Abstract
IL-1β is an important mediator of innate inflammatory responses and has been shown to contribute to liver injury in a number of etiologies. HIV patients have increased necroinflammation and more rapid fibrosis progression in chronic liver injury compared to non-HIV-infected patients. As the resident liver macrophage is critical to the IL-1β response to microbial translocation in chronic liver disease, we aim to examine the impact of HIV-1 and LPS stimulation on the IL-1β response of the resident hepatic macrophages. We isolated primary human liver macrophages from liver resection specimens, treated them with HIV-1BaL and/or LPS ex vivo, examined the IL-1β response, and then studied underlying mechanisms. Furthermore, we examined IL-1β expression in liver tissues derived from HIV-1 patients compared to those with no underlying liver disease. HIV-1 up-regulated TLR4 and CD14 expression on isolated primary CD68+ human liver macrophages and contributed to the IL-1β response to LPS stimulation as evidenced by TLR4 blocking. Nucleotide-binding domain, leucine-rich-containing family, pyrin domain-containing-3 (NLRP3) was shown to be involved in the IL-1β response of liver macrophages to HIV-1 infection and NLRP3 blocking experiments in primary CD68+ liver macrophages confirmed the contribution of the NLRP3-caspase 1 inflammatory signaling pathway in the IL-1β response. High in situ IL-1β expression was found in CD68+ cells in human liver tissues from HIV-1-infected patients, suggesting a critical role of IL-1β responses in patients infected by HIV. HIV infection sensitizes the IL-1β response of liver macrophages to LPS through up-regulation of CD14 and TLR4 expression and downstream activation of the NLRP3-caspase 1 pathway. These findings have implications for enhanced immune activation in HIV+ patients and mechanisms for rapid fibrosis progression in patients with chronic liver injury.
Keywords: HIV, IL-1β, liver macrophages, NLRP3 activation, TLR4
1 |. INTRODUCTION
As blood enters the liver, predominantly through the portal circulation, it is distributed through the hepatic sinusoids, which are lined by a uniquely fenestrated endothelium interspersed with resident liver macrophages, also known as Kupffer cells (KCs). KCs are long-lived and abundant, representing 15% to 20% of the total liver cell population.1,2 They are responsible for the clearance of translocated bacterial products and thus are tolerant to the proinflammatory effects of LPS under normal physiologic conditions. Therefore, function of these resident liver macrophages is critically important to the liver’s ability to clear translocated products, including endogenous TLR ligands that would otherwise promote hepatic inflammation. Furthermore, these cells may serve as the gatekeeper for systemic immune activation, which exists in HIV patients despite antiretroviral therapy.3 Although no surface marker clearly defines KCs, CD68 has been the most frequently used and therefore is used to define these resident liver macrophages in this study.
Liver disease is one of the leading causes of non-AIDS related mortality in HIV-1-infected patients receiving effective antiretroviral therapy (ART). While earlier initiation of ART and advances in the treatment of viral hepatitis have made a tremendous impact in improving liver outcomes in patients with HIV, there are HIV related alterations that are not restored by ART and may contribute to poor liver outcomes despite antiviral treatments.4–6 In HIV infection, CD4 gut depletion and increased gut permeability have been postulated to promote microbial translocation and drive immune activation. Though plasma LPS levels decrease in patients treated with ART, they never return to the level of HIV-uninfected individuals5 and microbial translocation is associated with persistent macrophage activation despite viral suppression or CD4 reconstitution.7 Therefore, HIV-1 infection and HIV-1 induced bacterial translocation have been implicated in accelerated liver disease progression in HIV-infected individuals. The importance of microbial translocation has been highlighted in many forms of chronic liver injury; thus HIV patients may be particularly at risk for the impact of the gut-liver axis in promoting chronic liver injury.
IL-1β, IL-6, and TNF-α are three classical proinflammatory cytokines implicated in the innate immune response of KCs. We have recently demonstrated that HIV infection of KCs results in an amplified IL-6 and TNF-α proinflammatory response to LPS.8,9 Inflammasome activation has been shown to be important in a number of liver diseases ranging from alcoholic steatohepatitis to nonalcoholic steatohepatitis (NASH) to chronic hepatitis C virus (HCV) infection. One of the main roles of inflammasomes in liver disease is to trigger the inflammatory cytokine IL-1β that synergizes with TLR signaling to amplify responses to LPS. IL-1β then also promotes the recruitment of inflammatory cells to the liver and activates hepatic stellate cells, which contribute to fibrosis.10 In general, IL-1β production is primed by the sensing of “danger signals,” either pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs), which then results in the production of mature IL-1β.11 In the liver, this inflammatory response is mainly mediated by the action of caspase-1, which is usually activated via the inflammasome, most notably by the nucleotide-binding domain leucine-rich-containing family pyrin domain-containing-3 (NLRP3) inflammasome. Although the underlying mechanisms for its contribution remain unclear, an NLRP3 inhibitory assay using a compound MCC950 has confirmed its importance in the caspase-1-IL-1β inflammatory axis.12,13
As a critical element in TLR signal transduction, NLRP3 plays an important role in viral-induced IL-1β maturation in macrophages. However, the pattern of IL-1β maturation in macrophages is different than that in monocytes. ATP, a potassium efflux inducer, is required for IL-1β maturation and secretion from LPS-primed macrophages.11 Several viral proteins influence IL-1β release.14 For example, influenza virus NS1 directly interacts with NLRP3 and inhibits NLRP3-mediated IL-1β production. In contrast, Influenza A virus M2 protein can activate NLRP3 and in turn promote IL-1β secretion.15 NLRP3 also contributes to the induction of the IL-1β response to Newcastle disease virus and HCV infection in THP1-derived macrophages. Furthermore, depletion of NLRP3 can dramatically reduce IL-1β production from the infected cells.16,17 Similarly, NLRP3 knockout significantly suppressed the IL-1β response of macrophages to influenza A virus infection in a mouse model.18 Our previous study has shown that HIV-1BaL infection induces an amplified inflammatory response through a TLR4-dependent pathway in primary human liver macrophages. However, the impact of HIV-1BaL infection, and the influence of concomitant microbial translocation, on the IL-1β response of liver macrophages remains incompletely described.
Here we show that HIV-1 infection primes the IL-1β response of CD68+ liver macrophages to LPS through up-regulation of CD14 and TLR4 expression and activation of the NLRP3-caspase 1 pathway. Specifically, in vitro/ex vivo HIV-1BaL infection of primary human hepatic CD68+ macrophages induced NLRP3 and caspase-1 activation and subsequently increased the IL-1β sensitivity of these cells to LPS stimulation. Small molecule NLRP3 inhibition significantly reduced the IL-1β response of CD68+ liver macrophages to LPS and HIV-1BaL stimulation. These results were supported by the demonstration of both increased IL-1β and NLRP3 in CD68+ cells in the livers of HIV-1-infected patients suggesting the up-regulation of this pathway and its potential role in clinical outcomes. Taken together, our findings suggest that HIV infection activates the TLR4-NLRP3-capsase-1 axis and consequently increases the IL-1β response of liver macrophages to translocated bacterial products, potentially leading to dysregulation of intrahepatic immune responses and progression of liver injury.
2 |. MATERIALS AND METHODS
2.1 |. Human liver tissue from HIV-1-infected patients
Tissues from HIV-infected patients were utilized for coimmunostaining of IL-1β, CD68, and NLRP3 and Western blot for IL-1β expression and co-immunostaining for CD68 and NLRP3 (Fig. 6). De-identified liver resection specimens from HIV-monoinfected infected patients (n = 7) were provided by Drs. Florman, Schwartz, Ganeskaran, and Fiel. In all cases, HIV was effectively suppressed with ART. Normal liver tissue obtained from the Mount Sinai Biobank served as controls. Tissues were formalin-fixed, frozen, or placed in DMEM. These studies have been approved by Mount Sinai IRB (GCO #06–0523 and #10–1211). In all other experiments, primary liver macrophages were isolated from non-HIV-infected livers and infected by HIV-1BaL ex vivo.
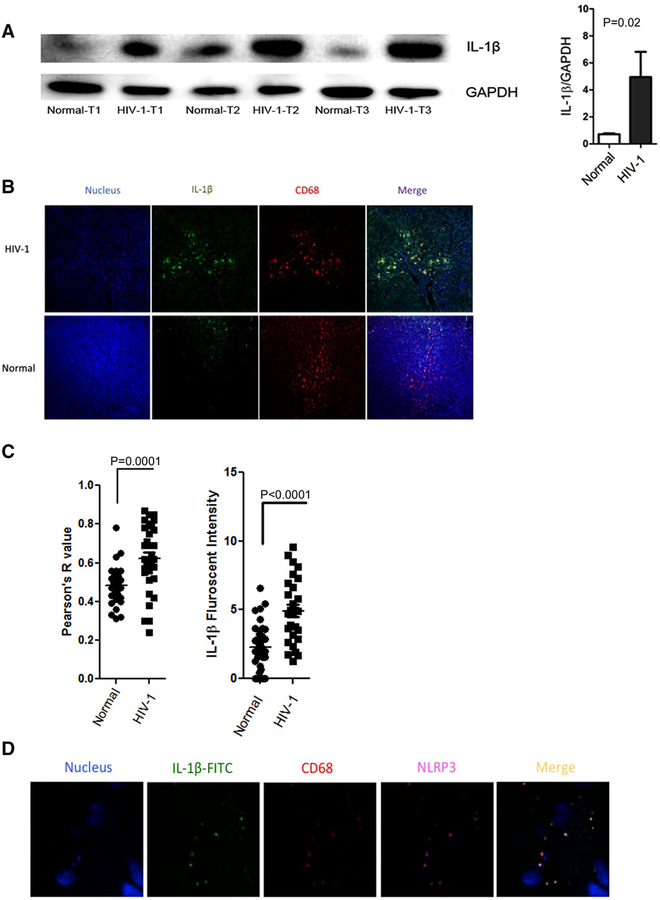
FIGURE 6. Immunoblot and Immunofluorescence staining reveals increased intrahepatic IL-1β expression in CD68+ macrophages in liver tissue from HIV-infected patients.
(A) IL-1β protein in human liver tissues was detected by Western blotting, GAPDH was used as internal control. Representative Western blot shown with densitometry from all specimens (P = 0.02) ((normal (n = 10) and HIV-1 monoinfected (n = 7)). (B) Confocal microscopic analysis of IL-1β and CD68 co-immunostaining in normal (n = 10) and HIV-1-infected liver tissues (n = 7). CD68 + liver macrophages (red), IL-1β (green) and nucleus (blue). (C) Colocalization analysis reveals increased co-localization of IL-1β and CD68 using image J in HIV-1 patients’ liver tissues compared to normal healthy livers (P = 0.0001). Increased IL-1β fluorescent intensity (= total intensity × Person’s R value) in HIV-1 liver tissue compared to normal livers (P < 0.0001). (D) Confocal microscopic analysis of IL-1β, NLRP3 and CD68 co-immunostaining in HIV-1-infected liver tissues. CD68 + liver macrophages (red), IL-1β (green), NLRP3 (magenta), and nucleus (blue). Panel A: Paired t test; Panel C: Welch’s test
2.2 |. Isolation of CD68+ primary human liver macrophages from liver resections
For all in vitro experiments examining the impact of HIV-1 on liver macrophage biology, primary liver macrophages were isolated from surgical resection specimens (metastatic rectal, colon, and parotid cancer, gall bladder cancer, cholangiocarcinomas, and hepatocellular carcinoma (HCC) (no HCV or hepatitis B virus or underlying cirrhosis) by performing collagenase perfusion and mincing of the liver tissue. Cells were washed with PBS 4 times, pelleted at 300 × g for 5 min, resuspended in DMEM followed by gradient centrifugation using 25% and 50% Percoll (Pharmacia, Uppsala, Sweden), as described.19 Liver macrophages were enriched by adherence to tissue culture plates. Cells (2 × 106 cells/well in a 24-well plate) were maintained in DMEM media with FBS and after 2 wk in culture, flow cytometry revealed that 82–98% of the cells were liver macrophages. Given the potential heterogeneity of intrahepatic macrophages, CD68+ cells, a common marker for macrophages, were used for subsequent characterization and experimentation.
2.3 |. Tissue immunofluorescence assay
The liver tissue sections were prepared in duplicate for each patient. Slides were rehydrated and treated for antigen retrieval. Then slides were washed with PBS and incubated with antibodies (please see the Supplemental Table 1 for antibodies) for overnight at 4°C in a humidified chamber; a secondary antibody was diluted at 1:500 and applied for secondary staining, respectively. After 2 hr incubation for each color at 37°C, the slides were stained with Hoechst 33342 and then mounted. Four random visual areas were selected for each tissue section. The images were captured with SP5 DM confocal microscope (magnification: 60×). Colocalization was analyzed by Fiji image J software (NIH, Bethesda, MD, USA) and presented by Pearson’s R value.
2.4 |. Immunostaining assay on isolated primary liver macrophages
Macrophages isolated from nontumorous noncirrhotic liver tissue were infected with HIV-1BaL +/−5nMATP in vitro as described, fixed with 2% paraformaldehyde at room temperature for 20 min. Then the cells were washed with PBS and incubated with first antibodies (please see the Supplemental Table 1 for antibodies) for overnight at 4°C in a humidified chamber. A secondary antibody was diluted at 1:500 and applied for secondary staining. After 2 hr incubation for each color at 37°C, the slides were stained with DAPI (Thermo Fisher, cat. no.: D1306, Waltham, MA). The images were captured with Zen software for LSM 880 laser Airscanning microscope (magnification: 100×). Colocalization was analyzed by Fiji image J software.
2.5 |. Flow cytometry analyses
Flow cytometry was used to determine impact of HIV-1BaL infection on CXCR4, CCR5, CD40, CD163, and CD11b expression. Furthermore, the impact of HIV-1BaL infection on TLR4, CD14, and IL-1b expression on CD68+ liver macrophages was examined. Cells were re-suspended in PBS containing 0.5% BSA at concentration 0.5–1 × 106 cells/ml. After addition of the Fc-R blocking reagent, cells were incubated with antihuman antibodies for 15 min. Cells were fixed and permeabilized at 40°C for 20 min, followed by intracellular staining with CD68-FITC and IL-1β-PB for 30 min on the ice. Flow cytometry analysis was performed on LSRII (BD Biosciences, Glendale, CA). Data were analyzed using FCS express 6 software (De Novo software).
2.6 |. Production and purification of HIV-1BaL virus
In order to obtain purified HIV-1BaL to utilize for infection of liver macrophages in vitro, we obtained Buffy coats from blood donors, differentiated primary human monocytes into macrophages and then infected with HIV-1BaL virus (NIH AIDS Reagent program, Bethesda, MD, USA). R5 virus HIV-1BaL was enriched from the supernatant of HIV-1BaL-infected macrophages and purified with 20% sucrose cushion. Viral titer was measured by serially diluted infection in ghost-CXCR4+-CCR5+ cells (NIH AIDS Reagent program, Bethesda, MD, USA).
2.7. HIV-1BaL infection in primary human liver macrophages
To determine replication of HIV-1BaL in primary liver macrophages, CD68+ liver macrophages were plated at concentration 2 × 105 cells/well in 24-well plates and cells were infected at a multiplicity of infection (MOI) of 0.10. The virus was washed off at 3 hr followed by the addition of fresh medium. Media was serially collected and HIV-1BaL p24 antigen production was measured by ELISA using the SAIC Frederick Kit (National Cancer Institute, Bethesda, MD, USA).
2.8. The impact of HIV-1BaL infection on primary human liver macrophage response to LPS
A total of 72 hr post -IV-1BaL infection CD68+ liver macrophages were challenged with TLR4 ligand ultra pure LPS (InvivoGen, San Diego, CA, USA; cat. no.: tlrl-ppglps) at a dose of 100 ng/ml for 4 hr, followed by RNA extraction and RT-qPCR at 8 hr posttreatment. Supernatants were collected for ELISA assay at 8 hr post-LPS treatment.
2.9 |. RNA extraction and RT-qPCR
Total RNA was extracted by using RNeasy spin columns (Qiagen, Chatsworth, CA, USA; cat. no.: 74136). After treatment with RNase-free DNase (Qiagen), RT was performed on RNA by using Clontech RNA to cDNA EcoDry kit according to the manufacturer’s protocol. RT-qPCR was performed on Roche Lightcycle II 480. Reactions were carried out in 10 μL by using SYBR Green PCR master mix according to the manufacturer’s protocol (Bio-Rad, cat. no.: 1708884, Hercules, CA). The concentration of primer pairs was 300 nM. All primer sequences are shown in the primer table (Supplemental Table 2). All reactions were performed in triplicate. To make comparisons between samples and controls, the CT (cycle threshold, defined as the cycle number at which the fluorescence is above the fixed threshold) values were normalized to the CT of α-actin in each sample.
2.10 |. Lactate dehydrogenase (LDH) cytotoxicity assay
Cell culture supernatant was collected from control cells, HIV-1BaL-infected cells, LPS treated cells, and HIV-1BaL+LPS treated cells. The cellular toxicity of different treatments was measured using an LDH cytotoxicity assay kit (Thermo Scientific, cat. no.: 88953, Waltham, MA) according to manufacturer’s protocols.
2.11 |. ELISA assay
Isolated macrophages were treated with different conditions and then incubated with 5 nM ATP for 8 hr. IL-1β production in the supernatant was collected at indicated time and measured by using human IL-1β ELISA kit (Affymetrix, cat. no.: 88-7261-86, Santa Clara, CA) according to the manufacturer’s protocol.
2.12 |. TLR4 signaling blockade
CD68+ liver macrophages isolated as described above were plated in 24-well plates, infected or noninfected with HIV were treated with either LPS (100 ng/ml) and/or TAK-242 (3 μM, Invivogen). TAK-242 also known as CLI-095, a small-molecule, has been shown to selectively inhibit TLR4 signaling to suppress LPS-induced inflammation.20 Eight hours after LPS treatment in the presence or absence of the TLR4 inhibitor, supernatants were collected for IL-1β ELISA.
2.13 |. NLRP3 inflammasome blockade
CD68+ liver macrophages isolated as described above were plated in 24-well plates, infected or noninfected with HIV for 3 hr. MCC950 (Sigma, San Jose, CA, USA; cat. no.: PZ0280–5MG) also known as CRID3, a small-molecule has been shown to selectively inhibit NLRP3 to suppress LPS-induced inflammation.20 Three hours postinfection, the cells were treated with 1 nM MCC950 for 30 min. Medium was aspirated at indicated time intervals and cells were then cultured with complete medium ±5 nM ATP for 8 hr. Supernatant was collected for IL-1β ELISA assay.
2.14 |. Western blot
Tissue proteins from normal liver and HIV-1 monoinfected liver were purified with T-PER tissue protein extraction reagent (Thermo, cat. no.: 78510, Waltham, MA). Equal amounts of protein were loaded in each lane for NuPAGE 4–12% Bis-tris gel and then transferred to polyvinylidene difluoride membranes. The membranes were washed with blotting buffer (1× PBS containing 0.1% Tween20) and then blocked for 60 min in blotting buffer containing 10% low-fat powdered milk. Membranes were washed 3 times with blotting buffer, incubated at 4°C overnight with primary antibody (1:1000) containing 5% lowfat powdered milk, and incubated with secondary antibody (1:1000) at room temperature for 60 min. Target proteins were developed with Tetramethylbenzidine-1 (TMB-1)-component membrane solution (SeraCare Life Sciences Inc., Milford, MA, USA). The blots were detected with Amersham Imager 600 system. The relative expression of proteins was normalized to β-actin and analyzed using Image J.
2.15 |. Statistical analysis
All data were analyzed using SigmaPlot14 software (Sigma). Data were first evaluated for normality using the Shapiro-Wilk test and then were tested for equal variance using the Brown-Forsythe test. Depending on the test results, either mean or median values were calculated. Comparisons between two groups were made using parametric (Student’s t test) or nonparametric (Wilcoxon signed ranks test) statistical tests, as appropriate. One-way ANOVA with Kruskal-Wallis test was used to compare multiple groups. For immunostaining, colocalization was analyzed by Fiji image J software; the Pearson’s R value was determined and unpaired t tests with Welch’s correction were used for statistical analysis. P values of <0.05 were considered statistically significant. Specific methods used for each figure panel provided in figure legends.
3. RESULTS
3.1 |. HIV infection of liver macrophages in vitro does not impact cell surface expression of HIV coreceptors or macrophage markers but does increaseIL-1β response
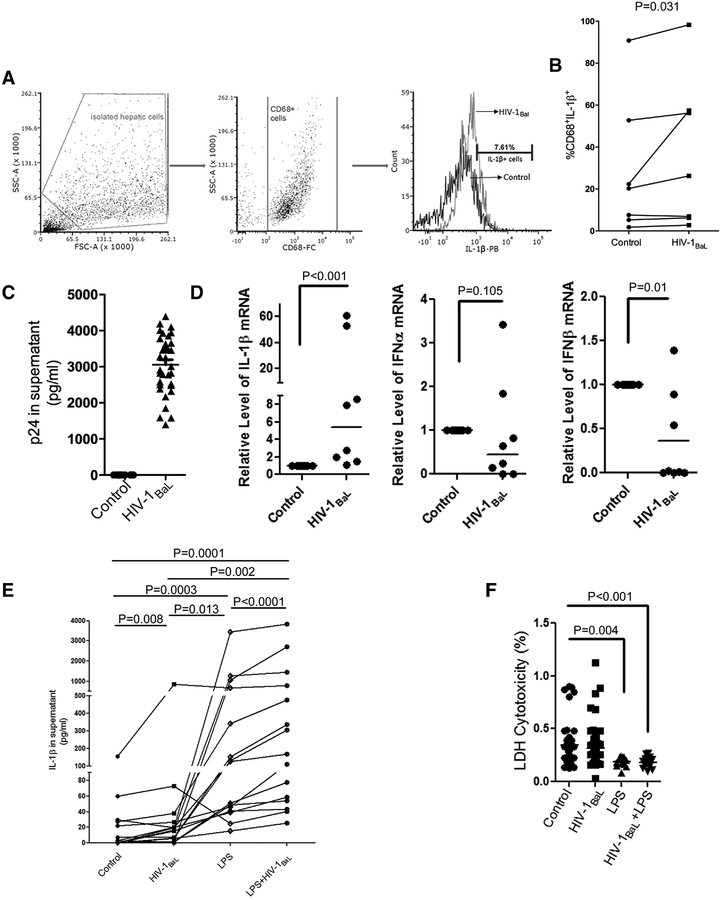
To study the contribution of HIV-1 infection to the IL-1β response in CD68+ liver macrophages, we isolated primary macrophages from liver tissues and first examined the impact of HIV infection on macrophage phenotypes and HIV coreceptors expression (CD68, CD163, CD40, CD11b, CCR5, and CXCR4) using flow cytometry. A high percentage of CD68+ macrophage cells (mean: 92.89%) were obtained. Nevertheless, no significant changes were found in relevant markers after HIV-1BaL infection (Fig. 1A and B). However, HIV-1BaL infection increased the percentage of intracellular IL-1β+ macrophages, suggesting that viral infection may promote IL-1β production in liver macrophages (median value: 20.24% vs. 26.27%) (Fig. 2A and B) that are productively infected by HIV-1BaL as reflected by p24 ELISA (Fig. 2C). RT-PCR results also revealed that whereas IL-1β mRNA was up-regulated by HIV-1BaL infection, no significant induction of IFNα or IFNβ mRNA was detected in HIV-1BaL-infected liver macrophages (Fig. 2D), indicating that HIV-1BaL infection employed different pathways to induce the IL-1β response and innate immune responses in liver macrophages, respectively. By quantifying IL-1β in the supernatant, we found that peak IL-1β secretion occurred 8 hr poststimulation with 5μM ATP (data not shown), but no detectable IL-1β induction was observed without ATP treatment (mean < 5 pg/ml), suggesting that ATP is required for the IL-1β release from HIV-1BaL-infected liver macrophages.21,22 ATP is known to be increased in chronic liver disease. Therefore, we used 5μM ATP to investigate IL-1β response in the following experiments. Compared with the control, a higher IL-1β secretion occurred in virus-infected cells after ATP addition (median value: 3.066 vs. 16.634 pg/ml, P = 0.008) (Fig. 2E). Liver macrophages play a major role in the response to microbial products and increased microbial translocation, which has been shown to be increased in HIV-1 infection.23 To determine whether HIV-1BaL infection alters IL-1β response of liver macrophages to LPS, we compared IL-1β secretion from macrophages treated with either LPS or HIV-1BaL alone versus LPS+HIV-1BaL. HIV-1BaL infection significantly enhanced IL-1β response to LPS in liver macrophages (median value: 40.91 pg/ml vs. 168.70 pg/ml, P < 0.0001) (Fig. 2E). In addition, LDH cytotoxicity assay further showed a low cytotoxicity of HIV-1BaL on cell proliferation, even after LPS stimulation (Fig. 2F), excluding a possibility of increased IL-1β due to the release from dead cells. These results suggest an important role of HIV-1BaL infection in activating the IL-1β response to LPS stimulation in intrahepatic macrophages.
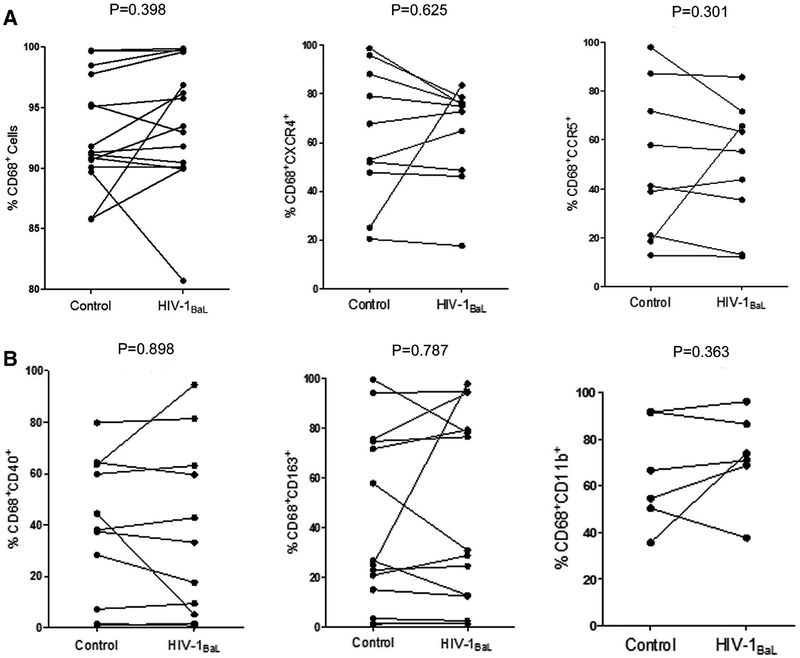
FIGURE 1. HIV infection does not significantly alter cell surface expression of HIV chemokine coreceptors or macrophage activation markers.
(A) Representative flow cytometry analyses of CD68 (n = 15, P = 0.398) expression and HIV-1 coreceptors, CCR5 (n = 10, P = 0.301) and CXCR4 (n = 9, P = 0.625), expression directly before and after HIV-1BaL infection. (B) Representative flow cytometry analyses of the expression CD40 (n = 11, P = 0.898), CD163 (n = 14, P = 0.787) and CD11b (n = 6, P = 0.363) in CD68+ macrophages directly before and after HIV-1BaL infection. Wilcoxon signed rank test was used for statistical analysis
FIGURE 2. HIV-1BaL infection induces an IL-1β response in primary CD68+ liver macrophages.
(A) Flow cytometry strategy to analyze IL-1β expression in isolated liver macrophages. (B) Flow cytometry analyses of IL-1β positive CD68+ macrophages directly before and after HIV-1BaL infection (n = 7; P = 0.031). (C) p24 was measured 48 hr postinfection with HIV-1BaL at an MOI of 0.10 (n = 8). (D) IL-1β mRNA (n = 8; P < 0.001), IFNα mRNA (n = 8; P = 0.105) and IFNβ mRNA (n = 8; P = 0.01) were measured by RT-PCR. (E) CD68+ liver macrophages were treated with HIV-1BaL, LPS, or LPS+HIV-1BaL and, IL-1β in culture supernatant was measured with ELISA (n = 15) HIV-1BaL vs. control, P = 0.008; LPS vs. control, P = 0.0003; LPS vs. LPS+ HIV-1BaL, P < 0.0001. (F) Cytotoxicity of control (n = 10), HIV-1BaL infected (n = 14), LPS treated (n = 4) or HIV-1BaL +LPS treated (n = 5) on isolated macrophages was examined by LDH assay to confirm that increases in IL-1β were not due to cell death (each sample quadruplicate). Panels B, D: Wilcoxon signed rank test; Panels E, F: Kruskal-Wallis one way analysis
3.2. HIV-1BaL induced TLR4 expression is critical for IL-1β response to LPS stimulation in CD68+ macrophages
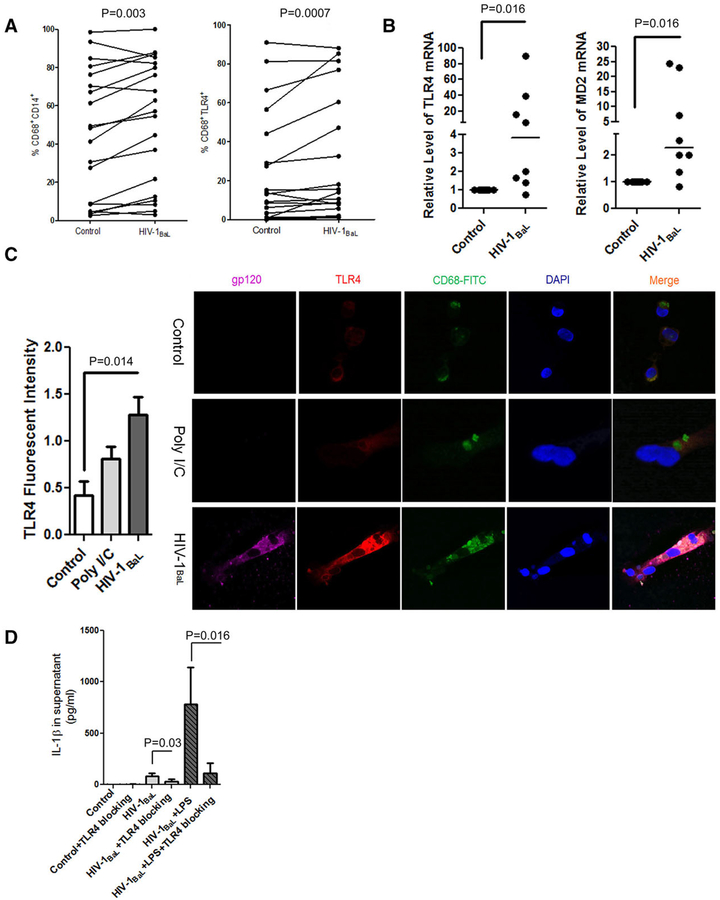
Because the TLR4 pathway contributes to the inflammatory response of macrophages to microbial products, we compared the expression of CD14 and TLR4 in liver macrophages before and after HIV-1BaL infection. As shown in Fig. 3A, HIV-1BaL infection increased CD14 by an average of 6.72% (P = 0.003) and TLR4 expression by 8.86% (P = 0.0007) on CD68+ macrophage cells. RT-PCR results also showed that HIV-1BaL infection up-regulated the levels of MD2 mRNA and TLR4 mRNA (P = 0.016) (Fig. 3B), suggesting that HIV-1BaL infection may up-regulate the IL-1β response to LPS stimulation in CD68+ liver macrophages via TLR4 signaling. To further investigate the impact of HIV-1BaL infection on the expression of TLR4, we examined the TLR4 expression in CD68+ liver macrophages after exposure to medium, polyI/C, and HIV-1BaL infection. A significant induction of TLR4 expression was observed after 24 hr of exposure to HIV-1BaL(P = 0.014), but not found in control or poly I/C treated cells (Fig. 3C), confirming the contribution of HIV-1BaL infection to the up-regulation of TLR4 expression. To further test the importance of TLR4 in the IL-1β response, we blocked TLR4 signaling with a small molecule inhibitor and examined the IL-1β response to LPS stimulation in HIV-1BaL-infected liver macrophages. A significant reduction of IL-1β (mean value: 192.281 pg/ml to 13.589 pg/ml, P = 0.016) was observed in HIV-1BaL plus LPS treated cells (Fig. 3D) in the presence of the TLR4 inhibitor, suggesting an amplifying effect of HIV-1BaL infection on IL-1β response to LPS through TLR4.
FIGURE 3. HIV-1BaL induced TLR4 expression is critical for the response of HIV-1BaL-infected CD68+ macrophages to LPS stimulation.
(A) Flow cytometry analyses of CD14 and TLR4 expression (n = 15; P = 0.003 and P = 0.0007) directly before and after HIV-1BaL infection; (B) TLR4 mRNA (n = 8; P = 0.016) and MD2 mRNA (n = 8; P = 0.016) were measured by RT-PCR; (C) Primary isolated CD68+ liver macrophages were treated with medium, poly I/C (2.5ug/ml), or HIV-1BaL infection (MOI = 0.1) for 24 hr without ATP treatment and TLR4 expression compared with TLR4 fluorescent intensity and examined by immunostaining. TLR4 (red), CD68 + liver macrophages (green), gp120 (magenta), and nucleus (blue). (D) CD68+ macrophages were treated by 0.1 MOI HIV-1BaL (n = 7) and or LPS stimulation (n = 7, P = 0.016) with or without 3 μM TAK-242. IL-1β was measured by ELISA. Panel A,C: Paired t test; Panel B: Wilcoxon signed rank test; Panel D: Kruskal-Wallis one way analysis (HIV vs. HIV+TLR4 blocking: Paired t test; LPS+HIV vs. HIV+LPS+TLR4 blocking: Wilcoxon signed rank test)
3.3 |. NLRP3 participates in IL-1β response to LPS stimulation in HIV-1BaL-infected CD68+macrophages
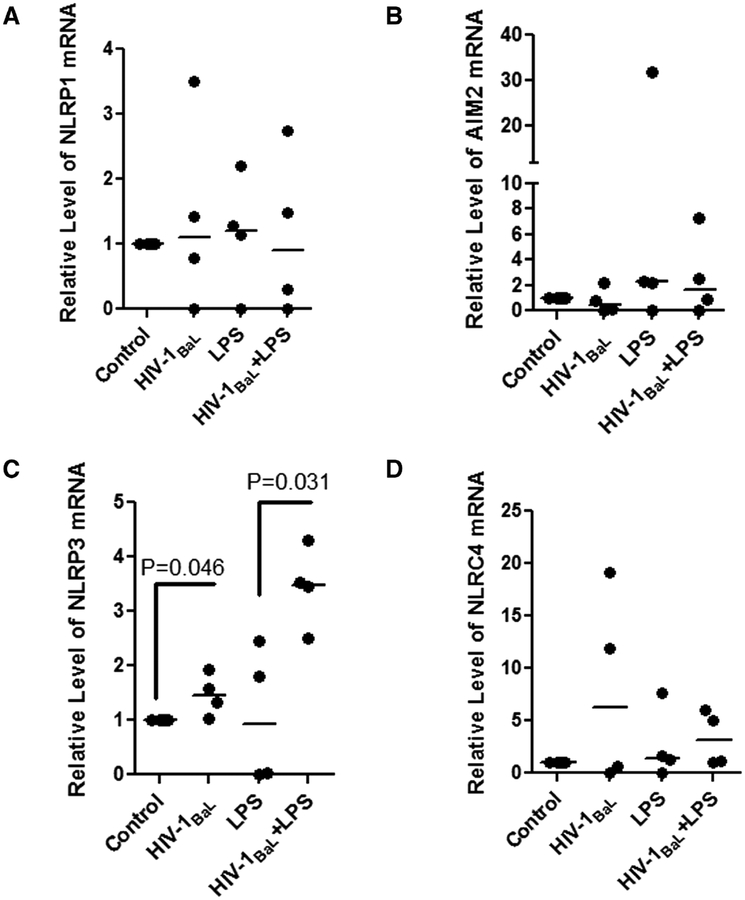
Caspase 1 inflammasomes have been implicated as important contributors to the activation of IL-1β signaling, and in the liver, NLRP3 has been additionally implicated in this signaling axis. To further investigate the underlying mechanisms and identify which inflammasome contributed to the sensitization of the IL-1β response to LPS stimulation in HIV-1BaL-infected liver macrophages, we examined the expression of absent in melanoma 2 (AIM2), NLRP1, NLRP3, and NLR family card domain containing 4 (NLRC4) between these two groups. The transcript level of AIM2, NLRP1, NLRC4 was not significantly changed by HIV-1BaL infection after LPS stimulation. However, the level of NLRP3 mRNA was up-regulated by HIV-1BaL infection and further increased in HIV-1BaL + LPS stimulated CD68+ liver macrophages (P = 0.046; P = 0.031, respectively) (Fig. 4C). These results suggest that NLRP3 may contribute to the IL-1β response to HIV-1BaL infection in KCs.
FIGURE 4. Increased NLRP3 is associated with the IL-1β response of HIV-1BaL-infected Liver macrophages to LPS stimulation.
The mRNA of (A) NLRP1, (B) AIM2, (C) NLRP3 (P = 0.045 and P = 0.031), and (D) NLRC4 were quantified by RT-PCR and compared among control, HIV-1BaL infection, LPS treated and LPS+HIV-1BaL treated macrophages, n = 4, each sample in triplicate; β-actin was used as internal control. Kruskal-Wallis one way analysis with paired t tests between groups
3.4. NLRP3 modulates IL-1β response to HIV-1BaL infection in liver macrophages
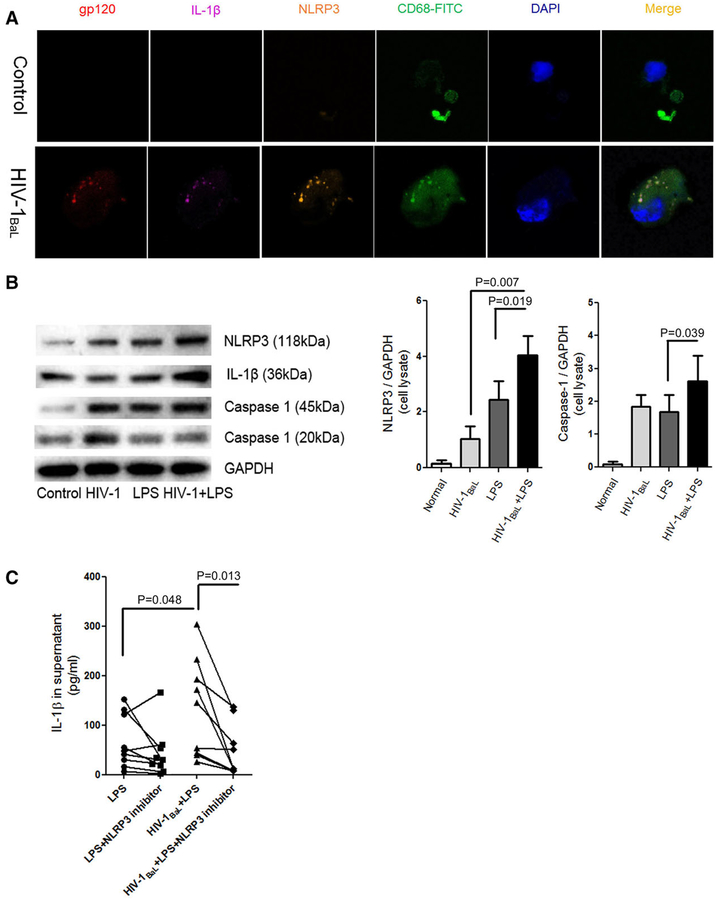
To determine whether NLRP3 modulates the IL-1β response in HIV-1BaL-infected CD68+ macrophages, we then infected CD68+ macrophages with HIV-1BaL and examined the expression/colocalization of IL-1β and NLRP3 expression in CD68+macrophages as assessed by confocal microscopy (Fig. 5A). Moreover, a higher level of caspase-1 and NLRP3 proteins were detected in HIV-1BaL+LPS treated cells compared to those in control cells and LPS stimulated cells (Fig. 5B). Consistent with the previous results, no induction of NLRP3 was found in normal control (medium + 5 μM ATP). In addition, no significant difference in cleaved caspase-1 was observed in supernatant between LPS treated and LPS plus HIV-1BaL stimulated liver macrophages. To further confirm the contribution of NLRP3 to the IL-1β response in HIV-1BaL-infected CD68+ liver macrophages, we blocked NLRP3 activation with a small molecule, MCC950. As expected, MCC950 significantly decreased the IL-1β response to LPS stimulation in HIV-1BaL-infected liver macrophages (median value:43.47 pg/ml vs. 10.84 pg/ml, P = 0.013) (Fig. 5C). Therefore, these results suggest that NLRP3 plays an important role in the modulation of IL-1β response to HIV-1BaL infection and LPS in liver macrophages.
FIGURE 5. NLRP3 modulates IL-1β response to LPS stimulation in HIV-1BaL-infected liver macrophages.
(A) NLRP3 confocal analysis in isolated liver macrophages with or without HIV-1BaL infection 8 hr posttreatment of 5 μM ATP. IL-1β (magenta), gp120 (red), CD68+ liver macrophages (green), NLRP3 (orange), and nucleus (blue). (B) NLRP3 (P = 0.019), intracellular caspase-1(P = 0.039) and cleaved caspase-1 proteins in the supernatant were measured after HIV-1, LPS or HIV-1 + LPS, GAPDH was taken as internal control. The results were obtained from 8 donors. (C) Liver macrophages were infected by HIV-1BaL at an MOI of 0.10 with or without 1.0 nM MCC9450. After 100 ng/ml LPS challenge, the supernatant was collected 8 hr posttreatment with 5 nM ATP and IL-1β was measured by ELISA (n = 9, duplicate, P = 0.013). Kruskal-Wallis one way analysis with paired t test between two groups
3.5. IL-1β expression is increased in intrahepatic CD68+ macrophages in liver tissue from HIV-infected patients
To assess IL-1β hepatic expression in patients with HIV-1 infection, protein lysates from frozen archived human liver sections from normal donors and HIV-1+ patients were purified and examined by Western blotting. As shown in Fig. 6A, a high expression of IL-1β was detected in HIV-1+ human liver tissues. To examine cellular localization, the liver tissues were co-immunostained for CD68 and IL-1β. HIV livers demonstrated an increased expression of IL-1β compared to normal livers with a high degree of colocalization in CD68+ macrophages (Fig. 6B and C). Furthermore, in higher power views, IL-1β and NLRP3 are colocalized (Fig. 6D). Consistent with previous data, these results further support the role of the IL-1β response of CD68+ liver macrophages in HIV-infected patients.
4 |. DISCUSSION
Overall our results suggest that HIV infection of CD68+ liver macrophages results in an amplified IL-1β response to LPS that is mediated through the TLR4-NLRP3 pathway. These findings are supported both ex vivo/in vitro HIV-1BaL infection of primary human liver macrophages and in vivo data derived from the livers of HIV-infected patients.
Macrophages are considered a major contributor to HIV-1 transmission and persistent infection.24 Although macrophages are not dividing cells, a high level of HIV-1 replication has been detected in the cells and well documented in a variety of studies.25,26 In the liver, KCs (resident macrophages) are the primary target of HIV-1. Infection of KCs by HIV-1 has been demonstrated by detection of HIV-1 RNA by in situ hybridization and by detection of proviral DNA by PCR in purified KCs derived from the livers of HIV-infected patients.27–29 The impact on the biology of KCs, however, has not been significantly studied. In addition, macrophages contribute to the first line defense against viral infection and microbial invasion. IL-1β is one of the key proinflammatory cytokines released from macrophages. It is generally primed by stimulation induced by microbial products and it is then processed into an activated form. ATP can rapidly promote caspase-1-dependent release of IL-1β. A high level of IL-1β expression has been found in the lymphoid tissues of HIV-1-infected patients with or without ART, suggesting that IL-1β may contribute to decreases in CD4+ T-cells.30 In addition, IL-1β can inhibit IL-7 production in epithelial cells, contributing to the poor survival of CD4+ T cells in the gut compartment.31 Supporting a role for IL-1β in disease progression, Paneth cells in SIV-infected macaques secrete IL-1β and induce gut inflammation, resulting in the disruption of the intestinal barrier.32 Also, Chattergoon et al. showed that HIV can drive a high IL-1β response through TLR activation and without induction of type I IFN responses.33 These studies indicate that IL-1β plays an important role in local innate and adaptive immunity in HIV-1 infection.
Innate immune responses with inflammasome activation plays a critical role in the progression of chronic liver disease and may be partially related to increases in gut permeability and microbial translocation into the portal vein, which delivers proinflamma-tory bacterial products to the liver.10 Fibrosis progression in HIV-infected patients is accelerated in patients with liver disease across a range of etiologies despite ART, and may also be associated with microbial translocation.34
The liver is an immunologically complex organ and serves as the gatekeeper for gut-derived pathogens/PAMPS to prevent them from promoting systemic immune activation. Dozens of DAMPS, including ATP, are constantly released from activated intrahepatic immune cells and injured parenchymal or nonparenchymal cells. Both PAMPS and DAMPS can result in high and persistent IL-1β inflammatory responses in liver macrophages.35 It has been shown that HIV-1 can infect resident hepatic macrophages both in vitro and in vivo. Our previous study8 also revealed that an amplified proinflammatory response, indicated by increased IL-6 and TNF-α can be induced by HIV-1 infection. In this study, IL-1 β expression was examined in HIV-1BaL-infected macrophages. We detected a high level of IL-1β expression in HIV-1BaL-infected human CD68+ macrophages. Given their key roles in the canonical IL-1β pathway in human liver macrophages, the NLRP3 inflammasome and caspase-1 were investigated to determine their contribution to the proinflammatory changes that resulted from HIV-1BaL infection. Caspase-1 is required for IL-1β release in HIV-1-infected human microglia.36 An association between a high NLRP3 expression and IL-1β in HIV-1 infection suggests an essential role of NLRP3 in IL-1β response to HIV-1 infection.37,38 Here we found that expression of NLRP3 in liver macrophages was up-regulated by HIV-1BaL infection. More importantly, the protein expression of caspase-1 and NLRP3 was significantly increased by LPS stimulation of HIV-1BaL-infected liver macrophages. Furthermore, inhibition of NLRP3 with a small chemical compound significantly reduced IL-1β release from HIV-1BaL plus LPS stimulated macrophages. Therefore, these results suggest that HIV-1BaL infection modulates the IL-1β response to LPS stimulation through increased NLRP3 expression in liver macrophages. To determine the potential clinical relevance of these in vitro findings, archived frozen liver tissue from HIV-1-infected patients was examined for expression of IL-1β and NLRP3. In line with our in ex vivo findings, our data revealed that IL-1β expression is increased in the livers from HIV-infected patients and IL-1β colocalizes with NLRP3 in intrahepatic CD68+ cells in these livers. We have previously shown that despite effective ART, KCs isolated from HIV-1+ patients maintain a dysregulated response to LPS. As archived tissue rather than fresh cells were available, it is not clear if direct HIV infection in vivo is contributing to these results but this will be explored in the future. Clearly the milieu created by HIV infection and potentially increased gut permeability results in increased IL-1β in the livers of HIV-monoinfected patients and is critical when considering the management of emerging liver diseases in this patient population.
Interestingly, IL-1β released from HIV-1-infected macrophages can disrupt the intestinal barrier and promote microbial translocation, and, in turn, microbial translocation has been found to produce a synergistic effect on the IL-1β response in HIV-1-infected macrophages.39 High levels of IL-1β production in HIV-1-infected macrophages have been associated with bacterial infection and could be reduced by antibacterial therapy.40,41 LPS stimulation in vitro also results in higher levels of IL-1β expression in PBMCs derived from HIV-1 exposed seronegative individuals compared to healthy controls.42 However, the interaction between microbial stimulation and inflammatory responses is complex in macrophages. The macrophages used in the current study were isolated from patients with cancer, though the cells themselves were isolated from neighboring nontumorous tissue. Studies have suggested that chemokines released from cancers may play an important role in the differentiation of infiltrating monocytes into M1- or M2-like macrophages. Moreover, clinical anticancer therapy targeting these chemokine receptors can result in a suppression of the M2-like tissue macrophages. Therefore, it is possible that endogenous pathway activation from the cancers or treatments of the cancers43,44 impacted the cells in our study. However, we examined cells from multiple different clinical specimens and had consistent results, supporting the validity of our findings. As shown in Fig. 1, macrophages enriched by the selection of rapidly adherent cells had an M1-like phenotype, with high expression of CD68. High levels of LPS in HIV-1 patients tend to drive the differentiation of monocytes into M1-like macrophages and augments proinflammatory responses. TLR signaling has been implicated in the initiation of the pro-IL-1β and NLRP3-caspase-1 pathway, leading to production of bioactive IL-1β in monocytes.45 Furthermore, TLR signaling has been shown to play an important role in chronic liver injury and liver fibrosis progression. HIV and HCV have been shown to activate the inflammasome in macrophages and monocytes via endosomal TLRs, independent of productive viral infection.33 TLR4, a major receptor of microbial products, is associated with liver injury, liver inflammation and a pro-fibrogenic signature in activated hepatic stellate cells.46 TLR4 signaling is important for hepatic fibrosis progression in multiple liver injury models.47 Moreover, HIV-1 related microbial translocation is associated with fibrosis progression in patients with chronic HCV though underlying mechanisms have not been defined.33
TLR4 and CD14 are major functional markers that are used to identify human KCs. Given the heterogeneity of macrophage populations, we used CD68 to identify liver macrophages. TLR4, MD2, and CD14 expression were up-regulated in HIV-1BaL-infected CD68+ macrophages in this study. Moreover, the expression of type I IFNs was not increased in HIV-1BaL-infected liver macrophages, suggesting that TLR4 is a major activator of NLRP3 expression. Inhibition of TLR4 and NLRP3 dramatically reduced the IL-1β response to LPS stimulation in HIV-1BaL-infected cells, but an NLRP3 inhibitor had no significant effects on LPS stimulated macrophages, suggesting that the IL-1β response to LPS stimulation in HIV-1BaL-infected CD68+ macrophages requires an intact TLR4-NLRP3 axis.
In conclusion, our data show that HIV-1BaL infection up-regulates the expression of IL-1β and increases the expression of NLRP3 and caspase-1 in CD68+ hepatic macrophages. Blocking assays confirmed that the NLRP3 inflammasome is a key transducer to amplify IL-1β response in HIV-1BaL-infected liver macrophages. Moreover, HIV-1BaL infection increases the expression of CD14 and TLR4 and results in a dysregulated IL-1β response to LPS. Blocking of a TLR4 signaling pathway can reverse this dysregulated IL-1β response in HIV-infected liver macrophages. Consistent with these findings, our data revealed that IL-1β expression was increased in liver tissues of HIV-monoinfected patients and IL-1β colocalized with CD68 and NLRP3. Whether these findings are directly due to direct infection of KCs by HIV or HIV-related microbial translocation or both remain to be determined. These findings may provide great insight in understanding the increasing prevalence of liver disease in HIV-infected patients in the era of effect ART and effective direct acting antivirals for HCV. The emergence of nonviral liver diseases such as NASH and ASH in this population and the role of inflammasome activation in promoting hepatic inflammation deserve greater attention and may inform unique therapeutic strategies.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by research grants NIH R56DK92128 and R01DK108364 to M.B.B. We appreciate support from the NIH AIDS Reagents Program.
Abbreviations:
- MOI
multiplicity of infection
- NLRC4
NLR family CARD domian containing
- NLRP
nucleotide-binding domain, leucine-rich-containing family, pyrin domain containing
Footnotes
DISCLOSURES
The authors declare no conflicts of interest.
SUPPORTING INFORMATION
Additional information may be found online in the Supporting Information section at the end of the article.
REFERENCES
- 1.Kolios G, Valatas V, Kouroumalis E. Role of Kupffer cells in the pathogenesis of liver disease. World J Gastroenterol. 2006;12:7413–7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Racanelli V, Rehermann B. The liver as an immunological organ. Hepatology. 2006;43:S54–S62. [DOI] [PubMed] [Google Scholar]
- 3.Klatt NR, Funderburg NT, Brenchley JM. Microbial translocation, immune activation, and HIV disease. Trends Microbiol. 2013;21:6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Price JC, Thio CL. Liver disease in the HIV-infected individual. Clin Gastroenterol Hepatol. 2010;8:1002–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brenchley JM, Price DA, Schacker TW, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–1371. [DOI] [PubMed] [Google Scholar]
- 6.Brenchley JM, Schacker TW, Ruff LE, et al. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med. 2004;200:749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wallet MA, Rodriguez CA, Yin L, et al. Microbial translocation induces persistent macrophage activation unrelated to HIV-1 levels or T-cell activation following therapy. AIDS. 2010;24:1281–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mosoian A, Zhang L, Hong F, et al. Frontline Science: HIV infection of Kupffer cells results in an amplified proinflammatory response to LPS. J Leukoc Biol. 2017;101:1083–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marchetti G, Tincati C, Silvestri G. Microbial translocation in the pathogenesis of HIV infection and AIDS. Clin Microbiol Rev. 2013;26:2–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szabo G, Petrasek J. Inflammasome activation and function in liver disease. Nat Rev Gastroenterol Hepatol. 2015;12:387–400. [DOI] [PubMed] [Google Scholar]
- 11.Netea MG, Simon A, van de Veerdonk F, Kullberg BJ, Van der Meer JW, Joosten LA. IL-1beta processing in host defense: beyond the inflammasomes. PLoS Pathog. 2010;6:e1000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tschopp J, Schroder K. NLRP3 inflammasome activation: the convergence of multiple signalling pathways on ROS production? Nat Rev Immunol. 2010;10:210–215. [DOI] [PubMed] [Google Scholar]
- 13.Coll RC, Robertson AA, Chae JJ, et al. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat Med. 2015;21:248–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin Z, Liang W, Kang K, Li H, Cao Z, Zhang Y. Classical swine fever virus and p7 protein induce secretion of IL-1beta in macrophages. J Gen Virol. 2014;95:2693–2699. [DOI] [PubMed] [Google Scholar]
- 15.Moriyama M, Chen IY, Kawaguchi A, et al. The RNA- and TRIM25-Binding domains of influenza virus NS1 protein are essential for suppression of NLRP3 inflammasome-mediated interleukin-1beta secretion. J Virol. 2016;90:4105–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang B, Zhu J, Li D, et al. Newcastle disease virus infection induces activation of the NLRP3 inflammasome. Virology. 2016;496: 90–96. [DOI] [PubMed] [Google Scholar]
- 17.Chen W, Xu Y, Li H, et al. HCV genomic RNA activates the NLRP3 inflammasome in human myeloid cells. PLoS One. 2014;9:e84953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen IC, Scull MA, Moore CB, et al. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. 2009;30:556–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alabraba EB, Curbishley SM, Lai WK, Wigmore SJ, Adams DH, Afford SC. A new approach to isolation and culture of human Kupffer cells. J Immunol Methods. 2007;326:139–144. [DOI] [PubMed] [Google Scholar]
- 20.Kawamoto T, Ii M, Kitazaki T, Iizawa Y, Kimura H. TAK-242 selectively suppresses Toll-like receptor 4-signaling mediated by the intracellular domain. Eur J Pharmacol. 2008;584:40–48. [DOI] [PubMed] [Google Scholar]
- 21.Pizzirani C, Ferrari D, Chiozzi P, et al. Stimulation of P2 receptors causes release of IL-1beta-loaded microvesicles from human dendritic cells. Blood. 2007;109:3856–3864. [DOI] [PubMed] [Google Scholar]
- 22.Lopez-Castejon G, Brough D. Understanding the mechanism of IL-1beta secretion. Cytokine Growth Factor Rev. 2011;22:189–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandler NG, Douek DC. Microbial translocation in HIV infection: causes, consequences and treatment opportunities. Nat Rev Microbiol. 2012;10:655–666. [DOI] [PubMed] [Google Scholar]
- 24.Koppensteiner H, Brack-Werner R, Schindler M. Macrophages and their relevance in human immunodeficiency virus type I infection. Retrovirology. 2012;9:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Honeycutt JB, Wahl A, Baker C, et al. Macrophages sustain HIV replication in vivo independently of T cells. J Clin Invest. 2016;126:1353–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saez-Cirion A, Hamimi C, Bergamaschi A, et al. Restriction of HIV-1 replication in macrophages and CD4+ T cells from HIV controllers. Blood. 2011;118:955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Housset C, Lamas E, Brechot C. Detection of HIV-1 RNA and p24 antigen in HIV-1 infected human liver. Res Virol. 1990;141:153–159. [DOI] [PubMed] [Google Scholar]
- 28.Housset C, Lamas E, Courgnaud V, et al. Presence of HIV-1 in human parenchymal and non-parenchymal liver cells in vivo. J Hepatol. 1993;19:252–258. [DOI] [PubMed] [Google Scholar]
- 29.Cao YZ, Dieterich D, Thomas PA, Huang YX, Mirabile M, Ho DD. Identification and quantitation of HIV-1 in the liver of patients with AIDS. AIDS. 1992;6:65–70. [DOI] [PubMed] [Google Scholar]
- 30.Shive CL, Mudd JC, Funderburg NT, et al. Inflammatory cytokines drive CD4+ T-cell cycling and impaired responsiveness to inter-leukin 7: implications for immune failure in HIV disease. J Infect Dis. 2014;210:619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thang PH, Ruffin N, Brodin D, et al. The role of IL-1beta in reduced IL-7 production by stromal and epithelial cells: a model for impaired T-cell numbers in the gut during HIV-1 infection. J Intern Med. 2010;268: 181–193. [DOI] [PubMed] [Google Scholar]
- 32.Hirao LA, Grishina I, Bourry O, et al. Early mucosal sensing of SIV infection by paneth cells induces IL-1beta production and initiates gut epithelial disruption. PLoS Pathog. 2014;10:e1004311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chattergoon MA, Latanich R, Quinn J, et al. HIV and HCV activate the inflammasome in monocytes and macrophages via endosomal Toll-like receptors without induction of type 1 interferon. PLoS Pathog. 2014;10:e1004082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balagopal A, Philp FH, Astemborski J, et al. Human immunodeficiency virus-related microbial translocation and progression of hepatitis C. Gastroenterology. 2008;135:226–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wree A, Marra F. The inflammasome in liver disease. J Hepatol. 2016;65:1055–1056. [DOI] [PubMed] [Google Scholar]
- 36.Walsh JG, Reinke SN, Mamik MK, et al. Rapid inflammasome activation in microglia contributes to brain disease in HIV/AIDS. Retrovirology. 2014;11:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pontillo A, Silva LT, Oshiro TM, Finazzo C, Crovella S, Duarte AJ. HIV-1 induces NALP3-inflammasome expression and interleukin-1beta secretion in dendritic cells from healthy individuals but not from HIV-positive patients. AIDS. 2012;26:11–18. [DOI] [PubMed] [Google Scholar]
- 38.Pontillo A, Oshiro TM, Girardelli M, Kamada AJ, Crovella S, Duarte AJ. Polymorphisms in inflammasome’ genes and susceptibility to HIV-1 infection. J Acquir Immune Defic Syndr. 2012;59:121–125. [DOI] [PubMed] [Google Scholar]
- 39.Chivero ET, Guo ML, Periyasamy P, Liao K, Callen SE, Buch S. HIV-1 Tat Primes and Activates Microglial NLRP3 Inflammasome-Mediated Neuroinflammation. J Neurosci. 2017;37:3599–3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sturm-Ramirez K, Gaye-Diallo A, Eisen G, Mboup S, Kanki PJ. High levels of tumor necrosis factor-alpha and interleukin-1beta in bacterial vaginosis may increase susceptibility to human immunodeficiency virus. J Infect Dis. 2000;182:467–473. [DOI] [PubMed] [Google Scholar]
- 41.Rebbapragada A, Howe K, Wachihi C, et al. Bacterial vaginosis in HIV-infected women induces reversible alterations in the cervical immune environment. J Acquir Immune Defic Syndr. 2008;49: 520–522. [DOI] [PubMed] [Google Scholar]
- 42.Biasin M, Piacentini L, Lo Caputo S, et al. TLR activation pathways in HIV-1-exposed seronegative individuals. J Immunol. 2010;184: 2710–2717. [DOI] [PubMed] [Google Scholar]
- 43.Zheng X, Turkowski K, Mora J, et al. Redirecting tumor-associated macrophages to become tumoricidal effectors as a novel strategy for cancer therapy. Oncotarget. 2017;8:48436–48452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silzle T, Kreutz M, Dobler MA, Brockhoff G, Knuechel R, Kunz-Schughart LA. Tumor-associated fibroblasts recruit blood monocytes into tumor tissue. Eur J Immunol. 2003;33:1311–1320. [DOI] [PubMed] [Google Scholar]
- 45.Guo H, Gao J, Taxman DJ, Ting JP, Su L. HIV-1 infection induces interleukin-1beta production via TLR8 protein-dependent and NLRP3 inflammasome mechanisms in human monocytes. J Biol Chem. 2014;289:21716–21726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo J, Friedman SL. Toll-like receptor 4 signaling in liver injury and hepatic fibrogenesis. Fibrogenesis Tissue Repair. 2010;3:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roh YS, Seki E. Toll-like receptors in alcoholic liver disease, non-alcoholic steatohepatitis and carcinogenesis. J Gastroenterol Hepatol. 2013;1:38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.