Abstract
Early parental loss is associated with social–emotional dysregulation and amygdala physiologic changes. Previously, we examined whole amygdala gene expression in infant monkeys exposed to early maternal deprivation. Here, we focus on an amygdala region with immature neurons at birth: the paralaminar nucleus (PL). We hypothesized that 1) the normal infant PL is enriched in a subset of neural maturation (NM) genes compared to a nearby amygdala subregion; and 2) maternal deprivation would downregulate expression of NM transcripts (mRNA). mRNAs for bcl2, doublecortin, neuroD1, and tbr1—genes expressed in post-mitotic neurons—were enriched in the normal PL. Maternal deprivation at either 1 week or 1 month of age resulted in PL-specific downregulation of tbr1—a transcription factor necessary for directing neuroblasts to a glutamatergic phenotype. tbr1 expression also correlated with typical social behaviors. We conclude that maternal deprivation influences glutamatergic neuronal development in the PL, possibly influencing circuits mediating social learning.
Keywords: ganglionic eminence, social behavior, tbr1, bcl2, dcx, neuroD1
1 |. INTRODUCTION
Inadequate caregiving has a lasting effect on social and emotional behaviors in primate species (Harlow & Zimmermann, 1959; Spitz, 1946). Young children placed in institutionalized orphanages are a stark example of this phenomenon (Bruce, Tarullo, & Gunnar, 2009; Sheridan, Fox, Zeanah, McLaughlin, & Nelson, 2012; Tottenham et al., 2010). Follow-up studies on several populations of these children show long-lasting behavioral changes, including difficulties with emotional regulation and increased anxiety (Bruce et al., 2009; Chisholm, 1998; Zeanah et al., 2009). In monkeys, maternal deprivation also results in behavioral changes that can be depressive or anxious in nature, and are dependent on the age at which maternal absence begins, similar to children with a history of institutionalized care (Cameron, 2001; Harlow & Zimmermann, 1959; Suomi, Collins, & Harlow, 1973).
Decreased sensory inputs to developing brain structures during specific developmental time-points may result in permanent alterations in brain function (Katz & Shatz, 1996). Thus, inadequate social and emotional stimulation such as occurs in institutional rearing in humans, or maternal deprivation in monkeys, may impart long-lasting changes to infant brain structures that are actively developing, with resulting effects on function. The amygdala is a key brain substrate that is critical for normal social–emotional development (Brothers, Ring, & Kling, 1990; Dicks, Myers, & Kling, 1969; Machado & Bachevalier, 2006; Machado et al., 2008; Prather et al., 2001; Shaw et al., 2004; Tranel & Hyman, 1990). It is dysregulated in many psychopathologic conditions (Burghy et al., 2012; Schumann, Bauman, & Amaral, 2011), as well as in the aftermath of inadequate caregiving in humans (Tottenham, 2012) and monkeys (Sabatini et al., 2007). We previously found that maternal deprivation in this cohort of monkeys and others, beginning at 1 week of age has markedly different effects on social-emotional development compared to maternal deprivation beginning at 1 month of age, with the former show long-lasting decreases in social behavior, and the latter showing increases in social behaviors (Bauer, Coleman, Kupfer, & Cameron, 2000; Cameron, 2001; Cameron et al., 1998; Cameron, Colemen, Dahl, Ryan, & Kupfer, 1999; Knudsen, Heckman, Cameron, & Shonkoff, 2006; Sabatini et al., 2007). Thus, input of age-appropriate sensory and social-related information to the amygdala in early childhood may be critical to later normative development and function.
Although the amygdala is frequently referred to as a unitary structure, it is a heterogeneous collection of nuclei. In both monkeys and humans, the amygdala subregion known as the paralaminar nucleus (PL) appears very late in gestation (Kordower, Piecinski, & Rakic, 1992), and is the only amygdalar region to contain immature cellular features at birth (Ulfig, Setzer, & Bohl, 2003) and postnatally (Bernier, Vinet, Cossette, & Parent, 2000; Chareyron, Lavenex, Amaral, & Lavenex, 2012; Fudge, 2004). Specifically, the PL contains a population of immature neuroblasts. These cells can be identified morphologically by their small size and singular cell process, as well as by histochemical (Chareyron et al., 2012) and immunocytological staining for neuroblast markers (Bernier et al., 2000; Fudge, 2004).
Importantly, the PL is not reliably detected in rodents. The PL is called a “nucleus,” but cells of the PL actually form a sheet-like layer that is most apparent in the ventral amygdala near the site of the former ganglionic eminence. From there, the PL stretches anteriorly to envelop the basal nucleus and to some extent, the lateral nucleus. The PL increases in size across primate phylogeny (accounting for body weight) and is largest in humans (Stephan & Andy, 1977; for review, deCampo & Fudge, 2012). Although the reason for this phylogenetic progression in monkeys, apes, and humans is not clear, it coincides with a relatively extended time-line for brain development, and a protracted juvenile state in these higher species (Joffe, 1997; Walker, Burger, Wagner, & Von Rueden, 2006).
Tract tracing studies indicate that the PL receives inputs from the ventral part of the lateral nucleus of the amygdala (Pitkanen & Amaral, 1998), and also from the hippocampus and entorhinal cortex (Fudge, deCampo, & Becoats, 2012; Saunders & Rosene, 1988; Saunders, Rosene, & Van Hoesen, 1988). On the efferent side, it has inputs to the lateral subregion of the central amygdala nucleus (Fudge & Tucker, 2009) and bed nucleus of the stria terminalis (Decampo & Fudge, 2013). These input/output paths place the PL at a crossroads where complex sensory and contextual information is integrated and transferred to key nodes of the “fear” system (Freese & Amaral, 2009). Although other nuclei of the amygdala also participate in “fear circuitry,” the PL is unique in containing neuroblasts—post-mitotic neurons that have not yet been incorporated into target circuits, based on markers such asdoublecortin (DCX) and class III beta tubulin (TUJI) (Bernier, Bedard, Vinet, Levesque, & Parent, 2002; Chareyron et al., 2012; deCampo & Fudge, 2012; Fudge, 2004; Ulfig, Setzer, & Bohl, 1998; Yachnis, Roper, Love, Fancey, & Muir, 2000). The percentage of immature neurons in the monkey PL begin to decline in the first year of life, whereas the proportion of mature neurons gradually increases (Chareyron etal.,2012),suggesting a period of postnatal neural development that may be particularly influenced by early life events.
Based on preliminary data demonstrating a subpopulation of immature neurons in the PL, we designed a custom gene set to probe genes involved in neural maturation (NM) to allow us to investigate the PL in both normally reared and maternally deprived infant monkeys at 3 months of age. We chose to examine early deprivation (with subsequent group-pen rearing), and the resulting effects on social behavior by 3 months of age, since this is the time period during which a secure base of attachment is forming for this species (see review by Suomi, 1997). Disruption of this primary parental social bond is therefore hypothesized to have important effects on the amygdala.
Beginning with the normal control cohort (Study 1), we first compared the expression of the NM gene set in the PL compared to the adjoining amygdalohippocampal area (AHA, which serves as a control region in having many similar connections, but few immature appearing neurons postnatally). We hypothesized that the NM gene set—or a subset of NM genes in the set—would have increased expression in the PL compared to the AHA. In Study 2, we tested the hypothesis that maternal deprivation would specifically affect genes associated with post-mitotic neural maturation in the PL.
2 |. MATERIALS AND METHODS
2.1. | Animals
The animals used in this study were of the same cohort that was previously behaviorally characterized in a study of the whole amygdala gene expression. Complete details of the maternal deprivation paradigm and behavioral assessment of these animals can be found in the previous paper (Sabatini et al., 2007). We briefly review the paradigm here (Figure 1). We used 12 female infant macaques (Macaco mulatta) born in the breeding colony at the University of Pittsburgh Primate Laboratory. Due to the sample size, which was constrained by the need to conserve animals and costs, we used only female animals so to avoid confounds on gene expression differences. However, it is important to note that this maternal deprivation paradigm has subsequently been conducted on cohorts of both females and males. These studies have shown no sex differences in social behavior parameters measured, even when tracked into adulthood (Bauer et al., 2000; Cameron et al., 1998, 1999). The experiments were reviewed and approved by the University of Pittsburgh Animal Care and Use Committee.
FIGURE 1.
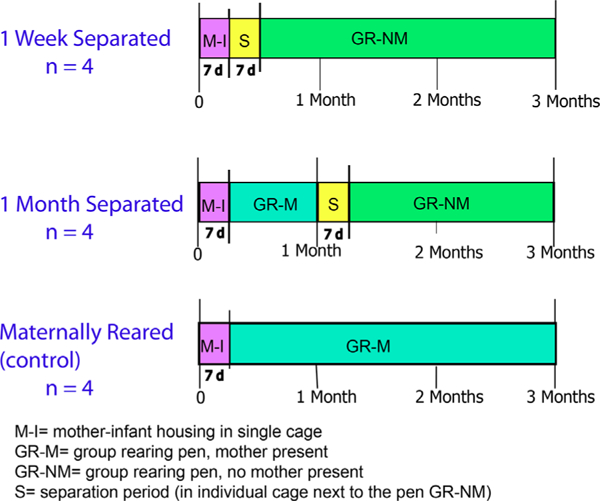
Timeline of maternal deprivation paradigm (Sabatini et al., 2007). Tissue was collected at 3 months of age
2.2 |. Experimental groups
Animals in all groups spent the first week of life in a single cage with their mother. After the first week of life, animals were arbitrarily designated as 1-week separated, 1-month separated, or control (maternally reared), and handled differently as described below. Although this paradigm is a “deprivation” rather than separation paradigm (see Section 2.3), we use the group names, “1-week separated” and “1-month separated” to reflect the age at which maternal deprivation began. Maternally reared monkeys stayed with their mother throughout the study duration.
2.3 |. Maternal deprivation paradigm
At 1 week of age, animals in the 1-week separated group were removed from their mothers and placed for a 5–7 days period into a single cage where they learned to bottle-feed. The cage was positioned immediately adjacent to a group-rearing pen that they would ultimately join. In the single cage, animals were provided with a soft cotton stuffed toy that could be held to provide contact comfort. After spending 5–7 days in the single cage, 1-week separated monkeys joined the group-rearing pen for weeks 3–12. Monkeys in the 1-month separated groups were introduced to the group-rearing pen with their mothers at 1 week of age. When infants turned 1 month of age, mothers were removed from the group-rearing pen, and infants were placed in the single cage adjacent to the group-rearing pen, as described for the 1-week separated monkeys. After the 5–7 days period in the single cage where they learned to bottle feed, 1-month separated monkeys were reintroduced into the group-rearing pen without their mother for weeks 6–12. After 1-weekof age, control (maternally reared) monkeys were placed into the group-rearing pen, which consisted of 4–5 monkeys, with their mother where they remained for weeks 2–12. The group-rearing pen also had a nursery enclosure where only infant monkeys could enter. Bottles containing standard human artificial formula were placed in the nursery and replaced three times each day. All infant monkeys were weighed weekly and had similar weight gains over the study. For details, see Supplementary Material S1.
2.4 |. Behavioral assessments
As noted, behavioral assessments on this cohort of animals were performed as part of an earlier study and took place at two time-points: 1) the week after maternal deprivation while infants were in a single cage; and 2) in the third month of life in the group-rearing pen (see methods and results section in Sabatini et al., 2007). The behaviors scored while infants were alone in the single cage included: general behaviors (e.g., locomotion, stationary, bottle drinking), where they were in the cage (e.g., adjacent to the pen) and self-comforting behaviors (e.g., toe/thumb-sucking, rocking). At the third month of life, all behaviors scored in the single cage were again scored, this time in the pen. In addition, at this time-point, time spent in social interactions was also recorded (e.g., social play, touch, groom, social huddle, aggression). In the present study, data collected only at the second time-point (3 months) was analyzed in conjunction with gene expression data. Brain tissue from the control monkeys was used for studies of the normal infant PL (Study 1) and tissue from all animals was examined in the deprivation study (Study 2).
2.5 |. Gene mRNA expression-behavior correlations
Following analysis of differential gene expression in Study 2, we found that the gene tbr1 showed significant differential expression between maternally deprived monkeys and control monkeys. Because mutations in tbr1 are associated with autism spectrum phenotypes (O’Roak et al., 2012), we chose to look at the correlation of tbr1 expression with the composite scores for typical social behavior (e.g., social play, touch, groom, social huddle, aggression) and self-comforting behaviors (e.g., toe/thumb sucking, rocking). Our rationale for looking at these behaviors was based on the altered social behavior, and stereotypies resembling self-comforting behaviors, that have been described in individuals with autism (Volkmar, Chawarska, & Klin, 2005). We examined the relation between tbr1 mRNA expression using silver grain counts from in situ hybridization studies (Supplementary Material S1). Pearson correlations were performed between the number of silver grains counted from the tbr1 in situ hybridization with either composite 1) social behaviors; or 2) self-comforting behaviors. A t-distribution table with n-2 degrees of freedom was used to determine the associated one-tailed p values. r values were considered significant when the associated p value was <0.05.
2.6 |. Tissue processing
At 12 weeks of age, brain tissue was collected from each monkey at the University of Pittsburgh. Monkeys were pre-anesthetized with ketamine HCl (10 mg kg–1, i.m.) and then deeply anesthetized with sodium pentobarbital (30 mg kg–1, i.v.). The chest was opened and a catheter placed in the heart with the tip in the ascending aorta and ~1 L of ice cold 0.9% NaCl, containing 5000 IU heparin and 2% sodium nitrite, was perfused transcardially. The left atrium was opened so perfusate could run out and when it ran clear the monkey was quickly decapitated. The brain was immediately removed from the skull and was hemisected by a midline sagittal cut. The right hemisphere, which was used in this study, was cut into 5-mm-thick coronal blocks, flash frozen in isopentane over dry ice, and stored at –80 °C until cryostat sectioning. For this study, tissue blocks containing the caudal amygdala at the level of the PL and amygdalohippocampal area (AHA) were selected and sectioned at 20 μm using a cryostat. One of the 1-month separated monkeys did not have enough tissue in the block for this study, resulting in three monkeys in the 1-month separated group for microarray studies. Ten adjacent sections of the caudal amygdala were thaw-mounted onto polyethylennaphtalate (PEN) foil slides (Leica Microsystems; Bannockburn, IL) for laser-capture microdissection, and frozen at –80 °C. The subsequent 10 adjacent sections of each block were used for in situ hybridization.
2.7 |. Laser-capture microdissection (LCM) and Microarray
We used LCM to isolate the PL and adjacent AHA on the same sections of each animal, and analyzed differential gene expression in each region using microarray (Figure 2). A custom-designed gene set enrichment approach was used to examine differential expression in the PL compared to the AHA in normal animals (Study 1) and in maternally deprived animals versus normal animals (Study 2) for the PL and AHA. We chose to use this approach to avoid the common problem in gene expression studies of identifying a list of significant, but unrelated genes. In gene set enrichment analysis, a set of genes with a common function or pathway are examined to see if there are significant differences in expression between two groups. Gene sets can be manually created, or obtained from Biocarta, KEGG, or GO databases. For this study, we manually created a list of genes that had a documented role in NM, and only examined changes in gene expression within this set. To create this custom-designed gene set, we included genes involved in NM based on 1) our previous observations of protein expression in immature neurons in the adult PL (Fudge, 2004; Fudge et al., 2012; Figure 3); and 2) primary literature searches of NM genes involved in the developing cortex and adult neurogenic zones. Genes were included in the custom-designed gene set if at least one published report showed a documented role in neural fate commitment (i.e., transition from a multi-potent precursor to committed neuronal precursor or involved in differentiation of a neural precursor into a specific neuronal cell type), migration, and/or neurite extension in post-mitotic neurons, regardless of whether specific genes had additional reported functions (Gupta, Tsai, & Wynshaw-Boris, 2002; Lie & Götz, 2009) (Supplementary Material S2). We reasoned that since most genes have more than one function, restricting the gene set to only one function might reduce discovery of important sets of genes that play a role in neural maturation in the PL.
FIGURE 2.
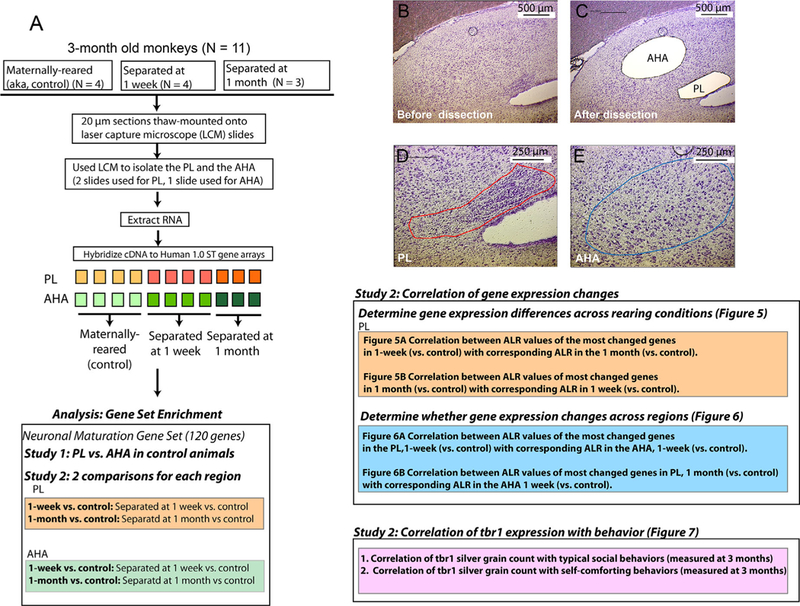
Overview of experimental methods. A) Schematic of methods used for this study (tissue from one animal deprived at 1 month was not used, n = 3). B and C) Coronal sections of the caudal amygdala, using a rapid thionin stain to identify the PL and AHA on the same section, before B), and after C), LCM. D and E) High-power image showing the PL D), and the AHA E), prior to dissection
FIGURE 3.
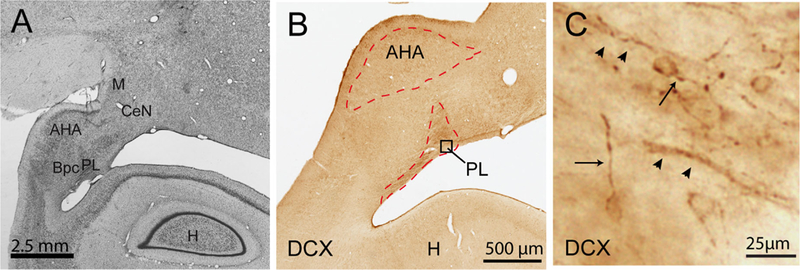
Photomicrographs of the PL and AHA. A) Coronal section of the caudal primate amygdala stained with cresyl violet. Scale bar = 2.5 mm. B) The caudal amygdala, at the level in B) seen at higher power in a section labeled for doublecortin (DCX) immunoreactivity. Laser capture microdissection of the PL and AHA was conducted at this amygdala level. C) Higher power view of DCX positive cells in the PL, boxed region in B). Morphologically, cells have a rounded soma and thin process (arrow). Cells are frequently found in close apposition to fibers of passage (arrowheads). CeN, central nucleus; Bpc, parvicellular subdivision of the basal nucleus; H, hippocampus; M, medial nucleus
PL and AHA boundaries were visualized using a rapid thionin staining protocol in which each section was placed in acetic acid, 0.5% thionin, and dehydrated through a series of alcohols. Total time for staining was 3–4 min. Immediately following staining, the PL and the AHA in each section (10 slides per animal), was rapidly dissected on a Leica A LMD microscope (Leica Microsystems, Bannockburn, IL). Using this method, the PL is distinguished by its small-sized cells and higher cell density (Price, Russchen, & Amaral, 1987). In separate studies, we noted that the PL visualized in this way, coincides with doublecortin (DCX)-immune reactive cells (Fudge et al., 2012) (Figure 3). The AHA is an elliptical shaped structure characterized by large cells and does not contain DCX-immuno reactive cells. A total area of 0.25 mm2 (PL) and 0.5 mm2 (AHA) was collected from the same section. Tissue from each region was collected into 30 μl RLT Lysis Buffer (Qiagen, Valencia, CA) with 1% β-mercaptoethanol (Sigma–Aldrich, St. Louis, MO) in under 10 min per slide, and rapidly frozen at –80 °C.
Total RNA was isolated using the RNeasy mini kit (Qiagen, Valencia, CA). RNA concentration and quality were measured using an RNA 6000 Pico Chip on an Agilent Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA). The average RNA integrity value (RIN) for the PL and the AHA was 3.0 and 5.7, respectively. Material captured from two slides for the PL and one slide for the AHA were used to acquire the 5 ng of RNA needed for amplification and hybridization. We chose samples whose 28:18 peak ratio was closest to 2. Amplification and synthesis of biotinylated cDNA was performed with the WT-Ovation PicoRNA Amplification kit (Nugen, San Carlos, CA). A total of 4.5 μg of cDNA was hybridized to Affymetrix Human Gene 1.0 ST arrays (Affymetrix Inc., Sunnyvale, CA). Following hybridization, samples were normalized using gene chip robust multi-array average (GC-RMA). Probe intensities were log2 transformed.
2.8 |. Microarray analyses
2.8.1 |. Study 1: Differential expression of NM genes in the PL versus AHA in normal infant animals
In Study 1, we hypothesized that the NM gene set as a whole, or a subset of genes within it, would be enriched (upregulated) in the PL compared to the AHA in control animals. To determine whether the gene set as a whole was differentially expressed in the two regions, the average log2 ratio (ALR) was computed between the PL and AHA for every gene on the microarray (|ALR| = 1 corresponds to a twofold increase or decrease, |ALR| = 0.585 represents a 1.5-fold change, | ALR| = 0.383 corresponds to a 1.3-fold change). These fold-changes are considered biologically relevant, and are detectable by quantitative polymerase chain reaction (qPCR) (Shelton et al., 2011). The “average ALR” for the NM gene set was computed by averaging the ALRs of each gene in this customized gene set. This average was then compared to the average of the ALRs for all genes on the chip using an unpaired one-tailed Student’s t-test with a p value threshold set at p < 0.05. We did not correct for multiple comparisons.
We then wanted to identify the individual genes within the NM gene set that were differentially expressed between the PL and the AHA. We used a two-criteria approach to determine differential expression (Mirnics, Levitt, & Lewis, 2006). First, an individual gene was considered to be differentially expressed if the ALR between the PL and the AHA was greater than 30% (|ALR| > 0.383). This criterion was chosen based on previous studies indicating that gene expression fold changes of 1.3 (ALR>30%) are biologically significant and can be verified with either real-time PCR or in situ hybridization at this level (Shelton et al., 2011). Genes that survived this first criterion were subjected to an unpaired 1-tailed Students t-test comparing expression levels between the two regions, with p value <0.05. Genes that met criteria were validated using qPCR and/or in situ hybridization (Supplementary Material S1) to determine whether they represented true biological changes.
2.8.2 |. Study 2: Maternal deprivation effects on gene expression in the PL and AHA
In Study 2, the effects of maternal deprivation on gene expression of the entire customized NM gene set in the PL was examined in two different comparisons: 1) 1-week separated monkeys relative to maternally reared monkeys (1-week separated vs. control); and 2) 1-month separated monkeys relative to maternally reared monkeys (1-month separated vs. control; Figure 2). As in Study 1, for every gene on the microarray the average log2 ratio (ALR) was calculated for each of the two comparisons (1-week separated vs. control, 1-month separated vs. control). The “average ALR” for the NM gene set as a whole was calculated by averaging the ALRs of all genes in the gene set. The average was then compared to the average of the ALRs for all genes on the chip using an unpaired one-tailed Student’s t-test with a p value threshold set at p < 0.05.
We then identified individual genes within the customized gene set that showed differential expression in the PL for each of the two comparisons (1-week separated vs. control, 1-month separated vs. control). We considered an individual gene within our customized gene set to be significant if it fulfilled our two criteria: 1) the gene’s ALR between the separated (separated at 1-week or at 1-month) and the maternally reared groups was greater than 30% (|ALR| > 0.383); and 2) the p-value for a one-way ANOVA (1-tailed) was p < 0.05.
Next, we wanted to determine whether genes that showed differential expression in the two comparisons were similar with respect to changes in direction and magnitude. We therefore plotted the ALR values of the most changed genes (|ALR| > 0.383) in the 1-week separated (vs. control) with corresponding values in the 1-month separated (vs. control). Pearson correlations were used to determine the r and p values. A t-distribution table with n-2 degrees of freedom was used to determine the one-tailed p values. r values were considered significant when the associated p was <0.05. Identical analyses were conducted on tissue collected from the AHA to identify the most changed genes in this region for each comparison. Differentially expressed genes are included in the results. However, as our focus was on the PL, AHA correlation plots are not shown.
To determine whether maternally deprived gene expression changes in the PL and AHA were region specific for the two comparisons, we correlated ALR values from one region with corresponding values in the other region, restricting our correlations to |ALR| > 0.383. This allowed us to see whether or not the most changed genes in the PL following maternal deprivation were correlated with most changed genes in the AHA. If a set of most changed genes in the PL were also changed in the same direction and of similar magnitude in the AHA, we interpreted this to mean that the effects of maternal deprivation on gene expression were non-specific to the PL. We used a Pearson correlation to determine r-values. A one-tailed p-value was used. r values were considered significant when the associated p < 0.05.
To validate some of the findings, qPCR and in situ hybridization were performed. qPCR was run for dcx, tbr1, and neuroD1, based on microarray results for both Study 1 and 2. In situ hybridization of tbr1 mRNA was done on frozen sections adjacent to those used for LCM (Additional Methods in Supplemenrtary Material S1).
3 |. RESULTS
3.1 |. Study 1: Differential expression of NM genes in the PL versus AHA in normal infant animals
In the normal infant amygdala, the NM gene set was decreased in the PL compared to the AHA (average ALR of all genes in the gene set compared to the average of the ALRs for all genes on the chip). This decrease (12.8%) (ALR = –0.174, p = 0.0001) in the NM gene set as a whole was in a direction contrary to our hypothesis. We reasoned that since many genes in the NM gene set had additional functions, a second level of analysis could be used to determine whether individual genes within the NM gene set had a higher expression in the PL relative to the AHA. From this analysis, four genes were found to have enriched expression in the PL based on the two criteria for differential expression (|ALR| > 0.383 and p < 0.05), and were confirmed by qPCR. They were: bc12 (ALR = 0.433, p = 0.01), dcx (ALR = 0.88, p = 0.01), neuroD1 (ALR = 0.84, p = 0.02), and tbr1 (ALR = 0.84, p = 0.01) (Figure 4A, red). qPCR confirmed the microarray findings, showing a significant enrichment in the expression of dcx of approximately fivefold (-ddCt 2.35, p = 0.02), tbr1 of approximately sixfold (-ddCt 2.62, p = 0.01), and neuroD1 approximately 11-fold (-ddCt 3.51, p = 0.001) in the PL relative to expression in the AHA (Figure 4B). Unfortunately, there was inadequate sample to confirm bcl2 expression differences by qPCR. Thus, a specific subset of NM genes expressed in post-mitotic neurons in the fetus, and in adult neuro-genetic zones (see Section 4), was enriched in the PL compared to the AHA, in normal 3-month old animals. Interestingly, a number of genes from the NM gene set were relatively enriched in the AHA versus the PL (Figure 4A, blue). These genes participate in axonal outgrowth and cytoskeletal stabilization, developmental processes that are somewhat different than those implicated by gene expression in the PL (see Section 4).
FIGURE 4.
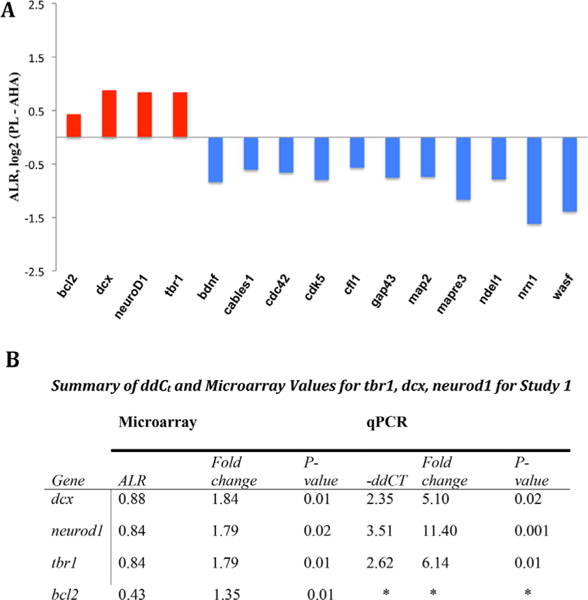
Subsets of differentially expressed genes in the NM gene set differ between the PL and AHA in control animals. A) Most changed genes in the PL versus the AHA based on our two criteria method: bcl2, dcx, neuroD1, tbr1 (red = increase). Eleven other genes were decreased in the PL relative to the AHA (blue = decreased). B) Microarray and qPCR values for most enriched genes in the PL relative to AHA in normal controls. *bcl2 was not validated by qPCR due to inadequate sample
3.2 |. Study 2: Maternal deprivation effects on gene expression in the PL and AHA
3.2.1 |. Maternal deprivation effects on gene expression in the PL
The effects of early maternal deprivation on PL gene expression were then examined. The NM gene set as a whole was examined first and was decreased in both maternal deprivation groups, although changes were small. For the 1-week separated versus control comparison, there was a 7.2% decrease (ALR = –0.10, p = 0.0001) and for the 1-month separated versus control comparison, there was a 4% decrease (ALR = –0.06, p = 0.0005) in the gene set in the PL compared to the ALR of all genes.
As in Study 1, we then identified a subset of NM genes with an |ALR| > 0.383 for each deprivation group within the PL (Figure 5). In the 1-week separated versus control comparison, we identified 13 genes meeting this criterion for fold-change. In the 1-month separated versus control comparison, we identified nine genes that met this criterion. Five of the genes in the 1-week versus control and 1-month versus control comparison were the same, displaying |ALR| > 0.383 in both cases (Figure 5, gray). The genes downregulated in both comparisons were: apc, ncam1, nrn1, rap1a, tbr1. Together these genes comprised a common core of genes involved in NM that were altered by early maternal deprivation in the PL.
FIGURE 5.
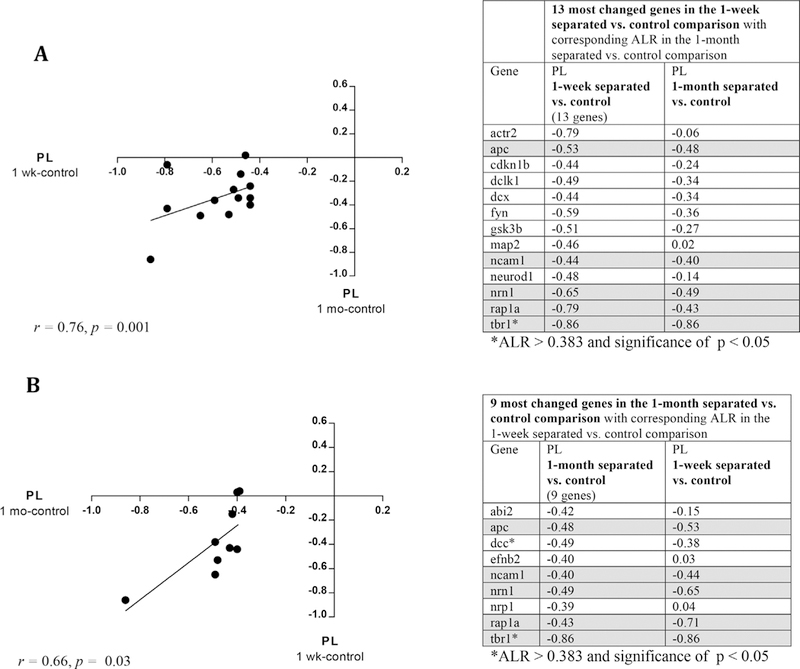
Most changed genes in PL following maternal deprivation, 1-week and 1-month separated subgroups. A) Plot of the most changed genes in the PL in 1-week separated animals (x-axis) versus their corresponding change in the PL of 1-month separated animals (y-axis) (r = 0.76, p = 0.001). B) Plot of the most changed genes in the PL in 1-month separated animals (x-axis) versus their corresponding change in the PL of 1-week separated animals (r = 0.66, p = 0.03). Tables of the individual genes are to the right of each plot. Gray boxes highlight gene changes common to both 1-week and 1-month separated groups (ALR > 0.383, with same directionality)
We then determined whether changes in gene expression in the two maternally deprived groups were similar (i.e., in the same direction and magnitude). Plotting the ALR values of the 13 most changed genes in the 1-week separated (vs. control) against their corresponding ALR values in the 1-month separated (vs. control), we found a correlation of r = 0.76, p = 0.001 (Figure 5A). Performing the reverse correlation (i.e., plotting values for the nine most changed genes in the 1-month separated (vs. control) comparison, against the corresponding ALR values in the 1-week separated (vs. control) comparison), gave a similar positive correlation r = 0.66, p = 0.03 (Figure 5B). These findings indicate that as a group, the most changed genes in the PL (relative to control values) had similar changes in direction and magnitude.
We then identified which genes within the 13 gene (1-week separated) and nine gene (1-month separated) subsets met additional criteria for significance (p < 0.05). In the PL, examination of differential gene expression values revealed that tbr1, a gene expressed in post-mitotic neurons destined to be projection neurons (Hodge, Kahoud, & Hevner, 2012), was significantly altered. tbr1 was also found to be relatively enriched in the normal PL in Study 1 (Figure 4). A one-way ANOVA showed that there was a significant effect of rearing condition on tbr1 expression in the PL at p < 0.05 [F(2,8) = 8.036, p = 0.0122]. Post hoc comparison indicated that tbr1 expression was significantly lower in monkeys deprived at 1 week and at 1 month of age compared to controls, with an ALR= –0.86 for each comparison. There was no significant difference in tbr1 expression between 1-week- and 1-month-separated comparisons. Expression levels of neuroD1 and dcx genes which are also expressed in post-mitotic neurons (Hodge et al., 2012), had meaningful decreased expression the 1-week separated versus control animals, but were not significant [neuroD1 ALR = –0.48, F(2,8) = 2.16, p = 0.18; dcx ALR = –0.86, F(2,8) = 1.21, p = 0.35]. Lack of statistical significance may mean that there is no true effect of group on neuroD1 and dcx expression. Alternately, the magnitude of the fold changes, and the fact that these genes are temporally and biologically linked (along with tbr1) in neuroblasts (Hodge et al., 2012), raises the strong possibility that there was insufficient power to detect statistically significant effects due to low sample size.
In addition to significant decreased expression of tbr1 in the 1-month separated versus control comparison, dcc expression was significantly lower in monkeys deprived at 1 month of age [F (2,8) = 14.56, p = 0.002, ALR = –0.49]. dcc encodes a membrane bound receptor that, when bound to netrin-1, is important in maintaining axonal growth along stereotyped paths during brain development (Fazeli et al., 1997; Keino-Masu et al., 1996). As yet, the developmental expression of dcc, or its ligands, in the immature primate brain have not been described.
3.2.2 |. Maternal deprivation also alters gene expression in the AHA
As a whole, the custom-designed NM gene set was downregulated in the AHA in the 1-week separated group (decreased by 12%; –0.16 ALR, p = 0.05), but upregulated in the 1-month separated group (increased by 3.5%; 0.05 ALR, p = 0.05) relative to maternally reared monkeys. Again, because many NM genes serve broad functions, to better evaluate specific gene expression changes, we identified the most changed genes in the AHA for each comparison as had been done for the PL. cables1, which plays a role in neurite outgrowth in the central nervous system (Zukerberg et al., 2000), was significantly altered in the AHA for both deprived groups compared to control animals [F(2,8) = 12.54, p = 0.01]. Although there were several other genes that were altered in either 1-week or 1-month separated comparisons, cables1 was the only gene to show a significant change in both comparisons, with its expression significantly lower in maternally deprived groups compared to controls (ALR= –0.49 for both comparisons). Cables1, which was also enriched in the AHA relative to the PL in normal animals (Study 1), is localized to growth cones where it modulates axonal outgrowth. Maternal deprivation at both 1 week and 1 month of age was associated with reduced cables1 expression in the AHA suggesting region-specific effects on axonal outgrowth. Maternal deprivation in the 1-week separated, but not 1-month separated comparison revealed several other significantly changed gene transcripts: bdnf (ALR = –0.51, p = 0.03) and cdk5 (ALR = –0.66, p = 0.01), which were enriched in the normal AHA, were downregulated in the 1-week separated monkeys. Additionally, some genes not found to be enriched in Study 1, including ncam1(ALR= −0.40, p = 0.02), rap1b (ALR= –0.90, p = 0.02), apc (ALR = –0.82, p = 0.01) and st8sia2 (ALR = +0.44, p = 0.01), were also altered in the 1-week separated comparison only.
3.2.3 |. Gene signatures in maternally deprived animals are distinct for the PL and AHA
Finally, to determine whether the most changed genes in the PL (|ALR| > 0.383) for each of the two comparisons were similar or different than those in the AHA (i.e., the specificity of gene expression changes in each region), we analyzed whether the ALR values from the PL (1-week separated vs. control, and 1-month separated vs. control) were similar in magnitude and direction compared to the values found in the AHA (Figure 6). For the 1-week separated versus control comparison, the correlation between the most changed genes in the PL with their corresponding values in the AHA was r = –0.011, p = 0.98; for the 1-month versus control comparison, the correlation was r = –0.3, p = 0.21. Therefore, in both comparisons, overall gene expression changes in the PL did not as a group, correspond to similar changes in the AHA. One striking difference was that tbr1 was downregulated in the PL of monkeys deprived at both 1 week and 1 month, but not the AHA (Figure 6, boxed). neuroD1 and dcx, genes which were also enriched in the PL of normal animals (Study 1), were uniquely downregulated in the PL compared to the AHA of 1-week separated monkeys, but did not reach significance (Figure 6A).
FIGURE 6.
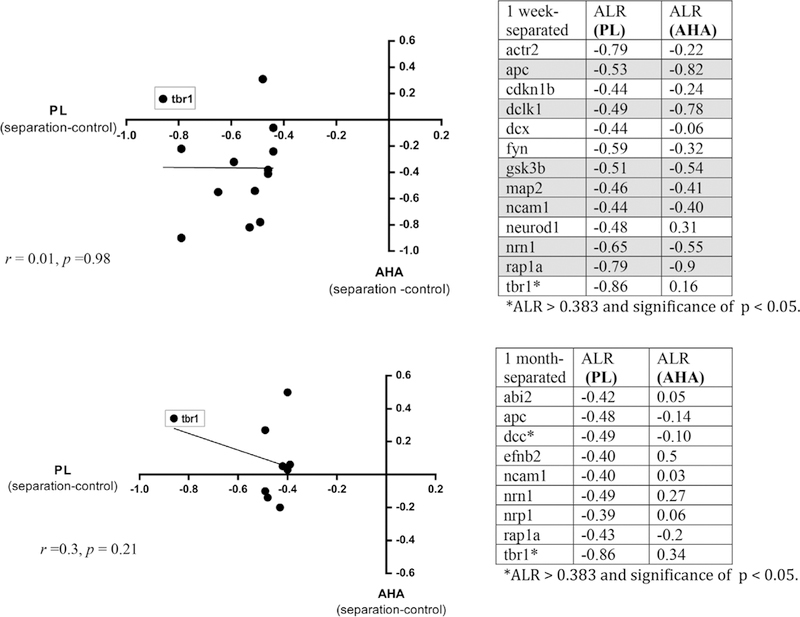
Most changed gene profiles in the PL versus AHA following maternal deprivation. Plots of the most changed genes (|ALR| > 0.383) in the PL (x-axis) versus their change in the AHA (y-axis). A). One-week versus control comparison (r = –0.011, p = 0.98). B) One-month versus control comparison (r = –0.3, p = 0.21). Gray boxes on graphs highlight the position of tbr1 for each comparison. Tables of the individual genes are to the right of each plot
Analysis of qPCR data for tbr1, dcx, and neuroD1 across rearing conditions for the PL and AHA was also performed. There was a significant effect of rearing condition on tbr1 expression [F(2,7) = 4.973, p = 0.045] (Figure 7A). Post hoc comparison using Fisher’s LSD test indicated that tbr1 expression was significantly lower in monkeys that were deprived at 1 week and 1 month compared with control animals, but not between 1-week and 1-month separated animals. Supporting microarray findings, tbr1 was not downregulated in the AHA in either the 1-week separated versus control or 1-month separated versus control comparisons. As with microarray data, dcx, and neuroD1 did not show significant changes in gene expression across group comparisons for the PL or the AHA.
FIGURE 7.
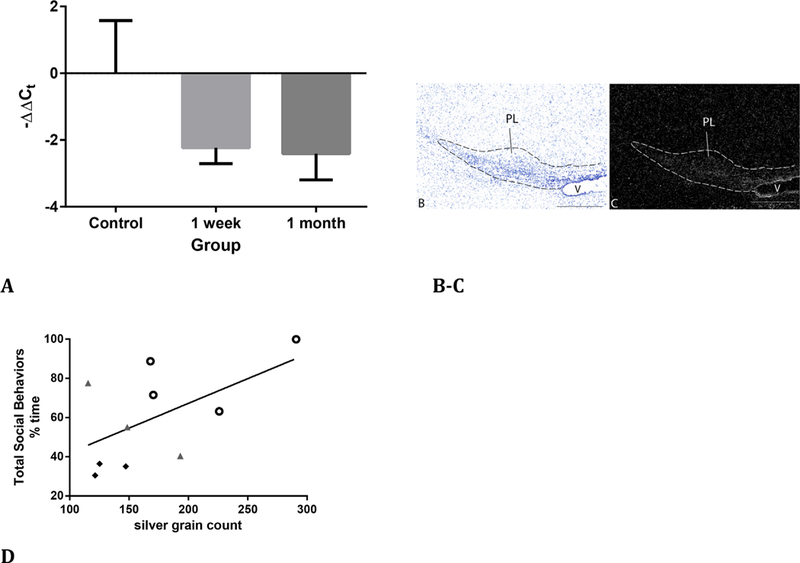
tbr1 mRNA and correlations with social behaviors at 3 months of age. A) qPCR of tbr1 transcript fold-changes in maternally deprived animals; 1-week separated versus control, p = 0.04 and, 1-month separated versus control, p = 0.03. B and C) Adjacent sections near the LCM site stained for cresyl violet, B) and processed for tbr1 mRNA. C) Dotted line indicates the PL. V, ventricle. Bar = 500 μm. D) Correlation between total social behaviors at 3 months and silver grain count (corrected for background) in the PL for the tbr1 probe across all animals. Black filled, diamonds 1-week separated monkeys; gray filled triangles, 1-month separated monkeys; open circles, maternally reared monkeys (r = 0.56, p = 0.045)
3.2.4 |. tbr1 mRNA expression in PL correlates with social behavior at 3 months of age
The maternal deprivation paradigm had no effects on the animals’ developmental trajectories of physical parameters such as weight and locomotor behavior. These were similar among all animals at the time brain tissue was collected (ANOVA, F = 0.284, p = 0.76; Sabatini et al., 2007). As previously reported for this cohort of monkeys, the percent time spent in typical social behaviors by 3 months of age was abnormal for all maternally deprived monkeys. Also as previously reported, 1-week separated animals demonstrated more contact-comforting behavior with a soft-cotton stuffed toy and decreased social contact initially (i.e., at time of deprivation), while later increasing self-comforting behaviors (thumb-/toe-sucking, rocking) and having decreased social-contact behaviors. One-month separated animals in contrast, initially showed increases in social-comforting behaviors and continued to engage in more social behavior (i.e., ventral contact, proximity, grooming) than 1-week separated or control groups (Cameron, 2001).
Spontaneous mutations in tbr1 are associated with autism, a condition that often presents with altered social behaviors and stereotypies that are thought to be self-comforting (O’Roak et al., 2012). We therefore examined correlations between tbr1 mRNA expression obtained from in situ hybridization methods with total typical social behaviors (e.g., play, grooming, touch, aggression, social huddle) and total self-comforting behaviors (e.g., toe/thumb sucking, rocking) measured at 3 months of age in the group-rearing pen. In situ hybridization confirmed that the density of silver grains was relatively higher over the PL than adjacent regions, including the AHA, in all animals (Figure 7B and C). Although there were no significant group differences in tbr1 mRNA expression by in situ hybridization [F(2,8) = 1.66, p = 0.25], these results showed a trend in the same direction as the significant changes detected by qPCR and microarray. The time spent in typical social behaviors by 3 months of age across all animals was significantly positively correlated with tbr1 PL expression (r = 0.56, p = 0.045; Figure 7D). Self-comforting behaviors and tbr1 expression were not significantly correlated (r = −0.47, p = 0.08).
4 |. DISCUSSION
4.1 |. Summary of main findings
In female infant monkeys, the pattern of NM gene enrichment (Study 1) indicates that in the first months of postnatal life, genes involved in the relatively early differentiation of neuroblasts predominate in the PL. Maternal deprivation (Study 2) during this time is associated with downregulation of tbr1, which regulates the production and differentiation of neuroblasts into glutamatergic neurons (Hodge et al., 2012) (Table 1). The percent time spent in typical social behaviors by 3 months was decreased in maternally deprived animals and correlated positively with tbr1 expression at this age.
TABLE 1.
Summary table of the main microarray findings that met dual-criteria for gene expression changes.
| Study 1 |
Study 2 |
|
|---|---|---|
| Maternally reared (control) PL versus AHA |
Maternal deprivation (1 week and 1 month combined) versus control PL |
AHA |
| neurod1 ⇑ | ||
| dcx ⇑ | ||
| bcl2 ⇑ | ||
| tbr1 ⇑ | tbr1 ⇓ |
cables 1⇓ |
Summary of microarray data meeting dual criteria for gene change (|ALR| > 0.383 and p < 0.05).
Gene expression changes shown, except cables 1 and bcl2, were additionally verified by qPCR.
4.2 |. Study 1: Differentially expressed genes identified in infant PL suggest postnatal maturation of glutamatergic neurons
In normal infant monkeys, we identified a subset of NM genes that distinguish the PL from its near-neighbor, the relatively developed AHA (Table 1). The particular gene signature of the PL supports the concept that it is a zone where maturational processes in post-mitotic neurons occur. As the caudal ganglionic eminence recedes in the last few months of human fetal life, the PL is progressively defined (Ulfig et al., 1998). Recent work in developing non-human primates has noted several immature neural characteristics of the PL, with changes over post-natal development. Chareyron et al. recently used histochemical and stereologic methods to show that the PL of monkeys between birth and 1 year of age contains only 15% of the total mature neurons present in young adulthood (5–9 years). In adulthood, there is an increase in the number of mature neurons paralleled by a decrease in the number of immature neurons. Importantly, the total number neurons in the PL does not differ between ages, suggesting that by adulthood, a significant number of immature neurons have matured (Chareyron et al., 2012).
Building on existing knowledge of the infant PL, we found that several transcripts involved in neuroblast fate determination, neurite extension, migration, terminal differentiation, and layer specification are relatively enriched in the PL of normal 3 month old animals (see below, this Section). Two of these gene products, bcl2 and dcx, have been previously described at the protein level in the PL of young adult monkeys (Bernier et al., 2002; Fudge, 2004; Fudge et al., 2012) and in humans (Yachnis et al., 2000). bcl2, while best-known for its anti-apoptotic activity (Zhong et al., 1993), is independently involved in differentiation of committed neurons and in neurite formation (Zhang, Westberg, Holtta, & Andersson, 1996). bcl2 is broadly expressed throughout human fetal brain (Merry, Veis, Hickey, & Korsmeyer, 1994), declines through gestation, and after birth is highly expressed only in restricted regions such as the hippocampal subgranular zone and PL in both humans and monkeys (Bernier et al., 2002; Fudge, 2004; Yachnis et al., 2000). dcx codes a microtubule-associated protein that is critically involved in neural migration (Gleeson, Lin, Flanagan, & Walsh, 1999), and its genetic mutation in humans results in severe disruption of cortical layering (des Portes et al., 1998; Gleeson et al., 1998). dcx is expressed in both radially and tangentially migrating neurons, possibly at different stages of migration or in different neuronal subclasses (i.e., interneurons and pyramidal cells) (Meyer, Perez-Garcia, & Gleeson, 2002).
neuroD1 and tbr1 transcripts, which have not previously been described in the postnatal primate brain, are involved in fate specification of post-mitotic neurons. neuroD1 encodes the neuronal differentiation 1 protein, which is required for terminal neuronal differentiation in the classic postnatal neurogenesis zones, namely the olfactory bulb (Boutin et al., 2010; Roybon, Deierborg, Brundin, & Li, 2009) and hippocampus (Miyata, Maeda, & Lee, 1999) based on work in the rodent. In these regions, as well as in the hippocampus and cortex of the fetal brain, the temporal expression of neuroD1 precedes and overlaps subsequent tbr1 expression (Hodge et al., 2012). tbr1 codes a transcription factor required for the development and laminar positioning of glutamatergic projection neurons in the cortex (Bulfone et al., 1995; Hevner et al., 2001; Hodge et al., 2012), cerebellum, and subgranular zone (Hodge et al., 2012) and marks precursors destined to become glutamatergic neurons. In fetal monkeys (Donoghue & Rakic, 1999), humans (Bulfone et al., 1995), and mice (Bulfone et al., 1995; Kolk, Whitman, Yun, Shete, & Donoghue, 2005), tbr1 expression is localized to the fetal subplate and deep cortical plate, both transient regions in which gene expression and transient efferent and afferent connections orchestrate formation of the mature cortex (Sidman & Rakic, 1973; Ulfig, Neudorfer, & Bohl, 2000). tbr1 expression in the PL may suggest a similar transient “way station” in the development of the amygdala in primates.
bcl2, dcx, neuroD1, and tbr1 transcripts are all closely temporally regulated during the final phases of neuronal differentiation and positioning in developing cortex and in adult neurogenic zones (Hodge et al., 2012), which is a specific phase of neural development prior to the integration into final circuits. The relative enrichment of these four transcripts in the postnatal PL in monkey suggests that continuing neural maturation processes in this subregion occur after birth, as the young animal begins its early interactions with the environment. Given the fact that the PL is minimal to non-existent in the rodent, and large in the human, our results highlight the importance of using non-human primate models to study normal and abnormal amygdala development that may pertain to humans, and human disease (deCampo & Fudge, 2012).
In contrast to genes involved in cell fate specification and differentiation (PL), genes enriched in AHA are more identified with molecular pathways involving neurite outgrowth and the cytoskeletal shifts that underlie neurite outgrowth (Figure 4A, blue). Several combinations of these genes participate in intracellular cascades including actin stabilization/polymerization (e.g., Hussain et al., 2001; Sumi, Matsumoto, Takai, & Nakamura, 1999) and neurite outgrowth (Cheung, Chin, Chen, Ng, & Ip, 2007; Fujino, Wu, Lin, Phillips, & Nedivi, 2008; Leemhuis et al., 2010; Meiri, Saffell, Walsh, & Doherty, 1998; Strittmatter, Fankhauser, Huang, Mashimo, & Fishman, 1995; Zuker-berg et al., 2000). Although these genes are critical in CNS development, they also participate in processes such as synaptic plasticity and learning (e.g., reviews by Dhavan & Tsai, 2001; Park & Poo, 2013).
4.3 |. Study 2: tbr1 is highly enriched in the normal infant PL and downregulated following maternal deprivation
The PL has a fundamentally different maturation profile compared to the rest of the amygdala, with PL neurons at a much earlier phase of development compared to the rest of the amygdala, particularly in the first few years of life (Bernier et al., 2002; Chareyron et al., 2012; Fudge, 2004). Our previous study of this cohort, using an exploratory approach to examine the entire amygdala found that differential expression of several genes distinguished the two separation groups, and that there were correlations with specific behavioral profiles typical of each group (Sabatini et al., 2007). In the present study, we dissected out the PL and found that the timing of deprivation (i.e., beginning at 1 week and 1 month) made no difference on tbr1 gene expression between 1-week and 1-month separated animals. We interpret this to mean that the very immature status of PL neurons in the first three months of life makes them sensitive to maternal deprivation at either time-point in the first month of life, specifically with respect to tbr1 expression. Decreased tbr1 expression implies an alteration in the maturation of glutamatergic neurons in the PL, which could affect these circuits, and/or their efferent and afferent contacts.
Although our focus was not on the AHA, in this nucleus, we found that cables1 (enriched in the AHA in normal animals in Study 1) was significantly downregulated in both maternal deprivation groups. Cables1 is a cdk5-interacting protein that is highly expressed in the brain and largely localized to growth cones (Zukerberg et al., 2000). Decreases in cables1 may suggest a negative influence on neurite outgrowth and circuit development in the AHA following early maternal deprivation. bdnf and cdk5—also relatively highly enriched in the AHA of normal animals—were additionally downregulated in monkeys deprived at 1 week of age. Although the downregulation of bdnf, cables 1, and cdk5 in 1-week separated monkeys may be entirely independent, it is intriguing that both bdnf and cables1 interact with cdk5 (Cheung et al., 2007; Zukerberg et al., 2000) to influence neurite growth.
4.3.1 |. Maternal deprivation, tbr1, and development of typical social behavior
We found that tbr1 expression in the PL was positively correlated with the frequency of typical social behaviors (e.g., ventral contact, proximity, grooming) at 3 months of age, suggesting a potential link between glutamatergic neuron development in the PL and the expression of social behavior. tbr1 has been identified as one of several genes whose disruption is associated with autism phenotypes (Huang & Hsueh, 2015; O’Roak et al., 2012), possibly through effects on developing glutamatergic cells. Autism spectrum disorders are heterogeneous disorders with varying phenotypic expression and severity, but a core feature is disrupted development of typical social behaviors (Volkmar et al., 2005). Similar deficits can be present in children raised in suboptimal caregiving environments in infancy (Rutter, Kreppner, & O’Connor, 2001).
Although it has long been known that normal amygdala development is critical for instantiation of appropriate social responses in monkeys (Bauman, Lavenex, Mason, Capitanio, & Amaral, 2004; Goursaud & Bachevalier, 2007; Prather et al., 2001; Schumann et al., 2011), the molecular-anatomic correlates of typical social learning and response in the infant have been unclear. Here, we find evidence that disrupted expression of tbr1 in the PL is associated with maternal deprivation and decreased time spent in typical social behavior by 3 months of life. Whether tbr1 is directly involved in the emergence of social behaviors, part of a larger gene network involved in social behavior, or a secondary effect of maternal deprivation and/or social impairment remains to be seen. We also do not yet know whether the gene-behavior correlation between tbr1 and time spent in age-appropriate social behavior persists over time. However, this association may be an important clue to determining how early maternal deprivation is translated to altered brain circuits with effects on social behavior. Whether disrupted gene expression in the PL is due to “stress-related” consequences of maternal deprivation such as cortisol activation (Feng et al., 2011) and inflammation (Cole et al., 2012)—or to deficits in social and sensory inputs required for normal PL development—also remain to be tested.
Finally, it is important to recognize that downregulation of tbr1 expression following early maternal loss may point to either regression or acceleration of young neurons along their developmental trajectory. Future studies will be needed to determine whether there are fewer or more mature neurons in the PL of monkeys undergoing early social stress. Several groups have suggested a hastening of development of neural growth and/or connectivity following a stressful early life experience (Gee et al., 2013; Schumann et al., 2011).
There are several limitations to this study. The most important is related to the sample size, necessitated by the value of the primate animals used and their extended gestational period. Larger animal numbers would clearly have enhanced statistical power to detect changes in other gene transcripts. In order to minimize variability in this small sample, the study was limited to female monkeys, and a larger future study should examine both sexes. Although we have not seen significant behavioral differences between the sexes in subsequent cohorts (Bauer et al., 2000; Cameron et al., 1998, 1999), it will be important to study gene expression and brain changes in both males and females, including into adulthood. In larger studies, the potential impact of nutritional differences between maternally deprived and maternally reared infants also needs to be examined, given growing recognition of nutritional differences between breast milk and standard human formula. Standard infant formula, such as used in this study, has decreased amounts of fatty acids, which are required for normal behavioral and neural development (Champoux et al., 2002). This fact has important implications for maternal deprivation paradigms in general and warrants further exploration.
On the microarray side, the relatively low RIN numbers for our samples indicate that our ability to detect other potentially important gene expression changes, particularly in the PL, is reduced (Schroeder et al., 2006). However, despite these challenges, several genes emerged in both Study 1 and Study 2 that were validated by qPCR. Although important genes were likely undetectable due to low sample size and lower than optimal RIN, we were able to show several robust subregion-specific gene expression changes between the PL and AHA in normal control animals, and altered gene expression of tbr1 in maternally deprived groups in the PL. Importantly, there was also differential expression of genes that are known to be temporally and/or biologically linked. Additionally, two of four genes in the NM gene set that were relatively highly expressed in normal PL in Study 1, bcl2, dcx, have also been demonstrated at the protein level, further validating those findings. Furthermore, the remaining two of the four genes were new discoveries: neuroD1 and tbr1. In a larger sample and with higher RIN, we would expect additional NM gene transcripts specific to the PL to be revealed. Despite limitations, both Study 1 and 2 highlight that the particular developmental “signature” of the amygdala varies by subregion, a point that needs to be considered in future studies of developmentally vulnerable circuitry.
Finally, we used a hypothesis-driven approach, focusing on genes involved in NM. The findings of this study do not diminish the possibility that other neurobiological processes in the PL are co-occurring in the 3-month animal, or are influenced by early maternal deprivation. In fact, we have previously published that maternal deprivation leads to time-dependent changes in gene expression in other non-NM genes in other subregions of the amygdala (Sabatini et al., 2007). Our future studies will utilize more broad pathway analysis to interrogate other biologic processes associated with these PL-specific findings.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Dana Helmreich (University of Rochester) for her assistance with in situ hybridization experiments, Dr. John Olschowka (University of Rochester) for assistance with qPCR, and Ms. Kelly Rogers (University of Pittsburgh) for her graciousness in providing technical and organizational support. We are especially grateful to Dr. Frank Middleton and Karen Gentile at the SUNY Upstate Microarray Facility for their support and use of laser capture microscope resources. The expert technical assistance in rearing infant monkeys by Sally Kuhn, Sally Acosta, Nathan Rockcastle, Erika Bauer, Denise Colosimo, Henry Lange, and Jennifer Dry-Henich was greatly appreciated. This work is supported by F30 MH096502 (DD), MH62931 (JF), MH41712, and the John D. and Catherine T. McArthur Foundation Network on Early Experience and Brain Development (JLC).
Funding information
John D. and Catherine T. McArthur Foundation Network on Early Experience and Brain Development (JLC), National Institute of Mental Health, Grant number: F30 MH096502(DD), R01 MH62931(JF), R01 MH41712(JLC)
Footnotes
Disclosures: The authors declare no competing financial interests.
SUPPORTING INFORMATION
Additional Supporting Information may be found online in the supporting information tab for this article.
REFERENCES
- Bauer EK, Coleman R, Kupfer DJ, & Cameron JL (2000). Social cognition in monkeys reared with maternal separation at one week of age. Abstract presented at the Society for Neuroscience. 17941. [Google Scholar]
- Bauman MD, Lavenex P, Mason WA, Capitanio JP, & Amaral DG (2004). The development of social behavior following neonatal amygdala lesions in rhesus monkeys. Journal of Cognitive Neuroscience, 16, 1388–1411. [DOI] [PubMed] [Google Scholar]
- Bernier PJ, Bedard A, Vinet J, Levesque M, & Parent A (2002). Newly generated neurons in the amygdala and adjoining cortex of adult primates. Proceedings of the National Academy of Sciences of the United States of America, 99, 11464–11469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier PJ, Vinet J, Cossette M, & Parent A (2000). Characterization of the subventricular zone of the adult human brain: Evidence for the involvement of Bcl-2. Neuroscience Research, 37, 67–78. [DOI] [PubMed] [Google Scholar]
- Boutin C, Hardt O, de Chevigny A, Core N, Goebbels S, Seidenfaden R, …Cremer H (2010). NeuroD1 induces terminal neuronal differentiation in olfactory neurogenesis. Proceedings of the National Academy of Sciences of the United States of America, 107, 1201–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brothers L, Ring B, & Kling A (1990). Response of neurons in the macaque amygdala to complex social stimuli. Behavioural Brain Research, 41, 199–213. [DOI] [PubMed] [Google Scholar]
- Bruce J, Tarullo AR, & Gunnar MR (2009). Disinhibited social behavior among internationally adopted children. Developmental Psychopathology, 21, 157–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulfone A, Smiga SM, Shimamura K, Peterson A, Puelles L, & Rubenstein JL (1995). T-brain-1: A homolog of Brachyury whose expression defines molecularly distinct domains within the cerebral cortex. Neuron, 15, 63–78. [DOI] [PubMed] [Google Scholar]
- Burghy CA, Stodola DE, Ruttle PL, Molloy EK, Armstrong JM, Oler JA,… Birn RM (2012). Developmental pathways to amygdala-prefrontal function and internalizing symptoms in adolescence. Nature Neuroscience, 15, 1736–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron JL (2001). Critical periods for social attachment: Deprivation and neural systems in rhesus monkeys. Paper presented at the Soc Res Child Development. 2–054. [Google Scholar]
- Cameron JL, Coleman K, Bench LM, Sabatini M, Owenby T, & Kupfer DJ (1998). Differential development of anxious and depressive behaviors in rhesus monkeys dependent on the timing of maternal separation. Abstract presented at the Society for Neuroscience. 2117. [Google Scholar]
- Cameron JL, Colemen K, Dahl RE, Ryan ND, & Kupfer DJ (1999). Disruption of the maternal-infant bond in early development leads to sustained alteratons in responsiveness to social stimuli. Abstract presented at the Society for Neuroscience. 2479. [Google Scholar]
- Champoux M, Hibbeln JR, Shannon C, Majchrzak S, Suomi SJ, Salem N, Higley JD (2002). Fatty acid supplementation and neuromotor development in Rhesus monkey neonates. Pediatric Research, 51, 273–281. [DOI] [PubMed] [Google Scholar]
- Chareyron LJ, Lavenex PB, Amaral DG, & Lavenex P (2012). Postnatal development of the amygdala: A stereological study in macaque monkeys. Journal of Comparative Neurology, 520, 1965–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung ZH, Chin WH, Chen Y, Ng YP, & Ip NY (2007). Cdk5 is involved in BDNF-stimulated dendritic growth in hippocampal neurons. PLoS Biology, 5, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm K (1998). A three year follow-up of attachment and indiscriminate friendliness in children adopted from Romanian orphanages. Child Development, 69, 1092–1106. [PubMed] [Google Scholar]
- Cole SW, Conti G, Arevalo JM, Ruggiero AM, Heckman JJ, & Suomi SJ (2012). Transcriptional modulation of the developing immune system by early life social adversity. Proceedings of the National Academy of Sciences of the United States of America, 109, 20578–20583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deCampo DM, & Fudge JL (2012). Where and what is the paralaminar nucleus? A review on a unique and frequently overlooked area of the primate amygdala. [Review]. Neuroscience and Biobehavioural Reviews, 36, 520–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decampo DM, & Fudge JL (2013). Amygdala projections to the lateral bed nucleus of the stria terminalis in the macaque: Comparison with ventral striatal afferents. Journal of Comparative Neurology, 521, 3191–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- des Portes V, Pinard JM, Billuart P, Vinet MC, Koulakoff A, Carrie A, …Chelly J (1998). A novel CNS gene required for neuronal migration and involved in X-linked subcortical laminar heterotopia and lissencephaly syndrome. Cell, 92, 51–61. [DOI] [PubMed] [Google Scholar]
- Dhavan R, & Tsai LH (2001). A decade of CDK5. [Review]. Nature Reviews Molecular Cell Biology, 2, 749–759. [DOI] [PubMed] [Google Scholar]
- Dicks D, Myers RE, & Kling A (1969). Uncus and amiygdala lesions: Effects on social behavior in the free-ranging rhesus monkey. Science, 165, 69–71. [DOI] [PubMed] [Google Scholar]
- Donoghue MJ, & Rakic P (1999). Molecular gradients and compartments in the embryonic primate cerebral cortex. Cerebral Cortex, 9, 586–600. [DOI] [PubMed] [Google Scholar]
- Fazeli A, Dickinson SL, Hermiston ML, Tighe RV, Steen RG, Small CG, …Weinberg RA (1997). Phenotype of mice lacking functional Deleted in colorectal cancer (Dcc) gene. Nature, 386, 796–804. [DOI] [PubMed] [Google Scholar]
- Feng X, Wang L, Yang S, Qin D, Wang J, Li C, … Hu X (2011). Maternal separation produces lasting changes in cortisol and behavior in rhesus monkeys. Proceedings of the National Academy of Sciences of the United States of America, 108, 14312–14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freese JL, & Amaral DG (2009). Neuroanatomy of the primate amygdala In Whalen PJ, & Phelps EA (Eds.), The human amygdala (pp. 3–42). New York: Guilford Press. [Google Scholar]
- Fudge JL (2004). Bcl-2 immunoreactive neurons are differentially distributed in subregions of the amygdala and hippocampus of the adult macaque. Neuroscience, 127, 539–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudge JL, deCampo DM, & Becoats KT (2012). Revisiting the hippocampal-amygdala pathway in primates: Association with immature-appearing neurons. Neuroscience, 212, 104–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudge JL, & Tucker T (2009). Amygdala projections to central amygdaloid nucleus subdivisions and transition zones in the primate. Neuroscience, 159, 819–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino T, Wu Z, Lin WC, Phillips MA, & Nedivi E (2008). Cpg15 and cpg15–2 constitute a family of activity-regulated ligands expressed differentially in the nervous system to promote neurite growth and neuronal survival. Journal of Comparative Neurology, 507, 1831–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee DG, Gabard-Durnam LJ, Flannery J, Goff B, Humphreys KL, Telzer EH, …Tottenham N (2013). Early developmental emergence of human amygdala-prefrontal connectivity after maternal deprivation. Proceedings of the National Academy of Sciences of the United States of America, 110, 15638–15643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson JG, Allen KM, Fox JW, Lamperti ED, Berkovic S, Scheffer I, … Walsh CA (1998). Doublecortin, a brain-specific gene mutated in human X-linked lissencephaly and double cortex syndrome, encodes a putative signaling protein. Cell, 92, 63–72. [DOI] [PubMed] [Google Scholar]
- Gleeson JG, Lin PT, Flanagan LA, & Walsh CA (1999). Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron, 23, 257–271. [DOI] [PubMed] [Google Scholar]
- Goursaud AP, & Bachevalier J (2007). Social attachment in juvenile monkeys with neonatal lesion of the hippocampus, amygdala and orbital frontal cortex. Behaviouraf Brain Research, 176, 75–93. [DOI] [PubMed] [Google Scholar]
- Gupta A, Tsai LH, & Wynshaw-Boris A (2002). Life is a journey: A genetic look at neocortical development. [Review]. Nature Reviews. Genetics, 3, 342–355. [DOI] [PubMed] [Google Scholar]
- Harlow HF, & Zimmermann RR (1959). Affectional responses in the infant monkey; orphaned baby monkeys develop a strong and persistent attachment to inanimate surrogate mothers. Science, 130, 421–432. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Shi L, Justice N, Hsueh Y, Sheng M, Smiga S … Rubenstein JL (2001). Tbr1 regulates differentiation of the preplate and layer 6. Neuron, 29, 353–366. [DOI] [PubMed] [Google Scholar]
- Hodge RD, Kahoud RJ, & Hevner RF (2012). Transcriptional control of glutamatergic differentiation during adult neurogenesis. Cellular and Molecular Life Sciences: CMLS, 69, 2125–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TN, & Hsueh YP (2015). Brain-specific transcriptional regulator T-brain-1 controls brain wiring and neuronal activity in autism spectrum disorders. [Review]. Frontiers in Neuroscience, 9, 406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain NK, Jenna S, Glogauer M, Quinn CC, Wasiak S, Guipponi M,… McPherson PS (2001). Endocytic protein intersectin-I regulates actin assembly via Cdc42 and N-WASP. Nature Cell Biology, 3, 927–932. [DOI] [PubMed] [Google Scholar]
- Joffe TH (1997). Social pressures have selected for an extended juvenile period in primates. Journal of Human Evolution, 32, 593–605. [DOI] [PubMed] [Google Scholar]
- Katz LC, & Shatz CJ (1996). Synaptic activity and the construction of cortical circuits. Science, 274, 1133–1138. [DOI] [PubMed] [Google Scholar]
- Keino-Masu K, Masu M, Hinck L, Leonardo ED, Chan SS, Culotti JG, & Tessier-Lavigne M (1996). Deleted in Colorectal Cancer (DCC) encodes a netrin receptor. Cell, 87, 175–185. [DOI] [PubMed] [Google Scholar]
- Knudsen EI, Heckman JJ, Cameron JL, & Shonkoff JP (2006). Economic, neurobiological, and behavioral perspectives on building America’s future workforce. Proceedings of the National Academy of Science of the United States of America, 103, 10155–10162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolk SM, Whitman MC, Yun ME, Shete P, & Donoghue MJ (2005). A unique subpopulation of Tbr1-expressing deep layer neurons in the developing cerebral cortex. Molecular Cell Neuroscience, 30, 538–551. [DOI] [PubMed] [Google Scholar]
- Kordower JH, Piecinski P, & Rakic P (1992). Neurogenesis of the amygdaloid nuclear complex in the rhesus monkey. Brain Research. Developmental Brain Research, 68, 9–15. [DOI] [PubMed] [Google Scholar]
- Leemhuis J, Bouche E, Frotscher M, Henle F, Hein L, Herz J, …Bock HH (2010). Reelin signals through apolipoprotein E receptor 2 and Cdc42 to increase growth cone motility and filopodia formation. Journal of Neuroscience, 30, 14759–14772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie DC, & Gotz M (2009). Adult neurogenesis: Similarities and differences in stem cell fate, proliferation, migration, and differentiation in distinct forebrain regions In Gage FH, Kempermann G, & Song H (Eds.), Adult neurogenesis (pp. 227–266). New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Machado CJ, & Bachevalier J (2006). The impact of selective amygdala, orbital frontal cortex, or hippocampal formation lesions on established social relationships in rhesus monkeys (Macaca mulatta). Behavioural Neuroscience, 120, 761–786. [DOI] [PubMed] [Google Scholar]
- Machado CJ, Emery NJ, Capitanio JP, Mason WA, Mendoza SP, & Amaral DG (2008). Bilateral neurotoxic amygdala lesions in rhesus monkeys (Macaca mulatta): Consistent pattern of behavior across different social contexts. Behavioral Neuroscience, 122, 251–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiri KF, Saffell JL, Walsh FS, & Doherty P (1998). Neurite outgrowth stimulated by neural cell adhesion molecules requires growth-associated protein-43 (GAP-43) function and is associated with GAP-43 phosphorylation in growth cones. Journal of Neuroscience, 18, 10429–10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merry DE, Veis DJ, Hickey WF, & Korsmeyer SJ (1994). Bcl-2 protein expression is widespread in the developing nervous system and retained in the adult PNS. Integrated Annual Indexes, 120, 301–311. [DOI] [PubMed] [Google Scholar]
- Meyer G, Perez-Garcia CG, & Gleeson JG (2002). Selective expression of doublecortin and LIS1 in developing human cortex suggests unique modes of neuronal movement. Cerebral Cortex, 12, 1225–1236. [DOI] [PubMed] [Google Scholar]
- Mirnics K, Levitt P, & Lewis DA (2006). Critical appraisal of DNA microarrays in psychiatric genomics. [Review]. Biological Psychiatry, 60, 163–176. [DOI] [PubMed] [Google Scholar]
- Miyata T, Maeda T, & Lee JE (1999). NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes & Development, 13, 1647–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG,… Shendure J (2012). Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science, 338, 1619–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, & Poo MM (2013). Neurotrophin regulation of neural circuit development and function. Nature Reviews Neuroscience, 14, 7–23. [DOI] [PubMed] [Google Scholar]
- Pitkanen A, & Amaral DG (1998). Organization of the intrinsic connections of the monkey amygdaloid complex: Projections originating in the lateral nucleus. Journal of Comparative Neurology, 398, 431–458. [DOI] [PubMed] [Google Scholar]
- Prather MD, Lavenex P, Mauldin-Jourdain ML, Mason WA, Capitanio JP, Mendoza SP, & Amaral DG (2001). Increased social fear and decreased fear of objects in monkeys with neonatal amygdala lesions. Neuroscience, 106, 653–658. [DOI] [PubMed] [Google Scholar]
- Price JL, Russchen FT, & Amaral DG (1987). The limbic region. II. The amygdaloid complex In Hokfelt BT, & Swanson LW (Eds.), Handbook of chemical neuroanatomy (pp. 279–381). Amsterdam: Elsevier. [Google Scholar]
- Roybon L, Deierborg T, Brundin P, & Li JY (2009). Involvement of Ngn2, Tbr and NeuroD proteins during postnatal olfactory bulb neurogenesis. European Journal of Neuroscience, 29, 232–243. [DOI] [PubMed] [Google Scholar]
- Rutter ML, Kreppner JM, & O’Connor TG (2001). Specificity and heterogeneity in children’s responses to profound institutional privation. British Journal of Psychiatry, 179, 97–103. [DOI] [PubMed] [Google Scholar]
- Sabatini MJ, Ebert P, Lewis DA, Levitt P, Cameron JL, & Mirnics K (2007). Amygdala gene expression correlates of social behavior in monkeys experiencing maternal separation. Journal of Neuroscience, 27, 3295–3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders RC, & Rosene DL (1988). A comparison of the efferents of the amygdala and the hippocampal formation in the rhesus monkey: I. Convergence in the entorhinal, prorhinal, and perirhinal cortices. Journal of Comparative Neurolology, 271, 153–184. [DOI] [PubMed] [Google Scholar]
- Saunders RC, Rosene DL, & Van Hoesen GW (1988). Comparison of the efferents of the amygdala and the hippocampal formation in the rhesus monkey: II. Reciprocal and non-reciprocal connections. Journal of Comparative Neurology, 271, 185–207. [DOI] [PubMed] [Google Scholar]
- Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, … Ragg T (2006). The RIN: An RNA integrity number for assigning integrity values to RNA measurements. [Evaluation Studies]. BMC Molecular Biology, 7, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann CM, Bauman MD, & Amaral DG (2011). Abnormal structure or function of the amygdala is a common component of neurodevelopmental disorders. Neuropsychologia, 49, 745–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P, Lawrence EJ, Radbourne C, Bramham J, Polkey CE, & David AS (2004). The impact of early and late damage to the human amygdala on ‘theory of mind’ reasoning. Brain, 127, 1535–1548. [DOI] [PubMed] [Google Scholar]
- Shelton RC, Claiborne J, Sidoryk-Wegrzynowicz M, Reddy R, Aschner M, Lewis DA, & Mirnics K (2011). Altered expression of genes involved in inflammation and apoptosis in frontal cortex in major depression. Molecular Psychiatry, 16, 751–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan MA, Fox NA, Zeanah CH, McLaughlin KA, & Nelson CA 3rd. (2012). Variation in neural development as a result of exposure to institutionalization early in childhood. Proceedings of the National Academy of Sciences of the United States of America, 109, 12927–12932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidman RL, & Rakic P (1973). Neuronal migration, with special reference to developing human brain: A review. Brain Research, 62, 1–35. [DOI] [PubMed] [Google Scholar]
- Spitz RA (1946). Anaclitic depression: An inquiry into the genesis of psychiatric conditions in early childhood. The Psychoanalytic Study of the Child, 2, 313–342. [PubMed] [Google Scholar]
- Stephan H, & Andy OJ (1977). Quantitative comparison of the amygdala in insectivores and primates. Acta Anatomica (Basel), 98, 130–153. [DOI] [PubMed] [Google Scholar]
- Strittmatter SM, Fankhauser C, Huang PL, Mashimo H, & Fishman MC (1995). Neuronal pathfinding is abnormal in mice lacking the neuronal growth cone protein GAP-43. Cell, 80, 445–452. [DOI] [PubMed] [Google Scholar]
- Sumi T, Matsumoto K, Takai Y, & Nakamura T (1999). Cofilin phosphorylation and actin cytoskeletal dynamics regulated by rho-and Cdc42-activated LIM-kinase 2. Journal of Cell Biology, 147, 1519–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomi SJ (1997). Early determinants of behaviour: Evidence from primate studies. British Medical Bulletin, 53, 170–184. [DOI] [PubMed] [Google Scholar]
- Suomi SJ, Collins ML, & Harlow HF (1973). Effects of permanent separation from mother on infant monkeys. Developmental Psychology, 9, 376–384. [Google Scholar]
- Tottenham N (2012). Human amygdala development in the absence of species-expected caregiving. Developmental Psychobiology, 54, 598–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tottenham N, Hare TA, Quinn BT, McCarry TW, Nurse M, Gilhooly T, … Casey, B. J. (2010). Prolonged institutional rearing is associated with atypically large amygdala volume and difficulties in emotion regulation. Developmental Science, 13, 46–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranel D, & Hyman BT (1990). Neuropsychological correlates of bilateral amygdala damage. Archives of Neurology, 47, 349–355. [DOI] [PubMed] [Google Scholar]
- Ulfig N, Neudorfer F, & Bohl J (2000). Transient structures of the human fetal brain: Subplate, thalamic reticular complex, ganglionic eminence. Histology Histopathology, 15, 771–790. [DOI] [PubMed] [Google Scholar]
- Ulfig N, Setzer M, & Bohl J (1998). Transient architectonic features in the basolateral amygdala of the human fetal brain. Acta Anatomica (Basel), 163, 99–112. [DOI] [PubMed] [Google Scholar]
- Ulfig N, Setzer M, & Bohl J (2003). Ontogeny of the human amygdala. Annals of the New York Academy of Sciences, 985, 22–33. [DOI] [PubMed] [Google Scholar]
- Volkmar F, Chawarska K, & Klin A (2005). Autism in infancy and early childhood. Annual Reviews in Psychology, 56, 315–336. [DOI] [PubMed] [Google Scholar]
- Walker R, Burger O, Wagner J, & Von Rueden CR (2006). Evolution of brain size and juvenile periods in primates. Journal of Human Evolution, 51, 480–489. [DOI] [PubMed] [Google Scholar]
- Yachnis AT, Roper SN, Love A, Fancey JT, & Muir D (2000). Bcl-2 immunoreactive cells with immature neuronal phenotype exist in the nonepileptic adult human brain. Journal of Neuropathology & Experimental Neurology, 59, 113–119. [DOI] [PubMed] [Google Scholar]
- Zeanah CH, Egger HL, Smyke AT, Nelson CA, Fox NA, Marshall PJ, & Guthrie D (2009). Institutional rearing and psychiatric disorders in Romanian preschool children. American Journal of Psychiatry, 166, 777–785. [DOI] [PubMed] [Google Scholar]
- Zhang KZ, Westberg JA, Holtta E, & Andersson LC (1996). BCL2 regulates neural differentiation. Proceedings of the National Academy of Sciences of the United States of America, 93, 4504–4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong LT, Sarafian T, Kane DJ, Charles AC, Mah SP, Edwards RH, & Bredesen DE (1993). Bcl-2 inhibits death of central neural cells induced by multiple agents. Proceedings of the National Academy of Sciences of the United States of America, 90, 4533–4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zukerberg LR, Patrick GN, Nikolic M, Humbert S, Wu CL, Lanier LM, … Tsai, L. H. (2000). Cables links Cdk5 and c-Abl and facilitates Cdk5 tyrosine phosphorylation, kinase upregulation, and neurite outgrowth. Neuron, 26, 633–646. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


