Abstract
The introgression of a small segment of wheat (Triticum aestivum L.) chromosome arm 1BS in the distal region of the rye (Secale cereale L.) 1RS.1BL arm translocation in wheat (henceforth 1RSRW) was previously associated with reduced grain yield, carbon isotope discrimination, and stomatal conductance, suggesting reduced access to soil moisture. Here we show that lines with the normal 1RS arm have longer roots than lines with the 1RSRW arm in both field and hydroponic experiments. In the 1RSRW lines, differences in seminal root length were associated with a developmentally regulated arrest of the root apical meristem (RAM). Approximately 10 d after germination, the seminal roots of the 1RSRW plants showed a gradual reduction in elongation rate, and stopped growing a week later. Seventeen days after germination, the roots of the 1RSRW plants showed altered gradients of reactive oxygen species and emergence of lateral roots close to the RAM, suggesting changes in the root meristem. The 1RSRW lines also showed reduced biomass (estimated by the normalized difference vegetation index) and grain yield relative to the 1RS lines, with larger differences under reduced or excessive irrigation than under normal irrigation. These results suggest that this genetic variation could be useful to modulate root architecture.
Keywords: Drought tolerance, lateral roots, reactive oxygen species (ROS), roots, rye, waterlogging, wheat
A wheat/rye polymorphism in chromosome 1B affects seminal root length and lateral root proliferation and is associated with differences in drought and waterlogging tolerance in the field.
Introduction
Approximately 750 Mt of wheat (Triticum aestivum L.) are produced worldwide every year (FAO, 2017), but further increases are required to feed a growing human population. One understudied area that can contribute to these yield increases is the role of different root architectures on wheat adaptation to different soils. Although some progress has been made in the understanding of root development and architecture in Arabidopsis (Yu and Luan, 2016; Liu et al., 2017), this knowledge is lacking in grass species (Voss-Fels et al., 2018). There have been some examples of phenotypic selection of root architecture in breeding programs (Richards et al., 2010; Wasson et al., 2012), but those methods are laborious and can be accelerated by a better understanding of the genes controlling wheat root architecture.
Rye (Secale cereal L.), a close relative of wheat, is more tolerant to water shortages than wheat, and has been reported to have a more robust root system. The translocation of the short arm of rye chromosome one (1RS) to wheat chromosome 1B (1RS.1BL) contributes to above-ground biomass (Shearman et al., 2005) and better performance under drought stress (Moreno-Sevilla et al., 1995; Ehdaie et al., 2003, 2012; Zarco-Hernandez et al., 2005; Hoffmann, 2008). To address bread-making quality problems associated with the 1RS.1BL translocation (Fenn et al., 1994), a recombinant 1RS chromosome including two wheat 1BS chromosome segment introgressions (henceforth 1RSWW) was developed to eliminate the two rye regions associated with the bread-making quality problems (Lukaszewski, 2000).
We introgressed the newly engineered chromosome into the spring wheat variety ‘Hahn’ and generated 1RS/1RSWW near isogenic lines (NILs). Previous field trials showed that the Hahn 1RS lines had significantly higher yield and better canopy water status than the 1RSWW NILs in both well-watered and water-stressed environments, although the differences were larger in the latter (Howell et al., 2014). From a cross between Hahn 1RSWW and Hahn 1RS, we generated two additional NILs, one carrying the distal (1RSRW) and the other the proximal wheat segment (1RSWR). The two NILs carrying the distal rye region (1RS and 1RSWR, henceforth 1RSxR) showed significant improvements in grain yield and canopy water status compared with NILs carrying the distal wheat segment (1RSWW and 1RSRW, henceforth 1RSxW). The 1RSxR NILs also showed higher carbon isotope discrimination and increased stomatal conductance, suggesting improved access to soil moisture relative to the 1RSxW NILs (Howell et al., 2014).
In the winter of 2013, heavy rains waterlogged a UC Davis experimental field which affected the four 1RS NILs at the early tillering stage. Although the affected areas were irregular, the 1RSxR NILs were less affected than the 1RSxW NILs. Based on this observation and previous results, we hypothesized that the 1RSxR lines might have a more extensive root system than the 1RSxW lines, which helped them tolerate both waterlogging in this experiment and water shortages in the previously published experiments (Howell et al., 2014).
The first objective of this study was to characterize the effect of the wheat/rye polymorphism in the distal region of the 1RS.1BL translocation on root architecture in the field, and on plant biomass and grain yield under normal, excessive, or reduced irrigation. After we observed that the lines with the distal wheat segment had shorter seminal roots than the lines with the distal rye segment in hydroponic conditions, we also decided to study the effect of these genotypes on seminal root growth rates, distribution of reactive oxygen species (ROS), and distribution of lateral roots. The implications of the observed differences in root development and architecture are discussed.
Materials and methods
Plant materials
In this study, we used four NILs that showed differences in grain yield in previous work (Howell et al., 2014). The recurrent common wheat parent of these NILs is the spring wheat cultivar ‘Hahn’ developed by the International Maize and Wheat Improvement Center (CIMMYT). The Hahn cultivar carries the complete 1RS translocation from rye, and the three NILs differed from Hahn in the presence of either a distal interstitial segment of wheat chromatin (1RSRW), a proximal interstitial segment of wheat chromatin (1RSWR), or both (1RSWW) (Howell et al., 2014). The interstitial wheat segments were introgressed from the common wheat cultivar ‘Pavon 76’ (also developed at CIMMYT) to eliminate the Sec-1 locus from 1RS and to incorporate the Glu-B3/Gli-B1 locus from 1BS into the 1RS chromosome to improve bread-making quality (Lukaszewski, 2000). The source of this 1RS arm was the rye cultivar ‘Petkus’, and the resulting 1RS.1BL translocation became widely distributed in wheat breeding programs around the world (Rabinovich, 1998; Schlegel and Korzun, 1997; Landjeva et al., 2006; Zhang et al., 2011).
All field experiments were conducted at the University of California field station in Davis, CA (38°32'N, 121°46'W), which has deep Yolo loam soils (fine-silty, mixed, superactive, non-acid, thermic Mollic Xerofluvent). Experiments were sown in plots at a density of 300 grains m–2 (3 million grains ha–1).
Waterlogging experiments
Controlled waterlogging experiments were conducted during the 2013–2014 and 2015–2016 growing seasons. An additional experiment was performed in 2014–2015, but it was not analyzed due to severe weed problems. The experiments were planted in November and harvested in June (experiments are designated by their harvest year).
The two waterlogging experiments were organized in a split-plot randomized complete block design (RCBD) with four blocks in 2014 and three blocks in 2016. Within each block, the main factor was irrigation treatment, and within each irrigation treatment–block combination, the Hahn 1RS, 1RSWW, 1RSRW, and 1RSWR genotypes were used as subplots. The average trait values of the 1RSxR and 1RSxW NILs were compared to determine the effect of the distal rye and wheat chromosome segments.
In the 2014 field experiment, each block included two different irrigation regimes as main plots. The first treatment was based on plant needs and normal practices in California’s Sacramento Valley and is designated hereafter as normal irrigation. The second treatment, referred to hereafter as waterlogging, consisted of artificial flooding twice a week starting in late January and ending in late March during the tillering stage, followed by normal irrigation. Water was applied via flood irrigation, and the soil profile remained saturated. While plants were not kept fully or partially submerged, there were persistent pools of water on the soil surface indicating a waterlogged environment. Each genotype was planted in three adjacent 1 m rows (experimental unit) with 30.5 cm spacing between rows at a rate of 30 grains per row. Genotypes were separated by an empty row (61 cm spacing), and treatments were separated by a minimum of a border row, an irrigation levee, and another border row, leaving >3 m between experimental units of different treatments. Experimental units were replicated six times within each of the four blocks in an RCBD pattern and were used as subsamples. At the end of the season, each set of three rows was harvested and grain yield was recorded. The average of the six subsamples was used as a single data point in the statistical analysis. Canopy spectral reflectance (CSR) measurements were taken for all subsamples on two days (17 April and 30 April, both post-anthesis). Subsamples were averaged within days, and day averages were used as repeated measures.
Canopy spectral reflectance measurements were taken with the ‘ASD HandHeld 2 Pro’ spectrometer from Malvern Panalytical. Measurements were taken using a ‘scanning’ method in which 50 measurements were taken on a single plot and averaged to give a single reflectance spectrum. From these measurements, differences in biomass between genotypes were estimated using the normalized difference vegetation index (NDVI), which was calculated using the formula (R900 nm–R680 nm)/(R900 nm+R680 nm), where R=reflectance at the specified wavelength.
In the 2016 field experiment, each block included three irrigation treatments. The first treatment was grown under normal irrigation as described above. The waterlogging treatment included flood irrigations three times a week, from the beginning of February (later than in 2014 due to a wet winter) to the end of February, followed by normal irrigation. The terminal drought treatment was grown under normal irrigation conditions until late March (before the booting stage), with no additional irrigations after that point. Within each block–treatment combination, each genotype was machine sown in 2.23 m2 plots (1.83×1.22 m), which were combine-harvested at maturity. In 2016, CSR measurements were taken as described above on 24 March (booting), 6 April (anthesis), and 13 and 28 April (both post-anthesis). Days were used as repeated measurements and were analyzed as sub-subplots in an RCBD split-split-plot design using conservative degrees of freedom for days and all their interactions (this was not necessary in 2014 because there were only two days and one degree of freedom). After the CSR measurements were completed, an irrigation pipe ruptured, flooding several sections of the experiment on 29 April, resulting in increased variability in the final yield measurements. Flooding was irregular and inconsistent across blocks, with major effects on replications two and three of the drought treatment and replication two of the waterlogging treatment.
Root depth experiments
The field experiment to estimate root length was conducted after a maize crop harvested in the summer of 2016. The field was organized in an RCBD with six blocks and four genotypes per block. Plots were machine sown in 4.5 m2 plots in November 2016 and were grown under normal irrigation conditions. To obtain soil core samples at specific depths and avoid differential soil compaction, we excavated ~2 m deep trenches cutting perpendicular across the middle of plots including complete blocks one (3 March, 2017, North side), three (20 March 2017, North side), and six (9 March, 2017, South side) to expose the root system. We took horizontal soil core samples from the center of each block at 20 cm intervals using a thin-walled copper pipe (5.08 cm diameter×35 cm long=709.4 cm3). Core samples were taken from 20 cm to 140 cm in the first block and from 20 cm to 180 cm in blocks three and six (see Supplementary Fig. S1 at JXB online) after we discovered the presence of roots at 140 cm in block one. Plants were at the tillering stage at the time of the root sampling.
Determination of root parameters in soil samples
Soil core samples were washed using a hydro-pneumatic elutriation system from Gillison’s Variety Fabrications, Inc. (Smucker et al., 1982). After washing and sorting white turgid roots from other organic matter and decayed roots of the previous maize crop (easily distinguished by color, texture, and diameter), we suspended the roots in water and scanned them using an EPSON Expression 11000XL flatbed scanner. Scanned root images were analyzed using the WinRhizo software package. Measurements of dry root biomass are not reported because they were too variable due to small biomass, stray soil contaminants, and changes in ambient moisture. The 20 cm sampling point was not used because the large amount of root biomass and organic matter present in these samples made them difficult to clean and measure.
Since all root measurements were performed using soil cores of identical volume (709.4 cm3), we refer to these measures as densities (except for average root diameter). Differences in total root length, surface and volume density, average root diameter, and root tip and fork densities were analyzed using a split-plot design with genotypes (1RSxR and 1RSxW) as main plots and depth as subplot. This is a conservative statistical analysis because it reduces the degrees of freedoms for genotype from three to one. Therefore, we also compared the same two pairs of genotypes using statistical contrasts in an ANOVA including all four genotypes. To account for the inability to randomize depths, we used a conservative estimate of the degrees of freedom for subplots and for the interaction between subplot and main plot. Conservative degrees of freedom were calculated by dividing their degrees of freedom by the number of subplots. This strategy is similar to that used for repeated measures in time and does not affect comparisons among main plots (genotypes), which are the main objective of this study. Homogeneity of variance and normality of the residuals was confirmed for all the individual ANOVAs performed at each depth for all parameters. When necessary, data were transformed using power transformations to satisfy ANOVA assumptions.
Hydroponic experiments
Hydroponic experiments were performed in growth chambers at 22–23 °C with a photoperiod of 16 h light/8 h dark provided by fluorescent lights supplemented with incandescent lighting. In all experiments, grains were imbibed at 4 °C for 4 d, after which they were placed at room temperature. Once most of the grains had germinated and the coleoptiles had emerged, healthy seedlings were transferred to a mesh suspended on water (UCD) or CaCl2 0.5 mM (Chascomús). Four days later, healthy seedlings were transferred to tanks with growth solutions [KH2PO4 0.2 mM, MgSO4·7H2O 1.0 mM, CaCl21.5 mM, KCl 1.5 mM, H3BO3 1 μM, (NH4)6Mo7O24·4H2O 0.05 μM, CuSO4·5H2O 0.5 μM, ZnSO4·7H2O 1 μM, MnSO4·H2O 1 μM, FeEDTA 0.1 mM, Ca(NO3)2 1.0 mM, (NH4)2SO4 1.0 mM]. After removing the grain, seedlings were wrapped at the crown with foam and inserted in holes pre-cut in a foam core board placed on top of the solution. Nutrient solution was changed 2–3 times a week for the duration of the experiment. Particular details of the methods used in the two laboratories are described below.
Davis, CA, USA
The experiments were performed in 13 liter hydroponic tanks containing the nutrient solution. Twenty-four seedlings were placed in each tank in a six by four pattern. All genotypes were included in one tank and, if necessary, multiple tanks were used as replications.
In the experiments to study the effect of different nitrogen sources (nitrate and ammonium) and concentrations (0.2 mM and 2 mM), seedlings were grown in normal growth solution for 7 d [from 5 to 12 days after germination (DAG)] and then transferred to four separate tanks with each of the four nitrogen sources for 10 d (12–22 DAG). Roots were measured at 22 DAG. Each tank included six replications of each of the four genotypes organized in a completely randomized design. The results were analyzed in a 2×2 factorial ANOVA with distal and proximal 1RS regions as factors and wheat or rye chromosome segments as levels.
For the analysis of distances between the first lateral roots and the root apical meristem (RAM), the four genotypes were grown in a tank with normal nitrogen conditions (2 mM nitrate) in a completely randomized design (n=12). Lines carrying distal rye (1RSxR) or wheat (1RSxW) segments were compared using a t-test.
Chascomús, Buenos Aires Argentina
The CaCl2 (0.5 mM) from the germination tank was replaced by nutrient solution on the fourth day. On the fifth day, plants were transferred to 350 ml pots containing nutrient solution, with each pot (plant) being a replicate. Pots were rotated every 2 d to ensure that they occupied different positions within the growth chamber. For the root elongation time course, the length of the second longest seminal root was measured daily 4 h after the start of the light period, beginning 6 DAG. Within each experiment, data were analyzed as repeated measures (split-plot in time with conservative degree of freedom). A combined ANOVA was performed using experiments as blocks.
Nitroblue tetrazolium (NBT) and 2',7'-dichlorofluorescein diacetate (DCF-DA) staining
The NBT staining experiments were performed using 5 cm root sections excised from the second longest root of 1RS and 1RSRW plants 17 DAG. This root segment was placed for 90 min in a 0.1 mg ml–1 NBT solution dissolved in 200 mM potassium phosphate buffer, pH 7.6, in darkness. For the DCF-DA staining, similar root segments were placed in the same buffer supplemented with 10 µM DCF-DA for 60 min, in darkness. Roots segments stained with NBT or DCF-DA were washed in the same buffer for 30 min and placed on a slide. Roots were observed using a Zeiss Discovery.V20 (Carl Zeiss MicroImaging, Germany) stereomicroscope equipped with a coaxial fluorescence mechanism. Pictures were obtained with an Axiocam 512 color (Carl Zeiss MicroImaging, Germany). The images were processed with the ImageJ software, obtaining a longitudinal profile of color and fluorescence intensities. These experiments were performed in Chascomús.
Results
1RSxW lines showed less tolerance to waterlogging than 1RSxR lines in the field
Controlled waterlogging experiments performed in 2014 and 2016 using the four Hahn NILs showed that the NDVI and yield responses of the two lines carrying the distal rye chromosome segment (1RSxR) were similar to each other and different from the two lines carrying the distal wheat chromosome segment (1RSxW), which were also similar to each other (Supplementary Fig. S2). These results were consistent across years and irrigation treatments, so for simplicity we grouped the genotypes with the same distal segment for the following statistical analyses and figures.
2014 field experiment
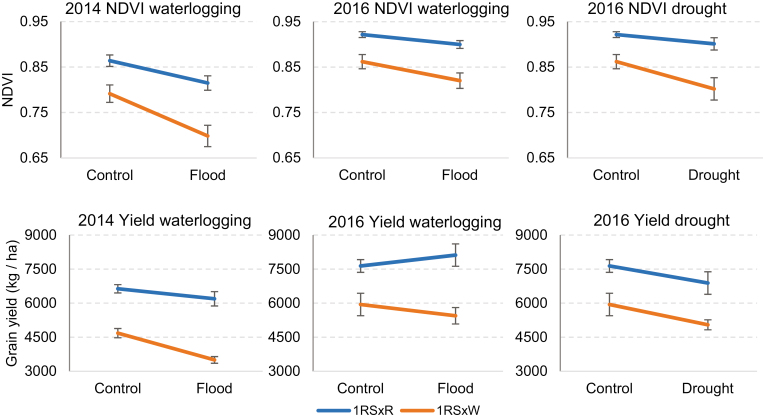
The repeated measures ANOVA (2 d) of the RCBD split-plot experiment harvested in 2014 showed significant differences between control and waterlogging conditions (main treatment) for NDVI (P=0.0025; Supplementary Table S1), but the effect of the treatment on yield was not significant (P=0.1556; Supplementary Table S3). For both traits, we detected highly significant differences between genotypes (subplots, P<0.0001), with the 1RSxR lines showing higher NDVI and grain yield than the 1RSxW lines (Fig. 1). The NDVI differences between genotypes under waterlogging conditions were larger than under normal irrigation. This was reflected in a significant interaction between waterlogging and genotype (P=0.0034; Supplementary Table S1) that can be visualized in the interaction graph as lack of parallelism between lines (Fig. 1). The same trend was observed for yield (Fig. 1), but the interaction was not significant (P=0.1101; Supplementary Table S3), probably due to the conservative statistical analyses used for these tests (see the Materials and methods) and the higher variability of the yield data.
Fig. 1.
NDVI (top row) and yield (bottom row) under normal irrigation (control), waterlogging, and drought conditions. Lines with the distal rye segment (1RSxR) showed significantly higher NDVI and yield values (P<0.0001) than the lines with the distal wheat segment (1RSxW) in all experiments. Differences between genotypes were generally larger under water stress conditions. Error bars are the SE of the means across blocks.
Since the genotype×day interaction for NDVI was significant, we also performed separate statistical analyses for each of the two days. Each of the two days showed similar results, with significant differences between treatments, genotypes, and genotype×treatment interactions (Supplementary Table S2).
2016 field experiment
This RCBD split-split-plot experiment included three irrigation treatments (control, waterlogging, and drought). To facilitate the comparisons with the 2104 experiment and to simplify the statistical analysis of the interactions, we present the ANOVA tables for waterlogging (Supplementary Tables S4–S6) and drought separately (Supplementary Tables S7–S9), although they share the same control treatment.
In the waterlogging experiment, we observed significant differences between treatments for NDVI (P=0.0010; Supplementary Table S4) but not for yield (P=0.9772; Supplementary Table S6). The differences between genotypes were highly significant for both NDVI (P<0.0001; Supplementary Table S4) and grain yield (P=0.0054; Supplementary Table S6), despite the rupture of an irrigation pipeline that affected the treatments and blocks in irregular patterns leading to increased variability of the grain yield results (all NDVI measurements were obtained before the incident). For both parameters, the 1RSxR lines showed higher values than the 1RSxW lines (Fig. 1). We observed a consistent trend for larger NDVI differences between genotypes in the waterlogging treatment than in the control (Fig. 1), but the waterlogging by genotype interaction was marginally non-significant in the overall NDVI analysis (P=0.0783; Supplementary Table S4) and not significant for yield (P=0.2888; Supplementary Table S6). For NDVI, the individual ANOVAs by day showed marginally non-significant interactions for the measurements taken in the middle of the waterlogging experiment (6 April P=0.06 and 16 April P=0.07), but not for those closer to the beginning and end of the measurement window (24 March P=0.91 and 28 April P=0.16; Supplementary Table S5). The differences between genotypes were highly significant for all four individual ANOVAs (Supplementary Table S5).
No significant differences were detected between the drought and control treatments for NDVI (P=0.1130) and grain yield (P=0.4587). In contrast, highly significant differences were detected between genotypes for both parameters (NDVI P=0.0002; Supplementary Table S7; grain yield P=0.0088; Supplementary Table S9). We also detected a significant treatment by genotype interaction for NDVI (P=0.0308; Supplementary Table S7) but not for yield (P=0.8561; Supplementary Table S9), probably due to the additional variability generated by the irrigation pipe incident. In the four individual ANOVAs by day, the 1RSxR lines showed higher NDVI and grain yield than the 1RSxW lines (Fig. 1; Supplementary Fig. S2, Supplementary Table S8).
Taken together, these results confirmed the better performance of the 1RSxR lines compared with the 1RSxW lines, with larger differences observed under excessive or insufficient irrigation than under control irrigation (Fig. 1).
1RSxW lines have shorter roots than 1RSxR lines in the field
Total root length density
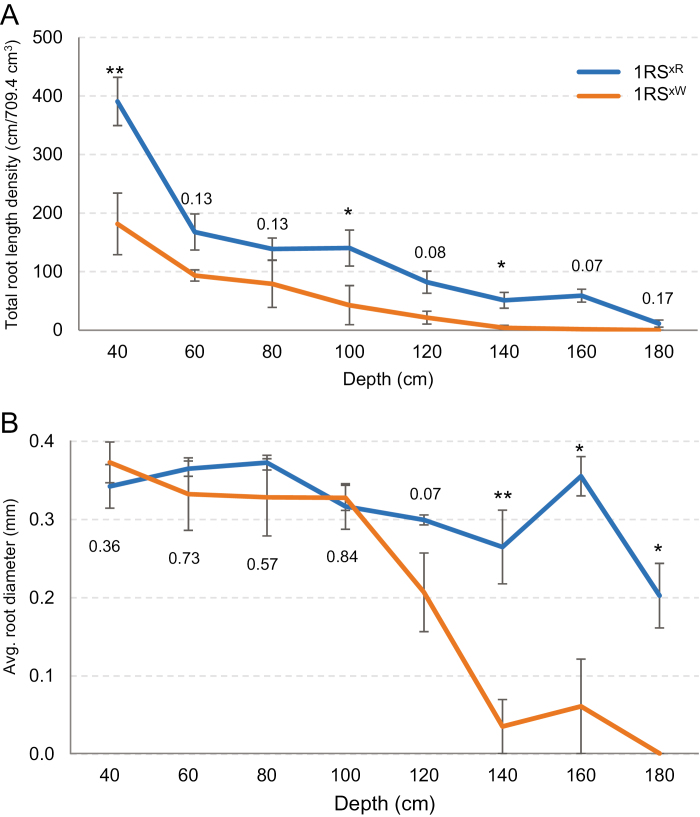
The graph for total root length density (total root length in a 709.4 cm3 soil volume) at different soil depths showed consistent differences between genotypes grouped by the distal rye or wheat segments (Fig. 2A). The total root length densities were consistently higher in the 1RSxR than in the 1RSxW NILs through the soil profile, with the largest absolute differences detected at 40 cm (Fig. 2A).
Fig. 2.
Change in total root length density and average root diameter in 1RSxW and 1RSxR. (A) Total root length density. Relative changes are presented in Supplementary Fig. S3A and B. (B) Average root diameter. P-values represent differences between 1RSxW and 1RSxR at individual depth (Supplementary Tables S12, S15). Error bars represent the SE of the means across blocks. Asterisks or numbers above error bars are P-values for the difference between genotypes at individual depths. *P<0.05, **P<0.01, ***P<0.001.
The overall ANOVA for total root length density showed significant differences between the 1RSxR and 1RSxW genotypes (P=0.0263; Supplementary Table S10). The limited degrees of freedom resulting from the averaged genotypes make this analysis very conservative. To increase the power of the analysis, we performed a statistical contrast between the two lines with the distal rye segment and the two lines with the distal wheat segment in a separate ANOVA including all four genotypes. In this analysis with a higher number of degrees of freedom, the difference between the 1RSxR and 1RSxW genotypes was highly significant (P=0.004; Supplementary Table S11).
The overall ANOVA showed a marginally significant interaction between genotype and depth (P=0.0497), but it became non-significant when adjusted using conservative degrees of freedom (P=0.190; Supplementary Table S10). The total root length density decreases with depth, which minimizes the absolute value of the differences between genotypes at more extreme depths. To address this limitation, we explored the differences between genotypes expressed as a percentage of the 1RSxR values {[(1RSxR–1RSxW)/1RSxR]×100}. This analysis showed that the relative difference in total root length between 1RSxW and 1RSxR plants was larger at the deepest sampling points (Supplementary Fig. S3A, regression between depth and percentage difference P=0.002). No roots were detected for 1RSxW at 180 cm.
Finally, we performed individual ANOVAs by depth and detected significant differences between genotypes at three depths and marginally non-significant differences at two (Fig. 2A; Supplementary Table S12). In summary, Fig. 2A and the statistical analyses in Supplementary Tables S10–S12 indicated that the 1RSxR lines have more extended and deeper root systems than the 1RSxW lines.
Average root diameter and combined root traits
In the overall split-plot ANOVA for average root diameter, the effect of depth was significant, even when using conservative degrees of freedom (P=0.02; Supplementary Table S13). The difference between the average 1RSxR and 1RSxW genotypes was marginally non-significant in the conservative analysis (P=0.0649; Supplementary Table S13), but was highly significant in the statistical contrast between the pairs of genotypes in the ANOVA including the four genotypes (P=0.006; Supplementary Table S14). In the analyses by individual depths, we observed similar average root diameters for the 1RSxR and 1RSxW genotypes between 40 cm and 100 cm, but those values started to differentiate at 120 cm (P=0.07) and were significant at 140, 160, and 180 cm. At all these depths, the average root diameter was larger in the 1RSxR than in the 1RSxW lines (Fig. 2B; Supplementary Table S15).
A similar result was also observed when the differences in root diameter were calculated as the decrease in 1RSxW relative to 1RSxR. In this graph, a sharp increase in the effect of genotype was observed below the 100 cm depth (Supplementary Fig. S3B; regression between depth and relative root diameter P=0.002). Taken together, these results suggest that the main seminal and/or adventitious roots from the 1RSxR lines reach deeper in the soil than those of the1RSxW lines, for which only thinner roots are found deeper in the soil profile.
The other root traits measured with WinRhizo showed a profile similar to that observed for total root length density (Supplementary Fig. S4). This is not surprising because root surface and volume density are a function of root length and diameter, and the number of root tips and forks is mainly affected by the abundance of roots. The contrasts between 1RSxW and 1RSxR in the ANOVAs using the four genotypes were significant for root surface (P=0.005), root volume (P=0.008), number of tips (P=0.02), and number of forks (P=0.003; Supplementary Fig. S4). Significant differences were also detected in 3–4 of the ANOVAs for individual depths in each of the traits (Supplementary Fig. S4).
1RSxW lines have shorter primary roots than 1RSxR lines in hydroponic cultures
The changes in total root length density and diameter with soil depth include both seminal and adventitious roots. To see if the differences observed between the 1RSxW and 1RSxR lines in the field could also be detected at early growth stages, when the root system is dominated by seminal roots, we performed hydroponic experiments.
Differences in root length
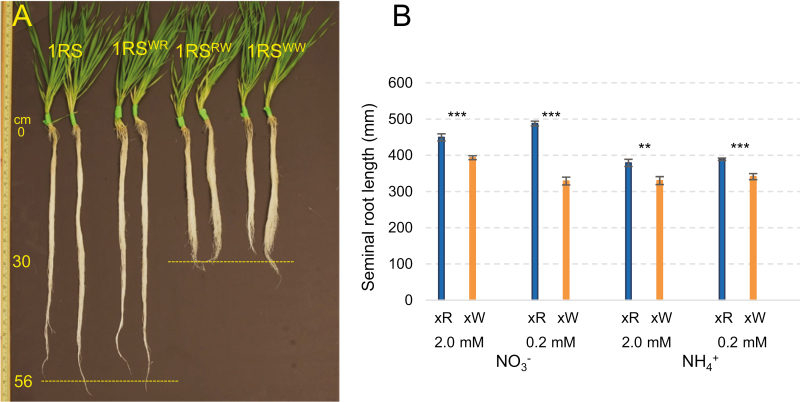
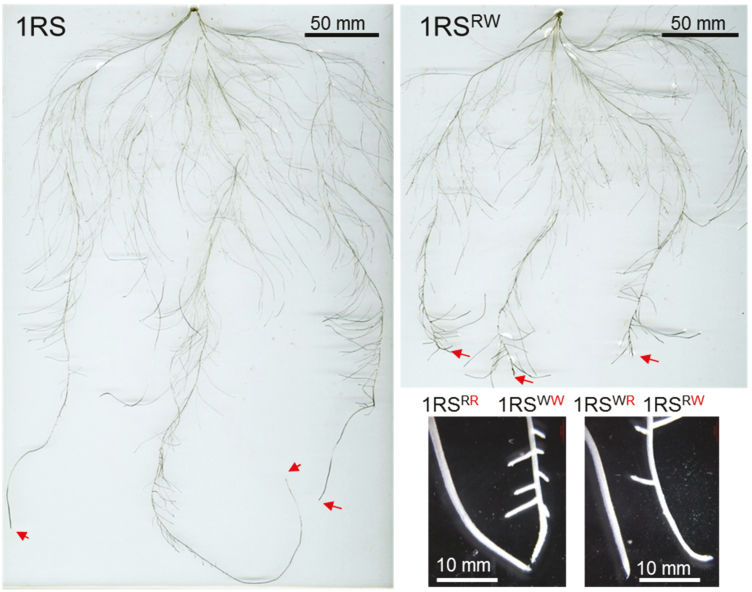
In the hydroponic experiments, the 1RSxR lines showed significantly longer seminal roots than the 1RSxW lines (Fig. 3A). Since root length was not significantly different between lines with the same distal chromosome segment, those lines were averaged for the statistical analyses (Fig. 3B). Highly significant differences in seminal root length were detected between the 1RSxW and 1RSxR plants in all four nitrogen treatments (Fig. 3B), which indicates that these differences are robust across different ionic environments.
Fig. 3.
Differences in primary root length between plants carrying the distal rye (1RS and 1RSWR) and wheat chromosome segments (1RSRW and 1RSWW) in hydroponic culture. (A) Representative plants in normal culture medium. (B) Seminal root length in hydroponic media using nitrate or ammonium at two different concentrations (2.0 mM and 0.2 mM). **P<0.01, ***P<0.001.
Root elongation time course
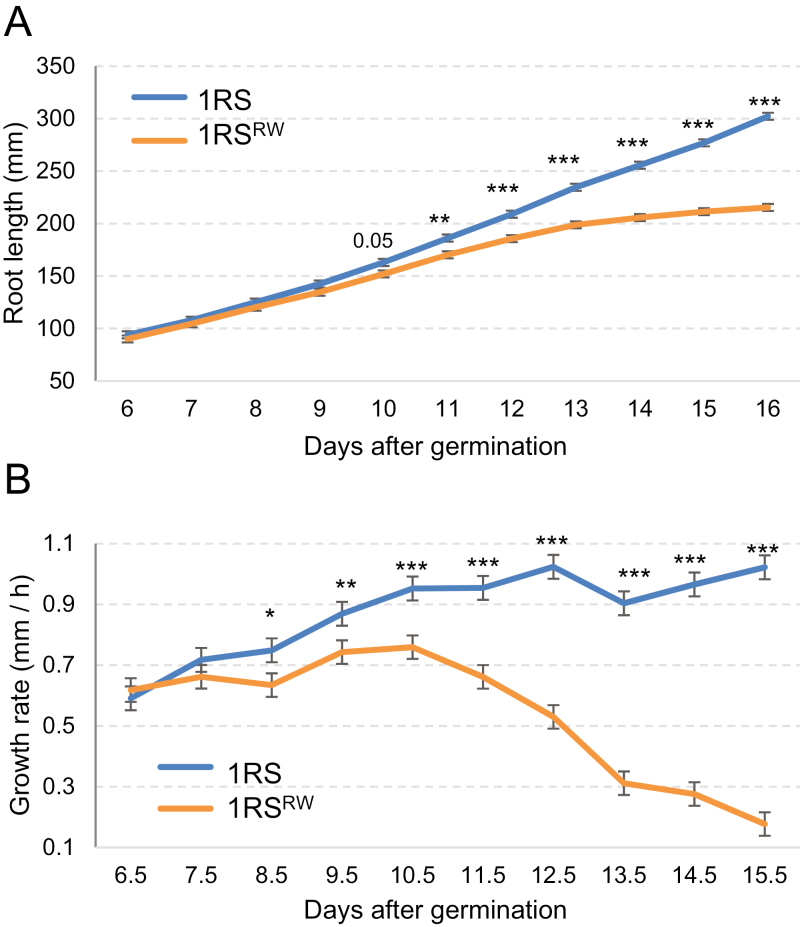
The previous results indicated that the differences in primary root length between the 1RSxR and 1RSxW plants start at an early stage of root development. These differences were consistent between the lines with the same distal chromosome segment, so root growth experiments were carried out using only the 1RS and 1RSRW lines (Supplementary Tables S16–S19). In the overall ANOVAs for accumulated root length, highly significant differences were detected between genotypes over time (P<0.0001; Supplementary Table S16; Fig. 4A) and elongation rate (Supplementary Table S18; Fig. 4B). The differences between days and the genotype × day interactions were also highly significant for both parameters (P<0.001; Supplementary Tables S16, S18). In separate ANOVAs performed for individual days, we detected significant differences in accumulated root length starting at 11 DAG, although differences were already close to significant at 10 DAG (P=0.053; Supplementary Table S17; Fig. 4A). In the same experiments, the differences in the rate of elongation were significant from day 8.5 (P=0.0328, Supplementary Table S19; Fig. 4B).
Fig. 4.
Time course of primary root growth of plants carrying the distal rye (1RS) or wheat chromosome segments (1RSRW) in hydroponic culture. Least square means and SEs are from the ANOVA of the three combined experiments. (A) Cumulative length of the second longest seminal root (mm; Supplementary Tables S16, S17). (B) Elongation rate of the second longest seminal root (mm h–1; Supplementary Tables S18, S19). *P<0.05, **P<0.01, ***P<0.001.
1RSxR and 1RSxW lines differ in the distribution of lateral roots and ROS
In seminal roots of the 1RSxR plants imaged at 22 DAG, lateral roots started to appear 10.42±0.47 cm from the root tip (Fig. 5; Supplementary Fig. S5). In contrast, the seminal roots of the 1RSxW plants showed lateral roots appearing significantly closer to the RAM (2.43±0.44 cm, P<0.0001; Supplementary Fig. S5). No significant differences were detected between lines differing in the proximal wheat and rye segments (Supplementary Fig. S5).
Fig. 5.
Development of laterals on the seminal roots of plants carrying the distal rye or wheat segments. Plants were grown for 22 d in hydroponics under low nitrogen conditions (0.2 mM nitrate). Arrows indicate the root apical meristem (RAM) of the seminal roots. The inset shows a detailed view of the seminal root tips for the four genotypes. 1RSxW lines show lateral roots close to the RAM.
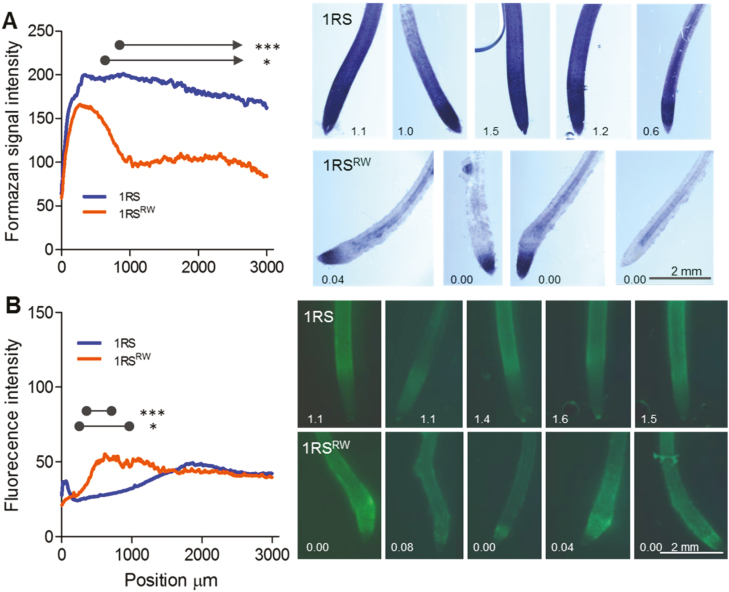
Since ROS gradients affect root elongation (Tyburski et al., 2009), we estimated their distribution in seminal roots 17 DAG by measuring the amount of formazan produced from the reduction of NBT and fluorescence in DCF-DA staining. Formazan intensity, associated with superoxide anions, was similar between genotypes near the root tips (distal region) but was significantly lower in the 1RSRW than in the 1RS roots starting at 650 μm from the root tip (P<0.05), and becoming even more pronounced after 860 μm (P<0.001; Fig. 6A). Fluorescence intensity from the DCF-DA staining, associated mainly with hydrogen peroxide, peroxynitrite, and hydroxyl radicals, showed a very different pattern, with significantly higher intensities in 1RSRW relative to the 1RS roots between 250 μm and 950 μm, and even higher differences between 350 μm and 640 μm measured from the root tip (P<0.001; Fig. 6B).
Fig. 6.
Distribution of ROS along the main roots of plants carrying the distal rye (1RS) or wheat chromosome segments (1RSRW) 17 d after germination. (A) Formazan signal intensity along the roots as determined with NBT (mainly detects superoxide anions). (B) Fluorescence signal intensity along the root as determined by DCF-DA (mainly detects hydrogen peroxide). Zero indicates the most distal point of the root (root tip). Representative NBT- and DCF-DA-stained roots from the corresponding experiment (number indicates growth rate in mm h–1 of the individual roots at the last sampling time). Circles connected with a line indicate the interval over which the indicated significance threshold is met; an arrow indicates that the interval is not bounded. *P<0.05, ***P<0.001, n=5.
Finally, we compared the distribution of formazan during lateral root development in 1RS and 1RSRW (Supplementary Fig. S6). No differences between genotypes were detected at 6 DAG, but at 9 DAG NBT staining revealed lateral root primordia close to the RAM in the 1RSRW line but not in the 1RS line. By 15 DAG, large lateral roots were formed close to the RAM in 1RSRW, but no lateral root primordia were observed in the same region in the 1RS line. These results confirmed the differences in the distribution of lateral roots between the two genotypes.
Discussion
1RSxR genotypes show deeper roots in the field and increased tolerance to waterlogging and terminal drought
Previous studies have shown an association between the introgression of the rye 1RS arm in wheat and improved resistance to water stress (Moreno-Sevilla et al., 1995; Ehdaie et al., 2003; Zarco-Hernandez et al., 2005; Hoffmann, 2008; Ehdaie et al., 2012). In three of these studies, the 1RS.1BL lines showed increased root biomass compared with the non-1RS control lines in large pot or sand-tube experiments. However, these differences were not validated in the field. In this study, we showed that differences in grain yield and biomass between plants carrying a complete 1RS translocation and NILs with an introgressed distal wheat chromosome segment (1BS) are associated with differences in total root length density and average root diameter in the field. Field excavations of the four different 1RS NILs provided an opportunity to visualize the differences in their root systems and to quantify these differences using horizontal soil cores at consistent depths.
This experiment confirmed the hypothesis that the 1RSxR lines have a higher root density throughout the soil profile, with roots that reach deeper in the soil than the 1RSxW lines (Fig. 2A). The more extensive root system of the 1RSxR lines relative to the 1RSxW lines may have contributed to their better tolerance to drought and waterlogging conditions in the experiments presented in this study (Fig. 1), and to the higher carbon isotope discrimination and increased stomatal conductance values detected in a previous study (Howell et al., 2014). Through their deeper root system, the 1RSxR plants can access more stored soil moisture and nutrients, keep their stomata open longer, and generate additional photosynthetic products and biomass compared with the 1RSxW plants. However, we cannot rule out the possibility that the genes in the distal 1BS introgression may have a more direct effect on aerial biomass or on other anatomical and/or physiological root differences known to impact tolerance to waterlogging and drought (Bailey-Serres and Voesenek, 2008; Berger et al., 2016).
1RSxW shows an earlier arrest of seminal root growth than 1RSxR in hydroponic culture
The differences in root depth observed between the Hahn 1RSxR and 1RSxW NILs in the field were paralleled by drastic changes in seminal root length in hydroponic cultures (Fig. 3A). These differences were robust across experiments and were detected with different nitrogen sources and concentrations (Fig. 3B). We hypothesize that these early differences in seminal root length may have contributed to the observed differences in total root length density observed in the deepest soil core samples in the field (Figs 2, 4B).
The early and consistent differences in root growth under controlled conditions provided the opportunity to study the process in detail. During the first week of development, root growth occurred at the same rate for both genotypes, suggesting that the differences were not primarily associated with embryonically determined differences in root elongation. Instead, differences in root growth consistently manifested during the second week across multiple experiments. The growth rate of the seminal roots of the 1RSxW plants gradually decreased during the second and third week, to come close to zero by the end of the third week, whereas growth continued in the 1RSxR plants (Fig. 4A). The consistent timing of these events suggests that these changes are developmentally regulated.
The growth arrest of the seminal roots in the 1RSxW plants was accompanied by the proliferation of lateral roots in close proximity to the RAM, suggesting important changes in the RAM. The RAM consists of a quiescent center (QC) surrounded by stem cells that generate new daughter cells, which undergo additional divisions in the proximal region of the meristem and differentiate in the transition zone (Heyman et al., 2014). At a cellular level, a balance between cell proliferation and cell elongation/differentiation determines root growth rate (Pacifici et al., 2015). The arrest of the growth of seminal roots in 1RSxW plants suggests a modification in cell proliferation and/or cell elongation/differentiation. Additional studies will be required to determine if this arrest involves changes in the QC and/or modifications in the root regions adjacent to the meristem. In any case, the dramatic reduction in seminal root growth and increased lateral root proliferation close to the RAM argues for an early developmental program switch in the regulation of the RAM in the 1RSxW plants.
The distribution of ROS along seminal roots differs between 1RSxW and 1RSxR lines
The transition from cell proliferation to cell elongation and differentiation and the subsequent development of lateral roots depend on the distribution of ROS along the root axis, specifically on the opposing gradients of superoxide and hydrogen peroxide. Superoxide is predominant in dividing cells in the meristematic zone, while hydrogen peroxide is predominant in elongated cells in the differentiation zone (Dunand et al., 2007; Tsukagoshi et al., 2010; Voothuluru and Sharp, 2013; Yamada et al., 2018, Preprint). The balance between these ROS modulates the transition between root proliferation and differentiation zones.
At 17 DAG, the apical region of 1RS seminal roots showed opposing gradients of superoxide and hydrogen peroxide characteristic of elongating roots (Fig. 6). A different ROS distribution was detected in the arrested 1RSxW roots, where superoxide was restricted to the distal ~700 μm and increased levels of DCF-DA fluorescence were detected between 250 μm and 950 μm in the cell proliferation zone (Fig. 6). The contrasting patterns of ROS distribution reflect the major developmental changes that differentiate the seminal roots of the 1RS and 1RSxW genotypes.
Studies in Arabidopsis have shown that changes in ROS distribution can be triggered by the altered expression of major genes that control the size of the meristematic zone. These genes include UPBEAT1 (UPB1), a basic helix–loop–helix (bHLH) transcription factor that regulates the meristematic zone size by restricting hydrogen peroxide distribution in the elongation zone (Tsukagoshi et al., 2010). In addition, ROOT MERISTEM GROWTH FACTOR 1 (RGF1) and the transcription factor gene RGF1 INDUCIBLE TRANSCRIPTION FACTOR 1 (RITF1) that mediates RGF1 signaling can modulate the distribution of ROS along the root developmental zones leading to enhanced stability of PLETHORA2 (PLT2) (Yamada et al., 2018, Preprint). Reduced expression of PLETHORA in the root apical region (Chen et al., 2011) or changes in its distribution (Ercoli et al., 2018) have been associated with impaired root growth. To test if these Arabidopsis results are applicable to wheat, we are initiating expression studies of these genes in the 1RSxR and 1RSxW lines.
It remains unknown whether the differential pattern of ROS distribution in the roots of the 1RSxW plants is the result of changes in the expression of wheat homologs of these central developmental genes or a more direct effect on genes affecting the redox balance in different developmental root zones. The differences in superoxide and hydrogen peroxide distribution between the seminal roots of the 1RSRW and 1RS plants were measured after the arrest in root growth (Fig. 6). Therefore, we currently do not know if the changes in ROS distribution are a cause or a consequence of the changes observed in root growth and lateral root proliferation close to the RAM.
Conclusions and future directions
Results presented here indicate that the differences in grain yield between the 1RSxW and 1RSxR lines were preceded by significant differences in aerial biomass. These differences were generally larger under restricted or excessive irrigation than under normal irrigation, suggesting that the introgression of the distal wheat segment in the 1RSxW lines resulted in a reduced tolerance to these stresses.
The hydroponic studies showed that the introgression of the distal wheat chromosome segment in 1RSxW resulted in a developmentally regulated arrest of seminal root growth associated with drastic changes in ROS gradients along the roots and the distribution of lateral roots close to the RAM. Since the 1RSxW and 1RSxR lines are highly isogenic (Howell et al., 2014), they provide a useful tool to identify the genes that regulate this major developmental change in the wheat RAM. The identification of these genes not only can contribute to our basic understanding of the regulation of root development in grasses, but also can provide tools to modulate wheat root architecture.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Excavation of the first block including the four genotypes.
Fig. S2. Plant biomass and grain yield.
Fig. S3. Percentage decrease in total root length density and diameter.
Fig. S4. Differences between 1RSxR and 1RSxW in different root parameters.
Fig. S5. Distance between the primary root apex and the first lateral root.
Fig. S6. Distribution of nitroblue tetrazolium (NBT) staining during root development.
Table S1. NDVI waterlogging 2013–2014.
Table S2. NDVI waterlogging 2013–2014 by day.
Table S3. Yield waterlogging 2013–2014.
Table S4. NDVI waterlogging 2015–2016.
Table S5. NDVI waterlogging 2015–2016 by day.
Table S6. Yield waterlogging 2015–2016.
Table S7. NDVI drought 2015–2016.
Table S8. NDVI drought 2015–2016 by day.
Table S9. Yield drought 2015–2016.
Table S10. Total root length density.
Table S11. Total root length density contrast.
Table S12. Total root length density contrast by depth.
Table S13. Average root diameter.
Table S14. Average root diameter contrast.
Table S15. Average root diameter contrast by depth.
Table S16. Repeated measures for root length
Table S17. Root length (mm) by day.
Table S18. Repeated measures for root elongation rate.
Table S19. Root elongation rate by day.
Acknowledgements
This project was supported by USA–Israel BARD grant UC-4916-16, the Agriculture and Food Research Initiative Competitive grant 2017-67007-25939 (WheatCAP) from the USDA National Institute of Food and Agriculture, by the International Wheat Yield Partnership (IWYP), and the Howard Hughes Medical Institute; and in Argentina by the CONICET and the ANPCYT (PICT 2014-1887).
References
- Bailey-Serres J, Voesenek LA. 2008. Flooding stress: acclimations and genetic diversity. Annual Review of Plant Biology 59, 313–339. [DOI] [PubMed] [Google Scholar]
- Berger J, Palta J, Vadez V. 2016. Review: an integrated framework for crop adaptation to dry environments: responses to transient and terminal drought. Plant Science 253, 58–67. [DOI] [PubMed] [Google Scholar]
- Chen Q, Sun J, Zhai Q, et al.. 2011. The basic helix–loop–helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. The Plant Cell 23, 3335–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunand C, Crèvecoeur M, Penel C. 2007. Distribution of superoxide and hydrogen peroxide in Arabidopsis root and their influence on root development: possible interaction with peroxidases. New Phytologist 174, 332–341. [DOI] [PubMed] [Google Scholar]
- Ehdaie B, Layne AP, Waines JG. 2012. Root system plasticity to drought influences grain yield in bread wheat. Euphytica 186, 219–232. [Google Scholar]
- Ehdaie B, Whitkus RW, Waines JG. 2003. Root biomass, water-use efficiency, and performance of wheat rye translocations of chromosomes 1 and 2 in spring bread wheat ‘Pavon’. Crop Science 43, 710–717. [Google Scholar]
- Ercoli MF, Ferela A, Debernardi JM, Perrone AP, Rodriguez RE, Palatnik JF. 2018. GIF transcriptional coregulators control root meristem homeostasis. The Plant Cell 30, 347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAOSTAT 2017. FAOSTAThttp://www.fao.org/faostat/en/#data. Rome: Food and Agriculture Organization (FAO) of the United Nations. [Google Scholar]
- Fenn D, Lukow OM, Bushuk W, Depauw RM. 1994. Milling and baking quality of 1BL/1RS translocation wheats. I. Effects of genotype and environment. Cereal Chemistry 71, 189–195. [Google Scholar]
- Heyman J, Kumpf RP, De Veylder L. 2014. A quiescent path to plant longevity. Trends in Cell Biology 24, 443–448. [DOI] [PubMed] [Google Scholar]
- Hoffmann B. 2008. Alteration of drought tolerance of winter wheat caused by translocation of rye chromosome segment 1RS. Cereal Research Communications 36, 269–278. [Google Scholar]
- Howell T, Hale I, Jankuloski L, Bonafede M, Gilbert M, Dubcovsky J. 2014. Mapping a region within the 1RS.1BL translocation in common wheat affecting grain yield and canopy water status. Theoretical and Applied Genetics 127, 2695–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landjeva S, Korzun V, Tsanev V, Vladova R, Ganeva G. 2006. Distribution of the wheat–rye translocation 1RS.1BL among bread wheat varieties of Bulgaria. Plant Breeding 125, 102–104. [Google Scholar]
- Liu J, Moore S, Chen C, Lindsey K. 2017. Crosstalk complexities between auxin, cytokinin, and ethylene in Arabidopsis root development: from experiments to systems modeling, and back again. Molecular Plant 10, 1480–1496. [DOI] [PubMed] [Google Scholar]
- Lukaszewski AJ. 2000. Manipulation of the 1RS.1BL translocation in wheat by induced homoeologous recombination. Crop Science 40, 216–225. [Google Scholar]
- Moreno-Sevilla B, Baenziger PS, Peterson CJ, Graybosch RA, Mcvey DV. 1995. The 1BL/1RS translocation—agronomic performance of F3-derived lines from a winter-wheat cross. Crop Science 35, 1051–1055. [Google Scholar]
- Pacifici E, Polverari L, Sabatini S. 2015. Plant hormone cross-talk: the pivot of root growth. Journal of Experimental Botany 66, 1113–1121. [DOI] [PubMed] [Google Scholar]
- Rabinovich SV. 1998. Importance of wheat–rye translocations for breeding modern cultivars of Triticum aestivum L. Euphytica 100, 323–340. [Google Scholar]
- Richards RA, Rebetzke GJ, Watt M, Condon AG, Spielmeyer W, Dolferus R. 2010. Breeding for improved water productivity in temperate cereals: phenotyping, quantitative trait loci, markers and the selection environment. Functional Plant Biology 37, 85–97. [Google Scholar]
- Schlegel R, Korzun V. 1997. About the origin of 1RS.1BL wheat–rye chromosome translocations from Germany. Plant Breeding 116, 537–540. [Google Scholar]
- Shearman VJ, Sylvester-Bradley R, Scott RK, Foulkes MJ. 2005. Physiological processes associated with wheat yield progress in the UK. Crop Science 45, 175–185. [Google Scholar]
- Smucker AJM, Mcburney SL, Srivastava AK. 1982. Quantitative separation of roots from compacted soil profiles by the hydropneumatic elutriation system. Agronomy Journal 74, 500–503. [Google Scholar]
- Tsukagoshi H, Busch W, Benfey PN. 2010. Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143, 606–616. [DOI] [PubMed] [Google Scholar]
- Tyburski J, Dunajska K, Tretyn A. 2009. Reactive oxygen species localization in roots of Arabidopsis thaliana seedlings grown under phosphate deficiency. Plant Growth Regulation 59, 27–36. [Google Scholar]
- Voothuluru P, Sharp RE. 2013. Apoplastic hydrogen peroxide in the growth zone of the maize primary root under water stress. I. Increased levels are specific to the apical region of growth maintenance. Journal of Experimental Botany 64, 1223–1233. [DOI] [PubMed] [Google Scholar]
- Voss-Fels KP, Snowdon RJ, Hickey LT. 2018. Designer roots for future crops. Trends in Plant Science 23, 957–960. [DOI] [PubMed] [Google Scholar]
- Wasson AP, Richards RA, Chatrath R, Misra SC, Prasad SV, Rebetzke GJ, Kirkegaard JA, Christopher J, Watt M. 2012. Traits and selection strategies to improve root systems and water uptake in water-limited wheat crops. Journal of Experimental Botany 63, 3485–3498. [DOI] [PubMed] [Google Scholar]
- Yamada M, Han X, Benfey PN. 2018. Root meristem growth factor 1 controls root meristem size through reactive oxygen species signaling. bioRxiv 244947. [Preprint]. [Google Scholar]
- Yu F, Luan S. 2016. Peptide signaling in plants: finding partners is the key. Cell Research 26, 755–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarco-Hernandez JA, Santiveri F, Michelena A, Pena RJ. 2005. Durum wheat (Triticum turgidum, L.) carrying the 1BL/1RS chromosomal translocation: agronomic performance and quality characteristics under Mediterranean conditions. European Journal of Agronomy 22, 33–43. [Google Scholar]
- Zhang LY, Liu DC, Guo XL, Yang WL, Sun JZ, Wang DW, Sourdille P, Zhang AM. 2011. Investigation of genetic diversity and population structure of common wheat cultivars in northern China using DArT markers. BMC Genetics 12, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.