ABSTRACT
Background
Proteins in human milk are essential and known to support the growth, development, protection, and health of the newborn. These proteins are highly modified by glycans that are currently being recognized as vital to protein structure, stability, function, and health of the intestinal mucosa. Although milk proteins have been studied, the quantitative changes in milk proteins and their respective site-specific glycosylation are unknown.
Objective
This study expanded the analytical tools for milk proteins and their site-specific glycosylation and applied these tools to a large cohort to determine changes in individual protein concentrations and their site-specific N-glycosylation across lactation.
Design
A tandem mass spectrometry method was applied to 231 breast-milk samples from 33 mothers in Davis, California, obtained during 7 different periods of lactation. Dynamic changes in the absolute abundances of milk proteins, as well as variation in site-specific N-glycosylation of individual proteins, were quantified.
Results
α-Lactalbumin, β-casein, k-casein, and α-antitrypsin were significantly increased from colostrum to transitional milk (4.37 ± 1.33 g/L to 6.41 ± 0.72 g/L, 2.25 ± 0.86 g/L to 2.59 ± 0.78 g/L, 1.33 ± 0.44 g/L to 1.60 ± 0.39 g/L, and 0.09 ± 0.10 g/L to 0.11 ± 0.04 g/L, respectively; P < 0.002). α-Lactalbumin (37%), β-casein (9%), and lysozyme (159%) were higher in mature milk than in colostrum. Glycans exhibited different behavior. Fucosylated glycans of lactoferrin and high-mannose, undecorated, fucosylated, sialylated, and combined fucosylated + sialylated glycans of secretory immunoglobulin A increased during lactation even when the concentrations of the parent proteins decreased.
Conclusions
Proteins in healthy mothers vary dynamically through lactation to support the development of infants. Individual milk proteins carried unique glycan modifications that varied systematically in structure even with site specificity. The role of glycosylation in human milk proteins will be important in understanding the functional components of human milk. This trial was registered at clinicaltrials.gov as NCT01817127.
Keywords: human milk proteins, site-specific N-glycosylation, lactation, mass spectrometry, fucosylation, sialylation
Introduction
Human milk is the remarkable product of the selective pressures on the mammary gland through evolution as the primary source of nutrients for the newborn. In the past few decades, human milk has been studied extensively to understand its nutritional value. In addition to delivering nutrients, milk provides many physiological benefits, such as the development of immunity and the maintenance of gut health for the newborn (1–4). Human milk contains a wide diversity of biologically active components, including carbohydrates, proteins, endogenous peptides, lipids, and minerals (5–8). Milk proteins are the fourth most abundant component in human milk, consisting of both caseins and whey proteins with a total concentration range of 10–20 mg/mL (9).
Variation in caseins and whey proteins in human milk is thought to be important because these proteins play many roles in the newborn's health and development. The three main types of caseins present in human milk are β-, k-, and α-casein. Caseins have been considered to be of predominantly nutritional value because they are completely digested by the infant's gastrointestinal proteases. However, functions of casein proteins and peptides continue to emerge with ongoing research. For example, caseins promote calcium absorption through the action of phosphopeptides, whose heavily phosphorylated groups can chelate Ca2+ (10). In addition, β-casein has shown antimicrobial properties toward streptococci (11) and Haemophilus influenza (12), and k-casein has been shown to inhibit the adhesion of Helicobacter pylori to the human gastric mucosa (13).
Whey proteins in milk consist of a wide variety of proteins. In addition to providing essential amino acids and energy, these proteins have exhibited a wide range of biological actions, including antimicrobial activity, prebiotic activity, and assisting the digestion of proteins, all of which promote healthy development of the newborn. α-Lactalbumin is responsible for lactose synthesis in the mammary gland and enhancing calcium, iron, and zinc absorption (14). Proteolytic fragments of α-lactalbumin can block harmful pathogens and encourage the growth of beneficial micro-organisms, such as Bifidobacteria (15, 16). Lactoferrin (LF), another abundant whey protein in milk that is known to facilitate the absorption of iron, has multiple physiological functions, including antiviral and antibacterial activities (17). Furthermore, immunoglobulins [secretory IgA (SIgA), IgM, and IgG], osteopontin, and lysozyme exert antipathogenic activities (18–22), whereas α-antitrypsin in milk guides digestion by functioning as a protease inhibitor (23).
A large number of milk proteins are known to be glycosylated. N-glycosylation is one of the most complicated but commonly occurring post-translational modifications of proteins. Glycosylation plays a variety of roles in the functions of proteins by stabilizing the three-dimensional structure and facilitating cell–cell recognition and cell signaling (24–26). It is not known precisely how, but the structure and functions of glycans on milk proteins may interact with the infant in various ways. There is evidence that glycosylated proteins act as an extra line of defense, where glycans attached to milk proteins have structural features that function as pathogen adhesion sites (9, 27, 28). Glycosylated proteins may also guide the community competition and functions of the intestinal microbiome. N-glycans are released through specialized enzymes from Bifidobacteria to provide free glycans as additional prebiotic compounds (29). To date, a very limited number of studies have focused on milk protein glycosylation during lactation. These studies release glycans and are therefore unable to capture many of the functions of glycosylated proteins because deglycosylation loses the information about the proteins of origin (27, 30).
In the current study, we measured the concentrations of >90% (by abundances) of the proteins in milk from healthy mothers during lactation. The proteins quantified include α-casein, β-casein, k-casein, α-lactalbumin, LF, SIgA, IgM, IgG, α-antitrypsin, osteopontin, and lysozyme. The respective glycosylations of the individual proteins were quantitatively monitored using dynamic multiple-reaction monitoring (MRM), yielding variations in site-specific glycosylation (31). The samples were obtained through the lactation study sponsored by the Foods for Health Institute at the University of California, Davis.
Methods
Materials and chemicals
Protein standards including α-lactalbumin, LF, IgA, IgG, IgM, and α-antitrypsin were purchased from Sigma-Aldrich. Human lysozyme was purchased from Lee Biosolutions. Standard peptides for the β-, k-, and α-caseins and osteopontin were purchased from Bionexus. Sequencing grade modified trypsin and dithiothreitol were purchased from Promega. Iodoacetamide was purchased from Sigma-Aldrich.
Milk samples
A total of 231 human milk samples were collected from 33 healthy mothers enrolled in the Food for Health Institute lactation study at the University of California, Davis. Milk samples were collected postnatally from mothers who gave birth to full-term infants (>37 weeks of gestation). Samples were binned into 7 groups to evaluate the changes in proteins and site-specific glycosylation across lactation: colostrum (days 2–5), week 2 (days 12–15), week 5 (days 28–42), week 10 (days 71–78), week 13 (days 85–92), week 17 (days 106–130), and week 24 (days 170–189). The University of California, Davis, institutional review board approved every aspect of this study. Written informed consent was obtained from each participant. Milk samples were collected in the morning using a modified, published method (32).
Trypsin digestion of a pooled milk sample for Q Exactive Plus Orbitrap LC-MS/MS analysis
A pooled milk sample from all the donors in this study was used for proteomic analysis. A 25-μL aliquot of the pooled milk sample was diluted in 175 μL of 50 mM ammonium bicarbonate. The sample was then reduced with 2 μL of 550 mM dithiothreitol at 60°C in a waterbath for 50 min and alkylated with 4 μL of 450 mM iodoacetamide for 30 min in the dark. Proteins were digested by adding 2 μg of trypsin and incubating for 18 h at 37°C.
Trypsin digestion of standards and human milk samples for ultraperformance LC–electrospray ionization–triple quadrupole analysis
For the quantification of milk proteins, accurate amounts of protein standards (200 g of LF and α-lactalbumin individually; 100 g of IgA; and 20 g of IgG, IgM, α-antitrypsin, and lysozyme individually) were used to make a standard protein mixture. For the quantification of caseins and osteopontin, accurate amounts of peptide standards, including 100 μg of β-casein peptide 1 (SPTIPFFDPQIPK), 50 μg of β-casein peptide 2 (VLPIPQQVVPYPQR), 20 μg of k-casein peptide 1 (RPAIAINNPYVPR), 20 μg of k-casein peptide 2 (TYYANPAVVRPHAQIPQR), 20 μg of α-casein peptide 1 (CAEQFCR), 20 μg of α-casein peptide 2 (NNVMLQW), and 20 μg of osteopontin peptide (GDSVVYGLR), were used to make a standard peptide mixture. Standard mixtures of proteins, peptides, and 25 μL of each milk sample were reduced, alkylated, and digested with trypsin in a 96-well plate under the same conditions as previously described (31). Digested proteins were purified with a C18 96-well cartridge plate. Peptides and glycopeptides were completely dried before MRM analysis.
Q Exactive Plus Orbitrap LC-MS/MS analysis
The tryptic digest of milk proteins was reconstituted with 2% (vol:vol) acetonitrile (ACN) in 0.1% formic acid (FA) prior to analysis with a Q Exactive Plus Orbitrap mass spectrometer in conjunction with an EASY-nLC II nano ultra-HPLC and Proxeon nanospray source. Peptides and glycopeptides were loaded on a 100 μm × 25 mm Magic C18 reverse-phase trap column prior to separation using a 75 μm × 150 mm Magic C18 reverse-phase column. A binary gradient containing 0.1% (vol:vol) FA in water (solvent A) and 100% ACN (solvent B) with a flow rate of 3 μL/min was used. MS spectra were obtained with an m/z of 300–1900. MS and MS/MS spectra for the tryptic peptides were acquired in the positive ion mode.
Ultraperformance LC–electrospray ionization–triple quadrupole analysis
For quantification, peptide and glycopeptide samples were analyzed using an Agilent 1290 Infinity ultraperformance LC system coupled with an Agilent 6490 triple quadrupole mass spectrometer. An Agilent Eclipse Plus C18 column (RRHD 1.8 μm, 2.1 mm × 100 mm) was used for LC separation. Standards and milk samples were reconstituted with 100 μL of nanopure water and 1.0 μL was injected for the analysis. To obtain protein concentrations, standard proteins and peptides were diluted serially to acquire calibration curves. Peptides and glycopeptides were separated using a 16-min binary gradient consisting of solvent A of 3% ACN and 0.1% FA and solvent B of 90% ACN and 0.1% FA in pure water (vol:vol) at a flow rate of 0.5 mL/min. The instrument was operated in the dynamic MRM mode with unit resolution, and peptide and glycopeptide ionization was performed in the positive ion mode.
Data processing and statistical analysis
Q Exactive Plus Orbitrap LC-MS/MS data for tryptic peptides were extracted using X! Tandem (www.thegpm.org), which is designed to search the SwissProt human proteome database. Masses allowed a 10-ppm error and 2 missed cleavages. Iodoacetamide derivative was chosen as a fix modification on cysteine. Phosphorylation in threonine, serine, and tyrosine, oxidation in methionine and tryptophan, and deamidation in asparagine and glutamine were selected as variable modification.
Triple quadrupole MS data for peak area and signal-to-noise ratio were analyzed using Agilent Mass-Hunter Quantitative Analysis B.6.0 software. To evaluate the sensitivity of this method, linear regression was applied in Microsoft Excel 2010, and the limits of quantification were determined as the concentrations of peptides with signal-to-noise ratio ≥10. Absolute protein concentrations were obtained for all 33 mothers at 7 lactation time points using calibration curves. Relative abundances of glycopeptides were calculated by normalizing the glycopeptide abundance to the quantifying peptide from the same glycoprotein.
Statistical analysis was performed using JMP Pro 13 (SAS Institute), and P ≤ 0.05 was considered statistically significant. Concentrations of the proteins and the relative abundances of the glycopeptides were transformed into natural log scale before any analysis. To observe the effect of the lactation time points on milk proteins and glycosylation, ANOVA and Wilcoxon's test were used. Student's t-test and nonparametric comparisons test were used to compare between two lactation stages. Means and SDs were calculated for the absolute abundances of proteins and relative abundances of glycoproteins and used to prepare graphical plots.
Results
Proteomic analysis was performed first to determine the identity of the proteins in human milk. A tryptic digest of pooled milk samples from all the donors was analyzed on a Q Exactive Plus Orbitrap LC-MS/MS instrument to identify milk proteins. We observed peptides corresponding to 639 milk proteins with a 99% confidence level (Supplemental Table 1). Among the identified proteins, 11 proteins represented >90% of the total abundances. This study thus focused efforts on the quantification of this most abundant group of proteins: α-lactalbumin, LF, β-casein, k-casein, α-casein, IgA, osteopontin, IgG, IgM, α-antitrypsin, and lysozyme.
Selection of peptides and glycopeptides for quantification
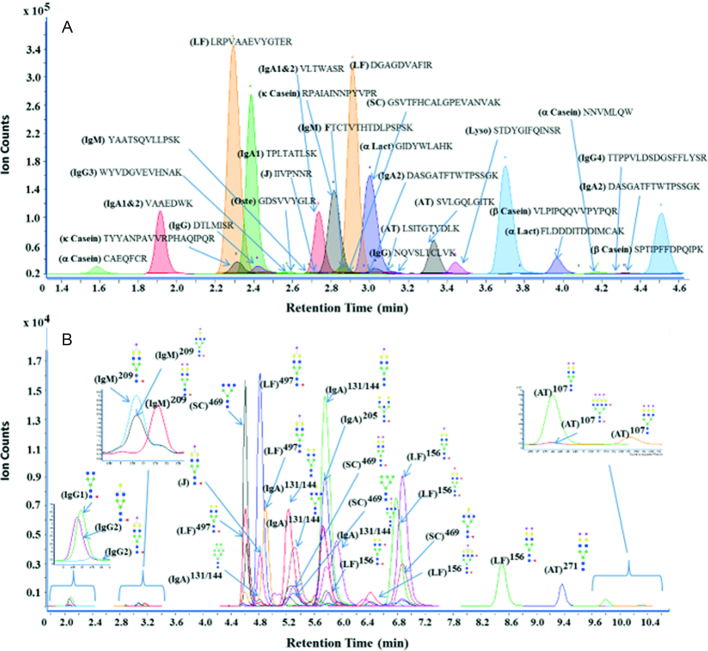
For the quantification of proteins, a novel strategy was developed to identify unique and unmodified peptides from each protein. A minimum of 2 unique peptides were selected for protein quantification. However, only 1 unique peptide for osteopontin was included because the majority of its tryptic peptides were post-translationally modified. The fragmentation pattern of each peptide was evaluated to identify the precursor ion and a minimum of 2 product ions (quantifier and qualifier) that were added to the MRM transitional list. For example, Supplemental Figure 1 shows the MS/MS spectrum of the α-casein peptide CAEQFCR and illustrates abundant b- and y- ions. For this peptide, the doubly charged quasimolecular ion ([M + 2H]+m/z 485.7) was chosen as the precursor ion, whereas the most abundant fragment ion m/z 739.3 was selected as a quantifier and m/z 610.3 was selected as a qualifier. For quantification, MRM transitions of tryptic peptides were optimized for their quantifier, qualifier, collision energy, and retention time. To increase the peak capacity, a dynamic MRM method was employed in which the analyte was monitored only when eluted. This reduced the number of concurrent transitions observed at a given time, rendering dynamic MRM to be very selective and sensitive. For example, the retention time for the α-casein peptide mentioned previously was 1.7 min with an optimized collision energy of 15 eV. MRM transitions for caseins and osteopontin peptides are listed with their precursor masses, product masses, retention times, and collision energies in Supplemental Table 2. MRM transitions for other whey proteins were previously optimized and reported (31). The chromatographic separation of peptides from α-casein, β-casein, k-casein, α-lactalbumin, LF, SIgA, IgM, IgG, α-antitrypsin, osteopontin, and lysozyme is shown in Figure 1A, with the separation of all peptides observed between 1.4 and 4.5 min.
FIGURE 1.
MRM chromatograms of (A) peptides and (B) glycopeptides in human milk. Blue squares, green circles, yellow circles, red triangles, and purple diamonds represent N-acetylglucosamine, mannose, galactose, fucose, and sialic acid, respectively. AT, α-antitrypsin; J, J chain; α-Lact, α-lactalbumin; LF, lactoferrin; Lyso, lysozyme; MRM, multiple reaction monitoring; Oste, osteopontin; SC, secretory component.
For glycopeptides, oxonium ions corresponding to m/z 204.1 for N-acetylhexosamine and m/z 366.1 for N-acetylhexosamine + hexose were universally the most abundant fragments observed under collision-induced dissociation (31). These ions were therefore used as the quantifiers for the glycopeptides. In the method, a total of 114 glycopeptides were monitored from LF, SIgA, IgM, IgG, and α-antitrypsin. Chromatographic separation of glycopeptides is shown in Figure 1B. Interestingly, glycopeptides containing the same peptide backbone with different glycans attached were eluted in similar retention time windows.
Quantification of milk proteins and glycopeptides
For absolute quantification, various concentrations of protein standards were used to prepare a standard mixture that was serially diluted to build the calibration curves. Because standard proteins of human milk caseins and osteopontin are not available, synthesized tryptic peptides were used for their absolute quantification. To quantify caseins and osteopontin, a single mixture with various concentrations of peptide standards was prepared. The resulting calibration curves for the previously mentioned peptides are shown in Supplemental Figure 2. All peptides exhibited very good linearity, with a regression coefficient >0.997. The linear ranges for the peptides spanned 3 orders of magnitude, which is sufficient to capture biological diversity. Abundances of the peptides in each protein were calculated using calibration curves, and the mass ratio between the peptide and the corresponding protein was used to calculate the absolute abundance of the protein present in milk. Limits of quantification for the proteins were observed at the femtomole level (Supplemental Table 2).
Absolute abundances of glycopeptides are complicated by the lack of commercial standards. To measure fold changes, the abundance of the glycopeptides was normalized to the protein abundance. This decoupled the degree of glycosylation to the variation in protein concentrations. A more extensive description of this method and its utility has been provided previously (31).
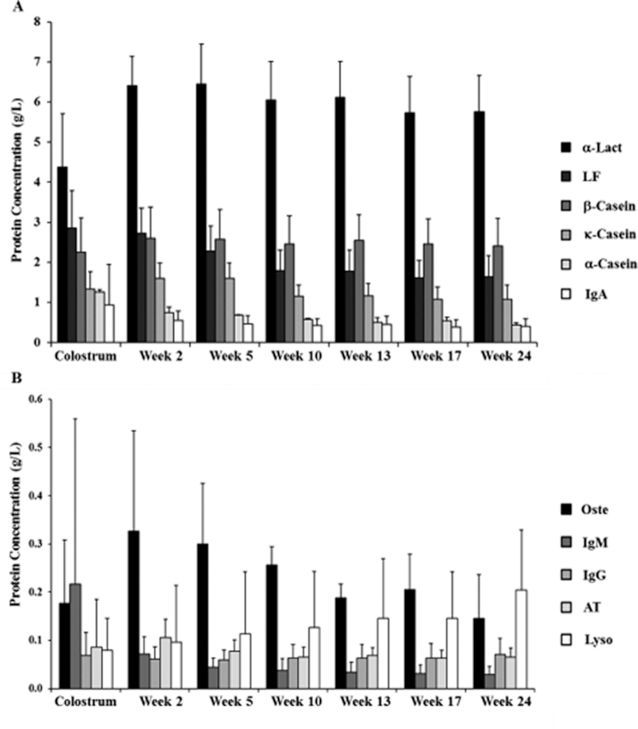
Dynamics of human milk proteins across lactation (colostrum to week 24)
The concentrations of the milk proteins throughout lactation are shown in Figure 2 and Supplemental Table 3. β-Casein was the most abundant casein in human milk. The concentration of β-casein showed a significant increase (P = 0.004) from colostrum to week 2 and gradually decreased until week 24. A similar trend was observed for k-casein across lactation, in which it exhibited a significant increase (P = 0.002) from colostrum to week 2 and gradually decreased until week 24. However, we observed a significant decrease (P = 0.011) in the concentration of k-casein from week 5 to week 10. In contrast to β- and k-casein, α-casein continuously decreased from colostrum to week 24, with a large decrease (P = 0.0004) from colostrum to week 2.
FIGURE 2.
Dynamics of human milk proteins across lactation in the order of decreasing concentration: (A) first 6 abundant proteins; (B) next 5 abundant proteins. Values are mean ± SDs, n = 33. AT, α-antitrypsin; α-Lact, α-lactalbumin; LF, lactoferrin; Lyso, lysozyme; Oste, osteopontin.
α-Lactalbumin exhibited a significant increase (P < 0.0001) in concentration from colostrum to week 2. It gradually decreased after week 2, and the concentration remained higher than in colostrum at week 24. After the first 2 time points, all the other subsequent days had no major changes. In contrast, the concentration of LF decreased throughout the lactation period from colostrum to week 24. A significant decrease was observed between week 2 and week 5 (P = 0.009) and also between week 5 and week 10 (P = 0.0007). Interestingly, the concentration of osteopontin increased from colostrum to week 2, although a gradual decrease was observed during the later stages of lactation. No significant differences were observed among lactation stages. A similar trend was observed for α-antitrypsin, in which its concentration significantly increased (P < 0.0001) from colostrum to week 2 and then significantly decreased (P < 0.0001) from week 2 to week 10. There was no significant change in concentration from week 10 to week 24. In contrast to all of the proteins observed, the concentration of lysozyme continuously increased from colostrum to week 24, with a significant increase (P < 0.029) from week 17 to week 24. The study also quantified immunoglobulins in human milk, including IgA, IgM, and IgG. IgA and IgM concentrations were higher in early lactation and gradually decreased in later lactation. In contrast, IgG remained mostly unchanged across lactation.
To evaluate the dynamic nature of milk proteins during lactation (colostrum to week 24), the percentage changes in protein concentrations were determined relative to the protein concentrations on day 3 (Figure 3). For α-casein, k-casein, LF, osteopontin, α-antitrypsin, IgA, and IgM, the concentrations decreased from colostrum to week 24. A group of proteins including osteopontin, k-casein, α-antitrypsin, and LF decreased <50% in abundances, whereas another group including IgA, α-casein, and IgM decreased even more. IgM had the largest decrease of 89% across lactation. In contrast, α-lactalbumin, lysozyme, and β-casein concentrations increased from colostrum to week 24. β-Casein and α-lactalbumin increased <50%, whereas lysozyme increased the most (159%).
FIGURE 3.
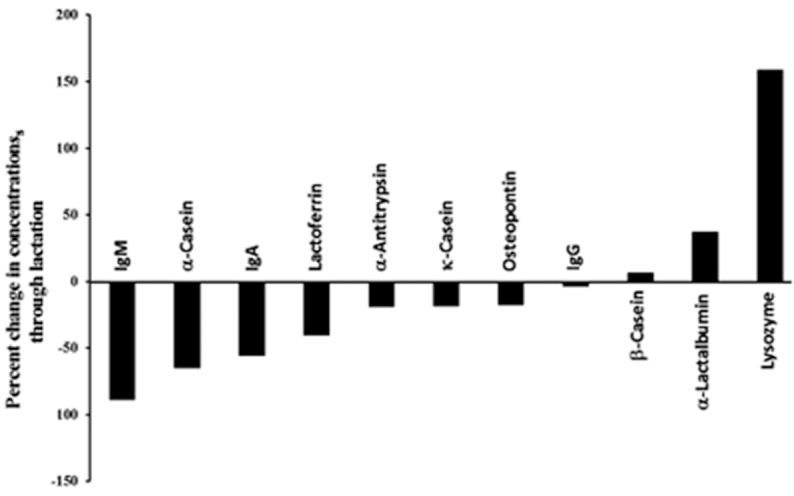
Percentage changes in protein concentrations across lactation (colostrum to week 24) for 11 proteins. Data are reported as mean values, n = 33.
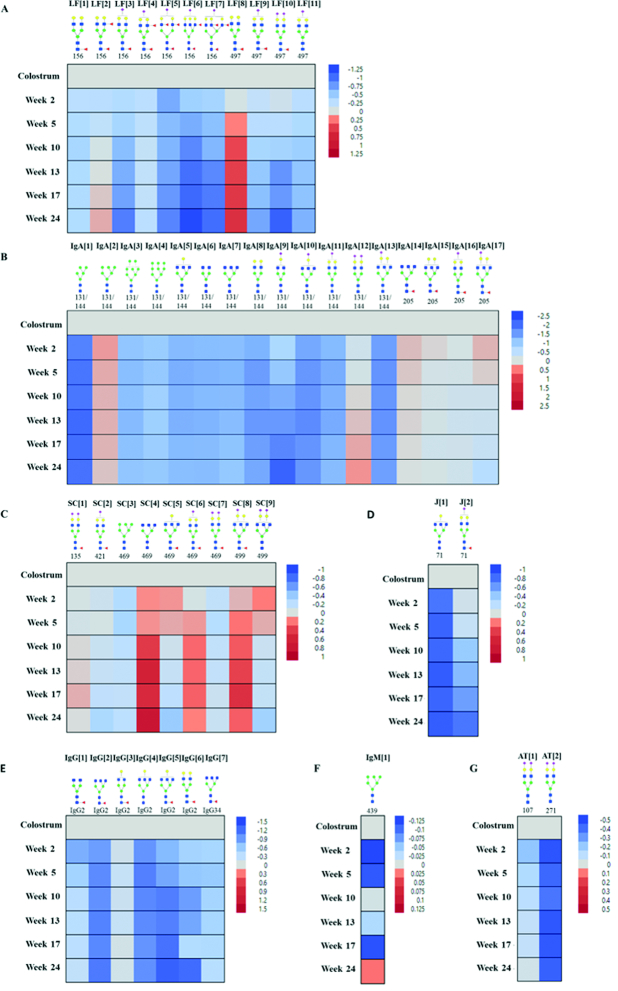
Dynamics of human milk site-specific N-glycosylation across lactation
Quantification of the site-specific glycosylation of the glycoproteins including LF, IgA, IgG, IgM, and α-antitrypsin during lactation was performed. Shown in Figure 4 are the fold changes associated with the more abundant glycans at specific glycosites relative to colostrum. In LF, 2 of the 3 sites are glycosylated, and the glycans on the left are on site N156 (asparagine 156), whereas most of those on the right belong to N497 (Figure 4A). Glycans at the two sites were mainly complex types that were fucosylated only (fucosylated), sialylated only (sialylated), and simultaneously fucosylated and sialylated (fucosylated + sialylated). Those that are neither fucosylated nor sialylated are termed undecorated. Fucosylated glycans increased at both sites during lactation. The monofucosylated biantennary LF[8] at site 497 and the difucosylated biantennary LF[2] at site 156 illustrate this behavior. Relative to colostrum, LF[8] increased slightly in week 2 (Figure 4A, slight pink) and increased more strongly and remained constant after week 10. Interestingly, LF[2] decreased immediately after colostrum (week 2) and increased in mature milk (week 10 to week 24). Sialylated and sialylated + fucosylated glycans generally decreased (Figure 4A, light blue; week 2) and more strongly across lactation (week 5 to week 24). They include LF[11] for sialylated glycans and LF[3], LF[5], LF[6], LF[7], and LF[10] for sialylated + fucosylated glycans.
FIGURE 4.
Variations in site-specific glycosylation across lactation of selected glycoproteins: (A) lactoferrin, (B) heavy chain of IgA, (C) secretory component of IgA, (D) J chain, (E) IgG, (F) IgM, and (G) α-antitrypsin. Blue squares, green circles, yellow circles, red triangles, and purple diamonds represent N-acetylglucosamine, mannose, galactose, fucose, and sialic acid, respectively. Data are reported as log value (mean) of fold change relative to colostrum, n = 33. AT, α-antitrypsin; J, J chain; LF, lactoferrin; SC, secretory component.
SIgA is the most abundant immunoglobulin in milk. The site-specific glycosylation of IgA1, IgA2, and secretory component (SC) and J chain was monitored and consisted of nearly 50 N-glycans across 8 glycosites. However, only 28 glycans were able to be quantified. IgA1 site N144 and IgA2 site N131 share the same tryptic backbone and cannot be separated in this method. The glycosites in SIgA were occupied with high-mannose and complex type glycans, including undecorated, fucosylated, sialylated, and fucosylated + sialylated glycans. In the heavy chain of SIgA (Figure 4B), we observed an increase in the abundance of Man6 IgA[2]. This behavior was accompanied by a decrease in Man5 IgA[1], Man8 IgA[3], and Man9 IgA[4]. Interestingly, the sialylation appeared to increase, as illustrated by IgA[12]; however, fucosylated + sialylated glycans, IgA[16] and IgA[17], increased at week 2, relative to colostrum, but decreased at week 5. The fucosylated glycans IgA[14] and IgA[15] increased at weeks 2 and 5 and decreased to the levels of colostrum thereafter.
Glycosylation in the SC of the IgA behaved dramatically, in that 3 species increased markedly compared to colostrum (Figure 4C): undecorated complex-type glycan SC[4] and fucosylated + sialylated glycan SC[6] on site 469 and SC[8] on site 499. Interestingly, the triantennary structures SC[4], SC[5], SC[6], SC[8], and SC[9] all increased in weeks 2 and 5. The J chain was found with 2 structures, but both decreased relative to colostrum (Figure 4D).
The glycosylation of IgG was monitored for all 4 subclasses (IgG1, IgG2, IgG3, and IgG4), with each subclass containing a single glycosylation site. The amino acid composition of the tryptic glycopeptides for IgG3 and IgG4 are identical and could not be distinguished with this method. A total of 25 glycan compositions from IgG were detected in the glycoproteomic map; however, only 7 structures had abundances suitable for quantification. All the glycans decreased immediately after colostrum and remained lower than colostrum in mature milk (Figure 4E). IgM had a single glycan that could be quantified, which had variable expression during lactation (Figure 4F).
α-Antitrypsin contained 3 N-glycosites that were mainly fucosylated and sialylated complex-type glycans. We observed 11 glycans in α-antitrypsin in all the sites; however, only 2 at different sites could be quantified. The abundance of the disialylated species at sites 107 AT[1] and 271 AT[2] decreased from colostrum to transitional milk and remained constant. AT[2] decreased more significantly (Figure 4G).
Discussion
This study represents the most extensive analysis of individual proteins during lactation and the first to examine variations in glycosylation on multiple glycoproteins. Total milk protein concentration is known to decrease during the lactation period (33). The results obtained in this study indicate that this trend is not general for all proteins, and the behavior is more complicated at the protein level. Nonetheless, the values obtained in this study are in general agreement with those of previous reports involving selected proteins, such as β-casein, k-casein, α-casein, LF, lysozyme, and α-lactalbumin (6, 34–36). Layered on top of the protein abundances is the glycan expression, which appears to be independent of protein expression. For example, the protein SIgA generally decreased across lactation, whereas several of the glycans increased during the same period.
A major role of milk is to develop the microbiome (37). In this regard, the glycans in milk proteins may play a complementary function, which includes pathogen defense and delivery for prebiotic compounds. In pathogen defense, the immunoglobulins represent the active immune system, and LF and lysozyme represent the passive immune system. The immunoglobulins have high concentrations early in lactation, and these decrease as the milk matures. IgG remains constant or slightly increases. As the infant's immune system develops, there is less of a need for the mother's immune system to protect the infant. Therefore, the immunoglobulins delivered to the infant decrease. Interestingly, the passive immune system appears to play a greater role as the concentration of lysozyme increases dramatically. Lysozyme is an enzyme that hydrolyzes the cell walls of gram-positive bacteria, and together with LF, lysozyme destroys gram-negative bacteria (38).
The understanding of the role of protein glycosylation is less developed, but the analytical methods employed in this study provide new insight. It is perhaps remarkable that with 33 mothers, the behavior of the glycans is generally correlated. This behavior is consistent throughout lactation for various types of glycans. Glycosylation in proteins may take on several roles. It can protect the peptide backbone from proteolysis (39). Bioactive peptides are produced in milk, and the extent of proteolysis is affected by the glycosylation on the parent protein. Glycans also play critical roles in the ability of proteins to block pathogens (9, 27, 28). The different glycoforms interact with specific bacteria more effectively. In LF, we found glycans with 1 or 2 fucose residues at sites 156 and 497. These glycans further increase in abundance in mature milk. The results are in agreement with those of a previous study performed in our laboratory that showed an increase in fucosylation of LF in mature milk (27). Core fucose is known to control biological activities including adhesion and cell–cell signaling (40, 41). Glycans with terminal fucose are Lewis family glycotopes and bind to lectin receptors of viruses and bacteria to block adhesion to epithelial cells, thereby preventing invasion and colonization (27, 42). Barboza et al. (27) showed that the enzymatic release of fucose from intact LF significantly increased the adhesion of Salmonella enterica typhimurium to host cells. Indeed, fucosylated glycans increased or at least remained constant during lactation.
SIgA proteins were found to be high in mono- and disialylated glycans. Increases in fucosylation and sialylation were found in the SC and the heavy chain of SIgA. SIgA is the predominant immunoglobulin that protects the surface mucosa against pathogen invasions. Sialylated glycans in the SIgA bind to S-fimbriated E. coli to prevent it from binding to epithelial cells. Escherichia coli causes meningitis and sepsis in newborns (43). The secretory component, also sialylated, is known to bind to toxin A (from Clostridium difficile) as well as E. coli to prevent them from interacting with epithelial cell surfaces (44). Abundant sialylated glycans are also found in antitrypsin, which suggests that it too may function as a pathogen block.
High-mannose glycans are found on SIgA and IgM glycoproteins. We observed Man5 to Man9 in various combinations on IgA and IgM. Both glycoproteins are richer in high-mannose glycans in colostrum. High-mannose-type glycans in IgA have been shown to aggregate Enterobacteriaceae-carrying type 1 fimbriae through mannose-specific lectins (45). Glycans with terminal N-acetylglucosamine were also observed in SIgA, with higher abundance in colostrum. These glycans are known to also bind to mannose-binding lectins in bacteria (46).
The intestinal microbiota of breastfed newborns has historically been dominated by Bifidobacterium species (37). The glycosidase present in Bifidobacteria longhum subsp. infantis releases N-glycans by cleavage of the chitobiose core. Protein glycosylation can therefore further affect the microbiome by releasing N-glycans as free oligosaccharides. These compounds would complement the existing free oligosaccharides that are abundant in human milk, providing nutrition for the gut microbiota.
In conclusion, through advanced analytical methods such as dynamic MRM, studies were performed to quantify human milk proteins and their site-specific glycosylation. Novel analytical protocols were developed that are capable of characterizing all the changes that occur in human milk, enabling more detailed understanding of the functions of individual components and how they work in unison and also a better understanding of the general role of food materials as they are consumed. Indeed, there is no single trend regarding protein concentration, and each protein behaves according to the function it provides. Although glycosylation increases the complexity of proteins, such post-translational modifications also provide new opportunities. The abundance of these proteins along with the changes in their glycosylation play roles in the development and protection of the newborn. The complexity of human milk suggests more than a single function for food. Nutrition will need to expand beyond essential nutrients, energy provision, and growth.
Supplementary Material
Acknowledgments
The authors’ responsibilities were as follows—EG, JH, JBG, and CBL: designed the research; EG: conducted the research and analyzed the data; EG, GX, LW, JTS, JBG, and CBL: wrote the manuscript; CBL: had primary responsibility for final content; and all authors: read and approved the final manuscript.
Notes
Funding for this study was provided by the NIH (R01GM049077 and AT007079), the Bill & Melinda Gates Foundation, and the Mills’ Group.
Author disclosures: EG, JH, GX, LW, JTS, JBG, and CBL, no conflicts of interest.
Supplemental Tables 1–3 and Supplemental Figures 1 and 2 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/jn/.
Abbreviations used: ACN, acetonitrile; FA, formic acid; LF, lactoferrin; MRM, multiple-reaction monitoring; SC, secretory component; SIgA, secretory IgA.
References
- 1. Kramer MS, Chalmers B, Hodnett ED, Sevkovskaya Z, Dzikovich I, Shapiro S, Collet JP, Vanilovich I, Mezen I, Ducruet T et al.. Promotion of breastfeeding intervention trial (PROBIT)—a randomized trial in the Republic of Belarus. J Am Med Assoc. 2001;285:413–20. [DOI] [PubMed] [Google Scholar]
- 2. Dewey KG, Heinig J, Nommsenrivers LA. Differences in morbidity between breast-fed and formula-fed infants. J Pediatr. 1995;126:696–702. [DOI] [PubMed] [Google Scholar]
- 3. Isolauri E. Development of healthy gut microbiota early in life. J Paediatr Child Health. 2012;48:1–6. [DOI] [PubMed] [Google Scholar]
- 4. Charbonneau MR, O'Donnell D, Blanton LV, Totten SM, Davis JCC, Barratt MJ, Cheng JY, Guruge J, Talcott M, Brain JR et al.. Sialylated milk oligosaccharides promote microbiota-dependent growth in models of infant undernutrition. Cell. 2016;164:859–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ruhaak LR, Lebrilla CB.. Advances in analysis of human milk oligosaccharides. Adv Nutr. 2012;3:406S–14S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lonnerdal B, Erdmann P, Thakkar SK, Sauser J, Destaillats F. Longitudinal evolution of true protein, amino acids and bioactive proteins in breast milk: a developmental perspective. J Nutr Biochem. 2017;41:1–11. [DOI] [PubMed] [Google Scholar]
- 7. Dallas DC, Guerrero A, Khaldi N, Castillo PA, Martin WF, Smilowitz JT, Bevins CL, Barile D, German JB, Lebrilla CB. Extensive in vivo human milk peptidomics reveals specific proteolysis yielding protective antimicrobial peptides. J Proteome Res. 2013;12:2295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee H, Lerno LA Jr, Choe Y, Chu CS, Gillies LA, Grimm R, Lebrilla CB, German JB. Multiple precursor ion scanning of gangliosides and sulfatides with a reversed-phase microfluidic chip and quadrupole time-of-flight mass spectrometry. Anal Chem. 2012;84:5905–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Froehlich JW, Dodds ED, Barboza M, McJimpsey EL, Seipert RR, Francis J, An HJ, Freeman S, German JB, Lebrilla CB. Glycoprotein expression in human milk during lactation. J Agric Food Chem. 2010;58:6440–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sato R, Noguchi T, Naito H. Casein phosphopeptide (Cpp) enhances calcium absorption from the ligated segment of rat small intestine. J Nutr Sci Vitaminol. 1986;32:67–76. [DOI] [PubMed] [Google Scholar]
- 11. Danielsson Niemi L, Hernell O, Johansson I. Human milk compounds inhibiting adhesion of mutans streptococci to host ligand-coated hydroxyapatite in vitro. Caries Res. 2009;43:171–8. [DOI] [PubMed] [Google Scholar]
- 12. Kroening TA, Baxter JH, Anderson SA, Hards RG, Harvey L, Mukerji P. Concentrations and anti-Haemophilusinfluenzae activities of β-casein phosphoforms in human milk. J Pediatr Gastroenterol Nutr. 1999;28:486–91. [DOI] [PubMed] [Google Scholar]
- 13. Stromqvist M, Falk P, Bergstrom S, Hansson L, Lonnerdal B, Normark S, Hernell O. Human-milk κ-casein and inhibition of Helicobacterpylori adhesion to human gastric mucosa. J Pediatr Gastroenterol Nutr. 1995;21:288–96. [DOI] [PubMed] [Google Scholar]
- 14. Lonnerdal B. Infant formula and infant nutrition: bioactive proteins of human milk and implications for composition of infant formulas. Am J Clin Nutr. 2014;99:712S–7S. [DOI] [PubMed] [Google Scholar]
- 15. Bruck WM, Graverholt G, Gibson GR. Use of batch culture and a two-stage continuous culture system to study the effect of supplemental α-lactalbumin and glycomacropeptide on mixed populations of human gut bacteria. FEMS Microbiol Ecol. 2002;41:231–7. [DOI] [PubMed] [Google Scholar]
- 16. Ebner K, Brodbeck U.. Biological role of α-lactalbumin: a review. J Dairy Sci. 1968;51:317–22. [DOI] [PubMed] [Google Scholar]
- 17. de Ferrer PAR, Baroni A, Sambucetti ME, Lopez NE, Cernadas JMC. Lactoferrin levels in term and preterm milk. J Am Coll Nutr. 2000;19:370–3. [DOI] [PubMed] [Google Scholar]
- 18. Goldman AS. The immune system of human milk: antimicrobial, antiinflammatory and immunomodulating properties. Pediatr Infect Dis J. 1993;12:664–71. [DOI] [PubMed] [Google Scholar]
- 19. Mantis NJ, Rol N, Corthesy B. Secretory IgA's complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 2011;4:603–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ashkar S, Weber GF, Panoutsakopoulou V, Sanchirico ME, Jansson M, Zawaideh S, Rittling SR, Denhardt DT, Glimcher MJ, Cantor H. η-1 (osteopontin): An early component of type-1 (cell-mediated) immunity. Science. 2000;287:860–4. [DOI] [PubMed] [Google Scholar]
- 21. Giachelli CM, Lombardi D, Johnson RJ, Murry CE, Almeida M. Evidence for a role of osteopontin in macrophage infiltration in response to pathological stimuli in vivo. Am J Pathol. 1998;152:353–8. [PMC free article] [PubMed] [Google Scholar]
- 22. Newburg DS, Walker WA.. Protection of the neonate by the innate immune system of developing gut and of human milk. Pediatr Res. 2007;61:2–8. [DOI] [PubMed] [Google Scholar]
- 23. Lonnerdal B. Nutritional and physiologic significance of human milk proteins. Am J Clin Nutr. 2003;77:1537s–43s. [DOI] [PubMed] [Google Scholar]
- 24. Shental-Bechor D, Levy Y.. Folding of glycoproteins: toward understanding the biophysics of the glycosylation code. Curr Opin Struct Biol. 2009;19:524–33. [DOI] [PubMed] [Google Scholar]
- 25. Fukuda MN, Sasaki H, Lopez L, Fukuda M. Survival of recombinant erythropoietin in the circulation: the role of carbohydrates. Blood. 1989;73:84–9. [PubMed] [Google Scholar]
- 26. Ohtsubo K, Marth JD.. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126:855–67. [DOI] [PubMed] [Google Scholar]
- 27. Barboza M, Pinzon J, Wickramasinghe S, Froehlich JW, Moeller I, Smilowitz JT, Ruhaak LR, Huang J, Lonnerdal B, German JB et al.. Glycosylation of human milk lactoferrin exhibits dynamic changes during early lactation enhancing its role in pathogenic bacteria–host interactions. Mol Cell Proteomics. 2012;11:M111015248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu B, Newburg DS. Human milk glycoproteins protect infants against human pathogens. Breastfeed Med. 2013;8:354–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kirmiz N, Robinson RC, Shah IM, Barile D, Mills DA. Milk glycans and their interaction with the infant gut microbiota. Annu Rev Food Sci Technol. 2018;9:429–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nwosu CC, Aldredge DL, Lee H, Lerno LA, Zivkovic AM, German JB, Lebrilla CB. Comparison of the human and bovine milk N-glycome via high-performance microfluidic chip liquid chromatography and tandem mass spectrometry. J Proteome Res. 2012;11:2912–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang J, Kailemia MJ, Goonatilleke E, Parker EA, Hong Q, Sabia R, Smilowitz JT, German JB, Lebrilla CB. Quantitation of human milk proteins and their glycoforms using multiple reaction monitoring (MRM). Anal Bioanal Chem. 2017;409:589–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ferris AM, Jensen RG.. Lipids in human milk: a review. 1: Sampling, determination, and content. J Pediatr Gastroenterol Nutr. 1984;3:108–22. [DOI] [PubMed] [Google Scholar]
- 33. Lonnerdal B. Nutritional and physiologic significance of human milk proteins. Am J Clin Nutr. 2003;77:1537S–43S. [DOI] [PubMed] [Google Scholar]
- 34. Liao Y, Weber D, Xu W, Durbin-Johnson BP, Phinney BS, Lonnerdal B. Absolute quantification of human milk caseins and the whey/casein ratio during the first year of lactation. J Proteome Res. 2017;16:4113–21. [DOI] [PubMed] [Google Scholar]
- 35. Montagne P, Cuilliere ML, Mole C, Bene MC, Faure G. Changes in lactoferrin and lysozyme levels in human milk during the first twelve weeks of lactation. Adv Exp Med Biol. 2001;501:241–7. [DOI] [PubMed] [Google Scholar]
- 36. Rai D, Adelman AS, Zhuang W, Rai GP, Boettcher J, Lonnerdal B. Longitudinal changes in lactoferrin concentrations in human milk: a global systematic review. Crit Rev Food Sci Nutr. 2014;54:1539–47. [DOI] [PubMed] [Google Scholar]
- 37. Yamada C, Gotoh A, Sakanaka M, Hattie M, Stubbs KA, Katayama-Ikegami A, Hirose J, Kurihara S, Arakawa T, Kitaoka M et al.. Molecular insight into evolution of symbiosis between breast-fed infants and a member of the human gut microbiome Bifidobacteriumlongum. Cell Chem Biol. 2017;24:515–24. [DOI] [PubMed] [Google Scholar]
- 38. Ellison RT 3rd, Giehl TJ. Killing of gram-negative bacteria by lactoferrin and lysozyme. J Clin Invest. 1991;88:1080–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Goettig P. Effects of glycosylation on the enzymatic activity and mechanisms of proteases. Int J Mol Sci. 2016;17(12):1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Becker DJ, Lowe JB.. Fucose: biosynthesis and biological function in mammals. Glycobiology. 2003;13:41R–53R. [DOI] [PubMed] [Google Scholar]
- 41. Niwa R, Natsume A, Uehara A, Wakitani M, Iida S, Uchida K, Satoh M, Shitara K. IgG subclass-independent improvement of antibody-dependent cellular cytotoxicity by fucose removal from Asn297-linked oligosaccharides. J Immunol Methods. 2005;306:151–60. [DOI] [PubMed] [Google Scholar]
- 42. Newburg DS. Glycobiology of human milk. Biochemistry (Mosc). 2013;78:771–85. [DOI] [PubMed] [Google Scholar]
- 43. Schroten H, Stapper C, Plogmann R, Kohler H, Hacker J, Hanisch FG. Fab-independent antiadhesion effects of secretory immunoglobulin A on S-fimbriated Escherichia coli are mediated by sialyloligosaccharides. Infect Immun. 1998;66:3971–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Royle L, Roos A, Harvey DJ, Wormald MR, van Gijlswijk-Janssen D, Redwan el RM, Wilson IA, Daha MR, Dwek RA, Rudd PM. Secretory IgA N- and O-glycans provide a link between the innate and adaptive immune systems. J Biol Chem. 2003;278:20140–53. [DOI] [PubMed] [Google Scholar]
- 45. Wold AE, Mestecky J, Tomana M, Kobata A, Ohbayashi H, Endo T, Eden CS. Secretory immunoglobulin-A carries oligosaccharide receptors for Escherichiacoli type 1 fimbrial lectin. Infect Immun. 1990;58:3073–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Roos A, Bouwman LH, van Gijlswijk-Janssen DJ, Faber-Krol MC, Stahl GL, Daha MR. Human IgA activates the complement system via the mannan-binding lectin pathway. J Immunol. 2001;167:2861–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.