ABSTRACT
The identification of effective biomarkers for early diagnosis, prognosis, and response to treatments remains a challenge in ovarian cancer (OC) research. Here, we present an unbiased high-throughput approach to profile ascitic fluid autoantibodies in order to obtain a tumor-specific antigen signature in OC.
We first reported the reactivity of immunoglobulins (Igs) purified from OC patient ascites towards two different OC cell lines. Using a discovery set of Igs, we selected tumor-specific antigens from a phage display cDNA library. After biopanning, 700 proteins were expressed as fusion protein and used in protein array to enable large-scale immunoscreening with independent sets of cancer and noncancerous control. Finally, the selected antigens were validated by ELISA.
The initial screening identified eight antigenic clones: CREB3, MRPL46, EXOSC10, BCOR, HMGN2, HIP1R, OLFM4, and KIAA1755. These antigens were all validated by ELISA in a study involving ascitic Igs from 153 patients (69 with OC, 34 with other cancers and 50 without cancer), with CREB3 showing the highest sensitivity (86.95%) and specificity (98%). Notably, we were able to identify an association between the tumor-associated (TA) antibody response and the response to a first-line tumor treatment (platinum-based chemotherapy). A stronger association was found by combining three antigens (BCOR, CREB3, and MRLP46) as a single antibody signature.
Measurement of an ascitic fluid antibody response to multiple TA antigens may aid in the identification of new prognostic signatures in OC patients and shift attention to new potentially relevant targets.
KEYWORDS: Ovarian cancer, biomarker, tumor-associated antigen, ascite, protein microarray
Introduction
Ovarian cancer (OC) is the primary cause of death among gynecologic cancers mainly because of diagnosis at late stage and development of chemoresistance.1 Most OC patients present with advanced stage disease due to a lack of specific symptoms, adequate diagnostic tools, and screening strategies.2 Standard treatment for advanced stage OC is tumor-debulking surgery followed by adjuvant platinum-based chemotherapy. Despite a good response rate to this front-line treatment, the majority of these patients eventually develop incurable platinum-resistant disease with a 5-y survival rate below 40%.3 Several new drugs have now become available for patients with OC; however, it is difficult to determine the effectiveness of a given treatment due to a lack of efficient biomarkers for patient selection, prognosis, and overall outcome.4 Cancer antigen-125 (CA-125) is the most extensively investigated OC biomarker,5 but it has a limited ability to detect the disease at an early stage and is mainly used for monitoring patients during follow-up procedures.6 A number of other biomarkers present in serum, plasma, and urine (such as HE4, mesothelin, prostasin, kallikreins, and osteopontin) have been identified.7 Although none of these markers meet the clinical standards when considered individually, the combination of multiple markers as a panel along with CA-125 has been demonstrated to improve diagnostic performances.8 Finding additional biomarkers is therefore needed not only for diagnosis but also for prognosis and response to treatment.9 Cancer cells induce a specific immune response to aberrantly expressed, mutated or posttranslationally modified proteins or other tumor-associated (TA) autoantigens and may be enhanced by TA inflammation or loss of immune self-tolerance, resulting in the production of TA autoantibodies.10,11 TA autoantibodies and TA antigens (TAAs) have emerged as clinically useful tools with both diagnostic and prognostic relevance.12,13 Measurement of TA autoantibodies by means of their corresponding antigens may be of clinical benefit in the early diagnosis of various solid tumors, including OC,14,15 breast cancer,16 lung cancer,17 hepatocellular carcinoma,18 gastric cancer,19 colorectal cancer,20 and prostate cancer.21 The mechanisms that lead to the production of antibodies as well as the exact pathological role of the different types of antibodies are largely undefined. The heterogeneous nature of TA antibodies underscore the importance of an innovative and unbiased approach to fully explore the complex information present in the antibody profile of cancer patients. By applying recent approaches in the field of biomarker discovery, it has been possible to perform so-called “seromic profiling”,22 which allows the identification of TAAs recognized by patients’ antibodies that are directed against aberrantly expressed, mutated or posttranslationally modified proteins. Using peptide/protein arrays,23 two-dimensional gel electrophoresis,24 and phage display technology,25 dozens of proteins have been identified as targets of the patient’s immune response in different tumors. Body fluids have been shown to be an excellent medium for biomarker discovery. Accumulation of ascitic fluid in the peritoneal cavity is an important feature of OC, and its accessibility over the course of treatment makes it an excellent alternative source for biomarker discovery and translational research.26,27 Malignant ascites typically comprise variable proportions of suspended cells that include tumor cells, mesothelial cells, fibroblasts, macrophages, white blood cells, red blood cells, and debris. In addition to the suspended cells, proteins secreted or leaked from tumor tissue, soluble growth factors that are associated with invasion and metastasis, factors of the complement system, chemokines and, above all, antibodies are contained with ascitic fluid. Ascitic fluids have received very little attention yet are an invaluable resource for prognostic and predictive biomarker identification, pharmacodynamic information, and molecular profiling of OC. Although some proteomics studies have been performed describing the thousands of proteins present within the fluid, only a few of the studies showed limited correlation of the proteins with clinical features of the disease.28–31 In this study, we present a systematic and in-depth profile of the antibody signature in OC ascites. We first showed that OC ascites contain antibodies targeting proteins expressed in OC cells. Then, using a high-throughput protein expression and screening platform that combines phage display and protein microarray, we assessed the ascitic fluid antibody response and identified eight new TA autoantibody-binding antigens. This tumor autoantibody signature is associated and correlated with response to chemotherapy.
Results
Project strategy
To identify the antigenic targets of antibodies detectable in OC ascites, we applied the research pipeline described in Figure 1. Initially, a total of 153 ascitic samples from OC patients and control subjects (without OC) were obtained, and immunoglobulins (Igs) were affinity purified. Validation of the hypothesis that OC ascites contain tumor antigen-targeting antibodies was verified by Western blot, immunofluorescence and ELISA performed with a smaller subset of the initial sample group ((i) 28 OC ascitic fluids collected from patients with stage I-VI diseases who were either platinum sensitive or platinum resistant (grade ranging from G1-G3), (ii) 18 ascitic fluids collected from patients with other types of cancer and (iii) 28 ascitic fluids from individuals without cancer). This step allows the identification of a “discovery set” of Igs that are used as bait for a phage display cDNA library on the basis of their reactivity on the OC cell lines. Specific biomarkers are then identified by screening protein microarrays, and the selected antigens are produced and purified as recombinant proteins before evaluation with ELISA on the complete sample set of ascite Igs to validate the protein antigenicity. Correlations between the ELISA signal, gene expression profile and clinical data were simultaneously explored.
Figure 1.
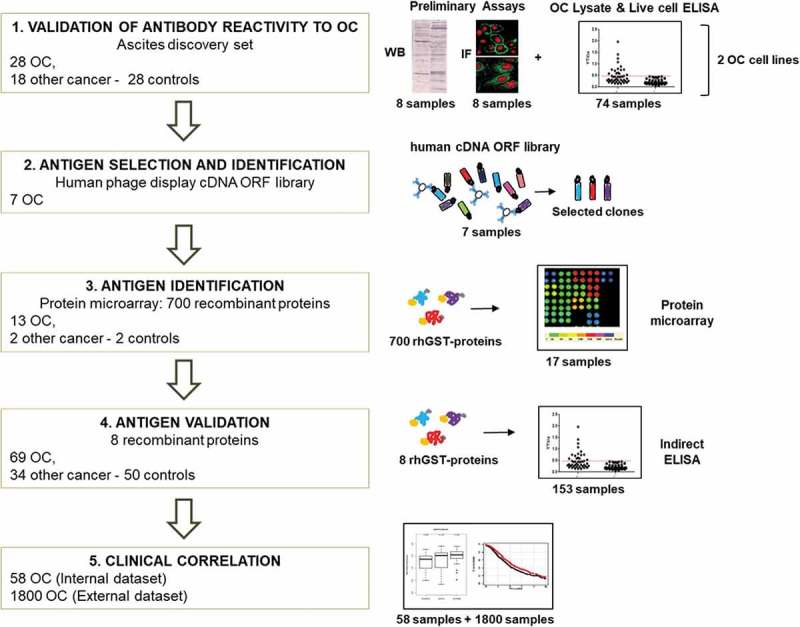
Workflow of the project.
Scheme of the project workflow and the main techniques used: 1) reactivity of antibodies from patients’ ascitic fluids against OC antigens was initially tested using a small group of samples, on two OC cell lines, by Western blot, ELISA, and immunofluorescence. 2) Antigens were identified by performing rounds of selection of an Open Reading Frame (ORF) fragments library displayed on the filamentous phage. Selection was performed with Igs from seven OC ascites. 3) Reactivity against selected putative antigens was further verified by protein microarray on 13 OC, two noncancerous, and two control samples. 4) Antigenicity validation was done by ELISA using the whole cohort of 153 ascites samples. 5) Clinical correlation of novel putative antigens to antibody titer and gene expression was performed, both on internal and external datasets.
Samples and controls used
In this study, 153 different ascitic fluids were collected from patients diagnosed with OC (N = 69), other cancers (N = 34) and noncancerous conditions (N = 50). The patients’ clinicopathological characteristics are summarized in Table 1. Igs from all samples were affinity purified with protein A agarose beads. Samples were checked by SDS-PAGE before and after purification, and the concentration of purified Igs was determined (Supplementary Figure 1). The median ascites Ig levels in samples from patients with OC, other types of cancer and noncancerous conditions were 2.97 µg/µl, 2.79 µg/µl and 2.86 µg/µl, respectively, which is in agreement with previous data.32
Table 1.
Clinicopathological characteristics of patients included in the study.
| Patient disease | No. | % | |
|---|---|---|---|
| Chemo-naïve ovarian cancer (69) | Age, y | ||
| Mean, median | 57, 60 | ||
| Range | 32–85 | ||
| Histology | |||
| Serous | 47 | 68 | |
| Nonserous | 22 | 32 | |
| Stage (FIGO) | |||
| II | 2 | 3 | |
| III | 45 | 65 | |
| IV | 21 | 31 | |
| Missing Information | 1 | 1 | |
| Grade | |||
| 1, Well differentiated | 1 | 1 | |
| 2, Moderately differentiated | 8 | 12 | |
| 3, Poorly differentiated | 60 | 87 | |
| Amount of residual disease | |||
| NED | 15 | 22 | |
| <1 cm, mRD | 11 | 16 | |
| >1 cm, GRD | 41 | 59 | |
| Missing information | 2 | 3 | |
| Response to Platinum treatment | |||
| Sensitive | 24 | 35 | |
| Partially sensitive | 16 | 23 | |
| Resistant | 28* | 41 | |
| Missing information | 1 | 1 | |
| Other cancers (34)# | Cancer of the lung, liver, colon, breast, gallbladder, pancreas and stomach rectum; lymphoma; mesothelioma | ||
| Noncancerous conditions (50) | Cirrhosis caused by various pathologies, pulmonary fibrosis, cholestatic liver diseases, pneumonia, side effects from transplantations. | ||
FIGO = International Federation of Gynecology and Obstetrics; NED = not evident disease; mRD = minimal residual disease; GRD = gross residual disease. * Includes four refractory patients. # These controls include ascites recovered from nonovarian tumors but in some cases presenting peritoneal diffusion similar to that observed in OC.
Ascites from OC contain antibodies targeting tumor cell antigens
To detect the presence of antibodies directed against antigens associated with OC, reactivity of purified Ig samples from ascitic fluids was tested on two different OC cell lines (OVCAR-3 and SKOV-3). This initial test was performed by Western blotting assay, in which normalized Ig samples from four distinct OC-derived ascites and four controls were used as a source of primary antibodies and challenged on the two cell lysates. We observed that the OC-derived samples generated distinct recognition patterns with a high number of proteins over a large range of molecular weights in lysates from both cell lines (Figure 2(a), upper and lower panels, lanes a-d). Notably, in the same experimental conditions, antibodies from noncancerous controls failed to clearly detect proteins with comparable frequency, specificity and intensity (Figure 2(a), upper and lower panels, lanes e-h). Equal loading of cell lysate was confirmed by probing the membrane strips with α-tubulin antibody. The presence of a specific response to cell antigens was further confirmed by immunofluorescence, the representative results of which are shown in Figure 2(b). As seen in Western blotting, a variable pattern of recognition was detected using different OC cell lines, and Igs from ascites of different OC patients were able to clearly stain cells in the different subcellular compartments of OVCAR-3 cells, including plasma membrane (panel a), nucleus (panel b), and cytoplasm (panel c). Cells stained with antibodies from noncancerous patients fail to produce detectable signals, confirming the Western blot results (panel d). The same pattern of recognition was obtained in a different OC cell line, SKOV-3; Igs purified from an OC ascite with cytoplasmic reactivity, clearly stained also SKOV-3 cells at cytoplasmic level (Figure 2(b) panel e) while Igs from a noncancerous control failed to give any signal (Figure 2(b) panel f).
Figure 2.
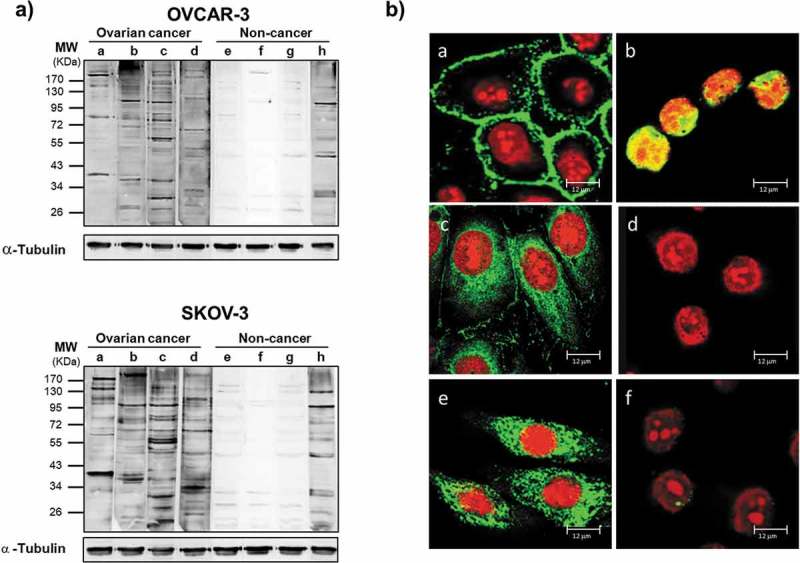
Antibodies from OC ascitic fluids react with tumor cells.
(a) Western blot of lysates from OVCAR-3 (upper panel) and SKOV-3 (lower panel) cells. Each sample was incubated with equal amount of Igs purified from ascites from OC patients (a-d) or from noncancerous controls (e-h). Immunoreactivity was detected with HRP-conjugated anti-human secondary antibody. After detection of ascitic IgG immunoreactivity, the membranes were stripped and probed with anti-α-tubulin antibody to assess the loading of lysate among the samples. (b) Immunofluorescence of OVCAR-3 and SKOV-3 cells with Igs purified from ascites. The cellular localization of TAAs was detected with Cy5-conjugated anti-human IgG (green). Nuclei were stained with propidium iodide (red). (a-d) Representative images of OVCAR-3 cells incubated with (a-c) Igs from OC ascites (a) without cell permeabilization, showing surface staining; (b) after cell fixation, showing nuclear staining; and (c) after permeabilization, showing cytoplasmic staining; (d) image of fixed and permeabilized OVCAR-3 cells incubated with Igs from noncancerous controls, showing a lack of any detectable signal. (e, f) Immunofluorescence analysis of fixed and permeabilized SKOV-3 cells incubated with Igs from ascites from patients with (e) OC and (f) noncancerous conditions, confirming the difference in reactivity.
To quantitatively confirm the presence of specific antibodies in OC-derived ascites, ELISA was performed using OVCAR-3 and SKOV-3 cells as a source of antigenic proteins. Purified antibodies from ascitic fluids from patients with OC (n = 28), other cancers (n = 18) or noncancerous (n = 28) were incubated either with fixed whole OVCAR-3 or SKOV-3 cells (Figure 3(a)) or with total cell lysate from OVCAR-3 or SKOV-3 cells (Figure 3(b)) to detect antibodies to cellular antigens. As expected, the ELISA results indicated the presence of a high level of tumor antigen-binding antibodies in OC ascites compared to that in ascites from patients with other cancers or with a noncancerous condition, confirming data obtained by immunofluorescence preliminary characterization.
Figure 3.
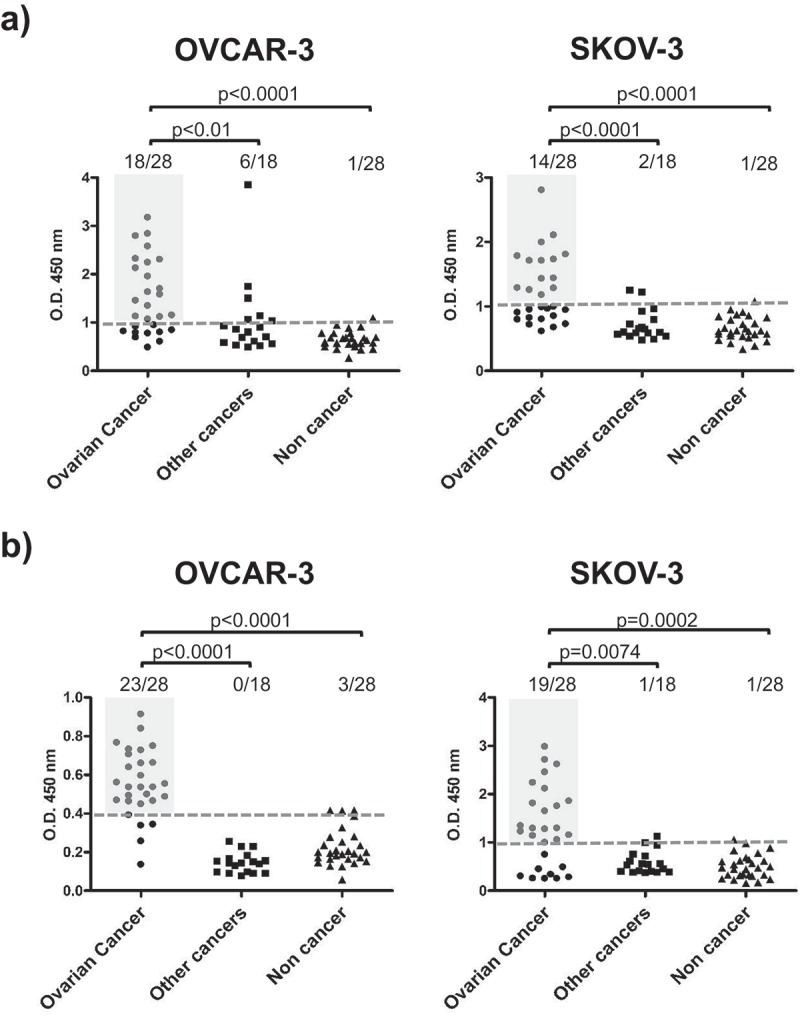
Antibodies from OC ascitic fluids react with tumor cells.
(a) OC whole-cell ELISA. Fixed and permeabilized OVCAR-3 or SKOV-3 cells at 80% confluence were incubated with 20 µg/ml Igs purified from ascites from patients with OC, other cancers or noncancerous conditions. (b) ELISA of OC cell lysate. OVCAR-3 or SKOV-3 cell lysates were prepared under nondenaturing conditions and used to coat the microtiter plates, which were subsequently incubated with 20 µg/ml Igs purified from ascites from patients with OC, other cancers or noncancerous conditions. Dotted lines represent cut-off levels calculated as the cumulative mean of the arbitrary units (AU) from noncancerous control wells. Light gray shaded areas indicate positive OC samples. Groups were compared by two-tailed unpaired t-test, and p-values are reported. p-Value<0.05 is considered significant.
Reactivity of OC ascites and that of noncancerous controls was further compared by indirect immunofluorescence assay on HEp-2 cells, a human laryngeal carcinoma cell line routinely used to evaluate autoantibody response in the context of different systemic autoimmune diseases. The assay was performed with purified antibodies from ascitic fluids from patients with OC (n = 15) and noncancerous (n = 14), results showed an overrepresentation of antibodies reacting against both nuclear and/or cytoplasmic antigens expressed by cancer cells (Supplementary Table 1). The data, supporting the presence of an immune response in OC patients, detectable in OC ascitic samples, were in line with what previously demonstrated by other group on OC using sera samples.33
Selection and identification of antigens by phage display and protein microarray
To identify the antigens recognized by the antibodies present in OC ascites, highly immunoreactive affinity-purified Igs were selected from the initially tested 28 OC-derived samples. This so-called “discovery set” was made by seven Ig samples purified from OC ascites of patients with different clinicopathological features and presents a distinct pattern of immunorecognition on both OVCAR-3 and SKOV-3 cellular antigens (Supplementary Table 2). Each of these seven samples was independently used as bait for two consecutive cycles of Open Reading Frame (ORF) cDNA phage display library selection46 to enrich for immunoreactive antigenic clones. Following biopanning, positive clones were identified using a protein microarray with the selected antigens expressed as GST-fusion proteins. One hundred clones from the final round of each of the seven selections were randomly picked, and recombinant proteins were expressed and affinity purified, yielding approximately 700 independent GST-fusion proteins. Protein microarrays were created by spotting purified proteins onto nitrocellulose slides along with controls and calibrator IgGs. Array quality control was performed with an anti-GST antibody (Supplementary Figure 2A), which indicated the correct printing and concentration range of more than 90% of the selected antigens. Each array was then challenged individually with purified antibodies from 17 different ascites representing patients with OC (n = 13, including the 7 used for selection), other cancer (n = 2) or noncancerous condition (n = 2). Antibodies from the noncancerous control subjects were given a limited number of positive staining spots (Supplementary Figure 2B), while arrays exhibited significantly increased immune recognition when incubated with Igs purified from OC ascites (Supplementary Figure 2C). Following spot signal quantification, 73 distinct clones were found to have increased immunogenicity towards one or more OC ascites IgG samples compared to noncancer ascites samples. Antigens were ranked according to the frequency of positive OC reactivity, and a subset of eight clones were identified as specifically recognized by all 13 tested OC samples but not from either type of control sample (i.e., other cancer and noncancerous). These clones were recovered, and proteins were identified by DNA sequencing. A total of eight different proteins were identified (Table 2): Cyclic AMP-responsive element-binding protein 3 (CREB3); Mitochondrial ribosomal protein L46 (MRPL46); Exosome component 10 (EXOSC10), BCL-6 corepressor (BCOR), High mobility group nucleosomal binding domain 2 (HMGN2), Huntingtin-interacting protein 1 related (HIP1R); Olfactomedin 4 (OLFM4) and the uncharacterized protein KIAA1755.
Table 2.
List of identified antigens.
| Protein Name | HUGO Name | Protein Length (aa) |
Protein Function | References |
|---|---|---|---|---|
| Cyclic AMP-responsive element-binding protein 3 | CREB3 | 395 | Transcription factor | 34,35 |
| 39S ribosomal protein L46, mitochondrial | MRPL46 | 279 | Structural constituent of ribosome | 36,37 |
| Exosome component 10 | EXOSC10 | 885 | rRNA Processing | 38 |
| BCL6 corepressor | BCOR | 1755 | Transcriptional corepressor | 39,40 |
| Non-histone chromosomal protein HMG-17 | HMGN2 | 90 | Nucleosomal DNA binding | 38,41 |
| Huntingtin-interacting protein 1-related protein | HIP1R | 1068 | Component of clathrin-coated pits and vesicles, | 42,43 |
| Olfactomedin-4 | OLFM4 | 510 | Cell adhesion | 44,45 |
| Uncharacterized protein KIAA1755 | KIAA1755 | 1200 | Unknown |
Identified tumor-associated antigens, their length and known main functions.
Validation of protein immunoreactivity by ELISA
To confirm the antigenicity of the selected proteins, recombinant proteins were produced to serve as antigens in an indirect ELISA to evaluate the reactivity of purified antibodies from patients and controls cohorts. Each selected ORF fragment was cloned in frame with the GST and the FLAG sequences at their N- and C-termini, respectively. The expected molecular weights of each recombinant GST-fusion product and the relative full-length protein are indicated in Supplementary Figure 3A. The quality of the affinity-purified GST-fused proteins was assessed by Coomassie staining (Supplementary Figure 3B) and by Western blotting performed with anti-GST and anti-FLAG antibodies (Supplementary Figure 3C and 3D). Preliminary set-up experiments were performed to establish the antigen coating concentration as well as the appropriate antibody dilution. Finally, each antigen was tested by ELISA against the full set of antibodies purified from the 153 ascites samples. As expected, ELISA reactivity towards the eight antigens was not homogenous, showing a wide range of immunorecognition (Figure 4). In general, antibodies from the control ascites samples showed no or very low reactivity towards all the antigens, resulting in a specificity higher than 94% for all proteins tested (see Supplementary Tables 3 and 4). Ascites samples from patients with other cancers showed a large degree of variability in recognition, while OC-derived ascites samples were highly reactive against all antigens tested. CREB3 showed a sensitivity of 87% (60 positive out of 69 total samples); MRPL46 and BCOR showed a sensitivity of approximately 73%, and EXOSC10 had a sensitivity of 56%. HMGN2 was the less sensitive among the antigenic panel, as only 13 of the 69 OC samples were positive (Supplementary Tables 3 and 4). When the antigens were used to identify differences in the three sample groups, the analyses showed that for all eight antigens, there was a statistically significant difference in immunoreactivity between the OC and noncancerous control ascites. Interestingly, three antigens, namely, CREB3, MRPL46, and EXOSC10, were able to significantly differentiate OC samples from samples derived from other cancers, thereby confirming the isolation of antigens specific for TA antibodies in OC ascites (Supplementary Table 4). When the immunoreactivity of each patient against the eight proteins was compared, we observed that 71% of the OC patients (49/69) showed reactivity against these three antigens, while positivity was observed for only 53% of patients with other cancers and none of noncancerous control subjects (Supplementary Figure 4).
Figure 4.
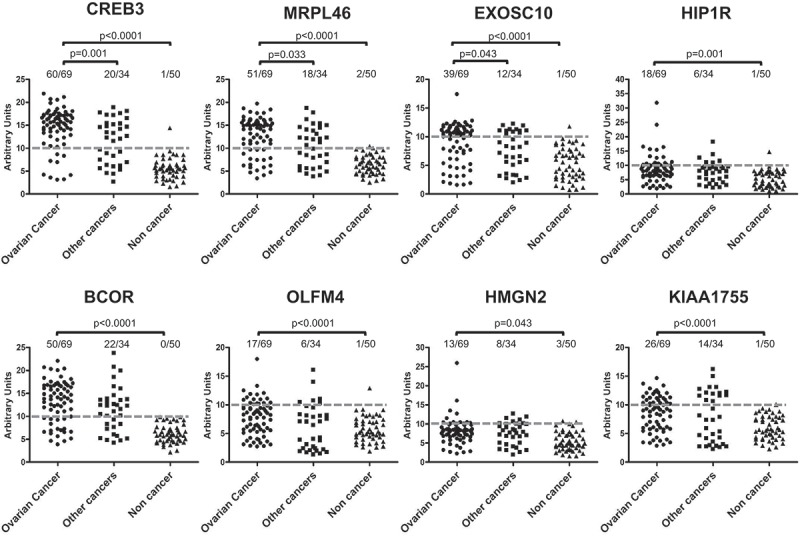
Validation of antigenic protein recognition by ELISA.
Each antigen was challenged with a 1:50 dilution of Igs purified from ascites from patients with OC, other cancers or noncancerous conditions. Immunoreactivity was assessed by measuring the amount of bound secondary anti-human IgG antibody and expressed as AU. Dotted lines represent cut-off levels. Differences between groups were calculated using Fisher’s exact test, and p-values are indicated. Difference was considered significant for a p-value<0.05.
Specific antibody response correlates with chemotherapy response in OC patients
We then analyzed whether the signal intensity of the TA antibody response to specific antigens correlated with clinical data. None of the clinicopathological characteristics except response to platinum-based therapy correlated with the antibody signal. Patients were stratified according to the response to treatment as follows: (i) ‘platinum-resistant’ (28 patients) with disease progression within 6 months from the end of therapy; (ii) ’partially platinum-sensitive’ (16 patients) with progression between 6 and 12 months; and (iii) ‘platinum-sensitive’ (24 patients) with progression after 12 months. The distribution of the ELISA signal values obtained from the individual antigens was then compared within the three classes of chemotherapy responses. For all eight antigens, there was no difference between the signals when the ‘platinum-resistant’ and ’partially platinum-sensitive’ classes were compared (Figure 5 and Supplementary Figure 5). Notably, when comparing ‘platinum-resistant’ and ‘platinum-sensitive’ patients, the BCOR antigen showed a statistically significant (p < .05) difference in TA autoantibody response with sensitive patients exhibiting a higher level of specific antibody (Figure 5(a)). No significant difference was observed for the other antigens (Figure 5(a) and Supplementary Figure 5); however, MRLP46 and CREB3 antigens showed a trend towards an increase in TA autoantibody response from resistant to sensitive patients, albeit not significant (Figure 5(a)). Furthermore, when BCOR, MRPL46, and CREB3 were combined as a single antibody signature (Figure 5(b,c)), a stronger association between higher levels of antibody and platinum sensitivity was observed, with a p-value reaching 6 × 10−3.
Figure 5.
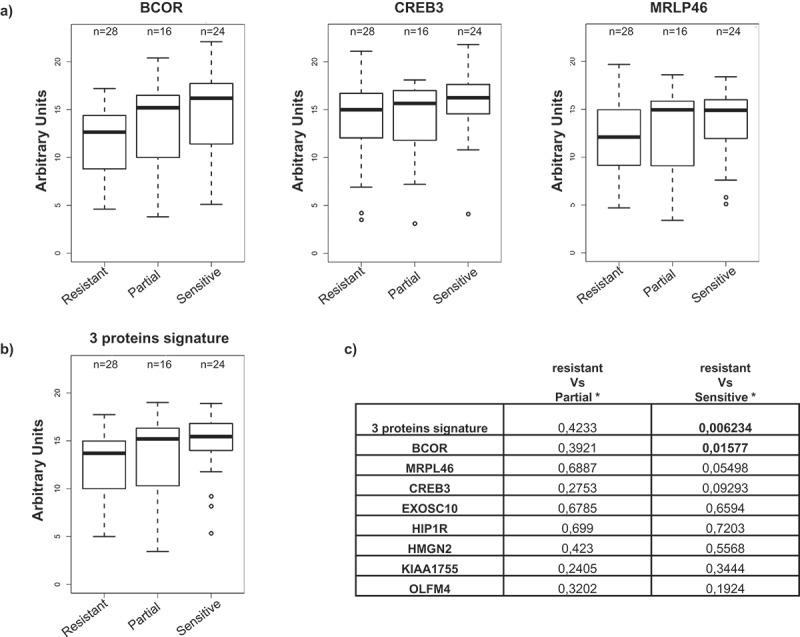
Antibody titer and correlation to clinical data.
OC patients were stratified in three groups according to their response to platinum-based treatment: ‘resistant’ (28 patients); ‘partially sensitive’ (16 patients) and ‘sensitive’ (24 patients). (a) Immunoreactivity in the different patients’ subgroups for BCOR, CREB3, and MRPL46; the values are expressed as AU. (b) immunoreactivity for the antibody signature comprising the combination of BCOR, CREB3 and MRPL46 signals in the different patient subgroups; the 3-gene signature is defined as the mean of the normalized values of the ELISA signal of the three genes. (c) p-values referring to ELISA signal comparisons for reactivity against the eight antigens or for the 3-protein signature between resistant vs. partially sensitive or resistant vs. sensitive patients (Mann–Whitney test). Statistically significance values are indicated in bold.
Antibodies to OC antigens only partially correlate with increased gene expression
The presence of high levels of TA autoantibodies to BCOR, CREB3, and MRLP46 in ascites from OC patients and the correlation of these high levels to platinum sensitivity prompted us to analyze the expression levels of the corresponding genes and their possible clinical impact; this was done with a curated collection of 23 datasets, including gene expression and clinical data from OC patients47 (see Supplementary Table 5).
More than 1800 OC patients with a 10-y follow-up were stratified for gene expression. We found that the correlation between gene expression of the identified antigens and patients’ overall survival (OS) was more complex than that seen with antibody levels and response to treatment. In fact, although the levels of anti-BCOR antibody had the most significant concordance between higher signal and platinum sensitivity, we found no significant correlation between high BCOR gene expression and longer OS (Figure 6(a)). By contrast, for both CREB3 (Figure 6(b)) and MRLP46 (Figure 6(c)), higher gene expression significantly correlated with longer OS. Interestingly, although the BCOR expression data alone did not show a significant correlation with OS, when the gene expression data of all three genes were combined into a novel gene signature (Figure 6(d)) (as done for the antibody), the level of significance was higher, reaching a p-value of 1.9 × 10−4 and thus identifying a novel prognostic gene signature, at least based on univariate analysis.
Figure 6.
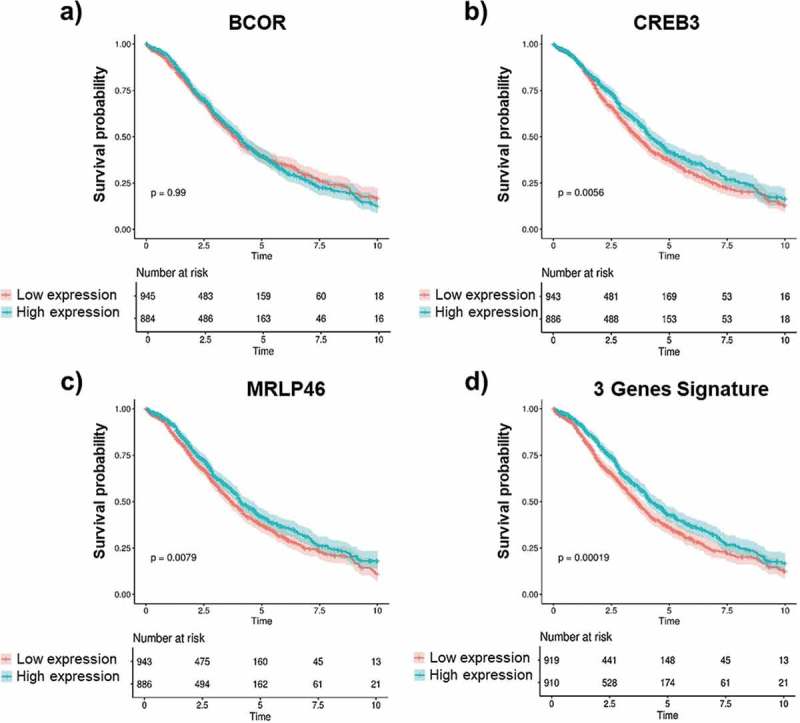
Gene expression and OS correlation.
Kaplan-Meier analysis of overall survival (OS) of OC patients according to the normalized mean expression of the genes of interest categorized as low (red lines) or high (blue lines). Gene expression levels of BCOR, CREB3, MRPL46 as well as the combined gene signature of these three antigens were measured in OC patients with 10 y of follow-up data. BCOR gene expression (a) shows no correlation with OS (y). Higher gene expression of CREB3 (b) and MRLP46 (c) is significantly associated with longer OS time (y). A much higher correlation was observed for combined signature of the expression data for all three genes as a novel gene signature (d). Log-rank p-values are reported. Shaded colors represent confidence intervals.
Discussion
Effective detection and monitoring of OC using biomarkers still holds great promise as an approach to reduce OC-related mortality. This study focuses on a systematic analysis of the OC antibody profile in ascites. We used a high-throughput proteomic-based technological platform that combines selection of a phage display cDNA library followed by a protein microarray,46 and we identified a final set of eight proteins whose recognition was unique in OC patients compared to control subjects. Ascitic fluids are accessible and an ideal source for analyzing both the cellular components of tumors and the secretome from tumor and nontumor cells. This unique specimen represents a still largely unexplored source for the identification of possible diagnostic, prognostic and predictive biomarkers.48 This study has the potential to enhance our insight into personalized cancer medicine and the development of effective targeted therapeutic strategies by providing an accurate molecular profile.26 Proteomic methods such as matrix-assisted laser desorption and ionization (MALDI), surface enhanced laser desorption and ionization (SELDI), and liquid chromatography followed by mass spectrometry (MS)49,50 have been used to elucidate the protein profile of malignant ascites and associated cells. In addition, automated high-throughput array-based proteomics techniques such as protein microarrays51 and reverse phase protein arrays (RPPAs) can be used to elucidate the differential expression of proteins in ascites from OC patients.52 Our initial strategy was aimed at identifying a unique antibody signature in ascitic fluid from OC patients. As a proximal fluid, ascites contain cancer-associated soluble factors, including TA antibodies, at concentrations much higher than those in serum. Ascites from various disease conditions were characterized for their antibody response against OC cellular antigens, and, although the level of antibody response was not homogenous within OC patients, it was considerably higher than that of patients with other types of cancer or that of patients with a noncancerous condition. The 8 proteins we identified following selection of an ORF cDNA phage display library and protein array screening produced a statistically significant difference in immunoreactivity when challenged with antibodies purified from ascites of OC patients compared to ascites from patients with noncancerous conditions and, to a lesser extent, to ascites from patients with other cancers. In fact, not surprisingly, some of the identified proteins have been previously associated with various cancer-related pathologies,38,39,42,53,54 suggesting that these antigens share common pathways in various disease conditions. A recent study found that most TA autoantigens are overexpressed or mutated, of which 42% were cytoplasmic, 26% were nuclear, 22% were membrane bound, and 10% were extracellular.13 Our results showed that although the first screening performed on OC cells identified a complex recognition pattern with immunofluorescence showing both intracellular (nuclear and cytoplasmic) and membrane staining, seven of the eight identified proteins were intracellular antigens. Tumor cell lysis and the induction of inflammation in the tumor microenvironment has been suggested to facilitate the release of intracellular antigens resulting in the abnormal levels of autologous antigens presentation to the immune system, which may explain a vast number of autoantibodies produced against intracellular antigens in cancer patients.11 We cannot exclude the possibility that other TA antibodies directed against surface antigens or glycosylated antigens are present in ascitic fluids from OC patients. Nevertheless, the autoantibody response to intracellular antigens is well known. A response to an intracellular antigen such as p53 has been detectable in OC patients as a result of a spontaneous and early humoral immune response of the host against the accumulation of an antigenic mutant p53 protein in tumor cells.55,56 In addition to their detection in serum, these antibodies can be detected in tissues, ascites, and other body fluids. We are aware that from a diagnostic point of view, assays relying on the detection of TA antibodies in serum rather than in ascites may be more useful for the early detection of OC. Indeed, the serum response of OC patients has been profiled to identify the signatures of proteins useful for disease detection. However, the added value of the selected TA antibodies as markers for the early detection of OC with the use of CA-125 remained limited.23,57
Tumor antibodies in OC have been explored as biomarkers not only for early diagnosis but also for predicting prognosis and recurrence as well as developing therapeutic approaches.58 The possibility of correlating the level of tumor autoantibodies with the clinical outcome of cancer patients might permit patient stratification into prognostic categories, which would help in the selection of second-line therapeutic treatments following primary debulking surgery and platinum-based first-line chemotherapy. The identification of patients most likely to respond to first-line treatment is still a clinically urgent need. In fact, after presenting an initial response, the majority of OC patients experience disease recurrence.57,59 We showed an association between the TA antibody signature in ascites from OC patients and their response to first-line treatment. OC patients who are sensitive to platinum treatment (either complete or partial) showed a higher production of anti-TA antibodies to the identified antigens than did patients who are resistant to platinum. Our data show a correlation between the presence of a treatment response with a higher immune response against BCOR, MRPL46 and CREB3 both individually and as a combined profile. This finding suggests higher levels of these antigens in particular subgroups of patients. However, the level of TA antibodies is the result of a complex and multistep activation of the immune system response, which does not always directly correlate with the gene expression level of the target antigens. Therefore, the level of gene expression of target antigens may not necessarily correlate with the patients’ prognosis.
To explore if the expression of genes, whose proteins we found to be able to induce an antibody response, was associated with a better prognosis, we analyzed publicly available OC gene expression profiles. The association of gene expression with patients’ prognosis was confirmed for CREB3 and MRPL46, but not for BCOR. However, as for the TA antibody, the combined signature of BCOR with CREB3 and MRPL46 gene expression levels had a greater prognostic impact than each of them individually.
The interest in relying on TA antibodies against TAAs as biomarkers for the diagnosis and prognosis of cancer is driven by the notion that these antibodies are generally absent or present in very low levels in healthy individuals and in people with noncancerous conditions.8,60
Furthermore, as antibodies represent biologically amplified markers, they circulate at higher concentrations than their corresponding antigen and demonstrate higher stability over time.15
The persistence and stability of anti-TAA antibodies in the body fluids of cancer patients is an advantage over other potential markers, including the TAAs themselves, which are released by tumors but rapidly degraded or cleared.61 Furthermore, the widespread availability of methods and reagents to detect autoantibodies facilitates their characterization in cancer patients. In conclusion, the strategy proposed in this work – systematically profiling OC ascites by coupling phage display and high-throughput screening technologies – could be a new valuable tool to select TAAs with potential diagnostic/prognostic relevance in any pathology involving autoantibodies and their corresponding antigens. The information obtained from these analyses can then be used to validate the impact of the identified molecules on patients’ sera.
Materials and methods
Patient samples and cell lines
Ascites from OC patients were collected at the Fondazione IRCCS Istituto Nazionale dei Tumori, Milan, while ascites from patients with nonovarian tumors or nononcological diseases were collected at the University of Torino. Ascites fluid was obtained with informed consent and approval from the institutional ethical committees. All our studies involving human subjects were conducted in compliance with the principles of the Declaration of Helsinki. Ascites were collected from 153 female patients with various disease conditions, including OC (n = 69), nonovarian tumors (n = 34), and noncancerous conditions (n = 50), the characteristics of whom are summarized in Table 1. All ascites from OC patients were collected during primary surgery. The amount of ascites collected was variable, ranging from centiliters to several liters. Fluids were transferred to 50 ml tubes and centrifuged at 2000 x g for 15 min at 4°C to remove cell debris, and the supernatant was stored at −80°C until use. The cellular composition of ascites from OC patients was determined by cytological evaluation. The presence of tumor cells in the 69 ascites samples from patients with OC was quantified as follows: absent (7/69), rare/some (19/69), and numerous (42/69); for one sample, the information was not available.
The human ovarian adenocarcinoma cell lines OVCAR-3 (ATCC® HTB-161™) and SKOV-3 (ATCC® HTB-77™) were grown in RPMI 1640 medium containing 10% fetal calf serum and 1% penicillin/streptomycin. Cells were maintained at 37°C in a humidified atmosphere containing CO2. Cells were routinely tested for mycoplasma contamination and confirmed by an STR profile.
Characterization of ascitic fluid based on autoantibody response
Igs were affinity purified using Protein A-agarose (Roche, Basel, Switzerland) according to the manufacturer’s instructions. To compare the antibody-mediated immune responses in the different samples, the Ig concentrations in all 153 purified ascitic fluids were set to 1 μg/μl and used as the standard starting concentrations for all experiments. Furthermore, we focused our analysis on the IgG-specific immune response by using specific anti-human IgG secondary antibodies. Affinity-purified Igs obtained from patients’ ascites were characterized for their reactivity using OVCAR-3 and SKOV-3 OC cell lines as described below.
Western blotting of cell extracts
Proteins from OVCAR-3 and SKOV-3 cells were extracted under denaturing conditions using 8M Urea, resolved by SDS-PAGE and transferred to nitrocellulose membranes, which were then blocked for 1 hr with 4% milk in PBS-Tween 20 buffer (MPBST) and incubated with Igs purified from ascites as the primary antibody source (diluted 1:50 in 2% milk in PBST) for 90 min. Immunoreactive bands were detected after the membranes were incubated with an HRP-conjugated anti-Human IgG (Thermo Fischer Scientific, Waltham, MA USA) for 1 hr at room temperature. Membrane strips were then stripped and reprobed with anti-tubulin antibody (Santa Cruz Biotechnology, CA, USA) followed by anti-mouse HRP-conjugated secondary antibody as protein loading control. Western blots were developed via chemiluminescence with a VersaDoc Imaging System (Biorad, Milano, Italy).
Immunofluorescence
OVCAR-3 and SKOV-3 cells were grown on glass coverslips to 80% confluence and subjected to immunofluorescence staining afterward. Images were obtained with a Leica DMIRE2 confocal fluorescence microscope (Leica Microsystems) equipped with Leica Confocal Software v.2.61. For intracellular staining, cells were washed twice with PBS, fixed with 4% paraformaldehyde (in PBS, 4% sucrose) and permeabilized with 0.2% Triton X-100. Fixed cells were blocked with 2% BSA and incubated for 2 hr at 37°C with a 1:10 dilution of Igs purified from ascites. Slides were then incubated with an anti-human IgG Cy5-conjugated secondary antibody (1:200 dilution; Jackson Immuno Research, Cambridgeshire, UK), and cell nuclei were counterstained with propidium iodide. For surface staining, cells grown on coverslips were washed with serum-free RPMI medium and incubated for 2 hr with the Igs purified from ascites (diluted 1:10 in blocking buffer (5% BSA in serum-free RPMI medium)). All incubation steps were carried out at 4°C to prevent cellular uptake of antibodies. After washing, bound Igs were revealed by incubation with the secondary antibody. The washed cells were fixed, permeabilized and counterstained as described above.
Whole-cell ELISA
OVCAR-3 and SKOV-3 cells were seeded in 96-well culture plates (104 cells/well) and allowed to adhere overnight (O/N). The cells were then washed with blocking buffer, fixed with methanol and incubated at 4°C for 90 min with Igs from ascites (diluted 1:50 in blocking buffer). Next, the cells were treated with a peroxidase-conjugated secondary antibody (Dako Cytomation) for 1 hr at 4°C. Immunocomplexes were revealed with 3,3ʹ,5,5ʹ-tetramethylbenzidine (TMB), and the plates were read at a wavelength of 450 nm. Experiments were performed in triplicate. The positive cut-off value for each antigen was calculated as the cumulative mean of the OD450 value obtained with Igs from ascites from noncancerous controls plus 2 standard deviations (SD).
Cell lysate ELISA
Confluent OVCAR-3 and SKOV-3 cells grown on 100 mm dishes were washed with ice-cold PBS and lysed with a nondenaturing extraction buffer (100 mM Tris, 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 1% IGEPAL CA-630, protease inhibitors; pH 7.4). Cell lysates were incubated on ice for 15–30 min and then briefly sonicated. The soluble material was recovered after centrifugation at 13000 rpm for 10 min at 4°C, and the protein concentration was determined with the BCA assay. To perform cell lysate ELISAs, microtiter plates were coated O/N with 3.2 µg of cell extract and then blocked with 2% BSA in PBST (0.05%) for 1 hr at 30°C. The plates were then incubated for 90 min at 30°C with Igs purified from ascites (1:50 dilution). Immunocomplexes were detected as described above.
Indirect immunofluorescence on HEp-2 cells
ANA reactivity was evaluated on HEp-2 ANA Slides (Inova Diagnostics, Inc., San Diego, CA, USA) following manufacturer’s instructions with Igs from OC (n = 15) and noncancerous controls (n = 14) ascites. Results were visually scored as negative or positive, and positive samples furthermore classified as cytoplasmic or nuclear or both nuclear and cytoplasmic by visual inspection of the fluorescence signal. Samples were acquired with a Leica DMIRE2 confocal fluorescence microscope (Leica Microsystems, Wetzlar, Germany) equipped with Leica Confocal Software v.2.61.
Construction and selection of the open reading frame cDNA phage display library
The human ORF cDNA phage display library was prepared from mRNA derived from different tissues as described by Di Niro et al.46 The biopanning of the library was performed with negative and positive selections using healthy control sera followed by independent selections using ascites from 7 different patients diagnosed with OC representing stage II-IV disease that had already been identified as the most reactive based on the initial immunological assays. Two successive rounds of selection using Igs purified from ascites and captured on protein G-coated magnetic beads (Dynabeads) (Thermo Fischer Scientific), were performed as described by Di Niro et al.62 Each cycle was preceded by a “pre-clearing step” done using antibodies from healthy donors to remove common or polyreactive clones.
ORF cDNA fragments from the second round of selection were subcloned from phagemid DNA into a modified pGEX-4T expression vector (GE-Healthcare, Chicago, IL, USA) to yield GST-fusion products with a C-terminal FLAG tag and transformed into the E. coli strain BL21 (DE3) RIPL for enhanced protein production. Approximately 700 individual clones were randomly picked (96 clones from each of the seven selections), and recombinant GST-fusion protein production was carried out in a 96-well format with the autoinducing medium ZYM5052 as described by Studier et al.63 Cells were grown under constant air supply using a custom-built air-well sparging minifermenter system.64 Proteins were affinity-purified using glutathione magnetic beads (Promega, Madison, WI, USA) and analyzed for their quantity and quality by Coomassie staining.
Protein microarrays and ORF sequencing
GST-fusion proteins were printed onto nitrocellulose-coated slides using a BioOdissey Calligrapher MiniArrayer (Biorad). The arrayed slides were processed as described by D’Angelo et al.65 The slides blocked with 3% milk in PBST were incubated individually with affinity-purified antibodies from 17 ascites samples (including OC, other cancer and noncancerous condition) as the source of primary antibodies (1:200 dilution in binding buffer (2% milk in PBST)). Bound antibodies were detected by incubating the slides with a Cy5-conjugated anti-human IgG secondary antibody (Jackson Immuno Research; 1:200 dilution) for 1 hr. Fluorescent signals were measured with a ScanArray Gx® (PerkinElmer, Waltham, MA, USA) and analyzed with ScanArray expression software (PerkinElmer). Arrays were normalized with a 2-step protocol. The background response was evaluated using the signal generated from each tested ascites sample against a reference GST protein, and a set of serial dilutions of purified IgG printed on each array was used to generate a calibration curve of arbitrary IgG units. Positive proteins were identified by comparing the frequency of reactive ascites samples. The proteins with a significant difference in reactivity between ascites samples from OC patients and those from noncancerous control subjects were identified as putative candidates for further analysis. The most immunoreactive clones in the microarray analysis were amplified by PCR using pGEX sense (5ʹ GGGCTGGCAAGCCACGTTTGGTG 3ʹ) and antisense primers (5ʹ GGTGAAAACCTCTGACACATGCAGCTCCCGG 3ʹ). The purified PCR products were sequenced in an ABI PRISM® 3100 Genetic Analyzer (Thermo Fischer Scientific) using BigDye Terminator v1.1 Cycle Sequencing Kit (Thermo Fischer Scientific).
Indirect ELISA
Recombinant GST-fused proteins were diluted in PBS to 10 μg/ml, and 100 μl incubated in the ELISA wells (Sigma Aldrich, St. Louis, MO, USA) O/N at 4°C. Wells were washed with PBS, and 200 μl of blocking solution (2% BSA in PBST) was added to each well for 1 hr at RT. Affinity-purified and normalized ascites Igs from 153 patients (1:50 dilution in blocking buffer) were used as primary antibodies and incubated for 90 min at 30°C. Extensive washes of the wells were performed with PBST and PBS to remove unbound primary antibodies. Then, the wells were incubated with a 1:3000 dilution of peroxidase-conjugated anti-human-IgG secondary antibody (Dako Cytomation) in blocking buffer at 30°C for 1 hr. After extensive washing, immunocomplexes were revealed with TMB, and the plate was read in a microplate reader at a wavelength of 450 nm. A cut-off value for positivity for each antigen was calculated independently as the mean OD450 value obtained with Igs from noncancerous ascites samples plus 2 SD.
Statistical analysis
The ELISA data were analyzed to test for differences in the presence of antibodies targeting the identified antigens among the three patient groups. Depending on the expected value of each cell in the cross-tabulation Chi-square or Fisher’s exact test was first performed to infer the statistical significance in the overall comparison among OC patients, other cancer patients and noncancerous control subjects and then when separately comparing OC vs. other tumors or vs. healthy subjects. ROC analyses were performed to evaluate the performances, including the AUC estimates with their 95% confidence limits, of each antigen in correctly classifying the disease condition. All tests were two-sided, and the level of statistical significance was set as p-values <0.05. All analyses were performed using Statistical Software Packages SPSS (IBM Corp. Released 2013. IBM SPSS Statistics for Windows, Version 22.0. Armonk, NY: IBM Corp).
Survival analysis of the internal dataset
The clinical correlation between the presence of a TA antibody and the patients’ response to treatment was assessed by the Mann–Whitney test; patients were stratified into three groups based on their treatment response (resistant, relapse occurring within 6 months after treatment completion; partially sensitive, relapse occurring between 6 and 12 months after treatment completion; and sensitive, relapse occurring after 12 months after treatment completion). All analyses were performed in the R/Bioconductor environment. For all analyses, differences were considered significant at p-values <0.05.
Survival analysis of the external dataset
The prognostic value of the expression levels of the genes of interest was analyzed in a dataset comprising more than 1800 OC patients from 23 studies with curated and documented clinical metadata.47 The curated Ovarian Data package provides data for gene expression analysis in patients with OC, and we selected samples for which OS information was available. Patients were classified as low or high according to the normalized mean expression of the genes of interest. A Mantel–Haenszel test was applied, and Kaplan-Meier survival curves were obtained and compared with the log-rank test. All analyses were performed in the R/Bioconductor environment using the survival package.
Supplementary Material
Funding Statement
This work was supported by Regione Piemonte (IMMONC Piattaforme Innovative), Cariplo Foundation (Milan) and AIRC under grant number AIRC IG-17475. FA is a recipient of an international PhD fellowship in Innovative Biomedical Technologies (IBT) funded by Cariplo Foundation. CY is supported by AIRC/FIRC 2015.
Disclosure of potential conflicts of interest
The authors report no conflicts of interest.
Author contribution
MP, MD, DC, SC, SD designed the study and wrote the paper; SMF, FA, CD, TO performed phage display selection and clone characterization; CY, PS, AF performed statistical analysis; MD, MP, RF collected and analyzed the sera from patients and control subjects. All authors analyzed the results and approved the final version of the manuscript.
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Tworoger SS, Shafrir AL, Hankinson SE.. Ovarian cancer. Schottenfeld Fraumeni Cancer Epidemiol. 2017;384(9951):889–908. Prev. Fourth Ed [Google Scholar]
- 2.Asai A, Okita T, Enzo A. Conflicting messages concerning current strategies against research misconduct in Japan: a call for ethical spontaneity. J Med Ethics. 2016;42(8):524–527. doi: 10.1001/jama.2017.21421. [DOI] [PubMed] [Google Scholar]
- 3.Vaughan S, Coward JI, Bast RC, Berchuck A, Berek JS, Brenton JD, Coukos G, Crum CC, Drapkin R, Etemadmoghadam D, et al. Rethinking ovarian cancer: recommendations for improving outcomes. Nat Rev Cancer. 2011;11(10):719–725. doi: 10.1038/nrc3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coleman RL, Monk BJ, Sood AK, Herzog TJ. Latest research and treatment of advanced-stage epithelial ovarian cancer. Nat Rev Clin Oncol. 2013;10(4):211–224. doi: 10.1038/nrclinonc.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zurawski VR, Orjaseter H, Andersen A, Jellum E. Elevated serum CA 125 levels prior to diagnosis of ovarian neoplasia: relevance for early detection of ovarian cancer. Int J Cancer. 1988;42(5):677–680. doi: 10.1002/ijc.2910420507. [DOI] [PubMed] [Google Scholar]
- 6.Skates SJ, Horick N, Yu Y, Xu FJ, Berchuck A, Havrilesky LJ, de Bruijn HW, van der Zee AG, Woolas RP, Jacobs IJ, et al. Preoperative sensitivity and specificity for early-stage ovarian cancer when combining cancer antigen CA-125II, CA 15-3, CA 72-4, and macrophage colony-stimulating factor using mixtures of multivariate normal distributions. J Clin Oncol. 2004;22(20):4059–4066. doi: 10.1200/JCO.2004.03.091JCO.2004.03.091. [DOI] [PubMed] [Google Scholar]
- 7.Leung F, Bernardini MQ, Brown MD, Zheng Y, Molina R, Bast RC, Davis G, Serra S, Diamandis EP, Kulasingam V. Validation of a novel biomarker panel for the detection of ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2016;25(9):1333–1340. doi: 10.1158/1055-9965.EPI-15-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore RG, Brown AK, Miller MC, Skates S, Allard WJ, Verch T, Steinhoff M, Messerlian G, DiSilvestro P, Granai CO, et al. The use of multiple novel tumor biomarkers for the detection of ovarian carcinoma in patients with a pelvic mass. Gynecol Oncol. 2008;108(2):402–408. doi: 10.1016/j.ygyno.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 9.Szajnik M, Czystowska-Kuźmicz M, Elishaev E, Whiteside TL. Biological markers of prognosis, response to therapy and outcome in ovarian carcinoma. Expert Rev Mol Diagn. 2016;16(8):811–826. doi: 10.1080/14737159.2016.1194758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Macdonald IK, Parsy-Kowalska CB, Chapman CJ. Autoantibodies: opportunities for early cancer detection. Trends Cancer. 2017;3(3):198–213. doi: 10.1016/j.trecan.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 11.Zaenker P, Gray ES, Ziman MR. Autoantibody production in cancer-the humoral immune response toward autologous antigens in cancer patients. Autoimmun Rev. 2016;15(5):477–483. doi: 10.1016/j.autrev.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 12.Kobold S, Lütkens T, Cao Y, Bokemeyer C, Atanackovic D. Autoantibodies against tumor-related antigens: incidence and biologic significance. Hum Immunol. 2010;71(7):643–651. doi: 10.1016/j.humimm.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 13.Reuschenbach M, Von Knebel Doeberitz M, Wentzensen N. A systematic review of humoral immune responses against tumor antigens. Cancer Immunol Immunother. 2009;58(10):1535–1544. doi: 10.1007/s00262-009-0733-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi JX, Qin JJ, Ye H, Wang P, Wang KJ, Zhang JY. Tumor associated antigens or anti-TAA autoantibodies as biomarkers in the diagnosis of ovarian cancer: A systematic review with meta-analysis. Expert Rev Mol Diagn. 2015;15(6):829–852. doi: 10.1586/14737159.2015.1035713. [DOI] [PubMed] [Google Scholar]
- 15.Fortner RT, Damms-Machado A, Kaaks R. Systematic review: tumor-associated antigen autoantibodies and ovarian cancer early detection. Gynecol Oncol. 2017;147(2):465–480. doi: 10.1016/j.ygyno.2017.07.138. [DOI] [PubMed] [Google Scholar]
- 16.Lacombe J, Mangé A, Jarlier M, Bascoul-Mollevi C, Rouanet P, Lamy PJ, Maudelonde T, Solassol J. Identification and validation of new autoantibodies for the diagnosis of DCIS and node negative early-stage breast cancers. Int J Cancer. 2013;132:1105–1113. doi: 10.1002/ijc.27766. [DOI] [PubMed] [Google Scholar]
- 17.Hirales Casillas CE, Flores Fernández JM, Camberos EP, Herrera López EJ, Pacheco GL, Velázquez MM. Current status of circulating protein biomarkers to aid the early detection of lung cancer. Futur Oncol. 2014;10(8):1501–1513. doi: 10.2217/fon.14.21. [DOI] [PubMed] [Google Scholar]
- 18.Hong Y, Huang J. Autoantibodies against tumor-associated antigens for detection of hepatocellular carcinoma. World J Hepatol. 2015;7(11):1581–1585. doi: 10.4254/wjh.v7.i11.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Werner S, Chen H, Tao S, Brenner H. Systematic review: serum autoantibodies in the early detection of gastric cancer. Int J Cancer. 2015;136(10):2243–2252. doi: 10.1002/ijc.28807. [DOI] [PubMed] [Google Scholar]
- 20.Chen H, Werner S, Tao S, Zörnig I, Brenner H. Blood autoantibodies against tumor-associated antigens as biomarkers in early detection of colorectal cancer. Cancer Lett. 2014;346(2):178–187. doi: 10.1016/j.canlet.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 21.Mintz PJ, Rietz AC, Cardó-Vila M, Ozawa MG, Dondossola E, Do K-A, Kim J, Troncoso P, Logothetis CJ, Sidman RL, et al. Discovery and horizontal follow-up of an autoantibody signature in human prostate cancer. Proc Natl Acad Sci. 2015;112(8):2515–2520. doi: 10.1073/pnas.1500097112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gnjatic S, Ritter E, Buchler MW, Giese NA, Brors B, Frei C, Murray A, Halama N, Zornig I, Chen Y-T, et al. Seromic profiling of ovarian and pancreatic cancer. Proc Natl Acad Sci. 2010;107(11):5088–5093. doi: 10.1073/pnas.0914213107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stafford P, Cichacz Z, Woodbury NW, Johnston SA. Immunosignature system for diagnosis of cancer. Proc Natl Acad Sci. 2014;111(30):E3072–E3080. doi: 10.1073/pnas.1409432111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gagnon A, Kim JH, Schorge JO, Ye B, Liu B, Hasselblatt K, Welch WR, Bandera CA, Mok SC. Use of a combination of approaches to identify and validate relevant tumor-associated antigens and their corresponding autoantibodies in ovarian cancer patients. Clin Cancer Res. 2008;14(3):764–771. doi: 10.1158/1078-0432.CCR-07-0856. [DOI] [PubMed] [Google Scholar]
- 25.Chatterjee M, Mohapatra S, Ionan A, Bawa G, Ali-Fehmi R, Wang X, Nowak J, Ye B, Nahhas FA, Lu K, et al. Diagnostic markers of ovarian cancer by high-throughput antigen cloning and detection on arrays. Cancer Res. 2006;66(2):1181–1190. doi: 10.1158/0008-5472.CAN-04-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kipps E, Tan DSP, Kaye SB. Meeting the challenge of ascites in ovarian cancer: new avenues for therapy and research. Nat Rev Cancer. 2013;13(4):273–282. doi: 10.1038/nrc3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuk C, Kulasingam V, Gunawardana CG, Smith CR, Batruch I, Diamandis EP. Mining the ovarian cancer ascites proteome for potential ovarian cancer biomarkers. Mol Cell Proteomics. 2009;8(4):661–669. doi: 10.1074/mcp.M800313-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin K, Ricciardelli C, Hoffmann P, Oehler MK. Exploring the immunoproteome for ovarian cancer biomarker discovery. Int J Mol Sci. 2011;12(1):410–428. doi: 10.3390/ijms12010410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang H, Li Y, Liu J, Zheng M, Feng Y, Hu K, Huang Y, Huang Q. Screening and identification of biomarkers in ascites related to intrinsic chemoresistance of serous epithelial ovarian cancers. PLoS One. 2012;7(12):e51256. doi: 10.1371/journal.pone.0051256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bery A, Leung F, Smith CR, Diamandis EP, Kulasingam V. Deciphering the ovarian cancer ascites fluid peptidome. Clin Proteomics. 2014;11(1):13. doi: 10.1186/1559-0275-11-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Xu B, Liu Y, Yao H, Lu N, Li B, Gao J, Guo S, Han N, Qi J, et al. The ovarian cancer-derived secretory/releasing proteome: A repertoire of tumor markers. Proteomics. 2012;12(11):1883–1891. doi: 10.1002/pmic.201100654. [DOI] [PubMed] [Google Scholar]
- 32.Bjørge L, Hakulinen J, Vintermyr OK, Jarva H, Jensen TS, Iversen OE, Meri S. Ascitic complement system in ovarian cancer. Br J Cancer. 2005;92(5):895–905. doi: 10.1038/sj.bjc.6602334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heegaard NHH, West-Nørager M, Tanassi JT, Houen G, Nedergaard L, Høgdall C, Høgdall E. Circulating antinuclear antibodies in patients with pelvic masses are associated with malignancy and decreased survival. PLoS One. 2012;7(2). doi: 10.1371/journal.pone.0030997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan CP, Kok KH, Jin DY. CREB3 subfamily transcription factors are not created equal: recent insights from global analyses and animal models. Cell Biosci. 2011;1(1):6. doi: 10.1186/2045-3701-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taniguchi M, Yoshida H. TFE3, HSP47, and CREB3 pathways of the mammalian golgi stress response. Cell Struct Funct. 2017;42(1):27–36. doi: 10.1247/csf.16023. [DOI] [PubMed] [Google Scholar]
- 36.Carim-Todd L, Sumoy L, Andreu N, Estivill X, Escarceller M. Cloning, mapping and expression analysis of C15 or f4, a novel human gene with homology to the yeast mitochondrial ribosomal protein Yml30 gene. Mitochondrial DNA. 2001;12(2):91–96. doi: 10.3109/10425170109047561. [DOI] [PubMed] [Google Scholar]
- 37.Brown A, Amunts A, Bai XC, Sugimoto Y, Edwards PC, Murshudov G, Scheres SHW, Ramakrishnan V. Structure of the large ribosomal subunit from human mitochondria. Science. 2014;346(6210):718–722. doi: 10.1126/science.1258026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Porkka K, Laakkonen P, Hoffman JA, Bernasconi M, Ruoslahti E. A fragment of the HMGN2 protein homes to the nuclei of tumor cells and tumor endothelial cells in vivo. Proc Natl Acad Sci U S A. 2002;99(11):7444–7449. doi: 10.1073/pnas.062189599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sturm D, Orr BA, Toprak UH, Hovestadt V, Jones DTW, Capper D, Sill M, Buchhalter I, Northcott PA, Leis I, et al. New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell. 2016;164(5):1060–1072. doi: 10.1016/j.cell.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huynh KD, Fischle W, Verdin E, Bardwell VJ. BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev. 2000;14(14):1810–1823. doi: 10.1111/j.1754-7121.1984.tb00653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martínez de Paz A, Ausió J; HMGNs . The enhancer charmers. BioEssays. 2016;38(3):226–231. doi: 10.1002/bies.201500157. [DOI] [PubMed] [Google Scholar]
- 42.Hsu CY, Lin CH, Jan YH, Su CY, Yao YC, Cheng HC, Hsu TI, Wang PS, Su WP, Yang CJ, et al. Huntingtin-interacting protein-1 is an early-stage prognostic biomarker of lung adenocarcinoma and suppresses metastasis via AKT-mediated epithelial-mesenchymal transition. Am J Respir Crit Care Med. 2016;193(8):869–880. doi: 10.1164/rccm.201412-2226OC. [DOI] [PubMed] [Google Scholar]
- 43.Porpaczy E, Bilban M, Heinze G, Gruber M, Vanura K, Schwarzinger I, Stilgenbauer S, Streubel B, Fonatsch C, Jaeger U. Gene expression signature of chronic lymphocytic leukaemia with Trisomy 12. Eur J Clin Invest. 2009;39(7):568–575. doi: 10.1111/j.1365-2362.2009.02146.x. [DOI] [PubMed] [Google Scholar]
- 44.Marimuthu A, Chavan S, Sathe G, Sahasrabuddhe NA, Srikanth SM, Renuse S, Ahmad S, Radhakrishnan A, Barbhuiya MA, Kumar RV, et al. Identification of head and neck squamous cell carcinoma biomarker candidates through proteomic analysis of cancer cell secretome. Biochim Biophys Acta - Proteins Proteomics. 2013;1834(11):2308–2316. doi: 10.1016/j.bbapap.2013.04.029. [DOI] [PubMed] [Google Scholar]
- 45.Yang Q, Bavi P, Wang JY, Roehrl MH. Immuno-proteomic discovery of tumor tissue autoantigens identifies olfactomedin 4, CD11b, and integrin alpha-2 as markers of colorectal cancer with liver metastases. J Proteomics. 2017;168:53–65. doi: 10.1016/j.jprot.2017.06.021. [DOI] [PubMed] [Google Scholar]
- 46.Di Niro R, Sulic AM, Mignone F, D’Angelo S, Bordoni R, Iacono M, Marzari R, Gaiotto T, Lavric M, Bradbury ARM, et al. Rapid interactome profiling by massive sequencing. Nucleic Acids Res. 2010;38(9). doi: 10.1093/nar/gkq052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ganzfried BF, Riester M, Haibe-Kains B, Risch T, Tyekucheva S, Jazic I, Wang XV, Ahmadifar M, Birrer MJ, Parmigiani G, et al. curatedOvarianData: clinically annotated data for the ovarian cancer transcriptome. Database. 2013;2013. doi: 10.1093/database/bat013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahmed N, Stenvers KL. Getting to know ovarian cancer ascites: opportunities for targeted therapy-based translational research. Front Oncol. 2013;3:256. doi: 10.3389/fonc.2013.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahmed N, Oliva KT, Barker G, Hoffmann P, Reeve S, Smith IA, Quinn MA, Rice GE. Proteomic tracking of serum protein isoforms as screening biomarkers of ovarian cancer. Proteomics. 2005;5(17):4625–4636. doi: 10.1002/pmic.200401321. [DOI] [PubMed] [Google Scholar]
- 50.Gortzak-Uzan L, Ignatchenko A, Evangelou AI, Agochiya M, Brown KA, St.Onge P, Kireeva I, Schmitt-Ulms G, Brown TJ, Murphy J, et al. A proteome resource of ovarian cancer ascites: integrated proteomic and bioinformatic analyses to identify putative biomarkers. J Proteome Res. 2008;7(1):339–351. doi: 10.1021/pr0703223. [DOI] [PubMed] [Google Scholar]
- 51.Gunawardana CG, Memari N, Diamandis EP. Identifying novel autoantibody signatures in ovarian cancer using high-density protein microarrays. Clin Biochem. 2009;42(4–5):426–429. doi: 10.1016/j.clinbiochem.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 52.Davidson B, Espina V, Steinberg SM, Flørenes VA, Liotta LA, Kristensen GB, Trope CG, Berner A, Ec K. Proteomic analysis of malignant ovarian cancer effusions as a tool for biologic and prognostic profiling. Clin Cancer Res. 2006;12(3I):791–799. doi: 10.1158/1078-0432.CCR-05-2516. [DOI] [PubMed] [Google Scholar]
- 53.Staals RH, Pruijn GJ. The human exosome and disease. Adv Exp Med Biol. 2011;702:132–142. [PubMed] [Google Scholar]
- 54.Shiba N, Yoshida K, Shiraishi Y, Okuno Y, Yamato G, Hara Y, Nagata Y, Chiba K, Tanaka H, Terui K, et al. Whole-exome sequencing reveals the spectrum of gene mutations and the clonal evolution patterns in paediatric acute myeloid leukaemia. Br J Haematol. 2016;175(3):476–489. doi: 10.1111/bjh.14247. [DOI] [PubMed] [Google Scholar]
- 55.Garziera M, Montico M, Bidoli E, Scalone S, Sorio R, Giorda G, Lucia E, Toffoli G. Prognostic role of serum antibody immunity to p53 oncogenic protein in ovarian cancer: a systematic review and a meta-analysis. PLoS One. 2015;10(10). doi: 10.1371/journal.pone.0140351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang WL, Gentry-Maharaj A, Simmons A, Ryan A, Fourkala EO, Lu Z, Baggerly KA, Zhao Y, Lu KH, Bowtell D, et al. Elevation of TP53 autoantibody before CA125 in preclinical invasive epithelial ovarian cancer. Clin Cancer Res. 2017;23(19):5912–5922. doi: 10.1158/1078-0432.CCR-17-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaaks R, Fortner RT, Hüsing A, Barrdahl M, Hopper M, Johnson T, Tjønneland A, Hansen L, Overvad K, Fournier A, et al. Tumor-associated autoantibodies as early detection markers for ovarian cancer? A prospective evaluation. Int J Cancer. 2018;143(3):515–526. doi: 10.1002/ijc.31335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chatterjee M, Tainsky MA. Autoantibodies as biomarkers for ovarian cancer. Cancer Biomarkers. 2010;8(4–5):187–201. doi: 10.3233/CBM-2011-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jayson GC, Kohn EC, Kitchener HC, Ledermann JA. Ovarian cancer. Lancet. 2014;384(9951):1376–1388. doi: 10.1016/S0140-6736(13)62146-7. [DOI] [PubMed] [Google Scholar]
- 60.Visintin I, Feng Z, Longton G, Ward DC, Alvero AB, Lai Y, Tenthorey J, Leiser A, Flores-Saaib R, Yu H, et al. Diagnostic markers for early detection of ovarian cancer. Clin Cancer Res. 2008;14(4):1065–1072. doi: 10.1158/1078-0432.CCR-07-1569. [DOI] [PubMed] [Google Scholar]
- 61.Zhang JY, Tan EM. Autoantibodies to tumor-associated antigens as diagnostic biomarkers in hepatocellular carcinoma and other solid tumors. Expert Rev Mol Diagn. 2010;10(3):321–328. doi: 10.1586/erm.10.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Di Niro R, D’Angelo S, Secco P, Marzari R, Santoro C, Sblattero D. Profiling the autoantibody repertoire by screening phage-displayed human cDNA libraries. Methods Mol Biol. 2009;570:353–369. doi: 10.1007/978-1-60327-394-7_20. [DOI] [PubMed] [Google Scholar]
- 63.Musgrove AJ, Austin GE, Hearn RD, Holt CA, Stroud DA, Wotton SR. Overwinter population estimates of British waterbirds. Br Birds. 2011;104(7):364–397. doi: 10.1016/j.jconrel.2016.07.053. [DOI] [Google Scholar]
- 64.Deantonio C, Sedini V, Cesaro P, Quasso F, Cotella D, Persichetti F, Santoro C, Sblattero D. An Air-well sparging minifermenter system for high-throughput protein production. Microb Cell Fact. 2014;13(1). doi: 10.1186/s12934-014-0132-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.D’Angelo S, Mignone F, Deantonio C, Di Niro R, Bordoni R, Marzari R, De Bellis G, Not T, Ferrara F, Bradbury A, et al. Profiling celiac disease antibody repertoire. Clin Immunol. 2013;148(1):99–109. doi: 10.1016/j.clim.2013.04.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


