ABSTRACT
Multiple reports have highlighted the importance of the local immunological cellular composition (i.e. the density of effector T cells and macrophage polarization state) in predicting clinical outcome in advanced metastatic stage of colorectal cancer. However, in spite of the general association between a high effector T cell density and improved outcome, our recent work has revealed a specific lymphocyte-driven cancer cell-supporting signal. Indeed, lymphocyte-derived CCL5 supports CCR5-positive tumor cell proliferation and thereby fosters tumor growth in metastatic liver lesions. Upon systematic analysis of CCR5 expression by tumor cells using immunohistochemistry, we observed that the intensity of CCR5 increases with primary tumor size and peaks in T4 tumors. In liver metastases however, though CCR5 expression intensity is globally heightened compared to primary tumors, alterations in the expression patterns appear, leading to “patchiness” of the stain. CCR5 patchiness is, therefore, a signature of liver metastases in our cohort (n = 97 specimens) and relates to globally decreased expression intensity, but does not influence the extent of the response to CCR5 inhibitor Maraviroc in patients. Moreover, CCR5 patchiness relates to a poor immune landscape characterized by a low cytotoxic-to-regulatory T cell ratio at the invasive margin and enriched cellular and molecular markers of macrophage M2 polarization. Finally, because higher numbers of PD-1- and CTLA-4-positive cells surround tumors with patchy CCR5 expression, one can speculate that these tumors potentially respond to immune checkpoint blockade. This hypothesis is corroborated by the prolonged disease-free survival and disease-specific survival observed in patients with low gene expression of CCR5 in metastases from two publically available cohorts. These observations highlight the complex role of the CCL5-CCR5 axis in CRC metastatic progression and warrant further investigations.
KEYWORDS: CCR5, colorectal cancer, microenvironment, immunotherapy, immune checkpoint, macrophages, T cells, CCL5, disease free survival
Introduction
The local immune infiltration in colorectal cancer (CRC) impacts the clinical course of the disease. 1–3 While the vast majority of research still focuses on the tumor microenvironment in primary colorectal cancer,4,5 the role of infiltrating immune cells is also clear for metastases of CRC.6,7 The composition of the different immune cells and their corresponding cytokines and chemokines shape the local microenvironment and the subsequent clinical course.8–11 The detailed analysis of the microenvironment revealed a plethora of modulating regulatory networks with intricate connections between the innate and the adaptive arm of the immune system.12
While surgical interventions in synergy with chemotherapy have improved the outcome for patients with a more limited disease, the situation for patients with advanced metastatic and non-resectable colorectal cancer is practically unchanged. Immunotherapy heralds new therapeutic options, but so far success has mainly been limited to CRC patients with microsatellite-instable tumors.13,14 In this setting, the analysis of the CCL5-CCR5 axis in metastatic colorectal cancer recently revealed a new therapeutic option: macrophage repolarization therapy.15
Also known as CD195, CCR5 (C-C motif chemokine receptor 5) is a β-chemokine receptor with seven transmembrane segments, the eighth containing an alpha-helix parallel to the plasma membrane and additionally with two disulfide bonds and palmitoylated cysteines.16 CCR5 is expressed by T cells, monocytes, dendritic cells and epithelial cells in response to inflammatory stimuli such as LPS. Its cognate ligands include CCL3 (C-C motif chemokine ligand 3), CCL4 and CCL5, which mediate potent chemotaxis functions. Because it was observed by Lusso and Gallo that these exact three chemokines were able to inhibit infection by macrophage-tropic HIV-1 strains,17 CCR5 was identified as one main co-receptor for HIV entry.18,19 CCR5 blockade has therefore mostly been studied in retrovirology and the CCR5 small molecule inhibitor Maraviroc has been approved and is vastly used in the clinics for highly active antiretroviral therapy (HAART) in AIDS patients.20–22 An important variant of CCR5 exists: the ∆32-CCR5 variant, which does not contain the extracellular domains for signaling23 and confers protection from HIV infection.24
So far, no relationship between the (homozygous) presence of allele encoding the non-functional ∆32-CCR5 variant and the occurrence of cancer could be confirmed in a large meta-analysis.25 However, data indicating potential tumor-supportive effects of CCR5 have appeared recently. These include the enhancement of the invasive properties of pancreatic,26 glioma27 or basal breast28 cancer cells in vitro, the promotion of tumor cell proliferation15,29 or the support of tumor growth and metastasis formation in xenografts models of colorectal30 or gastric cancer.31 Data from immunohistological analyses in primary CRC have indicated a relationship between stage and the CCR5 expression on tumor cells.32 In our previous study, we observed that inhibition of CCR5 in colorectal cancer liver metastases leads to the activation of an antiviral program in tumor-associated macrophages, followed by selective tumor cell death and improved responses in combination with chemotherapy.15
To better understand the expression and possible effects of CCL5-CCR5 in colorectal cancer, we analyzed the relationships between the observed patterns of CCR5 expression, CCR5 genomic status, and clinic-pathological features as well as immune landscape, utilizing different local cohorts of archived cryopreserved or FFPE tissue specimens of liver lesions from metastatic CRC patients as well as publically available gene expression datasets.
Material and methods
Patient material
Our total patient dataset originates from five different local cohorts: the MARACON cohort (n = 14 biopsies), HIPO34 (n = 20), Heilbronn (n = 30), Heidelberg HLM explants (n = 34) and Heidelberg primary tumors (n = 34). A summary of available data for each cohort is provided in Supplemental Table 1.
Samples were obtained from the Institute of Pathology and the Department of Surgery at the University of Heidelberg. All material was obtained after approval by the medical ethics committee of the University of Heidelberg (207–2005), written consent was obtained from all patients prior to analysis. Histopathologic and clinical findings were scored according to the Union for International Cancer Control (UICC)-TNM staging system.
Analyses of CCR5 status were performed on the samples from the MARACON trial (see hereafter), prior to the first dose of Maraviroc (n = 14). Additionally, the cohort for the analysis of ∆32-CCR5 expression was expanded with n = 20 metastatic lesion specimens from the HIPO-34 cohort, i.e. patients who underwent tumor resection between 2004 and 2009 at the Department of General, Visceral, and Transplantation Surgery, University of Heidelberg. Tissue collection was approved by the Ethics Committee of the University of Heidelberg (independent of the MARACON trial). A written informed consent was obtained preoperatively from all patients for (a) the tissue sampling and (b) the planned analyses. See supplemental Table 1 for patient characteristics.
For details regarding the MARACON trial, see Halama et al.15 Briefly, the MARACON-001 phase I trial („Treatment of Advanced Colorectal Cancer Patients with Hepatic Liver Metastases using the CCR5-Antagonist Maraviroc“, clinical trials.gov identifier NCT01736813, EudraCT 2012-000861-18) involved 14 patients with late-stage colorectal cancer, who received a monotherapy consisting of daily Maraviroc, a highly selective CCR5 inhibitor, for two months. Liver metastasis biopsies were sampled before and under treatment. The material was analyzed for CCR5 expression (using immunohistochemistry), CCR5 delta 32 mutation (PCR), immune cell distribution, density and activation (with immunostainings), tumor cell death, or cytokine and chemokine patterns. Safety and feasibility were the primary endpoints of this trial.
Tumor samples in the HIPO34 cohort were typed for MSI using BAT25, BAT26, and CAT25 as described earlier33 and no microsatellite instability was found. Pathological reports were available for all tissues. No sample from patients with inflammatory bowel disease was included in this analysis.
Immunohistochemistry, whole slide imaging and virtual image analysis
Tissue specimens were immunohistochemically analyzed for the presence and spatial distribution of specific surface antigens: CCR5 [MM0065-6H20], CD163 [EPR19518], pan-cytokeratin (polyclonal) from Dako, CD3 [Sp7], CD8 [Sp16], CD11b [EP1345Y], CD68 [KP1], CTLA4 [BNIS], PD-1 [NAT] and FoxP3 [236A/E7]. For negative control for CCR5 immunostain, a mouse IgG2b, κ isotype control from BioLegend was used. Tissue sections were either prepared from formalin-fixed, paraffin-embedded (FFPE) tissue (4 µm) or from cryopreserved specimens (7 µm). Cryosections were fixed either with 4% paraformaldehyde or 33% acetone in methanol prior staining. The complete staining procedure was carried out on a BOND-Max (Leica, Germany) using Bond Polymer Refine Detection kit (for DAB) or Bond Polymer Refine Red Detection kit from Leica Biosystems.
Whole slide images were acquired using an AT2 slide scanner (Leica Germany). The density and distribution of immune cells in complete microscopic images of full tissue sections were semi-automatically analyzed using QuPath open source software for digital pathology34 as previously described.35
CCR5 immunostains were scored by two independent-trained observers. CCR5 stain was named patchy when CCR5-positive and – negative cancer cells were observed in the same acini, leading to at least 10% of CCR5-negative cancer cells on the whole section. For staining intensity, 0 represented samples in which the immunoreactivity was undetectable whereas 1, 2 and 3 denoted samples with, respectively, a low, moderate and strong staining. For staining extent, 0, 1, 2 and 3 represented samples in which the immunoreactivity was detectable, respectively, in <5%, 6–33%, 34–66% and >67% of the tumor cells. In order to provide a global score for each case, the results obtained with the two scales were multiplied, yielding a single scale of 0, +1, +2, +3, +4, +6 or +9. All cases with divergent scoring between observers were reassessed.
PCR for identification of ∆32-CCR5
The procedure was performed according to the protocol described by Abdi and colleagues.36 The primer sequences were 5′-TGTTTGCGTCTCTCCCAG-3 (sense) and 5′-CACAGCCCTGTGCCTCTT-3′ (anti-sense). Wild-type and mutated CCR5 were discriminated by the size of the amplicon on an agarose gel using ethidium bromide (wild-type amplicon length: 233 bp, delta 32 amplicon length: 201 bp).
Sequence data analysis
Whole exome sequencing of DNA was conducted from the liver metastases and matched normal liver tissue in 20 patients (HIPO-34 cohort, supplemental Table 1). DNA was isolated from the fresh frozen tissue blocks and exon capture was performed using SureSelectXT Automation Reagent Kit and Human All Exon v5 (Agilent Technologies). Paired-end whole-exome sequencing was then performed on the HiSeq 2000 platform (Illumina) according to the manufacturer’s protocol, aiming for coverage of 60x for control and 80x for tumor samples (German Cancer Research Center Core Facility). Sequencing read pairs were mapped and aligned to the 1000 genomes phase 2 reference genome hs37d5 as previously described,37 using Burrows-Wheeler-Aligner (BWA) (version 0.6.2), and were processed with SAMtools (version 0.1.17) and Picard tools (version 1.61). All alignments were then visually inspected for the occurrence of the CCR5 deletion using the Integrative Genomics Viewer (IGV).38
Expression profile data analysis
Publically available datasets for gene expression in metastases from colorectal cancer (GSE17536 and GSE17537) were interrogated for the analysis of the CCR5 level’s impact on overall survival. The PROGgeneV2 algorithm was used and results were computed with adjustment for stage and the median value was used as group separator. The webpage can be accessed through the following link: http://watson.compbio.iupui.edu/chirayu/proggene/database/index.php .
Luminex analysis
Multiplex cytokine quantification was performed on tissue lysates as previously described.15 Briefly, sections of cryopreserved tissue were collected and lysed using the Bio-Plex cell lysis kit (Bio-Rad) and lysate concentration was adjusted to 300µg/ml. The protein concentration of soluble factors was determined using the Bio-Plex ProTM human cytokine assays, allowing the absolute measurement of 48 different soluble factors (Bio-Plex ProTM human cytokine, chemokine and growth factor assay, Bio-Rad) according to the manufacturer’s instructions.
Statistical analyses
Statistics were computed using GraphPad Prim 5.0 software. Univariate tests were used to compare groups two by two. For non-paired samples, unpaired T-tests or Mann Whitney tests (parametric and non-parametric, respectively) were used. Paired samples were compared using Wilcoxon matched pairs test or Student T-tests (non-parametric and parametric, respectively). Parametric tests were used in case of a normal distribution of values, as evaluated by D’Agostino and Pearson omnibus normality test. Differences were considered significant in case of a p-value < 0.05 and represented as follows: *: p-value < 0.05, **: p-value < 0.01 and ***: p-value < 0.001. For the analyses of Kaplan–Meyer curves, differences were assessed using the log rank test. For contingency table analyses, two-tailed Fisher’s exact tests were used in case of a 2 × 2 comparison. Chi-square tests were used otherwise.
Results
Patchy CCR5 expression patterns appear in liver metastases of colorectal cancer and relate to clinicopathological features
We conducted an immunohistochemistry analysis for CCR5 expression in liver lesions of metastatic colorectal cancer originating from four independent groups (from the “Maracon”, “HIPO-34”, “Heidelberg explants” and “Heilbronn” cohorts: for patients’ characteristics, see supplemental Table 1), as well as on an additional group of primary tumor samples. In stage IV primary tumors, CCR5 positive tumor cells were found in all (luminal/mucosal and invading/peritoneal) positions with comparable intensity (Figure 1(a)). The semi-quantitative scoring of CCR5 immunostaining showed that CCR5 expression increases and expands during tumor growth in primary tumors, peaking in T4 tumors (Figure 1(e)). CCR5 protein expression was further augmented in liver lesions, in which all specimens exhibited CCR5 positive tumor cells (Figure 1(b)). This finding was reproduced in one lung metastatic lesion (Figure 1(c)).
Figure 1.

CCR5 expression in colorectal cancer primary tumors and metastases. Representative micro-images of CCR5 immunostaining in (a) a primary colorectal cancer specimen, in the tumor core or at the invasive margin, (b) a liver metastasis and (c) a lung metastasis of colorectal cancer. (d) Negative control immunostaining with an isotype control antibody on a liver metastasis section. (e) Results of the semi-quantitative scoring of CCR5 expression by cancer cells in primary tumors (n = 31) according to their size classification (T2: n = 3; T3: n = 8, T4: n = 20) and in liver metastases (LM, n = 84), illustrating the increase of CCR5 expression by cancer cells during cancer progression. Groups were compared using the two-tailed Mann Withney test. *: p-value < 0.05; ***: p-value < 0.0001.
CCR5 was expressed by cancer cells (as illustrated by CCR5-cytokeratin double stains, Supplemental Figure 1(a-b)) and by specific subpopulations of tumor-associated macrophages, i.e. CD68-positive macrophages, but neither CD11b-nor CD163-positive macrophages (Supplemental Figure 1(c-H). Finally, CCR5 was expressed by CD3+ and CD8+ tumor-infiltrating T cells (TILs) (Supplemental Figure 2).
Surprisingly, when focusing on cancer cells, we noticed that the CCR5 expression pattern in liver metastases tended to differ from what was observed in primary tumors. Indeed, there was a significant proportion of liver metastases in which the CCR5 stain was “patchy”, i.e. at least 10% of tumor cells were negative for CCR5, in a patchwork-like configuration, with tumor cells in the very same acinus exhibiting positive and negative CCR5 stain (Figure 2(a-c)). Conversely, CCR5 expression patchiness was only rarely observed in T4 primary tumors (Figure 2(c)).
Figure 2.
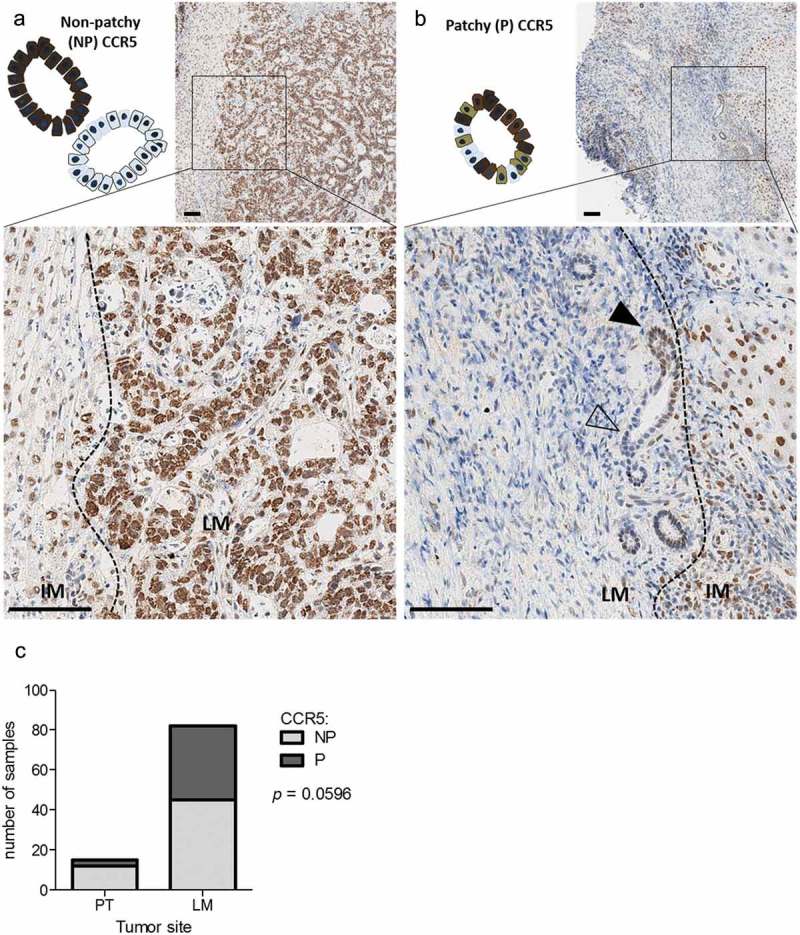
CCR5 expression patchiness in liver metastases of colorectal cancer. Representative micro-images and schematic representation of a patchy (a) and a non-patchy (b) CCR5 expression pattern in a liver metastasis. Black arrowheads indicate cancer cells positive for CCR5, empty arrowheads indicate negative cancer cells. Scale bar: 100 µm. (c) Contingency of tumors expressing CCR5 in a patchy or non-patchy fashion in stage IV primary tumors (PT, n = 15) or liver metastases (LM, n = 82), analyzed with a one-tailed Fisher’s exact test.
In an attempt to clarify, whether there is a relationship between CCR5 expression and other parameters specifically in liver lesions, CCR5 expression patterns were analyzed for association with clinicopathological variables. The patchy CCR5 expression on tumor cells in liver metastases of CRC was not associated with age, gender, tumor grade, K-Ras mutation status or neoadjuvant chemotherapy. However, it was associated to smaller tumors at diagnostic, and showed a trend for an association with the absence of lymph node metastasis at the time of diagnosis (Table 1). Finally, liver lesions originating from primary tumors that occurred in the sigmoid colon and the rectum expressed more frequently CCR5 in a patchy fashion, in contrast to liver lesions originating from a primary tumor occurring in the ascending or transverse colon (Table 1).
Table 1.
Clinicopathological features of liver metastases in function of CCR5 expression patterns.
| CCR5 expression pattern |
|||
|---|---|---|---|
| Non-patchy | Patchy | p-value | |
| Age | |||
| All three cohorts (<65/65+) | 18/13 | 12/11 | 0.7837 |
| MARACON (<65/65+) | 5/1 | 2/1 | 1.000 |
| HIPO34 (<65/65+) | 4/5 | 4/6 | 1.000 |
| Heilbronn (<65/65+) | 9/7 | 5/4 | 1.000 |
| Gender | |||
| All three cohorts (male/female) | 18/13 | 16/7 | 0.4115 |
| MARACON (male/female) | 4/2 | 2/2 | 1.000 |
| HIPO34 (male/female) | 6/3 | 7/3 | 1.000 |
| Heilbronn (male/female) | 8/8 | 7/2 | 0.2290 |
| Staging at diagnosis | |||
| T | |||
| All three cohorts (T2/T3/T4) | 1/24/5 | 3/7/8 | 0.0139 |
| MARACON (T2/T3/T4) | 0/4/3 | 0/1/1 | 1.000 |
| HIPO34 (T2/T3/T4) | 0/8/0 | 1/3/3 | 0.0443 |
| Heilbronn (T2/T3/T4) | 1/13/2 | 2/3/4 | 0.0567 |
| N | |||
| All three cohorts (N0/N+) | 6/24 | 6/13 | 0.4975 |
| MARACON (N0/N+) | 1/5 | 0/2 | 1.0000 |
| HIPO34 (N0/N+) | 1/5 | 3/4 | 0.5994 |
| Heilbronn (N0/N+) | 2/14 | 2/7 | 0.6016 |
| Synchronous metastases | |||
| All three cohorts (M0/M+) | 10/19 | 5/14 | 0.2666 |
| MARACON (M0/M+) | 3/3 | 0/2 | 0.4643 |
| HIPO34 (M0/M+) | 3/4 | 4/4 | 0.5952 |
| Heilbronn (M0/M+) | 4/12 | 1/8 | 0.6206 |
| Tumor Grade | |||
| All three cohorts (G2/G3) | 16/11 | 16/4 | 0.2025 |
| MARACON (G1/G2/G3) | 0/3/2 | 0/3/0 | 0.4643 |
| HIPO34 (G1/G2/G3) | 0/6/3 | 2/5/3 | 0.3599 |
| Heilbronn (G1/G2/G3) | 0/7/6 | 0/8/1 | 0.1649 |
| Site of the primary tumor | |||
| All groups (colon/rectum) | 23/6 | 11/10 | 0.0661 |
| MARACON (colon/rectum) | 4/1 | 3/0 | 1.000 |
| HIPO-34 (colon/rectum) | 6/2 | 5/4 | 0.6199 |
| Heilbronn (colon/rectum) | 13/3 | 3/6 | 0.0308 |
| Neoadjuvant chemotherapy | |||
| Both cohorts (yes/no) | 17/12 | 16/7 | 0.5635 |
| Heilbronn (yes/no) | 10/6 | 5/4 | 1.000 |
| Heidelberg explants (yes/no) | 7/6 | 11/3 | 0.2365 |
| K-Ras status | |||
| All two | 4/11 | 5/8 | 0.6891 |
| MARACON (mutated/WT) | 2/4 | 2/1 | 0.5238 |
| HIPO34 (mutated/WT) | 2/7 | 3/7 | 1.000 |
CCR5 patchiness relates to an unfavorable immune landscape
Metastatic liver lesions exhibiting patchy CCR5 expression did not display a different density or distribution of CD3+ TILs globally: most CD3+ TILs accumulated at the invasive margin and poorly infiltrated the tumor core, independently of CCR5 expression pattern (Figure 3(a) and supplemental Figure 3A, C, F, H). However, tumors with patchy CCR5 stains were characterized by a lesser CD8+ T cell accumulation (Figure 3(b) and supplemental Figure 3B, D, G, I) and a higher FoxP3+ T cell density (Figure 3(c) and supplemental Figure 3E) at the invasive margin. Moreover, these tumors were infiltrated by more PD-1+ and CTLA-4+ cells (Figure 3(c)). Finally, there is a (non-significant) trend towards higher infiltration of the CCR5 patchy metastases by macrophages in the tumor core (as assessed by pan-/CD68-staining and by CD163 staining for tumor-supporting macrophages) (Figure 3(d)). This suggests alternative polarization of tumor-associated macrophages in tumors that express patchy CCR5, which was confirmed by the observed enrichment of cytokines related to M2 polarization in corresponding tumor lysates (Figure 3(e)).
Figure 3.
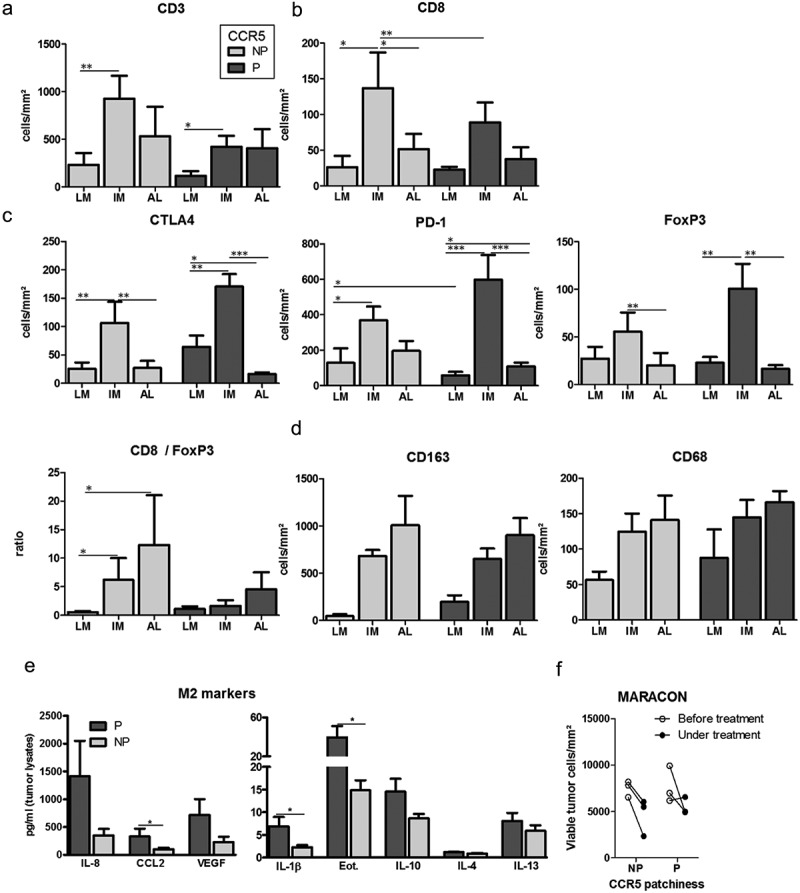
Immune landscape in liver metastases according to CCR5 expression patchiness. (a-d) Density of immune cells in tumor sections with patchy (P) or non-patchy (NP) CCR5 expression in the HIPO-34 cohort (n = 18). Results are expressed in cell numbers per mm2 in three regions of interest: liver metastasis core (LM), invasive margin (IM) and adjacent liver (AL). Histograms illustrate the density in CD3+ and CD8+ TILs (A-B), CTLA-4-, PD-1- and FoxP3-expressing cells (c), as well as the CD8/FoxP3 ratio (C), CD163- and CD68-positive macrophages (d). (e) Concentration in selected cytokines in whole tissue lysates of liver metastases, in the function of CCR5 expression patchiness (n = 28 samples: Heidelberg explants cohort). (f) Number of viable tumor cells per mm2 in liver lesion biopsies obtained before and under treatment during the MARACON trial (n = 6). In (a-d), paired samples (ex: AL versus IM in patchy tumors) were compared with a paired T-test. Samples from independent groups (ex; LM in patchy tumors versus LM in non-patchy tumors) were compared with a Student T-test. In (e), the samples were compared using the Mann Whitney test. *: p-value < 0.05; **: p-value < 0.01; ***: p-value < 0.001.
However, in our previously published data from the MARACON trial, where CCR5 inhibition with Maraviroc was used as a monotherapy and biopsies were obtained pre and post (or under) treatment,15 the initial patchiness of CCR5 expression was not related to the response to Maraviroc treatment, as measured by the magnitude of selective tumor cell death (Figure 3(f)).
Genetic variation of CCR5 as a natural tool to study the impact of CCR5 in human metastases
With the hypothesis that an abrogation or functional reduction of the CCL5-CCR5 axis would mimic patchiness and would also be reflected in metastatic progression, the CCR5 status was analyzed in the cohort of patients from the MARACON trial and in the HIPO-34 cohort. From 31 analyzed patients, 4 (13%) were heterozygous (WT/Δ32-CCR5) and 27 (87%) had homozygous wildtype CCR5 sequence. Though the limited size of the cohort and the low frequency of Δ32-CCR5 occurrence limit the potency of this analysis, the heterozygote patients tend to have a lower frequency of synchronous metastasis compared to the wild-type patients (Supplemental Table 2).
CCL5-CCR5 expression signature predicts OS in colorectal cancer
We analyzed the survival data of patients from the Heilbronn cohort and observed a trend towards a shorter overall survival associated with patchy CCR5 expression in the liver lesions (Supplemental Figure 4). To further confirm this in a bigger cohort from a publically available dataset, we translated CCR5 patchiness into expression levels of CCR5 using a semi-quantitative scoring system. CCR5 patchiness was associated to a slightly lower semi-quantitative score (Supplemental Figure 4). Therefore, we approximated a patchy CCR5 stain to a lower expression level for further analyses.
Metastatic CCR5 expression data from two independent publically available datasets (GSE17536 and GSE17537, n = 238 in total) were analyzed. Kaplan–Meyer curves were constructed, with correction for stage and revealed a deleterious effect of high CCR5 expression (assimilated to non-patchy stain) on disease-specific survival (HR = 1.51, p-value = 0.0206) and disease-free survival (HR = 1.42, p-value = 0.0293) (Figure 4(b–c)). Conversely, there was a trend towards longer overall survival in this cohort based on CCR5 expression (p = .246) (Figure 4(a)).
Figure 4.
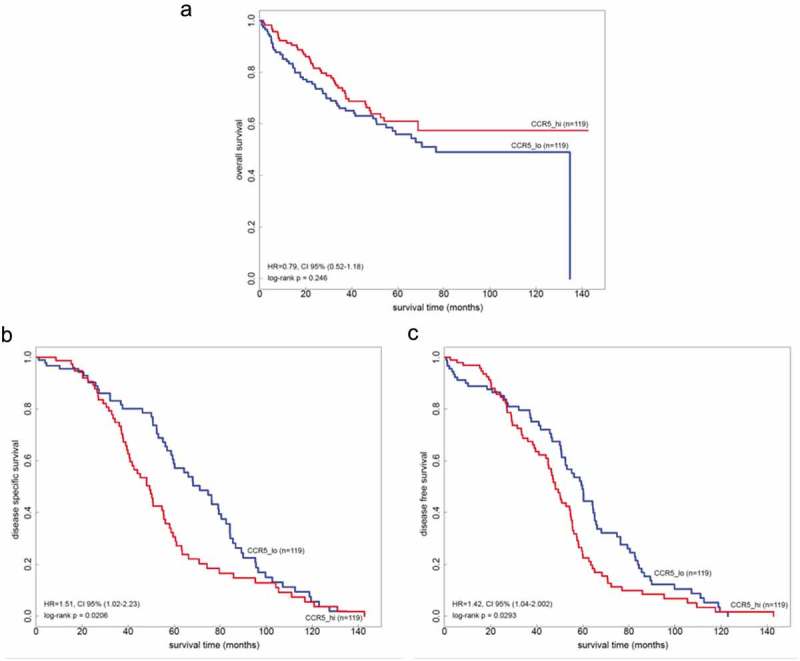
Overall, relapse-free and disease-specific survival in the function of intra-CCR5 gene expression in two independent cohorts of colorectal cancer patients. (a) Overall survival in n = 238 patients from two publically available cohorts (GSE17536 and GSE17537) adjusted for stage clustered according to the median mRNA expression of CCR5 in liver metastases. (b-c) Relapse-free and disease-specific survival in the same cohorts.
Discussion
Therapeutic options for patients with non-resectable metastatic CRC are limited and immunotherapy has not shown significant benefits for microsatellite stable cancers.13 Therefore, either prevention of the metastatic stage or expanded therapeutic – especially immunotherapeutic – options, are in the focus of current research.39
Our histological analysis of primary tumors showed an even distribution of CCR5 expression across the whole tissue sections, without predominance in invasive parts of the tumor, and is in accordance to previously published studies: CCR5 was expressed by cancer cells,40,41 by tumor-infiltrating lymphocytes32 as well as by macrophages.18 We additionally illustrate that CCR5 expression levels increase during primary tumor growth and peak in liver metastases, corroborating the hypothesis that CCR5 promotes metastasis. Our findings are supported by previously published data linking heightened CCR5 gene expression and metastasis in human specimens: CCR5 mRNA expression was indeed found elevated in liver and lung metastases (n = 10) compared to their matched primary tumors.42 Similarly, CCR5 gene expression has also been known to be enriched in human colorectal cancer specimens (n = 5) as compared to the non-neoplastic colon.32 The CCR5 expression on immune cells and on tumor cells has been investigated in vitro and in animal models mainly, underlying the impact of the CCL5-CCR5 axis on the acquisition by cancer cells of enhanced metastatic properties and on metastatic formation.28,29,31,42–46 As a hypothesis, the acquired CCR5 expression in progressing primary tumors allows tumor cells to follow the gradients of the respective chemokines, mostly CCL3, CCL4, and CCL5,47 and thereby contributes to the dissemination and metastatic spread.48 Finally, CCR5 expression in colorectal cancer primary tumors were recently found to be associated with a shorter overall survival.41
Not a single colorectal cancer liver metastasis in our hand contained only CCR5-negative tumor cells in all our analyzed samples so far. However, liver metastases exhibited patchiness in CCR5 immunostainings, i.e. some tumor cells did not express CCR5, leading to a patchwork-like stain, which was associated to a slightly lower semi-quantitative score of CCR5 (Figure 2 and Supplemental Figure 4). The mechanisms behind this phenomenon are unknown so far. The functionally inactive homozygous version of Δ32-CCR5 is present in a low number of patients (frequencies of homozygous individuals range between 1,1%49 and 3% in the general Caucasian population50,51) but the heterozygous situation is more likely (ranging from 10% to 20% according to studies).49,52 Interestingly, heterozygous individuals only express 20–30% of the wild-type protein on their cell surface.51 Nonetheless, our analysis has excluded that CCR5 patchiness might be accounted for by genetic variants of CCR5, namely the heterozygous Δ32-CCR5/wild-type situation: in the HIPO-34 cohort, for example, we identified 4/20 (20%) Δ32-CCR5/wild-type patients, but 10/20 (50%) tumors with patchy CCR5 expression.
If it appears that high CCR5 expression at the primary tumor is a poor prognosis indicator,41 is this also the case on the metastatic site? In liver lesions, patchy/decreased CCR5 expression relates to a poor immune landscape characterized by an unfavorable CD8+/FoxP3+ TIL ratio, elevated CTLA4+, and PD1+ cell numbers and elevated markers of alternatively activated macrophages.
The general notion that CCR5 inhibition can dampen T cell migration is supported by its well-described function as a T cell chemokine receptor. Maraviroc is indeed used in clinical settings to diminish T cell chemotaxis after allogeneic hematopoietic stem cell transplantation in case of visceral graft-versus-host disease.53 Our findings showing that patchy/decreased CCR5 expression is associated with lessened CD8+ T cell density specifically in the tumor microenvironment are moreover in agreement with data published by Zimmerman and collaborators, indicating an association, in primary colorectal tumors, between CCR5 expression and the infiltration of the tumor by CD8+ T cells.40
Our data also points to an increased regulatory T cell (Treg) density in tumors with patchy/decreased CCR5 expression, demonstrated in two independent cohorts (Figure 3(c) and supplemental Figure 3E). This differs from what has been illustrated in mouse models. Indeed, the blockade of CCR5 was shown to decrease the density of Tregs in the lung metastatic niche of mice following orthotopic breast cancer cell implantation,54 reducing the metastatic burden. The interspecies variations and the specificities of the lung and the liver microenvironment might account for those differences.
Finally, the association that we noticed between the patchy/decreased CCR5 expression and the increased density of CTLA4+ and PD-1+ cells at the invasive margin of liver lesions has never been shown to our knowledge. This observation has led us to speculate that patients bearing patchy CCR5-expressing tumors might respond differentially to immune checkpoint blockade. The currently running Luminescence trial (NCT03274804), in which CRC patients in the metastatic stage receive Maraviroc combined with Ipilimumab and Nivolumab, might shed some light on this potential effect.
The expression patterns of CCR5 in liver lesions are also associated with a specific cytokine profile linked to macrophage polarization. Indeed, patchy/decreased CCR5 expression is associated (i) with a trend to a higher density of CD163+ scavenger macrophages in the liver metastasis, inside the tumor core and (ii) with a significant increase of soluble factors reminiscent of a M2/scavenger phenotype of macrophages, i.e. Eotaxin, IL-8, CCL2, IL-1β and others. This is in line with our recent data from the MARACON trial, which have indicated for the first time a tightly interwoven T cell-macrophage network that is regulated by the CCL5-CCR5 axis specifically in liver lesions of CRC. Exhausted T cell-derived CCL5 was shown to enhance tumor cell survival and proliferation and to promote tumor-supportive functions of tumor-associated macrophages in cell culture models.15 Conversely, CCR5 inhibition using Maraviroc effectively polarized macrophages into ROS- and IFN-α-producing, tumor-killing macrophages. In the phase I clinical trial testing Maraviroc as a monotherapy in advanced colorectal cancer, this translated into selective necrosis of tumor cells in liver metastases.15 This association between CCR5 expression at the surface of macrophages, and the activation of a M2 polarization program upon its activation is supported by other reports55 and most compellingly, the functional (re-)activation of a M1 program in TAMs using Maraviroc has been shown as well by others in other tumor entities, such as non-Hodgkin lymphoma,56 breast cancer55 or gastric cancer (reviewed in57).
Also, it needs to be pointed out that in the MARACON trial, there was no direct relationship between patchy or non-patchy expression patterns and response to CCR5 inhibition in terms of the extent of selective tumor necrosis (Figure 2(f)), which also seems to indicate that the Maraviroc-mediated tumor cell death in this study did not exclusively rely on CCR5 expression by cancer cells but most likely centered on macrophages. This is line with our previously published results, where Maraviroc-mediated tumor cell death was relying on the presence of functional macrophages in tissue cultures of liver metastases.15
Low mRNA expression of CCR5 – that we approximate using patchiness (supplemental Figure 4) – is linked to a longer relapse-free survival and disease-specific survival (Figure 4). Interestingly, however, this effect only appears after a certain amount of time (around 2 years) during which low CCR5-expressing metastases are associated with a poorer profile of RFS (Figure 4(b-c)). Does this indicate that this is the timeframe for a CCR5-associated relapse to become visible? This timeframe of the first two years after curative resection nicely fits into the clinical observations.58 But this also brings up another important aspect: do we need a trial utilizing CCR5 inhibition in the adjuvant setting15,42? Can we prevent metastatic homing or at least delay the relapse and progression? Based on our data this could be considered, especially as this is reinforced by the trend towards overall survival advantage (Figure 4) in patients with high CCR5 expression.
Notably, the expression of CCR5 in liver lesions, albeit altered compared to primary tumors; directly relies on the primary tumor characteristics. CCR5 patchiness is indeed associated to liver lesions originating from bigger primary tumors arising in the rectum (and the sigmoid, data not shown), but rarely in the ascending and transverse colon, for unknown reasons so far. One might speculate on the mechanisms behind this and on the role of the microbiome in CCR5 expression by cancer cells, since colorectal cancers arising in the sigmoid and rectum are characterized by a specific microbiota.59
Finally, the prognosis impact of CCR5 expression in colorectal cancer diverges depending on the tumor site considered. When considering CCR5 expression in primary tumors, high expression relates to a shorter OS, shorter PFS, and DSS.41 High CCR5 expression in liver lesions, on the other hand, relates to lower PFS and DSS, but to longer OS as well.
During the MARACON trial, we did not see an association of clinical response to Maraviroc with the mutational status (e.g. k-RAS status), nor did we observe relevant toxicities, even for prolonged therapy periods. This is a good basis for considering this therapeutic option. Reducing the burden of relapse in adjuvant patients has to place exceptional emphasis on possible side effects and toxicities of prolonged periods. In this light, this possibility of CCR5 inhibition in addition to oxaliplatinum-based adjuvant therapy seems to be an option to be evaluated in clinical trials.
Funding Statement
This work was supported by the Wallonie-Bruxelles International.
Acknowledgments
The authors thank the DKFZ‐Heidelberg Center for Personalized Oncology (DKFZ‐HIPO) for technical support and funding through HIPO-034. The authors thank Rosa Eurich and Jana Wolf for technical support. Work by M.S-C was partly supported by a WBI-World Excellence grant of the WBI (Wallonie Brussels International) initiative (Belgium). The authors have no financial interest to disclose with the exception of intellectual property on CCR5 inhibition for cancer owned by University Heidelberg (NH, IZ, and DJ). MSC, NH, and DJ are supported by the Helmholtz-Gemeinschaft, Zukunftsthema “Immunology and Inflammation” (ZT-0027).
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, Tosolini M, Camus M, Berger A, Wind P, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 2.Fridman WH, Galon J, Dieu-Nosjean MC, Cremer I, Fisson S, Damotte D, Pagès F, Tartour E, Sautès-Fridman C.. Immune infiltration in human cancer: prognostic significance and disease control. Curr Top Microbiol Immunol. 2011;344:1–24. doi: 10.1007/82_2010_46. [DOI] [PubMed] [Google Scholar]
- 3.Bindea G, Mlecnik B, Fridman WH, Galon J.. The prognostic impact of anti-cancer immune response: a novel classification of cancer patients. Semin Immunopathol. 2011;33:335–340. doi: 10.1007/s00281-011-0264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becht E, Giraldo NA, Germain C, de Reynies A, Laurent-Puig P, Zucman-Rossi J, Dieu-Nosjean M-C, Sautès-Fridman C, Fridman WH. Immune contexture, immunoscore, and malignant cell molecular subgroups for prognostic and theranostic classifications of cancers. Adv Immunol. 2016;130:95–190. doi: 10.1016/bs.ai.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Pagès F, Mlecnik B, Marliot F, Bindea G, Ou F-S, Bifulco C, Lugli A, Zlobec I, Rau TT, Berger MD, et al. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. The Lancet. 2018;391:2128–2139. doi: 10.1016/S0140-6736(18)30789-X. [DOI] [PubMed] [Google Scholar]
- 6.Halama N, Michel S, Kloor M, Zoernig I, Benner A, Spille A, Pommerencke T, von Knebel DM, Folprecht G, Luber B, et al. Localization and density of immune cells in the invasive margin of human colorectal cancer liver metastases are prognostic for response to chemotherapy. Cancer Res. 2011;71:5670–5677. doi: 10.1158/0008-5472.CAN-11-0268. [DOI] [PubMed] [Google Scholar]
- 7.Tanis E, Julie C, Emile JF, Mauer M, Nordlinger B, Aust D, Roth A, Lutz MP, Gruenberger T, Wrba F, et al. Prognostic impact of immune response in resectable colorectal liver metastases treated by surgery alone or surgery with perioperative FOLFOX in the randomised EORTC study 40983. Eur J Cancer. 2015;51:2708–2717. doi: 10.1016/j.ejca.2015.08.014. [DOI] [PubMed] [Google Scholar]
- 8.Zitvogel L, Apetoh L, Ghiringhelli F, Andre F, Tesniere A, Kroemer G. The anticancer immune response: indispensable for therapeutic success?. J Clin Invest. 2008;118:1991–2001. doi: 10.1172/JCI35180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghiringhelli F, Menard C, Terme M, Flament C, Taieb J, Chaput N, Puig PE, Novault S, Escudier B, Vivier E, et al. CD4+CD25+ regulatory T cells inhibit natural killer cell functions in a transforming growth factor-beta-dependent manner. J Exp Med. 2005;202:1075–1085. doi: 10.1084/jem.20051511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dolcetti L, Peranzoni E, Ugel S, Marigo I, Fernandez Gomez A, Mesa C, Geilich M, Winkels G, Traggiai E, Casati A, et al. Hierarchy of immunosuppressive strength among myeloid-derived suppressor cell subsets is determined by GM-CSF. Eur J Immunol. 2010;40:22–35. doi: 10.1002/eji.200939903. [DOI] [PubMed] [Google Scholar]
- 11.Gallina G, Dolcetti L, Serafini P, De Santo C, Marigo I, Colombo MP, Basso G, Brombacher F, Borrello I, Zanovello P, et al. Tumors induce a subset of inflammatory monocytes with immunosuppressive activity on CD8+ T cells. J Clin Invest. 2006;116:2777–2790. doi: 10.1172/JCI28828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coussens LM, Zitvogel L, Palucka AK. Neutralizing tumor-promoting chronic inflammation: a magic bullet? Science. 2013;339:286–291. doi: 10.1126/science.1232227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kroemer G, Galluzzi L, Zitvogel L, Fridman WH. Colorectal cancer: the first neoplasia found to be under immunosurveillance and the last one to respond to immunotherapy? Oncoimmunology. 2015;4:e1058597. doi: 10.1080/2162402X.2015.1058597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asaoka Y, Ijichi H, Koike K. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;373:1979. doi: 10.1056/NEJMc1510353. [DOI] [PubMed] [Google Scholar]
- 15.Halama N, Zoernig I, Berthel A, Kahlert C, Klupp F, Suarez-Carmona M, Suetterlin T, Brand K, Krauss J, Lasitschka F, et al. Tumoral immune cell exploitation in colorectal cancer metastases can be targeted effectively by anti-CCR5 therapy in cancer patients. Cancer Cell. 2016;29:587–601. doi: 10.1016/j.ccell.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 16.Oppermann M. Chemokine receptor CCR5: insights into structure, function, and regulation. Cell Signal. 2004;16:1201–1210. doi: 10.1016/j.cellsig.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 17.Gallo RC, Lusso P. Chemokines and HIV infection. Curr Opin Infect Dis. 1997;10:12–17. doi: 10.1097/00001432-199702000-00004. [DOI] [Google Scholar]
- 18.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton RE, Hill CM, et al. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 19.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, Wu L, Mackay CR, LaRosa G, Newman W, et al. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. [DOI] [PubMed] [Google Scholar]
- 20.Kim MB, Giesler KE, Tahirovic YA, Truax VM, Liotta DC, Wilson LJ. CCR5 receptor antagonists in preclinical to phase II clinical development for treatment of HIV. Expert Opin Investig Drugs. 2016;25:1377–1392. doi: 10.1080/13543784.2016.1254615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Gao X, Martin J, Rosa B, Chen Z, Mitreva M, Henrich T, Kuritzkes D, Ratner L. Evolution of coreceptor utilization to escape CCR5 antagonist therapy. Virology. 2016;494:198–214. doi: 10.1016/j.virol.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McNiff T, Dezube BJ. CCR5 antagonists in the treatment of HIV-infected persons: is their cancer risk increased, decreased, or unchanged. AIDS Read. 2009;19:218–22, 24. [PubMed] [Google Scholar]
- 23.Henrich TJ, Hanhauser E, Hu Z, Stellbrink HJ, Noah C, Martin JN, Deeks SG, Kuritzkes DR, Pereyra F. Viremic control and viral coreceptor usage in two HIV-1-infected persons homozygous for CCR5 Delta32. Aids. 2015;29:867–876. doi: 10.1097/QAD.0000000000000629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quillent C, Oberlin E, Braun J, Rousset D, Gonzalez-Canali G, Métais P, Montagnier L, Virelizier J-L, Arenzana-Seisdedos F, Beretta A. HIV-1-resistance phenotype conferred by combination of two separate inherited mutations of CCR5 gene. The Lancet. 1998;351:14–18. doi: 10.1016/S0140-6736(97)09185-X. [DOI] [PubMed] [Google Scholar]
- 25.Ying H, Wang J, Gao X. CCL5-403, CCR5-59029, and Delta32 polymorphisms and cancer risk: a meta-analysis based on 20,625 subjects. Tumour Biol. 2014;35:5895–5904. doi: 10.1007/s13277-014-1780-9. [DOI] [PubMed] [Google Scholar]
- 26.Singh SK, Mishra MK, Eltoum I-EA, Bae S, Lillard JW, Singh R. CCR5/CCL5 axis interaction promotes migratory and invasiveness of pancreatic cancer cells. Sci Rep. 2018;8:1323. doi: 10.1038/s41598-018-19643-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Liu T, Yang N, Xu S, Li X, Wang D. Hypoxia and macrophages promote glioblastoma invasion by the CCL4-CCR5 axis. Oncol Rep. 2016;36:3522–3528. doi: 10.3892/or.2016.5171. [DOI] [PubMed] [Google Scholar]
- 28.Velasco-Velazquez M, Jiao X, De La Fuente M, Pestell TG, Ertel A, Lisanti MP, Pestell RG. CCR5 antagonist blocks metastasis of basal breast cancer cells. Cancer Res. 2012;72:3839–3850. doi: 10.1158/0008-5472.CAN-11-3917. [DOI] [PubMed] [Google Scholar]
- 29.Gao D, Rahbar R, Fish EN. CCL5 activation of CCR5 regulates cell metabolism to enhance proliferation of breast cancer cells. Open Biol. 2016;6:160122. doi: 10.1098/rsob.160122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanabe Y, Sasaki S, Mukaida N, Baba T. Blockade of the chemokine receptor, CCR5, reduces the growth of orthotopically injected colon cancer cells via limiting cancerassociated fibroblast accumulation. Oncotarget. 2016;7:48335. doi: 10.18632/oncotarget.v7i30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mencarelli A, Graziosi L, Renga B, Cipriani S, D’Amore C, Francisci D, Bruno A, Baldelli F, Donini A, Fiorucci S. CCR5 antagonism by maraviroc reduces the potential for gastric cancer cell dissemination. Transl Oncol. 2013;6:784–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Musha H, Ohtani H, Mizoi T, Kinouchi M, Nakayama T, Shiiba K, Miyagawa K, Nagura H, Yoshie O, Sasaki I. Selective infiltration of CCR5(+)CXCR3(+) T lymphocytes in human colorectal carcinoma. Int J Cancer. 2005;116:949–956. doi: 10.1002/ijc.21135. [DOI] [PubMed] [Google Scholar]
- 33.Findeisen P, Kloor M, Merx S, Sutter C, Woerner SM, Dostmann N, Benner A, Dondog B, Pawlita M, Dippold W, et al. T25 repeat in the 3’ untranslated region of the CASP2 gene: a sensitive and specific marker for microsatellite instability in colorectal cancer. Cancer Res. 2005;65:8072–8078. doi: 10.1158/0008-5472.CAN-04-4146. [DOI] [PubMed] [Google Scholar]
- 34.Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, McQuaid S, Gray RT, Murray LJ, Coleman HG, et al. QuPath: open source software for digital pathology image analysis. Sci Rep. 2017;7:16878. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kather JN, Suarez-Carmona M, Charoentong P, Weis CA, Hirsch D, Bankhead P. Topography of cancer-associated immune cells in human solid tumors. Elife. 2018 Sep 4;7:pii:e36967. doi: 10.7554/eLife.36967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abdi R, Tran TB, Sahagun-Ruiz A, Murphy PM, Brenner BM, Milford EL, McDermott DH. Chemokine receptor polymorphism and risk of acute rejection in human renal transplantation. J Am Soc Nephrol. 2002;13:754–758. [DOI] [PubMed] [Google Scholar]
- 37.Jones DT, Hutter B, Jager N, Korshunov A, Kool M, Warnatz HJ, Zichner T, Lambert SR, Ryzhova M, Quang DAK, et al. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet. 2013;45:927–932. doi: 10.1038/ng.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kather JN, Halama N. Harnessing the innate immune system and local immunological microenvironment to treat colorectal cancer. Br J Cancer. 2019;120:871–882. doi: 10.1038/s41416-019-0441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zimmermann T, Moehler M, Gockel I, Sgourakis GG, Biesterfeld S, Müller M, Berger MR, Lang H, Galle PR, Schimanski CC. Low expression of chemokine receptor CCR5 in human colorectal cancer correlates with lymphatic dissemination and reduced CD8+ T-cell infiltration. Int J Colorectal Dis. 2010;25:417–424. doi: 10.1007/s00384-009-0868-y. [DOI] [PubMed] [Google Scholar]
- 41.Nishikawa G, Kawada K, Nakagawa J, Toda K, Ogawa R, Inamoto S, Mizuno R, Itatani Y, Sakai Y. Bone marrow-derived mesenchymal stem cells promote colorectal cancer progression via CCR5. Cell Death Dis. 2019;10:264. doi: 10.1038/s41419-019-1508-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cambien B, Richard-Fiardo P, Karimdjee BF, Martini V, Ferrua B, Pitard B, Schmid-Antomarchi H, Schmid-Alliana A, Glinskii VV. CCL5 neutralization restricts cancer growth and potentiates the targeting of PDGFRβ in colorectal carcinoma. PLoS One. 2011;6:e28842. doi: 10.1371/journal.pone.0028842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, Richardson AL, Polyak K, Tubo R, Weinberg RA. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 44.Sicoli D, Jiao X, Ju X, Velasco-Velazquez M, Ertel A, Addya S, Li Z, Andò S, Fatatis A, Paudyal B, et al. CCR5 receptor antagonists block metastasis to bone of v-Src oncogene-transformed metastatic prostate cancer cell lines. Cancer Res. 2014;74:7103–7114. doi: 10.1158/0008-5472.CAN-14-0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Long H, Xiang T, Qi W, Huang J, Chen J, He L, Liang Z, Guo B, Li Y, Xie R, et al. CD133+ ovarian cancer stem-like cells promote non-stem cancer cell metastasis via CCL5 induced epithelial-mesenchymal transition. Oncotarget. 2015;6:5846–5859. doi: 10.18632/oncotarget.3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.D’Esposito V, Liguoro D, Ambrosio MR, Collina F, Cantile M, Spinelli R, Raciti GA, Miele C, Valentino R, Campiglia P, et al. Adipose microenvironment promotes triple negative breast cancer cell invasiveness and dissemination by producing CCL5. Oncotarget. 2016;7:24495–24509. doi: 10.18632/oncotarget.8336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alkhatib G. The biology of CCR5 and CXCR4. Curr Opin HIV AIDS. 2009;4:96–103. doi: 10.1097/COH.0b013e328324bbec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mantovani A, Savino B, Locati M, Zammataro L, Allavena P, Bonecchi R. The chemokine system in cancer biology and therapy. Cytokine Growth Factor Rev. 2010;21:27–39. doi: 10.1016/j.cytogfr.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 49.Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 50.Pereira RW, Pires ER, Duarte APM, Torloni H, Proietti AB, de Moura RP, Proietti AB, Simpson AJG, Pena SDJ. Frequency of the CCR5 32 allele in Brazilians: a study in colorectal cancer and in HTLV-I infection. Genet Mol Biol. 2000;23:4. doi: 10.1590/S1415-47572000000300003. [DOI] [Google Scholar]
- 51.O’Brien SJ, Moore JP. The effect of genetic variation in chemokines and their receptors on HIV transmission and progression to AIDS. Immunol Rev. 2000;177:99–111. [DOI] [PubMed] [Google Scholar]
- 52.Marmor M, Sheppard HW, Donnell D, Bozeman S, Celum C, Buchbinder S, Koblin B, Seage GR. Homozygous and heterozygous CCR5-Delta32 genotypes are associated with resistance to HIV infection. J Acquir Immune Defic Syndr. 2001;27:472–481. [DOI] [PubMed] [Google Scholar]
- 53.Reshef R, Luger SM, Hexner EO, Loren AW, Frey NV, Nasta SD, Goldstein SC, Stadtmauer EA, Smith J, Bailey S, et al. Blockade of lymphocyte chemotaxis in visceral graft-versus-host disease. N Engl J Med. 2012;367:135–145. doi: 10.1056/NEJMoa1201248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Halvorsen EC, Hamilton MJ, Young A, Wadsworth BJ, LePard NE, Lee HN, Firmino N, Collier JL, Bennewith KL. Maraviroc decreases CCL8-mediated migration of CCR5+ regulatory T cells and reduces metastatic tumor growth in the lungs. Oncoimmunology. 2016;5:e1150398. doi: 10.1080/2162402X.2016.1150398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nie Y, Huang H, Guo M, Chen J, Wu W, Li W, Xu X, Lin X, Fu W, Yao Y ,et al. Breast phyllodes tumors recruit and repolarize tumor-associated macrophages via secreting CCL5 to promote malignant progression, which can be inhibited by CCR5 inhibition therapy. Clin Cancer Res. 2019. [DOI] [PubMed] [Google Scholar]
- 56.Casagrande N, Borghese C, Visser L, Mongiat M, Colombatti A, Aldinucci D. CCR5 antagonism by maraviroc inhibits Hodgkin lymphoma microenvironment interactions and xenograft growth. Haematologica. 2019;104:564–575. doi: 10.3324/haematol.2018.196725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aldinucci D, Casagrande N. Inhibition of the CCL5/CCR5 Axis against the Progression of Gastric Cancer. Int J Mol Sci. 2018;19:1477. doi: 10.3390/ijms19051477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van Cutsem E, Borras JM, Castells A, Ciardiello F, Ducreux M, Haq A, Schmoll H-J, Tabernero J. Improving outcomes in colorectal cancer: where do we go from here?. Eur J Cancer. 2013;49:2476–2485. doi: 10.1016/j.ejca.2013.03.026. [DOI] [PubMed] [Google Scholar]
- 59.Flemer B, Lynch DB, Brown JMR, Jeffery IB, Ryan FJ, Claesson MJ, O’Riordain M, Shanahan F, O’Toole PW. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut. 2017;66:633–643. doi: 10.1136/gutjnl-2015-309595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


