The combination of cell sorting (FACS) and ATAC-seq enables the identification of cell type-specific chromatin changes that correlate with gene activity and effectively identifies regulatory elements in plant promoters.
Keywords: ATAC-seq, cell sorting, cell-type specification, chromatin, DORNRÖSCHEN-LIKE, lateral organ founder cells
Abstract
Fluorescence-activated cell sorting (FACS) and assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) were combined to analyse the chromatin state of lateral organ founder cells (LOFCs) in the peripheral zone of the Arabidopsis apetala1-1 cauliflower-1 double mutant inflorescence meristem. On a genome-wide level, we observed a striking correlation between transposase hypersensitive sites (THSs) detected by ATAC-seq and DNase I hypersensitive sites (DHSs). The mostly expanded DHSs were often substructured into several individual THSs, which correlated with phylogenetically conserved DNA sequences or enhancer elements. Comparing chromatin accessibility with available RNA-seq data, THS change configuration was reflected by gene activation or repression and chromatin regions acquired or lost transposase accessibility in direct correlation with gene expression levels in LOFCs. This was most pronounced immediately upstream of the transcription start, where genome-wide THSs were abundant in a complementary pattern to established H3K4me3 activation or H3K27me3 repression marks. At this resolution, the combined application of FACS/ATAC-seq is widely applicable to detect chromatin changes during cell-type specification and facilitates the detection of regulatory elements in plant promoters.
Introduction
The growth and architecture of the aerial plant body depends on the activity of the shoot apical meristem (SAM), a self-organizing array of small cells that maintains a stem cell pool in its central zone. In the SAM peripheral zone, lateral organs are initiated in a species-specific phyllotactic pattern, which in Arabidopsis is spiral for leaves and flowers or whorl-like for floral organs. An initial organogenesis step is the specification of groups of lateral organ founder cells (LOFCs) in the peripheral zone, where coordinated cell divisions generate a visible organ primordium whose fate is acquired according to the ontogenetic programme (Beveridge et al., 2007). Expression of the DORNRÖSCHEN-LIKE (DRNL) gene marks LOFCs in the Arabidopsis SAM; thus, the specification of LOFCs can be monitored by the confocal imaging of transgenic lines that express the green fluorescent protein (GFP) coding region from the DRNL promoter (DRNL::erGFP) (Chandler et al., 2011a). In the inflorescence meristem (IM), DRNL::erGFP expression depicts groups of LOFCs in a spiral phyllotaxy from close proximity to the central stem cell zone (Seeliger et al., 2016) to the IM periphery until a new floral bud becomes histologically evident (Chandler et al., 2011b). Likewise, in the developing Arabidopsis flower, DRNL::erGFP prepatterns organs in all four floral whorls (Chandler et al., 2011b). Deletion analysis of the DRNL promoter has revealed at least three enhancer elements, which redundantly or synergistically control qualitative and quantitative aspects of the dynamic DRNL transcription pattern in LOFCs in the IM and sequential floral meristems (FMs) (Comelli et al., 2016). The study of DRNL promoter activity raises two questions: (i) how is positional information perceived in the SAM peripheral zone? And (ii) what distinguishes LOFCs from surrounding meristematic cells?
The second question relates to DRNL function, which in Arabidopsis is masked by redundancy with its paralogue DORNRÖSCHEN (DRN) and the related PUCHI gene (Chandler and Werr, 2017). This redundancy is less pronounced in tomato, where mutation of the DRNL orthologue LEAFLESS (LFS) interferes with leaf development (Capua and Eshed, 2017). All four genes encode class VIIIb AP2-type transcription factors and their AP2 domain comprises a DNA binding motif common to ETHYLENE RESPONSE FACTORS (ERFs) or DROUGHT RESPONSE ELEMENT BINDING (DREB) factors. A point mutation at position 37 (A37V) of the AP2 domain abolishes DNA binding to the GCC box (Liu et al., 2006) and underlies the strong drnl-2 allele, identified as B class modifier 1 (bcm1) (Nag et al., 2007). As a transcription factor (TF), the DRNL protein potentially binds target gene promoters, which has been demonstrated for DRN and the GCC box (Matsuo et al., 2009). However, direct physical contact to the GCC box is increasingly a matter of debate, as the DRN activation of CLAVATA3 (CLV3) transcription in the SAM central stem cell zone requires a GCC box, whereas the A37V substitution in the DRN AP2 domain has no functional consequence and physical interactions between DRN and the GCC box could not be detected (Luo et al., 2018). Alternatively, DRN and DRNL may act as transcriptional co-activators, as the AP2 domains mediate protein interactions via the PAS-like C-terminal domain in class III homeodomain-zipper transcription factors such as PHAVOLUTA (PHV) or REVOLUTA (REV) (Chandler et al., 2007). This co-activator function for DRN or DRNL is supported by chromatin immunoprecipitation (ChIP) experiments that show they can enhance the precipitation of a SHOOT MERISTEMLESS (STM) promoter fragment by REVOLUTA (Zhang et al., 2018b).
In a previous study, we isolated LOFCs from apetala1-1 cauliflower-1 (ap1 cal) apices via fluorescence-activated cell sorting (FACS) based on expression of the DRNL::erGFP transgene for transcriptome analysis (Frerichs et al., 2016). The ap1 cal cauliflower phenotype results from a blocked acquisition of determinate FM identity and has been exploited by various other studies to determine cell type-specific gene expression, e.g. the stem cell transcriptome using the CLV3 promoter (Yadav et al., 2009; Yadav et al., 2014) or ChIP-seq, to determine the binding of MADS box TFs at successive stages of flower development (Pajoro et al., 2014), or histone modifications (Engelhorn et al., 2017). The DRNL::GFP expression pattern in the ap1 cal apex relies on transcriptional control and we wanted to assess whether LOFC specification is accompanied by changes in chromatin configuration.
Open chromatin can be monitored by its accessibility to endonuclease attack, initially by deoxynuclease I (DNase I) and micrococcal nuclease (MNase) methods, which have both been adapted to next-generation sequencing (NGS) techniques, but which require substantial cell numbers (Tsompana and Buck, 2014). More recent methodologies include formaldehyde-assisted isolation of regulatory elements (FAIRE) and assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq), which can effectively deal with input amounts as low as 500 cells (Tsompana and Buck, 2014). ATAC-seq is based on the ability of the hyperactive Tn5 transposase to fragment DNA while terminally adding adapters at cleavage sites in accessible chromatin regions, which serve for PCR amplification and paired-end NGS (Buenrostro et al., 2013). Recent examples of ATAC-seq in plants have identified conserved cis-regulatory elements across distantly related species, as well as chromatin changes during root-hair differentiation, or have compared stem cells in the SAM with differentiated leaf mesenchyma cells in Arabidopsis (Maher et al., 2018; Sijacic et al., 2018). For these three approaches, nuclei were enriched via isolation of nuclei tagged in specific cell types (INTACT) (Deal and Henikoff, 2010).
Here, we show that the combination of FACS and ATAC-seq can identify open chromatin regions at a resolution higher than that obtained with DNaseI, although both methods reveal a high congruence in open chromatin regions. Discrete transposase hypersensitive sites (THSs) distinguish LOFCs from surrounding meristematic cells and often show correlations with cell type-specific transcriptional activity. Substantial coincidence exists between THSs and phylogenetically conserved promoter regions, functional enhancer elements, or MADS domain TF binding sites.
Materials and methods
Fluorescence-activated cell sorting
Inflorescence apices of approximately 700–900 DRNL::erGFP ap1 cal plants were collected 4–5 weeks after sowing, and FACS was performed as described by Frerichs et al. (2016). The number of GFP+ protoplasts collected in 10 ml FACS medium typically varied between 95 000 and 300 000 per experiment. For comparison, a similar number of GFP− protoplasts was used as a control.
Assay for transposase-accessible chromatin
Protoplast treatment after FACS separation followed the protocol of Buenrostro et al. (2013) with some adaptations. The collected protoplasts were centrifuged twice (60 g, 10 min, 4 °C) and resuspended in 500 µl FACS medium before cells were lysed in 10 mM Tris–HCl pH 7,4, 10 mM NaCl, 3 mM MgCl2, 0.1% IGEPAL CA-630 for 30 min on ice. The nuclei were then pelleted (1000 g, 10 min, 4 °C) before resuspension in 15 µl nuclease-free H2O; 25 µl 2×TD Tagment DNA buffer and 10 µl TDE1 Tagment DNA Enzyme (TruSight One Kit) were added to a total reaction mix of 50 µl followed by incubation at 37 °C for 30 min with gentle mixing every 5 min. Subsequently, nuclear DNA was isolated with the MinElute PCR Purification Kit (28004, Qiagen, Hilden, Germany) and eluted in 10 µl elution buffer.
Sequencing
Sequencing was carried out at the Cologne centre for genomics. The purified tagmented DNA was amplified via a 12- or 15-cycle PCR programme using reagents from the Illumina TruSight One Library Preparation Kit. This procedure adds index 1 (i7) and index 2 (i5) information required for multiplexing, as well as common adapters (P5 and P7) for cluster generation and sequencing. To remove unwanted products, the final library was purified using Beckman Coulter Ampure XP Beads. After purification and validation (2200 TapeStation; Agilent Technologies), libraries were quantified by using the KAPA Library Quantification kit (Peqlab) and the 7900HT Sequence Detection System (Applied Biosystems) and subsequently sequenced on an Illumina HiSeq4000 sequencing system with a paired-end 2×75-nt sequencing protocol.
Bioinformatics
The quality of raw reads was assessed using FastQC (v0.11.5, www.bioinformatics.babraham.ac.uk/projects/fastqc). Paired-end reads were mapped onto the Arabidopsis genome (TAIR10) with the Burrows–Wheeler alignment tool (Li and Durbin, 2009). Duplicated pairs were removed using Picard (broadinstitute.github.io/picard). SAMtools (Li et al., 2009) were employed for further handling of the data and bedtools were used to generate bed files and map genomic regions to genes (Quinlan and Hall, 2010). All reads mapping to organelles were discarded from further analysis.
Visualization of sequencing data
Browser views, average patterns, and heat maps of assay for ATAC signals were generated as described in Engelhorn et al. (2017). To generate average views over replicates, reads per million were averaged at bedgraph level. To exclude bias from mapping artefacts in proximity to centromeric regions, signals higher than 20 reads per million where excluded from the visualization process. For accurate visualization of transposase binding sites in close-up views, only start positions of reads were displayed. Start positions were offset by +4 bases on the plus strand and −5 bp on the minus strand to match the centre of the transposase binding position as described in Buenrostro et al. (2013).
Identification of ATAC-seq peaks and quantitative differences
Since the ATAC peaks we observed resembled chromatin mark peaks, which can span relatively broad regions, we employed software designed for histone mark data analysis to analyse the ATAC-seq peaks and adjusted the parameters accordingly: we used the SICER-df-rb function of SICERV V1.1 (Zhang et al., 2018a) with no shifting of the reads and an effective genome size of 0.9. The e-value for detection of ATAC enriched regions was set to 100; no gaps were allowed, to maintain the resolution of separate peaks. The window size was 100 bases for THS and 25 bases for differential THS (dTHS) to obtain a very precise resolution. The P-value for the discovery of differentially enriched regions was chosen as 0.001. DNase data of Pajoro et al. (2014) and ATAC-seq data of Sijacic et al. (2018) were analysed using the same parameters. Regions were considered as THS/dTHS when reads were significantly enriched or differentially enriched after FACS in all three biological replicates (data in this study and those of Sijacic et al., 2018). Raw DNase data of Pajoro et al. (2014) were provided as a pool of two replicates; thus, the output of SICER was directly considered as DNase I hypersensitive sites (DHSs). Annotations were according to TAIR10 excluding pseudogenes and transposable elements; a gene was considered a THS gene or dTHS gene when it overlapped with the feature including the region 1 kb upstream or downstream of the gene; Venn diagrams were created using Venny 2.1 (bioinfogp.cnb.csic.es/tools/venny/). Spearman correlation coefficient graphs were generated using deep-tools2 (Ramírez et al., 2016).
GO analysis of differentially accessible genes
The Biological Networks Gene Ontology tool (v. 3.0.3; Maere et al., 2005) was used in Cytoscape (v. 3.6.0; Shannon et al., 2003) for the assignment of genes in specific gene ontology (GO) groups. As a reference, the gene association file for Arabidopsis was downloaded at http://geneontology.org (20 June 2015). The false discovery rate was amended by the Benjamin and Hochberg correction. Overrepresented GO groups are shown with a significance level of at least 0.01.
Phylogenetic shadowing
To determine sequence conservation outside of transcription units, we used the programme JBrowse (https://phytozome.jgi.doe.gov/jbrowse/) on the platform Phytozome 12 created by the department of Energy’s Joint Genome Institute (CA, USA). The Arabidopsis thaliana (TAIR10) genome was compared with the genomes of Arabidopsis halleri v1, Arabidopsis lyrata v1, Brassica rapa v1, Boechera stricta v1, Capsella grandiflora v1, Capsella rubella v1 and Eutrema salsugineum v1. Regions with a conservation level of at least 75% in four of the seven species compared with A. thaliana were considered as conserved.
Results
ATAC-seq depicts open chromatin with high resolution
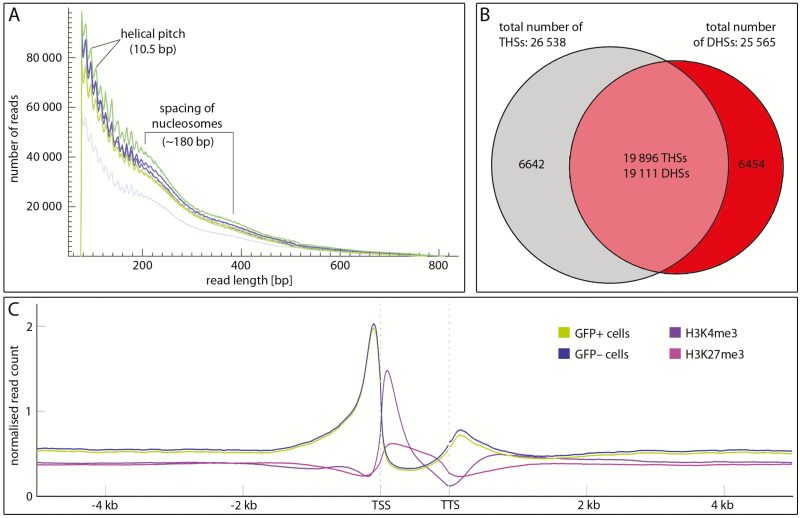
The separation of GFP+ and GFP– protoplasts of transgenic DRNL::erGFP ap1 cal Arabidopsis inflorescence apices via FACS resulted in about 95 000–300 000 protoplasts for ATAC-seq. Although ATAC-seq has been performed with as few as 500 cells in humans, we increased this number to obtain maximum resolution between the two similar cell types investigated. Despite up to a 3-fold difference in protoplast numbers, no significant differences in read numbers and signal quality were observed. Incubation with the hyperactive Tn5 transposase and NGS resulted in 118–142 million reads per sample in three independent biological replicates (Supplementary Table S1 at JXB online). Following deduplication, about 40 million reads in each sample mapped to the A. thaliana genome (TAIR10), approximately 40% of which were associated with organelle DNA, a comparable percentage to previous results (Maher et al., 2018). The distribution of fragment lengths (Fig. 1A) clearly depicted chromatin-dependent periodicities such as the characteristic 10.5-bp helical pitch pattern (Buenrostro et al., 2013) and weakly indicated the nucleosomal spacing of 180 bp characteristic for Arabidopsis flowers (Zhang et al., 2015).
Fig. 1.
Quality controls and distribution of ATAC-seq reads. (A) Absolute read numbers in a 50–820-bp size window in three biological replicates of GFP+ (green) and GFP– (blue) cells. The jagged line most prominent below 200 bp read length shows peaking read numbers at a spacing of 10.5 bp, which relates to the helical pitch, and the smooth curve between 200 and 400 indicates 180-bp spacing of nucleosomes. (B) Venn diagram comparing THSs (grey) with DHSs (red) in the ap1 cal IM; note that the majority of DHSs and THSs overlap (pale red). Similar fractions are unique to ATAC-seq or DNase-seq. (C) Mean ATAC-seq signal [green (GFP+) or blue (GFP–)] of all annotated Arabidopsis genes in 5 kb up- or downstream flanking sequences relative to the transcription unit compared with the distribution of H3K4me3 activation (purple) or H3K27me3 (pink) repression marks. Note the pronounced preference of THSs at the transcription start site (TSS) and at a lower frequency at the transcription termination site (TTS).
Read frequencies were highly reproducible within replicates, as supported genome-wide by the Spearman rank correlation (Supplementary Fig. S1A) as shown for individual genes in Fig. 2A and Supplementary Figs S2, S3. To validate ATAC-seq as a method to measure chromatin accessibility in ap1 cal inflorescences, we compared our data with DNase-seq data (Pajoro et al., 2014) originating from the same ap1 cal background. We therefore reanalysed the DNase-seq raw data with our analysis pipeline and found that THSs were commonly more pronounced than DHSs (Fig. 2A; Supplementary Fig. S2), although the chromosomal read numbers in the DNase-seq sample (two pooled replicates) well exceeded those obtained in each of six individual ATAC-seq samples (Supplementary Table S1). Genome-wide, 75% (19 111) of the DHSs overlapped with 19 896 THSs (Fig. 1B). The greater number of THSs resulted from the breakdown of individual DHSs into multiple THSs. Additionally, the size of accessible chromatin after ATAC-seq was smaller, calculated as 17.1 Mbp relative to 20.7 Mbp in the DNase-seq experiment (Supplementary Fig. S1B), although the total number of THSs exceeded that of DHSs by 3.8%.
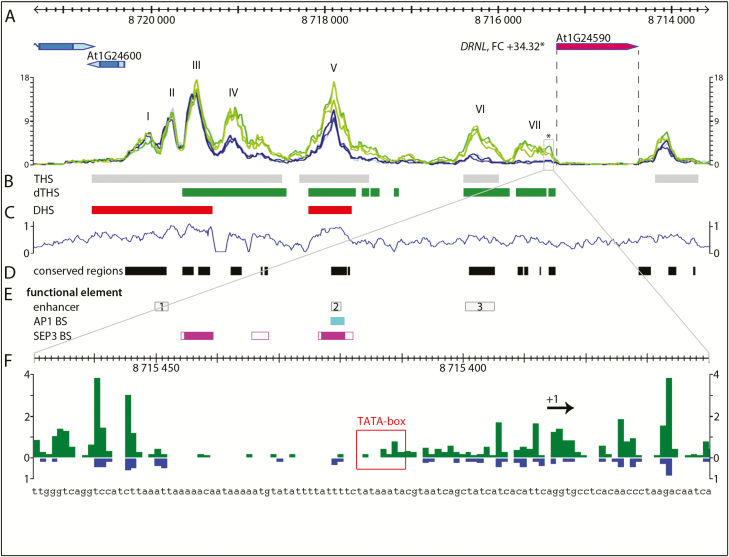
Fig. 2.
Open chromatin regions at the DRNL promoter correlate with functional elements. (A) Chromatin structure of the DRNL genomic region in three biological replicates of GFP+ (green) and GFP– (blue) cells. The DRNL gene is essentially untranscribed in GFP– cells but highly activated in GFP+ cells (fold change=+34.32; Frerichs et al., 2016). Individual THSs or peaks in the promoter upstream region are numbered (I–VII) and open chromatin regions in GFP+ and GFP– cells are marked as grey blocks (B). Regions with higher accessibility in LOFCs (dTHS-up) are indicated in green. (C) Comparison with DNase-seq data (Pajoro et al., 2014): red blocks mark significant DHSs and normalized read counts (y-axis) are depicted graphically to compare resolution to ATAC-seq. (D) DNA-sequence conservation among the Brassicaceae determined by phylogenetic shadowing of intergenic regions. (E) Position of functional elements in the DRNL promoter: DRNL enhancer elements En1–3 (top, Comelli et al., 2016), AP1 binding sites (centre) and SEP3 binding regions (bottom) (Pajoro et al., 2014). Peak calling in ChIP-seq data obtained at 2 d (filled boxes) or at 8 d (open boxes) following the onset of synchronized flower development by DEX application (Pajoro et al., 2014). (F) Enlargement of the DRNL transcription start region showing chromatin accessibility at the nucleotide level as a mean value of three biological replicates in GFP+ (green) and GFP– (blue) cells. Only start and stop positions of read pairs, i.e. sites of transposase cleavage, are shown. Note the substantial quantitative differences in read counts between the two cell types and LOFC-specific new read termini at the TATA box (red box) or at the transcription start site (+1).
THSs were highly enriched upstream of annotated transcriptional units and peaked prior to the transcription start site (TSS) in GFP+ and GFP– cells (Fig. 1C) and to a lesser degree, THSs flanked the transcription termination site (TTS). These preferences ceased beyond 1 kb distance from the TSS or TTS. Reads were under-represented within transcriptional units, where reduced densities exhibited a striking complementarity to the H3K4me3 activation or H3K27me3 repression marks (Engelhorn et al., 2017).
Chromatin accessibility at the DRNL promoter
The comparison between GFP+ LOFCs and surrounding GFP– tissue in the IM identified dTHSs, in which local open chromatin accessibility was significantly increased (dTHS-up) or decreased (dTHS-down) in LOFCs, and invariant THSs (iTHSs) that represented chromatin regions with the same Tn5 transposase accessibility in both GFP+ and GFP– protoplasts (Fig. 2B). A prominent LOFC-specific dTHS-up (peak VII in Fig. 2A) was located at the DRNL TSS, which correlates with a 34-fold higher DRNL expression in GFP+ relative to GFP– cells (Frerichs et al., 2016). This region is essentially inaccessible in GFP– cells, where the promoter is inactive, but open in GFP+ cells in which DRNL is expressed.
Further towards the At1g24600 gene, ATAC-seq reads form six additional, discrete and significant peaks (I–VI in Fig. 2A) that comprise either iTHS (I–III) or dTHS-up (IV–VI) (Fig. 2B). Although the read distribution from the DNase-seq experiment (Pajoro et al., 2014) showed a similar overall peak pattern, it failed to detect weaker THSs as DHSs (dTHS IV and VI), and depicted iTHS peaks I–III as a single peak. The absence of a corresponding signal proximal to the DRNL TSS probably relates to FACS sorting and increased accessibility of the dTHS-up in LOFCs. All seven ATAC-seq peaks in the DRNL promoter matched sequence elements conserved throughout the Brassicaceae (Fig. 2D), of which dTHS-up V and VI correspond to functional enhancer (En) elements En2 and En3 within the DRNL promoter (Comelli et al., 2016), which were both more accessible in GFP+ cells (Fig. 2E). The distal enhancer En1 bidirectionally regulates At1g24600 and DRNL transcription and resides between iTHS I and II (Fig. 2A). An AP1 MADS domain TF binding site is located in dTHS V, a region also bound by SEP3 in addition to distal promoter sequences in iTHS III and in the downstream shoulder of dTHS IV (Fig. 2E; Pajoro et al., 2014).
The DRNL TSS was further analysed at the nucleotide level and compared between GFP+ and GFP– nuclei (Fig. 2F). Few reads terminated in the TATA box (Comelli et al., 2016), which either relates to a preference of the Tn5 transposase for GC residues in proximity to the cleavage site (Madrigal, 2015), or the TATA box is occupied, e.g. by a TATA box binding protein (Heard et al., 1993). However, according to an increased number of read termini flanking the TATA box upstream region, the DRNL TSS acquires an open chromatin conformation in LOFCs, in contrast to in GFP– cells. Thus, ATAC-seq can detect chromatin changes associated with transcriptional activation of DRNL in LOFCs, which possibly correlate with the assembly of a pre-initiation complex for RNA polymerase entry.
Exemplary correlations between chromatin accessibility, transcriptional activity, and functional elements
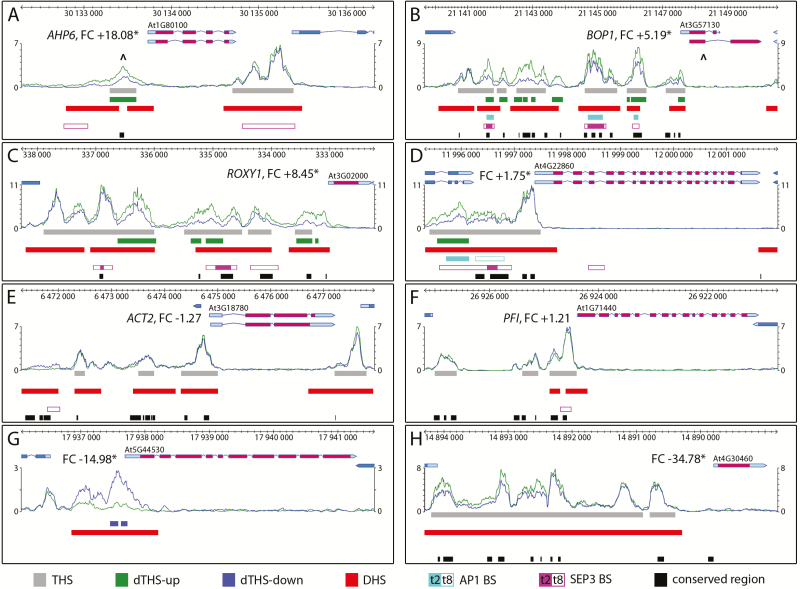
To assess further the correlation between changes in chromatin accessibility and transcription, we analysed six additional loci in detail that were up- or down-regulated in GFP+ protoplasts (Frerichs et al., 2016). Three out of a total of four promoters of up-regulated genes (Fig. 3A–D) were chosen for functional reasons. The first gene, ARABIDOPSIS HISTIDINE PHOSPHOTRANSFER PROTEIN 6 (AHP6, At1g801000) (Fig. 3A), is a potential target of DRNL (Ikeda et al., 2006), is co-expressed with DRNL at the IM periphery (Besnard et al., 2014; Chandler and Werr, 2014), and is almost as highly differentially expressed as DRNL in the RNA-seq comparison of GFP+/GFP– protoplasts (Frerichs et al., 2016). The dTHS-up in the AHP6 promoter correlated with a broad but fragmented DHS (see Supplementary Fig. S2A) and a small Brassicaceae-specific conserved sequence element. BLADE ON PETIOLE1 (BOP1, At3g57130) genetically interacts with DRNL (Chandler and Werr, 2017), whereas ROXY1 (At3g02000) relates to LOFC fate at the IM periphery, i.e. flower development. The transcription of BOP1 and ROXY1 was highly up-regulated in GFP+ protoplasts and THSs correlated with DHSs in the promoters of both genes (Fig. 3B, C). However, a better resolution was achieved by ATAC-seq and led to the identification of dTHSs-up (DHS details in Supplementary Fig. S2B, C).
Fig. 3.
Exemplary correlations between THSs/dTHSs and gene activity, DHSs, MADS box TF binding and conserved DNA elements. Chromatin accessibility is depicted as mean values of three biological replicates in GFP+ (green) and GFP– (blue) cells and compared between up-regulated (A–D), constitutively expressed (E, F) or down-regulated (G, H) genes. The genomic region with gene annotation and name, transcription unit, protein coding region and fold change (FC) is depicted above graphs that show the normalized read count (NRC) obtained by ATAC-seq. Significant THSs, dTHSs and DHSs, SEP3, or AP1 binding sites (BS) and phylogenetically conserved DNA sequence elements in the intergenic regions are indicated below the graph according to the colour code at the base of the figure. Asterisks mark a significant FC (≥1.5; P≤0.01), a criterion not met for the constitutively expressed ACT2 and PF1 genes. The position of a GCC box is marked in the APHP6 dTHS and the BOP1 first exon is indicated by the ^ symbol in (A, B).
Whereas AHP6, BOP1, and ROXY1 encode regulatory proteins, At4g22860 (Fig. 3D) encodes a mitotic spindle-associated protein and relates to cell-cycle control, the G1/S transition of which is affected by DRNL (Seeliger et al., 2016). The moderate increase in At4g22860 transcript levels [fold change (FC)=1.75] in GFP+ cells correlated with an invariant iTHS adjacent to the TSS, but a dTHS-up extended distally into the 3′-terminus of the flanking constitutively expressed At4g22850 gene. In the promoters of two uniformly expressed house-keeping genes, ACTIN 2 (ACT2, At3g18780; Fig. 3E) and PFIFFERLING (PFI, At1g71440; Fig. 3F), the ATAC-seq read distribution was similar in GFP+ LOFCs and GFP– meristematic cells. This contrasts with down-regulated genes, such as the subtilase At5g44530, where a dTHS-down extended from the TSS to more distal promoter regions (Fig. 3G) and correlated with a DHS. The down-regulated gene, At4g30460 (Fig. 3H), represents an exception; it encodes a glycine-rich protein of unknown function and despite high transcriptional repression in LOFCs (FC=−34), its chromatin was more accessible in GFP+ than in GFP– cells. Increased chromatin accessibility thus might not always relate to increased steady-state transcript levels, due to repressor binding or post-transcriptional mechanisms. In general, peak-calling in ATAC-seq resulted in substantially higher local read densities and thus more defined peaks than DNase-seq in the same ap1 cal background (compare y-axis scales in Supplementary Figs S2, S3), a resolution that relates to distinct THSs or dTHS at the position of AP1 or SEP3 binding sites and/or phylogenetically conserved sequence elements.
Genome-wide correlations
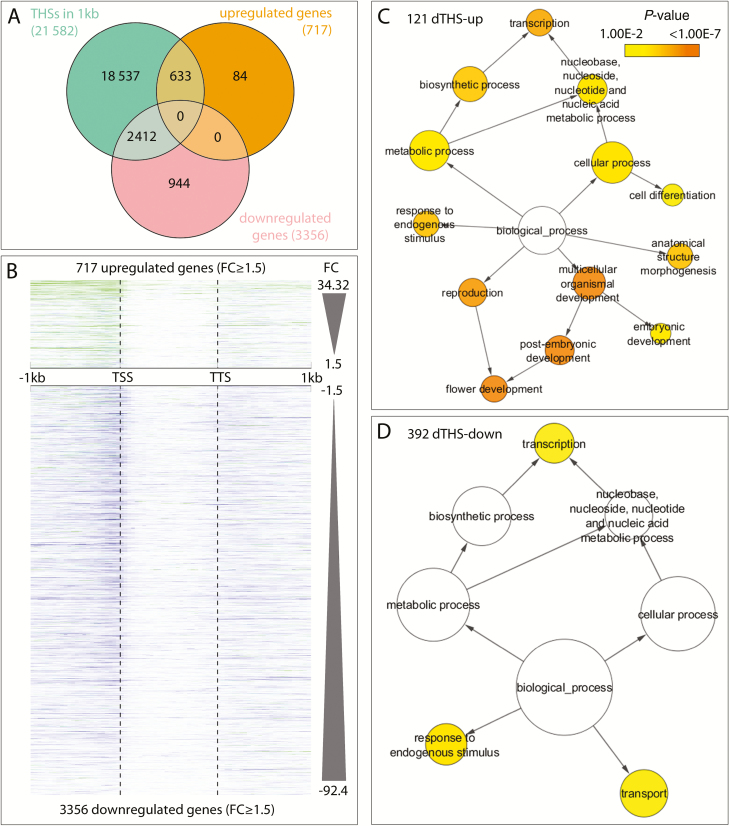
We further focused on THSs within transcriptional units and flanking 1 kb up- or downstream sequences. Comparison with RNA-seq data revealed THSs in the majority of transcribed Arabidopsis genes (28 496) and a positive correlation between chromatin accessibility and expression levels (Supplementary Fig. S4). We then analysed differentially expressed genes (DEGs) in the RNA-seq data (Frerichs et al., 2016) and confirmed that 88% (633 out of 717) of the up-regulated genes and 72% (2412 out of 3357) of the down-regulated genes in LOFCs contained a THS (Fig. 4A). Considering quantitative THS dynamics in the DEG chromatin, a strong correlation existed between the degree of chromatin accessibility and expression (Fig. 4B): almost all up-regulated genes displayed higher chromatin accessibility in LOFCs compared with IM cells, with the converse being true for down-regulated genes. Although the difference in chromatin openness between GFP+ and GFP– cells increased together with higher FC values for up-regulated genes in LOFCs, down-regulated genes displayed a lower relative change in chromatin accessibility with increasingly negative FCs.
Fig. 4.
Specific differences in chromatin structure between up- and down-regulated genes and between LOFCs and meristematic GFP– cells. (A) Distribution of THSs in up- and down-regulated DEGs (FC≥1.5; P≤0.01) (Frerichs et al., 2016); note the 88% (633 out of 717) overlap for up-regulated DEGs. (B) ATAC-seq signal intensity throughout the transcriptional unit and 1 kb flanking sequences in GFP+ relative to GFP– chromatin in up- or down-regulated DEGs sorted by FC value (Frerichs et al., 2016). Green and blue depict a higher signal in GFP+ or GFP– cells, respectively. Note the increasing frequency of green signals in the promoter upstream regions concomitant with increasingly positive FC values; no such correlation exists between ATAC-seq intensity and degree of DEG down-regulation. (C, D) Biological Network Gene Ontologies (BiNGO) of 121 genes with dTHSs-up (C) or 392 genes with dTHSs-down (D) within a maximum 1 kb distance from the transcription start in the GO-category biological processes. A significant over-representation of genes in a GO category is indicated according to the colour in the P-value bar.
Next, we analysed dTHS-up and dTHS-down and found 121 and 392 genes, respectively, that overlapped with dTHS sites (P=0.001). Among these, a significant over-representation was detected between the change in chromatin accessibility and gene expression; 37 up-regulated DEGs exhibited a dTHS-up and 189 down-regulated DEGs a dTHS-down, whereas reciprocal changes were rare (Supplementary Table S2). GO analysis of the 121 dTHS-up genes (Fig. 4C) revealed a significant preference in the GO categories flower development, cell differentiation, transcription, and anatomical structure morphogenesis. Known regulators of floral organ development among the dTHS-up genes in the GO category flower development included AHP6, ROXY1, PUCHI, CAL, ABNORMAL FLORAL ORGANS (AFO), MONOPTEROS (MP), and the potential DRNL target STYLISH1 (STY1) (Eklund et al., 2011). This contrasts with the 392 dTHS-down genes, which were only weakly over-represented in the GO categories transcription, responses to endogenous stimuli, and transport, the latter containing the auxin efflux carrier PIN-FORMED 7 (PIN7) (Fig. 4D).
LOFC versus stem cell chromatin
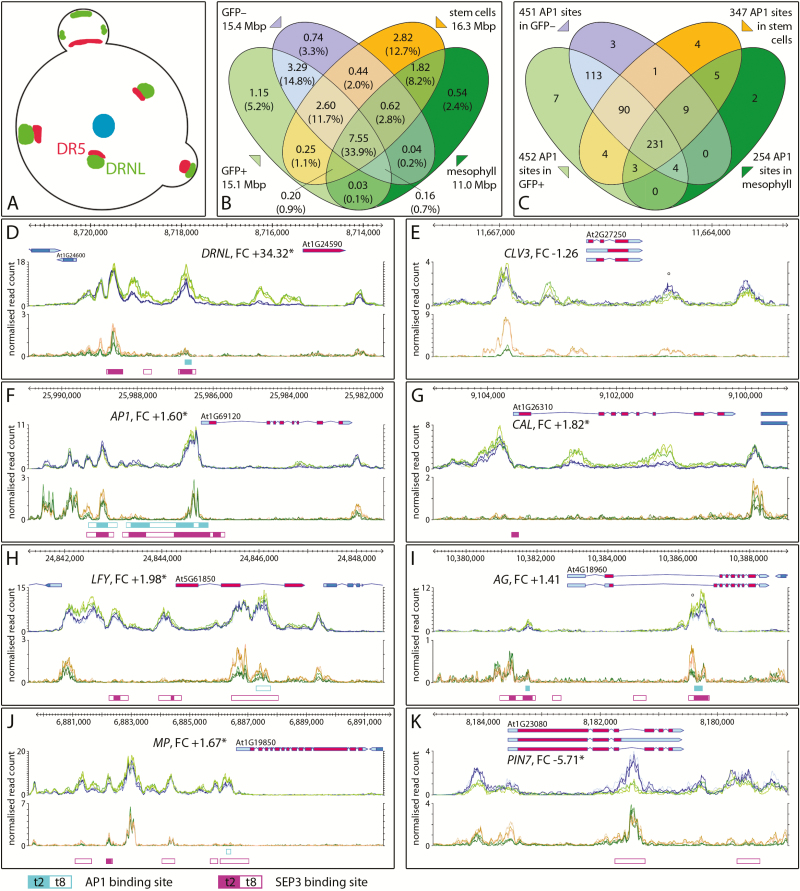
To assess differential chromatin accessibility between tissues, we compared the chromatin of the ap1 cal GFP+ LOFCs and GFP– meristematic cells with vegetative stem and mesophyll cell data from Sijacic et al. (2018). Stem cells and LOFCs occupy discrete domains in the IM (Fig. 5A) and in relation to auxin response maxima. The amount of open chromatin in stem and mesophyll cells or GFP+ and GFP– cells was similar, between 11 and 16 Mbp (Fig. 5B). All four cell types shared 33.9% open chromatin, and 11.7% was common to stem cells and cells of the ap1 cal IM (GFP+ and GFP–). Both ATAC-seq approaches rely on the cell type-specific promoter activity of DRNL to isolate LOFCs via FACS or of CLV3 to select stem cell nuclei via INTACT (Fig. 5D, E). The bilateral comparisons between each dataset revealed prominent cell type-specific dTHSs-up at the DRNL and CLV3 TSS that extended into the most proximal promoter upstream region and reflected promoter activity in LOFCs or stem cells, respectively. However, the DRNL promoter was inaccessible in vegetative stem cells and mesophyll cells, except for its distal iTHSs I–III (see Fig. 2A). In contrast, the CLV3 promoter mostly retained its characteristic THS pattern in the chromatin of the ap1 cal IM, except for the stem cell-specific THS at the TSS, which corresponded to low transcript levels in the ap1 cal IM. An iTHS downstream from the CLV3 transcriptional unit covers the enhancer where WUSCHEL (WUS) binding positively regulates CLV3 transcription (Perales et al., 2016). Both loci exemplify that cell type-specific dTHSs at the TSS directly relate to transcriptional activity, but these are combined with THSs stably detected in vegetative stem cells and the ap1 cal IM (CLV3) or restricted to the ap1 cal IM (DRNL).
Fig. 5.
Open chromatin regions in the ap1 cal inflorescence meristem relative to stem and mesophyll cells and MADS domain transcription factor binding. (A) Cartoon of the inflorescence meristem depicting the stem cell population (blue), DRNL-expressing cells (green), and auxin response maxima (red) (Chandler and Werr, 2014). (B) Venn diagram of open chromatin regions in GFP+ and GFP– cells in comparison with stem and mesophyll cells (Sijacic et al., 2018). (C) Distribution of AP1 binding sites in THSs of LOFCs or GFP– cells relative to AP1 binding sites in THSs of vegetative stem and leaf mesophyll cells. (D–K) Chromatin configuration of eight exemplary gene loci: graphs represent normalized ATAC-seq read counts of GFP+ (bright green) and GFP– (blue) cells of the ap1 cal IM (upper panel) or stem (orange) and mesophyll (dark green) cells (lower panel). (D) DRNL, (E) CLV3, (F) AP1, (G) CAL, (H) LFY, (I) AG, (J) MP, and (K) PIN7. The positions of AP1 and SEP3 binding sites (Pajoro et al., 2014) are depicted below the graphs according to the colour code below the figure with filled boxes representing stage 2 (t2) or open boxes stage 8 (t8). The position of WUS binding sites in the 3′-THS of CLV3 (E) and the intronic THS of AG (I) are marked by an open circle (°); all other gene descriptions are as in the legend of Fig. 3.
The floral meristem identity gene AP1 (Fig. 5F) was accessible from the early reproductive phase onwards and exhibited a characteristic THS pattern in stem cells, which remained the same throughout development as in ap1 cal LOFCs and GFP– cells or in leaf mesophyll cells. In contrast, the promoter of the AP1 paralogue, CAL, was accessible only in the reproductive IM. CAL is a transcriptionally activated dTHS-up gene in LOFCs (FC=+1.82), with two dTHS-up within its two largest introns. A similar ontogenetic change in chromatin openness was observed at the LEAFY (LFY) locus (Fig. 5H), where THSs in the proximal upstream promoter region were restricted to the IM, but an open chromatin region in the second intron was shared with vegetative stem cells and more weakly with mesophyll cells. Whereas AP1, CAL, and LFY affect meristem identity (Goslin et al., 2017), AGAMOUS (AG) functions later and controls stamens and carpel identity and contributes to FM determinacy (reviewed in Irish, 2017). In the arrested ap1 cal IM, THSs are weak in the AG promoter upstream region, but a prominent THS covers an essential enhancer element at the 3′-terminus of intron 2 (Hong et al., 2003). Open chromatin at this enhancer position was detected in vegetative stem cells and weakly in mesophyll cells, although read numbers were lower than in GFP+ and GFP– cells (compare the scale of the y-axes in Fig. 5I).
We also analysed two loci involved in auxin response or transport. MONOPTEROS (MP) encodes an auxin response factor and is transcriptionally up-regulated in LOFCs. The MP promoter was more accessible in the ap1 cal IM than in the vegetative SAM, where only a few of the THSs present in the IM were identified and the broad THS at the TSS (including a dTHS-up) was absent in both vegetative tissues. In contrast, the genomic region of the PIN7 auxin efflux carrier showed a similar THS pattern in bilateral comparisons of both datasets. Consistent with PIN7 transcriptional down-regulation in LOFCs, we observed two dTHSs-down in the upstream PIN7 promoter region and in intron 2, whereas the dTHS-up at the PIN7 TSS in stem cells suggests active transcription. Although directional auxin transport is effectively controlled on the protein level (Adamowski and Friml, 2015), the ATAC-seq data here indicate changes in the chromatin configuration at the PIN7 locus. PIN7 transcriptional activity in stem cells and its down-regulation in LOFCs supports the paradigm that auxin is actively exported from the central stem cell zone and is enriched in incipient primordia at the SAM periphery (Reinhardt et al., 2003).
THS and transcription factor binding
Concerted AP1 and CAL activity is essential for the IM/FM transition. Accordingly, we investigated the correlation between AP1 ChIP-seq data (Pajoro et al., 2014) and THSs in the ap1 cal IM and whether the physical AP1 binding sites resided in open chromatin regions in stem cells. We focused on the earliest data point (t2) of synchronized floral development, as this is closest to the specification of LOFCs at the IM periphery (Fig. 5C). In total, 231 (46.4%) out of all 498 AP1 target sites resided in THSs common to all four cell types: LOFCs, GFP–, vegetative stem, and mesophyll cells; a further 90 AP1 target sites residing in THSs shared between LOFCs, vegetative stem, and GFP– cells. The majority (64.5%) of t2 AP1 binding sites thus resided in open chromatin regions that are already established in the early seedling stem cell population and are possibly perpetuated from the vegetative to the reproductive phase. Unique to the ap1 cal IM, 113 AP1 target regions overlapped with THSs, whereas few binding sites uniquely overlapped with THSs in GFP– or GFP+ cells alone or in combination with vegetative stem cells. In the mutant ap1 cal IM, chromatin remodelling that leads to accessibility of these additional 113 binding sites was independent of AP1 activity. Thus, at least 87.1% (434) of the 498 early t2 AP1 binding sites (Pajoro et al., 2014) resided in open chromatin regions that existed prior to LOFC specification. On an individual gene basis, this coincidence is shown in Figs 2, 3, 5.
Because the GCC box is a potential DRNL target sequence, we queried its enrichment in THSs. The total number of 11 216 GCCGCC elements was 1.4-fold higher than expected (8104), when the GC-content of the Arabidopsis genome is considered. Given the cell type-specific DRNL expression pattern, one expectation for TF binding is that target sites are enriched in dTHSs-up. However, only five perfect GCCGCC elements resided in the 121 dTHSs-up genes, including one in the AHP6 promoter and one in the BOP1 first exon (Supplementary Table S3; Fig. 3A, B); the number of GCC boxes in the 392 dTHS-down genes was even lower (three). Predictably, the number of motifs increased to 15 in dTHS-up or 39 in dTHS-down, when stringency was relaxed at positions 2, 3, or 5 of the GCCGCC sequence, considering positional constraints or freedoms (Hao et al., 1998) (Supplementary Table S3). In contrast, more GCC boxes were present in genes carrying iTHSs, i.e. open chromatin regions, in both GFP+ and GFP– protoplasts (Supplementary Table S3). This distribution is incompatible with the assumption that binding of DRNL to the GCC box contributes to chromatin opening in LOFCs and to differential gene expression.
Discussion
ATAC-seq sensitivity
Because ATAC-seq has only recently been applied to plants, we compared our FACS/ATAC-seq data with existing DNase-seq data for the ap1 cal IM and to an ATAC-seq analysis comparing CLV3-expressing vegetative stem and leaf mesophyll cells (Sijacic et al., 2018). These comparisons revealed a similar THS (26 538) or DHS (25 565) frequency within the Arabidopsis genome, with 19 111 DHSs overlapping with 19 896 THSs and spanning 12.09 Mbp open chromatin detected by both methods (Fig. 1B; Supplementary Fig. S1B). Despite this congruence, a major difference was observed in local read densities in ATAC-seq that calculated to discrete peaks and structured open chromatin regions at a higher resolution than by DNase-seq (Fig. 2; Supplementary Figs S2, S3), despite the use of the ap1 cal IM in both analyses (Pajoro et al., 2014).
Secondly, we compared our FACS approach to separate DRNL::erGFP-expressing LOFC protoplasts from non-fluorescent protoplasts of the ap1 cal IM with the INTACT method that isolates epitope-tagged nuclei. Although the explant materials originated from different ontogenetic phases, i.e. stem and mesophyll cells from early vegetative seedling (Sijacic et al., 2018) and GFP+ and GFP– cells from reproductive IM tissue, substantial overlaps in open chromatin regions existed between all four cell types (Fig. 5B). A major advantage of the FACS relative to the INTACT method is that GFP+ and GFP− protoplasts originated from a single sorting experiment, whereas isolated nuclei via INTACT derived from different transgenic lines, tissues, and developmental windows, e.g. 6-day-old seedlings or leaves of 3-week-old plants. Thus, a GFP+/GFP– cell comparison addresses cell-type specification within a unique cell array, whereas vegetative stem and mesophyll cells are separated by a trajectory involving many developmental steps.
Despite methodological and cell-type differences, the transposase-accessible chromatin regions were remarkably congruent, but also exhibited differences (Fig. 5D–K: (i) dTHSs correlated with active transcription, such as at the TSS of CLV3 or DRNL in vegetative stem cells or LOFCs, respectively; (ii) specific THSs associated with a developmental window or cell type, as in the LFY or CAL promoter upstream regions; (iii) open chromatin regions were detectable in all four chromatin samples, e.g. generally in the AP1 promoter, and locally in the MP upstream region or in the AG second intron. The similar THS pattern at the AP1 locus in all four cell types strongly supports the conclusion that open chromatin regions are transmittable from vegetative stem to differentiated cells (Sijacic et al., 2018), but all other depicted genes showed pronounced dynamic THS patterns. The THS pattern in LFY chromatin contrasts with the stably transmitted AP1 pattern; although LFY acts upstream of AP1 in floral induction (Wagner et al., 2004), the LFY upstream promoter is only accessible in the ap1 cal IM, whereas a THS in intron 2 is accessible in stem cells, terminally differentiated mesophyll cells and in the ap1 cal IM. In contrast to the enhancer function in the AG intron (Hong et al., 2003), this intronic open chromatin region in the LFY transcriptional unit has to date not been reported to contribute to expression.
In summary, comparison of the ap1 cal IM chromatin state using DNase-seq and ATAC-seq revealed that the latter method is superior in terms of signal-to-noise ratio and the identification of discrete accessible chromatin regions. A comparative analysis of ATAC-seq data between four cell types at this higher resolution showed that up to 30% of the open chromatin or THSs are stably transmitted throughout ontogeny and cellular differentiation. On an individual gene basis, however, ATAC-seq detects substantial dynamics between cell types or related to transcriptional activity.
Transcription factor binding and evolutionary conservation
Besides DNase- and ATAC-seq, the ap1 cal IM has been used to synchronize flower development (Wellmer et al., 2006) and as a source for meristematic cells to study MADS domain TF binding during early flower development (Pajoro et al., 2014). From ChIP-seq data at successive floral stages (2–8 d), the earliest data point t2 is closest to LOFC specification and revealed 498 binding sites for AP1 and 1776 for SEPALATA3 (SEP3), another MADS domain TF activated by AP1 (Pajoro et al., 2014). In total, 321 of the t2 AP1 binding sites (231 + 90) resided within THSs identifiable in stem cells of 6-day-old seedlings (Fig. 5B) and an additional 113 AP1 binding sites were present in THSs unique to the ap1 cal IM. Thus in total, 434 (87.1%) out of 498 AP1 binding sites reside in open chromatin regions in the ap1 cal IM in the absence of AP1, a situation that questions whether AP1 binding specifically increases the chromatin accessibility of floral genes or rather relies on prepatterned accessible chromatin regions. This coincidence observed between THSs and t2 AP1 target sites remained unaffected in the late t8 or SEP3 ChIP-seq data sets, as is depicted for individual gene examples in Figs 3 and 5.
The binding of AP1 in iTHSs at its own promoter suggests autoregulation, whereas early and late developmental AP1 target sites are absent in CAL and LFY promoter upstream regions. This confirms that the LFY and AP1/CAL pathways are only partially redundant and show independent, synergistic or antagonistic contributions at individual target loci (Goslin et al., 2017). Intronic THSs observed for CAL, LFY, AG, and PIN7 are infrequent in the Arabidopsis ap1 cal IM (Fig. 1C) as well as in vegetative stem and mesophyll cells (Sijacic et al., 2018), which differs from animal ATAC-seq data, where such intronic THSs comprise a major fraction (Ackermann et al., 2016). However, intronic THSs can be functionally important, for example, at the AG locus, where AP1 and SEP3 bind an enhancer that is targeted by LFY during floral induction and by WUS later in flower development (Hong et al., 2003). This enhancer in the AG second intron is phylogenetically conserved, as are discrete ATAC-seq peaks covering enhancer elements in the DRNL promoter upstream region (Fig. 2E) or other genes (Fig. 3). In cases of such conservation, phylogenetic shadowing combined with ATAC-seq could effectively predict regulatory elements in the Arabidopsis genome and elucidate cell-type specificity by the integration of FACS or INTACT techniques. The correlation between THSs and evolutionarily conserved genomic regions could therefore assist the discovery of unknown functional plant enhancers (Weber et al., 2016) on a cell type-specific and genome-wide scale.
LOFC-specific gene expression
Three distinct chromatin states within the DRNL promoter have been identified by ATAC-seq: (i) a distal constitutively open chromatin region or iTHSs (I–III in Fig. 2A) that is shared with mesophyll and vegetative stem cells (Sijacic et al., 2018) and encompass enhancer element En1 (Fig. 2D); (ii) dTHS-up IV, which is not present in vegetative stem cells, or dTHS-up V, which covers En2 and is hardly accessible in vegetative stem cells; (iii) dTHS VI, which covers the DRNL TSS that includes En3 and is highly accessible only in LOFCs. All three DRNL enhancer elements are phylogenetically conserved. The DRNL TSS (dTHS VII in Fig. 2A) is unique, as promoter activity restricted to LOFCs was a condicio sine qua non for FACS, as was the CLV3 TSS in vegetative stem cells (Sijacic et al., 2018). This mutual exclusivity at the DRNL TSS (Fig. 5D) or CLV3 TSS (Fig. 5E) cannot be expected for other genes. The AHP6 promoter, which shows the highest FC increase after DRNL in LOFCs, showed no similar change in chromatin accessibility and although AHP6 shares a similar spatiotemporal expression pattern with DRNL (Besnard et al., 2014; Chandler and Werr, 2014), it is more highly expressed in GFP– cells than DRNL.
The AHP6 promoter is a potential target of DRNL (Ikeda et al., 2006) and contains a perfect GCC box in its dTHS-up. On the DNA level, the GCC box is required for gene expression in the STY1 and CLV3 promoters (Eklund et al., 2011; Luo et al., 2018). However, the failure of ChIP experiments (Eklund et al., 2011; Luo et al., 2018) and the undetectable effect of the A37V mutation in the DRN AP2 domain (Seeliger et al., 2016) increasingly argue against direct DRN protein–DNA contact. In contrast, ChIP experiments with DRNL and DRN at the STM promoter imply that they are indirectly recruited to target gene promoters via bridging proteins such as REV (Zhang et al., 2018b). Whether the DRNL AP2 domain is a DNA-binding or protein interaction domain (Chandler et al., 2007) relates to the distribution of GCC boxes in the Arabidopsis genome. The GCC box is normally distributed in genes possessing accessible chromatin regions in GFP+ and GFP– protoplasts. Furthermore, few GCC boxes reside in dTHS-up (five) or THS-down genes (three), which suggests that DRNL binding to GCC-target sites does not causally relate to chromatin changes detected by ATAC-seq in LOFCs (Supplementary Table S3).
However, dTHS-up genes, such as potential DRNL targets AHP6 and STY1, or ROXY1, PUCHI, CAL, PHV, SEP4, AFO, and MP, reside in the GO category flower development (Fig. 4C) and thus associate with increased chromatin accessibility in LOFCs at the IM periphery. The GO category flower development is under-represented in the 392 dTHS-down genes (Fig. 4D), which were only mildly enriched in the GO categories transcription, response to endogenous stimuli, and transport. In the transport group, PIN7 relates to auxin transport and indirectly to auxin response. Downregulation of PIN7 in LOFCs is accompanied by reduced chromatin accessibility in one promoter and one intronic dTHS-down (Fig. 5K). Concomitant with PIN7, the efflux carriers PIN2–5 are also transcriptionally down-regulated in LOFCs, although changes in chromatin accessibility here were small. In contrast, transcript levels of the auxin importers AUX1, LAX1, and LAX2 were unchanged in LOFCs (Frerichs et al., 2016). This implies less auxin efflux in LOFCs, whereas import remains unchanged, a scenario that conforms to auxin accumulation in incipient floral primordia (Reinhardt et al., 2003). Chromatin changes detected by FACS/ATAC-seq thus support the prospective floral fate of LOFCs with respect to dTHS-up and local changes in auxin efflux potential with respect to dTHS-down. With respect to dTHSs-down in PIN7, however, it should be recalled that the expression domains of DRNL only partially overlap with auxin response maxima (Chandler and Werr, 2014).
In summary, we have shown that ATAC-seq combined with FACS can identify cell type-specific chromatin changes, and together with RNA-seq data, genomic regions where chromatin accessibility correlates with transcriptional activation or repression. This feature is common to active or quiescent enhancer elements (Tsompana and Buck, 2014) and is here confirmed for functional elements within the DRNL promoter. Further support derives from established MADS domain TF binding sites, which mostly reside in open chromatin regions that are stably transmitted from vegetative stem cells into the inflorescence meristem and into LOFCs.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Comparison of biological replicates within the ATAC-seq analysis and overlap with available DNase-seq data.
Fig. S2. Comparison of the chromatin configurations of exemplary genes between ATAC-seq and DNase-seq, different cell types, and enrichment methods.
Fig. S3. DNase-seq accessibility in genes and promoters used for cell type-specific enrichment or relating to developmental decisions.
Fig. S4. Correlation between ATAC-signal and gene expression.
Table S1. ATAC-seq read numbers compared with DNase-seq data.
Table S2. Statistical evaluation of dTHS relative to differentially expressed genes (DEGs).
Table S3. Representation of GCC boxes in genes carrying dTHSs and THSs.
Acknowledgements
This work was supported by grants ERA-CAPS EVOREPRO to JG-M and DFG WE 1262/11–2 to WW; WW is a member of the Cluster of Excellence on Plant Science (CEPLAS; EXC 1028), which provided infrastructural support.
Glossary
Abbreviations:
- ATAC
assay for transposase-accessible chromatin
- ATAC-seq
assay for transposase-accessible chromatin with high-throughput sequencing
- DEG
differentially expressed gene
- DHS
DNase I hypersensitive site
- DNase-seq
DNase I hypersensitive sites sequencing
- dTHS
differential THS
- FACS
fluorescence-activated cell sorting
- FC
fold change
- GO
gene ontology
- iTHS
invariant THS
- LOFC
lateral organ founder cell
- RNA-seq
whole transcriptome shot-gun sequencing
- TF
transcription factor
- THS
transposase hypersensitive site
- TSS
transcription start site
- TTS
transcription termination site
Data deposition
The data discussed in this publication have been deposited in the NCBI Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE116972.
DOI: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE116972
References
- Ackermann AM, Wang Z, Schug J, Naji A, Kaestner KH. 2016. Integration of ATAC-seq and RNA-seq identifies human alpha cell and beta cell signature genes. Molecular Metabolism 5, 233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamowski M, Friml J. 2015. PIN-dependent auxin transport: action, regulation, and evolution. The Plant Cell 27, 20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard F, Refahi Y, Morin V, et al. . 2014. Cytokinin signalling inhibitory fields provide robustness to phyllotaxis. Nature 505, 417–421. [DOI] [PubMed] [Google Scholar]
- Beveridge CA, Mathesius U, Rose RJ, Gresshoff PM. 2007. Common regulatory themes in meristem development and whole-plant homeostasis. Current Opinion in Plant Biology 10, 44–51. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature Methods 10, 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capua Y, Eshed Y. 2017. Coordination of auxin-triggered leaf initiation by tomato LEAFLESS. Proceedings of the National Academy of Sciences, USA 114, 3246–3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler JW, Cole M, Flier A, Grewe B, Werr W. 2007. The AP2 transcription factors DORNRÖSCHEN and DORNRÖSCHEN-LIKE redundantly control Arabidopsis embryo patterning via interaction with PHAVOLUTA. Development 134, 1653–1662. [DOI] [PubMed] [Google Scholar]
- Chandler JW, Cole M, Jacobs B, Comelli P, Werr W. 2011a Genetic integration of DORNRÖSCHEN and DORNRÖSCHEN-LIKE reveals hierarchical interactions in auxin signalling and patterning of the Arabidopsis apical embryo. Plant Molecular Biology 75, 223–236. [DOI] [PubMed] [Google Scholar]
- Chandler JW, Jacobs B, Cole M, Comelli P, Werr W. 2011b DORNRÖSCHEN-LIKE expression marks Arabidopsis floral organ founder cells and precedes auxin response maxima. Plant Molecular Biology 76, 171–185. [DOI] [PubMed] [Google Scholar]
- Chandler JW, Werr W. 2014. Arabidopsis floral phytomer development: auxin response relative to biphasic modes of organ initiation. Journal of Experimental Botany 65, 3097–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler JW, Werr W. 2017. DORNRÖSCHEN, DORNRÖSCHEN-LIKE, and PUCHI redundantly control floral meristem identity and organ initiation in Arabidopsis. Journal of Experimental Botany 68, 3457–3472. [DOI] [PubMed] [Google Scholar]
- Comelli P, Glowa D, Chandler JW, Werr W. 2016. Founder-cell-specific transcription of the DORNRÖSCHEN-LIKE promoter and integration of the auxin response. Journal of Experimental Botany 67, 143–155. [DOI] [PubMed] [Google Scholar]
- Deal RB, Henikoff S. 2010. A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Developmental Cell 18, 1030–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund DM, Cierlik I, Ståldal V, Claes AR, Vestman D, Chandler J, Sundberg E. 2011. Expression of Arabidopsis SHORT INTERNODES/STYLISH family genes in auxin biosynthesis zones of aerial organs is dependent on a GCC box-like regulatory element. Plant Physiology 157, 2069–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhorn J, Blanvillain R, Kröner C, Parrinello H, Rohmer M, Posé D, Ott F, Schmid M, Carles C. 2017. Dynamics of H3K4me3 chromatin marks prevails over H3K27me3 for gene regulation during flower morphogenesis in Arabidopsis thaliana. Epigenomes 1, 8. [Google Scholar]
- Frerichs A, Thoma R, Abdallah AT, Frommolt P, Werr W, Chandler JW. 2016. The founder-cell transcriptome in the Arabidopsis apetala1 cauliflower inflorescence meristem. BMC Genomics 17, 855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goslin K, Zheng B, Serrano-Mislata A, et al. . 2017. Transcription factor interplay between LEAFY and APETALA1/CAULIFLOWER during floral initiation. Plant Physiology 174, 1097–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao D, Ohme-Takagi M, Sarai A. 1998. Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plant. The Journal of Biological Chemistry 273, 26857–26861. [DOI] [PubMed] [Google Scholar]
- Heard DJ, Kiss T, Filipowicz W. 1993. Both Arabidopsis TATA binding protein (TBP) isoforms are functionally identical in RNA polymerase II and III transcription in plant cells: evidence for gene-specific changes in DNA binding specificity of TBP. The EMBO Journal 12, 3519–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong RL, Hamaguchi L, Busch MA, Weigel D. 2003. Regulatory elements of the floral homeotic gene AGAMOUS identified by phylogenetic footprinting and shadowing. The Plant Cell 15, 1296–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y, Banno H, Niu QW, Howell SH, Chua NH. 2006. The ENHANCER OF SHOOT REGENERATION 2 gene in Arabidopsis regulates CUP-SHAPED COTYLEDON 1 at the transcriptional level and controls cotyledon development. Plant & Cell Physiology 47, 1443–1456. [DOI] [PubMed] [Google Scholar]
- Irish V. 2017. The ABC model of floral development. Current Biology 27, R887–R890. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhao TJ, Liu JM, Liu WQ, Liu Q, Yan YB, Zhou HM. 2006. The conserved Ala37 in the ERF/AP2 domain is essential for binding with the DRE element and the GCC box. FEBS Letters 580, 1303–1308. [DOI] [PubMed] [Google Scholar]
- Luo L, Zeng J, Wu H, Tian Z, Zhao Z. 2018. A molecular framework for auxin-controlled homeostasis of shoot stem cells in Arabidopsis. Molecular Plant 11, 899–913. [DOI] [PubMed] [Google Scholar]
- Madrigal P. 2015. On accounting for sequence-specific bias in genome-wide chromatin accessibility experiments: recent advances and contradictions. Frontiers in Bioengineering and Biotechnology 3, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. 2005. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449. [DOI] [PubMed] [Google Scholar]
- Maher KA, Bajic M, Kajala K, et al. . 2018. Profiling of accessible chromatin regions across multiple plant species and cell types reveals common gene regulatory principles and new control modules. The Plant Cell 30, 15–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo N, Mase H, Makino M, Takahashi H, Banno H. 2009. Identification of ENHANCER OF SHOOT REGENERATION 1-upregulated genes during in vitro shoot regeneration. Plant Biotechnology 26, 385–393. [Google Scholar]
- Nag A, Yang Y, Jack T. 2007. DORNRÖSCHEN-LIKE, an AP2 gene, is necessary for stamen emergence in Arabidopsis. Plant Molecular Biology 65, 219–232. [DOI] [PubMed] [Google Scholar]
- Pajoro A, Madrigal P, Muiño JM, et al. . 2014. Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biology 15, R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales M, Rodriguez K, Snipes S, Yadav RK, Diaz-Mendoza M, Reddy GV. 2016. Threshold-dependent transcriptional discrimination underlies stem cell homeostasis. Proceedings of the National Academy of Sciences, USA 113, E6298–E6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Research 44, W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Pesce ER, Stieger P, Mandel T, Baltensperger K, Bennett M, Traas J, Friml J, Kuhlemeier C. 2003. Regulation of phyllotaxis by polar auxin transport. Nature 426, 255–260. [DOI] [PubMed] [Google Scholar]
- Seeliger I, Frerichs A, Glowa D, Velo L, Comelli P, Chandler JW, Werr W. 2016. The AP2-type transcription factors DORNRÖSCHEN and DORNRÖSCHEN-LIKE promote G1/S transition. Molecular Genetics and Genomics 291, 1835–1849. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijacic P, Bajic M, McKinney EC, Meagher RB, Deal RB. 2018. Changes in chromatin accessibility between Arabidopsis stem cells and mesophyll cells illuminate cell type-specific transcription factor networks. The Plant Journal 94, 215–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsompana M, Buck MJ. 2014. Chromatin accessibility: a window into the genome. Epigenetics & Chromatin 7, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D, Wellmer F, Dilks K, William D, Smith MR, Kumar PP, Riechmann JL, Greenland AJ, Meyerowitz EM. 2004. Floral induction in tissue culture: a system for the analysis of LEAFY-dependent gene regulation. The Plant Journal 39, 273–282. [DOI] [PubMed] [Google Scholar]
- Weber B, Zicola J, Oka R, Stam M. 2016. Plant enhancers: a call for discovery. Trends in Plant Science 21, 974–987. [DOI] [PubMed] [Google Scholar]
- Wellmer F, Alves-Ferreira M, Dubois A, Riechmann JL, Meyerowitz EM. 2006. Genome-wide analysis of gene expression during early Arabidopsis flower development. PLoS Genetics 2, e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav RK, Girke T, Pasala S, Xie M, Reddy GV. 2009. Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proceedings of the National Academy of Sciences, USA 106, 4941–4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav RK, Tavakkoli M, Xie M, Girke T, Reddy GV. 2014. A high-resolution gene expression map of the Arabidopsis shoot meristem stem cell niche. Development 141, 2735–2744. [DOI] [PubMed] [Google Scholar]
- Zhang C, Du X, Tang K, et al. . 2018. a Arabidopsis AGDP1 links H3K9me2 to DNA methylation in heterochromatin. Nature Communications 9, 4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Wang J, Wenkel S, Chandler JW, Werr W, Jiao Y. 2018b Spatiotemporal control of axillary meristem formation by interacting transcriptional regulators. Development 145, dev158352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Zhang W, Jiang J. 2015. Genome-wide nucleosome occupancy and positioning and their impact on gene expression and evolution in plants. Plant Physiology 168, 1406–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.