Abstract
Vitamin C (ascorbic acid, AA) is a well-known regulator of bone and cartilage metabolism. However, the mechanisms of AA’s action in these tissues are only partly understood. In this study, we confirmed that AA contributes to bone and cartilage metabolism by showing decreased articular cartilage and trabecular bone in AA-deficient spontaneous fracture (sfx) mutant mice. In vitro, we found that AA exerts differential effects on chondrocyte and osteoblast differentiation. Since AA is known to increase levels of 5-hydroxymethylcytosine (5-hmC) and induce DNA demethylation via the ten-eleven translocases (TETs), and since prolyl hydroxylase domain-containing protein 2 (PHD2), a known mediator of AA’s effects in these tissues, is part of the same enzyme family as the TETs, we next investigated whether increases in 5-hmC might mediate some of these effects. All TETs and PHDs are expressed in chondrocytes and osteoblasts, and PHD2 is localized in both the cytoplasm and nucleus of the cell, lending plausibility to the hypothesis of altered 5-hmC content in these cells. We found that AA treatment increased levels of 5-hmC in both cell types globally, notably including promoter regions of osteoblast differentiation genes. Furthermore, inhibition of PHD2 decreased 5-hmC levels in chondrocyte differentiation gene promoters, and knockdown of Phd2 in chondrocytes reduced global 5-hmC levels, suggesting for the first time that PHD2 may itself directly mediate increases in 5-hmC in chondrocyte and osteoblast genes. Further investigation of this mechanism could lead to novel therapeutic approaches to treat debilitating diseases such as osteoarthritis and osteoporosis.
Introduction
Vitamin C (ascorbic acid, AA) is a known regulator of both bone and cartilage metabolism. Epidemiological evidence suggests links between vitamin C deficiency and risk of osteoporosis and osteoarthritis. For example, a recent systematic review and meta-analysis of observational studies found that greater dietary vitamin C intake was associated with decreased risks of hip fracture and osteoporosis and higher bone mineral density (BMD) at the femoral neck and lumbar spine, [1] and a case-control study showed that vitamin C is a possible risk factor for osteoarthritis in the knee. [2] Additionally, many studies have confirmed a key role for vitamin C in the regulation of osteoblasts [3–11] and chondrocytes. [12–14]
The importance of vitamin C for musculoskeletal tissues has been confirmed in animal models. We and others have previously published studies of AA-deficient rodents which had impaired osteoblast differentiation, reduced bone formation, and development of spontaneous fractures. [15–22] Studies on the mechanisms by which AA regulates gene expression in bone cells have identified both direct and indirect routes of vitamin C action. [23] For example, vitamin C is an antioxidant, and AA treatment of bone marrow stromal cells increases expression of osteoblast differentiation regulator osterix (Osx) by causing nuclear factor-E2-related factor-1 (Nrf1) to bind an antioxidant-responsive element (ARE) in the Osx promoter. [24] Another mechanism by which AA regulates gene expression is via its role as a cofactor of prolyl hydroxylase domain-containing protein 2 (PHD2). Our previous studies have shown that many of the skeletal and cartilage effects of AA are mediated via PHD2. [25] The most well studied mechanism of PHD2 involves the hypoxia signaling pathway. PHD2 hydroxylates HIF1α, thereby marking it for ubiquitin-mediated proteasomal degradation. In the absence of oxygen, PHD2 cannot function, leading to accumulation of HIF1α to transcriptionally regulate its downstream target genes. We have previously shown that proteasomal degradation is important for the ability of AA to induce osterix (Osx) expression in osteoblasts. [26] However, this mechanism may not explain all the effects of AA in bone and cartilage.
Another possible mechanism for AA regulation of gene expression involves DNA methylation status. When a gene’s promoter is enriched in methylated cytosine (5-methylcytosine, 5-mC), transcription of that gene is repressed. The ten-eleven translocation (TET) hydroxylase enzymes demethylate DNA; through a series of oxidation reactions, the 5-mC is converted to 5-hydroxymethylcytosine (5-hmC) and 5-formylcytosine (5-fC) and is eventually replaced by an unaltered cytosine, thus allowing the gene to be transcribed. The TETs are part of a family of 2-oxoglutarate- and iron-dependent dioxygenases which require AA to function. Indeed, AA has been shown to induce DNA demethylation via the TETs in embryonic stem cells and some cancers. [27–29] Furthermore, changes in 5-hmC levels have been observed in osteoarthritic human chondrocytes, [30] and CpG islands in promoter regions of chondrocyte differentiation genes were consistently hypomethylated in human synovium-derived mesenchymal stem cells, [31] suggesting that regulation of DNA demethylation may be important for musculoskeletal function.
Interestingly, the PHDs are part of the same enzyme family as the TETs, and a recent study has provided evidence that, in addition to its role as a reducing agent, L-ascorbate acts as a true co-substrate for PHD2 in the same way it does for the TETs. [32] Since AA can induce DNA demethylation, and since PHD2 mediates AA effects in bone and cartilage, we hypothesized that some of the AA effect in osteoblasts and chondrocytes may be due to TET- and/or PHD2-mediated increases in 5-hmC in the promoter regions of genes important for osteoblast and chondrocyte function. To test this hypothesis, we first examined the effects of AA on bone and cartilage at a phenotypic and gene expression level. Next, we measured levels of 5-hmC in osteoblasts and chondrocytes after AA treatment. Finally, we determined the levels of 5-hmC in the promoter regions of osteoblast and chondrocyte genes after AA treatment.
Materials and methods
Mice
In order to investigate the effect of AA deficiency in mice in vivo, we used mice which have a deletion in the GULO gene which encodes gulonolactone oxidase, a key enzyme in the AA synthesis pathway, and are thus AA-deficient. Since these mice develop spontaneous fractures, they are known as sfx mice. [18] AA-deficient sfx mice and their heterozygous, AA-replete littermates were fed an AA-free diet before skeletal and articular cartilage phenotype analysis via microCT and histology, respectively. In our previous studies, we have determined that there were no significant differences either in serum vitamin C levels or skeletal phenotype between heterozygous sfx/+ littermates and wild type littermates. Thus, using heterozygous littermates as controls allows us to use a breeding scheme (sfx/+ X sfx/sfx) which increases the yield of mutant mice to allow for higher sample sizes in experiments. Additionally, primary bone and cartilage cells were isolated from wild-type C57BL/6 mice and Phd2-floxed mice and grown in culture for in vitro experiments. Mice were housed at the VA Loma Linda Healthcare System Veterinary Medical Unit (Loma Linda, CA, USA) under standard approved laboratory conditions. All the procedures were performed with the approval of the Institutional Animal Care and Use Committee of the VA Loma Linda Healthcare System. For euthanasia, animals were exposed to CO2 prior to cervical dislocation.
Micro-computed tomography
The trabecular bone phenotype of 8-week-old sfx mice and their control littermates was evaluated at the primary and secondary femoral spongiosa via micro-computed tomography (microCT, Scanco vivaCT 40, Scanco Medical, Bruttisellen, Switzerland). The bones were scanned at a resolution of 10.5 μm with a 55kVp X-ray for measurement of trabecular bone microstructure. For primary spongiosa, a region of 0.36 mm starting from the end of the growth plate of the proximal femur was analyzed. For secondary spongiosa, a 1.2 mm region starting 0.36 mm distal to the growth plate was analyzed. The exact numbers and location of the slices used for analysis were corrected for bone length such that the analyzed regions were anatomically comparable between samples. Trabecular bone parameters were analyzed as described previously. [25]
Histology
The articular cartilage phenotype of 8-week-old sfx mice and their control littermates was evaluated at the femoral and tibial aspects of the knee joint. The knee joints were decalcified in 20% EDTA at 4°C for 4 weeks, dehydrated using alcohol, and embedded in paraffin. The paraffin-embedded knee joints were sectioned at a thickness of approximately 4 μm, and the sections were stained with Safranin-O using standard protocols. Articular cartilage area and width were quantitated (18–30 sections per knee joint) by a blinded observer using the OsteoMeasure system (OsteoMetrics, Atlanta, GA, USA). OARSI scoring was conducted as previously described. [33, 34]
Cell culture
Primary mouse calvarial osteoblasts and rib, growth plate, or epiphyseal chondrocytes were isolated from C57BL/6 mice or Phd2-floxed mice via enzymatic digestion with collagenase I (2 mg/mL) and hyaluronidase (1 mg/mL) or collagenase D (2 mg/mL), respectively. Primary cells and the ATDC5 (Abgent, San Diego, CA, USA) and MC3T3-E1 (ATCC, Manassas, VA, USA) cell lines were grown in AA-free alpha-MEM or DMEM/F-12 growth medium supplemented with 10% fetal bovine serum, penicillin (100 units/mL), and streptomycin (100 μg/mL) to 80% confluence before beginning experiments. ATDC5 cells were grown to confluence and treated with ascorbic acid and β-glycerophosphate; they were not induced to differentiate with, e.g., insulin and dexamethasone. Cells from Phd2-floxed mice were transduced according to previously published procedures [35] with adenoviral GFP or iCre vectors (produced by our colleague Weirong Xing) to knock down expression of Phd2. Knockdown was measured via real-time RT-PCR as described below. For each assay described below, cells were synchronized by serum starvation for 24 hours before 3-day treatment with 50 μg/mL (284 μM) AA and 10 mM β-glycerophosphate (unless other treatment durations and concentrations are specified). Chemical inhibition of PHD2 was achieved by treatment with PHD2-specific inhibitor Iox2 (20 μM; Cayman Chemical, Ann Arbor, MI, USA). To make sure that the effects of AA treatment are due to the AA we added as a treatment, all experiments were conducted in growth media which contained no AA.
Cell proliferation assay
Cells were plated at a density of 3,000 cells/well in a 96 well plate. One day later, a 24 hour period of serum-free synchronization was begun, followed by addition of media containing AA (50 μg/mL; 284 μM) and 10 mM β-glycerophosphate. Proliferation was measured 48 hours later using the CyQUANT kit according to the manufacturer’s instructions (ThermoFisher Scientific, Waltham, MA, USA).
Alkaline phosphatase (ALP) activity assay
Briefly, cells were plated at a density of 5,000 cells/well in a 96 well plate and allowed to proliferate for 24 hours before a 24 hour period of serum-free synchronization followed by addition of AA (doses ranging from 0–250 μg/mL, 0–1.42 mM) with 10 mM β-glycerophosphate. After 72 hours of treatment, cells were permeabilized with Triton X-100, PNPP substrate was added, and absorbance was read in a plate reader at intervals 0, 1, 3, and 5 hour time points. The difference in absorbance at 490 nm and 410 nm was used to calculate enzyme activity over time, which was normalized to total protein concentration as measured by a Pierce™ BCA kit according to manufacturer’s instructions (ThermoFisher Scientific, Waltham, MA, USA).
RNA extraction and quantitation
RNA samples were extracted from primary mouse osteoblasts and chondrocytes as well as the ATDC5 and MC3T3-E1 cell lines with TRI Reagent (MRC, Cincinnati, OH, USA) and the E.Z.N.A.® Total RNA Kit I (Omega Bio-tek, Norcross, GA, USA) according to the manufacturers’ instructions. An aliquot of RNA (300 ng) was reverse-transcribed using SuperScript II® reverse transcriptase (Invitrogen, Grand Island, NY, USA). Quantitative real-time RT-PCR was performed as previously described. The DDCT method was used to calculate relative gene expression with the housekeeping gene PPIA serving as an internal control as previously described. [36] Primer sequences can be found in Table 1.
Table 1. Primer sequences for real-time PCR.
| Gene | Forward | Reverse |
|---|---|---|
| Aggrecan | 5′-GACCAGGAAGGGAGGAGTAG-3′ | 5′-CAGCCGAGAAATGACACC-3′ |
| ALP | 5′-ATGGTAACGGGCCTGGCTACA-3′ | 5′-AGTTCTGCTCATGGACGCCGT-3′ |
| BiP | 5′-TTCAGCCAATTATCAGCAAACTCT-3′ | 5′-TTTTCTGATGTATCCTCTTCACCAGT-3′ |
| BSP | 5′-CTCTACTCCGTCTGCACAACA-3′ | 5′-TAAGCTCGGTAAGTGTCGCCA-3′ |
| CHOP | 5′-CCACCACACCTGAAAGCAGAA-3′ | 5′-AGGTGAAAGGCAGGGACTCA-3′ |
| Col10 | 5′-ACGGCACGCCTACGATGT-3′ | 5′-CCATGATTGCACTCCCTGAA-3′ |
| Col2 | 5′-TGGCTTCCACTTCAGCTATG-3′ | 5′-AGGTAGGCGATGCTGTTCTT-3′ |
| Gpx2 | 5′-ACCGATCCCAAGCTCATCAT-3′ | 5′-CAAAGTTCCAGGACACGTCTGA-3′ |
| GSR | 5′-GCTATGCAACATTCGCAGATG-3′ | 5′-AGCGGTAAACTTTTTCCCATTG-3′ |
| Gsta2 | 5′-CGTCCACCTGCTGGAACTTC-3′ | 5′-GCCTTCAGCAGAGGGAAAGG-3′ |
| Ihh | 5′-CCCCAACTACAATCCCGACATC-3′ | 5′-CGCCAGCAGTCCATACTTATTTCG-3′ |
| Jmjd1a | 5′-CCAGGAGAAGACTTCAGAGACATG-3′ | 5′-GGTGTACTCAGGCAGTGGAATG-3′ |
| Jmjd2b | 5′-GGCCAAGATCATTCCACCCA-3′ | 5′-CCCACAGTCATGGCCTTCTT-3′ |
| Osx | 5′-TCCTCTCTGCTTGAGGAAGAAG-3′ | 5′-GAGTCCATTGGTGCTTGAGAAG-3′ |
| Phd1 | 5′-GGAACCCACATGAGGTGAAG-3′ | 5′-AACACCTTTCTGTCCCGATG-3′ |
| Phd2 | 5′-GAAGCTGGGCAACTACAGGA-3′ | 5′-CATGTCACGCATCTTCCATC-3′ |
| Phd3 | 5′-AAGTTACACGGAGGGGTCCT-3′ | 5′-GGCTGGACTTCATGTGGATT-3′ |
| PPIA | 5′-CCATGGCAAATGCTGGACCA-3′ | 5′-TCCTGGACCCAAAACGCTCC-3′ |
| Tet1 | 5′-AGGAAATGCGAGGTGCTCAA-3′ | 5′-TCCCCATGACCACGTCTACT-3′ |
| Tet2 | 5′-AGCTCGAAAGCGTTCCTCTC-3′ | 5′-GAAGGTGCCTCTGGAGTGTT-3′ |
| Tet3 | 5′-GCATGTACTTCAACGGCTGC-3′ | 5′-TCGCCACATCCTCATTGGTC-3′ |
Protein extraction and quantitation
Cytoplasmic and nuclear protein samples were extracted from MC3T3-E1 pre-osteoblasts using a previously described method. [37] Briefly, cells were scraped, centrifuged and resuspended in PBS, and lysed with a buffer containing NP-40 on ice for 10 minutes. Nuclei were pelleted by centrifugation (2,500 RCF at 4°C for 10 minutes), and the supernatant containing cytosolic proteins was collected. Nuclei were washed and resuspended in a glycerol-containing buffer with 0.4 M NaCl followed by occasional vortexing at 4°C for 20 minutes. Following centrifugation (12,000 RCF at 4°C for 10 minutes), the supernatant containing nuclear proteins was collected. Protein samples were analyzed by immunoblotting of PVDF membranes with protein transferred from SDS-PAGE gels (antibodies used were as follows: EGLN1/PHD2, 48355, Cell Signaling, Danvers, MA, USA; β-actin, A1978, Sigma-Aldrich, St. Louis, MO, USA; histone 3, H0164, Sigma-Aldrich, St. Louis, MO, USA) followed by chemiluminescent imaging. Blots were quantitated using ImageJ.
Immunofluorescence microscopy
ATDC5 pre-chondrocytes and primary mouse rib chondrocytes were plated at a density of 20,000 cells/well on 8-well Lab-Tek™ Chamber Slides™ (177402, Thermo Fisher Scientific, Waltham, MA, USA), allowed to proliferate for 24 hours, and synchronized by serum starvation before treatment with AA. After three of days of treatment, cells were washed with PBS, fixed with 4% paraformaldehyde at room temperature for 10 minutes, washed again with PBS, and permeabilized with 0.5% Triton X-100 in 1X PBS for five minutes. Permeabilization buffer was washed off with PBS before blocking with 2.5% normal horse serum (from the VectaFluor™ R.T.U. Anti-Rabbit IgG kit, Vector Laboratories, Burlingame, CA, USA) for 1 hour at room temperature. Cells were incubated with primary antibody (PHD2, Cell Signaling; 1:200) in 1% normal horse serum overnight at 4°C. The next day, cells were washed with PBS three times for five minutes each, incubated with fluorophore-conjugated secondary antibodies (ATDC5: VectaFluor™ R.T.U. DyLight® 488 Anti-Rabbit IgG [DI-1788, Vector Laboratories]; rib chondrocytes: VectaFluor™ R.T.U. DyLight® 594 Anti-Rabbit IgG [DI-1794, Vector Laboratories]) for one hour at room temperature, washed again with PBS, counterstained with DAPI (286 nM), and rinsed again with PBS. Slide chambers and gaskets were removed, and slides were mounted with Fluoromount-G® (0100-01, SouthernBiotech, Birmingham, AL, USA). Cells were imaged using an Olympus Fluoview FV3000 confocal laser scanning microscope (Olumpus, Center Valley, PA, USA). Negative controls were processed in the same manner, excluding primary antibody in the primary antibody step. ATDC5 negative controls were processed under identical conditions but at a later time than the experimental samples.
5-hmC quantitation
After synchronization by serum starvation for 24 hours, chondrocytes and osteoblasts were treated with AA (50–100 μg/mL; 284–568 μM) for 3 days. DNA was extracted using the DNEasy Blood & Tissue DNA extraction kit (Qiagen, Germantown, MD, USA). 5-hmC was measured by dot blot as follows. Briefly, DNA was deposited on nitrocellulose membranes in 1 μL droplets containing a series of 2-fold dilutions beginning at 100 ng/μL, UV crosslinked to the membrane (120,000 μJ/cm2), dried at 80°C, rinsed with 2X saline-sodium citrate, and probed with a 1:1000 dilution of 5-hmC antibody (Diagenode, Denville, NJ, USA) for 1 hour before chemiluminescent imaging. Blots were quantitated using ImageJ. 5-hmC ELISA was performed using a kit (Epigentek, Farmingdale, NY, USA) according to manufacturer’s instructions.
Quantitation of promoters enriched in 5-hmC
Extracted DNA (20 μg/sample) was diluted in 300 μL 10 mM Tris-HCl (pH 8.5) and sonicated with 15 pulses of 20 seconds at power level 2 (Sonic Dismembrator Model 100, Fisher Scientific, PA, USA), with a 20-second pause on ice between each pulse. The sheared DNA samples were fragmented to 100–500 bp in size and visualized by ethidium bromide staining after electrophoresis on a 2% agarose gel.
5-hmC was enriched using a kit according to manufacturer’s instructions (Hydroxymethyl Collector, Active Motif, Carlsbad, CA). Briefly, 2.5 μg of fragmented DNA was spiked with 5-hmC positive control DNA (human APC gene) with a ratio of 1 to 20,000. 10% of the spiked DNA samples were set aside as input. The DNA samples were glycosylated with the presence of 0.15 mM UDP-azide-glucose and 8 units of β-glucosyltransferase in 50 μL reaction volume at 37°C for 1 hour. A negative control reaction omitting UDP-azide-glucose was also set up for each DNA sample. The reaction was further incubated with 20 μL biotin conjugation solution at 37°C for 1 hour. The reactions were purified with the columns provided by the kit. The hydroxymethylated DNA fragments were then captured by streptavidin beads and further purified with provided columns.
CpG islands in promoter regions of the genes of interest were retrieved from the UCSC genome browser, and primer pairs were designed within the CpG islands with an average Tm of 60°C and PCR products of 100–200 bp. 5 μL of the 5-hmC enriched DNA were used to perform real-time PCR with a 20 μL SYBR Green reaction mix. Enrichment of 5-hmC DNA was analyzed with DDCT method, and the APC control primers were used for normalization.
Statistical analysis
Results are expressed as mean ± SEM. Data were analyzed using Student’s t-tests or one-way ANOVA followed by pairwise t-tests with Bonferroni adjustments for multiple comparisons. Statistical software used were Microsoft Excel and R 3.5.3. [38].
Results
AA-deficient sfx mice had less articular cartilage
To evaluate the importance of vitamin C in articular cartilage, we compared histologic slides of knee joints stained with Safranin-O from AA-deficient sfx mice and control littermates at 8 weeks of age (Fig 1A). At the knee joint, AA-deficient sfx mice had significantly decreased articular cartilage area (21% at the femoral aspect and 15% at the tibial aspect, Fig 1B). Similarly, both the femoral and tibial aspects of the knee joint had decreased articular cartilage width (8%, Fig 1C). These reductions in articular cartilage area and width are characteristics of an osteoarthritis-like phenotype, a finding further confirmed by OARSI scoring (Fig 1D).
Fig 1. AA-deficient sfx mice had decreased articular cartilage.
(A) Representative sections of articular cartilage from control sfx/+ mice and AA-deficient sfx/sfx mice stained with Safranin-O. Articular cartilage was quantitated using the OsteoMeasure software, and sfx/sfx mice had less articular cartilage area (B) and width (C) at the femur and tibia. OARSI scores (D) are shown for sfx/+ and sfx/sfx mice. Results are presented as mean ± SEM. *P < 0.05 vs. control (Student’s t-test), n = 6 per group.
AA-deficient sfx mice had less trabecular bone
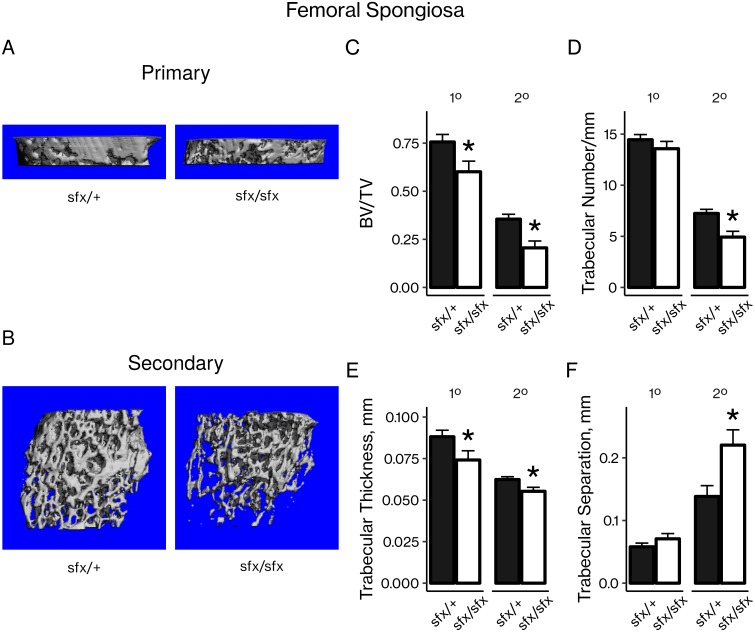
The primary and secondary spongiosa of 8-week-old AA-deficient sfx mice were analyzed by microCT (Fig 2A and 2B). AA-deficient mice had 20% and 42% less bone volume adjusted for tissue volume (BV/TV) at the primary and secondary spongiosa, respectively, compared to their control littermates (Fig 2C). These decreases were explained by a significant reduction in trabecular number at the secondary spongiosa (Fig 2D), significant reductions in trabecular thickness at both the primary and secondary spongiosa (Fig 2E), and a significant increase in trabecular separation at the secondary spongiosa (Fig 2F).
Fig 2. AA-deficient sfx mice had less trabecular bone at the femoral primary and secondary spongiosa.
Representative microCT images of (A) primary and (B) secondary spongiosa from control sfx/+ mice and AA-deficient sfx/sfx mice. Scans were quantitated by (C) bone volume adjusted for tissue volume (BV/TV), (D) trabecular number, (E) trabecular thickness, and (F) trabecular separation. Results are presented as mean ± SEM. *P < 0.05 vs. control (Student’s t-test), n = 6 per group.
AA had opposite effects on proliferation and differentiation in chondrocytes vs. osteoblasts
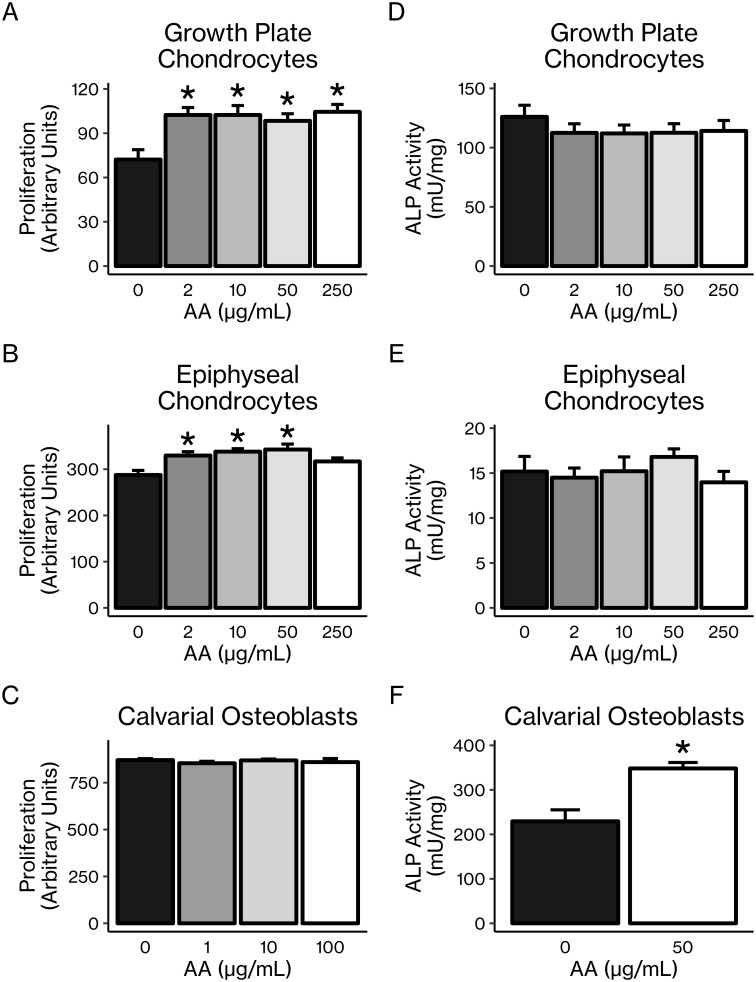
Treatment of chondrocytes in vitro with AA led to significant 20–40% (P < 0.05) increases in cellular proliferation of primary growth plate and epiphyseal chondrocytes (Fig 3A and 3B), although this effect was not seen in the ATDC5 chondrocytic cell line under the specific conditions we observed (S1A Fig). By contrast, in vitro AA treatment had no effect on proliferation of primary calvarial osteoblasts (Fig 3C) or the pre-osteoblastic MC3T3 cell line (S1B Fig). However, the opposite was true of AA’s effect on ALP activity, a marker of differentiation. While AA treatment had no effect on ALP activity in either primary chondrocytes (Fig 3D and 3E) or the ATDC5 cell line under the conditions we observed (S1C Fig), ALP activity was significantly increased by 30–160% (P < 0.05) in both primary calvarial osteoblasts (Fig 3F) and the MC3T3-E1 cell line (S1D Fig).
Fig 3. AA increased proliferation of primary chondrocytes and differentiation of osteoblasts in vitro.
Proliferation of primary (A) growth plate chondrocytes and (B) epiphyseal chondrocytes was increased by AA treatment. Proliferation of (C) primary calvarial osteoblasts was unaffected by AA treatment. ALP activity of (D) growth plate chondrocytes and (E) epiphyseal chondrocytes was not changed by AA treatment, while ALP activity of (F) calvarial osteoblasts was increased by AA treatment. Results are presented as mean ± SEM. *P < 0.05 vs. control (pairwise t-tests with Bonferroni corrections for multiple comparisons [A–E] or Student’s t-test [F]), n = 6–8 per group.
AA treatment regulated markers of osteoblast and chondrocyte differentiation and maturity
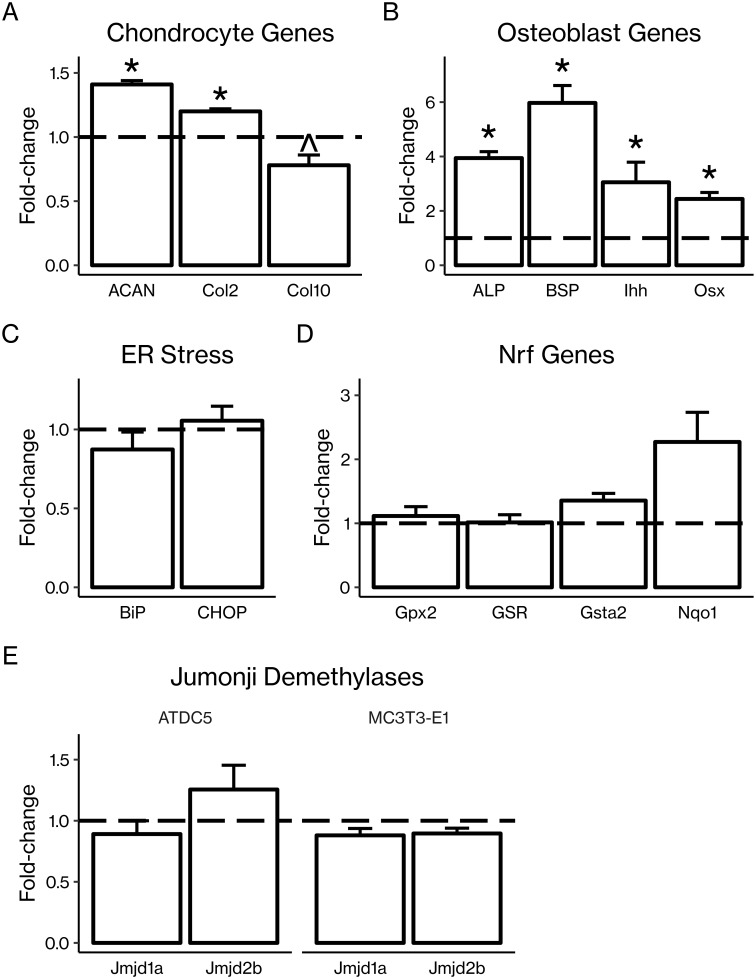
In primary mouse articular chondrocytes, AA treatment increased mRNA expression of immature chondrocyte markers Acan and Col2 1.2–1.4-fold (P < 0.05) while tending to decrease expression of mature chondrocyte marker Col10 0.8-fold (P < 0.1; Fig 4A). Consistent with the earlier finding that AA increased ALP activity in osteoblastic cells, we found that AA treatment in primary mouse calvarial osteoblasts produced 2.5–6-fold increases (P < 0.05) in mRNA expression of several markers of osteoblast differentiation including Alp, Bsp, Ihh, and Osx (Fig 4B).
Fig 4. AA regulated expression of chondrocyte and osteoblast genes at the mRNA level.
(A) Primary mouse articular chondrocytes were treated with AA (50 μg/mL) before RNA extraction and RT-qPCR. AA treatment significantly increased expression of immature chondrocyte genes aggrecan (AGCN) and type 2 collagen (Col2) but not mature chondrocyte differentiation marker type 10 collagen (Col10). (B) Primary mouse calvarial osteoblasts were treated with AA (50 μg/mL) before RNA extraction and RT-qPCR. AA treatment significantly increased expression of osteoblast differentiation genes alkaline phosphatase (ALP), bone sialoprotein (BSP), Indian hedgehog (Ihh), and osterix (Osx). ATDC5 chondrocytes were treated with AA before RNA extraction RT-qPCR, and no changes in (C) ER stress markers BiP or CHOP or (D) Nrf2 responsive genes Gpx2, GSR, Gsta2, or Nqo1 were observed. (E) AA treatment did not regulate expression of Jumonji family demethylases. demethylases. ATDC5 chondrocytes and MC3T3-E1 pre-osteoblasts were treated with AA before RNA extraction RT-qPCR. No significant changes in Jmjd1a or Jmjd2b were found. Results are presented as mean fold-change ± SEM. *P < 0.05 vs. control, ^P = 0.06 (Student’s t-test), n = 3–4 per group.
Differential regulation of TET and PHD expression in chondrocytes and osteoblasts
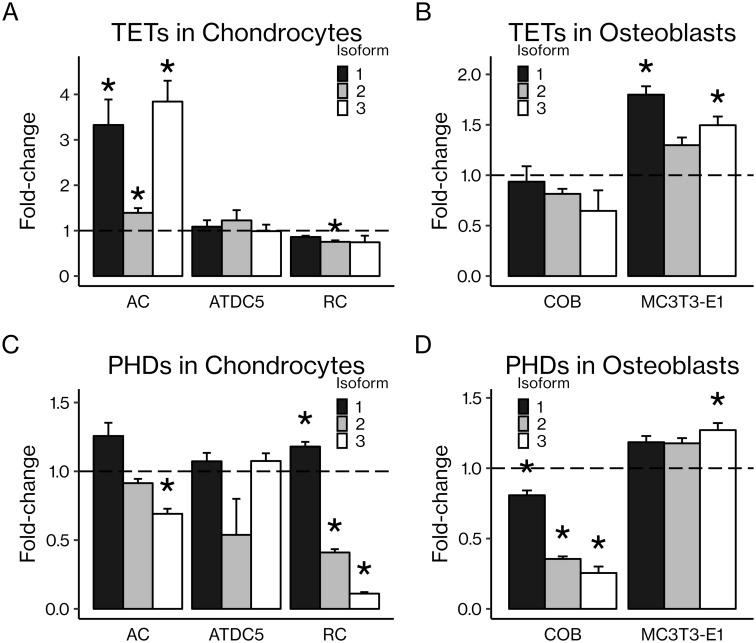
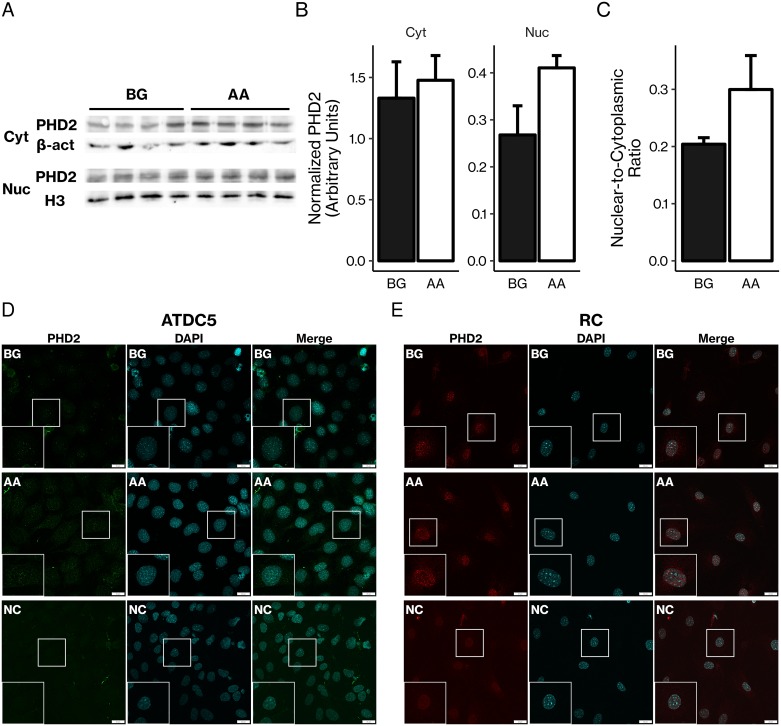
All isoforms of TETs and PHDs were detected at the mRNA level by real-time PCR in primary and cell line chondrocytes and osteoblasts (Fig 5). Absolute mRNA levels are provided in S1 Table. In vitro AA treatment had varying effects on each isoform; TETs tended to increase with AA treatment while PHDs tended to decrease. For example, AA increased expression of all three TET isoforms in articular chondrocytes and in the MC3T3-E1 pre-osteoblastic cell line. AA decreased expression of Phd3 in articular chondrocytes and all PHDs in calvarial osteoblasts. The varying effects of AA on TET and PHD mRNA expression appear to be dependent on cell type as well as stage of differentiation. Furthermore, PHD2 was expressed at the protein level in both the cytoplasm and nucleus of MC3T3-E1 calvarial osteoblasts, and AA treatment tended to increase the nuclear-to-cytoplasmic ratio of PHD2 expression (Fig 6A–6C), although this effect was not statistically significant. Additionally, immunofluorescent confocal micrographs of ATDC5 chondrocytes and rib chondrocytes stained for PHD2 show that PHD2 is present in both the cytoplasm and nucleus (Fig 6D and 6E).
Fig 5. All TETs and PHDs are expressed in chondrocytes and osteoblasts at the mRNA level, and expression of many TETs and PHDs is regulated by AA.
Primary articular chondrocytes (AC), ATDC5 chondrocytes, primary rib chondrocytes (RC), primary calvarial osteoblasts (COB), and MC3T3-E1 pre-osteoblasts were cultured in vitro and treated with AA (50 μg/mL) before RNA extraction and RT-qPCR. Expression of TETs in (A) chondrocytes and (B) osteoblasts as well as PHDs in (C) chondrocytes and (D) osteoblasts are reported. Results are presented as mean fold-change ± SEM. *P < 0.05 vs. control (Student’s t-test), n = 3–4 per group.
Fig 6. PHD2 is expressed in both the cytoplasm and nucleus at the protein level.
(A) MC3T3-E1 pre-osteoblasts were treated with β-glycerophosphate (BG) with or without AA before cytoplasmic (Cyt) and nuclear (Nuc) protein extraction and SDS-PAGE followed by immunoblotting. Loading controls were β-actin (β-act) for the cytoplasmic fraction and histone 3 (H3) for the nuclear fraction. (B) Immunoblots were quantitated with ImageJ, and signals from each fraction and treatment group were normalized to the respective loading control signals to produce quantitated, normalized PHD2 levels in the cytoplasmic and nuclear fractions. (C) The nuclear fraction signal was divided by the cytoplasmic fraction signal to give an indirect measure of the ratio of PHD2 levels in the nucleus vs. cytoplasm. AA treatment did not significantly change the nuclear-to-cytoplasmic ratio (P = 0.2, Student’s t-test). Results are presented as mean ± SEM. n = 4 per group. (D) ATDC5 chondrocytes immunostained for PHD2. Inserts are magnifications of indicated regions. NC = negative control. Green = PHD2; blue = DAPI. (E) Rib chondrocytes immunostained for PHD2. Inserts are magnifications of indicated regions. NC = negative control. Red = PHD2; blue = DAPI. Scale bars = 20 μm.
AA treatment did not regulate markers of ER stress or Nrf2 signaling
Vitamin C is known to modulate expression of antioxidant response element (ARE)-containing genes in part through the nuclear factor erythroid-derived 2-related factor (Nrf) proteins. [24, 39] In addition, crosstalk between Nrf2 and endoplasmic reticulum (ER) stress signaling has been established. [40] To determine whether AA’s effects may be mediated via regulation of ER stress or Nrf2 signaling, we measured expression of ER stress markers BiP and CHOP and Nrf responsive genes Gpx2, GSR, Gsta2, and Nqo1 in ATDC5 chondrocytes after AA treatment, and we found no significant changes (Fig 4C and 4D).
AA treatment increased 5-hmC levels in osteoblasts and chondrocytes
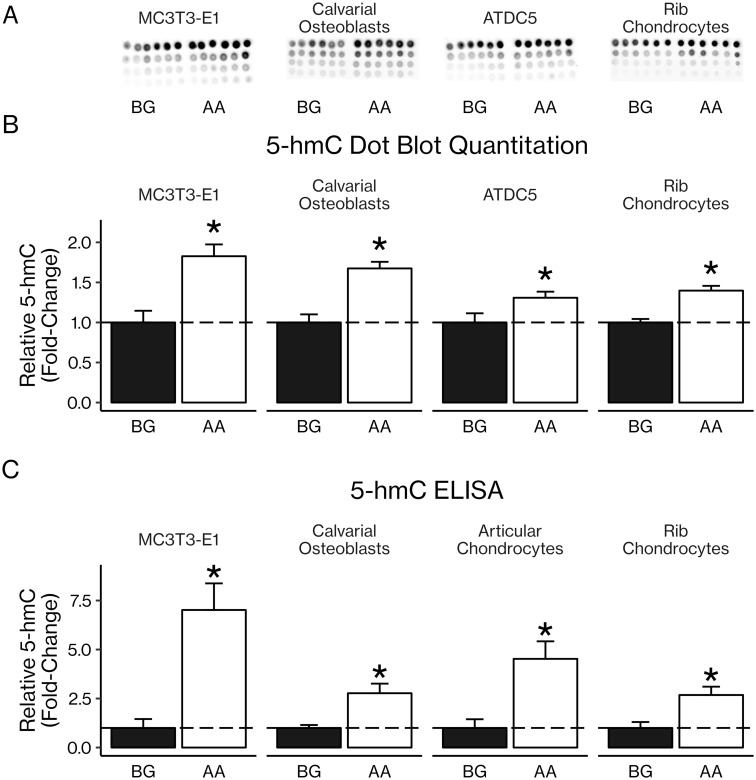
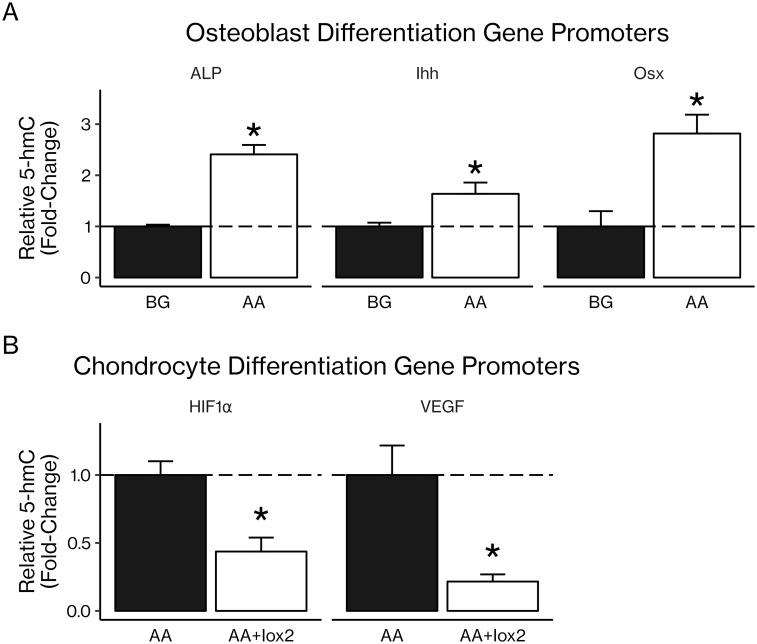
Three-day treatment of primary and cell line osteoblasts and chondrocytes with AA led to increased levels of 5-hmC, a potential marker of DNA demethylation, as measured by dot blot (30–90% increases, P < 0.05; Fig 7A and 7B). This result was confirmed using ELISA with an antibody specific for 5-hmC: AA treatment increased levels of 5-hmC (160–790% increases, P < 0.05; Fig 7C). To determine whether AA-induced increases in 5-hmC occurred in genes involved in osteoblast differentiation, we used a 5-hmC antibody to pull down DNA rich in 5-hmC and ran real-time PCR with primers specific for CpG-rich regions of promoters of interest. We found that AA treatment increased levels of 5-hmC in the promoter regions of osteoblast differentiation genes Alp, Ihh, and Osx (1.6–2.4-fold increases, P < 0.05; Fig 8A). In order to determine whether PHD2 could be involved in this increase in 5-hmC, we treated ATDC5 chondrocytes with AA and Iox2, a PHD2-specific inhibitor. We found that inhibition of PHD2 significantly reduced 5-hmC content in the promoter regions of chondrocyte differentiation genes Hif1α and Vegf (0.2–0.4-fold decreases, P < 0.05; Fig 8B).
Fig 7. AA increased 5-hmC levels in osteoblasts and chondrocytes.
(A) MC3T3-E1 pre-osteoblasts, primary calvarial osteoblasts, ATDC5 chondrocytes (not induced with, e.g., insulin and dexamethasone), and primary rib chondrocytes were treated with β-glycerophosphate (BG) with or without AA before DNA extraction and dot blotting using an antibody specific for 5-hmC. Each replicate is in a column on the dot blots, and each row indicates a serial two-fold dilution of the sample above it, beginning at 100 ng of DNA in each dot on the top row. (B) Dot blots were quantitated with ImageJ. (C) 5-hmC levels were also measured using an ELISA. Results are presented as mean fold-change ± SEM. *P < 0.05 (Student’s t-test), n = 6 per group for dot blot; technical issues during the ELISA led to several readings being invalidated, leaving the following n: MC3T3-E1 BG = 2, AA = 5; COB BG = 4, AA = 4; RC BG = 4, AA = 4; AC BG = 3, AA = 2.
Fig 8. Increased 5-hmC occurs in osteoblast and chondrocyte gene promoters.
After treatment with β-glycerophosphate (BG), BG with AA, or BG with AA and Iox2, a PHD2-specific inhibitor, DNA was extracted from MC3T3-E1 pre-osteoblasts and primary rib chondrocytes. An antibody specific for 5-hmC was used to pull down DNA rich in 5-hmC, and real-time PCR with primers for CpG-rich areas of osteoblast and chondrocyte gene promoters was used to determine the 5-hmC content of those promoters as an indirect measure of their demethylation status. (A) AA treatment significantly increased 5-hmC levels in the promoters of osteoblast differentiation genes alkaline phosphatase (ALP), Indian hedgehog (Ihh), and osterix (Osx). (B) Inhibition of PHD2 with Iox2 significantly decreased 5-hmC levels in the promoters of chondrocyte genes HIF1α and VEGF. Results are presented as mean fold-change ± SEM. *P < 0.05 (Student’s t-test), n = 3 per group.
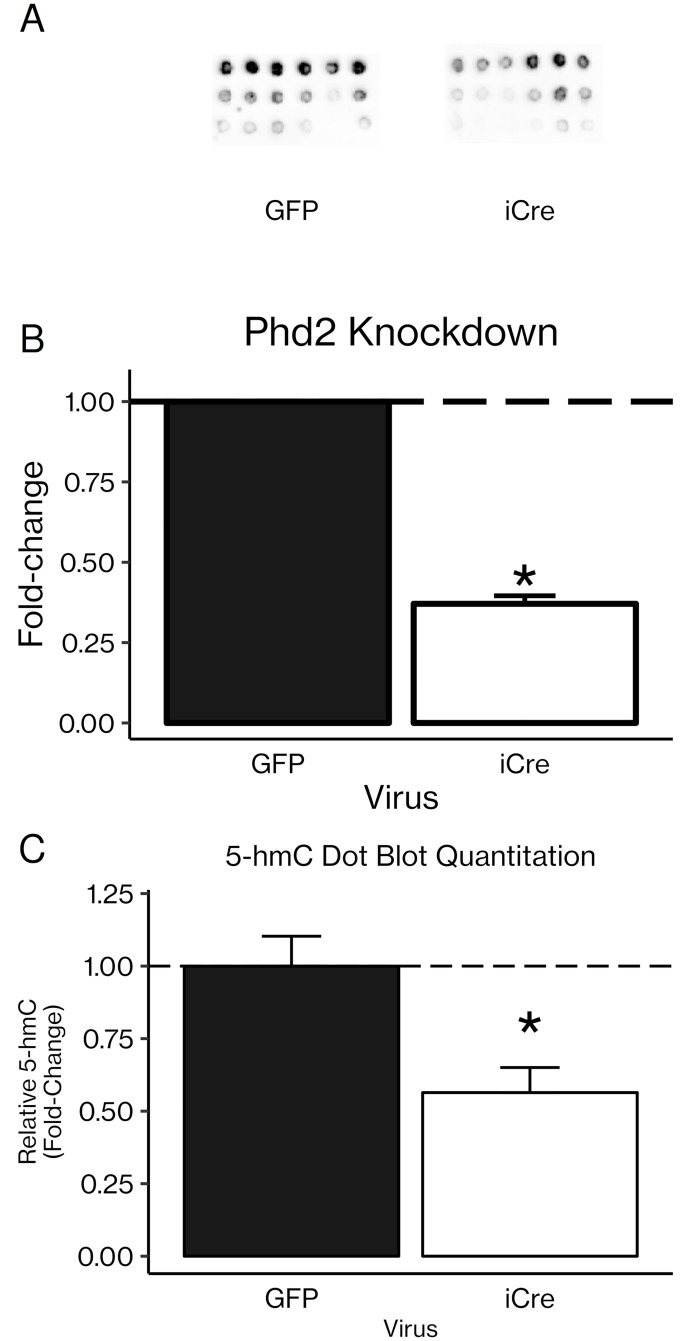
To further confirm whether PHD2 could itself mediate changes in 5-hmC levels, we used an adenoviral iCre vector to knock down Phd2 in rib chondrocytes from Phd2-floxed mice (60% knockdown, P < 0.001). Knocking down Phd2 led to a 0.56-fold decrease in 5-hmC (P < 0.05; Fig 9).
Fig 9. Knockdown of PHD2 significantly reduced 5-hmC levels in primary rib chondrocytes.
(A) Primary rib chondrocytes from Phd2-floxed mice were transduced with an adenoviral vector carrying iCre or GFP as a control. DNA was extracted and dot blotted as in Fig 7. (B) Phd2 gene expression after Phd2-floxed cells were transduced with adenoviral iCre. *P < 0.01 vs. adenoviral GFP (Student’s t-test), n = 3 per group. (C) Dot blots were quantitated with ImageJ. NB: The images shown in (A) are several exposures stacked to make differences more visible to the eye, while the quantitations in (C) were performed on single, non-stacked images. Results are presented as mean fold-change ± SEM. *P < 0.05 (Student’s t-test), n = 6 per group.
We and others have shown that vitamin C-mediated activation of PHD2 destabilizes HIF1α via proline hydroxylation-mediated proteasomal degradation. [26] Since HIF1α is a known regulator of Jumonji family demethylases, we evaluated whether AA regulates expression of Jumonji demethylases in osteoblasts or chondrocytes. Serum-free cultures of ATDC5 chondrocytes and MC3T3-E1 pre-osteoblasts were treated with vitamin C or vehicle, and RNA was extracted and used for real-time RT-PCR. We found expression of neither Jmjd1a nor Jmjd2b was affected by AA treatment in these cells (Fig 4E).
Discussion
The association between vitamin C and human health has a long history. Diseases resulting from vitamin C deficiencies have been observed as far back as 1500 BC, and one of the first documented clinical trials in the 18th century involved treating scurvy, a debilitating condition with musculoskeletal symptoms caused by a lack of vitamin C, with AA-rich citrus fruits. [23] More recently, epidemiological studies have examined the relationship between AA and bone health. Several cross-sectional studies have observed positive correlations between dietary vitamin C intake and bone mineral density (BMD) in postmenopausal women, [41–44] although in other studies this correlation was not significant, [45–48] and one study actually reported a significant negative correlation. [49] These inconsistencies are likely due to differences in study design and measurement techniques, however, and the fact the that the majority of studies have reported either significant positive correlations or positive but not significant correlations between vitamin C and skeletal outcomes suggest a likely complex but beneficial effect of vitamin C on bone. [23]
Similarly, several mouse or rat models have demonstrated the importance of AA for the skeleton. Unlike humans, who require dietary vitamin C, many animals including rodents are able to synthesize their own vitamin C. The skeletal phenotypes of several transgenic rodent models with mutations in the AA biosynthesis pathway and which are therefore AA-deficient have been analyzed. Vitamin C deficiency in these transgenic rodents has resulted in spontaneous fractures, missing trabeculae, and reduced cortical zones. [15–22] The fact that these skeletal phenotypes can be rescued by dietary supplementation with vitamin C suggests that it is vitamin C deficiency itself, not a secondary effect of the various mutations, which results in decreased BMD, trabecular number, and cortical thickness. Furthermore, these effects seem to be primarily due to decreased bone formation resulting from defective osteoblast function in AA-deficient animals. [23] Thus, vitamin C is an important regulator of bone health.
Consistent with these clinical observations, our findings show that AA-deficient sfx mice had decreased articular cartilage (Fig 1) and trabecular bone (Fig 2). However, the mechanisms for these changes have yet to be fully elucidated. In fact, our results suggest that vitamin C may have different effects in osteoblasts and chondrocytes. At the cellular level, AA is known to affect both chondrocytes and osteoblasts. In chondrocytes, AA has been shown to act via ERK signaling to increase chondrocyte proliferation and expression of chondrocyte differentiation genes. [12–14] Similarly, many studies demonstrate the ability of AA to stimulate osteoblast proliferation and differentiation, [3–11] and several of these studies implicate collagen synthesis as a mediator of these effects.
In this current study, we found that while in vitro AA treatment increased chondrocyte proliferation, AA had no significant effect on osteoblast proliferation (Fig 3A–3C). Conversely, AA had no effect on ALP activity in chondrocytes but significantly increased ALP activity in osteoblasts (Fig 3D–3F). The lack of significant effects of AA on proliferation in osteoblasts and differentiation in chondrocytes, which is in contrast to the findings of previous studies cited above, may be due to differences in the stage of cell maturation as well as treatment conditions and duration. Nevertheless, these results suggest that the primary effect of AA in chondrocytes is to maintain them in an immature, proliferative state while the primary effect of AA in osteoblasts is to induce differentiation.
Our mRNA expression data further support this explanation of the effects of AA on chondrocytes vs. osteoblasts. In chondrocytes, AA treatment increased expression of immature chondrocyte markers while tending to decrease expression of a marker of chondrocyte maturity (Fig 4A). In osteoblasts, AA treatment led to significant increases in markers of osteoblast differentiation (Fig 4B). Both of these effects would support the observed bone and cartilage phenotypes of sfx mice; the decrease in trabecular bone results from a lack of differentiated osteoblasts in the AA-deficient mice, and the decrease in articular cartilage results from a defect in chondrocyte differentiation and/or proliferation, potentially involving a lack of immature, proliferative chondrocytes to replace the cartilage as it is worn away in the AA-deficient mice.
In terms of molecular mechanisms of vitamin C actions, AA is a well-known antioxidant, and we have previously demonstrated the ability of AA to induce osteoblast differentiation by increasing binding of nuclear factor-E2-related factor-1 (Nrf1) to antioxidant response elements (AREs) in the osterix promoter. [24, 39] Both Nrf2 signaling and the induction of endoplasmic reticulum stress have been suggested as additional mechanisms of AA’s action. However, we did not find any significant changes in ER stress or Nrf genes in AA-treated ATDC5 chondrocytes (Fig 4C and 4D), suggesting that these are not primary mechanisms of AA’s action in these cells. An additional well established biological role of AA is as a cofactor for prolyl and lysyl hydroxylases. Hydroxylation of specific proline and lysine residues within the collagen molecule is required for collagen cross-linking, a crucial step in the collagen maturation process which contributes to building bone and cartilage. [3, 23, 50, 51] In addition to the regulation of collagen cross-linking, however, proline hydroxylation is known to play a role in regulation of other intracellular signaling pathways. AA is a cofactor for prolyl hydroxylase domain-containing (PHD) proteins, a family of dioxygenases which depend upon the presence of 2-oxoglutarate, Fe2+, and oxygen to function. [32] PHDs can hydroxylate proline residues in the hypoxia-inducible factors (HIFs), causing the HIFs to undergo ubiquitination followed by proteasomal degradation. [52] Indeed, we have demonstrated that PHD2-mediated proteasomal degradation of HIF1α is a mechanism of AA’s action in bone. [26]
Since AA is known to increase 5-hmC levels in DNA via the TETs, members of the same enzyme family as the PHDs, we decided to investigate TET- and/or PHD2-mediated increases in 5-hmC in osteoblasts and chondrocytes as a potential epigenetic mechanism for AA’s effects. Indeed, changes in 5-hmC levels have been reported in osteoarthritic human chondrocytes, [30] and hypomethylation of CpG islands in promoter regions of chondrocyte differentiation genes was observed in human synovium-derived mesenchymal stem cells, [31] suggesting that regulation of DNA demethylation may be important for musculoskeletal function. 5-hmC levels have been reported to rise in ATDC5 chondrocytic cells during differentiation, an effect mediated by TET1. [53] We confirmed that all three TET isoforms and all three PHD isoforms were expressed in both osteoblasts and chondrocytes at the mRNA level (Fig 5). The effect of AA treatment on TET and PHD expression varied by isoform, although TETs tended to increase while PHDs tended to decrease. However, the change in TET or PHD mRNA expression after AA treatment may be less important given that basal expression, of PHD2 at least, is quite high, and we are more interested in enzyme activity than changes in expression levels.
Furthermore, PHD2 is localized in both the cytoplasm and the nucleus, and AA treatment tended to increase the nuclear-to-cytoplasmic ratio of PHD2 expression (Fig 6). These findings confirm those of previous studies which have found that PHD2 is localized in both the cytoplasm and the nucleus and that PHD2 has significant hydroxylase activity in the nucleus. [54–57] Thus, PHD2 is present in the nucleus and could plausibly interact directly with DNA, contributing directly to AA-induced increases in DNA 5-hmC content, although this novel hypothesis remains to be investigated. Consistent with this possibility, our results show that AA increased 5-hmC levels in both osteoblasts and chondrocytes as measured by dot blot and ELISA (Fig 7). Since 5-hmC is the first product in the process of demethylating 5-mC, it is a possible indicator of DNA demethylation, although 5-hmC can exist as a stable epigenetic marker by itself.
Once we had confirmed that AA induced demethylation, we used a kit to pull down DNA rich in 5-hmC and then used real-time PCR with primers specific for CpG-rich regions of osteoblast gene promoters. This method gives us an indirect measure of how much the 5-hmC levels in those gene promoters have changed. We found significant increases in 5-hmC levels in osteoblast-specific genes Alp, Ihh, and Osx (Fig 8A), confirming that the global effect on 5-hmC does include osteoblast genes and supporting the hypothesis that some of AA’s effect in osteoblasts may in fact be due to increases in 5-hmC levels. Furthermore, inhibition of PHD2 using Iox2, a PHD2-specific inhibitor, resulted in significant decreases in 5-hmC in chondrocyte-specific genes Hif1α and Vegf (Fig 8B). In order to lend support to the hypothesis that the Iox2 effect on 5-hmC is mediated specifically via PHD2, we knocked down expression of the Phd2 gene in rib chondrocytes isolated from Phd2-floxed mice in vitro using an adenoviral iCre vector. Knockdown of Phd2 expression led to significantly decreased 5-hmC in the DNA isolated from these cells (Fig 9), suggesting for the first time that PHD2 may itself contribute to increased DNA 5-hmC content. However, there is not yet any evidence that PHD2 can directly modify DNA, and further mechanistic studies are needed to determine whether DNA can act as a substrate for PHD2.
While our findings provide evidence for the possibility of an epigenetic mechanism for AA’s effects in osteoblasts and chondrocytes, more work is needed to elucidate the specific TETs and/or PHDs which contribute to these changes in DNA 5-hmC content—particularly, to confirm the ability of PHD2 to directly increase 5-hmC levels—in addition to determining whether these changes in 5-hmC represent DNA demethylation and the extent to which this epigenetic mechanism contributes to AA effects in bone and cartilage in vivo. In particular, since PHD2 is known to mediate AA effects in bone and cartilage, and since we have reported here that knocking down Phd2 expression decreased levels of 5-hmC in rib chondrocytes, our future studies will investigate the novel hypothesis that PHD2 may be directly involved in DNA demethylation. Additionally, methylation sequencing and pathway analysis could provide a much deeper understanding of which genes are epigenetically regulated by AA and lead to novel targets for future therapies. Thus, we are planning a series of methylation sequencing studies to determine the specific effects of AA deficiency and PHD2 ablation on chondrocyte and osteoblast epigenomes. Further understanding of the mechanisms of bone and cartilage development and maintenance is crucial for the development of novel therapeutic approaches for diseases such as osteoporosis and osteoarthritis, and this epigenetic mechanism of DNA 5-hmC regulation and demethylation is an excellent candidate pathway for the discovery of new druggable targets.
Supporting information
Proliferation of (A) ATDC5 chondrocytic cells under the specific conditions we observed and of (B) MC3T3-E1 osteoblasts was not changed by AA treatment. ALP activity of (C) ATDC5 chondrocytic cells under the specific conditions we observed was not changed by AA treatment. However, ALP activity of (D) MC3T3-E1 osteoblasts was increased by AA treatment. Results are presented as mean ± SEM. *P < 0.05 vs. control (pairwise t-tests with Bonferroni corrections for multiple comparisons [A,C,D] or Student’s t-test [B]), n = 6–8 per group.
(TIF)
(PDF)
Acknowledgments
The authors would like to thank Catrina Godwin, Sheila Pourteymoor, and Heather Watt for technical assistance. We thank Dr. Weirong Xing for providing the adenoviral GFP and iCre vectors. All work was conducted in facilities provided by the US Department of Veterans Affairs.
This study was supported by funding from the US Department of Veterans Affairs BLR&D merit review grant (101-BX-001396) to SM and NIH grant 2 R25 GM060507 to the Loma Linda University Center for Health Disparities and Molecular Medicine. Salary support for SM is provided by a Senior Research Career Scientist Award from the US Department of Veterans Affairs.
Data Availability
All relevant data are within the manuscript.
Funding Statement
This study was supported by funding from the US Department of Veterans Affairs BLR&D merit review grant (101-BX-001396) to SM and NIH grant 2 R25 GM060507 to the Loma Linda University Center for Health Disparities and Molecular Medicine. Salary support for SM is provided by a Senior Research Career Scientist Award from the US Department of Veterans Affairs. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Malmir H, Shab-Bidar S, Djafarian K. Vitamin C Intake in Relation to Bone Mineral Density and Risk of Hip Fracture and Osteoporosis: A Systematic Review and Meta-Analysis of Observational Studies. Br J Nutr. 2018;119(8):847–858. 10.1017/S0007114518000430 [DOI] [PubMed] [Google Scholar]
- 2. Sanghi D, Mishra A, Sharma AC, Raj S, Mishra R, Kumari R, et al. Elucidation of Dietary Risk Factors in Osteoarthritis Knee—a Case-Control Study. J Am Coll Nutr. 2015;34(1):15–20. 10.1080/07315724.2013.875439 [DOI] [PubMed] [Google Scholar]
- 3. Franceschi RT, Iyer BS, Cui Y. Effects of Ascorbic Acid on Collagen Matrix Formation and Osteoblast Differentiation in Murine MC3T3-E1 Cells. J Bone Miner Res. 1994;9(6):843–854. 10.1002/jbmr.5650090610 [DOI] [PubMed] [Google Scholar]
- 4. Franceschi RT, Iyer BS. Relationship between Collagen Synthesis and Expression of the Osteoblast Phenotype in MC3T3-E1 Cells. J Bone Miner Res. 1992;7(2):235–246. 10.1002/jbmr.5650070216 [DOI] [PubMed] [Google Scholar]
- 5. Franceschi RT, Young J. Regulation of Alkaline Phosphatase by 1,25-Dihydroxyvitamin D3 and Ascorbic Acid in Bone-Derived Cells. J Bone Miner Res. 1990;5(11):1157–1167. 10.1002/jbmr.5650051111 [DOI] [PubMed] [Google Scholar]
- 6. Xiao G, Cui Y, Ducy P, Karsenty G, Franceschi RT. Ascorbic Acid-Dependent Activation of the Osteocalcin Promoter in MC3T3-E1 Preosteoblasts: Requirement for Collagen Matrix Synthesis and the Presence of an Intact OSE2 Sequence. Mol Endocrinol. 1997;11(8):1103–1113. 10.1210/mend.11.8.9955 [DOI] [PubMed] [Google Scholar]
- 7. Hadzir SN, Ibrahim SN, Abdul Wahab RM, Zainol Abidin IZ, Senafi S, Ariffin ZZ, et al. Ascorbic Acid Induces Osteoblast Differentiation of Human Suspension Mononuclear Cells. Cytotherapy. 2014;16(5):674–682. 10.1016/j.jcyt.2013.07.013 [DOI] [PubMed] [Google Scholar]
- 8. Buttery LD, Bourne S, Xynos JD, Wood H, Hughes FJ, Hughes SP, et al. Differentiation of Osteoblasts and in Vitro Bone Formation from Murine Embryonic Stem Cells. Tissue Eng. 2001;7(1):89–99. 10.1089/107632700300003323 [DOI] [PubMed] [Google Scholar]
- 9. Otsuka E, Yamaguchi A, Hirose S, Hagiwara H. Characterization of Osteoblastic Differentiation of Stromal Cell Line ST2 That Is Induced by Ascorbic Acid. Am J Physiol. 1999;277(1 Pt 1):C132–138. 10.1152/ajpcell.1999.277.1.C132 [DOI] [PubMed] [Google Scholar]
- 10. Harada S, Matsumoto T, Ogata E. Role of Ascorbic Acid in the Regulation of Proliferation in Osteoblast-like MC3T3-E1 Cells. J Bone Miner Res. 1991;6(9):903–908. 10.1002/jbmr.5650060902 [DOI] [PubMed] [Google Scholar]
- 11. Takamizawa S, Maehata Y, Imai K, Senoo H, Sato S, Hata RI. Effects of Ascorbic Acid and Ascorbic Acid 2-Phosphate, a Long-Acting Vitamin C Derivative, on the Proliferation and Differentiation of Human Osteoblast-like Cells. Cell Biol Int. 2004;28(4):255–265. 10.1016/j.cellbi.2004.01.010 [DOI] [PubMed] [Google Scholar]
- 12. Leboy PS, Vaias L, Uschmann B, Golub E, Adams SL, Pacifici M. Ascorbic Acid Induces Alkaline Phosphatase, Type X Collagen, and Calcium Deposition in Cultured Chick Chondrocytes. J Biol Chem. 1989;264(29):17281–17286. [PubMed] [Google Scholar]
- 13. Daniel JC, Pauli BU, Kuettner KE. Synthesis of Cartilage Matrix by Mammalian Chondrocytes in Vitro. III. Effects of Ascorbate. J Cell Biol. 1984;99(6):1960–1969. 10.1083/jcb.99.6.1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Temu TM, Wu KY, Gruppuso PA, Phornphutkul C. The Mechanism of Ascorbic Acid-Induced Differentiation of ATDC5 Chondrogenic Cells. Am J Physiol Endocrinol Metab. 2010;299(2):E325–334. 10.1152/ajpendo.00145.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kawai T, Nishikimi M, Ozawa T, Yagi K. A Missense Mutation of L-Gulono-Gamma-Lactone Oxidase Causes the Inability of Scurvy-Prone Osteogenic Disorder Rats to Synthesize L-Ascorbic Acid. J Biol Chem. 1992;267(30):21973–21976. [PubMed] [Google Scholar]
- 16. Mizushima Y, Harauchi T, Yoshizaki T, Makino S. A Rat Mutant Unable to Synthesize Vitamin C. Experientia. 1984;40(4):359–361. 10.1007/bf01952551 [DOI] [PubMed] [Google Scholar]
- 17. Maeda N, Hagihara H, Nakata Y, Hiller S, Wilder J, Reddick R. Aortic Wall Damage in Mice Unable to Synthesize Ascorbic Acid. Proc Natl Acad Sci USA. 2000;97(2):841–846. 10.1073/pnas.97.2.841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mohan S, Kapoor A, Singgih A, Zhang Z, Taylor T, Yu H, et al. Spontaneous Fractures in the Mouse Mutant Sfx Are Caused by Deletion of the Gulonolactone Oxidase Gene, Causing Vitamin C Deficiency. J Bone Miner Res. 2005;20(9):1597–1610. 10.1359/JBMR.050406 [DOI] [PubMed] [Google Scholar]
- 19. Jiao Y, Li X, Beamer WG, Yan J, Tong Y, Goldowitz D, et al. A Deletion Causing Spontaneous Fracture Identified from a Candidate Region of Mouse Chromosome 14. Mamm Genome. 2005;16(1):20–31. 10.1007/s00335-004-2414-0 [DOI] [PubMed] [Google Scholar]
- 20. Beamer WG, Rosen CJ, Bronson RT, Gu W, Donahue LR, Baylink DJ, et al. Spontaneous Fracture (Sfx): A Mouse Genetic Model of Defective Peripubertal Bone Formation. Bone. 2000;27(5):619–626. 10.1016/S8756-3282(00)00369-0 [DOI] [PubMed] [Google Scholar]
- 21. Gabbay KH, Bohren KM, Morello R, Bertin T, Liu J, Vogel P. Ascorbate Synthesis Pathway: Dual Role of Ascorbate in Bone Homeostasis. J Biol Chem. 2010;285(25):19510–19520. 10.1074/jbc.M110.110247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ishigami A, Fujita T, Handa S, Shirasawa T, Koseki H, Kitamura T, et al. Senescence Marker Protein-30 Knockout Mouse Liver Is Highly Susceptible to Tumor Necrosis Factor-Alpha- and Fas-Mediated Apoptosis. Am J Pathol. 2002;161(4):1273–1281. 10.1016/s0002-9440(10)64404-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Aghajanian P, Hall S, Wongworawat MD, Mohan S. The Roles and Mechanisms of Actions of Vitamin C in Bone: New Developments. J Bone Miner Res. 2015;30(11):1945–1955. 10.1002/jbmr.2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xing W, Singgih A, Kapoor A, Alarcon CM, Baylink DJ, Mohan S. Nuclear Factor-E2-Related Factor-1 Mediates Ascorbic Acid Induction of Osterix Expression via Interaction with Antioxidant-Responsive Element in Bone Cells. J Biol Chem. 2007;282(30):22052–22061. 10.1074/jbc.M702614200 [DOI] [PubMed] [Google Scholar]
- 25. Cheng S, Xing W, Pourteymoor S, Mohan S. Conditional Disruption of the Prolyl Hydroxylase Domain-Containing Protein 2 (Phd2) Gene Defines Its Key Role in Skeletal Development. J Bone Miner Res. 2014;29(10):2276–2286. 10.1002/jbmr.2258 [DOI] [PubMed] [Google Scholar]
- 26. Xing W, Pourteymoor S, Mohan S. Ascorbic Acid Regulates Osterix Expression in Osteoblasts by Activation of Prolyl Hydroxylase and Ubiquitination-Mediated Proteosomal Degradation Pathway. Physiological genomics. 2011;43(12):749–57. 10.1152/physiolgenomics.00229.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Blaschke K, Ebata KT, Karimi MM, Zepeda-Martinez JA, Goyal P, Mahapatra S, et al. Vitamin C Induces Tet-Dependent DNA Demethylation and a Blastocyst-like State in ES Cells. Nature. 2013;500(7461):222–6. 10.1038/nature12362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Minor EA, Court BL, Young JI, Wang G. Ascorbate Induces Ten-Eleven Translocation (Tet) Methylcytosine Dioxygenase-Mediated Generation of 5-Hydroxymethylcytosine. The Journal of biological chemistry. 2013;288(19):13669–74. 10.1074/jbc.C113.464800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Young JI, Züchner S, Wang G. Regulation of the Epigenome by Vitamin C. Annu Rev Nutr. 2015;35:545–564. 10.1146/annurev-nutr-071714-034228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Taylor SEB, Smeriglio P, Dhulipala L, Rath M, Bhutani N. A Global Increase in 5-Hydroxymethylcytosine Levels Marks Osteoarthritic Chondrocytes. Arthritis Rheumatol. 2014;66(1). 10.1002/art.38200 [DOI] [PubMed] [Google Scholar]
- 31. Ezura Y, Sekiya I, Koga H, Muneta T, Noda M. Methylation Status of CpG Islands in the Promoter Regions of Signature Genes during Chondrogenesis of Human Synovium-Derived Mesenchymal Stem Cells. Arthritis and rheumatism. 2009;60(5):1416–26. 10.1002/art.24472 [DOI] [PubMed] [Google Scholar]
- 32. Osipyants AI, Poloznikov AA, Smirnova NA, Hushpulian DM, Khristichenko AY, Chubar TA, et al. L-Ascorbic Acid: A True Substrate for HIF Prolyl Hydroxylase? Biochimie. 2018;147:46–54. 10.1016/j.biochi.2017.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cheng S, Pourteymoor S, Alarcon C, Mohan S. Conditional Deletion of the Phd2 Gene in Articular Chondrocytes Accelerates Differentiation and Reduces Articular Cartilage Thickness. Sci Rep. 2017;7:45408 10.1038/srep45408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Glasson SS, Blanchet TJ, Morris EA. The Surgical Destabilization of the Medial Meniscus (DMM) Model of Osteoarthritis in the 129/SvEv Mouse. Osteoarthr Cartil. 2007;15(9):1061–1069. 10.1016/j.joca.2007.03.006 [DOI] [PubMed] [Google Scholar]
- 35. Buo AM, Williams MS, Kerr JP, Stains JP. A Cost-Effective Method to Enhance Adenoviral Transduction of Primary Murine Osteoblasts and Bone Marrow Stromal Cells. Bone Res. 2016;4:16021 10.1038/boneres.2016.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cheng S, Zhao SL, Nelson B, Kesavan C, Qin X, Wergedal J, et al. Targeted Disruption of Ephrin B1 in Cells of Myeloid Lineage Increases Osteoclast Differentiation and Bone Resorption in Mice. PLoS ONE. 2012;7(3):e32887 10.1371/journal.pone.0032887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xing W, Archer TK. Upstream Stimulatory Factors Mediate Estrogen Receptor Activation of the Cathepsin D Promoter. Mol Endocrinol. 1998;12(9):1310–1321. 10.1210/mend.12.9.0159 [DOI] [PubMed] [Google Scholar]
- 38. R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2019. [Google Scholar]
- 39. Kim J, Xing W, Wergedal J, Chan JY, Mohan S. Targeted Disruption of Nuclear Factor Erythroid-Derived 2-like 1 in Osteoblasts Reduces Bone Size and Bone Formation in Mice. Physiol Genomics. 2010;40(2):100–110. 10.1152/physiolgenomics.00105.2009 [DOI] [PubMed] [Google Scholar]
- 40. Digaleh H, Kiaei M, Khodagholi F. Nrf2 and Nrf1 Signaling and ER Stress Crosstalk: Implication for Proteasomal Degradation and Autophagy. Cell Mol Life Sci. 2013;70(24):4681–4694. 10.1007/s00018-013-1409-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hall SL, Greendale GA. The Relation of Dietary Vitamin C Intake to Bone Mineral Density: Results from the PEPI Study. Calcif Tissue Int. 1998;63(3):183–189. 10.1007/s002239900512 [DOI] [PubMed] [Google Scholar]
- 42. New SA, Bolton-Smith C, Grubb DA, Reid DM. Nutritional Influences on Bone Mineral Density: A Cross-Sectional Study in Premenopausal Women. Am J Clin Nutr. 1997;65(6):1831–1839. 10.1093/ajcn/65.6.1831 [DOI] [PubMed] [Google Scholar]
- 43. Ilich JZ, Brownbill RA, Tamborini L. Bone and Nutrition in Elderly Women: Protein, Energy, and Calcium as Main Determinants of Bone Mineral Density. Eur J Clin Nutr. 2003;57(4):554–565. 10.1038/sj.ejcn.1601577 [DOI] [PubMed] [Google Scholar]
- 44. Sugiura M, Nakamura M, Ogawa K, Ikoma Y, Ando F, Shimokata H, et al. Dietary Patterns of Antioxidant Vitamin and Carotenoid Intake Associated with Bone Mineral Density: Findings from Post-Menopausal Japanese Female Subjects. Osteoporos Int. 2011;22(1):143–152. 10.1007/s00198-010-1239-9 [DOI] [PubMed] [Google Scholar]
- 45. New SA, Robins SP, Campbell MK, Martin JC, Garton MJ, Bolton-Smith C, et al. Dietary Influences on Bone Mass and Bone Metabolism: Further Evidence of a Positive Link between Fruit and Vegetable Consumption and Bone Health? Am J Clin Nutr. 2000;71(1):142–151. 10.1093/ajcn/71.1.142 [DOI] [PubMed] [Google Scholar]
- 46. Leveille SG, LaCroix AZ, Koepsell TD, Beresford SA, Van Belle G, Buchner DM. Dietary Vitamin C and Bone Mineral Density in Postmenopausal Women in Washington State, USA. J Epidemiol Community Health. 1997;51(5):479–485. 10.1136/jech.51.5.479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Simon JA, Hudes ES. Relation of Ascorbic Acid to Bone Mineral Density and Self-Reported Fractures among US Adults. Am J Epidemiol. 2001;154(5):427–433. 10.1093/aje/154.5.427 [DOI] [PubMed] [Google Scholar]
- 48. Wolf RL, Cauley JA, Pettinger M, Jackson R, Lacroix A, Leboff MS, et al. Lack of a Relation between Vitamin and Mineral Antioxidants and Bone Mineral Density: Results from the Women’s Health Initiative. Am J Clin Nutr. 2005;82(3):581–588. 10.1093/ajcn.82.3.581 [DOI] [PubMed] [Google Scholar]
- 49. Prynne CJ, Mishra GD, O’Connell MA, Muniz G, Laskey MA, Yan L, et al. Fruit and Vegetable Intakes and Bone Mineral Status: A Cross Sectional Study in 5 Age and Sex Cohorts. Am J Clin Nutr. 2006;83(6):1420–1428. 10.1093/ajcn/83.6.1420 [DOI] [PubMed] [Google Scholar]
- 50. Walmsley AR, Batten MR, Lad U, Bulleid NJ. Intracellular Retention of Procollagen within the Endoplasmic Reticulum Is Mediated by Prolyl 4-Hydroxylase. J Biol Chem. 1999;274(21):14884–14892. 10.1074/jbc.274.21.14884 [DOI] [PubMed] [Google Scholar]
- 51. Marini JC, Cabral WA, Barnes AM, Chang W. Components of the Collagen Prolyl 3-Hydroxylation Complex Are Crucial for Normal Bone Development. Cell Cycle. 2007;6(14):1675–1681. 10.4161/cc.6.14.4474 [DOI] [PubMed] [Google Scholar]
- 52. Fan L, Li J, Yu Z, Dang X, Wang K. The Hypoxia-Inducible Factor Pathway, Prolyl Hydroxylase Domain Protein Inhibitors, and Their Roles in Bone Repair and Regeneration. Biomed Res Int. 2014;2014:239356 10.1155/2014/239356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Taylor SEB, Li YH, Smeriglio P, Rath M, Wong WH, Bhutani N. Stable 5-Hydroxymethylcytosine (5hmC) Acquisition Marks Gene Activation During Chondrogenic Differentiation. J Bone Miner Res. 2016;31(3):524–534. 10.1002/jbmr.2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Depping R, Jelkmann W, Kosyna FK. Nuclear-Cytoplasmatic Shuttling of Proteins in Control of Cellular Oxygen Sensing. J Mol Med. 2015;93(6):599–608. 10.1007/s00109-015-1276-0 [DOI] [PubMed] [Google Scholar]
- 55. Berchner-Pfannschmidt U, Tug S, Trinidad B, Oehme F, Yamac H, Wotzlaw C, et al. Nuclear Oxygen Sensing: Induction of Endogenous Prolyl-Hydroxylase 2 Activity by Hypoxia and Nitric Oxide. J Biol Chem. 2008;283(46):31745–31753. 10.1074/jbc.M804390200 [DOI] [PubMed] [Google Scholar]
- 56. Metzen E, Berchner-Pfannschmidt U, Stengel P, Marxsen JH, Stolze I, Klinger M, et al. Intracellular Localisation of Human HIF-1 Alpha Hydroxylases: Implications for Oxygen Sensing. J Cell Sci. 2003;116(Pt 7):1319–1326. 10.1242/jcs.00318 [DOI] [PubMed] [Google Scholar]
- 57. Berra E, Roux D, Richard DE, Pouysségur J. Hypoxia-Inducible Factor-1 Alpha (HIF-1 Alpha) Escapes O(2)-Driven Proteasomal Degradation Irrespective of Its Subcellular Localization: Nucleus or Cytoplasm. EMBO Rep. 2001;2(7):615–620. 10.1093/embo-reports/kve130 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Proliferation of (A) ATDC5 chondrocytic cells under the specific conditions we observed and of (B) MC3T3-E1 osteoblasts was not changed by AA treatment. ALP activity of (C) ATDC5 chondrocytic cells under the specific conditions we observed was not changed by AA treatment. However, ALP activity of (D) MC3T3-E1 osteoblasts was increased by AA treatment. Results are presented as mean ± SEM. *P < 0.05 vs. control (pairwise t-tests with Bonferroni corrections for multiple comparisons [A,C,D] or Student’s t-test [B]), n = 6–8 per group.
(TIF)
(PDF)
Data Availability Statement
All relevant data are within the manuscript.