Abstract
Cys-loop ligand-gated ion channels mediate rapid neurotransmission throughout the central nervous system. They possess agonist recognition sites and allosteric sites where modulators regulate ion channel function. Using strychnine-sensitive glycine receptors, we identified a scaffold of hydrophobic residues enabling allosteric communication between glycine-agonist binding loops A and D, and the Zn2+ inhibition site. Mutating these hydrophobic residues disrupted Zn2+ inhibition, generating novel Zn2+ activated receptors and spontaneous channel activity. Homology modelling and electrophysiology revealed that these phenomena are caused by disruption to three residues on the ‘–’ loop face of the Zn2+ inhibition site, and to D84 and D86, on a neighbouring β3 strand, forming a Zn2+ activation site. We provide a new view for the activation of a Cys-loop receptor where, following agonist binding, the hydrophobic core and interfacial loops reorganise in a concerted fashion to induce downstream gating.
The Cys-loop ligand-gated ion channel superfamily includes nicotinic acetylcholine (nAChR), γ-aminobutyric acid type A (GABAA), glycine (GlyR) and serotonin type 3 (5HT3) receptors. Each subunit of these pentamers contains: a ligand-binding extracellular domain (ECD), formed by a sandwich of two β-sheets; a four α-helical membrane-spanning domain; and an intracellular region of unspecified quaternary structure 1. The interior of the ECD is hydrophobic2,3, and, as for most globular proteins, it is considered to be an entropic stabiliser of protein folding 4. Given the presumed stability of this hydrophobic core and its location between two sheets of rigid β-strands, it is usually regarded as a relatively inflexible structure. Thus, after agonist-induced activation, the core would move, if at all, as a rigid body 5. Accordingly, it would be the agonist-binding loops A-F, supported by the surrounding rigid β-strands, that would undergo a conformational change upon agonist binding to trigger rigid body movement and downstream channel opening2, 6, 7, 7–11. An alternative view, based on structural and modelling data, suggests that substantial portions of the inner and outer β-sheets of the ECD shift their orientation relative to one another upon receptor activation1, 12. Given the location of the hydrophobic core between the inner and outer β-sheets of each ECD it would then be expected that the core would reorganise, rather than move as a rigid body, to facilitate the reorientation of the β-sheets 13–17. This movement may induce separation of important charge interactions along neighbouring receptor subunit interfaces, allowing the ECDs to twist and induce downstream channel opening8, 16, 18, 19.
An ideal model system to investigate the role of the hydrophobic core in Cys-loop receptor activation is that involving Zn2+ inhibition of the GlyR. These receptors readily form homomers that are modulated by the physiological cation Zn2+ in a biphasic fashion. Zn2+ can be found in nanomolar concentrations in external medium and is also packaged into vesicles and released at synapses in sufficient amounts to endogenously modulate GlyRs, with low micromolar concentrations potentiating submaximal glycine responses, and higher concentrations causing inhibition 20–22. Two Zn2+ binding sites have been identified; the potentiation site is contained solely on the outer β sheet of the ECD 23, whereas the inhibition site spans neighbouring subunits on the inner β sheet of the ECDs 24,25,26 (Fig. 1a-c). Inhibition by Zn2+ of GlyR function involves the stabilisation of charge interactions between neighbouring subunit ECD interfaces, thereby hindering their movement. This supports the notion that charge separation of neighbouring ECD interfaces is necessary for receptor activation, and that agonist binding must transduce a signal near to the Zn2+ inhibition site to evoke a conformational change in this area leading to receptor activation. As the hydrophobic core is located between the glycine binding site and the Zn2+ inhibition site, identifying the molecular requirements for Zn2+ inhibition will elucidate the roles of the hydrophobic core and the subunit ECD interface in receptor function.
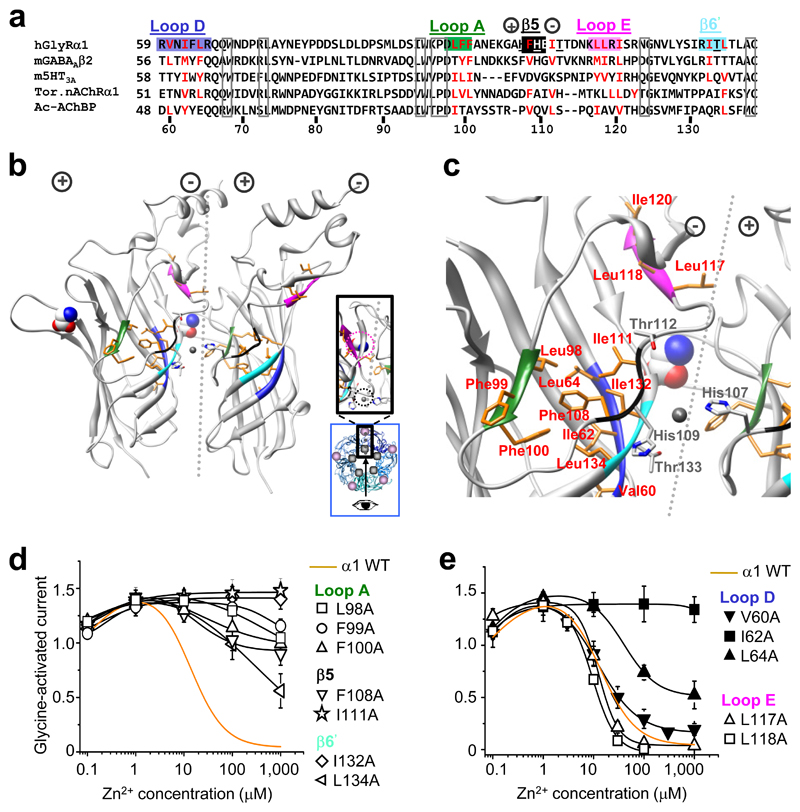
Figure 1. Hydrophobic determinants of Zn2+ inhibition.
(a) Partial protein alignment of Cys–loop receptor ECDs showing: GlyR α1 hydrophobic residues that are examined (red); where hydrophobicity is retained in other Cys–loop receptors (red); conserved residues (boxed); and the GlyR Zn2+ inhibition binding site residues (underlined). Hydrophobic motifs are labelled and colour–coded in accordance with (b, c), the GlyR α1 ECD homology model based on TornAChRα1 1. Clustered hydrophobic residues (orange) connect the Zn2+ inhibition site at the inner–subunit interface (black and light blue motifs – note, black motif is not a β–strand in the model but referred to as such in main text for clarity 2), to glycine binding loops A (green), D (dark blue) and E (mauve) of outer–subunit interface. Zn2+ inhibition site residues, His107, His109, Thr112 and Thr133, are in grey (nitrogen atoms are blue and oxygens are red). Dotted line depicts the interface with (+) and (–) subunit sides 28. Glycine, displayed in CPK, spacefill, was fitted manually in accordance with 35; Zn2+, fitted manually, in grey, spacefill. Exact side chain orientations presented here are speculative, based on best alignment and modelling estimations. Inset, blue box – pentamer plan view showing viewing angle (arrow) for main picture, and revealing the separation between the Zn2+ binding site (grey circle) and glycine binding site (mauve circle); enlarged plan view of interface shown in black box. (d, e) Zn2+ concentration response curves for modulation of EC50 glycine currents on wild–type (WT; dashed line) and alanine–substituted receptors. α1I120A did not traffic to cell surface so no recordings were made (Supplementary Figure 2). Curves fitted with Hill equation. All points are means ± s.e.m. (n = 3–6).
Here, we demonstrate that a cluster of residues forming a scaffold across the hydrophobic core are critical for Zn2+ inhibition and spontaneous opening of the human GlyR ion channel. Spontaneous opening was attributed to the apparent flexibility of a loop on the ‘–’ face of the Zn2+ inhibition site, which is exquisitely-sensitive to the molecular composition of the hydrophobic core. Disruption of this loop and the discovery of novel elements in a neighbouring β3 strand that are also important for receptor activation, demonstrate that charge redistribution at the ECD inner subunit interface is a key component of GlyR activation.
Results
Hydrophobic residues are required for Zn2+ inhibition
A GlyR homology model was constructed to guide our site-directed mutagenesis studies into receptor activation. The GlyR protein sequence was first aligned with other Cys-loop receptors, revealing that the region encompassing the GlyR Zn2+ inhibition site is not conserved across any of the Cys-loop receptors, for which atomic resolution ECD templates are available, and furthermore, it contains a 2 – 3 amino acid insertion. Structural alignment of three ECD template structures (conotoxin-bound Aplysia californica acetylcholine binding protein, (Ctx-Ac-AChBP 27); Torpedo (Tor) nAChR α1 1; and mouse nAChR α1 subunit 14), revealed substantial structural variation at the two loops of the ‘+’ and ‘–’ face 28 flanking the β5 strand (nomenclature of Brejc 2; Supplementary Fig. 1), which corresponds to the Zn2+ inhibition site. Using the variable loop regions as insertion points for the extra residues in our GlyR alignment (Fig. 1a) allowed us to generate a GlyR homology model (MODELLER-9.2 29; based on the TornAChR α1 template) with His107 and His109 exposed at the subunit interface 25 and Phe108 solvent accessible 30, in accord with published data.
To probe the link between Zn2+ inhibition and GlyR activation, hydrophobic residues positioned between the Zn2+ inhibition site, defined by His107, His109, Thr112 and Thr133 24, 26, 31, and the three closest agonist binding loops (A, D and E; Fig. 1b,c), were substituted with alanine. Substituted GlyRs were expressed in human embryonic kidney (HEK) cells and their sensitivities to Zn2+ potentiation and inhibition assessed by whole-cell recording of glycine (EC50) evoked responses in the presence of increasing concentrations of Zn2+. Substituting residues at the Zn2+ inhibition site ‘–‘ face (β5 F108A, I111A; β6’ I132A, L134A) and the loop A face (L98A, F99A, F100A), substantially reduced sensitivity to Zn2+ inhibition compared to wild-type, whilst Zn2+ potentiation remained the same (Fig. 1d,e). Substitutions in agonist binding loop D (V60A, I62A, L64A) also reduced Zn2+ inhibition with I62A causing ablation. This residue is orientated in the GlyR model towards other residues required for Zn2+ inhibition from loop A (Leu98, Phe99, Phe100) and those from β5 and β6’ adjoining the Zn2+ inhibition site (Phe108, Ile111, Ile132, Leu134). By contrast, substituting distally-located hydrophobic residues in agonist-binding loop E (Leu117 and Leu118; Fig. 1e and Table 1), and other hydrophobic residues located away from loops A/D and β5/6’ (Supplementary Fig. 3 and Supplementary Table 1), did not disrupt Zn2+ inhibition. All the substituted receptors with attenuated Zn2+ inhibition retained glycine EC50s within 10-fold of the wild-type (Table 1) and comparable maximal glycine currents (Imax) and Hill slopes, with the exception of α1F99A (Imax = 3.9 ± 0.4 nA; wild-type GlyR Imax = 6.8 ± 0.5 nA; P < 0.01).
Table 1.
Glycine activation, Zn2+ modulation and Zn2+ activation data from wild-type (WT) receptors and from GlyRs carrying alanine substitutions of hydrophobic residues at positions running from the Zn2+inhibition site to nearby agonist binding loops, expressed in HEK cells.
| Glycine | Zn2+ | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Inhibition | Activation | ||||||||
| EC50 (μM) |
nH |
Imax (nA) |
N | IC50 (μM) |
EC50 (μM) |
nH | Relative efficacy (% Gly Imax) |
N | |
| α1 WT | 35 ± 5 | 2.7 ± 0.2 | 6.8 ± 0.5 | 6 | 15 ± 2 | None | — | — | 4 |
| Agonist binding loop A | |||||||||
| α1L98A | 35 ± 6 | 1.9 ± 0.3 | 6.4 ± 0.5 | 6 | > 1000 | 0.26 ± 0.08 | 1.1 ± 0.2 | 49 ± 5 | 4 |
| α1F99A | 250 ± 40 | 2.3 ± 0.3 | 3.9 ± 0.4 | 4 | > 1000 | 0.13 ± 0.03 | 1.1 ± 0.1 | 86 ± 13 | 5 |
| α1F100A | 120 ± 20 | 1.8 ± 0.2 | 4.5 ± 0.7 | 5 | > 1000 | 9.2 ± 0.6 | 0.5 ± 0.1 | 7 ± 4 | 4 |
| Zn2+ binding site: β5 strand | |||||||||
| α1F108A | 15 ± 3 | 2.3 ± 0.1 | 4.5 ± 0.5 | 5 | > 1000 | 2.3 ± 0.6 | 0.7 ± 0.1 | 25 ± 6 | 6 |
| α1I111A | 39 ± 4 | 1.3 ± 0.2 | 4.6 ± 0.8 | 4 | > 1000 | 470 ± 130 | 0.7 ± 0.2 | 83 ± 3 | 3 |
| Zn2+ binding site: β6’ strand | |||||||||
| α1I132A | 95 ± 15 | 2.7 ± 0.4 | 6.1 ± 0.7 | 6 | > 1000 | 140 ± 30 | 1.2 ± 0.1 | 72 ± 5 | 5 |
| α1L134A | 58 ± 3 | 3.0 ± 0.4 | 6.0 ± 0.3 | 7 | > 1000 | 0.06 ± 0.01 | 1.3 ± 0.1 | 38 ± 9 | 5 |
| Agonist binding loop Δ | |||||||||
| α1V60A | 91 ± 15 | 3.5 ± 0.8 | 6.5 ± 1.3 | 4 | 10.1 ± 1.26 | None | — | — | 4 |
| α1I62A | 240 ± 7 | 2.5 ± 0.1 | 7.6 ± 0.7 | 3 | > 1000 | > 1000 | — | 3 ± 1 | 4 |
| α1L64A | 10 ± 3 | 2 ± 0.6 | 5.6 ± 0.8 | 4 | 70 ± 9 | 1.5 ± 1.3 | 1.1 ± 0.4 | 10 ± 3 | 3 |
| Agonist binding loop E | |||||||||
| α1L117A | 4200 ± 100 | 1.67 ± 0.1 | 4.4 ± 0.7 | 8 | 13.3 ± 3.3 | None | — | — | 3 |
| α1L118A | 1060 ± 110 | 3.00 ± 0.2 | 6.3 ± 0.7 | 3 | 10.3 ± 0.36 | None | — | — | 3 |
Removing Zn2+ inhibition creates Zn2+-activated GlyRs
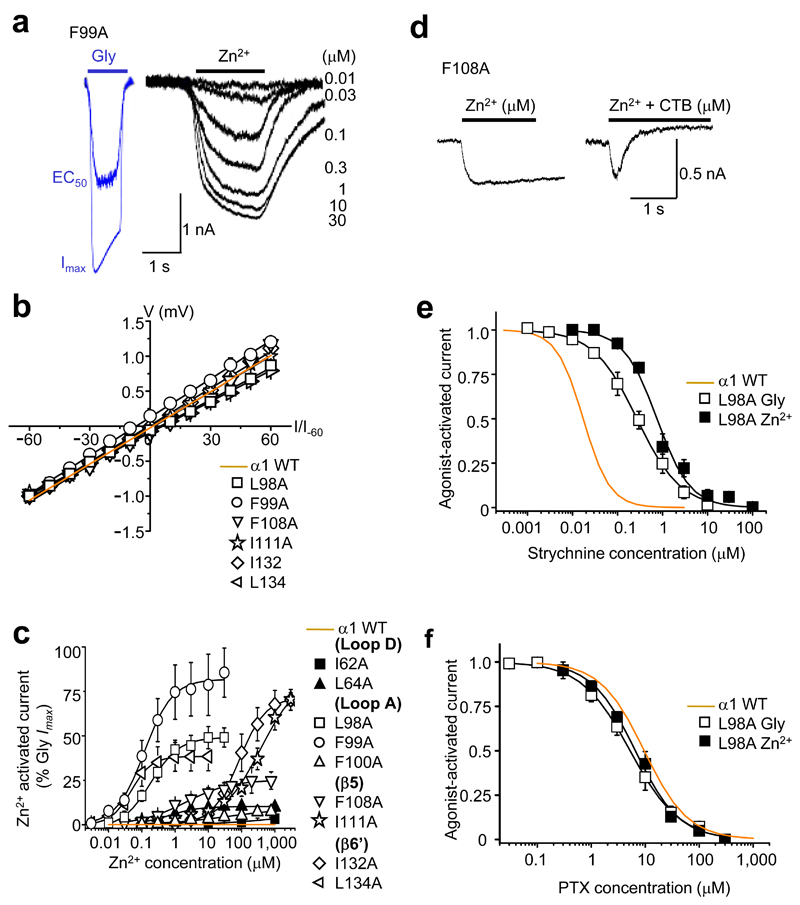
Although Zn2+ does not activate wild-type GABAA or glycine receptors, it generated inward currents at GlyRs with impaired Zn2+ inhibition, i.e., those with alanine substitutions in the glycine binding loops A/D and Zn2+ inhibition binding strands β5/6’ (L98A, F99A, F100A, F108A, I111A, I132A, L134A, I62A and I64A; Fig. 2a). The Zn2+-activated currents reversed close to ECl (0.4 ± 1.4 mV, n = 5), and the current-voltage relationships were comparable to those for glycine-activated Cl- currents at wild-type GlyRs (Fig. 2b). Zn2+ concentration response curves revealed that the potency and relative efficacy (maximal Zn2+ response as a percentage of maximal glycine response in the same cell), varied substantially between the substituted receptors (Fig. 2c and Table 1). α1L134A exhibited the highest sensitivity to Zn2+ (EC50 = 0.06 ± 0.01 μM, n = 5), whilst α1F99A exhibited the highest relative efficacy (86 ± 13%, n = 5). By contrast, α1I62A supported only 3 ± 1% maximal activation with 1 mM Zn2+ (Fig. 2c and Table 1). Entirely consistent with Zn2+ activating the substituted GlyRs, the anion-selective channel blocker, cyanotriphenylborate (CTB, 20 µM), abolished the Zn2+-activated currents (Fig. 2d). Furthermore, both strychnine, a selective GlyR competitive antagonist, and picrotoxin (PTX), a GABAAR and GlyR allosteric blocker, also inhibited Zn2+ activation (Fig. 2e,f). This prompted the classification of these substituted GlyRs as Zn2+-activated GlyRs (ZAGs).
Figure 2. Direct Zn2+ activation of substituted GlyRs.
(a) Representative glycine currents (blue traces) in the absence of Zn2+ (EC50 – 30 μM; Imax – 1000 μM) and Zn2+ currents (black traces) in absence of glycine (0.01 – 30 μM) for GlyR α1F99A. (b) Zn2+ current–voltage (I–V) relationships (normalised to the current recorded at –60 mV and fitted by linear regression) for the six most efficacious ZAGs, where EC50 Zn2+ responses were large enough to be recorded reliably, and for the wild–type (WT) receptor. (c) Zn2+ activation–response curves for alanine–substituted GlyRs; Zn2+ maxima are normalised to glycine maxima (10 mM) in same cell. (d) Representative α1F108A Zn2+ Imax (100 μM) current co–applied with and without 20 μM CTB. Inhibition greater than 100% is due to additional background leak (discussed later in Fig. 6). (e, f) α1L98A concentration inhibition curves for strychnine and picrotoxin, respectively on glycine or Zn2+ EC50–activated currents (n = 3 –6). Wild–type receptor activation sensitivities to the antagonists are shown in orange.
Hydrophobicity and sensitivity to Zn2+ inhibition
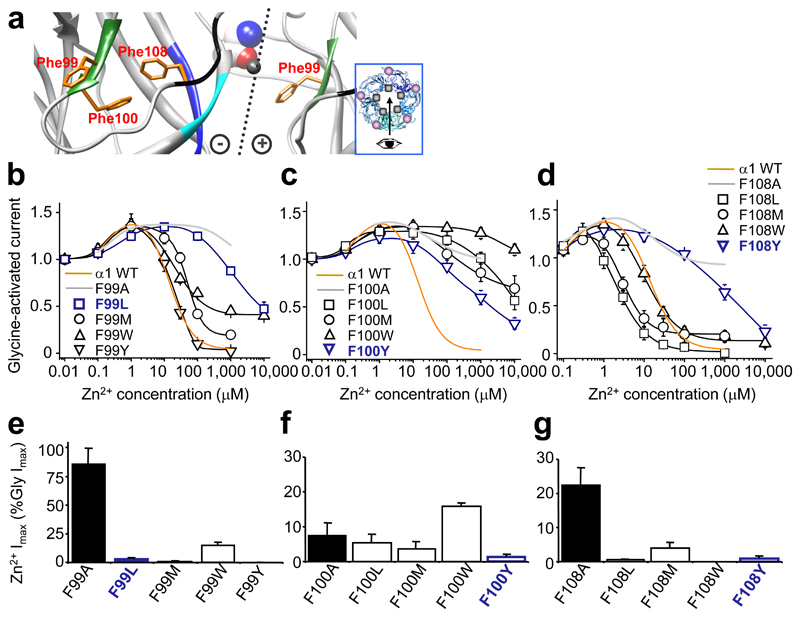
To investigate the ZAGs further, we substituted loop A, Phe99 and Phe100, and β5 Phe108 (Fig. 3a) individually with either: tyrosine or tryptophan – both aromatic like Phe; or leucine or methionine – aliphatic. Although many substitutions reduced Zn2+ inhibition (Fig. 3b-d), only some generated ZAG behaviour (Fig. 3e-g). Of the three Phe residues, Phe100 is the most critical for maintaining wild-type sensitivity to Zn2+ inhibition; however, there was no correlation between the properties of the substituting residue and the disruption to Zn2+ inhibition (Fig. 3b-d and Supplementary Table 2), suggesting each position has unique chemical and physical requirements.
Figure 3. Zn2+ inhibition and Zn2+ activation in GlyRs with conservative substitutions.
(a) Homology model showing Phe99 and Phe100 from glycine binding loop A (green) and Phe108 from β5 strand of Zn2+ inhibition site (black). Dotted line: interface with (+) and (–) subunit sides 28. Glycine (CPK, spacefill) was docked manually 35; Zn2+ (representation only, grey spacefill). Inset – pentamer plan viewing angle (arrow), Zn2+ binding site (grey spot), glycine binding site (mauve spot). (b–d) Zn2+ concentration response curves for modulation of EC50 glycine currents from GlyRs with conservative hydrophobic substitutions at positions Phe99 (b), Phe100 (c), and Phe108 (d). For comparison, wild–type (WT) inhibition profiles (orange) and alanine substituted receptor profiles (grey) are included. The same set of substituted GlyRs were assessed for maximal Zn2+ (1 mM)–activated currents (Imax), normalised to glycine Imax (10 mM) in the same cell: Phe99 (e), Phe100 (f) and Phe108 (g). Alanine substituted receptor Zn2+ Imax (black bars) is shown for comparison. Note: F99L, F100Y and F108Y had substantially attenuated Zn2+ inhibition (navy blue lines) but almost no Zn2+ activation (navy blue bars; < 2% Gly Imax). (n = 3 – 6)
Glycine EC50s for α1 Phe100 and Phe108 substituted receptors remained within two-fold of wild-type with the exceptions of α1F100Y and α1F108W (increased 15- and 25-fold, respectively; P<0.05; Supplementary Table 2). For Phe99 substituted receptors, glycine EC50s were significantly increased for α1F99L, α1F99W and α1F99Y (3- to 30-fold; P<0.05), but surprisingly the α1F99M receptor was 6-fold more sensitive to glycine (wild-type EC50 = 35 ± 5 μM; α1F99M EC50 = 5.5 ± 0.5 μM, n = 4 – 6; P<0.05), suggesting an important role for this residue in determining glycine binding. Interestingly, the GlyR model positions Phe99 facing into the glycine binding site (Fig. 1b,c).
Zn2+ activation originates from a novel binding site
The switch from Zn2+ inhibition to activation in ZAGs could have arisen if the function of an existing modulatory Zn2+ binding site was altered enabling activation in response to Zn2+ binding. However, substitution of Zn2+ binding residues with non-Zn2+ coordinating alanines at either the Zn2+ inhibition or potentiation sites revealed that neither site was required for Zn2+ activation (Supplementary Fig. 4). However, given that Zn2+ inhibition was severely compromised in ZAGs, we reasoned that regions bordering the inhibition site may have become structurally perturbed, sufficient to form a new Zn2+ activation site.
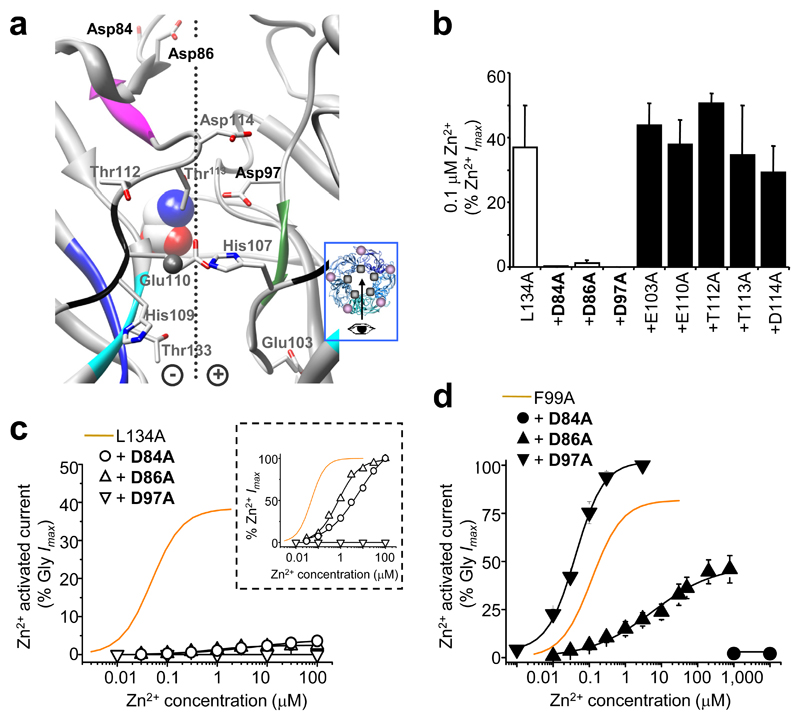
On an α1L134A ZAG background (most Zn2+-sensitive ZAG), potential Zn2+ coordinating residues neighbouring the Zn2+ inhibition site were substituted with alanine and assessed for activation by 0.1 μM Zn2+ (EC70 for α1L134A; Fig. 4a,b). Of these substitutions, Asp84, Asp86 (strand β3) and Asp97 (loop A) virtually abolished Zn2+ activation from 38 ± 9% (α1L134A) to 5 ± 1% (α1L134A, D84A) and 2.4 ± 1.2% (α1L134A, D86A), with no detectable activation for α1LL134A, D97A (Fig. 4c). The Zn2+ EC50s for α1L134A, D84A and α1L134A, D86A were increased 120- and 15-fold, respectively (Fig. 4c inset); whilst glycine EC50s were only shifted 2-fold (Supplementary Table 3). Notably, using a wild-type receptor background, the substitutions D84A, D86A or D97A, caused only modest (< 2-fold) changes in GlyR sensitivity to Zn2+ inhibition and potentiation (Supplementary Fig. 5 and Supplementary Table 4).
Figure 4. Identifying potential residues for the Zn2+ activation binding site in ZAGs.
(a) Homology model showing potential Zn2+ coordinating residues at the subunit interface around the His107 /His109 /Thr112 /Thr133 Zn2+ inhibition site. Side chains in grey, nitrogens in blue, oxygens in red. Dotted line: interface with (+) and (–) sides 28. Glycine (CPK, spacefill) was docked manually 35; Zn2+ (representation only, grey, spacefill). Inset shows the viewing angle (arrow), with Zn2+ (grey spot) and glycine binding sites (mauve spot). (b) 0.1μM Zn2+–activation of alanine–substituted α1L134A background GlyRs, expressed as a percentage Zn2+ Imax in the same cell. (c) Zn2+ activation concentration response curves for α1L134A, D84A, α1L134A, D86A and α1L134A, D97A receptors normalised to the Gly Imax and also normalised to the Zn2+ Imax (inset). (d) Zn2+ activation response curves for the equivalent mutations on an α1F99A background showing that although Asp84 and Asp86 are still required for Zn2+ activation, Asp97 is not. (n = 3–6).
According to the GlyR homology model (Fig. 4a), Asp84 and Asp86 are positioned approximately 17 Å from Asp97, which is too far for the three residues to coordinate a single Zn2+ ion 32. Furthermore, Asp97, which is conserved across the Cys-loop receptor family, probably supports loop B via the carboxyl side chain 33, precluding its involvement in Zn2+ binding. To establish the importance of Asp84, Asp86 and Asp97 for Zn2+ activation, alanine substitutions were also made on an alternative ZAG background, α1F99A (most efficacious ZAG). Substituting Asp84 or Asp86 again substantially reduced the sensitivity to Zn2+ activation, but substituting Asp97 was ineffective (Fig. 4d). Thus, the role of Asp97 in Zn2+ activation is more complex than just directly binding Zn2+.
Zn2+ activation site is not a potentiation site
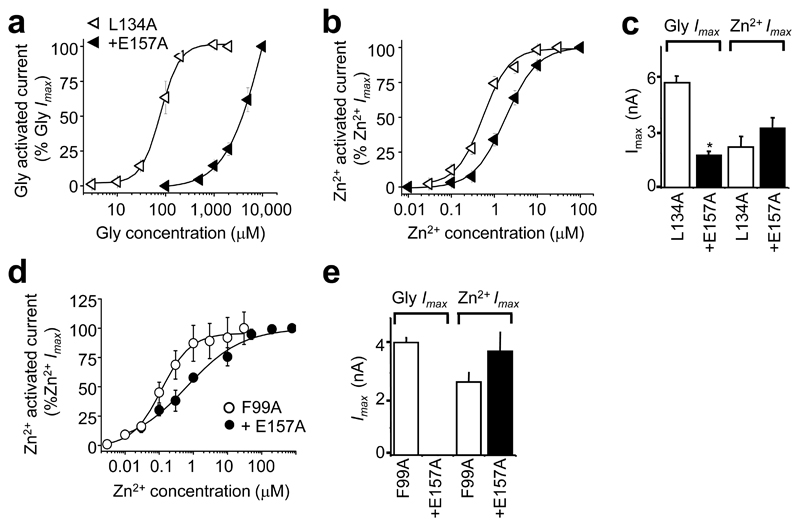
As reagents and water are ubiquitously contaminated with glycine (~ 50 nM 34), it is feasible that Asp84/Asp86 may actually form part of a (second) Zn2+ potentiation site, rather than an activation site. Occupancy of this site by Zn2+ would then enhance the receptor’s sensitivity to glycine allowing activation by very low contaminating glycine concentrations. To address this, an α1L134A ZAG background was used with an extra mutation, E157A on glycine binding loop B35, to produce a receptor with 50-fold reduced sensitivity to glycine. The threshold concentration for glycine was now >100 μM (Fig. 5a) and 2000-fold higher than the predicted level of glycine contamination. Nevertheless, the α1L134,E157A ZAG showed only a modest 3-fold reduced sensitivity to Zn2+ activation (Fig. 5b) and retained comparable maximal responses to Zn2+ (Fig. 5c). Using an alternative F99A background, α1F99A,E157A yielded identical Zn2+ sensitivity to α1F99A, despite being insensitive up to 10 mM glycine (Fig. 5d,e). Thus, in the absence of glycine-mediated activation, Zn2+ activation is still apparent, suggesting it originates from a pure activation site, not an additional Zn2+ potentiation site.
Figure 5. Zn2+–activation of GlyRs lacking a high affinity glycine binding site.
Glycine (a) and Zn2+ (b) concentration response curves on an α1L134A background with an extra mutation in the glycine binding site (E157A). (c) Maximum currents evoked by glycine (10 mM) and Zn2+ (1 mM) for α1L134A and α1L134A, E157A. Incorporating E157A on an α1F99A background, ablated activation by up to 10 mM glycine, but induced only a modest, 3–fold decrease in sensitivity to Zn2+ activation (d) and no reduction in Zn2+ Imax (1 mM) (e) (n = 3–4).
ZAGs exhibit spontaneous channel activity
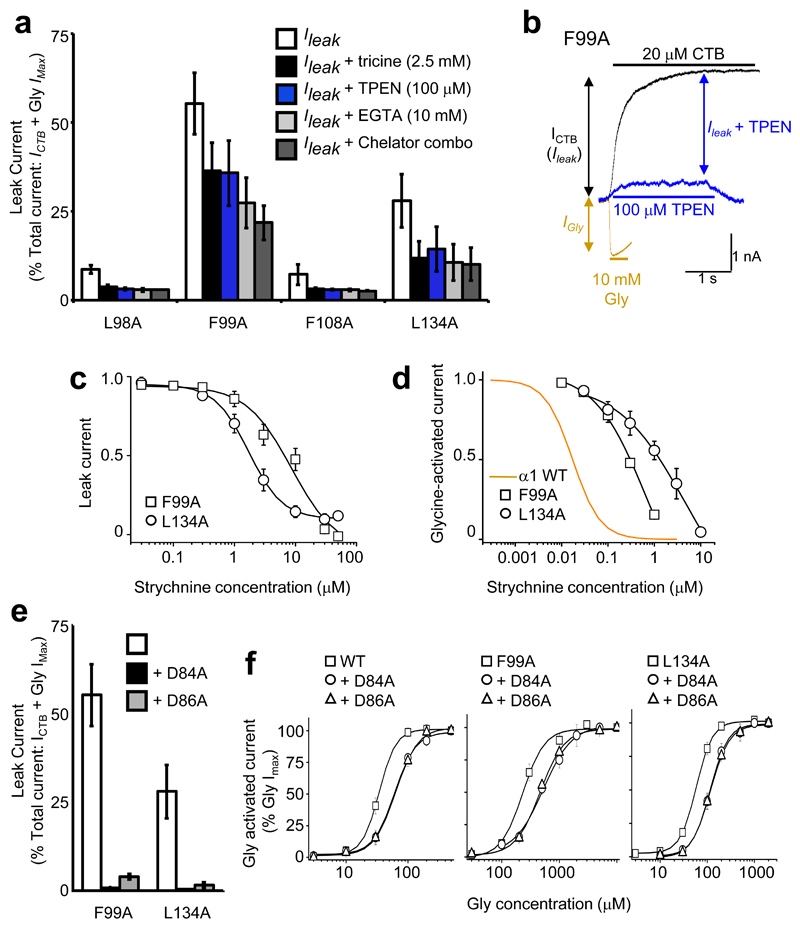
HEK cells expressing the most sensitive ZAGs, α1L98A, α1F99A, α1F108A and α1L134A, all exhibited sizable (0.5 – 3 nA) leak currents. A minor component was caused by Zn2+ contamination of the external solution (~200 nM 36) activating the ZAGs, as this was reduced by the Zn2+ chelators, tricine (2.5 mM) or N,N,N’,N’-tetrakis-(2-pyridylmethyl)-ethylenediamine (TPEN; 100 μM; Fig. 6a, b). The remaining component depended on spontaneous GlyR channel activity since it was abolished by CTB (20 μM) to less than 50 pA standing current (considered full abolition of receptor-mediated leak). Strychnine also attenuated the leak, by 100 % for ZAG α1F99A and by 90 ± 4% for α1L134A. Strychnine was 10-fold less potent inhibiting the leak current compared to glycine-activated currents (Fig. 6c, d).
Figure 6. Spontaneous activation of alanine–substituted receptors.
(a) Persistent leak currents for alanine substituted GlyRs (= % CTB blockable current (ICTB)/(ICTB + Gly IMax)) in presence of individual and combined ion chelators. (b) Membrane currents for α1F99A ZAG in the presence of either glycine (orange), CTB (black) or TPEN (blue), overlaid to demonstrate their relative contributions to the total current (c) Strychnine concentration curves for inhibiting leak currents for α1F99A and α1L134A ZAGs (c) and EC50 glycine–activated current (d). Orange line indicates strychnine concentration inhibition curve for WT GlyRs, which are notably more sensitive as the agonist binding loops A and D are not perturbed. (e) Persistent leak current absent from receptors with alanine substitutions at Asp84 or Asp86 on α1F99A and α1L134A backgrounds. (f) Glycine sensitivity is modestly reduced in alanine–substituted Asp84 or Asp86 receptors, as compared to wild–type, α1F99A and α1L134A backgrounds. n = 3 –6.
Interestingly, Asp84 and Asp86, which are important for Zn2+ activation, were also required for spontaneous activation with α1F99A, D84A, α1F99A, D86A, α1L134A, D84A, and α1L134A, D86A failing to exhibit spontaneous channel activity (Fig. 6e). Furthermore, Asp84 and Asp86 also influenced glycine-induced receptor activation, as alanine substitutions induced a modest but consistent 2-fold reduction in glycine sensitivity of wild-type, α1F99A and α1L134A backgrounds (Fig. 6d and Supplementary Table 4). A double substituted receptor, α1D84A, D86A, was non-functional.
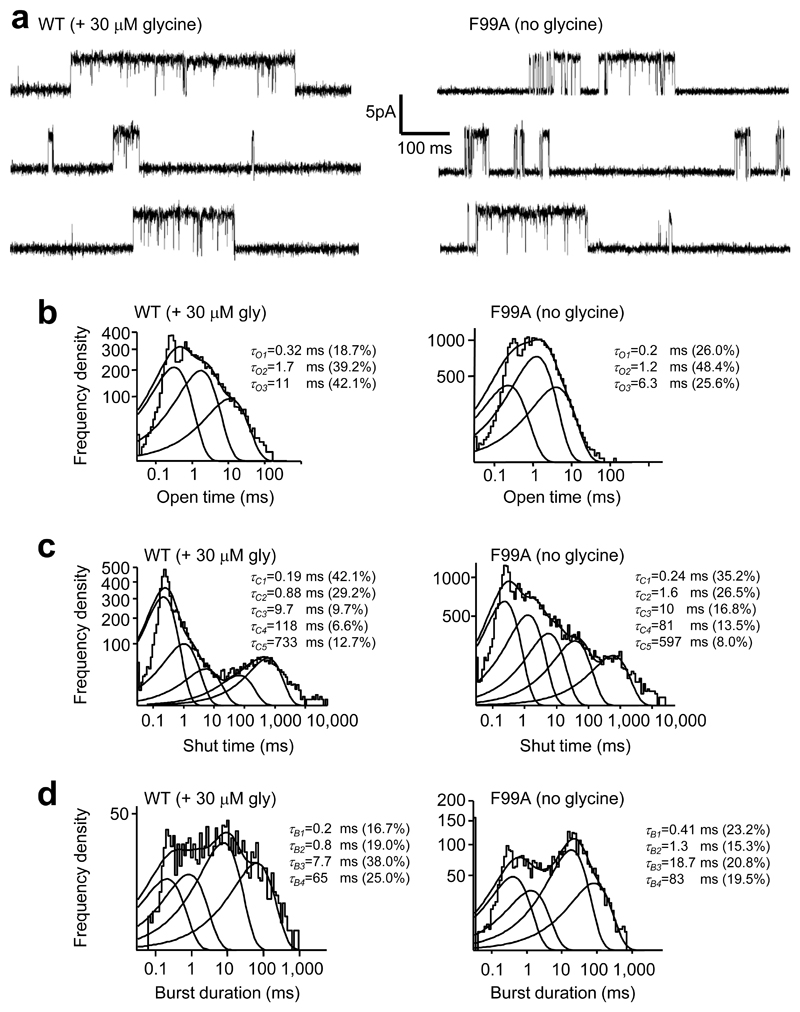
To investigate whether spontaneous activity mimics agonist-induced activity, single channel currents were recorded in cell-attached mode (pipette potential +60 mV) for wild-type channels activated by glycine (20 μM; EC30) and for spontaneously-gating α1F99A ZAGs without glycine. TPEN (100µM) was present throughout to remove any activation by contaminating Zn2+. The single channel currents for each receptor population were comparable at 4 – 5 pA with estimated conductances of ~ 60 pS 37 (Fig. 7a). The corresponding open time distributions were best fit by three Gaussian components with similar mean time constants and relative areas (P > 0.05; Fig. 7b and Table 2). The shut time distributions required five Gaussian components giving similar time constants for both receptors, with the exception of τC2 and τC3, which were 2-fold higher for α1F99A receptors (Fig. 7c and Table 2). These changes will contribute to the lower open probability (PO) for clusters of openings at α1F99A (0.53 ± 0.07) compared to wild type (0.9 ± 0.03, n = 3) channels. With regard to the burst duration distributions, four Gaussian components were required with comparable time constants and relative areas, except for τB3, which was 2-fold longer for α1F99A (Fig. 7d).
Figure 7. Spontaneous channel activation mimics agonist–induced activation.
(a) Single channel currents from cell–attached recordings of HEK cells expressing either WT α1 GlyRs or GlyR α1F99A, at a pipette potential +60 mV. Burst activity was recorded in the presence of glycine (EC30) for WT and in its absence for spontaneously–activated α1F99A. TPEN (100 μM) was present in the pipette solution to remove contaminating Zn2+. (b–d) Dwell time distributions for (b) open times, (c) shut times, and (d) burst durations.
Table 2.
Data from cell-attached single-channel recordings made from α1 wild-type (WT) or α1F99A receptors expressed in HEK 293 cells. Includes: average durations of open, closed and burst time constants and areas of exponential components that fitted the distributions; single channel amplitudes; PO for openings within bursts. n = 3. * denotes significant variations between the two receptor populations (P<0.05).
| α1 WT (+30 μM Gly) |
α1F99A (no glycine) |
|||
|---|---|---|---|---|
| Open times | τO (ms) | Area (%) | τO (ms) | Area (%) |
| 1 | 0.28 ± 0.029 | 36 ± 11 | 0.4 ± 0.1 | 36.9 ± 9.3 |
| 2 | 1.5 ± 0.2 | 39 ± 3 | 1.7 ± 0.3 | 41.9 ± 4.0 |
| 3 | 8.9 ± 2.1 | 26 ± 10 | 5.2 ± 0.8 | 21.2 ± 7.2 |
| Closed times | τC (ms) | Area (%) | τC (ms) | Area (%) |
| 1 | 0.22 ± 0.03 | 44 ± 16 | 0.26 ± 0.026 | 37 ± 2 |
| 2 | 0.90 ± 0.24 | 35 ± 15 | 1.6 ± 0.17 * | 30 ± 3 |
| 3 | 4.3 ± 1.6 | 8 ± 1 | 10.0 ± 2.9 | 18 ± 1 * |
| 4 | 109 ± 6.2 | 10 ± 2 | 89 ± 39 | 12 ± 1 |
| 5 | 2200 ± 1000 | 4 ± 1 | 1200 ± 400 | 3 ± 3 |
| Burst durations | τB (ms) | Area (%) | τB (ms) | Area (%) |
| 1 | 0.3 ± 0.06 | 28 ± 7 | 0.3 ± 0.06 | 26 ± 2 |
| 2 | 1.4 ± 0.6 | 22 ± 4 | 1.4 ± 0.1 | 22 ± 8 |
| 3 | 6.9 ± 2.2 | 35 ± 5 | 17.6 ± 0.8 * | 32 ± 8 |
| 4 | 63.4 ± 14.8 | 19 ± 7 | 77.1 ± 10.7 | 20 ± 1 |
| Amplitude (pA) | 4.5 ± 0.7 | 4.1 ± 0.6 | ||
| PO | 0.9 ± 0.03 | 0.53 ± 0.07 | ||
The ‘–’ face affects Zn2+ activation and spontaneous activity
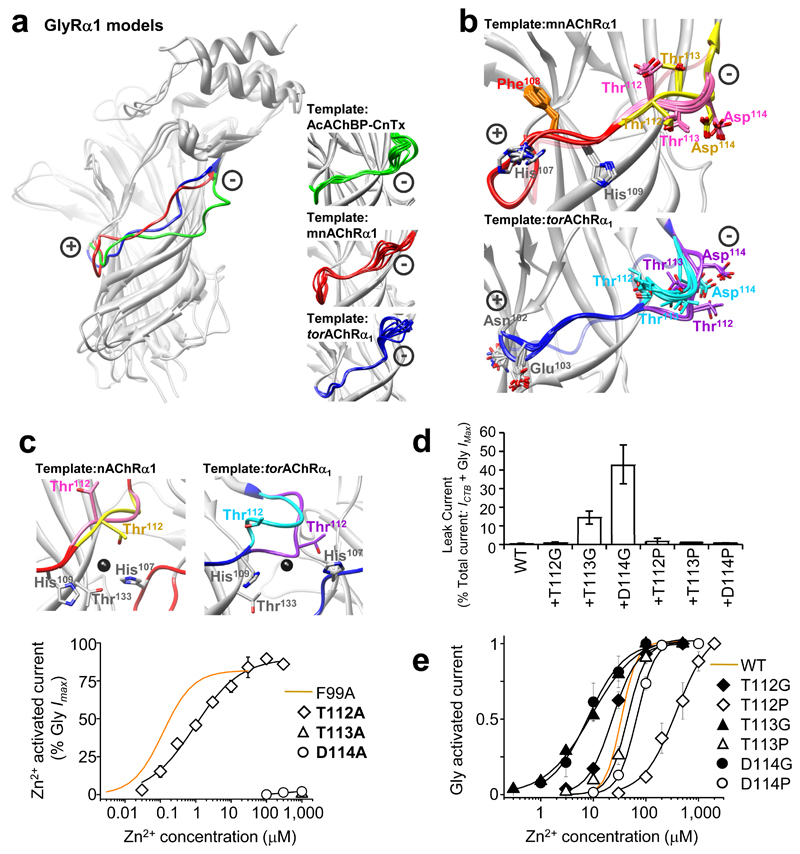
Conceivably, Zn2+ activation and spontaneous channel activity may arise if the substitutions of hydrophobic residues exert a common conformational effect on a region that undergoes critical movement during channel gating. As the Zn2+ inhibition site is perturbed in ZAGs, it is the ideal region to examine for conformational flexibility. The top ten GlyR homology models (lowest distant-dependent atomic statistical potential (DOPE)38 score from 100 models run in MODELLER-9.2) based on three related template structures, conotoxin-bound Ac-AChBP, TornAChR α1 and mnAChR α1 1, 14, 27, showed much greater structural variability at the ‘+’ and ‘–’ loop faces surrounding the β5 strand of the Zn2+ inhibition site, compared to other more rigid β-strands and across the structure as a whole (Fig. 8a, insets and Supplementary Table 5). By using DOPE loop modelling to optimise the structures of the GlyR ‘+’ and ‘–’ face loops, using TornAChR α1 and mnAChR α1 as templates, the ten best conformations for the ‘+’ loops were all comparable (Fig. 8b), whereas for the ‘–’ face loops, variable conformations were equally favoured with residues exhibiting multiple orientations at this location (Fig. 8b and Supplementary Table 6).
Figure 8. Zn2+ and spontaneous channel activation originates via the Zn2+ inhibition site ‘–’ loop face.
(a) CE structural alignments 55 of GlyR α1 homology models based on three predicted ‘closed’ conformation templates. Note, structural variability at the Zn2+ inhibition site (+) and (–) faces between the conotoxin–bound Ac–AChBP 27 (green), mnAChR α1 14 (red) and TornAChR α1 1 (blue) templates. (a – insets) Overlays of 10 lowest energy models for each template reveal particular uncertainty over the fitting of the (–) face but less uncertainty for the (+) face. (b) 10 lowest energy loop conformations resulting from DOPE loop refinement 38 of the (+) and (–) Zn2+ inhibition site loops starting from the best GlyR α1 model based on two different templates. This revealed one favourable conformation for the (+) face loop for each template–derived model, with only minor deviations in the peptide backbone (red) and side chain positions between models. In contrast, multiple peptide backbone conformations were equally preferred for the (–) face loop (pink or yellow for mnAChR α1; aquamarine or purple for TornAChR α1) with clear divergence in side chain positions. Original model backbone conformations before loop refinement are shown in translucent red (mnAChR α1) and translucent blue (TornAChR α1). (c) Zn2+ activation–response curves from GlyRs with potential Zn2+ coordinating residues on the (+) and (–) inhibition site faces substituted with alanines; Zn2+ maxima normalised to glycine maximal current (10 mM) in same cell. Only α1T112A exhibited highly–sensitive and efficacious ZAG activity. ZAG α1F99A (orange line; originally from Fig. 2c) included for comparison. Insets: DOPE loop refinement of GlyR α1 models from mnAChR α1 and TornAChR α1 templates show how alternate favourable conformations on the (–) face loop could have a substantive impact on the organisation of Thr112 in the Zn2+ binding site. Zn2+ (fitted manually, dark grey, spacefill). (d) Significant spontaneous activity, measured as persistent leak currents (ICTB/(ICTB + Gly IMax)) in the presence of 100 μM TPEN, was observed for GlyR α1T113G and α1D114G where ‘–’ loop flexibility was increased, but not for GlyR α1T113P and α1D114P where flexibility was reduced. (e) Glycine concentration response curves for receptors with glycine or proline substitutions at positions 112, 113 or 114 in the ‘–’ loop. Glycine increased agonist sensitivity compared to proline substitution at the equivalent position. Glycine curve for WT receptors included for comparison (orange line; n = 3–7).
To corroborate the modelling data, polar residues Thr112, Thr113 and Asp114 on the apex of the ‘–’ face, the point of greatest variability between DOPE loop-fitted structures, were individually substituted with alanine and examined for Zn2+ activation and spontaneous activity. Although T112A yielded a highly-sensitive and efficacious ZAG (Fig. 8c), no single alanine substitution generated a spontaneously-active receptor (data not shown). We altered the apex flexibility of the ‘–’ face by individually substituting Thr112, Thr113 and Asp114 with either glycine to increase, or proline to reduce, backbone flexibility 39, 40. Whilst the proline-substituted receptors lacked spontaneous activity, two glycine-substituted receptors, α1T113G and α1D114G, exhibited 15 ± 4% and 43 ± 10% (n = 4 – 6) spontaneous activity, respectively (Fig. 8d). Furthermore, there was a 3–fold increase in glycine sensitivity for α1T113G (EC50 = 9 ± 2 μM) and α1D114G (EC50 = 10 ± 3 μM) compared to WT (EC50 = 35 ± 5 μM; n = 4–6); whereas the proline substituted receptors all exhibited reduced sensitivities to glycine (Fig. 8e). Thus, increasing the flexibility of the ‘–’ face around Thr112–Asp114 increased the propensity of GlyR to open spontaneously, and in response to agonist binding, while decreasing flexibility by proline substitution had the opposite effect.
Discussion
This study identifies a scaffold of hydrophobic residues in the GlyR that functionally link glycine binding loops A and D with the Zn2+ binding β5 and β6’ strands of the Zn2+ inhibition site. Exchanging the hydrophobic residues, but not others outside the scaffold, initiated spontaneous channel opening, severely attenuated Zn2+ inhibition, and enabled Zn2+ to act as a novel activator of GlyRs. This suggests the hydrophobic scaffold is pivotally involved in receptor activation by stabilising one or more closed GlyR conformations. This is achieved by regulating the ‘–’ loop face of the Zn2+ inhibition site, as specific substitutions of polar residues in the ‘–’ face produced receptors with the same properties to those generated by alanine substitutions in the hydrophobic scaffold.
Structurally linking two discrete ligand binding sites
The current view of Cys–loop receptor activation is that agonist binding at the interface between subunits induces a rearrangement of interacting residues allowing the ECDs to twist relative to one another. The newly–orientated loops at the bases of the ECDs then promote rearrangement of opposing transmembrane domains to open the channel 6, 16, 18, 41–44. By binding to its interfacial inhibitory site on GlyRs, Zn2+ stabilises subunit interfaces, preventing the ECDs from twisting and initiating activation. It is therefore plausible that by distorting the Zn2+ binding ‘–’ loop interface we will not only ablate Zn2+ inhibition, but also enable spontaneous channel activity, particularly if the distortion mimics the conformation that occurs in the activated receptor state. Thus, the attenuation of Zn2+ inhibition and appearance of spontaneous channel activation are intrinsically linked. The extent to which both properties are seen in mutated receptors, will depend on the degree by which each substitution perturbs the ‘–’ loop away from a closed towards an activated conformation.
Notably, the molecular pathway by which Zn2+ causes inhibition is entirely different to that for Zn2+ potentiation at GlyRs. The potentiation site resides very close to the Cys–loop where it may interact directly with Thr151 to facilitate channel gating 23. This negates the need for any interaction with the hydrophobic scaffold identified here, explaining why Zn2+ potentiation was unaffected in this study.
The molecular pathway identified here is the first to be described in a Cys–loop ligand–gated ion channel that functionally connects two distinct binding sites, linking the agonist binding site to downstream activation. The importance of the hydrophobic scaffold in mediating GlyR activation is emphasised by the common kinetics of spontaneously–active α1F99A and agonist–activated wild–type GlyRs. Specifically, for α1F99A, it is the alanine substitution that artificially perturbs loop A to induce activation, whilst for wild–type GlyRs it is presumably agonist binding that similarly perturbs loop A to cause activation. Although a critical role for loop A in directing receptor activation is evident for GABAARs 45 and nAChRs 46, loop C, possibly via transmission of a conformational change along the outside of the ECDs (β7, 9 and 10 strands), has also been suggested to mediate activation upon agonist binding 8, 10. Our data does not preclude this scenario, but advocates loop A as an important contributor to the conformational wave that precedes channel opening 47.
At the glycine binding site, Phe99 appears ideally positioned to directly influence the receptor’s sensitivity to glycine, possibly via a cation–π interaction 48. The action of Phe99 to induce GlyR activation in response to agonist binding may then be mediated via the hydrophobic scaffold and subsequent ‘–’ loop face of the Zn2+ inhibition site. Indeed, Phe99 probably does this via Leu98 and Phe100, which are predicted to face, opposite to Phe99, into the hydrophobic scaffold towards the residues supporting the ‘–’ face loop. Such an interaction with Phe99 would explain why Phe100 could also influence the receptor’s sensitivity to glycine (Supplementary Figure 6; Supplementary Table 2). The ability of Phe99 to influence important residues within the hydrophobic scaffold may explain why it produces the most efficacious ZAG and the most spontaneously–active receptors when substituted with alanine.
From the perspective of the polar residues at the Zn2+ site’s ‘–’ loop face, substituting Thr112 or Ile111 produced a receptor that was insensitive to Zn2+ inhibition (cf 25, 31) and capable of Zn2+ activation. Isoleucine 111 faces into the core, in close proximity with the other residues comprising the hydrophobic scaffold. Thus Thr112, via Ile111, is ideally located to act as a relay following perturbation of the hydrophobic scaffold. Sequential substitution of Thr113 and Asp114 within the ‘–‘ loop by glycine, but not by alanine or proline, also yielded spontaneously–active receptors. These residues must also be ideally located to respond to perturbations of the hydrophobic scaffold, with increased loop flexibility enabling the receptor to shift to an activated state, while imposed rigidity (e.g., proline insertion or when Zn2+ binds to stabilise this region) hinders receptor activation.
β5 loop movement during GlyR activation
Although we propose that the ‘–’ face loop undergoes a conformational change to facilitate receptor activation, comparative structural evidence does not, so far, support this idea. Overlaying crystal structures of Aplysia californica AChBP bound to either α–conotoxin PnIA (‘inactive conformation’) or HEPES (‘active conformation’ 27) does not reveal any variation around the corresponding ‘–’ face loop region in the GlyR model (Supplementary Figure 7). Furthermore, structurally aligning Torpedo nAChR α1 and α2 subunits (presumed closed conformation), compared to β, δ and γ subunits (presumed open) for the pentamer, reveals only a small degree of variability around the corresponding ‘–’ face loop region (Supplementary Figure 8). Of course, as static structures, it is possible that neither the HEPES–bound AChBP nor βδγ TorAChR subunits represent fully–activated receptors. Alternatively, they might undergo different conformational rearrangements after activation compared to GlyRs. Simulation studies on nAChRs also do not support movements in the ‘–’ face region 49, although the nanosecond timescales for these studies are as yet too short to encompass all conformational rearrangements in pentameric Cys–loop receptors.
Despite the caveats, the ‘–’ loop face of the GlyR Zn2+ inhibition site was predicted to adopt multiple conformations and side chain orientations with the potential to influence receptor function. Moreover, previous functional studies support a role for the ‘–’ face loop in the GlyR activation process, notably: Thr112 is important in determining partial agonist efficacy 50; it is accessible to Cys–scanning mutagenesis, resulting in dynamic disruption to agonist–evoked responses 30; and Zn2+ binds between subunits at the ‘–’ loop face to stabilise the GlyR closed conformation, suggesting this interface is mobile during receptor activation 25, 31.
Creating a Zn2+ activation site
The Zn2+ activation site was localised to Asp84/Asp86 on strand β3, directly above the ‘–’ face loop. Structural perturbation of the ‘–’ loop face may therefore have a knock–on effect on the β3 region, allowing Asp84/Asp86 to form a novel Zn2+ binding site that aids movement of the subunit interfaces, rather than hinders them, so inducing activation. This provides further evidence that charge dispersal at subunit interfaces plays an important role in regulating Cys–loop receptor excitability 16, 19, 46, 51, and also indicates that a dynamic interaction occurs between the ‘–’ loop face and the β3 strand to facilitate activation of wild–type receptors. The variable potency and efficacy of Zn2+ at different ZAGs further indicates that hydrophobic residues within the scaffold differentially affect the ‘–’ loop face, and consequently the juxtaposed β3 strand, so determining the efficiency with which Asp84/Asp86 forms a new Zn2+ activation site.
The general activation mechanism presented here for the GlyR is in accord with the hydrophobic scaffold and ‘–’ face loop dynamically responding to agonist binding. This provides a new vista on Cys–loop receptor activation whereby, during activation, the reorganisation of the hydrophobic scaffold and ‘–’ face facilitate the re–alignment of the inner and outer β–sheets relative to one another 12, 13, 16, 18. This then initiates movement of subunit interfaces, which is subsequently transmitted to the transmembrane domains for receptor activation 42.
Methods
cDNA constructs and mutagenesis
We used human (h) GlyR α1L cDNA constructs and the mutant cDNAs were prepared using the Stratagene Quikchange kit. Mutated cDNAs were sequenced using an ABI sequencer.
Cell culture, transfection and electrophysiology
By using a calcium phosphate transfection method (3 GlyR α1:1 eGFP) we expressed GlyR in HEK cells (ATCC CRL1573) grown on poly–L–Lysine–coated coverslips at 10% confluence. Whole–cell membrane currents were recorded after 24 h at 20–22°C from single HEK cells held at –40 mV using the patch clamp technique (Axopatch 200B, Molecular Devices). For rapid drug applications (exchange rate ~50–100 ms), we used a Y–tube. Patch electrodes (4 – 5 MΩ) were filled with (mM): 140 KCl, 2 MgCl2, 1 CaCl2, 10 HEPES, 11 EGTA, and 2 ATP, pH 7.2 (≈ 300 mOsm). External solution contained (mM): 140 NaCl, 4.7 KCl, 1.2 MgCl2, 2.5 CaCl2, 10 HEPES, and 11 D–Glucose, pH 7.4 (≈ 300 mOsm). For GlyRs exhibiting nanomolar sensitivities to Zn2+ activation, the tricine (2.5 mM; Zn2+ complexation KD = 10–5 M, ref. 52), was added to the saline to remove Zn2+ contamination 36. For single channel analysis, thick–walled electrodes were used (10 – 20 MΩ) and filled with external saline solution containing 100 μM TPEN, and 10 mM TEA to block endogenous potassium channel activity. Single–channel recordings were made in cell–attached mode at +60 mV pipette potential.
Data acquisition and analysis
Membrane currents were filtered using a high–pass Bessel filter at 3 kHz (–36dB per octave) and series resistance compensation was routinely achieved up to 70%. Data were recorded in 20 s epochs directly to a Pentium IV, 3.5 GHz computer into Clampex 8.0 via a Digidata 1322A (Axon instruments) sampling at 200 μs intervals. Due to Zn2+ activation in many of the receptors, Zn2+ inhibition profiles were measured by prolonged (4 s) co–application of Zn2+ with glycine, with response measurements being taken at the 4 s time point (to allow Zn2+ inhibition sufficient time to reach equilibrium 26). The digitised membrane current records were analysed offline using Axoscope 8.2. The concentration response relationships for glycine and Zn2+ were fitted with modified Hill equations as previously described 26.
For the single channel data analyses, stored pre–filtered (2.7 kHz Bessel) single channel data were digitised at 33 kHz prior to analysis. A fixed time resolution based on the dead time of the system was set at 80 μs. The analysis of the single channel current amplitudes was performed by fitting Gaussian components to the amplitude distributions to determine the mean single channel current, standard deviation and the total area of the component using a non–linear least–squares fitting routine. Single–channel conductances were calculated from the mean unitary current and the difference between the patch potential and glycine response reversal potential. The patch potential was estimated in cell–attached recordings, by estimating the cell membrane potential.
All open and shut durations were measured with a 50% threshold cursor applied to the main single channel current amplitude (WinEDR v2.8.9). The duration of events that were included in the analysis was not less than 200 μs before fitting the dwell time histograms. Frequency distributions were constructed from the measured individual open and shut durations and analysed by fitting a mixture of exponentials, defined by:
where Ai represents the area of the ith component to the distribution and τi represents the respective time constant. The areas, time constants and standard errors of the individual components of the distribution were determined. The burst duration analysis required the determination of a critical shut time (τcrit) 53 determined between the shut time constants, τC3 and τC4 by solving:
Channel open probability (PO) was calculated as the percentage of time that the channel spent in the open state within a cluster. All statistical comparisons used an unpaired t test and P<0.05 was considered significant.
Homology and loop modelling
We used ClustalW 54 to produce protein sequence alignments. Aplysia californica acetylcholine binding protein, Ac–AChBP (2br8 27; conotoxin–bound form), Torpedo (Tor)nAChR α1βδα2γ (2BG9) 1, and mouse (m)nAChR α1 (2QC1 14) were used for the Combinatorial Extension (CE) structural alignment method 55, which helped identify divergent regions in the GlyR α1 model. The final alignment reflected both alignment strategies. The Torpedo nAChR α1 subunit was selected as the final template structure to guide the homology modelling of the GlyR α subunit, as it has only two less residues around the β5 strand (the GlyR Zn2+ inhibition site), compared to three less residues for Ac–AChBP; and also, the structure of TornAChR α1 was determined as part of a pentamer, whereas mnAChR α1 was crystallized as a non–physiological monomer with several artificial point mutations 14. The TornAChR α1 pentamer was built by overlaying a second α1 subunit over the α2 subunit and then using Chimera 56, to build a five–fold symmetric pentamer. Using MODELLER–9.2 29, 100 hGlyR α1 models were generated with Cys bridges added into the agonist binding loop C (C198–C209), for the principle TornAChR α1 pentamer template, and also for the Ac–AChBP–conotoxin–bound pentamer and mnAChR α1 monomer. Side chain configurations were generated using SCWRL3 57. Fifty loops were generated using DOPE loop modelling in Modeller 9.2, for each loop before (‘+’) and after (‘–’) the β5 strand for each of the subunit templates (‘+’ loop residues 102NEKGAH107; ‘–’ loop residues 110EITTDN115). Models were evaluated using MolProbity 58 and gave good general agreement with each other. Uncertainty regarding the short β5 strand, ascribed as a β–strand in Ac–AChBP (1UW6) and nAChR α1 (2QC1), but not in TornAChR α1 (2BG9), was considered unimportant, as it had little effect on side chain positioning, and a PSIPRED 59 secondary structure prediction of the GlyR sequence gave low confidence for the presence of a β–strand, suggesting neither one nor other template was more likely to be correct. All 3–D images were prepared and rendered using Chimera 56.
Supplementary Material
Acknowledgements
This work was supported by the MRC, BBSRC and the Wellcome Trust. We thank Alastair Hosie, Philip Thomas and Megan Wilkins for helpful comments and Helena Da Silva for technical assistance.
References
- 1.Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4A resolution. J Mol Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 2.Brejc K, et al. Crystal structure of an ACh–binding protein reveals the ligand–binding domain of nicotinic receptors. Nature. 2001;411:269–276. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y, Reilly K, Chang Y. Evolutionarily conserved allosteric network in the Cys loop family of ligand–gated ion channels revealed by statistical covariance analyses. J Biol Chem. 2006;281:18184–18192. doi: 10.1074/jbc.M600349200. [DOI] [PubMed] [Google Scholar]
- 4.Chandler D. Interfaces and the driving force of hydrophobic assembly. Nature. 2005;437:640–647. doi: 10.1038/nature04162. [DOI] [PubMed] [Google Scholar]
- 5.Corringer PJ, Le NN, Changeux JP. Nicotinic receptors at the amino acid level. Annu Rev Pharmacol Toxicol. 2000;40:431–458. doi: 10.1146/annurev.pharmtox.40.1.431. [DOI] [PubMed] [Google Scholar]
- 6.Lester HA, Dibas MI, Dahan DS, Leite JF, Dougherty DA. Cys–loop receptors: new twists and turns. Trends Neurosci. 2004;27:329–336. doi: 10.1016/j.tins.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Sine SM, Engel AG. Recent advances in Cys–loop receptor structure and function. Nature. 2006;440:448–455. doi: 10.1038/nature04708. [DOI] [PubMed] [Google Scholar]
- 8.Law RJ, Henchman RH, McCammon JA. A gating mechanism proposed from a simulation of a human alpha7 nicotinic acetylcholine receptor. Proc Natl Acad Sci U S A. 2005;102:6813–6818. doi: 10.1073/pnas.0407739102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Celie PH, et al. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron. 2004;41:907–914. doi: 10.1016/s0896-6273(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 10.Lyford LK, Sproul AD, Eddins D, McLaughlin JT, Rosenberg RL. Agonist–induced conformational changes in the extracellular domain of alpha 7 nicotinic acetylcholine receptors. Mol Pharmacol. 2003;64:650–658. doi: 10.1124/mol.64.3.650. [DOI] [PubMed] [Google Scholar]
- 11.McLaughlin JT, Fu J, Sproul AD, Rosenberg RL. Role of the Outer beta–Sheet in Divalent Cation Modulation of {alpha}7 Nicotinic Receptors. Mol Pharmacol. 2006;70:16–22. doi: 10.1124/mol.106.023259. [DOI] [PubMed] [Google Scholar]
- 12.McLaughlin JT, Fu J, Rosenberg RL. Agonist–driven conformational changes in the inner beta–sheet of alpha7 nicotinic receptors. Mol Pharmacol. 2007;71:1312–1318. doi: 10.1124/mol.106.033092. [DOI] [PubMed] [Google Scholar]
- 13.Purohit P, Auerbach A. Acetylcholine receptor gating: movement in the alpha–subunit extracellular domain. J Gen Physiol. 2007;130:569–579. doi: 10.1085/jgp.200709858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dellisanti CD, Yao Y, Stroud JC, Wang ZZ, Chen L. Crystal structure of the extracellular domain of nAChR alpha1 bound to alpha–bungarotoxin at 1.94 A resolution. Nat Neurosci. 2007;10:953–962. doi: 10.1038/nn1942. [DOI] [PubMed] [Google Scholar]
- 15.Chakrapani S, Bailey TD, Auerbach A. Gating dynamics of the acetylcholine receptor extracellular domain. J Gen Physiol. 2004;123:341–356. doi: 10.1085/jgp.200309004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Unwin N, Miyazawa A, Li J, Fujiyoshi Y. Activation of the nicotinic acetylcholine receptor involves a switch in conformation of the alpha subunits. J Mol Biol. 2002;319:1165–1176. doi: 10.1016/S0022-2836(02)00381-9. [DOI] [PubMed] [Google Scholar]
- 17.McLaughlin JT, Fu J, Rosenberg RL. Agonist–driven conformational changes in the inner beta–sheet of alpha7 nicotinic receptors. Mol Pharmacol. 2007;71:1312–1318. doi: 10.1124/mol.106.033092. [DOI] [PubMed] [Google Scholar]
- 18.Taly A, et al. Implications of the quaternary twist allosteric model for the physiology and pathology of nicotinic acetylcholine receptors. Proc Natl Acad Sci U S A. 2006;103:16965–16970. doi: 10.1073/pnas.0607477103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukhtasimova N, Sine SM. An intersubunit trigger of channel gating in the muscle nicotinic receptor. J Neurosci. 2007;27:4110–4119. doi: 10.1523/JNEUROSCI.0025-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirzel K, et al. Hyperekplexia phenotype of glycine receptor alpha1 subunit mutant mice identifies Zn(2+) as an essential endogenous modulator of glycinergic neurotransmission. Neuron. 2006;52:679–690. doi: 10.1016/j.neuron.2006.09.035. [DOI] [PubMed] [Google Scholar]
- 21.Bloomenthal AB, Goldwater E, Pritchett DB, Harrison NL. Biphasic modulation of the strychnine–sensitive glycine receptor by Zn2+ Mol Pharmacol. 1994;46:1156–1159. [PubMed] [Google Scholar]
- 22.Laube B, et al. Modulation by zinc ions of native rat and recombinant human inhibitory glycine receptors. J Physiol. 1995;483(Pt 3):613–619. doi: 10.1113/jphysiol.1995.sp020610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller PS, Da Silva HM, Smart TG. Molecular basis for zinc potentiation at strychnine–sensitive glycine receptors. J Biol Chem. 2005;280:37877–37884. doi: 10.1074/jbc.M508303200. [DOI] [PubMed] [Google Scholar]
- 24.Harvey RJ, Thomas P, James CH, Wilderspin A, Smart TG. Identification of an inhibitory Zn2+ binding site on the human glycine receptor alpha1 subunit. J Physiol. 1999;520:53–64. doi: 10.1111/j.1469-7793.1999.00053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nevin ST, et al. Insights into the structural basis for zinc inhibition of the glycine receptor. J Biol Chem. 2003;278:28985–28992. doi: 10.1074/jbc.M300097200. [DOI] [PubMed] [Google Scholar]
- 26.Miller PS, Beato M, Harvey RJ, Smart TG. Molecular determinants of Glycine receptor {alpha}{beta}subunit sensitivities to Zn2+ inhibition. J Physiol. 2005 doi: 10.1113/jphysiol.2005.088575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Celie PH, et al. Crystal structure of nicotinic acetylcholine receptor homolog AChBP in complex with an alpha–conotoxin PnIA variant. Nat Struct Mol Biol. 2005;12:582–588. doi: 10.1038/nsmb951. [DOI] [PubMed] [Google Scholar]
- 28.Fu DX, Sine SM. Asymmetric contribution of the conserved disulfide loop to subunit oligomerization and assembly of the nicotinic acetylcholine receptor. J Biol Chem. 1996;271:31479–31484. doi: 10.1074/jbc.271.49.31479. [DOI] [PubMed] [Google Scholar]
- 29.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 30.Han NL, Haddrill JL, Lynch JW. Characterization of a glycine receptor domain that controls the binding and gating mechanisms of the beta–amino acid agonist, taurine. J Neurochem. 2001;79:636–647. doi: 10.1046/j.1471-4159.2001.00601.x. [DOI] [PubMed] [Google Scholar]
- 31.Laube B, Kuhse J, Betz H. Kinetic and mutational analysis of Zn2+ modulation of recombinant human inhibitory glycine receptors. J Physiol. 2000;522(Pt 2):215–230. doi: 10.1111/j.1469-7793.2000.t01-1-00215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Auld DS. Zinc coordination sphere in biochemical zinc sites. Biometals. 2001;14:271–313. doi: 10.1023/a:1012976615056. [DOI] [PubMed] [Google Scholar]
- 33.Cashin AL, Torrice MM, McMenimen KA, Lester HA, Dougherty DA. Chemical–scale studies on the role of a conserved aspartate in preorganizing the agonist binding site of the nicotinic acetylcholine receptor. Biochemistry. 2007;46:630–639. doi: 10.1021/bi061638b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lerma J, Zukin RS, Bennett MV. Glycine decreases desensitization of N–methyl–D–aspartate (NMDA) receptors expressed in Xenopus oocytes and is required for NMDA responses. Proc Natl Acad Sci U S A. 1990;87:2354–2358. doi: 10.1073/pnas.87.6.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grudzinska J, et al. The beta subunit determines the ligand binding properties of synaptic glycine receptors. Neuron. 2005;45:727–739. doi: 10.1016/j.neuron.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 36.Wilkins ME, Smart TG. Redox modulation of GABAA receptors obscured by Zn2+ complexation. Neuropharmacology. 2002;43:938–944. doi: 10.1016/s0028-3908(02)00238-1. [DOI] [PubMed] [Google Scholar]
- 37.Beato M, Groot–Kormelink PJ, Colquhoun D, Sivilotti LG. Openings of the rat recombinant alpha 1 homomeric glycine receptor as a function of the number of agonist molecules bound. J Gen Physiol. 2002;119:443–466. doi: 10.1085/jgp.20028530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Creighton TE. The energetic ups and downs of protein folding. Nat Struct Biol. 1994;1:135–138. doi: 10.1038/nsb0394-135. [DOI] [PubMed] [Google Scholar]
- 40.Yaron A, Naider F. Proline–dependent structural and biological properties of peptides and proteins. Crit Rev Biochem Mol Biol. 1993;28:31–81. doi: 10.3109/10409239309082572. [DOI] [PubMed] [Google Scholar]
- 41.Mukhtasimova N, Sine SM. An intersubunit trigger of channel gating in the muscle nicotinic receptor. J Neurosci. 2007;27:4110–4119. doi: 10.1523/JNEUROSCI.0025-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kash TL, Jenkins A, Kelley JC, Trudell JR, Harrison NL. Coupling of agonist binding to channel gating in the GABA(A) receptor. Nature. 2003;421:272–275. doi: 10.1038/nature01280. [DOI] [PubMed] [Google Scholar]
- 43.Lee WY, Sine SM. Principal pathway coupling agonist binding to channel gating in nicotinic receptors. Nature. 2005;438:243–247. doi: 10.1038/nature04156. [DOI] [PubMed] [Google Scholar]
- 44.Lummis SC, et al. Cis–trans isomerization at a proline opens the pore of a neurotransmitter–gated ion channel. Nature. 2005;438:248–252. doi: 10.1038/nature04130. [DOI] [PubMed] [Google Scholar]
- 45.Boileau AJ, Newell JG, Czajkowski C. GABA(A) receptor beta 2 Tyr97 and Leu99 line the GABA–binding site. Insights into mechanisms of agonist and antagonist actions. J Biol Chem. 2002;277:2931–2937. doi: 10.1074/jbc.M109334200. [DOI] [PubMed] [Google Scholar]
- 46.Chakrapani S, Bailey TD, Auerbach A. The role of loop 5 in acetylcholine receptor channel gating. J Gen Physiol. 2003;122:521–539. doi: 10.1085/jgp.200308885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grosman C, Zhou M, Auerbach A. Mapping the conformational wave of acetylcholine receptor channel gating. Nature. 2000;403:773–776. doi: 10.1038/35001586. [DOI] [PubMed] [Google Scholar]
- 48.Padgett CL, Hanek AP, Lester HA, Dougherty DA, Lummis SC. Unnatural amino acid mutagenesis of the GABA(A) receptor binding site residues reveals a novel cation–pi interaction between GABA and beta 2Tyr97. J Neurosci. 2007;27:886–892. doi: 10.1523/JNEUROSCI.4791-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Henchman RH, Wang HL, Sine SM, Taylor P, McCammon JA. Ligand–induced conformational change in the alpha7 nicotinic receptor ligand binding domain. Biophys J. 2005;88:2564–2576. doi: 10.1529/biophysj.104.053934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmieden V, Kuhse J, Betz H. A novel domain of the inhibitory glycine receptor determining antagonist efficacies: further evidence for partial agonism resulting from self–inhibition. Mol Pharmacol. 1999;56:464–472. doi: 10.1124/mol.56.3.464. [DOI] [PubMed] [Google Scholar]
- 51.Chang Y, Weiss DS. Site–specific fluorescence reveals distinct structural changes with GABA receptor activation and antagonism. Nat Neurosci. 2002;5:1163–1168. doi: 10.1038/nn926. [DOI] [PubMed] [Google Scholar]
- 52.Paoletti P, Ascher P, Neyton J. High–affinity zinc inhibition of NMDA NR1–NR2A receptors. J Neurosci. 1997;17:5711–5725. doi: 10.1523/JNEUROSCI.17-15-05711.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Colquhoun D, Sakmann B. Fast events in single–channel currents activated by acetylcholine and its analogues at the frog muscle end–plate. J Physiol. 1985;369:501–557. doi: 10.1113/jphysiol.1985.sp015912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position–specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 56.Pettersen EF, et al. UCSF Chimera––a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 57.Canutescu AA, Shelenkov AA, Dunbrack RL., Jr A graph–theory algorithm for rapid protein side–chain prediction. Protein Sci. 2003;12:2001–2014. doi: 10.1110/ps.03154503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davis IW, et al. MolProbity: all–atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jones DT. Protein secondary structure prediction based on position–specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 60.Miesenbock G, De Angelis DA, Rothman JE. Visualizing secretion and synaptic transmission with pH–sensitive green fluorescent proteins. Nature. 1998;394:192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.