SUMMARY
Anabolic resistance and impaired myocellular quality contribute to age-related sarcopenia, which exacerbates with obesity. Diet-induced muscle mass loss is attenuated by resistance or aerobic plus resistance exercise compared to aerobic exercise in obese elderly. We assessed chronic effects of weight loss plus different exercise modalities on muscle protein synthesis response to feeding and myocellular quality. Obese older adults were randomized to weight-management program plus aerobic, resistance or combined aerobic and resistance exercise or to control. Participants underwent vastus lateralis biopsies at baseline and 6 months. Muscle protein synthesis rate increased more in resistance and combined than in control. Autophagy mediators’ expression decreased more in combined than in aerobic, which experienced a higher increase in inflammation and mitochondrial regulators’ expression. In obese elderly, combined aerobic and resistance exercise is superior to either mode independently for improving muscle protein synthesis and myocellular quality, thereby maintaining muscle mass during weight-loss therapy.
Keywords: Aging, Sarcopenia, Diet, Calorie Restriction, Weight Loss, Exercise, Lifestyle Intervention, Muscle Protein Synthesis, Muscle Quality
Graphical Abstract
In Brief
Anabolic resistance and impaired myocellular quality contribute to age-related sarcopenia, which worsens with obesity. However, weight loss programs can exacerbate sarcopenia. Colleluori et al., show that during weight-loss therapy, aerobic plus resistance exercise is more effective than aerobic or resistance exercise alone in improving muscle protein synthesis and myocellular quality, thereby preserving muscle mass in dieting, obese older adults.
INTRODUCTION
Frailty is an age-related condition of impaired homeostatic reserve and reduced capacity of the organism to withstand stress, resulting in increased vulnerability to adverse health outcomes (Fried et al., 2001). One of the main features of frailty is sarcopenia, a complex multifactorial process defined as a loss of muscle mass and strength or function (Fielding et al., 2011). In older adults, loss of muscle mass was reported to be a result of blunted muscle protein synthesis (MPS) response to anabolic stimuli (e.g. insulin and amino acids) as opposed to an increased muscle protein breakdown (Volpi et al., 2000). However, since age-related loss in muscle strength is three times faster than muscle mass loss, understanding the pathways defining muscle quality is critical (Goodpaster et al., 2006). Dysfunctional neuromuscular junction, myogenesis, protein quality control, mitochondrial function, along with increased inflammation and fat infiltration, are all considered age-related mechanisms impairing muscle quality and triggering muscle wasting (Bell et al., 2016; Cartee et al., 2016; Egan and Zierath, 2013; Romanello and Sandri, 2015).
Sarcopenia during aging is exacerbated by obesity (Batsis and Villareal, 2018; Roubenoff, 2004) which has developed into an epidemic in the Western world. By 2030, 20% of the population will be represented by older adults (defined as age ≥65 years) half of whom will be obese (Flegal et al., 2016). Lifestyle modifications consisting of diet and regular exercise are cornerstones of treatment for obesity; however, their adoption in elderly frail is still under debate because of the weight loss-induced reduction of muscle and bone mass (Jensen et al., 2014; Villareal et al., 2005). We demonstrated that the combination of diet and exercise in obese elderly improves outcomes of frailty (Villareal et al., 2011a) and that ~10% diet-induced weight loss (WL) accompanied by a combination of aerobic and resistance exercise is the most effective strategy in improving functional status of obese older adults while attenuating the negative effects of WL (Villareal et al., 2017a). Specifically, we compared the effect of WL plus aerobic, resistance exercise, or both, and we observed that, while resistance and combined exercise were the best interventions attenuating the WL-induced reduction in muscle mass, combined exercise resulted in the greatest improvement in physical function (Villareal et al., 2017a). However, the mechanisms underlying our observations are not clear, and insights into key aspects of specific exercise protocols designed to improve overall physical capacity, are considered an important topic of investigation (Cartee et al., 2016). Therefore, our objective was to compare the chronic (6-months) effects of WL plus aerobic or resistance or the combination of both exercise modalities on changes in 1) MPS response to feeding, 2) expression of regulators of myocellular quality (e.g. proteostasis, mitochondrial function, myogenesis and inflammation) and 3) circulating C-terminal agrin fragment (CAF), a marker of neuromuscular junction integrity and muscle wasting (Drey et al., 2013). We hypothesized that resistance and combined aerobic and resistance exercise would improve MPS more than aerobic exercise, thereby preserving lean mass despite energy deficit from WL. Based on the observed improvements in physical function, we further hypothesized that the combination of aerobic and resistance exercise would improve myocellular quality more than aerobic or resistance exercise alone.
RESULTS
Study overview and participants
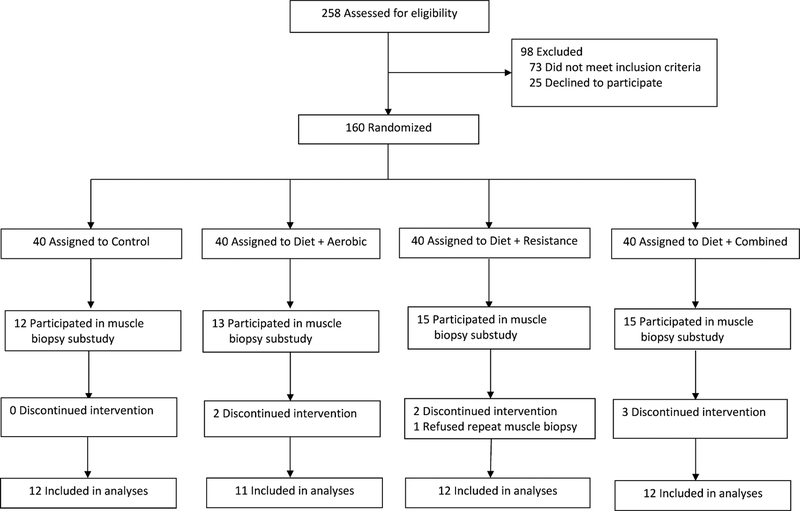
Forty-seven individuals from the randomized controlled trial (RCT) “Lifestyle Intervention Trial in Obese Elderly” (LITOE ClinicalTrials.gov number, ) (Villareal et al., 2017a) completed the muscle metabolism substudy for analyses of MPS rate and gene expression before and after the interventions and are included in this report (Figure 1). The study was conducted at the University of New Mexico School of Medicine and New Mexico Veterans Affairs Health Care System, after approval by the local institutional review board; participants were recruited using advertisements and underwent a comprehensive medical screening after providing written informed consent. Inclusion criteria were: older age (age ≥ 65 years), obesity (BMI ≥ 30 kg/m2), sedentary life-style (regular exercise <1 hour per week), stable body weight (loss or gain of no greater than 2 kg), stable medication use for 6 months before enrollment, and mild-to-moderate frailty (Brown et al., 2000; Villareal et al., 2004). Exclusion criteria included severe cardiopulmonary disease, musculoskeletal or neuromuscular impairments that precluded exercise training, or cognitive impairments. Participants were randomly assigned, with stratification according to sex, to one of four groups: 1) a control group that included neither a weight-management nor an exercise intervention (CON, n=12), 2) an aerobic group that participated in a weight-management program and aerobic exercise training (AEX, n=11), 3) a resistance group that participated in a weight-management program and resistance exercise training (REX, n=12), 4) and a combination group that participated in a weight management program and combined aerobic and resistance exercise training (COMB, n=12). We did not include a WL alone group (without exercise) for ethical reasons because WL would cause further muscle and bone loss in the elderly (Villareal et al., 2011a). The goal of the current study was not to distinguish the effect of calorie restriction versus different exercise modalities, but to compare different exercise regimens in combination with matched calorie restriction in obese elderly. Average age, average BMI, and number of males and females for the entire sample were: 70.5 ± 0.7 years, 35.9 ± 0.6 kg/m2 and 20 males and 27 females, respectively (Table 1). Baseline characteristics were not different among groups (Table 1). Intervention lasted 6 months. Details regarding participants’ characteristics, dietary and behavioral therapy, specific aerobic, resistance, and combined exercise training, as well as compliance to the diet therapy and exercise sessions and adverse events have been previously reported (Villareal et al., 2017a). For the subset who participated in the current study, exercise adherence was 97±1% (performed at 2.9±0 days per week) in AEX group, 96±2% (performed at 2.9±1 days per week) in REX group, and 93±2% (performed at 2.8±1 days per week) in COMB group.
Figure 1. Screening, Randomization, and Follow-up.
Study groups included a Control group that participated in neither a weight-management nor an exercise intervention and three exercise groups: a group that participated in aerobic exercise training (Aerobic group), a group that participated in resistance exercise training (Resistance group), and a group that received combined aerobic and resistance exercise training (Combined group); all three exercise groups also participated in a weight-management program.
Table 1:
Baseline Characteristics of the Subset who Participated in the Muscle Metabolism Study
| Control (n = 12) |
Aerobic (n = 11) |
Resistance (n = 12) |
Combined (n = 12) |
|
|---|---|---|---|---|
| Age, years | 70 ± 1 | 71 ± 1 | 72 ± 2 | 69 ± 1 |
| Sex, number (%) | ||||
| Male | 4 (33) | 4 (36) | 6 (50) | 6 (50) |
| Female | 8 (67) | 7 (64) | 6 (50) | 6 (50) |
| Race, number (%) | ||||
| White | 11 (92) | 10 (91) | 11 (92) | 10 (84) |
| Black | 0 (0) | 0 (0) | 0 (0) | 1 (8) |
| Other | 1 (8) | 1 (9) | 1 (8) | 1 (8) |
| Ethnicity, number (%) | ||||
| Hispanic or Latino | 7 (58) | 7 (64) | 8 (67) | 7 (58) |
| Not Hispanic or Latino | 5 (42) | 4 (36) | 3 (25) | 5 (42) |
| Unknown | 0 (0) | 0 (0) | 1 (8) | 0 (0) |
| Marital status, number (%) | ||||
| Single | 3 (25) | 1 (9) | 3 (25) | 3 (25) |
| Married | 2 (17) | 7 (64) | 6 (50) | 6 (50) |
| Divorced | 3 (25) | 2 (18) | 3 (25) | 2 (17) |
| Widowed | 4 (33) | 1 (9) | 0 (0) | 1 (8) |
| Education, number (%) | ||||
| Less than college degree | 5 (42) | 6 (55) | 4 (33) | 6 (50) |
| College degree | 5 (42) | 3 (27) | 6 (50) | 4 (33) |
| Graduate school | 2 (18) | 2 (18) | 2 (17) | 2 (16) |
| Body mass index (kg/m2) | 35.0 ± 1.0 | 35.5 ± 1.0 | 37.3 ± 1.7 | 36.1 ± 1.2 |
Values are means ± SE. There were no significant group differences in baseline characteristics (all p>0.05).
Intervention groups: Aerobic = weight management and aerobic training, Resistance = weight management and resistance training, Combined = weight management and combined aerobic and resistance training.
Physical function improved most in COMB group
The primary aim of the LITOE trial was to determine the effectiveness of several exercise modes in reversing frailty and preventing reduction of muscle and bone mass induced by weight loss (Villareal et al., 2017a). The primary results showed that WL plus combined aerobic and resistance exercise was most effective in improving functional status of obese older adults and was associated with relative preservation of lean mass (Villareal et al., 2017a). The effects of the interventions on these outcomes are reported here for the subset of individuals who participated in the present substudy (Table 2). There were no significant differences in body weight reduction among the three intervention groups, with all losing an average of ~9% of their body weight from baseline. CON group’s body weight did not significantly change during the intervention period (1% decrease). Physical Performance Test score increased more in COMB group than in AEX and REX groups (5.6±0.5 [20% increase], 3.8±0.6 [13% increase], and 3.8±5 [13% increase], respectively), while scores in all three exercise groups increased more than those in the CON group (0.6±0.5 [2% increase]). Peak oxygen consumption (VO2peak, measured as milliliters per kilogram of body weight per minute) increased more in COMB and AEX groups than in REX group (3.0±0.4 [16% increase], 2.9±0.4 [16% increase], and 1.3±0.4 [7% increase], respectively). Lean mass decreased less in COMB (−1.7±0.3 kg [3% decrease]) and REX (−1.1±0.3 kg [2% decrease]) groups than in AEX group (−2.8±0.3 kg [5% decrease]). Fat mass decreased by −6.5±0.9 kg (16% decrease) in AEX, −8.7±0.9 kg (21% decrease) in REX group and −7.5±0.8 kg (18% decrease) in COMB group. Changes in thigh muscle and thigh fat among the exercise groups were similar to those observed for lean mass and fat mass, respectively. Total one-repetition maximum (1-RM) strength increased in REX group (56±8 kg [16% increase]) and COMB group (54±8 kg [17% increase]), whereas it was maintained in AEX group (7±8 kg [3% increase]). Therefore, although weight loss was similar among the intervention groups, COMB and REX preserved muscle mass more than AEX while increasing muscle strength, whereas COMB experienced the most improvement in overall functional status (Villareal et al., 2017a).
Table 2.
Effect of Exercise Modes added to Diet-induced Weight Loss on Body Composition and Physical Function in the Subset who Participated in the Muscle Metabolism Study
| Outcome Variables | Control (n = 12) |
Aerobic (n = 11) |
Resistance (n = 12) |
Combined (n = 12) |
|---|---|---|---|---|
| Body weight (kg) | ||||
| Baseline | 97.0 ± 6.1 | 95.5 ± 3.2 | 105.3 ± 1.9 | 98.4 ± 2.1 |
| Change at 6 months | −0.4 ± 1.0 | −9.1 ± 1.0* | −9.7 ± 1.1* | −9.5 ± 0.9* |
| Physical Performance Test (score) | ||||
| Baseline | 28.0 ± 1.1 | 29.8 ± 0.4 | 29.7 ± 0.3 | 27.8 ± .0.7 |
| Change at 6 months | 0.6 ± 0.5 | 3.8 ± 0.6* | 3.8 ± 0.5* | 5.6 ± 0.5*†‡ |
| VO2peak (ml/kg/min) | ||||
| Baseline | 17.7 ± 1.0 | 18.6 ± 1.1 | 17.3 ± 1.4 | 17.8 ± 1.1 |
| Change at 6 months | −0.2 ± 0.4 | 3.0 ± 0.4* | 1.3 ± 0.4 | 2.9 ± 0.4*‡ |
| Functional Status Questionnaire Test (score) | ||||
| Baseline | 29.9 ± 0.7 | 29.8 ± 1.2 | 28.9 ± 0.7 | 28.1 ± 1.0 |
| Change at 6 months | 0.5 ± 0.5 | 2.2 ± 0.5 | 2.0 ± 0.5 | 3.8 ± 0.5*‡ |
| Lean mass (g/cm2) | ||||
| Baseline | 55.8 ± 4.5 | 55.5 ± 3.0 | 63.4 ± 2.5 | 57.2 ± 2.3 |
| Change at 6 months | 0.4 ± 0.2 | −2.8 ± 0.3* | −l.l ± 0.3*† | −1.7 ± 0.3*† |
| Fat mass (kg) | ||||
| Baseline | 41.2 ± 2.7 | 39.9 ± 1.9 | 41.8 ± 2.8 | 41.3 ± 1.6 |
| Change at 6 months | −0.7 ± 0.8 | −6.5 ± 0.9* | −8.7 ± 0.9* | −7.5 ± 0.8* |
| Thigh muscle (cm3) | ||||
| Baseline | 1236 ± 118 | 1346 ± 136 | 1279 ± 62 | 1170 ± 141 |
| Change at 6 months | 31 ± 15 | −80 ± 15* | −23 ± 14† | −41 ± 14* |
| Thigh fat (cm3) | ||||
| Baseline | 1665 ± 155 | 1695 ± 127 | 1687 ± 273 | 1725 ± 191 |
| Change at 6 months | −23 ± 69 | −268 ± 74* | −265 ± 62* | −263 ± 62* |
| Total 1-repetition muscle strength (kg) | ||||
| Baseline | 267 ± 35 | 269 ± 27 | 350 ± 36 | 322 ± 33 |
| Change at 6 months | −1 ± 8 | 7 ± 8 | 56 ± 8*† | 54 ± 8*† |
Values are means ± SE.
p<0.05 compared to Control group,
p<0.05 compared to Aerobic group (superscript for resistance and/or combined groups),
p<0.05 compared to Resistance group (superscript only for combined group).
Insulin sensitivity improved in AEX, REX, and COMB groups but not in CON group
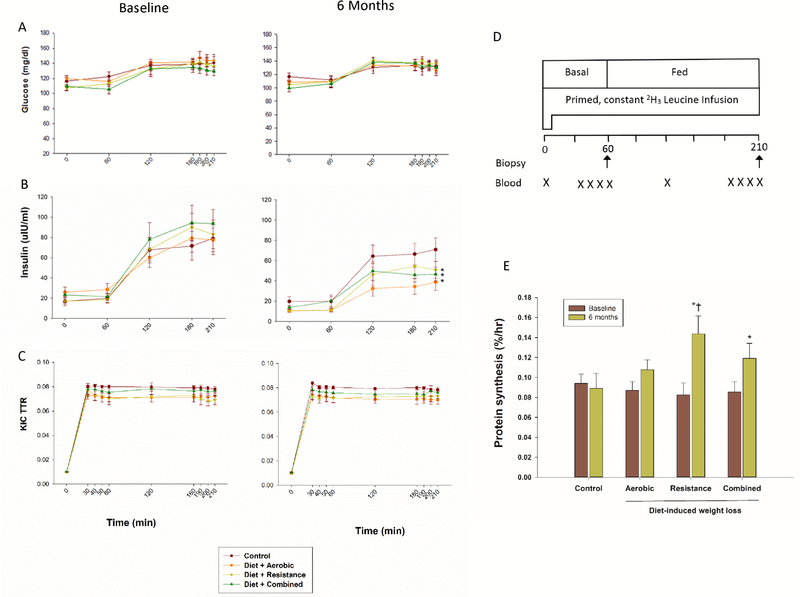
We measured plasma glucose concentrations, insulin concentrations, and α-ketoisocaproic acid (KIC) enrichments during basal (postabsorptive) state and in response to a mixed meal during a stable isotope infusion with [5,5,5-2H3] leucine (Figure 2). Glucose concentrations during the stable isotope infusion were not different before and after the interventions, neither during basal (postabsorptive) conditions nor during feeding, which increased glucose concentrations by ~25 mg/dl above basal levels in all groups (Figure 2A, Table S1). In contrast, insulin concentrations, which were greater in the fed state, decreased similarly in AEX, REX, and COMB groups, while they remained unchanged in CON group (Figure 2B, Tables S1 and S2) at 6 months. Accordingly, insulin sensitivity index, as assessed by the HOMA-IR, improved similarly in AEX (by −2.8 ± 0.6 [41% decrease]), REX (by −2.4 ± 0.9 [36% decrease]), and COMB (by −2.6 ± 0.6 [41% decrease]) groups, while it did not change in CON (0.4 ± 0.7 [7% increase]) group (Table S2). Plasma leucine concentrations were not different before and after the interventions, neither during basal (postabsorptive) conditions, nor during feeding, which increased leucine concentration by ~20 μM above basal values in all groups (Table S1).
Figure 2. Muscle Protein Metabolism Studies.
Glucose concentrations (Panel A), insulin concentrations (Panel B), and α-ketoisocaproic acid (KIC) enrichments (Panel C) during the stable isotope infusion in basal (post-absorptive) conditions and mixed meal feeding before and at 6 months of diet-induced weight loss plus different types of exercise interventions in obese older adults. Outline of the protocol (Panel D). Muscle protein synthesis rate during mixed meal feeding before and at 6 months of interventions (Panel E). Muscle protein synthesis rate was calculated using muscle tissue fluid as precursor pool for protein synthesis. Between-group differences in changes from baseline: * p <0.05 for comparison with changes in Control group, † p<0.05 for comparison with changes in Aerobic group. I bars indicate standard errors, n = 11–12/group. Please see also Tables S1 and S2, and Figure S1.
Although insulin stimulates net MPS and anabolism in young adults, several studies have shown that aging is associated with impaired ability of insulin to stimulate MPS in response to a mixed meal, typical of many meals ingested by older adults throughout the day (Guillet et al., 2004; Rasmussen et al., 2006; Volpi et al., 2000). Accordingly, since it is well known that meal intake has an anabolic effect on MPS, a chronic reduction in energy and protein intake related to WL might be of particular concern in older adults who already have reduced anabolic response to nutrient stimuli (i.e. mixed meal). In fact, this problem could be exacerbated in frail, obese older adults due to a further decrease in anabolic stimuli from a lack of regular exercise (i.e. exercise deficiency) (Aguirre and Villareal, 2015; Evans, 2002). The present findings, based on the first head-head comparison of different exercise types added to WL, have important implications for WL therapy of obese older adults. They demonstrate that lifestyle intervention by means of AEX, REX, or COMB are equally effective in improving insulin sensitivity and insulin response to a mixed meal, supporting the importance of combining WL with regular exercise to help preserve muscle mass during negative energy balance (Villareal et al., 2017a; Villareal et al., 2011a).
MPS rate in response to feeding improved most in COMB and REX groups
Since muscle loss during aging is related to a lower sensitivity of MPS to feeding (Guillet et al., 2004; Rasmussen et al., 2006; Volpi et al., 2000), which might be exacerbated by chronic energy deficit but countered by exercise training, we examined the differential effects of AEX, REX, or COMB on MPS rate in response to a mixed meal before and after WL (Figure 2). We measured MPS ~24 h after the last bout of exercise to minimize acute effects of exercise. We also provided a small meal, comparable to a typical breakfast of Americans, containing an amount of protein (~15 g) that would substantially stimulate the rate of MPS while avoiding a possible ceiling effect. In all groups, plasma α-KIC and leucine enrichments were steady between 30 and 60 min during the basal state (prior to the first muscle biopsy) and between 180 min and 210 min during the fed state (prior to the second muscle biopsy) before and after the interventions (Figure 2C, Table S1). Accordingly, muscle-free leucine enrichments were also not different between these time points during stable isotope infusions over the study period, consistent with steady state precursor enrichment (Table S1). At the beginning of the study, MPS rate during the fed state was not different among groups (Table S1). However, after 6 months of interventions, MPS in response to mixed meal (calculated using muscle tissue fluid enrichment as precursor pool) increased more in REX (0.084 ± 0.009%/hr to 0.154 ± 0.015%/hr [114% increase]) and COMB groups (0.093 ± 0.006%/hr to 0.131 ± 0.013%/hr [45% increase]) than in CON group (0.094 ± 0.010%/hr to 0.089 ± 0.016%/hr [7% decrease]); the increase in MPS experienced by REX group was significantly higher than that experienced by AEX group (0.087 ± 0.009%/hr to 0.108 ± 0.010 %/hr [28% increase]) (Figure 2E). Similar results were observed when MPS rate was calculated using integrated plasma leucine enrichment as precursor pool (Figure S1). These findings demonstrate that, despite negative energy balance from calorierestriction induced WL, obese older adults can respond and adapt to exercise training by increasing MPS rate, thus helping to minimize loss of lean mass induced by WL. These data also indicate that among the exercise types to combine with WL, exercises that include a resistance component are particularly important to stimulate MPS and thereby preserve lean mass, supporting our prior report that muscle mass decreased less in COMB and REX groups than in AEX group (Villareal et al., 2017a).
Previous studies of the effects of exercise on MPS were performed after acute or relatively short-term training (≤ 12 weeks), mostly conducted in the basal (post-absorptive) state. An isolated bout of resistance exercise increased MPS by ~100% within 24 h which declined towards baseline by ~72 h in both young and old adults (Balagopal et al., 2001; Drummond et al., 2008; Phillips et al., 1997; Welle et al., 1999; Yarasheski et al., 1993). Eight weeks of resistance training increased the initial MPS response to feeding by ~160% while reducing the duration of the MPS response to ~30 h (Tang et al., 2008). In the present study, the 114% increase in MPS after 24 h in the REX group could represent an additive effect of resistance training and WL because we previously reported that WL alone increased MPS, indicating no adverse effect of WL on MPS in obese older adults (Villareal et al., 2012). On the other hand, aerobic exercise increased MPS rate to a lesser degree (<50%) both acutely and after training in young and old adults (Carraro et al., 1990; Pikosky et al., 2006; Sheffield-Moore et al., 2004; Short et al., 2004), and thus, the ~28% increase in MPS in AEX group in the present study is consistent with these reports. We did not observe a greater increase in MPS in COMB group than in REX group probably because concurrent aerobic training interferes with the resistance training-induced adaptation of muscle protein metabolism (Cadore et al., 2010; Hickson, 1980; Wilson et al., 2012). Nevertheless, because both COMB and REX increased MPS more than CON, and were associated with relative preservation of lean mass, both exercise types can be recommended when frail, obese older adults undertake WL therapy to improve physical function (Villareal et al., 2005). On the other hand, aerobic exercise is known to increase mitochondrial MPS (Robinson et al., 2017), while resistance exercise increases contractile MPS (Wilkinson et al., 2008). We measured ‘mixed’ muscle protein synthesis rate which did not allow us to specifically distinguish which muscle protein compartment we altered. It is thus possible that our AEX group improved mitochondrial MPS, an increase that contributed to, but cannot be distinguished from, the measurement of mixed muscle protein synthesis rate. Moreover, although it is possible that the improved MPS responses to feeding observed in our study could be, in part due to the WL and exercise-induced improvements in insulin sensitivity, these effects alone would not be sufficient to account for the differential MPS responses (i.e. REX > AEX), given that insulin sensitivity improved equally (by ~40%) in the three intervention groups relative to CON (Table S2). Moreover, besides the positive effects of exercise on MPS, it is likely that our exercise interventions helped preserve muscle mass and function through effects on myocellular quality, as described below.
Myogenesis regulator expression was preserved in COMB and REX but reduced in the AEX group
Alteration of muscle growth and regeneration as well as MPS may contribute to the etiology of age-related sarcopenia. Compared to other myocyte enhancer factors that mediate muscle growth, MEF2A is absolutely required for myoblast differentiation and regeneration (Estrella et al., 2015). MEF2A expression decreased more in the AEX group than in COMB and REX groups, in which its expression was preserved (Table 3, Section 1.1; Figures 3A and 3B). Interestingly, in the AEX group, which had the greatest reduction in muscle mass, changes in MEF2A expression correlated positively with changes in muscle volume (r=0.91, p=0.03). It is thus possible that the worse preservation of muscle mass experienced by the AEX group could be due to reduction in MEF2A expression. MEF2A knockdown in C2C12 myotube impairs cell differentiation, whereas MEF2 -B, -C, or -D deficiency does not block myotube formation (Estrella et al., 2015). Myotubes’ MEF2A depletion leads to a unique transcriptional profile, affecting myogenesis, sarcolemmal repair mechanisms and actin-signaling required for myotubes fusion and differentiation. In contrast, since MEF2B, -C, and -D regulated genes extensively overlap, the lack of one these isoforms is compensated by the presence of the other one (Estrella et al., 2015). Altogether these evidences support the unique role played by MEF2A in myoblast differentiation and the implication of the reduced expression observed in our AEX group. We did not observe significant differences in changes in MEF2A protein levels (Figure 3C), likely because of the small sample size available for this analysis. Changes in the expression of most contractile proteins and regulators of anabolism did not differ among the intervention groups, except for MYLK (myosin light chain kinase) which increased more in AEX compared to CON and REX, and STAT5B (GH pathway, activated after exercise) which decreased more in AEX compared to CON and REX (Table 3, Section 1.2).
Table 3.
Differences in Changes in Gene expression Between Groups
| Symbol | Function | Control | Aerobic | Resistance | Combined | P |
|---|---|---|---|---|---|---|
| 1. PROTEOSTASIS, APOPTOSIS & MYOGENESIS | ||||||
| 1.1 MYOGENESIS & MUSCLE REGENERATION | ||||||
| MEF2A | Myocyte enhancer factor, transcription factor involved in the regulation of muscle regeneration. It activates muscle specific growth factors. | 0.98 | 0.83* | 1.00† | 0.93† | <0.01 |
| 1.2 REGULATORS OF ANABOLISM & STRUCTURAL PROTEINS | ||||||
| STAT5B | Involved in the induction of IGF 1 expression. | 1.01 | 0.76* | 1.07† | 0.82‡ | 0.01 |
| MYLK | This gene encodes myosin light chain kinase which phosphorylates myosin regulatory light chains to facilitate myosin interaction with actin filaments to produce contractile activity. | 0.88 | 1.49* | 0.83† | 1.08‡ | <0.01 |
| 1.3 CASPASE: APOPTOSIS & DIFFERENTIATION | ||||||
| IAPS | This gene encodes for an apoptosis inhibitor, it ubiquitinates caspase reducing the time of activation and promoting myocellular differentiation vs apoptosis. | 1.01 | 0.87* | 1.08† | 0.95‡ | <0.01 |
| CASPASE7 | Regulator of apoptosis and myogenic differentiation: sustained activation leads to the first, whereas transient activation leads to the last. | 0.93 | 0.89 | 1.07*† | 0.93‡ | 0.01 |
| CASPASE3 | Regulator of apoptosis and myogenic differentiation: sustained activation leads to the first, whereas transient activation leads to the last. | 0.91 | 0.96 | 0.93 | 1.18*†‡ | <0.01 |
| XRCC1 | The protein encoded by this gene is involved in the efficient repair of DNA single-strand breaks. | 1.06 | 0.98 | 0.85* | 0.94*† | 0.03 |
| 1.4 AUTOPHAGY & HEAT SHOCK PROTEINS | ||||||
|
FIP200 (ATG17) |
The protein encoded by this gene interacts with other autophagic regulators promoting the formation of the autophagosome. | 0.98 | 0.83 | 0.99† | 0.82*‡ | 0.02 |
| ATG101 | Protects ATG13 from proteosomal degradation, therefore stabilizing levels of ATG13 found in cells and regulating levels of macroautophagy. | 1.03 | 0.95 | 1.00 | 0.79*†‡ | 0.02 |
| LAMP2 | Responsible for the fusion of autophagosome with lysosome, thus the formation of the autophagolysosome. | 0.95 | 0.88 | 0.91 | 0.71*†‡ | 0.01 |
| HSP704_2 | Cytosolic constitutive heat shock protein. | 1.08 | 0.67* | 0.96† | 0.68*‡ | 0.01 |
| 2. MITOCHONDRIA | ||||||
| 2.1 MITOCHONDRIA BIOGENESIS | ||||||
| TFAM | Mitochondrial biogenesis master regulator. | 1.00 | 1.05 | 0.92† | 0.87*† | <0.01 |
| 2.2 MITOCHONDRIA FISSION & FUSION | ||||||
| FIS1 | Mitochondrial fission mediator. | 1.00 | 0.99 | 1.03 | 0.86*†‡ | 0.001 |
| DRP | Mitochondrial fission mediator. | 0.98 | 1.17* | 0.96† | 1.02†‡ | 0.02 |
| MFF | Mitochondrial fission mediator. | 0.95 | 1.14* | 0.98† | 0.82†‡ | <0.001 |
| OPA1 | Controls fusion, cristae remodeling and assembly of the respiratory chain. It affects mitochondrial morphology and function. | 0.91 | 1.15* | 1.01 | 0.88† | 0.02 |
| 2.3 MITOCHONDRIA PROTEOSTASIS | ||||||
|
AFG3L2 mAAA |
Degrade misfolded proteins, first line of defense against mitochondrial damage with the capability to control mitochondrial fate. | 1.02 | 1.03 | 1.01 | 0.68*†‡ | O.001 |
| LONP1 | This gene encodes a mitochondrial matrix protein that mediates the selective degradation of misfolded, unassembled or oxidatively damaged polypeptides in the mitochondrial matrix. | 1.01 | 0.93 | 1.02 | 0.85*‡ | <0.01 |
| LONP2 | The protein encoded by this gene plays a role in maintaining overall peroxisome homeostasis as well as in proteolytically degrading peroxisomal proteins damaged by oxidation. | 1.00 | 0.99 | 1.00 | 0.88*†‡ | 0.01 |
| PARL | This gene encodes a mitochondrial matrix protein involved in maintenance of proteostasis and mitochondrial function. | 1.01 | 0.95 | 1.02 | 0.87*‡ | 0.04 |
| HSP704_9 | Mitochondrial constitutive heat shock protein. | 0.98 | 1.23* | 1.11 | 0.99† | 0.02 |
| 2.4 OXPHOS & FATTY ACIDS UTILIZATION | ||||||
| FABP5 | This gene encodes for fatty acid binding protein which binds fatty acids and increases the efficiency of their oxidation. | 0.84 | 1.42* | 0.97† | 1.07† | 0.02 |
| SUCLA2 | Succinyl-CoA synthetase is a mitochondrial matrix enzyme involved in the oxidative phosphorylation. | 0.93 | 1.25* | 1.03† | 0.90† | <0.01 |
| CKMT2 | Mitochondrial creatine kinase is responsible for the transfer of high energy phosphate from mitochondria to the cytosolic carrier, creatine | 0.90 | 1.91* | 1.12† | 0.99† | <0.01 |
| COX5B | Cytochrome c oxidase subunit Vb, oxidative protein | 0.95 | 1.34* | 1.05† | 0.96† | <0.01 |
| C0X7C | This gene encodes for cytochrome c oxidase subunit 7C | 0.92 | 1.18* | 1.04 | 0.89†‡ | <0.01 |
| 3. INFLAMMATION | ||||||
| TGFβ1 | Immunosuppressor, produced by T-reg | 1.03 | 1.11 | 1.06 | 1.42*†‡ | 0.04 |
| TLR2 | Transmembrane proteins involved in the detection of pathogens and of endogenous danger signals released following tissue damage, such as HSP | 0.80 | 1.25* | 0.87† | 0.78† | O.001 |
| CD68 | Marker of macrophage infiltration | 0.76 | 1.31* | 1.14* | 0.94† | 0.02 |
| CCL2 | Chemoattractant to the tissue for activation of inflammatory response | 0.85 | 1.05 | 1.19* | 0.82†‡ | <0.01 |
Fold changes (FC) are calculated as the ratio between gene expression at the end of the study and gene expression at baseline. Values >1 indicate a positive regulation, while values <1 reflect a downregulation. P value column indicates overall between group differences P<0.05.
p<0.05 compared to Control group,
p<0.05 compared to Aerobic group (superscript for resistance and/or combined groups),
p<0.05 compared to Resistance group (superscript only for combined group).
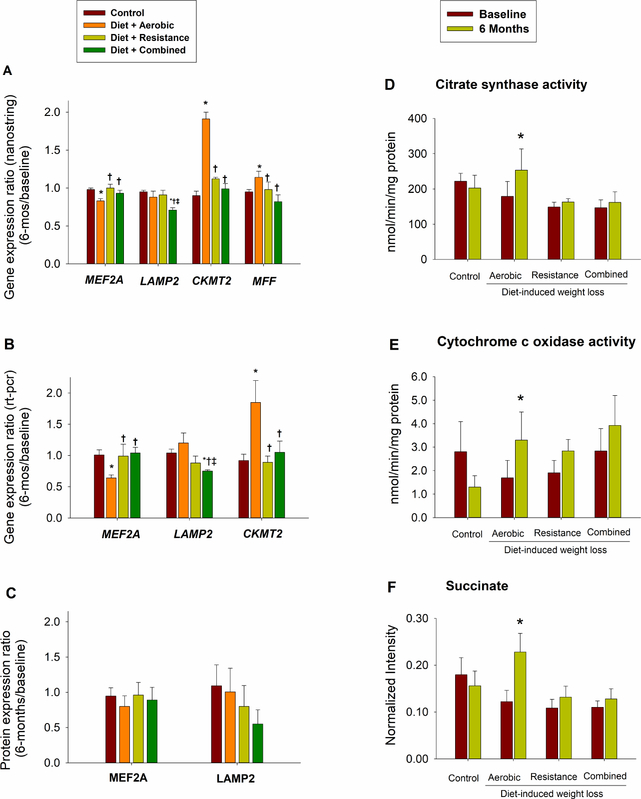
Figure 3. Gene Expression, Enzyme Activities, and Metabolites Studies.
Gene expression analyses by nanostring (Panel A) and RT-qPCR (Panel B) and protein expression analyses by western blot (Panel C) of representative targets. Fold changes are calculated as the ratio between gene expression or protein levels at 6 months and at baseline. Citrate synthase (Panel D) and Cytochrome c oxidase (Panel E) activities expressed in nmol/min/mg of proteins at baseline and 6 months. Succinate metabolite (Panel F) expressed as normalized intensity at baseline and 6 months. Between-group comparisons in changes from baseline: * p<0.05 for comparison with Control group, † p<0.05 for comparison with Aerobic group, ‡ p<0.05 for comparison with Resistance group. I bars indicate standard errors. n = 11–12/group (Panels A and B); n= 4–7/group (Panel C); n=6/group (Panels D and E) and n = 8/group (Panel F). Please see also Tables S3 and S4.
Higher reduction in expression of inhibitors of apoptosis in AEX group
Besides initiating apoptosis, caspase are proteolytic enzymes able to promote muscle growth and regeneration in conditions of transient activation (Bell et al., 2016). For this purpose, inhibitors of apoptosis, IAPs, are crucial (Bell et al., 2016). Among the intervention groups, REX had the highest increase in the expression of the member of the IAPs family, XIAP, whereas AEX experienced the highest reduction (Table 3, Section 1.3). On the other hand, XIAP targets expression, CASP7 and CASP3, increased more in REX and COMB, respectively (Table 3, section 1.3). Changes in XIAP expression were positively correlated with changes in CASP3 in both REX and COMB groups (r=0.75, p=0.004 and r=0.89, r<0.0001, respectively). Furthermore, compared to CON, REX and COMB groups reduced XRCC1 expression (Table 3, section 1.3), a regulator of DNA repair mechanisms that follows caspase activated DNase (CAD) activity whose expression did not change. Satellite cell commitment into cell lineage is pivotal for muscle growth and regeneration, and caspases 3/7 play a crucial role, allowing the acquisition of differentiation competences and activating several promyogenic kinases (Bell et al., 2016). Transient caspases activation promotes differentiation as opposed to apoptosis, and the short duration of their activity is ensured by IAPs (Bell et al., 2016). The opposite direction of the changes in XIAPs expression between the AEX and REX groups could be linked to the different capability of both groups to preserve muscle mass.
Higher Reduction in Atrogenes Expression in COMB group
Autophagy and ubiquitin-proteasome system (UPS) are protein quality control mechanisms activated in response to stress and their excessive up or downregulation lead to muscle wasting and cellular malfunction, respectively (Cohen et al., 2015). Therefore, mediators of both pathways are also known as “atrogenes” (atrophy-related genes) (Cohen et al., 2015). Stress induced by diet and exercise promotes acute atrogenes activation, responsible for myocellular quality improvement as a result of the degradation of old damaged proteins (Cartee et al., 2016). In this context, chronic consequences of lifestyle intervention remain poorly understood. Sustained atrogene activation reflects persistent cellular stress, and could be a health threat for muscle loss, especially in the context of older-frail (Cartee et al., 2016; Cohen et al., 2015). Compared to normal weight individuals, overweight individuals had higher expression of autophagy-related genes because of higher inflammation and oxidative stress (Potes et al., 2017). In addition, young and life-long trained senior subjects had similarly low expression of autophagy-related genes as compared to sedentary old individuals (Zampieri et al., 2015). In our study conducted in obese, frail elderly, COMB group experienced a chronic higher reduction in the expression of autophagy related genes (ATG101, LAMP-2 and FIP200 - also known as ATG17) as compared to all other groups and, together with AEX, had a significant higher downregulation of HSPA704_2 (stress-induced cytosolic chaperone, ensuring correct protein folding), as compared to REX and CON (Table 3, section 1.4). We did not observe significant differences in changes in LAMP2 protein levels (Figure 3, panel C), possibly because of the lower sample size available for this analysis. On the other hand, changes in the expression of UPS-related genes (e.g. MURF1, MAFbx also known as atrogen-1) did not differ significantly between groups (data not shown). These results suggest a greater chronic reduction in cellular stress experienced by COMB group. To assess cellular stress status, we studied the expression of inflammatory and mitochondrial stress markers, considered upstream autophagic activators triggering muscle wasting (Cohen et al., 2015).
Mitochondrial stress markers were reduced in COMB group but increased in AEX group
Mitochondrial fission segregates dysfunctional components of the mitochondrial network allowing their removal by mitophagy (Romanello and Sandri, 2015). Age-related muscle atrophy results in mitochondrial fission which in turn promotes the activation of proteolytic pathways (Cartee et al., 2016). Consistently, fission inhibition prevents muscle loss in atrophying conditions (Romanello and Sandri, 2015). In COMB group, expression of regulators of mitochondrial fission, mitophagy and proteostasis (FIS1, LONP1, LONP2, AFG3L2, PARL) was reduced more compared to all other groups (Table 3, sections 2.2 and 2.3), suggesting a lower presence of dysfunctional mitochondrial structures. On the other hand, AEX group increased more the expression of mito-fusion/fission and protein chaperones (OPA1/DRP-MFF and HSP704_9) likely as a result of the observed higher increase in mitochondrial bioenergetic regulators (SUCLA2, COX5B, COX7C, CKMT2, FABP5; Table 3, sections 2.3 and 2.4), and mitochondrial activity (citrate synthase and cytochrome c oxidase activities, Figures 3D and 3E). Interestingly, according to our results, AEX overcame citrate activity reduction induced by calorie restriction (Gouspillou and Hepple, 2013). In addition, although we did not detect any differences in the expression of the mitochondrial master regulator, PGC1α, AEX group increased TFAM expression (mitochondrial biogenesis) more than REX and COMB, the latter of which experienced a large reduction in expression (Table 3, section 2.1). AEX group experienced a higher increase in succinate, intermediate in the tricarboxylic acid cycle (TCA), cell’s metabolic hub (Figure 3F, Table S4). As the product of succinate dehydrogenase activity which couples TCA with electron transport chain and OXPHOS, succinate concentrations were positively correlated with changes in VO2peak and metabolic fitness in overweight and obese individuals undergoing a 6-month exercise intervention (Huffman et al., 2014). Consistent with our results, succinate was the only TCA metabolite that increased significantly in obese performing high amount/vigorous intensity aerobic exercise compared to controls, and did not change in the individuals performing resistance exercise training (Huffman et al., 2014). Importantly, succinate increases in conditions of ketone body utilization or branched chain amino acid catabolism (Tretter et al., 2016). It is thus possible that our AEX group experienced a comparatively higher muscle catabolism, hypothesis also supported by the higher lean mass loss in this group (Villareal et al., 2017a). Elderly obese require a degree of autophagy that aging does not support (Potes et al., 2017) and show a blunted adaptation to exercise training compared to young adults (Cartee et al., 2016). Although endurance exercise was reported to prevent the decline in mitochondrial respiratory capacity experienced by elderly (Cartee et al., 2016), it is possible that a higher activation of the oxidative network in the context of calorie restriction, aging, and sarcopenia could lead to higher muscle loss (as that experienced by our AEX group) due to greater catabolic stimulation.
Higher reduction in expression of inflammatory macrophage markers in COMB group
Obesity-related chronic inflammation, responsible for muscle atrophy, is reduced after diet and exercise interventions (Gleeson et al., 2011). Whether one exercise modality is more effective than the other is not known. In our population, we did not detect any difference in changes in the expression of cytokines or downstream effectors (e.g. IL6, TNFα, NFkβ, data not shown). However, mechanisms proposed for the anti-inflammatory effect of exercise include reduced expression of toll like receptors (TLRs) on monocytes and macrophages (Gleeson et al., 2011). Loss of 4% of leg lean mass was accompanied by an increase in vastus lateralis TLR4 expression in older adults undergoing bed rest (Drummond et al., 2013). Skeletal muscle of obese, diabetic individuals expressed higher pro-inflammatory (M1) macrophage infiltration markers and lower TGFβ1 compared to non-obese (Fink et al., 2013). Our obese frail individuals assigned to the COMB group not only reduced more the expression of markers of macrophage infiltration (TLR2, CD68, and CCL2) compared to those assigned to the AEX and REX groups (for which the same markers changed in the opposite direction), but also increased more the expression of the immunosuppressor TGFβ1 (Table 3, section 3). These data suggest a possible increase in M2 (anti-inflammatory), as opposed to M1 macrophages, in the COMB group which may in part explain the best phenotype of this group.
C-Terminal Agrin Fragment changes did not differ between groups
Agrin is a proteoglycan synthesized by motoneurons which promotes neuromuscular junction maintenance. An increase in circulating levels of the product of its degradation, CAF, was observed in sarcopenic individuals, indicating that it could be a valid marker of muscle wasting (Drey et al., 2013). According to data collected on 333 elderly participating in the LIFE-P trial, circulating CAF changes did not differ between elderly exercising and “healthy aging” controls (Bondoc et al., 2015). Our objective was to evaluate the effect of different exercise protocols on CAF changes in the context of weight loss. Similar to the findings in the LIFE-P trial, we did not detect differences in changes in serum CAF in our population: CON (104 ± 68 to 95 ± 62 pM [change of −3 ± 54%]); AEX (142 ± 105 to 134 ± 119 pM [change of −0.1 ± 69%]); REX (139 ± 141 to 131 ± 139 pM [change of 8 ± 32%]); COMB (143 ± 97 to 129 ± 89 pM [change of −11 ± 32%]; p=0.64 for between-group comparisons). This finding suggests that improvements in physical function observed in response to our interventions may not be primarily due to changes in neuromuscular junction efficiency.
SUMMARY
This is the first RCT investigating the chronic effect of different types of exercise training accompanied by ~10% diet-induced weight loss on the response of MPS rate to feeding and changes in the expression of regulators of myocellular quality in frail, obese older adults. Main strength of the study: unique but prevalent population, study design and successful performance of the protocol. The findings revealed different possible mechanisms responsible for the better preservation of muscle mass experienced by COMB and REX groups compared to AEX group after 6 months of lifestyle intervention. Among the outcomes investigated, it is likely that the least improvement in MPS response to feeding is primarily responsible for the observed phenotype in the AEX group, as opposed to COMB and REX which experienced both, greater increases in MPS and muscle mass preservation. Additionally, the highest reduction in expression of muscle growth regulators and the highest increase in mitochondrial and inflammatory markers of stress could contribute to the worse preservation of muscle mass in the AEX group, possibly exacerbating the catabolic state associated with calorie restriction. Moreover, the best preservation of muscle growth regulators and reduction of atrogenes expression could be important elements promoting muscle mass preservation and improvement in physical function in COMB group, which underwent what we demonstrated to be the most effective intervention for dieting obese frail elderly. Our data strongly support that resistance plus aerobic exercise training is needed to preserve lean mass by improving MPS response to anabolic stimuli and preserve myocellular quality in dieting elderly obese, and that therefore, it should be prescribed as part of a lifestyle intervention in this population. Additional studies further exploring changes in the expression of mediators of the investigated pathways are needed to expand our observation.
Limitations of Study
Small sample size and relatively short duration of the trial are among our study limitations. It was not possible to distinguish between the effect of calorie restriction and different exercise modalities because of the lack of a “weight loss alone” group. Furthermore, we could not identify the specific muscle protein compartment affected by our intervention because of the use of mixed muscle protein to assess muscle protein synthesis rate. Lastly, given the limitation of human intervention studies, we could not perform all experiments on the full set of samples because of the small number of muscle specimens available for each analysis.
STAR METHODS
Contact for Reagents and Resources Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dennis T. Villareal (dennis.villareal@bcm.edu).
Experimental Model and Subjects Details
Below is a brief description of study population, intervention, and protocol which has been published elsewhere (Villareal et al., 2017). Forty-seven obese (BMI ≥ 30 kg/m2) older (age ≥ 65 yrs) adults were enrolled in the LITOE Muscle Metabolism Substudy after signing a written informed consent. The study was approved by the institutional review board of the University of New Mexico School of Medicine and monitored by an independent data and safety monitoring board.
Human Subjects and Study Design
Sample size was based on our preliminary studies and published data (Parise et al., 2001; Villareal et al., 2012; Villareal et al., 2011b; Yarasheski et al., 1993) showing that 12 subjects per group were needed to provide 80% power to detect a clinically important difference between groups of 20% in the change in MPS, at an alpha level of 0.05. Total duration of the interventions was 26 weeks. CON attended group educational classes about a healthful diet during monthly visits and was asked not to participate in external weight-loss or exercise programs. All intervention groups participated in a weight management program and met weekly with an experienced dietitian for dietary adjustment and behavioral therapy to achieve the goal of approximately 10% weight loss. Participants were prescribed a balanced diet that provided an energy deficit of 500 to 750 kcal per day and contained ~ 1 g of high-quality protein per kg body weight per day (Villareal et al., 2005). In addition to the WL, the intervention groups performed exercise sessions three times weekly: AEX group 60 minutes long aerobic training, REX 60 minutes long resistance training and COMB group performed both aerobic and resistance 75 to 90 minutes long exercise training (sessions of longer duration to test the interference effect) (Cadore et al., 2010; Hickson, 1980; Villareal et al., 2017a; Villareal et al., 2017b; Wilson et al., 2012). The aerobic training consisted of treadmill walking, stationary cycling, and stair climbing, in which participants exercised at approximately 65% of their peak heart rate progressively increased to 70 to 85% (Villareal et al., 2017a). The resistance training consisted of nine upper-body and lower-body exercising using weight-lifting machines, in which the initial sessions were 1 to 2 sets of 8 to 12 repetitions at 65% of the one-repetition maximum (1-RM), increased progressively to 2 to 3 sets at approximately 85% of the 1-RM. Exercise sessions were supervised by exercise trainers (Villareal et al., 2017a). All participants received supplements to ensure an intake of approximately 1500 mg of calcium per day and approximately 1000 IU of vitamin D per day (Villareal et al., 2005).
Methods Details
All assessments were done at baseline and 6 months. Assessors were unaware of the study group assignments.
Physical Function
Frailty was assessed objectively with the Physical Performance Test, which is a performance-based global measure of physical performance, (Brown et al., 2000) and with the peak oxygen consumption (VO2peak) during graded treadmill walking, as described previously (Villareal et al., 2004). Frailty was also assessed subjectively with the Functional Status Questionnaire that assesses ability to perform activities of daily living (Jette and Cleary, 1987). Muscle strength was assessed using total 1-RM (the maximum weight lifted in one attempt, in the biceps curl, bench press, seated row, knee extension, knee flexion and one leg press).
Body Composition
Lean mass and fat mass of the whole body were measured using dual energy x-ray absorptiometry (Lunar DPX [General Electric] or Discovery A [Hologic] scanner), as described previously (Villareal et al., 2004; Villareal et al., 2006). We also measured thigh muscle and fat volumes by magnetic resonance imaging (Magnetom Avanto [Siemens]), as described previously (Weiss et al., 2007).
Insulin Sensitivity
The Homeostatic model assessment of insulin resistance (HOMA-IR) was calculated as fasting glucose (mg/dL) multiplied by fasting insulin (μIU/mL) divided by 22.5 (Matthews et al., 1985). The areas under the curve (AUC) for glucose and insulin in response to mixed meal during the stable isotope infusions were calculated using the trapezoid method (Allison et al., 1995).
Muscle Protein Metabolism Study
Tracer infusions protocol.
Participants were instructed to adhere to their regular diet and to refrain from vigorous exercise (before intervention only) for 3 days before the study. They were admitted to the Clinical Research Unit of the University of New Mexico Hospital the evening before the procedure and consumed a standardized meal containing 12 kcal/kg adjusted body weight which was calculated as ideal body weight (the midpoint of the medium frame of the Metropolitan Life Insurance Company Table172) + 0.25 × (actual body weight - ideal body weight). Meal was consumed at 2000 and contained 55% of total energy carbohydrates, 15% protein and 30% fat. Participants then rested in bed and fasted until the completion of the study the next day. Stable labeled isotope tracer technique was performed in fed conditions to assess MPS. The isotope infusion protocol is outlined in Figure 2 Panel D. At 0600 h, on the day after admission, one catheter was inserted into an antecubital vein for the infusion of a stable isotope labeled leucine tracer (purchased from Isotec Inc, Miamisburg, OH); another one was inserted into a hand vein for blood sampling; the hand was heated to 55°C by using a thermostatically controlled box 15 min before collection of each blood sample to obtain arterialized blood samples (Jensen and Heiling, 1991). At 0800 h, after baseline blood samples were obtained, a constant infusion of [5,5,5-2H3] leucine (priming dose: 7.2 μmol/kg lean mass, infusion rate: 0.12 μmol/kg/lean mass/min) was started. Sixty minutes after the start of the tracer infusion, a liquid meal (Ensure, Abbott Laboratories, Abbott Park, IL, containing 15% of energy as protein, 55% as carbohydrate and 30% as fat) was given as small boluses every 10 minutes for 150 min to achieve steady state plasma amino acid concentrations and leucine enrichment (Table S1). The dose of the meal was equivalent to 70 mg protein/kg/lean mass/h (priming dose: 23 mg/kg FFM), which results in ~50% higher plasma AA concentration compared to basal values and ~ half maximal stimulation of MPS according to previous publications (Bohe et al., 2003). Muscle biopsies from the vastus lateralis were obtained at 60 min (on one leg) and at 210 min (on the contralateral leg) after the start of the tracer infusion to determine the MPS rate in response to increased amino acid availability (Bohe et al., 2001) Muscle tissue was obtained under local anesthesia using a Tilley-Henkel forceps. Lidocaine (2%) was injected at the site of the biopsy, a small incision (~1 cm) was made to ease passage of the forceps (5 mm) into the vastus lateralis, and ~150 mg of tissue collected. After collection, muscle specimens were cleaned of blood and fat and immediately snap-frozen in liquid nitrogen for biochemical analysis. Separate pieces of tissue from the 60-min time point were collected and frozen in liquid nitrogen for gene expression (immersed in RNAlater -Ambion, Austin, TX-) and protein quantification analysis. Blood samples were obtained at several time points during the isotope infusion to measure leucine, glucose, and insulin concentrations and the leucine and α-ketoisocaproic acid (KIC) enrichments in plasma. Samples were collected in pre-chilled tubes containing EDTA to determine the isotopic enrichment and substrate concentration.
Sample processing and analyses.
Plasma glucose concentration was determined on an automated glucose analyzer (Yellow Spring Instruments, Yellow Springs, OH). Plasma insulin concentration was determined by radioimmunoassay (Linco Research, St Louis, MO). Plasma leucine concentration, plasma α-KIC tracer-to-tracee ratios (TTR), and the leucine TTR in muscle proteins and muscle tissue fluid were determined using gas-chromatography/mass-spectrometry (Agilent 6890N Gas Chromatograph and Agilent 5973N Mass Selective Detector (GC-MS); Agilent, Palo Alto, CA) and standards of known isotope enrichment as previously described (Patterson et al., 1997; Reeds et al., 2006; Smith et al., 2007; Villareal et al., 2011b). Briefly, a known amount of norleucine was added to the plasma, proteins were precipitated, and the supernatant, containing free amino and keto acids, was collected to prepare the N-heptafluorobutyryl n-propyl ester (HFBPr) and O-t-butyldimethylsilyl quinoxalinols derivative of leucine and α-KIC, respectively. Muscle samples (~20 mg) were homogenized in 1 ml trichloroacetic acid solution (3% wt/vol), proteins were precipitated by centrifugation, and the supernatant, containing free amino acids, was collected. The pellet containing muscle proteins was washed and then hydrolyzed in 6 N HCl at 110°C for 24 h. Amino acids in the protein hydrolysate and supernatant samples were purified on cation-exchange columns (Dowex 50WX8–200; Bio-Rad Laboratories, Richmond, CA) and leucine converted to its HFBPr derivative to determine the TTR by gas-chromatography/mass spectrometry (Agilent 6890N Gas Chromatograph and Agilent 5973N Mass Selective Detector (GC-MS); Agilent, Palo Alto, CA).
Calculations.
The fractional synthesis rate (FSR) of mixed muscle protein was calculated based on the incorporation rate of tracer into muscle proteins using the precursor-product model: (Chinkes et al., 1993) FSR = [(Ep/Eic) × 1/t × 100 where ΔEp is the increment of protein-bound leucine enrichment between two sequential biopsies, t is the time period between the two sequential biopsies, Eic is the mean enrichment of the precursor for protein synthesis (during the time between the two biopsies) (Wolfe, 1992). We used the free leucine enrichment in muscle tissue fluid as well as the average plasma leucine enrichments as surrogates for the immediate precursor for muscle protein synthesis (i.e., aminoacyl t-RNA) (Watt et al., 1991). In addition, we calculated the muscle protein FSR by using the average plasma α-KIC enrichments and this did not affect the results from our study. Therefore, data from this analysis are not included in this article. The post-intervention protein metabolism study was performed in the morning (i.e. approximately 24 h) after the last bout of exercise in the intervention groups.
Gene Expression Study
We studied chronic changes in the expression of regulators of myogenesis, proteostasis, mitochondrial function and inflammation (Table S3). In order to identify regulators of muscle mass and quality affected by aging, obesity, diet and exercise, we conducted a research in pubmed including the words “sarcopenia” OR “muscle quality” OR “muscle mass” AND “aging”, “frailty”, “life-style”, “caloric restriction”, “diet”, “exercise”. We then selected clinical and review articles reporting molecular aspects of age-related sarcopenia. Based on the available data, we selected two-hundred and fifteen targets for our gene expression analysis by nanostring. Data obtained were subjected to rigorous statistical analysis to exclude type one errors (please see Statistical Methods section). Results were then validated by RT-qPCR and western blotting.
Total RNA was isolated from vastus lateralis biopsies obtained at baseline and after 6 months using RNeasy Plus Universal Mini Kit (QIAGEN, Valencia, CA, USA) and FastPrep 24–5G homogenizer (MP Biomedicals, Santa Ana, CA, USA) following the manufacturer’s instructions. The quality and quantity of total RNA were analyzed by using both, nanodrop and Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). Gene expression profiles were analyzed with NanoString nCounter Gene Expression assay (NanoString Technologies) following the manufacturer’s protocols. The digital multiplexed NanoString Technologies were performed using a custom designed set of probes (CodeSet). Data quality control was performed using nSolver analysis software (NanoString Technologies). Reporter counts were normalized to each sample using positive control and housekeeping genes. The values are shown as the mean of the ratio between the gene expression at 6 months (end of the study) and the gene expression at baseline, with values >1 and <1 showing an increase and decrease in the expression, respectively. RT-qPCR was also performed for data validation. Two-hundred ng of RNA were used for retrotranscription into cDNA which was performed using SuperScript VILO Master Mix (Invitrogen, Carlsbad, CA, USA) as per protocol instruction. Relative quantification (ΔΔCT gene expression at 6 months vs gene expression at baseline adjusted for housekeeping gene) and data analysis were performed using Real Time PCR system QuantStudio5 and QuantStudio Design & Analysis Software 1.3.1, respectively. FAM labeled TaqMan Gene expression assays (Applied Biosystem, College Station, TX, USA) for LAMP2 (assay ID: Hs00174474_m1), MEF2A (assay ID: Hs01050406_g1), CKMT2 (assay ID: Hs00176502_m1) and VIC labeled TaqMan gene expression assay for B2M (housekeeping gene, assay ID: Hs00187842_m1) and TaqMan Universal Master Mix were used following the manufacturer’s protocol.
Immunoblotting
Vastus lateralis biopsies obtained baseline and 6 months were used for the immunoblotting. Samples were homogenized in 1 ml of cold MSD-Tris lysis buffer (Mesoscale Discovery, R60TX-3) supplemented with phosphatase inhibitor and protease inhibitor cocktail tablets, using a FastPrep 24 instrument (MP Biomedicals). Protein concentration was determined by BCA assay kit (Thermo Scientific). 20 micrograms of purified proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) on either a 4–20% or a 4–15% gradient gel (BioRad) and transferred onto PVDF membranes (Thermo Fisher Scientific). Membranes were incubated overnight at 4 °C with the following primary antibodies: MEF2A (Thermo Fisher Scientific, PA5–27380), and LAMP2 (Thermo Fisher Scientific #PA1–655). Anti-rabbit horseradish peroxidase-conjugated antibodies (Santa Cruz) were used as secondary antibodies. Densitometric analyses of western blot images were performed by using Image Studio Lite v5.2 (LiCOR) and normalized to Revert Total Protein Stain (LiCOR; #926–11011). Because muscle samples were used up for the MPS and gene expression analyses, only 4–7 subjects per group had remaining samples available for immunoblotting.
Enzymatic activity assays
Cells were homogenized using FastPrep 24–5G homogenizer (MP Biomedicals, Santa Ana, CA, USA) and the protein concentration was determined by Bradford assay. ETC enzyme activity was analyzed in duplicate using Tecan Infinite M200, as previously described (May Y et al., 2010). ETC complexes and citrate synthase activity measurements were normalized to the protein concentration.
Muscle Metabolites
Reagents and internal standards-
High-performance liquid chromatography (HPLC)-grade acetonitrile, methanol, and water were procured from Burdick & Jackson (Morristown, NJ). Mass spectrometry-grade formic acid was purchased from Sigma-Aldrich (St Louis, MO). Calibration solution containing multiple calibrants in a solution of acetonitrile, trifluroacetic acid, and water was purchased from Agilent Technologies (Santa Clara, CA). Metabolites and internal standards, including N-acetyl Aspartic acid-2H3, Tryptophan-15N2, Sarcosine-2H3, Glutamic acid-2H5, Thymine-2H4, Gibberellic acid, Trans-Zeatine, Jasmonic acid, Anthranilic acid 15N, and Testosterone-2H3, were purchased from Sigma-Aldrich (St. Louis, MO).
Internal Standard Solution and Quality controls -
Aliquots (200 L) of 10 mM solutions of N-acetyl Aspartic acid-2H3, Tryptophan-15N2, Sarcosine-2H3, Glutamic acid-2H5, Thymine-2H4, Gibberellic acid, Trans-Zeatine, Jasmonic acid, Anthranilic acid 15N, and Testosterone-2H3, were mixed and diluted up to 8 ml (final concentration 0.25 mM) and aliquoted into a 20 L portions. The aliquots were dried and stored at −80°C. Two kinds of controls were used to monitor the sample preparation and mass spectrometry. To monitor instrument performance, 20 L of a matrix-free mixture of the internal standards described above, reconstituted in 100 L of methanol: water (50:50) and analyzed by SRM.
Sample Preparation-
25mg of each tissue sample was taken. To this 750 μL ice-cold methanol:water (4:1) containing 20 μL spiked internal standards was added to each tissue sample. Ice-cold chloroform and water were added in a 3:1 ratio for a final proportion of 4:3:2 methanol:chloroform:water. The organic (methanol and chloroform) and aqueous layers were mixed, dried and resuspended with 50:50 methanol: water. The extract was deproteinized using a 3kDa molecular filter (Amicon ultracel-3K Membrane; Millipore Corporation, Billerica, MA) and the filtrate was dried under vacuum (Genevac EZ-2plus; Gardiner, Stone Ridge, NY). Prior to mass spectrometry, the dried extracts were re-suspended in identical volumes of injection solvent composed of 1:1 water: methanol (100ul) and were subjected to liquid chromatography-mass spectrometry using a 6495-triple quadrupole mass spectrometer (Agilent Technologies, Santa Clara, CA) coupled to a HPLC system (Agilent Technologies, Santa Clara, CA).
Measurement of glycolytic intermediates, TCA cycle:
To target the glycolysis, TCA, the samples were delivered to the mass spectrometer via normal phase chromatography using a Luna 3 um NH2 100 A (150 × 2mm) (Phenomenex, Torrance, CA) in ESI/MS negative ion mode. Source parameters were as follows: Gas temperature was 250 °C; Gas flow was 11 l/min; Nebulizer was 20psi; Sheath gas temperature was 300 °C; Sheath gas flow was 11 l/min; Capillary was 3000 V positive and 3500 V negative; Nozzle voltage was 1500 V positive and 1500 V negative. The mobile phase was water solvent A modified by the addition of 5mM ammonium acetate (pH 9.9), and 100% acetonitrile (ACN) solvent B. The binary pump flow rate was 0.2 ml/min with a gradient spanning 80% B to 2% A over a 20-minute period followed by 2% solvent B to 80% solvent A for a 5 min period and followed by 80% B for 13-minute. The flow rate was gradually increased during the separation from 0.2 mL/min (0–20 mins), 0.3 mL/min (20.1–25 min), 0.35 mL/min (25–30 min), 0.4 mL/min (30–37.99 min) and finally set at 0.2 mL/min (5 min). The injection volume was 10ul.
Circulating CAF detection
Fasting blood samples were collected at baseline and after 6 months of intervention; serum samples were extracted and stored at −80°C until analysis. Baseline and 6 month’s serum samples belonging to each individual were analyzed in duplicate in the same ELISA plate (#NT1001, Neurotune, Switzerland) to reduce inter-assay variability. The coefficient of variability for this assay is <10%.
Quantification and Statistical Analysis
Randomization of participants was performed using a computer-generated block random permutation procedure, stratified for ethnicity (Hispanic vs. Non-Hispanic), body mass index (BMI <35 kg/m2 vs. ≥ 35 kg/m2), and gender (women vs. men). Differences in baseline characteristics between groups were assessed by analysis of variance or Fisher’s exact test. For analyses of data from the muscle protein metabolism study, gatekeeping strategy (described in detail below) (Dmitrienko et al., 2011; Villareal et al., 2017a) was used to control for multiple comparisons while satisfying the family-wise error rate (FWER) < 0.05. Accordingly, longitudinal changes between groups were first tested with the use of mixed-model repeated-measures analyses of covariance, with adjustment for baseline values. Thereafter, a significant group-by-time interaction (<0.05) for the change in outcome (e.g. MPS rate) was required in order to continue with posthoc testing; comparisons of the exercise groups with the control group were performed with Dunnett’s test (Dunnett, 1980) while comparisons among the intervention groups were performed with the Fisher-Hayter test (Hayter, 1986). Our study sample size was not large enough to analyze a potential effect of gender on our outcomes; however, we controlled for the effect of gender by including gender in the block stratification of our study design.
Gene expression data by nanostring were assessed for the 215 genes measured before and after the intervention on the 47 subjects grouped into the 4 intervention groups. Six of the selected targets were housekeeping genes that were considered not to be affected by exercise; the mean of these were used to exclude data for any subject with sum >1.5. The remainder of 207 genes were grouped into 6 established pathways: proteostasis (autophagy, UPS, apoptosis), mitochondria, glycolysis, myogenesis and inflammation (Table S3). The before/after gene expression differences due to the interventions were expressed as ratios (6 months/baseline). Statistical significant before/after differences were determined by one-sample t-test of the ratio compared to 1 or the fold difference compared to 0 for each gene for all subjects by intervention group; this is descriptive only since the number of t-tests precluded robust conclusions. Thus, as in the analyses of the muscle protein metabolism study above, we applied a gatekeeping strategy (Dmitrienko and Tamhane, 2011) that we have used in the NEJM clinical trial paper (Villareal et al., 2017a).
We applied a Bonferroni correction for the 6 pathways using an alpha=.05/6 (approximately equal to 0.01). If no gene in a given pathway showed any difference among the 4 groups by a one-way ANOVA then that pathway was excluded from further statistical testing. N= 5 pathways survived this gate.
For the genes in surviving pathways, we applied Dunnett’s test to compare each intervention group to the control group. This gate required at least one significant finding.
For the genes that survived Dunnett’s gate, we applied the Fisher-Hayter test for the comparisons among the 3 intervention groups.
Note that both the Dunnett’s multiple comparison tests and the Fisher-Hayter multiple comparison tests satisfy the FWER criteria.
Since our measures of gene expression were often not normally distributed in box plots, we used a 2-step method of transformation to normality similar to that of Templeton (Templeton, 2011). A nonparametric center and variability of each distribution were computed as the median and the pseudo standard deviation (PSD= IQR /1.35, IQR=interquartile range). Then the raw data was transformed to a standard normal distribution (mean=0, SD=1) as normal scores computed from their ranks and represented by a random variable labeled Z. The data in each exercise group used in the ANOVA procedures were obtained as X = median + PSD*Z. The analysis of nanostring data was validated by using RT-qPCR assays for selected genes of the banked samples from the same subjects and exercise groups. Pearson correlation test was performed to assess the degree of the relationship between changes in linearly-related variables.
Data on changes in mitochondrial enzyme activities and TCA metabolites were considered tertiary outcomes and were also analyzed by using repeated-measures analyses of covariance, with adjustments for baseline values. Within the framework of the mixed model, when the P value for an interaction was significant, the appropriate contrast was used to test the null hypothesis that changes between 2 specific time points were equal to corresponding changes in another group. Statistical analysis for the TCA metabolites were performed on log-transformed data.
All statistical tests were 2-tailed, and P < .05 was considered statistically significant for the between-group comparisons. All data are presented as mean (±SE) unless otherwise indicated. Analyses were performed with SAS software, version 9.4 (SAS Institute).
Data Availability
Nanostring dataset are provided in the file entitled “Data S1”. Descriptive Title: Gene Expression Data by Nanostring Related to Table 3. Data S1 can be also found at the following URL: https://data.mendeley.com/datasets/ry4dmc24n6/draft?a=075d9c6d-8b8a-4c0f-b140-ae88a46e04fc Nanostring dataset accession number is provided in Table S3.
Supplementary Material
Data S1 Gene Expression Data by Nanostring Related to Table 3. Data were analyzed with the use of NanoString nCounter Gene Expression assay (NanoString Technologies). Baseline (Sheet 1) and 6 months (Sheet 2).
Table S3 Gene Selection List with Accession Numbers Related to Table 3. A: Proteostasis; B: Mitochondria; C: Myogenesis; D: Inflammation; E: Glycolysis; F: Housekeeping Genes and Controls.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-LAMP2 | ThermoFisher | Cat# PA1–655 RRID:AB_2134625 |
| Mouse anti-rabbit monoclonal IgG-HRP | Santa Cruz Biotech | Cat# sc-2357 RRID:AB_2714189 |
| Rabbit polyclonal anti-MEF2A | ThermoFisher | Cat# PA5–27380 RRID:AB_2544856 |
| Chemicals, Peptides, Metabolites and Recombinant Proteins | ||
| [5,5,5-2H3] leucine tracer | Sigma Aldrich | Cat#486825–1G |
| Liquid meal | Ensure, Abbott Laboratories | N/A |
| L-Norleucine, ≥98% (TLC) | Sigma-Aldrich | Cat# N6877 |
| 2-Mercaptoethanol | Sigma-Aldrich | Cat# M6250 |
| 4X Laemmli Sample Buffer | Bio-Rad | Cat# 1610747 |
| MSD-Tris lysis buffer | Mesoscale Discovery | Cat# R60TX-3 |
| Halt™ Protease Inhibitor Cocktail (100X) | ThermoFisher | Cat# 78430 |
| Ponceau S | Fisher Scientific | Cat# BP103–10 |
| REVERT total protein stain | LiCOR | Cat# 926–11011 |
| RNALater | ThermoFisher | Cat# AM7020 |
| HPLC-grade acetonitrile | ThermoFisher | Cat#A955–500 |
| HPLC-grade methanol | ThermoFisher on | Cat#A456–500 |
| HPLC-grade water | ThermoFisher | Cat#W64 |
| MS-grade formic acid | Sigma-Aldrich | Cat#399388 |
| Calibration solution | Agilent Technologies | Cat#G1969–85000 |
| N-acetyl Aspartic acid-2H3 | Sigma-Aldrich | Cat#616060 |
| Tryptophan-15N2 | Sigma-Aldrich | Cat#574600 |
| Sarcosine-2H3 | Sigma-Aldrich | Cat#493120 |
| Glutamic acid-2H5 | Sigma-Aldrich | Cat#616281 |
| Thymine-2H4 | Sigma-Aldrich | Cat# 487066 |
| Gibberellic acid | Sigma-Aldrich | Cat# G7645 |
| Trans-Zeatine | Sigma-Aldrich | Cat# Z0876 |
| Anthranilic acid 15N | Cambridge isotopes | Cat#NLM-3294 |
| Testosterone-2H3 | Sigma-Aldrich | Cat# T2655 |
| Jasmonic acid | Sigma-Aldrich | Cat# J2500 |
| Critical Commercial Assays | ||
| Insulin Detection Assay | Siemens | Cat# LKIN1 |
| CAF ELISA kit | Neurotune | Cat# NT1001 |
| RNeasy Plus Universal Mini Kit | QIAGEN | Cat# 73404 |
| TaqMan Universal PCR Master Mix | ThermoFisher | Cat# 4304437 |
| SuperScript VILO Master Mix | ThermoFisher | Cat# 11755250 |
| Bradford Assay, Bio-Rad Assay Dye | Bio-Rad | Cat#5000006 |
| NanoString nCounter Customized Gene Expression Assay | NanoString Technologies | N/A |
| Pierce™ Rapid Gold BCA Protein Assay Kit | ThermoFisher | Cat# A53225 |
| Oligonucleotides | ||
| Taqman FAM Probe MEF2A | ThermoFisher | Hs01050406_g1 |
| Taqman FAM Probe CKMT2 | ThermoFisher | Hs00176502_m1 |
| Taqman VIC Probe B2M | ThermoFisher | Hs00187842_m1 |
| Taqman FAM Probe LAMP2 | ThermoFisher | Hs00174474_m1 |
| Software and Algorithms | ||
| nSolver analysis software | Nanostring Tech | N/A |
| Quant Studio Design and Analysis Software 1.3.1 | Thermofisher | N/A |
| Image Studio Lite v5.2 | LI-COR Biosciences | N/A |
| Sigma Plot Version 13 | Systat Software, Inc | N/A |
| EndNotes Version 8.2 | Clarivate Analytics | N/A |
| SAS Version 9.4 | Sas Institute Inc | N/A |
| Others | ||
| Amicon ultracel-3K Membrane; | Millipore Corporation, | Cat#UFC5003BK |
| Vacuum | Genevac EZ-2plus | Cat#EZ-2.3 |
| Data generated with Nanostring technology can be found in Data S1 at the following link https://data.mendeley.com/datasets/ry4dmc24n6/draft?a=075d9c6d-8b8a-4c0f-b140-ae88a46e04fc | ||
Highlights.
Diet-induced weight loss leads to muscle mass reduction in obese older adults
Diet-induced muscle loss is attenuated by addition of resistance exercise
Muscle protein synthesis following a meal improves with resistance exercise
Myocellular quality improves with aerobic plus resistance exercise
Context and Significance.
Age-related loss of muscle mass and strength (sarcopenia) is exacerbated by obesity, and significantly contributes to loss of functional independence. Weight-loss induced muscle mass loss may exacerbate sarcopenia in the obese elderly population. In dieting, obese older adults, the combination of aerobic and resistance exercise improves muscle protein synthesis following a meal (anabolic stimulus) and myocellular quality compared to either aerobic or resistance exercise alone, thus preserving muscle mass. Health care providers should prescribe such specific lifestyle intervention therapies to obese older adults in order to ensure muscle mass preservation, muscle quality improvements and reverse frailty.
ACKNOWLEDGMENTS
We thank the participants for their cooperation, Nancy Morgan for assistance with preparation of stable isotopes, and Robert Scott, Vasanta Putluri, and Mylinh Bernardi for technical assistance. Supported by grants RO1-AG031176, CX-00096, UL1-TR00041, UL1-TR000135, P41-GM103422, P30-DK056341, RO1-DK111436, P30-DK020579, RP170005, P30-CA125123, P30-DK56338, R01-CA234479, and R03-CA212816. We also thank the members of the Alkek Foundation for their support. The findings reported in this article are the result of work supported with resources and the use of facilities at the New Mexico VA Health Care System and Michael E. DeBakey VA Medical Center.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health and the Department of Veterans Affairs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could aect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTEREST
The authors declare no conflicts of interest.
Additional Resources
Clinical Trial Registry Number: .
Description: https://clinicaltrials.gov/ct2/show/NCT01065636
REFERENCES
- Aguirre LE, and Villareal DT (2015). Physical Exercise as Therapy for Frailty. Nestle. Nutr. Inst. Workshop Ser 83, 83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison DB, Paultre F, Maggio C, Mezzitis N, and Pi-Sunyer FX (1995). The use of areas under curves in diabetes research. Diabetes Care 18, 245–250. [DOI] [PubMed] [Google Scholar]
- Balagopal P, Schimke JC, Ades P, Adey D, and Nair KS (2001). Age effect on transcript levels and synthesis rate of muscle MHC and response to resistance exercise. Am. J. Physiol Endocrinol. Metab 280, E203–E208. [DOI] [PubMed] [Google Scholar]
- Batsis JA, and Villareal DT (2018). Sarcopenic obesity in older adults: aetiology, epidemiology and treatment strategies. Nat Rev Endocrinol 14, 513–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RA, Al-Khalaf M, and Megeney LA (2016). The beneficial role of proteolysis in skeletal muscle growth and stress adaptation. Skelet Muscle 6, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohe J, Low A, Wolfe RR, and Rennie MJ (2003). Human muscle protein synthesis is modulated by extracellular, not intramuscular amino acid availability: a dose-response study. J. Physiol 552, 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondoc I, Cochrane SK, Church TS, Dahinden P, Hettwer S, Hsu FC, Stafford RS, Pahor M, Buford TW, and Life Study, I. (2015). Effects of a One-Year Physical Activity Program on Serum C-Terminal Agrin Fragment (CAF) Concentrations among Mobility-Limited Older Adults. J Nutr Health Aging 19, 922–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M, Sinacore DR, Binder EF, and Kohrt WM (2000). Physical and performance measures for the identification of mild to moderate frailty. J. Gerontol. A Biol. Sci. Med. Sci 55, M350–M355. [DOI] [PubMed] [Google Scholar]
- Cadore EL, Pinto RS, Lhullier FL, Correa CS, Alberton CL, Pinto SS, Almeida AP, Tartaruga MP, Silva EM, and Kruel LF (2010). Physiological effects of concurrent training in elderly men. Int. J. Sports Med 31, 689–697. [DOI] [PubMed] [Google Scholar]
- Carraro F, Stuart CA, Hartl WH, Rosenblatt J, and Wolfe RR (1990). Effect of exercise and recovery on muscle protein synthesis in human subjects. Am. J Physiol 259, E470–E476. [DOI] [PubMed] [Google Scholar]
- Cartee GD, Hepple RT, Bamman MM, and Zierath JR (2016). Exercise Promotes Healthy Aging of Skeletal Muscle. Cell Metab 23, 1034–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinkes DL, Rosenblatt J, and Wolfe RR (1993). Assessment of the mathematical issues involved in measuring the fractional synthesis rate of protein using the flooding dose technique. Clin. Sci. (Lond) 84, 177–183. [DOI] [PubMed] [Google Scholar]
- Cohen S, Nathan JA, and Goldberg AL (2015). Muscle wasting in disease: molecular mechanisms and promising therapies. Nat Rev Drug Discov 14, 58–74. [DOI] [PubMed] [Google Scholar]
- Dmitrienko A, Millen BA, Brechenmacher T, and Paux G (2011). Development of gatekeeping strategies in confirmatory clinical trials. Biom. J 53, 875–893. [DOI] [PubMed] [Google Scholar]
- Drey M, Sieber CC, Bauer JM, Uter W, Dahinden P, Fariello RG, Vrijbloed JW, and Fi, A.T.i.g. (2013). C-terminal Agrin Fragment as a potential marker for sarcopenia caused by degeneration of the neuromuscular junction. Exp Gerontol 48, 76–80. [DOI] [PubMed] [Google Scholar]
- Drummond MJ, Dreyer HC, Pennings B, Fry CS, Dhanani S, Dillon EL, Sheffield-Moore M, Volpi E, and Rasmussen BB (2008). Skeletal muscle protein anabolic response to resistance exercise and essential amino acids is delayed with aging. J Appl. Physiol 104, 1452–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond MJ, Timmerman KL, Markofski MM, Walker DK, Dickinson JM, Jamaluddin M, Brasier AR, Rasmussen BB, and Volpi E (2013). Short-term bed rest increases TLR4 and IL-6 expression in skeletal muscle of older adults. Am J Physiol Regul Integr Comp Physiol 305, R216–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnett CW (1980). Pairwise multiple comparisons in the homogeneous vriance, unequal sample size case. Journal of the American Statistical Association 75, 789–795. [Google Scholar]
- Egan B, and Zierath JR (2013). Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab 17, 162–184. [DOI] [PubMed] [Google Scholar]
- Estrella NL, Desjardins CA, Nocco SE, Clark AL, Maksimenko Y, and Naya FJ (2015). MEF2 transcription factors regulate distinct gene programs in mammalian skeletal muscle differentiation. J Biol Chem 290, 1256–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans WJ (2002). Exercise as the standard of care for elderly people. J Gerontol. A Biol. Sci. Med. Sci 57, M260–M261. [DOI] [PubMed] [Google Scholar]
- Fielding RA, Vellas B, Evans WJ, Bhasin S, Morley JE, Newman AB, Abellan van KG, Andrieu S, Bauer J, Breuille D, et al. (2011). Sarcopenia: an undiagnosed condition in older adults. Current consensus definition: prevalence, etiology, and consequences. International working group on sarcopenia. J Am Med Dir. Assoc 12, 249–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink LN, Oberbach A, Costford SR, Chan KL, Sams A, Bluher M, and Klip A (2013). Expression of anti-inflammatory macrophage genes within skeletal muscle correlates with insulin sensitivity in human obesity and type 2 diabetes. Diabetologia 56, 1623–1628. [DOI] [PubMed] [Google Scholar]
- Flegal KM, Kruszon-Moran D, Carroll MD, Fryar CD, and Ogden CL (2016). Trends in Obesity Among Adults in the United States, 2005 to 2014. JAMA 315, 2284–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried LP, Tangen CM, Walston J, Newman AB, Hirsch C, Gottdiener J, Seeman T, Tracy R, Kop WJ, Burke G, et al. (2001). Frailty in older adults: evidence for a phenotype. J Gerontol. A Biol. Sci. Med. Sci 56, M146–M156. [DOI] [PubMed] [Google Scholar]
- Gleeson M, Bishop NC, Stensel DJ, Lindley MR, Mastana SS, and Nimmo MA (2011). The anti-inflammatory effects of exercise: mechanisms and implications for the prevention and treatment of disease. Nat Rev Immunol 11, 607–615. [DOI] [PubMed] [Google Scholar]
- Goodpaster BH, Park SW, Harris TB, Kritchevsky SB, Nevitt M, Schwartz AV, Simonsick EM, Tylavsky FA, Visser M, and Newman AB (2006). The loss of skeletal muscle strength, mass, and quality in older adults: the health, aging and body composition study. J Gerontol. A Biol. Sci. Med. Sci 61, 1059–1064. [DOI] [PubMed] [Google Scholar]
- Gouspillou G, and Hepple RT (2013). Facts and controversies in our understanding of how caloric restriction impacts the mitochondrion. Exp Gerontol 48, 1075–1084. [DOI] [PubMed] [Google Scholar]
- Guillet C, Prod’homme M, Balage M, Gachon P, Giraudet C, Morin L, Grizard J, and Boirie Y (2004). Impaired anabolic response of muscle protein synthesis is associated with S6K1 dysregulation in elderly humans. FASEB J 18, 1586–1587. [DOI] [PubMed] [Google Scholar]
- Hayter AJ (1986). The maximum familywise error rate of Fisher’s least significant difference test. Biometrics 12, 1000–1004. [Google Scholar]
- Hickson RC (1980). Interference of strength development by simultaneously training for strength and endurance. Eur. J Appl. Physiol Occup. Physiol 45, 255–263. [DOI] [PubMed] [Google Scholar]
- Huffman KM, Koves TR, Hubal MJ, Abouassi H, Beri N, Bateman LA, Stevens RD, Ilkayeva OR, Hoffman EP, Muoio DM, et al. (2014). Metabolite signatures of exercise training in human skeletal muscle relate to mitochondrial remodelling and cardiometabolic fitness. Diabetologia 57, 2282–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen MD, and Heiling VJ (1991). Heated hand vein blood is satisfactory for measurements during free fatty acid kinetic studies. Metabolism 40, 406–409. [DOI] [PubMed] [Google Scholar]
- Jensen MD, Ryan DH, Apovian CM, Ard JD, Comuzzie AG, Donato KA, Hu FB, Hubbard VS, Jakicic JM, Kushner RF, et al. (2014). 2013 AHA/ACC/TOS guideline for the management of overweight and obesity in adults: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines and The Obesity Society. J. Am. Coll. Cardiol 63, 2985–3023. [DOI] [PubMed] [Google Scholar]
- Jette AM, and Cleary PD (1987). Functional disability assessment. Phys. Ther 67, 1854–1859. [DOI] [PubMed] [Google Scholar]
- Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, and Turner RC (1985). Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 28, 412–419. [DOI] [PubMed] [Google Scholar]
- Parise G, Mihic S, MacLennan D, Yarasheski KE, and Tarnopolsky MA (2001). Effects of acute creatine monohydrate supplementation on leucine kinetics and mixed-muscle protein synthesis. J Appl. Physiol 91, 1041–1047. [DOI] [PubMed] [Google Scholar]
- Patterson BW, Zhang XJ, Chen Y, Klein S, and Wolfe RR (1997). Measurement of very low stable isotope enrichments by gas chromatography/mass spectrometry: application to measurement of muscle protein synthesis. Metabolism 46, 943–948. [DOI] [PubMed] [Google Scholar]
- Phillips SM, Tipton KD, Aarsland A, Wolf SE, and Wolfe RR (1997). Mixed muscle protein synthesis and breakdown after resistance exercise in humans. Am J Physiol 273, E99–107. [DOI] [PubMed] [Google Scholar]
- Pikosky MA, Gaine PC, Martin WF, Grabarz KC, Ferrando AA, Wolfe RR, and Rodriguez NR (2006). Aerobic exercise training increases skeletal muscle protein turnover in healthy adults at rest. J Nutr 136, 379–383. [DOI] [PubMed] [Google Scholar]
- Potes Y, de Luxan-Delgado B, Rodriguez-Gonzalez S, Guimaraes MRM, Solano JJ, Fernandez-Fernandez M, Bermudez M, Boga JA, Vega-Naredo I, and Coto-Montes A (2017). Overweight in elderly people induces impaired autophagy in skeletal muscle. Free Radic Biol Med 110, 31–41. [DOI] [PubMed] [Google Scholar]
- Rasmussen BB, Fujita S, Wolfe RR, Mittendorfer B, Roy M, Rowe VL, and Volpi E (2006). Insulin resistance of muscle protein metabolism in aging. FASEB J 20, 768–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeds DN, Yarasheski KE, Fontana L, Cade WT, Laciny E, DeMoss A, Patterson BW, Powderly WG, and Klein S (2006). Alterations in liver, muscle, and adipose tissue insulin sensitivity in men with HIV infection and dyslipidemia. Am. J Physiol Endocrinol. Metab 290, E47–E53. [DOI] [PubMed] [Google Scholar]
- Robinson MM, Dasari S, Konopka AR, Johnson ML, Manjunatha S, Esponda RR, Carter RE, Lanza IR, and Nair KS (2017). Enhanced Protein Translation Underlies Improved Metabolic and Physical Adaptations to Different Exercise Training Modes in Young and Old Humans. Cell Metab 25, 581–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanello V, and Sandri M (2015). Mitochondrial Quality Control and Muscle Mass Maintenance. Front Physiol 6, 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roubenoff R (2004). Sarcopenic obesity: the confluence of two epidemics. Obes Res 12, 887–888. [DOI] [PubMed] [Google Scholar]
- Sheffield-Moore M, Yeckel CW, Volpi E, Wolf SE, Morio B, Chinkes DL, Paddon-Jones D, and Wolfe RR (2004). Postexercise protein metabolism in older and younger men following moderate-intensity aerobic exercise. Am. J Physiol Endocrinol. Metab 287, E513–E522. [DOI] [PubMed] [Google Scholar]
- Short KR, Vittone JL, Bigelow ML, Proctor DN, and Nair KS (2004). Age and aerobic exercise training effects on whole body and muscle protein metabolism. Am J Physiol Endocrinol Metab 286, E92–101. [DOI] [PubMed] [Google Scholar]
- Smith GI, Villareal DT, and Mittendorfer B (2007). Measurement of human mixed muscle protein fractional synthesis rate depends on the choice of amino acid tracer. Am. J. Physiol Endocrinol. Metab 293, E666–E671. [DOI] [PubMed] [Google Scholar]
- Tang JE, Perco JG, Moore DR, Wilkinson SB, and Phillips SM (2008). Resistance training alters the response of fed state mixed muscle protein synthesis in young men. Am. J Physiol Regul. Integr. Comp Physiol 294, R172–R178. [DOI] [PubMed] [Google Scholar]
- Templeton G (2011). A Two-Step Approach for Transforming Continuous Variables to Normal: Implications and Recommendations for IS Research. Communications of the Association for Information Systems. 28, 41–48. [Google Scholar]
- Tretter L, Patocs A, and Chinopoulos C (2016). Succinate, an intermediate in metabolism, signal transduction, ROS, hypoxia, and tumorigenesis. Biochim Biophys Acta 1857, 1086–1101. [DOI] [PubMed] [Google Scholar]
- Villareal DT, Aguirre L, Gurney AB, Waters DL, Sinacore DR, Colombo E, Armamento-Villareal R, and Qualls C (2017a). Aerobic or Resistance Exercise, or Both, in Dieting Obese Older Adults. N. Engl. J. Med 376, 1943–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villareal DT, Apovian CM, Kushner RF, and Klein S (2005). Obesity in older adults: technical review and position statement of the American Society for Nutrition and NAASO, The Obesity Society. Am J Clin Nutr 82, 923–934 [also published in: Obes Res. 2005: 2013:1849–1863]. [DOI] [PubMed] [Google Scholar]
- Villareal DT, Banks M, Siener C, Sinacore DR, and Klein S (2004). Physical Frailty and Body Composition in Obese Elderly Men and Women. Obesity Research 12, 913–920. [DOI] [PubMed] [Google Scholar]
- Villareal DT, Chode S, Parimi N, Sinacore DR, Hilton T, Armamento-Villareal R, Napoli N, Qualls C, and Shah K (2011a). Weight loss, exercise, or both and physical function in obese older adults. N Engl J Med 364, 1218–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villareal DT, Fontana L, Weiss EP, Racette SB, Steger-May K, Schechtman KB, Klein S, and Holloszy JO (2006). Bone Mineral Density Response to Caloric Restriction-Induced Weight Loss or Exercise-Induced Weight Loss: A Randomized Controlled Trial. Archives of Internal Medicine 166, 2502–2510. [DOI] [PubMed] [Google Scholar]
- Villareal DT, Smith GI, Shah K, and Mittendorfer B (2012). Effect of weight loss on the rate of muscle protein synthesis during fasted and fed conditions in obese older adults. Obesity. (Silver. Spring) 20, 1780–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villareal DT, Smith GI, Sinacore DR, Shah K, and Mittendorfer B (2011b). Regular multicomponent exercise increases physical fitness and muscle protein anabolism in frail, obese, older adults. Obesity. (Silver. Spring) 19, 312–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villareal DT, Waters DL, and Qualls C (2017b). Exercise Type in Dieting Obese Older Adults. N. Engl. J. Med 377, 599–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpi E, Mittendorfer B, Rasmussen BB, and Wolfe RR (2000). The response of muscle protein anabolism to combined hyperaminoacidemia and glucose-induced hyperinsulinemia is impaired in the elderly. J. Clin. Endocrinol. Metab 85, 4481–4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss EP, Racette SB, Villareal DT, Fontana L, Steger-May K, Schechtman KB, Klein S, Ehsani AA, and Holloszy JO (2007). Lower extremity muscle size and strength and aerobic capacity decrease with caloric restriction but not with exercise-induced weight loss. J Appl. Physiol 102, 634–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welle S, Bhatt K, and Thornton CA (1999). Stimulation of myofibrillar synthesis by exercise is mediated by more efficient translation of mRNA. J Appl. Physiol 86, 1220–1225. [DOI] [PubMed] [Google Scholar]
- Wilkinson SB, Phillips SM, Atherton PJ, Patel R, Yarasheski KE, Tarnopolsky MA, and Rennie MJ (2008). Differential effects of resistance and endurance exercise in the fed state on signalling molecule phosphorylation and protein synthesis in human muscle. J Physiol 586, 3701–3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JM, Marin PJ, Rhea MR, Wilson SM, Loenneke JP, and Anderson JC (2012). Concurrent training: a meta-analysis examining interference of aerobic and resistance exercises. J. Strength. Cond. Res 26, 2293–2307. [DOI] [PubMed] [Google Scholar]
- Wolfe RR (1992). Radioactive and Stable Isotope Tracers in Biomedicine: Principles and Practice of Kinetic Analysis. (New York,: Wiley-Liss; ), pp. 377–416. [Google Scholar]
- Yarasheski KE, Zachwieja JJ, and Bier DM (1993). Acute effects of resistance exercise on muscle protein synthesis rate in young and elderly men and women. Am. J. Physiol 265, E210–E214. [DOI] [PubMed] [Google Scholar]
- Zampieri S, Pietrangelo L, Loefler S, Fruhmann H, Vogelauer M, Burggraf S, Pond A, Grim-Stieger M, Cvecka J, Sedliak M, et al. (2015). Lifelong physical exercise delays age-associated skeletal muscle decline. J Gerontol A Biol Sci Med Sci 70, 163–173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1 Gene Expression Data by Nanostring Related to Table 3. Data were analyzed with the use of NanoString nCounter Gene Expression assay (NanoString Technologies). Baseline (Sheet 1) and 6 months (Sheet 2).
Table S3 Gene Selection List with Accession Numbers Related to Table 3. A: Proteostasis; B: Mitochondria; C: Myogenesis; D: Inflammation; E: Glycolysis; F: Housekeeping Genes and Controls.
Data Availability Statement
Nanostring dataset are provided in the file entitled “Data S1”. Descriptive Title: Gene Expression Data by Nanostring Related to Table 3. Data S1 can be also found at the following URL: https://data.mendeley.com/datasets/ry4dmc24n6/draft?a=075d9c6d-8b8a-4c0f-b140-ae88a46e04fc Nanostring dataset accession number is provided in Table S3.