Abstract
Adoptive cell therapy (ACT) with T cells targeting neoantigens can mediate durable responses in patients with metastatic cancer. Cell therapies targeting common shared antigens for epithelial cancers are not yet broadly available. Here, we report the identification and characterization in one patient of T-cell receptors (TCRs) recognizing mutated p53 p.R175H, which is shared among a subset of patients with cancer. Tumor-infiltrating lymphocytes were screened for recognition of mutated neoantigens in a patient with metastatic colorectal cancer. HLA-A*0201-restricted recognition of mutated p53 p.R175H was identified, and the minimal peptide epitope was HMTEVVRHC. Reactive T cells were isolated by tetramer sorting, and three TCRs were identified. These TCRs mediated recognition of commercially available ovarian cancer, uterine carcinoma, and myeloma cell lines, as well as an NIH patient–derived esophageal adenocarcinoma line that endogenously expressed p53 p.R175H and HLA-A*0201. They also mediated recognition of p53 p.R175H+ colon, breast, and leukemia cell lines after transduction with a retrovirus encoding HLAA*0201. This work demonstrates that common shared mutated epitopes such as those found in p53 can elicit immunogenic responses and that the application of ACT may be extended to patients with any cancer histology that expresses both HLA-A*0201 and the p53 p.R175H mutation.
Introduction
Adoptive cell therapy (ACT) with tumor-infiltrating lymphocytes (TILs) can induce complete, durable cancer regression in patients with metastatic melanoma (1). Single patient reports have shown that ACT can target mutated antigens and mediate durable responses in patients with metastatic cholangiocarcinoma, colon, and cervical cancers (2–4). One of several challenges in translating neoantigen-targeted therapies to patients with cancer is the unique neoantigen repertoire of each patient. There are few shared mutated targets among patients, even among patients with similar histologic cancer types. However, the identification of shared immunogenic neoantigens would facilitate the development of therapies that could be more broadly applied to patients with cancer.
The TP53 gene is frequently mutated in cancer; TP53 mutations are found in 40% to 50% of cancer patients (5–8). TP53 mutations affect most of the hallmarks of cancer cells, including genome instability, increased invasion, metastasis, apoptosis, and proliferation. Moreover, cancers with TP53 mutations frequently have single-nucleotide variants, including “hotspot” mutations at amino acid positions R175, G245, R248, R249, R273, and R282. A substantial proportion of TP53 mutations are found at one of these six different hotspot locations across all cancers (9). Mutations in TP53 have been associated with conferring growth advantage to tumor cells, making these mutations desirable as a neoantigen target (9). Despite the phenotypic effects, no current pharmacotherapies exist that target mutated TP53 in cancer patients. Here, we describe T-cell–mediated recognition of mutated TP53 in the context of a common HLA allele and characterize multiple T-cell receptors that can be of use in the ACT of cancer patients.
Materials and Methods
Patient and treatment characteristics
A 36-year-old woman presented to the Surgery Branch, NCI with metastatic colorectal cancer (KRAS wild-type, microsatellite stable) involving bilateral lungs, liver, and lymph nodes. Disease had demonstrated progression through treatment with capecitabine, oxaliplatin, and bevacizumab. Pulmonary metastases (tumors 4196–1, 4196–2) were resected via video-assisted thoracoscopic surgery for generation of TIL and genetic analysis following an NCI IRB-approved tissue procurement protocol. Upon identification of mutation-reactive lymphocyte cultures, she was enrolled on the NCI IRB-approved phase I/II protocol 10-C-0166, the purpose of which is to evaluate the safety and efficacy of the adoptive transfer of autologous, ex vivo–expanded TIL in patients with advanced gastrointestinal cancers. Treatment consisted of nonmyeloablative lymphodepleting chemotherapy (cyclophosphamide and fludarabine), a single dose of pembrolizumab administered two days prior to cell infusion, infusion of selected autologous TIL, and three additional doses of pembrolizumab given at 3-week intervals. Posttreatment, a newly identified adrenal metastasis (tumor 4249) was resected for additional evaluation. We obtained informed written consent for every aspect of her treatment, and all studies were conducted in accordance with The Declaration of Helsinki, The Belmont Report, and the U.S. Common Rule.
Whole-exome sequencing and RNA-seq
Portions of both lung tumors were retained for the isolation and purification of genomic DNA and RNA for whole-exome sequencing and RNA-seq as previously described (2–4). Genomic DNA (gDNA) and RNA were isolated and purified from fresh tumor and patient-matched normal apheresis samples using the QIAGEN DNA/RNA AllPrep MiniKit (cat. # 80204) per manufacturer instructions. Whole-exome library construction and exon capture of approximately 20,000 coding genes were prepared using Agilent Technologies SureSelectXT Target Enrichment System (cat. #5190–8863). Whole-exome sequencing (WES) was performed using a NextSeq 500 desktop sequencer (Illumina). RNA-seq libraries were prepared using 200 ng of total RNA with the Illumina TruSeq RNA Stranded library prep kit following the manufacturer’s protocol. RNA-seq libraries were paired-end sequenced on our NextSeq 500/550 desktop sequencer (Illumina) again using the same mechanism.
Mutation identification and copy-number aberration
WES was performed on tumor tissue and normal peripheral blood cells. Alignments were performed using NovoAlign MPI (Novocraft, http://www.novocraft.com/) to human genome build hg19. Duplicates were marked and sorted using Picard’s MarkDuplicates tool. Reads were then split and trimmed using GATK SplitNTrim tool, after which In/Del realignment and base recalibration were performed using GATK toolbox. Four different variant calling tools were applied including Varscan2 (http://varscan.sourceforge.net), SomaticSniper (http://gmt.genome.wustl.edu/packages/somatic-sniper/), Strelka (https://sites.google.com/site/strelkasomaticvariantcaller/), and Mutect (https://www.broadinstitute.org/gatk/). After the variant calling, we used the following filters to identify putative mutations: a tumor and normal coverage of greater than 10, a variant allele frequency (vaf) of 7% or above, variant read counts of 4 or above, and two of the four callers (Varscan 2, Somatic Sniper, Strelka, and Mutect) used to identify single-nucleotide variants (snvs), although indels were only required to be identified using one of the two callers capable of returning indels (VarScan 2 and Strelka). The tumor purity estimates (10) for the two resected lesions 4196–1and 4196–2 were 30% and 49%, respectively, and the mean target coverage for 4196–1 and 4196–2 were 202 and 224, respectively. The minimum variant allele frequency was lowered from 10%, which has typically been used as a cutoff, to 7% to facilitate the identification of subclonal mutations in tumors with greater than 50% normal contamination. The requirement for 4 or more variant calls, coupled with the high read depth and the requirement for single-nucleotide variants (SNV) to be identified using multiple callers, helped to minimize the number of false positives identified using these filters. In addition, variant reads were manually curated using the Integrative Genomics Viewer. Sequenza, R package, was applied to determine aberrations in tumor chromosome copy numbers, with normal samples as reference and hg19 coordinates.
HLA typing and calculation of HLA-A allele frequency
Both PHLAT (v1.0) and HLA sequencing were used for HLA typing. Chromosome 6 was extracted from the WES data and aligned to HLA genome reference from the IMGT/HLA database using Novocraft (v3.02.10). After sorting using SAMtools, reads that perfectly matched to HLA-A*0201 and HLA-A*2402 alleles were identified. The mismatched sequence nucleotides were identified at the genomic level. Only reads that contained nucleotides unique for a given HLA-A allele were counted to determine the frequency of HLA-A*0201 and HLA-A*2402 in tumor. The frequency of one HLA-A allele was calculated by averaging the frequency at each of the locations. The following equation was used to calculate the frequency:
[Number of one unique HLA-A allele reads at the given location] × 100/[Sum of both HLA-A allele reads at the give location]. Wilcoxon signed-rank test was used to calculate the significance of the comparison between the HLA-A*0201 and HLA-A*2402 allele.
Generation of TIL for screening
TILs were generated using previously described methods (11). Briefly, resected tumors were trimmed of normal tissue immediately after surgical excision. Areas of firm, solid tumor were selected for processing and sized to about 1 to 3 mm per section. Individual fragments were placed in wells of a 24-well plate in 2 mL of complete media containing high-dose IL2 (6000 IU/mL, Chiron). Fragments were cultured at 37°C at 5% CO2 for 5 days. On day 5, culture media were replenished with fresh media and IL2 (6000 IU/mL, Prometheus Laboratories) and reassessed every 2 to 3 days. When cultures exceeded 1e6 cells/mL or were nearly confluent, the wells were split 1:1. Each fragment was maintained as a separate culture. Complete media consisted of RPMI-1640 supplemented with 10% in-house serum, 2 mmol/L L-glutamine, 25 mmol/L HEPES, and 10 μg/mL gentamicin.
Rapid expansion of TIL for treatment
G-Rex 100 Gas Permeable Cell Culture Flasks (Wilson Wolf Manufacturing) were used to expand selected fragments for patient treatment in 50/50 media, which consisted of 50% AIM-V media (Thermo Fisher Scientific) and 50% complete media (described above) containing IL2 (500 CU/mL).
Frozen, irradiated donor PBMC (feeder cells) were thawed, and 5e8 feeder cells were added per G-Rex 100 flask, in addition to 400 mL 50/50 media, OKT3 (30 ng/mL), 5e6 patient TIL, and cultured at 37° C at 5% CO2 for 5 days. Media were replenished on day 5 and split on day 7. Media were replenished again between 2 and 5 days prior to infusion (days 9–12 of REP), depending upon degree of cell growth.
Generation of autologous dendritic cells (DCs)
PBMCs were thawed in warmed AIM-V media and centrifuged at 900 rpm for 10 minutes. Cell pellets were washed with AIM-V and allowed to rest on tissue culture-treated 162 cm2 flasks for 90 minutes. Adherent cells were washed twice with RPMI-1640, once with AIM-V, and then allowed to culture for 3 days in DC media (RPMI-1640 þ L-glutamine, 5% heat-inactivated commercial human serum albumin, 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mmol/L L-glutamine, 200 U/mL IL4 [PeproTech], and 800 IU/mL GM-CSF [PeproTech]). Media were replenished with fresh DC media (200 U/mL IL4 and 800 IU/mL GM-CSF) on day 3. Cells were harvested for screening or frozen for future use on day 5.
Construction of tandem minigene (TMG) constructs and in vitro transcription of TMG RNA
One hundred and seventy-one mutations were identified by whole-exome and transcriptome sequencing of the 4196 tumors (Supplementary Table S1). For each mutation, a minigene encoding the mutated amino acid flanked by 12 amino acids on either side was generated and synthesized in tandem to create TMG constructs as previously described (12). Briefly, candidate tumor neoepitopes were synthesized into minigenes containing the sequence encoding the mutated amino acid flanked by 12 amino acids from the wild-type protein sequence. Sixteen minigenes were constructed in succession to create a TMG; 11 total TMGs were synthesized for this patient (Supplementary Table S1). Plasmids encoding the TMGs were linearized with the restriction enzyme Sac II. A control pcDNA3.1/V5-His-TOPO vector encoding GFP was linearized with NotI. Restriction digests were terminated with EDTA, sodium acetate, and ethanol precipitation. Complete plasmid digestion was verified by standard agarose gel electrophoresis. Approximately 1 μg of linearized plasmid was used for the generation of IVT RNA using the mmessage mmachine T7 Ultra kit (Life Technologies) as directed by the manufacturer. RNA was precipitated using the LiCl2 method, and RNA purity and concentrations were assessed using a NanoDrop spectrophotometer. RNA was then aliquoted into microtubes and stored at −80°aC until use.
Peptide pool synthesis
Candidate tumor neoepitopes were synthesized into 25 amino acid crude peptides. The mutant amino acid was flanked with 12 amino acids from the wild-type protein sequence. Peptides were synthesized by GenScript and resuspended at 10 mg/mL in DMSO. One microliter aliquots was taken from each stock solution and pooled with a total of 18 peptides to create a peptide pool to screen for TIL reactivity via coculture assay. Ten total peptide pools (PP) were created (Supplementary Table S2).
Identification of neoantigen-reactive T cells
Immature autologous DCs were generated for TIL screening as described above. DCs were either electroporated with TMG-derived RNA or pulsed with PPs. Electroporation was performed by suspending DCs at 1 × 107 cells/mL in Opti-Mem Reduced Serum Medium (Thermo Fisher Scientific) and mixed gently with 8 μg IVT RNA before being transferred to a 2-mm cuvette. Electroporation was performed with a BTX ECM 830 Electroporation Generator (Fisher Scientific) at 150 V for 10 milliseconds. Cells were harvested and immediately plated in DC media to rest overnight. For DCs pulsed with peptides, ~5 × 104 cells were plated in a 96-well plate and pulsed with peptides at a final total peptide concentration of 50 μg/mL. Transfected and peptide-pulsed cells were rested overnight in DC media at 37° C at 5% CO2 for 18 to 20 hours. On the day of coculture, DCs were centrifuged at 1,300 rpm for 5 minutes and supernatant was removed. TIL was plated on IFNγ ELISPOT plates and allowed to coculture with transferred electroporated or pulsed DCs (E:T ratio 2:5) overnight in media at 37° C at 5% CO2 for 18 to 20 hours. TIL were then harvested and assayed using flow cytometry (BD FACSCanto I or II, BD Biosciences). Data were analyzed using FlowJo software (BD Biosciences).
Reactivity was assessed by measuring upregulated expression of 4-1BB (CD137) and OX40 (CD134), or generation of IFNγ (IFNγ ELISPOT) at least 2.5 times background expression. Both of these assays are described more completely below. Fragments selected for treatment were then expanded in an institution-standardized rapid expansion protocol in G-Rex flasks (as described above in the section entitled “Rapid expansion of TIL for treatment”). Cells were cultured at 37°C at 5% CO2 and replenished with fresh 50/50 media with IL2 on day 5 and during days 9 to 12, depending upon TIL growth. Cells were harvested on day 14 for processing for inclusion in the patient’s therapeutic infusion bag (Rx1 TIL infusion bag).
Flow cytometry antibodies
The following mouse anti-human flow cytometry antibodies were used in this report:
CD3 APC-H7 (clone SK7) cat no. 641406 (BD Biosciences)
TCR beta monoclonal antibody—FITC (H57–597) cat no. 11-5961-86 (Thermo Fisher Scientific)
CD8 Pe-Cy7 (clone SK1, RUO GMP) cat no. 335787 (BD Biosciences)
CD4 PE (clone SK3) cat no. 347327 (BD Biosciences)
CD137 APC (clone 4B4–1, RUO) cat no. 550890 (BD Biosciences)
CD134 FITC (clone ACT35, RUO) cat no. 555837 (BD Biosciences)
Flow-cytometric assessment of 4–1BB (CD137), OX40 (CD134)
On the day of assessment, cells were gently harvested from 96-well ELISPOT plates and centrifuged at 1300 rpm for 5 minutes. Supernatant was flicked out of the plate, and remaining cell pellet was aliquoted with 10 μL staining antibody cocktail:
Staining antibody cocktail
CD3 (APC-Cy7) 0.4 μL/well
CD8 (Pe-Cy7) 0.2 μL/well
CD4 (PE) 0.5μL/well
4–1BB (CD137, APC) 0.5 μL/well
OX40 (CD134, FITC) 0.5 μL/well or mTCRb (FITC) at 0.5 μL/well for TCR-transduced cells
FACS buffer (up to 10 μL/well)—comprised of PBS (5% heat-inactivated FCS)
Cells were stained at 4°C for 30 minutes. After incubation, the plate was washed with 200 μL/well FACS buffer and centrifuged for 5 minutes at 1,300 rpm. Supernatant was removed, and cell pellets resuspended with FACS buffer/PI (1:60 dilution) for total volume of 120 μL/well. Flow cytometry was performed using the BD FACSCanto I (BD Biosciences) or Canto II and BD FACSDIVA software (BD Biosciences).
IFNγ ELISPOT assay
Nitrocellulose-backed multiscreen (PVDF) ELISPOT plates (Millipore; cat no. MAIPSWU10) were pre-wet with 70% ethanol for 90 to 120 seconds. Plates were washed 3 × with 200 μL PBS. IFNy capture antibody (1-D1K, stock 1 mg/mL, Mabtech, Inc.; cat no. 3420-3-1000) was diluted to 10 μg/mL (1:100) in PBS and plated at 50 μL/well. Plates were covered with plastic wrap and incubated at 4°C overnight.
On the day of coculture, plates were washed again with PBS (3 × with 200 μL/well) and then plated with 200 μL/well 50/50 media. Plates were allowed to incubate for 2 hours at 37°C. Media were then discarded, and effector cells were plated at 2e4 cells/well and target cells transferred, into a total volume of 200 μL/well. Plates were then incubated for 16 to 21 hours at 37°C at 5% CO2.
On day 3, cells were harvested from ELISPOT plates. ELISPOT plates were then washed 5 × in PBS/0.05% Tween-20 and plated with 100 μL/well of biotinylated anti-human IFNg detection antibody (7-B6–1-biotin, stock 1 mg/mL, Mabtech, Inc.; cat no. 3420-6-1000) after dilution to 1 μg/mL and filtration through a0.22 mm 50 mL filter (MilliporeSigma; PES membrane, cat no. SLGP033RS). Plates were incubated for 2 hours at room temperature, then washed in previous wash buffer. Streptavidin–ALP was diluted (1:3,000) with PBS/0.05% FCS, then added to each well (100 μL/well). Plates were incubated for 1 hour at room temperature and then washed 5 × in PBS. BCIP/NBT substrate (VWR; cat no. 50-81-10) was filtered with a 0.45 μm filter (Millipore). One hundred microliters was plated per well and allowed to incubate in the dark at room temperature for 13 minutes. Development was stopped by washing the plate thoroughly under running tap water. Excess water was flicked vigorously off, and the plates were allowed to dry. Spots were scanned and counted using an ImmunoSpot plate reader and associated software (Cellular Technologies Ltd.).
Additional characterization of mutant p53-reactive cells
To evaluate the HLA restriction of mutant p53-reactive cells, Cos7 cells were transfected with the DNA plasmid for the TMG encoding TP53 (TMG1) and DNA plasmids for each of the patient’s class I HLA alleles (A*0201, A*2402, B13, B15, C*03,C*06) using Lipofectamine 2000 (Thermo Fisher Scientific). Approximately 1 × 105 Cos7 cells were cocultured with 2 × 104 T cells from the patient’s Rx1 TIL infusion bag (E:T ratio 1:5) in 50/50 media without added cytokines on IFNγ ELISPOT membranes. Cells were harvested for flow-cytometric evaluation of CD137 expression, and the membrane was processed to evaluate IFNγ secretion.
netMHC4.0 was used to predict candidate minimal epitopes for evaluation, based on evidence that the mutation was HLA-A*0201 restricted. The 25mer mutated peptide (YKQSQHMTEVVRHCPHHERCSDSDG) was input for prediction of 9mer, 10mer, and 11mer peptides. The top five predicted candidate peptides containing the mutated amino acid were synthesized (4 mg, crude synthesis; GenScript). Candidate peptides were suspended in DMSO to 10 mg/mL, diluted sequentially with 10-fold dilutions, and pulsed onto T2 cells for 2 hours. Approximately 1 ×105 T2 cells were washed and cocultured with 2 ×104 T cells from the patient’s Rx1 TIL infusion bag. Reactivity was evaluated by IFNγ ELISPOT and upregulation of CD137 as described above.
Isolation and identification of mutant p53-reactive TCRs
To isolate mutant p53-reactive TCRs, we used a single-cell RNA-seq method that has been described previously (11) with some modifications. Briefly, an HLA-A*0201 tetramer conjugated with the HLA-A*0201–restricted minimal epitope (HMTEVVRHC) was constructed using the QuickSwitch custom tetramer kit (MBL). The tetramer was then used to stain T cells from the patient’s Rx1 TIL infusion bag; the top 5% staining cells were single-cell sorted into a 96-well plate using the BD FACSAria III (BD). Sorted T cells were then subjected to automated single-cell RNA-seq sample preparation using the Fluidigm C1 platform following the manufacturer’s instructions (Fluidigm). Single-cell RNA-seq analysis was performed using the Illumina MiSeq sequencer (Illumina) at NCI/CCR Genomics Core. The paired full-length TCRα/β sequences were identified using an in-house software (13).
Candidate TCR vectors were synthesized by linking TCRα V–J regions with the mouse TCRα-constant chain and TCRβ V–D–J region linked to the mouse TCRβ–constant chain to facilitate identification of transduced cells. The full-length TCRα and TCRβ were cloned into a pMSGV1 vector with Nco1 and EcoRI sites (GenScript).
293 cells expressing gag and pol proteins (293 GP cells) were transfected with the desired retroviral vector plasmid and RD114 envelope protein plasmid using Lipofectamine 2000 per manufacturer instructions. Retroviral supernatant was harvested the day after transfection and either used fresh or added to polybrene and frozen for storage at −80°C.
Donor PBMC was thawed, counted, and resuspended at 3.75e6 cells/mL in 50/50 media with 300 IU/mL IL2 and 50 ng/mL OKT3. A total of 7.5e6 cells were plated per well in a 24-well plate 2 days prior to transduction.
The day prior to transduction, non–tissue-culture–coated plates were plated with Retronectin (20 μg/mL in PBS) with 2 mL/well in a 6-well plate. Plates were wrapped with plastic wrap and stored overnight at 4°C.
The day of transduction, Retronectin was aspirated off the non–tissue culture plates, and wells were subsequently blocked with sterile 2% bovine serum albumin (in PBS) for 30 minutes at room temperature (2 mL/well for a 6-well plate). After blocking, wells were washed with PBS twice and then plated with 4 mL/well viral supernatant. Plates were wrapped with plastic wrap and centrifuged at 2,000 × g for 2 hours at 32°C. At the end of 2 hours, viral supernatant was aspirated. Stimulated donor PBMC were harvested, washed, and resuspended in 50/50 media with 300 IU/mL IL2 at 5e5 cells/mL, then plated at 4 mL/well. Plates were wrapped in plastic wrap and spun for 10 minutes at 1,500 rpm at room temperature. Plates were then incubated at 37°C overnight. GFP and mock-transduced cells were used as controls. Transduced cells were cryopreserved 2 to 3 weeks after transduction for future use.
Evaluation of specificity and sensitivity of mutant p53-reactive TCRs
TCR bulk-transduced cells were tested for recognition of mutant p53-expressing tumors. Commercially available tumor lines expressing p.R175H-mutated p53 including colon adenocarcinoma (LS123; ATCC), breast adenocarcinomas (AU-565 and SKBr3; ATCC), ovarian cancer (TYKNU and TYKNU CpR; Japanese Cell Bank), uterine carcinoma (KLE; Japanese Cell Bank), myeloma (KMS26; Japanese Cell Bank), primary ductal carcinoma (HCC1395; ATCC), leukemias (CEM/C1 and CCRF-CEM; ATCC), leiomyosarcoma (SKUT1; ATCC), and lung adenocarcinoma (VMRCLCD) were obtained and passaged several times before testing. All cell lines were purchased in 2016, 2017, or 2018 and were used within 6 months of the original thaw dates. An NIH patient esophageal adenocarcinoma tumor line, which was HLAA*0201+ by HLA typing and p53 p.R175H+ by genotyping, was also evaluated. Mycoplasma testing was not performed, but HLAA2 expression was evaluated by FACS, and expression of the TP53 R175H mutation was evaluated using quantitative RT-PCR specific for the mutation as described below.
HLA-A*0201 was retrovirally transduced into the LS123, AU565, and CCRF-CEM cell lines using retroviral supernatant. Frozen retroviral supernatant encoding HLA-A*0201 was obtained from Paul Robbins (Surgery Branch), thawed, and diluted to 4 mL before being plated onto tumor cells at 5e5 cells/mL in a 6-well plate. Plates were wrapped in plastic wrap, spun for 10 minutes at 1500 rpm at room temperature, then incubated at 37°C. Tumor cells were selected for A°02:01-transduced cells using puromycin.
We also genetically introduced the mutant TP53 R175H gene into the HLA-A°0201+ bone osteosarcoma cell line SAOS-2. The full-length TP53 gene with R175H codon change was fused to a RAKR–SGSG–P2A linker, and truncated CD19 was then synthesized de novo and cloned into the Sleeping Beauty transposon for nonviral stable transgene expression. SAOS2 cells were cotransfected with TP53 transposon and SB11 transposase DNA plasmids with Lipofectamine 2000 and were selected the following day with CD19 microbeads (Miltenyi Biotec). A second CD19 microbead enrichment was performed 14 days later to create the cell lines for T-cell coculture.
To evaluate the specificity and sensitivity of the transduced T cells, they were washed thoroughly and plated at ~2 ×104 cells per ~1 × 105 target tumor cells per well (200 μL/well in a 96-well plate) for 18 to 21 hours in plain 50/50 media without exogenous cytokines. In some experiments, transduced and nontransduced tumor lines were either pulsed with 1 μL DMSO or 1 μL A*02:01-restricted TP53 minimal epitope (stock concentration 1 mg/mL) for 2 hours at 37°C. Cells were washed three times with 50/50 media before coculture with TCR-transduced cells (E:T ratio 1:5) for 16 to 21 hours. Transduced T cells were harvested for flowcytometric analysis for mTCRβ and cell-surface CD137 (4–1BB) expression; IFNγ generation was evaluated by IFNγ ELISPOT assay.
TCR-transduced cells were sorted based on mouse TCRb and either CD4+ or CD8+ using the BD FACSAria III, then rapidly expanded per institution protocol as described above in the section entitled “Rapid expansion of TIL for treatment”). HLA-matched, allogeneic immature DCs were pulsed with serial 10-fold dilutions of minimal epitope peptide and allowed to incubate for 2 hours; DCs were then washed and coincubated with CD4+- or CD8+-sorted TCR-transduced cells for 18 to 20 hours at 37°C and 5% CO2. Reactivity was assessed by CD137 expression by flow cytometry and IFNγ ELISPOT assay.
Evaluation of expression of p53 R175H in tumor cell lines by quantitative RT-PCR
Mutant-specific reverse primer (T175H-RP: 5′-GAGCAGCGCTCATGGTGGGGGCAGT-3′) and a competitive WT-specific reverse DNA oligo (WT-specific blocker 3′-phosphorylated oligo: 5′-GCTCATGGTGGGGGCAGC-3′phospho) plus WT cDNA-specific forward primer (5′-CTTGCATTCTGGGACAGCCAAGTC-3′) were used to amplify mutant-specific amplicons using an SYBR-green qPCR assay kit (iQ SYBR-green supermix, Bio-Rad). The ratio of reverse primer to phosphor-blocker oligo was 50:50 in all tumor samples. A standard qPCR protocol for 40 cycles was used to obtain amplification curves and PCR data were analyzed by Bio-Rad CFX manager (PCR machine CFX96). Copy numbers of TP53-mutant amplicons were measured using a standard curve drawn from known copy numbers of TP53 that were amplified from serially diluted plasmids. Similarly, GAPDH expression for each sample was quantified using a standard curve drawn from known copy numbers of GAPDH that were amplified from serially diluted plasmids. All DNA oligos including primers for GAPDH (FP: 5′-CGACCACTTTGT CAAGCTCA-3′, RP: 5′-AGGGGTCTACATGGCAACT-3′) and TP53 (FP: 5′-CTTGCATTCTGGGACAGCCAAGTC-3′ and RP: 5′-GAGCAGCGCTCATGGTGGGGGCAGC-3′) were obtained from IDT.
Results
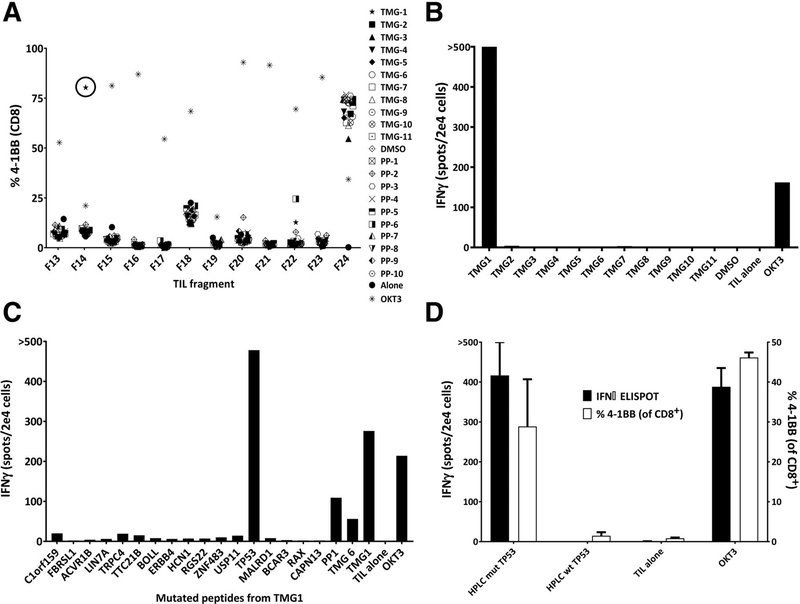
WES of two pulmonary metastases from patient 4196 yielded 171 nonsynonymous mutations for evaluation. These mutations were encoded onto 11 TMGs, with 16 minigenes in each TMG, using previously described methods (10). 25mer peptides encoding each mutation were also synthesized for screening. Eighteen peptides were used to create each peptide pool, totaling in 10 PPs. Lymphocytes grown from 24 tumor fragments were screened for reactivity against the TMGs and PPs presenting all of the patient’s neoepitopes. This identified reactivities in fragment 14 to TMG1, in fragment 20 to peptide pool 2 (PP2), and in fragment 22 to peptide pool 6 (PP6; Fig. 1A and B; Supplementary Fig. S1). Individual peptide testing identified that CD8+ T cells in fragment 14 were reactive with p53 p.R175H (Fig. 1C and D). CD4+ T cells in fragment 20 were reactive with GTF2E1 p.S334F (Supplementary Fig. S2), and CD8+ T cells in fragment 22 were reactive to ARMCX5-GPRASP2 p.G722W, although we have not been able to determine if this reactivity was truly mutation specific (Supplementary Fig. S3). The remainder of the 21 tested fragments were either nonreactive or contained lower percentages of reactive cells (Supplementary Fig. S1). Based on these results, fragments 14, 20, and 22 were selected for expansion and inclusion in the patient’s treatment TIL (Rx1 TIL) per institution protocol.
Figure 1.
Identification of mutation-reactive T cells from TIL. A, Representative plot of the TIL fragment screen for 41BB+ cells following coculture with TMG-transfected and peptide-pulsed autologous DCs. B, IFNγ ELISPOT after coculture of fragment 14 TIL with TMG-transfected autologous DCs. C, Coculture of fragment 14 TIL with autologous DCs pulsed with individual peptides encoding for each mutation on TMG1 (41BB%, IFNγ). D, Fragment 14 TIL reactivity to HPLC-purified p53 peptide (n = 3); error bars, standard deviation.
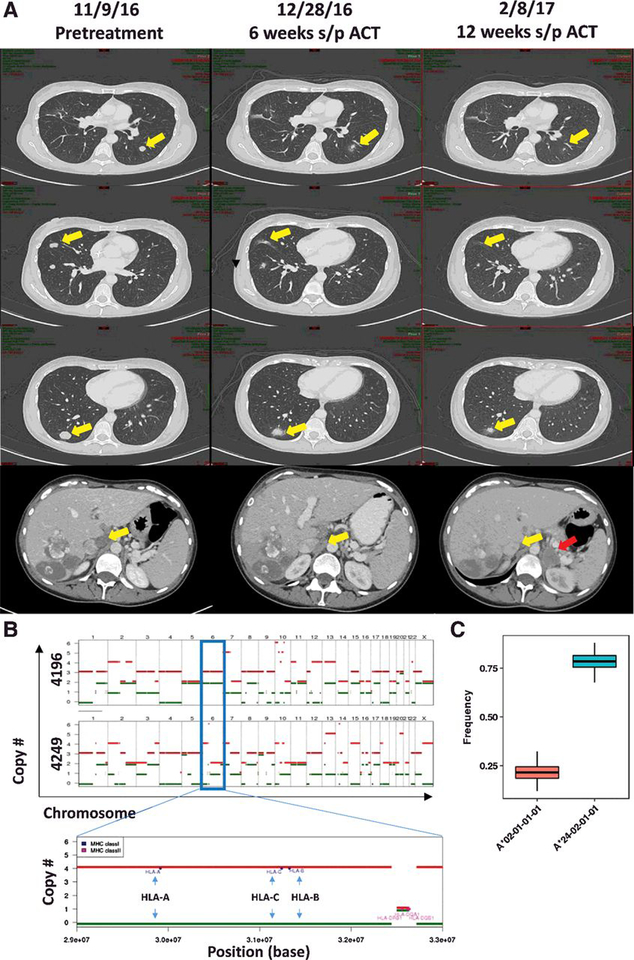
The patient underwent preparative chemotherapy including a pre-cell dose of pembrolizumab followed by infusion of 91.8 ×109 cells. Respiratory distress following infusion precluded the customary post-cell administration of high-dose IL2. She recovered and was discharged on day 8. At her first protocol evaluation (6 weeks after TIL), her target lesions had decreased 11% by RECIST criteria and were associated with similar regression in nontarget liver lesions and aortocaval lymph nodes (Fig. 2A). She received third and fourth doses (final) of pembrolizumab per protocol. At her second protocol evaluation (12 weeks after TIL), her target and off-target lesions continued to decrease in size. However, computed tomography at that evaluation identified a new solitary left adrenal mass, indicating progressive disease (Fig. 2A, bottom, red arrow). This adrenal mass continued to grow and was laparoscopically resected. Chromosome copy-number analysis of the resected lung tumors (4196) and adrenal tumor (4249) demonstrated evolution of loss of heterozygosity (LOH) in a portion of chromosome 6 known to encode the HLA allele. The patient’s adrenal mass did not have the A*0201 allele (Fig. 2B and C; Supplementary Fig. S4). The adrenal tumor also had LOH of the X chromosome, leading to loss of the ARMCX5-GPRASP2 gene. After 7 months, the patient had developed large bowel obstruction and further progressive disease with new bilateral pulmonary lesions, peritoneal carcinomatosis, and ascites. She was taken off study and referred for systemic chemotherapy.
Figure 2.
Treatment of a patient with metastatic colon cancer with TILs. A, Serial cross-sectional imaging indicating initial regression in the patient’s target lesions (lungs, top three panels; aortocaval lymph node, bottom) and evolution of a new left adrenal mass by 12 weeks after ACT (bottom, red arrow).B, Chromosome copy-number analysis comparing the pulmonary lesions (4196) with the adrenal tumor (4249). Red and green bars represent the relative numbers of homologs for each chromosome with relative height on the chart indicating the number of copies for that homolog (top two panels). The HLA locus is located on chromosome 6 (blue outlined box). A magnified view of the earlier portion of chromosome 6 is shown (bottom). Here, the relative locations of HLA-A, HLA-B, and HLA-C alleles are indicated (blue arrows), suggesting that LOH had contributed to the loss of a complete set of class I alleles. C, HLA allele reads were used to estimate the relative frequency of each HLA allele in 4249 (the subsequently resected adrenal lesion). A*0201 is denoted in the red box plot, whereas A 2402 is denoted in the blue box plot. Normal anticipated frequency would be 0.50.
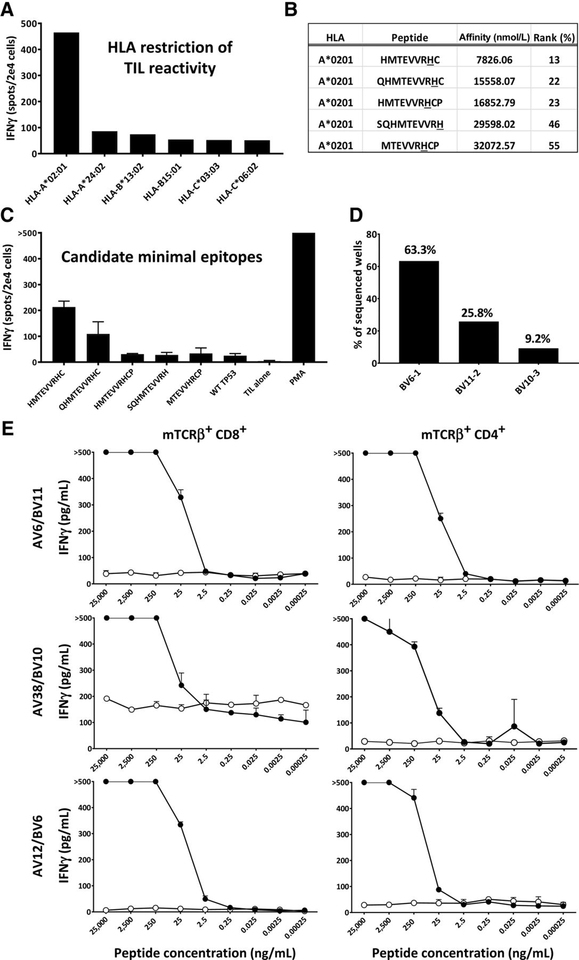
After identifying TIL reactivity to mutated p53, we sought to characterize the HLA restriction and minimal epitope required for T-cell recognition. To assess HLA restriction, the patient’s TIL were cocultured with COS7 cells transfected with the DNA plasmids corresponding to each of the patient’s individual class I HLA alleles as well as the TMG encoding mutant TP53. The patient’s TIL were reactive only when mutated TP53 was presented by HLA-A*0201–transfected COS7 cells (Fig. 3A). The online, open-source netMHC4.0 was used to predict 9mer, 10mer, and 11mer candidate minimal epitopes for evaluation, based on evidence that the mutation was HLA-A*0201 restricted. We selected five candidate peptides containing the mutated amino acid with the highest predicted binding affinities for evaluation. None of the epitopes were predicted to bind to HLA-A*0201 with high-affinity (<50 nmol/L; Fig. 3B). This may be due to the presence of a cysteine residue at the C terminal anchor position of the top two peptides. T2 cells pulsed with candidate minimal epitopes were cocultured with TIL used to treat the patient (Rx1). TIL generated the most IFNγ when presented with the 9mer HMTEVVRHC (Fig. 3C).
Figure 3.
Isolation and characterization of A*0201-restricted TP53 TCR.A, IFNγ generated by TIL after coculture with COS7 cells transfected with TMG1 (containing mutated TP53) and each of the patient’s class I HLA alleles. B, netMHC4.0 prediction of top candidate minimal epitopes based on HLA-A*0201 restriction and mutated 25mer. C, IFNγ generated after TIL coculture with autologous DCs stimulated with each candidate minimal epitope (n = 3); data shown are representative of 3 different experiments. Error bars denote standard deviation. D, Frequencies of three unique candidate TCRs identified from tetramer+ cells. E, CD8+- and CD4+-sorted transduced cells were cocultured with autologous DCs pulsed with decreasing concentrations of mutated p53 peptide (n = 3); error bars, standard deviation.
To isolate p53-reactive TCRs, HLA-A 0201 tetramers loaded with HMTEVVRHC were synthesized and used to stain Rx1 TIL. The top 5% staining cells were sorted into a 96-well plate, and single-cell RNA-seq samples were prepared using the Fluidigm C1 platform (Fluidigm). Single-cell RNA-seq was performed using the Illumina MiSeq with subsequent identification of paired full-length TCRα and TCRβ chain sequences using in-house software. Four possible TCRs were identified using this method. Three unique TCRβ chains were identified, one of which was paired with two possible TCRα chains, with BV6–1 comprising 63.3%, BV11–2 comprising 25.8%, and BV10–3 comprising 9.2% of all productive sequences identified (Fig. 3D). Candidate TCR plasmids were cloned into p.MSGV1 vectors and retrovirally introduced into allogeneic donor lymphocytes. Three receptors exhibited mutation-specific reactivity against the mutant p53 peptide at concentrations as low as 25 ng/mL (Fig. 3E). The amino acid sequences of these three TCRs can be found in Supplementary Table S3. Further, antigen titration with CD4 sorted, transduced cells demonstrated that antigen reactivity was also seen in the CD4+ selected population (purity range, 86.5%–93.3%), suggesting that these TCRs might be coreceptor independent.
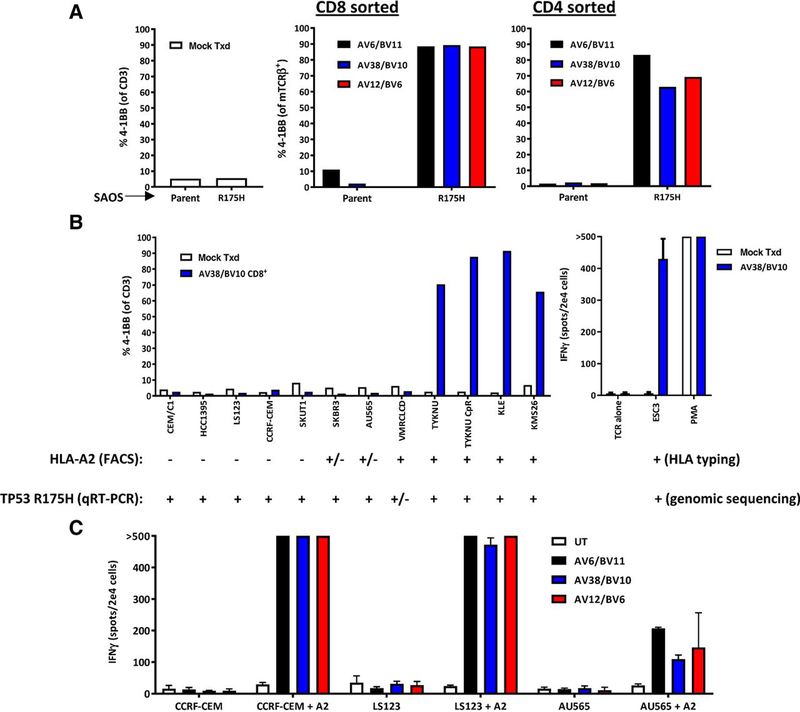
We also evaluated whether these TCR-transduced T cells would recognize tumor lines endogenously expressing mutated p53. We genetically introduced the mutant TP53 gene into the HLAA*0201+ bone osteosarcoma cell line SAOS-2 and evaluated recognition by CD8+ and CD4+ T-cell populations expressing each of the three TCRs (Fig. 4A). Specific recognition of SAOS-2 cells expressing p53 p.R175H in comparison with the parental cell line was mediated by all three TCRs, although recognition appeared slightly better by CD8+ T cells in comparison with CD4+ T cells. In addition, we assembled a set of commercially available cell lines reported to contain themutant TP53 gene including colon adenocarcinoma (LS123; ATCC), breast adenocarcinomas (AU-565 and SKBr3; ATCC), ovarian cancer (TYKNU and TYKNU CpR; Japanese Cell Bank), uterine carcinoma (KLE; Japanese Cell Bank), myeloma (KMS26; Japanese Cell Bank), primary ductal carcinoma (HCC1395; ATCC), leukemias (CEM/C1 and CCRF-CEM; ATCC), leiomyosarcoma (SKUT1; ATCC), and lung adenocarcinoma (VMRCLCD). All cell lines that were positive for HLA-A2 expression by FACS (Supplementary Fig. S5) and expressed greater than 1,000 copies of TP53 p.R175H by quantitative RT-PCR (Supplementary Table S1) were recognized by T cells expressing each of the three TCRs (Fig. 4B; Supplementary Fig. S6A and S6B). An NIH patient-derived esophageal adenocarcinoma line (ESC3) naturally expressing A*0201 and mutated p53 p.R175H was also recognized by all three TCR-transduced cell populations (Fig. 3B, left; Supplementary Fig. S6C). Three cell lines that expressed little or no HLA-A2, as determined by FACS (CCRF-CEM, LS123, and AU565), were transduced with HLA-A*0201 retroviral particles and passaged using puromycin selection. All three TCR-transduced T-cell populations were cocultured with each tumor line overnight and evaluated for reactivity. TCR-transduced cells recognized all three A*0201-transduced tumor lines (Fig. 4C).
Figure 4.
TCR-transduced cell recognition of p53 p.R175H tumors. A, Recognition of genetically modified SAOS2 cells expressing p53 p.R175H was compared with that of the parental cell line based on expression of 4–1BB by TCR-transduced T cells. B, Left, recognition of commercially available cell lines expressing p53 p.R175H was evaluated based on expression of 4–1BB by TCR-transduced T cells. Expression of HLA-A2 on cell lines was evaluated by FACS (Supplementary Fig. S5), and expression of p53 p.R175H was evaluated by quantitative RT-PCR (Supplementary Table S4). HLA-A2 expression was considered weak (+/−) if less than 10% of the cells stained positively compared with isotype, and the MFI with the anti–HLA-A2 antibody divided by that of the isotype was < 2.5. p53+ p.R175H expression was considered weak (+/−) if the copy number was less than 1,000 per 1e5 copies of the house keeping gene GAPDH. B. Right, recognition of an NIH patient–derived tumor line that was HLA typed to be HLA-A*0201+ and was genotyped to contain mutated TP53 was evaluated based on IFNγ secretion by TCR-transduced T cells via ELISPOT. C, HLA-A*0201 was genetically introduced into three cell lines that did not express, or only weakly expressed, HLA-A2 by FACS, and recognition was evaluated using IFNγ ELISPOT assays.
The patient’s pre- and post-TIL infusion blood was assessed for persistence of antigen-reactive TCRs using deep sequencing. The prevalence in the Rx1 TIL infusion bag of the three p53-reactive TCRs was 1.866%, 0.545%, and 0.431%, respectively. None of these were detectable in the patient’s pretreatment pheresis sample. Of the reactive TCRs, one (BV10–3) was undetectable in blood by day +42 after treatment, and another (BV6–1) was undetectable by day +60. The remaining p53-reactive TCR (BV11–2) remained detectable at low levels (0.0027%) through day +120, at which point the patient was taken off protocol (Supplementary Fig. S7).
Discussion
Reports have suggested that treating patients with metastatic GI epithelial cancers using neoantigen-reactive TIL can mediate durable objective responses (2–4). However, this approach is limited by the unique mutational profile of each patient’s tumor. Further, common epithelial cancers exhibit a paucity of shared mutated neoepitopes.
Mutated TP53 represents an ideal shared antigen target. In an assessment of four different pan-cancer sequencing studies, TP53 was found to be mutated in 40% to 50% of sequenced cases (12,520 total) representing a broad range of cancer histologies, including lung cancer (89% of 82 cases), colorectal cancer (72.7% of 1,014 cases), and esophageal cancer (70.7% of 341 cases; refs. 8, 14). Amino acid location R175 is one of the most frequently mutated and comprises approximately 5% of all identified mutations in TP53. Ninety-four percent result in a histidine substitution (5, 6). These specific hotspot mutations can be therapeutic targets for multiple cancer cellular pathologies. Further, the identification of a p53-reactive TCR with a common HLA-restriction element has broad implications. HLA-A*0201 is present in approximately 16% of African Americans and 48% of Caucasian American patients, making it one of the most dominant MHC class I HLAs in the United States (15). Large numbers of patients could thus potentially be treated using a TCR reactive to the mutated p53 p.R175H epitope.
In this patient, several factors appeared to contribute to unsuccessful TIL therapy. First, less than 3% of the adoptively transferred cells in the therapeutic infusion bag were reactive to p53. Copy-number analysis of the patient’s adrenal mass demonstrated LOH in chromosome 6, resulting in the loss of the A*0201 allele. Although mutated p53 was identified in each resected metastatic lesion, loss of the HLA-restriction element precluded TIL recognition of the target neoantigen. Clonal diversity between primary and metastatic tumors has been demonstrated in non–small cell lung cancers and continues to be a challenge with respect to targeting all tumor subpopulations in a given patient (16). Although we presume that a well-characterized tumor suppressor gene such as TP53 is likely to be clonal, the potential loss of TP53 from metastatic lesions may preclude the immunogenic recognition of those lesions. Although the number of neoantigen-reactive cells can be manipulated in TCR-transduced cells, the persistence of infused cells could affect the efficacy of the cell product and the duration of response.
Here, we describe the identification and characterization of three HLA-A*0201–restricted TCRs that recognize p53 p.R175H, thereby demonstrating that mutated TP53 can be an immunogenic antigen. Although reactivity was demonstrated at a single hotspot location (amino acid position 175), this opens the possibility that other hotspots in TP53 may also elicit an immunogenic response. Our results demonstrate that these TCRs can exhibit recognition of the target epitope at low peptide concentrations and when endogenously presented by A*0201-restricted tumor lines. This latter finding suggests that an off-the-shelf TCR—transduced at high frequency into a patient’s PBL for infusion—may be a valuable reagent for the treatment of patients whose tumors express HLA-A*0201 and the TP53 p.R175H mutation.
Supplementary Material
Acknowledgments
We gratefully acknowledge the assistance of Julie Hong, for provision of the ESC3 tumor line; Zhili Zheng, for assistance in TCR sequencing; Robert Somerville and the Surgery Branch Cell Production Facility, for TIL culture and infusion product generation; and Stephanie Goff and the Immunotherapy team, for participation in patient evaluation, treatment, and follow-up.
Footnotes
Note: Supplementary data for this article are available at Cancer Immunology Research Online (http://cancerimmunolres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
E. Tran is a consultant/advisory board member for PACT Pharma and Genocea Biosciences. L. Jia is a senior bioinformatician at NCI. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Rosenberg SA, Restifo NP. Adoptive cell transfer as personalized immunotherapy for human cancer. Science 2015;348:62–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tran E, Turcotte S, Gros A, Robbins PF, Lu YC, Dudley ME, et al. Cancer immunotherapy based on mutation-specific CD4þ T cells in a patient with epithelial cancer. Science 2014;344:641–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stevanovic S, Pasetto A, Helman SR, Gartner JJ, Prickett TD, Howie B, et al. Landscape of immunogenic tumor antigens in successful immunotherapy of virally induced epithelial cancer. Science 2017;356:200–05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tran E, Robbins PF, Lu YC, Prickett TD, Gartner JJ, Jia L, et al. T-cell transfer therapy targeting mutant KRAS in cancer. N Engl J Med 2016;375:2255–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Erratum: mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23:1004. [DOI] [PubMed] [Google Scholar]
- 8.Tran E, Ahmadzadeh M, Lu YC, Gros A, Turcotte S, Robbins PF, et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science 2015;350:1387–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabapathy K, Lane DP. Therapeutic targeting of p53: all mutants are equal, but some mutants are more equal than others. Nat Rev Clin Oncol 2018. Jan;15(1):13–30. [DOI] [PubMed] [Google Scholar]
- 10.Favero F, Joshi T, Marquard AM, Birkbak NJ, Krzystanek M, Li Q, et al. Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data. Ann Oncol 2015;26:64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dudley ME, Wunderlich JR, Shelton TE, Even J, Rosenberg SA. Generation of tumor-infiltrating lymphocyte cultures for use in adoptive transfer therapy for melanoma patients. Journal of Immunotherapy 2003;26: 332–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu YC, Yao X, Crystal JS, Li YF, El-Gamil M, Gross C, et al. Efficient identification of mutated cancer antigens recognized by T cells associated with durable tumor regressions. Clin Cancer Res 2014;20: 3401–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu YC, Zheng Z, Robbins PF, Tran E, Prickett TD, Gartner JJ, et al. An efficient single-cell RNA-Seq approach to identify neoantigen-specific T cell receptors. Mol Ther 2018;26:379–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23:703–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez-Galarza FF, Takeshita LY, Santos EJ, Kempson F, Maia MH, da Silva AL, et al. Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res 2015;43(Database issue): D784–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, et al. Tracking the evolution of non–small-cell lung cancer. N Engl J Med 2017;376:2109–21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.