Abstract.
Tanzania is one of the sub-Saharan countries that have experienced a number of Rift Valley fever (RVF) outbreaks at intervals of 10–20 years since the first isolation of the virus during the early 1930s. Recent studies have reported serological evidence of inter epizootic/epidemic period circulation of RVF virus (RVFV) in livestock and humans. The aim of this study was to conduct a cross-sectional survey in Tanzania during 2015/16 to further explore the possibility that RVFV was circulating among cattle during the Inter epizootic/epidemic period. A total of 443 cattle samples were collected in Manyara, Dodoma, Singida, and Mbeya regions of Tanzania. The samples were tested for RVFV antibodies using a commercial ELISA kit and a plaque reduction neutralization test. Serum samples were also tested for RVFV viral RNA by an reverse transcription polymerase chain reaction (RT-PCR) assay. An overall RVFV seroprevalence rate of 7.7% (34/443) was detected by ELISA among cattle in all four regions. The Mbeya region cattle had the highest seroprevalence of 26.4% (23/87), followed by Dodoma 5.9% (10/171) and lastly Singida 0.9% (1/101). Of 33 ELISA antibody-positive samples, only 0.2% (1/443) had IgM antibody. Of 36 ELISA antibody-positive and doubtful samples, 32 were positive for neutralizing antibody with titers between 10 and > 10,240. None of the samples were positive for RVFV viral RNA by RT-PCR. The detection of RVFV antibodies in cattle suggested that these animals were involved in an enzootic cycle during the interepidemic period and that the high antibody titers may confer protection of cattle against RVFV.
INTRODUCTION
Rift Valley fever virus (RVFV) is the most common pathogen of genus Phlebovirus, family Phenuividae, order Bunyavirales known to infect and cause disease among ruminants as well as humans in Africa.1 Rift Valley fever virus is an enveloped RNA virus that contains a tripartite genome consisting of S (small), M (medium), and L (large) segment. The segments contain genes coding for structural and nonstructural proteins responsible for viral pathogenesis and replication.2,3 Rift Valley fever virus is mainly transmitted by mosquitoes of the genera Culex and Aedes.4,5 Rift Valley fever virus has also been isolated from many other mosquito species, including Anopheles spp., as well as Simulium blackflies, sand flies, and Amblyomma ticks.5–7
The virus is enzootic in most African countries and has been reported to cause significant morbidity and mortality in livestock and humans. Also, the virus has been reported to cause outbreaks in some countries outside Africa, including Saudi Arabia and Yemen.8–12
Rift Valley fever virus is known to cause devastating epizootics characterized by abortions storms, neonatal animal mortality approaching 100% and significant mortality (10–20%) among adult’s ruminants, especially sheep and cattle.13–17 Outbreaks of RVF occur at an interval of 5–20 years and are generally associated with explosions of mosquito populations following periods exceptionally high rainfall.18
Tanzania has experienced a number of RVF epizootics/epidemics since the first reported outbreak of RVF in Kenya in 1930–1931.19 However, subsequent epizootics/epidemics were very patchy with little awareness and, therefore, lack of documentation in most areas in the country.20 The latest outbreak during 2006/7 in Tanzania caused thousands of cases in ruminants and several hundred human fatalities primarily in the central and northern zones of the country.20–23 The outbreak in 2006/7 initially affected ruminants in the northern part of Tanzania bordering Kenya and later spread to 52% of all 21 regions by the end of the outbreak.21 The regions affected included Manyara, Dodoma, and Singida which are the target sites of this study. Of 194,750 domestic ruminants cases reported during the past 10 minor and major outbreaks in Tanzania (1930–2007), 54.01% were cattle, of which 0.4% of approximately 12 million cattle were affected in the latest outbreak in all 11 regions.21,24 Livestock of pastoral and agro-pastoral communities was most affected in the latest outbreak.22 Several studies reported suspected inter-epizootic/epidemic transmission of RVFV among human and livestock in areas with/without a history of outbreaks based mainly on the detection of RVFV antibodies.23–25 In Tanzania, RVF IgG and IgM antibodies have been detected in human and livestock in different regions, including Mbeya, Dodoma, Arusha, Morogoro, Kigoma, Tanga, Mara, and Kagera.26–29
In addition to the possible low-level circulation between mosquitoes and livestock, one study revealed that the virus could be maintained during inter-epizootic/epidemic periods by transovarial transmission in Aedes mosquitoes.4 Moreover, mosquitoes of Aedes and Culex species are known to amplify and transmit the virus in two distinct cycles of RVFV; the enzootic and epizootic/epidemic cycles.22,23,27 The enzootic cycle has been reported to possibly involve Aedes species mosquitoes as the vectors that serve to circulate the virus at a very low incidence without noticeable clinical manifestation in both humans and animals.23 During heavy rainfall and flooding, the population density of Aedes species increases rapidly, resulting in transmission and amplification of the virus by infected vertebrate hosts, leading to further infection of other mosquito species that are capable of transmitting the virus.25,28 The epizootic/epidemic cycle is driven by the involvement of secondary vectors, including Culex species capable of transmitting the virus to both humans and animals.4 Tanzania is known to have the third largest cattle population in Africa, about 21.3 million cattle, after Ethiopia and Sudan (National Bureau of Statistics, 2011), which are at risk of infection by RVFV. The control of RVF in Africa is critically needed to improve food security, economic status, household nutrition, and the well-being of livestock keepers as well as to improve the agricultural sector, which contributes 44.5% of the Tanzania’s national gross domestic product. An understanding of the inter-epizootic/epidemics mechanism(s) of maintenance of RVFV is a priority requirement for the development of improved surveillance and control strategies for this devastating disease. Therefore, the primary aim of this study was to determine the prevalence of RVFV antibody in cattle in selected regions of Tanzania as a possible vertebrate host for sustaining the RVFV cycle during the inter-epidemic/epizootic period (IEP).
MATERIAL AND METHODS
Study area.
The study was conducted in four regions, namely, Manyara, Dodoma, Singida, and Mbeya, which are located in the northern, central, and southwestern zones of Tanzania (Figure 1). All four regions selected are located in the eastern wing of the Rift Valley in Tanzania, which was most affected in the previous outbreaks. Three of the four regions selected had a history of RVF outbreaks, including the more recent one during 2006/2007. In the fourth, or the Mbeya, region selected, there was a recent report of transmission of RVFV to humans; however, there was no evidence of clinical disease in either humans or animals.30
Figure 1.
A map of Tanzania showing the four regions where blood samples were obtained from cattle.
The specific districts in each of the four regions selected for obtaining blood samples from cattle included the Kyela district located at 9°35′0″S/33°51′0″E and with an elevation of 1,637 ft in the Mbeya region, southwest Tanzania.30 Kyela has a flat topography with an average temperature of 59–93°F and heavy rainfall (0.5 in.) between May and October. Kyela is mainly an agricultural district known for growing rice, with 61% of its vegetation covered with cropland, herbaceous vegetation (20%), and shrubs (15%). Ikungi district is located (4°91′20″S, 4°94′80″E and with an elevation of 4,957 ft in the Singida region, most of its vegetation is shrubs (75%), cropland (25%), and trees (32%). The climate is semi-arid with heavy rains from September to November. Yearly temperature varies between 55°F and 84°F. It is located in the central zone of Tanzania, which was affected most during the 2006/2007 RVF outbreak. Agro-pastoralism is the major economic activity in this area.
Simanjiro district is located 3°52′0.01″S, 36°36′0.00″E and with an elevation of 4,882 ft in the northern Tanzania where pastoralist keep a large number of ruminants for their livelihood.
Bahi district is located 5.1359°S, 34.7699°E, and with an elevation of 2,749 ft in the central zone of Tanzania. Bahi receives annual average rainfall of about 500 to 700 mm and an average temperature of about 22.6°C. It is one of the seven districts of the Dodoma region with a semi-arid climate. The major economic activity is agro-pastoralism and one of the two communities that were most affected during the 2006/2007 RVF outbreak.
Study design.
An observational cross-sectional study was conducted in cattle of the four districts of Tanzania. Blood samples were obtained from cattle during September 2015, before the onset of rainy season in Mbeya and Manyara and during May 2016 after the short rainy season in Dodoma and Singida. This was approximately 8–9 years after the 2006/2007 RVF outbreak in Tanzania. Oral consent was obtained from a herd owner to collect blood samples from healthy RVF-unvaccinated cattle born after the last RVF outbreak (2006/7) and more than 6 months (6 months–8/9 years) of age. Convenience sampling was used to select wards in each of the district included in the study. Ten percent of each herd was sampled in selected wards of each district. Information was obtained on each of the animals sampled from household head/owners and veterinary officers, including cattle vaccination history, age, sex, breed, herd composition, type of grazing and animal history. Animals above 6 months of age, and below 1 year were recorded as young, whereas animals aged 1 year and above but below 8/9 years were considered as an adult. In this study, animals that were confined and fed in an enclosed building were classified as indoors, whereas those taken for a free range were grouped as extensive grazing.
Sample collection.
Blood samples were aseptically collected using 5-mL vacutainer tubes from the jugular vein with an 18-gauge needle attached to the tube. Then the vacutainer tubes were kept in a cool box containing ice packs to keep the samples chilled and to allow serum separation. Some sera were separated by centrifugation at 8,500 × g for 15 minutes. The samples were transported to the virology laboratory at the Sokoine University of Agriculture (SUA) where the serum was transferred in duplicates to 1.8-mL cryovial tubes and stored at −20°C until tested for RVFV viral RNA and/or antibody.
Assay of samples for RVFV antibody.
cELISA.
An indirect ELISA was used to test cattle sera samples for RVFV antibodies. Primary detection of RVFV antibody was performed using an ID Screen® Rift Valley Fever Competition Multi-species Kit (Innovative Diagnostics, Montpellier, France) which detects all RVFV antibodies present in a sample. The samples were analyzed in pools of five samples each as described previously. For every antibody-positive pool, individual samples were retested to determine the exact number of positive samples in a respective pool. RVFV antibody-positive and antibody-negative control samples were included in each test run. ELISA procedures, as well as validation, were performed as per the manufacturer’s protocol and the optical density (OD) values were recorded at 405 nm. For each sample, the competition percentage (S/N%) was calculated, and if any value was equal to or less than 40%, the sample was considered positive. A value greater than 50% was a negative result and the values between 40% and 50% indicated doubtful results.
IgM ELISA.
All RVFV IgG antibody–positive samples were tested for RVFV IgM antibody to determine evidence of a recent RVFV infection. Samples were tested using ID Screen® Rift Valley Fever IgM Capture ELISA Kit (Innovative Diagnostics, Montpellier, France). ELISA procedures, validation, and interpretation were performed as per the manufacturer’s protocol. The OD values were recorded at 450 nm. Each sample was tested in duplicate in even and odd wells. The net OD of each sample and controls was calculated by subtracting OD of odd wells from even wells. Then the ratio of a sample (S) and positive control (P) (S/P%) was calculated by taking the net OD sample or net OD-positive control × 100 was used to interpret results. Samples with the ratios (S/P%) less than 40% were considered negative, between 40% and 50% as doubtful, and above 50% were positive.
Virus neutralization test.
Confirmatory virus neutralization test, the plaque reduction neutralization test (PRNT) assay was performed to test all ELISA RVFV antibody-positive and antibody-doubtful samples. Plaque reduction neutralization test was performed using Vero E6 cell lines and RVF vaccine virus strain, MP12 as described in the following paragraphs.
Cell culture.
The Vero E6 cell lines were received from Multi-Chemical Industry Sante Animale, Mohammedia, Morocco, and propagated at SUA in a biosafety level (BSL) 2 virology laboratory. The cells were grown in Eagle’s Minimum Essential Medium with Earle’s balanced salts, L-glutamine, and sodium bicarbonate supplemented with calcium, 2× penicillin/streptomycin, 2× vitamins, and 8% gamma-irradiated fetal bovine serum. Cells at a concentration of 4 × 108 were cultured in 24-well plates for 4–5 days to obtain confluent monolayer for testing sera samples for RVFV and for testing samples for RVFV antibody by the plaque reduction neutralization assays.
Virus propagation and quantification.
The RVF vaccine virus strain (MP-12), which was developed during the 1980s from an Egyptian RVFV virulent isolate, ZH548 strain, by 12 serial passages in human diploid lung Medical Research Council-5 cells in the presence of a chemical mutagen, 5-fluorouracil,31 was selected to be used in the neutralization assay. The MP 12 vaccine is considered to be safe for use in BSL level 2 laboratories. Although the vaccine has attenuations in all three segments of the genome, the growth and multiplication characteristics in cell culture are similar to that of the wild-type virus. The MP 12 virus was obtained from the University of Texas-El Paso and was stored at −80°C until replicated in confluent Vero E6 cell lines at SUA virology laboratory to prepare a working stock virus.
The infectivity titer of the MP12 virus was determined by preparing and testing log10 dilutions by the plaque assay technique in Vero E6 cells. Tenfold virus dilutions were prepared in Hanks’ balanced salt solution (HBSS) and incubated at 37°C for 1 hour. Following 1 hour of incubation, 100 uL of the diluted virus was then inoculated in wells of a confluent 24-well plates and incubated at 37°C for 1 hour. Each culture was overlaid with a mixture of equal volumes of essential basal media with Earle’s salt (EBME) and 1% agarose and then incubated at 37°C for 2 days. The cells were then stained using an overlay made by mixing equal parts of one percent agarose and 5% neutral red stained with EBME. Each culture received 0.5 mL of the stain overlay and incubated at 37°C. On the next day, the cultures were observed using a light box to enumerate the number of plaque forming units (PFU) for each virus dilution. The titer of the MP-12 virus was 108 PFU/mL as determined by plaque assay. Therefore, a dilution of 1/4,000 would yield about 40 PFU, which was selected for use as the virus dose in the PRNT.
Plaque reduction neutralization test analysis.
All RVFV antibody-positive and antibody-doubtful ELISA samples were serially diluted fourfold to a maximum dilution of 10,240 in HBSS. The MP-12 virus was diluted to yield about 40 PFUs in HBSS for use as the dose in the PRNT. Thereafter, equal parts of the diluted virus and serum dilutions (75 uL each) were mixed and incubated at 37°C for 1 hour. After 1 hour of incubation, 1 mL of the mixture was added to each well of a 24-well plate. The overlay and staining procedures were carried out as described previously. On day 3 post-inoculation, the cultures were observed and the number of PFUs was recorded per each dilution. The antibody titer was the dilution of sera samples that reduced 80% of the virus dose (PRNT 80%). Any sample that had an antibody titer of 1:10 and above was considered positive, whereas < 1:10 was considered negative.
Sample Analysis and Testing for RVFV.
Nucleic acid detection.
RNA extraction.
The RNA was extracted from the cattle serum samples using a Qiagen viral RNA extraction kit. A 50-uL aliquot of each of five samples from the same farm/herd was pooled for a total of 250 uL and mixed by vortexing. RNA was extracted as per the manufacturer’s protocol and stored at −80°C until tested for RVFV.
Polymerase chain reaction (PCR).
The RNA samples were analyzed by one-step RT-PCR for RVFV and other phlebovirus. Samples collected from farms that had IgM antibody–positive and cELISA-doubtful animals were re-tested individually using RVFV-specific primers. Two sets of primers designed to amplify glycoprotein (M-segment) of RVFV and nucleoprotein (N) of phlebovirus were used.32 In phlebovirus and RVF PCR reactions, the MP-12 vaccine was used as the positive control giving amplicons of 550 bp and 370 bp by species-specific and generic primers, respectively, and Rift Valley fever RNA detection was performed as per the QIAGEN One-Step RT-PCR Kit manufacturer’s protocol. Initially, detection of RVFV RNA was done using genus-specific primers (phlebovirus), F1-TTTGCTTATCAAGGATTTGACC, F2-TTTGCTTATCAAGGATTTGATGC, and R1-TCAATCAGTCCAGCAAAGCTGGGATGCATCAT in a reaction set of 25-uL reaction mix with 5 uL of the RNA sample and 0.1 uM of the primers. Negative samples collected in ELISA-positive and ELISA-doubtful farms were re-tested using an RVFV-specific RT-PCR reaction set in a 12.5-uL reaction mix with 3 uL of RNA and 0.1 uM of RVF1: TGTGAACAATAGGCATTGG and RVF2: GACTACCAGTCAGCTCATTACC. All the primers were run at Reverse Transcription at 55°C for 30 minutes; initial denaturation at 95°C for 15 minutes; 40 cycles of denaturation 95°C for 30 seconds, annealing at 55°C for 1 minute, and extension at 72°C for 2 minutes; and final extension 72°C for 10 minutes.
Gel electrophoresis.
Polymerase chain reaction amplicons were visualized in 1% E-Z Vision–stained gel electrophoresis, prepared in 1× TBE and run at 100 V for 30 minutes. A DNA ladder of 50 bp was used to estimate the band size of the PCR amplicons.
Data analysis.
Seroprevalence data were compiled in a Microsoft Excel 2007 spreadsheet and analyzed using R statistical program version 3.4.1, and a map was drawn using ArcGIS. The chi-squared test at 99% CI was used to determine the significant difference between and within different variables.
RESULTS
Rift Valley fever virus seroprevalence by cELISA.
A total of 443 of cattle sera samples were tested in approximately 89 pools by competitive ELISA kit for RVFV antibody. The overall seroprevalence for all cattle tested was 7.7% (34/443). There was a significant difference between regions sampled with the Mbeya region having the highest seroprevalence rate of 26.4% (23/87), followed by Dodoma 5.9% (10/171) and at last by Singida 0.9% (1/101) (Table 1). None of the cattle samples from Manyara were positive for RVFV antibody. However, the animal’s age, grazing mode, breed, and sex were not significantly associated with the seropositivity in three regions at 99% confidence. Seropositive cattle aged below 1 year were collected in Mbeya and Dodoma.
Table 1.
A total number of cattle samples tested and seroprevalence for anti-nucleoprotein Rift Valley fever virus antibody in the four regions of Tanzania
| Variables | Levels | Total animals | Negative animals | Positive animals | Doubtful animals | Overall seroprevalence | P-Value |
|---|---|---|---|---|---|---|---|
| Regions | Dodoma | 171 | 158 | 10 | 3 | 5.9 | 0.001* |
| Manyara | 84 | 84 | 0 | 0 | 0 | ||
| Mbeya | 87 | 64 | 23 | 0 | 26.4 | ||
| Singida | 101 | 99 | 1 | 1 | 0.9 | ||
| Sex | Male | 167 | 156 | 11 | 0 | 6.6 | 0.2288 |
| Female | 276 | 249 | 23 | 4 | 8.3 | ||
| Grazing | Indoors | 24 | 17 | 3 | 0 | 15 | 0.4173 |
| Extensive | 419 | 388 | 31 | 4 | 7.4 | ||
| Breed | Indigenous | 387 | 358 | 25 | 4 | 6.5 | 0.03237 |
| Exotic | 56 | 47 | 9 | 0 | 16.1 | ||
| Age | Young | 164 | 157 | 6 | 3 | 3.7 | 0.04393 |
| Adult | 279 | 248 | 28 | 1 | 10.1 |
Analysis performed by chi-squared test.
* Shows variables with a significant difference by chi-squared test at a CI of 99%.
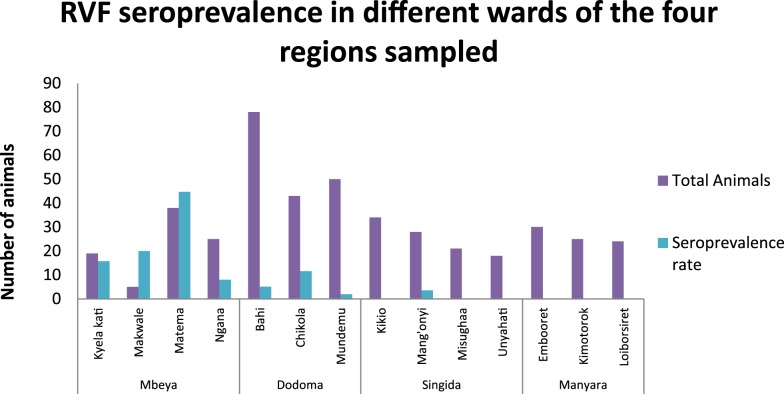
Statistical analysis within regions revealed a significant difference in seropositivity between wards sampled in Mbeya. Matema ward, which is located near Lake Nyasa, had the highest prevalence of 44.7% (17/38) followed by Makwale 20% (1/5), Kyela Kati 15.8% (3/19), and at last Ngana ward 8% (2/25) (Figure 2). Although there was no significant difference between wards in Dodoma, the highest seroprevalence rate was observed in Chikola 11.6% (5/43), followed by Bahi 5.1% (4/78) and at last by Mundemu 2% (1/50) (Figure 2). In Singida, only Mang’onyi ward had one seropositive cattle sample at a rate of 3.6% (1/28) (Figure 2).
Figure 2.
Seroprevalence rates for Rift Valley fever virus (RVFV) antibodies determined by cELISA among cattle in different wards of different regions of Tanzania. This figure appears in color at www.ajtmh.org.
Rift Valley fever virus seroprevalence by IgM ELISA.
Of 33 cELISA-positive samples tested by IgM ELISA, only one sample had IgM antibody giving an overall IgM seroprevalence of 0.2%. One of the ELISA-positive samples was not analyzed by IgM ELISA because of the lower amount of sera that was depleted in cELISA. The cattle sample with RVF IgM antibody at a titer of 160 was an adult female animal sampled in the Mbeya region. None of the cattle samples from the other three regions had IgM antibody.
Rift Valley fever virus seroprevalence by PRNT.
A total of 33 ELISA-positive and three ELISA-doubtful samples were tested by the PRNT to confirm and determine the antibody titer. Of 36 samples tested, 32 samples (96%) were positive by the PRNT, giving an overall seroprevalence of 6.9%. The slight changes in seroprevalence were also observed in individual regions as shown in Table 2. All cELISA-doubtful samples were negative with titers less than 1:10 (Table 3). Antibody titers ranged from a low of 1:10 and to a high of 1:10,240 and above (Table 3). Some ELISA-positive cattle samples gave titers below 1:10, which was considered negative. The highest titer of > 1:10,240 was obtained from an adult female cow collected in Kyela district, Mbeya. The IgM-positive cattle sample had a PRNT titer of 160. There was no correlation between ELISA OD and PRNT titers (data not shown).
Table 2.
Distribution of seropositive cattle samples at different variables as tested by ELISA and PRNT
| Variables | Levels | Total cattle | ELISA positive | ELISA doubtful | PRNT positive | Seropositivity by PRNT | Seropositivity by ELISA |
|---|---|---|---|---|---|---|---|
| Regions | Dodoma | 171 | 9 | 2 | 8 | 4.7 | 5.8 |
| Manyara | 84 | 0 | 0 | 0 | 0 | 0 | |
| Mbeya | 87 | 23 | 0 | 22 | 25.3 | 24.7 | |
| Singida | 101 | 1 | 1 | 1 | 0.9 | 0.9 | |
| Sex | Female | 276 | 11 | 0 | 21 | 7.9 | 8.3 |
| Male | 167 | 23 | 4 | 10 | 5.9 | 6.6 | |
| Age | Adult | 30 | 27 | 3 | 25 | 9.4 | 10.1 |
| Young | 419 | 6 | 0 | 6 | 3.7 | 3.7 | |
| Grazing | Extensive | 387 | 30 | 3 | 28 | 6.9 | 7.4 |
| Indoors | 56 | 3 | 6 | 3 | 15 | 15 | |
| Breed | Exotic | 164 | 9 | 0 | 9 | 16.1 | 16.7 |
| Indigenous | 276 | 23 | 3 | 22 | 5.9 | 6.5 |
PRNT = plaque reduction neutralization test.
Table 3.
Distribution of neutralization titers obtained on 33 ELISA-positive and three ELISA-doubtful samples tested by PRNT 80%
| Variables | < 1:10 | 1:10 | 1:40 | 1:160 | 1:640 | 1:2,560 | 1:10,240 | > 1:10,240 | Total samples animal tested | ELISA positive | ELISA doubtful | PRNT positive | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Region | Dodoma | 3 | 0 | 1 | 4 | 3 | 0 | 0 | 0 | 11 | 9 | 2 | 8 |
| Manyara | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Mbeya | 1 | 5 | 4 | 3 | 6 | 2 | 1 | 1 | 23 | 23 | 0 | 22 | |
| Singida | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 1 | 1 | 1 | |
| Sex | Female | 4 | 3 | 4 | 5 | 7 | 1 | 1 | 1 | 26 | 23 | 3 | 21 |
| Male | 0 | 3 | 1 | 2 | 3 | 1 | 0 | 0 | 10 | 10 | 0 | 10 | |
| Age | Adult | 4 | 6 | 4 | 3 | 9 | 2 | 1 | 1 | 30 | 27 | 3 | 25 |
| Young | 0 | 0 | 1 | 4 | 1 | 0 | 0 | 0 | 6 | 6 | 0 | 6 | |
| Grazing | Extensive | 4 | 6 | 5 | 6 | 9 | 1 | 1 | 1 | 33 | 30 | 3 | 28 |
| Indoors | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 3 | 3 | 0 | 3 | |
| Breed | Exotic | 0 | 1 | 1 | 2 | 3 | 2 | 0 | 0 | 9 | 9 | 0 | 9 |
| Indigenous | 4 | 5 | 4 | 5 | 7 | 0 | 1 | 1 | 27 | 23 | 3 | 22 | |
Samples with titers below 1:10 are considered negative, whereas the titers above 1:10 are positive.
Rift Valley fever virus prevalence by PCR.
Of the 445 RNA samples analyzed in 89 pools, none of the samples were positive using both of the genus-specific primers. Also, none of the samples from the four farms were positive except for the positive control (MP12), which gave amplicons of 550 bp and 400 bp by phlebovirus and RVF species-specific primers, respectively.
DISCUSSION
The overall RVFV seroprevalence for cattle observed in this study was slightly lower than that reported for cattle sampled in Kilombero, Morogoro, during the inter-epizootic/epidemic period in 2013.26 The difference may have reflected the study area, sampling criteria, and the distance from the latest outbreak. Other factors which have been associated with seropositivity include climate, number of ruminants, proximity to perennial water bodies, and animal trading activities.24–27,30,36 The present study did not reveal any significant difference in seropositivity between sex despite higher seroprevalence in female animals, which was also observed in other RVFV serological studies.24,27,28,37,38 Insignificant association of seropositivity among animal breed, herd composition, sex, and grazing practices as observed in the present study was also reported in ruminants in previous studies.24 Contrary to what was observed during the latest RVF outbreak in 2006/2007, in this study, animals kept indoors had higher RVFV antibodies than those under an extensive farming system.22
Of the four regions sampled, the highest seroprevalence was in Mbeya region, which is consistent with the results of other studies that documented Kyela, in the Mbeya region, as one of the active foci for the enzootic circulation of RVFV.27,30 This could have reflected the presence of water-retaining soil for rice plantations, flat topology temperate climate, a large number of cattle, and live animal trading activities in Kyela.24,27 These factors were previously associated with the occurrence of previous RVF outbreaks.37 Another difference was observed between the sampled wards of Mbeya regions, whereby the Matema ward, which is located closer to Lake Nyasa, had the highest seroprevalence rate. Closeness to water bodies was previously associated with seropositivity among cattle in Tanzania, Senegal, and Saudi Arabia.37,39,40 Detection of the highest antibody titers (> 1:10,240) in Mbeya suggested recent and re-exposure of animals to RVFV infection. The presence of neutralizing antibodies confers strong protection in cattle and, hence, decreases chances of an epizootic/clinical case to occur when animals are exposed to the virus.16,40 Despite a large number of cattle in Mbeya and a favorable climate for vector mosquitoes, epizootic may not occur because of the presence of RVFV-neutralizing antibodies in these animals.
Although there was no evidence of active infection, this study highlights exposure of animals to wild-type RVFV after the last outbreak in Tanzania because all animals tested are those born after the latest RVF outbreak. Previous studies have reported a number of RVF outbreaks in Dodoma and Singida and hereby IEP transmission, therefore confirming the circulation of RVFV during both enzootic and epidemic cycles in these regions. Results obtained in the present study suggested the circulation of RVFV at very low levels without noticeable clinical manifestation in animals as was postulated in the previous study.23 A number of studies have reported seropositivity in animals without clinical manifestations in other countries, including Kenya, Mozambique, and Madagascar.36,41,42
ELISA kits used in this study had 99.51% specificity and 99.02% negative predictive value. Only the antibody-positive ELISA samples were confirmed by neutralization test and all of the ELISA-negative samples were negative by the PRNT. Slight discrepancies between ELISA and PRNT results observed in the present study were also reported in previous studies.36 Although it is laborious, expensive, and requires 5–7 days for completion, the PRNT overcomes the possibility of cross-reactivity, which is a major shortcoming of ELISA assays.43–45
Evidence for RVFV transmission during inter-epizootic/epidemic periods has also been reported among in humans, livestock, and wild animals.23,24,27,28,37,39,41,46 Seroprevalence results obtained for RVFV during several studies in Tanzania were based solely on different ELISA assays, whereas the results of the present study are based on both the ELISA and confirmatory virus neutralization assays. The results of our study, therefore, confirmed the specificity and neutralizing ability of RVFV antibodies circulating during the inter-epizootic/epidemic period in the study areas. PCR could not detect RVFV or any other phlebovirus during RVFV inter-epizootic/epidemic period in the study area. The negative PCR and IgM results obtained in Dodoma, Singida, and Manyara suggested the absence of active/acute RVFV transmission in these areas.
The use of genus-specific primers for screening cattle sera aimed to determine circulation of other phlebovirus in ruminants.47
The presence of other viruses of Phenuiviridae family, which might cross-react with RVFV antibody in ELISA, was another possibility of misdiagnosis by this assay which is mostly used in Tanzania. A more effective study design could be to monitor a cohort of the same animals’ overtime with sampling at intervals to maximize the chances of possibility of detecting an RVFV during the inter-epizootic/epidemic period.
CONCLUSION
In conclusion, the findings of this study confirmed the circulation of RVF antibodies in an area with and without a history of RVF during inter-epizootic/epidemic periods. This is among the first study that has documented RVFV-neutralizing antibodies circulating in cattle in Tanzania during inter-epizootic/epidemic periods. Strong herd immunity as a result of high neutralizing antibody titers in cattle confers protection of cattle against RVFV, thus reducing the possibility of epizootics/epidemics. Likewise, PRNT confirmed the specificity and titers of RVFV antibodies circulating in the study area during inter-epizootic/epidemic periods. Last, phleboviruses were not detected by RT-PCR in cattle sampled in the study areas.
RECOMMENDATIONS
The approach used in this study provided evidence that supported the transmission of RVFV among cattle during the RVF IEP in the selected region of Tanzania. On the basis of these findings, it is recommended that a longitudinal study be implemented using a cohort of cattle to obtain repeated observations, including blood samples to gain a better understanding and documentation of active RVFV transmission during the inter-epizootic/epidemic period in this region. The scope of such a study should also be expanded to include other livestock, such as sheep and goats, and to include the sampling and testing of mosquitoes for RVFV to gain a possible understanding of the mechanism of enzootic transmission of RVFV to cattle and possibly other domestic animals.
LIMITATION OF THE STUDY
The study did not use reliable ways of detecting the age, for example, dentition due to the absence of expert in the sampling team. Instead, we relied on livestock owner information which can be misleading. Because RVFV is known to infect all domestic animals, this study could be carried out simultaneously in cattle, sheep, and goats.
Acknowledgments:
We convey our sincere gratitude to USAID for providing the support required to implement this study and to the research team of the Feed the Future Innovation Laboratory for Rift Valley Fever Control in Agriculture, including the Sokoine University of Agriculture (SUA), University of Texas at El Paso (UTEP), and Multi-Chemical Industry (MCI) Santé Animale for their invaluable support throughout this study. Also, we sincerely thank Jessica Rowland of the University of Texas at El Paso and district veterinary officers and livestock owners of Ikungi, Kyela, Simanjiro, and Bahi districts for their contributions to this study.
REFERENCES
- 1.Adams MJ, et al. 2017. Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2017). Arch Virol 162: 2505–2538. [DOI] [PubMed] [Google Scholar]
- 2.Collett MS, 1986. Messenger RNA of the M segment RNA of Rift Valley fever virus. Virology 151: 151–156. [DOI] [PubMed] [Google Scholar]
- 3.Collett MS, Purchio AF, Keegan K, Frazier S, Hays W, Anderson DK, Parker MD, Schmaljohn C, Schmidt J, Dalrymple JM, 1985. Complete nucleotide sequence of the M RNA segment of Rift Valley fever virus. Virology 144: 228–245. [DOI] [PubMed] [Google Scholar]
- 4.Linthicum KJ, Davies FG, Kairo A, Bailey CL, 1985. Rift Valley fever virus (family Bunyaviridae, genus Phlebovirus). Isolations from Diptera collected during an inter-epizootic period in Kenya. Epidemiol Infect 95: 197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jupp PG, Kemp A, Grobbelaar A, Lema P, Burt FJ, Alahmed AM, Al Mujalli D, Al Khamees M, Swanepoel R, 2002. The 2000 epidemic of Rift Valley fever in Saudi Arabia: mosquito vector studies. Med Vet Entomol 16: 245–252. [DOI] [PubMed] [Google Scholar]
- 6.Pepin M, Bouloy M, Bird BH, Kemp A, Paweska J, 2010. Rift Valley fever virus (Bunyaviridae: Phlebovirus): an update on pathogenesis, molecular epidemiology, vectors, diagnostics and prevention. Vet Res 41: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sang R, et al. 2010. Prediction, assessment of the Rift Valley fever activity in east and southern Africa 2006–2008 and possible vector control strategies. Am J Trop Med Hyg 83 (Suppl 2): 43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abdo-Salem S, Gerbier G, Bonnet P, Al-Qadasi M, Tran A, Thiry E, Al-Eryni G, Roger F, 2006. Descriptive and spatial epidemiology of Rift Valley fever outbreak in Yemen 2000–2001. Ann N Y Acad Sci 1081: 240–242. [DOI] [PubMed] [Google Scholar]
- 9.Al-Afaleq AI, Hussein MF, 2011. The status of Rift Valley fever in animals in Saudi Arabia: a mini review. Vector Borne Zoonotic Dis 11: 1513–1520. [DOI] [PubMed] [Google Scholar]
- 10.Balkhy HH, Memish ZA, 2003. Rift Valley fever: an uninvited zoonosis in the Arabian peninsula. Int J Antimicrob Agents 21: 153–157. [DOI] [PubMed] [Google Scholar]
- 11.Shoemaker T, et al. 2002. Genetic analysis of viruses associated with emergence of Rift Valley fever in Saudi Arabia and Yemen, 2000–01. Emerg Infect Dis 8: 1415–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sissoko D, Giry C, Gabrie P, Tarantola A, Pettinelli F, Collet L, D’Ortenzio E, Renault P, Pierre V, 2009. Rift Valley fever, Mayotte, 2007–2008. Emerg Infect Dis 15: 568–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bird BH, Ksiazek TG, Nichol ST, MacLachlan NJ, 2009. Rift Valley fever virus. J Am Vet Med Assoc 234: 883–893. [DOI] [PubMed] [Google Scholar]
- 14.Coetzer JA, 1982. The pathology of Rift Valley fever. II. Lesions occurring in field cases in adult cattle, calves and aborted foetuses. Onderstepoort J Vet Res 49: 11–17. [PubMed] [Google Scholar]
- 15.Coetzer JA, 1977. The pathology of Rift Valley fever. I. Lesions occurring in natural cases in new-born lambs. Onderstepoort J Vet Res 44: 205–211. [PubMed] [Google Scholar]
- 16.Munyua P, et al. 2010. Rift Valley fever outbreak in livestock in Kenya, 2006–2007. Am J Trop Med Hyg 83 (Suppl 2): 58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swanepoel R, 1976. Studies on the epidemiology of Rift Valley fever. J S Afr Vet Assoc 47: 93–94. [PubMed] [Google Scholar]
- 18.Turell MJ, Rossi CA, 1991. Potential for mosquito transmission of attenuated strains of Rift Valley fever virus. Am J Trop Med Hyg 44: 278–282. [DOI] [PubMed] [Google Scholar]
- 19.Daubney R, Hudson JR, Garnham PC, 1931. Enzootic hepatitis or Rift Valley fever. An undescribed virus disease of sheep cattle and man from east Africa. J Pathol Bacteriol 34: 545–579. [Google Scholar]
- 20.Sindato C, Karimuribo E, Mboera LE, 2011. The epidemiology and socio-economic impact of Rift Valley fever in Tanzania: a review. Tanzan J Health Res 13: 305–318. [DOI] [PubMed] [Google Scholar]
- 21.Sindato C, Karimuribo ED, Pfeiffer DU, Mboera LE, Kivaria F, Dautu G, Bernard B, Paweska JT, 2014. Spatial and temporal pattern of Rift Valley fever outbreaks in Tanzania; 1930 to 2007. PLoS One 9: e88897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mlozi MRS, Mtambo MMA, 2008. Socio-Economic Impact Analysis of the Recent Rift Valley Fever Outbreak in Tanzania. FAO Consultancy Report.
- 23.Anyamba A, et al. 2010. Prediction, assessment of the Rift Valley fever activity in east and southern Africa 2006–2008 and possible vector control strategies. Am J Trop Med Hyg 83 (Suppl 2): 43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sumaye RD, Geubbels E, Mbeyela E, Berkvens D, 2013. Inter-epidemic transmission of Rift Valley fever in livestock in the Kilombero river valley, Tanzania: a cross-sectional survey. PLoS Negl Trop Dis 7: e2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McIntosh BM, 1972. Rift Valley fever: 1. Vector studies in the field. J S Afr Vet Assoc 43: 391–395. [PubMed] [Google Scholar]
- 26.Kifaro EG, Nkangaga J, Joshua G, Sallu R, Yongolo M, Dautu G, Kasanga CJ, 2014. Epidemiological study of Rift Valley Fever virus in Kigoma, Tanzania. Onderstepoort J Vet Res 81: E1–E5. [DOI] [PubMed] [Google Scholar]
- 27.Sindato C, Pfeiffer DU, Karimuribo ED, Mboera LE, Rweyemamu MM, Paweska JT, 2015. A spatial analysis of Rift Valley fever virus seropositivity in domestic ruminants in Tanzania. PLoS One 10: e0131873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sumaye RD, Abatih EN, Thiry E, Amuri M, Berkvens D, Geubbels E, 2015. Inter-epidemic acquisition of Rift Valley fever virus in humans in Tanzania. PLoS Negl Trop Dis 9: e0003536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swai ES, Schoonman L, 2009. Prevalence of Rift Valley fever immunoglobulin G antibody in various occupational groups before the 2007 outbreak in Tanzania. Vector Borne Zoonotic Dis 9: 579–582. [DOI] [PubMed] [Google Scholar]
- 30.Heinrich N, et al. 2012. High seroprevalence of Rift Valley fever and evidence for endemic circulation in Mbeya region, Tanzania, in a cross-sectional study. PLoS Negl Trop Dis 6: e1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caplen H, Peters CJ, Bishop DH, 1985. Mutagen-directed attenuation of Rift Valley fever virus as a method for vaccine development. J Gen Virol 66: 2271–2277. [DOI] [PubMed] [Google Scholar]
- 32.Ibrahim MS, Turell MJ, Knauert FK, Lofts RS, 1997. Detection of Rift Valley fever virus in mosquitoes by RT-PCR. Mol Cell Probes 11: 49–53. [DOI] [PubMed] [Google Scholar]
- 33.Meegan JM, Yedloutschnig RJ, Peleg BA, Shy J, Peters CJ, Walker JS, Shope RE, 1987. Enzyme-linked immunosorbent assay for detection of antibodies to Rift Valley fever virus in ovine and bovine sera. Am J Vet Res 48: 1138–1141. [PubMed] [Google Scholar]
- 34.Niklasson BS, Meadors GF, Peters CJ, 1984. Active and passive immunization against Rift Valley fever virus infection in Syrian hamsters. Acta Pathol Microbiol Immunol Scand C 92: 197–200. [DOI] [PubMed] [Google Scholar]
- 35.Paweska JT, Mortimer E, Leman PA, Swanepoel R, 2005. An inhibition enzyme-linked immunosorbent assay for the detection of antibody to Rift Valley fever virus in humans, domestic and wild ruminants. J Virol Methods 127: 10–18. [DOI] [PubMed] [Google Scholar]
- 36.Swanepoel R, Struthers JK, Erasmus MJ, Shepherd SP, McGillivray GM, Erasmus BJ, Barnard BJH, 1986. Comparison of techniques for demonstrating antibodies to Rift Valley fever virus. Epidemiol Infect 97: 317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.LaBeaud AD, Muchiri EM, Ndzovu M, Mwanje MT, Muiruri S, Peters CJ, King CH, 2008. Interepidemic Rift Valley fever virus seropositivity, northeastern Kenya. Emerg Infect Dis 14: 1240–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguku PM, et al. 2010. An investigation of a major outbreak of Rift Valley fever in Kenya: 2006–2007. Am J Trop Med Hyg 83 (Suppl 2): 5–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ringot D, Durand J-P, Tolou H, Boutin J-P, Davoust B, 2004. Rift Valley fever in Chad. Emerg Infect Dis 10: 945–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elfadil AA, Hasab-Allah KA, Dafa-Allah OM, Elmanea AA, 2006. The persistence of Rift Valley fever in the Jazan region of Saudi Arabia. Rev Sci Tech 25: 1131–1136. [PubMed] [Google Scholar]
- 41.Thiongane Y, Gonzalez J-P, Fati A, Akakpo JA, 1991. Changes in Rift Valley fever neutralizing antibody prevalence among small domestic ruminants following the 1987 outbreak in the Senegal River basin. Res Virol 142: 67–70. [DOI] [PubMed] [Google Scholar]
- 42.Fafetine J, Neves L, Thompson PN, Paweska JT, Rutten VP, Coetzer JA, 2013. Serological evidence of Rift Valley fever virus circulation in sheep and goats in Zambézia Province, Mozambique. PLoS Negl Trop Dis 7: e2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeanmaire EM, et al. 2011. Prevalence of Rift Valley fever infection in ruminants in Madagascar after the 2008 outbreak. Vector Borne Zoonotic Dis 11: 395–402. [DOI] [PubMed] [Google Scholar]
- 44.Paweska JT, van Vuren PJ, Kemp A, Buss P, Bengis RG, Gakuya F, Breiman RF, Njenga MK, Swanepoel R, 2008. Recombinant nucleocapsid-based ELISA for detection of IgG antibody to Rift Valley fever virus in African buffalo. Vet Microbiol 127: 21–28. [DOI] [PubMed] [Google Scholar]
- 45.Paweska JT, Smith SJ, Wright IM, Williams R, Cohen AS, Van Dijk AA, Grobbelaar AA, Croft JE, Swanepoel R, Gerdes GH, 2003. Indirect enzyme-linked immunosorbent assay for the detection of antibody against Rift Valley fever virus in domestic and wild ruminant sera. Onderstepoort J Vet Res 70: 49–64. [PubMed] [Google Scholar]
- 46.Matiko MK, Salekwa LP, Kasanga CJ, Kimera SI, Evander M, Nyangi WP, 2018. Serological evidence of inter-epizootic/inter-epidemic circulation of Rift Valley fever virus in domestic cattle in Kyela and Morogoro, Tanzania. PLoS Negl Trop Dis 12: e0006931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Montgomery E, 1917. On a tick-borne gastroenteritis of sheep and goats occurring in British east Africa. J Comp Pathol Ther 30: 28–57. [Google Scholar]