A high-throughput, convenient, reliable, and cost-efficient sequencing method to identify transgene insertion positions in the genome is described.
Keywords: Complex genome, cost efficient, high throughput, mapping insertion position, MinNON sequencing, transgene
Abstract
Transgenic technology was developed to introduce transgenes into various organisms to validate gene function and add genetic variations >40 years ago. However, the identification of the transgene insertion position is still challenging in organisms with complex genomes. Here, we report a nanopore-based method to map the insertion position of a Ds transposable element originating in maize in the soybean genome. In this method, an oligo probe is used to capture the DNA fragments containing the Ds element from pooled DNA samples of transgenic soybean plants. The Ds element-enriched DNAs are then sequenced using the MinION-based platform of Nanopore. This method allowed us to rapidly map the Ds insertion positions in 51 transgenic soybean lines through a single sequencing run. This strategy is high throughput, convenient, reliable, and cost-efficient. The transgenic allele mapping protocol can be easily translated to other eukaryotes with complex genomes.
Introduction
Transgenic technologies that introduce genetic variation into bacteria, animals, and plants were developed in 1972 (Cohen et al., 1972), 1974 (Jaenisch and Mintz, 1974), and 1982 (Barton et al., 1983), respectively. Since then, transgenic technologies have become great tools to improve the traits of various organisms and to analyze gene functions. In higher plants, a single copy or multiple copies of transgenes are randomly inserted into the genome (Weising et al., 1988; Kim et al., 2007). Expression levels of a transgene are often influenced by the genomic context surrounding the transgenic allele and the complexity of the genome (Weising et al., 1988; Day et al., 2000; van Leeuwen et al., 2001; Butaye et al., 2004). Moreover, the transgene insertion position may also affect the function of surrounding genes (Weising et al., 1988; Azpiroz-Leehan and Feldmann, 1997). Importantly, prior knowledge of the insertion position of a transgene is beneficial when breeding programs begin to introgress the allele into other varieties. Consequently, there is a need to characterize transgenic alleles in higher plants efficiently and accurately.
Strategies have been developed for mapping of transgenic alleles (Lepage et al., 2013; Guo et al., 2016). The complexity of the transgenic locus can be estimated through multiple approaches including Southern blot analysis (Southern, 1975), quantitative PCR (qPCR; Ingham et al., 2001), and droplet PCR (Glowacka et al., 2016). One of the first methods used to successfully map a transgenic allele in higher plants was plasmid rescue. This strategy involves restriction enzyme digestion of the host genome containing the transgenic allele, cloning the cleavage products into a plasmid, and selection of the plasmid containing the transgene fragment (Nan and Walbot, 2009). Subsequent methods for mapping transgenic alleles are primarily PCR based, include thermal asymmetric interlaced PCR (TAIL-PCR) (Liu et al., 1995; Liu and Chen, 2007), adaptor PCR which is sometimes referred to as anchored PCR (Singer and Burke, 2003; Thole et al., 2009), and T-linker PCR that utilizes a specific T/A ligation (Yuanxin et al., 2003). However, these methods are often challenging to scale-up for high throughput (Guo et al., 2016; Ji and Braam, 2010). Moreover, failure to map transgenes can happen due to the complexity of the transgenic locus and/or issues associated with the genomic context of the transgenic allele (Wahler et al., 2013). Next-generation Illumina sequencing technology can be used to map transgenic alleles in plants due to its depth of sequencing capacity (Polko et al., 2012; Lepage et al., 2013; Guo et al., 2016). However, because this method produces short reads, a high degree of sequencing depth is needed, especially in crops that have large genomes that are rich in repetitive sequence. This, in turn, impacts the cost per transgenic locus mapped. In addition, short-read sequencing data are challenging to resolve transgene insertion position in many plant species, such as soybean, due to issues related to genome rearrangements and copy number variations, which may lead to inaccurate mapping locations.
Recently, single molecule real-time (SMRT) sequencing technologies have been developed to provide long-read sequencing data sets. These SMRT platforms developed by Pacific Biosciences (PacBio®) and Oxford Nanopore Technologies® (ONT) offer significant attributes for genotyping plant species. The most significant benefit is read lengths, with the PacBio® platform generating up to 60 kb reads, and Nanopore® reads being up to ~1 Mb (Lu et al., 2016; Jain et al., 2018). Both technologies have been used in genome assembly (Rhoads and Au, 2015; Badouin et al., 2017; Schmidt et al., 2017; Jain et al., 2018; Michael et al., 2018). The MinION device, which was developed by ONT and entered the market in 2014, is a portable apparatus of <100 g in weight. Furthermore, it is compatible with a PC or laptop with USB 3.0 ports (Jain et al., 2016) giving it a flexibility attribute permitting use outside of a laboratory setting (Castro-Wallace et al., 2017). In addition, compared with PacBio®, the Nanopore® technology apparatus is affordable in most laboratories. Thus, the MinION platform provides potential for a high-throughput, cost-effective strategy to map transgenic alleles in plant species with complex genomes.
Described herein is a MinION-based pipeline designed for high-throughput mapping of transgenic alleles in plant species. Through employing a target enrichment approach using an oligo probe to capture the transgene-containing DNA fragments, this method was able to rapidly map the transgene insertion positions in 51 transgenic soybean plants in a single 1D sequencing run. The cost is estimated to be US$1360 for mapping all 51 events, and the results were generated within 1 week. Moreover, the cost per sample can be further reduced because the numbers of transgenic plants in each sequencing run can be increased. These results demonstrate that this Nanopore®-based sequencing method is rapid, convenient, reliable, cost efficient, and high throughput.
Materials and methods
Soybean growth condition
The soybean plants were grown in controlled greenhouse conditions with a 14 h photoperiod and 28/26 °C day/night temperature. The soybean plants harboring the Ds element are in the Thorne genetic background.
DNA extraction and shearing
DNAs of soybean leaves were extracted using the cetyltrimethylammonium bromide (CTAB) method (Healey et al., 2014) and purified with the DNeasy Plant Mini Kit (69104, QIAGEN). A 6 μg aliquot of genomic DNA in a total of 150 μl of nuclease-free water was sheared into ~8 kb fragments with g-TUBEs (520079, Covaris) by following the manufacturer’s instructions.
DNA barcodes and enrichment of the Ds element-containing fragments
A 1 μg aliquot of sheared DNA fragments was end-repaired with Ultra II End-prep enzyme mix (E7546L, NEB) for 5 min at 20 °C and 5 min at 65 °C using a thermal cycler, followed by purification with AMPure XP beads in a 1.5 ml DNA LoBind Eppendorf tube. After end-repair, DNA fragments were ligated to the Barcode Adapter from the barcode Kit 1D (EXP-PBC001, Nanopore) using Blunt/TA Ligase Master Mix (M0367L, NEB). Following purification with AMPure XP beads, the DNAs were ligated to the Barcode (EXP-PBC001, Nanopore) using LongAmp Taq (M0287S, NEB). The barcoded DNA library was then purified with AMPure XP beads. After barcoding, the library was purified by extraction with Tris-buffered phenolchloroform pH 8. Then, the enrichment of Ds element-containing DNA fragments was performed in a hybridization reaction containing 4.8 μl of H2O, 8.5 μl of xGen 2× hybridization buffer, 2.7 μl of xGen hybridization enhancer (1072281, Integrated DNA Technologies, IDT), and 1 μl of probe. The hybridization was performed at 65 °C for 4 h in a thermal cycler. After hybridization, the targets were captured by the Dynabeads M-270 streptavidin beads (65-305, Thermo Fisher Scientific) that recognize the dual-biotinylated probe. After washing with stringent wash buffer and wash buffer I, II, and III by following the manufacture’s protocol, the captured target fragments were amplified for 12 cycles with primers recognizing the barcode using LongAmp Taq under the PCR conditions: 15 s at 98 °C, 30 s at 60 °C, and 6 min at 72 °C. The resulting PCR products were purified with AMPure XP beads, which were subjected to second-round enrichment (steps 3 and 4), or library construction following the manufacturer’s instruction. The 5'-dual-biotinylated probe was synthesized by IDT and its sequence is shown in Supplementary Table S1 at JXB online).
Library construction and sequencing
Following target enrichment, barcoded libraries were pooled and 1 μg samples were end-repaired with the Ultra II End-prep enzyme, purified with the AMPure XP beads, and then ligated to the sequencing adaptor (SQK-LSK108, Nanopore) with the Blunt/TA Ligation Master Mix. After purification with the AMPure beads, the adapted DNA libraries were sequenced in the flow cells (R9.4 version, FLC-MIN106, Nanopore). After 20–24 h, the sequencing was stopped.
Assessment of target enrichment efficiency
To assess the target enrichment, 2% of samples were used as templates to perform qPCR using SYBR Green PCR Master Mix (Bio-Rad) with primers recognizing the Ds element or an unrelated intergenic region in soybean chromosome 7. The primer sequences are shown in Supplementary Table S1.
PCR validation
PCR was performed with the primers listed in Supplementary Table S1 using the condition: 95 °C 2 min; 95 °C 30 s, 50 °C 30 s, 72 °C 1 h 20 min for 34 cycles; and 72 °C 5 min. The PCR products were isolated on a 1% agarose gel and visualized by ethidium bromide staining.
Bioinformatics analysis
All barcoded reads were de-multiplexed and adaptors were trimmed off using Porechop version 0.2.1 (https://github.com/rrwick/Porechop) with default parameters. To identify reads with the Ds target sequence, the Ds target sequence was searched against trimmed reads for each sample with E-values ≤10–3. For all hits with the Ds target sequence, the 5' end and 3' end sequences of the Ds target sequence were scanned on each read to identify long reads with one or two complete ends of the Ds target sequence. Sequences on 5' end and/or 3' end sequences of long reads beyond the Ds target sequence, if of length >20 bp, were recorded as flanking sequences, which come from the soybean genome. The flanking sequences underwent blast searches against the soybean genome (v1.0). Uniquely aligned hits with aligned length >200 bp and >80% sequence identity were kept. The genomic location for each flanking sequence was determined based on its alignment. The insertion sites were determined based on statistically enriched flanking sequences. The zero-inflated Poisson regression was used to model count data that have an excess of zero counts. All read counts were fitted into the zero-inflated Poisson regression model with the R package, ZIM. For each peak of read counts, to determine if it was a significant peak, a P-value was calculated as the probability of observing a count value equally as extreme as, or more extreme than, the given read count based on the fitted distribution.
Results
Mapping of maize Ds transpositions in the soybean genome through MinION sequencing without target enrichment
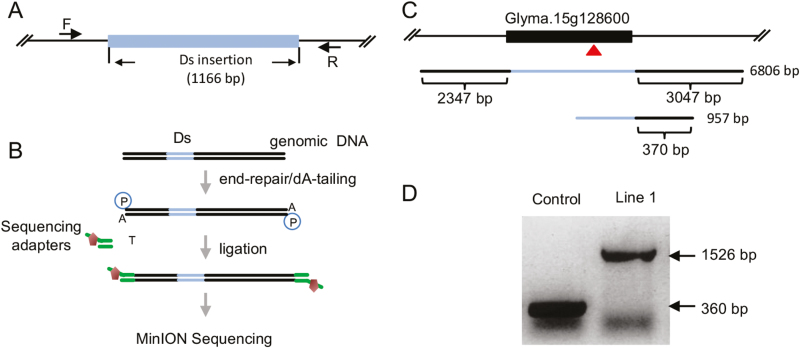
To evaluate the potential application of MinION sequencing to map transgenic alleles, soybean lines, which contain a transgene stack harboring the maize Activator (Ac)/Dissociation (Ds) transposon system, were used. The Ac transposase is controlled by the Cauliflower mosaic virus (CaMV) 35S promoter, and the Ds element harbors the Cassava vein mosaic virus promoter (CsVMV) as an activation tag. The selected soybean lines were previously genotyped via Southern blot analysis to ascertain the presence of the Ds loci and the absence of the Ac allele, along with mapping of the Ds allele using TAIL-PCR (Fig. 1A; Supplementary Fig. S1). To assess the power of MinION sequencing to map transgenic alleles, genomic DNA isolated from one of the selected genotyped soybean lines carrying the Ds activation tag was sequenced on the FLO-MIN106 flow cell following the 1D sequencing protocol without DNA fragmentation (Fig. 1B). A 24 h sequencing run produced ~1 million reads, resulting in ~2.8 Gb of sequence data (Table 1). Mining the sequence data for the Ds element revealed two reads containing the Ds element (Table 1). One read was 957 bp covering a partial Ds element flanked by a 370 bp sequence at the 3' end, and the other was 6806 bp, containing the full-length Ds element flanked by 2347 bp of 5' upstream sequence and 3047 bp of downstream sequence flanking the Ds sequence (Fig. 1C). The identified Ds junction fragment sequences were mapped to the soybean Glyma.15g128600 gene (Fig. 1C), in agreement with the TAIL-PCR results. To further validate the sequencing and TAIL-PCR outcomes, PCRs were carried out with a primer set designed to span the Ds/junction around the insertion site (Fig. 1A). The data revealed a 360 bp PCR product amplified from the endogenous Glyma.15g128600 gene when control DNAs were used as templates, and a 1526 bp fragment predicted to carry the Ds/junction target sequence amplified from DNAs of the transgenic soybean plants (Fig. 1D). These results demonstrate the potential of MinION sequencing to map a transgenic allele in the soybean genome. However, given the few reads that contain the Ds, refinement in the genomic DNA processing steps would be required for a high-throughput/cost-effective mapping pipeline with this technology.
Fig. 1.
MinION sequencing without Ds enrichment. (A) A schematic diagram of Ds insertion in the soybean genome. The length of the Ds insertion is 1166 bp. The positions of forward (F) and reverse (R) primers used for PCR genotyping are shown. (B) Workflow of direct genome sequencing without target enrichment. Genomic DNA was end-repaired and dA-tailed, ligated with sequencing adaptors, and sequenced on the FLO-MIN106 flow cell. (C) A schematic diagram of the Ds insertion in the Glyma.15g128600 gene. Two reads are shown. The first one covers 2347 bp in the 5'-flanking region and 3047 bp in the 3'-flanking region. The second one contains 370 bp flanking sequence in the 3' region. (D) PCR validation of the Ds insertion in Line 1. Thorne was used as control plant. The length of the DNA fragment without the Ds element in control plant is 360 bp, while the fragment length from Ds-containing Line 1 is 1526 bp.
Table 1.
Sequencing result of one line without enrichment
| Total read number | Longest read (bp) | Target read number | Percentage of target reads | Longest read with targets (bp) | |
|---|---|---|---|---|---|
| Line 1 | 1 061 117 | 351 899 | 2 | 0.00019 | 6806 |
Longest read indicates the longest read in all readings.
Target enrichment of the transgenic allele to improve mapping throughput with MinION sequencing
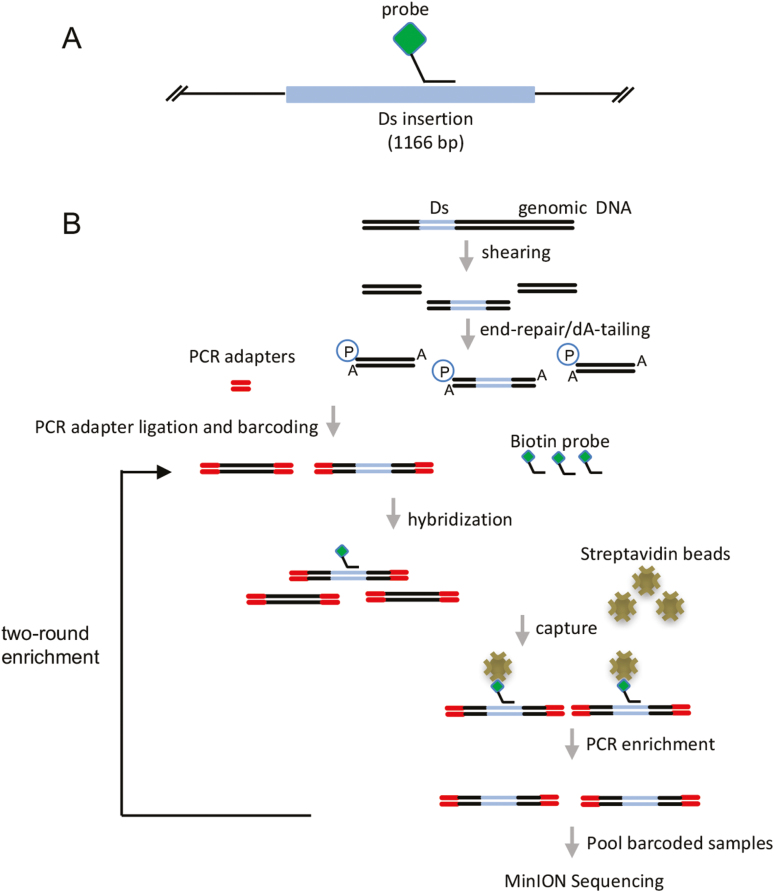
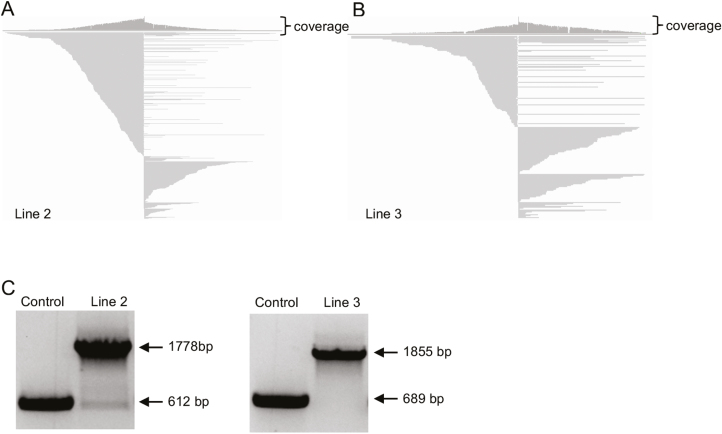
To improve read counts around the junction of a transgenic allele, a PCR-based method to enrich the target sequences in the DNA library (Fig. 2) was developed. To test the enrichment protocol, DNAs from two soybean lines carrying a Ds activation tag allele, previously characterized via Southern blot and mapped by TAIL-PCR, were used. The enrichment protocol incorporated steps for fragmentation of DNA to ~8 kb, end-repairing and dT tailing, with subsequent ligation to barcode adaptors and PCR barcoding (Fig. 2). The resultant reaction products were subjected to a 120 nucleotide 5'-dual-biotinylated probe designed to capture the transgenic Ds allele (Fig. 2). Following the probe capture step, the probe-captured fraction was re-amplified by PCR and products were pooled for sequencing (Fig. 2). Total reads obtained were 357 765 and 326 189 for Lines 2 and 3, respectively (Table 2). The average read length of Line 2 was 2426 bp, with the longest read of 20 453 bp (Table 2), while the average read length of Line 3 was 2445 bp, with the longest read of 48 971 bp (Table 2). Among the reads obtained implementing the enrichment steps, 203 and 438 contained the Ds allele sequence, for Lines 2 and 3, respectively, which correctly mapped to gene calls, Glyma.19g105100 and Glyma.11g247400, respectively (Fig. 3A, B; Table 2). The map positions were re-confirmed using PCR analyses incorporating a primer set designed to amplify the Ds/junction fragment region (Fig. 3C).
Fig. 2.
The workflow of the enrichment of Ds-containing fragments in DNA libraries. (A) Schematic diagram of the oligo probe used to capture the Ds element. The probe is dual biotinylated at the 5' end (diamond). (B) The workflow of sequencing the enriched Ds-containing DNA fragments. Genomic DNA was sheared and ligated to PCR barcode adaptors. The Ds-containing fragments were enriched for one or two rounds. The enriched fragments were pooled and sequenced.
Table 2.
Sequencing result of two lines with one-round enrichment
| Total read number | Longest read (bp) | Target read number | Percentage of target reads | Longest read with targets (bp) | |
|---|---|---|---|---|---|
| Line 2 | 357 765 | 20 453 | 203 | 0.057 | 6524 |
| Line 3 | 326 189 | 48 971 | 438 | 0.134 | 6725 |
Longest reads indicate the longest reads in the individual barcoded lines.
Fig. 3.
Sequencing results after one-round enrichment of the Ds-containing fragments. (A and B) Schematic diagram of the flanking sequences of Line 2 (A) and Line 3 (B). Partial sequences of reads are shown. (C) PCR validation of the Ds insertion in Lines 2 and 3. Thorne was used as the control plant. The lengths of the DNA fragment without the Ds element are 612 bp for Line 2 and 689 bp for Line 3. With the Ds elements, the lengths of the DNA fragments are 1778 bp for Line 2 and 1855 bp for Line 3.
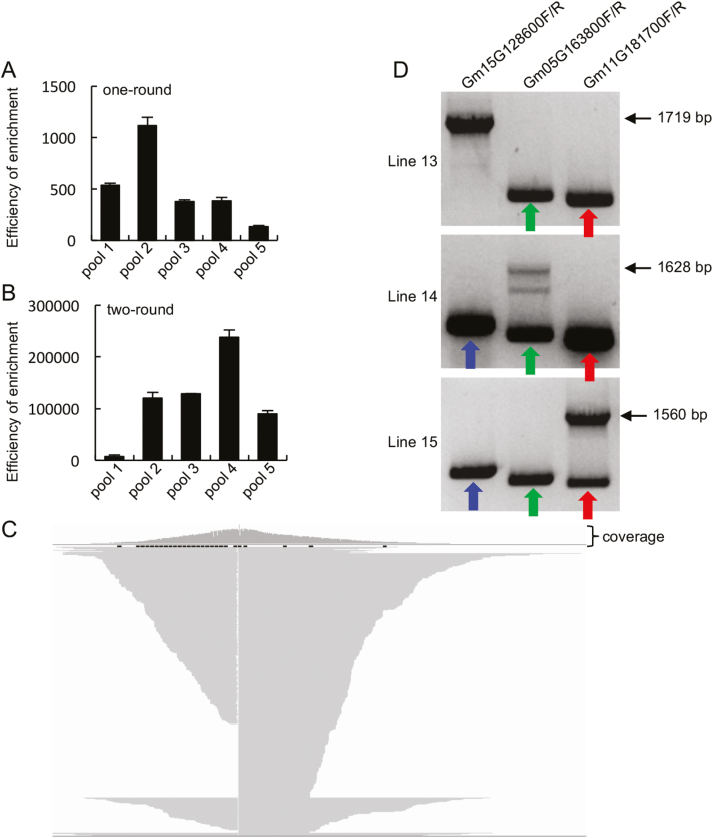
Given the high number of reads containing the Ds element, following the targeted enrichment approach, the method appeared to be amenable for higher throughput by increasing the sample pool size. To this end, 15 soybean lines previously ascertained to harbor a single Ds element (Lines 4–18) were selected for integrating a pooling strategy with the targeted enrichment method. Here five DNA pools, each of which contained DNAs from three soybean lines (Table 3; Supplementary Fig. S2), were prepared. Following the first target enrichment step, the pools were subjected to an additional round of purification to increase coverage of the Ds-containing DNA fragments (Fig. 2). Subsequent to each purification step, qPCR was used to estimate the relative enrichment level of the target fragment compared with an unrelated DNA region that served as an internal control. After one round of enrichment, the ratio of target fragments to the unrelated region was enriched 132–1120× across all DNA pools (Fig. 4A). Following two rounds of purification, the enrichment ratio ranged from 7469 to 238 193 times in the pools (Fig. 4B). MinION sequencing of the double enriched products resulted in a total number of reads ranging from 117 266 to 523 192 across the pools (Table 3), with reads containing the Ds sequence ranging from 1856 to 36 388 in the pools (Table 3). These results were translated to ratios of reads containing the target sequence per total read counts for each pool in the range of 0.53–6.95% (Table 3). The average length of these Ds-containing reads was longer than 2 kb and the majority (>99%) of these reads were longer than 1.2 kb (Supplementary Fig. S3). The Ds-containing reads of each DNA pool were successfully mapped to three positions in the soybean genome (Supplementary Table S2; Fig. 4C showing a position of reads at the soybean genome from pool 4), reflecting that the pools each contained three independently integrated Ds elements within the soybean genome. The predicted mapped locations identified in pool 4 were subsequently verified by PCR using primer sets that spanned the Ds element/soybean genome junction (Lines 13–15; Fig. 4D).
Table 3.
Sequencing result of the 15 sample pools
| Line number | Total read number | Longest read (bp) | Target read number | Percentage of target reads | Longest read with targets (bp) | |
|---|---|---|---|---|---|---|
| DNA pool 1 | Lines 4–6 | 351 722 | 16 352 | 1856 | 0.53 | 5100 |
| DNA pool 2 | Lines 7–9 | 490 852 | 21 457 | 30 937 | 6.30 | 9317 |
| DNA pool 3 | Lines 10–12 | 117 266 | 8000 | 3165 | 2.70 | 5770 |
| DNA pool 4 | Lines 13–15 | 523 192 | 25 983 | 36 388 | 6.95 | 13 213 |
| DNA pool 5 | Lines 16–18 | 234 809 | 14 215 | 5412 | 2.30 | 6008 |
Line number indicates the pooled Ds-containing lines.
Longest reads indicate the longest reads in each pool.
Fig. 4.
Sequencing results after two-round enrichment of the Ds-containing fragments. (A and B) Efficiency of one-round (A) and two-round (B) enrichment of the Ds element-containing fragments; 2% of samples before and after probe enriching were used to perform qPCR. The amount of target fragments was normalized to that of the internal control. (C) Schematic diagram of the flanking sequences of Line 15. Partial sequences of reads are shown. (D) PCR validation of the Ds insertion in Lines 13, 14, and 15. The three individual lines were examined with three pairs of primers each. Each primer pair (labeled above the picture) recognizes a potential insertion position of the Ds element, identified by sequencing. Line 13 containing a Ds insertion in the Glyma15G128600 gene produced a 1719 bp fragment, while Lines 14 and 15 without insertions in this gene generated ~559 bp fragments (indicated with arrows). Line 14 containing a Ds insertion in the Glyma05G163800 gene produced a 1628 bp fragment, while Lines 13 and 15 without insertions in this gene generated 462 bp fragments (indicated with arrows). Line 15 containing a Ds insertion in the Glyma11G181700 gene produced a 1560 bp fragment, while Lines 13 and 14 without insertions in this gene generated 394 bp fragments (indicated with arrows).
MinION sequencing provides a platform for a high-throughput method to identify the map position of transgenic alleles in plants
The numbers of reads containing the Ds element averaged in the hundreds (Table 3), suggesting that MinION sequencing technology is a cost-effective tool that could be translated to a high-throughput method to map a transgenic allele in the soybean genome. To test its throughput further, an expanded pooling was performed with the enrichment steps, wherein 51 independent soybean lines containing a single Ds element were divided into six pools, each of which contained 8–10 lines (Table 4), for MinION sequencing. This expanded throughput evaluation resulted in total read counts ranging from 19 758 to 282 690 across the pools, with reads containing the Ds sequence ranging from 212 to 16 146 (Table 4). These data were sufficient to successfully map the transgenic allele in each of the 51 soybean lines analyzed (Supplementary Table S3).
Table 4.
Sequencing result of the 51 sample pools
| Line number | Total read number | Longest read (bp) | Target read number | Percentae of target reads | Longest read with targets (bp) | |
|---|---|---|---|---|---|---|
| DNA pool 1 | Lines 19–26 | 20 317 | 8512 | 2104 | 10.36 | 6096 |
| DNA pool 2 | Lines 27–34 | 47 257 | 9995 | 212 | 0.45 | 6042 |
| DNA pool 3 | Lines 35–42 | 19 758 | 6953 | 1569 | 7.94 | 5476 |
| DNA pool 4 | Lines 43–50 | 181 763 | 10 577 | 14 485 | 7.97 | 8178 |
| DNA pool 5 | Lines 51–59 | 63 227 | 10 137 | 5698 | 9.01 | 7007 |
| DNA pool 6 | Lines 60–69 | 282 690 | 10 397 | 16 146 | 5.71 | 8691 |
Line number indicates the pooled Ds-containing lines.
Longest reads indicate the longest reads in each pool.
To further validate whether this method is suitable to map potential multiple transgene insertions, we selected 18 transgenic soybean lines harboring 1–3 copies of the original Ds transgene (~5 kb), which were determined by Southern blot (Supplementary Fig. S4; Supplementary Table S4). We divided these plants into four pools, and performed MinION sequencing after target enrichment. We were able to identify 29 transgenic insertion loci (Supplementary Table S4), agreeing with the Southern blot result. This result suggests that our method can be used to map transgene insertions pooled from transgenic plants with multiple transgene insertions.
Discussion
Here, we present a Nanopore®-based method to map transgene insertion positions in the soybean genome. This method has at least five advantages. First, it provides reliable information on sequences flanking the insertion position. In most scenarios, >100 reads contain the target transgenic allele and associated junction sequences. Secondly, the method is scalable. We currently are able to determine the insertion position for 51 samples in a single sequencing event with this method. Sample numbers can be further increased since the Ds-containing reads are much more than needed for determining the Ds insertion. Moreover, the target enrichment method still has potential for additional refinement given that the ratios of reads containing the target sequences per total read count are still low (ranging from ~0.5% to 10%). The current enrichment step only incorporates one probe to the target allele. A refinement in the enrichment step might include the use of multiple probes that recognize different regions of the target allele, thereby improving specificity and efficiency of capture. Thirdly, the cost per mapped event is extremely low, estimated at US$1360 per 51 samples, excluding labor. If sample numbers per single sequencing run can be increased, the cost can be further reduced. In addition, after one-round purification, we may also use primers that recognize the target and adaptor to amplify the target-containing fragments, which will eliminate second-round purification and improve specificity, and thereby further reduce the cost and allow pooling of more samples. Fourthly, it is rapid, with the time frame from DNA fragmentation to mapped transgenic allele being ~1 week. Lastly, it is convenient and has broad usability, given the fact that MinION is a portable device that can run on a laptop or computer, which allows most laboratories to use it.
The introduction of novel genetic variation into higher plants offers a powerful way to complement plant breeding programs. Prior knowledge of transgene insertion position facilitates breeding decisions. The MinION-based sequencing strategy outlined here is a powerful, high-throughput tool to determine the insertion position of transgenic alleles in higher plants. The average length of reads containing the Ds element here was ~2.1 kb and that of the longest reads was ~10 kb in the 51 sample sequencing. This length should be sufficient to cover a portion of a longer transgene with flanking sequencing at one end. Indeed, we used this method to determine a population of soybean lines containing an ~5 kb transgene. The average numbers of reads containing the Ds elements are >100, which should be sufficient to identify multiple insertion events in the genome. However, it may still be a challenge to identify transgene copy numbers with the current target enrichment method when multiple copies of the transgene exist in the same location on the genome. In this scenario, the read length needs to be improved. A possible solution is to perform size selection after each round of target enrichment or after adaptor addition to eliminate the short DNA fragments, and thereby to improve the read length, although this may reduce the numbers of reads containing the transgene. We also noticed variations of reading within a barcode. This may due to the difference of DNAs surrounding the insertion positions, which results in variations in efficiency of ligation or PCR. Moreover, there are variations in the numbers of Ds-containing reads among different pools. This is likely to be due to the fact that different barcodes may have different optimal PCR conditions and the same PCR amplification condition was used in the study. Recently, MinION-based de novo genome assembly was used to identify both homozygous and hemizygous T-DNA insertions in the Arabidopsis genome (Jupe et al., 2019). However, the method described here cannot determine whether a transgenic insertion is in a hemizyous or homozygous state, since the target enrichment step will eliminate DNA fragments that do not contain the transgene insertion.
Although this method was optimized using a population of soybean lines containing the same transgene, it can be adapted to map the transgene insertions in any other organisms. Moreover, this method can also be used to map transgenes from transgenic lines containing different transgenes that do not share common fragments using probes targeting individual transgenes. Thus, we expect that this method will have broad applications.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Diagram of the Ds system.
Fig. S2. Agarose gel electrophoresis of sheared DNAs.
Fig. S3. Size distribution of readings containing the Ds elements.
Fig. S4. Copy numbers of the transgenes in various soybean transgenic lines determined by Southern blot.
Table S1. Oligo DNAs used in this study.
Table S2. Positions of Ds insertions identified in the 15 sample sequencing.
Table S3. Positions of Ds insertions identified in the 51 sample sequencing.
Table S4. Sequencing result of T-DNA insertion lines.
Acknowledgements
This work was supported by the National Institute of Health (GM127414 to BY), the National Science Foundation (awards OIA-1557417 to BY, CZ, EC, and TC, IOS-1444581 to TC, and MCB-1808182 to BY), the Nebraska Soybean Board (to BY, CZ, EC, and TC), the Agricultural Research Division of University of Nebraska (to DH), National Natural Science Foundation of China (31872816 to SL), and Taishan Scholars (tsqn201812114 to S.L). The authors declare no conflict of interest.
References
- Azpiroz-Leehan R, Feldmann KA. 1997. T-DNA insertion mutagenesis in Arabidopsis: going back and forth. Trends in Genetics 13, 152–156. [DOI] [PubMed] [Google Scholar]
- Badouin H, Gouzy J, Grassa CJ, et al. 2017. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 546, 148–152. [DOI] [PubMed] [Google Scholar]
- Barton KA, Binns AN, Matzke AJ, Chilton MD. 1983. Regeneration of intact tobacco plants containing full length copies of genetically engineered T-DNA, and transmission of T-DNA to R1 progeny. Cell 32, 1033–1043. [DOI] [PubMed] [Google Scholar]
- Butaye KM, Goderis IJ, Wouters PF, Pues JM, Delauré SL, Broekaert WF, Depicker A, Cammue BP, De Bolle MF. 2004. Stable high-level transgene expression in Arabidopsis thaliana using gene silencing mutants and matrix attachment regions. The Plant Journal 39, 440–449. [DOI] [PubMed] [Google Scholar]
- Castro-Wallace SL, Chiu CY, John KK, et al. 2017. Nanopore DNA sequencing and genome assembly on the international space station. Scientific Reports 7, 18022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SN, Chang AC, Hsu L. 1972. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proceedings of the National Academy of Sciences, USA 69, 2110–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day CD, Lee E, Kobayashi J, Holappa LD, Albert H, Ow DW. 2000. Transgene integration into the same chromosome location can produce alleles that express at a predictable level, or alleles that are differentially silenced. Genes & Development 14, 2869–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Głowacka K, Kromdijk J, Leonelli L, Niyogi KK, Clemente TE, Long SP. 2016. An evaluation of new and established methods to determine T-DNA copy number and homozygosity in transgenic plants. Plant, Cell & Environment 39, 908–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Guo Y, Hong H, Qiu LJ. 2016. Identification of genomic insertion and flanking sequence of G2-EPSPS and GAT transgenes in soybean using whole genome sequencing method. Frontiers in Plant Science 7, 1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healey A, Furtado A, Cooper T, Henry RJ. 2014. Protocol: a simple method for extracting next-generation sequencing quality genomic DNA from recalcitrant plant species. Plant Methods 10, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham DJ, Beer S, Money S, Hansen G. 2001. Quantitative real-time PCR assay for determining transgene copy number in transformed plants. Biotechniques 31, 132–140. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Mintz B. 1974. Simian virus 40 DNA sequences in DNA of healthy adult mice derived from preimplantation blastocysts injected with viral DNA. Proceedings of the National Academy of Sciences, USA 71, 1250–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Koren S, Miga KH, et al. 2018. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nature Biotechnology 36, 338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Olsen HE, Paten B, Akeson M. 2016. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biology 17, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J, Braam J. 2010. Restriction site extension PCR: a novel method for high-throughput characterization of tagged DNA fragments and genome walking. PLoS One 5, e10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jupe F, Rivkin AC, Michael TP, et al. 2019. The complex architecture and epigenomic impact of plant T-DNA insertions. PLoS Genetics 15, e1007819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IH, Nagel J, Otten S, Knerr B, Eils R, Rohr K, Dietzel S. 2007. Quantitative comparison of DNA detection by GFP-lac repressor tagging, fluorescence in situ hybridization and immunostaining. BMC Biotechnology 7, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepage E, Zampini E, Boyle B, Brisson N. 2013. Time- and cost-efficient identification of T-DNA insertion sites through targeted genomic sequencing. PLoS One 8, e70912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Chen Y. 2007. High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques 43, 649–650, 652, 654 passim. [DOI] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF. 1995. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. The Plant Journal 8, 457–463. [DOI] [PubMed] [Google Scholar]
- Lu H, Giordano F, Ning Z. 2016. Oxford nanopore MinION sequencing and genome assembly. Genomics, Proteomics & Bioinformatics 14, 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Jupe F, Bemm F, Motley ST, Sandoval JP, Lanz C, Loudet O, Weigel D, Ecker JR. 2018. High contiguity Arabidopsis thaliana genome assembly with a single nanopore flow cell. Nature Communications 9, 541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan GL, Walbot V. 2009. Plasmid rescue: recovery of flanking genomic sequences from transgenic transposon insertion sites. Methods in Molecular Biology 526, 101–109. [DOI] [PubMed] [Google Scholar]
- Polko JK, Temanni MR, van Zanten M, van Workum W, Iburg S, Pierik R, Voesenek LA, Peeters AJ. 2012. Illumina sequencing technology as a method of identifying T-DNA insertion loci in activation-tagged Arabidopsis thaliana plants. Molecular Plant 5, 948–950. [DOI] [PubMed] [Google Scholar]
- Rhoads A, Au KF. 2015. PacBio sequencing and its applications. Genomics, Proteomics & Bioinformatics 13, 278–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MH, Vogel A, Denton AK, et al. 2017. De novo assembly of a new Solanum pennellii accession using nanopore sequencing. The Plant Cell 29, 2336–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer T, Burke E. 2003. High-throughput TAIL-PCR as a tool to identify DNA flanking insertions. Methods in Molecular Biology 236, 241–272. [DOI] [PubMed] [Google Scholar]
- Southern EM. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. Journal of Molecular Biology 98, 503–517. [DOI] [PubMed] [Google Scholar]
- Thole V, Alves SC, Worland B, Bevan MW, Vain P. 2009. A protocol for efficiently retrieving and characterizing flanking sequence tags (FSTs) in Brachypodium distachyon T-DNA insertional mutants. Nature Protocols 4, 650–661. [DOI] [PubMed] [Google Scholar]
- van Leeuwen W, Ruttink T, Borst-Vrenssen AW, van der Plas LH, van der Krol AR. 2001. Characterization of position-induced spatial and temporal regulation of transgene promoter activity in plants. Journal of Experimental Botany 52, 949–959. [DOI] [PubMed] [Google Scholar]
- Wahler D, Schauser L, Bendiek J, Grohmann L. 2013. Next-generation sequencing as a tool for detailed molecular characterisation of genomic insertions and flanking regions in genetically modified plants: a pilot study using a rice event unauthorised in the EU. Food Analytical Methods 6, 1718–1727. [Google Scholar]
- Weising K, Schell J, Kahl G. 1988. Foreign genes in plants: transfer, structure, expression, and applications. Annual Review of Genetics 22, 421–477. [DOI] [PubMed] [Google Scholar]
- Yuanxin Y, Chengcai A, Li L, Jiayu G, Guihong T, Zhangliang C. 2003. T-linker-specific ligation PCR (T-linker PCR): an advanced PCR technique for chromosome walking or for isolation of tagged DNA ends. Nucleic Acids Research 31, e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.