Abstract
With this review, we aimed to provide a synopsis of recently proposed applications of machine learning (ML) in radiology focusing on prostate magnetic resonance imaging (MRI). After defining the difference between ML and classical rule-based algorithms and the distinction among supervised, unsupervised and reinforcement learning, we explain the characteristic of deep learning (DL), a particular new type of ML, including its structure mimicking human neural networks and its ‘black box’ nature. Differences in the pipeline for applying ML and DL to prostate MRI are highlighted. The following potential clinical applications in different settings are outlined, many of them based only on MRI-unenhanced sequences: gland segmentation; assessment of lesion aggressiveness to distinguish between clinically significant and indolent cancers, allowing for active surveillance; cancer detection/diagnosis and localisation (transition versus peripheral zone, use of prostate imaging reporting and data system (PI-RADS) version 2), reading reproducibility, differentiation of cancers from prostatitis benign hyperplasia; local staging and pre-treatment assessment (detection of extraprostatic disease extension, planning of radiation therapy); and prediction of biochemical recurrence. Results are promising, but clinical applicability still requires more robust validation across scanner vendors, field strengths and institutions.
Keywords: Machine learning, Magnetic resonance imaging, Prostate, Prostatic neoplasms, Radiomics
Key points
Machine/deep learning is a powerful tool to analyse large amounts of data, also applied to prostate magnetic resonance imaging (MRI).
Differences in the pipelines for applying machine and deep learning to prostate MRI exist.
Applications of machine/deep learning to prostate MRI regarding gland segmentation, cancer detection and localisation, assessment of lesion aggressiveness, local staging and pre-treatment assessment, and prediction of biochemical recurrence.
Many of these applications are based only on MRI-unenhanced sequences.
Background
Prostate cancer (PCa) represents the most common cancer in the male population, and its early detection is fundamental to reduce mortality [1]. Since only a part of PCa cases are clinically significant (csPCa), risk stratification is of crucial importance in order to avoid overdiagnosis and overtreatment. While for biopsy-naïve patients this has been performed employing clinical and laboratory parameters, imaging, especially magnetic resonance imaging (MRI), has acquired an increasingly important role in this task [2].
In the past, transrectal ultrasound was the main imaging technique for assessment of patients with suspected PCa, but it presents numerous limitations, with both low sensitivity and specificity rates, especially for transition zone lesions [2]. More recently, multiparametric MRI (mpMRI) has demonstrated a better diagnostic accuracy and is becoming a clinical routine examination for patients at risk of having csPCa [3–5]. The second version of the Prostate Imaging Reporting and Data System (PI-RADS) was recently updated both in regard to minimum technical acquisition parameters and image interpretation [6]. It describes a standard prostate mpMRI protocol that combines anatomical T2-weighted images with one or more functional sequences, i.e., diffusion-weighted imaging (DWI) and/or dynamic contrast-enhanced (DCE) sequences (Fig. 1). In short, DWI, together with apparent diffusion coefficient (ADC) maps, is the dominant sequence to detect and establish the aggressiveness of peripheral zone (PZ) lesions. On the other hand, T2-weighted images are the most useful tool for diagnosing tumours of the transition zone. DCE has a relatively minor role as it is mainly used for the characterisation of PZ lesions in conjunction with DWI and ADC maps [7–9]. It has also been found that the use of mpMRI-targeted biopsies increases the accuracy of diagnosing csPCa and reduces the number of patients requiring repeat biopsies when compared to transrectal ultrasound-guided biopsies [4, 10].
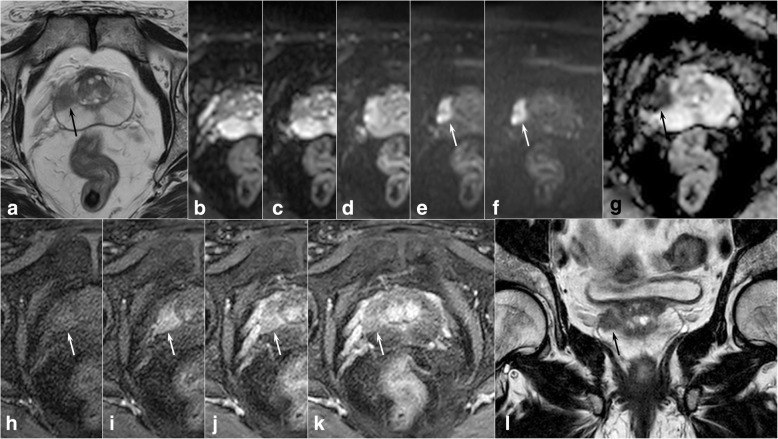
Fig. 1.
Prostate multiparametric magnetic resonance imaging showing a neoplastic lesion of the right peripheral zone (arrows). The lesion is hypointense on axial (a) and coronal (l) T2-weighted images and demonstrates diffusion restriction on diffusion-weighted images (b values 0, 100, 500, 1000, and 1400 s/mm2, from b to f, respectively), confirmed by the apparent diffusion coefficient map (g). Lesion enhancement is also evident on dynamic contrast-enhanced perfusion-weighted imaging (from h to k). PI-RADSv2 diagnostic category: 5
Nonetheless, mpMRI still presents some limitations. In particular, variability is reported in terms of inter-reader agreement and diagnostic accuracy, mainly dependent on reader experience [11–14].
Machine learning (ML) is a branch of data science, and in particular of artificial intelligence, based on the development and training of algorithms, by which computers may learn from data and perform predictions without previous specific programming. The main difference with classical rule-based algorithms is represented by their ability to take advantage of increased exposure to large and new data as well as to improve and learn over time. We can identify three different types of ML algorithms [15]:
Supervised learning, the most used in radiology, which depends on train data labelling prior to the learning process
Unsupervised learning, characterised by the absence of preliminary human division of data in categories
Reinforcement learning, in which the algorithm learns from both its mistakes and successes, thanks to a continuous feedback
The main strength of ML is its ability to analyse and employ an enormous quantity of data, much more efficiently than possible for humans through classical statistical analyses. Therefore, it is not surprising that its increased role in radiology has followed the growing role and potential shown in research by radiomics. This is another expanding field that allows the extraction of great volumes of quantitative data from medical images [16]. These large datasets have been analysed to obtain useful clinical information such as correlation to other biomarkers, patient prognosis or treatment outcome [17–19].
A particular application in the field of ML is that of deep learning. This method exploits algorithms, also called networks, that are structured in order to somewhat mimic human neural structure [20]. Briefly, for its application in the field of medical imaging analysis, data is transformed in feature vectors, derived from its voxels, which then constitute the input neurons of the network. Between the input and output strata of the algorithm, a variable number of hidden layers, also made up by neural nodes, with various structures can be implemented. Each node, represented by a numerical value, is connected to those in other layers with different strengths (or weights), leading to the output neurons that encode the final outcome [21].
In recent years, ML has been proposed for a wide range of applications in medical imaging. The most common are the detection and characterisation of neoplastic lesions in different anatomical regions [22–25]. On the other hand, it also has many other possible uses including for example acquisition time reduction, organ and lesion automated segmentation and early detection of neurodegenerative disorders [26–30]. Unfortunately, ML algorithms are still far from a widespread application in clinical practice, mainly due to the current unavailability of the large quantity of data that would be necessary for their validation. Some solutions to this issue have already been proposed. For example, Pinto dos Santos et al. [31] focused on the essential need to create structured reports which could greatly improve the quality and the reproducibility of data available to refine artificial intelligence algorithms.
There is a broad interest in the applications of ML to prostate imaging. The purpose of this article is to review the various approaches proposed in the recent literature for gland segmentation, PCa detection, lesion aggressiveness assessment, local staging, pre-treatment assessment and follow-up.
Machine learning pipelines for prostate MRI
A typical ML post-processing pipeline applied to prostate MRI for radiomic analysis may be constituted by:
mpMRI examination: T2-weighted sequences, diffusion-weighted imaging (DWI) with apparent diffusion coefficient (ADC) maps and dynamic contrast-enhanced (DCE) sequences
Image segmentation through the delineation of regions of interest (ROIs), which can include whole gland volume, a specific zone or one or multiple lesions
Image pre-processing: voxel grey value normalisation (when using non-quantitative images, i.e., T2-weighted, DWI, or DCE sequences), decomposition filtering for the creation of additional mineable data (e.g., Laplacian of Gaussian)
Feature extraction from the ROI: shape, histogram, and texture (second-order features) parameters
Integration of radiomic data with clinical, laboratory, prognostic, and/or genomic data
Feature selection in relation to the class of interest
Algorithm training and testing
Validation on an external population
Alternatively, a deep learning approach would only require:
mpMRI examination: T2-weighted sequences, DWI with ADC maps, and DCE sequences
Annotation of the ROI or of the whole image, according to the desired classification output
Algorithm training and testing
Validation on an external population
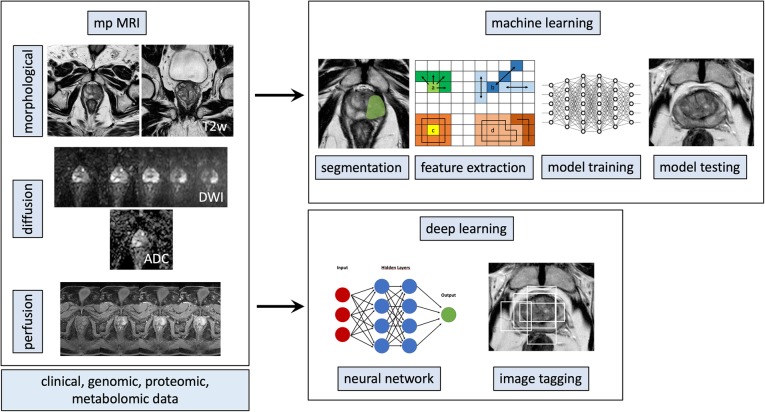
An overview of both approaches is shown in Fig. 2. While the deep learning approach may appear simpler and more flexible, it does require much larger quantities of data for algorithm training and its structure is usually more complex and less transparent. These latter characteristics contribute to the ‘black box’ nature of deep learning algorithms, one of the main limitations preventing their widespread adoption. Finally, the two approaches can be variously mixed and matched combining for example radiomic data with deep learning algorithms.
Fig. 2.
Radiomic workflow pipeline for both machine learning and deep learning approaches for prostate magnetic resonance imaging. See the text for details
Applications
Segmentation
The segmentation of the prostate gland is often a necessary step in clinical settings as well as for further image analysis. Therefore, the possibility of automating whole prostate as well as lesion segmentation is of great interest for the potential time-saving and increased reproducibility. A robust automated segmentation could in turn lead to a fully automated post-processing pipeline.
There are interesting studies in the literature that describe the potentials of deep learning to achieve this goal. For example, Wang et al. [32] compared manual segmentation to a novel deep learning-based one. They were able to demonstrate the feasibility of three-dimensional fully convolutional networks and subsequently validate their findings on a public dataset. Another research group [33] proposed a model based on a propagation deep neuronal network by which data from different levels of complexity were extracted and combined so as to obtain a more trustworthy segmentation of the gland and its boundaries. The propagation deep neuronal network was able to outperform baseline deep neural networks, resulting competitive with the reference standard.
Finally, Alkadi et al. [34] applied a unimodal deep learning-based system using only T2-weighted images for the automated segmentation of both the prostate and PCa lesions. Their algorithm was able to achieve an area under the receiver operating characteristic curve (AUC) of 0.995, comparable to that of other multimodal systems in lesion detection while outperforming other proposed two-dimensional and three-dimensional prostate segmentation approaches.
Cancer detection
There is a high interest in analysing the usefulness of ML-based computer-assisted diagnosis (CAD) software in the field of PCa, as it could improve radiologists’ diagnostic performances and reproducibility. For example, already in 2012, PZ maps created by a decision support system model based on endorectal mpMRI appeared to be a promising tool in correctly localising PZ tumours [35].
More recently, Kwak et al. [36] have demonstrated that it is possible to employ radiomics and ML for the analysis of different tissues and cellular densities in the prostate gland to aid in PCa detection. In order to do so, they developed an MRI-based patient-specific prostate mould in order to ensure correspondence of prostatectomy specimens to the images. A significant difference was found in ML-determined prostatic tissue composition between benign and malignant areas. In 2018, McGarry et al. [37] found that the data derived from ten patients was sufficient to obtain a stable fit for ML MRI detection of increased epithelium and decreased lumen density areas, indicative of high-grade PCa. These authors were therefore able to generate lesion detection maps, also validated against whole-mount histopathological specimens oriented with three-dimensional printed slicing moulds.
Another proposal has been based on volumetric ROI analysis of index lesions on mpMRI [38]. These authors evaluated the usefulness of histogram parameters obtained from T2-weighted, DWI and DCE images in combination with a support vector machine (SVM) ML approach for the improvement of PI-RADSv2 scores, significantly increasing the radiologist’s performance. In a recent study [39], comparing the analysis of ADC radiomics with ML analysis and the performance of mean ADC values alone for differential diagnosis of benign and malignant prostate lesions, the resulting accuracy was similar without a statistically significant difference between approaches.
Ginsburg et al. [40] have suggested that different ML predictive models should be developed for the transition zone and PZ, as lesions and normal prostatic tissue have different imaging characteristics in these zones. In particular, they compared two zone-specific algorithms, trained on different radiomic feature datasets, with a zone-ignorant one. Interestingly, they found that while a significant difference in performance was found between the PZ-specific algorithm and the zone-ignorant one, no differences were found for the transition zone-specific algorithm.
A further use case for ML is the differentiation of stromal benign prostatic hyperplasia from PCa in the transition zone. This diagnosis can be quite challenging, especially with small lesions. Statistical analysis of previously established quantitative features (ADC maps, shape, and image texture) demonstrated a high accuracy in the differentiation of small neoplastic lesions from benign ones using either linear regression and SVM classifiers [41].
Another potential advantage of ML CAD-assisted mpMRI is the improvement of inter-reader agreement. Greer et al. [42] compared index lesion sensitivity, specificity and agreement between eight radiologists from different institutions and with various levels of experience. The readers, after a first qualitative examination, reanalysed the same images using CAD: agreement and sensitivity were higher in the CAD-assisted mpMRI, but specificity worsened. Furthermore, the system was more useful for PZ lesions.
Another CAD system was developed by Ishioka et al. [43], although their proposal was based on a convolutional neural network deep learning algorithm. While the accuracy reported in two validation sets was not very high, with AUC respectively of 0.645 and 0.636, further development could lead to a reproducible, automated CAD system. On the other hand, the research unit lead by Wang [44] has shown better results for a deep learning-based fully automated segmentation when compared to a non-deep learning model. The authors assessed their ability to distinguish PCa from benign pathologies such as prostatitis or prostate benign hyperplasia. Their results showed a better accuracy and reliability of deep learning, with a statistically significantly higher AUC than that of the non-deep learning model (0.84 vs. 0.70). Interestingly, their approach was conducted without any need for segmentation of the training set, potentially easing further evolution of their software with added data.
Assessment of lesion aggressiveness
As PCa is frequently indolent, it is of great importance to assess the aggressiveness of detected lesions, and therefore their clinical significance, for patient management [2]. The standard approach of MRI and subsequent targeted biopsy have improved identification of csPCa foci, but significant disease is still missed, as shown in a recent meta-analysis [45]. Texture features have shown in the past potential as biomarkers of PCa aggressiveness [46, 47].
An interesting study was performed on 56 PCa patients in active surveillance who underwent MRI-guided biopsies [48]. The authors assessed differences between patients who were biopsy- and MRI-negative and patients who were biopsy- and MRI-positive, in radiomic features extracted from T2-weighted and ADC images. They subsequently employed the ten selected parameters to construct models to identify subjects who were biopsy-positive while MRI-negative and those biopsy-negative and MRI-positive. Quadratic discriminant analysis enabled to obtain the best accuracy improving, compared to PI-RADS alone, by 80% for the first classification and 60% in the second. While focusing only on the central gland, Li et al. [49] have shown that an SVM approach, trained on six features extracted from mpMRI exams depicting 152 prostate lesions, was able to consistently predict Gleason score. This is especially important as it is one of the main determinants of PCa clinical significance.
For the task of distinguishing indolent PCas and csPCas, deep learning methods have shown some potential for future applications. Zhong et al. [50] compared both a deep learning and a deep transfer-learning algorithm to the performance of PI-RADSv2. In their validation cohort of 30 patients with 47 lesions, it was comparable to the radiologist’s assessment (AUC 0.73 and 0.71, respectively), while outperforming deep learning alone (AUC 0.69). Similarly, Yuan et al. [51] have shown that transfer-learning, in their case applied to the AlexNet neural network, was able to achieve an overall accuracy of 87% for the prediction of lesion Gleason score.
Finally, a novel approach for the detection of csPCa, as defined in the National Comprehensive Cancer Network guidelines, using radiomics in combination with ML, was recently performed by Varghese et al. [52]. A framework for the robust testing of seven ML algorithms with fivefold cross-validation showed that the best classifier was a quadratic kernel-based SVM with an overall accuracy of 0.92 in the validation cohort.
Local staging and pre-treatment assessment
mpMRI also gives important information for pre-treatment local staging. In this setting, proposals for artificial intelligence applications have been more limited. A recent preliminary report has shown that a radiomics-based Bayesian network achieved a high accuracy (AUC 0.88) in the detection of extraprostatic extension of disease in preoperative MRI, using radical prostatectomy as the reference standard [53]. This finding is promising for further development of ML in PCa local staging.
Some novel applications of ML to prostate MRI have been proposed in the setting of treatment planning. In particular, Sun et al. [3] have developed a new type of focal radiotherapy (bio-focused therapy) that requires prior evaluation of some biological features of the tumour, such as cell density and aggressiveness, which relate to therapy response. These have been assessed by the authors non-invasively, with a voxel-wise approach: patients underwent mpMRI before radical prostatectomy, and imaging data were combined with histopathological information to extract tissue features (cell density). They were then able to create models of prostate tissue cell density, used for the targeting of bio-focused therapy in order to achieve better response and lower toxicity. Shafai-Erfani et al. [54] trained a ML algorithm with paired CT and MRI datasets in order to generate synthetic CT images to be used for patient radiation therapy setup and dose calculation. ML proved capable of producing reliable CT images, comparable to the ground truth for both tasks.
Biochemical recurrence
About 30% of PCa relapse after radical prostatectomy [55]. Also, radiation therapy-resistant PCa or recurrent PCa is not uncommon. The elevation of serum levels of prostate-specific antigen after treatment is currently the most employed biomarker of this condition and is known as ‘biochemical recurrence’ [56]. Very complex altered molecular networks lie behind it and, more extensively, PCa recurrence [55]. ML applied to mpMRI has been proposed as a viable tool for early detection of recurrence or prediction of treatment outcome.
Abdollahi et al. [57] investigated how mpMRI ML models could predict the response to an intensity-modulated radiation therapy. Their findings suggest that both pre- and post-treatment radiomic features can give a reliable prediction of therapeutic success, especially if compared to prostate-specific antigen dosage. A recent study [58] has shown that a SVM could accurately predict biochemical recurrence within 3 years of radical prostatectomy, outperforming a linear regression model based on both MRI and the D’Amico patient risk classification. In fact, ML showed an overall accuracy of 92.2% compared to the 79% achieved by linear regression. The potential of ML for this task was confirmed in 2018 by another study [59] that employed SVM, linear discriminant analysis and random forest on radiomic features extracted from T2-weighted and ADC images.
Conclusions
A number of studies showed that ML, with or without radiomic feature extraction, has a great potential to improve the diagnostic performance and to expand the clinical role of prostate MRI. Its applications range from segmentation, lesion detection and aggressiveness prediction to local staging and assessment prior to and following treatment. It is interesting to note that many of the cited studies do not employ DCE images. Thus, ML could help in avoiding the systematic use of contrast agents for prostate imaging, as suggested by current guidelines, a development to monitor as gadolinium administration has come under scrutiny for the growing evidences of its accumulation in the body [60].
Clinical applicability still requires more robust validation across scanner vendors, field strengths and institutions, as for ML in all fields of medical imaging. As highlighted in a recent letter by Peter L. Choyke, the current limitations of ML also prevent them from gaining the trust of the ideal end-users, radiologists [61]. On the other hand, the growth in quantity and quality of research in the years is also undeniable, and as ML software obtain approval for clinical use in other settings, it is difficult to imagine a future for prostate MRI without it.
Abbreviations
- ADC
Apparent diffusion coefficient
- AUC
Area under the curve
- CAD
Computer-assisted diagnosis
- csPCa
Clinically significant prostate cancer
- DCE
Dynamic contrast-enhanced
- DWI
Diffusion-weighted imaging
- ML
Machine learning
- mpMRI
Multiparametric MRI
- MRI
Magnetic resonance imaging
- PCa
Prostate cancer
- PI-RADS
Prostate Imaging Reporting and Data System
- PZ
Peripheral zone
- ROI
Region of interest
- SVM
Support vector machine
Authors’ contributions
RC, MBC, VR, and AS contributed to the conception and design, interpretation of the literature data, manuscript preparation and editing. LR, AB, and MI revised critically the intellectual content of the manuscript and contributed to the interpretation of the data, manuscript preparation, and editing. LU prepared the figures. All authors read and approved the final manuscript.
Funding
The authors declare that this work has not received any funding.
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Mottet N, Bellmunt J, Bolla M, et al. EAU-ESTRO-SIOG Guidelines on prostate cancer. Part 1: screening, diagnosis, and local treatment with curative intent. Eur Urol. 2017;71:618–629. doi: 10.1016/j.eururo.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Sun Y, Reynolds HM, Parameswaran B, et al. Multiparametric MRI and radiomics in prostate cancer: a review. Australas Phys Eng Sci Med. 2019;42:3–25. doi: 10.1007/s13246-019-00730-z. [DOI] [PubMed] [Google Scholar]
- 4.van der Leest M, Cornel E, Israël B, et al. Head-to-head comparison of transrectal ultrasound-guided prostate biopsy versus multiparametric prostate resonance imaging with subsequent magnetic resonance-guided biopsy in biopsy-naïve men with elevated prostate-specific antigen: a large prospective multicenter clinical study. Eur Urol. 2019;75:570–578. doi: 10.1016/j.eururo.2018.11.023. [DOI] [PubMed] [Google Scholar]
- 5.Winoker JS, Pinto PA, Rastinehad AR. MRI to guide biopsies or avoid biopsies? Curr Opin Urol. 2018;28:522–528. doi: 10.1097/MOU.0000000000000555. [DOI] [PubMed] [Google Scholar]
- 6.Turkbey B, Rosenkrantz AB, Haider MA et al (2019) Prostate imaging reporting and data system version 2.1: 2019 update of prostate imaging reporting and data system version 2. Eur Urol. pii: S0302-2838(19)30180-0 [Epub ahead of print] 10.1016/j.eururo.2019.02.033 [DOI] [PubMed]
- 7.Barentsz JO, Weinreb JC, Verma S, et al. Synopsis of the PI-RADS v2 guidelines for multiparametric prostate magnetic resonance imaging and recommendations for use. Eur Urol. 2016;69:41–49. doi: 10.1016/j.eururo.2015.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stanzione A, Imbriaco M, Cocozza S, et al. Biparametric 3T magnetic resonance imaging for prostatic cancer detection in a biopsy-naïve patient population: a further improvement of PI-RADS v2? Eur J Radiol. 2016;85:2269–2274. doi: 10.1016/j.ejrad.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 9.Cuocolo R, Stanzione A, Rusconi G, et al. PSA-density does not improve bi-parametric prostate MR detection of prostate cancer in a biopsy naïve patient population. Eur J Radiol. 2018;104:64–70. doi: 10.1016/j.ejrad.2018.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Boesen Lars, Nørgaard Nis, Løgager Vibeke, Balslev Ingegerd, Thomsen Henrik S. A Prospective Comparison of Selective Multiparametric Magnetic Resonance Imaging Fusion-Targeted and Systematic Transrectal Ultrasound-Guided Biopsies for Detecting Prostate Cancer in Men Undergoing Repeated Biopsies. Urologia Internationalis. 2017;99(4):384–391. doi: 10.1159/000477214. [DOI] [PubMed] [Google Scholar]
- 11.Rosenkrantz AB, Ayoola A, Hoffman D, et al. The learning curve in prostate MRI interpretation: self-directed learning versus continual reader feedback. AJR Am J Roentgenol. 2017;208:W92–W100. doi: 10.2214/AJR.16.16876. [DOI] [PubMed] [Google Scholar]
- 12.Gatti Marco, Faletti Riccardo, Calleris Giorgio, Giglio Jacopo, Berzovini Claudio, Gentile Francesco, Marra Giancarlo, Misischi Francesca, Molinaro Luca, Bergamasco Laura, Gontero Paolo, Papotti Mauro, Fonio Paolo. Prostate cancer detection with biparametric magnetic resonance imaging (bpMRI) by readers with different experience: performance and comparison with multiparametric (mpMRI) Abdominal Radiology. 2019;44(5):1883–1893. doi: 10.1007/s00261-019-01934-3. [DOI] [PubMed] [Google Scholar]
- 13.Glazer DI, Mayo-Smith WW, Sainani NI, et al. Interreader agreement of Prostate Imaging Reporting and Data System version 2 using an in-bore MRI-guided prostate biopsy cohort: a single institution’s initial experience. AJR Am J Roentgenol. 2017;209:W145–W151. doi: 10.2214/AJR.16.17551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Li, Tang Min, Chen Sipan, Lei Xiaoyan, Zhang Xiaoling, Huan Yi. A meta-analysis of use of Prostate Imaging Reporting and Data System Version 2 (PI-RADS V2) with multiparametric MR imaging for the detection of prostate cancer. European Radiology. 2017;27(12):5204–5214. doi: 10.1007/s00330-017-4843-7. [DOI] [PubMed] [Google Scholar]
- 15.Choy G, Khalilzadeh O, Michalski M, et al. Current applications and future impact of machine learning in radiology. Radiology. 2018;288:318–328. doi: 10.1148/radiol.2018171820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aerts HJ, Velazquez ER, Leijenaar RT et al (2014) Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 5:4006 10.1038/ncomms5006 [DOI] [PMC free article] [PubMed]
- 18.Trebeschi S, Drago S G, Birkbak N J, Kurilova I, Cǎlin A M, Delli Pizzi A, Lalezari F, Lambregts D M J, Rohaan M W, Parmar C, Rozeman E A, Hartemink K J, Swanton C, Haanen J B A G, Blank C U, Smit E F, Beets-Tan R G H, Aerts H J W L. Predicting response to cancer immunotherapy using noninvasive radiomic biomarkers. Annals of Oncology. 2019;30(6):998–1004. doi: 10.1093/annonc/mdz108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Oikonomou A, Wong A, Haider MA, Khalvati F (2017) Radiomics-based prognosis analysis for non-small cell lung cancer. Sci Rep 7:46349 10.1038/srep46349 [DOI] [PMC free article] [PubMed]
- 20.Zaharchuk G., Gong E., Wintermark M., Rubin D., Langlotz C.P. Deep Learning in Neuroradiology. American Journal of Neuroradiology. 2018;39(10):1776–1784. doi: 10.3174/ajnr.A5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 22.Chang P., Grinband J., Weinberg B.D., Bardis M., Khy M., Cadena G., Su M.-Y., Cha S., Filippi C.G., Bota D., Baldi P., Poisson L.M., Jain R., Chow D. Deep-Learning Convolutional Neural Networks Accurately Classify Genetic Mutations in Gliomas. American Journal of Neuroradiology. 2018;39(7):1201–1207. doi: 10.3174/ajnr.A5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romeo V, Maurea S, Cuocolo R, et al. Characterization of adrenal lesions on unenhanced MRI using texture analysis: a machine-learning approach. J Magn Reson Imaging. 2018;48:198–204. doi: 10.1002/jmri.25954. [DOI] [PubMed] [Google Scholar]
- 24.Ciompi F, Chung K, van Riel SJ, et al. Towards automatic pulmonary nodule management in lung cancer screening with deep learning. Sci Rep. 2017;7:46479. doi: 10.1038/srep46479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le E.P.V., Wang Y., Huang Y., Hickman S., Gilbert F.J. Artificial intelligence in breast imaging. Clinical Radiology. 2019;74(5):357–366. doi: 10.1016/j.crad.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Golkov V, Dosovitskiy A, Sperl JI, et al. q-space deep learning: twelve-fold shorter and model-free diffusion MRI scans. IEEE Trans Med Imaging. 2016;35:1344–1351. doi: 10.1109/TMI.2016.2551324. [DOI] [PubMed] [Google Scholar]
- 27.Ding Y, Sohn JH, Kawczynski MG, et al. A deep learning model to predict a diagnosis of Alzheimer disease by using 18F-FDG PET of the Brain. Radiology. 2019;290:456–464. doi: 10.1148/radiol.2018180958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bai W, Sinclair M, Tarroni G, et al. Automated cardiovascular magnetic resonance image analysis with fully convolutional networks. J Cardiovasc Magn Reson. 2018;20:65. doi: 10.1186/s12968-018-0471-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burström Gustav, Buerger Christian, Hoppenbrouwers Jurgen, Nachabe Rami, Lorenz Cristian, Babic Drazenko, Homan Robert, Racadio John M., Grass Michael, Persson Oscar, Edström Erik, Elmi Terander Adrian. Machine learning for automated 3-dimensional segmentation of the spine and suggested placement of pedicle screws based on intraoperative cone-beam computer tomography. Journal of Neurosurgery: Spine. 2019;31(1):147–154. doi: 10.3171/2018.12.SPINE181397. [DOI] [PubMed] [Google Scholar]
- 30.Comelli A, Stefano A, Bignardi S, et al. Active contour algorithm with discriminant analysis for delineating tumors in positron emission tomography. Artif Intell Med. 2019;94:67–78. doi: 10.1016/j.artmed.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 31.Pinto dos Santos D, Baeßler B. Big data, artificial intelligence, and structured reporting. Eur Radiol Exp. 2018;2:42. doi: 10.1186/s41747-018-0071-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang B, Lei Y, Tian S, et al. Deeply supervised 3D fully convolutional networks with group dilated convolution for automatic MRI prostate segmentation. Med Phys. 2019;46:1707–1718. doi: 10.1002/mp.13416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan Ke, Wang Xiuying, Kim Jinman, Khadra Mohamed, Fulham Michael, Feng Dagan. A propagation-DNN: Deep combination learning of multi-level features for MR prostate segmentation. Computer Methods and Programs in Biomedicine. 2019;170:11–21. doi: 10.1016/j.cmpb.2018.12.031. [DOI] [PubMed] [Google Scholar]
- 34.Alkadi R, Taher F, El-Baz A, Werghi N (2018) A deep learning-based approach for the detection and localization of prostate cancer in T2 magnetic resonance images. J Digit Imaging, [Epub ahead of print]. 10.1007/s10278-018-0160-1 [DOI] [PMC free article] [PubMed]
- 35.Shah V, Turkbey B, Mani H, et al. Decision support system for localizing prostate cancer based on multiparametric magnetic resonance imaging. Med Phys. 2012;39:4093–4103. doi: 10.1118/1.4722753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwak JT, Sankineni S, Xu S, et al. Correlation of magnetic resonance imaging with digital histopathology in prostate. Int J Comput Assist Radiol Surg. 2016;11:657–666. doi: 10.1007/s11548-015-1287-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGarry Sean D., Hurrell Sarah L., Iczkowski Kenneth A., Hall William, Kaczmarowski Amy L., Banerjee Anjishnu, Keuter Tucker, Jacobsohn Kenneth, Bukowy John D., Nevalainen Marja T., Hohenwalter Mark D., See William A., LaViolette Peter S. Radio-pathomic Maps of Epithelium and Lumen Density Predict the Location of High-Grade Prostate Cancer. International Journal of Radiation Oncology*Biology*Physics. 2018;101(5):1179–1187. doi: 10.1016/j.ijrobp.2018.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Jing, Wu Chen-Jiang, Bao Mei-Ling, Zhang Jing, Wang Xiao-Ning, Zhang Yu-Dong. Machine learning-based analysis of MR radiomics can help to improve the diagnostic performance of PI-RADS v2 in clinically relevant prostate cancer. European Radiology. 2017;27(10):4082–4090. doi: 10.1007/s00330-017-4800-5. [DOI] [PubMed] [Google Scholar]
- 39.Bonekamp D, Kohl S, Wiesenfarth M, et al. Radiomic machine learning for characterization of prostate lesions with MRI: comparison to ADC values. Radiology. 2018;289:128–137. doi: 10.1148/radiol.2018173064. [DOI] [PubMed] [Google Scholar]
- 40.Ginsburg SB, Algohary A, Pahwa S, et al. Radiomic features for prostate cancer detection on MRI differ between the transition and peripheral zones: preliminary findings from a multi-institutional study. J Magn Reson Imaging. 2017;46:184–193. doi: 10.1002/jmri.25562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu M, Krishna S, Thornhill RE, Flood TA, McInnes MDF, Schieda N (2019) Transition zone prostate cancer: Logistic regression and machine-learning models of quantitative ADC, shape and texture features are highly accurate for diagnosis. J Magn Reson Imaging, [Epub ahead of print. 10.1002/jmri.26674 [DOI] [PubMed]
- 42.Greer MD, Lay N, Shih JH, et al. Computer-aided diagnosis prior to conventional interpretation of prostate mpMRI: an international multi-reader study. Eur Radiol. 2018;28:4407–4417. doi: 10.1007/s00330-018-5374-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ishioka J, Matsuoka Y, Uehara S, et al. Computer-aided diagnosis of prostate cancer on magnetic resonance imaging using a convolutional neural network algorithm. BJU Int. 2018;122:411–417. doi: 10.1111/bju.14397. [DOI] [PubMed] [Google Scholar]
- 44.Wang X, Yang W, Weinreb J, et al. Searching for prostate cancer by fully automated magnetic resonance imaging classification: deep learning versus non-deep learning. Sci Rep. 2017;7:15415. doi: 10.1038/s41598-017-15720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sathianathen Niranjan J., Butaney Mohit, Bongiorno Connie, Konety Badrinath R., Bolton Damien M., Lawrentschuk Nathan. Accuracy of the magnetic resonance imaging pathway in the detection of prostate cancer: a systematic review and meta-analysis. Prostate Cancer and Prostatic Diseases. 2018;22(1):39–48. doi: 10.1038/s41391-018-0075-4. [DOI] [PubMed] [Google Scholar]
- 46.Vignati A, Mazzetti S, Giannini V, et al. Texture features on T2-weighted magnetic resonance imaging: new potential biomarkers for prostate cancer aggressiveness. Phys Med Biol. 2015;60:2685–2701. doi: 10.1088/0031-9155/60/7/2685. [DOI] [PubMed] [Google Scholar]
- 47.Wibmer A, Hricak H, Gondo T, et al. Haralick texture analysis of prostate MRI: utility for differentiating non-cancerous prostate from prostate cancer and differentiating prostate cancers with different Gleason scores. Eur Radiol. 2015;25:2840–2850. doi: 10.1007/s00330-015-3701-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Algohary A, Viswanath S, Shiradkar R, et al. Radiomic features on MRI enable risk categorization of prostate cancer patients on active surveillance: preliminary findings. J Magn Reson Imaging. 2018;48:818–828. doi: 10.1002/jmri.25983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J, Weng Z, Xu H, et al. Support vector machines (SVM) classification of prostate cancer Gleason score in central gland using multiparametric magnetic resonance images: a cross-validated study. Eur J Radiol. 2018;98:61–67. doi: 10.1016/j.ejrad.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 50.Zhong X, Cao R, Shakeri S, et al. Deep transfer learning-based prostate cancer classification using 3 Tesla multi-parametric MRI. Abdom Radiol (NY) 2018;44:2030–2039. doi: 10.1007/s00261-018-1824-5. [DOI] [PubMed] [Google Scholar]
- 51.Yuan Y, Qin W, Buyyounouski M, et al. Prostate cancer classification with multiparametric MRI transfer learning model. Med Phys. 2019;46:756–765. doi: 10.1002/mp.13367. [DOI] [PubMed] [Google Scholar]
- 52.Varghese B, Chen F, Hwang D, et al. Objective risk stratification of prostate cancer using machine learning and radiomics applied to multiparametric magnetic resonance images. Sci Rep. 2019;9:1570. doi: 10.1038/s41598-018-38381-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stanzione A, Cuocolo R, Cocozza S et al (2019) Detection of extraprostatic extension of cancer on biparametric MRI combining texture analysis and machine learning: preliminary results. Acad Radiol:1–7. 10.1016/j.acra.2018.12.025 [DOI] [PubMed]
- 54.Shafai-Erfani G, Wang T, Lei Y et al (2019) Dose evaluation of MRI-based synthetic CT generated using a machine learning method for prostate cancer radiotherapy. Med Dosim, pii: S0958-3947(19)30011-1. 10.1016/j.meddos.2019.01.002 [DOI] [PMC free article] [PubMed]
- 55.Gu Yan, Lin Xiaozeng, Kapoor Anil, Mei Wenjuan, Tang Damu. Attempt to predict early recurrence of prostate cancer following prostatectomy through machine learning. AME Medical Journal. 2018;3:96–96. doi: 10.21037/amj.2018.09.06. [DOI] [Google Scholar]
- 56.Zaorsky Nicholas G., Raj Ganesh V., Trabulsi Edouard J., Lin Jianqing, Den Robert B. The Dilemma of a Rising Prostate-Specific Antigen Level After Local Therapy: What Are Our Options? Seminars in Oncology. 2013;40(3):322–336. doi: 10.1053/j.seminoncol.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 57.Abdollahi H, Mofid B, Shiri I, et al. Machine learning-based radiomic models to predict intensity-modulated radiation therapy response, Gleason score and stage in prostate cancer. Radiol Med. 2019;124:555–567. doi: 10.1007/s11547-018-0966-4. [DOI] [PubMed] [Google Scholar]
- 58.Zhang YD, Wang J, Wu CJ et al (2016) An imaging-based approach predicts clinical outcomes in prostate cancer through a novel support vector machine classification. Oncotarget 7:78140–78151 10.18632/oncotarget.11293 [DOI] [PMC free article] [PubMed]
- 59.Shiradkar R, Ghose S, Jambor I, et al. Radiomic features from pretreatment biparametric MRI predict prostate cancer biochemical recurrence: preliminary findings. J Magn Reson Imaging. 2018;48:1626–1636. doi: 10.1002/jmri.26178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Layne KA, Dargan PI, Archer JRH, Wood DM. Gadolinium deposition and the potential for toxicological sequelae – a literature review of issues surrounding gadolinium-based contrast agents. Br J Clin Pharmacol. 2018;84:2522–2534. doi: 10.1111/bcp.13718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Choyke PL. Quantitative MRI or machine learning for prostate MRI: which should you use? Radiology. 2018;289:138–139. doi: 10.1148/radiol.2018181304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.