Abstract
As one of the most commonly reported malignancies of the urinary system, clear cell renal cell carcinoma (ccRCC) is an advanced metastatic tumor with high mortality rates. The Rac family small GTPase 2 (RAC2) is a member of the Rho GTPases. Although Rho GTPases play an important role in numerous different types of tumor, whether they have functions in ccRCC remains uncertain. The present study utilized bioinformatics analyses in order to compare the expression levels of RAC2 in ccRCC tumors vs. adjacent tissues, and assessed the association between RAC2 expression and clinicopathological parameters. Furthermore, reverse transcription-quantitative PCR, western blotting and immunohistochemistry assays were performed to validate RAC2 expression levels in human ccRCC tissues and cell lines. Functional experiments were also conducted in order to identify the roles of RAC2 in vitro. The results revealed that RAC2 was upregulated in ccRCC tissues and cell lines. In addition, elevated expression levels of RAC2 were significantly associated with a poor overall survival (P=0.0061), higher Tumor-Node-Metastasis stage and worse G grade. Receiver operating characteristic analysis indicated that high expression levels of RAC2 could be a diagnostic index for ccRCC (area under the curve, 0.9095; P<0.0001). Furthermore, knockdown of RAC2 in vitro attenuated the proliferation, migration and invasion of renal carcinoma cells. In conclusion, the results of the present study demonstrated that RAC2 may act as a promising prognostic and diagnostic biomarker of ccRCC, and could be considered as a potential therapeutic target for treating ccRCC.
Keywords: clear cell renal cell carcinoma, Rac family small GTPase 2, biomarker, prognosis, therapy
Introduction
Renal cell carcinoma (RCC) is a malignant tumor originating in the renal parenchymal urothelial system. It is one of the most frequently observed malignant neoplasms in the urinary system, forming ~85-90% of primary renal malignancies (1). At present, the number of new cases of renal cell cancer has been estimated to be 73,820; a total of 14,770 mortalities have been estimated in the USA (2). Despite the various pathological types of renal cell carcinoma, >80% of RCCs are clear cell RCC (ccRCC) (1,3), which has high risk of metastasis and poor response to chemoradiotherapy (4). Due to the relatively mild initial symptoms (low backache, hematuria and masses), certain patients with ccRCC have experienced local progression and distant metastasis at the time of diagnosis (5). Despite progress having been made in the diagnosis and treatment of ccRCC in recent years, the prognosis of patients with renal cancer remains poor. Postoperative metastasis or local recurrence occurred in ~20-40% of patients (6,7). Targeted drugs have been applied to patients with ccRCC; however, the overall survival (OS) of patients at the terminal stage of disease is unsatisfactory (8). As a result, efficient biomarkers are urgently required for early diagnosis and to determine the molecular mechanisms underlying ccRCC progression and metastasis.
Rac proteins (ras-associated C3 botulinum toxin substrate) are a subfamily of Ras homology (Rho) small GTPases that consist of RAC1, RAC2, RAC3 and RhoG (9). They are activated by guanine nucleotide exchange factors, which exchange GDP for GTP (10). Studies have demonstrated various functions of Rho small GTPases, including regulating actin cytoskeleton rearrangement, regulating cell growth and maintaining stem cells during development (11-13). Furthermore, a number of reports have demonstrated the roles of this family in tumor progression (9,14-16). Pei et al (15) demonstrated that RAC2 and transcription factor jun-B serve as oncogenes in the tumorigenesis of NSCLC. In addition, in a study focusing on glioblastoma, Lai et al (9) revealed that not just RAC1, but RAC2 and RAC3 are also essential for glioblastoma tumorigenesis. However, few studies have investigated these proteins in RCC.
RAC2 is a GTPase with a molecular weight of 21 kDa, which contains the catalytic subunit of NADPH oxidase (17). It is expressed primarily in hematopoietic cells, such as the lymph nodes, bone marrow and spleen, but not in normal kidney tissue (17). Numerous studies that have reported RAC2 as important in cytoskeleton remodeling (18), host defense responses (19) and the expression of oncogenes (20). To the best of our knowledge, there are no studies focusing on RAC2 and ccRCC currently published. Therefore, the present study investigated whether RAC2 expression was associated with clinicopathological characteristics and prognosis of ccRCC. Furthermore, the role of RAC2 in renal cell carcinoma was investigated in vitro.
Materials and methods
ccRCC tissue samples
A total of 120 pairs of ccRCC and adjacent normal renal tissues, as well as 10 benign renal angiomyolipoma tissues and renal cyst tissues were collected between May 2015 and May 2018 at the Department of Urology, Union Hospital, Tongji Medical College (Wuhan, China). The age range of these patients was 22-79 years old. The adjacent normal renal tissues were collected ≥2 cm away from the edge of the tumor site. The proteins and RNA extracted from 50 pairs of these resected samples were analyzed via western blotting and reverse transcription-quantitative PCR (RT-qPCR). The remaining tissues were analyzed via immunohistochemistry (IHC). The basic clinical characteristics (age, sex, tumor size, tumor location and tumor stage) of the patients are presented in Table I. Fuhrman and TNM grading was conducted (21,22). No patients had received any adjuvant anticancer therapy prior to or following surgery. The present study was approved by the Human Research Ethics Committee of Huazhong University of Science and Technology. Written informed consent was provided by the patients or the patients' family. The study methodologies conformed to the standards set by the Declaration of Helsinki.
Table I.
Clinical characteristics of patients with ccRCC.
| Characteristic | N (%) |
|---|---|
| Age | |
| Mean ± SEM, years | 53±13 |
| Sex | |
| Male/female | 72/48 |
| Tumor size | |
| Mean ± SEM, cm | 5.8±3.2 |
| Location | |
| Right/left | 63/57 |
| T stage | |
| T1a | 26 (21.67) |
| T1b | 53 (44.16) |
| T2a | 16 (13.33) |
| T2b | 11 (9.17) |
| T3 | 5 (4.17) |
| T4 | 2 (1.67) |
| Unknown | 7 (5.83) |
| N stage | |
| N0 | 106 (88.33) |
| N1 | 14 (11.67) |
| M stage | |
| M0 | 109 (90.83) |
| M1 | 11 (9.17) |
| Fuhrman grade | |
| 1 | 31 (25.83) |
| 2 | 54 (45.00) |
| 3 | 18 (15.00) |
| 4 | 9 (7.50) |
| Unknown | 8 (6.67) |
SEM, standard error of the mean.
Cell culture
The human renal proximal tubular epithelial cell line HK-2, and the following human renal cell carcinoma cell lines: 786-O, OSRC-2, ACHN, A498 and CAKI-1, were employed in the present study, and were purchased from the American Type Culture Collection. HK-2, 786-O, OSRC-2, ACHN, A498 and CAKI-1 cells were used for RT-qPCR and western blotting. 786-O and ACHN cells were used for transient transfection, cell proliferation, migration and invasion assays. The cells were cultured in high glucose Dulbecco's Modified Eagle's medium (DMEM; Servicbio, Inc.) containing 10% fetal bovine serum (Biomart) and 1% penicillin-streptomycin solution, and maintained in a humidified atmosphere with 5% CO2 at 37°C.
Western blotting
Cells and tissues were lysed in radioimmunoprecipitation assay lysis (Servicbio, Inc.) buffer containing protease inhibitors. The protein concentration of each sample was measured using a BCA Protein Assay kit (Beyotime Institute of Biotechnology). For western blotting, 30 µg proteins were separated via SDS-PAGE (12% gel) and transferred to a polyvinylidene difluoride (PVDF) membrane (EMD Millipore) at 300 mA for 90 min. The PVDF membranes were blocked with 5% non-fat milk for 2 h at room temperature, and then incubated with specific primary antibodies against RAC2 (1:1,000; ProteinTech Group, Inc., cat. no. 60077-1-Ig) and β-actin (1:3,000; Abcam, ab8226) overnight at 4°C. Following incubation with the primary antibodies, the membranes were incubated with the corresponding secondary antibodies (1:3,000; Servicebio, Inc.) for 2 h at room temperature following washing with TBST (3×10 min). Finally, the protein bands were visualized with Pierce™ ECL Western Blotting Substrate (Thermo Fisher Scientific, Inc., cat. no. 32106) using ChemiDoc-XRS+ (Bio-Rad Laboratories, Inc.).
RNA extraction and RT-qPCR
Total RNA was isolated from tissues or cells using TRIzol® reagent (Thermo Fisher Scientific, Inc.). The concentration and purity of the RNA solution were detected using a NanoDrop 2000 spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Inc.). Extracted RNA was then reverse transcribed into cDNA using a Superscript II reverse transcription kit (Takara Bio, Inc.) according to the manufacturer's protocols. The reaction conditions were as follows: 37°C for 15 min; 85°C for 5 sec. Subsequently, the cDNA was subjected to qPCR using a SYBR-Green master kit (ChamQ; Vazyme) on a LightCycler 480 II (Roche Diagnostics) according to the manufacturer's protocols. The qPCR conditions were as follows: Pre-denaturation at 95°C for 3 min; 40 cycles of denaturation at 95°C for 3 sec; annealing and extension at 60°C for 20 sec. The housekeeping gene, GAPDH, was used to normalize the relative expression of RAC2 as an endogenous control by the comparative Cq (threshold cycle) method (2−ΔΔCq) (23). All RT-qPCR reactions were performed in duplicate. The primers used to amplify RAC1, RAC2, RAC3, RhoG and GAPDH were chemically synthesized by TSINGKE. The primer sequences were as follows: RAC1: 5′-ATG CAG GCC ATC AAG TGT GTG GTG-3′ (forward) and 5′-TTA CAA CAG CAG GCA TTT TCT CTT CC-3′ (reverse); RAC2: 5′-CGT CAG CCC AGC CTC TTA TG-3′ (forward) and 5′-TCA GGC CTC TCT GGG TGA G-3′ (reverse); RAC3: 5′-CTC CTA CCC CCA AAC TGA CG-3′ (forward) and 5′-TCA CAG AGC CCA CCA ATC TC-3′ (reverse); RhoG: 5′-ACT ACA GCA ACT GCA CCC ACG A-3′ (forward) and 5′-ACC ACC ACG CAC TTG ATG CTC T-3′ (reverse); and GAPDH: 5′-AAA AGC ATC ACC CGG AGG AGA A-3′ (forward) and 5′-AAG GAA ATG AAT GGG CAG CCG-3′ (reverse).
Immunohistochemistry (IHC) assay
The IHC assay was performed as previously described (24). Briefly, ccRCC tissues and adjacent normal tissues were sequentially fixed in formalin at room temperature for 12 h, dehydrated and embedded in paraffin. Tissue sections were then incubated with a rabbit antibody against RAC2 (1:100; ProteinTech Group, Inc., cat. no. 60077-1-Ig) overnight at 4°C. They were then rinsed three times with PBS and incubated with secondary antibodies that were conjugated to horseradish peroxidase (1:200; GB23303; Servicebio, Inc.) at room temperaturefor 2 h. Finally, tissues were observed in three randomly selected fields under a light microscope (Olympus CX41-32C02; Olympus Corporation) at 40, 100, and 200x magnification.
Knockdown of RAC2
Small interfering RNA (siRNA) oligonucleotide sequences specifically targeting RAC2 (si-RAC2) and a negative control (si-NC) siRNA (cat. no. siN0000002-1-5) were obtained from Guangzhou RiboBio Co., Ltd. BLAST alignment of all siRNA sequences was performed to ensure off-target effects were not exhibited within the sequences (https://blast.ncbi.nlm.nih.gov/Blast.cgi). For transient transfection, RCC cells were incubated in 6-well plates until they reached 50% confluence. si-RAC2 and si-negative control (NC) with a final concentration of 50 nM were transfected with Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Cells were collected for subsequent experiments 48 h post-transfection. The si-RAC2 sequence was as follows: 5′-GCC AAT GTG ATG GTG GAC A-3′.
Cell proliferation assay
ACHN and 786-O cells had been transfected with si-RAC2 or si-NC 48 h before subsequent experimentation. Cells were inoculated on 96-well plates at a cell density of 1×103 cells per well with 100 µl of medium. A cell proliferation assay was performed using a Cell Counting Kit-8 (CCK8; Dojindo Molceular Technologies, Inc.) every 24 h for a total of 96 h, according to the manufacturer's protocols. CCK8 solution (10 µl) was added to each well. After incubation for 3 h at 37°C, the optical density of each well was measured at 450 nm with a spectrophotometer to evaluate the quantity of living cells. Finally, the number of cells over 4 days were plotted in a graph that reflected the rate of cell proliferation.
Cell migration and invasion assays
Migration and invasion assays were performed as previously described (25). ACHN and 786-O cells had been transfected with si-RAC2 or si-NC 48 h before subsequent experimentation. Prior to the migration and invasion assays, cells were incubated in DMEM without serum for 6-8 h. Boyden Transwell chambers and 24-well plates (Corning Inc.) with 8-µm membrane filters were used in the migration and invasion assays. Serum-starved cells (1×105) were seeded into the upper chambers in serum-free medium, and the lower chambers were filled with DMEM containing 10% FBS. After incubation for 24 h at 37°C, the lower chamber was washed twice with PBS and fixed with 100% methanol for 15 min at room temperature, and stained with 0.1% crystal violet dye for 20 min at room temperature. Following washing of the chamber again three times with PBS, non-migrated and non-invaded cells were removed from the upper chamber with a cotton bud. Migrated cells in lower chambers were observed in five randomly selected fields under a light microscope (Olympus CX41-32C02; Olympus Corporation) at 100x magnification. Based on the migration assay, a cell invasion assay was performed in Matrigel-coated Transwell insert chambers (BD Biosciences), which had already been incubated at 37°C for 6-8 h. The remaining procedure was performed as described for the cell migration assays.
Bioinformatics analysis
RAC2 mRNA expression levels and clinical data, including sex, age, Tumor-Node-Metastasis (TNM) stage, G stage and OS of The Cancer Genome Atlas (TCGA) clear cell renal cell carcinoma dataset (TCGA_KIRC) were downloaded from the Xena Functional Genomics Explorer of University of California Santa Cruz (https://xenabrowser.net/heatmap/). The median of RAC2 expression was set as the cutoff point for dividing patients into high and low expression groups. Gumz renal, Lenburg renal, Jones renal and Yusenko renal datasets (26-29) were obtained from the Oncomine database (https://www.oncomine.org). In order to determine which RAC2 signaling pathways were involved in the pathogenesis of ccRCC, a gene set enrichment analysis (GSEA; http://www.broadinstitute.org/gsea) was used with the Kyoto Encylopedia of Genes and Genomes and Gene Ontology databases (c2.all.v6.2.symbols.gmt). For the enriched gene sets, after performing 1,000 permutations, the false discovery rate (FDR) value <0.25 and the P<0.05 were considered to indicate statistically significant enriched pathways (30). The protein expression level of RAC2 in ccRCC tissues was also obtained from The Human Protein Atlas (http://www.proteinatlas.org).
Statistical analysis
Statistical analyses were performed using GraphPad Prism (version 7.0; GraphPad Software) and SPSS Statistics (version 22.0; IBM Corp.). The Numerical data of each group are presented as the mean ± standard deviation. The significant differences in RAC2 expression between each ccRCC subgroup was analyzed using a Student's t-test. A paired Student's t-test was used to analyze RAC2 expression in tumor tissues and matched normal kidney tissues. The associations between RAC2 expression and clinicopathological characteristics in patients with ccRCC were evaluated using Pearson's χ2 test. Receiver operator characteristic (ROC) curves and areas under the curve (AUC) were used to calculate the diagnostic values of RAC2 expression in patients with ccRCC. The association between RAC2 expression and OS was investigated using Kaplan-Meier curves with log-rank tests. P<0.05 was considered to indicate a statistically significant difference.
Results
RAC2 expression is upregulated and associated with numerous types of clinicopathological parameters in ccRCC
In order to determine whether Rac proteins are associated with the development of ccRCC, the mRNA expression values of the four members of this family (RAC1, RAC2, RAC3 and RhoG) were downloaded from TCGA database. In the heatmap constructed using these data, RAC2 was highlighted as having the most significantly different expression levels between ccRCC and normal tissues (Fig. 1). Furthermore, the mRNA expression levels of RAC1, RAC2, RAC3 and RhoG were determined in ccRCC tissues and adjacent normal tissues. The results revealed that the RAC2 was upregulated in the majority of tumor tissues than the other Rac proteins (Fig. S1A-D); thus, RAC2 was selected for subsequent investigation.
Figure 1.
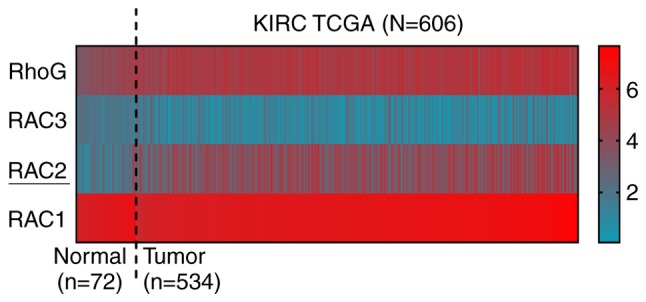
Heatmap of Rac protein family expression levels in samples from TCGA combined human clear cell renal cell carcinoma microarray dataset (n=606). Red indicates high expression; blue indicates low expression. KIRC, kidney renal clear cell carcinoma; RAC, Rac family small GTPase; RhoG, Rho family member G; TCGA, The Cancer Genome Atlas.
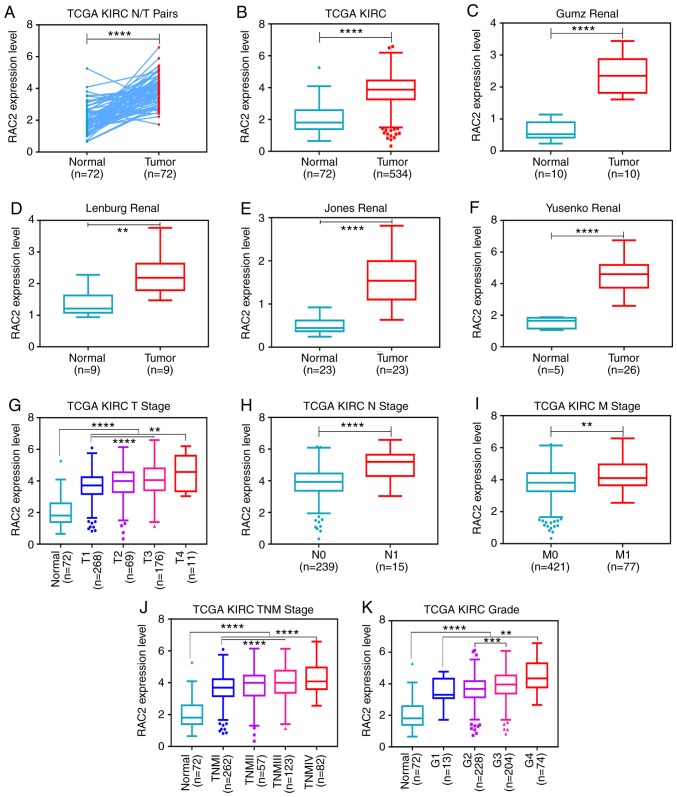
RAC2 mRNA expression levels in ccRCC tissues were then compared with those in adjacent normal tissues downloaded from TCGA. RAC2 expression levels in tumor tissues were significantly higher than that in normal tissues (Fig. 2A and B). To confirm these results, the Oncomine database was searched for RAC2 expression levels in ccRCC. The data from studies by Gumz et al, Lenburg et al, Jones et al and Yusenko et al supported our findings (Fig. 2C-F) (26-29). In addition, the present study analyzed the expression levels of RAC2 using 524 cases from TCGA database with different clinicopathological parameters. It was revealed that high expression levels of RAC2 were significantly associated with higher tumor T stage, distant metastasis, tumor lymph node metastasis (N stage), pathological TNM stage and advanced histological grade (G stage) in ccRCC (Fig. 2G-K; Table II). However, no notable differences were observed between the RAC2 expression levels in patients aged ≥60 years and in those aged <60 years or between males and females. These results indicated that RAC2 expression levels are upregulated and significantly associated with T stage, N stage, M stage, TNM stage and G grade in ccRCC.
Figure 2.
RAC2 expression is upregulated in ccRCC, and is associated with various clinicopathological parameters in ccRCC tissues. The mRNA expression levels of RAC2 were obtained from TCGA dataset, which contained 72 adjacent normal tissues and 534 ccRCC tissues. (A) RAC2 expression was higher in the ccRCC tissues than in the adjacent normal tissues in the 72 paired tissues from patients with ccRCC. The mRNA expression levels of RAC2 were increased in (B) ccRCC tissues than in normal tissues in TCGA, (C) Gumz renal database, (D) Lenburg renal dataset, (E) Jones renal dataset and (F) Yusenko renal dataset. The high expression of RAC2 mRNA was associated with various clinicopathological factors: (G) T stage, (H) lymph node metastasis, (I) distant metastases, (J) TNM stage and (K) G grade. ****P<0.0001; ***P<0.001; **P<0.01. RAC2, Rac family small GTPase 2; ccRCC, clear cell renal cell carcinoma; KIRC, kidney renal clear cell carcinoma; TCGA, The Cancer Genome Atlas; TNM, Tumor-Node-Metastasis.
Table II.
Association between RAC2 mRNA expression and clinicopathological parameters of patients with clear cell renal cell carcinoma.
| Parameter | Number | RAC2 mRNA expression
|
P-value | |
|---|---|---|---|---|
| Low (n=262) | High (n=262) | |||
| Age (years) | 0.793 | |||
| <60 | 244 | 124 | 120 | |
| ≥60 | 280 | 138 | 142 | |
| Sex | 0.462 | |||
| Female | 181 | 95 | 86 | |
| Male | 343 | 167 | 176 | |
| T stage | 0.001a | |||
| T1 or T2 | 337 | 187 | 150 | |
| T3 or T4 | 187 | 75 | 112 | |
| N stage | 0.007 | |||
| N0 or NX | 509 | 260 | 249 | |
| N1 | 15 | 2 | 13 | |
| M stage | 0.006 | |||
| M0 or MX | 447 | 235 | 212 | |
| M1 | 77 | 27 | 50 | |
| G grade | <0.001a | |||
| G1 or G2orGx | 246 | 149 | 97 | |
| G3 or G4 | 278 | 113 | 165 | |
| TNM stage | <0.001a | |||
| I + II | 319 | 180 | 139 | |
| III + IV | 205 | 82 | 123 | |
P<0.05.
Upregulation of RAC2 expression indicates a poor clinical prognosis
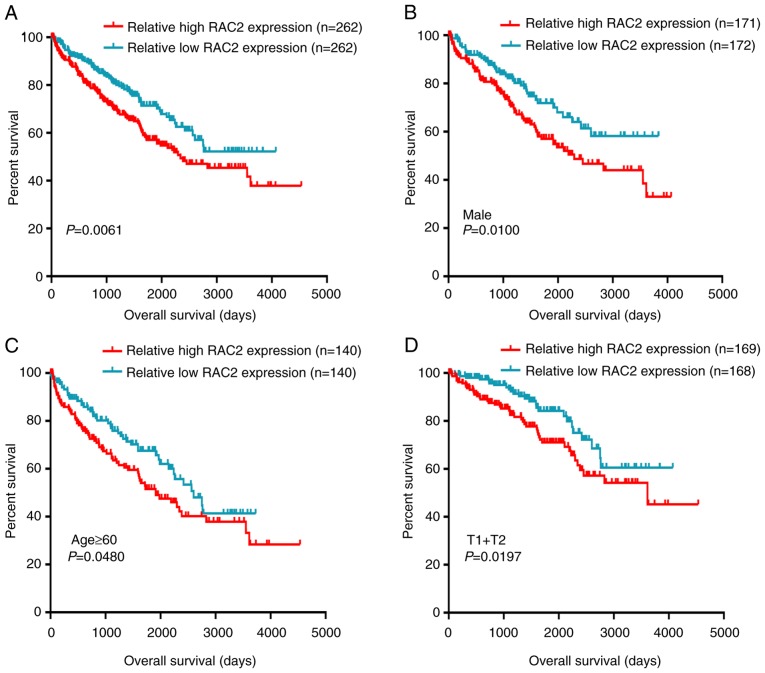
To estimate the prognostic value of RAC2, a Kaplan-Meier survival analysis with log-rank test was applied in order to determine whether OS was associated with RAC2 expression. First, the 524 patients with ccRCC from TCGA database were divided into a relatively high RAC2 expression group and a relatively low RAC2 expression group using the median value of RAC2 mRNA expression level. The results revealed that the OS of the high RAC2 expression group was poorer (P=0.006; Fig. 3A). Further Kaplan-Meier survival analyses regarding RAC2 expression were performed to assess the patients with ccRCC with different clinicopathological characteristics. According to the results from the present study, low RAC2 expression levels tended to indicate a good prognosis for male patients with ccRCC (P=0.0100; Fig. 3B) or those aged ≥60 years (P=0.0480; Fig. 3C) or in T1 + T2 stage (P=0.0197; Fig. 3D). These results indicated that RAC2 may be a potential prognostic biomarker in patients with ccRCC and that elevated RAC2 expression levels indicated relatively poor patient outcome.
Figure 3.
High RAC2 mRNA expression is associated with poor OS in patients with ccRCC. Patient samples from The Cancer Genome Atlas were separated into two groups: Those with low RAC2 expression and those with high RAC2 expression. (A) OS of patients with ccRCC was associated with RAC2 expression. OS subanalysis in regards to RAC2 expression was conducted in subgroups of patients with ccRCC: (B) Male, (C) age ≥60 years and (D) T1 + T2 stage. RAC2, Rac family small GTPase 2; ccRCC, clear cell renal cell carcinoma; OS, overall survival.
RAC2 expression levels could be valuable for the clinical diagnosis of patients with ccRCC
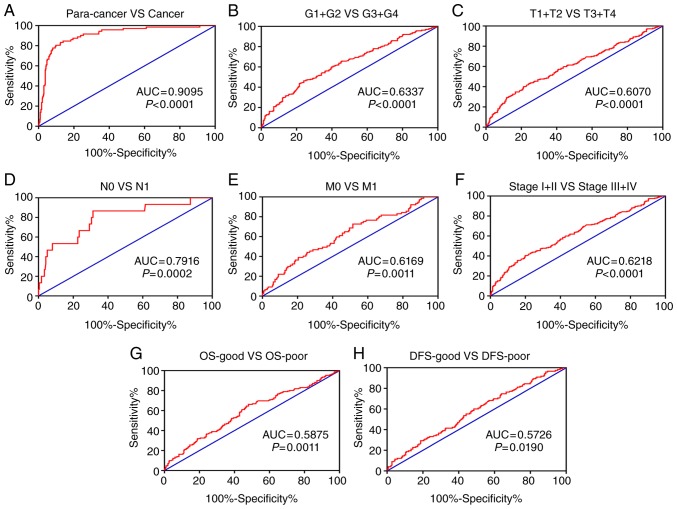
In order to investigate the diagnostic role of RAC2 in ccRCC, a ROC curve was used to assess the clinicopathological factors of the patients. With the AUC, the diagnostic efficiency of RAC2 expression was presented. Generally, ccRCC could be sufficiently distinguished from normal tissues by RAC2 expression with an AUC of 0.9095 (P<0.0001; Fig. 4A). Furthermore, the diagnostic values of RAC2 expression levels were also analyzed in the subgroups as follows: G1 + G2 vs. G3 + G4 stage (AUC, 0.6337; P<0.0001; Fig. 4B); T1 + T2 vs. T3 + T4 stage (AUC, 0.6070; P<0.0001; Fig. 4C); N0 vs. N1 stage (AUC, 0.7916; P=0.0002; Fig. 4D); M0 vs. M1 stage (AUC, 0.6169; P=0.0011; Fig. 4C); stage I + II vs. stage III + IV (AUC, 0.6218; P<0.0001; Fig. 4F); OS-good vs. OS-poor (AUC, 0.5875; P=0.0011; Fig. 4G); and disease-free survival (DFS)-good vs. DFS-poor (AUC, 0.5726; P=0.0190; Fig. 4H). Therefore, RAC2 may be a potential diagnostic biomarker for clear cell renal cell carcinoma.
Figure 4.
RAC2 expression may be a diagnostic biomarker in patients with ccRCC. (A) RAC2 effectively discriminated between ccRCC and paired normal tissues (AUC 0.9095; P<0.0001). Receiver operating characteristic curve subanalysis was performed with respect to the following subgroups of patients with ccRCC: (B) G grade, (C) T stage, (D) lymph node metastasis, (E) distant metastases, (F) Tumor-Node-Metastasis stage, (G) overal survival and (H) disease free survive. RAC2, Rac family small GTPase 2; ccRCC, clear cell renal cell carcinoma; AUC, area under the curve; OS, overall survival; DFS, disease-free survival.
RAC2 is highly expressed in ccRCC cells and tissues
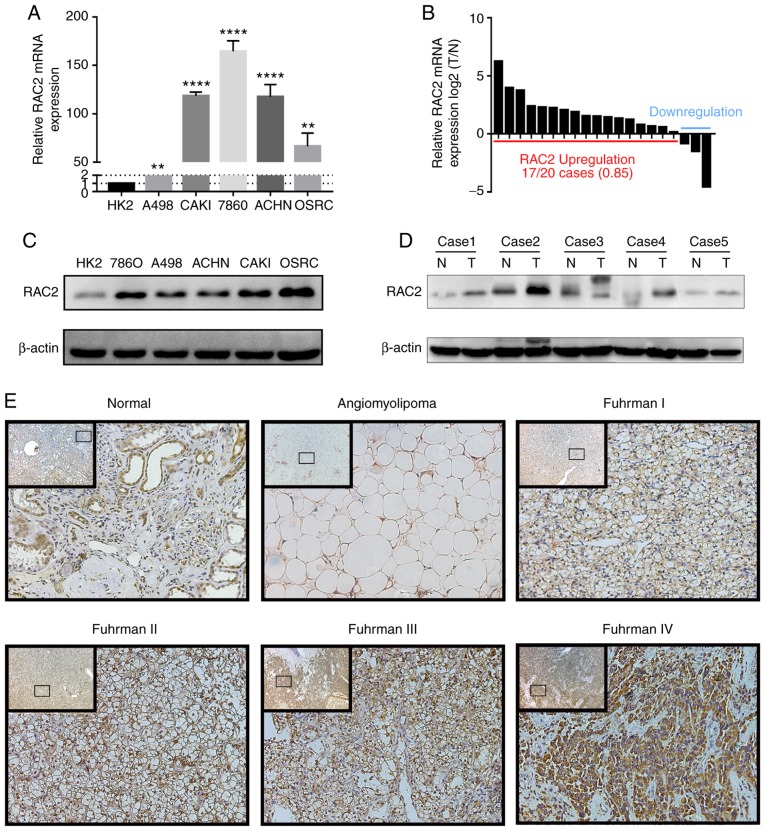
In order to verify the expression levels of RAC2 in RCC cells, RT-qPCR and western blot assays were performed. Compared with that of HK-2, whether at the mRNA or protein levels, RAC2 expression levels in RCC cell lines, including 786-O, OSRC-2, CAKI-1, A498 and ACHN, were upregulated (Fig. 5A-C). As for the ccRCC tissues, similarly, total RNA and protein were isolated and it was observed that RAC2 expression levels were markedly higher in tumor tissues when compared with the adjacent normal tissues (Fig. 5D). Furthermore, IHC was performed in ccRCC tissues, adjacent normal tissues and benign renal tumor tissues. RAC2 was primarily located in the cytoplasm and membranes of cancer cells and renal tubular epithelial cells. These results demonstrated that, with the increase of Fuhrman grade, the expression levels of RAC2 appeared to increase (Fig. 5E). The IHC data of RAC2 in ccRCC was also collected from The Human Protein Atlas (Fig. S1E).
Figure 5.
RAC2 was up-regulated in RCC cells and tissues. (A and B) Reverse transcription-quantitative PCR assays of RAC2 mRNA expression in normal renal tubular epithelial cells (HK-2) and renal cancer cell lines (A498, ACHN, CAKI-1, OSRC-2, 786-O), and in 50 paired tissue samples of patients with ccRCC. (C and D) Western blot assays of RAC2 expression in normal renal tubular epithelial cells (HK-2) and in renal cancer cell lines (A498, ACHN, CAKI-1, OSRC-2, 786-O) and in 50 paired tissue samples of ccRCC patients. (E) Immunohistochemistry for RAC2 expression in ccRCC tissues, adjacent normal tissues and renal angiomyolipoma tissues. The inset images are the lower magnification of the same tissue as that presented in the larger image of each set. Magnification, ×40 and ×200. RAC2 expression was normalized to β-actin expression. The values of each group are presented as the mean ± standard deviation. Bars represented the means of three independent experiments. ****P<0.0001; **P<0.01 vs. HK-2. RAC2, Rac family small GTPase 2; ccRCC, clear cell renal cell carcinoma.
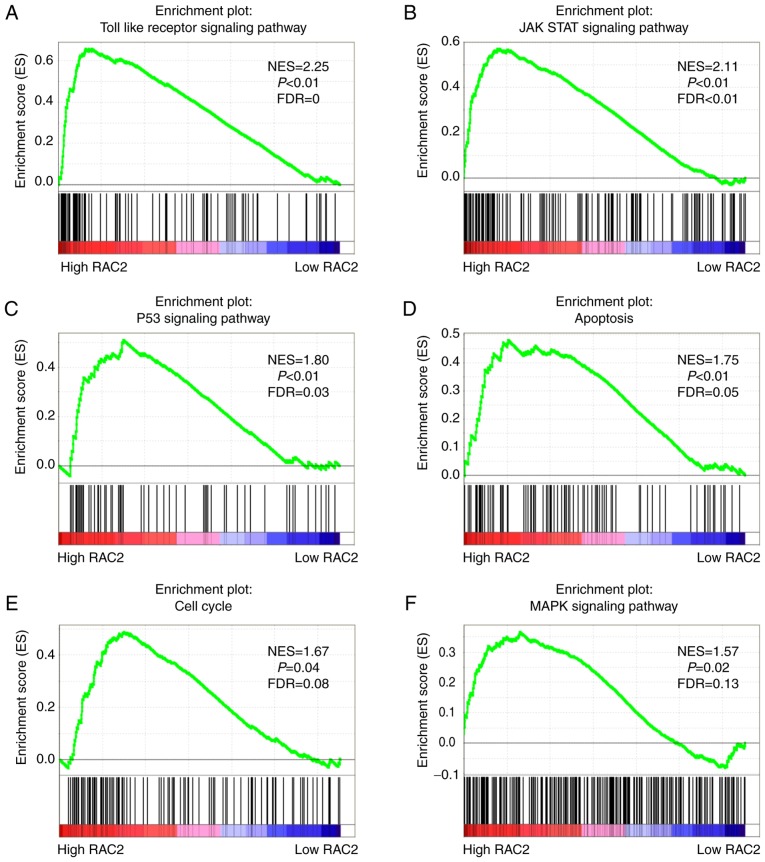
RAC2 is involved in biological pathways associated with the pathogenesis ccRCC
In order to determine how RAC2 is involved in ccRCC pathogenesis, GSEA was performed using TCGA database with the aim of obtaining more information regarding the biological pathways associated with this disease. As demonstrated by the results of the present study, activated gene sets associated with the Toll-like receptor signaling pathway normalized enrichment score (NES), 2.25; P<0.01; FDR=0; Fig. 6A), Janus kinase/signal transducer and activator of transcription (JAK/STAT) signaling pathway (NES, 2.11; P<0.01; FDR<0.01; Fig. 6B), p53 signaling pathway (NES, 1.80; P<0.01; FDR=0.03; Fig. 6C), apoptosis (NES, 1.75; P<0.01; FDR=0.05; Fig. 6D), cell cycle (NES, 1.67; P=0.04; FDR=0.08; Fig. 6E) and the MAPK signaling pathway (NES, 1.57; P=0.02; FDR=0.13; Fig. 6F) were more relevant to patients with higher RAC2 expression than those with lower expression.
Figure 6.
RAC2 regulates certain tumor-related pathways. Enrichment curves are shown for activated gene sets related to the (A) Toll like receptor, (B) JAK STAT and (C) P53 signaling pathways, (D) apoptosis, and the (E) cell cycle and (F) MAPK signal pathways. RAC2, Rac family small GTPase 2; NES, normalized enrichment score; FDR, false discovery rate; JAK, Janus kinases; STAT, signal transducer and activator of transcription proteins; MAPK, mitogen-activated protein kinase.
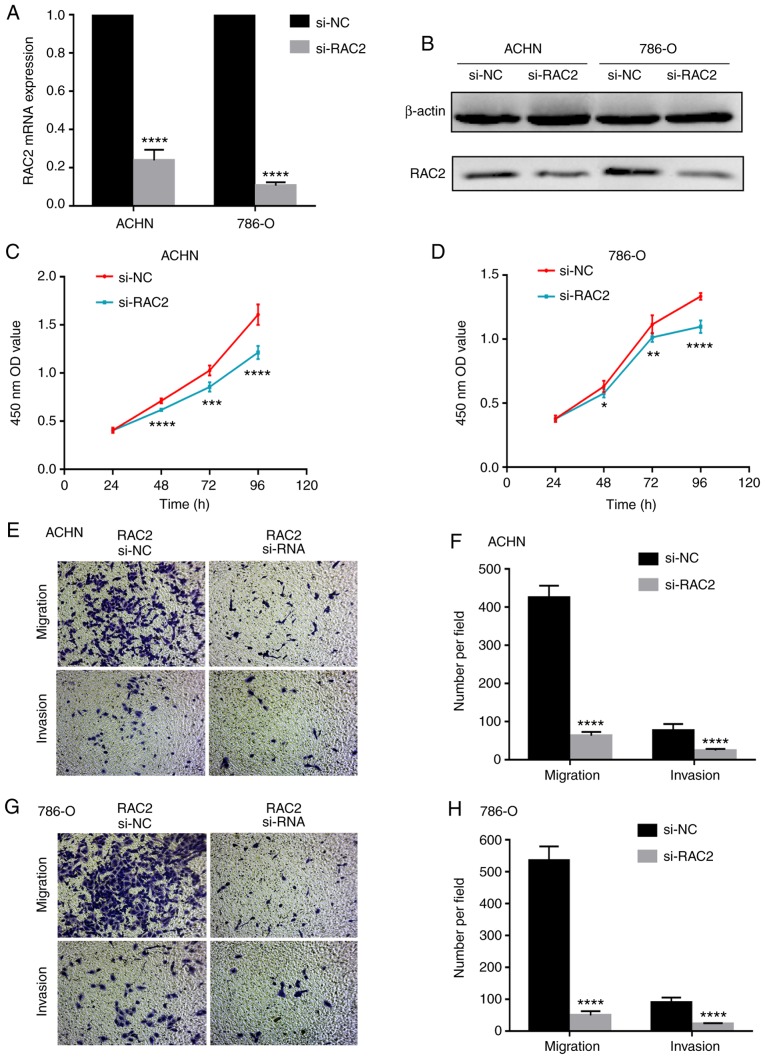
RAC2 facilitates the proliferation, invasion and migration of RCC cells in vitro
In order to investigate the function of RAC2 on the biological behaviors of renal cancer, RCC cell lines were transfected with si-RAC2 to downregulate the expression of RAC2. Significantly decreased RAC2 mRNA and protein expression levels were detected in ACHN cells and 786-O cells compared with the corresponding control (Fig. 7A and B). Following transfection, cell proliferation assays, which were performed with the prior-transfected ACHN and 786-O cells, demonstrated that cell proliferation was significantly inhibited with RAC2 knockdown compared with the corresponding control (Fig. 7C and D). In addition, cell migration and invasion assays were conducted, which verified that downregulation of RAC2 significantly attenuated the cell migration and invasive abilities of the cells (Fig. 7E-H). Furthermore, we compared cell proliferative, migration and invasive abilities between untransfected 786-O cells and transfected si-NC 786-O cells. The results (Fig. S2A-D) revealed no significant cytotoxicity of the siRNA delivery method. These results indicated that RAC2 may contribute to the proliferation, migration and invasion of RCC cells.
Figure 7.
RAC2 promotes the proliferation, invasion and migration of RCC cell lines in vitro. (A and B) Reverse transcription-qauntitative PCR and western blotting assays of RAC2 knockdown in ACHN and 786-O cells; β-actin was used as a loading control. (C and D) Cell counting kit-8 assays detected the effects of RAC2 knockdown on the proliferation of ACHN and 786-O cells. (E-H) Representative images of migration and invasion assays performed using ACHN and 786-O cells (magnification, ×100). Data are presented as the mean ± standard deviation from three independent experiments. ****P<0.0001; ***P<0.001; **P<0.01 and *P<0.05 vs. si-NC. RAC2, Rac family small GTPase 2; ccRCC, clear cell renal cell carcinoma; NC, negative control; si-RNA, small interfering RNA; OD value, optical density value.
Discussion
ccRCC is an advanced metastatic tumor with high mortality. Currently, <6-10% of patients with ccRCC have characteristic symptoms, such as lower backache, hematuria and masses. This can result in patients being diagnosed at a more advanced stage of disease, whereby metastases have already developed (31,32). Partial or total nephrectomy is currently the main treatment method for ccRCC; however, ~35% of patients with clinical T1 stage ccRCC still experience recurrence or metastasis following surgery (33). The therapeutic effects of other treatment methods, such as radiotherapy, chemotherapy or targeted drug therapy remain unsatisfactory for those with advanced stage renal cancer. Although immunotherapy is considered to be very promising for treating RCC currently, anti-cytotoxic T-lymphocyte protein 4 drugs and anti-PD-L1 (programmed cell death 1 ligand 1) drugs are still being evaluated in clinical trials (34,35). It is critical to further analyze potential tumor-associated proteins and develop personalized precision-based care for ccRCC (36). According to recent studies, perilipin (PLIN)2, PLIN3, regulator of calcinerurin 1 and certain other biomarkers may be useful in the diagnosis and prognosis of renal cancer (5,23,37). In addition, the results of the present study provide evidence to suggest that RAC2 may also be a potential biomarker.
The Rho GTPases family consists of Rac, Rho, Cdc42 subfamilies and other less studied GTPases (38). They serves important roles in a diverse range of fundamental cellular functions through the regulation of actin contraction and peripheral actin structures, including cell morphology, locomotion and polarity (39). As a result, certain physiological process, such as cell growth, cell division and cell survival, are associated with the Rho GTPases. Recently, an increasing number of studies have suggested that the Rho GTPases, particularly Rac GTPases, are primarily involved in the signaling networks of malignant cell transformation and metastasis, and thus, may be potential therapeutic targets in cancer (40,41). These findings are consistent with the GSEA of the present study regarding RAC2. The Toll-like receptor signaling pathway, JAK/STAT signaling pathway, p53 signaling pathway, cell cycle, apoptosis pathway and MAPK signaling pathway are all classical pathways that are involved in cell proliferation, cell migration, cell death, tumorigenesis or drug resistance (http://software.broadinsti-tute.org/gsea/index.jsp). These findings suggested that RAC2, as a member of the Rac subfamily, may be a critical element in oncogenesis and tumor development.
Previous studies have suggested that Rac proteins were upregulated in various different types of tumor and could serve as a prognostic biomarker, including in prostate (42), testicular (43), ovarian (44), lung (45) and gastric cancer (46). RAC1 was reported to accumulate in the nucleolus and participate in the synthesis of ribosomal RNA in lung cancer cells (47,48). Furthermore, particular attention has been paid to RAC1-targeting drugs. Rac1 inhibitors, such as NSC23766, which specifically disrupts RAC1 interaction with Tiam1 and Trio (39,40,49), have been developed. However, few studies have focused on Rac proteins and RCC. In the present study, the heatmap depicting RAC1, RAC2, RAC3 and RoG expression levels in normal tissues and ccRCC tissues suggested that the differences in expression existed most frequently in RAC2 instead of RAC1; current understanding of the specific function of RAC2 remains limited. Several studies have suggested that RAC2 is associated with myeloid cell dysfunction and myeloid leukemia (50-52).
In the present study, the clinical data downloaded from TCGA KIRC database and Oncomine database were assessed, and it was revealed that RAC2 was upregulated in ccRCC tissues compared with in normal tissues. Analysis of clinical characteristics demonstrated that higher expression levels of RAC2 were associated with higher clinical and pathological grade and poorer OS in patients with ccRCC. However, with the ROC analyses, it was hypothesized that RAC2 is a diagnostic index for patients with different clinicopathological parameters. The differential expression levels of RAC2 were verified in 50 pairs of ccRCC tissues and matched normal tissues collected in our hospital, as well as in RCC cell lines. In order to validate the hypothesis that RAC2 promoted renal cancer progression, GSEA analyses and functional experiments were performed in the present study. The results demonstrated that RAC2 participated in a number of tumor-associated pathways, and that downregulated RAC2 suppressed the proliferative, migration and invasive abilities of RCC cells.
To the best of our knowledge, the present study is the first to report that RAC2 may be a potential diagnostic and prognostic biomarker that promotes tumor progression in ccRCC. The results from the present study also indicated that downregulated expression of RAC2 attenuated the proliferation, migration and invasion of RCC cells, providing further evidence that RAC2 may serve as a therapeutic target for ccRCC. However, there are limitations to our study. First, the association between RAC2 expression and the ccRCC biological behaviors was not confirmed in vivo. Furthermore, the underlying molecular mechanism by which RAC2 facilitates renal cell carcinoma was not investigated thoroughly. Hence, further research is required in order to overcome these issues.
In conclusion, the results from the present study demonstrated that high expression levels of RAC2 were associated with poor overall survival and higher tumor grade in patients with ccRCC, which, to the best of our knowledge, is the first time this has been reported. In addition, low expression levels of RAC2 may decrease the proliferation, migration and invasion ability of RCC cells in vitro. The aforementioned results indicated that RAC2 may be a promising prognostic and diagnostic biomarker for renal cancer, and thus, be considered as a potential therapeutic target for treating ccRCC.
Supplementary Materials
Acknowledgments
Not applicable.
Funding
This study was supported by the National Natural Science Foundation of China (grant nos. 81672524, 81672528 and 81874090), the National Key R&D Program of China (grant no. 017YFB1303100), the Hubei Provincial Natural Science Foundation of China (grant no. 2018CFA038), the Independent Innovation Foundation of Huazhong University of Science and Technology (grant no. 118530309), the Clinical Research Physician Program of Tongji Medical College, Huazhong University of Science and Technology (grant no. 5001530015) and the Integrated Innovation Team for Major Human Disease Program of Tongji Medical College, Huazhong University of Science and Technology.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XZ and GC designed the study. YL, ZS, TX and HR carried out data acquisition and analysis. YL, GC, ZS and DL performed the majority of the experiments. YL and KC wrote the manuscript. LZ conducted immunohistochemistry analyses. QC and KW collected the clinical samples and managed the clinical data. JL and LB contributed to bioinformatics analysis. HY and KC were involved in project management, and contributed to preparing and making figures and tables. HY and XZ supervised the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study and experimental procedures were approved by the Human Research Ethics Committee of Huazhong University of Science and Technology (Wuhan, China). Written informed consent was obtained from the patients/patients' families. The study was conducted according to the principles outlined in the Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Moch H, Cubilla AL, Humphrey PA, Reuter VE, Ulbright TM. The 2016 WHO classification of tumours of the urinary system and male genital organs-Part A: Renal, penile, and testicular tumours. Eur Urol. 2016;70:93–105. doi: 10.1016/j.eururo.2016.02.029. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Rini BI, Atkins MB. Resistance to targeted therapy in renal-cell carcinoma. Lancet Oncol. 2009;10:992–1000. doi: 10.1016/S1470-2045(09)70240-2. [DOI] [PubMed] [Google Scholar]
- 4.Gong J, Maia MC, Dizman N, Govindarajan A, Pal SK. Metastasis in renal cell carcinoma: Biology and implications for therapy. Asian J Urol. 2016;3:286–292. doi: 10.1016/j.ajur.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song Z, Cao Q, Ruan H, Yang H, Wang K, Bao L, Cheng G, Xu T, Xiao H, Wang C, et al. RCAN1.4 acts as a suppressor of cancer progression and sunitinib resistance in clear cell renal cell carcinoma. Exp Cell Res. 2018;372:118–128. doi: 10.1016/j.yexcr.2018.09.017. [DOI] [PubMed] [Google Scholar]
- 6.Pichler M, Hutterer GC, Chromecki TF, Jesche J, Kampel-Kettner K, Rehak P, Pummer K, Zigeuner R. External validation of the leibovich prognosis score for nonmetastatic clear cell renal cell carcinoma at a single European center applying routine pathology. J Urology. 2011;186:1773–1777. doi: 10.1016/j.juro.2011.07.034. [DOI] [PubMed] [Google Scholar]
- 7.Novara G, Ficarra V, Antonelli A, Artibani W, Bertini R, Carini M, Cosciani Cunico S, Imbimbo C, Longo N, Martignoni G, et al. Validation of the 2009 TNM version in a large multi-institutional cohort of patients treated for renal cell carcinoma: Are further improvements needed? Eur Urol. 2010;58:588–595. doi: 10.1016/j.eururo.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 8.Bergers G, Hanahan D. Modes of resistance to anti-angio-genic therapy. Nat Rev Cancer. 2008;8:592–603. doi: 10.1038/nrc2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lai YJ, Tsai JC, Tseng YT, Wu MS, Liu WS, Lam HI, Yu JH, Nozell SE, Benveniste EN. Small G protein Rac GTPases regulate the maintenance of glioblastoma stem-like cells in vitro and in vivo. Oncotarget. 2017;8:1803–18049. doi: 10.18632/oncotarget.14949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bustelo XR, Sauzeau V, Berenjeno IM. GTP-binding proteins of the Rho/Rac family: Regulation, effectors and functions in vivo. Bioessays. 2007;29:356–370. doi: 10.1002/bies.20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancelas JA. On how Rac controls hematopoietic stem cell activity. Transfusion. 2011;51(Suppl 4):153S–159S. doi: 10.1111/j.1537-2995.2011.03378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aspenstrom P, Fransson A, Saras J. Rho GTPases have diverse effects on the organization of the actin filament system. Biochem J. 2004;377:327–337. doi: 10.1042/bj20031041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 14.Chen X, Zhang JX, Luo JH, Wu S, Yuan GJ, Ma NF, Feng Y, Cai MY, Chen RX, Lu J, et al. CSTF2-induced shortening of the RAC1 3'UTR promotes the pathogenesis of urothelial carcinoma of the bladder. Cancer Res. 2018;78:5848–5862. doi: 10.1158/0008-5472.CAN-18-0822. [DOI] [PubMed] [Google Scholar]
- 15.Pei H, Guo Z, Wang Z, Dai Y, Zheng L, Zhu L, Zhang J, Hu W, Nie J, Mao W, et al. RAC2 promotes abnormal proliferation of quiescent cells by enhanced JUNB expression via the MAL-SRF pathway. Cell Cycle. 2018;17:1115–1123. doi: 10.1080/15384101.2018.1480217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mack NA, Whalley HJ, Castillo-Lluva S, Malliri A. The diverse roles of Rac signaling in tumorigenesis. Cell Cycle. 2011;10:1571–1581. doi: 10.4161/cc.10.10.15612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.NCBI; 2019. RAC2 Rac family small GTPase 2 [Homo sapiens (human)] (Gene ID: 5880). https://www.ncbi.nlm.nih.gov/gene?Db=gene&Cmd=DetailsSearch&Term=5880 Accessed July 7 , 2019. [Google Scholar]
- 18.Filippi MD, Harris CE, Meller J, Gu Y, Zheng Y, Williams DA. Localization of Rac2 via the C terminus and aspartic acid 150 specifies superoxide generation, actin polarity and chemotaxis in neutrophils. Nat Immunol. 2004;5:744–751. doi: 10.1038/ni1081. [DOI] [PubMed] [Google Scholar]
- 19.Croker BA, Handman E, Hayball JD, Baldwin TM, Voigt V, Cluse LA, Yang FC, Williams DA, Roberts AW. Rac2-deficient mice display perturbed T-cell distribution and chemotaxis, but only minor abnormalities in T(H)1 responses. Immunol Cell Biol. 2002;80:231–240. doi: 10.1046/j.1440-1711.2002.01077.x. [DOI] [PubMed] [Google Scholar]
- 20.Yang FC, Kapur R, King AJ, Tao W, Kim C, Borneo J, Breese R, Marshall M, Dinauer MC, Williams DA. Rac2 stimulates Akt activation affecting BAD/Bcl-XL expression while mediating survival and actin function in primary mast cells. Immunity. 2000;12:557–568. doi: 10.1016/S1074-7613(00)80207-1. [DOI] [PubMed] [Google Scholar]
- 21.Fuhrman SA, Lasky LC, Limas C. Prognostic significance of morphologic parameters in renal cell carcinoma. Am J Surg Pathol. 1982;6:655–663. doi: 10.1097/00000478-198210000-00007. [DOI] [PubMed] [Google Scholar]
- 22.Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A, editors. AJCC cancer staging manual. 7th edition. Springer Verlag; New York, NY: 2009. pp. 547–560. [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Cao Q, Ruan H, Wang K, Song Z, Bao L, Xu T, Xiao H, Wang C, Cheng G, Tong J, et al. Overexpression of PLIN2 is a prognostic marker and attenuates tumor progression in clear cell renal cell carcinoma. Int J Oncol. 2018;53:137–147. doi: 10.3892/ijo.2018.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu R, Qin X, Ji C, Zeng W, Yang Y, Tan W. Pygopus 2 promotes kidney cancer OS-RC-2 cells proliferation and invasion in vitro and in vivo. Asian J Urol. 2015;2:151–157. doi: 10.1016/j.ajur.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yusenko MV, Zubakov D, Kovacs G. Gene expression profiling of chromophobe renal cell carcinomas and renal oncocytomas by Affymetrix GeneChip using pooled and individual tumours. Int J Biol Sci. 2009;5:517–527. doi: 10.7150/ijbs.5.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones J, Otu H, Spentzos D, Kolia S, Inan M, Beecken WD, Fellbaum C, Gu X, Joseph M, Pantuck AJ, et al. Gene signatures of progression and metastasis in renal cell cancer. Clin Cancer Res. 2005;11:5730–5739. doi: 10.1158/1078-0432.CCR-04-2225. [DOI] [PubMed] [Google Scholar]
- 28.Lenburg ME, Liou LS, Gerry NP, Frampton GM, Cohen HT, Christman MF. Previously unidentified changes in renal cell carcinoma gene expression identified by parametric analysis of microarray data. BMC Cancer. 2003;3:31. doi: 10.1186/1471-2407-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gumz ML, Zou H, Kreinest PA, Childs AC, Belmonte LS, LeGrand SN, Wu KJ, Luxon BA, Sinha M, Parker AS, et al. Secreted frizzled-related protein 1 loss contributes to tumor phenotype of clear cell renal cell carcinoma. Clin Cancer Res. 2007;13:4740–4749. doi: 10.1158/1078-0432.CCR-07-0143. [DOI] [PubMed] [Google Scholar]
- 30.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patard JJ, Leray E, Rodriguez A, Rioux-Leclercq N, Guillé F, Lobel B. Correlation between symptom graduation, tumor characteristics and survival in renal cell carcinoma. Eur Urol. 2003;44:226–232. doi: 10.1016/S0302-2838(03)00216-1. [DOI] [PubMed] [Google Scholar]
- 32.Lee CT, Katz J, Fearn PA, Russo P. Mode of presentation of renal cell carcinoma provides prognostic information. Urol Oncol. 2002;7:135–140. doi: 10.1016/S1078-1439(01)00185-5. [DOI] [PubMed] [Google Scholar]
- 33.Campbell SC, Novick AC, Belldegrun A, Blute ML, Chow GK, Derweesh IH, Faraday MM, Kaouk JH, Leveillee RJ, Matin SF, et al. Guideline for management of the clinical T1 renal mass. J Urol. 2009;182:1271–1279. doi: 10.1016/j.juro.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 34.Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, Tykodi SS, Sosman JA, Procopio G, Plimack ER, et al. Nivolumab versus Everolimus in advanced renal-cell carcinoma. New Engl J Med. 2015;373:1803–1813. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dempke WCM, Fenchel K, Uciechowski P, Dale SP. Second- and third-generation drugs for immuno-oncology treatment-the more the better? Eur J Cancer. 2017;74:55–72. doi: 10.1016/j.ejca.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 36.Li QK, Pavlovich CP, Zhang H, Kinsinger CR, Chan DW. Challenges and opportunities in the proteomic characterization of clear cell renal cell carcinoma (ccRCC): A critical step towards the personalized care of renal cancers. Semin Cancer Biol. 2019;55:8–15. doi: 10.1016/j.semcancer.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang K, Ruan H, Song Z, Cao Q, Bao L, Liu D, Xu T, Xiao H, Wang C, Cheng G, et al. PLIN3 is up-regulated and correlates with poor prognosis in clear cell renal cell carcinoma. Urol Oncol. 2018;36:343.e9–343.e19. doi: 10.1016/j.urolonc.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 38.Olson MF. Rho GTPases, their post-translational modifications, disease-associated mutations and pharmacological inhibitors. Small GTPases. 2018;9:203–215. doi: 10.1080/21541248.2016.1218407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kazanietz MG, Caloca MJ. The Rac GTPase in cancer: From old concepts to new paradigms. Cancer Res. 2017;77:5445–5451. doi: 10.1158/0008-5472.CAN-17-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cardama GA, Gonzalez N, Maggio J, Menna PL, Gomez DE. Rho GTPases as therapeutic targets in cancer (Review) Int J Oncol. 2017;51:1025–1034. doi: 10.3892/ijo.2017.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Casado-Medrano V, Baker MJ, Lopez-Haber C, Cooke M, Wang S, Caloca MJ, Kazanietz MG. The role of Rac in tumor susceptibility and disease progression: From biochemistry to the clinic. Biochem Soc Trans. 2018;46:1003–1012. doi: 10.1042/BST20170519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kobayashi T, Inoue T, Shimizu Y, Terada N, Maeno A, Kajita Y, Yamasaki T, Kamba T, Toda Y, Mikami Y, et al. Activation of Rac1 is closely related to androgen-independent cell proliferation of prostate cancer cells both in vitro and in vivo. Mol Endocrinol. 2010;24:722–734. doi: 10.1210/me.2009-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamai T, Yamanishi T, Shirataki H, Takagi K, Asami H, Ito Y, Yoshida K. Overexpression of RhoA, Rac1, and Cdc42 GTPases is associated with progression in testicular cancer. Clin Cancer Res. 2004;10:4799–4805. doi: 10.1158/1078-0432.CCR-0436-03. [DOI] [PubMed] [Google Scholar]
- 44.Leng R, Liao G, Wang H, Kuang J, Tang L. Rac1 expression in epithelial ovarian cancer: Effect on cell EMT and clinical outcome. Med Oncol. 2015;32:329. doi: 10.1007/s12032-014-0329-5. [DOI] [PubMed] [Google Scholar]
- 45.Zhou Y, Liao Q, Han Y, Chen J, Liu Z, Ling H, Zhang J, Yang W, Oyang L, Xia L, et al. Rac1 overexpression is correlated with epithelial mesenchymal transition and predicts poor prognosis in non-small cell lung cancer. J Cancer. 2016;7:2100–2109. doi: 10.7150/jca.16198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ji J, Feng X, Shi M, Cai Q, Yu Y, Zhu Z, Zhang J. Rac1 is correlated with aggressiveness and a potential therapeutic target for gastric cancer. Int J Oncol. 2015;46:1343–1353. doi: 10.3892/ijo.2015.2836. [DOI] [PubMed] [Google Scholar]
- 47.Justilien V, Ali SA, Jamieson L, Yin N, Cox AD, Der CJ, Murray NR, Fields AP. Ect2-Dependent rRNA synthesis is required for KRAS-TRP53-driven lung adenocarcinoma. Cancer Cell. 2017;31:256–269. doi: 10.1016/j.ccell.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baker MJ, Cooke M, Kazanietz MG. Nuclear PKC iota-ECT2-Rac1 and ribosome biogenesis: A novel axis in lung tumorigenesis. Cancer Cell. 2017;31:167–169. doi: 10.1016/j.ccell.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 49.Gao Y, Dickerson JB, Guo F, Zheng J, Zheng Y. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc Natl Acad Sci USA. 2004;101:7618–7623. doi: 10.1073/pnas.0307512101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Troeger A, Williams DA. Hematopoietic-specific Rho GTPases Rac2 and RhoH and human blood disorders. Exp Cell Res. 2013;319:2375–2383. doi: 10.1016/j.yexcr.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mizukawa B, Wei J, Shrestha M, Wunderlich M, Chou FS, Griesinger A, Harris CE, Kumar AR, Zheng Y, Williams DA, Mulloy JC. Inhibition of Rac GTPase signaling and downstream prosurvival Bcl-2 proteins as combination targeted therapy in MLL-AF9 leukemia. Blood. 2011;118:5235–5245. doi: 10.1182/blood-2011-04-351817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gu Y, Williams DA. RAC2 GTPase deficiency and myeloid cell dysfunction in human and mouse. J Pediat Hematol Oncol. 2002;24:791–794. doi: 10.1097/00043426-200212000-00027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.