A mutation in Large Grain Size 1 results in increases in grain length, grain width, grain weight, and endosperm cell number, as well as enhancement of cold tolerance in rice.
Keywords: Cold tolerance, grain size, map-based gene cloning, microRNA, OsGRF4, RNA sequencing
Abstract
Grain shape is controlled by quantitative trait loci (QTLs) in rice (Oryza sativa L.). A rice mutant (JF178) with long and large grains has been used in a breeding program for over a decade, but its genetic basis has been unclear. Here, a semi-dominant QTL, designated Large Grain Size 1 (LGS1), was cloned and the potential molecular mechanism of LGS1 function was studied. Near-isogenic lines (NILs) and a map-based approach were employed to clone the LGS1 locus. LGS1 encodes the OsGRF4 transcription factor and contains a 2 bp missense mutation in the coding region that coincides with the putative pairing site of miRNA396. The LGS1 transcript levels in the mutant line were found to be higher than the lgs1 transcript levels in the control plants, suggesting that the mutation might disrupt the pairing of the LGS1 mRNA with miR396. In addition to producing larger grains, LGS1 also enhanced cold tolerance at the seedling stage and increased the survival rate of seedlings after cold stress treatment. These findings indicate that the mutation in LGS1 appears to disturb the GRF4–miR396 stress response network and results in the development of enlarged grains and enhancement of cold tolerance in rice.
Introduction
Rice (Oryza sativa L.) can grow in a wide range of environments, and provides the main calorific intake for more than half of the world’s population (Huang et al., 2013). As the world’s population is predicted to reach 9 billion by the middle of this century, requiring a 70–100% increase in food production relative to the current levels (Godfray et al., 2010; Abe et al., 2012), it is a great challenge to ensure sustainable food production and supplies worldwide. Among the yield-associated traits, the thousand-grain weight is restricted mainly by the grain shape, which can be further characterized by the grain length, grain width, and grain thickness. In addition, rice grain shape is known to strongly affect grain yield, grain quality, and market values (Huang et al., 2013; S. Wang et al., 2015). Thus, grain shape is an important trait in rice breeding programs.
More than 400 quantitative trait loci (QTLs) associated with the rice grain shape trait have been identified, and nearly 30 genes have been cloned and demonstrated to regulate the traits of grain shape and grain weight in various genetic studies (Huang et al., 2011, 2013; Zhao et al., 2011; Liu et al., 2015; S. Wang et al., 2015; Y. Wang et al., 2015; Wu et al., 2017). The identification and functional characterization of these genes have provided an important theoretical basis for the enrichment of genetic resources and development of new breeding and cultivation strategies in rice.
Growth-regulating factors (GRFs) are a family of plant-specific proteins that regulate the size and number of tissues, organs, and cells in plants (Van der Knapp et al., 2000). GRFs contain two highly conserved regions, the QLQ (Gln, Leu, Gln) and the WRC (Trp, Arg, Cys) domains, both of which are present in the N-terminal regions of the protein (Van der Knapp et al., 2000; Kim et al., 2003; Choi et al., 2004; Jones-Rhoades and Bartel, 2004; Kim and Kende, 2004; Zhang et al., 2008; Kim et al., 2012). The expression of GRFs is regulated at the transcriptional level by endogenous miRNAs, which base-pair with the corresponding region of mRNA coding for the WRC domain, resulting in target mRNA cleavage (Jones-Rhoades and Bartel, 2004; Jones-Rhoades et al., 2006; Gao et al., 2010; Liu et al., 2014; Duan et al., 2015; Hu et al., 2015; Li et al., 2016).
In this study, we took the map-based approach to clone a semi-dominant grain shape locus, designated as Large Grain Size 1 (LGS1). LGS1 encodes a GRF4 transcription factor and contains a 2 bp missense mutation corresponding to the WRC domain. The identification of LGS1 will greatly facilitate the improvement of grain shape in rice through molecular marker-assisted selection approaches.
Materials and methods
Identification of LGS1
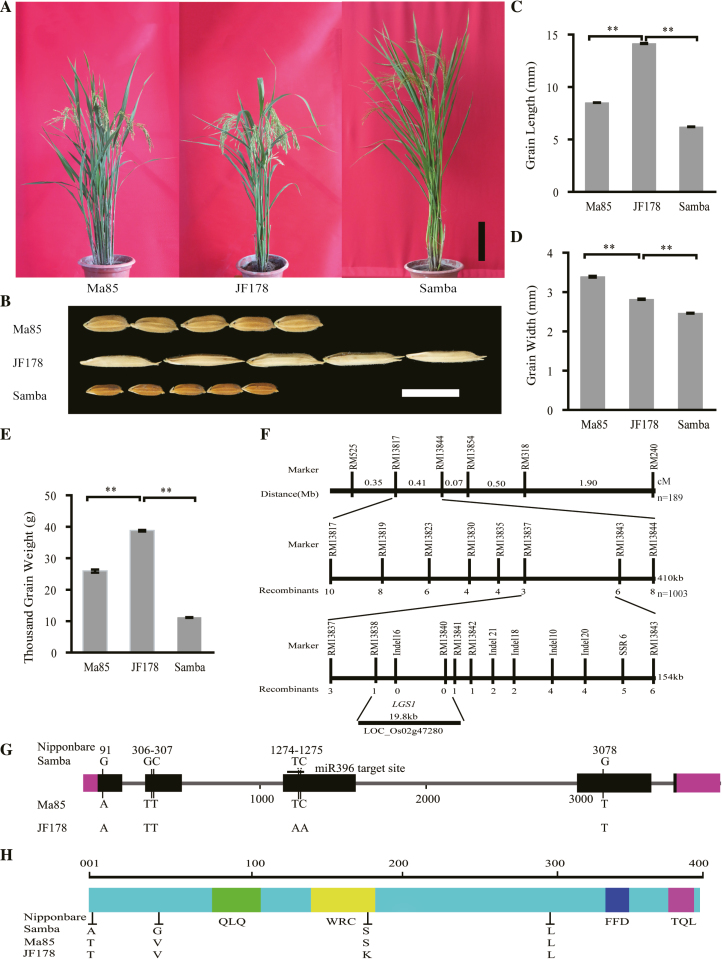
The mutant line JF178 (Oryza sativa L. ssp. indica cv) that produced larger and longer grains (Fig. 1A–E; Supplementary Table S2 at JXB online) was selected from the progeny of the rice cultivar Ma85 (O. sativa L. ssp. indica cv) that was subjected to mutagenesis by γ-Co60 irradiation in the 1990s. JF178 and Samba (O. sativa L. ssp. japonica cv) were chosen as parents for mapping QTLs for grain length (Fig. 1F; Supplementary Fig. S1). A set of LGS1 near-isogenic lines (NILs) was developed by crossing the small grain cultivar Samba (maternal parent) with JF178 (paternal parent). F1 plants that contained the LGS1 locus as identified through marker-assisted selection were chosen for backcross using Samba as the recurrent parent. NIL-LGS1JF178 was finally selected from the BC10F3 generation in the Samba genetic background (Supplementary Fig. S1). For phylogenetic tree analysis of LGS1, a total of 47 rice cultivars collected in China and the JF series of breeding lines from our research group were used for PCR amplification utilizing the marker-assisted selection primers and LGS1 PCR primers (Supplementary Tables S1, S2).
Fig. 1.
Map-based cloning of LGS1. (A) Plant phenotype of Ma85, JF178, and Samba. Scale bar=20 cm. (B) Grain phenotype of Ma85, JF178, and Samba (n=5). Scale bar=10 mm. (C–E) Comparisons of grain length (C), grain width (D), and thousand-grain weight (E) among Ma85, JF178, and Samba (n=90). Values are given as the mean ±SE. **P<0.01, Student’s t-test. (F) Fine mapping of LGS1. The numbers under each marker indicate the number of recombinants between LGS1 and the molecular markers. (G) Gene structure and sequence variations of LGS1 among JF178, Ma85, Samba, and Nipponbare. The blocks represent exons, and the 5'- and 3'-UTRs, the lines represent introns, and the vertical lines indicate the nucleotide sequence variations in exons. (H) Domain structure of LGS1 protein and amino acid variations among JF178, Ma85, Samba, and Nipponbare. The blocks indicate different conserved domains. The vertical lines indicate amino acid variations.
Growing conditions and traits measurement
Rice plants were cultivated twice a year in the experimental field of Xiamen University, Fujian Province, China, and were planted with an interplant spacing of 20×20 cm2 for transplanting. The plants were arranged in a completely randomized block design with three repeats. The traits of thousand-grain weight, grain length, grain width, grain number per panicle, plant height, spike length of the main stem panicle, and of the primary, secondary, and tertiary branch panicles were measured using 30 plants. Fully filled grains were chosen for measurement at the harvest stage. Grain yield per unit area was measured from 100 plants for each variety.
Tissue section and microscopy
Rice spikelets were sampled at the heading stage and fixed in the formalin–acetic acid–alcohol (FAA) solution (5% formaldehyde, 5% glacial acetic acid, and 50% ethanol), followed by degassing under a vacuum pump for 25 min, and incubation in the FAA solution for at least 24 h. Fixed tissue samples were dehydrated in an ethanol solution series, destained in xylene solution, and embedded in paraffin. Tissue sections (7 μm thick) were cut with a rotary microtome (Leica, Germany) and mounted on a glass slide, followed by treatment with xylene and staining with 0.2% toluidine blue (Sigma, Germany). Sections were photographed under a Nikon 50i microscope (Nikon, Japan).
Scanning electron microscopy
The entire spikelet hulls and endosperm were collected at the harvest stage and used for SEM. Gold-plated outer surface cells of the lemma and palea from mature seeds and endosperm slices (2 mm thick) were observed under a JSM-6390LV scanning electron microscope (JEOL, Japan) at 5 kV. The cell length and width, and the endosperm cell number per unit area were measured using Image IS Capture software (TuSimple, China). The cell number along both the longitudinal axis and transverse axis was recorded.
Plasmid construction and genetic transformation
For construction of pLGS1::LGS1 for genetic complementation (Supplementary Fig. S3A), a DNA fragment of 7.4 kb containing the 2.7 kb promoter, the 3.825 kb coding region, and the 0.9 kb termination site was amplified from Ma85 and JF178, and cloned into binary vector pCAMBIA1301. The resulting vectors, plgs1Ma85::lgs1Ma85 and pLGS1JF178::LGS1JF178, were introduced into JF178 and Ma85, respectively.
To construct the LGS1 RNAi plasmid, two DNA fragments unique to LGS1 [a 194 bp fragment containing the 84 bp 5'-untranslated region (UTR) and the other fragment 110 bp downstream from the LGS1 start codon; Supplementary Fig. S3B] were amplified from JF178 and Samba, and cloned into pCUbi1390 (Okuley et al., 1994; Smith et al., 2000; Wesley et al., 2001; Stoutjesdijk et al., 2002). The expression of the LGS1 RNAi was driven by the ubiquitin promoter. The resulting LGS1JF178 and lgs1Samba RNAi vectors were introduced into JF178 and Samba, respectively.
The lgs1 promoter::GUS (β-glucuronidase) reporter plasmid (Supplementary Fig. S3C) was constructed by fusion of the 2.8 kb lgs1 promoter (including 36 bp downstream from the lgs1 start codon) from Ma85 to the GUS reporter gene. The construct was subcloned into pCAMBIA1381Xa to generate plgs1::GUS, which was used to transform the rice cultivar Zhonghua 11 (O. sativa L. ssp. japonica cv).
Plasmids were confirmed by DNA sequencing and introduced into Agrobacterium tumefaciens strain EHA105 for rice transformation as described previously (Hiei et al., 1994). Positive transgenic plants of the T2 generation were used for recording of grain shape data.
Histochemical GUS assays
Specimens were stained for GUS activity as described by Jefferson et al. (1987), and photographed under a BX50 OLYPUS microscope (Olympus, Japan).
Quantitative RT–PCR of mRNA, and miRNA analysis
Total RNA and miRNA were extracted from various rice tissues using the Wolact® Plant RNA Isolation Kit (Wolact, Hong Kong, China). cDNAs were reverse transcribed using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA). miRNAs were reverse transcribed using the Taqman® miRNA Reverse Transcriptase Kit (Thermo Fisher Scientific, Foster, CA, USA) and stem–loop reverse transcription primers (Sangon, Shanghai, China). Quantitative RT–PCR was carried out using TransStart Top Green qPCR SuperMix (Transgene Biotech, Beijing, China) with OsActin1 (for mRNA) or the U6 (for miRNA) gene as an endogenous control for fold enrichment. Quantitative RT–PCRs of mRNA and miRNA analyses were carried out using the CFX96 Real-Time PCR System (Agilent, California, USA) in accordance with the manufacturer’s instructions. The qPCR cycling conditions were 2 min at 95 °C followed by 38 cycles of amplification (95 °C for 15 s, 60 °C for 20 s, and 72 °C for 25 s). All assays were repeated at least three times using independent RNA samples and performed with three technical repetitions. The sequences of primers are given in Supplementary Table S1.
Subcellular localization
For subcellular localization experiments (Supplementary Fig. S3D; Supplementary Table S1), the coding sequence of LGS1 was amplified from JF178, and inserted into pDONR201-GFP-LGS1. The plasmid was used to react with pK2GW7 (Invitrogen, Carlsbad, CA, USA) to generate PCAMV35S::GFP-LGS1. The vector was introduced into A. tumefaciens strain GV1301 for transformation into Arabidopsis thaliana. PCAMV35S::Collin-RFP was used as a nuclear localization marker (Cui et al., 2015). All Arabidopsis plants were grown in soil at 22 °C under long days (16 h light/8 h dark) with white fluorescent light (120 μmol m−2 s−1). Green and red fluorescence signals of 2-week-old transgenic plants were observed under a confocal laser scanning microscope (Zeiss LSM 780/Carl Zeiss Meditec AG, Jena, Germany).
Transactivation activity assays in yeast
Various segments of LGS1 with truncated cDNA were amplified from JF178 and Ma85, and cloned into the pGBKT7 vector for transcriptional activation activity analysis by the yeast two-hybrid system according to the manufacturer’s protocol (Clontech, USA). The empty pGBKT7 vector was used as control. The prey and bait plasmids were co-transformed into the yeast Y2H Gold strain, which was grown in SC/DDO-Trp/-Leu (synthetic complete medium with dropout of Trp and Leu), SC/DDO-Trp/-Leu+100 ng ml–1 Aureobasidin A (AbA) medium for 2 d at 30 °C. The interactions between GRF and GIF (GRF-interacting factor) were assayed in the Matchmaker GAL4 Two-Hybrid System (Clontech, USA). LGS1JF178 and lgs1Ma85 proteins were expressed as bait from pGBKT7. The coding sequences of OsGIF1, OsGIF2, and OsGIF3 were amplified from Ma85 and expressed as prey from pGADT7. The interaction assays were based on the growth ability of the co-transformants on medium supplemented with AbA. The experiments were repeated three times.
RNA-seq and data analysis
Tissue samples of 5–6 cm long young panicles, corresponding to the differentiation stage of meiotic division of pollen mother cells, were taken from rice plants grown under field conditions, and collected for total RNA extraction. To obtain a comprehensive profile of the LGS1 transcriptome, library construction was performed with a NEBNext® Multiplex RNA Library Prep Set (NEB), which used the Illumina Hiseq 2000 to perform high-throughput RNA sequencing (RNA-seq) of the recurrent parent (Samba) and the NIL line (NIL-LGS1JF178). In total, 30.68 Gb of clean RNA-seq reads were generated from the six libraries, and mapped to the rice genome using TopHat2 (Trapnell et al., 2010; Kim et al., 2013). The transcriptome data were collected from three replicas of the wild-type control and three replicas of the LGS1 NIL line. The number of fragments per kilobase of transcript per million mapped reads (FPKM) was analyzed for gene expression. Differentially expressed genes (DEGs) were defined by a 2-fold change expression difference at a false discovery rate (FDR) of <0.01. The genome data of Nipponbare (O. sativa L. ssp. japonica cv) were used as a reference.
Abiotic stress treatment and cold treatment
NIL-LGS1JF178 and Samba were used for analysis of the response to plant hormone treatment. Ma85 and JF178 were used for stress treatment. Rice seeds were germinated and seedlings were grown for 15 d under normal growth conditions (14 h light, 28 °C/10 h dark, 20 °C, 20000LX; Yiheng, Shanghai, China) with daily watering. Seedlings were then treated for 12 h with normal growth conditions (the control), 35 °C, 4 °C, 20 mg ml–1 GA3 (gibberellic acid 3), 1 mg ml–1 IAA (indole-3-acetic acid), 25 mg ml–1 ABA (abscisic acid), 20 mg ml–1 BL (brassinolide), 0.25 g ml–1 NaCl, and 0.1 g ml–1 NaHCO3 (pH 9.0), or UV-B (TL20W/01RS; Philips, Eindhoven, The Netherlands) for 1 h. Seedlings were then sampled for quantitative real-time PCR (qRT-PCR) analysis of LGS1 expression.
For cold stress treatment, 15-day-old seedlings were held at 4 °C for different periods of time as indicated, and then sampled for qRT-PCR analysis. After the cold treatment, seedlings were grown under normal growth conditions for 5 d to recover from the cold stress. The survival rates after the recovery from cold treatment were recorded, and the seedlings were sampled for qRT-PCR analysis of expression of lgs1 and miR396.
Accession codes
Accession codes for the DNA Data Bank of Japan (DDBJ) are as follows: LGS1_JF178, LC333009; lgs1_Ma85, LC333010; and lgs1_Samba, LC333011.
Results
Mapping of LGS1 and polymorphisms in rice germplasm
JF178 and Samba were selected as parents to map QTLs for grain length. JF178 is a larger grain indica mutant line (thousand-grain weight, 39.0±0.6 g; grain length, 14.16±0.05 mm), whereas Samba is a small grain japonica variety (thousand-grain weight, 11.9±0.1 g; grain length, 6.21±0.02 mm; Fig. 1A–E). Using 189 random subpopulations of the BC5F2 segregating population, we were able to perform primary mapping of LGS1, the major QTL locus affecting grain shape in JF178, to a region between markers RM13817 and RM13844 on chromosome 2 (Supplementary Fig. S1A). Using 1003 recessive plants (short grain, grain length <6.2 mm) from the BC5F3 population, we fine-mapped LGS1 to a 154 kb interval between markers RM13838 and RM13841 on chromosome 2 (Supplementary Fig. S1A, B). From the 1003 recessive plants of the BC5F3 population, we identified 18 recombinant plants, which allowed us to narrow down the LGS1 locus to a 19.8 kb region between markers RM13838 and RM13840 on the long arm of chromosome 2 (Fig. 1F; Supplementary Fig. S1A, B). This region contains only one gene, LOC_Os02g47280, the only candidate for LGS1 (Fig. 1F). Comparison between the genomic DNA and cDNA sequences of LGS1 revealed the presence of five exons and four introns for LGS1, with the full-length DNA of 3825 bp encoding 394 amino acids (Fig. 1G, H). Comparison of the nucleotide sequences of LGS1 alleles from Ma85 and JF178 revealed two nucleotide substitutions (TC to AA) in exon 3 that resulted in the amino acid substitution of serine to lysine (S163K) in the LGS1 protein (Fig. 1G, H; Supplementary Table S2). We then analyzed the nucleotide sequences of LGS1 from the irradiated mutant lines of our collection and the natural germplasm resources. Multiple polymorphisms in the LGS1 coding region and non-coding region were detected. These polymorphisms can be grouped into three types: haplotype I (two nucleotide substitutions), haplotype II (natural nucleotide sequence of indica subspecies), and haplotype III (natural nucleotide sequence of japonica subspecies). There are three single nucleotide polymorphisms (SNPs) between haplotype II and III. SNP1 has a one nucleotide difference, resulting in an amino acid change to T2A in the japonica subspecies. SNP2 contains a variation of two nucleotides, resulting in an amino acid change to V41G in the japonica subspecies. SNP3 has a one nucleotide difference that does not alter the amino acid residue between indica and japonica subspecies (Fig. 1G, H; Supplementary Tables S2, S3). These data indicate that the two nucleotide substitution at position 477–488 of LGS1 is the cause of the large grain shape trait observed in JF178 and other mutant lines from our experimental station.
LGS1 is a semi-dominant gene regulating grain shape and panicle traits
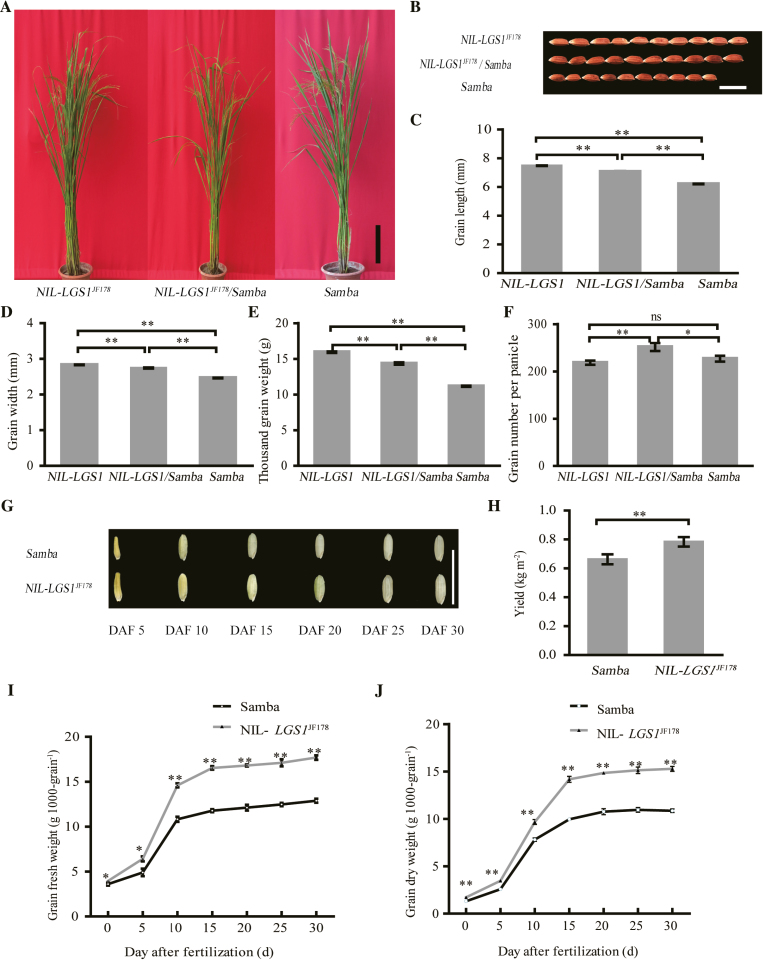
We performed reciprocal crosses of JF178 and Samba, and assayed the grain traits of F1 plants. Our data showed that there was no significant difference between the reciprocal crosses in grain length, grain width, thousand-grain weight, grain thickness, and grain number of the main panicle (Supplementary Fig. S2A–F), suggesting that LGS1 is a typical nuclear inherited locus with no maternal effect on grain traits. Analysis of grain trait data showed that grains of the homozygous and heterozygous NILs (NIL-LGS1JF178 and NIL-LGS1JF178/lgs1Samba) were 20.47% and 13.83% longer, 14.95% and 11.35% wider, and 42.81% and 28.53% heavier, respectively, than those of Samba (Fig. 2A–E; Supplementary Fig. S1). This finding suggests that LGS1 regulates grain length, grain width, and thousand-grain weight in rice. The grain filling rate was also greatly increased in NIL-LGS1JF178 as compared with Samba (Fig. 2G, I, J), resulting in a significant increase (15.72%) in grain yield in NIL-LGS1JF178 (Fig. 2H). In addition, analysis of plant height and panicle traits in NILs revealed that LGS1 had pleiotropic effects on the spike length, effective panicles (tillers), and tertiary branches per panicle (Fig. 2F; Supplementary Fig. S2G–L). Taken together, these analyses suggest that LGS1 is a semi-dominant gene with pleiotropic effects on grain shape and panicle traits in rice.
Fig. 2.
Grain size phenotype of near-isogenic lines (NILs). (A) Morphological characteristics of LGS1 NILs. Scale bar=20 cm. (B) Grain shape phenotype of LGS1 NILs (n=10). Scale bar=10 mm. (C–F) Comparisons of grain length (C), grain width (D), thousand-grain weight (E), and grain number per panicle (F) in LGS1 NILs (n=90–3000). (G) Characterization of grain filling in LGS1 NILs (DAF, day after fertilization). Scale bar=10 mm. (H) Yield per unit area at the harvest stage (n=3). (I) Time-course of grain fresh weight increase (n=300). (J) Time-course of grain dry weight increase (n=300). LGS1 from JF178 is a semi-dominant allele. NIL-LGS1JF178 (NIL-LGS1) contains the LGS1 allele from JF178 in the Samba genetic background. Samba carries the lgs1 allele and serves as a control. Values are given as the mean ±SE. ns P>0.05, *P<0.05, **P<0.01 were calculated by Student’s t-test.
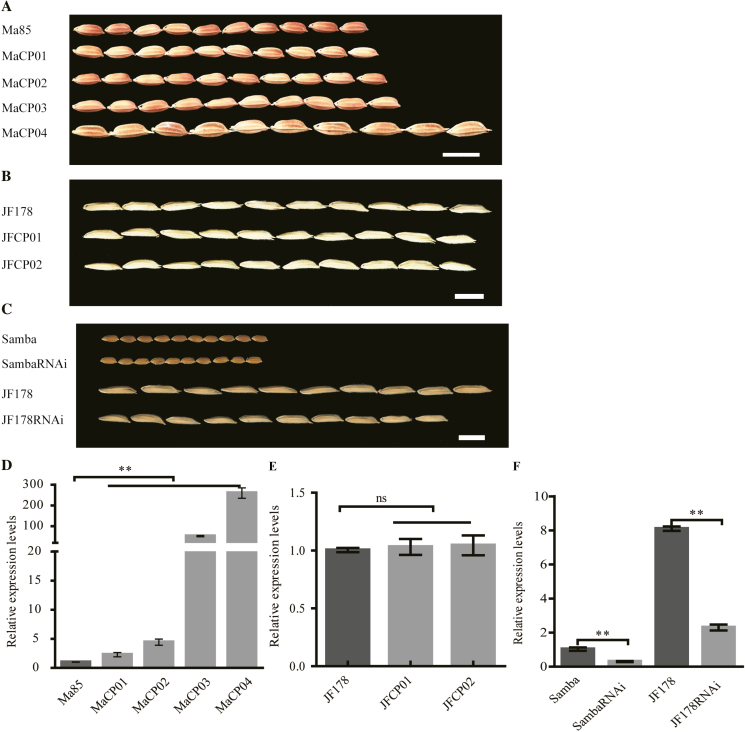
To verify the LGS1 function on grain traits, we introduced the pLGS1::LGS1 vector (Supplementary Fig. S3A) containing the LGS1 allele from JF178 into the small-grain cultivar Ma85. Transgenic plants were found to have increased transcript levels of LGS1 at the young spike differentiation stage, the critical time of reproductive development in rice. We selected four transgenic lines expressing the LGS1 allele and compared them with Ma85. Grains produced by the four transgenic lines (MaCP01–MaCP04) were longer (6.75, 5.31, 10.02, and 37.08%, respectively), wider (1.89, 3.05, 4.79, and 19.34%), and heavier (6.96, 15.66, 21.33, and 33.93%) than those of the control Ma85 (Fig. 3A, D; Supplementary Fig. S4A–C). We also performed the reciprocal experiment by introducing the plgs1::lgs1 vector (Supplementary Fig. S3A) containing the Ma85 allele into JF178. Transgenic plants of JF178 expressing the Ma85 lgs1 allele did not change the transcript level of its native LGS1 (Fig. 3E). Grains of the two transgenic lines (JFCP01 and JFCP02) were shorter (2.05% and 5.14% reduction, respectively), not significantly different in width (only 1.10% and 2.80% reduction in grain width), and considerably lighter in thousand-grain weight (26.50% and 30.81% reduction) (Fig. 3B; Supplementary Fig. S4D–F). These results further suggest that LGS1 is a semi-dominant gene with pleiotropic effects on multiple grain traits, which is consistent with the conclusion from genetic observations described above.
Fig. 3.
Grain phenotype of transgenic plants and LGS1-RNAi plants. (A–C) Grain phenotype of Ma85 and four transgenic lines of Ma85 expressing the genetic complementation vector pLGS1::LGS1 (A), JF178 and two lines of JF178 expressing plgs1::lgs1 (B), and Samba and JF178 expressing the LGS1-RNAi construct (C). Scale bars=1 cm in (A–C). (D–F) Transcript levels of LGS1 in Ma85 and four transgenic lines of Ma85 expressing pLGS1::LGS1 (D), JF178 and two transgenic lines of JF178 expressing plgs1::lgs1 (E), and Samba, JF178, and transgenic RNAi plants (F). RNA was isolated from young panicles (5 cm long, n=4). MaCP, transgenic Ma85 expressing the complementation vector pLGS1::LGS1; JFCP, transgenic JF178 expressing plgs1::lgs1. Ma85, JF178, and Samba and JF178 were used as controls in (A) and (D), (B) and (E), and (C) and (F), respectively. Values are given as the mean ±SE. ns P>0.05, **P<0.01 were calculated by Student’s t-test.
LGS1 is highly expressed in young vegetative tissues and developing grains
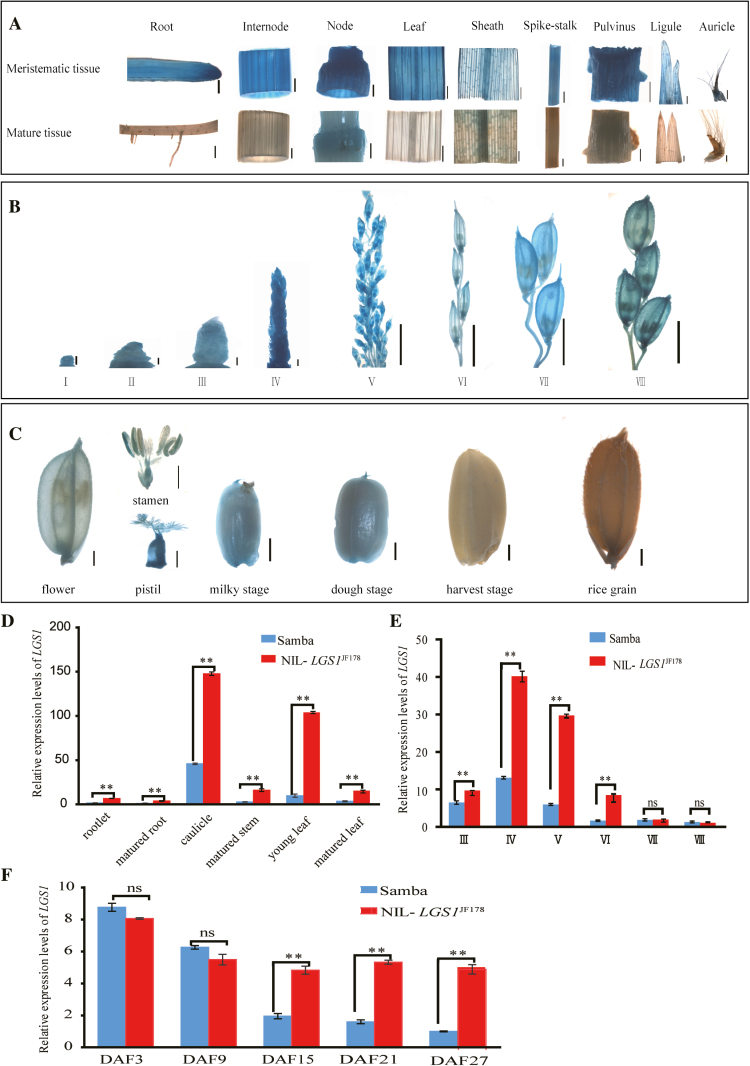
To investigate the temporal and spatial expression patterns of lgs1, we constructed the plgs1Ma85::GUS reporter vector (Supplementary Fig. S3C) and used it to transform Zhonghua 11. The GUS signals were considerably stronger in the growing tissues of root, internode, node, leaf, sheath, spike-stalk, pulvinus, ligule, and auricle than in the mature tissues (Fig. 4A). The GUS signal remained strong in the nodes of mature plants (Fig. 4A), where there was still active meristematic activity. During panicle development, the GUS activity was markedly stronger in the developing flowers, pistils, stamens, hulls, and fruits than in ripened grains (Fig. 4B, C; Supplementary Table S4).
Fig. 4.
Temporal and spatial expression patterns of LGS1. (A–C) Expression patterns of LGS1 in vegetative tissues (A), panicle meristems and developing young panicles (B; Supplementary Table S4), and developing fruits (C). Scale bars=1 mm in (A) (root, internode, node, leaf, sheath, and spike-stalk) and (C); 100 μm in (A) (pulvinus and ligule); 500 μm in (A) (auricle) and (B) (I, II, III, IV), and 5 mm in (B) (V, VI, VII, VIII). (D–F) Relative expression levels of LGS1 in vegetative tissues (D), developing young panicles (E), and developing fruits (F). DAF, day after fertilization. The expression levels of LGS1 were measured by qRT-PCR (n=6). The expression values of mature leaves (D), stage VIII (E), and DAF 27 in Samba (F) are set as 1. Values are given as the mean ±SE. ns P>0.05, **P<0.01 were calculated by Student’s t-test.
Transcript levels of LGS1 are higher in large grain NILs
We performed qRT-PCR to detect the expression of LGS1 in NILs. Our results showed that the transcript levels of LGS1 in the large grain NIL-LGS1JF178 were significantly higher in roots, stems, leaves, young panicles (stages III–VI), and developing fruits (15, 21, and 27 d after fertilization) than those of the small grain Samba. The differences in transcript levels between NIL-LGS1 and Samba were highly significant at stages IV–V (~5 cm young spikes) (Fig. 4D–F; Supplementary Table S4). Based on these observations, we conclude that LGS1 is expressed widely in various developing tissues and organs in rice and that its expression levels positively correlate with grain size.
Grain size is reduced in transgenic plants expressing LGS1 RNAi
To verify the hypothesis that the LGS1 mRNA level correlated with its function in grain development, we constructed the ubiquitin promoter-driven RNAi vectors pUbi::lgs1Samba and pUbi::LGS1JF178 (Supplementary Fig. S3B), and used them to transform Samba and JF178, respectively. The expression level of LGS1 was found to be strongly reduced in young panicles of transgenic Samba and JF178 plants expressing the corresponding RNAi construct. As a result, grains of RNAi-expressing Samba and JF178 were not significantly different and there was a 4.75% reduction in grain length, 4.09% and 12.90% reductions in grain width, and 11.28% and 17.72% reductions in thousand-grain weight, respectively (Fig. 3C, F; Supplementary S4G–I). Taken together, these observations indicate that the LGS1 allele from JF178 causes the LGS1 mRNA to accumulate and increases grain length, grain width, and thousand-grain weight in JF178 mutant plants, NIL-LGS1JF178 plants, and transgenic Ma85 plants expressing LGS1JF178.
LGS1 encodes rice growth-regulating factor 4
The deduced peptide sequence of LGS1 encodes the putative transcription factor OsGRF4 (rice growth-regulating factor 4; Fig. 1H), one of the 12 members of the GRF family. Similar to most other GRF proteins, LGS1 contains a QLQ domain that may be involved in chromatin remodeling, a WRC domain that may participate in transcription regulation, and two other domains (FFD and TQL) with unknown functions (Fig. 1H). The mutation in JF178 occurs in the WRC domain with a serine to lysine substitution at position 163 of LGS1 (Fig. 1H). The LGS1 protein was localized to the cell nucleus (Supplementary Fig. S3D, I), and had transcription activation activity when expressed in yeast cells (Supplementary Fig. S3E–G, J). Using the yeast one-hybrid system, we were able to further delimitate the activation domain of LGS1 to its C-terminus (Supplementary Fig. S3E–G , J).
It has been reported that GRF proteins interact with GIF family members, and form GRF–GIF complexes in Arabidopsis, maize, and rice (Kim and Kende, 2004; Zhang et al., 2008; Duan et al., 2015; Kim and Tsukaya, 2015; Li et al., 2016). To test if any GIF may interact with LGS1, we cloned OsGIF1 (Os11g0615200), OsGIF2 (Os12g0496900), and OsGIF3 (Os03g0733600), and expressed them as prey proteins in the yeast two-hybrid system (Supplementary Fig. S3E, H). LGS1JF178 and lgs1Ma85 were expressed as bait. The results showed that both LGS1 and lgs1 (OsGRF4) interacted with the three OsGIFs tested. The interaction with OsGIF3 was stronger than that with OsGIF1 or OsGIF2 (Supplementary Fig. S3K). These results demonstrate that LGS1 interacts with OsGIFs and functions as a transcription factor.
LGS1 controls grain size and milk filling rate
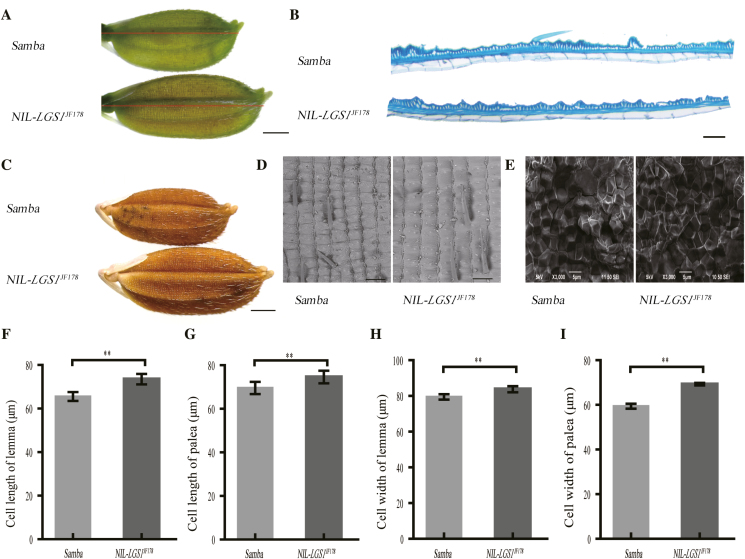
Given that the NIL-LGS1 spikelet hull was longer and wider than that of Samba at the flowering and harvest stages, we asked whether the increase in the hull size was caused by the increase in cell number, cell size, or both. We cut sections of paraffin-embedded spikelets and observed the longitudinal sections of the central spikelet hull, the surface sections of the lemma and palea, and cross-sections of endosperm under a scanning electron microscope (Fig. 5A–E). The length of lemma inner epidermal cells was significantly greater in NIL-LGS1JF178 than in Samba (Fig. 5A, B). Specifically, the epidermal cells of the lemma and palea in NIL-LGS1JF178 were longer (12.20% and 7.19% increase, respectively) and wider (5.44% and 16.44% increase, respectively) than those of Samba. Interestingly, there was also a significant increase in the longitudinal cell number of the lemma and palea (15.03% and 13.10% increase, respectively) in NIL-LGS1JF178 compared with that in Samba. The transverse cell numbers of lemma and palea in NIL-LGS1JF178 were not significantly different from those in Samba (Fig. 5C–I; Supplementary S5A–D). There was also no significant difference in endosperm cell size between NIL-LGS1JF178 and Samba (Fig. 5E; Supplementary Fig. S5E). However, NIL-LGS1 and Samba differed significantly in endosperm dry weight (Supplementary Fig. S5F), which resulted from the increased endosperm cell number and increased grain filling rate. Thus, our data demonstrate a significant increase in grain length, grain width, longitudinal cell number, and endosperm cell number in NIL-LGS1JF178, thereby suggesting that LGS1 may play a role in the regulation of cell division during grain development in rice.
Fig. 5.
Anatomic analyses of spikelet hulls at the heading stage and endosperm at the harvest stage in LGS1-NILs. (A) Spikelets. Scale bar=1 mm. The line indicates the position of the longitudinal section. (B) Magnified view of the spikelet hull longitudinal section of the lemma. Scale bar=100 μm. (C) Grain phenotype at the harvest stage. Scale bar=1 mm. (D) SEM analysis of the lemma surface in NILs. Scale bar=100 μm. (E) SEM analysis of the endosperm cross-section at the centre of the grain. Scale bar=5 μm. (F–I) Comparison of cell length of the lemma (F) and palea (G), and cell width of the lemma (H) and palea (I) in NILs (n=100). Samba as a control. Values are given as the mean ±SE. **P<0.01 was calculated by Student’s t-test.
LGS1 regulates cell division and hormone response pathways
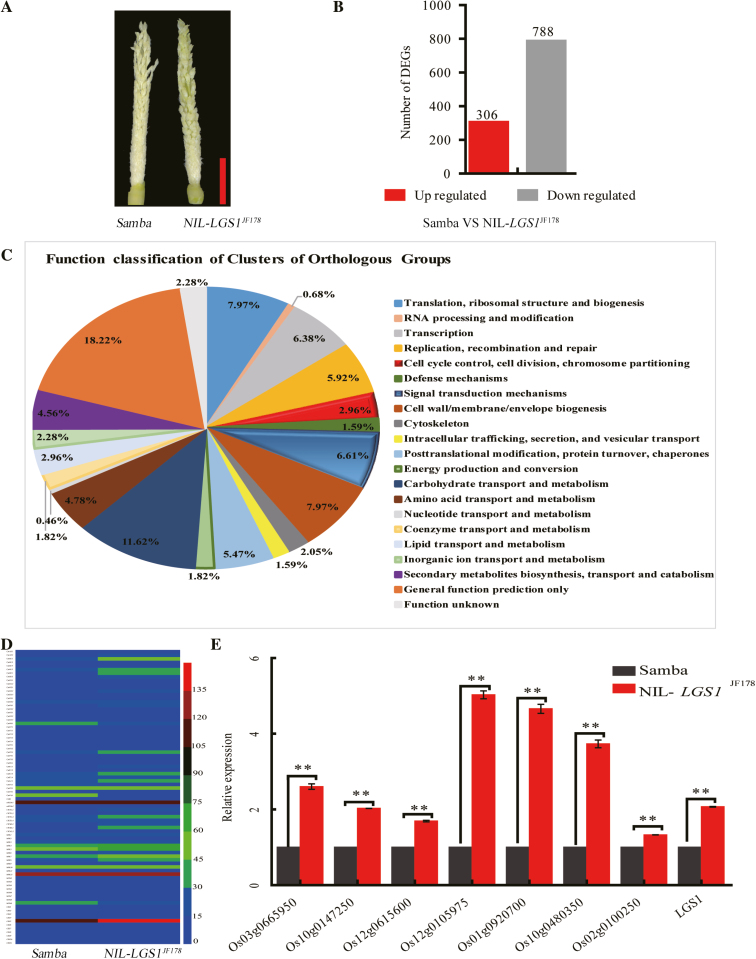
To identify genes that are regulated by LGS1, we performed RNA-seq analyses in young panicles (~4–5 cm long, Fig. 6A) and compared the whole transcriptome profiles between NIL-LGS1JF178 and Samba. A total of 1094 DEGs (ratio ≥2) were identified in the young panicles, among which 306 genes were up-regulated and 788 down-regulated (Fig. 6B). This observation indicates that the number of down-regulated genes was ~2.57 times that of the up-regulated genes (Fig. 6B). Enrichment analysis of function classification of clusters of orthologous groups (COGs) identified the ‘cell cycle control, cell division, chromosome partitioning’ (2.96%) and ‘signal transduction mechanisms’ (6.61%) as the annotated DEG pathways (Fig. 6C). These analyses infer that LGS1 may control grain development through regulation of cell division, and participate in hormone responses.
Fig. 6.
RNA-seq analysis of LGS1-NILs. (A) Phenotype of young panicles. Scale bar=1 cm. (B) Number of differentially expressed genes (DEGs, >2-fold change) of NIL-LGS1JF178 as compared with Samba. (C) DEG analysis of COG annotation. (D) Heat map of cell cycle-related DEGs (n=3). The scale bar, horizontal bars, and numbers indicate the FPKM (fragments per kilobase million) values. (E) Expression of the uncharacterized cell cycle-related genes by qRT-PCR (n=3). Samba as a control. Values are given as the mean ±SE. **P<0.01 was calculated by Student’s t-test.
We analyzed the 82 identified cell cycle genes that may affect the grain size (Fig. 6D; Supplementary Table S5). Analysis of annotated DEGs revealed that 25 DEGs encode cell cycle proteins, among which 11 were up-regulated and 14 down-regulated. Five DEGs encode expansin-like proteins, all of which were down-regulated. Nineteen DEGs encode leucine-rich repeat extensin-like proteins, among which seven were up-regulated and 12 down-regulated (Supplementary Table S6). We performed qRT-PCR to verify the up-regulated cell cycle genes (fold change >2) in NIL-LGS1JF178, and our results confirmed that the expression levels of all genes tested were significantly higher in NIL-LGS1JF178 than in Samba (Fig. 6E). These results demonstrate that LGS1 may alter the expression of cell cycle genes to promote cell division, extension, and expansion, resulting in increased grain size.
We also noticed that there were significant increases in seedling fresh weight, dry weight, and seedling length in NIL-LGS1JF178 as compared with Samba, whereas no significant difference in plant height at the harvest stage was observed between NIL-LGS1 and Samba (Supplementary Figs S2G, S6A–H). These features indicate that LGS1 may be involved in the hormone response pathways, which is consistent with the detection of DEGs in signal transduction pathways revealed by RNA-seq analysis (Fig. 6C). To explore this possibility, we treated NILs with different plant hormones (BL, ABA, IAA, and GA3) and measured seedling length and lamina inclination. We observed that there were significant increases in seedling length in NIL-LGS1JF178 in response to BL, ABA, and IAA as compared with the seedling length in Samba. The sensitivity of lamina inclination to BL treatment was impaired in NIL-LGS1JF178, and no significant change in seedling height was observed in NIL-LGS1JF178 treated with GA3 as compared with that in Samba (Supplementary Fig. S7A–H). In summary, these crosstalk responses of plant hormones imply that LGS1 may promote the growth of vegetative organs, resulting in more efficient nutrient supply for the later development of spikelets and endosperm. Whether and how the plant hormone signaling pathways are linked to the increased grain size in NIL-LGS1JF178 remains to be studied in the future.
LGS1 transcript levels are regulated by miR396
It has been documented that the GRF genes are targets of miRNA 396 (miR396), contain the miR396 recognition sequence in Arabidopsis, tobacco, and rice (Lan et al., 2012; Baucher et al., 2013; Liang et al., 2014; Rodriguez et al., 2015), and are responsive to stress (Gao et al., 2010). Nine miRNA precursor sequences of the miR396 family in rice (osa-miR396a–osa-miR396i) were predicted from the miRbase and psRNA target databases (Supplementary Fig. S8A; Supplementary Table S7). Sequence analysis showed that the mutation site occurred within the complementary sequence of OsmiR396 (Fig. 1G). We investigated whether OsmiR396 plays a role in regulating the LGS1 mRNA level in rice. qRT-PCR analysis of young panicles revealed that the expression pattern of lgs1 was opposite to that of OsmiR396; that is, the decrease in the LGS1 mRNA level during stages IV–VIII coincided with the increase in the expression levels of OsmiR396 members (Supplementary Fig. S8B). These results suggest that the transcript abundance of lgs1 is severely altered by osa-miR396a–osa-miR396i and further demonstrate that the expression of miR396 members down-regulates the transcript levels of lgs1.
LGS1 enhances cold tolerance at the seedling stage
To investigate the role of miR396a– osa-miR396i in the regulation of LGS1 during stress, we grew Ma85 and JF178 plants under different abiotic stress conditions or treated them with plant hormones. We found that LGS1 expression was up-regulated by treatment with IAA and NaHCO3, and slightly down-regulated by treatment with cold (4 °C), UV, ABA, and NaCl in JF178 as compared with the lgs1 levels in Ma85. It was striking to note that LGS1 expression was severely down-regulated by treatment with GA3 and BL in JF178 as compared with the lgs1 levels in Ma85 (Supplementary Fig. S9A). After treatment with abiotic stress and hormones, the seedlings were allowed to recover for a further 5 d under normal growth conditions. We found that there was a highly significant increase in survival rate in JF178 from cold (4 °C) stress (Fig. 7A, B). We further investigated the time-course of expression patterns of LGS1 in response to cold treatment, and found that lgs1 expression was significantly down-regulated (Supplementary Fig. S9B), which was correlated with the up-regulation of osa-miR396 members except miR396f, which was not affected by cold treatment (Supplementary Fig. S9C). After recovery from cold treatment, the expression of lgs1 and miR396 members returned to normal levels (Supplementary Fig. S9D, E). After cold treatment for 40 min, the expression of osa-miR396a–osa-miR396e and miR396g–osa-miR396i in Ma85, JF178, and JF178 RNAi plants was enhanced, and the expression of osa-miR396a–osa-miR396c in Ma85-expressing LGS1JF178 (MaCP) plants was significantly up-regulated, but osa-miR396f was significantly down-regulated (Supplementary Fig. S10A–C). Based on these data, we conclude that the expression of miR396 members is up-regulated in response to cold stress, which in turn strongly down-regulates LGS1 transcript levels.
Fig. 7.
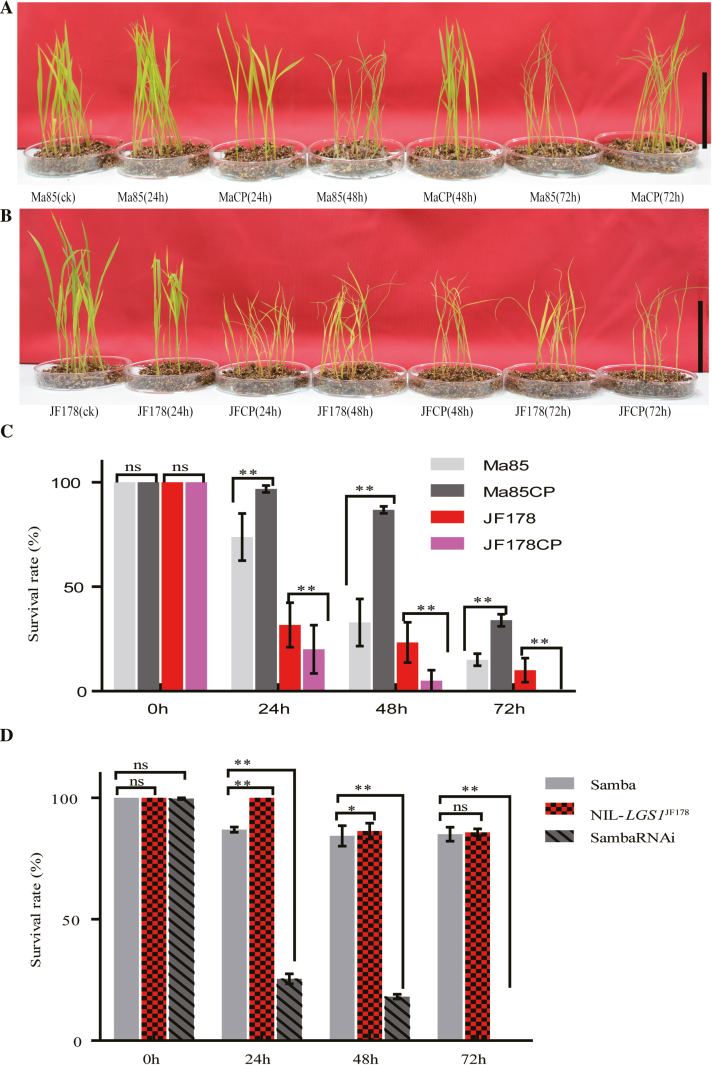
Abiotic stress response of LGS1. (A and B) Phenotype of Ma85 and MaCP (A), and JF178 and JFCP (B) after cold (4 °C) treatment (n=30). Ma85(ck) and JF178(ck), unstressed controls; MaCP and JFCP, transgenic plants expressing the complementation vector pLGS1::LGS1 in Ma85 and plgs1::lgs1 in JF178, respectively. Plants were treated for 24, 48, and 72 h, respectively. Scale bar=10 cm in (A and B). (C and D) Comparison of survival rates after cold treatment in Ma85, JF178, and transgenic plants expressing the complementation vectors pLGS1::LGS1 in Ma85 and plgs1::lgs1 in JF178, respectively (C), and Samba, NIL-LGS1JF178, and Samba RNAi lines (D). Ma85, JF178, and Samba serve as controls in (C) and (D). Values are given as the mean ±SE. ns P>0.05, *P<0.05, **P<0.01 were calculated by Student’s t-test.
We also treated seedlings of Ma85, JF178, Samba, NIL-LGS1JF178, and SambaRNAi transgenic plants with 4 °C, and then allowed them to grow for a further 5 d under normal growth conditions. We found that the survival rates of the Ma85 seedlings expressing LGS1JF178 (MaCP) after 4 °C treatment for 24, 48, and 72 h were 31.2, 164.1, and 126.2%, respectively, higher than those of Ma85 (Fig. 7C). In contrast, the survival rates of the JF178 seedlings expressing lgs1Ma85 (JFCP) were 36.8, 78.6, and 99.7%, respectively, lower than those of JF178 (Fig. 7C). Furthermore, the survival rates after 4 °C treatment for 24, 48, and 72 h were 15.1, 2.3, and 1.0%, respectively, higher in NIL-LGS1JF178 seedlings, but 70.7, 78.5, and 100% lower in SambaRNAi seedlings, than those in the control Samba (Fig. 7D). Our data also showed that the expression levels of lgs1 and miR396 members returned to normal levels (Supplementary Fig. S11A–E) after the removal of cold stress, suggesting that these seedlings had recovered from the cold treatment. Taken together, these data demonstrate that the LGS1 allele enhances cold tolerance at the seedling stage of rice.
Discussion
A missense mutation in LGS1 results in the development of enlarged grains
In this study, we cloned and characterized a gain-of-function mutant LGS1, and our data demonstrate that a 2 bp substitution (TC to AA) in LGS1 results in increases in cell length and cell numbers in the longitudinal direction of developing grains and promotes cell expansion in the latitudinal direction. As a result, there is an increase in glume volume and endosperm size in the JF178 mutant that contains the LGS1 allele. The increase in glume volume was found to be correlated with increases in grain length, grain width, thousand-grain weight, and grain yield in JF178. Analysis of the genetic transformation tests reveals that the elevated LGS1 mRNA is the cause of the enlarged grains in JF178, indicating that the LGS1 mRNA level serves as a positive regulator of grain size in rice.
OsGRF4 regulates grain formation through interaction with OsGIF co-activator
LGS1 encodes a plant-specific growth-regulating factor, an allele of OsGRF4, also known as GS2 (Duan et al., 2015; Hu et al., 2015), GL2 (Che et al., 2015), PT2 (Sun et al., 2016), and GLW2 (Li et al., 2016). Similar to LGS1, these alleles of OsGRF4 also positively control grain length, grain width, grain weight, and other traits. The GRF family of transcription factors comprises nine members in A. thaliana, and has been shown to regulate leaf cell proliferation and expansion, stress responses, and pistil development (Kim et al., 2003; Rodriguez et al., 2010; Kim et al., 2012; Liang et al., 2014). There are 14 GRF family members in Zea mays, some of which have been shown to control the bolting stage and inflorescence stem growth (Zhang et al., 2008). Both the indica (Supplementary Fig. S12) and japonica subspecies of rice (Choi et al., 2004) have 12 GRF members. Previous observations have shown that GRF family members regulate stem elongation, inflorescence development, and floral organogenesis in plants (Van der Knopp et al., 2000; Choi et al., 2004; Liu et al., 2014; Gao et al., 2015). In general, GRF transcription factor proteins typically contain a QLQ domain and a WRC domain, which are implicated in DNA remodelling and transcription activation. In addition to having these two highly conserved domains, rice LGS1 also includes a transcription activation domain at its C-terminus (Supplementary Fig. S3D–G, I, J). The QLQ domain of GRFs has been shown to bind to the SNH domain of GIFs to form a transcriptional co-activator, which promotes continuous differentiation and generation of new tissues throughout the plant life cycle (Kim and Kende, 2004; Zhang et al., 2008; Liang et al., 2014; Kim and Tsukaya, 2015). Consistent with these reports, our analysis using the yeast system showed that both LGS1 and lgs1 proteins interacted strongly with OsGIF3 (note that OsGIF3 of Supplementary Fig. S12 is designated as OsGIF1 in Duan et al., 2015 and Li et al., 2016). Interestingly, overexpression of OsGIF1 has previously been found to lead to development of larger and heavier grains than the wild-type variety (Che et al., 2015; Duan et al., 2015; Li et al., 2016). Whether other OsGIF genes also regulate grain size remains to be studied in the future.
miR396 members regulate the transcript level of LGS1
The WRC domain of GRF proteins is highly conserved, and its corresponding mRNA has been believed to be the target of miR396 in plants (Baucher et al., 2013; Kim and Tsukaya, 2015). Most miRNAs exert their functions to down-regulate the expression of target genes, and have been implicated in the regulation of cell differentiation and organogenesis in plants (Liu et al., 2009; Rodriguez et al., 2010; Ercoli et al., 2016; Rodriguez et al., 2016). miR396b has been found to mediate OsGRF6 and regulate rice yield by shaping inflorescence architecture (Gao et al., 2015). miR396c, and g/h/i may mediate GS2 (Duan et al., 2015; Hu et al., 2015), miR396c mediates GLW2 (Li et al., 2016), and miR396d mediates GL2 (Che et al., 2015). The role of miR396 in mediation of GRF transcripts has also been shown by miR396c overexpression, which markedly decreases rice grain size and weight (Li et al., 2016). GRF4NGR2 prevents miR396-mediated cleavage of GRF4 mRNA, thereby increasing GRF4 mRNA and GRF4 abundance, and promoting NH4+ uptake (Li et al., 2018). Analysis of the lgs1 (OsGRF4) sequence suggested that it could be targeted by all nine miR396 precursors (Supplementary Table S7; Supplementary Fig. S8A). The expression levels of OsmiR396 were also found to correlate with a regulatory network of OsGRF4. Thus, the expression of OsmiR396 members could significantly attenuate the transcript level of lgs1 (Supplementary Fig. S8B). The mutation in LGS1 appeared to prevent cleavage by miR396, which led to an elevated transcript level of LGS1, resulting in the formation of enlarged grains in the mutant line JF178. Taken together, these findings indicate that miR396 regulates the transcript level of OsGRF4 (lgs1) in an antagonistic manner and the 2 bp mutation of LGS1 impedes the down-regulation of LGS1 transcripts by miR396.
LGS1 improves cold tolerance of rice seedlings
Transcription factors have been implicated widely in the regulation of signaling and metabolic pathways in response to adversity stress, including biotic stress (bacterial pathogens, fungi, and insects) and abiotic stress (heavy metals, high temperatures, low temperatures, drought, waterlogging, high salinity, and high alkalinity). The pathways of stress responses overlap extensively in plants (Singh et al., 2002; Nakashima et al., 2009; Brotman et al., 2012; Ambavaram et al., 2014). The regulation of gene expression via miRNAs is one of the mechanisms involved in the different stress responses. Plants produce crosstalk signal responses that regulate cellular physiology and metabolism, and ultimately yield formation in crops (Jones-Rhoades and Bartel, 2004; Ding et al., 2011; Ambavaram et al., 2014). Previous studies on the OsGRF gene–miR396 stress networks in rice have shown that OsGRF4 is down-regulated by drought, overexpression of miR396c leads to a decrease in salt and alkali stress tolerance, and the expression of miR396d is down-regulated by cadmium stress (Gao et al., 2010; Ding et al., 2011; Tang and Chu, 2017). OsGRF4 and GL2 transcription activation activity is known to be down-regulated by GSK2 to a large extent and functions in the brassinosteroid signaling pathway (Che et al., 2015), which has been shown to be cold stress responsive in Arabidopsis (Li et al., 2017). In this study, we observed that OsGRF4 may be involved in the signaling pathways of hormone and stress responses (Supplementary Fig. S9A). Although the stress response patterns were highly complex, the mutation in LGS1 significantly enhanced tolerance to low temperatures and increased the survival rates of seedlings after recovery (Fig. 7). This conclusion is consistent with the observation that the transcript levels of lgs1 were down-regulated by cold treatment, whereas OsmiR396 expression was up-regulated by low temperatures (Supplementary Figs S9B, C, S10). Taken together, these results indicate that miR396 acts as a negative regulator to target OsGRF4 for cleavage and mediate plant stress responses. This study identified the missense mutation of LGS1 as the interfering site that breaks the antagonistic regulation pattern of OsGRF4–miR396 stress response networks, resulting in the development of large grains and the enhancement of cold tolerance in rice.
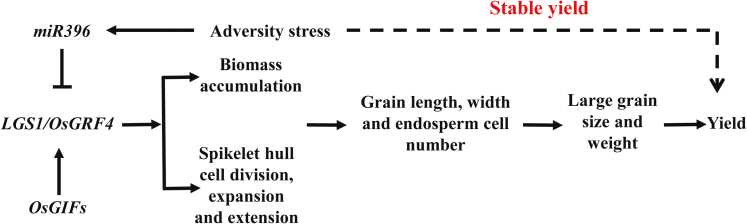
In summary, this work has identified an important gene that regulates grain development and stress tolerance in rice. We propose a model to elucidate the possible mechanism of LGS1 in the regulation of rice growth and development, including grain and yield formation and adaptability to stress environments (Fig. 8). The results presented in this report provide new insight into a new genetic regulation mechanism of the OsGRF4–OsGIF–OsmiR396 stress response network. This model contributes to the understanding of the complex regulatory mechanisms underlying seed development and yield formation in rice and other crops, and could be useful for applications in molecular breeding and molecular marker-assisted selection breeding in rice.
Fig. 8.
Schematic model for the role of LGS1 in the regulation of grain length, grain width, and grain weight. LGS1 forms a protein complex with OsGIF to regulate gene expression. The transcript level of LGS1 is regulated by miR396. LGS1 promotes cell division, cell extension, and cell expansion, resulting in increases in vegetative biomass and spikelet glume volume. During grain development, LGS1 enhances the grain filling rate and increases grain length and endosperm cell numbers, leading to the production of enlarged grains and higher yield in rice. Furthermore, the expression of miR396 is up-regulated by stress, which in turn reduces the transcript level of lgs1. The nucleotide substitution in LGS1 prevents regulation of LGS1 by miR396 and allows the LGS1 transcript to accumulate during adversity stress, which improves stress tolerance in rice seedlings.
Supplementary data
Fig. S1. Construction of the near-isogenic line NIL-LGS1.
Fig. S2. Panicle phenotype analysis of LGS1.
Fig. S3. LGS1 expression vector constructs, subcellular localization, and interaction partner.
Fig. S4. Grain shape traits of transgenic plants.
Fig. S5. Glume cell number, endosperm size, and grain filling dynamics of NILs.
Fig. S6. Biomass accumulation of NILs.
Fig. S7. Response of NILs to plant hormone treatments.
Fig. S8. Targeting of the LGS1 and lgs1 mRNAs by miR396.
Fig. S9. Expression levels of LGS1 and miR396 in response to low temperature stress.
Fig. S10. Expression levels of miR396 members in Ma85, JF178, Samba, and transgenic lines in response to cold treatment.
Fig. S11. Expression levels of LGS1 and miR396 in rice seedlings after recovery from cold treatment.
Fig. S12. Phylogenetic tree of the GRF family members in rice.
Table S1. Sequences of primers for vector construction and PCR analysis.
Table S2. Polymorphisms of the LGS1 coding sequence in different accessions.
Table S3. Polymorphisms of the LGS1 non-coding sequence in different accessions.
Table S4. Young panicle differentiation stage of rice.
Table S5. RNA-seq analysis of cell cycle genes in NILs.
Table S6. RNA-seq analysis of unknown cell cycle, expansin, and extensin genes in NILs.
Table S7. Alignment sites of the miR396 target in OsGRF4.
Acknowledgements
We thank Dr Tao Huang, Dr Luming Yao, Dr Caiming Wu (School of Life Sciences, Xiamen University), and Dr Jiwai He (Hunan Agricultural University) for generously providing technical assistance. This work was supported by grants from the Seed Industry Innovation and Industrialization Project of Fujian Province (fjzycxny2017004), the Fundamental Research Funds for the Central Universities (2013121040), the Program on Technology of Fujian Province (2017N2010290), the Open Program of State Key Laboratory of Rice Biology of China (170101), and the USDA-Hatch project (IDA-01423).
Glossary
Abbreviations
- DEG
differentially expressed gene
- GIF
GRF-interacting factor
- GRF
growth-regulating factor
- miR396
microRNA 396
- LGS1
Large grain size 1
- NIL
near-isogenic line
- QLQ
glutamine–leucine–glutamine domain
- QTL
quantitative trait locus
- SC/DO
yeast synthetic complete medium with dropouts
- WRC
tryptophan–arginine–cysteine domain
References
- Abe A, Kosugi S, Yoshida K, et al. . 2012. Genome sequencing reveals agronomically important loci in rice using MutMap. Nature Biotechnology 30, 174–178. [DOI] [PubMed] [Google Scholar]
- Ambavaram MM, Basu S, Krishnan A, Ramegowda V, Batlang U, Rahman L, Baisakh N, Pereira A. 2014. Coordinated regulation of photosynthesis in rice increases yield and tolerance to environmental stress. Nature Communications 5, 5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucher M, Moussawi J, Vandeputte OM, Monteyne D, Mol A, Pérez-Morga D, El Jaziri M. 2013. A role for the miR396/GRF network in specification of organ type during flower development, as supported by ectopic expression of Populus trichocarpa miR396c in transgenic tobacco. Plant Biology 15, 892–898. [DOI] [PubMed] [Google Scholar]
- Brotman Y, Lisec J, Méret M, Chet I, Willmitzer L, Viterbo A. 2012. Transcript and metabolite analysis of the Trichoderma-induced systemic resistance response to Pseudomonas syringae in Arabidopsis thaliana. Microbiology 158, 139–146. [DOI] [PubMed] [Google Scholar]
- Che R, Tong H, Shi B, et al. . 2015. Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nature Plants 2, 15195. [DOI] [PubMed] [Google Scholar]
- Choi D, Kim JH, Kende H. 2004. Whole genome analysis of the OsGRF gene family encoding plant-specific putative transcription activators in rice (Oryza sativa L.). Plant & Cell Physiology 45, 897–904. [DOI] [PubMed] [Google Scholar]
- Cui Y, Rao S, Chang B, et al. . 2015. AtLa1 protein initiates IRES-dependent translation of WUSCHEL mRNA and regulates the stem cell homeostasis of Arabidopsis in response to environmental hazards. Plant, Cell & Environment 38, 2098–2114. [DOI] [PubMed] [Google Scholar]
- Ding Y, Chen Z, Zhu C. 2011. Microarray-based analysis of cadmium-responsive microRNAs in rice (Oryza sativa). Journal of Experimental Botany 62, 3563–3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan P, Ni S, Wang J, Zhang B, Xu R, Wang Y, Chen H, Zhu X, Li Y. 2015. Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nature Plants 2, 15203. [DOI] [PubMed] [Google Scholar]
- Ercoli MF, Rojas AM, Debernardi JM, Palatnik JF, Rodriguez RE. 2016. Control of cell proliferation and elongation by miR396. Plant Signaling & Behavior 11, e1184809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Wang K, Liu Y, et al. . 2015. Blocking miR396 increases rice yield by shaping inflorescence architecture. Nature Plants 2, 15196. [DOI] [PubMed] [Google Scholar]
- Gao P, Bai X, Yang L, Lv D, Li Y, Cai H, Ji W, Guo D, Zhu Y. 2010. Over-expression of osa-MIR396c decreases salt and alkali stress tolerance. Planta 231, 991–1001. [DOI] [PubMed] [Google Scholar]
- Godfray HC, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF, Pretty J, Robinson S, Thomas SM, Toulmin C. 2010. Food security: the challenge of feeding 9 billion people. Science 327, 812–818. [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. 1994. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. The Plant Journal 6, 271–282. [DOI] [PubMed] [Google Scholar]
- Hu J, Wang Y, Fang Y, et al. . 2015. A rare allele of GS2 enhances grain size and grain yield in rice. Molecular Plant 8, 1455–1465. [DOI] [PubMed] [Google Scholar]
- Huang R, Jiang L, Zheng J, Wang T, Wang H, Huang Y, Hong Z. 2013. Genetic bases of rice grain shape: so many genes, so little known. Trends in Plant Science 18, 218–226. [DOI] [PubMed] [Google Scholar]
- Huang X, Zhao Y, Wei X, et al. . 2011. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nature Genetics 44, 32–39. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO Journal 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP. 2004. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Molecular Cell 14, 787–799. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. 2006. MicroRNAs and their regulatory roles in plants. Annual Review of Plant Biology 57, 19–53. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Choi D, Kende H. 2003. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. The Plant Journal 36, 94–104. [DOI] [PubMed] [Google Scholar]
- Kim JH, Kende H. 2004. A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proceedings of the National Academy of Sciences, USA 101, 13374–13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Tsukaya H. 2015. Regulation of plant growth and development by the GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR duo. Journal of Experimental Botany 66, 6093–6107. [DOI] [PubMed] [Google Scholar]
- Kim JS, Mizoi J, Kidokoro S, et al. . 2012. Arabidopsis growth-regulating factor7 functions as a transcriptional repressor of abscisic acid- and osmotic stress-responsive genes, including DREB2A. The Plant Cell 24, 3393–3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y, Su N, Shen Y, et al. . 2012. Identification of novel miRNAs and miRNA expression profiling during grain development in indica rice. BMC Genomics 13, 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ye K, Shi Y, Cheng J, Zhang X, Yang S. 2017. BZR1 positively regulates freezing tolerance via CBF-dependent and CBF-independent pathways in Arabidopsis. Molecular Plant 10, 545–559. [DOI] [PubMed] [Google Scholar]
- Li S, Gao F, Xie K, et al. . 2016. The OsmiR396c–OsGRF4–OsGIF1 regulatory module determines grain size and yield in rice. Plant Biotechnology Journal 14, 2134–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Tian Y, Wu K, et al. . 2018. Modulating plant growth–metabolism coordination for sustainable agriculture. Nature 560, 595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, He H, Li Y, Wang F, Yu D. 2014. Molecular mechanism of microRNA396 mediating pistil development in Arabidopsis. Plant Physiology 164, 249–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Song Y, Chen Z, Yu D. 2009. Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiologia Plantarum 136, 223–236. [DOI] [PubMed] [Google Scholar]
- Liu H, Guo S, Xu Y, Li C, Zhang Z, Zhang D, Xu S, Zhang C, Chong K. 2014. OsmiR396d-regulated OsGRFs function in floral organogenesis in rice through binding to their targets OsJMJ706 and OsCR4. Plant Physiology 165, 160–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Hua L, Dong S, Chen H, Zhu X, Jiang J, Zhang F, Li Y, Fang X, Chen F. 2015. OsMAPK6, a mitogen-activated protein kinase, influences rice grain size and biomass production. The Plant Journal 84, 672–681. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Ito Y, Yamaguchi-Shinozaki K. 2009. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiology 149, 88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuley J, Lightner J, Feldmann K, Yadav N, Lark E, Browse J. 1994. Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. The Plant Cell 6, 147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez RE, Ercoli MF, Debernardi JM, Breakfield NW, Mecchia MA, Sabatini M, Cools T, De Veylder L, Benfey PN, Palatnik JF. 2015. MicroRNA miR396 regulates the switch between stem cells and transit-amplifying cells in Arabidopsis roots. The Plant Cell 27, 3354–3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez RE, Mecchia MA, Debernardi JM, Schommer C, Weigel D, Palatnik JF. 2010. Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137, 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez RE, Schommer C, Palatnik JF. 2016. Control of cell proliferation by microRNAs in plants. Current Opinion in Plant Biology 34, 68–76. [DOI] [PubMed] [Google Scholar]
- Singh K, Foley RC, Oñate-Sánchez L. 2002. Transcription factors in plant defense and stress responses. Current Opinion in Plant Biology 5, 430–436. [DOI] [PubMed] [Google Scholar]
- Smith NA, Singh SP, Wang MB, Stoutjesdijk PA, Green AG, Waterhouse PM. 2000. Total silencing by intron-spliced hairpin RNAs. Nature 407, 319–320. [DOI] [PubMed] [Google Scholar]
- Stoutjesdijk PA, Singh SP, Liu Q, Hurlstone CJ, Waterhouse PA, Green AG. 2002. hpRNA-mediated targeting of the Arabidopsis FAD2 gene gives highly efficient and stable silencing. Plant Physiology 129, 1723–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P, Zhang W, Wang Y, He Q, Shu F, Liu H, Wang J, Wang J, Yuan L, Deng H. 2016. OsGRF4 controls grain shape, panicle length and seed shattering in rice. Journal of Integrative Plant Biology 58, 836–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Chu C. 2017. MicroRNAs in crop improvement: fine-tuners for complex traits. Nature Plants 3, 17077. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Knapp E, Kim JH, Kende H. 2000. A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiology 122, 695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Li S, Liu Q, et al. . 2015. The OsSPL16–GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nature Genetics 47, 949–954. [DOI] [PubMed] [Google Scholar]
- Wang Y, Xiong G, Hu J, et al. . 2015. Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nature Genetics 47, 944–948. [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, et al. . 2001. Construct design for efficient, effective and high-throughput gene silencing in plants. The Plant Journal 27, 581–590. [DOI] [PubMed] [Google Scholar]
- Wu W, Liu X, Wang M, et al. . 2017. A single-nucleotide polymorphism causes smaller grain size and loss of seed shattering during African rice domestication. Nature Plants 3, 17064. [DOI] [PubMed] [Google Scholar]
- Zhang D, Li B, Jia G, Zhang T, Dai J, Li J, Wang S. 2008. Isolation and characterization of genes encoding GRF transcription factors and GIF transcriptional coactivators in maize (Zea mays L.). Plant Science 175, 809–817. [Google Scholar]
- Zhao K, Tung CW, Eizenga GC, et al. . 2011. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nature Communications 2, 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.