REPLY
To quantitatively measure the beta diversities between microbiomes, Microbiome Search Engine (MSE) (1) calculates phylogeny similarity using operational taxonomy unit (OTU) profiles; for both query and database samples, all 16S rRNA gene sequences are mapped to the Greengenes database (version 13-8) (2) for reference-based OTU picking with a 97% cutoff. Thus, in MSE, the comparison between query and database samples is approximately at the species level (3), although the actual taxonomic resolution varies according to taxon, due to differences in the evolutionary rates of the 16S rRNAs. Moreover, in MSE, both the relative abundance (with 16S rRNA gene copy number normalization [4]) and the phylogenetic structures of OTUs are utilized for similarity calculation (as in UniFrac [5, 6]), yet the speed is optimized by nonrecursive computing to enable real-time responses (7).
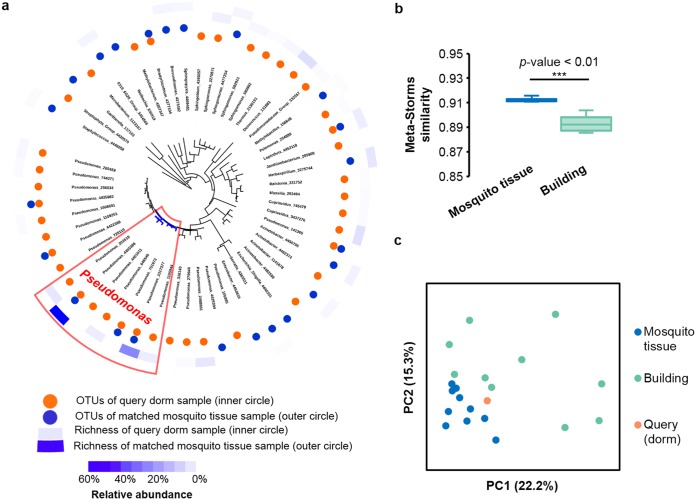
By comparing the query sample (i.e., dust from university dormitories) provided by Sun et al. (8) and the MSE top-hit samples, which are from mosquito tissues, we found that although abundant sequences of the two (query and the top-hit) samples are distributed among different OTUs (species) within the Pseudomonas genus, they are still very close in the common OTU-based phylogenetic tree (extracted from the Greengenes tree) (Fig. 1a), resulting in a high similarity of 0.916. To test whether this match is significant, we ranked this value in pairwise similarity calculation among all microbiomes (n = 177,022) in MSE [in total, (n · n – 1)/2 = 15,668,305,731 times). The resulting P value of the permutation test is 0.0009, suggesting a highly significant match. This might have revealed potential interaction or transmission between mosquitos and dust, as these mosquitos were collected from residential properties and buildings (samples for generating 16S rRNA amplicon libraries were prepared by grinding one insect or a pool of individual insects [9]) (Table 1), or it might have highlighted communities that are distinct yet still dominated by microbes that are similar to one another when the overall picture of the bacterial tree is considered.
FIG 1.
Comparison between the query microbiome (dorm dust) and the top hits reported by MSE-based searches. (a) Distribution of OTUs in the common phylogeny tree between the query and the top hit from the full MSE reference database. Those abundant OTUs from the Pseudomonas genus are marked in the red box, and the shared subbranches of the query and the hits are indicated in blue. (b) The similarities between the query sample and each of the top 10 hits against the building reference samples are significantly lower than those between the query and each of the 10 hits against the entire database, as suggested by both t test (b) and PCoA (c). PC1 and PC2, principal components 1 and 2, respectively.
TABLE 1.
Details for the top 10 hits for the query microbiome, dorm dust
| MSE database ID of top 10 hit | Habitat | Similarity | Sampling location | Sampling date (yr/mo/day) |
Reference |
|---|---|---|---|---|---|
| IDs from entire MSE database | |||||
| S_10815.C1OtvW34TOR2012 | Mosquito tissue | 0.91586 | Toronto, Canada | 2012/8/21 | 9 |
| S_10815.NOjW34MSL2012 | Mosquito tissue | 0.91350 | Toronto, Canada | 2012/7/24 | 9 |
| S_10815.3A081OjW32LAM2012 | Mosquito tissue | 0.91291 | Toronto, Canada | 2012/8/7 | 9 |
| S_10815.Can2CxW32MSL2012 | Mosquito tissue | 0.91283 | Toronto, Canada | 2012/8/22 | 9 |
| S_10815.O3AvW34TOR2012 | Mosquito tissue | 0.91260 | Toronto, Canada | 2012/6/12 | 9 |
| S_10815.Y12A2AnpW31PEE2012 | Mosquito tissue | 0.91183 | Toronto, Canada | 2012/8/1 | 9 |
| S_10815.C1AvW30TOR2012 | Mosquito tissue | 0.91134 | Toronto, Canada | 2012/7/24 | 9 |
| S_10815.Can10AvW32MSL2012 | Mosquito tissue | 0.91097 | Toronto, Canada | 2012/8/15 | 9 |
| S_10815.M1AvW32WEC2012 | Mosquito tissue | 0.91095 | Toronto, Canada | 2012/7/31 | 9 |
| S_10815.B4AvW25TOR2013 | Mosquito tissue | 0.91088 | Toronto, Canada | 2013/6/18 | 9 |
| IDs from “Building” subset of reference microbiomes in MSE database |
|||||
| S_10172.815 | Room surface dust | 0.90388 | Chicago, IL, USA | 2017/5/24 | 10 |
| S_10172.828 | Nurse station surface dust |
0.90063 | Chicago, IL, USA | 2017/5/24 | 10 |
| S_1772.H23Cb | Kitchen cutting board | 0.89745 | Raleigh-Durham, NC, USA | 2013/5/22 | 11 |
| S_10172.286 | Cold tap water | 0.89666 | Chicago, IL, USA | 2017/5/24 | 10 |
| S_10172.830 | Nurse station surface dust |
0.89300 | Chicago, IL, USA | 2017/5/24 | 10 |
| S_SRR5574403 | Kitchen dust | 0.89109 | Oakland, CA, USA | 2017/5/17 | 12 |
| S_10423.34E7LN0ZRJUQB | Carpet dust | 0.88931 | Toronto, Canada | 2004/7/14 | 13 |
| S_10172.10456 | Cold tap water | 0.88743 | Chicago, IL, USA | 2017/5/24 | 10 |
| S_10172.8331 | Glove | 0.88592 | Chicago, IL, USA | 2017/5/24 | 10 |
| S_10172.291 | Room surface dust | 0.88534 | Chicago, IL, USA | 2017/5/24 | 10 |
To test whether microbiomes from similar environments are more similar to each other than those from distinct environments, we next searched the query sample (which is dust collected inside a building) against all “building” samples in the reference database of MSE (a subset that includes 11,248 samples that were labeled as “building” from 35 studies). The similarities between the query and each of the top 10 hits (10–13) (Table 1) against the building reference samples are significantly lower than those between the query and each of the top 10 hits against the entire database (Fig. 1b) (t test P value = 2.75E–08). Findings from principal-component analysis (PCoA) support this conclusion, because the query sample is closer to the mosquito samples (i.e., to hits from the entire database) than to the building sample hits (i.e., hits from the building database) (Fig. 1c). These results suggest that microbiomes from similar environments can indeed be more different from each other than from certain samples from other environments that would intuitively be considered distinct.
In our current MSE implementation (1), the microbiome novelty score (MNS) is calculated based on the top hits against the whole reference database in MSE, rather than against only a subset of the reference microbiomes or those from a specific environment. We are grateful to Sun et al.’s suggestion of allowing the choice of reference databases when using MSE. In the upcoming release of MSE (http://mse.ac.cn), we plan to allow the selection of a specific environment or ecosystem as the reference database to search against, although we caution strongly that such restricted searches may lead to incorrect interpretation of results when the databases are not comprehensive.
Recently, amplicon sequence variant (ASV)-based approaches have been developed to improve the resolution of classifying 16S rRNA genes (14–16), but they require a unified sequencing platform and identical gene amplicon regions among the data sets. At present, the majority of historical microbiome samples were produced via a variety of platforms and amplicon regions; e.g., the V1-V3 and V3-V5 regions of 16S rRNA gene were sequenced via Roche 454 in the Human Microbiome Project (17), while the V4 region was sequenced via Illumina HiSeq and MiSeq in the Earth Microbiome Project (18). This reality limits the prospect of adopting the ASV scheme in MSE for searching against the current 16S rRNA-based microbiome data space. On the other hand, with the rapid accumulation of shotgun metagenomic data sets, we expect MSE to accommodate such data sets and eventually allow microbiome searches at the strain level, as Sun et al. have suggested.
Footnotes
This is a response to a letter by Sun et al. (https://doi.org/10.1128/mBio.00892-19).
Citation Su X, Jing G, McDonald D, Wang H, Wang Z, Gonzalez A, Sun Z, Huang S, Navas J, Knight R, Xu J. 2019. Reply to Sun et al., “Identifying composition novelty in microbiome studies: improvement of prediction accuracy.” mBio 10:e01234-19. https://doi.org/10.1128/mBio.01234-19.
Contributor Information
Keith A. Crandall, George Washington University.
Paul Keim, Northern Arizona University.
REFERENCES
- 1.Su X, Jing G, McDonald D, Wang H, Wang Z, Gonzalez A, Sun Z, Huang S, Navas J, Knight R, Xu J. 2018. Identifying and predicting novelty in microbiome studies. mBio 9:e02099-18. doi: 10.1128/mBio.02099-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Su X, Xu J, Ning K. 2012. Meta-Storms: efficient search for similar microbial communities based on a novel indexing scheme and similarity score for metagenomic data. Bioinformatics 28:2493. doi: 10.1093/bioinformatics/bts470. [DOI] [PubMed] [Google Scholar]
- 4.Jing G, Sun Z, Wang H, Gong Y, Huang S, Ning K, Xu J, Su X. 2017. Parallel-META 3: comprehensive taxonomical and functional analysis platform for efficient comparison of microbial communities. Sci Rep 7:40371. doi: 10.1038/srep40371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamady M, Lozupone C, Knight R. 2010. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J 4:17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Su X, Wang X, Jing G, Ning K. 2014. GPU-Meta-Storms: computing the structure similarities among massive amount of microbial community samples using GPU. Bioinformatics 30:1031–1033. doi: 10.1093/bioinformatics/btt736. [DOI] [PubMed] [Google Scholar]
- 8.Sun Y, Li Y, Yuan Q, Fu X. 2019. Identifying composition novelty in microbiome studies: improvement for prediction accuracy. mBio 10:e00892-19. doi: 10.1128/mBio.00892-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Novakova E, Woodhams DC, Rodríguez-Ruano SM, Brucker RM, Leff JW, Maharaj A, Amir A, Knight R, Scott J. 2017. Mosquito microbiome dynamics, a background for prevalence and seasonality of West Nile virus. Front Microbiol 8:526. doi: 10.3389/fmicb.2017.00526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lax S. 2017. Bacterial colonization and succession in a newly opened hospital. Sci Transl Med 9:eaah6500. doi: 10.1126/scitranslmed.aah6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunn RR, Fierer N, Henley JB, Leff JW, Menninger HL. 2013. Home life: factors structuring the bacterial diversity found within and between homes. PLoS One 8:e64133. doi: 10.1371/journal.pone.0064133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adams RI, Lymperopoulou DS, Misztal PK, De Cassia Pessotti R, Behie SW, Tian Y, Goldstein AH, Lindow SE, Nazaroff WW, Taylor JW, Traxler MF, Bruns TD. 2017. Microbes and associated soluble and volatile chemicals on periodically wet household surfaces. Microbiome 5:128. doi: 10.1186/s40168-017-0347-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chase J, Fouquier J, Zare M, Sonderegger DL, Knight R, Kelley ST, Siegel J, Caporaso JG. 2016. Geography and location are the primary drivers of office microbiome composition. mSystems 1:e00022-16. doi: 10.1128/mSystems.00022-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Callahan BJ, McMurdie PJ, Holmes SP. 2017. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J 11:2639–2643. doi: 10.1038/ismej.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caruso V, Song X, Asquith M, Karstens L. 2019. Performance of microbiome sequence inference methods in environments with varying biomass. mSystems 4:e00163-18. doi: 10.1128/mSystems.00163-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amir A, McDonald D, Navas-Molina JA, Kopylova E, Morton JT, Zech Xu Z, Kightley EP, Thompson LR, Hyde ER, Gonzalez A, Knight R. 2017. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2:e00191-16. doi: 10.1128/mSystems.00191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson LR, Sanders JG, McDonald D, Amir A, Ladau J, Locey KJ, Prill RJ, Tripathi A, Gibbons SM, Ackermann G, Navas-Molina JA, Janssen S, Kopylova E, Vázquez-Baeza Y, González A, Morton JT, Mirarab S, Zech Xu Z, Jiang L, Haroon MF, Kanbar J, Zhu Q, Jin Song S, Kosciolek T, Bokulich NA, Lefler J, Brislawn CJ, Humphrey G, Owens SM, Hampton-Marcell J, Berg-Lyons D, McKenzie V, Fierer N, Fuhrman JA, Clauset A, Stevens RL, Shade A, Pollard KS, Goodwin KD, Jansson JK, Gilbert JA, Knight R. 2017. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551:457–463. doi: 10.1038/nature24621. [DOI] [PMC free article] [PubMed] [Google Scholar]