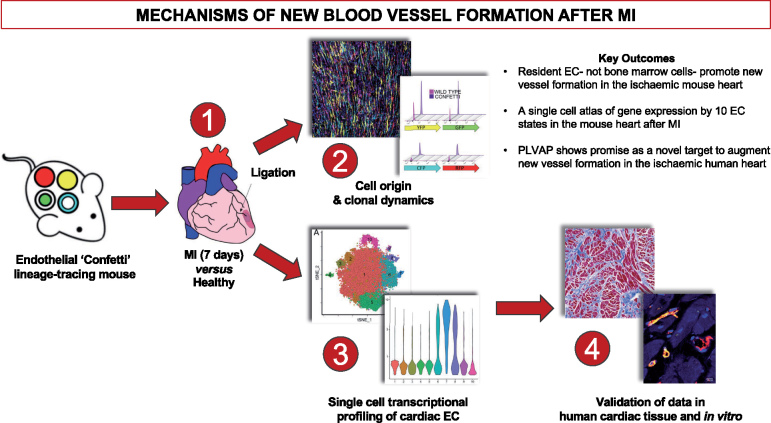
Abstract
Aims
A better understanding of the pathways that regulate regeneration of the coronary vasculature is of fundamental importance for the advancement of strategies to treat patients with heart disease. Here, we aimed to investigate the origin and clonal dynamics of endothelial cells (ECs) associated with neovascularization in the adult mouse heart following myocardial infarction (MI). Furthermore, we sought to define murine cardiac endothelial heterogeneity and to characterize the transcriptional profiles of pro-angiogenic resident ECs in the adult mouse heart, at single-cell resolution.
Methods and results
An EC-specific multispectral lineage-tracing mouse (Pdgfb-iCreERT2-R26R-Brainbow2.1) was used to demonstrate that structural integrity of adult cardiac endothelium following MI was maintained through clonal proliferation by resident ECs in the infarct border region, without significant contributions from bone marrow cells or endothelial-to-mesenchymal transition. Ten transcriptionally discrete heterogeneous EC states, as well as the pathways through which each endothelial state is likely to enhance neovasculogenesis and tissue regeneration following ischaemic injury were defined. Plasmalemma vesicle-associated protein (Plvap) was selected for further study, which showed an endothelial-specific and increased expression in both the ischaemic mouse and human heart, and played a direct role in regulating human endothelial proliferation in vitro.
Conclusion
We present a single-cell gene expression atlas of cardiac specific resident ECs, and the transcriptional hierarchy underpinning endogenous vascular repair following MI. These data provide a rich resource that could assist in the development of new therapeutic interventions to augment endogenous myocardial perfusion and enhance regeneration in the injured heart.
Keywords: Myocardial infarction, Endothelial cells, Lineage tracing, Single-cell RNA sequencing, Therapeutic angiogenesis, Cell proliferation
Translational perspective
Chronic heart failure following acute myocardial infarction (MI) has reached global epidemic proportions, and new therapeutic approaches are vital. Functional neovascularization in the infarct border may delay progression to heart failure and improve outcome, although the underpinning mechanisms remain unclear. We show that resident cardiac endothelial cells (ECs), and not bone marrow cells, mediate therapeutic angiogenesis post-MI, and provide a unique single-cell atlas of the transcriptional signature of these regenerative cardiac EC. We predict that these data will inform future design of clinical strategies aimed at promoting vascular perfusion and long-term functional viability of the ischaemic heart.
Introduction
Chronic heart failure as a consequence of left ventricular impairment following acute myocardial infarction (MI) has reached epidemic proportions affecting more than 23 million patients worldwide.1–4 Patients have a poor prognosis with a mortality of 45–60% by 5 years.5 Early reperfusion following MI is important to limit infarct size, however, patients who present late with extensive myocardial injury benefit little from these treatments. In this high-risk group, new therapeutic approaches are required to enhance myocardial perfusion, limit infarct expansion, and promote cardiac repair and regeneration. As such, extensive research has focused on harnessing the regenerative value of exogenous cell transplantation, and on elucidating the endogenous mechanisms that promote structural repair in the injured heart.
Experimental studies have produced disparate results regarding the regenerative potential of transplanted bone marrow cells in the setting of MI, including their contribution to vascular structures in vivo (reviewed in Refs6,7). Moreover, clinical trials using autologous bone marrow-derived cell transplantation aimed at promoting recovery following acute MI have largely failed to show sustained clinical benefit.6,8 Recently, endothelial cell (EC) clonal expansion in the post-ischaemic myocardium was shown using a Cdh5-CreERT2 lineage-tracing mouse.9 However, Cre-recombinase can be detected in bone marrow cells as well as endothelium in Cdh5-Cre lines and therefore the origin of these proliferative EC remains somewhat inconclusive.10 In the same study, clonally expanded EC co-expressed endothelial and mesenchymal markers, leading to the conjecture that ‘partial’ endothelial-to-mesenchymal transition (EndMT) may be associated with new vessel growth post-MI.9 However, a direct role for EndMT in therapeutic angiogenesis has not been conclusively shown, to date. Therefore, the cellular origin and mechanisms of neovascularization following MI remain unresolved, including whether reparative EC derive from a bone marrow niche or reside locally within the cardiac microvasculature.
Here, we have used an EC-specific multispectral lineage-tracing mouse (Pdgfb-iCreERT2-R26R-Brainbow2.1) coupled with single-cell RNA sequencing to collectively investigate the origin, proliferative dynamics and transcriptional profile of EC that mediate neovasculogenesis in the adult mouse heart. We show that a subpopulation of resident EC with progenitor-like functional properties directly contribute to new blood vessel formation at 7 days following MI, with evidence that bone marrow-derived cells and EndMT are unlikely to contribute. We have identified multiple discrete heterogeneous EC states in the healthy and post-ischaemic heart, and characterized gene expression profiles of each EC state at the single-cell level. These data identified plasmalemma vesicle-associated protein (Plvap) as a novel endothelial-specific marker of cardiac neovasculogenesis, which was further validated in cardiac samples from patients with ischaemic heart disease and following gene silencing in vitro. Together, these data provide a comprehensive single-cell index of gene expression by resident cardiac EC that contribute to endogenous neovascularization in the infarcted myocardium and will assist identification of new therapeutic targets for heart disease.
Methods
See Supplementary material online for extended experimental procedures.
Results
Brainbow2.1 (Confetti) reporter fluorophore expression is specific to endothelial cells in adult Pdgfb-iCreERT2-R26R-Brainbow2.1 mouse hearts
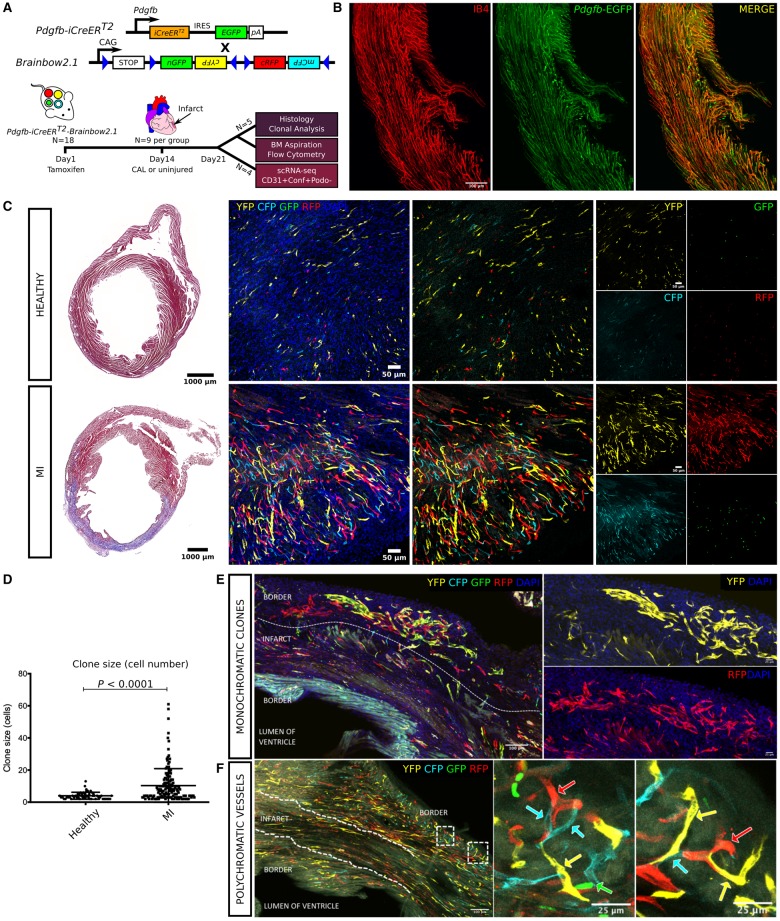
We first evaluated Pdgfb expression in the adult mouse coronary vasculature, which has not been widely characterized. We used the presence of a sequence coding for enhanced green fluorescent protein (EGFP) located downstream of iCreERT2 in Pdgfb-iCreERT2 mice11 (Figure 1A, Supplementary material online, Figure S1A–C) to show that Pdgfb-EGFP expression was widespread and endothelial-specific, confirmed by co-localization of EGFP with isolectin B4 (Figure 1B). We further showed using flow cytometry that Pdgfb was expressed by 94.5 ± 5.6% of CD31+ vascular EC in the adult Pdgfb-iCreERT2-R26R-Brainbow2.1 mouse heart (Supplementary material online, Figure S1C). Expression of the four fluorophores encoded by the Brainbow2.1 transgene (YFP, RFP, nGFP, mCFP) was specific to Pdgfb-EGFP+ cells (Figures 1C; Supplementary material online, Figure S1D and E). Cre-recombination efficiency was 46.6 (±9.3) % of total cardiac Pdgfb-EGFP+ cells, and no reporter expression was observed in mice administered peanut oil as a vehicle control for tamoxifen or in Cre-negative mice administered tamoxifen (Supplementary material online, Figure S1D–G).
Figure 1.
Genetic labelling strategy used for study of Pdgfb-iCreERT2 expressing endothelial cells using the R26R-Brainbow2.1 ‘Confetti’ reporter mouse (A). Tamoxifen was administered by intraperitoneal injection followed by coronary artery ligation 14 days later. Injured and healthy hearts) were collected 7 days post-surgery to quantify clonal proliferation and bone marrow reporter expression, or for single-cell RNA sequencing (scRNA-seq) of vascular cardiac Pdgfb-lineage endothelial cells (CD31+ podoplanin− Confetti+) (A). Isolectin B4 (IB4) was injected intravenously 15 min prior to cull to identify perfused vessels in the vasculature of Pdgfb-iCreERT2-R26R-Brainbow2.1 adult mouse hearts. Co-expression of isolectin B4 (IB4, red) with Pdgfb-EGFP endothelial cells (green, with co-localized expression shown in orange) demonstrated widespread patency in vessels formed by Pdgfb-lineage endothelial cell clonal proliferation (B). Masson’s trichrome staining confirmed healthy myocardial tissue or the presence of an infarct in each group (C). Clonal proliferation was quantified in 100 μm tissue wholemounts in the infarct border and equivalent healthy region, where a clone was defined as two or more adjacent cells expressing YFP, RFP, nGFP, or mCFP. Clones of each colour were observed throughout the healthy heart with a significant increase in fluorophore expression and clone size in the infarcted (MI) hearts (cells per clone = 4.0 ± 2.1 vs. 10.3 ± 10.6, P < 0.0001) (C, D). Clones of >50 cells were commonly observed in the infarct border (E; 2 large multicellular clones in the infarct border expressing RFP or YFP). Vessels with a polychromatic endothelium were observed in healthy and infarcted hearts but were significantly less abundant than monoclonal vessels in both groups (healthy hearts = 25.5 ± 4.1% vs. 74.5 ± 4.1%, P < 0.001; MI hearts = 9.3 ± 15.3% vs. 90.7 ± 15.3%, P < 0.001) (F; with arrows showing contiguous vessels composed of Pdgfb-lineage endothelial cells expressing different Confetti reporter fluorophores in the infarct border).
Cardiac Pdgfb-lineage endothelial cells undergo clonal proliferation to form new perfused blood vessels following myocardial infarction
Clonal proliferation by EC was observed in the healthy heart at 21 days post-tamoxifen, highlighting that Pdgfb-lineage cardiac EC are not quiescent in physiological conditions (Figure 1C). Clone size was significantly increased in the infarct border region at 7 days post-MI, with clones of >50 cells frequently present (cells per clone = 4.0 ± 2.1 vs. 10.3 ± 10.6, P < 0.0001; Figure 1C–E; Supplementary material online, Movie S1). Representative histological images of injured and healthy hearts are shown in Supplementary material online, Figure S2. Perfusion of vessels formed by clonal proliferation of Pdgfb-EC was confirmed by fluorophore co-expression with isolectin-B4, infused intravenously just prior to cull (Figure 1B). Consistent with other studies using the R26R-Brainbow2.1 mouse,12 we observed a bias in fluorophore distribution in both groups with YFP, RFP, nGFP, and mCFP expressed by 51.9 ± 13.5, 25.7 ± 8.7, 11.3 ± 6.2, and 11.8 ± 5.8% of total reporter expressing cells in the healthy heart, and by 52.9 ± 26.5, 18.8 ± 15.0, 26.5 ± 29.8, and 8.8 ± 7.8% of total reporter expressing cells in the injured heart (Supplementary material online, Figure S1H and I). However, clone size did not differ between each fluorophore in either group (Supplementary material online, Figure S1J and K). Therefore, patches were likely to be formed through clonal expansion, and not by the merger of multiple smaller clones of the same colour (see Supplementary material online file for detailed methods used for quantitative analysis of clonal proliferation).
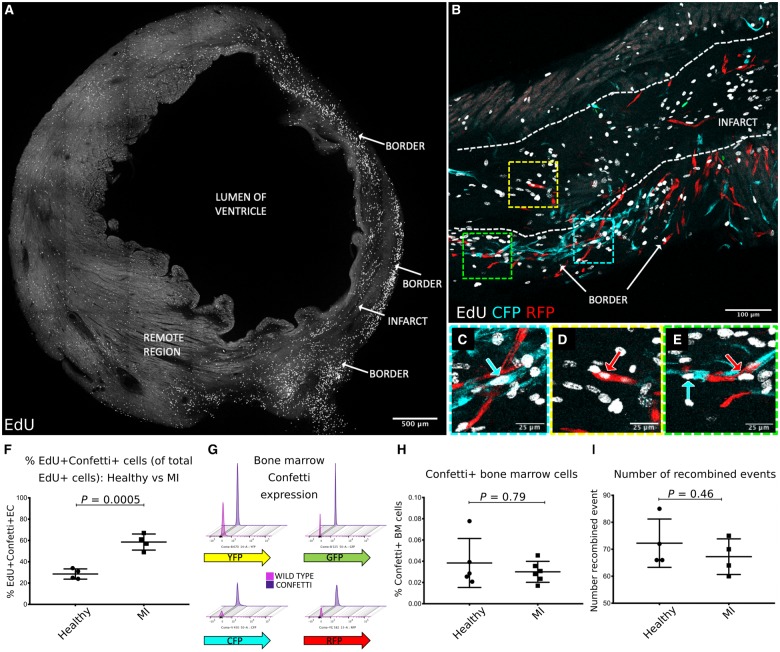
Vessels with a polychromatic endothelium were frequently observed in healthy and infarcted hearts and most likely represented pre-existing vessels composed of labelled EC that had not undergone proliferation within the study timeframe, or new vessels formed from the merger of non-proliferative labelled EC (Figure 1F). Polychromatic vessels were significantly less abundant than monoclonal vessels in both groups (healthy hearts = 25.5 ± 4.1% vs. 74.5 ± 4.1%, P < 0.001; MI hearts = 9.3 ± 15.3% vs. 90.7 ± 15.3%, P < 0.001), implying that clonal proliferation is a dominant mechanism of cardiac EC turnover in homeostasis and in response to injury. The proportion of Confetti+ EC present as single cells, i.e. that had not undergone division during the preceding 21 days, was similar in healthy and MI groups (54.6 ± 6.6% vs. 63.4 ± 4.7%, P = 0.28). Therefore, vascular turnover and neovascularization by clonal proliferation appears be restricted to a subpopulation of Pdgfb-lineage EC, presumably with progenitor-like properties. The thymidine analogue 5-ethynyl-2′-deoxyuridine (EdU) was administered 1 h prior to cull to label cells in S phase. EdU+ total cardiac cells were significantly increased in the infarct border at 7 days post-MI compared with the healthy left ventricle, as expected (0.09 ± 0.04% vs. 1.9 ± 0.6%, P < 0.0001). The percentage of Confetti+ EC that co-expressed EdU was also increased in the infarct border compared with the healthy left ventricle (28.5 ± 4.8% vs. 58.5 ± 7.6%, P = 0.0005), underlining our evidence that the infarct border is a region of dense neovasculogenesis with maximal clonal proliferation by Pdgfb-EC (Figure 2A–F).
Figure 2.
A complete transverse section of the left ventricle at 7 days post-MI showing increased density of EdU expressing cells in the border region compared with the infarct and remote region (A). Dense neovascularization due to clonal proliferation of Pdgfb-lineage endothelial cells was observed in the infarct border region (B, with high power inserts in C–E). Confetti+Pdgfb-lineage endothelial cells frequently co-expressed EdU (B–E) and were significantly increased in the infarct border at 7 days post-MI compared with the Confetti+ EdU+ Pdgfb-lineage endothelial cells in the left ventricle of healthy uninjured mice (28.5 ± 4.8% vs. 58.5 ± 7.6%, P = 0.0005) (F). Representative flow cytometry plots showing very low reporter fluorophore expression in femoral bone marrow cells from adult Pdgfb-iCreERT2-R26R-Brainbow2.1 mice are shown in (G) with threshold gates set for each fluorophore using C57Bl6 wild type mice bone marrow cells as a negative control. No change in fluorophore expression by bone marrow cells was observed between healthy and MI groups (0.04 ± 0.02% vs. 0.03 ± 0.009%, P = 0.79) (H). The founding number of recombined events (where a Confetti+ cell or clone was counted as one event) was unchanged between healthy and MI groups (72.3 ± 9.0 vs. 67.3 ± 6.6 events per section, P = 0.46) (I).
Brainbow2.1 reporter fluorophore expression is minimal in bone marrow cells in Pdgfb-iCreERT2-R26R-Brainbow2.1 mice
We observed minimal Brainbow2.1 reporter expression in femoral bone marrow cells in healthy and MI groups (0.04 ± 0.02% vs. 0.03 ± 0.009%, P = 0.79) (Figure 2F and G). Therefore, we postulated that Pdgfb-lineage EC reside locally within the cardiac vasculature. This was corroborated by our observation that the founding number of recombined events (i.e. a Confetti+ cell or clone) did not differ significantly between groups (P = 0.46) (Figure 2H) and thus migration of Pdgfb-lineage Brainbow2.1 fluorophore expressing EC from outside the heart was likely to be rare.
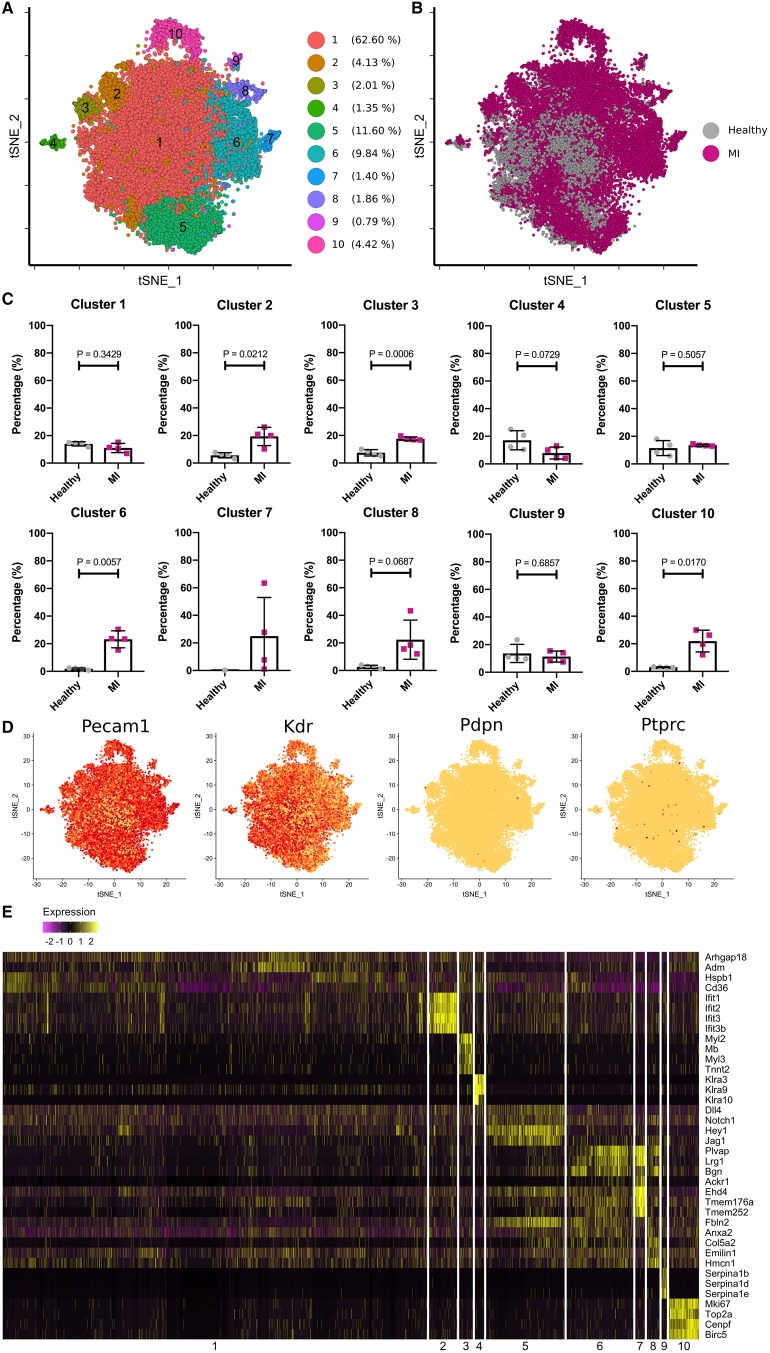
Single-cell RNA sequencing defines the transcriptional signature of 10 heterogeneous cardiac endothelial cell states in the adult mouse heart
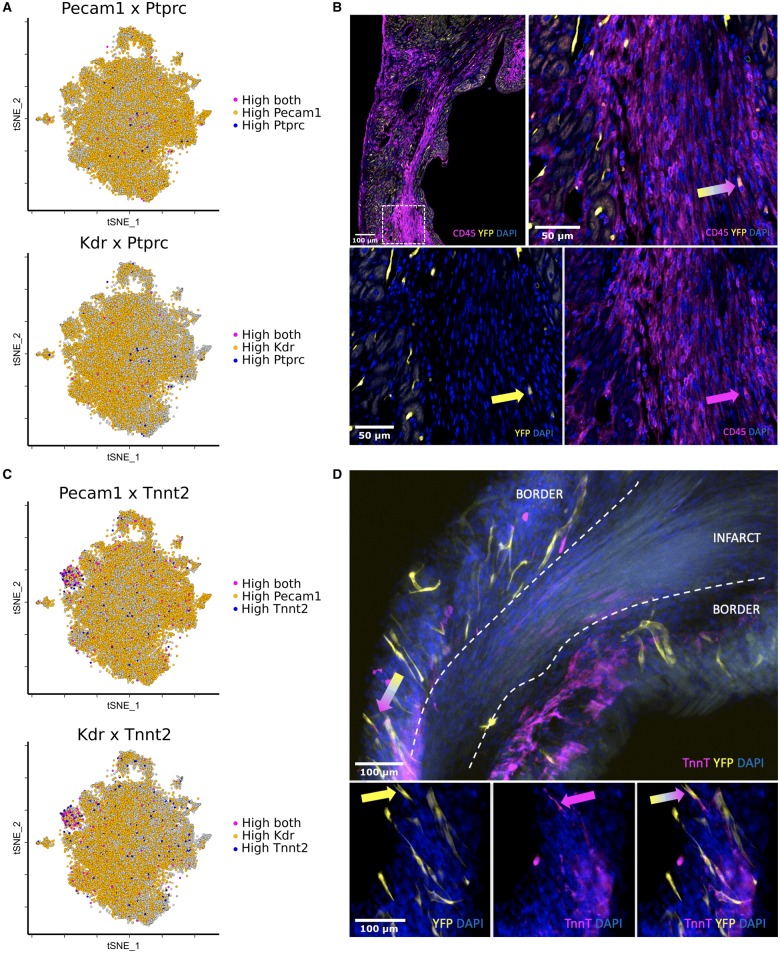
Pdgfb-lineage vascular EC (Confetti+ CD31+ podoplanin−) were isolated as single cells from dissociated ventricles of healthy and ischaemic hearts at 7 days post-MI for single-cell RNA sequencing (Supplementary material online, Figure S3A). Mean 3195 (range 2637–3535) and 3955 (range 3326–4744) cells were retained for analysis from the healthy and MI groups, respectively (P = 0.11) after filtering of cells with a low gene count (<400 genes per cell) and a high mitochondrial transcript ratio (threshold of 20%) (Supplementary material online, Figure S3B). Mean number of unique molecular identifiers (UMIs) was 2232 (range 2035–2398) and 5041 (range 4165–5604) (P = 0.03), and median gene number per cell was 1180 (range 1046–1217) and 2133 (range 1807–2251) (P = 0.03) for the healthy and MI groups (Supplementary material online, Figure S3B). Dimension reduction by principal component analysis of the whole transcriptome gene signature followed by 3D t-distributed stochastic neighbour embedding (t-SNE) projection assigned cells into 10 transcriptionally distinct clusters, i.e. discrete heterogeneous EC states (Figure 3A). Cluster distributions were conserved in each biological replicate animal in each group (Supplementary material online, Figure S3C). The proportion of cells in each cluster, the contribution of cells from both groups to individual clusters, as well as a heat map of top differentially expressed genes in each cluster are shown in Figure 3B, C, and E. An isolated cluster of cells that expressed the haematopoietic marker Ptprc (CD45) but did not express endothelial markers Pecam-1 (CD31) or Kdr (VEGFR2) was removed from the analysis, as it was likely due to cellular contamination during FACS. Following this, we confirmed enrichment of Pecam-1 and Kdr in all clusters and a minimal presence of lymphatic markers in our analysis (Figure 3D). A very small proportion of Ptprc+ cells from the MI group that co-expressed Pecam-1 (0.29% total cells) or KDR (0.19% total cells) was dispersed throughout several clusters (Figure 4A). Correspondingly, we observed rare in situ expression of CD45 by Pdgfb-lineage EC post-MI (Figure 4B). These CD45+ cells may be EC that have transitioned towards an inflammatory phenotype in response to injury or, more likely, scarce infiltrating bone marrow cells, described above.
Figure 3.
Ten discrete heterogeneous endothelial cell populations were identified in healthy and infarcted adult mouse hearts (A and B). Contributions of cells from both healthy and myocardial infarction hearts to each cluster were compared, and cluster 2, 3, 6, 8, and 10 were largely constituted by the cells from the myocardial infarction hearts. Cluster 7 was composed exclusively of cells from the myocardial infarction hearts (C). Broad expression of endothelial cell markers, such as Pecam1 and Kdr, and the rare presence of Pdpn and Ptprc expressing cells showed purity of the studied endothelial cell population (D). Top differentially expressed marker genes were shown for each of the 10 clusters in the heatmap (E).
Figure 4.
Rare Ptprc+ (CD45) cells that co-expressed high levels of Pecam-1 (0.29% total cells) or KDR (0.19% total cells) were dispersed throughout several clusters in the MI group (A). Cells that co-expressed CD45 and a Confetti reporter fluorophore were observed at the infarct border in 100 μm tissue wholemounts (B, arrows). Cluster 3 showed co-expression of endothelial markers, Pecam-1 and Kdr, with genes encoding cardiac muscle markers including troponin-T (Tnnt2) (C). Cells that co-expressed a Brainbow2.1 fluorophore and the cardiomyocyte marker, troponin-T, were identified in the infarct border region in 100 μm tissue wholemounts (D, arrows). These cells were scarce and resembled vascular structures in morphology, rather than cardiomyocytes.
We used Gene Ontology (GO) term enrichment analysis (PANTHER, GOrilla and GeneMANIA) to reveal significant terms associated with the top differentially expressed genes in each cluster, which allowed us to extrapolate information regarding the functional identity of each EC state (Table 1). For example, cluster 10 predominantly comprised cells from the MI group with a high and restricted expression of cell cycle and proliferation markers (e.g. Mki67, Top2a, Cenpf, Cks2, Birc5, Cenpa, Ube2c, Cdc2029,30). Our aforementioned observations of heightened clonal proliferation post-MI by Pdgfb-lineage EC compared with the healthy heart, substantiated this gene expression profile (Figure 1D). Cluster 3 showed selective expression of genes encoding ventricular cardiac muscle morphogenesis (Myl2, Mb, Myl3, Tnnt2, Tnni3, Actc1), suggesting that resident adult cardiac EC may switch on cardiomyocyte lineage genes in response to MI. This was supported by a study showing cardiac muscle cells derived from embryonic EC following their direct injection into the ischaemic adult mouse heart.19 We confirmed rare in situ existence of cells that co-expressed a Brainbow2.1 fluorophore and the cardiomyocyte marker, troponin-T, in the infarct border region, but not in the healthy heart (Figure 4C and D). Therefore, an endothelial to cardiomyocyte fate shift may occur post-MI, although did not contribute significantly to myocardial regeneration at the studied timepoint.
Table 1.
Top differentially expressed markers and predicted functions of each cluster
| Cluster number | Enrichment in EC from healthy or MI groups | Top differentially expressed genes | Specific to cluster | Predicted function in Pdgfb-EC | References to support predicted function |
|---|---|---|---|---|---|
| 1 | NA (P = 0.34) | Arhgap18, Adm, Hspb1, CD36 | N | Cellular homeostasis | (13–16) |
| 2 | MI (P = 0.02) | Ifit1, Ifit2, Ifit3, Ifit3b, Usp18, Cxcl10 | Y | Interferon signalling | (17) IFN signalling in CVD including MI; (18) Induction of EC proliferation by IFN |
| 3 | MI (P = 0.0006) | Myl2, Mb, Myl3, Tnnt2, Tnni3, Actc1 | Y | Ventricular cardiac muscle remodelling | (19) Embryonic EC trans-differentiation to cardiac muscle cells in MI |
| 4 | NA (P = 0.07) | Klra3, Klra9, Klra10 | Y | Killer cell lectin-like receptor signalling | (20) NK cell interaction with EC driving neovasculogenesis post-MI; (21) EC expression of Klra family genes; (22) CD31 expression by NK cells |
| 5 | NA (P = 0.51) | Dll4, Notch1, Hey1, Jag1, Gja4 | Y (Hey1, Jag1, Gja4) | Endothelial cell regulation via Notch signalling | (23) Notch regulation of EC; (24,25) Gja4 encodes Connexin37 in EC, which regulates arterial-venous specification; (26) Gja4-deficient mice have abnormal vascular regeneration in ischaemic limb |
| 6 | MI (P = 0.006) | Plvap, Lrg1, Pbp1, Bgn, vWF | N | Ventricular remodelling [via retinoic acid (RA) signalling] | (27,28) Regulation of ventricular remodelling by Rbp1 and Bgn via RA signalling post-MI |
| 7 | MI (100%) | Ackr1, Ehd4, Tmem176a, Tmem252, Tmem176b, Selp | Y (Ackr1, Tmem252, Selp) | Stalk cell markers. Tip and stalk cell-mediated neovasculogenesis | (29,30) EC tip and stalk cell signalling; (31) Fate switching between tip and stalk cells, enhanced proliferation by stalk versus tip cells |
| 8 | MI (P = 0.07) | Fbln2, Anxa2, Col5a2, Emilin1, Hmcn1, Bgn, Mgp | Y (Col5a2, Mgp) | Endothelial ECM proteins, cardiac remodelling post-MI | (32) Cardiac remodelling in human and pigs post-MI; (33) Endothelial ECM critical for blood vessel network stabilization and maturation |
| 9 | NA (P = 0.69) | Serpina1b, Serpina1d, Serpina1e | Y | Serine protease inhibitor alpha-1 antitrypsin (AAT) signalling | (34,35) AAT therapy in ischaemic disease; (36,37) AAT is cytoprotective following ischaemic injury |
| 10 | MI (P = 0.02) | Mki67, Top2a, Cenpf, Cks2, Birc5, Cenpa, Ube2c, Cdc20 | Y | Proliferation and cell cycle regulation | (38) |
Endothelial-to-mesenchymal transition does not contribute to cardiac neovasculogenesis at 7 days post-myocardial infarction
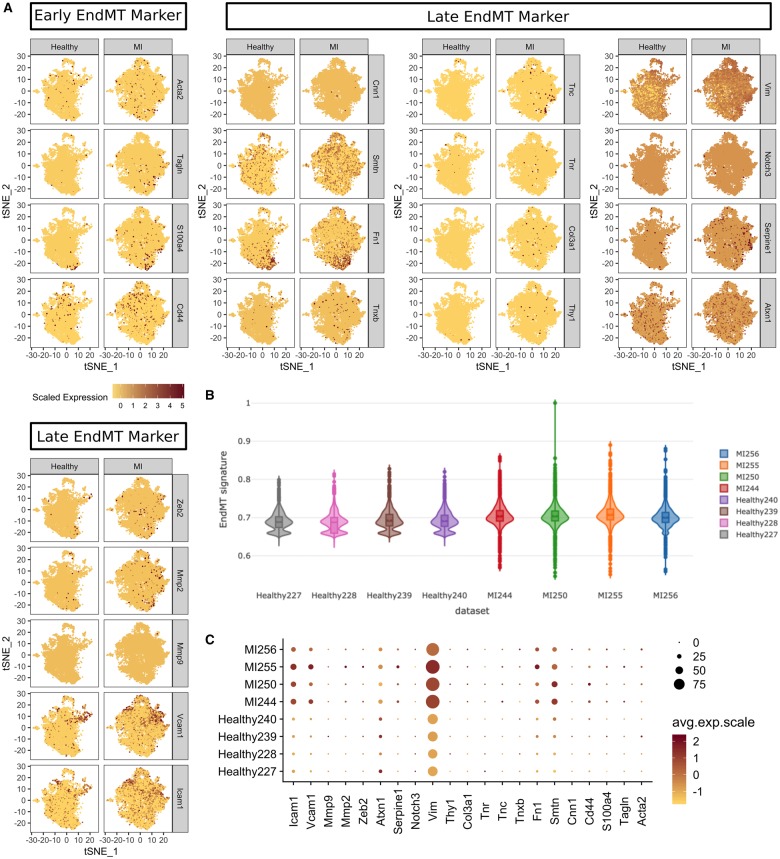
Endothelial markers were not reduced after MI in our data (Figure 3D), and therefore, we investigated the role of ‘partial EndMT’, recently hypothesized to play a role in neovascularization in the ischaemic mouse heart.9 However, the overall EndMT signature was unchanged in both groups (Figure 5A and B) despite an increase in some EndMT markers after MI (Icam1, Vcam1, Vim, Fn1 and Smtn) (Figure 5C). This indicates that EndMT and partial EndMT are unlikely to participate significantly in our model at the studied timepoint.
Figure 5.
Endothelial-to-mesenchymal transition/partial endothelial-to-mesenchymal transition was investigated in our data using a comprehensive panel of markers of early and late endothelial-to-mesenchymal transition (A). The change in an endothelial-to-mesenchymal transition gene signature between healthy and myocardial infarction groups was minimal and not specific to any cluster (B) despite an increase in some markers (C).
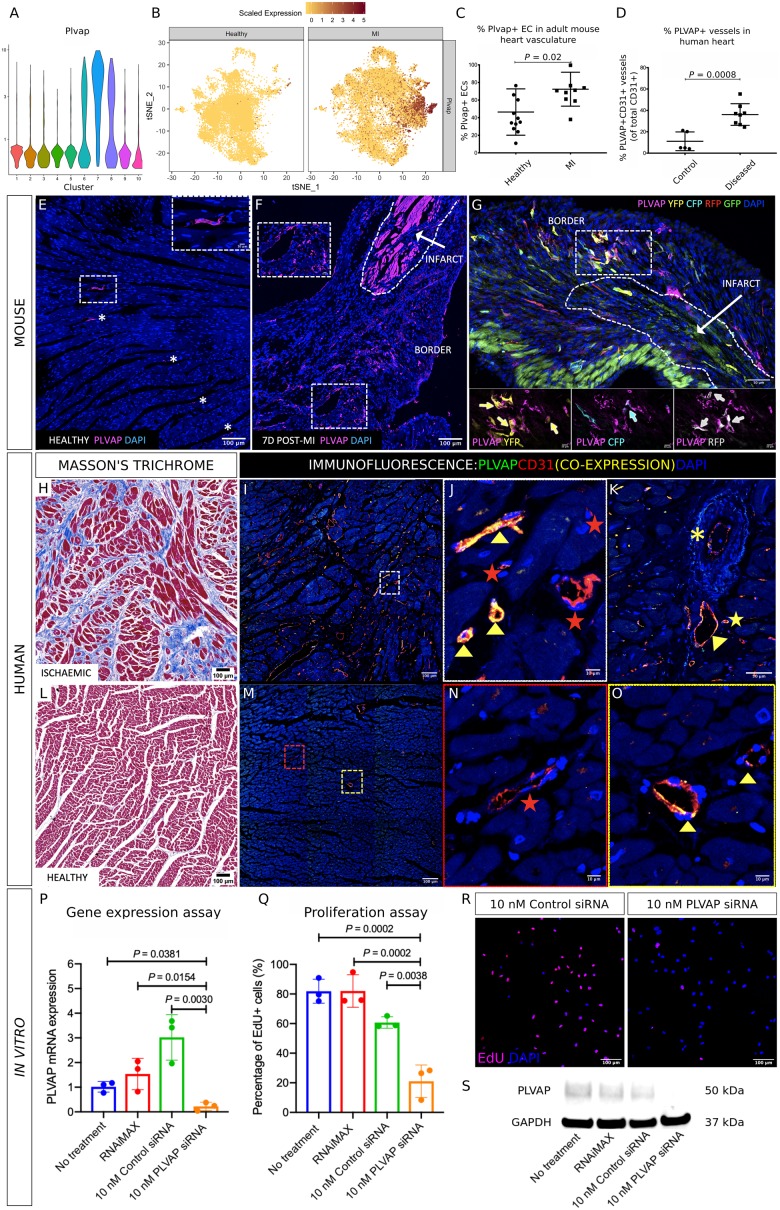
Plasmalemma vesicle-associated protein represents a novel potential therapeutic target that mediates neovasculogenesis in the ischaemic mouse and human heart
Clusters 6, 7, and 8 predominantly comprised cells from the MI group (Figure 3B), indicating that their gene expression profiles may be relevant to neovasculogenic pathways. Gene expression of Plvap was highest in clusters 6, 7, and 8 (Figure 6A and B). Plasmalemma vesicle-associated protein encodes an EC-specific marker,39 although its role in neovasculogenesis in the adult heart is unknown. We quantified Plvap expression in the healthy and infarcted mouse heart, as well as in healthy and ischaemic human cardiac tissue sections. Plvap expression was specific to CD31+ EC and was increased in the infarct border compared with the healthy left ventricle, thus validating our RNA sequencing data at the protein level (% Plvap+ EC = 70.5 ± 19.9% vs. 38.7 ± 28.2%, P = 0.002, Figure 6C, E–G). Plvap expression was also increased in EC adjacent to regions of fibrosis and scarring in the ischaemic human heart, compared with healthy human hearts (% Plvap+ EC = 36.9 ± 10.1% vs. 11.1 ± 8.8%, P = 0.002, Figure 6D, H–O). In order to better understand whether Plvap can directly modulate neovascularization we carried out in vitro siRNA gene silencing on human umbilical venous EC (HUVEC) where Plvap expression was significantly reduced at the mRNA level compared with control siRNA (RQ = 3.0 ± 0.9 vs. 0.2 ± 0.2, P = 0.003) and confirmed at the protein level by western blot (Figure 6P, S). Proliferation was assessed using an EdU incorporation assay and was significantly inhibited following Plvap gene silencing compared with control siRNA treatment (% EdU+ HUVEC = 60.7 ± 3.9% vs. 21.1 ± 11.0%, P = 0.0038; Figure 6Q and R), providing strong evidence for a direct functional role of PLVAP in endothelial proliferation.
Figure 6.
Violin plots showing heightened expression of Plvap in clusters 6, 7, and 8 (A). t-SNE plots highlighting an increased Plvap gene expression in cardiac endothelial cells from the myocardial infarction group compared with the healthy group (B). Plvap was significantly increased in the infarct border region at 7 days post-myocardial infarction, compared with the healthy mouse heart (70.5 ± 19.9% vs. 38.7 ± 28.2%, P = 0.002) (C). Plvap expression was also increased in endothelial cells adjacent to regions of fibrosis and scarring in the ischaemic human heart, compared with healthy human hearts (% Plvap+ endothelial cells = 36.9 ± 10.1% vs. 11.1 ± 8.8%, P = 0.002) (D). Representative images showing immunofluoresence staining for Plvap accompanying the above data showing the change in expression in cardiac endothelial cells in the healthy (E) and injured Pdgfb-iCreERT2-R26R-Brainbow2.1 mouse heart in the infarct border at 7 days post-myocardial infarction (F), with co-expression of Pdgfb-EC Confetti fluorophore-expressing clones (G with high power inserts and arrows highlighting Plvap+ Confetti+ EC). Consecutive sections from ischaemic and healthy human cardiac tissue were stained using Masson’s Trichrome (H = healthy, L = ischaemic) and by immunofluorescence for PLVAP and CD31 (green and red, respectively, with co-expression in yellow; I–K = ischaemic, M–O = healthy). PLVAP expression was specific to CD31+ cells and was increased in regions of fibrosis and scarring in the diseased compared with the healthy heart. Plvap+ CD31+ vessels are indicated by yellow arrowheads (J, O), CD31+ Plvap− vessels indicated by red stars (J, N). Plvap+ CD31+ EC expression in a venule (yellow arrowhead), arteriole (yellow asterisk) and capillary (yellow star) is shown in (K). siRNA gene silencing of Plvap in human umbilical venous endothelial cells gave a significant reduction at the mRNA level compared with control siRNA (RQ = 3.0 ± 0.9 vs. 0.2 ± 0.2, P = 0.003) (P) and was confirmed at the protein level by western blot (S). Proliferation assessed using an EdU incorporation assay was significantly inhibited following Plvap gene silencing compared with control siRNA treatment (% EdU+ human umbilical venous endothelial cells = 60.7 ± 3.9% vs. 21.1 ± 11.0%, P = 0.0038; Q, R).
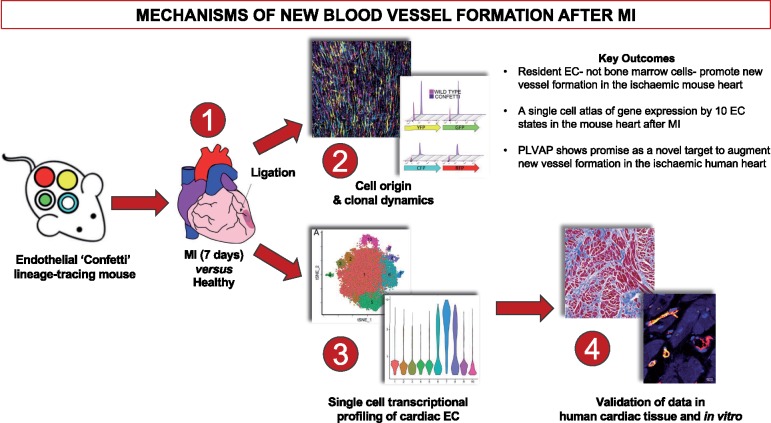
Take home figure.
Single cell transcriptome analyses provide a single cell gene expression atlas of resident cardiac endothelial cells that promote neovascularisation following ischaemic injury by undergoing clonal proliferation.
Discussion
In this study, we demonstrate that structural integrity of the endothelium of the adult heart during physiological conditions and following MI is maintained through clonal proliferation of a subset of resident EC with progenitor-like functional properties with minimal, if any, contribution by bone marrow cells. We further present the first comprehensive single-cell atlas of gene expression by 10 individual EC subpopulations in the adult mouse heart after MI. Finally, we show that Plvap is likely to represent a novel target to augment new vessel formation in the ischaemic human heart.
Our observation that clonal expansion of resident cardiac EC is the predominant mechanism driving neovasculogenesis expand on recent findings of He et al.40 who used genetic lineage tracing to show that new cardiac blood vessels are formed from pre-existing EC. This may, in part, explain the disappointing outcomes of clinical studies using autologous bone marrow-derived cells for therapeutic neovascularization in patients with ischaemic heart disease.6,8 It is important to note that a contribution of Pdgfb-lineage EC to cardiac neovasculogenesis post-MI cannot be discounted, i.e. due to an inactive Cre-driver or the migration of non-recombined cells. However, despite a 46.6% Cre-recombination efficiency in cardiac ECs we observed Brainbow2.1 reporter fluorophore expression in fewer than 0.05% of bone marrow cells. Furthermore, the founding number of recombined events in the cardiac vasculature did not change before or after ischaemic injury. This led us to postulate that Pdgfb-lineage EC contributing to neovasculogenesis via clonal proliferation reside locally within the cardiac vasculature with minimal, if any, contribution from bone marrow cells. Therefore, future research and clinical focus should aim to enhance the regenerative capacity of endogenous resident cardiac EC to promote therapeutic neovasculogenesis in the injured heart.
Using single-cell RNA sequencing, we have defined endothelial heterogeneity in the healthy and injured mouse heart through characterization of 10 EC states with distinct gene expression signatures, and by mapping the transcriptional changes arising in each subset following injury, at the level of a single cell. Transcriptional profiling of the non-myocyte cardiac cellulome of healthy mice was recently reported,41 although EC were strategically depleted prior to sequencing to enable focused analysis of fibroblasts and rarer cardiac lineages such as glia and mural cells. Single-cell RNA sequencing was also recently applied to map cell populations in the healthy and injured mouse heart 3 days after ischaemia–reperfusion surgery.42 This study identified two EC clusters, although detailed EC gene expression analysis was not provided as this study focused on changing cardiomyocyte subpopulations. Therefore, we believe our data is the first in-depth characterization of the molecular profiles that demarcate cardiac endothelial heterogeneity and plasticity in response to ischaemic injury.
We defined changes in EndMT gene signatures between healthy and injured cardiac EC using a broad panel of markers.9,43 However, as we found no inhibition of endothelial genes nor a significant change in the EndMT gene signature, we concluded that EndMT does not play an obvious role in our model at the timepoint studied. This contrasts with a recent report suggesting that partial induction of EndMT may support cardiac neovascularization after ischaemia.9 This discrepancy may be due to a transient nature of EndMT, as the authors did not clarify whether partial EndMT genes were enriched at 7 or 14-days post-MI. Therefore, further temporal analysis of EndMT gene signatures following injury, as well as investigation into whether EndMT directly facilitates cardiac neovasculogenesis are warranted.
Plasmalemma vesicle-associated protein was selected for further study, due to its cluster-restricted increase in MI, and the lack of understanding of its role in endothelial regulation in the adult heart. Plasmalemma vesicle-associated protein is specifically expressed by EC and increased in some inflammatory states44 as well as in pathogenic angiogenesis.45–47 Here, we showed increased Plvap expression specifically within EC in clonal neovessels in the ischaemic border region of the mouse heart, and in regions of fibrosis and scarring in the ischaemic human heart, compared with non-ischaemic hearts. We further demonstrated that targeted silencing of Plvap in vitro directly inhibits EC proliferation. Therefore, Plvap shows early promise as a gene of interest for therapeutic strategies to promote cardiac regeneration. Future studies must now systematically evaluate the molecular pathways and regenerative significance of enhanced Plvap expression in EC in the infarct border region at the time of injury. However, at present, methods for cardiac endothelial-specific gene delivery to increase expression are challenging, and therefore, genetic mouse models and/or a pharmacological approach would be required.
The current study provides a high-resolution single cell gene expression atlas of resident cardiac EC in both physiological conditions and following permanent ligation of the left anterior descending coronary artery at 7 days post-MI. This model results in massive cardiomyocyte death, coagulative necrosis and scarring, with evidence of significant neovasculogenesis in the infarct border region. However, this model is unlikely to stimulate EC-mediated myocardial regeneration, and therefore, molecular targets identified from the analyses presented herein should be rigorously interrogated to investigate whether they promote myocardial regeneration and prevent cardiomyocyte death through enhanced tissue perfusion, rather than merely supporting scar formation. One could venture that a potential limitation of this study lies within the vast wealth of data generated from a single snap-shot in time, and care must be taken to extract and validate future targets that most effectively tackle the clinical problem. Therefore, consulting publicly available EC transcriptomics data, such as the endothelial database, EndoDB,48 will assist the construction of novel hypotheses for future studies.
In summary, this study provides a rich resource of targets and pathways that can be interrogated to reveal their role in promoting neovascularization by endogenous cardiac EC and thus may guide novel therapeutic strategies aimed at enhancing myocardial repair and regeneration.
Supplementary Material
Acknowledgements
We would like to acknowledge Edinburgh Genomics for single-cell RNA sequencing, the University of Edinburgh SuRF facility for H&E staining, and the University of Edinburgh MRC Tissue Brain Bank for providing and sectioning human cardiac tissue samples. We would like to thank Dr Julie Rodor (University of Edinburgh, Baker laboratory) for assistance with analysis of the siRNA study.
Funding
M.B. and Z.L. are supported by the British Heart Foundation (FS/16/4/31831). M.B. is further supported by the British Heart Foundation Centre for Vascular Regeneration (RM/17/3/33381) and the University of Edinburgh BHF Centre of Research Excellence (RE/13/3/30183). A.H.B. is supported by the British Heart Foundation Chair of Translational Cardiovascular Sciences (CH/11/2/28733). N.C.H. is supported by a Wellcome Trust Senior Research Fellowship in Clinical Science (ref. 103749).
Declaration of Helsinki
This study was performed in accordance with the Declaration of Helsinki.
Conflict of interest: none declared.
See page 2521 for the editorial comment on this article (doi: 10.1093/eurheartj/ehz375)
References
- 1. Dunlay SM, Roger VL.. Understanding the epidemic of heart failure: past, present, and future. Curr Heart Fail Rep 2014;11:404–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ponikowski P, Voors AA, Anker SD, Bueno H, Cleland JGF, Coats AJS, Falk V, Gonzalez-Juanatey JR, Harjola VP, Jankowska EA, Jessup M, Linde C, Nihoyannopoulos P, Parissis JT, Pieske B, Riley JP, Rosano GMC, Ruilope LM, Ruschitzka F, Rutten FH, van der Meer P; Group Scientific Document Group. 2016 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure: the task force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC)Developed with the special contribution of the Heart Failure Association (HFA) of the ESC. Eur Heart J 2016;37:2129–2200. [DOI] [PubMed] [Google Scholar]
- 3. Thygesen K, Alpert JS, Jaffe AS, Chaitman BR, Bax JJ, Morrow DA, White HD, Group E.. Fourth universal definition of myocardial infarction (2018). Eur Heart J 2019;40:237–269. [DOI] [PubMed] [Google Scholar]
- 4. Madonna R, Van Laake LW, Davidson SM, Engel FB, Hausenloy DJ, Lecour S, Leor J, Perrino C, Schulz R, Ytrehus K, Landmesser U, Mummery CL, Janssens S, Willerson J, Eschenhagen T, Ferdinandy P, Sluijter JP.. Position Paper of the European Society of Cardiology Working Group Cellular Biology of the Heart: cell-based therapies for myocardial repair and regeneration in ischemic heart disease and heart failure. Eur Heart J 2016;37:1789–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Levy D, Kenchaiah S, Larson MG, Benjamin EJ, Kupka MJ, Ho KK, Murabito JM, Vasan RS.. Long-term trends in the incidence of and survival with heart failure. N Engl J Med 2002;347:1397–1402. [DOI] [PubMed] [Google Scholar]
- 6. Boudoulas KD, Hatzopoulos AK.. Cardiac repair and regeneration: the Rubik's cube of cell therapy for heart disease. Dis Model Mech 2009;2:344–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Madigan M, Atoui R.. Therapeutic use of stem cells for myocardial infarction. Bioengineering (Basel) 2018;5:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Henry TD, Moye L, Traverse JH.. Consistently inconsistent-bone marrow mononuclear stem cell therapy following acute myocardial infarction: a decade later. Circ Res 2016;119:404–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Manavski Y, Lucas T, Glaser SF, Dorsheimer L, Gunther S, Braun T, Rieger MA, Zeiher AM, Boon RA, Dimmeler S.. Clonal expansion of endothelial cells contributes to ischemia-induced neovascularization. Circ Res 2018;122:670–677. [DOI] [PubMed] [Google Scholar]
- 10. Joseph C, Quach JM, Walkley CR, Lane SW, Lo Celso C, Purton LE.. Deciphering hematopoietic stem cells in their niches: a critical appraisal of genetic models, lineage tracing, and imaging strategies. Cell Stem Cell 2013;13:520–533. [DOI] [PubMed] [Google Scholar]
- 11. Claxton S, Kostourou V, Jadeja S, Chambon P, Hodivala-Dilke K, Fruttiger M.. Efficient, inducible Cre-recombinase activation in vascular endothelium. Genesis 2008;46:74–80. [DOI] [PubMed] [Google Scholar]
- 12. Chappell J, Harman JL, Narasimhan VM, Yu H, Foote K, Simons BD, Bennett MR, Jorgensen HF.. Extensive proliferation of a subset of differentiated, yet plastic, medial vascular smooth muscle cells contributes to neointimal formation in mouse injury and atherosclerosis models. Circ Res 2016;119:1313–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lovelace MD, Powter EE, Coleman PR, Zhao Y, Parker A, Chang GH, Lay AJ, Hunter J, McGrath AP, Jormakka M, Bertolino P, McCaughan G, Kavallaris M, Vadas MA, Gamble JR.. The RhoGAP protein ARHGAP18/SENEX localizes to microtubules and regulates their stability in endothelial cells. Mol Biol Cell 2017;28:1066–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marcovecchio PM, Thomas GD, Mikulski Z, Ehinger E, Mueller KAL, Blatchley A, Wu R, Miller YI, Nguyen AT, Taylor AM, McNamara CA, Ley K, Hedrick CC.. Scavenger receptor CD36 directs nonclassical monocyte patrolling along the endothelium during early atherogenesis. Arterioscler Thromb Vasc Biol 2017;37:2043–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Batulan Z, Pulakazhi Venu VK, Li Y, Koumbadinga G, Alvarez-Olmedo DG, Shi C, O'Brien ER.. Extracellular release and signaling by heat shock protein 27: role in modifying vascular inflammation. Front Immunol 2016;7:285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Beygui F, Wild PS, Zeller T, Germain M, Castagne R, Lackner KJ, Munzel T, Montalescot G, Mitchell GF, Verwoert GC, Tarasov KV, Tregouet DA, Cambien F, Blankenberg S, Tiret L.. Adrenomedullin and arterial stiffness: integrative approach combining monocyte ADM expression, plasma MR-Pro-ADM, and genome-wide association study. Circ Cardiovasc Genet 2014;7:634–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van den Borne P, Quax PH, Hoefer IE, Pasterkamp G.. The multifaceted functions of CXCL10 in cardiovascular disease. Biomed Res Int 2014;2014:893106.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gomez D, Reich NC.. Stimulation of primary human endothelial cell proliferation by IFN. J Immunol 2003;170:5373–5381. [DOI] [PubMed] [Google Scholar]
- 19. Condorelli G, Borello U, De Angelis L, Latronico M, Sirabella D, Coletta M, Galli R, Balconi G, Follenzi A, Frati G, Cusella De Angelis MG, Gioglio L, Amuchastegui S, Adorini L, Naldini L, Vescovi A, Dejana E, Cossu G.. Cardiomyocytes induce endothelial cells to trans-differentiate into cardiac muscle: implications for myocardium regeneration. Proc Natl Acad Sci USA 2001;98:10733–10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bouchentouf M, Forner KA, Cuerquis J, Michaud V, Zheng J, Paradis P, Schiffrin EL, Galipeau J.. Induction of cardiac angiogenesis requires killer cell lectin-like receptor 1 and alpha4beta7 integrin expression by NK cells. J Immunol 2010;185:7014–7025. [DOI] [PubMed] [Google Scholar]
- 21. Sobanov Y, Bernreiter A, Derdak S, Mechtcheriakova D, Schweighofer B, Duchler M, Kalthoff F, Hofer E.. A novel cluster of lectin-like receptor genes expressed in monocytic, dendritic and endothelial cells maps close to the NK receptor genes in the human NK gene complex. Eur J Immunol 2001;31:3493–3503. [DOI] [PubMed] [Google Scholar]
- 22. Marelli-Berg FM, Clement M, Mauro C, Caligiuri G.. An immunologist's guide to CD31 function in T-cells. J Cell Sci 2013;126:2343–2352. [DOI] [PubMed] [Google Scholar]
- 23. Mack JJ, Iruela-Arispe ML.. NOTCH regulation of the endothelial cell phenotype. Curr Opin Hematol 2018;25:212–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fang JS, Coon BG, Gillis N, Chen Z, Qiu J, Chittenden TW, Burt JM, Schwartz MA, Hirschi KK.. Shear-induced Notch-Cx37-p27 axis arrests endothelial cell cycle to enable arterial specification. Nat Commun 2017;8:2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Meens MJ, Pfenniger A, Kwak BR, Delmar M.. Regulation of cardiovascular connexins by mechanical forces and junctions. Cardiovasc Res 2013;99:304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fang JS, Angelov SN, Simon AM, Burt JM.. Cx37 deletion enhances vascular growth and facilitates ischemic limb recovery. Am J Physiol Heart Circ Physiol 2011;301:H1872–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bilbija D, Haugen F, Sagave J, Baysa A, Bastani N, Levy FO, Sirsjo A, Blomhoff R, Valen G.. Retinoic acid signalling is activated in the postischemic heart and may influence remodelling. PLoS One 2012;7:e44740.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ahmed MS, Oie E, Vinge LE, Yndestad A, Andersen GG, Andersson Y, Attramadal T, Attramadal H.. Induction of myocardial biglycan in heart failure in rats–an extracellular matrix component targeted by AT(1) receptor antagonism. Cardiovasc Res 2003;60:557–568. [DOI] [PubMed] [Google Scholar]
- 29. Moya IM, Umans L, Maas E, Pereira PN, Beets K, Francis A, Sents W, Robertson EJ, Mummery CL, Huylebroeck D, Zwijsen A.. Stalk cell phenotype depends on integration of Notch and Smad1/5 signaling cascades. Dev Cell 2012;22:501–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hellstrom M, Phng LK, Hofmann JJ, Wallgard E, Coultas L, Lindblom P, Alva J, Nilsson AK, Karlsson L, Gaiano N, Yoon K, Rossant J, Iruela-Arispe ML, Kalen M, Gerhardt H, Betsholtz C.. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature 2007;445:776–780. [DOI] [PubMed] [Google Scholar]
- 31. Jakobsson L, Franco CA, Bentley K, Collins RT, Ponsioen B, Aspalter IM, Rosewell I, Busse M, Thurston G, Medvinsky A, Schulte-Merker S, Gerhardt H.. Endothelial cells dynamically compete for the tip cell position during angiogenic sprouting. Nat Cell Biol 2010;12:943–953. [DOI] [PubMed] [Google Scholar]
- 32. Barallobre-Barreiro J, Didangelos A, Schoendube FA, Drozdov I, Yin X, Fernández-Caggiano M, Willeit P, Puntmann VO, Aldama-López G, Shah AM, Doménech N, Mayr M.. Proteomics analysis of cardiac extracellular matrix remodeling in a porcine model of ischemia/reperfusion injury. Circulation 2012;125:789–802. [DOI] [PubMed] [Google Scholar]
- 33. Davis GE, Senger DR.. Endothelial extracellular matrix: biosynthesis, remodeling, and functions during vascular morphogenesis and neovessel stabilization. Circ Res 2005;97:1093–1107. [DOI] [PubMed] [Google Scholar]
- 34. Lomas DA, Mahadeva R.. Alpha1-antitrypsin polymerization and the serpinopathies: pathobiology and prospects for therapy. J Clin Invest 2002;110:1585–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lewis EC. Expanding the clinical indications for alpha(1)-antitrypsin therapy. Mol Med 2012;18:957–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Toldo S, Seropian IM, Mezzaroma E, Van Tassell BW, Salloum FN, Lewis EC, Voelkel N, Dinarello CA, Abbate A.. Alpha-1 antitrypsin inhibits caspase-1 and protects from acute myocardial ischemia-reperfusion injury. J Mol Cell Cardiol 2011;51:244–251. [DOI] [PubMed] [Google Scholar]
- 37. Daemen MARC, Heemskerk VH, van ’t Veer C, Denecker G, Wolfs TGAM, Vandenabeele P, Buurman WA.. Functional protection by acute phase proteins α1-acid glycoprotein and α1-antitrypsin against ischemia/reperfusion injury by preventing apoptosis and inflammation. Circulation 2000;102:1420–1426. [DOI] [PubMed] [Google Scholar]
- 38. Scott RE, Ghule PN, Stein JL, Stein GS.. Cell cycle gene expression networks discovered using systems biology: significance in carcinogenesis. J Cell Physiol 2015;230:2533–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Guo L, Zhang H, Hou Y, Wei T, Liu J.. Plasmalemma vesicle-associated protein: a crucial component of vascular homeostasis. Exp Ther Med 2016;12:1639–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. He L, Huang X, Kanisicak O, Li Y, Wang Y, Li Y, Pu W, Liu Q, Zhang H, Tian X, Zhao H, Liu X, Zhang S, Nie Y, Hu S, Miao X, Wang QD, Wang F, Chen T, Xu Q, Lui KO, Molkentin JD, Zhou B.. Preexisting endothelial cells mediate cardiac neovascularization after injury. J Clin Invest 2017;127:2968–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Skelly DA, Squiers GT, McLellan MA, Bolisetty MT, Robson P, Rosenthal NA, Pinto AR.. Single-cell transcriptional profiling reveals cellular diversity and intercommunication in the mouse heart. Cell Rep 2018;22:600–610. [DOI] [PubMed] [Google Scholar]
- 42. Gladka MM, Molenaar B, de Ruiter H, van der Elst S, Tsui H, Versteeg D, Lacraz GPA, Huibers MMH, van Oudenaarden A, van Rooij E.. Single-cell sequencing of the healthy and diseased heart reveals cytoskeleton-associated protein 4 as a new modulator of fibroblasts activation. Circulation 2018;138:166–180. [DOI] [PubMed] [Google Scholar]
- 43. Dejana E, Hirschi KK, Simons M.. The molecular basis of endothelial cell plasticity. Nat Commun 2017;8:14361.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ichimura K, Stan RV, Kurihara H, Sakai T.. Glomerular endothelial cells form diaphragms during development and pathologic conditions. J Am Soc Nephrol 2008;19:1463–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Madden SL, Cook BP, Nacht M, Weber WD, Callahan MR, Jiang Y, Dufault MR, Zhang X, Zhang W, Walter-Yohrling J, Rouleau C, Akmaev VR, Wang CJ, Cao X, St Martin TB, Roberts BL, Teicher BA, Klinger KW, Stan RV, Lucey B, Carson-Walter EB, Laterra J, Walter KA.. Vascular gene expression in nonneoplastic and malignant brain. Am J Pathol 2004;165:601–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stan RV. Endothelial stomatal and fenestral diaphragms in normal vessels and angiogenesis. J Cell Mol Med 2007;11:621–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Strickland LA, Jubb AM, Hongo JA, Zhong F, Burwick J, Fu L, Frantz GD, Koeppen H.. Plasmalemmal vesicle-associated protein (PLVAP) is expressed by tumour endothelium and is upregulated by vascular endothelial growth factor-A (VEGF). J Pathol 2005;206:466–475. [DOI] [PubMed] [Google Scholar]
- 48. Khan S, Taverna F, Rohlenova K, Treps L, Geldhof V, de Rooij L, Sokol L, Pircher A, Conradi LC, Kalucka J, Schoonjans L, Eelen G, Dewerchin M, Karakach T, Li X, Goveia J, Carmeliet P.. EndoDB: a database of endothelial cell transcriptomics data. Nucleic Acids Res 2018;47:D736–D744. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.