Abstract
The function and evolution of eukaryotic cells depend upon direct molecular interactions between gene products encoded in nuclear and cytoplasmic genomes. Understanding how these cytonuclear interactions drive molecular evolution and generate genetic incompatibilities between isolated populations and species is of central importance to eukaryotic biology. Plants are an outstanding system to investigate such effects because of their two different genomic compartments present in the cytoplasm (mitochondria and plastids) and the extensive resources detailing subcellular targeting of nuclear-encoded proteins. However, the field lacks a consistent classification scheme for mitochondrial- and plastid-targeted proteins based on their molecular interactions with cytoplasmic genomes and gene products, which hinders efforts to standardize and compare results across studies. Here, we take advantage of detailed knowledge about the model angiosperm Arabidopsis thaliana to provide a curated database of plant cytonuclear interactions at the molecular level. CyMIRA (Cytonuclear Molecular Interactions Reference for Arabidopsis) is available at http://cymira.colostate.edu/ and https://github.com/dbsloan/cymira and will serve as a resource to aid researchers in partitioning evolutionary genomic data into functional gene classes based on organelle targeting and direct molecular interaction with cytoplasmic genomes and gene products. It includes 11 categories (and 27 subcategories) of different cytonuclear complexes and types of molecular interactions, and it reports residue-level information for cytonuclear contact sites. We hope that this framework will make it easier to standardize, interpret, and compare studies testing the functional and evolutionary consequences of cytonuclear interactions.
Keywords: chloroplast, cytonuclear coevolution, mitochondria, OXPHOS, photosynthesis
Introduction
The endosymbiotic history of eukaryotes has resulted in cells that are operated under divided genetic control between nuclear and cytoplasmic (i.e., mitochondrial and plastid) genomes. Core eukaryotic functions depend on integration and coevolution between these genomic compartments. The level of integration extends down to direct molecular interactions within multisubunit enzyme complexes (Rand et al. 2004). For example, the major enzymes in mitochondria and plastids such as oxidative phosphorylation (OXPHOS) complexes, photosynthetic machinery, and ribosomes are “chimeric” in the sense that they are composed of gene products from both nuclear and cytoplasmic genomes. This organization reflects an evolutionary history in which many of the genes ancestrally present in cytoplasmic genomes have been replaced by a combination of gene transfer to the nucleus and substitution by existing nuclear genes (Sloan et al. 2018). There are also extensive interactions between cytoplasmic RNAs and nuclear-encoded proteins that are responsible for posttranscriptional processes, such as transcript end-processing, intron splicing, RNA editing, base modifications, and tRNA aminoacylation (Germain et al. 2013; Salinas-Giegé et al. 2015). Furthermore, many nuclear-encoded proteins must directly interact with the cytoplasmic genomes themselves to mediate processes of DNA replication, repair, recombination, and transcription (Zhang et al. 2016; Gualberto and Newton 2017).
The intimacy of these interactions has made them an attractive arena for studying molecular coevolution, especially because they can elucidate the consequences of genes evolving in very different genomic contexts (e.g., differences in mutation rates, replication and expression mechanisms, frequency of recombination, effective population sizes, and modes of inheritance). Not surprisingly, disruption of cytonuclear interactions can have significant functional consequences, and genetic incompatibilities between nuclear and cytoplasmic genomes contribute to reproductive isolation in many systems (Burton et al. 2013; Hill 2015; Sloan et al. 2017). It is possible that cytonuclear incompatibilities evolve at a faster pace than nuclear–nuclear incompatibilities because of the differences in genome evolution and the conflicting genealogical histories that can often distinguish these compartments (Burton and Barreto 2012; Toews and Brelsford 2012).
To test such hypotheses, it is often useful to compare nuclear-encoded proteins that are involved in direct cytonuclear molecular interactions against relevant “control” proteins. For example, classic studies in animals have taken advantage of OXPHOS complex II (succinate dehydrogenase), which is entirely nuclear-encoded, in order to make comparisons with the other OXPHOS complexes, which are all chimeric (Ellison and Burton 2006). In the current genomic era, it has become increasingly popular for evolutionary studies to partition nuclear gene content into categories based on whether they are targeted to mitochondria/plastids and whether they are involved in direct molecular interactions with cytoplasmic genomes and gene products within these organelles (Barreto and Burton 2013; Rogell et al. 2014; Pett and Lavrov 2015; Sloan et al. 2015; Zhang et al. 2015, 2016; Adrion et al. 2016; Rockenbach et al. 2016; Weng et al. 2016; Eslamieh et al. 2017; Havird et al. 2017; Sharbrough et al. 2017; Barreto et al. 2018; Forsythe et al. 2018; Morales et al. 2018; Ferreira et al. 2019; Li et al. 2019; Yan et al. 2019; Zaidi and Makova 2019). Such approaches are an effective means to investigate the evolutionary effect of organelle targeting and molecular interactions. Because plants contain two endosymbiotically derived organelles, they are an especially appealing system in which to study such questions. However, comparing across studies can be challenging because of the variable ways in which authors classify and partition gene sets. Although there are many excellent databases with gene-specific information on subcellular targeting in plants (table 1), none of these provide comprehensive information about direct cytonuclear interactions at the level of protein subunits and amino-acid residues. To address this limitation, we have taken advantage of the extensive work on cytonuclear biology in the model angiosperm Arabidopsis thaliana to create the Cytonuclear Molecular Interactions Reference for Arabidopsis (CyMIRA).
Table 1.
Set of Nine Databases with Information on Subcellular Localization of Proteins in Plants That Were Used for Automated Curations of Targeting Predictions
| Database | Targeting Predictions |
References | ||
|---|---|---|---|---|
| Mito | Plastid | Dual | ||
| SUBA predicted | 2,370 | 2,644 | 97 | Hooper et al. (2017) |
| eSLDB | 848 | 4,427 | 69 | Pierleoni et al. (2007) |
| PA-GOSUB | 985 | 730 | 14 | Lu et al. (2005) |
| SUBA experimental | 1,217 | 2,128 | 785 | Hooper et al. (2017) |
| SWISS PROT | 311 | 657 | 20 | Boutet et al. (2007) |
| TAIR | 397 | 1,598 | 266 | Reiser et al. (2017) |
| LocDB | 446 | 1,527 | 234 | Rastogi and Rost (2011) |
| PPDB | 327 | 1,570 | 73 | Sun et al. (2009) |
| Organelle DB | 512 | 276 | 11 | Wiwatwattana et al. (2007) |
Note.—Counts reflect number of genes in each targeting category.
Results and Discussion
CyMIRA is a detailed curation of A. thaliana cytonuclear interactions at the molecular level, which is available as Supplementary Material online with this article (supplementary file S1, Supplementary Material online). Future updates will be disseminated via GitHub (https://github.com/dbsloan/cymira), and we have also generated a queryable web interface to extract specific subsets of the data: http://cymira.colostate.edu/.
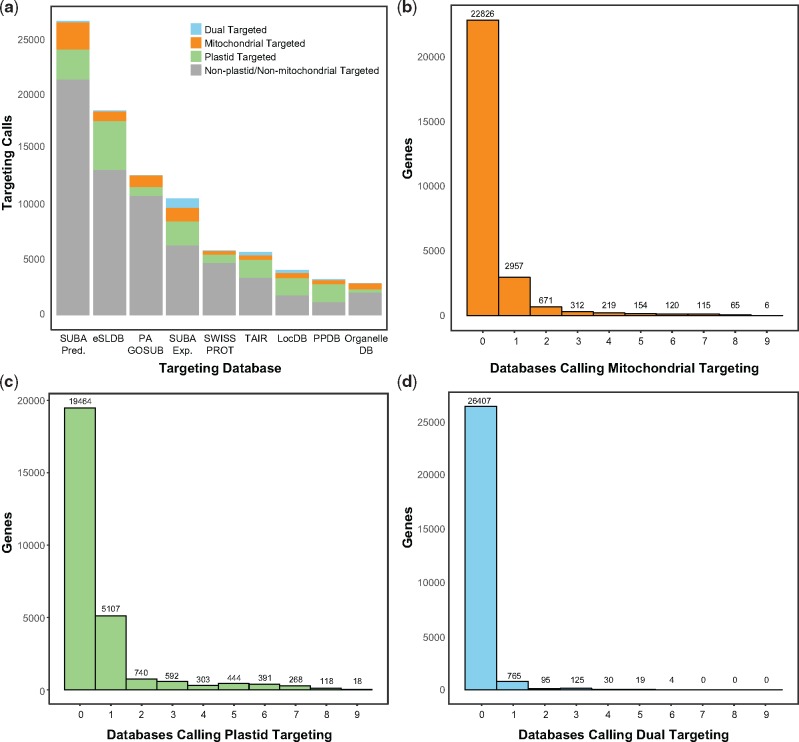
Our initial automated predictions of organelle targeting based on nine existing databases (table 1) identified a total of 4,130 nuclear-encoded protein-coding genes (1,256 mitochondrial-localized, 2,468 plastid-localized, and 406 dual-localized). The sampled databases differed greatly in their number of organelle-targeting predictions, and very few genes shared the same prediction across all nine databases (fig. 1 and supplementary table S1, Supplementary Material online). Because we limited our classification to predictions shared by at least two databases, there were thousands of genes that were excluded because they had a mitochondrial or plastid targeting prediction in only a single database (fig. 1). As such, taking the full union of predictions across all nine databases would have massively exceeded typical estimates of mitochondrial and plastid proteome content. There are likely multiple factors that contribute to the substantial differences in predictions among databases. First, many databases utilize distinct methods for inferring subcellular localization (e.g., fluorescent protein activity vs. mass spectrometry vs. targeting peptide identification), each of which may differ in sensitivity and/or bias. Second, many of the proteins targeted to the organelles also serve other functions inside the cell, resulting in ambiguities in how to apply a classification scheme. Finally, some of the databases include a relatively small number of organelle-targeting predictions and, therefore, may be applying a conservative approach that is likely to miss many genes with mitochondrial and/or plastid localization (supplementary text, Supplementary Material online).
Fig. 1.
—Summary of nine existing databases on subcellular protein targeting plants that were used to generate our automated targeting predictions.
Subsequent manual curation of proteins with direct cytonuclear interactions led to the inclusion of 138 new genes and changed the prediction for six genes that were initially identified as targeting just one organelle to dual targeting. As a result, our final organelle targeting count was 4,268 with 1,337 mitochondrial, 2,495 plastid, and 436 dual. Of these, 910 were classified as being involved in direct cytonuclear molecular interactions, meaning that they are components of chimeric cytonuclear enzyme complexes or directly interact with cytoplasmic DNA and/or RNA transcripts (table 2). The majority of genes involved in these direct cytonuclear interactions were characterized as exclusively mitochondrial (535) or plastid (293), but there are also 82 dual targeted genes in this group, many of which are involved in DNA recombination/replication/repair, tRNA aminoacylation, and posttranscriptional RNA modifications (supplementary file S1, Supplementary Material online).
Table 2.
List of Functional Categories Used in Manual Curation of Direct Cytonuclear Interactions
| Category | Subcategory | Mito | Plastid | Dual | Key Reference(s) |
|---|---|---|---|---|---|
| ACCase | 0 | 4 | 0 | Sasaki and Nagano (2004) | |
| Chlororibosome | 0 | 42 | 0 | Bonen and Calixte (2006); Sloan et al. (2014) | |
| Large subunit | 0 | 31 | 0 | Bieri et al. (2017) | |
| Small subunit | 0 | 11 | 0 | Tiller et al. (2012) | |
| Clp protease | 0 | 15 | 0 | Nishimura and van Wijk (2015) | |
| DNA-RRR | 11 | 8 | 17 | Zhang et al. (2016); Gualberto and Newton (2017) | |
| TAT complex | 1 | 0 | 0 | Carrie et al. (2016) | |
| Mitoribosome | 88 | 0 | 0 | Waltz et al. (2019) | |
| Large subunit | 41 | 0 | 0 | ||
| Small subunit | 47 | 0 | 0 | ||
| OXPHOS | 91 | 0 | 0 | Senkler et al. (2017) | |
| Complex I | 48 | 0 | 0 | ||
| Complex III | 14 | 0 | 0 | ||
| Complex IV | 14 | 0 | 0 | ||
| Complex V | 15 | 0 | 0 | ||
| Photosynthesis | 0 | 67 | 0 | ||
| ATP synthase | 0 | 3 | 0 | Friso et al. (2004) | |
| Cytochrome b6f | 0 | 2 | 0 | Friso et al. (2004) | |
| NDH | 0 | 18 | 0 | Shikanai (2016) | |
| PSI | 0 | 18 | 0 | Jensen et al. (2007) | |
| PSII | 0 | 22 | 0 | van Bezouwen et al. (2017) | |
| Rubisco | 0 | 4 | 0 | Izumi et al. (2012) | |
| PPR | 308 | 110 | 36 | Cheng et al. (2016) | |
| Transcription and transcript maturation | 33 | 46 | 5 | ||
| Intron splicing | 7 | 7 | 1 | de Longevialle et al. (2010) | |
| mTERF | 17 | 11 | 0 | Shevtsov et al. (2018) | |
| RNA polymerase | 1 | 1 | 1 | Kühn et al. (2007) | |
| rRNA base modification | 1 | 2 | 0 | Yu et al. (2008) | |
| Sigma factor | 0 | 6 | 0 | Zhang et al. (2015) | |
| Transcript end processing | 5 | 5 | 3 | Perrin et al. (2004); Stoll and Binder (2016) | |
| tRNA base modification | 2 | 14 | 0 | Chen et al. (2010) | |
| tRNA aminoacylation | 3 | 1 | 24 | Duchêne et al. (2005) | |
| Total | 535 | 293 | 82 |
Note.—Counts reflect number of genes in each targeting category. Key references are listed by category. More extensive literature references are provided in supplementary table S2, Supplementary Material online.
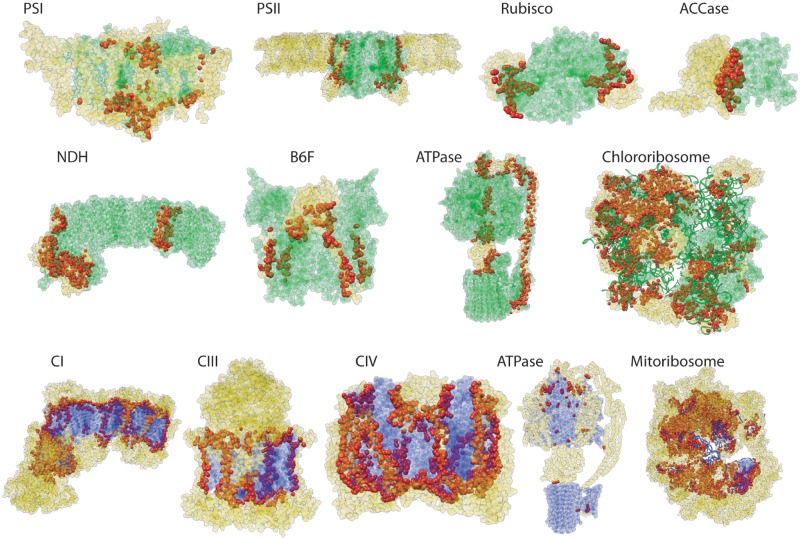
Many studies have begun taking advantage of protein structural data to specifically investigate molecular evolution at the physical interface between contacting cytoplasmic and nuclear gene products (Osada and Akashi 2012; Havird et al. 2015; Zhang et al. 2015; Havird and McConie 2019; Yan et al. 2019). We therefore used structural data from 13 protein complexes (fig. 2) to identify which nuclear subunits actually contact cytoplasmically encoded subunits within these complexes and their specific interacting amino acid positions (supplementary file S1, Supplementary Material online). However, the efficacy of this structural mapping approach varied greatly depending on the completeness and phylogenetic relatedness of the reference structures. For many photosynthetic complexes, reference structures are available from angiosperms or even A. thaliana itself, but other complexes required use of structures from anciently divergent species, including bacteria and mammals (supplementary file S1, Supplementary Material online), making inference of residue homology tenuous. Furthermore, even when structures from close relatives were available, they were sometimes known to be missing certain subunits (van Bezouwen et al. 2017; Laughlin et al. 2019). Therefore, we did not analyze many subunits within these complexes because of their absence from reference structures or low level of sequence similarity, designating them simply as not available (“NA”). Some additional subunits were classified only as “likely” or “not likely” to be involved in direct cytonuclear interactions because of low confidence in the reference mapping. Despite these limitations, structural data suggest that most nuclear-encoded proteins within these chimeric complexes do physically contact cytoplasmic gene products (91% of those for which assignments were made).
Fig. 2.
—Chimeric cytonuclear protein complexes showing cytoplasmic-encoded, nuclear-encoded, and nuclear contact residues. Plastid-encoded residues are in green, mitochondrial-encoded residues are in purple, nuclear-encoded noncontact residues are in yellow, and nuclear-encoded contact residues are in red. Amino acids are shown as spheres, RNA is shown as ribbons. PDB accessions for reference structures: PSI: 2O01, PSII: 5MDX, rubisco: 5IU0, ACCase: 2F9Y, NDH: 6NBY, B6F: 1VF5, plastid ATPase: 6FKF, chlororibosome: 5MMM, CI: 5LNK, CIII: 1BGY, CIV: 1V54, mitochondrial ATPase: 5ARA, and mitoribosome: 3J9M.
Our goal in generating CyMIRA is to provide a standardized partitioning of plant nuclear gene content based on cytonuclear interactions at a molecular level to improve consistency across evolutionary genomic studies. One obvious need that will arise is to extend this A. thaliana annotation to genomic data sets from nonmodel plant species that lack the same level of functional data. Because of the extensive history of gene and whole-genome duplication and the associated process of neofunctionalization in plants (Panchy et al. 2016), we recommend against relying solely on homology searches when porting the CyMIRA annotations to other species. Instead, we suggest combining such information with tools that perform in silico predictions of organelle targeting to increase confidence in assignments (Bannai et al. 2002; Small et al. 2004; Emanuelsson et al. 2007; Sperschneider et al. 2017).
A further complication in expanding to evolutionary studies across species is that the landscape of cytonuclear integration and interactions is rapidly shifting in plants. Unlike many eukaryotes in which the gene content in cytoplasmic genomes has reached a period of long-term stasis (Johnston and Williams 2016; Janouškovec et al. 2017), flowering plants remain highly active in the process of endosymbiotic gene transfer to the nucleus (Timmis et al. 2004). For example, our CyMIRA annotations do not include OXPHOS complex II because this is entirely nuclear-encoded in A. thaliana. In contrast, many other angiosperms have retained functional complex II genes (sdh3 and/or sdh4) in their mitochondrial genomes. Ribosomal subunits are also subject to ongoing functional transfers to the nucleus, resulting in substantial heterogeneity in cytoplasmic gene content across angiosperms (Adams et al. 2002). Therefore, species-specific additions and deletions to this data set, even at the whole complex level, should be considered based on the retained cytoplasmic gene content in each lineage. Although this continued need for refinement across phylogenetic scales undoubtedly poses a challenge for future studies, the dynamic nature of cytoplasmic genomes in plants is also one of the strongest motivations for studying cytonuclear interactions in these systems.
In summary, the proliferation of plant genomic resources makes this an exciting time to take studies of cytonuclear biology to a genome-wide level, and methodological consistency will be key to the efficacy of such efforts. We hope that CyMIRA will serve as useful community resource in this respect.
Materials and Methods
Curation of Mitochondrial and Plastid Targeting Databases
To identify mitochondrial- and plastid-targeted genes, we integrated predictions from nine existing databases (table 1 and supplementary text, Supplementary Material online). Based on these data sets, we classified all nuclear-encoded proteins in the A. thaliana Araport11 genome annotation into five targeting categories: mitochondrial, plastid, dual (both mitochondrial and plastid), other, or unknown. Because of the cytonuclear focus of this project, in cases where organelle-targeted proteins were known to have additional subcellular localizations, we still classified them based on their mitochondrial/plastid targeting status alone. To classify a protein as having an organellar localization, we required it to be identified as such in at least two different databases. Because it is well documented that many plant proteins play a dual functional role in both the mitochondria and plastids (Carrie and Small 2013), we assigned genes to the dual-targeted category as long as there were at least two databases supporting targeting to the mitochondria and at least two supporting targeting to the plastids. It was possible (although not required) for these to be the same two databases because the selected databases explicitly classify some genes as dual targeted. Some of these automated database classifications were subsequently refined based on manual curation of direct molecular interactions as described below.
Curation of Direct Cytonuclear Molecular Interactions
We conducted a literature-based curation to generate a resource that could distinguish nuclear proteins that are simply targeted to mitochondria and plastids from those that are involved in direct and intimate interactions with cytoplasmic genomes or their gene products. We assigned genes to 11 types of cytonuclear enzyme complexes and molecular interactions, which are further divided into 27 subcategories (table 2).
Because of the manual nature of this curation, our classifications often required judgment calls and special considerations. With respect to major multisubunit enzymes, we aimed to restrict our classification to the core complex, excluding proteins such as assembly factors involved in more transient interactions (e.g., Lu 2016; Ligas et al. 2019).
One of the largest classes of genes involved in plant cytonuclear interactions is the RNA-binding pentatricopeptide repeat (PPR) family (Schmitz-Linneweber and Small 2008). These proteins are overwhelmingly targeted to the mitochondria and plastids where they play diverse roles in RNA processing and maturation. We classified six specialized PPRs as components of the mitochondrial ribosome (Waltz et al. 2019) or as functioning in tRNA end processing (Gobert et al. 2010). The remaining PPRs were assigned to their own category. Even though many PPRs still lack detailed functional characterization, we considered these examples of direct cytonuclear interactions because of their near universal role in binding cytoplasmic transcripts. A total of 109 PPRs (24%) were not identified as mitochondrial or plastid targeted based on our automated database curation. In these cases, we reassigned their targeting classification using The Arabidopsis Information Resource (TAIR) Gene Ontology (GO) cellular component designations (Berardini et al. 2015). As a result, all PPRs were assigned as mitochondrial and/or plastid targeted, with the exception of only nine genes (AT1G06150, AT1G77150, AT2G20720, AT3G13150, AT3G47530, AT3G58590, AT5G09320, AT5G15300, AT5G44230), which we excluded from the direct-interaction data set. A large portion of PPR genes function as specificity factors in C-to-U RNA editing of organellar transcripts. Therefore, RNA editing interactions are effectively subsumed within the PPR category. Although other types of nuclear proteins have been found to function in RNA editing (Sun et al. 2016), we are not aware of any evidence that these directly bind to organellar transcripts, so they were not classified as directly interacting.
Mitochondrial transcription termination factors (mTERFs) are another sizeable family of organelle-targeted nucleic-acid binding proteins (Shevtsov et al. 2018). Similar to how we handled PPRs, we defined mTERFs as their own subcategory within the transcription and transcript maturation category, even though many individual mTERF genes await functional characterization.
Although our manual curation of direct cytonuclear interactions overwhelmingly agreed with general subcellular targeting predictions from our database summary, there were 189 genes (21%, including 100 PPRs; see above) for which the automated targeting predictions did not include the organelle(s) found in our manual analysis. In such cases, we updated the original automated targeting call by adding the location of direct cytonuclear interactions (but we did not remove other predicted localizations from the automated call set).
As a companion to this curated interaction data set, we also made use of the TAIR Interactome v 2.0 (Geisler-Lee et al. 2007), which identifies proteins with direct physical interactions. We used all pairwise interactions to create a list of partners for each Araport11 protein (supplementary file S1, Supplementary Material online). For organelle-targeted proteins, lists were further refined to include interacting partners that are targeted to the same subcellular compartment.
Identification of Direct Cytonuclear Contact Sites within Multisubunit Enzyme Complexes
In some cases, nuclear-encoded proteins may form part of a cytonuclear enzyme complex but still not physically contact a cytoplasmic gene product within the complex. Therefore, to identify direct cytonuclear interactions at the level of subunits and amino-acid residues, we mapped A. thaliana protein sequences to reference structures of 13 multisubunit enzyme complexes that are involved in OXPHOS, photosynthesis, protein translation, and fatty acid biosynthesis. Reference structures for these complexes were searched in the Protein Data Bank (PDB) and were chosen based on their completeness and relatedness to A. thaliana (supplementary file S1, Supplementary Material online). We identified cytonuclear contact residues in these structures using the “find clashes/contacts” tool in Chimera version 1.12 (Pettersen et al. 2004) with default contact settings except that the van der Waals (VDW) overlap was changed to ≤−1 Å. VDW overlap is essentially a measure of the distance between two atoms, and changing this value to ≤−1 Å allows for detecting more weakly interacting residues than by default. We determined homologous genes and residues in A. thaliana by querying the structural reference sequences with TAIR BLAST 2.2.8, and we aligned the resulting hits with MUSCLE as implemented in MEGA 7 (Kumar et al. 2016) to identify the corresponding contact residues in A. thaliana genes.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Stephane Bentolila, Hans-Peter Braun, José Gualberto, Oren Ostersetzer-Biran, Mareike Schallenberg-Rüdinger, and Ian Small for helpful discussion on different types of cytonuclear molecular interactions. This work was supported by NSF grants IOS-1829176 and MCB-1733227, an NSF GAUSSI graduate research fellowship (DGE-1450032), and start-up funds from the University of Texas.
Literature Cited
- Adams KL, Qiu YL, Stoutemyer M, Palmer JD.. 2002. Punctuated evolution of mitochondrial gene content: high and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc Natl Acad Sci U S A. 99(15):9905–9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrion JR, White PS, Montooth KL.. 2016. The roles of compensatory evolution and constraint in aminoacyl tRNA synthetase evolution. Mol Biol Evol. 33(1):152.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannai H, Tamada Y, Maruyama O, Nakai K, Miyano S.. 2002. Extensive feature detection of N-terminal protein sorting signals. Bioinformatics 18(2):298–305. [DOI] [PubMed] [Google Scholar]
- Barreto F, et al. 2018. Genomic signatures of mitonuclear coevolution across populations of Tigriopus californicus. Nat Ecol Evol. 2(8):1250–1257. [DOI] [PubMed] [Google Scholar]
- Barreto FS, Burton RS.. 2013. Evidence for compensatory evolution of ribosomal proteins in response to rapid divergence of mitochondrial rRNA. Mol Biol Evol. 30(2):310–314. [DOI] [PubMed] [Google Scholar]
- Berardini TZ, et al. 2015. The Arabidopsis information resource: making and mining the “gold standard” annotated reference plant genome. Genesis 53(8):474–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieri P, Leibundgut M, Saurer M, Boehringer D, Ban N.. 2017. The complete structure of the chloroplast 70S ribosome in complex with translation factor pY. EMBO J. 36(4):475–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonen L, Calixte S.. 2006. Comparative analysis of bacterial-origin genes for plant mitochondrial ribosomal proteins. Mol Biol Evol. 23(3):701–712. [DOI] [PubMed] [Google Scholar]
- Boutet E, Lieberherr D, Tognolli M, Schneider M, Bairoch A.. 2007. UniProtKB/Swiss-Prot. Methods Mol Biol. 406:89–112. [DOI] [PubMed] [Google Scholar]
- Burton RS, Barreto FS.. 2012. A disproportionate role for mtDNA in Dobzhansky-Muller incompatibilities? Mol Ecol. 21(20):4942–4957. [DOI] [PubMed] [Google Scholar]
- Burton RS, Pereira RJ, Barreto FS.. 2013. Cytonuclear genomic interactions and hybrid breakdown. Annu Rev Ecol Evol Syst. 44(1):281–302. [Google Scholar]
- Carrie C, Small I.. 2013. A reevaluation of dual-targeting of proteins to mitochondria and chloroplasts. Biochim Biophys Acta. 1833(2):253–259. [DOI] [PubMed] [Google Scholar]
- Carrie C, Weißenberger S, Soll J.. 2016. Plant mitochondria contain the protein translocase subunits TatB and TatC. J Cell Sci. 129(20):3935–3947. [DOI] [PubMed] [Google Scholar]
- Chen P, Jäger G, Zheng B.. 2010. Transfer RNA modifications and genes for modifying enzymes in Arabidopsis thaliana. BMC Plant Biol. 10:201.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, et al. 2016. Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J. 85(4):532–547. [DOI] [PubMed] [Google Scholar]
- de Longevialle AF, Small ID, Lurin C.. 2010. Nuclearly encoded splicing factors implicated in RNA splicing in higher plant organelles. Mol Plant. 3(4):691–705. [DOI] [PubMed] [Google Scholar]
- Duchêne A-M, et al. 2005. Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 102:16484–16489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison CK, Burton RS.. 2006. Disruption of mitochondrial function in interpopulation hybrids of Tigriopus californicus. Evolution 60(7):1382–1391. [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H.. 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2(4):953–971. [DOI] [PubMed] [Google Scholar]
- Eslamieh M, Williford A, Betrán E.. 2017. Few nuclear-encoded mitochondrial gene duplicates contribute to male germline-specific functions in humans. Genome Biol Evol. 9(10):2782–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira dCJ, et al. 2019. Cytonuclear interactions remain stable during allopolyploid evolution despite repeated whole-genome duplications in Brassica. Plant J. 98:443–447. [DOI] [PubMed] [Google Scholar]
- Forsythe ES, Nelson ADL, Beilstein MA.. 2018. Biased gene retention in the face of massive nuclear introgression obscures species relationships. bioRxiv. 10.1101/197087 [DOI] [PMC free article] [PubMed]
- Friso G, et al. 2004. In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. Plant Cell. 16(2):478–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler-Lee J, et al. 2007. A predicted interactome for Arabidopsis. Plant Physiol. 145(2):317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain A, Hotto AM, Barkan A, Stern DB.. 2013. RNA processing and decay in plastids. Wiley Interdiscip Rev RNA. 4(3):295–316. [DOI] [PubMed] [Google Scholar]
- Gobert A, et al. 2010. A single Arabidopsis organellar protein has RNase P activity. Nat Struct Mol Biol. 17(6):740–744. [DOI] [PubMed] [Google Scholar]
- Gualberto JM, Newton KJ.. 2017. Plant mitochondrial genomes: dynamics and mechanisms of mutation. Annu Rev Plant Biol. 68:225–252. [DOI] [PubMed] [Google Scholar]
- Havird JC, McConie HJ.. 2019. Sexually antagonistic mitonuclear coevolution in duplicate oxidative phosphorylation genes. Integrative & Comparative Biology; 10.1093/icb/icz021. [DOI] [PubMed] [Google Scholar]
- Havird JC, Trapp P, Miller C, Bazos I, Sloan DB.. 2017. Causes and consequences of rapidly evolving mtDNA in a plant lineage. Genome Biol Evol. 9(2):323–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havird JC, Whitehill NS, Snow CD, Sloan DB.. 2015. Conservative and compensatory evolution in oxidative phosphorylation complexes of angiosperms with highly divergent rates of mitochondrial genome evolution. Evolution 69(12):3069–3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill GE. 2015. Mitonuclear ecology. Mol Biol Evol. 32(8):1917–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper CM, Castleden IR, Tanz SK, Aryamanesh N, Millar AH.. 2017. SUBA4: the interactive data analysis centre for Arabidopsis subcellular protein locations. Nucleic Acids Res. 45(D1):D1064–D1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M, Tsunoda H, Suzuki Y, Makino A, Ishida H.. 2012. RBCS1A and RBCS3B, two major members within the Arabidopsis RBCS multigene family, function to yield sufficient Rubisco content for leaf photosynthetic capacity. J Exp Bot. 63(5):2159–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janouškovec J, et al. 2017. A new lineage of eukaryotes illuminates early mitochondrial genome reduction. Curr Biol. 27(23):3717–3724. [DOI] [PubMed] [Google Scholar]
- Jensen PE, et al. 2007. Structure, function and regulation of plant photosystem I. Biochim Biophys Acta. 1767(5):335–352. [DOI] [PubMed] [Google Scholar]
- Johnston IG, Williams BP.. 2016. Evolutionary inference across eukaryotes identifies specific pressures favoring mitochondrial gene retention. Cell Syst. 2(2):101–111. [DOI] [PubMed] [Google Scholar]
- Kühn K, Bohne A, Liere K, Weihe A, Börner T.. 2007. Arabidopsis phage-type RNA polymerases: accurate in vitro transcription of organellar genes. Plant Cell. 19(3):959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughlin T, Bayne A, Trempe J, Savage D, Davies K.. 2019. Structure of the complex I-like molecule NDH of oxygenic photosynthesis. Nature 566(7744):411–414. [DOI] [PubMed] [Google Scholar]
- Li C, et al. 2019. Cytonuclear coevolution following homoploid hybrid speciation in Aegilops tauschii. Mol Biol Evol. 36(2):341–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligas J, Pineau E, Bock R, Huynen M, Meyer E.. 2019. The assembly pathway of complex I in Arabidopsis thaliana. Plant J. 97(3):447–459. [DOI] [PubMed] [Google Scholar]
- Lu P, et al. 2005. PA-GOSUB: a searchable database of model organism protein sequences with their predicted Gene Ontology molecular function and subcellular localization. Nucleic Acids Res. 33(Database issue):D147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y. 2016. Identification and roles of photosystem II assembly, stability, and repair factors in Arabidopsis. Front Plant Sci. 7:168.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales H, et al. 2018. Concordant divergence of mitogenomes and a mitonuclear gene cluster in bird lineages inhabiting different climates. Nat Ecol Evol. 2:1258–1267. [DOI] [PubMed] [Google Scholar]
- Nishimura K, van Wijk KJ.. 2015. Organization, function and substrates of the essential Clp protease system in plastids. Biochim Biophys Acta. 1847(9):915–930. [DOI] [PubMed] [Google Scholar]
- Osada N, Akashi H.. 2012. Mitochondrial-nuclear interactions and accelerated compensatory evolution: evidence from the primate cytochrome C oxidase complex. Mol Biol Evol. 29(1):337.. [DOI] [PubMed] [Google Scholar]
- Panchy N, Lehti-Shiu M, Shiu S.. 2016. Evolution of gene duplication in plants. Plant Physiol. 171(4):2294–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin R, Lange H, Grienenberger JM, Gagliardi D.. 2004. AtmtPNPase is required for multiple aspects of the 18S rRNA metabolism in Arabidopsis thaliana mitochondria. Nucleic Acids Res. 32(17):5174–5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pett W, Lavrov DV.. 2015. Cytonuclear interactions in the evolution of animal mitochondrial tRNA metabolism. Genome Biol Evol. 7(8):2089–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, et al. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 25(13):1605–1612. [DOI] [PubMed] [Google Scholar]
- Pierleoni A, Martelli PL, Fariselli P, Casadio R.. 2007. eSLDB: eukaryotic subcellular localization database. Nucleic Acids Res. 35(Database issue):D208–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand DM, Haney RA, Fry AJ.. 2004. Cytonuclear coevolution: the genomics of cooperation. Trends Ecol Evol. 19(12):645–653. [DOI] [PubMed] [Google Scholar]
- Rastogi S, Rost B.. 2011. LocDB: experimental annotations of localization for Homo sapiens and Arabidopsis thaliana. Nucleic Acids Res. 39(Database issue):D230–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser L, Subramaniam S, Li D, Huala E.. 2017. Using the Arabidopsis Information Resource (TAIR) to find information about Arabidopsis genes. Curr Protoc Bioinformatics 60:1.11.11–1.11.45. [DOI] [PubMed] [Google Scholar]
- Rockenbach KD, et al. 2016. Positive selection in rapidly evolving plastid-nuclear enzyme complexes. Genetics 204(4):1507–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogell B, Dean R, Lemos B, Dowling DK.. 2014. Mito-nuclear interactions as drivers of gene movement on and off the X-chromosome. BMC Genomics 15:330.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas-Giegé T, Giegé R, Giegé P.. 2015. tRNA biology in mitochondria. Int J Mol Sci. 16(3):4518–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Nagano Y.. 2004. Plant acetyl-CoA carboxylase: structure, biosynthesis, regulation, and gene manipulation for plant breeding. Biosci Biotechnol Biochem. 68(6):1175–1184. [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Small I.. 2008. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 13(12):663–670. [DOI] [PubMed] [Google Scholar]
- Senkler J, Senkler M, Braun HP.. 2017. Structure and function of complex I in animals and plants – a comparative view. Physiol Plant. 161(1):6–15. [DOI] [PubMed] [Google Scholar]
- Sharbrough J, Havird JC, Noe GR, Warren JM, Sloan DB.. 2017. The mitonuclear dimension of Neanderthal and Denisovan ancestry in modern human genomes. Genome Biol Evol. 9(6):1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevtsov S, et al. 2018. Control of organelle gene expression by the mitochondrial transcription termination factor mTERF22 in Arabidopsis thaliana plants. PLoS One 13(7):e0201631.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T. 2016. Chloroplast NDH: a different enzyme with a structure similar to that of respiratory NADH dehydrogenase. Biochim Biophys Acta. 1857(7):1015–1022. [DOI] [PubMed] [Google Scholar]
- Sloan DB, et al. 2018. Cytonuclear integration and co-evolution. Nat Rev Genet. 19(10):635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Fields PD, Havird JC.. 2015. Mitonuclear linkage disequilibrium in human populations. Proc R Soc B. 282(1815):20151704.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Havird JC, Sharbrough J.. 2017. The on-again, off-again relationship between mitochondrial genomes and species boundaries. Mol Ecol. 26(8):2212–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Triant DA, Wu M, Taylor DR.. 2014. Cytonuclear interactions and relaxed selection accelerate sequence evolution in organelle ribosomes. Mol Biol Evol. 31(3):673–682. [DOI] [PubMed] [Google Scholar]
- Small I, Peeters N, Legeai F, Lurin C.. 2004. Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4(6):1581–1590. [DOI] [PubMed] [Google Scholar]
- Sperschneider J, et al. 2017. LOCALIZER: subcellular localization prediction of both plant and effector proteins in the plant cell. Sci Rep. 7:44598.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoll B, Binder S.. 2016. Two NYN domain containing putative nucleases are involved in transcript maturation in Arabidopsis mitochondria. Plant J. 85(2):278–288. [DOI] [PubMed] [Google Scholar]
- Sun Q, et al. 2009. PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Res. 37(Database issue):D969–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Bentolila S, Hanson M.. 2016. The unexpected diversity of plant organelle RNA editosomes. Trends Plant Sci. 21(11):962–973. [DOI] [PubMed] [Google Scholar]
- Tiller N, et al. 2012. The plastid-specific ribosomal proteins of Arabidopsis thaliana can be divided into non-essential proteins and genuine ribosomal proteins. Plant J. 69(2):302–316. [DOI] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W.. 2004. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 5(2):123–135. [DOI] [PubMed] [Google Scholar]
- Toews DP, Brelsford A.. 2012. The biogeography of mitochondrial and nuclear discordance in animals. Mol Ecol. 21(16):3907–3930. [DOI] [PubMed] [Google Scholar]
- van Bezouwen L, et al. 2017. Subunit and chlorophyll organization of the plant photosystem II supercomplex. Nat Plants. 3:17080.. [DOI] [PubMed] [Google Scholar]
- Waltz F, et al. 2019. Small is big in Arabidopsis mitochondrial ribosome. Nat Plants. 5(1):106–117. [DOI] [PubMed] [Google Scholar]
- Weng ML, Ruhlman TA, Jansen RK.. 2016. Plastid-nuclear interaction and accelerated coevolution in plastid ribosomal genes in Geraniaceae. Genome Biol Evol. 8(6):1824–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiwatwattana N, Landau CM, Cope GJ, Harp GA, Kumar A.. 2007. Organelle DB: an updated resource of eukaryotic protein localization and function. Nucleic Acids Res. 35(Database issue):D810–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Ye G, Werren J.. 2019. Evolutionary rate correlation between mitochondrial-encoded and mitochondria-associated nuclear-encoded proteins in insects. Mol Biol Evol. 36(5):1022.–1036. [DOI] [PubMed] [Google Scholar]
- Yu F, Liu X, Alsheikh M, Park S, Rodermel S.. 2008. Mutations in SUPPRESSOR OF VARIEGATION1, a factor required for normal chloroplast translation, suppress var2-mediated leaf variegation in Arabidopsis. Plant Cell. 20(7):1786–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi A, Makova K.. 2019. Investigating mitonuclear interactions in human admixed populations. Nat Ecol Evol. 3(2):213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, et al. 2016. Coevolution between nuclear-encoded DNA replication, recombination, and repair genes and plastid genome complexity. Genome Biol Evol. 8(3):622–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ruhlman TA, Sabir J, Blazier JC, Jansen RK.. 2015. Coordinated rates of evolution between interacting plastid and nuclear genes in Geraniaceae. Plant Cell. 27(3):563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.