Abstract
Starch and storage proteins, the primary storage substances of cereal endosperm, are a major source of food for humans. However, the transcriptional regulatory networks of the synthesis and accumulation of storage substances remain largely unknown. Here, we identified a rice endosperm-specific gene, NF-YC12, that encodes a putative nuclear factor-Y transcription factor subunit C. NF-YC12 is expressed in the aleurone layer and starchy endosperm during grain development. Knockout of NF-YC12 significantly decreased grain weight as well as altering starch and protein accumulation and starch granule formation. RNA-sequencing analysis revealed that in the nf-yc12 mutant genes related to starch biosynthesis and the metabolism of energy reserves were enriched in the down-regulated category. In addition, starch and protein contents in seeds differed between NF-YC12-overexpression lines and the wild-type. NF-YC12 was found to interact with NF-YB1. ChIP-qPCR and yeast one-hybrid assays showed that NF-YC12 regulated the rice sucrose transporter OsSUT1 in coordination with NF-YB1 in the aleurone layer. In addition, NF-YC12 was directly bound to the promoters of FLO6 (FLOURY ENDOSPERM6) and OsGS1;3 (glutamine synthetase1) in developing endosperm. This study demonstrates a transcriptional regulatory network involving NF-YC12, which coordinates multiple pathways to regulate endosperm development and the accumulation of storage substances in rice seeds.
Keywords: Grain filling, NF-Y factor, rice (Oryza sativa), seed specific expression, storage substance accumulation, transcriptional regulation
NF-YC12, which encodes a rice nuclear factor-Y transcription factor subunit C, plays a key regulatory role in the accumulation of storage substances during grain filling in rice.
Introduction
Rice is the foremost food source for nearly half of the global population (Zuo and Li, 2014). The rice endosperm, which is the main storage tissue, is the triploid product of the fertilization of two polar nuclei in the central cell of the embryo sac with one sperm cell nucleus (Sabelli and Larkins, 2009). A fully developed endosperm, which occupies most of the seed space, has accumulated large amounts of storage substances. The endosperm consists of an outer aleurone layer (AL) inside which is the starchy endosperm (SE) (Becraft, 2001; Krishnan and Dayanandan, 2003; Zuo and Li, 2014; Ishimaru et al., 2015; Wu et al., 2016). Proteins, lipids, and minerals are accumulated in the AL, whereas starch and small amounts of storage proteins are mainly stored in the SE (Olsen, 2004; Zheng and Wang, 2014). Among these primary nutrient components, starch and storage proteins account for ~90% and 5–8% of the dry weight of rice seeds, respectively, and 60–80% and 20–30% of the proteins belong to the glutelins and prolamins, respectively (Zhou et al., 2013).
Starch is synthesized from sucrose and is rapidly stored in specialized amyloplasts via a complex process involving multiple subunits or isoforms of five major enzymes, namely ADP-glucose pyrophosphorylase (AGPase), soluble starch synthases (SSs), starch-branching enzymes (BEs), debranching enzymes (DBEs), and granule-bound starch synthase (GBSS) (Hannah and James, 2008; Sabelli and Larkins, 2009; Nakamura, 2018). In rice, mutants defective in some of these key enzymes show abnormal starch synthesis, resulting in floury or chalky phenotypes of the endosperm. Loss of function of SSs causes chalky endosperm, in which starch granules are irregularly shaped and loosely packed (Hirose and Terao, 2004; Ryoo et al., 2007; Zhang et al., 2011). Mutations in AGPase cause shrunken endosperms and reduced starch content (Lee et al., 2007; Tang et al., 2016; Wei et al., 2017). Glutelins, the predominant storage proteins in rice, are encoded by a multigene family consisting of GluA, GluB, GluC, and GluD subfamilies (Okita et al., 1989; Kawakatsu et al., 2008). Prolamins are encoded by 34 genes in rice (Xu and Messing, 2009). Suppressed expression of several storage protein genes can change the seed weight, starch content, and protein accumulation in rice (Kawakatsu et al., 2010).
In addition to biosynthesis enzymes, other factors indirectly related to starch synthesis and storage protein accumulation during endosperm development have also been identified. For example, FLOURY ENDOSPERM2 (FLO2), which encodes a protein with a tetratricopeptide repeat (TPR) motif, can regulate starch synthesis. The flo2 mutation results in decreases in grain weight and in accumulation of storage substances (She et al., 2010). FLO6, a protein containing the C-terminal carbohydrate-binding module 48 (CBM48) domain, modulates starch synthesis and starch granule formation (Peng et al., 2014). FLO7 is required for starch synthesis and amyloplast development within the peripheral endosperm in rice (Zhang et al., 2016). The basic leucine zipper factor RISBZ1 and the rice prolamin box binding factor (RPBF) are seed-specific transcription factors, and suppression of their expression results in a significant reduction of storage protein accumulation in seeds (Yamamoto et al., 2006; Kawakatsu et al., 2009). In addition, RISBZ1/OsbZIP58 has been shown to directly bind to the promoters of six genes related to starch synthesis, namely OsAGPL3, Wx, OsSSIIa, SBE1, OsBEIIb, and ISA2, and to regulate starch biosynthesis in rice seeds (Wang et al., 2013). However, the synthesis and accumulation of seed storage substances are quite complex, and the related transcriptional regulatory networks remain largely unknown.
Nuclear factor-Y (NF-Y), also known as Heme activator protein (HAP) or CCAAT-binding factor (CBF), is a class of transcription factors that bind to the CCAAT box in eukaryote promoter regions. NF-Y is composed of three subunits: NF-YA (CBF-B or HAP2), NF-YB (CBF-A or HAP3), and NF-YC (CBF-C or HAP5) (Laloum et al., 2013). NF-YB can interact with NF-YC, forming a tight heterodimer via their conserved histone fold motifs (HFMs) in the cytoplasm. This heterodimer is then translocated to the nucleus, where it interacts with NF-YA to form a mature NF-Y complex (Mantovani, 1999; Petroni et al., 2012; Laloum et al., 2013). In mammals and yeast, there is a single gene for each NF-Y subunit, while in plants each subunit is encoded by multiple genes belonging to a family (Siefers et al., 2009; Petroni et al., 2012). Genome-wide analysis in rice has resulted in the identification of 11 NF-YA, 11 NF-YB, and 12 NF-YC genes (Li et al., 2016; Yang et al., 2017).
The NF-Y subunits play important roles in multiple plant developmental processes. Arabidopsis NF-YB9 (LEC1, LEAFY COTYLEDON1) and its homolog NF-YB6 (L1L, LEC1-like) are required for embryo development (Kwong et al., 2003; Lee et al., 2003). In rice, NF-YB2 and its close homologs NF-YB3 and NF-YB4 control chloroplast biogenesis (Miyoshi et al., 2003). OsNF-YB7/OsHAP3E function in both vegetative and reproductive development (Ito et al., 2011; Zhang and Xue, 2013). NF-YB11 (also called DTH8/Ghd8/LHD1) is involved in the control of photoperiodic flowering time, plant height, and yield (Yan et al., 2011). NF-YC2 and NF-YC4 regulate the photoperiodic flowering response via their effects on three flowering-time genes in rice. Repression of OsNF-YC2 and OsNF-YC4 leads to earlier flowering under long-day conditions (Kim et al., 2016b). Rice endosperm-specific NF-YB1 has been identified to regulate cell proliferation and sucrose loading during endosperm development (Sun et al., 2014; Bai et al., 2016). In addition, NF-YB1 can form a protein complex with NF-YC members and the ERF transcription factor OsERF115 to regulate the transcription of downstream genes and hence to mediate grain filling and endosperm development (Xu et al., 2016). In a recent publication, it was demonstrated that NF-YB1 plays an important role in the transcription of Wx by forming a heteotrimer complex with NF-YC12 and bHLH144 (Bello et al., 2019).
In our previous work, we identified five rice endosperm-specific NF-Y genes, namely NF-YA8, NF-YB1, NF-YB9, NF-YC10, and NF-YC12 (Nie et al., 2013; Yang et al., 2017). NF-YC10 encodes a seed-specific NF-YC transcription factor that regulates grain size by influencing the cell proliferation (Jia et al., 2019). In recent years, NF-YB1 has been thoroughly studied in relation to various aspects of rice endosperm development and grain filling (Sun et al., 2014; Bai et al., 2016; Xu et al., 2016; Bello et al., 2019; E et al., 2018). However, it remains unclear whether other NF-Y members also function in endosperm development and how they synergistically regulate the grain-filling process. In this study, we demonstrate that NF-YC12 is involved in regulating endosperm development and the accumulation of storage substances in rice. Consistent with the phenotype, NF-YC12 is preferentially expressed in the AL and SE of the seed. RNA-seq and genome-wide identification of NF-YC12 binding targets reveal a regulatory network involving multiple pathways for starch and storage protein accumulation. The interaction between NF-YC12 and NF-YB1 is confirmed, and NF-YC12 can regulate endosperm development and the accumulation of storage substances with or without NF-YB1. This study reveals a universal regulatory role of NF-YC12 in rice endosperm development.
Materials and methods
Generation of constructs and transgenic plants
For generation of rice (Oryza sativa) nf-yc12 mutants, specific single-guide RNA (sgRNA) targeting the NF-YC12 gene was designed using the web-based tool CRISPR-P (http://crispr.hzau.edu.cn/CRISPR/) and assembled into the binary expression vector pCXUN-Cas9 (sgRNA was driven by the OsU3 promoter), as previously described (He et al., 2017). To generate the NF-YC12-overexpressing plants, a 1005-bp cDNA fragment without a stop codon and encoding full-length NF-YC12 was amplified, and then cloned into the KpnI and BamHI sites of the binary expression vector pCAMBIA2301-FLAG (driven by a maize ubiquitin promoter). The recombinant constructs were transferred into calli of the cultivar ‘zhonghua11’ (ZH11; O. sativa ssp. japonica) by Agrobacterium tumefaciens-mediated transformation using the strain EHA105 (Lin and Zhang, 2005). The transgenic lines were grown in paddy fields in Huazhong Agricultural University, Wuhan, China, during the normal rice growing seasons. The phenotypes were detected in the homozygous T2 generation of the transgenic plants. The sequences of the primers are listed in Supplementary Table S1 at JXB online.
RNA isolation and quantitative RT-PCR analysis
Total RNA was extracted using TRIzol reagent (Invitrogen) and reverse-transcribed using SuperScript III reverse transcriptase (Invitrogen) to obtain cDNA according to the manufacturer’s instructions. Gene expression levels were measured by quantitative real-time PCR (qRT-PCR) using the Ubiquitin gene (LOC_Os03g13170) as the internal control. Relevant primer sequences are listed in Supplementary Table S1. qRT-PCR was performed on a CFX96 Real-time system (Bio-Rad). Changes in gene expression were calculated using the 2-ΔΔCT method. Three technical replicates were performed for each sample.
mRNA in situ hybridization
Fresh tissues from ZH11 were collected and fixed in FAA solution (50 ml ethanol, 5 ml acetic acid, 10 ml 37% formaldehyde, and 35 ml DEPC-H2O) for 24 h at 4 °C, and the solution was then replaced with 70% ethanol twice. The samples were then dehydrated with an ethanol series, infiltrated by xylene from 50% to 100%, embedded in paraffin (Sigma-Aldrich), and sectioned to a thickness of 8–10 μm with a microtome (Leica RM2145). The sections were mounted on RNase-free glass slides. The 138-bp specific 3´-region of NF-YC12 FL-cDNA was amplified by PCR (primer sequences are listed in Supplementary Table S1), and subcloned into the pGM-T vector (TaKaRa). Sense and antisense RNA probes were synthesized using SP6 and T7 RNA polymerase, respectively, with digoxigenin-UTP as a label. RNA hybridization and immunologic detection of the hybridized probes were performed on sections as described previously (Wang et al., 2015). Slides were observed and photographed using a BX53 microscope (Olympus).
Yeast two-hybrid and one-hybrid analysis
The coding sequences of NF-YA8, NF-YB1, NF-YC10, and NF-YC12 were amplified by PCR and subcloned into either the pGADT7 or pGBKT7 vector (Clontech). The prey and bait plasmids were verified by sequencing and subsequently transformed into yeast strain AH109. pGADT7-T was co-transformed with pGBKT7-53 as a positive control. The yeast cells were grown on SD lacking Leu and Trp (DDO) selection media at 30 °C for 3 d. Interactions were tested using SD/–Leu/–Trp/–His/–Ade (QDO) medium. QDO with X-α-Gal was used to detect the α-galactosidase activity of the yeast strains. Images were taken 5 d after the incubation.
In the yeast one-hybrid analysis, DNA fragments corresponding to the promoters of target genes were independently inserted into the pHIS2.0 plasmid (Clontech). NF-YC12 was fused to GAL4 transcriptional activation domain (AD). These constructs were transformed into the yeast strain AH109. A one-hybrid assay was performed following the manufacturer’s instructions (Clontech). Primers used for cloning are listed in Supplementary Table S1.
In vitro pull-down assays
For glutathione S-transferase (GST)-tagged NF-YB1 protein expression, pGEX4T-1-NF-YB1 was constructed and expressed in the Escherichia coli BL21 strain (primers are listed in Supplementary Table S1). For His-tagged NF-YC12 protein expression, pET28a-NF-YC12 was constructed and expressed in the E. coli BL21 strain. For GST pull-down assays, GST or GST-NF-YB1 and His-NF-YC12 recombinant proteins were incubated in binding buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 0.6% Triton X-100) for 2 h at 4 °C. GST resin was added and incubated for 1 h. After washing five times with GST pull-down buffer, bound proteins were eluted in loading buffer. The proteins were separated on a 10% SDS-PAGE gel and immunoblotted with anti-GST and anti-His antibodies (Abcam).
BiFC and subcellular localization analysis
For bimolecular fluorescence complementation (BiFC) assays, the full-length coding sequence of NF-YB1 was cloned in-frame into the pSAT4-cCFP-N (pE3451) vector (Lee et al., 2008). The coding sequences of NF-YC12 were cloned in-frame into the pSAT6-nCerulean-N (pE3248) vector (Lee et al., 2008). For the observation of subcellular localization, the transient expression vectors 35S::NF-YC12-YFP and 35S::NF-YB1-GFP were constructed. 35S::Ghd7-CFP was used as a nuclear localization marker (Xue et al., 2008). Pairs of the constructed vector plasmids were used to co-transform rice protoplast cells. Fluorescence signals were detected with a confocal laser-scanning microscope (Leica SP8). The channel specifications followed those of Lee et al., (2008) and Wang et al. (2014).
Microscopy
SEM was performed as described previously (Zhou et al., 2017). The brown rice seeds of the wild-type and the nf-yc12 mutant were cut transversely with a knife and coated with gold under vacuum conditions. Samples were observed using a SEM (JSM-6390LV), and the analysis was based on at least three biological replicates. Observation of the ultrastructure of developing seeds at 7–14 d after pollination (DAP) was performed as previously described (Wang et al., 2010) using a TEM (Hitachi H7650).
Analysis of grain quality and physicochemical properties of seeds
Harvested rice grains were air-dried and stored at room temperature for at least 3 months before testing. Fully filled grains were used for measuring grain size (length and width) and 1000-grain weight as previously described (Li et al., 2014). The percentage of grains with chalkiness (PGWC) was determined as previously described (Xu et al., 2016). Dehulled rice grains were ground to powder and samples of 50 mg, 10 mg, and 0.5 g powder were used to measure the total starch, amylose, and protein contents, respectively. Total starch content and the glutelin and prolamin contents were measured as previously described by Wang et al. (2015), and the amylose content was determined as described by Li et al. (2014). The gelatinization properties of the endosperm starch were examined by measuring the solubility in urea solutions according to previous studies (Nishi et al., 2001; Peng et al., 2014).
RNA-seq and GO analysis
Total RNAs were extracted from the endosperm of nf-yc12 and the wild-type at 7 DAP using Trizol. For each library, three independent replicated RNA samples were prepared. The RNA samples were sequenced on an Illumina HiSeq 2000 system. The raw reads were filtered for adaptors and low-quality reads. Clean reads were mapped to the reference genome of rice (RGAP v. 7.0) using HISAT2 (v.2.0.5) (Kim et al., 2015). The gene expression levels were calculated using the FPKM method (expected number of fragments per kilobase of transcript sequence per million base pairs sequenced) (Trapnell et al., 2010). DESeq in R was used to select the differentially expressed genes (DEGs). DEGs were defined as those with an expression change >1.5-fold and corrected P-value <0.05. Gene ontology (GO) analysis was performed using the GOseq software (Young et al., 2010) to identify the pathways enriched among the DEGs.
ChIP-seq and data analysis
Transgenic lines expressing pUbi::NF-YC12-FLAG were used for ChIP-seq analysis. Expression of the transformed target protein was verified by western blot analysis using anti-FLAG M2 monoclonal antibodies (Sigma, F3165; 1:2000 dilution). ChIP assays were performed as described previously (Bowler et al., 2004) with some modifications. Briefly, endosperm at 7 DAP was harvested and immediately crosslinked in 1% formaldehyde under vacuum for 30 min, and 3 g of tissues for each sample was used for chromatin isolation. Chromatin was fragmented to 200–500 bp by sonication. For ChIP-seq, the DNA was immunoprecipitated by anti-FLAG® M2 magnetic beads (Sigma, M8823) according to the manufacturer’s instructions, and the precipitated DNA was purified and dissolved in distilled water. The immunoprecipitated DNA and input DNA were then subjected to sequencing using the Illumina HiSeq 2000 platform.
ChIP-seq reads were aligned to the rice reference genome (RGAP v. 7.0) using BWA (Li and Durbin, 2009). Only uniquely mapped reads were used for peak identification. MACS2 (Zhang et al., 2008) was used for peak calling. Peaks were identified as significantly enriched (corrected P-value <0.05) in the IP libraries compared with input DNA. NF-YC12-bound genes were defined when peaks appeared on their genic or promoter region (including 2 kb upstream of the TTS). Motif enrichment analysis was performed using DREME (Bailey, 2011) with default parameters.
ChIP-quantitative PCR
To detect the specific DNA targets, the precipitated DNA and input DNA were applied for qPCR analysis (specific primers are listed in Supplementary Table S1). ChIP assays were carried out with two biological replicates with each including three technical replicates, and the enrichment values were normalized to the input sample. The significance of differences was estimated using Student’s t-test.
Transient transcription dual-luciferase (LUC) assays
Dual-LUC assays using rice protoplasts were performed as described previously (Zong et al., 2016). The luciferase activity of the transformed protoplasts was analysed with a luminometer (Promega) using commercial LUC reaction reagents according to the manufacturer’s instructions (Promega). Three independent experiments were performed at different times (three biological replicates). For the effectors used in this study, the full-length CDS of NF-YB1 or NF-YC12 was fused into a ‘none’ vector. For the reporters, the promoters of NF-YC12-potential targets were cloned into 190-LUC as previously described (Zong et al., 2016). The primers used are listed in Supplementary Table S1. The relative luciferase activity was calculated as the ratio between fLUC and rLUC.
Statistical analysis
Significant differences between individual means were established using a two-tailed Student’s t-test in Microsoft Office Excel 2010.
Accession numbers
Genes from this paper can be found in the RAP-DB databases under the following accession numbers: NF-YB1 (Os02g0725900), NF-YC10 (Os01g0346900), NF-YC12 (Os10g0191900), FLO6 (Os03g0686900), OsSUT1 (Os03g0170900), OsGS1;3 (Os03g0712800), OsMST4 (Os03g0218400), OsMST6 (Os07g0559700), OsHXK7 (Os05g0187100), OsAGPS1 (Os09g0298200), OsAGPS2 (Os08g0345800), OsAGPL2 (Os03g0735000), OsAGPL3 (Os03g0735000), Wx (Os06g0133000), OsSSIIa (Os06g0229800), OsSSIIb (Os02g0744700), OsSSIIIa (Os08g019 1433), OsSSIIIb (Os04g0624600), GluA3 (Os03g0427300), GluB1 (Os02g0249800), GluB4 (Os02g0242600), GluD1 (Os02g0249000). The RNA-seq and ChIP-seq data are deposited in the NCBI Gene Expression Omnibus with accession codes GSE119576 and GSE119575.
Results
NF-YC12 interacts with NF-YB1 in vitro and in vivo
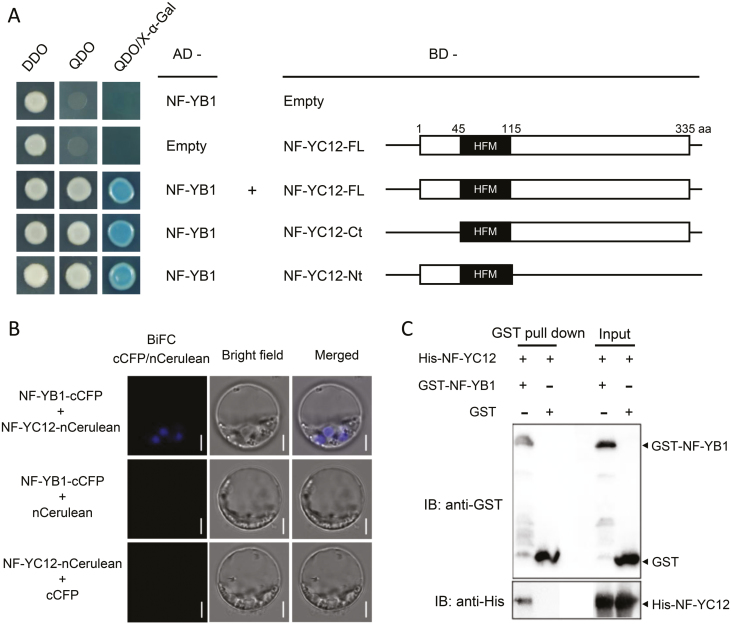
It has been reported that NF-YB can interact with NF-YA or NF-YC subunits (Petroni et al., 2012; Xing et al., 2015). NF-YA8, NF-YC10, and NF-YC12 were selected to identify the endosperm-specific NF-Y proteins interacting with NF-YB1 in yeast. The results confirmed the interaction of NF-YC12 with NF-YB1 (Fig. 1A), while NF-YA8 and NF-YC10 did not interact with NF-YB1 (Supplementary Fig. S1). Two deletion constructs of NF-YC12 were then used to map the region required for the interaction. As shown in Fig. 1A, NF-YC12-Ct (without N-terminus) and NF-YC12-Nt (without C-terminus), both of which contained a conserved HFM domain, interacted with NF-YB1, indicating that NF-YC12 can interact with NF-YB1 through its HFM domain.
Fig. 1.
Interaction between rice NF-YB1 and NF-YC12. (A) Yeast two-hybrid assay. The full-length and truncated NF-YC12 cDNAs were cloned into a vector bearing the DNA binding domain (BD), and the full length cDNAs of NF-YB1 were cloned into a vector bearing an activation domain (AD). The transformants were grown on DDO (SD/–Leu/–Trp), QDO (SD/–Leu/–Trp/–His/–Ade), and QDO with X-α-Gal plates. (B) BiFC assays of NF-YC12 and NF-YB1. NF-YB1-cCFP and NF-YC12-nCerulean interacted to form a functional CFP in rice protoplast cells. Scale bars are 5 μm. (C). Pull-down assays Showing that there was a direct interaction between GST-NF-YB1 and His-NF-YC12 in vitro. IB, immunoblotting.
We next performed BiFC analysis to examine the interaction between NF-YC12 and NF-YB1 in rice protoplasts. Blue fluorescence generated from the interaction between NF-YC12-nCerulean and NF-YB1-cCFP in the same cell was detected (Fig. 1B). In contrast, no fluorescence signal was produced from the co-expression of NF-YB1-cCFP and empty nCerulean or empty cCFP and NF-YC12-nCerulean. We then examined subcellular localization. The transient expression vectors 35S::NF-YC12-YFP and 35S::NF-YB1-GFP were each co-transformed into rice protoplasts with another transient expression vector, 35S:Ghd7-CFP. Ghd7 was used as a marker of nucleus localization (Xue et al., 2008). The fluorescent signals showed that both the GFP-tagged NF-YB1 and YFP-tagged NF-YC12 proteins were localized in the nucleus and cytoplasm (Supplementary Fig. S2A, B). Co-localization of NF-YC12 and NF-YB1 and their overlapping signals that occurred predominantly in the nucleus (Supplementary Fig. S2C) indicated that they could form a heterodimer in the nucleus.
To further confirm the direct interaction of NF-YC12 with NF-YB1, a pull-down assay was carried out. NF-YB1 was fused to a GST tag, which was then incubated with His-tagged NF-YC12, with GST used as a negative control. After the pull-down assay, the NF-YC12 protein was detected by His-tag antibodies in the sample containing GST-NF-YB1, but not in the control (Fig. 1C). These results confirmed the interaction between NF-YC12 and NF-YB1 in vitro.
Functional loss of NF-YC12 reduces grain weight and causes chalky endosperm
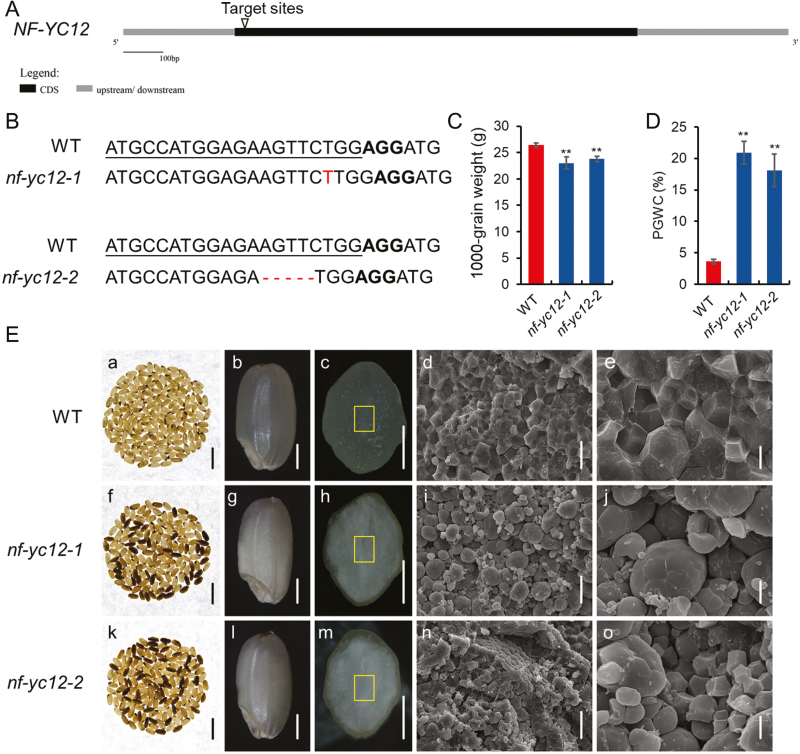
To investigate the biological roles of NF-YC12 in rice endosperm development, the CRISPR/Cas9 genome editing system was used to specifically knockout NF-YC12 in the Zhonghua11 (ZH11, japonica) background. The sgRNA target site was designed at the exon of the NF-YC12 gene (86–105 bp from the ATG codon) using the web-based tool CRISPR-P, and this was expected to generate a mutation in the coding region of the gene (Fig. 2A), thereby ensuring the generation of a loss-of-function mutant. After introduction of the construct into rice embryogenic calli by Agrobacterium-mediated transformation, 32 independent T0 transgenic plants were regenerated. We then examined the mutation efficiency by PCR with the CRISPR/Cas9 constructs. A very high mutagenesis rate of 71.9% was observed for the T0 transformants (Supplementary Table S2). Six T0 homozygous plants were found by decoding the sequencing chromatograms. Sequencing of the mutated region revealed that various mutations had been obtained, including insertion and deletion. To test for possible off-target effects, we identified the locus with the highest probability based on the target site used in this study. No off-target mutations were found by sequencing in T0 plants (Supplementary Table S3). The six T0 homozygous mutant lines and the wild-type (WT) controls were grown in the field and the T2 plants were investigated.
Fig. 2.
Rice nf-yc12 knockout mutants generated by CRISPR/Cas9 and their phenotypes. (A) Schematic diagram of the genomic region of NF-YC12 and the sgRNA target site. (B) Mutation sites in nf-yc12-1 and nf-yc12-2, as compared with wild-type (WT) sequences. The target sites are underlined, protospacer-adjacent motif sequences are shown in bold, and inserted or deleted nucleotides are indicated in red. (C) Thousand-grain weights and (D) percentage of grains with chalkiness (PGWC) of WT and nf-yc12 seeds. Data are means (±SD) from three replicates, each of which included at least 200 seeds. Significant differences between the WT and the mutants were determined using Student’s t-test (**P<0.01). (E) Phenotypes of seeds of the WT and nf-yc12 mutants. (a, f, k) Images of 200 grains of mature seeds; scale bars are 10 mm. (b, g, l) Appearance of mature seeds; scale bars are 1 mm. (c, h, m) Cross-sections of mature seeds; scale bars are 1 mm. (d, e, i, j, n, o) SEM images of the central area of mature endosperm at different magnifications: the areas are indicated by the squares in (c, h, m). Scale bars are 20 μm (d, i, n), 5 μm (e, j, o).
Sequencing of PCR-amplified NF-YC12 genomic DNA from transgenic T2 homozygous plants resulted in the identification of two knockout mutants: nf-yc12-1 (with a 1-bp insertion) and nf-yc12-2 (with a 5-bp deletion) (Fig. 2B, Supplementary Fig. S3A, C), which were predicted to produce truncated polypeptides with only 73 and 71 amino acids, respectively. Both mutations resulted in premature stop codons, and produced mutant proteins (Supplementary Fig. S3B).
No visible defects were found during the vegetative stage, including plant height, flowering time, and panicle number per plant, while the homozygous nf-yc12-1 and nf-yc12-2 plants showed a significant reduction in 1000-grain weight (Fig. 2C). In addition, nf-yc12 grains displayed a higher rate of chalkiness than those of the WT control (Fig. 2D, E-a, b, f, g, k, l). Notably, cross-section analysis showed that the inner starchy endosperm of nf-yc12 appeared to be floury-white, while that of the WT was translucent (Fig. 2E-c, h, m). Furthermore, SEM images of transverse sections of nf-yc12 grains indicated that starch granules (SGs) were small, round, and loosely packed, while they were regularly polyhedral and densely packed in WT endosperm cells (Fig. 2E-d, e, i, j, n, o). In contrast to the defective endosperm and decreased grain weight, the germination rate of homozygous nf-yc12 mutants was not affected (Supplementary Fig. S4). Together, these data suggested that functional loss of NF-YC12 caused a decrease in grain weight and chalkiness of the endosperm.
Seed storage substances and compound starch granules are altered in nf-yc12 endosperm
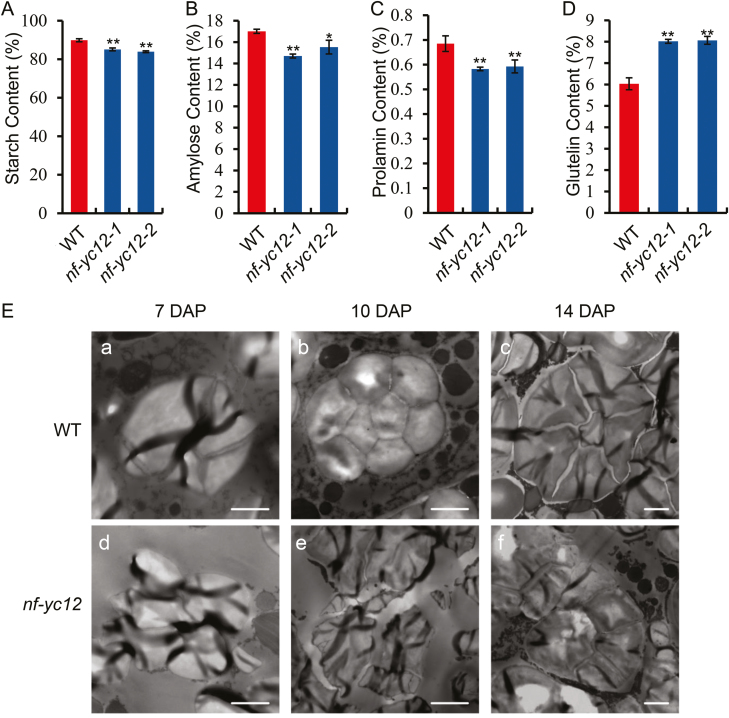
To investigate the intrinsic changes of grains in nf-yc12, we examined the contents of storage substances in mature seeds. The total starch and amylose contents were significantly lower than those in the WT (Fig. 3A, B). The prolamin content was decreased, while the glutelin content was higher than that in the WT (Fig. 3C, D). To further examine the starch physicochemical properties, the gelatinization properties of the SGs were determined by measuring the solubility of starch in urea solutions (Peng et al., 2014). Various concentrations (0–9 M) of urea solutions were added to powdered starch, and the results showed that the gelatinization characteristics of nf-yc12 were significantly different from those of the WT in solutions of 4–5 M (Supplementary Fig. S5).
Fig. 3.
Seed properties and amyloplast development in wild-type rice and nf-yc12 mutants. (A–D) Quality trait parameters of mature seeds from wild-type (WT) and nf-yc12. Data are means (±SD) of n=3 replicates. Significant differences between the WT and the mutant were determined using Student’s t-test (*P<0.05; **P<0.01). (E) TEM images of the compound starch granules of the WT and nf-yc12 mutant at (a, d) 7 d after pollination (DAP), (b, e) 10 DAP, and (c, f) 14 DAP. Scale bars are 2 μm.
To examine whether the SGs in nf-yc12 were morphologically defective, TEM was carried out to observe the development of starchy endosperm cells at early developmental stages (7, 10, and 14 DAP). In the WT, smaller SGs were initially formed in amyloplasts and gradually filled their interior regions, eventually forming regularly shaped polyhedrons (Fig. 3E). No significant differences were detected between the WT and mutant SGs at 7 DAP (Fig. 3E-a, d). However, at 10 DAP and 14 DAP, in contrast to the dense arrangement in the WT, the SGs in nf-yc12 showed a loose and irregular arrangement (Fig. 3E-b, c, e, f), implying an arrest of amyloplast growth in the mutant. Together, these results suggested that NF-YC12 might play a vital role in accumulation of storage substances and in SG development during the grain-filling stage.
Overexpression of NF-YC12 increases grain size and grain weight
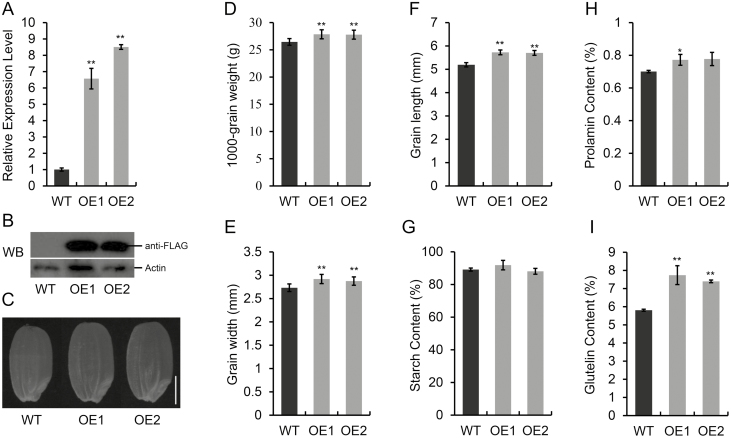
To further explore the function of NF-YC12 in rice seed development, we generated transgenic lines expressing pUbi::NF-YC12-FLAG. qRT-PCR analysis showed that the expression of NF-YC12 was greatly elevated in the overexpression (OE) lines, with OE1 and OE2 having 6.5- and 8.5-fold greater transcripts, respectively, compared with the WT (Fig. 4A). Expression of the transformed target NF-YC12-FLAG fusion protein was verified by western blot analysis (Fig. 4B), and indicated the effectiveness of the FLAG tags. We then selected these two independent and homozygous OE lines for further analysis.
Fig. 4.
Transgenic lines of rice overexpressing NF-YC12. (A) Expression levels of NF-YC12 in the developing seeds at 7 d after pollination (DAP) of the wild-type (WT) and two overexpression lines (OE1 and OE2), as determined by real-time quantitative RT-PCR. Data are means (±SD) for n=3 replicates. Significant differences between the WT and OE lines were determined using two-tailed Student’s t-tests (*P<0.05; **P<0.01). (B) Expression levels of representative protein in the WT and OE lines. Total protein extracted from developing seeds at 10 DAP was used for western blot analysis with an anti-FLAG antibody. (C) Representative images of the shapes of the grains of the WT and OE lines; the scale bar is 2 mm. (D–F) Thousand-grain weight (D), grain width (E), grain length (F), and contents of (G) starch, (H) prolamin, and (I) glutelin of the WT and OE lines. Data are means (±SD) of n=20 replicates (D–F) and n=3 replicates (G–I). Significant differences between the WT and OE lines were determined using Student’s t-test (*P<0.05; **P<0.01).
Under normal field conditions, the OE lines of NF-YC12 displayed no significant differences from the WT plants during seeding and the vegetative growth stages. At the mature grain stage, we measured the length, width, and weight of the grains and found that the OE lines produced larger and heavier grains compared with the WT (Fig. 4C–F). At the same time, the contents of prolamin and glutelin were clearly increased in the OE lines (Fig. 4H, I) but the total starch content was not significantly altered compared with that of WT (Fig. 4G). The results suggested that overexpression of NF-YC12 may enlarge grain size by increasing the length and width, and result in increased grain weight through promoting grain filling in rice.
NF-YC12 is predominantly expressed in the starchy endosperm and the aleurone layer
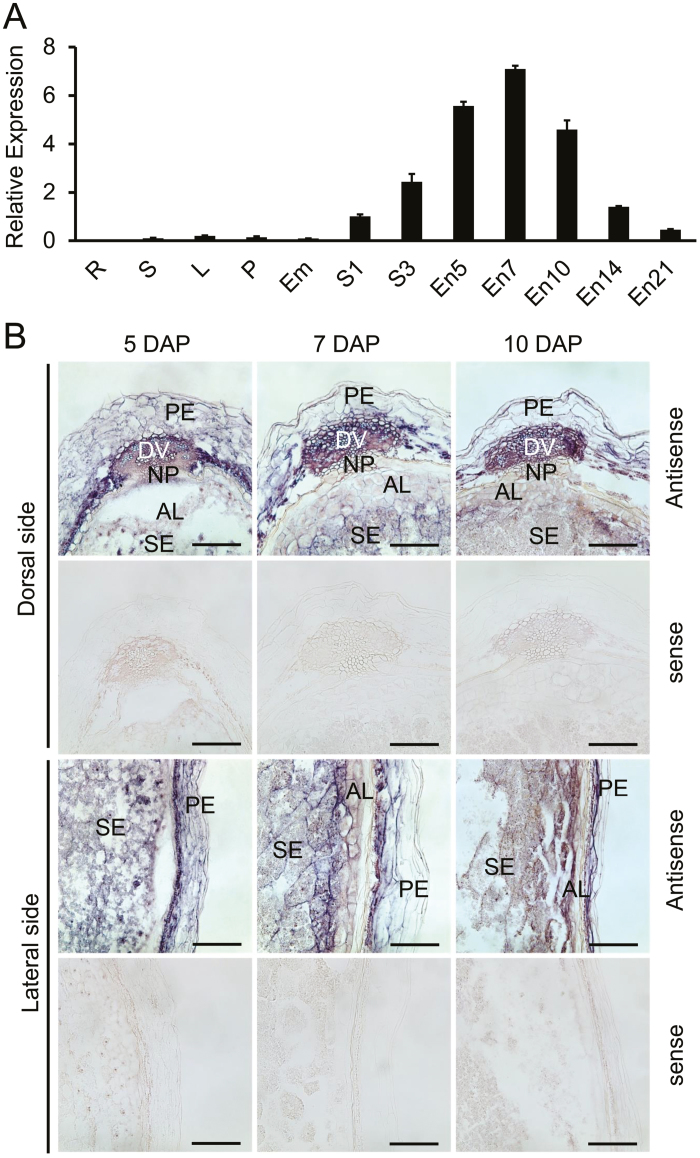
The expression patterns of NF-YC12 in different tissues were firstly analysed using qRT-PCR. The results showed that NF-YC12 was highly expressed in developing caryopses, while its expression was almost negligible in vegetative tissues (Fig. 5A). During seed development, the expression of NF-YC12 increased and peaked at 7 DAP and then gradually declined with seed maturation (Fig. 5A). These results suggested that NF-YC12 is specifically expressed in developing seeds.
Fig. 5.
Transcriptional characterization of rice NF-YC12. (A) qRT-PCR analysis of NF-YC12 In different organs: R, roots; S, stems; L, leaves; P, panicles; Em, embryo; S1, caryopses collected at 1 d after pollination (DAP); S3, caryopses collected at 3 DAP; En5–En21, endosperm collected at 5–21 DAP. (B) In situ hybridization of sectioned caryopses collected at 5, 7, and 10 DAP using antisense and sense probes. NF-YC12 was highly expressed in the aleurone layer (AL) and the starchy endosperm (SE). DV, dorsal vascular bundle; NP, nucellar projection; PE, pericarp. Scale bars are 100 μm.
To more precisely determine the spatial and temporal expression patterns of NF-YC12, mRNA in situ hybridization analysis was performed with immature seeds. The results showed that during seed development, NF-YC12 was mainly expressed in the starchy endosperm (SE) and pericarp, and weakly expressed in the aleurone layer (AL) at the dorsal side at 5 DAP. As endosperm differentiation and the accumulation of storage substances progresses, NF-YC12 was moderately expressed in the AL, and showed high expression in the SE at 7–10 DAP (Fig. 5B). No signals were observed in the sense-probe control, indicating the reliability of this experiment. NF-YC12 expression was also detected in the dorsal vascular bundle and nucellar epidermis in developing seeds (Fig. 5B). No expression was detected in leaves and panicles (Supplementary Fig. S6). These results indicated that the specific expression patterns of NF-YC12 were consistent with the defective phenotypes of nf-yc12 observed in endosperm development and in the grain-filling process.
To further examine the specific expression distributions of NF-YC12 and NF-YB1, the starchy endosperm and a mixture of the AL and the testa were mechanically isolated at 10 DAP and used for qRT-PCR analysis. This indicated that the expression of NF-YB1 was specific in the AL and it was undetectable in the SE (Supplementary Fig. S7), which was consistent with previous findings (Bai et al., 2016; Xu et al., 2016). Expression of NF-YC12 was observed in both tissues, but more preferentially in the SE (Supplementary Fig. S7).
NF-YC12 affects the transcription of genes related to starch biosynthesis and the metabolism of energy reserves
To study the functional mechanisms of NF-YC12, we identified significantly differentially expressed genes (DEGs) by RNA-seq analysis. The samples consisted of three independent biological replicates from pooled WT and nf-yc12 endosperms at 7 DAP. The data showed that the mutation of NF-YC12 significantly altered the expression of 4038 genes (corrected P-value <0.05, fold-change >1.5), of which 2000 were down-regulated and 2038 were up-regulated (Supplementary Dataset S1).
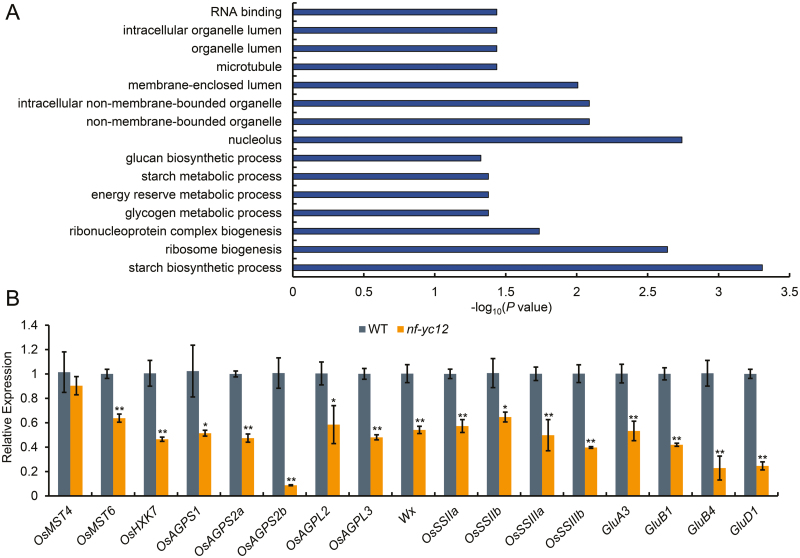
Gene Ontology (GO) enrichment analysis was performed to examine the biological roles of the DEGs in the endosperm. The significantly enriched GO terms were associated with ‘starch biosynthetic process’ (corrected P-value=4.92×10–4), ‘ribosome biogenesis’ (corrected P=2.29×10–3), ‘glycogen metabolic process’ (P=0.0419), and ‘energy reserve metabolic process’ (P=0.0419E) (Fig. 6A). These results were consistent with the accumulation of seed storage substances and associated metabolic processes, indicating a key transcriptional regulatory role of NF-YC12 in the rice endosperm. We also performed separate GO analyses of the down- and up-regulated genes. The most significantly enriched categories were different between these two groups of DEGs, with ‘starch biosynthetic process’ being predominant for the down-regulated genes and ‘ribosome biogenesis’ being predominant for the up-regulated genes (Supplementary Fig. S8), indicating that the down-regulated DEGs may have the more significant effects on the process of accumulation of storage substances. Notably, 20 genes previously identified as being critical for sugar transport and storage-substance accumulation were found to be down-regulated in the nf-yc12 RNA-seq data (Table 1), including monosaccharide transporter gene OsMST6 (Wang et al., 2008), cytosolic hexokinase encoding gene OsHXK7 (Kim et al., 2016a), ADP-glucose pyrophosphorylase encoding genes OsAGPS1, OsAGPS2, and OsAGPL2 (Lee et al., 2007; Tang et al., 2016; Wei et al., 2017), starch synthase encoding gene OsSSIIIa (Ryoo et al., 2007), storage protein-related gene GluB1 (Kawakatsu et al., 2009), and GluD1 (Kawakatsu et al., 2008).
Fig. 6.
Transcriptomic analyses of the rice nf-yc12 mutant. (A) A selection of enriched gene ontology (GO) terms of the differentially expressed genes (DEGs) as determined by RNA-seq using endosperm at 7 d after pollination (DAP). Wallenius’ non-central hyper-geometric distribution was implemented using the R package GOseq (Young et al., 2010). Only GO terms with a corrected P-value <0.05 and including at least five annotated genes were kept. The length of the bars represents the negative logarithm (base 10) of the corrected P-value. (B) qRT-PCR analysis confirming the down-regulated genes in the endosperm of the nf-yc12 mutant. The relative expressions of genes involved in starch biosynthesis and metabolic process were calculated. The expression of each gene in the wild-type (WT) endosperm at 7 DAP was set as a reference value of 1. Data are means (±SD) from n=3 replicates. Significant differences between the WT and the mutant were determined using Student’s t-test (*P<0.05; **P<0.01).
Table 1.
A selection of DEGs involved in accumulation of storage substances in nf-yc12 endosperm as identified by RNA-seq analysis.
| Gene ID | Gene Annotation | Gene Name | Fold-change | Corrected P-value |
|---|---|---|---|---|
| Sugar transport/synthesis | ||||
| Os01g0919400 | Sucrose-phosphate synthase | SPS1 | –27.17 | 2.73×10–5 |
| Os03g0218400 | Monosaccharide transporter | OsMST4 | –2.91 | 8.55×10–12 |
| Os07g0559700 | Monosaccharide transporter | OsMST6 | –2.50 | 2.66×10–67 |
| Os05g0187100 | Hexokinase | OsHXK7 | –2.35 | 3.54×10–41 |
| Starch biosynthesis | ||||
| Os09g0298200 | ADP-glucose pyrophosphorylase | OsAGPS1 | –2.12 | 8.03×10–62 |
| Os08g0345800 | ADP-glucose pyrophosphorylase | OsAGPS2 | –3.67 | 0.00 |
| Os01g0633100 | ADP-glucose pyrophosphorylase | OsAGPL2 | –1.50 | 1.49×10–42 |
| Os03g0735000 | ADP-glucose pyrophosphorylase | OsAGPL3 | –1.83 | 3.96×10–36 |
| Os06g0133000 | Starch synthase | Wx | –1.83 | 5.63×10–93 |
| Os06g0229800 | Starch synthase | OsSSIIa | –2.04 | 6.77×10–41 |
| Os02g0744700 | Starch synthase | OsSSIIb | –2.51 | 3.13×10–4 |
| Os08g0191433 | Starch synthase | OsSSIIIa | –2.88 | 6.14×10–202 |
| Os04g0624600 | Starch synthase | OsSSIIIb | –1.54 | 1.59×10–6 |
| Protein biosynthesis | ||||
| Os03g0427300 | Seed storage protein | GluA3 | –1.58 | 4.34×10–55 |
| Os02g0249800 | Seed storage protein | GluB1 | –2.56 | 1.18×10–67 |
| Os02g0242600 | Seed storage protein | GluB4 | –2.65 | 1.07×10–241 |
| Os02g0249000 | Seed storage protein | GluD1 | –2.47 | 3.61×10–201 |
| Os02g0248800 | Seed storage protein | Unnamed | –2.53 | 1.77×10–191 |
| Os02g0268300 | Seed storage protein | Unnamed | –1.57 | 6.60×10–53 |
| Os02g0249600 | Seed storage protein | Unnamed | –1.52 | 2.89×10–46 |
qRT-PCR was carried out to verify the decreases in the expression levels of the selected genes associated with starch and protein synthesis in endosperm of nf-yc12 at 7 DAP (Fig. 6B), and the results demonstrated the reliability of the differences in gene expression that were determined by RNA-seq. These data further indicated that the transcriptional regulatory function of NF-YC12 is associated with the accumulation of storage substances in rice seeds.
NF-YC12 directly regulates OsSUT1, OsGS1;3, and FLO6 in storage-substance accumulation
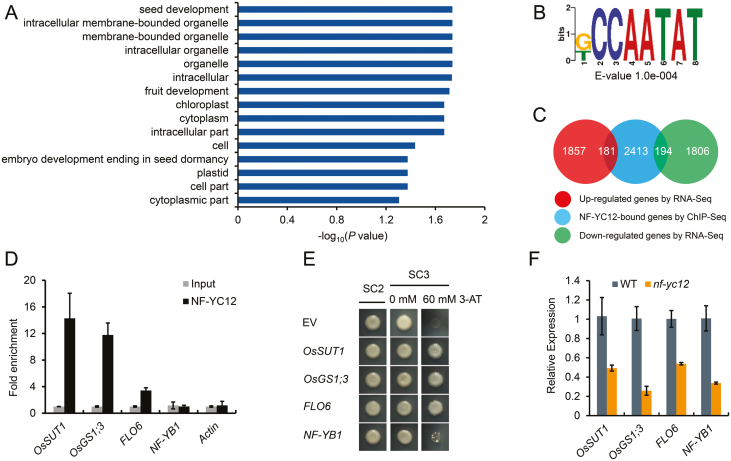
To further elucidate its regulatory mechanisms, the genes bound by NF-YC12 in the endosperm were identified using ChIP experiments with an anti-FLAG antibody followed by DNA-sequencing analysis (ChIP-seq). The ChIP-seq data were analysed with the statistical software MACS2 (Zhang et al., 2008) and 4188 peaks were finally identified (Supplementary Dataset S2). These peaks were linked to 2788 neighbor genes, which were considered as the predicted NF-YC12-bound genes. GO annotation analysis showed that the terms for the biological processes related to seed and fruit development were significantly enriched (Fig. 7A). We first attempted to identify the binding motifs of the NF-YC12 protein using DREME (Bailey, 2011) and found that one of the top five significantly enriched motifs was the typical one of NF-Y factors, CCAAT-box (CCAATAT) (Fig. 7B). Among the 4188 NF-YC12 binding peaks, the CCAAT box was found in 1176 peaks, while the GCC box (GCCGCC), the binding motif of NF-YB1 (Xu et al., 2016), was not enriched. These results implied that NF-YC12 probably binds to the promoters of its downstream targets through the CCAAT-box motif.
Fig. 7.
Overview of ChIP-seq data and identification of NF-YC12 direct target genes in rice. (A) Enriched gene ontology (GO) terms of the genes bound by NF-YC12 as determined by ChIP-seq analysis. Only GO terms with a corrected P-value <0.05 and including at least five annotated genes were kept. The length of the bars represents the negative logarithm (base 10) of the corrected P-value. (B) Motif analysis of NF-YC12 binding peaks by DREME. The ‘CCAATA’ (CCAAT-box) motif was identified as one of the top five enriched motifs. The E-value is the enrichment P-value multiplied by the number of candidate motifs tested. The enrichment P-value was calculated using Fisher’s exact test for enrichment of the motif in the positive sequences. (C) Venn diagram showing the number of overlapping genes between the NF-YC12-bound gene set (ChIP-seq data) and the NF-YC12-regulated gene set (RNA-seq). (D) ChIP-PCR verification of NF-YC12-bound regions. The data are the mean values (±SD) of fold-enrichment from n=3 technical replicates. (E) The interaction between NF-YC12 and the promoters of target genes as determined by yeast one-hybrid analysis. EV, empty vector; SC2,: SD/–Leu/–Trp; SC3, SD/–Leu/–Trp/–His. (F) qRT-PCR analysis of expression levels of the target genes in nf-yc12 Compared with the wild-type (WT). Ubiquitin was used as the reference gene.
To further explore the target genes regulated by NF-YC12 at the transcript level, we combined the data sets of DEGs from RNA-seq and the NF-YC12-bound genes from ChIP-seq. The results showed that 181 up-regulated genes and 194 down-regulated genes were bound by NF-YC12 in the endosperm at 7 DAP (Fig. 7C). The potential NF-YC12 targets included several known synthesis genes of starch and transcription factors, such as OsAGPS2, OsSSIIIb, OsGS1;3, and NF-YB1. Based on the RNA-seq and ChIP-seq analysis, we then selected OsGS1;3 and NF-YB1 as potential targets of NF-YC12 for validation of the protein–DNA interactions. In addition, given the targets of NF-YB1 and the floury endosperm phenotype, OsSUT1, 3, 4, and FLO6 were also selected for ChIP-qPCR testing. The results showed that NF-YC12 binds to the promoters of OsSUT1, OsGS1;3, and FLO6, while the promoter region of NF-YB1, which showed enrichment in the ChIP-seq data, was not enriched (Fig. 7D). In addition, a yeast one-hybrid assay was performed to further confirm the interactions between NF-YC12 and the promoters of target genes, and it showed that the promoters of OsSUT1, OsGS1;3, and FLO6 were specifically recognized by the NF-YC12 protein (Fig 7E). Loss of function of NF-YC12 significantly down-regulated OsSUT1, OsGS1;3, and FLO6 (Fig. 7F). qRT-PCR results indicated that NF-YC12 positively regulated the expression of OsSUT1, OsGS1;3, and FLO6 in the NF-YC12 overexpression lines (Supplementary Fig. S9). These results indicated that OsSUT1, OsGS1;3, and FLO6 are the direct targets of NF-YC12 in rice during endosperm development. LUC transient transcriptional activity assays in protoplasts were performed, and the showed that NF-YC12 specifically activated the OsSUT1 and OsGS1;3 promoters in vivo, although the NF-YC12 protein showed no significant activation of FLO6 transcription (Supplementary Fig. S10).
In addition, OsGS1;3, which encodes a cytosolic glutamine synthetase (GS), was abundantly expressed in developing endosperm, and the expression reached a maximum at 10 DAP (Supplementary Fig. S11). A similar expression pattern was observed for NF-YC12. OsSUT1, which encodes a sucrose transporter protein, is one of the direct targets of NF-YB1 (Bai et al., 2016). Loss of function of FLO6 results in a similar chalky endosperm phenotype and alters the accumulation of storage substances in rice seeds (Peng et al., 2014). Taken together, the results suggested that NF-YC12 functions cooperatively with NF-YB1 to regulate SUTs, while it directly regulates OsGS1;3 and FLO6 in the endosperm for accumulation of storage substances.
Discussion
In this study, we identified the function of NF-YC12, an endosperm-specific NF-Y transcription factor. Our genetic analysis indicated that loss of function of NF-YC12 resulted in significantly decreased grain weight and starch content as well as an obvious chalky endosperm phenotype (Figs 2, 3). In addition, the prolamin and glutelin contents were also significantly altered in the seeds of nf-yc12 (Fig. 3). Previous studies have shown that there are compensatory effects between different storage proteins (Kawakatsu et al., 2009; Kawakatsu and Takaiwa, 2010). The percentage of storage substances is constant, and an increase or decrease in one component leads to a change in content of another component (Kawakatsu and Takaiwa, 2010; Zhou et al., 2017). It is known that overexpression of RAG2 increases the content of storage proteins and decreases that of starch, and it enlarges the size and weight of grains significantly by influencing the grain filling (Zhou et al., 2017). Our results showed that a change in the contents of storage proteins was directly linked to the level of NF-YC12 expression. The contents of prolamin and glutelin were clearly increased in the overexpression (OE) lines (Fig. 4). This suggests that overexpression of NF-YC12 in rice possibly promotes grain filling and improves the accumulation of storage proteins, hence increasing the grain size and weight. NF-YC12 is therefore a potential useful gene in cereal breeding programs.
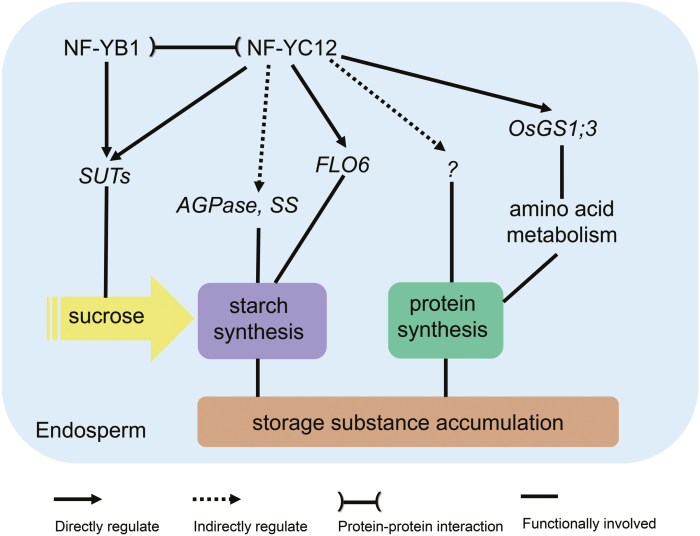
Comprehensive transcriptome and DNA-binding analysis showed that genes related to ‘starch biosynthesis’ and ‘energy reserve metabolic process’ were enriched in the down-regulated category in the nf-yc12 mutant (Fig. 6). Moreover, we also demonstrated that NF-YC12 not only regulates the genes for sucrose transport in the AL through interacting with NF-YB1, but also controls the key gene related to the starch synthesis process (FLO6) and the amino acid synthetase gene OsGS1;3 in the endosperm (Fig. 8). Taken together, this indicates a broad regulatory function of NF-YC12, involving multiple pathways for the accumulation of storage substances in the rice endosperm.
Fig. 8.
Schematic diagram of the regulatory network of NF-YC12 in rice endosperm. NF-YC12 plays upstream regulatory roles in sucrose loading, endosperm development, and the accumulation of storage substances. It modulates starch synthesis through direct regulation of FLO6, which is a key regulator involved in starch synthesis, and through indirectly regulating other starch synthesis genes, including AGPase and SS. At the same time, NF-YC12 also influences accumulation of storage proteins through directly regulating the amino acid metabolic enzyme OsGS1;3 and other as yet undetermined seed storage-protein synthases. In addition, NF-YC12 interacts with NF-YB1, and they co-regulate sucrose loading through directly regulating SUTs in the aleurone layer.
NF-YC12 functions cooperatively with NF-YB1 to regulate SUTs in the aleurone layer
Previous studies have shown that OsNF-YB1 is specifically expressed in the AL of the endosperm, and not in the SE (Bai et al., 2016; Xu et al., 2016). Consistent with this, we also found that the expression of NF-YB1 was AL-specific (Supplementary Fig. S7). mRNA in situ hybridization and qRT-PCR analysis indicated that NF-YC12 was highly expressed in both the AL and SE (Fig. 5, Supplementary Fig. S7). Comparison of the expression patterns between NF-YC12 and NF-YB1 in the endosperm showed that they were co-expressed in the AL.
In plants, the subcellular localization of NF-YB is variable due to the different interacting NF-YCs (Hackenberg et al., 2012). NF-YB1 and NF-YC12 were predominantly located in the nucleus when co-expressed in rice protoplasts (Supplementary Fig. S2), which is in agreement with their nuclear translocation mechanism (Hackenberg et al., 2012; Xu et al., 2016). During our studies, two other groups reported the interaction between NF-YB1 and NF-YC12 (Xu et al., 2016; Bello et al., 2019). A series of experiments in our study suggested that NF-YC12 interacts with NF-YB1 in vitro and vivo (Fig. 1). NF-YBs and NF-YCs are characterized by their core domain HFM motif, which is involved in both protein–DNA and protein–protein interactions (Laloum et al., 2013). NF-YC12 is a typical NF-YC subunit, and can interact with NF-YB1 through its HFM domain, suggesting the possibility that NF-YC12 and NF-YB1 form a NF-YB/C dimer in the AL to regulate endosperm development.
OsSUT1 is the direct target of NF-YB1 in the AL (Bai et al., 2016). We found that its expression was markedly decreased in nf-yc12 mutants (Fig. 7). ChIP-qPCR and yeast one-hybrid assays confirmed that NF-YC12 directly binds to the promoter of OsSUT1. Thus, OsSUT1 is a common target of both NF-YC12 and NF-YB1. Furthermore, the same defective endosperm phenotype was observed in both nf-yb1 and nf-yc12 mutants (Fig. 2). Previous studies have shown that suppressed OsSUT1 expression leads to impaired grain filling and reduces the final grain weight (Ishimaru et al., 2001; Ishibashi et al., 2014). Our work further supports the view that the AL of the endosperm is important for sucrose translocation during the grain-filling stage. Thus, the NF-YB1–NF-YC12 dimer is likely to regulate the expression of SUTs in the AL for the loading of sugar to the rice endosperm. Previous studies have indicated that the NF-YB/YC transcriptional complex can coordinately regulate the common pathway (Xu et al., 2016; Bello et al., 2019). It has been reported that NF-YC2 and NF-YC4 interact with three NF-YB proteins (NF-YB8, NF-YB10, and NF-YB11) in the regulation of flowering time in rice (Kim et al., 2016b). Similarly, NF-YC12 functions cooperatively with NF-YB1 to regulate SUT1 in the AL for rice endosperm development (Fig. 8). However, further studies are needed to clarify the regulatory network of the NF-YB1 and NF-YC12 complex in the AL.
The functional mechanism of NF-YC12 in regulating the accumulation of storage substances in the endosperm
RNA-seq data and GO analysis showed that the DEGs were involved in cellular carbohydrate metabolic processes and glucan synthase activity (Fig. 6). Furthermore, GO annotation analysis of the NF-YC12-bound genes showed significant enrichment of terms for biological processes related to seed and fruit development. These results reveal a broad regulatory function of NF-YC12 in the developing rice endosperm. The expression levels of 16 genes related to starch synthesis and seed storage proteins were reduced in the nf-yc12 mutant (Fig. 6). Intriguingly, several well-characterized genes encoding starch synthases (OsSSIIIa/FLO5, OsAGPL2) and genes related to protein synthesis (GluB1 and GluD1) were significantly down-regulated in the nf-yc12 endosperm. Mutant lines of OsSSIIIa/FLO5 show chalky endosperm and decreased starch contents (Ryoo et al., 2007). A loss-of-function mutation of OsAGPL2 results in floury endosperm and severe defects in starch and storage protein synthesis (Tang et al., 2016; Wei et al., 2017). The endosperm-specific glutelin gene GluD1 is predominantly expressed in the inner SE, and the promoter of GluD1 is specifically recognized by RISBZ1 and RPBF (Kawakatsu et al., 2008, 2009). Another glutelin gene, GluB1, has been shown to be involved in storage protein synthesis, and the core motifs in its promoter for seed-specific expression have been identified (Wu et al., 2000; Chen et al., 2014). Similarly, nf-yc12 mutants showed floury endosperm and abnormal storage-substance accumulation (Figs 2, 3). This suggest that NF-YC12 modulates the process of storage-substance accumulation by regulating the expression of multiple genes associated with starch and protein biosynthesis, and hence influences seed-related phenotypes of rice. However, further studies are required to determine whether NF-YC12 regulates these synthesis genes directly or indirectly during grain filling.
We confirmed that FLO6 is one of the targets of NF-YC12, and its expression was significantly reduced in nf-yc12 (Fig. 7). It has been reported that FLO6 encodes a protein containing a CBM domain that acts as a starch-binding protein involved in starch synthesis (Peng et al., 2014). The flo6 mutant displays chalky endosperm and reduced grain weight, and the contents of starch and proteins are also altered in its seeds (Peng et al., 2014). The nf-yc12 exhibited the same phenotype as flo6 in terms of synthesis of storage substances and grain traits (Figs 2, 3). Taken together, NF-YC12 affects the synthesis of endosperm storage substances by directly regulating FLO6 expression.
Our ChIP-seq and RNA-seq analysis provided clues to the potential targets of NF-YC12. OsGS1;3 was verified to be a direct downstream target of NF-YC12 (Fig. 7). Plant glutamine synthetase (GS, EC 6.3.1.2) catalyses an ATP-dependent conversion of glutamate to glutamine for amino acid interconversion. Cytosolic glutamine synthetase (GS1) has three homologous genes (OsGS1;1, OsGS1;2, and OsGS1;3). Homozygous mutants lacking OsGS1;1 show severe retardation in growth and grain filling under normal conditions (Tabuchi et al., 2005; Kusano et al., 2011). Previous studies have shown that OsGS1;3 is mainly expressed in spikelets (Tabuchi et al., 2005). Microarray data in CREP (http://crep.ncpgr.cn; microarray data sets: GSE19024) show that OsGS1;3 is preferentially expressed in the spikelets and seeds (Wang et al., 2010). In our study, qRT-PCR results revealed that OsGS1;3 was predominantly expressed in the endosperm, overlapping with the expression of NF-YC12 (Supplementary Fig. S11). Therefore, NF-YC12 may directly regulate OsGS1;3, which is related to amino acid metabolism for protein accumulation in the rice endosperm.
It is notable that the expression of NF-YC12 was more extensive in the endosperm than that of NF-YB1, and was higher in the SE than in the AL (Supplementary Fig. S7), which is consistent with a previous report that NF-YCs are probably highly expressed in the SE (E et al., 2018). It has been reported that NF-YC proteins (NF-YC11 and NF-YC12) do not show any transactivation activities in yeast (E et al., 2018). NF-YC10 has transcriptional activation ability in yeast (Jia et al., 2019), and NF-YC12 shows a certain degree of transcriptional activation in vivo (Bello et al., 2019). We found transactivation of NF-YC12 on OsSUT1 and OsGS1;3 (Supplementary Fig. S10), suggesting that it directly activates them. Although NF-YC12 has not been shown to activate FLO6 in vivo, more experiments need to be undertaken to examine this. We provide direct evidence to demonstrate NF-YC12-mediated transcriptional regulation of FLO6, and we believe that FLO6 is a direct target of NF-YC12. A model was proposed for the function of NF-YC12 in the gene network that regulates sucrose loading and the accumulation of storage substances in the rice endosperm (Fig. 8). NF-YC12 may not only work in coordination with NF-YB1 to regulate the expression of SUTs in the AL, but also act as a direct activator of the downstream genes FLO6 and OsGS1;3 and other as yet undetermined targets to regulate the accumulation of storage substances during endosperm development.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Interactions between selected rice endosperm- specific NF-Ys.
Fig. S2. Subcellular localization of NF-YB1 and NF-YC12 in rice protoplasts.
Fig. S3. Identification of CRISPR/Cas9-induced target mutations.
Fig. S4. Seed germination rates of mature seeds of the wild-type and nf-yc12.
Fig. S5. Gelatinization characteristics of starch from nf-yc12 mutant seeds.
Fig. S6. In situ hybridization of NF-YC12 in vegetative organs.
Fig. S7. Expression levels of NF-YB1 and NF-YC12 in different endosperm tissues.
Fig. S8. GO analysis of DEGs that were down- and up-regulated in nf-yc12.
Fig. S9. Expression levels of NF-YC12 potential targets in the developing seeds of the wild-type and overexpression lines at 7 DAP.
Fig. S10. LUC transient transcriptional activity assays in rice protoplast.
Fig. S11. Real-time PCR analysis of the expression pattern of OsGS1;3 in the endosperm.
Table S1. Primers used in this study.
Table S2. Percentage of T0 plants with mutation in the target sequence of NF-YC12.
Table S3. Mutations detected in putative CRISPR/Cas9 off-target sites.
Dataset S1. Differentially expressed genes between the wild-type and the nf-yc12 mutant.
Dataset S2. NF-YC12 binding sites identified by ChIP-seq.
Acknowledgements
We thank Prof. Yidan Ouyang (Huazhong Agricultural University, China) for helping revise the manuscript and for English language editing. We thank Prof. Meizhong Luo (Huazhong Agricultural University, China) for providing the plasmids pSAT4-cCFP-N and pSAT6-nCerulean-N. This research was supported by grants from the National Natural Science Foundation of China (no. 31570321 and no. 31660046). The funders had no role in the study design, data collection and analysis, the decision to publish, or in the preparation of the manuscript.
References
- Bai AN, Lu XD, Li DQ, Liu JX, Liu CM. 2016. NF-YB1-regulated expression of sucrose transporters in aleurone facilitates sugar loading to rice endosperm. Cell Research 26, 384–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. 2011. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 27, 1653–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becraft PW. 2001. Cell fate specification in the cereal endosperm. Seminars in Cell & Developmental Biology 12, 387–394. [DOI] [PubMed] [Google Scholar]
- Bello BK, Hou Y, Zhao J, et al. . 2019. NF-YB1-YC12-bHLH144 complex directly activates Wx to regulate grain quality in rice (Oryza sativa L.). Plant Biotechnology Journal. In press, doi: 10.1111/pbi.13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C, Benvenuto G, Laflamme P, Molino D, Probst AV, Tariq M, Paszkowski J. 2004. Chromatin techniques for plant cells. The Plant Journal 39, 776–789. [DOI] [PubMed] [Google Scholar]
- Chen Y, Sun A, Wang M, Zhu Z, Ouwerkerk PB. 2014. Functions of the CCCH type zinc finger protein OsGZF1 in regulation of the seed storage protein GluB-1 from rice. Plant Molecular Biology 84, 621–634. [DOI] [PubMed] [Google Scholar]
- E Z, Li T, Zhang H, Liu Z, Deng H, Sharma S, Wei X, Wang L, Niu B, Chen C. 2018. A group of nuclear factor Y transcription factors are sub-functionalized during endosperm development in monocots. Journal of Experimental Botany 69, 2495–2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenberg D, Wu Y, Voigt A, Adams R, Schramm P, Grimm B. 2012. Studies on differential nuclear translocation mechanism and assembly of the three subunits of the Arabidopsis thaliana transcription factor NF-Y. Molecular Plant 5, 876–888. [DOI] [PubMed] [Google Scholar]
- Hannah LC, James M. 2008. The complexities of starch biosynthesis in cereal endosperms. Current Opinion in Biotechnology 19, 160–165. [DOI] [PubMed] [Google Scholar]
- He Y, Zhang T, Yang N, Xu M, Yan L, Wang L, Wang R, Zhao Y. 2017. Self-cleaving ribozymes enable the production of guide RNAs from unlimited choices of promoters for CRISPR/Cas9 mediated genome editing. Journal of Genetics and Genomics 44, 469–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T, Terao T. 2004. A comprehensive expression analysis of the starch synthase gene family in rice (Oryza sativa L.). Planta 220, 9–16. [DOI] [PubMed] [Google Scholar]
- Ishibashi Y, Okamura K, Miyazaki M, Phan T, Yuasa T, Iwaya-Inoue M. 2014. Expression of rice sucrose transporter gene OsSUT1 in sink and source organs shaded during grain filling may affect grain yield and quality. Environmental and Experimental Botany 97, 49–54. [Google Scholar]
- Ishimaru K, Hirose T, Aoki N, et al. . 2001. Antisense expression of a rice sucrose transporter OsSUT1 in rice (Oryza sativa L.). Plant & Cell Physiology 42, 1181–1185. [DOI] [PubMed] [Google Scholar]
- Ishimaru T, Ida M, Hirose S, Shimamura S, Masumura T, Nishizawa NK, Nakazono M, Kondo M. 2015. Laser microdissection-based gene expression analysis in the aleurone layer and starchy endosperm of developing rice caryopses in the early storage phase. Rice 8, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Thirumurugan T, Serizawa A, Hiratsu K, Ohme-Takagi M, Kurata N. 2011. Aberrant vegetative and reproductive development by overexpression and lethality by silencing of OsHAP3E in rice. Plant Science 181, 105–110. [DOI] [PubMed] [Google Scholar]
- Jia S, Xiong Y, Xiao P, Wang X, Yao J. 2019. OsNF-YC10, a seed preferentially expressed gene regulates grain width by affecting cell proliferation in rice. Plant Science 280, 219–227. [DOI] [PubMed] [Google Scholar]
- Kawakatsu T, Hirose S, Yasuda H, Takaiwa F. 2010. Reducing rice seed storage protein accumulation leads to changes in nutrient quality and storage organelle formation. Plant Physiology 154, 1842–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T, Takaiwa F. 2010. Differences in transcriptional regulatory mechanisms functioning for free lysine content and seed storage protein accumulation in rice grain. Plant & Cell Physiology 51, 1964–1974. [DOI] [PubMed] [Google Scholar]
- Kawakatsu T, Yamamoto MP, Hirose S, Yano M, Takaiwa F. 2008. Characterization of a new rice glutelin gene GluD-1 expressed in the starchy endosperm. Journal of Experimental Botany 59, 4233–4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T, Yamamoto MP, Touno SM, Yasuda H, Takaiwa F. 2009. Compensation and interaction between RISBZ1 and RPBF during grain filling in rice. The Plant Journal 59, 908–920. [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nature Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HB, Cho JI, Ryoo N, Shin DH, Park YI, Hwang YS, Lee SK, An G, Jeon JS. 2016a. Role of rice cytosolic hexokinase OsHXK7 in sugar signaling and metabolism. Journal of Integrative Plant Biology 58, 127–135. [DOI] [PubMed] [Google Scholar]
- Kim SK, Park HY, Jang YH, Lee KC, Chung YS, Lee JH, Kim JK. 2016b. OsNF-YC2 and OsNF-YC4 proteins inhibit flowering under long-day conditions in rice. Planta 243, 563–576. [DOI] [PubMed] [Google Scholar]
- Krishnan S, Dayanandan P. 2003. Structural and histochemical studies on grain-filling in the caryopsis of rice (Oryza sativa L.). Journal of Biosciences 28, 455–469. [DOI] [PubMed] [Google Scholar]
- Kusano M, Tabuchi M, Fukushima A, et al. . 2011. Metabolomics data reveal a crucial role of cytosolic glutamine synthetase 1;1 in coordinating metabolic balance in rice. The Plant Journal 66, 456–466. [DOI] [PubMed] [Google Scholar]
- Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, Harada JJ. 2003. LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. The Plant Cell 15, 5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloum T, De Mita S, Gamas P, Baudin M, Niebel A. 2013. CCAAT-box binding transcription factors in plants: Y so many? Trends in Plant Science 18, 157–166. [DOI] [PubMed] [Google Scholar]
- Lee H, Fischer RL, Goldberg RB, Harada JJ. 2003. Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proceedings of the National Academy of Sciences, USA 100, 2152–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LY, Fang MJ, Kuang LY, Gelvin SB. 2008. Vectors for multi-color bimolecular fluorescence complementation to investigate protein–protein interactions in living plant cells. Plant Methods 4, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Hwang SK, Han M, et al. . 2007. Identification of the ADP-glucose pyrophosphorylase isoforms essential for starch synthesis in the leaf and seed endosperm of rice (Oryza sativa L.). Plant Molecular Biology 65, 531–546. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Yan W, Chen H, Tan C, Han Z, Yao W, Li G, Yuan M, Xing Y. 2016. Duplication of OsHAP family genes and their association with heading date in rice. Journal of Experimental Botany 67, 1759–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Fan C, Xing Y, et al. . 2014. Chalk5 encodes a vacuolar H+-translocating pyrophosphatase influencing grain chalkiness in rice. Nature Genetics 46, 398–404. [DOI] [PubMed] [Google Scholar]
- Lin YJ, Zhang Q. 2005. Optimising the tissue culture conditions for high efficiency transformation of indica rice. Plant Cell Reports 23, 540–547. [DOI] [PubMed] [Google Scholar]
- Mantovani R. 1999. The molecular biology of the CCAAT-binding factor NF-Y. Gene 239, 15–27. [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Ito Y, Serizawa A, Kurata N. 2003. OsHAP3 genes regulate chloroplast biogenesis in rice. The Plant Journal 36, 532–540. [DOI] [PubMed] [Google Scholar]
- Nakamura Y. 2018. Rice starch biotechnology: rice endosperm as a model of cereal endosperms. Starch 70, 1600375. [Google Scholar]
- Nie DM, Ouyang YD, Wang X, Zhou W, Hu CG, Yao J. 2013. Genome-wide analysis of endosperm-specific genes in rice. Gene 530, 236–247. [DOI] [PubMed] [Google Scholar]
- Nishi A, Nakamura Y, Tanaka N, Satoh H. 2001. Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. Plant Physiology 127, 459–472. [PMC free article] [PubMed] [Google Scholar]
- Okita TW, Hwang YS, Hnilo J, Kim WT, Aryan AP, Larson R, Krishnan HB. 1989. Structure and expression of the rice glutelin multigene family. The Journal of Biological Chemistry 264, 12573–12581. [PubMed] [Google Scholar]
- Olsen OA. 2004. Nuclear endosperm development in cereals and Arabidopsis thaliana. The Plant Cell 16, S214–S227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C, Wang Y, Liu F, et al. . 2014. FLOURY ENDOSPERM6 encodes a CBM48 domain-containing protein involved in compound granule formation and starch synthesis in rice endosperm. The Plant Journal 77, 917–930. [DOI] [PubMed] [Google Scholar]
- Petroni K, Kumimoto RW, Gnesutta N, Calvenzani V, Fornari M, Tonelli C, Holt BF 3rd, Mantovani R. 2012. The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. The Plant Cell 24, 4777–4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo N, Yu C, Park CS, Baik MY, Park IM, Cho MH, Bhoo SH, An G, Hahn TR, Jeon JS. 2007. Knockout of a starch synthase gene OsSSIIIa/Flo5 causes white-core floury endosperm in rice (Oryza sativa L.). Plant Cell Reports 26, 1083–1095. [DOI] [PubMed] [Google Scholar]
- Sabelli PA, Larkins BA. 2009. The development of endosperm in grasses. Plant Physiology 149, 14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She KC, Kusano H, Koizumi K, et al. . 2010. A novel factor FLOURY ENDOSPERM2 is involved in regulation of rice grain size and starch quality. The Plant Cell 22, 3280–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siefers N, Dang KK, Kumimoto RW, Bynum WE 4th, Tayrose G, Holt BF 3rd. 2009. Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiology 149, 625–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Ling S, Lu Z, Ouyang YD, Liu S, Yao J. 2014. OsNF-YB1, a rice endosperm-specific gene, is essential for cell proliferation in endosperm development. Gene 551, 214–221. [DOI] [PubMed] [Google Scholar]
- Tabuchi M, Sugiyama K, Ishiyama K, Inoue E, Sato T, Takahashi H, Yamaya T. 2005. Severe reduction in growth rate and grain filling of rice mutants lacking OsGS1;1, a cytosolic glutamine synthetase1;1. The Plant Journal 42, 641–651. [DOI] [PubMed] [Google Scholar]
- Tang XJ, Peng C, Zhang J, et al. . 2016. ADP-glucose pyrophosphorylase large subunit 2 is essential for storage substance accumulation and subunit interactions in rice endosperm. Plant Science 249, 70–83. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Xu H, Zhu Y, Liu QQ, Cai XL. 2013. OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm. Journal of Experimental Botany 64, 3453–3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xie W, Chen Y, . et al 2010. A dynamic gene expression atlas covering the entire life cycle of rice. The Plant Journal 61, 752–766. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhou W, Lu Z, Ouyang Y, Su OC, Yao J. 2015. A lipid transfer protein, OsLTPL36, is essential for seed development and seed quality in rice. Plant Science 239, 200–208. [DOI] [PubMed] [Google Scholar]
- Wang Y, Peng W, Zhou X, Huang F, Shao L, Luo M. 2014. The putative Agrobacterium transcriptional activator-like virulence protein VirD5 may target T-complex to prevent the degradation of coat proteins in the plant cell nucleus. New Phytologist 203, 1266–1281. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ren Y, Liu X, et al. . 2010. OsRab5a regulates endomembrane organization and storage protein trafficking in rice endosperm cells. The Plant Journal 64, 812–824. [DOI] [PubMed] [Google Scholar]
- Wang Y, Xiao Y, Zhang Y, Chai C, Wei G, Wei X, Xu H, Wang M, Ouwerkerk PB, Zhu Z. 2008. Molecular cloning, functional characterization and expression analysis of a novel monosaccharide transporter gene OsMST6 from rice (Oryza sativa L.). Planta 228, 525–535. [DOI] [PubMed] [Google Scholar]
- Wei X, Jiao G, Lin H, Sheng Z, Shao G, Xie L, Tang S, Xu Q, Hu P. 2017. GRAIN INCOMPLETE FILLING 2 regulates grain filling and starch synthesis during rice caryopsis development. Journal of Integrative Plant Biology 59, 134–153. [DOI] [PubMed] [Google Scholar]
- Wu C, Washida H, Onodera Y, Harada K, Takaiwa F. 2000. Quantitative nature of the Prolamin-box, ACGT and AACA motifs in a rice glutelin gene promoter: minimal cis-element requirements for endosperm-specific gene expression. The Plant Journal 23, 415–421. [DOI] [PubMed] [Google Scholar]
- Wu X, Liu J, Li D, Liu CM. 2016. Rice caryopsis development II: Dynamic changes in the endosperm. Journal of Integrative Plant Biology 58, 786–798. [DOI] [PubMed] [Google Scholar]
- Xing Q, Zheng Z, Zhou X, Chen X, Guo Z. 2015. Ds9 was isolated encoding as OsHAP3H and its C-terminus was required for interaction with HAP2 and HAP5. Journal of Plant Biology 58, 26–37. [Google Scholar]
- Xu J, Messing J. 2009. Amplification of prolamin storage protein genes in different subfamilies of the Poaceae. Theoretical and Applied Genetics 119, 1397–1412. [DOI] [PubMed] [Google Scholar]
- Xu JJ, Zhang XF, Xue HW. 2016. Rice aleurone layer specific OsNF-YB1 regulates grain filling and endosperm development by interacting with an ERF transcription factor. Journal of Experimental Botany 67, 6399–6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Xing Y, Weng X, et al. . 2008. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nature Genetics 40, 761–767. [DOI] [PubMed] [Google Scholar]
- Yamamoto MP, Onodera Y, Touno SM, Takaiwa F. 2006. Synergism between RPBF Dof and RISBZ1 bZIP activators in the regulation of rice seed expression genes. Plant Physiology 141, 1694–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan WH, Wang P, Chen HX, et al. . 2011. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Molecular Plant 4, 319–330. [DOI] [PubMed] [Google Scholar]
- Yang W, Lu Z, Xiong Y, Yao J. 2017. Genome-wide identification and co-expression network analysis of the OsNF-Y gene family in rice. Crop Journal 5, 21–31. [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A. 2010. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biology 11, R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Cheng Z, Zhang X, Guo X, Su N, Jiang L, Mao L, Wan J. 2011. Double repression of soluble starch synthase genes SSIIa and SSIIIa in rice (Oryza sativa L.) uncovers interactive effects on the physicochemical properties of starch. Genome 54, 448–459. [DOI] [PubMed] [Google Scholar]
- Zhang JJ, Xue HW. 2013. OsLEC1/OsHAP3E participates in the determination of meristem identity in both vegetative and reproductive developments of rice. Journal of Integrative Plant Biology 55, 232–249. [DOI] [PubMed] [Google Scholar]
- Zhang L, Ren Y, Lu B, et al. . 2016. FLOURY ENDOSPERM7 encodes a regulator of starch synthesis and amyloplast development essential for peripheral endosperm development in rice. Journal of Experimental Botany 67, 633–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, et al. . 2008. Model-based analysis of ChIP-seq (MACS). Genome Biology 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Wang Z. 2014. Protein accumulation in aleurone cells, sub-aleurone cells and the center starch endosperm of cereals. Plant Cell Reports 33, 1607–1615. [DOI] [PubMed] [Google Scholar]
- Zhou SR, Yin LL, Xue HW. 2013. Functional genomics based understanding of rice endosperm development. Current Opinion in Plant Biology 16, 236–246. [DOI] [PubMed] [Google Scholar]
- Zhou W, Wang X, Zhou D, Ouyang Y, Yao J. 2017. Overexpression of the 16-kDa α-amylase/trypsin inhibitor RAG2 improves grain yield and quality of rice. Plant Biotechnology Journal 15, 568–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong W, Tang N, Yang J, Peng L, Ma S, Xu Y, Li G, Xiong L. 2016. Feedback regulation of ABA signaling and biosynthesis by a bZIP transcription factor targets drought-resistance-related genes. Plant Physiology 171, 2810–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Li J. 2014. Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annual Review of Genetics 48, 99–118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.