Abstract
Aberrant activation of Wnt/β-catenin signaling is observed in >90% of colorectal cancer cases. microRNAs (miRNAs) regulate the expression of key genes in Wnt/β-catenin signaling. As a result, abnormal expression of miRNAs regulates the activation of Wnt/β-catenin signaling in several types of cancer. In the current study, it was demonstrated that miR-501-3p was overexpressed in colorectal tumor tissues compared to the adjacent normal tissues. Downregulation of miR-501-3p inhibited cell proliferation and sphere formation, while it induced cell cycle arrest at the G1 phase in colorectal cancer cells. Bioinformatics analysis results predicted that adenomatous polyposis coli (APC), a negative regulator of Wnt/β-catenin signaling, was a potential target gene of miR-501-3p. Inhibition of miR-501-3p increased APC expression in colorectal cancer cells. Additionally, β-catenin was destabilized following miR-501-3p inhibition; immunofluorescence analysis revealed that β-catenin translocated from nucleus to cytoplasm. In addition, cyclin D1 and c-Myc, two well-characterized target genes of Wnt/β-catenin signaling, were downregulated following miR-501-3p inhibition. Transfection of APC small interfering RNA re-activated β-catenin and stimulated the expression of cyclin D1 and c-Myc. Furthermore, silencing of APC reversed the miR-501-3p inhibitor-induced cell cycle disruption, and the inhibition of cell proliferation and sphere formation in colorectal cancer cells. In conclusion, the present study identified miR-501-3p as a novel regulator of Wnt/β-catenin signaling in colorectal cancer cells via targeting APC, suggesting that miR-501-3p may act as a novel oncogenic miRNA in colorectal cancer.
Keywords: microRNA-501-3p, Wnt/β-catenin signaling, colorectal cancer
Introduction
Colorectal cancer (CRC) is the fourth most commonly diagnosed cancer type worldwide, with estimated 1.1 million new cases and 551,000 deaths in 2018 (1). Despite the observation that the incidence of CRC has been gradually decreased and integrated therapy has greatly improved the prognosis of patients with CRC in the recent years, the overall survival of patients with CRC remains poor, and the five-year survival is ~60% (2,3). Therefore, there is an urgent need to develop innovative detection and therapeutic approaches for patients with CRC.
β-catenin-dependent Wnt signaling, also known as canonical Wnt signaling, has a pivotal role in mediating cancer cell proliferation, metastasis and stemness maintenance (4-6). Upon activation, such as following Wnt ligand binding, β-catenin, the major effector of Wnt/β-catenin signaling, translocates into the nucleus of cells to form a complex with co-regulators of transcription factors, thus promoting the transcription of multiple oncogenes (7). Overactivation of Wnt/β-catenin signaling has been observed in several types of cancer, including CRC (8). Activation of Wnt/β-catenin signaling is essential for the initiation of colorectal cancer, as well as promotes chemoradiotherapy resistance (9,10). The activity of Wnt/β-catenin signaling is tightly controlled by several positive and negative regulators (11). Adenomatous polyposis coli (APC) is one of the well-studied negative regulators of Wnt/β-catenin signaling (12). A APC truncating mutation has been observed in a large proportion of patients with CRC (13), suggesting that Wnt/β-catenin signaling may be a promising target for the treatment of patients with CRC. Altered expression of non-coding RNA and transcription factors also contributes to the downregulation of APC in cancer cells (14,15).
microRNAs (miRNAs) are small, non-coding RNA molecules which were first discovered in Caenorhabditis elegans (16). Mechanistically, miRNAs function via base-pairing with sequences on the 3′-untranslated region (3′-UTR) of target gene mRNAs (17). Dysregulation of miRNAs leads to aberrant expression of target genes, resulting in the disruption of signaling networks in cells. Accumulating evidence suggests that miRNAs are involved in almost all physiological and pathological conditions (18,19). In CRC, multiple miRNAs have been identified as oncogenes or tumor suppressors via regulating key genes in oncogenic signaling pathways (20). For example, miR-144 was reported as a regulator of cell proliferation and rapamycin sensitivity in CRC through directly targeting mTOR (21). miRNA microarray analysis of normal, adenoma and carcinoma tissues discovered several dysregulated miRNAs (22), however, their roles have not been studied yet. Through bioinformatic analysis, miR-501-3p was discovered as a differentially expressed miRNA in CRC. miR-501-3p has been previously confirmed as an oncogenic miRNA in cervical cancer and hepatocellular carcinoma (23,24). However, the role of miR-501-3p in CRC remains unknown.
The current study focused on the role of this differentially expressed miRNA, miR-501-3p, in CRC cells. Reverse transcription-quantitative PCR (RT-qPCR) suggested that miR-501-3p was overexpressed in tumor tissues of patients with CRC. In addition, downregulation of miR-501-3p inhibited cell proliferation, cell cycle progression and sphere formation in CRC cells. Furthermore, miR-501-3p regulated CRC progression via targeting APC to activate Wnt/β-catenin signaling.
Materials and methods
Patient samples
A total of 30 patients with CRC who underwent surgical resection at the Fourth People's Hospital of Shanxi from September 2015 to January 2018 were included in the present study. None of the patients received chemotherapy or radiotherapy prior to surgery. Written consents were provided by all participants and all patients agreed to the use of their tissue samples in the current study. The experiments were performed under the supervision of the Ethics Committee of the Fourth People's Hospital of Shanxi. There were 19 males and 11 females, aged 35-68 years. Exclusion criteria were as follows: Patients with a history of other malignancies or other serious active diseases recently; and patients with drug hepatitis, alcoholic liver disease or autoimmune liver disease. The normal and tumor tissues were immediately stored at −80°C, until use for the following experiments.
Cell culture
CRC cell lines HCT116, Caco2, LOVO, HT29 and the fetal colon epithelial cell line FHC were purchased from the Cell Center in Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Shanghai, China). All cells were cultured in DMEM (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin-streptomycin (Sigma-Aldrich; Merck KGaA) in a 95% humid incubator with 5% CO2.
RNA extraction and RT-qPCR
Total RNA was extracted from tissues and cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's protocol. The concentration of RNA was determined using a NanoDrop 2000 (Thermo Fisher Scientific, Inc.). RNA was reverse transcribed into cDNA using SMART MMLV Reverse Transcriptase (Takara Bio, Inc.). qPCR was performed on a CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, Inc.) with TB Green Premix Ex Taq (Takara Bio, Inc.). The qPCR conditions were as follows: Predenature at 98°C for 30 sec, followed by 35 cycles of denature at 98°C for 5 sec and elongation and annealing at 60°C for 30 sec. GAPDH and U6 were used as internal controls for mRNA and miRNA, respectively. Relative gene expression was calculated using the 2−ΔΔCq method (25). The primer sequences were as follows: APC, forward 5′-AAG CAT GAA ACC GGC TCA CAT-3′ and reverse 5′-CAT TCG TGT AGT TGA ACC CTG A-3′; β-catenin, forward 5′-ATG GAC AGT ATG CAA TGA CTC G-3′ and reverse 5′-TAG CAG ACA CCA TCT GAG GAG A-3′; cyclin D1, forward 5′-GCT GCG AAG TGG AAA CCA TC-3′ and reverse 5′-CCT CCT TCT GCA CAC ATT TGA A-3′; c-Myc, forward 5′-GGC TCC TGG CAA AAG GTC A-3′ and reverse 5′-CTG CGT AGT TGT GCT GAT GT-3′; GAPDH, forward 5′-GGA GCG AGA TCC CTC CAA AAT-3′ and reverse 5′-GGC TGT TGT CAT ACT TCT CAT GG-3′; stem-loop primer, 5′-CTC AAC TGG TGT CGT GGA GTC GGC AAT TCA GTT GAG AGA ATC-3′; miR-501-3p, forward 5′-GCC GAG AAT GCA CCC GGG CA-3′ and reverse 5′-CTC AAC TGG TGT CGT GGA-3′; U6, forward 5′-CTC GCT TCG GCA GCA C-3′ and reverse 5′-AAC GCT TCA CGA ATT TGC G-3′.
Protein extraction and western blotting
Primary antibodies against APC (cat. no. 2504; 1:1,000), β-catenin (cat. no. 8480; 1:1,000), phosphorylated (p-) β-catenin (cat. no. 9561; 1:1,000), cyclin D1 (cat. no. 2978; 1:1,000) and c-Myc (cat. no. 9402; 1:1,000) were purchased from Cell Signaling Technology, Inc. The antibodies targeting histone H1 (H1; cat. no. ab71594; 1:10,000) and GAPDH (cat. no. ab8245; 1:5,000) were purchased from Abcam. Horseradish peroxidase-conjugated mouse (cat. no. SA00001-1; 1:10,000) and rabbit (cat. no. SA00001-2; 1:10,000) secondary antibodies were products of ProteinTech Group, Inc. Protein lysates were prepared with RIPA lysis buffer (Thermo Fisher Scientific, Inc.) following the manufacturer's protocol. The concentration of lysates was determined with BCA Protein Assay kit (Thermo Fisher Scientific, Inc.). A total of 20 µg lysate sample was loaded in each well of 8% SDS gels and separated by electrophoresis. The proteins were transferred to PVDF membranes and incubated with 5% non-fat milk for 30 min at room temperature. Afterwards, the membrane was incubated with primary antibody and secondary antibody for 1 h at room temperature. The blots were developed with enhanced chemiluminescence ECL Western Blotting Substrate (Thermo Fisher Scientific, Inc.). The relative protein expression was analyzed with Image J software version 1.51J8 (National Institutes of Health).
Transfection of miR-501-3p mimic and inhibitor
miR-501-3p mimic (5′-AAU CCU UUG UCC CUG GGU GAG A-3′), miR-negative control (NC) mimic (5′-UCA CAA CCU CCU AGA AAG AGU AGA-3′), miR-501-3p inhibitor (5′-UCU CAC CCA GGG ACA AAG GAU U-3′) and miR-NC inhibitor (5′-CAG UAC UUU UGU GUA GUA CAA-3′) were synthesized and purchased from Shanghai GenePharma Co., Ltd. For transfection, 50 nM miR-501-3p mimic or miR-NC mimic or miR-501-3p inhibitor or miR-NC inhibitor was transfected into the indicated cells using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's protocol. At 48 h post-transfection, the cultured cells were harvested for subsequent experiments.
Cell proliferation assay
To determine the cell proliferation ability, a Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.) was used. Briefly, the cells were seeded in 6-well plates and transfected with miR-501-3p inhibitor or miR-NC inhibitor or miR-NC inhibitor + APC small interfering (si)RNA. On the next day, the cells were harvested and re-seeded in 96-well plates at a concentration of 10,000 cells per well. Thereafter, 10 µl CCK8 solution was added for 2 h into each well at 0, 24, 48, 72 h post-cell adhesion. The medium was then transferred into new 96-well plates and the absorbance of each well was determined using a microplate reader.
Cell cycle assay
At 48 h post-transfection, the cells were fixed in 75% ethanol at −20°C overnight. On the next day, the cells were washed in cold PBS and stained with propidium iodide (Invitrogen; Thermo Fisher Scientific, Inc.) for 30 min at room temperature. Finally, the cells were analyzed using a FACSCalibur flow cytometer (BD Biosciences) and FlowJo software version 10.4 (FlowJo LLC).
Sphere formation assay
Briefly, after transfection, the cells were harvested by trypsinization, and the cell suspensions were washed with PBS three times. The cells were resuspended with sphere condition media (DMEM/F12; Thermo Fisher Scientific, Inc.) supplemented with B27 (Thermo Fisher Scientific, Inc.) and recombinant epidermal growth factor (20 ng/ml; PeproTech, Inc.) at a final concentration of 1,000 cells/ml. These cell suspensions (1 ml) were seeded in a 96-well ultra-low attachment plate (Corning Inc.). At day 7 of cell culture, the number of spheres (diameter >50 µm) was counted in three random fields using an inverted light microscope (magnification, ×20).
Immunofluorescence
HCT116 cells were plated on a slide glass in 96-well plates. Following transfection with miR-501-3p inhibitor or miR-NC inhibitor in combination with control siRNA or APC siRNA, HCT116 cells were fixed with 4% formaldehyde for 1 h at room temperature, washed with PBS containing 0.5% triton, and blocked with 5% bovine serum (Sigma Aldrich; Merck KGaA) for 1 h at room temperature. The cells were then incubated with β-catenin antibody (cat. no. 8480; 1:100; Cell Signaling Technology, Inc.) at 4°C overnight. On the next day, the cells were washed with PBS and incubated with Alexa Fluor 488-conjugated secondary antibody (cat. no. A-11034; 1:100,000; Thermo Fisher Scientific, Inc.) at room temperature for 2 h. After washing with PBS, Vectashield antifade mounting medium with DAPI (Vector Laboratories, Inc.) was used. Cells were observed and images were captured with a fluorescence microscope (magnification, ×40; Leica Microsystems GmbH).
Cycloheximide (CHX) chase assay
To detect the stability of proteins, a CHX chase assay was performed. Following transfection with miR-501-3p inhibitor or miR-NC inhibitor for 48 h, the cells were treated with CHX (10 µg/µl) for 0, 2 or 4 h, respectively. The cells were then harvested for western blotting, as aforementioned.
Extraction of nuclear and cytoplasmic protein
The NE-PER™ Nuclear and Cytoplasmic Extraction Reagents kit (Thermo Fisher Scientific, Inc.) was used to extract nuclear and cytoplasmic proteins from cells, following the manufacturer's protocol. H1 and GAPDH were used as controls for the nuclear and cytoplasmic extracts, respectively.
Dual luciferase reporter assay
For construction of the pGL3-APC 3'UTR-WT reporter plasmid, the 3'UTR sequence of the APC gene containing the putative binding sites for miR-501-3p was amplified from cDNA generated from HCT116 cells and ligated into the pGL3 plasmid (Promega Corporation) with T4 DNA ligase (New England Biolabs, Inc.). Briefly, HCT116 cells were seeded in 24-well plates. On the next day, miR-501-3p mimic or miR-NC mimic was co-transfected with pGL3-APC 3'UTR-WT or pGL3-APC 3'UTR-Mut into HCT116 cells using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.). After 24 h, the relative luciferase activity of HCT116 cells was determined with a Dual Luciferase Reporter System Assay kit (Promega Corporation) via normalization of the firefly luciferase activity to Renilla luciferase activity.
Silencing of APC
APC siRNA (5′-GGA TCA GCC TAT TGA TTA T-3′) and control siRNA (5′-TTC TCC GAA CGT GTC ACG T-3′) were synthesized and purchased from Shanghai GenePharma Co., Ltd. For silencing of APC, the cells were transfected with 50 nM APC siRNA using Lipofectamine RNAiMax (Invitrogen; Thermo Fisher Scientific, Inc.). At 48 h post-transfection, the cells were subjected to subsequent experiments.
Statistical analysis
All experiments were repeated at least three times. Data were analyzed with GraphPad Prism 5.0 (GraphPad Software, Inc.) and presented as mean ± SD. The correlation between miR-501-3p and APC expression levels was analyzed by Pearson's correlation analysis. The patients were divided into high and low expression groups according to the median expression value of miR-501-3p, and Chi-square test was used to compare the pathological data from the collected samples. Two groups were compared with Student's t-test. Three groups were analyzed with one-way ANOVA followed by Newman-Keuls test. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-501-3p is upregulated in CRC tissues and cells
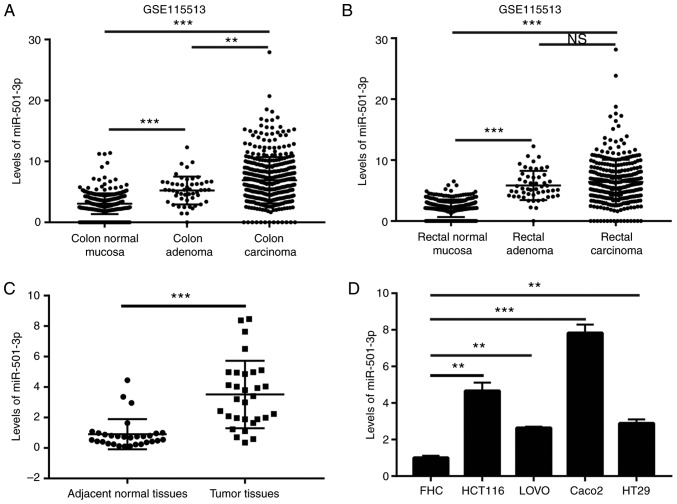
To investigate the potential role of miR-501-3p in CRC, the expression levels of miR-501-3p in colon normal mucosa, colon adenoma, colon carcinoma, rectal normal mucosa, rectal adenoma and rectal carcinoma were analyzed from a GEO dataset (GSE115513) (22). The results indicated that miR-501-3p levels were elevated in adenoma and carcinoma compared with normal tissues (Fig. 1A and B). To confirm this observation, RT-qPCR was used to detect the miR-501-3p expression levels in 30 pairs of normal and tumor tissues from patients with CRC collected for the present study. The results demonstrated that miR-501-3p was significantly overexpressed in tumor tissues compared to normal tissues (Fig. 1C). Additionally, high expression of miR-501-3p was associated with increased TNM stage (III/IV) in patients with CRC (Table I). In vitro, miR-501-3p was significantly overexpressed in CRC cell lines (HCT116, LOVO, Caco2 and HT29) compared with a colon epithelial cell line FHC (Fig. 1D). These data suggested that miR-501-3p might be involved in the progression of CRC.
Figure 1.
Overexpression of miR-501-3p in colorectal cancer tissues and cell lines. (A) Analysis of expression data in GSE115513 revealed that miR-501-3p was overexpressed in colon adenoma and colon carcinoma compared with colon normal mucosa. (B) Analysis of expression data in GSE115513 revealed that miR-501-3p was overexpressed in rectal adenoma and rectal carcinoma compared with rectal normal mucosa. (C) RT-qPCR analysis revealed that miR-501-3p was overexpressed in tumor tissues compared to normal tissues from 30 patients with colorectal cancer. (D) RT-qPCR analysis revealed that miR-501-3p was overexpressed in colorectal cancer cell lines (HCT116, LOVO, Caco2, HT29) compared with the epithelial colorectal cell line FHC. **P<0.01 and ***P<0.001, with comparisons indicated by lines. miR, microRNA; RT-qPCR, reverse transcription-quantitative PCR; n.s., not significant.
Table I.
Association between miR-501-3p expression and clinicopathological features of patients with colorectal cancer.
| Clinicopathological features | Number of cases | miR-501 -3p expression
|
P-value | |
|---|---|---|---|---|
| High (n=15) | Low (n=15) | |||
| Age | 1.000 | |||
| >50 | 17 | 9 | 8 | |
| ≤50 | 13 | 6 | 7 | |
| Sex | 0.427 | |||
| Male | 21 | 9 | 12 | |
| Female | 9 | 6 | 3 | |
| Tumor size (mm) | 0.272 | |||
| >5 | 14 | 9 | 5 | |
| ≤5 | 16 | 6 | 10 | |
| TNM stage | 0.008 | |||
| I-II | 12 | 2 | 10 | |
| III-IV | 18 | 13 | 5 | |
Downregulation of miR-501-3p inhibits CRC cell proliferation and stemness
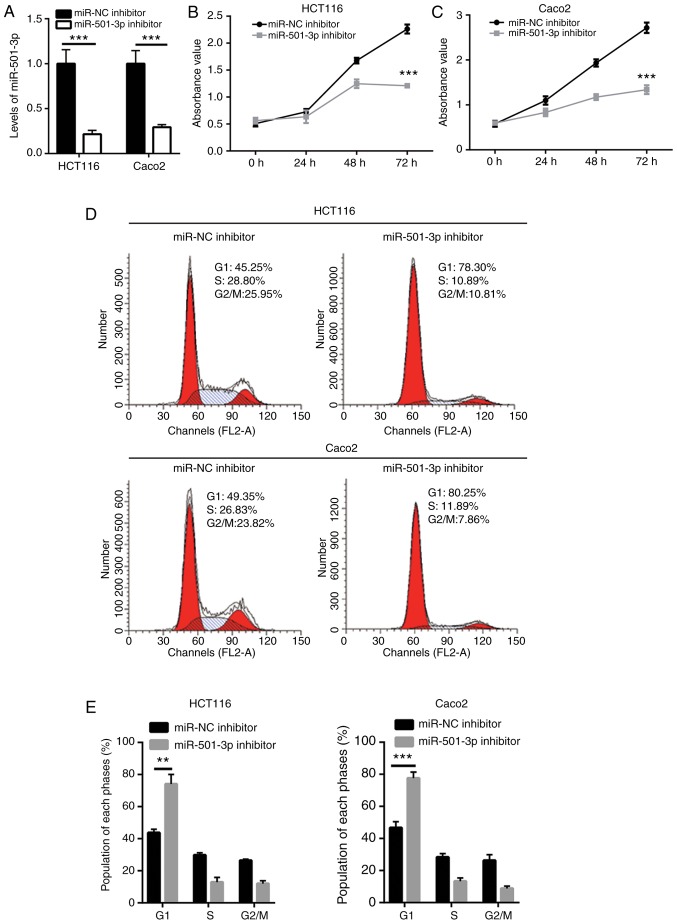
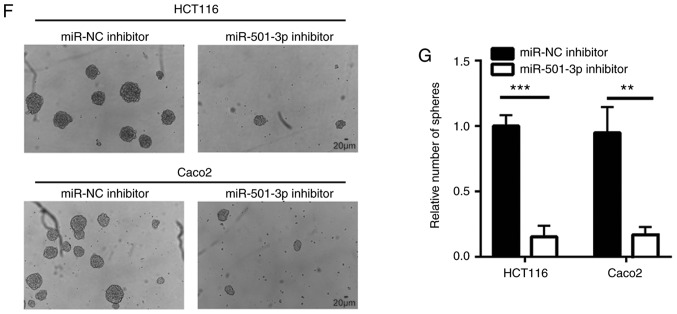
To explore the function of miR-501-3p in CRC, CRC cells were transfected in vitro with a miR-501-3p inhibitor. Transfection of mR-501-3p inhibitor effectively decreased the miR-501-3p expression levels in HCT116 and Caco2 cells (Fig. 2A). The cell proliferation assay indicated that miR-501-3p inhibitor significantly suppressed cell proliferation in both HCT116 and Caco2 cells (Fig. 2B and C). Uncontrolled cell cycle progression leads to fast cell proliferation in cancer cells. Results from flow cytometry analysis revealed that downregulation of miR-501-3p induced an accumulation of cells in the G1 phase in HCT116 and Caco2 cells (Fig. 2D and E), suggesting that miR-501-3p might regulate cell proliferation via cell cycle control. Cancer stem cells are cancer cells that exhibit strong self-renewing ability and are associated with cancer initiation and drug resistance. The sphere formation assay revealed that downregulation of miR-501-3p significantly reduced the number of spheres formed by HCT116 and Caco2 cells (Fig. 2F and G), indicating that miR-501-3p could maintain the stemness of CRC cells.
Figure 2.
miR-501-3p regulates colorectal cancer cell proliferation, cell cycle progression and sphere formation. (A) Transfection with miR-501-3p inhibitor decreased miR-501-3p expression levels in HCT116 and Caco2 cells. (B) The cell proliferation ability was inhibited following miR-501-3p downregulation in HCT116 cells. (C) The cell proliferation ability was inhibited following miR-501-3p downregulation in Caco2 cells. (D) Representative images of cell cycle distribution analysis in HCT116 and Caco2 cells transfected with miR-NC inhibitor or miR-501-3p inhibitor. (E) Quantification of cell cycle distribution analysis revealed that miR-501-3p downregulation accumulated cells in the G1 phase. (F) Representative images of sphere formation in HCT116 and Caco2 cells transfected with miR-NC inhibitor or miR-501-3p inhibitor. Scale bar, 20 µm. (G) Quantification of sphere formation assay revealed that miR-501-3p downregulation reduced the sphere formation ability of HCT116 and Caco2 cells. **P<0.01 and ***P<0.001, with comparisons indicated by lines. miR, microRNA; NC, negative control.
APC is a direct target gene of miR-501-3p in CRC cells
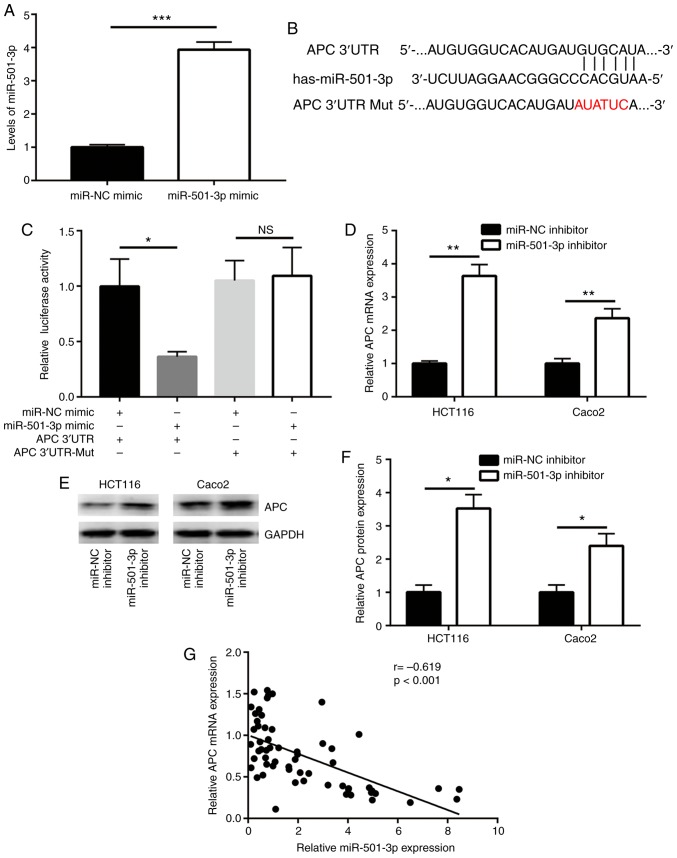
miRNAs function via directly binding to the 3'UTR of target mRNAs to downregulate their expression. Using TargetScan software (26), several mRNAs were predicted to be potential target genes of miR-501-3p. Among them, the 3'UTR of APC, a well-known negative regulator of Wnt/β-catenin signaling, was found to harbor a complementary binding site for miR-501-3p (Fig. 3B). To further investigate whether APC is a direct target of miR-501-3p, mimics transfection and a dual luciferase assay were employed. First, the efficacy of miR-501-3p overexpression by mimic transfection was confirmed (Fig. 3A). In the dual luciferase reporter assay, overexpression of miR-501-3p significantly reduced the relative luciferase activity in HCT116 cells transfected with APC 3'UTR, but not the mutant APC 3'UTR (Fig. 3C). In addition, inhibition of miR-501-3p significantly increased the APC mRNA expression levels in HCT116 and Caco2 cells (Fig. 3D). Western blot analysis revealed that miR-501-3p inhibition also significantly increased the APC protein expression levels in these two cell lines (Fig. 3E and F). Of note, using RT-qPCR to examine the mRNA expression levels of APC, a strong negative correlation was observed between miR-501-3p and APC mRNA expression in tumor and normal tissues from the 30 patients with CRC (Fig. 3G).
Figure 3.
miR-501-3p directly targets APC in colorectal cancer cells. (A) Confirmation of successful miR-501-3p overexpression in HCT116 cells following mimics transfection. (B) Bioinformatic analysis revealed an miR-501-3p binding site within the 3'UTR of APC. The nucleotides that were mutated to generate the APC 3'UTR-Mut reporter are shown in red. (C) Overexpression of miR-501-3p specifically repressed relative luciferase activity in cells transfected with APC 3'UTR. (D) Downregulation of miR-501-3p increased APC mRNA expression levels in HCT116 and Caco2 cells. (E) Western blotting revealed that transfection with the miR-501-3p inhibitor increased the APC protein expression levels in HCT116 and Caco2 cells. (F) Quantitative analysis of APC protein expression levels in D. (G) Reverse transcription-quantitative PCR was used to detect APC mRNA expression levels in 30 tumors from patients with colorectal cancer. Pearson correlation analysis was applied to examine the correlation between miR-501-3p and APC expression levels in the 30 pairs of tumors and normal tissues. *P<0.05, **P<0.01 and ***P<0.001, with comparisons indicated by lines. miR, microRNA; APC, adenomatous polyposis coli; UTR, untranslated region; NC, negative control; Mut, mutant.
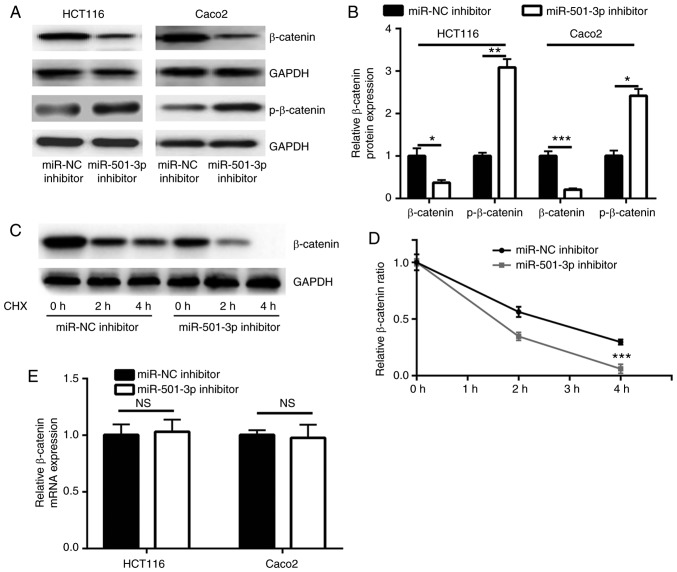
miR-501-3p inhibition downregulates β-catenin expression via targeting APC in CRC cells
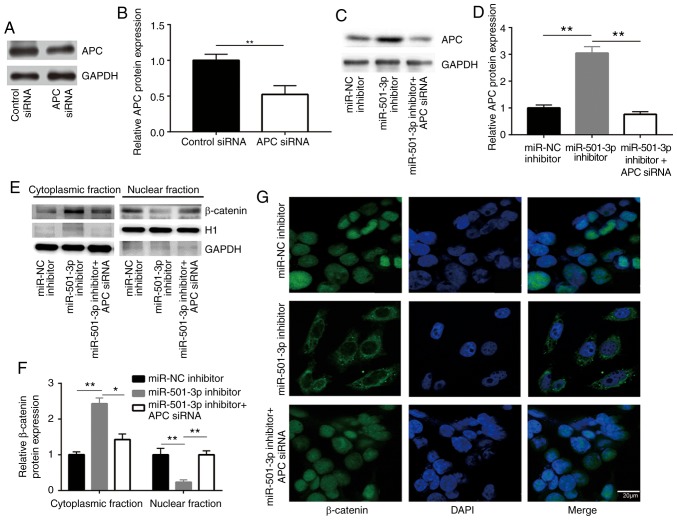
Consistent with the upregulation of APC, miR-501-3p inhibition significantly decreased β-catenin protein expression levels and increased p-β-catenin levels, in both HCT116 and Caco2 cells (Fig. 4A and B). APC promotes the degradation of β-catenin protein via forming a complex with glycogen synthase kinase 3β and other proteins to phosphorylate β-catenin (28). In a CHX chase assay, inhibition of miR-501-3p significantly destabilized the β-catenin protein in HCT116 cells (Fig. 4C and D). RT-qPCR results indicated that the mRNA expression levels of β-catenin were not altered following miR-501-3p inhibition (Fig. 4E), suggesting that miR-501-3p regulated the stability of the β-catenin protein rather than its gene transcription. The subcellular localization of β-catenin is tightly controlled by APC and associated with its transcriptional activity (28). To investigate whether APC was involved in the regulation of β-catenin by miR-501-3p, APC siRNA was transfected into HCT116 cells to downregulate APC expression (Fig. 5A and B). While the miR-501-3p inhibitor increased the APC protein expression levels, APC silencing by siRNA decreased the APC protein expression levels in HCT116 cells (Fig. 5C and D). To explore whether miR-501-3p regulated β-catenin cellular localization, cytoplasmic and nuclear protein extracts were separately prepared. Western blotting demonstrated that transfection with the miR-501-3p inhibitor significantly increased the proportion of β-catenin in the cytoplasm and decreased the proportion of β-catenin in the nucleus, compared with the cells transfected with the negative control (Fig. 5E and F). By contrast, APC silencing reversed the translocation of β-catenin in HCT116 cells (Fig. 5E and F), suggesting the inactivation of Wnt/β-catenin signaling. The translocation of β-catenin from the nucleus to the cytoplasm following miR-501-3p inhibition was further confirmed by immunofluorescence (Fig. 5G); APC silencing reversed this effect (Fig. 5G). These results indicated that miR-501-3p regulated Wnt/β-catenin signaling via targeting APC in CRC cells.
Figure 4.
miR-501-3p stabilizes the β-catenin protein in colorectal cancer cells. (A) Downregulation of miR-501-3p decreased the β-catenin total protein expression levels and increased the p-β-catenin protein expression levels in HCT116 and Caco2 cells. (B) Quantification of β-catenin and p-β-catenin protein expression levels in A. (C) The CHX chase assay suggested that downregulation of miR-501-3p led to an increase of β-catenin degradation in HCT116 cells. (D) Analysis of β-catenin protein degradation in HCT116 cells transfected with miR-NC inhibitor or miR-501-3p inhibitor. (E) Reverse transcription-quantitative qPCR analysis revealed that miR-501-3p downregulation had no effect on β-catenin mRNA expression levels in HCT116 and Caco2 cells. *P<0.05, **P<0.01 and ***P<0.001, with comparisons indicated by lines. miR, microRNA; p-, phosphorylated; CHX, cycloheximide; NC, negative control; n.s., not significant.
Figure 5.
Downregulation of miR-501-3p promotes translocation of β-catenin via targeting APC in colorectal cancer cells. (A) Western blotting confirmed that transfection of APC siRNA significantly decreased APC protein expression levels in HCT116 cells. (B) Quantitative analysis of APC protein expression in A. (C) Western blotting confirmed that miR-501-3p inhibitor increased APC protein expression, and this was effectively reversed following APC siRNA transfection in HCT116. (D) Quantitative analysis of APC protein expression levels in C. (E) Western blotting revealed that miR-501-3p downregulation resulted in an increase of β-catenin protein in the cytoplasmic fraction and a decrease of β-catenin in the nuclear fraction, while silencing of APC reversed the altered subcellular localization of β-catenin in HCT116 cells. H1 and GAPDH were used as internal controls for the nuclear and the cytoplasmic protein extracts, respectively. (F) Quantitative analysis of β-catenin protein expression in E. (G) Immunofluorescence analysis revealed that miR-501-3p downregulation induced translocation of β-catenin from the nucleus to the cytoplasm, and silencing of APC reversed this translocation in HCT116 cells. Scale bar, 20 µm. *P<0.05 and **P<0.01, with comparisons indicated by lines. miR, microRNA; APC, adenomatous polyposis coli; si, small interfering; H1, histone H1.
miR-501-3p regulates Wnt/β-catenin signaling-related gene expression via targeting APC in CRC cells
Wnt/β-catenin signaling directly activates several key genes which are involved in cell proliferation, cell cycle and stemness to control cancer progression (29). In HCT116 and Caco2 cells, transfection with miR-501-3p inhibitor significantly decreased the mRNA expression levels of cyclinD1 and c-Myc, two Wnt/β-catenin pathway target genes; this effect was effectively reversed by APC silencing (Fig. 6A and B). In addition, the protein expression levels of cyclinD1 and c-Myc were also reduced with miR-501-3p inhibition, which was again reversed following APC silencing (Fig. 6C-F).
Figure 6.
Downregulation of miR-501-3p increases cyclinD1 and c-Myc expression via targeting APC in colorectal cancer cells. (A) Downregulation of miR-501-3p decreased cyclinD1 and c-Myc mRNA expression levels, while APC silencing reversed the downregulation of cyclinD1 and c-Myc in HCT116 cells and in (B) Caco2 cells. (C) Downregulation of miR-501-3p decreased cyclinD1 and c-Myc protein expression levels, while APC silencing reversed the downregulation of cyclinD1 and c-Myc in HCT116 cells. (D) Quantitative analysis of cyclinD1 and c-Myc protein expression levels in C. (E) Downregulation of miR-501-3p decreased cyclinD1 and c-Myc protein expression levels, while APC silencing reversed downregulation of cyclinD1 and c-Myc in Caco2 cells. (F) Quantitative analysis of cyclinD1 and c-Myc protein expression levels in E. **P<0.01 and ***P<0.001, with comparisons indicated by lines. miR, microRNA; APC, adenomatous polyposis coli; NC, negative control; si, small interfering.
APC is pivotal for miR-501-3p-mediated CRC cell proliferation and stemness
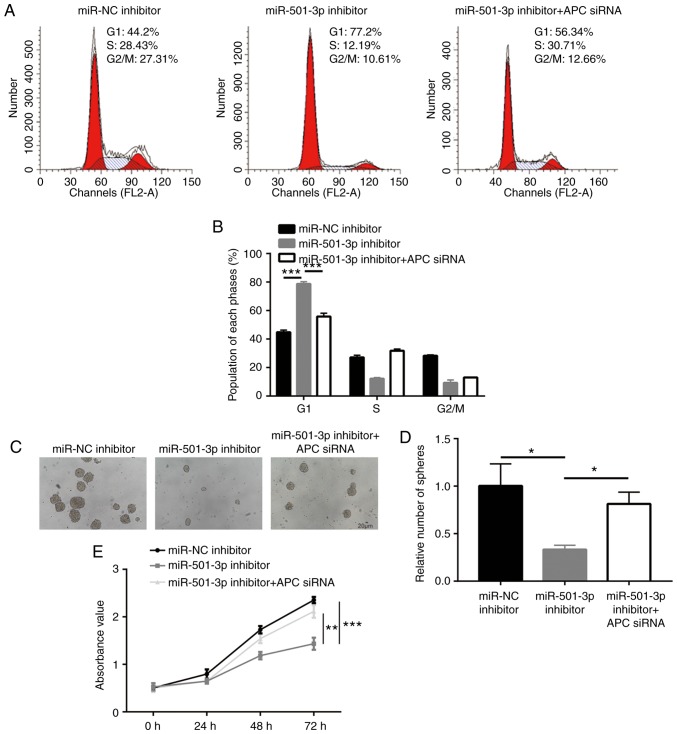
Next, the present study aimed to confirm whether APC was critical for the function of miR-501-3p in CRC cells. Analysis of cell cycle phase distribution revealed that an accumulation of cells in the G1 phase induced by the miR-501-3p inhibitor was also rescued after APC silencing (Fig. 7A and B). In addition, transfection with the miR-501-3p inhibitor induced a significant decrease of sphere formation ability, which was recovered following APC silencing (Fig. 7C and D). Furthermore, the cell proliferation assay suggested that APC silencing reversed the miR-501-3p inhibitor-induced cell growth arrest in HCT116 cells (Fig. 7E). The present results demonstrated that miR-501-3p regulated CRC cell proliferation and stemness via regulation of APC expression.
Figure 7.
miR-501-3p/APC regulate colorectal cancer cell proliferation, cell cycle progression and sphere formation. (A) Representative images of cell cycle distribution in HCT116 cells transfected with miR-NC inhibitor or miR-501-3p inhibitor or miR-501-3p inhibitor + APC siRNA. (B) Quantitative analysis of cell cycle distribution indicated that miR-501-3p downregulation induced cell accumulation in the G1 phase, and this effect was reversed after APC silencing. (C) Representative images of sphere formation in HCT116 cells transfected with miR-NC inhibitor or miR-501-3p inhibitor or miR-501-3p inhibitor + APC siRNA. Scale bar, 20 µm. (D) Quantitative analysis of the sphere formation assay. (E) Downregulation of miR-501-3p induced cell growth arrest, and this was reversed after APC silencing. *P<0.05, **P<0.01 and ***P<0.001, with comparisons indicated by lines. miR, microRNA; APC, adenomatous polyposis coli; NC, negative control; si, small interfering.
Discussion
Accumulating evidence has suggested that miRNAs are involved in the initiation and development of cancer (30). Several miRNAs have been discovered as differentially expressed between tumor and normal tissues, or serum from healthy volunteers and from patients with cancer (31-33). Several have been experimentally validated as oncogenes or tumor suppressors via targeting key genes that are involved in cancer progression (34,35). Compared with normal colonic mucosa, miR-501-3p is one of the most significantly upregulated miRNAs in colorectal carcinomas, as reported by Slattery et al (22). The current study further confirmed the upregulation of miR-501-3p in CRC tumors and cell lines compared with normal. Notably, the present results demonstrated that miR-501-3p promoted CRC cell proliferation and stemness via targeting APC to activate Wnt/β-catenin signaling.
miR-501-3p is involved in carcinogenesis and Alzheimer's disease (23,36). In cervical cancer and hepatocellular carcinoma, miR-501-3p is overexpressed in tumor tissues compared with normal tissues, and promotes cell proliferation, migration and invasion via targeting CYLD lysine 63 deubiquitinase (23,24). The present study demonstrated that downregulation of miR-501-3p inhibited CRC cell proliferation, cell cycle progression and sphere formation in CRC cells. The results of functional assays suggested that miR-501-3p might accelerate cell cycle progression to promote cell proliferation; in addition, they emphasized a role of miR-501-3p in mediating cancer cell stemness for the first time. Cancer stem cells are pivotal for tumor initiation and drug resistance (37,38); therefore, it would be interesting to examine the potential role of miR-501-3p in regulating drug sensitivity in the future. Overall, in agreement with previous studies, the present data support an oncogenic role of miR-510-3p in CRC.
Among several oncogenic signaling pathways, the Wnt/β-catenin signaling pathway is the most well-characterized driver of CRC (13). Several negative regulators of the Wnt/β-catenin signaling pathway, such as APC and axin 1/2, are frequently mutated, resulting in loss of function in CRC (39). In addition, almost all the key components of Wnt/β-catenin signaling are reported to be regulated by miRNAs in CRC. For example, miR-7 targets the transcription factor YY1 to inactivate Wnt/β-catenin signaling and to inhibit CRC cell proliferation (40). miR-23b inhibits CRC cell proliferation via targeting Frizzled class receptor 7, a receptor of Wnt/β-catenin signaling (41). By contrast, miR-135a/b targets APC to promote CRC progression (42). Several miRNAs, such as miR-3607, have been shown to target APC in other types of cancer (43). In the present study, it was predicted and confirmed that APC was a target gene of miR-501-3p in CRC cells. Inhibition of miR-501-3p resulted in downregulation of β-catenin protein expression, as well as its translocation from the nucleus to the cytoplasm, inactivating Wnt/β-catenin signaling in CRC cells. As target genes of Wnt/β-catenin signaling, CyclinD1 and c-Myc are major regulators of the G0/G1 cell cycle checkpoint and stemness (44,45). The present data showing that cyclinD1 and c-Myc were downregulated following miR-501-3p inhibition, suggested that miR-501-3p might regulate cell proliferation, cell cycle and sphere formation via APC. In addition, it was demonstrated that the inhibition of cell proliferation, cell cycle and sphere formation induced by miR-501-3p downregulation were reversed by APC silencing. The current data revealed miR-501-3p as a novel regulator of Wnt/β-catenin signaling in CRC.
In conclusion, the present results demonstrated that miR-501-3p promoted the proliferation and stemness of CRC cells through targeting APC and activating Wnt/β-catenin signaling. These findings enrich the current understanding on the molecular mechanism of miR-501-3p function and the activity of Wnt/β-catenin signaling in CRC, which may facilitate the research and development of novel treatment approaches. However, the association between miR-501-3p and the prognosis of patients with CRC was not included in the current study; further studies will be needed to assess the prognostic potential of miR-501-3p.
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
FL participated in the design and performance of the experiments. XG and TX contributed to the collection of samples and clinical data. TX supervised the study and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All procedures performed in the present study involving human specimens were approved by the Ethics Committee of The Fourth People's Hospital of Shanxi. Written informed consent was provided by all patients prior to surgery.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 3.Araghi M, Soerjomataram I, Jenkins M, Brierley J, Morris E, Bray F, Arnold M. Global trends in colorectal cancer mortality: Projections to the year 2035. Int J Cancer. 2019;144:2992–3000. doi: 10.1002/ijc.32055. [DOI] [PubMed] [Google Scholar]
- 4.Jiang S, Miao D, Wang M, Lv J, Wang Y, Tong J. MiR-30-5p suppresses cell chemoresistance and stemness in colorectal cancer through USP22/Wnt/β-catenin signaling axis. J Cell Mol Med. 2019;23:630–640. doi: 10.1111/jcmm.13968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Claessen MM, Schipper ME, Oldenburg B, Siersema PD, Offerhaus GJ, Vleggaar FP. WNT-pathway activation in IBD-associated colorectal carcinogenesis: Potential biomarkers for colonic surveillance. Cell Oncol. 2010;32:303–310. doi: 10.3233/CLO-2009-0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng H, Sun X, Li J, He P, Liu W, Meng X. Knockdown of Uba2 inhibits colorectal cancer cell invasion and migration through downregulation of the Wnt/β-catenin signaling pathway. J Cell Biochem. 2018;119:6914–6925. doi: 10.1002/jcb.26890. [DOI] [PubMed] [Google Scholar]
- 7.Bienz M, Clevers H. Linking colorectal cancer to Wnt signaling. Cell. 2000;103:311–320. doi: 10.1016/S0092-8674(00)00122-7. [DOI] [PubMed] [Google Scholar]
- 8.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 9.Emons G, Spitzner M, Reineke S, Möller J, Auslander N, Kramer F, Hu Y, Beissbarth T, Wolff HA, Rave-Fränk M, et al. Chemoradiotherapy resistance in colorectal cancer cells is mediated by Wnt/β-catenin signaling. Mol Cancer Res. 2017;15:1481–1490. doi: 10.1158/1541-7786.MCR-17-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Myant KB, Cammareri P, McGhee EJ, Ridgway RA, Huels DJ, Cordero JB, Schwitalla S, Kalna G, Ogg EL, Athineos D, et al. ROS production and NF-κB activation triggered by RAC1 facilitate WNT-driven intestinal stem cell proliferation and colorectal cancer initiation. Cell Stem Cell. 2013;12:761–773. doi: 10.1016/j.stem.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peifer M, Polakis P. Wnt signaling in oncogenesis and embryogenesis-a look outside the nucleus. Science. 2000;287:1606–1609. doi: 10.1126/science.287.5458.1606. [DOI] [PubMed] [Google Scholar]
- 12.Sparks AB, Morin PJ, Vogelstein B, Kinzler KW. Mutational analysis of the APC/beta-catenin/Tcf pathway in colorectal cancer. Cancer Res. 1998;58:1130–1134. [PubMed] [Google Scholar]
- 13.Sawa M, Masuda M, Yamada T. Targeting the Wnt signaling pathway in colorectal cancer. Expert Opin Ther Targets. 2016;20:419–429. doi: 10.1517/14728222.2016.1098619. [DOI] [PubMed] [Google Scholar]
- 14.Olsen AK, Coskun M, Bzorek M, Kristensen MH, Danielsen ET, Jørgensen S, Olsen J, Engel U, Holck S, Troelsen JT. Regulation of APC and AXIN2 expression by intestinal tumor suppressor CDX2 in colon cancer cells. Carcinogenesis. 2013;34:1361–1369. doi: 10.1093/carcin/bgt037. [DOI] [PubMed] [Google Scholar]
- 15.Shu Z, Chen L, Ding D. miR-582-5P induces colorectal cancer cell proliferation by targeting adenomatous polyposis coli. World J Surg Oncol. 2016;14:239. doi: 10.1186/s12957-016-0984-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee RC, Feinbaum RL, Ambros V. The C. elegans heter-ochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 17.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 19.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 20.Thomas J, Ohtsuka M, Pichler M, Ling H. MicroRNAs: Clinical relevance in colorectal cancer. Int J Mol Sci. 2015;16:28063–28076. doi: 10.3390/ijms161226080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iwaya T, Yokobori T, Nishida N, Kogo R, Sudo T, Tanaka F, Shibata K, Sawada G, Takahashi Y, Ishibashi M, et al. Downregulation of miR-144 is associated with colorectal cancer progression via activation of mTOR signaling pathway. Carcinogenesis. 2012;33:2391–2397. doi: 10.1093/carcin/bgs288. [DOI] [PubMed] [Google Scholar]
- 22.Slattery ML, Herrick JS, Pellatt DF, Stevens JR, Mullany LE, Wolff E, Hoffman MD, Samowitz WS, Wolff RK. MicroRNA profiles in colorectal carcinomas, adenomas and normal colonic mucosa: Variations in miRNA expression and disease progression. Carcinogenesis. 2016;37:245–261. doi: 10.1093/carcin/bgv249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanches JGP, Xu Y, Yabasin IB, Li M, Lu Y, Xiu X, Wang L, Mao L, Shen J, Wang B, et al. miR-501 is upregulated in cervical cancer and promotes cell proliferation, migration and invasion by targeting CYLD. Chem Biol Interact. 2018;285:85–95. doi: 10.1016/j.cbi.2018.02.024. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y, Chai Y, Zhang J, Tang J. A Function variant at miR-501 alters susceptibility to hepatocellular carcinoma in a Chinese Han population. Cell Physiol Biochem. 2016;38:2500–2508. doi: 10.1159/000445600. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreno-Bueno G, Hardisson D, Sánchez C, Sarrió D, Cassia R, García-Rostán G, Prat J, Guo M, Herman JG, Matías-Guiu X, et al. Abnormalities of the APC/beta-catenin pathway in endometrial cancer. Oncogene. 2002;21:7981–7990. doi: 10.1038/sj.onc.1205924. [DOI] [PubMed] [Google Scholar]
- 28.Gumbiner BM. Carcinogenesis: A balance between beta-catenin and APC. Curr Biol. 1997;7:R443–R446. doi: 10.1016/S0960-9822(06)00214-4. [DOI] [PubMed] [Google Scholar]
- 29.Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 30.Gomase VS, Parundekar AN. MicroRNA: Human disease and development. Int J Bioinform Res Appl. 2009;5:479–500. doi: 10.1504/IJBRA.2009.028678. [DOI] [PubMed] [Google Scholar]
- 31.Bailey ST, Westerling T, Brown M. Loss of estrogen-regulated microRNA expression increases HER2 signaling and is prognostic of poor outcome in luminal breast cancer. Cancer Res. 2015;75:436–445. doi: 10.1158/0008-5472.CAN-14-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Gu J, Roth JA, Hildebrandt MA, Lippman SM, Ye Y, Minna JD, Wu X. Pathway-based serum microRNA profiling and survival in patients with advanced stage non-small cell lung cancer. Cancer Res. 2013;73:4801–4809. doi: 10.1158/0008-5472.CAN-12-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yokoi A, Matsuzaki J, Yamamoto Y, Yoneoka Y, Takahashi K, Shimizu H, Uehara T, Ishikawa M, Ikeda SI, Sonoda T, et al. Integrated extracellular microRNA profiling for ovarian cancer screening. Nat Commun. 2018;9:4319. doi: 10.1038/s41467-018-06434-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu C, Liu R, Zhang D, Deng Q, Liu B, Chao HP, Rycaj K, Takata Y, Lin K, Lu Y, et al. MicroRNA-141 suppresses prostate cancer stem cells and metastasis by targeting a cohort of pro-metastasis genes. Nat Commun. 2017;8:14270. doi: 10.1038/ncomms14270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu J, Lei R, Zhuang X, Li X, Li G, Lev S, Segura MF, Zhang X, Hu G. MicroRNA-182 targets SMAD7 to potentiate TGFβ-induced epithelial-mesenchymal transition and metastasis of cancer cells. Nat Commun. 2016;7:13884. doi: 10.1038/ncomms13884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hara N, Kikuchi M, Miyashita A, Hatsuta H, Saito Y, Kasuga K, Murayama S, Ikeuchi T, Kuwano R. Serum microRNA miR-501-3p as a potential biomarker related to the progression of Alzheimer's disease. Acta Neuropathol Commun. 2017;5:10. doi: 10.1186/s40478-017-0414-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.James MI, Iwuji C, Irving G, Karmokar A, Higgins JA, Griffin-Teal N, Thomas A, Greaves P, Cai H, Patel SR, et al. Curcumin inhibits cancer stem cell phenotypes in ex vivo models of colorectal liver metastases, and is clinically safe and tolerable in combination with FOLFOX chemotherapy. Cancer Lett. 2015;364:135–141. doi: 10.1016/j.canlet.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boman BM, Fields JZ, Bonham-Carter O, Runquist OA. Computer modeling implicates stem cell overproduction in colon cancer initiation. Cancer Res. 2001;61:8408–8411. [PubMed] [Google Scholar]
- 39.Novellasdemunt L, Antas P, Li VS. Targeting Wnt signaling in colorectal cancer. A review in the theme: Cell signaling: Proteins, pathways and mechanisms. Am J Physiol Cell Physiol. 2015;309:C511–C521. doi: 10.1152/ajpcell.00117.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang N, Li X, Wu CW, Dong Y, Cai M, Mok MT, Wang H, Chen J, Ng SS, Chen M, et al. microRNA-7 is a novel inhibitor of YY1 contributing to colorectal tumorigenesis. Oncogene. 2013;32:5078–5088. doi: 10.1038/onc.2012.526. [DOI] [PubMed] [Google Scholar]
- 41.Zhang H, Hao Y, Yang J, Zhou Y, Li J, Yin S, Sun C, Ma M, Huang Y, Xi JJ. Genome-wide functional screening of miR-23b as a pleiotropic modulator suppressing cancer metastasis. Nat Commun. 2011;2:554. doi: 10.1038/ncomms1555. [DOI] [PubMed] [Google Scholar]
- 42.Nagel R, le Sage C, Diosdado B, van der Waal M, Oude Vrielink JA, Bolijn A, Meijer GA, Agami R. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008;68:5795–5802. doi: 10.1158/0008-5472.CAN-08-0951. [DOI] [PubMed] [Google Scholar]
- 43.Lin Y, Gu Q, Sun Z, Sheng B, Qi C, Liu B, Fu T, Liu C, Zhang Y. Upregulation of miR-3607 promotes lung adenocar-cinoma proliferation by suppressing APC expression. Biomed Pharmacother. 2017;95:497–503. doi: 10.1016/j.biopha.2017.08.052. [DOI] [PubMed] [Google Scholar]
- 44.Liu L, Zhang H, Shi L, Zhang W, Yuan J, Chen X, Liu J, Zhang Y, Wang Z. Inhibition of Rac1 activity induces G1/S phase arrest through the GSK3/cyclin D1 pathway in human cancer cells. Oncol Rep. 2014;32:1395–1400. doi: 10.3892/or.2014.3388. [DOI] [PubMed] [Google Scholar]
- 45.Lee SH, Chen TY, Dhar SS, Gu B, Chen K, Kim YZ, Li W, Lee MG. A feedback loop comprising PRMT7 and miR-24-2 interplays with Oct4, Nanog, Klf4 and c-Myc to regulate stemness. Nucleic Acids Res. 2016;44:10603–10618. doi: 10.1093/nar/gkw788. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.