Abstract
The developmental biology of neural crest cells in humans remains unexplored due to technical and ethical challenges. The use of pluripotent stem cells to model human neural crest development has gained momentum. We recently introduced a rapid chemically defined approach to induce robust neural crest by WNT/ β-CATENIN activation. Here we investigate the temporal requirements of ectopic WNT activation needed to induce neural crest cells. By altering the temporal activation of canonical WNT/ β-CATENIN with a GSK3 inhibitor we find that a 2 Day pulse of WNT/β-CATENIN activation via GSK3 inhibition is optimal to generate bona fide neural crest cells, as shown by their capacity to differentiate to neural crest specific fates including peripheral neurons, glia, melanoblasts and ectomesenchymal osteocytes, chondrocytes and adipocytes. Although a 2 Day pulse can impart neural crest character when GSK3 is inhibited days after seeding, optimal results are obtained when WNT is activated from the beginning, and we find that the window of competence to induce NCs from non-neural ectodermal/placodal precursors closes by day 3 of culture. The reduced requirement for exogenous WNT activation offers an approach that is cost effective, and we show that this adherent 2-dimensional approach is efficient in a broad range of culture platforms ranging from 96-well vessels to 10 cm dishes.
Keywords: human embryonic stem cells, pluripotent stem cells, neural crest, signaling, WNT, CHIR99021, CHIR, GSK3, osteoblast, adipocyte, chondrocyte, smooth muscle, peripheral neurons, glia, melanocytes
Introduction
The specification of neural crest cells has received much attention in model organisms (Pla and Monsoro-Burq, 2018; Stuhlmiller and García-Castro, 2012), and although some molecular players involved in NC biology have conserved expression in human embryos (Betters et al., 2010), both technical and ethical hurdles have precluded our understanding of human neural crest (hNC) embryonic development. Human pluripotent stem cells (hPSC) which include human Embryonic Stem Cells (hESC) and induced Pluripotent Stem Cells (iPSC) offer a great opportunity to advance our understanding of hNC development, and substantial progress has been made since the initial derivation of hNCs from hPSCs co-cultured with stromal cells (Pomp et al., 2005). Subsequent approaches relied on the generation of neurospheres, neural rosettes and embryoid bodies (Bajpai et al., 2010; Brokhman et al., 2008; Curchoe et al., 2010; Jiang et al., 2009; Lee et al., 2007; Liu et al., 2012; Rada-Iglesias et al., 2012; Sparks et al., 2018). But given the expression of paracrine signals of unknown identities elicited under such conditions, the precise contributions of specific molecules required to generate hNCs in these approaches remain enigmatic.
Common to several approaches of hNC induction in chemically defined conditions is the inhibition of TGF-β molecules (either dual BMP and TGF-β inhibition or just TGF-β), and more recently, the inclusion of WNT activation (Chambers et al., 2012; Menendez et al., 2011; Mica et al., 2013). However, these protocols retain the use of serum or serum replacement cocktails with undisclosed components, preventing a thorough understanding of significant molecular contributions during NC formation. Also, the time required to generate hNCs under these protocols varies from 11 days to 3 weeks, with some requiring multiple passages, and thus leading to longer culture times. Finally, the levels of efficiency varied greatly, and some of these approaches actually require enrichment or purification approaches.
Our group recently reported a novel model of hNC formation from hPSCs. This approach provides high efficiencies, takes only 5 days and is based on WNT induction achieved with either WNT3A or the GSK3β inhibitor CHIR99021 (Adachi et al.) (Leung et al., 2016). With the use of this model we investigated the ontogeny of anterior hNC, and suggest that a pre-border state defined by a synexpressed group of early markers precedes the expression of typical neural plate border markers associated with NC (Leung et al., 2016).
Altogether, these models offer a great opportunity to explore basic principles of human NC development and promise to facilitate clinical translational efforts to ameliorate hNC pathologies. Yet a complete understanding of the precise contributions of the multiple components suggested to direct hNC induction is still missing, and improvements to make the methods more effective would be appreciated.
Here we further analyze the temporal dynamics of WNT activation during hNC formation and demonstrate that a pulse, from 0–2 days after hESC dissociation, leads to a strong induction with similar developmental progression. This improved protocol also produces hNCs able to differentiate into expected NC terminal derivatives and is scalable from 96 well plates to 10 cm dishes.
Results
Temporal regimens of CHIR treatment and hNC induction
The inhibition of GSK3β via CHIR is known to elicit WNT signaling by preventing β-CATENIN destruction, and thus enabling its nuclear translocation, where it functions as a transcription factor (Aberle et al., 1997; Ring et al., 2003). Indeed, in our previous study, we demonstrated that hPSCs can be efficiently directed towards a NC fate when cultured with CHIR during just 5 days; and that canonical WNT activation is fundamental in this process, as β-CATENIN knockdown prevents hNC formation (Leung et al., 2016). Yet the optimal temporal dynamics of GSK3β inhibition for hNC induction remain unexplored. Is exogenous WNT signaling required for the full 5 days? Or could a shorter activation render similar or better results? Which time-points are optimal to translate the WNT signal into NC formation?
To address these questions, we started by assessing whether continuous GSK3β inhibition through CHIR99021 was required to induce hNCs. Therefore, we assessed hNC formation from hESCs treated with CHIR for different numbers of consecutive days, resulting in regimens of 0–1, 0–2, 0–3, 0–4, and 0–5 days (D) of CHIR treatment. Regimens other than 0–5D, received medium without CHIR for the rest of the culture until reaching day 5, when all regimens were fixed and evaluated by immunofluorescence for the expression of SOX10 and PAX7, NC markers expressed in human embryos (Betters et al., 2010). As shown in Figure 1A, 0–1D CHIR did not lead to hNC formation. Instead, all other regimens (0–2D to 0–5D CHIR) lead to robust NC formation, showing ≥ 60% SOX10/PAX7 positive cells. Intriguingly, immunofluorescence of 0–2D CHIR seemed slightly better than all other regimens, while comparison of cell counts show minimal differences between 0–2D and 0–5D CHIR treatment for these markers (Figure 1C), and becomes more apparent after separation of the SOX10, PAX7 and DAPI immunofluroescence signals (Figure 1B). Real-time quantitative PCR (RT-qPCR) analysis for several NC markers including SOX10, PAX7, PAX3 and FOXD3 from these samples confirm that unlike 0–1D CHIR (which does not lead to NC formation), 0–2D to 0–5D CHIR treatments rendered robust NC marker expression (Figure 1D), and here the 2D CHIR regimen produced a consistently stronger expression of the NC markers tested. This result suggests that 0–1D treatment is insufficient for NC formation, and that a 2D CHIR treatment is enough to promote robust NC formation, while CHIR treatment longer than 2Ds does not improve the efficiency of NC formation (Figure 1C). With this in mind, we aimed to test if deployment of 2D CHIR treatment at different time points during the 5 Day culture would improve NC formation. Dissociated hESCs were seeded as before, but 2D CHIR treatments were provided on 0–2, 1–3, or 2–4 days after seeding (See schematic, Figure 1E). PAX7 and SOX10 expression was tested after fixation at the end of the fifth day. We found PAX7 and SOX10 expression in all three conditions (Figure 1F, top row). However, quantification of both SOX10+ and PAX7+ nuclei shows a significantly reduced yield after 1–3 and 2–4 D treatments (Figure 1G). In our previous experiments, robust PAX7 and SOX10 appear 5 days after the initial treatment with CHIR, and here we only tested the expression of NC markers on Day 5, but 1–3D and 2–4D regimens only progressed 3 and 4 days after the initiation of WNT treatment respectively. To compensate for this difference, we extended cultures to allow for a full 5-Day period from the initiating time of WNT treatment. To this end, cells were treated with 2D-CHIR on days 1–3, 2–4, or 3–5, and analyzed on days 6, 7, and 8 respectively. Despite the extended culture after CHIR treatment, the best outcome was still the original 0–2 Day treatment (Figure 1F, bottom row and Figure 1G).
Figure1. A 2 Day pulse of CHIR is sufficient to induce hNCs.
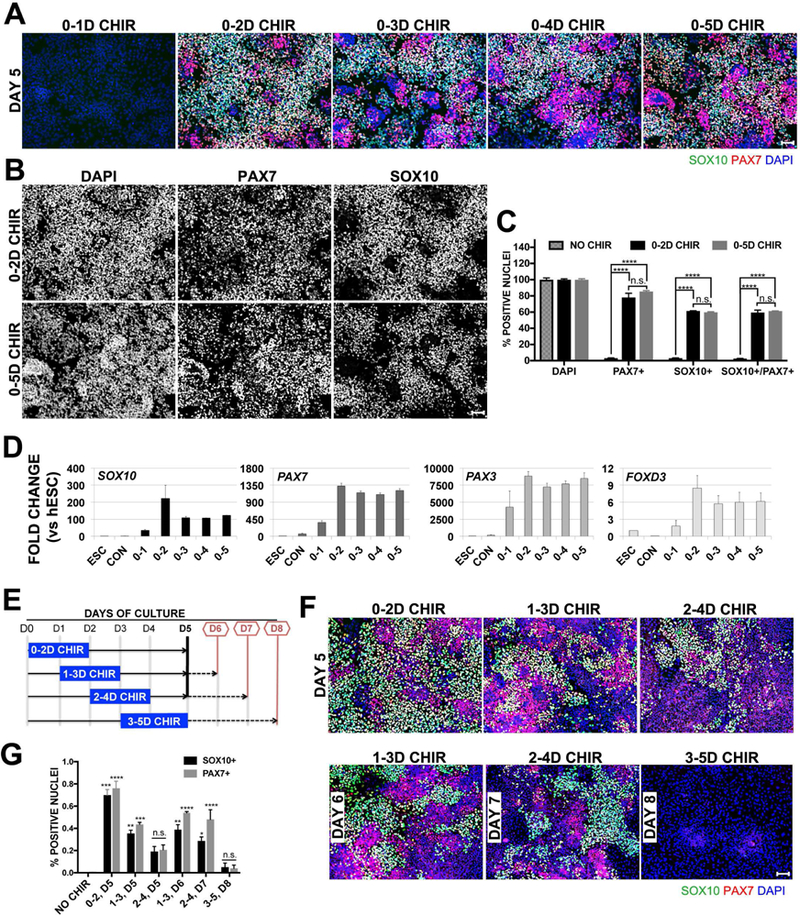
(A) Immunofluroescence, IF, on day 5 for neural crest markers SOX10 (green), PAX7 (red) and 4,6-diamidino-2-phenylindole, DAPI (blue) treated with 3uM CHIR for durations indicated above images. (B) IF channels of 0–2D CHIR and 0–5D CHIR shown in panel A separated for clarity (C) Quantification of IF data for NO CHIR, 0–2D CHIR, or 0–5D CHIR treatment on Day 5 represented as a percentage normalized by total cells counted via DAPI stain. Error bars are ±SEM, Statistical analysis performed by one–way ANOVA. n.s. no significance, **p<0.05, ***p<0.005, ****p<0.0005. Scale bars are 100um. Data are representative of 3 independent experiments. (D) RT-qPCR of NC markers SOX10, PAX7, PAX3 and FOXD3 on day 5. Conditions in x-axis are hESC, NO CHIR (CON), or after 3uM CHIR treatment at (Days) indicated. Fold change is relative to hESCs, error bars are ± SEM (E) Schematic for data presented in panel F. 3uM CHIR was added on days indicated by blue rectangle. First set was examined on Day 5 (solid black line, top row in panel F), second set was evaluated 5 days after CHIR addition (red lines, bottom row in panel F). (F) IF for SOX10 (green), PAX7 (red) and DAPI (blue). Top row are conditions evaluated on day 5, and bottom row are conditions evaluated 5 days after initial addition of CHIR on days 6, 7, or 8. (G) Quantification of IF data shown in panel F. Error bars are ±SEM, Statistical analysis performed by one–way ANOVA, significance is relative to NO CHIR controls on D5. n.s. no significance, * p<0.05, **p<0.05, ***p<0.005, ****p<0.0005. Scale bars are 100um. Data are representative of 3 independent experiments, and RT-qPCR values reflect data from pooled replicates.
Acquisition of NC markers upon 2D and 5D CHIR treatments
In 2D CHIR treatment, GSK3β inhibition is removed 3 days earlier than in 5D treatment, which leads to an apparently slightly stronger NC induction. To better characterize the acquisition of NC character under this culture condition we explored the daily expression of known Canonical WNT targets AXIN2 and SP5 (Jho et al., 2002; Park et al., 2013), during hNC formation under 2D CHIR treatment. The expression of both direct targets increases during the first 2 days of CHIR treatment (Figure 2A). Surprisingly, on the third day, after a whole 24 hours of culture in the absence of CHIR, we found even stronger expression of both genes, which continue to increase at day 4, and was only slightly reduced at day 5 for AXIN2, and more strongly for SP5. These results suggest that the brief pulse of WNT activation during the first 2 days is enough to initiate a sustained activation of endogenous WNT signaling.
Figure 2. The temporal dynamics of hNC differentiation following a short pulse of CHIR administration.
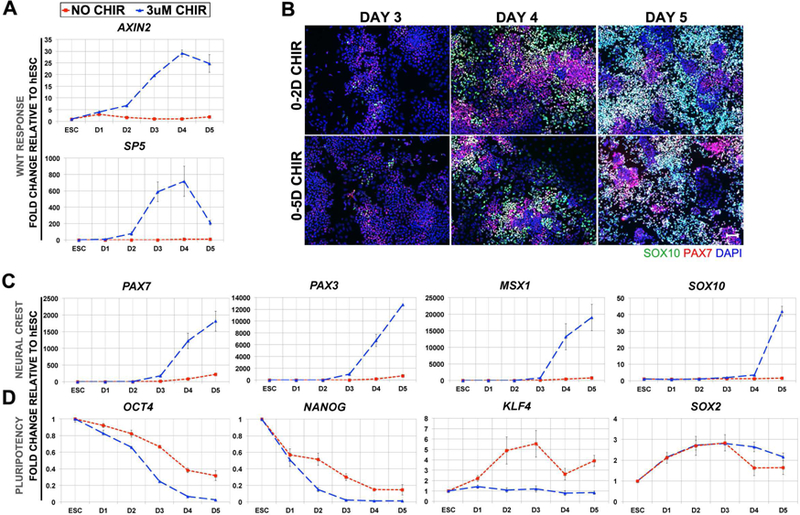
(A) Daily temporal RT-qPCR for WNT-response genes AXIN2 and SP5. (B) Time-course immunofluorescence for SOX10 (green), PAX7 (red) and DAPI (blue) in cells treated with 3uM CHIR for either 0–2 days (top row) or 0–5 days (bottom row) and evaluated on D3, D4, or D5. Scale is 100um. Daily temporal analysis of (C) RT-qPCR for NC genes PAX7, PAX3, MSX1, and SOX10 and (D) RT-qPCR for pluripotency genes OCT4, NANOG, KLF4 and SOX2. Fold changes were calculated relative to hESCs and normalized by BACTIN. NO CHIR controls are represented by red dashed line and 3uM CHIR by blue dashed line. Error bars are ± SEM. IF results are representative of 2 independent experiments and RT-qPCR data are pooled from 3 independent experiments.
As a next step, we assessed possible changes in the progressive acquisition of hNC markers in the 2D CHIR treatment. Here we simultaneously performed 2D- and 5D-CHIR treatments of hESCs and collected cells daily until the end of day 5 and monitored expression of NC markers via immunofluorescence. We previously reported that under the 5D CHIR treatment, a few cells express PAX7 and SOX10 on day 3. We detected a similar acquisition of these markers by immunofluorescence in both conditions through the 5 Day culture period. Robust PAX7 and SOX10 appear in a few cells on day 3, and increasing number of cells express these markers thereafter. From these results, it appears that reducing the temporal treatment of GSK3β inhibition to 2 days does not modify the temporal acquisition of NC markers (Figure 2B). Next, we evaluated the transcriptional levels of NC markers in 2D CHIR treated cells via RT-qPCR. Although there is slight activation of PAX3 and PAX7 in NO CHIR controls by the fifth day, the levels are minimal compared with the robust expression detected in the CHIR treated counterparts (Figure 2C). Furthermore, the levels of all NC markers examined are increased by day 3 in the 2D CHIR treated group, with levels rising gradually as seen by immunofluorescence for PAX7 and SOX10. These trends concur with what we have observed when CHIR is added for the entire 5 days (Leung et al., 2016).
Continuous treatment of hESCs with CHIR for 5 days results in a gradual loss of pluripotency markers OCT4 and NANOG (Leung et al., 2016). Here we extended the analysis to include KLF4 and SOX2, in addition to OCT4 and NANOG. Similar to our reported results with the 5D CHIR treatment, we found with the 2D CHIR treatment, that by the first day of culture there is already a transcriptional decrease of the pluripotency markers OCT4 and NANOG, and that their expression levels continue to be progressively reduced throughout the 5 days of culture. Interestingly, KLF4 expression remains unaltered throughout while SOX2 levels increase gradually but clearly from day 1 onwards, reaching a maximum level by day 3, and then its expression is slightly reduced but remains higher than levels in hESCs by day 5. In addition to other roles in stemness and neural fate, SOX2 expression and function has been characterized in neural crest (Wakamatsu et al., 2004; Wang et al., 2012). It has been reported that Sox2 gets downregulated in neural crest cells (NCs) prior to emigration away from the dorsal neural tube, and overexpression of Sox2 reduces maintenance of NCs, as revealed by reduced expression of Slug/Snai2 and reduction in HNK1 migrating NCs (Wakamatsu et al., 2004). In agreement with these model organism studies, our previous report on human NC formation from hESC confirmed the transient increase of expression of SOX2 (Leung et al., 2016), similar to our current findings. Overall, these results suggest that our treatment conditions result in an altered pluripotent transcriptional profile within the first day (Figure 2D).
Finally, we previously reported that NC cells generated via the 5D CHIR treatment emerge independently from neural ectoderm, with minimal expression of PAX6, the earliest definitive neuroectoderm marker in humans and ESCs (Chambers et al., 2009; Gerrard et al., 2005; Surmacz et al., 2012; Zhang et al., 2010). Specifically, we pointed out that only a very small subset of cells express PAX6 in our cultures, and that these do not overlap with SOX10+ cells. Furthermore, via short hairpin RNAs, we generated PAX6 knock-downs that did not reduce NC formation or expression of NC markers (Leung et al.). To asses if NC treated with 2D CHIR also emerge independently of PAX6 expression, we performed immunostaining of PAX6 and SOX10. As before, only a small number of cells express PAX6, and no overlap was detected between SOX10 and PAX6 in NC cultures generated by 2D CHIR treatment (data not shown). This result confirms our previous findings that early NC independently from neural ectoderm.
hNCs induced with 2D CHIR treatment retain the potential to differentiate to expected terminal derivatives
In order to validate the “NC status” of the cells generated with 2D CHIR treatment, it is fundamental to test their differentiation potential towards NC-terminal derivatives, and because transcriptional profiles of NC markers suggest increased values, we compared their differentiation potential with NCs induced by 5D CHIR treatment. We induced hNCs with either 2D CHIR or 5D CHIR treatment, and at the end of day 5 exposed these cells to defined protocols to evoke terminal differentiation of NC derivatives including osteoblasts, chondrocytes, adipocytes, smooth muscle, peripheral neurons, glia and melanoblasts. As shown in figure 3, hNC generated by both 2D CHIR and 5D CHIR treatment efficiently differentiate into all of the derivatives tested, without any notable difference between these populations. To better establish if the differentiation capacity of the NC induced by either 2D or 5D CHIR treatment was equivalent, we scored the proportion of cells expressing specific markers associated with neural crest terminal derivatives. The percentage of cells expressing differentiation markers (marker/DAPI nuclear counts) for 2D vs 5D CHIR treatment reveal that smooth muscle (SMA: 98.31%; 99.16 %), peripheral neurons (Peripherin: 98.58%; 96.77. HuD: 20.46%; 17.15%), Glia (S100β: 96.62;95.81), and melanoblast (MITF: 85.04%;84.25%. SOX10: 85.97%; 86.99%) arise at similar rates under both NC induction regimes. This strongly suggests that the shortened 2D CHIR treatment generates hNCs with the same multipotent capacity exhibited by NCs formed following 5D CHIR treatment (Leung et al., 2016).
Figure 3. hNCs induced by short pulse of CHIR retain potential for terminal differentiation.
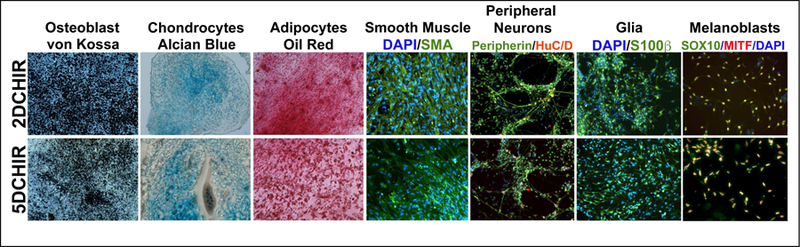
hNCs induced on day 5 following either 2D CHIR or 5D CHIR treatment were subject to terminal differentiation with distinct protocols ranging from 2–6 weeks and assessed histologically by alizarin red for osteoblasts, alcian blue for chondrocytes, and oil red for adipocytes. Smooth muscle was evaluated by Immunofluorescence for smooth muscle actin, SMA (green); peripheral neurons for Peripherin (green) and HuC/D (red); glia by S100B (green), and Melanoblasts by SOX10 (green) and MITF (red).
hNC induction via 2D CHIR treatment is effective in multiple culture platforms
Whether our human NC induction approach is scalable is an important issue worth addressing in order to provide flexibility across multiple platforms and avenues of research. We therefore examined whether it is possible to obtain hNC induction in vessels of different dimensions following 2D CHIR treatment. We tested whether we could induce hNCs in different volumes with our fixed seeding densities and 3µM CHIR concentration in culture vessels with surface areas as low as 0.32 cm2 in 96-well plates through 78.5 cm2 in 10cm dishes. Evaluation by immunofluorescence on day 5 of culture with SOX10 and PAX7 reveal the successful induction of NCs in hESCs treated with CHIR in all formats tested (Figure 4). Similar experiments with other NC markers like ETS1 further support our results (data not shown). This demonstrates the amenability of these culture conditions to produce scalable quantities of human neural crest. Moreover, the capacity of this method to generate hNCs in 96-wells will facilitate high throughput screens for basic research and translational efforts alike.
Figure 4. hNCs can be generated via 2D-CHIR in platforms of various dimensions.
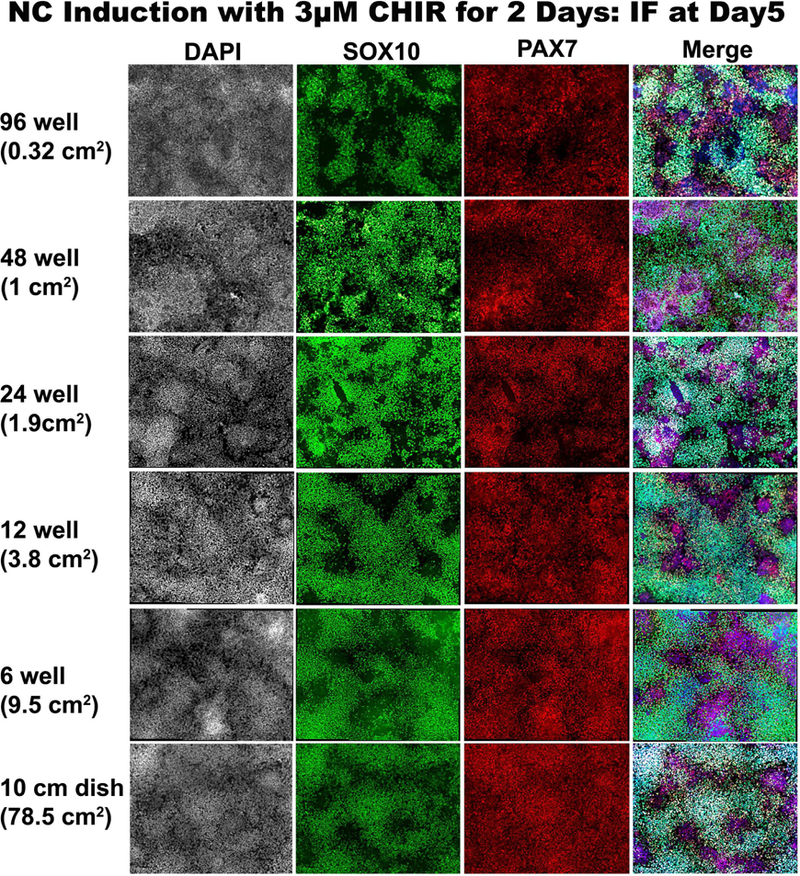
SOX10 (green), and PAX7 (red) immunofluorescence of day 5 cultures treated with 2D 3uM CHIR on days 0–2. Images are from 96-well, 48-well, 24-well, 12-well, 6-well and 10cm dish formats. DAPI counterstain is grey on left column and blue in right merged column.
Discussion
We have found here that a 48-hour pulse of WNT activation between 0 and 48 hours is sufficient to direct hESCs toward the NC fate, suggesting that the critical period for the induction of hNCs from hESCs lies within the first 2 days of differentiation. Under the 2D-CHIR protocol, hNCs express known NC genes like PAX3, PAX7, FOXD3, and SOX10. It is intriguing that while we removed ectopic WNT activation after 2 days under this new regimen, the expression of SOX10 and PAX7 appear in a similar time frame as we reported under the 5D-CHIR regimen (Leung et al., 2016). We note that in the 2D CHIR protocol, direct WNT/β-CATENIN targets like AXIN2 and SP5 continued to increase during the following days, suggesting that endogenous WNT activation is sustained after CHIR removal. This may very well explain why the 2D and 5D CHIR regimens perform in a similar fashion. Furthermore, the hNC we obtain after 5 days not only express NC markers, they are also able to adopt NC-terminal derivative fates of both neurogenic and mesenchymal character including peripheral neurons, glia, melanocytes, osteoblasts, smooth muscle, chondrocytes and adipocytes, in agreement with their proposed NC status.
The 2D-CHIR regimen is also sufficient to trigger hNC formation when applied to cells on the 1st and 2nd days of culture, although at reduced efficiencies. This posits that the cells remain responsive to WNT signaling to induce the expression of SOX10 and PAX7 in adherent cells until the 2nd day of culture. These cells have been dissociated and plated in DMEM with B27 media, but without CHIR, and we previously showed that these conditions trigger hESC differentiation towards a non-neural/placodal ectoderm fate. Instead, when 2D-CHIR treatment was deployed on the 3rd or 4th day of culture, no NC where formed. It seems possible then, that non-neural ectoderm precursors retain the plasticity to form NC under WNT induction during the early days of their differentiation.
What factors enable the responsiveness of these cells to form NC upon WNT activation is not yet known. OCT4- β-CATENIN interaction is fundamental for mesoderm formation (Funa et al., 2015), and it is appealing to suggest a possible role during NC formation as well, given that the hESCs express high levels of OCT4. Interestingly, the expression of OCT4 is gradually reduced from the hESC levels through the differentiation of both NC and control cells (non-neural ectoderm precursors) and it is still detectable until the 3rd and 4th day of culture (Leung et al., 2016). However, as we stated above, CHIR treatment is unable to elicit NC at these later time-points and thus we suggest that the reduced expression levels of OCT4 found at days 3 and 4 is insufficient to provide “responsiveness” to CHIR towards the NC lineage.
Interestingly, within the dual SMAD inhibition protocol proposed by the Studer group, they also found beneficial effects of WNT activation. Furthermore, while testing for optimal temporal deployment of WNT activation, they found that a short pulse of WNT was sufficient to induce NCs, but a longer exposure to WNT lead to better outcomes. Surprisingly, in their approach CHIR activation from the beginning prevents NC formation (Mica et al., 2013).
Our hNC protocol requires less culture time than those reported by other groups, cutting down on expensive culture reagents. Furthermore, we have demonstrated that our 2D-CHIR protocol can effectively lead to hNC formation in a wide range of culture scales, rendering above 60% PAX7/ SOX10 double positive cells from large 10cm2 plates, to small 96 well plates. These ranges are ideal for protein characterization and enable high-throughput approaches for chemical and genomic screens alike.
In conclusion, here we show that 48hr inhibition of GSK3 leads to robust NC induction from hESCs in just 5 days with high efficiency (>60% of SOX10, PAX7 double positive cells), that progressively acquire NC markers, and effectively differentiate to expected NC terminal derivatives. Furthermore, we show that this approach allows a range of culture dimensions from high-throughput 96 well plates to 10 cm dishes. This more streamlined and cost-effective protocol is well suited for a variety of applications to advance our understanding of human NC development and NC associated pathologies.
Methods
RT-qPCR
Total RNA was collected in TRIzol (Thermo Fisher Scientific), cDNA was prepared with Applied Biosystems High Capacity cDNA Reverse Transcription Kit (Fisher Scientific), and RT-qPCR was performed with SYBR Premix Ex Taq II (Clontech) on an Applied Biosystems Step One Plus system. Fold-Changes were evaluated by the ∆∆-Ct method and plotted on Excel or Graphpad prism. qPCR primers used: BACTIN:(F)TGAACCCCAAGGCCAACCGC (R)GACCCCGTCACCGGAGTCCA
SOX10:(F)CCTTCATGGTGTGGGCTC (R)CGCTTGTCACTTTCGTTCAG
PAX7:(F)TGACAGCTTCATGAATCCGG (R)GATGGAGAAGTCAGCCTGTG
PAX3:(F)GGCTTTCAACCATCTCATTCCCG (R)GTTGAGGTCTGTGAACGGTGCT
FOXD3:(F)GCATCTGCGAGTTCATCAGC (R)CGTTGAGTGAGAGGTTGTGG
MSX1:(F)GCTCGTCAAAGCCGAGAG (R)ACGGTTCGTCTTGTGTTTGC
OCT4:(F)GAGAAGGAGAAGCTGGAGCA (R)CTTCTGCTTCAGGAGCTTGG
NANOG:(F)GATTTGTGGGCCTGAAGAAA (R)CAGATCCATGGAGGAAGGAA
KLF4:(F)ACCCACACAGGTGAGAAACC (R)ATGCTCGGTCGCATTTTTGG
SOX2:(F)TCAAGCGGCCCATGAATGCC (R)AGCCGCTTAGCCTCGTCGAT
AXIN2:(F)CGGGAGCCACACCCTTCT (R)TGGACACCTGCCAGTTTCTTT
SP5:(F)CTTCGGGTGTCCATGCCTC (R)GTGCGGTCCTGGAGAAAGG
Immunohistochemistry
Cells were fixed in 4% paraformaldehyde, rinsed in PBS, permeabilized with 0.4% triton X100, blocked with 4% fetal bovine serum and incubated with primaries overnight. Alexa Fluor conjugated secondary antibodies (Invitrogen) were used at 1:2000 dilution. Mouse anti-SOX10 antibody, SC271163 (Santa Cruz Biotechnology) was used at 1:200. Mouse anti-PAX7 antibody from Developmental Studies Hybridoma Bank was used at 1:200.
Cell Counts
Immunofluorescence images taken on NIKON eclipse 80i were converted to nd2 format and counted on NIS-Elements AR Analysis software (version 4.6). Nuclei were counted by clustered method at 5px/spot, at a typical diameter of 10um, with variable contrast levels and dark objects removed. Statistical analysis of cell counts and graph was performed on Graphpad Prism7.
Microscopy
Images were captured on a Nikon eclipse 80i microscope on a Spot SE camera and software. 96-well images were captured on an inverted Nikon Eclipse Ti microscope with NIS Elements software. All images were compiled and adjusted in Adobe Photoshop CS5.
Neural Crest Differentiation
Human embryonic stem cells line H1 (WA01) obtained from WiCell Research Institute, Inc. (Madison WI, USA), was maintained in mTeSR1 (Stem Cell Technologies) on matrigel coated dishes at 37°C in 5% CO 2, 5% O2 and passaged regularly with Dispase (Stem Cell Technologies) or Versene (Thermo Fisher Scientific). For hNC differentiation, hESCs were collected 4 to 5 days after last passage at 80% to 90% confluency. Cells were first rinsed 3 times in 1X PBS Ca+ and Mg+ free (Thermo Scientific, Cat. No. 14190144), then dissociated in Accutase (Stem Cell Technologies) for 4 minutes 30 seconds at 37°C in 5% CO 2, 5% O2. Accutase was immediately quenched with warmed 1X DMEM/F12 (Invitrogen, Cat. No. 17504–044) containing 10uM Rock Inhibitor, Y-27632 (Tocris, Cat. No. 1254), then cells were centrifuged 1200 RPM for 4 minutes at room temperature. 1X DMEM/F12 was discarded leaving cell pellet in-tact, then replaced with NC Induction media [1X DMEM/F12 (Invitrogen Cat. No. 11320), 1X serum-free B27 supplement (Invitrogen, Cat. No. 17504–044), 1X Glutamax (Thermo fisher scientific, Cat. No. 35050061), 0.5% BSA (Sigma, A7979)] containing 10uM Y-27632. hESC clusters were further dissociated to single cells mechanically by trituration through a 5mL serological pipette a total of 20 times. Cells were counted on a hemocytometer, diluted to an optimal seeding density of 20 X 103 cells/cm2 per culture vessel, then seeded on culture vessels pre-coated with Matrigel (BD Matrigel hESC-qualified Matrix Cat. No. 354277). CHIR99021 (Tocris, Cat. No. 4423) was added to cells in induction media during trituration to a final concentration of 3uM prior to seeding. 10uM Rock Inhibitor, Y-27632, was added from days 0–2 in all cases and left out of induction media during remainder of culture. Induction media was changed daily until day of collection for further analysis. Cells were cultured at 37°C in 5% CO2, 5% O2 throughout the procedure. Note, the CHIR99021 concentration required to elicit optimal NC induction efficiency is sensitive to the concentration of CHIR used, which varies depending on the different lots of CHIR99021, resuspension, pipetting, etc., We obtained optimal inductions at ranges ~2.5uM – 3uM, but report all results at 3uM for consistency. This parameter needs to be tested for each lot prepared.
Terminal differentiation protocols for neural crest derivatives
All protocols were adapted and derived from Studer lab group, except chondrocyte differentiation. (Chambers et al., 2012; Lee et al., 2007; Mica et al., 2013). Osteogenic differentiation: Neural crest cells are plated at 300K/cm2 in osteogenic differentiation media (10 mM β-glycerol phosphate, 0.1 µM dexamethasone and 200 µM AA in α-MEM medium containing 10% FBS) and media is changed every 2–3 days for 21 days when calcification can be seen. Cells were stained with alizarin red for calcification. Chondrogenic differentiation: Mesencult-Chondrogenic Differentiation media (Stem cell technologies) was used as per the supplier’s prescribed protocol. Chondrocyte pellets at the end of the protocol were paraffin sectioned and stained with Alcian blue. Adipogenic differentiation: Neural crest cells at day 5 were dissociated with accutase and plated at 50K/cm2 density in adipogenic media (1 mM dexamethasone, 10 µg/ml Insulin and 0.5 mM IBMX in α-MEM medium containing 10% FBS) with media changes every 2–4 days for 21 days and then stained with oil red. Myofibroblast differentiation: Neural crest cells at day 5 were dissociated and plated at 50K/cm2 density in alpha-MEM with 10%FBS for 14 days. Cells were stained with smooth muscle actin (Sigma, A2547 at 1:500) and S100β (abcam, ab52642 at 1:200). Peripheral neurons differentiation: On day 5 of NC induction, nociceptor induction was initiated with the addition of perineural media (3 µM CHIR99021, 10 µM SU5402 and 10 µM DAPT, in N2/DMEM/F12). Cells were harvested after 5 days to test for Peripherin (Millipore AB1530 at 1:200) and HuC/D (Molecular Probes, A-21271 at 1:300). Glial cell differentiation: NC cells on day 5 were plated on to PO/Lamini/Fibronectin coated wells in DMEM/F12/N2 medium supplemented with 10 ng/ml of FGF2 and 10 ng/ml of EGF for 14 days. After FGF2/EGF culture, cells are differentiated towards Glia in N2 medium supplemented with ciliary neurotrophic factor (10 ng/ml), neuregulin (20 ng/ml), FGF2 (10 ng/ml) and cyclic AMP (0.5 mM) for 21 days. Cells were stained with S100β. Melanocyte differentiation: On day5 of NC induction, melanocyte differentiation media was added (25 ng/ml BMP4 and 100 nM EDN3 in DMEM/F12/N2) for 5 days. Cells were further passed on to PO/Lamini/Fibronectin coated wells in Mel Maturation media (Neurobasal/Mel Media as described in Mica et al., 2013) for another 2–3 passage then stained for Sox10/MITF (R&D systems, AF5769 at 1:100). Differentiation percentages were calculated by measuring the number of cells expressing specific terminal differentiation markers, comparing positive cells vs DAPI nuclear counts, for smooth muscle, peripheral neurons, glia and melanoblasts. A minimum of 3 frames were scored per marker, from 2 biological replicas of terminal differentiation experiments from NC induced under either 2D or 5D CHIR regimes.
Highlights.
A pulse of exogenous WNT activation for 2 days is sufficient to induce human neural crest formation from human pluripotent stem cells in chemically defined conditions
Human neural crest induced by this protocol can be differentiated to derivatives expected of cranial neural crest cells
This protocol is efficient in a broad range of culture platforms
Acknowledgements
We thank Jacqueline Cely and Man Wong for technical assistance, and the reviewers for their helpful questions. This work was funded by National Institute of Health (grant R01DE017914 to M.I.G.C)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aberle H, Bauer A, Stappert J, Kispert A, Kemler R, 1997. beta-catenin is a target for the ubiquitin-proteasome pathway. The EMBO Journal 16, 3797–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi K, Suemori H, Yasuda S-Y, Nakatsuji N, Kawase E, 2010. Role of SOX2 in maintaining pluripotency of human embryonic stem cells. Genes to cells : devoted to molecular & cellular mechanisms 15, 455–470. [DOI] [PubMed] [Google Scholar]
- Bajpai R, Chen DA, Rada-Iglesias A, Zhang J, Xiong Y, Helms J, Chang C-P, Zhao Y, Swigut T, Wysocka J, 2010. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature 463, 958–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betters E, Liu Y, Kjaeldgaard A, Sundström E, García-Castro MI, 2010. Analysis of early human neural crest development. Developmental Biology 344, 578–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brokhman I, Lina G-Z, Oz P, Aharonowiz M, Reubbinoff BE, Goldstein RS, 2008. Peripheral sensory neurons differentiate from neural precursors derived from human embryonic stem cells. Differentiation 76, 145–155. [DOI] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L, 2009. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nature Biotechnology 27, 275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Qi Y, Mica Y, Lee G, Zhang X-J, Niu L, Bilsland J, Cao L, Stevens E, Whiting P, Shi S-H, Studer L, 2012. Combined small-molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nature Biotechnology 30, 715–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curchoe CL, Maurer J, McKeown SJ, Cattarossi G, Cimadamore F, Nilbratt M, Snyder EY, Bronner-Fraser M, Terskikh AV, 2010. Early acquisition of neural crest competence during hESCs neuralization. PLoS ONE 5, e13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funa NS, Schachter KA, Lerdrup M, Ekberg J, Hess K, Dietrich N, Honoré C, Hansen K, Semb H, 2015. β-Catenin Regulates Primitive Streak Induction through Collaborative Interactions with SMAD2/SMAD3 and OCT4. Cell Stem Cell 16, 639–652. [DOI] [PubMed] [Google Scholar]
- Gerrard L, Rodgers L, Cui W, 2005. Differentiation of human embryonic stem cells to neural lineages in adherent culture by blocking bone morphogenetic protein signaling. Stem cells (Dayton, Ohio) 23, 1234–1241. [DOI] [PubMed] [Google Scholar]
- Jho E.-h., Zhang T, Domon C, Joo C-K, Freund J-N, Costantini F, 2002. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Molecular and Cellular Biology 22, 1172–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Gwye Y, McKeown SJ, Bronner-Fraser M, Lutzko C, Lawlor ER, 2009. Isolation and characterization of neural crest stem cells derived from in vitro-differentiated human embryonic stem cells. Stem cells and development 18, 1059–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Kim H, Elkabetz Y, Al Shamy G, Panagiotakos G, Barberi T, Tabar V, Studer L, 2007. Isolation and directed differentiation of neural crest stem cells derived from human embryonic stem cells. Nature Biotechnology 25, 1468–1475. [DOI] [PubMed] [Google Scholar]
- Leung AW, Murdoch B, Salem AF, Prasad MS, Gomez GA, García-Castro MI, 2016. WNT/β-catenin signaling mediates human neural crest induction via a pre-neural border intermediate. Development 143, 398–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Spusta SC, Mi R, Lassiter RNT, Stark MR, Höke A, Rao MS, Zeng X, 2012. Human neural crest stem cells derived from human ESCs and induced pluripotent stem cells: induction, maintenance, and differentiation into functional schwann cells. Stem cells translational medicine 1, 266–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez L, Yatskievych TA, Antin PB, Dalton S, 2011. Wnt signaling and a Smad pathway blockade direct the differentiation of human pluripotent stem cells to multipotent neural crest cells. Proceedings of the National Academy of Sciences of the United States of America 108, 19240–19245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mica Y, Lee G, Chambers SM, Tomishima MJ, Studer L, 2013. Modeling neural crest induction, melanocyte specification, and disease-related pigmentation defects in hESCs and patient-specific iPSCs. CellReports 3, 1140–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D-S, Seo J-H, Hong M, Bang W, Han J-K, Choi S-C, 2013. Role of Sp5 as an essential early regulator of neural crest specification in xenopus. Developmental Dynamics 242, 1382–1394. [DOI] [PubMed] [Google Scholar]
- Pla P, Monsoro-Burq AH, 2018. The Neural Border: Induction, Specification and Maturation of the territory that generates Neural Crest cells. Developmental Biology [DOI] [PubMed] [Google Scholar]
- Pomp O, Brokhman I, Ben-Dor I, Reubinoff B, Goldstein RS, 2005. Generation of peripheral sensory and sympathetic neurons and neural crest cells from human embryonic stem cells. Stem cells (Dayton, Ohio) 23, 923–930. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Prescott S, Brugmann SA, Swigut T, Wysocka J, 2012. Epigenomic annotation of enhancers predicts transcriptional regulators of human neural crest. Cell Stem Cell 11, 633–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring DB, Johnson KW, Henriksen EJ, Nuss JM, Goff D, Kinnick TR, Ma ST, Reeder JW, Samuels I, Slabiak T, Wagman AS, Hammond M-EW, Harrison SD, 2003. Selective Glycogen Synthase Kinase 3 Inhibitors Potentiate Insulin Activation of Glucose Transport and Utilization In Vitro and In Vivo. Diabetes 52, 588–595. [DOI] [PubMed] [Google Scholar]
- Sparks NRL, Martinez IKC, Soto CH, Zur Nieden NI, 2018. Low Osteogenic Yield in Human Pluripotent Stem Cells Associates with Differential Neural Crest Promoter Methylation. Stem cells (Dayton, Ohio) 36, 349–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuhlmiller TJ, García-Castro MI, 2012. Current perspectives of the signaling pathways directing neural crest induction. Cellular and molecular life sciences : CMLS 69, 3715–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surmacz B, Fox H, Gutteridge A, Fish P, Lubitz S, Whiting P, 2012. Directing differentiation of human embryonic stem cells toward anterior neural ectoderm using small molecules. Stem cells (Dayton, Ohio) 30, 1875–1884. [DOI] [PubMed] [Google Scholar]
- Wakamatsu Y, Endo Y, Osumi N, Weston JA, 2004. Multiple roles of Sox2, an HMG-box transcription factor in avian neural crest development. Developmental Dynamics 229, 74–86. [DOI] [PubMed] [Google Scholar]
- Wang Z, Oron E, Nelson B, Razis S, Ivanova N, 2012. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell 10, 440–454. [DOI] [PubMed] [Google Scholar]
- Zhang X, Huang CT, Chen J, Pankratz MT, Xi J, Li J, Yang Y, Lavaute TM, Li X-J, Ayala M, Bondarenko GI, Du Z-W, Jin Y, Golos TG, Zhang SC, 2010. Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell 7, 90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]


