Abstract
As obligate parasites, plant viruses usually hijack host cytoskeletons for replication and movement. Rhabdoviruses are enveloped, negative-stranded RNA viruses that infect vertebrates, invertebrates, and plants, but the mechanisms of intracellular trafficking of plant rhabdovirus proteins are largely unknown. Here, we used Barley yellow striate mosaic virus (BYSMV), a plant cytorhabdovirus, as a model to investigate the effects of the actin cytoskeleton on viral intracellular movement and viral RNA synthesis in a mini-replicon (MR) system. The BYSMV P protein forms mobile inclusion bodies that are trafficked along the actin/endoplasmic reticulum network, and recruit the N and L proteins into viroplasm-like structures. Deletion analysis showed that the N terminal region (aa 43–55) and the remaining region (aa 56–295) of BYSMV P are essential for the mobility and formation of inclusions, respectively. Overexpression of myosin XI-K tails completely abolishes the trafficking activity of P bodies, and is accompanied by a significant reduction of viral MR RNA synthesis. These results suggest that BYSMV P contributes to the formation and trafficking of viroplasm-like structures along the ER/actin network driven by myosin XI-K. Thus, rhabdovirus P appears to be a dynamic hub protein for efficient recruitment of viral proteins, thereby promoting viral RNA synthesis.
Keywords: Actin cytoskeleton, Barley yellow striate mosaic virus, intracellular movement, myosin motor, P bodies, rhabdovirus
A cytorhabdovirus phosphoprotein forms mobile inclusion bodies that traffic along the actin/endoplasmic reticulum network in a process that is important for viral RNA synthesis.
Introduction
As obligate parasites, plant viruses are heavily reliant on host cell functions for replication and movement, and there is accumulating evidence to suggest that microtubules and microfilaments are implicated in the intracellular trafficking of viral proteins (Laporte et al., 2003; Sambade et al., 2008; Amari et al., 2011; Yuan et al., 2011; Agbeci et al., 2013; Pitzalis and Heinlein, 2017). In plant cells, the endoplasmic reticulum (ER) runs along actin filaments and forms a cytoplasmic actin/ER network that induces rapid streaming of the ER in the cortical cytoplasm (Boevink et al., 1998). Myosins play the main role in driving actin-based cytoplasmic streaming within plant cells (Tominaga et al., 2003; Avisar et al., 2008b; Kurth et al., 2017). A number of movement proteins of plant viruses have been reported to make use of the actomyosin cytoskeleton for transportation targeted at the plasmodesmata (PD). For instance, class VIII myosins are required for PD targeting of the closterovirus Hsp70 homolog, the NSvc4 protein of Rice stripe virus (RSV), and the movement protein (MP) of Tobacco mosaic virus (TMV) (Avisar et al., 2008a; Yuan et al., 2011; Amari et al., 2014). The cell-to-cell movement of Grapevine fanleaf virus (GFLV) requires class XI myosins for tubule-guided transport between plant cells (Amari et al., 2011). In addition, several viral movement-independent proteins also form mobile inclusions, and their trafficking along the ER/actin network is required for optimal replication (Kawakami et al., 2004; Liu et al., 2005; Harries et al., 2009a; Cui et al., 2010; Amari et al., 2014; Ishikawa et al., 2015; Feng et al., 2016). This suggests that trafficking of inclusions along the ER/actin network is a general mechanism that is exploited by various plant viruses to support viral propagation and inter- and intracellular movement.
The intracellular transport of diverse viral proteins from non-enveloped plant viruses has been well studied, whereas in contrast our understanding of intracellular trafficking of enveloped plant virus proteins is limited. Most animal viruses are enveloped and require the ER–Golgi secretory pathway for the formation of viral replication complexes (VRCs), and for intra- and intercellular movement (Brandenburg and Zhuang, 2007). However, it has been shown that nucleocapsid proteins of two plant enveloped viruses, Tomato spotted wilt tospovirus (TSWV) and Fig mosaic virus (FMV), form cytoplasmic inclusions that are trafficked along the ER/actin network in a manner that is dependent on class XI myosins (Feng et al., 2013; Ishikawa et al., 2015). It remains to be determined whether the movement pathways of viral nucleocapsids are conserved in other enveloped plant viruses.
Rhabdoviruses are bullet-shaped or bacilliform enveloped virions containing single-stranded, negative-sense RNA genomes and they have a broad host range, including vertebrates, invertebrates, and plants. Rhabdovirus genomes encode five structural proteins, namely nucleoprotein (N), phosphoprotein (P), matrix protein (M), glycoprotein (G), and polymerase (L), in the conserved order 3′–N–P–M–G–L–5′. The N, P, and L proteins are required for viral RNA replication and mRNA transcription, which occurs in a highly regulated manner (Ivanov et al., 2011). In the nucleocapsid (NC) complex, a minimal infectious unit, the RNA genome is encapsulated by N proteins into an N–RNA complex that serves as a template for viral replication and transcription by the polymerase complex, which is comprised of the L and P proteins (Ivanov et al., 2011; Leyrat et al., 2011). The rhabdovirus P proteins mainly serve as non-catalytic L co-factors and chaperone RNA-free N proteins (N0) (Ivanov et al., 2011; Leyrat et al., 2011). The rhabdovirus L proteins exhibit several enzymatic activities, including RNA synthesis, mRNA capping, and polyadenylation (Ivanov et al., 2011). Although rhabdovirus NC complexes have been the subject of numerous studies, it is not known whether the plant intracellular trafficking pathways are implicated in their assembly.
Barley yellow striate mosaic virus (BYSMV) is a member of the genus Cytorhabdovirus, and is transmitted to cereal hosts by the small brown planthopper (SBPH, Laodelphax striatellus) in a persistent-propagative manner (Cao et al., 2018). The BYSMV genome encodes 10 proteins in the order 3′–N–P–P3–P4/P5–P6–M–G–P9–L–5′ (Yan et al., 2015). Here, we used BYSMV as a model cytorhabdovirus to investigate localization and mobility of plant rhabdovirus proteins, and found that the P protein forms mobile inclusion bodies along the ER/actin network driven by class XI myosins. We further demonstrated that mobile P bodies recruit the N and L proteins into viroplasm-like structures. Our results reveal an entirely new role for the P protein in the assembly of plant rhabdovirus N and L proteins. We further examined the potential role of P movement in a mini-genome system in Nicotiana benthamiana leaves.
Materials and methods
Plasmid constructs
For construction of BYSMV mini-replicon (MR) reporter cassettes, total RNA was extracted from BYSMV-infected Laodelphax striatellus with TRIzol reagent (Invitrogen) and used as a template for reverse transcription (RT) to synthesize the first-strand full-length cDNA of BYSMV with SuperScript III Reverse Transcriptase (Invitrogen). The full-length cDNA of the BYSMV anti-genomic (ag) RNA was amplified using Phanta Max Super-Fidelity DNA Polymerase (Vazyme Biotech) with the primer pair BYS-F and BYS-R (see Supplementary Table S1 at JXB online for a list of all primers used in this study), and then engineered into a binary plasmid pCB301-2×35S-Nos (Shen et al., 2014). For construction of pBYSMV-agMR, the pBYSMV-NP was generated by reverse PCR using the full-length cDNA clone of the BYSMV agRNA as a template with primers MR1-F and MR1-R. Then, a PCR fragment containing the pBYSMV-NP sequence up to the N protein ORF was amplified by reverse PCR with the MR2-F and MR2-R primers, and fused with the green fluorescent protein (GFP) ORF amplified from pGDG (Goodin et al., 2002) with the primer pair GFP-F and GFP-R. Using a similar strategy, the P protein ORF was replaced with the red fluorescent protein (RFP) reporter gene from pGD-RFP (Goodin et al., 2002) with two pairs of primers (MR1-F and MR3-R, RFP-F and RFP-R). The pGD-N and pGD-P plasmids were generated by insertion of the BYSMV N, P, and L ORF cDNA sequences into the pGD vector (Goodin et al., 2002). pGD-L was generated by insertion of the BYSMV L ORF cDNA fused with a 6×Myc tag into the pGD vector (Goodin et al., 2002). To generate the pGD-VSRs plasmid, the ORF sequences of three viral suppressors of RNA silencing (VSRs), namely the Tomato bushy stunt virus p19, the Tobacco etch virus HC-Pro, and the Barley stripe mosaic virus γb, were individually inserted into the pGD vector. The pGD expression cassettes were then amplified, ligated, and integrated into another pGD vector to simultaneously express the three VSRs.
For subcellular localization assays, the N and P genes of BYSMV or NCMV, as well as their relative mutants, were fused in-frame with cyan fluorescent protein (CFP) or GFP at either the N- or C-terminus and were introduced into the pGD vector (Goodin et al., 2002) with different primers. The L gene of BYSMV was inserted into pSuper1300-mCherry (Jin et al., 2018) and fused in-frame with the mCherry N-terminus. All of these constructs were confirmed by sequencing. The ER marker mCherry-HDEL and Golgi marker STtmd-RFP were as described previously (Boevink et al., 1998; Nelson et al., 2007). For the actin marker CFP-ABD2-CFP, the ABD2 sequence (At4G26700) was amplified from cDNA of Arabidopsis, fused with CFP at both the N- and C-terminus, and cloned into the pGD vector (Goodin et al., 2002). The dominant-negative myosin constructs of VIII-1, VIII-2, VIII-B, XI-2, XI-F, and XI-K were as described previously (Avisar et al., 2008b). The GFP-ABD2-GFP and mCherry-MAP65-1 plasmids were also as described previously (Smertenko et al., 2004; Wang et al., 2008; Feng et al., 2013).
Plant material and transient expression
Four-week-old H2B-RFP transgenic (Martin et al., 2009) and wild-type plants of N icotiana benthamiana were agro-infiltrated for transient expression assays as described previously (Zhang et al., 2017). Briefly, Agrobacterium tumefaciens cells containing various constructs were resuspended in infiltration buffer (10 mM MES, pH5.6, 10 mM MgCl2, 150 μM Acetosyringone) at 25 °C at least 3 h before infiltration. For mini-replicon assays, Agrobacterium harboring pBYSMV-agMR (OD600 0.3), pGD-VSRs (OD600 0.1), pGD-N (OD600 0.1), pGD-P (OD600 0.1), and pGD-L (OD600 0.3) were mixed and infiltrated into N. benthamiana leaves. For subcellular localization assays, Agrobacterium containing different constructs encoding the N, P, L, and organelle marker-fused protein genes and dominant-negative myosin tails were diluted to OD600 0.3 and infiltrated into N. benthamiana leaves.
Western blotting analysis
Agro-infiltrated N. benthamiana leaves were harvested at 2 to 6 d post infiltration (dpi), and total proteins were extracted in SDS buffer [100 mM Tris (pH 6.8), 20% glycerol, 4% SDS, and 0.2% bromophenol blue, 10% β-mercaptoethanol] for western blotting analyses. Proteins were separated by SDS-PAGE gels followed by either staining with Coomassie blue for equal loading control or transfer to nitrocellulose membranes to detect accumulation using polyclonal antiserum specific to the BYSMV N (1:3000), P (1:3000), GFP (1:2000), RFP (1:2000), or HA (1:3000) proteins, followed by treatment with goat anti-rabbit/mouse IgG horseradish peroxidase conjugate (Bio-Rad, 1:3000), Pierce ECL Plus chemiluminescent substrate, and exposure to X-ray films (Fujifilm).
RNA analysis by northern blotting
Northern blotting was performed as described previously (Dong et al., 2016). For analysis of RNA synthesis in the BYSMV MR system, total RNA was extracted from agro-infiltrated N. benthamiana leaves using TRIzol reagent (Sigma). Northern blot detection of BYSMV MR gRNA, GFP mRNA, and RFP mRNA was carried out using 5 μg of total RNA following the methods described above. For gRNA detection, the agMR fragment under the control of the T7 promoter was transcribed in vitro with T7 RNA polymerase in the presence of [32P] UTP. For mRNA detection, randomly labeled cDNA probes of GFP and RFP were used for GFP and RFP mRNA detection, respectively.
Real-time quantitative RT-PCR (qRT-PCR)
Total RNA isolated from MR-infiltrated leaves was treated with DNase I (Takara) to eliminate DNA contamination. The treated RNA (2 μg) was then used as a template for reverse-transcription with the oligo(dT) primer to detect mRNA or the B-RT-F primers to detect mini-genome RNA using M-MLV Reverse Transcriptase (Promega). Real-time quantitative RT-PCR reactions were performed by amplifying GFP and RFP fragments to detect transcription levels and trailer fragments to monitor replication levels using SsoFast EvaGreen Supermix (Bio-Rad). All qRT-PCR data were normalized to the EF1A gene. Three independent biological replicates were performed.
Drug treatments and virus detection
Leaves of N. benthamiana were treated with 10 μM Latrunculin B (LatB; Abcam) to disrupt actin assembly at 40 h after agro-infiltration. DMSO was used for negative controls at equivalent dilutions. Confocal images were taken at 3–6 h after drug treatments. For systemic infection assays, barley plants at the single-leaf stage were infiltrated with 5 μM LatB or DMSO control solutions. At 3 h after the treatments, more than 20 viruliferous L. striatellus were transferred onto each treated barley leaf for a single day of inoculation feeding. The stems were loosely wrapped with paper to keep the L. striatellus on the drug-treated leaves. Nine plants were used in each treatment. Inoculated leaves and the systemically infected leaves were harvested at 3 dpi or 6 dpi for detection of viral accumulation using anti-N antibodies (1:3000).
Confocal laser-scanning microscopy
For subcellular localization, images were captured at 2–3 d post agro-infiltration using a Leica TCS-SP8 or Zeiss LSM710 confocal laser-scanning microscope (CLSM) or at 6 dpi for mini-replicon assays. CFP was excited with 440 nm and emissions were captured between 450–490 nm. GFP was excited with 488 nm and emissions were captured between 500–540 nm. RFP and mCherry were excited with 568 nm and emissions were captured between 580–620 nm. Time-series programs in the microscopes were used to obtain videos. For each video, one frame was obtained over 3 s or 4 s and 50 frames were obtained from a single optical plane. After acquisition, the confocal images were processed using the software provided with the CLSMs. The videos were edited to play seven frames per second using the ImageJ software. Imaris Software Surpass modes were used to plot tracks and to calculate the velocities of GFP-P bodies. For the mean velocities, 50 frames were captured at 4 s intervals and more than 90 bodies were recorded for each treatment.
Statistical analysis
One-way ANOVA was used to analyse the overall statistical differences between the different group means. Significant differences were determined by Tukey’s multiple comparison test.
Results
Construction of a BYSMV mini-replicon in N. benthamiana leaves
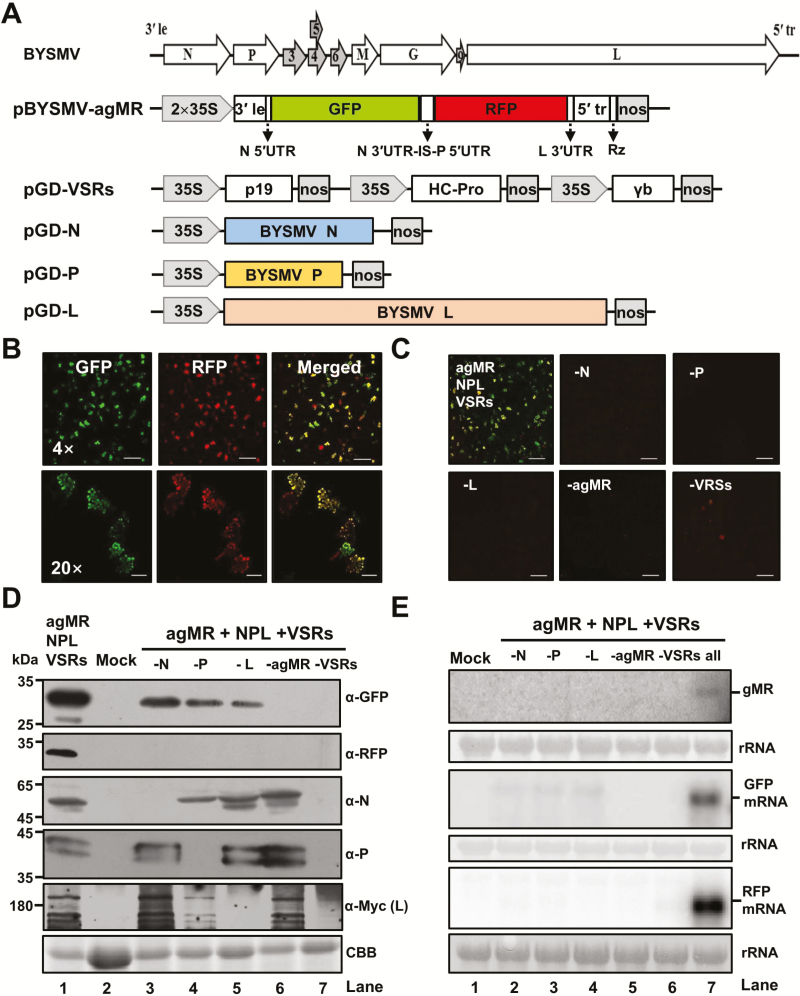
To investigate the functions of the BYSMV core proteins and cis-acting sequences in vivo, we first developed a BYSMV mini-replicon (MR) system in N. benthamiana leaves. We generated a reporter cassette of anti-genomic-sense BYSMV derivative (pBYSMV-agMR), in which the BYSMV N and P ORFs were substituted with red-shifted GFP and RFP, respectively, and were flanked by the two terminal sequences outside the N gene and the L gene (Fig. 1A). The ORFs of the BYSMV N, P, and L genes were engineered into pGD expression vectors for ectopic expression. Three VSRs, namely the Tomato bushy stunt virus p19, the Tobacco etch virus HC-Pro, and the Barley stripe mosaic virus γb, were individually inserted into pGD, and then three expression cassettes were combined into one plasmid.
Fig. 1.
Construction of a BYSMV antigenomic-sense mini-replicon. (A) Illustration of binary Agrobacterium vectors designed to generate antigenomic-sense mini-replicon (agMR) RNA and to express the BYSMV N, P, and L proteins in vivo. In the pBYSMV-agMR plasmid, a reporter cassette of antigenomic-sense BYSMV derivative was inserted between the truncated CaMV double 35S promoter (2×35S) and the hepatitis delta (HDV) ribozyme sequence (RZ). Le, leader; tr, trailer; UTR, untranslated region; N 3′UTR-IS-P 5′UTR, intergenic sequences including N 3´UTR, intergenic sequence (IS), and P 5´UTR; nos, nopaline synthase terminator. (B) GFP and RFP foci in N. benthamiana leaves agro-infiltrated with BYSMV-agMR combinations at 5 d post inoculation (dpi). Scale bars in the top panels are 500 μm and the bars in the bottom panels are 100 μm. (C) Requirement of N, P, L, and viral suppressors of RNA silencing (VSRs) for expression of MR reporter genes. GFP and RFP fluorescence were observed at 5 dpi. Scale bars are 500 μm. (D) Western blot analysis showing accumulation of the GFP, RFP, N, P, and L proteins in the leaves shown in (C) as determined using rabbit antibodies against GFP, RFP, N, P, and a mouse Myc antibody for BYSMV L. The mock sample was infiltrated with Agrobacterium harboring the pGD vector. Coomassie brilliant blue (CBB) staining was used for protein loading controls. (E) Northern blot analysis of BYSMV agMR transcription and replication. Total RNA extracted from leaves shown in panel (C) was hybridized with an in vitro transcript of a positive-sense GFP probe to detect the genomic-sense MR (gMR) RNA. Randomly labeled GFP and RFP cDNA probes were used for detection of GFP and RFP mRNA, respectively. rRNAs stained with methylene blue were used as loading controls.
To assess the BYSMV MR system, Agrobacterium harboring plasmids for expression of agMR, N, P, L, and VSRs were mixed and co-infiltrated into N. benthamiana leaves. At 5 dpi, GFP and RFP fluorescence were observed throughout the infiltrated leaves (Fig. 1B). Confocal microscopy at high magnification indicated that the fluorescence originated from single cells. To determine the requirement for reporter gene expression of each component, Agrobacterium combinations without either N, P, L, agMR, or VSRs were tested in N. benthamiana leaves. The results showed that a combination of all the components was essential for detectable reporter gene expression in vivo (Fig. 1C).
Western blotting analyses were performed to assess the expression of reporter genes and viral core proteins (Fig. 1D). In line with the fluorescence observations, GFP accumulation was significantly higher in the presence of the complete combination of BYSMV-agMR systems than for any other combination (Fig. 1D, lane 1), indicating that active transcription of GFP mRNA from replicating agMR had occurred. Unexpectedly, a residual GFP signal accumulated in leaves without expression of N, P, or L proteins (Fig. 1D, lanes 3–5), which resulted from direct translation from the GFP ORF at the 5´-end of the agRNA transcripts. RFP at the second ORF was absolutely reliant upon co-expression of N, P, L, agMR, and VSRs in combination (Fig. 1D, lane 1).
Northern blotting was performed to assess the MR and mRNA accumulation in co-infiltrated N. benthamiana leaves at 5 dpi. The genome-sense MR (gMR) that represented replication products was only detected in those tissues infiltrated together with the combination of the N, P, L, MR, and VSRs (Fig. 1E, top panel, lane 7). Highly abundant GFP and RFP mRNAs were detected with the complete combination of BYSMV-agMR systems (Fig. 1E, middle and bottom panels, lane 7). Taken together, these results demonstrated that the co-expressed N, P, and L proteins were functionally active in replication of the agMR derivatives and in transcription of reporter mRNAs.
Functional BYSMV GFP-P forms motile cytoplasmic inclusion bodies
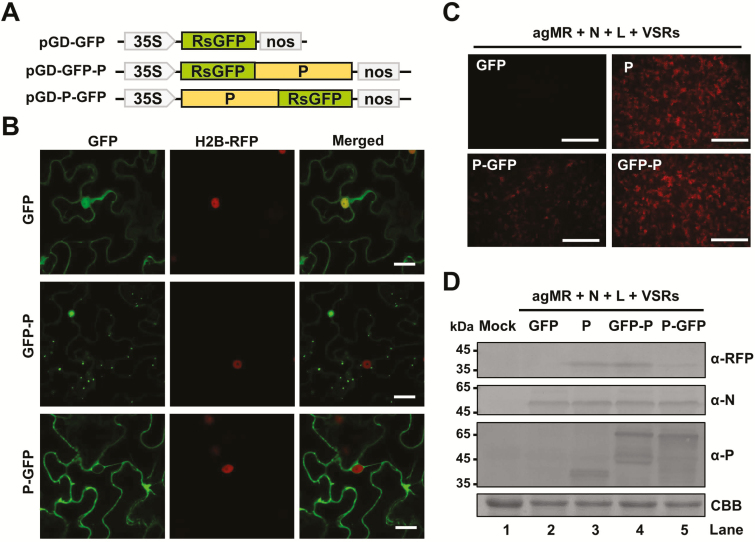
We next examined the localization of the BYSMV N and P proteins and their functions in BYSMV mini-genome infections. First, we fused the ORFs of N and P to the N or C termini of GFP under the control of the Cauliflower mosaic virus (CaMV) 35S promoter (Fig. 2A, Supplementary Fig. S1A). Agrobacterium harboring plasmids for expression of GFP-tagged proteins or free GFP were infiltrated into the leaves of H2B‐RFP transgenic N. benthamiana plants (Martin et al., 2009), and fluorescence was then monitored by confocal microscopy at 2 dpi. Free GFP localized to both the cytoplasm and nuclei (Fig. 2B, Supplementary Fig. S1B). GFP-N and N-GFP localized evenly in the cytoplasm, but not in the nuclei (Supplementary Fig. S1B). In contrast, GFP-P formed small punctate structures, which we designated BYSMV-P bodies, that were trafficked throughout the cytoplasm but failed to move between cells (Fig. 2B, Supplementary Video S1).P-GFP only exhibited diffuse fluorescence or very small inclusion bodies in the cytoplasm (Fig. 2B).
Fig. 2.
Subcellular localization and mobility of the BYSMV P protein. (A) Schematic diagram of pGD vectors for expression of free GFP, GFP-P, and P-GFP. (B) Confocal micrographs showing the subcellular distribution of free GFP, GFP-P, and P-GFP in epidermal cells of agro-infiltrated leaves of H2B-RFP transgenic N. benthamiana at 2 d post inoculation (dpi). Scale bars are 20 μm. (C) RFP foci at 6 dpi in leaves infiltrated with Agrobacterium engineered for expression of free GFP, P, GFP-P, or P-GFP in antigenomic-sense mini-replicon (agMR) combinations. Scale bars are 1 mm. (D) Western blotting analysis showing accumulation of RFP, and N and P proteins in infiltrated leaves with anti-RFP, anti-N, or anti-P polyclonal antibodies, respectively. Buffer-infiltrated leaves were used as a negative control (mock). Coomassie brilliant blue (CBB) staining was used for protein loading controls.
To examine whether GFP-P or P-GFP supported mini-genome RNA replication, GFP-P, P-GFP, or wild-type P proteins were individually co-expressed with the N, L, VSRs, and agMR in N. benthamiana leaves by agro-infiltration. The fluorescent RFP foci supported by GFP-P were comparable to those of the wild-type P protein, whereas much less intensity was observed in P-GFP samples (Fig. 2C). Western blotting consistently showed increased RFP protein levels in the P and GFP-P samples compared with that of P-GFP despite similar N and P protein levels (Fig. 2D, compare lanes 3, 4, and 5). These results therefore suggested that GFP-P, rather than P-GFP, exhibited a function similar to wild-type P in the BYSMV MR infection.
Northern cereal mosaic virus (NCMV) and BYSMV are closely related cytorhabdoviruses, and share similar genome organization and high sequence identity of major structural proteins (Yan et al., 2015). Therefore, we next investigated whether the NCMV P protein had similar features of formation and trafficking of inclusion bodies. To this end, the ORFs of NCMV N and P were fused with the N- or C-terminus of GFP, and were transiently expressed in N. benthamiana leaves. In line with the BYSMV protein localization, GFP-P, but not P-GFP, of NCMV formed trafficking inclusion bodies, whereas both GFP-N and N-GFP of NCMV were distributed evenly in the cytoplasm of the agro-infiltrated epidermal cells (Supplementary Fig. S2, Supplementary Video S2). Thus, the trafficking of BYSMV-P bodies might represent a general feature among closely related cytorhabdoviruses.
Identification of BYSMV P domains responsible for inclusion body formation and trafficking
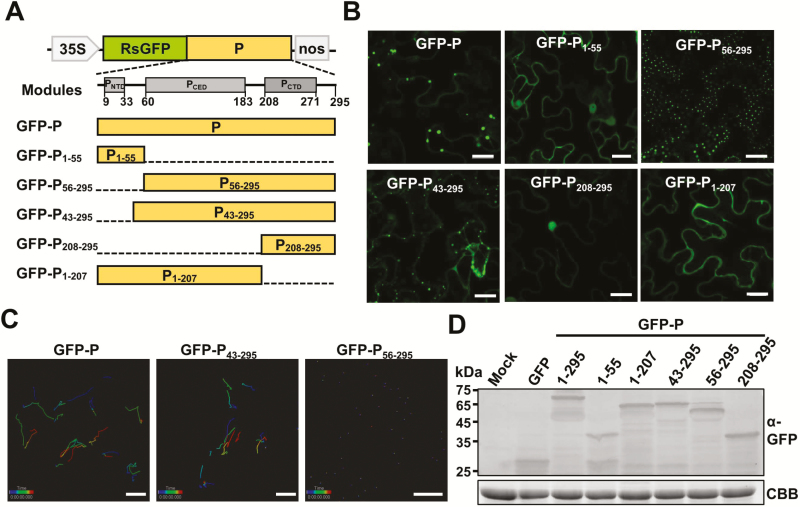
The rabies virus P protein has several truncated forms in infected cells in addition to full-length P due to alternate translation initiation at downstream AUG codons (Chenik et al., 1995). Likewise, the N-terminus of the BYSMV P protein contains an internal AUG codon that is probably used for translation of truncated forms, including P2 (aa 56–295; Supplementary Fig. S3A). Using deletion analysis, we found that P2 did indeed exist in BYSMV-infected barley plants (Supplementary Fig. S3B). To further investigate the localization of BYSMV P2, we fused the N terminal region (aa 1–55, named P1–55), or the remaining P2 region (aa 56–295, named P56–295) to the C-terminus of GFP (Fig. 3A). At 2 dpi, observation of epidermal cells revealed that GFP-P1–55 localized evenly in both the cytoplasm and nucleus, while GFP-P56–295 formed small and stationary cytoplasmic inclusion bodies throughout the cytoplasm (Fig. 3B, upper panels, and Supplementary Video S3).
Fig. 3.
Identification of BYSMV P protein domains responsible for trafficking and formation of inclusion bodies. (A) Modular organization of the BYSMV P protein and schematic presentation of the designs of deletion mutants. Normalized disorder scores (D-scores) were calculated from 15 different predictors, and structured regions (grey boxes) were defined as locations within a D-score above a threshold of 0.5 and disordered regions (lines) were defined as having a D-score below 0.5. Numbers below the boxes indicate the limits of the structured regions. Deleted regions of BYSMV P are indicated by dashed lines. (B) Fluorescence micrographs showing localizations of GFP-fused P and deletion mutants. Images were taken at 2 d post inoculation. Scale bars are 20 μm. (C) Tracking of individual GFP-P, GFP-P43–295, and GFP-P56–295 bodies using Imaris software. Individual particles in epidermal cells of agro-infiltrated N. benthamiana leaves were tracked in 1-s bursts separated by 4-s dark intervals, and 50 images were collected. Different time intervals are indicated by the different colors. Scale bars are 20 μm. (D) Detection of GFP-tagged P protein and P mutant proteins by western blotting with anti-GFP polyclonal antibodies. Coomassie brilliant blue (CBB) staining was used as the protein loading control. Healthy N. benthamiana leaves served as mock samples.
To further identify the domains required for the formation of the P bodies, we used 15 independent predictors available through web servers to analyse the structural domains of BYSMV P, including PNTD (aa 9–33), PCED (aa 60–183), and PCTD (aa 208–270) (Fig. 3B). Two BYSMV P mutants, P208–295 and P1–207, were individually fused with the C-terminus of GFP and transiently expressed in N. benthamiana leaves, and both failed to form inclusion bodies (Fig. 3B, lower panels), indicating that both the PCED and PCTD domains are essential for BYSMV-P body formation.
To further identify the domain required for mobility of the BYSMV-P bodies, we fused P43–295 to the C-terminus of GFP and expressed GFP-P43–295 in N. benthamiana leaves. The results showed that, in contrast to GFP-P56–295, GFP-P43–295 maintained movement ability (Fig. 3B and Supplementary Video S4). We then used time-lapse confocal images to monitor trafficking of GFP-P, GFP-P43–295, and GFP-P56–295 over time in leaf epidermal cells. As shown in Fig. 3C, where individual bodies are indicated by the colored lines, the GFP-P and GFP-P43–295 bodies were highly dynamic in the cells, whereas none of the GFP-P56–295 bodies exhibited appreciable movement. It should be noted that the full-length and deleted mutants of BYSMV P were expressed at similar levels (Fig. 3D), suggesting that the deletions did not affect the stability of P proteins but did change their localization and mobility.
Collectively, these results demonstrated that the PCED and PCTD domains are essential for BYSMV-P body formation, and the N-terminal aa 43–55 domain is required for cytoplasmic trafficking.
The BYSMV-P bodies are trafficked along the actin/ER network
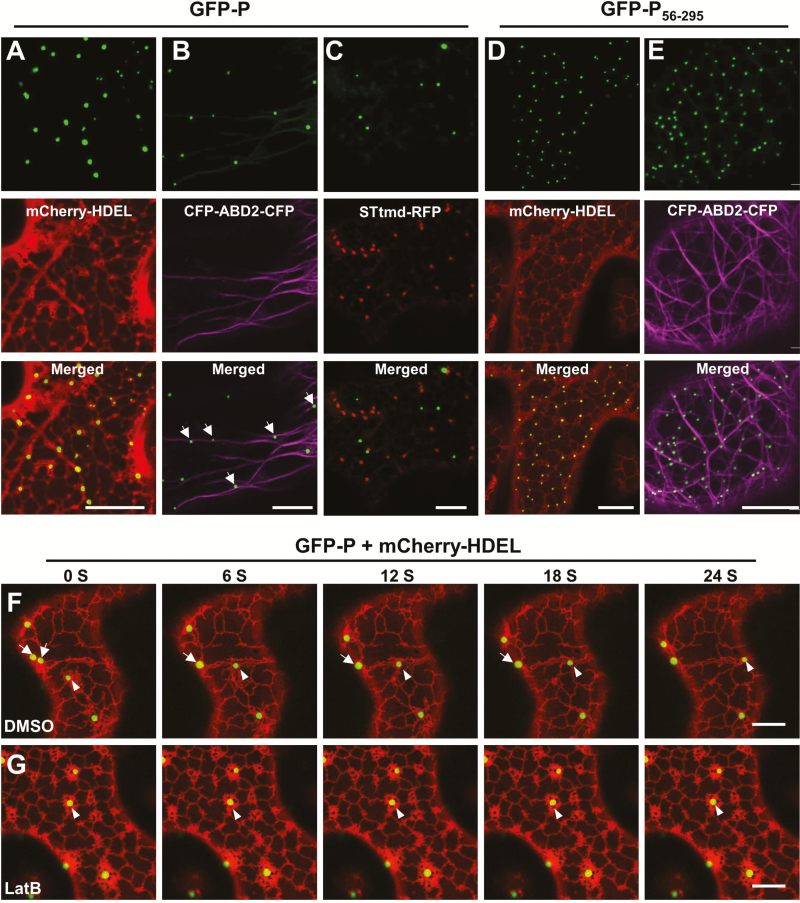
We next examined the possibility that the movement of the BYSMV-P bodies was dependent on the actin/ER network, which has been shown to be essential for intracellular movement of various virus proteins (Kurth et al., 2017; Pitzalis and Heinlein, 2017). GFP-P was therefore co-expressed transiently with a cortical ER marker (mCherry-HDEL) in N. benthamiana leaves (Boevink et al., 1996). At 2 dpi, the GFP-P bodies remained closely associated with the cortical ER network labeled with mCherry-HDEL, and individual P bodies were motile and remained closely associated with the ER tubules (Fig. 4A and Supplementary Video S5). We further generated a construct for transient expression of an actin-binding domain 2 (ABD2) with CFP fusions at both termini. Co-expression of GFP-P with CFP-ABD2-CFP resulted in close association between the P bodies and individual microfilament bundles (Fig. 4B). We demonstrated that GFP-P bodies were trafficked along actin filaments that were labelled with GFP-ABD2-GFP (Supplementary Fig. S4A, B and Supplementary Video S6). To examine whether GFP-P associated with microtubules, a microtubule-specific marker, mCherry-MAP65-1 (Feng et al., 2013), was co-expressed with GFP-P in N. benthamiana leaves. The results showed that GFP-P bodies were not trafficked along the microtubules (Supplementary Fig. S4C and Supplementary Video S7).
Fig. 4.
Confocal micrographs showing the localization of BYSMV P protein bodies in relation to the ER/actin network. (A–E) Co-expression of GFP-P with mCherry-HDEL (A), CFP-ABD2-CFP (B), and STtmd-RFP (C), and co-expression of GFP-P56–295 with mCherry-HDEL (D) and CFP-ABD2-CFP (E) were monitored at 2 d post inoculation (dpi). Scale bars are 20 μm. (F, G) Time-lapse confocal images of GFP-P and mCherry-HDEL at 2 dpi in co-infiltrated N.benthamiana leaves treated with DMSO (F) or latrunculin B (LatB) to disrupt actin assembly (G) at 2 dpi. Images were taken 3 h after DMSO or 10 μM LatB treatments. Scale bars are 10 μm. In the DMSO-treated leaves (F), arrows indicate fusion of two GFP-P inclusion bodies, and arrowheads indicate movement of a GFP-P body along the ER network. In the LatB-treated leaves (G), arrowheads indicate an immobile GFP-P body.
Since the morphology and motion of the BYSMV-P bodies resembled those of the plant Golgi apparatus (Boevink et al., 1998; Avisar et al., 2008b), we next determined whether the P bodies co-localized with the ER/actin network. To this end, the Golgi apparatus was visualized using an RFP fusion with a Golgi marker, rat 2, 6-sialyl transferase (STtmd-RFP). When GFP-P and STtmd-RFP were co-expressed in infiltrated N. benthamiana leaves, the GFP-P fluorescent signal was clearly separate from that of STtmd-RFP (Fig. 4C). Nonetheless, GFP-P bodies and STtmd-RFP bodies exhibited similar intracellular movement along the ER/actin network (Supplementary Video S8). In contrast, movement of GFP-P56–295 bodies was difficult to detect and was closely associated with ER tubules, especially in the three-way junctions of the ER network (Fig. 4D, E).
To further verify actin-mediated movement of the BYSMV-P bodies, N. benthamiana leaves expressing GFP-P and mCherry-HDEL were incubated in DMSO buffer with or without the actin-depolymerizing agent LatB. Using time-lapse confocal microscopy, we found that movement of BYSMV-P bodies was completely abolished by treatment with 10 μM LatB, in contrast to highly dynamic movement along ER tubules observed in the DMSO control (Fig. 4F, G, arrow heads, Supplementary Videos S9, S10). In addition, we noticed that two small BYSMV-P bodies were fused together along the ER tubules in the DMSO-treated samples (Fig. 4F, arrows), indicating the bodies can fuse into larger bodies during movement. Collectively, these results indicated that the BYSMV-P bodies were trafficked along the ER/actin network.
Overexpression of class XI myosin tails interferes with the movement of BYSMV-P bodies
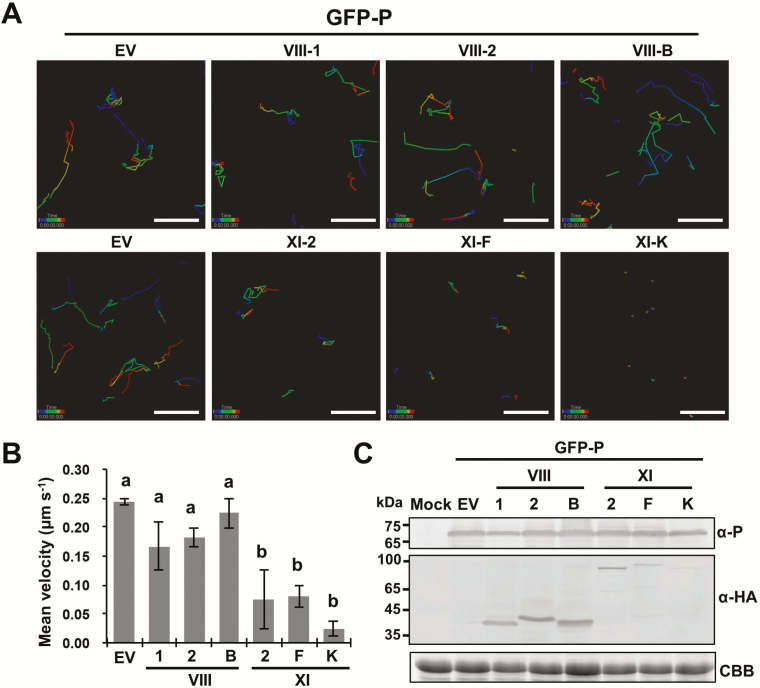
Cytoplasmic trafficking along the ER/actin network is primarily dependent on actomyosin mobility. We therefore next examined the myosin motor protein(s) responsible for trafficking of the BYSMV-P bodies. Because myosin tails are required for attachment of organelles on actin microfilaments, overexpression of headless tails can compete for organelles and inhibit their trafficking in dominant negative inhibition assays (Avisar et al., 2008a, 2008b; Harries et al., 2009a; Feng et al., 2016). To gain insights into the specific contributions of myosins to the movement of BYSMV-P bodies, GFP-P was co-expressed with the myosin tails XI-2, XI-F, XI-K, VIII-1, VIII-2, and VIII-B in N. benthamiana leaves. Based on time-lapse observations, the trafficking of the BYSMV-P bodies was nearly halted by the overexpression of the myosin XI-K tail (Fig. 5A and Supplementary Video S11), and was substantially compromised by the overexpression of the XI-2 and XI-F tails (Fig. 5A, Supplementary Videos S12, S13). In contrast with the pGD empty vector (Fig. 5A, Supplementary Video S14), none of the myosin VIII tails had obvious effects on the movement of BYSMV-P bodies (Fig. 5A, Supplementary Videos S15, S16, S17).
Fig. 5.
Effects of myosin tail overexpression on trafficking of BYSMV-P protein bodies in co-infiltrated N. benthamiana. (A) Representative confocal images of GFP-P bodies in epidermal cells overexpressing myosin tails VIII-1, VIII-2, VIII-B, XI-2, XI-F, and XI-K at 2 d post inoculation. Individual bodies were recorded in a time-series and are indicated by connecting lines. Different time intervals are represented by the different colors. (B) Mean velocities of the BYSMV-P bodies in the infiltrated leaves shown in (A). The mean velocities were calculated from the velocities of at least 90 bodies over 4 min. Data are means (±SE) of three independent experiments. Different letters indicate significant differences as determined by ANOVA followed by Turkey’s multiple comparison test (P<0.05). (C) Accumulation of GFP-P and myosin tails in those N. benthamiana leaves shown in (A). Anti-P, and anti-HA antibodies were used to detect the accumulation of GFP-P and HA-tagged myosin tails, respectively. Uninfiltrated N. benthamiana leaves were used as mock controls. Coomassie brilliant blue (CBB) staining was used for loading controls. EV, empty vector.
To determine the statistical significance of the differences in the movements of the BYSMV-P bodies, mean velocities were calculated based on two-dimensional movement. Overexpression of the VIII-1, VIII-2, and VIII-B tails did not significantly interfere with the mean velocity (0.15–0.25 μm s−1) (Fig. 5B). Overexpression of myosin XI-2 and XI-F tails resulted in ~2-fold reduction of the mean velocity. In contrast, overexpression of the XI-K tail reduced the mean velocity by ~6-fold; indeed, the residual velocity resulted from oscillations rather than effective movement of the BYSMV-P bodies (Fig. 5A and Supplementary Video S11). Western blotting analysis showed that BYSMV-P and myosin tails were expressed in co-infiltrated leaves (Fig. 5C). Taken together, these results demonstrated that the movement of BYSMV-P bodies was mainly mediated by myosin XI-K, and partially mediated by XI-2 and XI-F.
The BYSMV-P bodies recruit the N and L proteins into viroplasm-like complexes
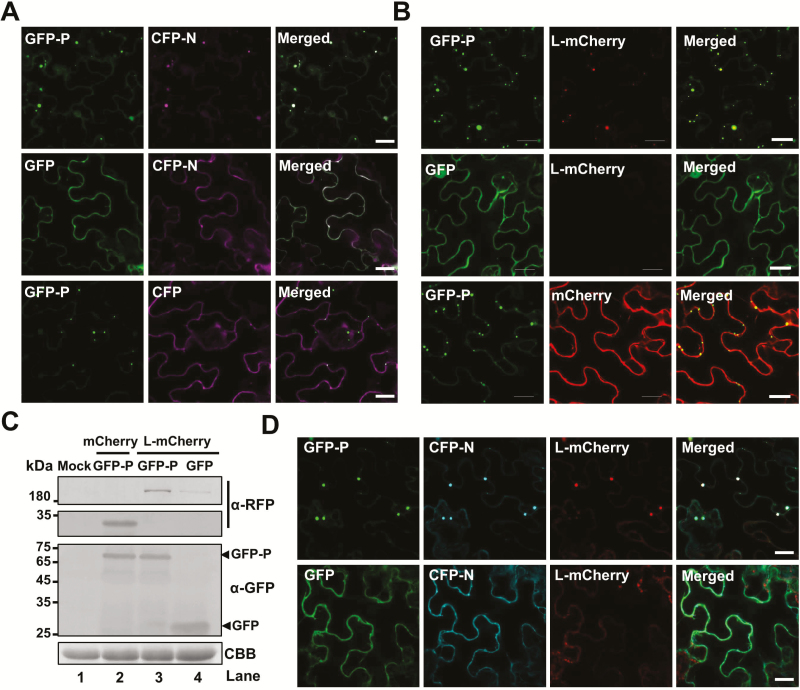
Because rhabdovirus P acts as a flexible ‘hub’ protein that anchors the polymerase complexes to the N–RNA templates, we assumed that trafficking of BYSMV-P bodies might facilitate recruitment of the N and L proteins for optimal RNA synthesis. To test this hypothesis, the CFP-N or CFP proteins were co-expressed with the GFP-P or GFP proteins in N. benthamiana leaf cells by agro-infiltration. At 3 dpi, the CFP-N and GFP-P proteins co-localized in the trafficking bodies (Fig. 6A and Supplementary Video S18), whereas CFP-N and GFP did not form bodies (Fig. 6A). When co-expressed with free CFP, GFP-P formed trafficking bodies but did not recruit CFP (Fig. 6A). To rule out the effects of the GFP fusion, the BYSMV P protein was co-expressed with GFP-N or free GFP. This showed that GFP-N rather than free GFP could form inclusion bodies when co-expressed with the BYSMV P protein (Supplementary Fig. S5A). These results suggested that GFP-P formed trafficking bodies and then recruited CFP-N into complexes. Using the same strategy, we found that the GFP-P bodies also recruited L-mCherry, rather than mCherry (Fig. 6B and Supplementary Video S19). It should be noted that higher levels of L-mCherry accumulated when it was co-expressed with GFP-P compared with those of free GFP samples, as shown by the fluorescence intensity and by western blotting (Fig. 6B, C). This was in agreement with a previous study that showed that the vesicular stomatitis virus P protein could stabilize the L protein (Canter and Perrault, 1996).
Fig. 6.
Recruitment of the BYSMV N and L proteins into BYSMV-P viroplasm-like inclusion bodies. (A) Co-localization of CFP-N, but not CFP, with GFP-P bodies. GFP-P, GFP, CFP-N, and CFP proteins were expressed in different combinations via agro-infiltration in N. benthamiana leaves, and fluorescence images were observed at 2 d post inoculation (dpi). Scale bars are 20 μm. (B) Co-localization of L-mCherry, but not mCherry, with GFP-P bodies. Co-expression of GFP-P/L-mCherry, GFP-P/mCherry, and GFP/L-mCherry in N. benthamiana epidermal cells in fluorescence images taken at 2 dpi. Scale bars are 20 μm. (C) Accumulation of GFP-P and L-mCherry in the N. benthamiana leaves shown in (B). Anti-GFP and anti-RFP antibodies were used to detect accumulation of GFP-P and L-mCherry, respectively. Coomassie brilliant blue (CBB) staining was used as loading protein control. Healthy N. benthamiana leaves served as mock samples. (D) Accumulation of CFP-N and L-mCherry in GFP-P bodies, and lack of association with GFP. CFP-N and L-mCherry were expressed with GFP-P or GFP in agro-infiltrated N.benthamiana leaves. Fluorescence images were taken at 2 dpi. Scale bars are 20 μm.
We further co-expressed CFP-N and L-mCherry with GFP-P or GFP in N. benthamiana leaf cells. The results showed that GFP-P, CFP-N, and L-mCherry formed trafficking bodies (Fig. 6D and Supplementary Video S20). By contrast, CFP-N and L-mCherry were evenly distributed in the cytoplasm when co-expressed with GFP (Fig. 6D). Furthermore, GFP-N and L-mCherry could form inclusion bodies when co-expressed with BYSMV P (Supplementary Fig. S5B, upper panels). However, GFP-N and L-mCherry distributed evenly in the cytoplasm when co-expressed with the empty vector (Supplementary Fig. S5B, lower panels). Collectively, these results suggested that BYSMV P first forms trafficking bodies and then recruits the N and L proteins into viroplasm-like complexes, probably for optimal replication and transcription.
Trafficking of BYSMV-P bodies driven by actin/myosin is required for optimal viral RNA synthesis
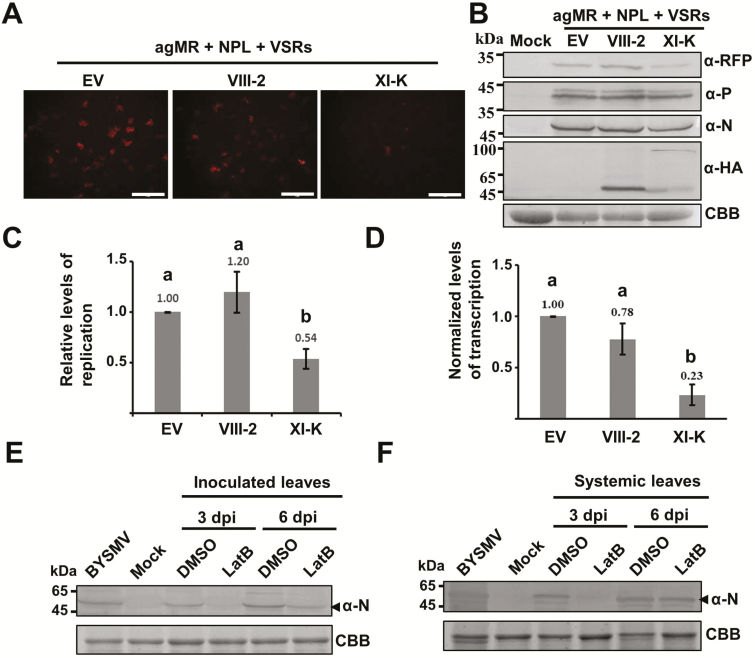
The leading role of myosin XI-K in the movement of BYSMV-P bodies prompted us to determine whether the trafficking of the BYSMV-P/N/L complex was required for optimal viral RNA synthesis. To this end, we overexpressed the XI-K or VIII-2 myosin tails with mini-replicons containing the agMR, N, P, L, and VSRs in N. benthamiana leaves. Compared to the empty vector and VIII-2 tail, overexpression of the XI-K tail resulted in dramatic reductions in the numbers of RFP fluorescence foci at 6 dpi (Fig. 7A). Western blotting consistently showed that overexpression of the XI-K tail substantially reduced RFP accumulation, despite having negligible effects on the accumulation of the N and P proteins (Fig. 7B). Quantitative RT-PCR analysis was carried out to determine the levels of mini-genome RNA replication and transcription at 6 dpi, representing RNA replication activities. To compare accurate transcription activities, we normalized the transcribed RFP mRNA levels relative to the gMR template levels. As shown in Fig. 7C, D, overexpression of the XI-K tail resulted in a reduction of replication of ~50% and a reduction of transcription accumulation of 75% compared to those of the empty control and the VIII-2 tail. These results therefore suggested that trafficking of the BYSMV P protein was required for optimal viral RNA synthesis, including replication and transcription.
Fig. 7.
Effects of BYSMV-P protein body trafficking on mini-genome RNA synthesis. (A) Representative confocal images of RFP fluorescence expressed from antigenomic-sense mini-replicon (agMR) combinations in epidermal cells of N. benthamiana treated with either the empty vector (EV) or with overexpressed myosin tails VIII-2 and XI-K at 6 d post inoculation (dpi). Scale bars are 1 mm. (B) Accumulation of RFP, P, N, and myosin tails shown in (A), as detected by western blotting with antibodies against RFP, P, N, and HA, respectively. Coomassie brilliant blue (CBB) staining was used as a loading control. Healthy N. benthamiana leaves served as mock samples. (C) Quantitative real-time PCR analysis of mini-genome RNA replication levels and (D) real-time quantitative PCR analysis of the relative levels of RFP mRNA versus mini-genome RNA in the samples shown in in (A). The EF1A housekeeping gene served as an internal control. The values of viral replication and relative transcription in the samples treated with the empty vector (EV) were set to 1. Data are means (±SE) of three independent experiments, and the mean values are indicated. Different letters indicate significant differences as determined by ANOVA followed by Turkey’s multiple comparison test (P<0.05). (E) Accumulation of BYSMV N protein in the inoculated leaves after treatment with DMSO or latrunculin B (LatB, 5 μM) to disrupt actin assembly, as analysed by western blotting. (F) Accumulation of BYSMV N protein in systemically infected leaves that emerged after viruliferous planthoppers had been given access to leaves that had been treated as described in (E). BYSMV-infected barley plants and healthy plants (mock) provided positive and negative controls, respectively. CBB) staining was used as loading controls.
We further evaluated the effects of LatB treatment on BYSMV infection of barley plants. Barley leaves were first treated with 5 μM LatB or DMSO control solutions for 3 h, and were then exposed to BYSMV-infected SBPHs (20 insects per leaf) for a 2-d inoculation period. Western blot analysis indicated that at 3 dpi and 6 dpi, BYSMV N protein accumulation in LatB-treated leaves exposed to viruliferous SBPHs was substantially lower than that of control leaves (Fig. 7E). Compared with the DMSO control, LatB treatment resulted in a reduced accumulation of the BYSMV N protein in systemically infected leaves that emerged after the planthopper access period at 3 dpi (Fig. 7F). By 6 dpi, the BYSMV N protein had accumulated to similar levels in all systemically infected leaves. The results therefore suggested that the actin-driven trafficking of BYSMV-P bodies was required for efficient infection of BYSMV in barley plants.
Discussion
The reverse genetic systems of viruses make it possible to study the functions of viral proteins and their associated host factors through manipulation of the complementary DNA (cDNA) of the viral RNA genome. Recently, reverse genetic applications with plant nucleorhabdoviruses have been established through co-infiltration of N. benthamiana leaves with Agrobacterium harboring plasmids encoding core proteins and mini-genome derivatives or full-length cDNA of Sonchus yellow net virus (SYNV) (Ganesan et al., 2013; Wang et al., 2015). However, reverse genetic systems for plant cytorhabdoviruses remain to be developed for studying the distinct features of their infection cycles. In our current study, we developed a BYSMV cytorhabdovirus mini-replicon (MR) system using the co-infiltration strategy in N. benthamiana leaves (Fig. 1) to permit functional analysis of BYSMV core proteins in viral transcription and replication.
Using the BYSMV MR system, we attempted to evaluate the effects of localization and mobility of the BYSMV core proteins on MR infections. In previous studies, the N and P proteins of the nucleorhabdoviruses SYNV, Potato yellow dwarf virus (PYDV), and Maize fine streak virus (MFSV) were found to co-localize to subnuclear loci that were distinct from the localizations of either protein alone (Goodin et al., 2001; Tsai et al., 2005; Deng et al., 2007; Bandyopadhyay et al., 2010). Likewise, the N and P proteins of Lettuce necrotic yellows virus (LNYV), a type cytorhabdovirus, localize to the cytoplasm at the cell periphery when expressed individually, whereas they form aggregates near the nucleus upon co-expression (Martin et al., 2012). In our current study, we found that the BYSMV P protein formed inclusion bodies when expressed alone and moved rapidly along the ER/actin network (Figs 2, 3). Given that the rhabdovirus P proteins have hub roles connecting the N and L proteins (Ivanov et al., 2011), it is not surprising to find that they are the best candidates in the inclusion body for direct recruitment of the N and L proteins (Fig. 6D). The resulting viroplasm-like complexes are then trafficked along the ER/actin network where they probably function in the assembly of nascent viral and host components for virus RNA synthesis. Indeed, our results showed that mobile viroplasm-like complexes are required for efficient MR replication and transcription (Fig. 7).
A growing number of animal and plant viruses are known to hijack the actin cytoskeleton for replication, and intra- and intercellular movement (Harries et al., 2009b, 2010; Sattentau, 2011; Pitzalis and Heinlein, 2017). In plants, the class VIII and class XI myosins are important molecular motors that drive the trafficking of cargos along actin filaments. For instance, the N protein of TSWV is dependent on myosin XI-K for ER streaming (Feng et al., 2013). The nucleocapsid protein (NP) of FMV requires XI-1, XI-2, and XI-K for formation of VRCs and supports efficient ER streaming (Ishikawa et al., 2015). By contrast, the TMV 126-k protein complex hijacks myosin XI-2 for formation of VRCs and supports efficient replication (Amari et al., 2014). Here, we found that the BYSMV P protein was trafficked along the actin/ER network, and was mainly driven by myosin XI-K, and partially by myosin XI-2 and XI-F (Fig. 5). Collectively, our results showed that enveloped and non-enveloped viruses have evolved distinct proteins that hijack different myosin motors for their life cycles.
In summary, our work provides a mechanistic model of the interplay between BYSMV and the actin filament. The middle and the C-terminal domains of BYSMV P associate with the ER to form P bodies that bind actin filaments through the N termini of the P protein. Myosin XI-K mainly drives the dynamic BYSMV P bodies along the ER/actin network to recruit the N and L proteins. Thus, it seems plausible that myosin XI-K contributes to the formation of large viroplasm structures that support optimal virus replication and transcription.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Primers used in this study.
Fig. S1. Confocal micrographs showing the subcellular localization of the BYSMV N protein in the epidermal cells of agro-infiltrated N. benthamiana leaves.
Fig. S2. Subcellular localization of the NCMV N and P proteins in epidermal cells of agro-infiltrated N. benthamiana leaves.
Fig. S3. Detection of BYSMV P56–295 in Hordeum vulgare infected with BYSMV and in agro-infiltrated N. benthamiana.
Fig. S4. Subcellular localization of the GFP-P inclusion bodies with actin filaments and microtubule in the epidermal cells of agro-infiltrated N. benthamiana leaves.
Fig. S5. GFP-N and L-mCherry form inclusion bodies with the BYSMV P protein.
Video. S1. High mobility of BYSMV GFP-P inclusion bodies in epidermal cells of agro-infiltrated N. benthamiana leaves.
Video. S2. High mobility of NCMV GFP-P inclusion bodies in epidermal cells of agro-infiltrated N. benthamiana leaves.
Video. S3. Immobile BYSMV GFP-P56–295 inclusion bodies in epidermal cells of agro-infiltrated N. benthamiana leaves.
Video. S4. High mobility of BYSMV GFP-P43–295 inclusion bodies in epidermal cells of agro-infiltrated N. benthamiana leaves.
Video. S5. Close association of BYSMV GFP-P inclusion bodies and the mCherry-HDEL labeled cortical ER network.
Video. S6. Mobility of BYSMV GFP-P inclusion bodies along the GFP-ABD2-GFP labeled actin filaments.
Video. S7. Mobility of BYSMV GFP-P inclusion bodies and mCherry-MAP65-1-labeled microtubules.
Video. S8. High mobility of BYSMV GFP-P inclusion bodies and Golgi labeled with STtmd-RFP in N. benthamiana leaf epidermal cells.
Video. S9. Effects of DMSO on the intracellular movement of BYSMV GFP-P inclusion bodies in N. benthamiana leaf epidermal cells expressing BYSMV GFP-P and mCherry-HDEL.
Video. S10. Effects of LatB on the intracellular movement of BYSMV GFP-P inclusion bodies in N. benthamiana leaf epidermal cells expressing BYSMV GFP-P and mCherry-HDEL.
Video. S11. Intracellular movement of BYSMV GFP-P inclusion bodies in N. benthamiana leaf epidermal cells co-expressing the myosin tail of XI-K.
Video. S12. Intracellular movement of BYSMV GFP-P inclusion bodies in N. benthamiana leaf epidermal cells co-expressing the myosin tail of XI-2.
Video. S13. Intracellular movement of BYSMV GFP-P inclusion bodies in N. benthamiana leaf epidermal cells co-expressing the myosin tail of XI-F.
Video. S14. Intracellular movement of BYSMV GFP-P inclusion bodies in N. benthamiana leaf epidermal cells co-expressing the empty vector (EV).
Video. S15. Intracellular movement of BYSMV GFP-P inclusion bodies in N. benthamiana leaf epidermal cells co-expressing the myosin tail of VIII-1.
Video. S16. Intracellular movement of BYSMV GFP-P inclusion bodies in N. benthamiana leaf epidermal cells co-expressing the myosin tail of VIII-2.
Video. S17. Intracellular movement of BYSMV GFP-P inclusion bodies in N. benthamiana leaf epidermal cells co-expressing the myosin tail of VIII-B.
Video. S18. BYSMV CFP-N co-localization with GFP-P and trafficking bodies formed when co-expressed in N. benthamiana leaf epidermal cells.
Video. S19. BYSMV L-mCherry trafficking bodies formed with GFP-P in N. benthamiana leaf epidermal cells.
Video. S20. BYSMV CFP-N and L-mCherry trafficking bodies formed with GFP-P in N. benthamiana leaf epidermal cells.
Acknowledgements
We thank our colleagues Jialin Yu, Dawei Li, Chenggui Han, and Yongliang Zhang for their helpful suggestions and constructive criticism. We thank Prof. Xiaorong Tao at the Department of Plant Pathology of Nanjing Agricultural University for his gift of the pCB301-2×35S-Nos plasmid. We thank Prof. V. V. Dolja at Oregon State University for providing the dominant negative myosin constructs. We would also like to thank Prof. Michael M. Goodin (University of Kentucky, Lexington, KY, USA) for the H2B-RFP transgenic N. benthamiana seeds. This work was supported by grants from the National Key Research and Development Program (2016YFC1200605), the Natural Science Foundation of China (31872920, 31571978), and the Beijing Nova Program (Z171100001117038).
Glossary
Abbreviations:
- agMR
anti-genome minireplicon
- BYSMV
Barley yellow striate mosaic virus
- ER
endoplasmic reticulum
- MR
mini-replicon
References
- Agbeci M, Grangeon R, Nelson RS, Zheng H, Laliberté JF. 2013. Contribution of host intracellular transport machineries to intercellular movement of turnip mosaic virus. PLoS Pathogens 9, e1003683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amari K, Di Donato M, Dolja VV, Heinlein M. 2014. Myosins VIII and XI play distinct roles in reproduction and transport of tobacco mosaic virus. PLoS Pathogens 10, e1004448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amari K, Lerich A, Schmitt-Keichinger C, Dolja VV, Ritzenthaler C. 2011. Tubule-guided cell-to-cell movement of a plant virus requires class XI myosin motors. PLoS Pathogens 7, e1002327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avisar D, Prokhnevsky AI, Dolja VV. 2008a. Class VIII myosins are required for plasmodesmatal localization of a closterovirus Hsp70 homolog. Journal of Virology 82, 2836–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avisar D, Prokhnevsky AI, Makarova KS, Koonin EV, Dolja VV. 2008b. Myosin XI-K Is required for rapid trafficking of Golgi stacks, peroxisomes, and mitochondria in leaf cells of Nicotiana benthamiana. Plant Physiology 146, 1098–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay A, Kopperud K, Anderson G, Martin K, Goodin M. 2010. An integrated protein localization and interaction map for Potato yellow dwarf virus, type species of the genus Nucleorhabdovirus. Virology 402, 61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boevink P, Oparka K, Cruz SS, Martin B, Betteridge A, Hawes C. 1998. Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. The Plant Journal 15, 441–447. [DOI] [PubMed] [Google Scholar]
- Boevink P, Santa Cruz S, Hawes C, Harris N, Oparka K. 1996. Virus‐mediated delivery of the green fluorescent protein to the endoplasmic reticulum of plant cells. The Plant Journal 10, 935–941. [Google Scholar]
- Brandenburg B, Zhuang X. 2007. Virus trafficking – learning from single-virus tracking. Nature Reviews Microbiology 5, 197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canter DM, Perrault J. 1996. Stabilization of vesicular stomatitis virus L polymerase protein by P protein binding: a small deletion in the C-terminal domain of L abrogates binding. Virology 219, 376–386. [DOI] [PubMed] [Google Scholar]
- Cao Q, Xu WY, Gao Q, Jiang ZH, Liu SY, Fang XD, Gao DM, Wang Y, Wang XB. 2018. Transmission characteristics of Barley yellow striate mosaic virus in its planthopper vector Laodelphax striatellus. Frontiers in Microbiology 9, 1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenik M, Chebli K, Blondel D. 1995. Translation initiation at alternate in-frame AUG codons in the rabies virus phosphoprotein mRNA is mediated by a ribosomal leaky scanning mechanism. Journal of Virology 69, 707–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Wei T, Chowda-Reddy RV, Sun G, Wang A. 2010. The Tobacco etch virus P3 protein forms mobile inclusions via the early secretory pathway and traffics along actin microfilaments. Virology 397, 56–63. [DOI] [PubMed] [Google Scholar]
- Deng M, Bragg JN, Ruzin S, Schichnes D, King D, Goodin MM, Jackson AO. 2007. Role of the Sonchus yellow net virus N protein in formation of nuclear viroplasms. Journal of Virology 81, 5362–5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong K, Wang Y, Zhang Z, Chai LX, Tong X, Xu J, Li D, Wang XB. 2016. Two amino acids near the N-terminus of Cucumber mosaic virus 2b play critical roles in the suppression of RNA silencing and viral infectivity. Molecular Plant Pathology 17, 173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Chen X, Bao Y, Dong J, Zhang Z, Tao X. 2013. Nucleocapsid of Tomato spotted wilt tospovirus forms mobile particles that traffic on an actin/endoplasmic reticulum network driven by myosin XI-K. New Phytologist 200, 1212–1224. [DOI] [PubMed] [Google Scholar]
- Feng Z, Xue F, Xu M, et al. 2016. The ER-membrane transport system is critical for intercellular trafficking of the NSm movement protein and Tomato spotted wilt tospovirus. PLoS Pathogens 12, e1005443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan U, Bragg JN, Deng M, et al. 2013. Construction of a Sonchus yellow net virus minireplicon: a step toward reverse genetic analysis of plant negative-strand RNA viruses. Journal of Virology 87, 10598–10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodin MM, Austin J, Tobias R, Fujita M, Morales C, Jackson AO. 2001. Interactions and nuclear import of the N and P proteins of Sonchus yellow net virus, a plant nucleorhabdovirus. Journal of Virology 75, 9393–9406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO. 2002. pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. The Plant Journal 31, 375–383. [DOI] [PubMed] [Google Scholar]
- Harries PA, Palanichelvam K, Yu W, Schoelz JE, Nelson RS. 2009a. The Cauliflower mosaic virus protein P6 forms motile inclusions that traffic along actin microfilaments and stabilize microtubules. Plant Physiology 149, 1005–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harries PA, Park J-W, Sasaki N, Ballard KD, Maule AJ, Nelson RS. 2009b. Differing requirements for actin and myosin by plant viruses for sustained intercellular movement. Proceedings of the National Academy of Sciences, USA 106, 17594–17599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harries PA, Schoelz JE, Nelson RS. 2010. Intracellular transport of viruses and their components: utilizing the cytoskeleton and membrane highways. Molecular Plant-Microbe Interactions 23, 1381–1393. [DOI] [PubMed] [Google Scholar]
- Ishikawa K, Miura C, Maejima K, Komatsu K, Hashimoto M, Tomomitsu T, Fukuoka M, Yusa A, Yamaji Y, Namba S. 2015. Nucleocapsid protein from Fig mosaic virus forms cytoplasmic agglomerates that are hauled by endoplasmic reticulum streaming. Journal of Virology 89, 480–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I, Yabukarski F, Ruigrok RW, Jamin M. 2011. Structural insights into the rhabdovirus transcription/replication complex. Virus Research 162, 126–137. [DOI] [PubMed] [Google Scholar]
- Jin X, Jiang Z, Zhang K, et al. 2018. Three-dimensional analysis of chloroplast structures associated with virus infection. Plant Physiology 176, 282–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami S, Watanabe Y, Beachy RN. 2004. Tobacco mosaic virus infection spreads cell to cell as intact replication complexes. Proceedings of the National Academy of Sciences, USA 101, 6291–6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth EG, Peremyslov VV, Turner HL, Makarova KS, Iranzo J, Mekhedov SL, Koonin EV, Dolja VV. 2017. Myosin-driven transport network in plants. Proceedings of the National Academy of Sciences, USA 114, E1385–E1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte C, Vetter G, Loudes AM, Robinson DG, Hillmer S, Stussi-Garaud C, Ritzenthaler C. 2003. Involvement of the secretory pathway and the cytoskeleton in intracellular targeting and tubule assembly of Grapevine fanleaf virus movement protein in tobacco BY-2 cells. The Plant Cell 15, 2058–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyrat C, Ribeiro EA, Gérard FC, Ivanov I, Ruigrok RW, Jamin M. 2011. Structure, interactions with host cell and functions of rhabdovirus phosphoprotein. Future Virology 6, 465–481. [Google Scholar]
- Liu JZ, Blancaflor EB, Nelson RS. 2005. The Tobacco mosaic virus 126-kilodalton protein, a constituent of the virus replication complex, alone or within the complex aligns with and traffics along microfilaments. Plant Physiology 138, 1853–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, Goodin MM. 2009. Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. The Plant Journal 59, 150–162. [DOI] [PubMed] [Google Scholar]
- Martin KM, Dietzgen RG, Wang R, Goodin MM. 2012. Lettuce necrotic yellows cytorhabdovirus protein localization and interaction map, and comparison with nucleorhabdoviruses. The Journal of General Virology 93, 906–914. [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenfuhr A. 2007. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. The Plant Journal 51, 1126–1136. [DOI] [PubMed] [Google Scholar]
- Pitzalis N, Heinlein M. 2017. The roles of membranes and associated cytoskeleton in plant virus replication and cell-to-cell movement. Journal of Experimental Botany 69, 117–132. [DOI] [PubMed] [Google Scholar]
- Sambade A, Brandner K, Hofmann C, Seemanpillai M, Mutterer J, Heinlein M. 2008. Transport of TMV movement protein particles associated with the targeting of RNA to plasmodesmata. Traffic 9, 2073–2088. [DOI] [PubMed] [Google Scholar]
- Sattentau QJ. 2011. The direct passage of animal viruses between cells. Current Opinion in Virology 1, 396–402. [DOI] [PubMed] [Google Scholar]
- Shen Y, Zhao X, Yao M, Li C, Miriam K, Zhang X, Tao X. 2014. A versatile complementation assay for cell-to-cell and long distance movements by cucumber mosaic virus based agro-infiltration. Virus Research 190, 25–33. [DOI] [PubMed] [Google Scholar]
- Smertenko AP, Chang HY, Wagner V, Kaloriti D, Fenyk S, Sonobe S, Lloyd C, Hauser MT, Hussey PJ. 2004. The Arabidopsis microtubule-associated protein AtMAP65-1: molecular analysis of its microtubule bundling activity. The Plant Cell 16, 2035–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga M, Kojima H, Yokota E, Orii H, Nakamori R, Katayama E, Anson M, Shimmen T, Oiwa K. 2003. Higher plant myosin XI moves processively on actin with 35 nm steps at high velocity. The EMBO Journal 22, 1263–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CW, Redinbaugh MG, Willie KJ, Reed S, Goodin M, Hogenhout SA. 2005. Complete genome sequence and in planta subcellular localization of Maize fine streak virus proteins. Journal of Virology 79, 5304–5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Ma X, Qian S, Zhou X, Sun K, Chen X, Zhou X, Jackson AO, Li Z. 2015. Rescue of a plant negative-strand RNA virus from cloned cDNA: insights into enveloped plant virus movement and morphogenesis. PLoS Pathogens 11, e1005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YS, Yoo CM, Blancaflor EB. 2008. Improved imaging of actin filaments in transgenic Arabidopsis plants expressing a green fluorescent protein fusion to the C- and N-termini of the fimbrin actin-binding domain 2. New Phytologist 177, 525–536. [DOI] [PubMed] [Google Scholar]
- Yan T, Zhu JR, Di D, Gao Q, Zhang Y, Zhang A, Yan C, Miao H, Wang XB. 2015. Characterization of the complete genome of Barley yellow striate mosaic virus reveals a nested gene encoding a small hydrophobic protein. Virology 478, 112–122. [DOI] [PubMed] [Google Scholar]
- Yuan Z, Chen H, Chen Q, Omura T, Xie L, Wu Z, Wei T. 2011. The early secretory pathway and an actin–myosin VIII motility system are required for plasmodesmatal localization of the NSvc4 protein of Rice stripe virus. Virus Research 159, 62–68. [DOI] [PubMed] [Google Scholar]
- Zhang XP, Liu DS, Yan T, Fang XD, Dong K, Xu J, Wang Y, Yu JL, Wang XB. 2017. Cucumber mosaic virus coat protein modulates the accumulation of 2b protein and antiviral silencing that causes symptom recovery in planta. PLoS Pathogens 13, e1006522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.