Abstract
Background:
Experimental autoimmune encephalomyelitis (EAE) in the common marmoset is a nonhuman primate model of multiple sclerosis (MS) that shares numerous clinical, radiological, and pathological features with MS. Among the clinical features are motor and sensory deficits that are highly suggestive of spinal cord (SC) damage.
Objective:
To characterize the extent and nature of SC damage in symptomatic marmosets with EAE using a combined magnetic resonance imaging (MRI) and histopathology approach.
Materials and Methods:
SC tissues from five animals were scanned using 7-tesla MRI to collect high-resolution ex vivo images. Lesions were segmented and classified based on shape, size, and distribution along the SC. Tissues were processed for histopathological characterization (myelin and microglia/macrophages). Statistical analysis, using linear mixed-effects models, evaluated the association between MRI and histopathology.
Results:
Marmosets with EAE displayed two types of SC lesions: focal and subpial lesions. Both lesion types were heterogeneous in size and configuration, and corresponded to areas of marked demyelination with high density of inflammatory cells. Inside the lesions, the MRI signal was significantly correlated with myelin content (p<0.001).
Conclusion:
Our findings underscore the relevance of this nonhuman primate EAE model for better understanding mechanisms of MS lesion formation in the SC.
Keywords: Multiple sclerosis, Spinal cord, EAE, MRI, Marmoset
Introduction
Experimental autoimmune encephalomyelitis (EAE) is the most commonly used animal model for studying multiple sclerosis (MS)1,2. While mouse EAE models are a vital component of preclinical research in MS, the marmoset EAE model has recently gained some attention due to marmosets’ closer genetic and immunological proximity to humans3,4. This nonhuman primate model of MS also bears strong pathological and radiological similarities to MS,5 while still maintaining practicality as a model, making it an excellent system for studying the mechanisms of lesion formation in the brain6.
Among the various clinical symptoms experienced by marmosets with EAE, motor and sensory deficits are often present, which strongly suggest the presence of demyelinated lesions in the spinal cord (SC). A few histopathological studies have reported the presence of pathology in SC tissues from marmosets with EAE3,7,8,9,10. However, these studies did not characterize in a comprehensive manner the extent of the SC pathology.
Magnetic resonance imaging (MRI) is a noninvasive and sensitive tool for detecting pathology in the central nervous system (CNS). However, MRI of the SC is challenging, especially in small animals like the common marmoset, due to the extremely small dimensions of the cord. Therefore, we investigated the spinal cord pathology by postmortem MRI, where high signal-to-noise ratio can be obtained to produce high-quality and high-resolution images. These ex vivo MRI images can then be used to guide precisely any subsequent tissue sampling for histology, and to evaluate pathologically specific areas of the CNS affected by EAE12.
In this study, we used an experimental approach combining ex vivo MRI with histopathology, with the goal of characterizing the extent and nature of the pathology affecting the SC in the marmoset model of MS.
Materials and Methods
EAE induction
All common marmosets (Callithrix jacchus) used in this study were housed at the NIH Intramural Research facilities, in accordance with the standards of the American Association for Accreditation of Laboratory Animal Care and the NINDS Animal Care and Use Committee. Five adult marmosets [mean age 3.5 years (range: 1.5–4.9), 4 females] were included in this study. Four animals were induced with EAE using white matter homogenate in complete Freund’s adjuvant13 which in our hands produces a disease that bears strong radiological similarities to MS5. The fifth animal was used as a healthy, non-EAE control. Animals were frequently weighed and scored according to an EAE scale based on clinical symptoms14. Table 1 provides the demographics and clinical summary of these animals. Once the animals showed paraplegia and/or 20% weight loss, the experiment was terminated by performing transcardial perfusion fixation with cold 4% paraformaldehyde under anesthesia. The spine was removed and stored in 4% neutral buffered formalin.
Table 1:
Demographic and clinical information of the five marmosets included in this study.
| Animal | Immunization | Sex | Age (years) | Disease duration (days) | EAE score at termination | Clinical symptoms |
|---|---|---|---|---|---|---|
| M#1 | WMH | M | 3.6 | 134 | 3 | Weight loss, lethargy, ataxia, severe weakness, sensory loss, vision loss |
| M#2 | WMH | F | 4.6 | 159 | 3 | Apathy, tetraparesis, sensory loss, vision loss |
| M#3 | WMH | F | 2.9 | 123 | N/A | N/A |
| M#4 | WMH | F | 1.5 | 89 | N/A | N/A |
| M#5 | None | F | 4.9 | - | - | - |
Table subheading: Note that clinical information for M#3 and M#4 were not tracked over the course of the disease. Disease duration is measured as time from immunization to termination. EAE scores are defined in Kap YS et al., 20084. WMH: white matter homogenate, M: male, F: female, N/A: not available.
MRI acquisition
After extraction and preparation of the SC samples (see supplementary Figure 1), MRI was performed on a 7T/30 cm MRI scanner (Bruker BioSpin Corp., Ettlingen, Germany) equipped with a 15 cm gradient set of 450 mT/m strength (Resonance Research Inc., Billerica, MA, USA). A proton-only transmit-receive volume coil (25 mm inner diameter) (Bruker) was used for imaging the tissue samples loaded in the plastic tubes. During each scanning session, a 3-dimensional (3D) T2*-weighted gradient-echo sequence with an in-plane resolution of 70 μm and a slice thickness of 200 μm was acquired 4 times consecutively for a total acquisition time of 12 hours (see Supplementary Table 1 for acquisition parameters).
Image analysis
All four acquisitions were first averaged using the MIPAV image analysis software (National Institutes of Health, Bethesda, Maryland; http://mipav.cit.nih.gov). After a N4-bias-field correction15 was applied to correct for intensity inhomogeneity, images were upsampled to an in-plane axial resolution of 35 μm × 35 μm and a 70 μm slice thickness and concatenated to generate a single image volume of the entire SC using ImageJ software (National Institutes of Health, Bethesda, Maryland; https://imagej.nih.gov/ij/) and MIPAV. Finally, the different spinal level segments were annotated based on the location of the nerve roots16.
Focal lesions (areas of abnormal signal intensity within the white matter (WM)) were manually segmented (ITK-SNAP version 3.2.0) using triplanar viewing mode. Lesions were classified according to their volume using the following arbitrary categories: small (0.009-0.06 mm3), medium (0.06-0.4 mm3), and large (>0.4 mm3). The specific position of the focal lesions was also recorded as anterior, posterior, and lateral (left/right). Subpial lesions were also manually segmented on select images at three different levels of the cord (C7, T7, and L3). Regions-of-interest (ROIs) of the entire cross-sectional cord, normal appearing white matter (NAWM), and gray matter (GM) were drawn manually (Supplementary Figure 2) using a semi-automated tool in JIM V.7.0 (Xinapse Systems, Leicester, UK). The relative thickness of subpial lesions (areas of abnormal signal intensity along the edge of the WM) was characterized as a percentage of total WM using the following formula: Area ROIsubpial lesion / (Area ROIcross-sectional cord – Area ROIGM) × 100. The degree of severity for the subpial lesions was arbitrarily defined as: low if < 20%; intermediate if between 20–50%; and high if > 50%.
Histopathology
Focal and subpial lesions with various sizes, shapes, and locations identified by ex vivo MRI were targeted during the cutting process (transaxial cuts of 3 mm-thick slabs). Slabs from the control animal were selected at similar cord levels for comparison purposes. A total of 32 slabs were cut, and then embedded in paraffin wax. These paraffin-embedded slabs were sectioned in 5 μm-thick slices to find the lesions detected by MRI. Luxol fast blue with periodic acid-Schiff (LFB-PAS) staining, ionized calcium-binding adapter molecule 1 (Iba-1) immunohistochemistry, and 3,3’-diaminobenzidine (DAB)-enhanced Turnbull staining were performed on a subset of slices containing lesions. Details of the staining protocols are provided in the supplementary material. Digital pictures of the stained slides were generated using a Zeiss Observer 1 microscope (ZEN blue software (Zeiss, Thomwood, NY) at 10× optical zoom with a pixel resolution 0.7 μm, and exported as TIFF files.
MRI-histopathological comparison
A semi-quantitative comparison was performed by selecting 11 different lesions (9 focal and 2 subpial) on MRI and rating their corresponding histological images. A contour line for each lesion was first drawn on each histological image using the following procedure. MRI images were first coregistered to the corresponding LFB-PAS histology images using thin-plate splines with control points initialized manually (MIPAV, NIH). Lesions were then segmented on MRI images by using an intensity level threshold. The corresponding contour line was defined using the ROI tool and propagated to the histological images. The contrast between lesion and GM (Contrastlesion-to-GM = Signal intensitylesion/Signal intensityGM) was computed and reported for these selected lesions. The level of demyelination inside the contoured lesions was rated on LFB-PAS using the following grading system: 0 = normal myelin density; 1 = reduced myelin density; 2 = completely demyelinated. The level of inflammation was rated on Iba-1 using the following grading system: 0 = absent/few inflammatory cells; 1 = moderate density of inflammatory cells; 2 = high density of inflammatory cells.
A more quantitative comparison between MRI and histopathology was performed using regression models (see “Statistical analysis”) on the coregistered ex vivo MRI, LFB-PAS, and Iba-1 images. To generate data for fitting the statistical models, a total of 92 ROIs were drawn manually on LFB-PAS images using ImageJ at different cross-sectional cord locations (WM, GM, and lesion) of several sections (8–14 ROIs per section) (Supplementary Figure 3). ROIs were then propagated to the corresponding MRI images and Iba-1 staining. From each staining, the function “color deconvolution” (ImageJ) was applied to extract the channel corresponding to the stain of interest (LFB channel for the myelin component of LFB-PAS and DAB channel for the inflammatory cells of Iba-1). Mean signal intensity for each ROI was computed on both MRI and grayscale-deconvoluted staining images.
Statistical analysis
The nonparametric two-tailed Mann Whitney test (Prism v7.0b) was performed to test any association between the Contrastlesion-to-GM values and histopathology scores. To assess the association between MRI and histopathology signals and identify which variables influenced the MRI signal, we performed regression models and evaluated different statistical models, summarized in Table 2. In all models, the independent variable was the histopathology and the dependent variable was the MRI signal intensity. Two linear mixed-effects models (M1 and M2) were used with random effect of section, to account for the correlated observations taken from the same section. A simple linear model M0 was run for completeness. M2, the most complex model, included an interaction term to test the hypothesis that the relationship between MRI and pathology depends on the tissue type (NAWM, GM, lesions). We evaluated these different regression models for both LFB (called the myelin model) and Iba-1 (called the inflammation model) by comparing their Akaike Information Criteria (AIC)17 values; the best model was considered to have the lowest AIC value. Because AIC values are ordinal and give a rank of the model goodness-of-fit, the ΔAIC values were reported as well: ΔAIC(n) = AIC(n) – AICmin where n denotes the model of interest and AICmin the minimum AIC of all the models tested. Regression results were considered significant for p≤0.05. The coefficient of determination, R2, was reported to evaluate the goodness-of-fit using the method of Nakagawa et al. adapted for the linear mixed-effects models18. Statistical analysis and graphical visualization were performed using R version 3.3.3 and Rstudio (1.0.143). The lme4 package version 1.1-16 was used to fit the mixed-effects model using restricted maximum likelihood estimation.
Table 2:
Summary of the different regression models applied to the data with their respective Akaike information criteria results for the different myelin models (using the LFB-PAS histology) and inflammation models (using the Iba-1).
| Models | Dependent Variable | Fixed-Effects | Interaction Term | Random Effect | Myelin model | Inflammation model | ||
|---|---|---|---|---|---|---|---|---|
| AIC | ΔAIC | AIC | ΔAIC | |||||
| M0 | MRI | Histopathology, tissue type | No | No | 1640.6 | 91.6 | 1618.4 | 20.1 |
| M1 | MRI | Histopathology, tissue type | No | Section | 1557.8 | 8.8 | 1598.3 | 0 |
| M2 | MRI | Histopathology, tissue type | Yes | Section | 1549 | 0 | 1599.3 | 1 |
Table subheading: AIC: Akaike information criteria
Results
All immunized animals developed clinical signs between 65 and 100 days post-immunization and reached EAE scores ≥2.5 at the time of sacrifice. Disease duration (from immunization to death) ranged from 71 to 159 days (mean: 113 days) (Table 1). All four EAE animals displayed SC abnormalities on postmortem MRI images (Figure 1). Focal WM lesions with various sizes and shapes were observed at different cord levels (Figure 1, red arrows). A total of 85 focal WM lesions were segmented on the MRI images across the four EAE animals. Thirty-four lesions (40%) were found at the cervical level, 40 (47%) at the thoracic level, and 11 (13%) at the lumbar level (Table 3). Fifty-seven lesions (67%) were classified as small (average volume: 0.028 mm3), 24 lesions (28%) as medium-sized (0.11 mm3), and 4 lesions (5%) as large (2.54 mm3) (Table 3). Sixty-nine focal WM lesions (81%) were located in the lateral WM, fifteen lesions (18%) were located in the posterior WM, and only 1 lesion was found in the anterior WM (Table 3). Subpial WM lesions were also observed along the entire cord, with variable thickness (Figure 1, green arrows). Three animals (M#1, M#2, and M#3) displayed thin subpial WM lesions (relative thickness <21%) throughout their entire cords, whereas one animal (M#4) showed thicker subpial WM lesions (relative thickness >39%) in the lumbar portion of the cord (Table 4).
Figure 1.
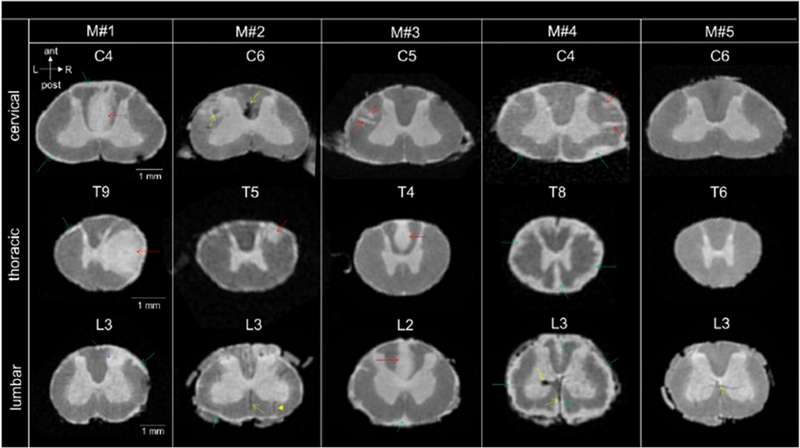
High-resolution postmortem MRI scans of four marmoset spinal cords with EAE (M#1, M#2, M#3, M#4) and one healthy control (M#5) showing cervical, thoracic, and lumbar spinal cord levels. Anatomical orientation axes are included (white arrows): anterior (ant), posterior (post), left (L) and right (R). The heterogeneity of the focal lesions (red arrows) and subpial lesions (green arrows) in terms of size, shape and location can be observed across the marmosets with EAE. Areas of hypointense signal (yellow arrows) were sometimes observed in these animals (M#2, C6 and M#4, L3), most likely originating from blood deposits (see Supplementary Figure 4). Parenchymal vessels with deoxygenated blood could also be visualized as dark thin lines (yellow dotted arrows, L3 for M#2, M#4 and M#5), sometimes running centrally through a focal lesion (plain yellow arrow, M#2, L3). Note the mass effect produced by a large tumefactive lesion (M#1, T9, red arrow) that involves most of the right lateral column and some of the GM. Another lesion affecting the GM (purple arrow) coud also be found within the dorsal horn at the lumbar level (M#1, L3). Various degree of confluence for the subpial lesions can be appreciated. For three animals (M#1, M#2, M#3), subpial lesions were subtle and not confluent compared to animal M#4 that displayed partially confluent (C4) and completelely confluent at the lower levels of the SC (T8 and L3).
Table 3:
Characterization of the focal lesions by spinal cord level, size, and white matter location in the four marmosets with EAE.
| Animal | M#1 | M#2 | M#3 | M#4 | Average across marmosets | |
|---|---|---|---|---|---|---|
| Number of focal WM lesions by cord level | cervical | 11 | 5 | 10 | 8 | 8.5 |
| thoracic | 9 | 10 | 10 | 11 | 10 | |
| lumbar | 1 | 2 | 5 | 3 | 2.8 | |
| Number of focal lesions classified by size and corresponding average volume [STD] in mm3 | small | 10 ; 0.025 [0.013] | 9 ; 0.033 [0.014] | 21 ; 0.028 [0.009] | 17 ; 0.026 [0.013] | 14 ; 0.028 [0.01] |
| medium | 9 ; 0.14 [0.1] | 8 ; 0.13 [0.089] | 2 ; 0.078 [0.003] | 5 ; 0.086 [0.022] | 6 ; 0.12 [0.08] | |
| large | 2 ; 4.18 [0.59] | 0 | 2 ; 0.9 [0.68] | 0 | 1 ; 2.54 [1.96] | |
| Number of focal WM lesions by location | Posterior | 9 | 1 | 3 | 2 | 3.8 |
| Left lateral | 8 | 10 | 20 | 10 | 6.9 | |
| Right lateral | 3 | 6 | 2 | 10 | 5.3 | |
| Anterior | 1 | 0 | 0 | 0 | 0.25 | |
Table subheading: WM: white matter; STD: standard deviation
Table 4:
Characterization of the subpial lesions as a percentage of the total white matter for selected spinal levels in the four marmosets with EAE.
| Animal | M#1 | M#2 | M#3 | M#4 | Average across marmosets | |
|---|---|---|---|---|---|---|
| Relative thickness (% of white matter) of subpial lesions by cord level | Cervical C7 | 18 | 15 | 5 | 30 | 17 |
| Thoracic T7 | 16 | 20 | 11 | 54 | 25 | |
| Lumbar L3 | 19 | 14 | 10 | 39 | 21 |
Anatomical features observed on MRI were visually matched on histological sections in control and EAE animals (Figure 2). In the control animal, both MRI and histology depicted normal WM with dark gray (denoted here as “isointense”) MRI signal (Figure 2A1), normal LFB staining (Figure 2B1), and absence of “reactive” inflammatory cells on Iba-1 (Figure 2C1). In the EAE animals, focal and subpial WM cord lesions detected as areas of hyperintense MRI signal (Figure 2A2-A4) corresponded to areas of clear demyelination characterized by low LFB (Figure 2B2-B4) staining, as well as intense inflammation demonstrated by strong Iba-1 staining (Figure 2C2-C4).
Figure 2.
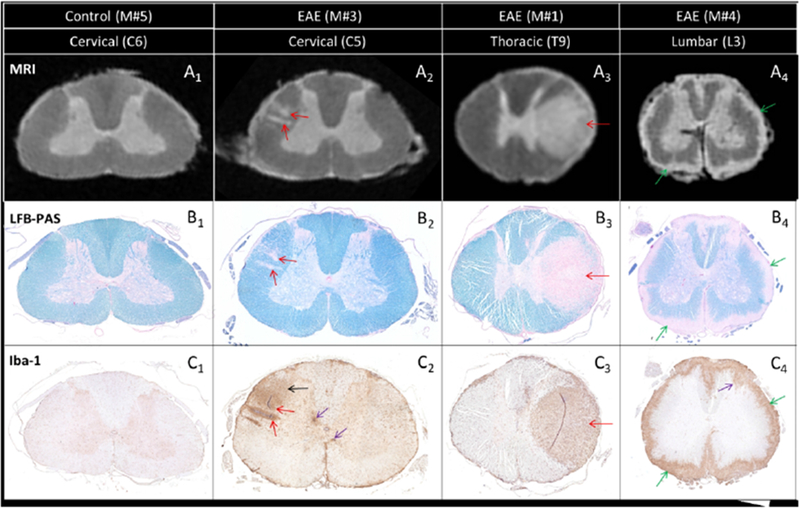
Comparison between MRI scans (A) and histopathology sections (B and C) from a control animal (M#5) and three EAE animals (M#3, M#1, and M#4). Two small focal lesions detected by MRI in the cervical section (A2, red arrows) correspond to two small demyelinated areas (B2, red arrows) with intense inflammatory cell infiltration (C2, red arrows). Note also the more diffuse inflammatory cell infiltration located above the two lesions, not captured by MRI (C2, black arrow). The large tumefactive focal lesion detected by MRI in the thoracic section (A3, red arrow) matches an area of demyelination (B3, red arrow) and inflammatory cell infiltration (C3, red arrow). A similar correspondence between MRI and histopathology is observed at the lumbar level (A4, B4, C4) for the subpial lesion (green arrows). Although not visible on the MRI, inflammation can be found within the GM (purple arrows) either infiltrating the dorsal horn (C4) or seen as small foci (C2).
The semi-quantitative analysis performed on a subset of 11 lesions revealed heterogeneity in myelin content and density of inflammation (Supplementary Table 2). This heterogeneity is illustrated by representative examples of a completely demyelinated lesion (Figure 3A2-3) with intense inflammation (Figure 3A4) and a partially demyelinated lesion (Figure 3B2-3) with moderate inflammation (Figure 3B4). The semi-quantitative analysis also indicated an association between MRI signal and myelin content/density of inflammatory cells (Supplementary Table 2). Completely demyelinated lesions (score = 2) showed a significantly higher Contrastlesion-to-GM value than lesions with partial demyelination (score = 1) (Figure 4). A similar trend was observed when comparing lesions with high density of Iba-1 cells (score = 2) against lesions with a moderate density of cells (score = 1), although the difference in Contrastlesion-to-GM values was not significant in this case (Figure 4).
Figure 3.
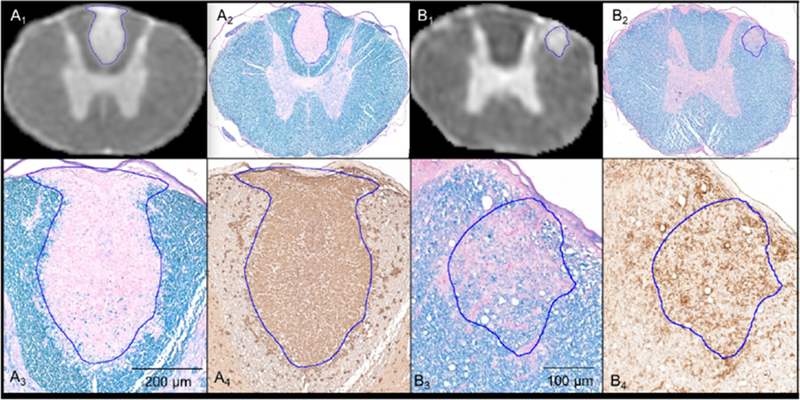
MRI scans (A1 and B1) coregistered to the corresponding LFB-PAS images (A2 and B2) for defining the lesion contour ROI (blue lines). Magnified views of the lesions for LFB-PAS (A3 and B3) and Iba-1 (A4 and B4). A3 and A4 show complete myelin loss and uniform intensity of inflammatory cells throughout the entire lesion. B3 and B4 show a partially demyelinated lesion with moderate inflammation.
Figure 4.
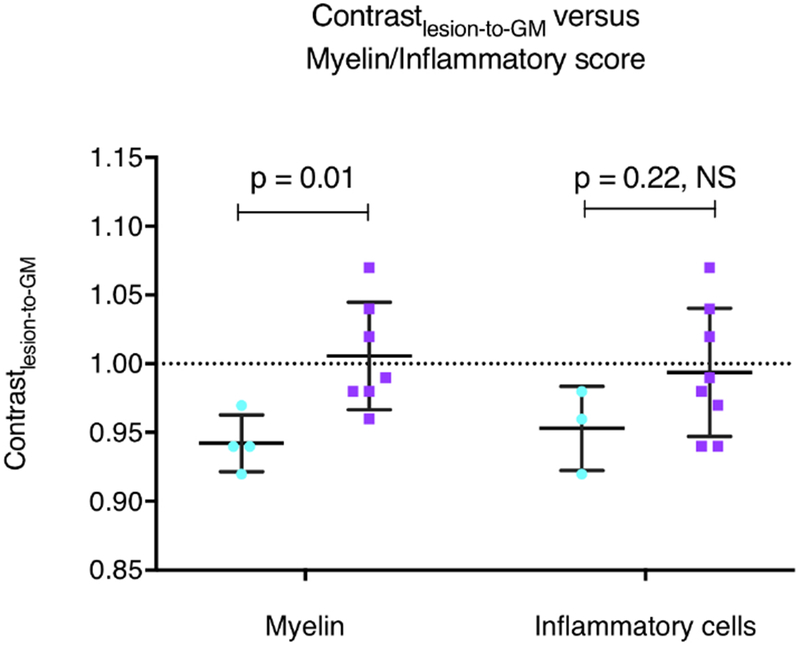
Contrastlesion-to-GM values for each white matter lesion (n=11) grouped according to histopathological scores for myelin and inflammatory cells (light blue: score = 1; purple: score = 2).
The quantitative analysis performed on the individual 92 ROIs revealed a lower AIC value when an interaction term for tissue type and random effect for sections were added in the myelin model (M2) (see Table 2). Figure 5 illustrates the importance of the random effect when predicting MRI values. In the M2 regression model, R2 of 0.96 indicated that 96% of the variability in the MRI data were explained by the predictors (fixed and random effects) in this model (Supplementary Figure 5). P-values and mean slope coefficients from the linear mixed-effects model M2 are summarized in Supplementary Table 3. Significant and strong positive associations were found between LFB and MRI values for both lesional and GM tissues. No significant association was seen between the LFB and MRI values in the NAWM, but the variation in MRI signal intensity within NAWM was comparatively small. A negligible difference in AIC values was observed when adding an interaction term in the inflammation models (Table 2). The lowest AIC value was obtained for M1, which includes random effects but no interaction term. No significant association was found between the Iba-1 and the MRI signal (slope [standard error] = −10 [10.9], p=0.35).
Figure 5.
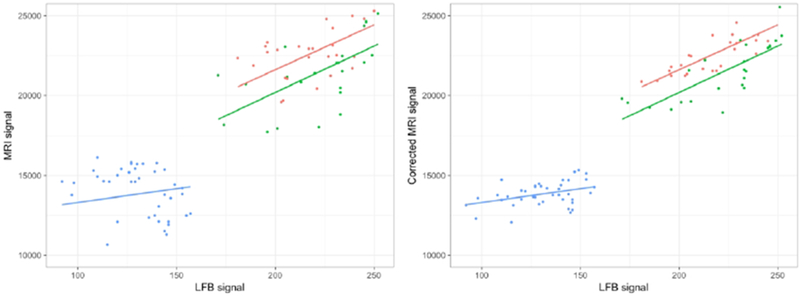
Left: relationship between the observed MRI signal intensity values and LFB signal intensity using myelin model M2. Each dot represents the individual ROI values according to their tissue type. The fitted line represents the estimated MRI values for fixed effect only. Right: representation of the random effect correction on the MRI value estimations. The corrected MRI signal was obtained by removing the estimated random effect value from the mixed effect model for each section. The residual variation, which is neither explained by fixed effect nor random effect, is shown by the distance between the individual dot and its corresponding value within the fitted line. Green: lesion; red: gray matter; blue: NAWM.
Discussion
In this study, we investigated the SC pathology in marmosets with EAE using a combined MRI-histopathological approach. By acquiring high-resolution ex vivo MRI, we first identified two types of lesions: (1) focal lesions located within the WM of the SC, (2) subpial lesions along the WM abutting the subarachnoid space. By comparing our ex vivo MRI images with histology, we then demonstrated that these lesions corresponded to areas of demyelination and marked inflammation.
Similar to focal MS lesions19,20, focal EAE lesions were found throughout the SC of the marmoset, with a prevalent localization in the lateral and posterior columns. We observed a heterogeneous distribution in terms of number and size of the focal lesions, which is also consistent with the lesion heterogeneity described in MS patients21. To our knowledge, only one histopathological study has reported the presence of diffuse subpial demyelination22. In our model, subpial EAE lesions were distributed throughout the animal cords and showed variable thicknesses between animals. Note that these subpial lesions were difficult to visualize by postmortem MRI without performing first a laminectomy. Indeed, the tissues (bones, fat, and muscles) surrounding the SC generated magnetic susceptibility-related artifacts that hid these subtle lesions located at the edge of the cord (Supplementary Figure 6). Thus, we hypothesize that the technical issues described above combined with comparatively low resolution MRI might explain why subpial cord lesions have not been yet reported by MRI in MS patients. A recent in vivo imaging study of the cervical cord in MS patients found a decrease in magnetization transfer ratio values for the outermost voxels of the SC,23 which could correspond to the subpial pathology observed in our study, but this would need to be verified histopathologically.
For lesion types, our histopathological evaluation revealed variable degrees of demyelination and inflammatory cell densities, which most likely reflects different stages of lesion development, as recently demonstrated for marmoset brain lesions6. Note that the small focal inflammatory demyelinated lesions appeared to follow parenchymal blood vessels, whereas the larger subpial inflamed demyelinated lesions detected in our model are consistent with a leptomeningeal inflammatory origin, as is postulated to be the case for subpial cortical lesions24,25. Interestingly, meningeal inflammation has already been reported in MS SC22,26.
By performing a quantitative comparison between MRI and histopathology using regression models, we established a strong association between the (T2*-weighted) MRI signal intensity and the amount of myelin derived from the LFB-PAS stain intensity level within the GM and lesional areas. By contrast, the association between the MRI signal and the density of inflammatory cells was weaker (Figure 4). The use of a linear mixed-effects model allowed us to improve the prediction of MRI values by correcting for the correlation within section (Figure 5). These results highlight the fact that T2*-weighted MRI signal can be highly sensitive to demyelination (although not specific) and corroborate previous postmortem studies investigating MRI and its pathological correlates in the MS SC27.
Important limitations of this study were the use of a single, postmortem timepoint with a single MRI contrast (T2*-weighted). The postmortem timepoint illuminates the SC lesion load when symptoms are severe (20% weight loss or paraplegia), but cannot provide spatiotemporal context of the pathology. Additionally, because we were not able to scan these spinal cords in vivo, we have no information regarding the age and the blood-brain-barrier integrity of the lesions analyzed here. Both of these processes can play a role in the development of vasogenic edema and expansion of the interstitial space, which can also impact signal intensity on postmortem MRI. Finally, our MRI sequence was optimized to provide the best delineation between lesions and NAWM. Therefore, lesions affecting the GM of the SC could not be easily identified by MRI in this study, although we detected GM pathology by histology in some of our sections (Figures 2 and 3, purple arrows).
Conclusions
In this study, we investigated SC tissues from marmosets with EAE showing symptoms suggestive of SC damage. Using a combined MRI-histopathological approach, we established the existence of two types of WM SC lesions: focal WM lesions with characteristics (location, distribution, and size) overlapping those found in human MS SC, and subpial WM lesions, which so far have not been reported in MS. We found that both focal and subpial EAE lesions detected by MRI corresponded to areas of demyelination and marked inflammation. Our findings underscore the relevance of this nonhuman primate model of MS for understanding mechanisms of lesion formation in the SC. Future work is needed to investigate the existence of SC subpial lesions in MS as well as in vivo marmoset SC imaging to unravel the spatiotemporal development of SC pathology.
Supplementary Material
Acknowledgments
The authors wish to thank Cecil Chern-Chyi Yen who helped with troubleshooting the scanner used in this study; Sam Antonio and James O’Malley in the NIH animal facility for marmoset care and project support; and Gina Norato, NINDS statistician, for her help to apply the linear mixed-effect models.
This research was supported by the Intramural Research Program of the NIH, NINDS. This research was presented in part at the 24th annual meeting of the International Society for Magnetic Resonance in Medicine (Singapore, 2016).
Funding
This research was supported by the Intramural Research Program of the National Institute of Neurological Disorders and Stroke.
Abbreviations:
- EAE
experimental autoimmune encephalomyelitis
- MS
multiple sclerosis
- SC
spinal cord
- LFB
Luxol fast blue
- Iba-1
ionized calcium-binding adapter molecule 1
- 7T
7-tesla
- T2*w
T2*-weighted
Footnotes
Declaration of conflicting interest
J.A.L, J.R.G, N.J.L., S.K.H., E.L., M.D.S, A.C.S., S.J., S.L, D.S.R, P.S, declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Baxter AG. The origin and application of experimental autoimmune encephalomyelitis. Nat Rev Immunol. 2007; 7: 904–12. [DOI] [PubMed] [Google Scholar]
- 2.Linker RA and Lee DH. Models of autoimmune demyelination in the central nervous system: on the way to translational medicine. Exp Transl Stroke Med. 2009; 1: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Massacesi L, Genain CP, Lee-Parritz D, et al. Active and passively induced experimental autoimmune encephalomyelitis in common marmosets: a new model for multiple sclerosis. Ann Neurol. 1995; 37: 519–30. [DOI] [PubMed] [Google Scholar]
- 4.Kap YS, Laman JD, ‘t Hart BA. Experimental autoimmune encephalomyelitis in the common marmoset, a bridge between rodent EAE and multiple sclerosis for immunotherapy development. JNeuroimmune Pharmacol. 2010;5(2):220–30. [DOI] [PubMed] [Google Scholar]
- 5.Absinta M, Sati P, Reich DS. Advanced MRI and staging of multiple sclerosis lesions. Nat Rev Neurol. 2016;12(6):358–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maggi P, Macri SM, Gaitan MI, et al. The formation of inflammatory demyelinated lesions in cerebral white matter. Ann Neurol. 2014;76(4):594–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hart BA, Bauer J, Muller HJ, et al. Histopathological characterization of magnetic resonance imaging-detectable brain white matter lesions in a primate model of multiple sclerosis: a correlative study in the experimental autoimmune encephalomyelitis model in common marmosets (Callithrix jacchus). Am J Pathol. 1998;153(2):649–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.‘t Hart BA, Massacesi L Clinical, pathological, and immunologic aspects of the multiple sclerosis model in common marmosets (Callithrix jacchus). J Neuropathol Exp Neurol. 2009;68(4):341–55. [DOI] [PubMed] [Google Scholar]
- 9.Laman JD, van Meurs M, Schellekens MM, et al. Expression of accessory molecules and cytokines in acute EAE in marmoset monkeys (Callithrix jacchus). J Neuroimmunol. 1998;86(1):30–45. [DOI] [PubMed] [Google Scholar]
- 10.Villoslada P, Hauser SL, Bartke I, et al. Human nerve growth factor protects common marmosets against autoimmune encephalomyelitis by switching the balance of T helper cell type 1 and 2 cytokines within the central nervous system. J Exp Med. 2000;191(10):1799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stroman PW, Wheeler-Kingshott C, Bacon M, et al. The current state-of-the-art of spinal cord imaging: methods. Neuroimage. 2014;84:1070–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guy JR, Sati P, Leibovitch E, et al. Custom fit 3D-printed brain holders for comparison of histology with MRI in marmosets. J Neurosci Methods. 2016;257:55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee NJ, Ha SK, Sati P, et al. Spatiotemporal distribution of fibrinogen in marmoset and human inflammatory demyelination. Brain. 2018;141(6):1637–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kap YS, Smith P, Jagessar SA, et al. Fast progression of recombinant human myelin/oligodendrocyte glycoprotein (MOG)-induced experimental autoimmune encephalomyelitis in marmosets is associated with the activation of MOG34-56-specific cytotoxic T cells. J Immunol. 2008;180(3):1326–37. [DOI] [PubMed] [Google Scholar]
- 15.Tustison NJ, Avants BB, Cook PA, et al. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging. 2010;29(6): 1310–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watson C, Sengul G, Tanaka I, et al. The spinal cord of the common marmoset (Callithrix jacchus). Neurosci Res. 2015; 93:164–75. [DOI] [PubMed] [Google Scholar]
- 17.AKAIKE H (1973). Information theory and an extension of the maximum likelihood principle In 2nd International Symposium on Information Theory, Ed. Petrov BN and Csaki F, pp. 267–81. Budapest: Akademia Kiado. [Google Scholar]
- 18.Nakagawa S and Schielzeth H A general and simple method for obtaining R2 from generalized linear mixed-effects models. Methods Ecol Evol, 2013. February 1;4(2):133–42 [Google Scholar]
- 19.Oppenheimer DR. The cervical cord in multiple sclerosis. Nenropathol Appl Neurobiol. 1978;4(2):151–162. [DOI] [PubMed] [Google Scholar]
- 20.Weier K, Mazraeh J, Naegelin Y, et al. Biplanar MRI for the assessment of the spinal cord in multiple sclerosis. Mult Scler. 2012; 18(ll):1560–9. [DOI] [PubMed] [Google Scholar]
- 21.Nijeholt GJ, Bergers E, Kamphorst W, et al. Post-mortem high-resolution MRI of the spinal cord in multiple sclerosis: a correlative study with conventional MRI, histopathology and clinical phenotype. Brain. 2001;124(Pt 1): 154–166. [DOI] [PubMed] [Google Scholar]
- 22.Androdias G, Reynolds R, Chanal M, et al. Meningeal T cells associate with diffuse axonal loss in multiple sclerosis spinal cords. Ann Neurol. 2010;68(4):465–76. [DOI] [PubMed] [Google Scholar]
- 23.Kearney H, Yiannakas MC, Samson RS, et al. Investigation of magnetization transfer ratio-derived pial and subpial abnormalities in the multiple sclerosis spinal cord. Brain. 2014;137(Pt 9):2456–2468. [DOI] [PubMed] [Google Scholar]
- 24.Reich DS, Lucchinetti CF, Calabresi PA. Multiple Sclerosis. N Engl J Med. 2018;378(2):169–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi SR, Howell OW, Carassiti D, Magliozzi R, Gveric D, Muraro PA, et al. Meningeal inflammation plays a role in the pathology of primary progressive multiple sclerosis. Brain. 2012;135(Pt 10):2925–37. [DOI] [PubMed] [Google Scholar]
- 26.DeLuca GC, Alterman R, Martin JL, et al. Casting light on multiple sclerosis heterogeneity: the role of HLA-DRB1 on spinal cord pathology. Brain. 2013;136(Pt 4):1025–34. [DOI] [PubMed] [Google Scholar]
- 27.Schmierer K, McDowell A, Petrova N, et al. Quantifying multiple sclerosis pathology in post mortem spinal cord using MRI. Neuroimage. 2018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


