Abstract
Artemisinin is a sesquiterpene lactone produced by the Chinese traditional herb Artemisia annua and is used for the treatment of malaria. It is known that salicylic acid (SA) can enhance artemisinin content but the mechanism by which it does so is not known. In this study, we systematically investigated a basic leucine zipper family transcription factor, AaTGA6, involved in SA signaling to regulate artemisinin biosynthesis. We found specific in vivo and in vitro binding of the AaTGA6 protein to a ‘TGACG’ element in the AaERF1 promoter. Moreover, we demonstrated that AaNPR1 can interact with AaTGA6 and enhance its DNA-binding activity to its cognate promoter element ‘TGACG’ in the promoter of AaERF1, thus enhancing artemisinin biosynthesis. The artemisinin contents in AaTGA6-overexpressing and RNAi transgenic plants were increased by 90–120% and decreased by 20–60%, respectively, indicating that AaTGA6 plays a positive role in artemisinin biosynthesis. Importantly, heterodimerization with AaTGA3 significantly inhibits the DNA-binding activity of AaTGA6 and plays a negative role in target gene activation. In conclusion, we demonstrate that binding of AaTGA6 to the promoter of the artemisinin-regulatory gene AaERF1 is enhanced by AaNPR1 and inhibited by AaTGA3. Based on these findings, AaTGA6 has potential value in the genetic engineering of artemisinin production.
Keywords: Artemisinin, Artemisia annua, salicylic acid, transcription factor
Binding of the bZIP transcription factor AaTGA6 to the promoter of the artemisinin-regulatory gene AaERF1 is inhibited by AaTGA3 and enhanced by AaNPR1, which acts in the salicylic acid signaling pathway.
Introduction
Malaria is a serious disease that threatens the health of more than 212 million people (World Health Organization, 2016). The sesquiterpene artemisinin is produced in the glandular trichomes of the Chinese medicinal plant Artemisia annua (Duke et al., 1994) and in recent decades artemisinin combination therapy (ACT) has been recommended by the World Health Organization (WHO) to treat malaria caused by the Plasmodium falciparum parasite. Artemisinin has also been shown to have additional potential roles in the treatment of several human cancers and viral diseases (Ma et al., 2014; O’Brien et al., 2015). It may also inhibit Mycobacterium tuberculosis and has great potential to cure diabetes (Li et al., 2016; Zheng et al., 2017).
The artemisinin biosynthetic pathway has been determined (Bouwmeester et al., 1999; Mercke et al., 2000; Teoh et al., 2006, 2009; Zhang et al., 2008) (Supplementary Fig. S1 at JXB online) and the small-scale production of an artemisinin precursor in engineered Saccharomyces cerevisiae has been developed; however, this is only enough to supplement, rather than replace, the agricultural production of artemisinin (Westfall et al., 2012). The price of artemisinin remains relatively high in developing countries, and increasing its yield is an effective way to reduce the cost of production. In this context, the genetic engineering of artemisinin production is a promising method to improve the availability of this compound (Tang et al., 2014).
Salicylic acid (SA), a phenolic compound, plays an important role in plant defense responses against pathogens (Raskin et al., 1990). Many studies have indicated that NON-EXPRESSOR OF PATHOGENESIS-RELATED GENES 1 (NPR1) is an important modulator of plant immunity in SA signal transduction (Després et al., 2000; Kinkema et al., 2000; Zhou et al., 2000; Mukhtarv et al., 2009; Spoel et al., 2009; Wu et al., 2012). NPR1 contains ankyrin repeats and a BTB/POZ (broad-complex, tramtrack, and bric-a-brac/pox virus and zinc finger) domain (Rochon et al., 2006; Boyle et al., 2009). The ankyrin repeat domain is thought to be involved in interactions with TGA transcription factors (Després et al., 2003) and the BTB/POZ domain is necessary for the DNA binding activity of the TGA transcription factors. Under the stimulation of SA, TGA2 can be incorporated into a transactivating complex with the BTB/POZ domain (residues 80–91) of NPR1 and form an enhanceosome (Fan and Dong, 2002; Rochon et al., 2006). There are 10 TGA transcription factors in Arabidopsis, and three TGAs (TGA2/5/6, termed class II TGAs) interact with NPR1 via strong binding (Zhang et al., 1999; Després et al., 2000; Zhou et al., 2000).
TGA factors are the basic region/leucine zipper motif (bZIP) transcription factors involved in the regulation of pathogen defense, stress signaling, flower development, and seed maturation by binding to the as-1 element (Lam and Lam, 1995; Xiang et al., 1997; Zhang et al., 1999; Després et al., 2000; Zhou et al., 2000). The class II TGA proteins are the essential molecular linkers of the SA and jasmonic acid/ethylene signaling networks for defense responses (Zander et al., 2010; Van der Does et al., 2013). Moreover, class II TGA factors can interact with NPR1 to form an enhanceosome complex at the PR-1 promoter, indicating that they have redundant roles in modulating gene expression in the process of defense (Rochon et al., 2006). In addition, in vivo assays indicate that some homodimers and heterodimers are formed between TGA factors (Foster et al., 1994; Lam and Lam, 1995).
Much research has shown that SA regulates the biosynthesis of secondary metabolites (Pu et al., 2009; Okada et al., 2009). SA can improve the artemisinin content (Pu et al., 2009; Yu et al., 2012), but the mechanism of the process is still unknown. In general, AP2/ERF transcription factors are targets of SA (Van der Does et al., 2013; Zander et al., 2014). AP2/ERF transcription factors have a conserved AP2 DNA-binding domain of ~60 amino acid residues. In plants, the AP2/ERF family plays important roles in the modulation of secondary metabolite biosynthesis. The AP2/ERF proteins AaERF1 and AaERF2 are involved in enhancing transcription levels of the artemisinin biosynthetic pathway genes ADS and CYP71AV1. Artemisinin levels are increased by 19–67% and 24–51% in AaERF1- and AaERF2-overexpressing plants, respectively, suggesting that AaERF1/AaERF2 play important roles in sesquiterpenoid biosynthesis (Yu et al., 2012). Several other AP2/ERF transcription factors have also been shown to regulate artemisinin production, including AaORA (Lu et al., 2013) and AaTAR1 (Tan et al., 2015).
In this study, we examined the SA-mediated molecular mechanisms that modulate artemisinin biosynthesis and found that a bZIP transcription factor AaTGA6 specifically targets the promoter of AaERF1. In addition, we demonstrated that AaNPR1 acts as a transcriptional co-activator and interacts with AaTGA6, forming an enhanceosome at the AaERF1 promoter. AaTGA3 interacts with AaTGA6 to form a heterodimer and this exerts a negative effect on the DNA-binding activity of AaTGA6. Finally, we provided evidence of AaTGA6-mediated artemisinin synthesis by the AaNPR1-AaTGA6-AaTGA3 complex, which modulates AaERF1 to influence the accumulation of artemisinin. This network may provide a reference for other medicinal plants.
Materials and methods
Plant material
Seeds of Artemisia annua L. (Qinghao) ‘Huhao 1’ from Chongqing, China, were surface-sterilized with 75% ethanol for 1 min and then sterilized using 20% (v/v) NaOCl (sodium hypochlorite) for 20 min (Shen, et al., 2016). All seeds were washed three times for 5 min in sterilized water, planted on MS medium (Murashige and Skoog, 1962), and incubated with a photoperiod of 16/8 h light/dark with 7500 lux at 26 °C.
Seeds of Nicotiana benthamiana were sown in a soil mixture (vermiculite:perlite:peat moss, 7:0.5:2) and incubated under the same conditions. The tobacco leaves were used for infiltration experiments after 5 weeks.
Plant hormone treatments
For plant hormonal treatment, 1-month-old A. annua seedlings were sprayed with 1 mM SA (Sigma-Aldrich; adjusted to pH 7.0 with NaOH) (Pu et al., 2009) and then sampled at 0, 0.5, 1, 3, 6, 12, and 24 h for RNA isolation.
RNA isolation and qPCR analysis
Total RNA was extracted using an RNAprep Pure Plant Kit (Tiangen Biotech) (Lv et al., 2016). Samples of 500 ng of total RNA were used for first-strand cDNA synthesis using a cDNA synthesis kit (TaKaRa Biotech) according to the manufacturer’s instructions. For qPCR, the diluted cDNA was used as a template, and SYBR Green (Kapa Biosystems) was used to detect transcript levels. The β-ACTIN gene of A. annua was used as an internal control (Zhang, et al., 2015; Lv et al., 2016; Shen, et al., 2016). The primers used for qPCR are listed in Supplementary Table S1.
Dual-LUC assay
AaTGA6, AaTGA3, AaNPR1, N-AaNPR1, C-AaNPR1, N-AaTGA6, and C-AaTGA6 were individually subcloned into the PHB vector to generate the effector. The promoters of AaERF1 and ADS were fused to the vector pGreenⅡ0800 to generate a reporter. The reporter and effector constructs were then separately transformed into Agrobacterium tumefaciens strain GV3101. The bacterial cells were resuspended in MS medium with 10 mM methylester sulfonate and 150 μM acetosyringone to OD600=0.6 and then incubated at room temperature for 3 h. The bacteria-harboring constructs were infiltrated into tobacco leaves according to Zhang, et al. (2015). The leaves were collected after 48 h for dual-LUC assays using a Dual-Luciferase Reporter Assay System according to the manufacturer’s instructions (Promega). Three independent biological replicates were measured for each sample.
Y2H, BiFC, and pull-down analysis
For yeast two-hybrid (Y2H) assays, the ORF of AaTGA6 was cloned into the yeast GAL4 DNA-binding domain vector GBKT7 (Clontech) and used as bait. The ORFs of AaTGA3 and AaNPR1 were fused to the vector GADT7 as prey. The plasmids of bait and prey were co-transformed into the yeast strain AH109 (Clontech) by the LiAC-polyethylene glycol method (Gietz and Schiestl, 2007). The transformants were reconfirmed on Leu- and Trp-deficient synthetic dropout (SD) medium plates. The interactions of AaTGA6 with AaNPR1 and AaTGA3 were tested on SD–Leu–Trp–His or SD–Ade–Leu–Trp–His plates with 3-amino-1,2,4-triazole. At least five individual clones were analysed.
For the bimolecular fluorescence complementation (BIFC) assays, AaNPR1 and AaTGA3 were subcloned into the vector pEG202 to generate AaNPR1-pEG202 and AaTGA3-pEG202; AaTGA6 was subcloned into the vector pEG201 to generate AaTGA6-pEG201. Plasmids of AaNPR1-pEG202, AaTGA3-pEG202, and AaTGA6-pEG201 were transformed into A. tumefaciens strain GV3101. The A. tumefaciens-harboring sets of constructs were resuspended in MS liquid medium buffer with 10 mM methylester sulfonate and 150 μM acetosyringone at OD600=0.6 and then incubated at room temperature for 3 h. Tobacco leaves were infiltrated with the A. tumefaciens suspensions. Yellow fluorescent protein (YFP) was observed by confocal laser microscopy.
For pull-down assays, His, AaTGA6-His, GST, AaTGA3-GST, and AaNPR1-GST were expressed E. coli Rosetta (DE3) (Novagen). The recombinant proteins were purified using glutathione sepharose beads (Amersham Biosciences) and Ni-NTA agarose beads (QIAGEN). The pull-down assays were conducted according to Bratzel et al. (2010).
Y1H assays
To detect whether AaTGA6 could interact with the ‘TGACG’ box in yeast, yeast one-hybrid (Y1H) assay were used and examined according to Zhang et al. (2015).
EMSA analysis
To test whether AaTGA6 could interact with the ‘TGACG’ box, an electrophoretic mobility shift assay (EMSA) was performed. The ORF regions of AaTGA6, AaNPR1, and AaTGA3 were cloned into the PET28A vector (Novagen) and the resulting constructs (AaTGA6-PET28, AaNPR1-PET28, and AaTGA3-PET28) were introduced into E. coli strain BL21(DE3). The purification of the AaTGA6-PET28, AaNPR1-PET28, and AaTGA3-PET28 fusion proteins was performed by Ni-NTA agarose, according to the manufacturer’s instructions (Qiagen). The 40-bp oligonucleotides were the recognition sites of AaTGA6, labeled by digoxin as probes for the EMSA studies, and the sequences are shown in Supplementary Table S1. The EMSA reactions were performed using a DIG Gel Shift Kit, 2nd Generation, according to the manufacturer’s instructions (Roche). After the EMSA reaction, the products were loaded on a 5% polyacrylamide gel (acrylamide:bisacrylamide, 37.5:1; Bio-Rad) by applying a voltage of 80 V for electrophoresis. The DNA–protein complex was transferred to a nylon membrane for 120 min at 30 V. The membrane was then cross-linked at 120 MJ. The detection was performed according to instructions from Roche.
RNA-seq analysis
total RNA samples were isolated from 1-month-old plants of A. annua and sequenced on a HiSeq 4000 (Illumina Inc.). RNA-seq analysis was performed according to Shi et al. (2018).
Quantification of artemisinin using HPLC-ELSD
Artemisinin was quantified using HPLC-ELSD (evaporative light scattering detection). Young leaves at the upper levels of secondary branches of A. annua were collected and dried at 50 °C for 24 h and ground into powder (Nair et al., 2013). The powdered samples (0.1 g) were used for artemisinin detection with a Waters Alliance 2695 HPLC system (Zhang et al., 2009).
Accession Numbers
Sequence data from this article are deposited in the GenBank databases under the following accession numbers: AaNPR1 (MH201457), AaNPR2 (MH201458), AaNPR3 (MH201459), AaNPR4 (MH201460), AaNPR5 (MH201461), AaTGA1 (MH201462), AaTGA2 (MH201463), AaTGA3 (MH201464), AaTGA4 (MH201465), AaTGA5 (MH201466), AaTGA6 (MH201467). The accession number of Raw RNA-seq data is available from the NCBI Sequence Read Archive (PRJNA524494).
Results
Characterization of AaTGA6
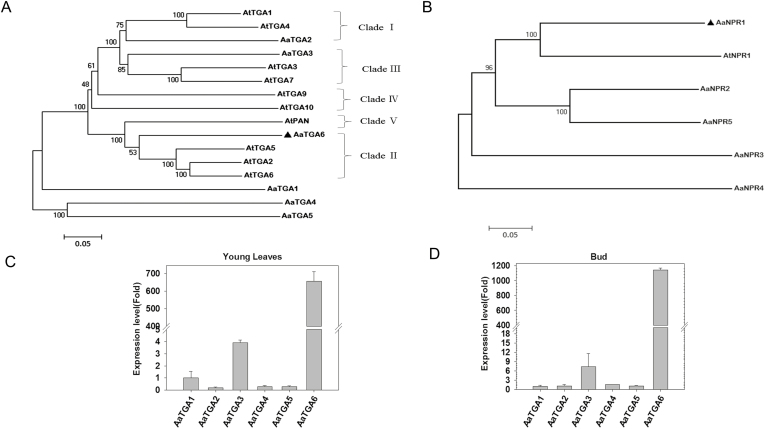
SA regulation of artemisinin content has previously been shown to increase the transcript level of ADS in the artemisinin biosynthetic pathway in A. annua (Supplementary Fig. S2) (Pu et al., 2009; Yu et al., 2012). We hypothesized that the TGA transcription factors might modulate the expression level of ADS, either directly or indirectly. According to transcriptome sequencing, we first analysed the data and obtained the gene sequences of NPRs and TGAs in A. annua. There were six basic leucine zipper (bZIP) transcription factors of TGA (AaTGA1, AaTGA2, AaTGA3, AaTGA4, AaTGA5, and AaTGA6) and five NPR genes (AaNPR1, AaNPR2, AaNPR3, AaNPR4, and AaNPR5) in the transcriptome database of A. annua. We constructed a phylogenetic tree to identify TGA transcription factors in A. annua, which showed that AaTGA6 clustered with AtTGA2/AtTGA5/AtTGA6 (Fig. 1A). Previous studies have indicated that monocot and dicot plants share a conserved sequence and function in NPR1 (Kuai and Desprã, 2016). AaNPR1 had a close phylogenetic relationship with AtNPR1 (Fig. 1B), indicating that AaNPR1 may play a co-activator role in A. annua. To investigate which TGAs had the potential to affect the accumulation of artemisinin, we used qPCR to monitor mRNA levels of AaTGA1–6 in the different tissues (roots, stems, leaves, and buds) and different leaves (leaf 0–leaf 7; Ma et al., 2018). Most of TGA factors were mainly expressed in the roots, buds, and in leaves 4–7 (Supplementary Fig. S3).
Fig. 1.
Characterizations of NPRs and TGAs in Artemisia annua. Phylogenetic trees of (A) TGAs and (B) NPRs in A. annua (Aa) and Arabidopsis (At). The trees were constructed using MEGA 5 with the neighbor-joining method (1000 bootstrap replicates). The scale bars indicate the evolutionary distance of the sequences examined. AaTGA6 and AaNPR1 are indicated with black triangles. Expression of TGAs in (C) young leaves and (D) buds. Expression values are relative to those of TGA1. Data are means (±SE) of n=3 biologically independent samples.
The artemisinin biosynthetic pathway genes ADS, CYP71AV1, DBR2, and ALDH1 are mainly expressed in young leaves and buds (Lommen et al., 2006; Lu et al., 2013), and our qPCR assays showed a significantly higher expression level of AaTGA6 in these tissues (Fig. 1C, D), indicating that AaTGA6 may be the principle TGA that regulates artemisinin biosynthesis. In addition, AaTGA3 had similar expression patterns to AaTGA6, although at much lower levels.
AaTGA6 was localized to the nucleus in A. annua (Supplementary Fig. S4A) and may have been involved in artemisinin biosynthesis (Fig. 1C). We performed dual-LUC assays and these indicated that AaTGA6 was active in the transcription of ADS (Supplementary Fig. S4C).
AaTGA6 activates the transcription of AaERF1 in vivo
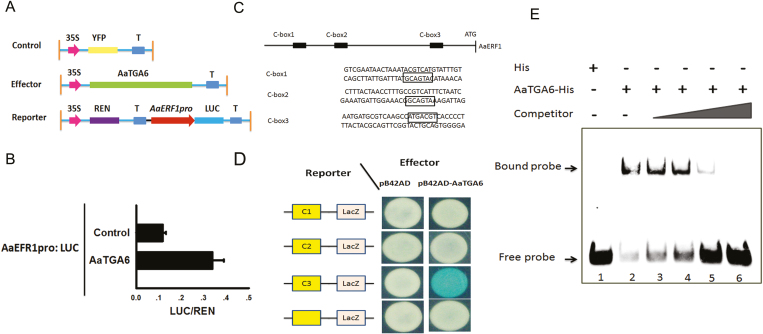
Previous studies have indicated that the AP2/ERF transcription factor ORA59 is the direct target of class II TGA transcription factors (Zander et al., 2014), and we hypothesized that AaTGA6 could directly modulate AP2/ERF transcription factor expression levels. Three AP2/ERF transcription factors (AaERF1, AaERF2, and AaORA) involved in artemisinin biosynthesis have been identified (Yu et al., 2012; Lu et al., 2013). We scanned the promoter region of AaERF1/AaERF2/AaORA and found ‘TGACG’ elements existing in the promoter of AaERF1 (Supplementary Fig. S5).
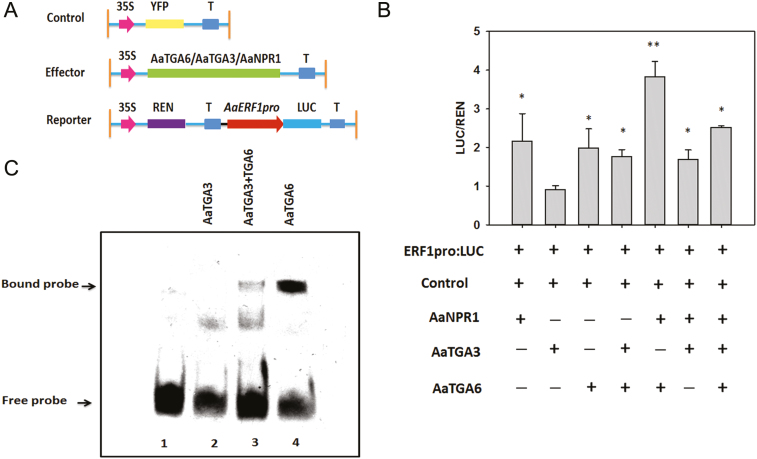
To survey the target transcription factors of AaTGA6, we performed deep RNA sequencing. One-month-old A. annua seedlings were treated with 1 mM SA, sampled at 0, 1, 5, and 7 h, and sequenced using the Illumina platform. According to the resulting heat map the expression levels of 101 genes were significantly enhanced under SA treatment, (P<0.01; Supplementary Fig. S6). Thus, all the available evidence indicated that AaERF1 may be a candidate target for modulation by AaTGA6. Further studies were therefore focused on the relationship between AaTGA6 and AaERF1. Using a dual-LUC assay, we examined whether AaTGA6 activated the transcription of AaERF1 in vivo. The results showed that AaTGA6 activated the expression of AaERF1 promoters, as evidenced by a higher value of LUC/REN than the control (Fig. 2B). AaTGA6 may therefore have great potential for increasing the artemisinin content in A. annua.
Fig. 2.
AaTGA6 is a transactivator that binds to the C-‘TGACG’ box in the promoter of AaERF1. (A) Schematic diagram of the reporter and effector constructs used in the transient dual-LUC assays. (B) Transient dual-LUC analysis showing AaTGA6 activation of the transcription of AaERF1 in N. benthamiana leaves. LUC/REN represents the luciferase/Renilla ratio of n=3 independent experiments; P<0.05 as determined using Student’s t-test. (C) Schematic diagram of the cis-element in the sequences of the AaERF1 promoter. C-box1, C-box2, and C-box3 were used for the yeast one-hybrid (Y1h) assays. The ‘TGACG’ boxes are indicated. (D) Y1H assays showing the interaction between AaTGA6 and ‘TGACG’ boxes in the promoter of AaERF1. Yeast transformants were diluted and plated on synthetic dropout medium without Ura and Trp plus 20 mM X-gal. (E) EMSA analysis to test the binding of AaTGA6-His to the ‘TGACG’ motifs in the promoter of AaERF1. Unlabeled ‘TGACG’ was used as competitor DNA at molar ratios of 1:1; 1:6; 1:50; and 1:100.
A Y1H assay was performed and demonstrated that AaTGA6 bound directly to the AaERF1 promoter in vivo (Fig. 2D). In addition, an EMSA analysis was performed to demonstrate the DNA-binding activity of AaTGA6. A retarded band of AaTGA6 was observed when AaTGA6 was added to the binding mixture (Fig. 2E, lane 2). To further test the binding specificity, the competition probe concentration was increased and this increased the number of retarded bands (Fig. 2E, lanes 3–6). Certain sequences that flank the conserved region ‘TGACG’ also played key roles in AaTGA6 binding. When the flanking sequences were mutated, the DNA-binding effects of AaTGA6 were changed (Supplementary Fig. S7). These results indicated that AaTGA6 specifically recognized the ‘TGACG’ sequence within the AaERF1 promoter.
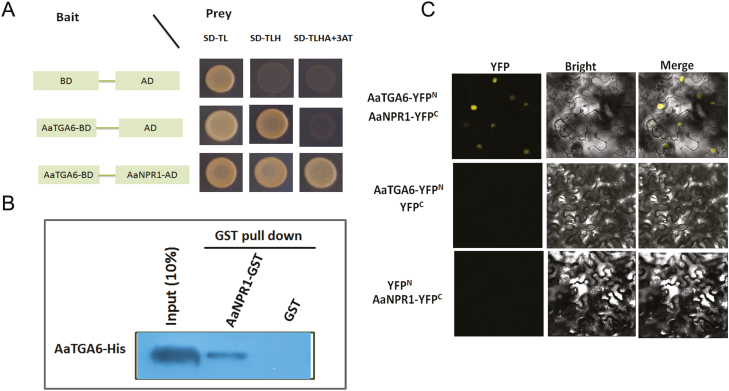
AaTGA6 interacts with AaNPR1
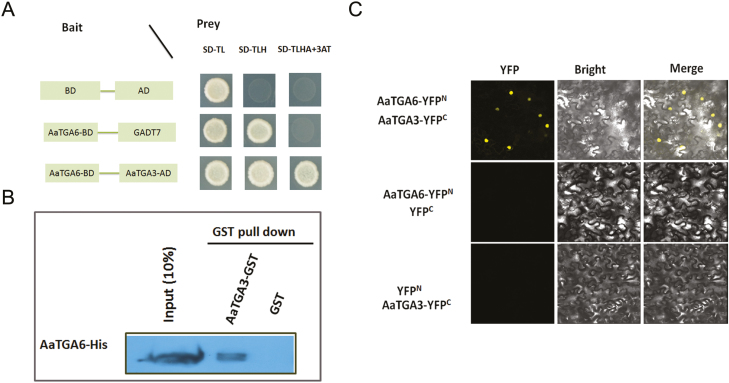
The class II TGA transcription factors (AtTGA2/5/6) can interact with AtNPR1, which is known as a mediator protein between TGA2/5/6 and the PR1 gene in Arabidopsis (Fan and Dong, 2002). We hypothesized that AaNPR1 might interact with the class II TGA transcription factor AaTGA6 in A. annua, and therefore performed a yeast two-hybrid (Y2H) assay. As shown in Fig. 3A, the interaction between AaNPR1 and AaTGA6 was confirmed. In addition, we performed YFP BiFC assays and pull-down assays, which also confirmed the interaction (Fig. 3B, C).
Fig. 3.
Interactions between AaNPR1 and AaTGA6. (A) Yeast two-hybrid assays. Yeast cells were plated on synthetic dropout media either without Leu and Trp (SD-TL), without Leu, Trp, and His (SD-TLH), or without Leu, Trp, His, and Ade but containing 80 mM 3-AT (SD-TLHA+3AT). (B) Interactions between AaNPR1 and AaTGA6 examined by BiFC in N. benthamiana leaves. AaNPR1 and AaTGA6 were fused in the vectors pEarleyagte202 and pEarleyagte201, respectively. (C) Pull-down assays showing the interaction of AaTGA6 and AaNPR1. Prokaryotic expression of AaTGA6 and AaNPR1 proteins fused to His or a GST tag were subjected to GST pull-down, and the proteins were detected by immunoblotting using a GST antibody.
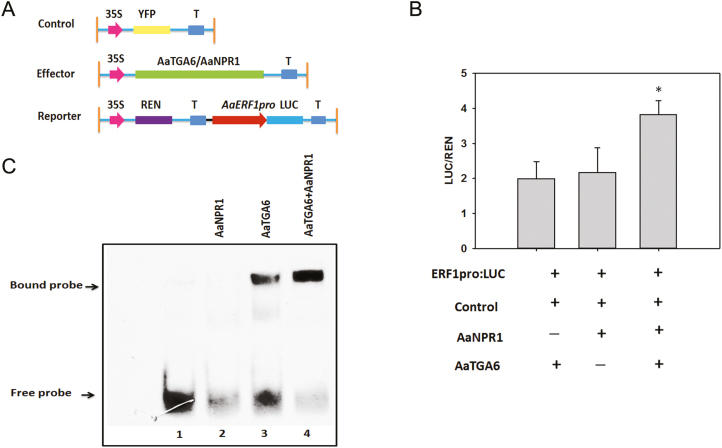
AaNPR1 enhances the binding of AaTGA6 to the ‘TGACG’ sequence
Previous studies have shown that NPR1 increases the binding of TGA2 to its cognate promoter element, as-1 (Després et al., 2000). The transcription factor ABF2 substantially enhances the binding of ANAC096 to the NACRSelements at the RD29A promoter (Xu et al., 2013). To determine whether it behaved in a similar way, we investigated the effect of AaNPR1 on the binding of AaTGA6 factors to the ‘TGACG’ sequence within the AaERF1 promoters. Dual-LUC assays were performed in the presence or absence of in vitro-translated AaNPR1, AaTGA6, or both. We found that the relative reporter activity of LUC/REN was low in the presence of AaNPR1 or AaTGA6, but in the presence of both the activation of LUC/REN significantly increased (Fig. 4B). In addition, AaNPR1 also activated the promoter of AaERF1. Surprisingly, the amino-terminal regions of AaNPR1 alone (N-AaNPR1,1–351 residues) seemed to have a high level of activation of the AaERF1 promoters (Supplementary Fig. S8). Since full-length AaNPR1 could activate the transcription of AaERF1, we hypothesized that N-AaNPR1 or C-AaNPR1 might also activate it. A dual-LUC assay showed that N-AaNPR1 exhibited high transcriptional activities at the AaERF1 promoters (Supplementary Fig. S8).
Fig. 4.
AaNPR1 enhances the DNA-binding activity of AaTGA6. (A) Schematic diagram of the reporter and effector constructs used in the transient dual-LUC assays. (B) Transient dual-LUC analysis showing that AaNPR1 increases AaTGA6 activation of the transcription of AaERF1. LUC/REN represents the luciferase/Renilla ratio of n=3 independent experiments; Significant differences were determined using Student’s t-test: *P<0.05. (C) EMSA analysis showing that AaNPR1 enhances the DNA-binding activity of AaTGA6.
To gain further insights into the role of AaNPR1, EMSA assays were performed in the presence or absence of AaTGA6, AaNPR1, or both. No migrating band was observed when AaNPR1 was present in the binding mixture (Fig. 4C, lane 2) and a retarded band was detected when AaTGA6 was present (Fig. 4C, lane 3). However, the addition of AaNPR1 to the AaTGA6 binding mixture substantially increased the number of migrating bands (Fig. 4C, lane 4). These results confirmed that AaNPR1 enhanced the binding of AaTGA6 to the ‘TGACG’ sequence within the AaERF1 promoters.
AaTGA3 interacts with AaTGA6 and inhibits its binding to the ‘TGACG’ sequence
Previous studies have indicated that heterodimers are preferentially formed in bZIP transcription factors (Ehlert et al., 2006). We had found that AaTGA3 had similar expression patterns to AaTGA6 in young leaves and buds (Fig. 1C, D) and we hypothesized that AaTGA6 might form heterodimers with other TGAs. Y2H, BiFC, and pull-down assays demonstrated that AaTGA3 could interact with AaTGA6 (Fig. 5A–C). To verify the biological significance of the interaction, dual-LUC assays were performed and indicated that AaTGA3 substantially repressed the binding of AaTGA6 to its cognate promoter element ‘TGACG’ in the promoter of AaERF1 (Fig. 6B). We also confirmed the inhibition using EMSA analysis, which showed that co-incubation of AaTGA3 with AaTGA6 resulted in a weak retarded band, indicating that AaTGA3 inhibition was functional in the EMSA assay (Fig. 6C, lane 3).
Fig. 5.
Interactions between AaTGA3 and AaTGA6. (A) Yeast two-hybrid assays with AaTGA3 and AaTGA6 fused to GAL4 activation domain (AD) or GAL4-binding domain (BD). Interactions were examined on synthetic dropout media either without Leu and Trp (SD-TL), without His, Leu, and Trp (SD-TLH), or without Ade, Leu Trp, His but containing 80 mM 3 AT (SD-ALTH). 3 AT inhibited the self-transcription activity of AaTGA6-BD. (B) Interactions between AaTGA3 and AaTGA6 were examined by BiFC in N. benthamiana leaves. AaTGA3 and AaTGA6 were fused in the vectors pEarleyagte201 and pEarleyagte202, respectively. (C) Pull-down assay showing the interaction of AaTGA6 and AaTGA3. Prokaryotic expression of AaTGA6 and AaTGA3 proteins fused to His or a GST tag were subjected to a GST pull-down, and the proteins were detected by immunoblotting using a GST antibody.
Fig. 6.
AaTGA3 represses the DNA-binding activity of AaTGA6. (A) Schematic diagram of the reporter and effector constructs in the transient dual-LUC assays. (B) Transient dual-LUC analysis showing that AaTGA3 inhibits AaTGA6 activation of the transcription of AaERF1. Luc/Ren represents the luciferase/Renilla ratio of n=3 independent experiments; Significant differences were determined using Student’s t-test: *P<0.05; **P<0.01. (C) EMSA analysis showing that AaTGA3 inhibits the DNA-binding activity of AaTGA6.
Molecular and chemical characterization of AaTGA6-overexpression and -RNAi knockdown transgenic lines
The data presented above indicated that AaTGA6 was predominantly modulated at the mRNA level of AaERF1, the positive regulator of artemisinin. We therefore overexpressed AaTGA6 in A. annua. Driven by the double-35S promoter, the AaTGA6 was infused into the plasmid PHB to generate the overexpression vector AaTGA6-PHB. There was no significant difference in the phenotype between the transgenic lines and the wild-type under normal growth conditions (Supplementary Fig. S9).
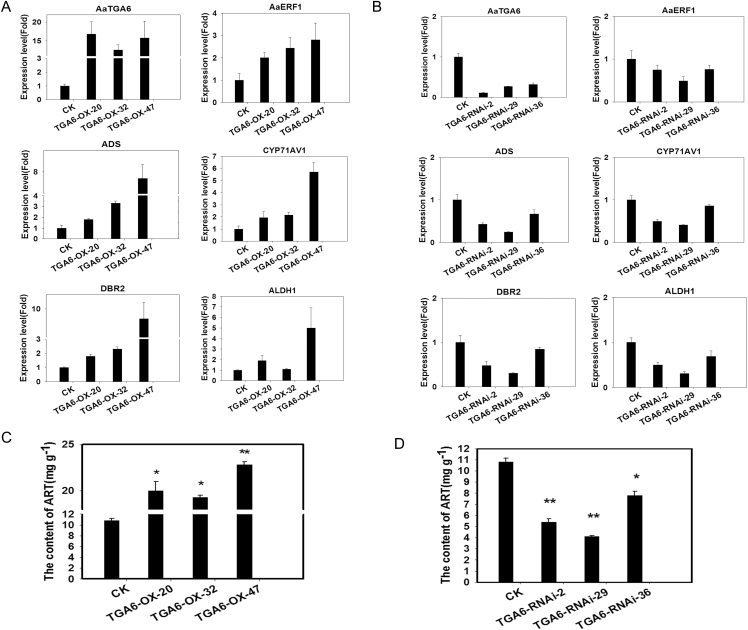
Three overexpression and three RNAi lines of transgenic plants were selected for further study and qPCR was employed to determine the expression levels of AaTGA6. The results showed that the relative mRNA levels o were increased by 11- to 15-fold in the overexpression plants and decreased by 70–90% in the RNAi plants (Fig. 7A, B). As key catalysis genes involved in the artemisinin biosynthetic pathway, the expression levels of ADS, CYP71AV1, DBR2, and ALDH1 were also evaluated and were found to be increased in the AaTGA6 overexpression lines by between 1- to 7-fold (Fig. 7A).The artemisinin content in the overexpression plants was increased by 90–120% compared with the wild-type (Fig. 7C). In the AaTGA6-RNAi plants, the expression levels of ADS, CYP71AV1, DBR2, and ALDH1were reduced by between 20–70% (Fig. 7B) and the artemisinin content was decreased by 20–60% compared with the wild-type (Fig. 7D).
Fig. 7.
AaTGA6 is a positive modulator of artemisinin biosynthesis. (A, B) Expression levels of AaTGA3, AaERF1, ADS, CYP71AV1, DBR2, and ALDH1 in (A) AaTGA6-overexpression (OX) and (B) AaTGA6-RNAi plants, as determined by qPCR analysis. β-ACTIN was used as the endogenous reference gene. Data are means (±SE) of n=3 biologically independent samples. (C, D) Artemisinin (ART) contents of (C) OX and (D) RNAi plants, as detected by HPLC. CK, wild-type control.
Discussion
AaTGA6 modulates artemisinin content by directly binding to the promoter of AaERF1
The AP2/ERF transcription factors are often targeted by TGA factors, such as TGA2/5/6, which directly bind to the cis-element ‘TGACG’ of the promoter of ORA59 (Zander et al., 2014). Previous studies have indicated that three AP2/ERF transcription factors, namely, AaERF1, AaERF2 and AaORA, play important roles in the regulation of artemisinin biosynthesis (Yu et al., 2012; Lu et al., 2013). Our promoter sequence analysis indicated that only the that of AaERF1 harbored the ‘TGACG’ cis-element. Since we found that AaTGA6 was an important modulator for the regulation of artemisinin content in A. annua (Fig. 7C), we first established its target gene by transcriptome sequencing. Six highly significantly different transcription factors were found in the transcriptome, including AaERF1 (Supplementary Fig. S6). In AaTGA6-RNAi transgenic plants, the levels of AaERF1 mRNA decreased, which coincided with decreased artemisinin content (Fig. 7D), so we postulated that AaERF1 was the direct target of AaTGA6, and Fig. 2 shows that AaTGA6 directly activated the promoter of AaERF1. Therefore, the results are consistent with previous studies showing that AP2/ERF transcription factors are direct targets of class II TGA factors.
N-AaNPR1 activates the promoter of AaERF1
Previous studies have indicated that NPR1 may act with a receptor for SA and play important roles in immune responses by interacting with NPR3/ NPR4 (Fu et al., 2012; Wu et al., 2012; Ding et al., 2018), and thus NPR1 plays an important role in SA signaling. There were five NPR genes in A. annua. NPR1 contains two domains: an ankyrin repeat domain and a BTB/POZ domain, which was first cloned by Cao et al. (1997). Ankyrin repeats play important roles in the interaction with TGA transcription factors, and the BTB/POZ domain is the enhancer for the transactivation function of the TGA2–NPR1 compound (Boyle et al., 2009). To investigate the functions of the different domains of AaNPR1 in A. annua, we analysed the activation of N-AaNPR1 (1–351 residues) and C-AaNPR1 (352–570 residues) using dual-LUC assays. N-AaNPR1 showed a high level of AaERF1 transactivation activity, while C-AaNPR1 had a low level of activity (Supplementary Fig. S8).
The N-terminus of AaNPR1 had a SKP1/BTB/POZ domain, which might be involved in the transactivation activity of AaERF1 (Sedgwick and Smerdon, 1999). Previous studies have indicated that the BTB/POZ domain may play a specific role in protein–protein interactions (Bardwell and Treisman, 1994). We assume that N-AaNPR1, which acted as a modulator of AaERF1 and enhanced its transactivation activity, may interact with other proteins. The reason for this may be that the BTB/POZ domain of N-AaNPR1 represses the C-terminal transactivation domain (Wu et al., 2012).
AaNPR1 increases the DNA-binding activity of AaTGA6
TGA factors act as regulatory components of the SA signaling pathway. The class II TGA transcription factors, such as TGA2, TGA5, and TGA6 in Arabidopsis, have been demonstrated to regulate the as-1 element of PR-1 (Zhang et al., 1999; Després et al., 2000). AP2/ERF transcription factors are potential targets of class II TGA transcription factors (Van der Does et al., 2013; Zander et al., 2014). We determined that AaTGA6 belongs to the class II TGA transcription factors (Fig. 1A) and can activate the promoter of AaERF1 (Fig. 4D). It has previously been shown that AaERF1 is an important modulator of artemisinin biosynthesis (Yu et al., 2012). We postulated that AaTGA6 may bind to the promoter of AaERF1 directly, and this was supported by the results of the Y1H assays and EMSAs (Fig 2D, E). Hence, AaTGA6 enhances artemisinin content by directly binding to the promoter of AaERF1.
Y2H and BiFC assays indicated that AaTGA6 appeared to interact with AaNPR1 (Fig 3A–C), which is consistent with previous results in Arabidopsis (Zhang et al., 1999; Zhou et al., 2000). However, it was unclear whether AaNPR1 enhanced the DNA-binding activity of AaTGA6. Dual-LUC assays and EMSAs were employed and clearly demonstrated that AaNPR1 was required for more efficient binding of AaTGA6 to the ‘TGACG’ element in the AaERF1 promoter (Fig. 4). These results are consistent with previous findings that NPR1 causes an increase in DNA-binding activity in Arabidopsis (Zhang et al., 1999; Després et al., 2000).
AaTGA3 plays a negative role in artemisinin biosynthesis
There are many kinds of highly concentrated secondary metabolites in glandular trichomes with pharmaceutical, pesticidal, flavor- and fragrance-related activities (Duke, 1994). Artemisinin is an important product of the glandular trichome of A. annua, and it has a phytotoxic effect on most plants, including itself (Duke, 1994). To avoid autotoxicity hazards, plants may limit the production of artemisinin by a putative feedback mechanism (Arsenault et al., 2010).
AaERF1 has previously been shown to be a positive regulator of artemisinin biosynthesis (Yu et al., 2012). We found that AaTGA6 activated the promoter of AaERF1, while AaTGA3 behaved in the opposite manner (Fig. 6B). The interaction of AaTGA3 with AaTGA6 (Fig. 5A–C) reflected the fact that heterodimerization between them occurs (Llorca et al., 2014). Thus, AaTGA3 inhibits the binding of AaTGA6 to its cognate promoter element ‘TGACG’ in the promoter of AaERF1 and participates in a negative feedback loop of artemisinin biosynthesis. This may be a mechanism by which A. annua protects itself from autotoxicity.
Mode of regulation of artemisinin by the AaNPR1-AaTGA6-AaTGA3 complex
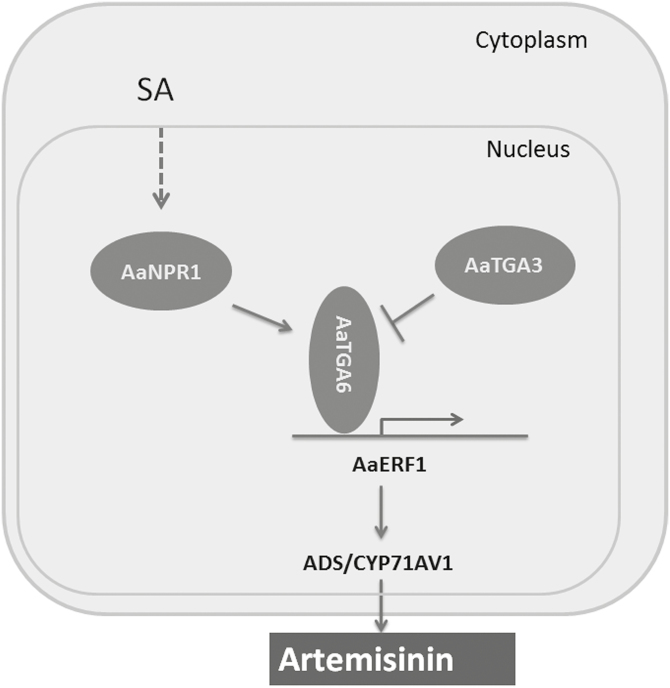
Our results showed that AaTGA6 can increase artemisinin content by binding to the promoter of AaERF1, which encodes a transcription factor that plays an important role in the modulation of artemisinin content by directly binding to the promoters of ADS and CYP71AV1 (Yu et al., 2012). In addition, we found that AaNPR1 interacts with AaTGA6 to enhance its DNA-binding activity. However, there may be feedback inhibition of artemisinin biosynthesis in A. annua, as we showed that another transcription factor, AaTGA3, suppresses the transcription activity of AaERF1 by interacting with AaTGA6. Thus, SA modulation of artemisinin content may be mediated by the AaNPR1-AaTGA6-AaTGA3 complex (Fig. 8).
Fig. 8.
Proposed molecular regulatory network for modulation of artemisinin biosynthesis by AaTGA6 in Artemisia annua. AaERF1 directly regulates artemisinin biosynthesis by activating transcription of artemisinin-related genes such as ADS (Yu et al., 2012). AaNPR1 interacts with AaTGA6 and activates transcription of artemisinin-related genes such as AaERF1. AaTGA3 repression of the binding of AaTGA6 to the promoter of AaERF1 may play a feedback-inhibition role in artemisinin biosynthesis. Arrows indicate positive regulation and blocked lines indicate negative regulation; the dashed line indicates an unidentified pathway.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. The artemisinin biosynthesis pathway in A. annua.
Fig. S2. Expression levels of ADS, CYP71AV1, DBR2, and ALDH1 in A. annua under SA treatment.
Fig. S3. Expression levels of AaTGA1–6 in different organs in A. annua.
Fig. S4. Subcelluar location and transient dual-LUC assays of AaTGA6 expressed in tobacco leaves.
Fig. S5. Promoter sequences of AaERF1, AaERF2, and AaORA.
Fig. S6. Heat map of expression of SA-related genes in A. annua.
Fig. S7. Results of EMSAs to test the binding of AaTGA6-His to mutant ‘TGACG’ motifs in the promoter of AaERF1.
Fig. S8. Dual-LUC assays showing that N-AaNPR1 can activate the promoter of AaERF1.
Fig. S9. Phenotypes of transgenic A. annua plants with either overexpression or knockdown of AaTGA6, compared with the wild-type.
Table S1. List of primers used in this study.
Acknowledgements
This work was funded by the China Transgenic Research Program (grant no. 2016ZX08002001-006), the National Science and Technology Major Project (2017ZX09101002-003-002), the Shanghai Leading Academic Discipline Project (Horticulture),and the State Key Laboratory of Subtropical Silviculture, Zhejiang A & F University (Grant no. 2018FR003, ZY20180206). The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Author contributions: ZL, KT, WC, and Le.Z conceived and designed the research; ZL, ZG, FZ, WJ, QS, XF, and TY performed most of the experiments; PS, LL, XH, YM, MC, and WC provided technical assistance; ZL, KT, WC, and Li.Z wrote the manuscript; and LL, KT, WC, and Le.Z helped with the organization and editing.
References
- Arsenault PR, Vail D, Wobbe KK, Erickson K, Weathers PJ. 2010. Reproductive development modulates gene expression and metabolite levels with possible feedback inhibition of artemisinin in Artemisia annua. Plant Physiology 154, 958–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell VJ, Treisman R. 1994. The POZ domain: a conserved protein–protein interaction motif. Genes & Development 8, 1664–1677. [DOI] [PubMed] [Google Scholar]
- Bouwmeester HJ, Wallaart TE, Janssen MH, van Loo B, Jansen BJ, Posthumus MA, Schmidt CO, De Kraker JW, König WA, Franssen MC. 1999. Amorpha-4,11-diene synthase catalyses the first probable step in artemisinin biosynthesis. Phytochemistry 52, 843–854. [DOI] [PubMed] [Google Scholar]
- Boyle P, Le Su E, Rochon A, Shearer HL, Murmu J, Chu JY, Fobert PR, Després C. 2009. The BTB/POZ domain of the Arabidopsis disease resistance protein NPR1 interacts with the repression domain of TGA2 to negate its function. The Plant Cell 21, 3700–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratzel F, López-Torrejón G, Koch M, Del Pozo JC, Calonje M. 2010. Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Current Biology 20, 1853–1859. [DOI] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X. 1997. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63. [DOI] [PubMed] [Google Scholar]
- Després C, Chubak C, Rochon A, Clark R, Bethune T, Desveaux D, Fobert PR. 2003. The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. The Plant Cell 15, 2181–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després C, DeLong C, Glaze S, Liu E, Fobert PR. 2000. The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. The Plant Cell 12, 279–290. [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Sun T, Ao K, Peng Y, Zhang Y, Li X, Zhang Y. 2018. Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell 173, 1454–1467.e15. [DOI] [PubMed] [Google Scholar]
- Duke MV, Paul RN, Elsohly HN, Sturtz G, Duke SO. 1994. Localization of artemisinin and artemisitene in foliar tissues of glanded and glandless biotypes of Artemisia annua L. International Journal of Plant Sciences 155, 365–372. [Google Scholar]
- Duke SO. 1994. Glandular trichomes-a focal point of chemical and structural interactions. International Journal of Plant Sciences 155, 617–620. [Google Scholar]
- Ehlert A, Weltmeier F, Wang X, Mayer CS, Smeekens S, Vicente-Carbajosa J, Dröge-Laser W. 2006. Two-hybrid protein–protein interaction analysis in Arabidopsis protoplasts: establishment of a heterodimerization map of group C and group S bZIP transcription factors. The Plant Journal 46, 890–900. [DOI] [PubMed] [Google Scholar]
- Fan W, Dong X. 2002. In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis. The Plant Cell 14, 1377–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster R, Izawa T, Chua NH. 1994. Plant bZIP proteins gather at ACGT elements. FASEB Journal 8, 192–200. [DOI] [PubMed] [Google Scholar]
- Fu ZQ, Yan S, Saleh A, et al. . 2012. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486, 228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH. 2007. Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nature Protocols 2, 38–41. [DOI] [PubMed] [Google Scholar]
- Kinkema M, Fan W, Dong X. 2000. Nuclear localization of NPR1 is required for activation of PR gene expression. The Plant Cell 12, 2339–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuai X, Desprã SC. 2016. Defining Arabidopsis NPR1 orthologues in crops for translational plant immunity. Canadian Journal of Plant Pathology 38, 25–30. [Google Scholar]
- Lam E, Lam YK. 1995. Binding site requirements and differential representation of TGF factors in nuclear ASF-1 activity. Nucleic Acids Research 23, 3778–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Casteels T, Frogne T, et al. . 2016. Artemisinins target GABAA receptor signaling and impair alpha cell identity. Cell 168, 86–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorca CM, Potschin M, Zentgraf U. 2014. bZIPs and WRKYs: two large transcription factor families executing two different functional strategies. Frontiers in Plant Science 5, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommen WJ, Schenk E, Bouwmeester HJ, Verstappen FW. 2006. Trichome dynamics and artemisinin accumulation during development and senescence of Artemisia annua leaves. Planta Medica 72, 336–345. [DOI] [PubMed] [Google Scholar]
- Lu X, Zhang L, Zhang F, Jiang W, Shen Q, Zhang L, Lv Z, Wang G, Tang K. 2013. AaORA, a trichome-specific AP2/ERF transcription factor of Artemisia annua, is a positive regulator in the artemisinin biosynthetic pathway and in disease resistance to Botrytis cinerea. New Phytologist 198, 1191–1202. [DOI] [PubMed] [Google Scholar]
- Lv Z, Zhang F, Pan Q, et al. . 2016. Branch pathway blocking in Artemisia annua is a useful method for obtaining high yield Artemisinin. Plant & Cell Physiology 57, 588–602. [DOI] [PubMed] [Google Scholar]
- Ma G, Park K, Chung W. 2014. The effects of artemisinin, dihydroartemisinin and artemisinin-glycolipid on non-small cell lung cancer induced bone destruction. Cancer Research 74, 2147. [Google Scholar]
- Ma YN, Xu DB, Li L, et al. . 2018. Jasmonate promotes artemisinin biosynthesis by activating the TCP14-ORA complex in Artemisia annua. Science Advances 4, eaas9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercke P, Bengtsson M, Bouwmeester HJ, Posthumus MA, Brodelius PE. 2000. Molecular cloning, expression, and characterization of amorpha-4,11-diene synthase, a key enzyme of artemisinin biosynthesis in Artemisia annua L. Archives of Biochemistry and Biophysics 381, 173–180. [DOI] [PubMed] [Google Scholar]
- Mukhtarv MS, Nishimura MT, Danglv J. 2009. NPR1 in plant defense: it’s not over ’til it’s turned over. Cell 137, 804–806. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. 1962. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia Plantarum 15, 473–497. [Google Scholar]
- Nair P, Misra A, Singh A, Shukla AK, Gupta MM, Gupta AK, Gupta V, Khanuja SP, Shasany AK. 2013. Differentially expressed genes during contrasting growth stages of Artemisia annua for artemisinin content. PloS ONE 8, e60375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien KL, Chandran A, Weatherholtz R, et al. . 2015. Efficacy of motavizumab for the prevention of respiratory syncytial virus disease in healthy Native American infants: a phase 3 randomised double-blind placebo-controlled trial. The Lancet Infectious Diseases 15, 1398–1408. [DOI] [PubMed] [Google Scholar]
- Okada A, Okada K, Miyamoto K, Koga J, Shibuya N, Nojiri H, Yamane H. 2009. OsTGAP1, a bZIP transcription factor, coordinately regulates the inductive production of diterpenoid phytoalexins in rice. The Journal of Biological Chemistry 284, 26510–26518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu GB, Ma DM, Chen JL, Ma LQ, Wang H, Li GF, Ye HC, Liu BY. 2009. Salicylic acid activates artemisinin biosynthesis in Artemisia annua L. Plant Cell Reports 28, 1127–1135. [DOI] [PubMed] [Google Scholar]
- Raskin I, Skubatz H, Tang W, Meeuse BJ. 1990. Salicylic acid levels in thermogenic and non-thermogenic plants. Annals of Botany 66, 369–373. [Google Scholar]
- Rochon A, Boyle P, Wignes T, Fobert PR, Després C. 2006. The coactivator function of Arabidopsis NPR1 requires the core of its BTB/POZ domain and the oxidation of C-terminal cysteines. The Plant Cell 18, 3670–3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick SG, Smerdon SJ.. 1999. The ankyrin repeat: a diversity of interactions on a common structural framework. Trends in Biochemical Sciences 24, 311–316. [DOI] [PubMed] [Google Scholar]
- Shen Q, Lu X, Yan T, Fu X, Lv Z, Zhang F, Pan Q, Wang G, Sun X, Tang K. 2016. The jasmonate-responsive AaMYC2 transcription factor positively regulates artemisinin biosynthesis in Artemisia annua. New Phytologist 210, 1269–1281. [DOI] [PubMed] [Google Scholar]
- Shi P, Fu X, Shen Q, et al. . 2018. The roles of AaMIXTA1 in regulating the initiation of glandular trichomes and cuticle biosynthesis in Artemisia annua. New Phytologist 217, 261–276. [DOI] [PubMed] [Google Scholar]
- Spoel SH, Mou Z, Tada Y, Spivey NW, Genschik P, Dong X. 2009. Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 137, 860–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H, Xiao L, Gao S, Li Q, Chen J, Xiao Y, Ji Q, Chen R, Chen W, Zhang L. 2015. TRICHOME AND ARTEMISININ REGULATOR 1 is required for trichome development and artemisinin biosynthesis in Artemisia annua. Molecular Plant 8, 1396–1411. [DOI] [PubMed] [Google Scholar]
- Tang K, Shen Q, Yan T, Fu X. 2014. Transgenic approach to increase artemisinin content in Artemisia annua L. Plant Cell Reports 33, 605–615. [DOI] [PubMed] [Google Scholar]
- Teoh KH, Polichuk DR, Reed DW, Covello PS. 2009. Molecular cloning of an aldehyde dehydrogenase implicated in artemisinin biosynthesis in Artemisia annua. Botany 87, 635–642. [Google Scholar]
- Teoh KH, Polichuk DR, Reed DW, Nowak G, Covello PS. 2006. Artemisia annua L. (Asteraceae) trichome-specific cDNAs reveal CYP71AV1, a cytochrome P450 with a key role in the biosynthesis of the antimalarial sesquiterpene lactone artemisinin. FEBS Letters 580, 1411–1416. [DOI] [PubMed] [Google Scholar]
- Van der Does D, Leon-Reyes A, Koornneef A, et al. . 2013. Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. The Plant Cell 25, 744–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall PJ, Pitera DJ, Lenihan JR, Eng D, Woolard FX, Regentin R, Horning T, Tsuruta H, Melis DJ, Owens A. 2012. Production of amorphadiene in yeast, and its conversion to dihydroartemisinic acid, precursor to the antimalarial agent artemisinin. Proceedings of the National Academy of Sciences, USA 109, E111–E118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. 2016. World malaria report. Geneva: World Health Organization. [Google Scholar]
- Wu Y, Zhang D, Chu JY, Boyle P, Wang Y, Brindle ID, De Luca V, Després C. 2012. The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Reports 1, 639–647. [DOI] [PubMed] [Google Scholar]
- Xiang C, Miao Z, Lam E. 1997. DNA-binding properties, genomic organization and expression pattern of TGA6, a new member of the TGA family of bZIP transcription factors in Arabidopsis thaliana. Plant Molecular Biology 34, 403–415. [DOI] [PubMed] [Google Scholar]
- Xu ZY, Kim SY, Hyeon do Y, et al. . 2013. The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. The Plant Cell 25, 4708–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu ZX, Li JX, Yang CQ, Hu WL, Wang LJ, Chen XY. 2012. The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Molecular Plant 5, 353–365. [DOI] [PubMed] [Google Scholar]
- Zander M, La Camera S, Lamotte O, Métraux JP, Gatz C. 2010. Arabidopsis thaliana class-II TGA transcription factors are essential activators of jasmonic acid/ethylene-induced defense responses. The Plant Journal 61, 200–210. [DOI] [PubMed] [Google Scholar]
- Zander M, Thurow C, Gatz C. 2014. TGA transcription factors activate the salicylic acid-suppressible branch of the ethylene-induced defense program by regulating ORA59 expression. Plant Physiology 165, 1671–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Fu X, Lv Z, et al. . 2015. A basic leucine zipper transcription factor, AabZIP1, connects abscisic acid signaling with artemisinin biosynthesis in Artemisia annua. Molecular Plant 8, 163–175. [DOI] [PubMed] [Google Scholar]
- Zhang L, Jing F, Li F, Li M, Wang Y, Wang G, Sun X, Tang K. 2009. Development of transgenic Artemisia annua (Chinese wormwood) plants with an enhanced content of artemisinin, an effective anti-malarial drug, by hairpin-RNA-mediated gene silencing. Biotechnology and Applied Biochemistry 52, 199–207. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Fan W, Kinkema M, Li X, Dong X. 1999. Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proceedings of the National Academy of Sciences, USA 96, 6523–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Teoh KH, Reed DW, Maes L, Goossens A, Olson DJ, Ross AR, Covello PS. 2008. The molecular cloning of artemisinic aldehyde Δ11(13) reductase and its role in glandular trichome-dependent biosynthesis of artemisinin in Artemisia annua. The Journal of Biological Chemistry 283, 21501–21508. [DOI] [PubMed] [Google Scholar]
- Zheng H, Colvin CJ, Johnson BK, Kirchhoff PD, Wilson M, Jorgensen-Muga K, Larsen SD, Abramovitch RB. 2017. Inhibitors of Mycobacterium tuberculosis DosRST signaling and persistence. Nature Chemical Biology 13, 218–225. [DOI] [PubMed] [Google Scholar]
- Zhou JM, Trifa Y, Silva H, Pontier D, Lam E, Shah J, Klessig DF. 2000. NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Molecular Plant-Microbe Interactions 13, 191–202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.