Abstract
Glycosylation is a ubiquitous post-translational modification responsible for a multitude of crucial biological roles. As obligate parasites, viruses exploit host-cell machinery to glycosylate their own proteins during replication. Viral envelope proteins from a variety of human pathogens including HIV-1, influenza virus, Lassa virus, SARS, Zika virus, dengue virus, and Ebola virus have evolved to be extensively glycosylated. These host-cell derived glycans facilitate diverse structural and functional roles during the viral life-cycle, ranging from immune evasion by glycan shielding to enhancement of immune cell infection. In this review, we highlight the imperative and auxiliary roles glycans play, and how specific oligosaccharide structures facilitate these functions during viral pathogenesis. We discuss the growing efforts to exploit viral glycobiology in the development of anti-viral vaccines and therapies.
Keywords: Virus-host interactions, Glycosylation, Virus, Glycoprotein, Structure, Glycan shielding
1. Introduction
The increasing emergence of infectious diseases, particularly those caused by viral infections, constitutes a significant threat to global health [1]. Numerous enveloped viral pathogens, have evolved to take advantage of host-cell processes, including glycosylation pathways, to decorate the surface of their proteins with “self” glycan moieties. The analysis of viral glycosylation across different families has revealed significant variation in the structure, density and conservation of glycans. Understanding the endogenous functionality and the immunological basis driving viral diversity has the promise of guiding the development of the next generation of vaccines and anti-virals. Here, we will discuss the varying roles that glycosylation plays in the viral lifecycle and host immune responses to infection.
Viral glycosylation has been investigated in the context of a number of envelope glycoproteins, including the envelope glycoprotein (Env) of human immunodeficiency virus-1 (HIV-1) [[2], [3], [4]], hemagglutinin glycoprotein (HA) of influenza virus [[5], [6], [7]], coronavirus glycoprotein spike (S) [[8], [9], [10]], glycoprotein (GP) of Ebola virus [11,12], glycoprotein complex (GPC) of Lassa virus [13], and envelope (E) glycoprotein of dengue, Zika, and other flaviviruses [14,15]. However, glycosylation on viruses is not limited to envelope glycoproteins. Many secreted viral proteins present glycans which are necessary for their respective functions, such as the non-structural protein-1 (NS1) of flaviviruses [16], the secreted GP of Ebola [17], and the secreted glycoprotein G (sgG) of herpes simplex virus (HSV) [18].
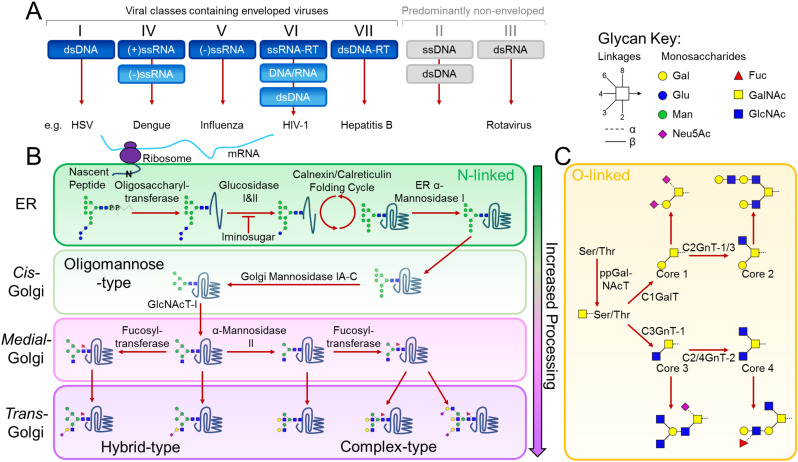
Protein-displayed glycosylation is fundamental to a wide range of molecular and cellular processes, which can be divided according to intrinsic and extrinsic functions [19,20]. Intrinsic functions are based upon the intramolecular properties of glycoproteins, while extrinsic functions result from modulation of intermolecular interactions of glycoproteins with glycan-binding partners, such as lectins and antibodies. While several forms of glycosylation occur in nature [21,22], here, we focus on the intrinsic and extrinsic functions of N-linked and mucin-type O-linked glycosylation in viral pathogenesis (Fig. 1 ). N-linked glycosylation occurs on asparagine residues of Asn-X-Ser/Thr sequons, where X is any amino acid except proline. Viral proteins are glycosylated by the host-cell as viruses are able to hijack cellular glycosylation. In addition, as glycans are genetically encoded, glycosylation can be under significant selective pressure from factors such as immune evasion, and functions in glycoprotein folding and assembly [23]. The initial step of N-linked glycan biosynthesis involves the covalent en bloc attachment of the N-linked glycan precursor onto an asparagine residue of a nascent polypeptide chain by oligosaccharyltransferase (OST) early in the endoplasmic reticulum (ER). From this point, other processing enzymes trim down and subsequently build up the glycans, resulting in a potentially vast assortment of different classes of glycans namely: oligomannose, hybrid, and complex-type N-glycan structures (Fig. 1B). It is important to note that in the context of viruses, glycosylation pathways may not be strictly linear and it is evident that some viral particles bud off early/translocate in the glycosylation pathway, which may account for the presence of unusual glycans on some viral glycoproteins [24,25]. Furthermore, it is known that many viral glycoproteins do not follow the classical secretion pathway but can, for example, traffic directly from the ER to the plasma membrane, bypassing glycan maturation in the Golgi apparatus. This particular route is exhibited by hepatitis C virus and yields viral populations that are dominated by oligomannose-type glycans [26].
Fig. 1.
Glycosylation of a viral glycoprotein in the classical mammalian N-linked and mucin-type O-linked pathways. (A) Viral classes containing predominantly enveloped and non-enveloped viruses are coloured in blue and grey, respectively. Although not enveloped, viruses such as rotaviruses can also exploit the host-glycosylation pathways to modify their proteins. (B) Following mRNA synthesis, the mature tri-glucosylated N-linked glycan precursor, dolichol-P-P-glycan, is co-translationally transferred en bloc by oligosaccharyltransferase to the asparagine residue of an Asn-X-Ser/Thr sequon on a nascent polypeptide chain. Following transfer of the precursor glycan to the protein, glucosidases in the ER remove the three glucose residues while the protein folds in the Calnexin/Calreticulin cycle. A series of ER and Golgi mannosidases subsequently cleave mannose residues down to the Man5GlcNAc2 glycan. The action of GlcNAc transferase-I (GlcNAcT-I) initiates the first branch of the N-glycan. Once α-Mannosidase II removes the two remaining outer mannose residues, other glycan processing enzymes such as galactosyl-, fucosyl- and sialyl-transferases, can act to construct a huge assortment of complex-type glycans. (C) The mucin-type O-linked glycosylation pathway is initiated by a family of ppGalNAc transferases that covalently link a N-acetylgalactosamine (GalNAc) monosaccharide to any serine, threonine and tyrosine residue. Following this conjugation, a series of glycosyltransferases can act upon the primary GalNAc residue to generate the four common O-linked glycan cores. Each of these cores can be extended and processed further to generate numerous mucin-type O-linked glycans. Glycans are presented using Consortium for Functional Glycomics symbolic nomenclature and Oxford system linkages [31], as per the key.
The existence of non-classical secretion routes also has important experimental implications. For example, the Env glycoprotein from HIV-1 pseudoviruses exhibit unusually elevated levels of oligomannose-type glycans compared to those of infectious virus produced in peripheral blood mononuclear cells [27]. This is likely due to deviations away from a native-like virus budding pathway in the pseudoviral system [3,28,29]. The cellular basis of these effects are not entirely clear; however, it is known that virus budding can compromise native membrane integrity [30]. If variations in budding routes are apparent in experimental systems, it is likely that virus biology, and its associated glycosylation, is heterogeneous in the setting of a native infection.
In mucin-type O-linked glycosylation, serine, threonine and tyrosine [32] residues are covalently linked to a GalNAc residue by polypeptide-N-acetylgalactosaminetransferases (ppGalNAcT) to form the so called 'Tn antigen'. There is a total of 8 mucin-type O-glycan cores, with 4 common core structures (cores 1-4), each of which can be processed further as proteins pass through the Golgi apparatus (Fig. 1C). Unlike N-linked glycosylation, which is identifiable by a specific amino acid sequon, no such simplicity has been identified for O-linked glycosylation, most likely due to differential peptide substrate preferences of multiple ppGalNAcTs. However, O-linked glycosylation in mucin-like domains can be detected by their high levels of serine, threonine and proline residues [33,34], and more recent developments have improved prediction of individual O-linked glycosylation sites in non-mucin-like domains [33,[35], [36], [37]]. The heterogeneous occupancy and composition of O-linked glycosylation constitutes a major challenge in studies of glycan structure and function. A number of viral proteins, notably Ebola GP and sGP [38], herpes simplex virus gC protein [39], and respiratory syncytial virus G protein [40] contain mucin-like domains with high populations of serine and threonine residues, that are heavily modified by mucin-type O-linked glycans.
Whilst most known viruses typically utilise the N- and O-linked host-cell glycosylation pathways to adorn their glycoproteins with carbohydrate modifications, it is important to mention known exceptions. There are a few notable examples of viruses that encode some, if not all, of the enzymes involved in the glycosylation of their proteins and other macromolecules such as, chloroviruses and mimiviruses [[41], [42], [43], [44]].
The characterization of N- and O-linked glycans has presented a particular analytical challenge due to the extraordinary heterogeneity and complexity arising from the number of glycan processing enzymes residing in the ER and Golgi apparatus. However, coincident with the advent of potent mass spectrometric and chromatographic techniques and the commercial availability of recombinant glycosidases, investigation of these post-translational modifications has become more feasible [45]. These advances have enabled experimental approaches including matrix assisted laser desorption ionisation (MALDI) and electrospray ionisation (ESI) mass spectrometry, allowing characterization of viral glycome (i.e. the carbohydrate repertoire of a specific viral protein in isolation or as presented by the pathogen). More recently, with the augmentation of biophysical techniques, namely in-line liquid chromatography mass spectrometry (LC-MS) [46], it has been possible to define the exact glycan structures presented at individual glycosylation sites [47]. Additionally, investigation of carbohydrate-protein interactions has been aided by the development of glycan microarrays capable of screening the carbohydrate specificity of glycan-binding proteins to immobilized and pre-defined glycan structures [48,49]. Furthermore, advances in computational biology and sequencing technologies have also provided crucial insights into the evolution of viral glycosylation [50,51]. These bioinformatics approaches can allow greater prediction and understanding of how viruses evolve to utilise glycosylation.
Given that any particular cell exhibits a specific portfolio of glycan processing enzymes, the cellular origin of viral replication has the potential to significantly influence viral glycosylation [52]. Similarly, any multicellular target host will display a range of tissue-specific glycosylation that will potentially influence viral tropism [53]. At one extreme, glycosylation can be compositionally different across different species and this is an important feature of inter-species transmission potential. Indeed, many human pathogens are zoonotic viruses with animal reservoirs and although the mammalian N-linked glycan pathway is largely well-conserved [54], several key differences can impact the viral antigenicity [55]. For example, humans lack the galactose-α-1,3-galactose epitope that is a common structure at the terminals of mammalian glycans [56] and antibody-based immunity against these epitopes can limit cross-species viral infectivity [57]. Similarly, O-linked glycans and glycolipids can exhibit pronounced compositional differences across species. At the other extreme, these immunological effects of host-specific protein and lipid glycosylation can also influence the spread of viruses within a species. Such glycan differences are evident within the human population as the ABO blood group system, with people of blood group A and blood group B expressing a fucosylglycoprotein α-N-acetylgalactosaminyltransferase and fucosylglycoprotein galactosyltransferase, respectively [58]. The presence of antibodies against the glycan structures of foreign blood groups, for example anti-A present in a blood group B individual, can limit viral transmission from a blood group B to blood group A individual. These effects have been documented in the infectivity of HIV-1 [59] and other viruses [60], and the distribution of blood groups may be a selective pressure shaping the presence and distribution of viral spread [60]. Similarly, the targeting of specific blood groups as attachment factors in both viral and bacterial entry may also influence prevalence of particular viral strains based on the frequency of blood groups within a population [61].
Tissue-specific glycosylation is also a key determinant of interspecies viral transmission potential and can lead to targeting of vulnerable tissues within a host [61]. For example, the capacity of the avian influenza HA to interact with both α2,3- and α2,6-linked sialic acid has been shown to facilitate spill-over of the virus from birds to humans [53,62,63]. In addition, differential expression of these sialic acid structures between the upper and lower respiratory tracts in humans can shape the distribution of influenza infection within an individual [64,65].
The presence and distribution of glycan specific cell surface lectins, together with the host-specific viral glycosylation, also influence transmission. For example, in the transmission of insect-borne viruses to mammals, the founder virus will display features specific to invertebrate glycosylation whilst subsequent virions manufactured in the new host will contain mammalian glycosylation. Thus, viral glycosylation undergoes a compositional switch upon inter-species transmission that will influence the interaction with immunological receptors and responses which will also influence viral tropism. Invertebrates display chiefly paucimannosidic structures [66], thus for insect-borne diseases such as Dengue and Zika, these structures may affect lectin-utilisation, infectivity, and tropism depending on the origin of viral replication.
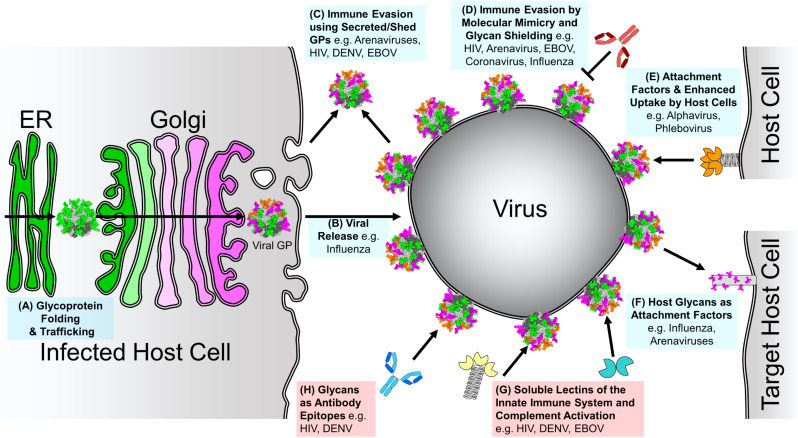
Overall, it can be extremely important to consider the cellular biosynthetic origins of viruses, both naturally and in experimental systems. In this review, we survey the wide-ranging factors shaping the presence and processing of viral glycosylation and the role glycosylation plays in viral pathogenesis (Fig. 2 ).
Fig. 2.
Roles of Glycosylation in Viral Pathogenesis. The roles contributing to viral pathogenesis and host-cell strategies used to respond to viral infection are coloured blue and red, respectively. Green and pink indicate oligomannose and complex-type N-linked glycan processing states, respectively. (A) Glycoprotein Folding and Trafficking. As with all glycoproteins, glycans on viral glycoproteins aid in folding and trafficking through the host secretory pathway. (B) Glycosylation in Viral Release. Glycosylation of infected host cell proteins can influence viral spread. (C) Immune Evasion Using Secreted/Shed Glycoproteins. Viruses can shed or secrete glycoproteins to act as immune decoys. (D) Immune Evasion by Molecular Mimicry and Glycan Shielding. Extensively glycosylated viral proteins shield themselves from the host immune response by occluding the immunogenic proteinous surface with a dense coat of host-derived glycans. (E) Glycans as Attachment Factors and Enhanced Uptake by Immune Cells. Some virus envelope-displayed glycoproteins contain under-processed oligomannose-type glycans that function as host-cell attachment factors to augment or facilitate infection of immune cells. (F) Host Glycans as Attachment Factors. Viruses can recognise glycans presented on host cell-surface proteins to facilitate host cell attachment. (G) Soluble Lectins of the Innate Immune System and Complement Activation. As under-processed glycans are rarely presented on mature host cell glycoproteins [67,68], the innate immune system is able to recognise these glycans as pathogen-associated molecular patterns (PAMPs) using soluble lectins. (H) Glycans as Antibody Epitopes. Where glycan shielding is conserved on viral glycoproteins, it is possible for the humoral immune response, in rare cases, to elicit neutralising antibodies that target sugars as part of their epitopes.
2. Glycoprotein folding and trafficking
One of the most crucial roles of protein glycosylation is its effect on glycoprotein folding, structure, sorting, trafficking, and stability [69]. Glycans can be structurally integrated into the protein fold and exhibit significant glycan-protein interactions to stabilise the protein, for example by increasing the solubility of proteins and interacting with surface hydrophobic residues. In addition, glycans can also assist glycoprotein folding by mediating chaperone interactions during biosynthesis [70].
Glycoprotein maturation in the ER requires the stepwise removal of the two terminal glucose residues of the N-linked glycan precursor, by ER α-glucosidases I and II, to leave the monoglucosylated precursor. ER chaperones, calnexin and calreticulin, bind to this substrate glycan to sequester the glycoprotein in the ER such that the protein is able to properly fold. Once correctly folded, the chaperones are released and alpha-glucosidase II cleaves off the final glucose residue; if the glycoprotein at this stage is properly folded, it is processed further and exported to the Golgi apparatus for further modification. Conversely, if the glycoprotein is incorrectly assembled, it is re-glucosylated by UDP-glucose:glycoprotein glucosyltransferase (UGGT), resulting in the glycoprotein being chaperoned once more. This feedback loop acts as a quality control mechanism ensuring that solely correctly folded glycoproteins exit the ER.
Due to this dual effect on protein folding, it can be difficult to determine the contribution of any single glycan to glycoprotein folding and assembly. However, comprehensive mutagenesis studies of individual N-linked glycosylation site knockouts of several viral glycoproteins, and direct biophysical analysis of glycan-protein interactions, have demonstrated their reliance on the host's translational machinery for correct protein folding, and thereby pathogenesis [14,16,[71], [72], [73], [74], [75], [76], [77], [78]]. For example, the importance of N-linked glycosylation on HIV-1 Env protein folding and stabilization has been investigated thoroughly, including a study that systematically dissected the requirement of each N-linked glycosylation site, which highlighted integral glycans necessary for correct gp160 incorporation into virions [79,80]. The lack of these glycans seemingly causes misfolding of trimeric Env, preventing efficient secretion and display on the viral surface, highlighting crucial roles for specific N-linked glycans [[79], [80], [81], [82]].
Due to the importance of protein folding for viral protein function, certain sugar mimetics, namely iminosugars, have been investigated for their antiviral activity. Iminosugars, such as N-butyl-deoxynojirimycin (NB-DNJ), inhibit ER α-glucosidase activity, resulting in the generation of unfolded, non-functional viral glycoproteins (Fig. 1B). The antiviral activity of iminosugars has been tested on a diverse spectrum of viruses including HIV-1 [[83], [84], [85]], influenza viruses [86,87], Ebola virus [88], arenaviruses [89], and flaviviruses [[90], [91], [92]]. A variety of iminosugar compounds have been shown to potently inhibit the release of infectious virion particles and the number of infected cells [93]. The inhibition of Dengue pathogeny by these compounds demonstrates the importance of glycosylation in Dengue glycoprotein folding and virulence. Despite these promising antiviral properties, iminosugars have yet to be clinically approved for antiviral indications. One barrier to their clinical application at the concentrations required to treat viral infections, such as HIV-1, appears to be the off-target effects of the drug which impede host gut glucosidases leading to diarrhoea and abdominal pains [85]. However, further derivatives are being investigated that may increase anti-viral specificity [94].
As per N-linked glycosylation, O-linked glycosylation has also been shown to be crucial for efficient viral particle formation and infectivity. In the case of HSV-1, which possess eight structural glycoproteins featuring O-linked glycosylation, viral expression in a cell line that only displays truncated Tn and sialylated Tn antigens resulted in severely reduced viral titres [35]. Whilst the exact mechanisms are yet to be elucidated, it is conceivable that correct processing and elongation of O-linked glycosylation is integral for either viral glycoprotein trafficking, stability and viral particle formation, or infectivity [35]. N-linked glycosylation of HSV-1 glycoproteins has also been shown to play a significant role in intracellular trafficking and folding [78]. We direct readers to the review by Bagdonaite et al. which covers glycosylation of herpesviruses thoroughly [95].
3. Glycosylation in viral release
Some viruses exploit the glycosylation pathways not only to modify their own proteins, but also utilise host glycans as receptors and attachment factors (Fig. 2F), however, these same interactions can impede viral budding from the infected cell. Consequently such viruses have evolved carbohydrate-cleaving enzymes to facilitate efficient viral budding and release. The most notable example is the ability of neuraminidases of influenza viruses to cleave sialic acid residues on the host cell surface to negate the ability of hemagglutinins (HA) to bind the host-cell receptor sugars [96,97]. During virus budding at the cell membrane, HA spikes continue to bind virion particles to sialic acid residues on the cell surface until the sialidase activity of NA removes the terminal sugar residue. Additionally, NA cleaves sialic acid residues from the glycoproteins on the virus envelope itself and enhances infectivity by preventing aggregation of virus particles [98]. Furthermore, NA is thought to act upon sialic acid residues on mucins to gain access to epithelial cells, potentially playing a secondary role in aiding virus entry to host cells [99]. Additionally, a variety of paramyxoviruses, including mumps [100], parainfluenza [101], and Newcastle disease [102] viruses possess hemagglutinin-neuraminidase (HN) glycoproteins that both bind and cleave sialic acid residues, as such these viral glycoproteins are integral for facilitating host cell entry and egress [103].
A variety of therapeutics have been developed to mimic sugars that bind at the relatively deep sugar binding site of NA (compared to HA) to combat influenza virus infection. These NA-inhibitors are most commonly N-Acetylneuraminic acid (Neu5Ac) derivatives, which competitively inhibit NA activity [104]. Additionally structure-based development of competitive inhibitors against the glycan binding sites of HN glycoproteins have been explored [102,105].
Beyond the role of NA in viral release, this viral glycoprotein has emerged as a focus in vaccine development. Current influenza vaccines poorly display NA epitopes and do not produce robust anti-NA antibodies [106], whilst during infection many NA-reactive B cells and antibodies are produced. The development of recombinant influenza vaccines, which improve the display of influenza NA, may provide a method of vaccine optimisation to improve targeting of NA for broader protection against divergent influenza strains circulating in human populations [[106], [107], [108]].
4. Immune evasion using secreted and shed viral glycoproteins
Shedding or secretion of viral glycoproteins constitutes an important stratagem employed by some viruses to misdirect the humoral immune response (Fig. 2C). This can either occur by absorbing neutralising antibodies or by modulating the immune response to target non-neutralising epitopes. The GP gene of Ebola virus contains a poly-U stutter region that drives transcriptional frame-shifting that generates three glycoproteins: a dimeric secreted GP (sGP) that accounts for approximately 75% of transcripts, the full-length membrane-bound fusion protein (GP) accounting for ~25% of transcripts, and trace levels of a small soluble GP (ssGP) [17,109]. Whilst the function of ssGP is yet unknown, sGP that is secreted by infected cells has been associated with viral immune evasion by acting as an antibody decoy [17]. Ebola sGP has been found to “antigenically subvert” the immune system, wherein sGP redirects the humoral immune response to target epitopes shared with the full-length GP. The study by Mohan et al. also demonstrated that sGP also subverts the host immune response of mice previously immunised with Ebola GP [17]. As the major antigenic transcript, most antibodies isolated from Ebola survivors and macaques are directed towards sGP rather than GP [110,111]; importantly these antibodies are likely to be non-neutralising and the neutralising antibodies elicited against GP may cross-react and be absorbed by the more abundant sGP [112]. The extent to which mimicry of the glycan component of GP by sGP contributes to the overall antigenic mimicry in immune decoy functions is unknown. However, given both that glycans have been shown to contribute to epitope formation on sGP/GP [113], and that there is congruence in their glycan processing [11], glycans are likely to be an important component of these functions.
Shedding of attachment glycoprotein subunit (GP1) of Lassa virus has also been observed during acute Lassa fever in humans [114,115]. Whilst the exact function of shed GP1 is yet to be elucidated, it has been proposed that it may act as an immunological decoy in a manner akin to Ebola sGP. In the case of Lassa virus, the soluble GP1 structure [116] is conformationally distinct to that presented on the trimeric GPC [117], further highlighting its potential function as an immune decoy, since shed GP1 will likely present a non-functional conformation, and thereby elicit non-neutralising antibodies. The conservation of such substantial structural rearrangements of isolated GP1 subunits from other Old World arenaviruses, such as Loei River [118] and Morogoro virus [119] may highlight a conserved mechanism of immune evasion amongst these pathogens.
Dengue virus non-structural protein-1 (NS1) is an intriguing glycoprotein that is secreted from infected cells. NS1 exhibits two N-linked glycosylation sites that have been shown, via mutagenesis studies, to be essential for a number of crucial roles, including hexamer formation, facilitating secretion and modulating protein stability [120]. NS1 binds to heparin sulphate and chondroitin sulphate glycosaminoglycans on the surface of various cells [121]. NS1 has been shown to play a role in both viral replication [122] and immune evasion [123,124], and it has been hypothesised that immune recognition of NS1 on endothelial cell surfaces may facilitate vascular leakage during severe Dengue infection [125]. The use of secreted/shed GPs in order to subvert the immune system and/or enhance infectivity appears to be a common phenomenon utilised by viruses. Other examples include the secreted form of respiratory syncytial virus G glycoprotein, which distracts the antibody-immune response by acting as a decoy [126], secreted BARF1 and soluble gp42 glycoproteins of Epstein-Barr virus, which interfere with macrophage differentiation and activation, and MHC class II-mediated antigen presentation respectively [127,128].
Untethered monomeric gp120 subunits from HIV-1 Env can also arise due to improper processing of the spike or by gp120 shedding [129]. These viral envelope fragments can divert the humoral immune response by exposing immunodominant protein epitopes not presented on functional Env proteins, thereby eliciting undesirable non-neutralising antibodies during HIV-1 infection [[129], [130], [131]]. The role of non-neutralising epitopes in subverting the host immune response, away from neutralising epitopes, is an important consideration in vaccine development, and has stimulated the development of immunogens that closely mimic the native states of the HIV-1 viral spike [131,132].
5. Immune evasion by molecular mimicry and glycan shielding
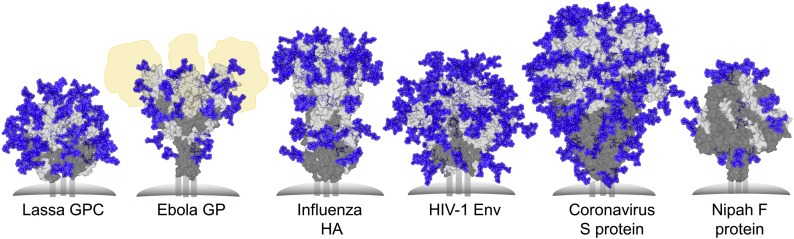
A number of viruses, including HIV-1, influenza, Lassa, coronaviruses, and Ebola have evolved to shield their respective envelope glycoproteins with host-derived glycans to prevent antibody recognition of the underlying protein surface. The importance of N-linked glycosylation of envelope proteins, with respect to immune evasion, has been observed across many viruses including: Hepatitis C [133,134], Hepatitis B [135], Hendra [136], Newcastle disease [137] and Herpes simplex virus [138,139]. Some class I fusion proteins exhibit particularly high densities of glycans, consistent with their role in shielding. Here, we focus on the glycan shields of the trimeric class I fusion proteins from HIV-1, Influenza, Coronavirus, Lassa, Ebola, and Nipah viruses (Fig. 3 ).
Fig. 3.
Glycan Shielding of Viral Class I Fusion Proteins. Left to right: Glycan shield models of Lassa virus GPC (PDB ID: 5VK2) [117,140], Ebola GP (PDB ID: 5JQ3) [141], A/H3N2/361/Victoria/2011 H3N2 Influenza virus hemagglutinin (PDB ID: 4O5N) [142,143], BG505 SOSIP.664 HIV-1 Env (PDB ID: 4ZMJ) [3,144], human coronavirus-NL63 (HCoV-NL63) S protein (PDB ID: 5SZS) [8], Nipah F protein (PDB ID: 5EVM) [145]. Glycans and proteins are shown in blue and grey, respectively. The fusion protein subunit is shown in dark grey. The positions of mucin-like domains of Ebola GP are shown in yellow. Most predominant sugar compositions were modelled onto each N-linked glycan site, using pre-existing GlcNAc residues if possible, with Man5GlcNAc2 modelled on if compositional information was lacking.
5.1. HIV-1 envelope glycoprotein
One of the best characterised glycan shields is that of HIV-1 Env, which is the sole glycoprotein found on the virus surface [146]. Mature Env exists as a trimer of non-covalently associated gp120-gp41 heterodimers that are generated by furin cleavage of a gp160 polypeptide precursor [147]. The gp120 receptor binding subunit helps dictate host cell tropism and facilitate attachment to CD4 receptor and CXCR4/CCR5 co-receptors, while gp41 facilitates viral and host cell membrane fusion upon receptor binding by gp120, and the extensive conformational changes of Env that follow. HIV-1 Env presents between 18-33 glycans per gp120 monomer, with a median of 25 glycan sites on gp120 and 4 glycans on the gp41 subunit [23]. The extent of these post-translational modifications, combined with the intrinsic flexibility of the trimeric Env [148,149] and constant evolution of the glycan shield, renders HIV-1 Env an immunologically challenging target [150,151]. The result of the dense glycosylation on the viral protein surface leads to the formation of the so-called intrinsic and trimer associated mannose-patches (IMP and TAMP, respectively) [[152], [153], [154], [155], [156]], which arise due to the steric blocking of glycan-processing enzyme in the ER and Golgi apparatus by neighbouring glycans [157]. These often under-processed glycans are weakly immunogenic, since they are produced by the host-processing pathway, thereby limiting the response to deviating glycosylation [158]. Astoundingly, in the case of HIV-1, the conservation of these under-processed glycans can result in the generation of broadly neutralising antibodies (bnAbs) that actually target these immunoquiescent host-derived glycans as part of their epitope [[159], [160], [161], [162], [163], [164]]. As more bnAbs have been discovered, it has become clear that they can not only recognise the viral mannose patches, but some have also evolved to recognise complex and hybrid-type glycans [165]. However, the observation that bnAbs only develop in a subset of patients, and only after several years, underscores the fact that the glycan shield of HIV-1 has evolved under sustained immunological pressure, and represents an extreme challenge for the immune system and an important consideration in vaccination strategies [146,159,[166], [167], [168], [169]].
5.2. Influenza hemagglutinin
Considerable antigenic evolution enables the persistence and spread of influenza virus in human populations [170]. The surface glycoprotein HA exists as a trimer of hetero-dimers that is synthesised as a single polypeptide (HA0) that is subsequently cleaved into the receptor binding HA1 domain and the membrane-fusion mediating HA2 subunit by host cell proteases. Similarly to HIV-1 Env, HA is a class I viral fusion protein (Fig. 3) and plays a vital role in viral entry by binding to host receptors via the receptor binding site (RBS) located on HA1. As the main focus of the immune response, HA evolves at the highest rate compared to other influenza viral proteins [171], and the presence and acquisition of N-linked glycosylation sites on HA plays a crucial role in immune evasion [172]. The evolution of the HA glycan shield constitutes an important defense mechanism by masking neutralising epitopes [173]. In 1968, H3N2 HAs possessed an average of 6 N-linked glycosylation sites per protomer. Since then, H3N2 HAs have acquired and retained over twice as many sites, with current strains exhibiting approximately 13 PNGs per HA1-HA2 protomer [7,172]. Similarly, from the Spanish flu in 1918 to its temporary departure from human circulation in 1957, H1N1 strains have incorporated between 1 to 3 N-linked glycan sites per HA1-HA2 protomer [50,174]. Interestingly, the seasonal accumulation of glycans on HAs are most-commonly incorporated at the globular head domain (HA1), in regions of variability where the majority of antigenic sites lie [[175], [176], [177], [178], [179], [180], [181], [182]], seemingly due to the preference of neutralising antibodies to target the RBS-proximal regions [172].
The accretion of N-linked glycans that occlude antigenic sites on HA can, however, affect HA binding to its receptor, particularly when they neighbour the RBS [183]. This functional constraint renders HAs not able to be completely shielded by glycans. Indeed, there appears to be a maximum number of N-linked glycosylation sites allowed on HAs at any one time, resulting in the replacement of non-conserved glycosylation sites with an alternative N-linked glycan sequon [172]. In other words, the plateauing of the number of glycosylation sites on HAs appears to derive from a balance between functionality and immune evasion.
Despite the potential fitness cost of increased glycosylation sites, their presence has been shown to compensate for the fitness costs associated with other antibody escape mutations [184]. Compositionally, although the total number of glycans present on HAs is significantly less than on HIV-1 Env, glycosylation analyses has revealed the abundance of oligomannose-type glycans at specific sites [143,185,186], indicating a restriction in glycan processing in potential areas of higher glycan density and shielding.
5.3. Coronavirus spike protein
Epitope masking by glycosylation has also been observed on coronavirus spike (S) proteins [8,187]. As with HIV-1 Env and influenza HAs, which use N-linked glycosylation to shield conserved CD4 [188] and sialic acid binding sites [189,190], respectively, coronaviruses appear to also occlude receptor binding domains by utilising N-linked glycans [8]. The S proteins of coronaviruses are large glycoproteins with between 23-38 potential N-linked glycan sites per protomer. Glycan analysis performed by Ritchie et al. of severe acute respiratory syndrome (SARS) S protein revealed a significant population of oligomannose-type glycans (30%) [10]. Furthermore, compositional analysis of the glycan shield of alpha- and delta-coronaviruses, expressed in Drosophila S2 cells, also displayed a wide variety of oligomannose-type glycans in addition to fly-type paucimannosidic glycans [8,9]. Whilst it is extremely likely that N-linked glycan sites on coronavirus spikes have been incorporated to occlude certain epitopes, it remains unclear how effective the glycan shield is at facilitating immune evasion, since antibodies that target S proteins are readily elicited [[191], [192], [193]]. The sparsity of the glycan shield may be reflected in the overall oligomannose-type glycan proportions which is low (30%) [10] compared to the other “evasion strong” [166,194] viruses such as HIV-1, Influenza, and Lassa (>50%) [3,140,143] indicating less dense glycosylation across the surface, permitting the processing of glycans by enzymes in the ER and Golgi apparatus, and allowing antibodies to target the protein surface. As with other viral glycoproteins, it appears that glycans surrounding the receptor binding sites can negatively impact functionality [195], suggesting that the slightly limited glycan shield of coronaviruses may result from the necessity to ensure efficient receptor binding.
5.4. Lassa virus glycoprotein complex
Lassa virus is a highly pathogenic arenavirus that causes severe haemorrhagic fever in humans. Arenaviruses are generally categorised as Old and New World pathogens, based on geographical, genetic, and antigenic properties [196]. Both Old and New World Arenavirus GPCs are extensively glycosylated with a remarkable conservation of the glycosylation sequons [13,140,[197], [198], [199]]. Arenaviral GPCs are expressed as a single polyprotein precursor that is proteolytically processed and displayed as a trimer of tripartite protomers, with each protomer consisting of a receptor-binding GP1 domain, a class I membrane fusion GP2 domain, and a retained myristoylated stable signal peptide. The oft-inadequate antibody immune response elicited against arenaviruses has been, in part, attributed to the shielding of would-be antigenic epitopes by conserved N-linked glycans [13]. We previously reported the composition and model of the Lassa virus glycan shield, which comprises approximately 25% of the glycoprotein mass, revealing the extent of shielding across GPC, which consisted of a dense cluster of oligomannose-type glycans on the side of trimer [140]. These N-linked glycans protect vulnerable regions of the GPC, including the putative α-dystroglycan [200] and lysosomal-associated membrane protein-1 (LAMP-1) [116,119,201] binding sites, and regions surrounding the fusion loop [117]. Further evolutionary analysis of Lassa virus GPC revealed that residues with elevated nonsynonymous to synonymous substitutions (dN/dS) ratios co-localise to regions on Lassa virus GPC with lower levels of glycan density. This provided a rationale for how the Lassa virus glycan shield protects the proteinous surface from the humoral antibody response of the host [140]. Additionally, we have hypothesized that the Old World Loei River arenavirus GPC, which encodes two additional N-linked glycans on the GP1 subunit that are not observed on most other Old World arenaviruses may contribute to a more expanded oligomannose-type glycan cluster than that on Lassa virus GPC [118]. Nevertheless, with the conservation of N-linked glycosylation sites on arenaviral GPCs it seems extremely likely that the glycan shielding of the protein surface is conserved across the family to facilitate immune evasion.
5.5. Ebolavirus glycoprotein
Ebola virus is a highly virulent pathogen capable of causing severe haemorrhagic fever, in humans. The Ebola virus envelope surface glycoprotein (GP) also exhibits dense glycosylation, suggesting a role in immune evasion. Ebola GP is initially expressed as pre-GP that is cleaved by furin into two subunits, GP1 and GP2, which are associated by a disulphide bridge. This heterodimer assembles into a trimer, mediated by extensive inter-subunit interactions, on the surface of nascent virions to form mature Ebola GP that is solely responsible for receptor attachment, fusion and entry into target cells [202]. Ebola GP presents approximately 17 N-linked glycosylation sequons and an extensive mucin-like domain, per protomer (Fig. 3). Both N-linked glycosylation [112,203,204] and the extensive O-linked glycosylation of the mucin-like domain [38] have been shown to negatively impact immunogenicity by hiding immunogenic protein epitopes [112]. Interestingly, antibodies elicited against the so called GP-1 “glycan cap” and the mucin-like domain of Ebola GP have been characterised, although compared to antibodies that bind other epitope classes, such as the GP base, GP2-resident fusion-loop, and the GP1-head domain, neutralisation was limited [113]. It has been hypothesised that variation in neutralising potential within “glycan cap” and mucin antibodies may be due to heterogeneity in both N-and O-linked glycosylation processing states [113].
5.6. Nipah virus fusion protein
Glycan shielding of paramyxoviruses, such as Nipah virus, is influenced by the different protein architecture of this family (Fig. 3), although a similar theme in immune evasion has been reported. Nipah virus is a deadly emerging paramyxovirus that enters host cells using two envelope glycoproteins; the attachment (G) protein that binds to cell receptors [103,205], and the fusion (F) glycoprotein that subsequently implements membrane fusion (PMID: 21511478). Paramyxovirus F proteins, unlike the other class I fusion proteins, described above, do not participate in receptor binding and exhibit significantly varied protein architectures compared to other class I fusion proteins [206]. However, they all share similarities in the utilisation of N-linked glycans to protect against the humoral immune response. Nipah virus F protein possesses 5 PNGs per protomer, which does not indicate the presence of an extensive shield that occludes much of the protein surface, as exemplified in Fig. 3. The lack of a dense glycan shield is likely related to the fact that the N-linked glycans on Nipah F modulates the efficiency of membrane fusion and viral entry [207]. Nevertheless, these carbohydrate moieties have been shown to protect virions from neutralisation by antibodies, suggesting a trade-off between the evolutionary advantage of increased protection against the immune system and the effects on protein functionality. As a surface-displayed protein, Nipah virus G protein is also under immunological pressure and N-linked glycans on the G protein have been shown to play a role in shielding virions against antibody neutralization [208].
Overall, immune evasion by glycan shielding and molecular mimicry by exploiting the host glycosylation pathway is a common feature observed on viral class I fusion proteins. One important consideration is understanding the potentially varied roles and efficiencies of glycan shields in the context of immune evasion. Regardless of these potential differences, the widespread use of glycosylation as a tactic to thwart the response of the humoral-immune system underscores the vulnerability of protein epitopes to immune pressure. Some of these viruses such as HIV-1, Influenza, and Lassa appear to be “evasion strong” pathogens [166] that utilise varied mechanisms to change how immunogenic epitopes are presented on their respective envelope proteins. However, shielding of the protein surface of the viral glycoprotein with glycans is not fool-proof, and can lead to the evolution of anti-glycan antibodies and recognition by the host immune response; both innate and humoral.
6. Glycans as attachment factors and enhanced uptake by immune cells
Oligomannose-type glycans can both impede and facilitate viral propagation. On one hand, these glycans can trigger a sterilising innate immune response, for example by the activation of the complement cascade. On the other hand, viruses can exploit innate immune receptors such as dendritic cell-specific ICAM-3-grabbing nonintegrin (DC-SIGN) [[209], [210], [211], [212], [213], [214], [215]] to facilitate and/or augment infection. DC-SIGN possesses a C-type carbohydrate recognition domain (CRD) that co-ordinates a calcium ion in the primary subsite to facilitate the binding of oligomannose-type glycans found on viruses (Fig. 4C). DC-SIGN binds to the Manα1-3(Manα1-6)Manα tri-mannose structure found on oligomannose type glycans [216,217]. Importantly, not all glycoforms can be bound by DC-SIGN. Specificity to oligomannose-type glycans is conferred by the presence of a phenylalanine side chain in the CRD that acts as a steric block obstructing the GlcNAc residues of the N-linked glycan core, when a core mannose residue of a complex-type glycan attempts to bind [217]. This selectivity to oligomannose-type glycans ensures that DC-SIGN interactions are predominantly with foreign pathogens, due to the rarity of oligomannose-type glycans expressed on correctly processed host glycoproteins [67,68].
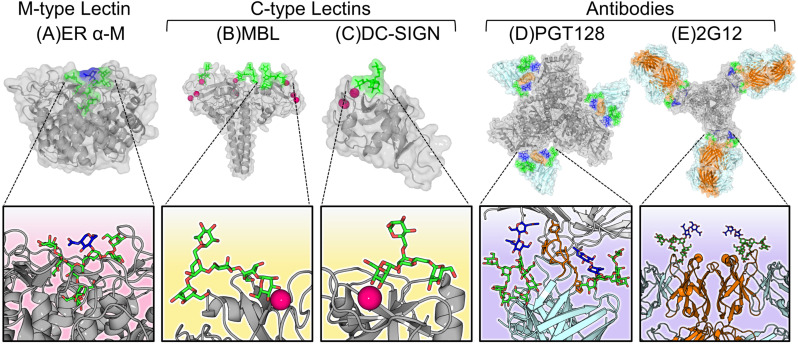
Fig. 4.
Diverse modes of oligomannose-type glycan recognition. (A) The glycan processing enzyme, ER α-mannosidase. Notice the deep binding pocket of the protein that nearly encapsulates the entire Man9GlcNAc2 glycan (PDB ID: 5KIJ) [242]. (B) The collagen-containing C-type lectin, mannose binding lectin (MBL), that recognises oligomannose-type glycans via a carbohydrate recognition domain containing a calcium ion (pink), which is responsible for mediating the primary interactions to bind the sugar (PDB ID: 1KX1) [243]. (C) The membrane bound C-type lectin receptor, DC-SIGN recognises oligomannose-type glycans via a Ca2+ ion and initiates phagocytosis of the pathogen (PDB ID: 1SL4) [216]. (D) An anti-HIV-1 broadly neutralising antibody, PGT128, that recognises conserved oligomannose-type glycans on Env using an extended complementarity-determining region (CDR) loop (orange) that penetrates the glycan shield (PDB ID: 5ACO) [244]. (E) The unique anti-oligomannose antibody, 2G12, featuring its unique “domain-exchanged” architecture where the heavy chains (orange) of the opposite Fab domain are swapped over (PDB ID: 6N2X) [245,246].
The relative pro- and anti-viral effects of the presence of oligomannose-type glycans are specific to the viral pathogen. It is also important to note that the necessity of glycans as host-cell invasion factors can vary greatly, and whilst a wide spectrum of viruses have been shown to interact with lectins, the majority of viruses, for example, HIV-1 [211], arenaviruses [209], influenza [218], hepatitis C [219], and coronaviruses [212,220], are recognised by host lectins on immune cells but do not utilise lectins as obligate receptors. However, as in the case of HIV, these lectin interactions have been shown to expand and enhance infectivity routes. A comprehensive table referencing viruses and associated lectin interactions can be found in the review by Van Breedam et al. [221].
Although many viruses utilise both lectins and other host proteins during infection, some viruses use lectins as obligate receptors. Alphaviruses and Phleboviruses have been reported to employ DC-SIGN as the primary receptor [214,215] that recognises oligomannose-type glycans on the viral surfaces [52,222]. Glycosylation analysis of the envelope glycoproteins of a prototypic alphavirus, Semliki Forest virus (SFV), revealed near-exclusive oligomannose-type glycan populations on the E2 attachment glycoprotein, but more complex-type glycosylation on the membrane-proximal E1 fusion protein [52]. Similarly, phleboviruses of the Phenuiviridae family, such as Rift Valley fever virus and other members of the Bunyavirales order, such as severe fever with thrombocytopenia syndrome virus (SFTSV) [223], display oligomannose-type glycans [224], which are required for DC-SIGN dependent cellular attachment and entry into mammalian dendritic cells [225]. Although the exact composition of the glycans at each of the sites on the Gn and Gc glycoproteins is not understood, it has been shown that more than one site can be used for attachment [224,226]. A glycomic study into understanding the post-translational modifications on the Gn and Gc glycoproteins of Uukuniemi virus, a relative of Rift Valley fever virus, revealed significant populations of oligomannose-type glycans, predominantly on the Gc glycoprotein of Uukuniemi virus [222]. However, the dependency of DC-SIGN and other C-type lectins [227] for infection across phleboviruses is still not fully understood, especially considering that the positions of N-linked glycosylation sites are not well-conserved across the family [228].
Dengue virus is another pathogen that utilises both the macrophage mannose receptor [229] and DC-SIGN [210,230,231] as putative receptors to infect macrophages and dendritic cells, respectively. Structural data using cryo-EM of Dengue in complex with the CRD of DC-SIGN revealed binding of N-linked glycan residues at N67 on two neighbouring E proteins by a single CRD [230]. Binding of DC-SIGN CRD does not result in any significant conformational changes, suggesting that DC-SIGN is an attachment receptor that mediates initial binding, prior to membrane fusion, which is triggered by pH-dependent rearrangements [232]. Nevertheless, DC-SIGN has been shown to be indispensable for Dengue infection of dendritic cells [210,231,233]. C-type lectin usage by flaviviruses is not limited to infection of mammals; both West Nile virus and Dengue appear to employ C-type lectins in Aedes aegypti called mosquito galactose specific lectins (mosGCTL) as ligands to promote infection of these invertebrate vectors [234,235]. An important question remains with regards to flavivirus E protein glycosylation and their usage of lectins as receptors during viral pathogeny. Does the compositional variation in glycosylation between insect and mammalian expression systems determine the variable interactions of these lectins during different stages of infection?
The dense glycan shield of Lassa virus that contributes to its immunological resistance against the immune system [13], leads to the generation of specific oligomannose-type glycan clusters across the side of the GPC spike [140]. These glycans have been shown to play an advantageous role for the virus in infecting human dendritic cells, via DC-SIGN mediated recognition [209]. Lassa infection of macrophages and dendritic cells is thought to be crucial to viral pathogenesis, as the virus can be propagated in these cells while also not inducing their activation, suggesting a mechanism of avoiding immune surveillance [236]. Thus, whilst DC-SIGN may not be the primary receptor for Lassa virus, the oligomannose-type glycans on the GPC do act as attachment factors and positively influence infection of immune cells. A similar phenomenon has been observed for HIV-1 Env, where recognition of oligomannose-type glycans by DC-SIGN has been shown to augment infection of T cells [237]. This DC-SIGN-mediated enhancement of infection of target cells via trans-infection has also been observed across other viruses, including SARS [238]. A wide range of other membrane-associated lectins besides DC-SIGN and mannose receptor such as, hepatic asialoglycoprotein receptor, liver/lymph node sinusoidal endothelial cell C-type lectin, siglecs, and macrophage Gal/GalNAc-specific C-type lectin have been implicated to have pro-viral effects [221].
A growing body of research indicates that membrane-associated lectin binding of many viruses augments infection and is advantageous for pathogenesis. Whether these paradoxical consequences are due to lectin functions to tackle non-viral pathogens such as fungi and bacteria, it would seem likely that immune cells exhibiting these lectins, can utilise them to combat viral infection using downstream functions, such as antigen presentation [239]. A prominent example of anti-viral lectin activity includes oligomannose-type glycan recognition on HIV-1 Env by langerin, a C-type lectin expressed on Langerhans cells, which results in virus internalisation and degradation [240]. As such, it seems feasible that membrane-bound lectins constitute an important part of the innate immune response against other oligomannose glycan-bearing viruses, perhaps by also aiding in antigen presentation [241].
7. Host glycans as attachment factors
Glycan binding properties are not limited to host proteins, viral proteins can also possess glycan-binding properties. Some viral glycoproteins are capable of specifically recognizing distinct glycan motifs on the host cell surface and thereby mediate viral entry, infection, and tropism. Depending on the virus, host cell glycans can act as primary receptors, co-receptors, and/or attachment factors.
Perhaps the most well-characterised example is the hemagglutinin of influenza A viruses [247,248]. The ability of HA to recognise and bind specific glycan moieties via the receptor binding site has led to it being regarded as a viral lectin [249]. The binding event between HA and its glycan receptor leads to the endocytic internalisation of the virion, subsequent pH-dependent viral-endosomal membrane fusion, and the infection of the target cell. Remarkably, cross-species transmission is determined by preferential binding of HAs to glycan receptors with a single change in sialic acid linkage to the penultimate galactose residue; Neu5Ac-α(2,3)-Gal that is primarily expressed in bird and pig gastro-intestinal and respiratory tract cells or Neu5Ac-α(2,6)-Gal that is the predominant structure found in human upper respiratory epithelial cells [[250], [251], [252], [253], [254]]. Sialic acid and host glycan recognition is not limited to influenza viruses; many pathogens such as paramyxoviruses [102], rotaviruses [255], noroviruses [256], polyomaviruses [257], and coronaviruses [258,259] also possess glycoproteins with sialic acid binding capabilities that facilitate host cell entry. Glycans recognised by viruses are not solely presented on glycoproteins. These glycan epitopes can also be found on glycolipids, which are composed of oligosaccharides linked to ceramide lipid molecules, and widely found on host-cell surfaces [260]. Furthermore, interactions between glycolipids on the viral envelope and cellular receptors have been implicated to contribute to viral pathogenesis, for example sialyllactose-containing glycolipids on the HIV-1 viral membrane can be bound by siglec-1, a lectin found on dendritic cells, to mediate trans-infection of T cells [261].
GPCs of Old World arenaviruses, including the important human pathogens, lymphocytic choriomeningitis virus, Lassa virus, and Clade C New World arenaviruses also utilise a specific host glycan motif to mediate infection. Initial host cell entry is mediated by GPC-mediated attachment to an unusual O-mannosyl class glycan composed of a tri-saccharide core with xylose-glucuronic acid repeats, presented on the cell surface receptor, α-dystroglycan [[262], [263], [264], [265]]. Once the GPC recognises this so called matriglycan, these arenaviruses can enter the endocytic pathway, where the virions are internalised into endosomal compartments [266]. In the case of Lassa virus, the associated lowered pH induces a conformational change of the GPC that leads to binding to lysosomal-associated membrane protein 1 (LAMP1) [201,264]. The subsequent dissociation of GP1 from the GPC triggers viral envelope-endosomal host membrane fusion. It has been suggested that glycosylation is key not only in the primary binding event of GPC but also in the following step of arenavirus viral fusion. Specifically, the sialylated N-linked glycan at position Asn76 on human LAMP1 has been demonstrated to be crucial for Lassa virus GPC recognition [119,264]. In any case, glycosylation of specific host proteins is critical for the attachment of many arenaviruses.
Aside from specific host-glycan binding capabilities of viral envelope proteins, broader interactions between host glycosaminoglycans (GAGs) present on the target cell surface, and viruses, including HIV [267], herpes simplex virus-1 [268], and Dengue [269] have been shown to occur and promote viral attachment independent of cognate receptor binding. Contact with GAGs often constitute the initial interaction between a virus and the target host cell. In the case of herpesviruses, glycoproteins C (gC) and B (gB) are known to initially bind to the GAG, heparan sulphate (HS), which is a proteoglycan consisting of glucuronic acid (GlcA) and GlcNAc disaccharide repeats with various sulphate groups on the saccharide residue [268,270]. This primary interaction of herpes simplex virus-1 to HS proteoglycans is critical and removal of HS from the cell surface, or the use of HS-deficient mutant cell lines renders the cells resistant to infection [268]. Herpes simplex virus-1 interaction with GAGs is not limited to HS; chondroitin sulfate another GAG found in the extracellular matrix, which is composed of alternating repeats of GalNAc and GlcA carbohydrates with sulphate groups, was revealed to be efficiently bound by herpes simplex virus-1, indicating multiple binding specificities to sulphated GAGs [271]. Interestingly, the mucin-like domain of HSV-1 Gc has been shown to modulate binding to GAGs, highlighting the complex interplay between viral and host glycosylation, which can be an important determinant in viral infectivity [272].
8. Soluble lectins of the innate immune system and complement activation
The presence of oligomannose-type glycans is an uncommon feature on mature secreted and cell-surface host glycoproteins, and as such readily triggers the host innate immune system [273]. As pathogen-associated molecular patterns (PAMPs), they can be recognised by a variety of soluble lectins, for example, surfactant protein A and D (SPA and SPD), and mannose-binding lectin (MBL) (Fig. 4B). MBL is a trimeric, soluble C-type lectin of the collectin family that constitutively circulates in the bloodstream [274]. Each protomer of the trimer possesses a C-terminal CRD, a coiled coil neck domain, a collagen-like domain, and a cysteine-rich N-terminal domain. MBL specifically binds to equatorial 3- and 4- hydroxyl groups on terminal mannose sugars and GlcNAc residues. Whilst individual CRD binding is relatively weak (KD of approximately 1 mM), the ability of collectins to form multi-trimeric oligomers significantly enhances glycan binding affinity via avidity effects resulting in KD values in the nM range [275,276]. Once MBL binds to viral glycoproteins, it recruits mannose-binding lectin-associated serine proteases (MASPs), which trigger the lectin pathway of the complement system [[277], [278], [279]]. Furthermore, collagen-like domains can interact with collectin receptors found on phagocytic immune cells to promote clearance of pathogens. Ficolins, another member of collagenous soluble lectin family found in serum specifically recognize GlcNAc moieties [280]. These lectins contribute toward a functional complement system that is vital in combating viruses. MBL has been shown to recognise N-linked glycans on Dengue [281], HIV-1 [282], Marburg virus [283], Ebola [283], influenza [284], and SARS coronavirus [285] and neutralize them by mechanisms including recruitment of complement, promoting clearance, blocking viral fusion, and interfering with DC-SIGN mediated trans-infection.
In addition to these soluble lectins found in sera, SPA and SPD, which share the basic structure of other collectins, recognize oligomannose-type glycans on viral particles in the lungs [286]. The mechanisms of SPA and SPD activity in combating influenza virus infection, whether by inhibition of hemagglutinin activity [287,288], neuraminidase activity [289], or promoting opsonisation of virion by alveolar macrophages [290] have been investigated. Interestingly, the gradual addition of N-linked glycan sites on HAs of H3N2 influenza viruses has been thought to attenuate viral pathogenicity, partly due to increased levels of SPD mediated clearance from the lungs [291]. Other respiratory viruses including coronaviruses [292], and respiratory syncytial virus [293] have been shown to be targeted by these surfactant proteins.
Galectins are a class of small soluble, so called S-type, lectins that bind to galactose and galactose-containing glycoconjugates. Unlike C-type lectins, glycan binding by the CRD is independent of calcium binding. Galectin-1 has been shown to bind to the envelope fusion glycoprotein (F) of Nipah [[294], [295], [296]], Hendra [294], and HA of influenza [297] viruses and inhibit membrane fusion. However, galectin-1 has also been shown to promote HIV infection of T cells by cross-linking the virus to host cells [298,299]. The varied pro- and anti-viral consequences of soluble lectin interactions highlights the diverse, at times, paradoxical roles that they play in the innate immune system.
9. Glycans as antibody epitopes
Self-glycans are by their very nature, poorly immunogenic, which is an important feature for how glycan shields function. There are very few examples where a host antibody response readily recognises a glycan component of viral glycoproteins. However, there are contexts where such phenomena have been observed. The two most characterised examples, HIV-1 and Dengue, reveal varied mechanisms to how these glycan-binding antibodies can arise. In the case of HIV-1 the chronic nature of infection enables, the eventual evolution of antibodies, capable of recognising conserved glycan features on the envelope spike. Whereas in Dengue virus, antibodies have been isolated that target structurally integral glycans. In both cases the challenge when exploiting these potential vulnerabilities in vaccine design is that precursor B-cells exhibiting the required glycan specificities are negatively selected.
9.1. HIV-1 envelope protein
A subset of patients infected with HIV-1 develop broadly neutralising antibodies capable of recognising glycan structures on Env. Due to the immense genetic diversity across HIV-1, many broadly neutralising antibodies target conserved features across the envelope spike, namely the glycan shield. Many such antibodies have been characterised for HIV-1, that not only target regions of dense of glycosylation that generate mannose patches [146,160,161,163,164,245,[300], [301], [302], [303]], but also hybrid-type [304], and tri- and tetra-antennary complex type glycans [301].
Due to the intrinsically weak immunogenic nature of host-cell derived glycans, many anti-HIV bnAbs that target glycans often adopt unusual architectures to overcome epitope masking. For example, bnAbs PGT128, 145, and 151 display long heavy chain complementarity determining region-3 (HCDR3) loops that penetrate the glycan shield to make protein-protein contacts [161] (Fig. 4D), and 2G12 that features a unique domain-exchange phenomenon [245,305], where the variable domains from the two heavy chains are exchanged, giving rise to a compact dual Fab architecture (Fig. 4E). Since numerous anti-HIV-1 Env bnAbs specifically recognise areas of the glycan shield, the compositional conservation of glycans on HIV-1 Env has significant implications in immunogen design as any vaccine candidate would have to emulate the native glycosylation of infectious virions [169], in order to elicit these bnAbs.
9.2. Dengue virus envelope protein
Broadly neutralising antibodies with epitopes comprised of glycans have also been characterised against Dengue virus E proteins. Specifically, these bnAbs are able to cross-neutralise all four Dengue virus serotypes by binding to the conserved so-called E dimer epitope (EDE) [306]. Dengue E proteins are presented on the mature virion surface as 90 repeating dimers, each of which contains the EDE that bridges the two monomeric subunits with two conserved N-linked glycosylation sites (N67 and N153). These EDE antibodies are divided into two sub-classes called EDE1, which do not require the conserved glycan at N153 for binding, and EDE2, which utilises the sugar at N153 for stronger binding as part of the neutralising epitope. Beyond the fact that these EDE antibodies possess broadly neutralising capabilities across multiple serotypes [307], these glycan-binding EDE2 antibodies are able to also bind partially immature viral particles as well as the fully mature virions [306]. As such, the use of immunogens that elicit EDE antibodies has been proposed as an alternative route for next-generation vaccines due to their broadly neutralising nature [308]. An important consideration of these vaccines will be the effect of antibody-dependent enhancement of infection due to the cross-reactive nature of these antibodies [307,309]. In any case, it is likely that glycan compositions of any immunogen to combat Dengue virus will have to be carefully considered, especially when trying to elicit antibodies that target the EDE.
10. Concluding remarks
The ability of enveloped viruses to hijack host cell glycosylation machinery and adorn their own glycoproteins with host-derived glycans is vital for multiple facets of viral pathogenesis. The recent advances in the field of glycosylation analysis have provided greater insights into the roles that glycans play in protein trafficking and folding, viral attachment, and immune responses to infection. Often these viral glycoproteins are sole antigens expressed on the viral surface, and as such constitute crucial vaccine targets. Since antigenic mimicry is fundamental to most licensed vaccines [169]; it is important that the glycosylation of immunogens is representative of that observed on the virus, as the elicitation of antibodies against epitopes that are occluded on the virus, and against non-native glycoforms is undesirable. Moreover, glycosylation of immunogens can play central roles in innate immune recognition to enhance humoral immunity [310]. The understanding of glycan structures, modes of recognition, and their functionality have also resulted in the development of several therapeutics to combat a wide array of deadly pathogens [93,105,[311], [312], [313], [314], [315]]. We suggest that a growing understanding of viral glycobiology will provide further opportunities to rationally develop novel therapeutics and vaccines.
Acknowledgements
The Wellcome Centre for Human Genetics is supported by grant 203141/Z/16/Z. We thank the Medical Research Council (MR/L009528/1 to T.A.B.), the NIH (R56 AI127371 to I.A.W.), the Bill and Melinda Gates Foundation (grant OPP1196345 to I.A.W.), the International AIDS Vaccine Initiative, the Bill and Melinda Gates Foundation through the Collaboration for AIDS Discovery (grants OPP1084519 and OPP1115782 to M.C.), and the Scripps Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery (CHAVI-ID) (1UM1AI100663 to M.C.). This project has received funding from the European Union's Horizon 2020 for Research & Innovation program under grant agreement No. 681137 (to M.C.).
References
- 1.Jones K.E., Patel N.G., Levy M.A., Storeygard A., Balk D., Gittleman J.L., Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao L., Pauthner M., Andrabi R., Rantalainen K., Berndsen Z., Diedrich J.K., Menis S., Sok D., Bastidas R., Park S.-K.R., Delahunty C.M., He L., Guenaga J., Wyatt R.T., Schief W.R., Ward A.B., Yates J.R., Burton D.R., Paulson J.C. Differential processing of HIV envelope glycans on the virus and soluble recombinant trimer. Nat. Commun. 2018;9:3693. doi: 10.1038/s41467-018-06121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Struwe W.B., Chertova E., Allen J.D., Seabright G.E., Watanabe Y., Harvey D.J., Medina-Ramirez M., Roser J.D., Smith R., Westcott D., Keele B.F., Bess J.W., Sanders R.W., Lifson J.D., Moore J.P., Crispin M. Site-specific glycosylation of virion-derived HIV-1 Env Is mimicked by a soluble trimeric immunogen. Cell Rep. 2018;24:1958–1966.e5. doi: 10.1016/j.celrep.2018.07.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Panico M., Bouché L., Binet D., O'Connor M.-J., Rahman D., Pang P.-C., Canis K., North S.J., Desrosiers R.C., Chertova E., Keele B.F., Bess J.W., Lifson J.D., Haslam S.M., Dell A., Morris H.R. Mapping the complete glycoproteome of virion-derived HIV-1 gp120 provides insights into broadly neutralizing antibody binding. Sci. Rep. 2016;6:32956. doi: 10.1038/srep32956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kobayashi Y., Suzuki Y. Evidence for N-glycan shielding of antigenic sites during evolution of human influenza A virus hemagglutinin. J. Virol. 2012;86:3446–3451. doi: 10.1128/JVI.06147-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wanzeck K., Boyd K.L., McCullers J.A. Glycan shielding of the Influenza virus hemagglutinin contributes to immunopathology in mice. Am. J. Respir. Crit. Care Med. 2011;183:767–773. doi: 10.1164/rccm.201007-1184OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee P.S., Ohshima N., Stanfield R.L., Yu W., Iba Y., Okuno Y., Kurosawa Y., Wilson I.A. Receptor mimicry by antibody F045-092 facilitates universal binding to the H3 subtype of influenza virus. Nat. Commun. 2014;5:3614. doi: 10.1038/ncomms4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walls A.C., Tortorici M.A., Frenz B., Snijder J., Li W., Rey F.A., DiMaio F., Bosch B.-J., Veesler D. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat. Struct. Mol. Biol. 2016;23:899–905. doi: 10.1038/nsmb.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiong X., Tortorici M.A., Snijder J., Yoshioka C., Walls A.C., Li W., McGuire A.T., Rey F.A., Bosch B.-J., Veesler D. Glycan shield and fusion activation of a deltacoronavirus spike glycoprotein fine-tuned for enteric infections. J. Virol. 2018;92 doi: 10.1128/JVI.01628-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ritchie G., Harvey D.J., Feldmann F., Stroeher U., Feldmann H., Royle L., Dwek R.A., Rudd P.M. Identification of N-linked carbohydrates from severe acute respiratory syndrome (SARS) spike glycoprotein. Virology. 2010;399:257–269. doi: 10.1016/j.virol.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ritchie G., Harvey D.J., Stroeher U., Feldmann F., Feldmann H., Wahl-Jensen V., Royle L., Dwek R.A., Rudd P.M. Identification of N-glycans from Ebola virus glycoproteins by matrix-assisted laser desorption/ionisation time-of-flight and negative ion electrospray tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2010;24:571–585. doi: 10.1002/rcm.4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J.E., Fusco M.L., Hessell A.J., Oswald W.B., Burton D.R., Saphire E.O. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sommerstein R., Flatz L., Remy M.M., Malinge P., Magistrelli G., Fischer N., Sahin M., Bergthaler A., Igonet S., ter Meulen J., Rigo D., Meda P., Rabah N., Coutard B., Bowden T.A., Lambert P.-H., Siegrist C.-A., Pinschewer D.D. Arenavirus glycan shield promotes neutralizing antibody evasion and protracted infection. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005276. e1005276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mondotte J.A., Lozach P.-Y., Amara A., Gamarnik A.V. Essential role of dengue virus envelope protein N glycosylation at asparagine-67 during viral propagation. J. Virol. 2007;81:7136–7148. doi: 10.1128/JVI.00116-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fontes-Garfias C.R., Shan C., Luo H., Muruato A.E., Medeiros D.B.A., Mays E., Xie X., Zou J., Roundy C.M., Wakamiya M., Rossi S.L., Wang T., Weaver S.C., Shi P.-Y. Functional analysis of glycosylation of Zika virus envelope protein. Cell Rep. 2017;21:1180–1190. doi: 10.1016/j.celrep.2017.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flamand M., Megret F., Mathieu M., Lepault J., Rey F.A., Deubel V. Dengue virus type 1 nonstructural glycoprotein NS1 is secreted from mammalian cells as a soluble hexamer in a glycosylation-dependent fashion. J. Virol. 1999;73:6104–6110. doi: 10.1128/jvi.73.7.6104-6110.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohan G.S., Li W., Ye L., Compans R.W., Yang C. Antigenic subversion: a novel mechanism of host immune evasion by Ebola virus. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1003065. e1003065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorander S., Mbwana J., Lyamuya E., Lagergard T., Liljeqvist J.-A. Mature glycoprotein G Presents high performance in diagnosing Herpes Simplex Virus type 2 infection in sera of different Tanzanian cohorts. Clin. Vaccine Immunol. 2006;13:633–639. doi: 10.1128/CVI.00051-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor M.E., Drickamer K. Oxford University Press; 2011. Introduction to Glycobiology. [Google Scholar]
- 20.Varki A., Gagneux P. Cold Spring Harbor Laboratory Press; 2015. Biological Functions of Glycans. [DOI] [PubMed] [Google Scholar]
- 21.Spiro R.G. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology. 2002;12:43R–56R. doi: 10.1093/glycob/12.4.43R. [DOI] [PubMed] [Google Scholar]
- 22.Varki A. Cold Spring Harbor Laboratory Press; 2009. Essentials of Glycobiology. [PubMed] [Google Scholar]
- 23.Korber B., Gaschen B., Yusim K., Thakallapally R., Kesmir C., Detours V. Evolutionary and immunological implications of contemporary HIV-1 variation. Br. Med. Bull. 2001;58:19–42. doi: 10.1093/bmb/58.1.19. [DOI] [PubMed] [Google Scholar]
- 24.Jäntti J., Hildén P., Rönkä H., Mäkiranta V., Keränen S., Kuismanen E. Immunocytochemical analysis of Uukuniemi virus budding compartments: role of the intermediate compartment and the Golgi stack in virus maturation. J. Virol. 1997;71:1162–1172. doi: 10.1128/jvi.71.2.1162-1172.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stertz S., Reichelt M., Spiegel M., Kuri T., Martínez-Sobrido L., García-Sastre A., Weber F., Kochs G. The intracellular sites of early replication and budding of SARS-coronavirus. Virology. 2007;361:304–315. doi: 10.1016/J.VIROL.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bayer K., Banning C., Bruss V., Wiltzer-Bach L., Schindler M. Hepatitis C Virus is released via a noncanonical secretory route. J. Virol. 2016;90:10558–10573. doi: 10.1128/JVI.01615-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doores K.J., Bonomelli C., Harvey D.J., Vasiljevic S., Dwek R.A., Burton D.R., Crispin M., Scanlan C.N. Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. Proc. Natl. Acad. Sci. U. S. A. 2010;107:13800–13805. doi: 10.1073/pnas.1006498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonomelli C., Doores K.J., Dunlop D.C., Thaney V., Dwek R.A., Burton D.R., Crispin M., Scanlan C.N. The glycan shield of HIV is predominantly oligomannose independently of production system or viral clade. PLoS One. 2011;6 doi: 10.1371/journal.pone.0023521. e23521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pritchard L.K., Harvey D.J., Bonomelli C., Crispin M., Doores K.J. Cell- and protein-directed glycosylation of native cleaved HIV-1 envelope. J. Virol. 2015;89:8932–8944. doi: 10.1128/JVI.01190-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gluschankof P., Mondor I., Gelderblom H.R., Sattentau Q.J. Cell membrane vesicles are a major contaminant of gradient-enriched human immunodeficiency virus type-1 preparations. Virology. 1997;230:125–133. doi: 10.1006/viro.1997.8453. [DOI] [PubMed] [Google Scholar]
- 31.Harvey D.J., Merry A.H., Royle L., P. Campbell M., Dwek R.A., Rudd P.M. Proposal for a standard system for drawing structural diagrams of N- and O-linked carbohydrates and related compounds. Proteomics. 2009;9:3796–3801. doi: 10.1002/pmic.200900096. [DOI] [PubMed] [Google Scholar]
- 32.Halim A., Brinkmalm G., Rüetschi U., Westman-Brinkmalm A., Portelius E., Zetterberg H., Blennow K., Larson G., Nilsson J. Site-specific characterization of threonine, serine, and tyrosine glycosylations of amyloid precursor protein/amyloid beta-peptides in human cerebrospinal fluid. Proc. Natl. Acad. Sci. U. S. A. 2011;108:11848–11853. doi: 10.1073/pnas.1102664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steentoft C., Vakhrushev S.Y., Joshi H.J., Kong Y., Vester-Christensen M.B., Schjoldager K.T.-B.G., Lavrsen K., Dabelsteen S., Pedersen N.B., Marcos-Silva L., Gupta R., Paul Bennett E., Mandel U., Brunak S., Wandall H.H., Levery S.B., Clausen H. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013;32:1478–1488. doi: 10.1038/emboj.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishikawa I., Nakajima Y., Ito M., Fukuchi S., Homma K., Nishikawa K. Computational prediction of O-linked glycosylation sites that preferentially map on intrinsically disordered regions of extracellular proteins. Int. J. Mol. Sci. 2010;11:4991–5008. doi: 10.3390/ijms11124991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bagdonaite I., Nordén R., Joshi H.J., Dabelsteen S., Nyström K., Vakhrushev S.Y., Olofsson S., Wandall H.H. A strategy for O-glycoproteomics of enveloped viruses—the O-glycoproteome of Herpes Simplex virus type 1. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004784. e1004784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bagdonaite I., Nordén R., Joshi H.J., King S.L., Vakhrushev S.Y., Olofsson S., Wandall H.H. Global mapping of O-glycosylation of Varicella Zoster virus, human Cytomegalovirus, and Epstein-Barr virus. J. Biol. Chem. 2016;291:12014–12028. doi: 10.1074/jbc.M116.721746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iversen M.B., Reinert L.S., Thomsen M.K., Bagdonaite I., Nandakumar R., Cheshenko N., Prabakaran T., Vakhrushev S.Y., Krzyzowska M., Kratholm S.K., Ruiz-Perez F., Petersen S.V., Goriely S., Bibby B.M., Eriksson K., Ruland J., Thomsen A.R., Herold B.C., Wandall H.H., Frische S., Holm C.K., Paludan S.R. An innate antiviral pathway acting before interferons at epithelial surfaces. Nat. Immunol. 2016;17:150–158. doi: 10.1038/ni.3319. [DOI] [PubMed] [Google Scholar]
- 38.Martinez O., Tantral L., Mulherkar N., Chandran K., Basler C.F. Impact of Ebola mucin-like domain on antiglycoprotein antibody responses induced by Ebola virus-like particles. J. Infect. Dis. 2011;204:S825–S832. doi: 10.1093/infdis/jir295. Suppl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nordén R., Halim A., Nyström K., Bennett E.P., Mandel U., Olofsson S., Nilsson J., Larson G. O-Linked glycosylation of the mucin domain of the Herpes Simplex virus type 1-specific glycoprotein gC-1 is temporally regulated in a seed-and-spread manner. J. Biol. Chem. 2015;290:5078–5091. doi: 10.1074/jbc.M114.616409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McLellan J.S., Ray W.C., Peeples M.E. Structure and function of respiratory syncytial virus surface glycoproteins. Curr. Top. Microbiol. Immunol. 2013;372:83–104. doi: 10.1007/978-3-642-38919-1_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luther K.B., Hülsmeier A.J., Schegg B., Deuber S.A., Raoult D., Hennet T. Mimivirus collagen is modified by bifunctional lysyl hydroxylase and glycosyltransferase enzyme. J. Biol. Chem. 2011;286:43701–43709. doi: 10.1074/jbc.M111.309096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiang Y., Baxa U., Zhang Y., Steven A.C., Lewis G.L., Van Etten J.L., Rossmann M.G. Crystal structure of a virus-encoded putative glycosyltransferase. J. Virol. 2010;84:12265–12273. doi: 10.1128/JVI.01303-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Markine-Goriaynoff N., Gillet L., Van Etten J.L., Korres H., Verma N., Vanderplasschen A. Glycosyltransferases encoded by viruses. J. Gen. Virol. 2004;85:2741–2754. doi: 10.1099/vir.0.80320-0. [DOI] [PubMed] [Google Scholar]
- 44.Piacente F., De Castro C., Jeudy S., Gaglianone M., Laugieri M.E., Notaro A., Salis A., Damonte G., Abergel C., Tonetti M.G. The rare sugar N-acetylated viosamine is a major component of Mimivirus fibers. J. Biol. Chem. 2017;292:7385–7394. doi: 10.1074/jbc.M117.783217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Flaherty R., Trbojević-Akmačić I., Greville G., Rudd P.M., Lauc G. The sweet spot for biologics: recent advances in characterization of biotherapeutic glycoproteins. Expert Rev. Proteomics. 2018;15:13–29. doi: 10.1080/14789450.2018.1404907. [DOI] [PubMed] [Google Scholar]
- 46.Thaysen-Andersen M., Packer N.H. Advances in LC–MS/MS-based glycoproteomics: Getting closer to system-wide site-specific mapping of the N- and O-glycoproteome. Biochim. Biophys. Acta, Proteins Proteomics. 2014;1844:1437–1452. doi: 10.1016/J.BBAPAP.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 47.Xiao H., Sun F., Suttapitugsakul S., Wu R. Global and site-specific analysis of protein glycosylation in complex biological systems with mass spectrometry. Mass Spectrom. Rev. 2019 doi: 10.1002/mas.21586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith D.F., Song X., Cummings R.D. Use of glycan microarrays to explore specificity of glycan-binding proteins. Methods Enzymol. 2010;480:417–444. doi: 10.1016/S0076-6879(10)80033-3. [DOI] [PubMed] [Google Scholar]
- 49.Rillahan C.D., Paulson J.C. Glycan microarrays for decoding the glycome. Annu. Rev. Biochem. 2011;80:797–823. doi: 10.1146/annurev-biochem-061809-152236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang M., Gaschen B., Blay W., Foley B., Haigwood N., Kuiken C., Korber B. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology. 2004;14:1229–1246. doi: 10.1093/glycob/cwh106. [DOI] [PubMed] [Google Scholar]
- 51.Wagh K., Kreider E.F., Li Y., Barbian H.J., Learn G.H., Giorgi E., Hraber P.T., Decker T.G., Smith A.G., Gondim M.V., Gillis L., Wandzilak J., Chuang G.-Y., Rawi R., Cai F., Pellegrino P., Williams I., Overbaugh J., Gao F., Kwong P.D., Haynes B.F., Shaw G.M., Borrow P., Seaman M.S., Hahn B.H., Korber B. Completeness of HIV-1 envelope glycan shield at transmission determines neutralization breadth. Cell Rep. 2018;25:893–908.e7. doi: 10.1016/j.celrep.2018.09.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crispin M., Harvey D.J., Bitto D., Bonomelli C., Edgeworth M., Scrivens J.H., Huiskonen J.T., Bowden T.A. Structural plasticity of the Semliki Forest Virus glycome upon interspecies transmission. J. Proteome Res. 2014;13:1702–1712. doi: 10.1021/pr401162k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shelton H., Ayora-Talavera G., Ren J., Loureiro S., Pickles R.J., Barclay W.S., Jones I.M. Receptor binding profiles of avian influenza virus hemagglutinin subtypes on human cells as a predictor of pandemic potential. J. Virol. 2011;85:1875–1880. doi: 10.1128/JVI.01822-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kornfeld R., Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 55.Kim N.Y., Jung W.-W., Oh Y.-K., Chun T., Park H.-Y., Lee H.-T., Han I.-K., Yang J.M., Kim Y.B. Natural protection from zoonosis by alpha-gal epitopes on virus particles in xenotransmission. Xenotransplantation. 2007;14:104–111. doi: 10.1111/j.1399-3089.2007.00377.x. [DOI] [PubMed] [Google Scholar]
- 56.Antonopoulos A., North S.J., Haslam S.M., Dell A. Glycosylation of mouse and human immune cells: insights emerging from N-glycomics analyses. Biochem. Soc. Trans. 2011;39:1334–1340. doi: 10.1042/BST0391334. [DOI] [PubMed] [Google Scholar]
- 57.Galili U. Natural anti-carbohydrate antibodies contributing to evolutionary survival of primates in viral epidemics? Glycobiology. 2016;26:1140–1150. doi: 10.1093/glycob/cww088. [DOI] [PubMed] [Google Scholar]
- 58.Brockhausen I. Pathways of O-glycan biosynthesis in cancer cells. Biochim. Biophys. Acta, Gen. Subj. 1999;1473:67–95. doi: 10.1016/S0304-4165(99)00170-1. [DOI] [PubMed] [Google Scholar]
- 59.Neil S.J.D., McKnight A., Gustafsson K., Weiss R.A. HIV-1 incorporates ABO histo-blood group antigens that sensitize virions to complement-mediated inactivation. Blood. 2005;105:4693–4699. doi: 10.1182/blood-2004-11-4267. [DOI] [PubMed] [Google Scholar]
- 60.Cooling L. Blood groups in infection and host susceptibility. Clin. Microbiol. Rev. 2015;28:801–870. doi: 10.1128/CMR.00109-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seymour R.M., Allan M.J., Pomiankowski A., Gustafsson K. Evolution of the human ABO polymorphism by two complementary selective pressures. Proc. Biol. Sci. 2004;271:1065–1072. doi: 10.1098/rspb.2004.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matrosovich M., Tuzikov A., Bovin N., Gambaryan A., Klimov A., Castrucci M.R., Donatelli I., Kawaoka Y. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J. Virol. 2000;74:8502–8512. doi: 10.1128/jvi.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shinya K., Ebina M., Yamada S., Ono M., Kasai N., Kawaoka Y. Influenza virus receptors in the human airway. Nature. 2006;440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 64.Nicholls J.M., Bourne A.J., Chen H., Guan Y., Peiris J.S.M. Sialic acid receptor detection in the human respiratory tract: evidence for widespread distribution of potential binding sites for human and avian influenza viruses. Respir. Res. 2007;8:73. doi: 10.1186/1465-9921-8-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kumlin U., Olofsson S., Dimock K., Arnberg N. Sialic acid tissue distribution and influenza virus tropism. Influenza Other Respir. Viruses. 2008;2:147–154. doi: 10.1111/j.1750-2659.2008.00051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shi X., Jarvis D.L. Protein N-glycosylation in the baculovirus-insect cell system. Curr. Drug Targets. 2007;8:1116–1125. doi: 10.2174/138945007782151360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Max Crispin M.D., Ritchie G.E., Critchley A.J., Paul Morgan B., Wilson I.A., Dwek R.A., Sim R.B., Rudd P.M. Monoglucosylated glycans in the secreted human complement component C3: implications for protein biosynthesis and structure. FEBS Lett. 2004;566:270–274. doi: 10.1016/j.febslet.2004.04.045. [DOI] [PubMed] [Google Scholar]
- 68.Loke I., Kolarich D., Packer N.H., Thaysen-Andersen M. Emerging roles of protein mannosylation in inflammation and infection. Mol. Asp. Med. 2016;51:31–55. doi: 10.1016/j.mam.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 69.Hebert D.N., Lamriben L., Powers E.T., Kelly J.W. The intrinsic and extrinsic effects of N-linked glycans on glycoproteostasis. Nat. Chem. Biol. 2014;10:902–910. doi: 10.1038/nchembio.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wormald M.R., Dwek R.A. Glycoproteins: glycan presentation and protein-fold stability. Structure. 1999;7:R155–R160. doi: 10.1016/s0969-2126(99)80095-1. [DOI] [PubMed] [Google Scholar]
- 71.Shi X., Brauburger K., Elliott R.M. Role of N-linked glycans on bunyamwera virus glycoproteins in intracellular trafficking, protein folding, and virus infectivity. J. Virol. 2005;79:13725–13734. doi: 10.1128/JVI.79.21.13725-13734.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shi X., Elliott R.M. Analysis of N-linked glycosylation of hantaan virus glycoproteins and the role of oligosaccharide side chains in protein folding and intracellular trafficking. J. Virol. 2004;78:5414–5422. doi: 10.1128/JVI.78.10.5414-5422.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li S., van Oers M.M., Vlak J.M., Shen S., Hu Z., Wang M., Braakman I., Wang H., Li X., Deng F. Mutational and functional analysis of N-linked glycosylation of envelope fusion protein F of Helicoverpa armigera nucleopolyhedrovirus. J. Gen. Virol. 2016;97:988–999. doi: 10.1099/jgv.0.000404. [DOI] [PubMed] [Google Scholar]
- 74.Lee E., Weir R.C., Dalgarno L. Changes in the Dengue virus major envelope protein on passaging and their localization on the three-dimensional structure of the protein. Virology. 1997;232:281–290. doi: 10.1006/viro.1997.8570. [DOI] [PubMed] [Google Scholar]
- 75.Guirakhoo F., Hunt A.R., Lewis J.G., Roehrig J.T. Selection and partial characterization of Dengue 2 virus mutants that induce fusion at elevated pH. Virology. 1993;194:219–223. doi: 10.1006/viro.1993.1252. [DOI] [PubMed] [Google Scholar]
- 76.Dubuisson J., Cocquerel L., Fournillier A., Meunier J.C., Cahour A., Choukhi A., Wychowski C. Analysis of the glycosylation sites of hepatitis C virus (HCV) glycoprotein E1 and the influence of E1 glycans on the formation of the HCV glycoprotein complex. J. Gen. Virol. 1999;80:887–896. doi: 10.1099/0022-1317-80-4-887. [DOI] [PubMed] [Google Scholar]
- 77.Moll M., Kaufmann A., Maisner A. Influence of N-glycans on processing and biological activity of the Nipah virus fusion protein. J. Virol. 2004;78:7274–7278. doi: 10.1128/JVI.78.13.7274-7278.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Luo S., Hu K., He S., Wang P., Zhang M., Huang X., Du T., Zheng C., Liu Y., Hu Q. Contribution of N-linked glycans on HSV-2 gB to cell–cell fusion and viral entry. Virology. 2015;483:72–82. doi: 10.1016/J.VIROL.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 79.Wang W., Nie J., Prochnow C., Truong C., Jia Z., Wang S., Chen X.S., Wang Y. A systematic study of the N-glycosylation sites of HIV-1 envelope protein on infectivity and antibody-mediated neutralization. Retrovirology. 2013;10:14. doi: 10.1186/1742-4690-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.François K.O., Balzarini J. The highly conserved glycan at asparagine 260 of HIV-1 gp120 is indispensable for viral entry. J. Biol. Chem. 2011;286:42900–42910. doi: 10.1074/jbc.M111.274456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kong L., Wilson I.A., Kwong P.D. Crystal structure of a fully glycosylated HIV-1 gp120 core reveals a stabilizing role for the glycan at Asn262. Proteins. 2015;83:590–596. doi: 10.1002/prot.24747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang X., Liu X., Yang D., Chen S., Peng D., Zhang X., Zhu J. Role of stem glycans attached to haemagglutinin in the biological characteristics of H5N1 avian influenza virus. J. Gen. Virol. 2015;96:1248–1257. doi: 10.1099/vir.0.000082. [DOI] [PubMed] [Google Scholar]
- 83.Gruters R.A., Neefjes J.J., Tersmette M., de Goede R.E.Y., Tulp A., Huisman H.G., Miedema F., Ploegh H.L. Interference with HIV-induced syncytium formation and viral infectivity by inhibitors of trimming glucosidase. Nature. 1987;330:74–77. doi: 10.1038/330074a0. [DOI] [PubMed] [Google Scholar]
- 84.Fleet G.W.J., Karpas A., Dwek R.A., Fellows L.E., Tyms A.S., Petursson S., Namgoong S.K., Ramsden N.G., Smith P.W., Son J.C., Wilson F., Witty D.R., Jacob G.S., Rademacher T.W. Inhibition of HIV replication by amino-sugar derivatives. FEBS Lett. 1988;237:128–132. doi: 10.1016/0014-5793(88)80185-6. [DOI] [PubMed] [Google Scholar]
- 85.Fischl M.A., Resnick L., Coombs R., Kremer A.B., Pottage J.C., Fass R.J., Fife K.H., Powderly W.G., Collier A.C., Aspinall R.L., Smith S.L., Kowalski K.G., Wallemark C.-B. The safety and efficacy of combination N-butyl-deoxynojirimycin (SC-48334) and zidovudine in patients with HIV-1 infection and 200-500 CD4 cells/mm3. J. Acquir. Immune Defic. Syndr. 1994;7:139–147. [PubMed] [Google Scholar]
- 86.Hussain S., Miller J.L., Harvey D.J., Gu Y., Rosenthal P.B., Zitzmann N., McCauley J.W. Strain-specific antiviral activity of iminosugars against human influenza A viruses. J. Antimicrob. Chemother. 2015;70:136–152. doi: 10.1093/jac/dku349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Datema R., Romero P.A., Legler G., Schwarz R.T. Inhibition of formation of complex oligosaccharides by the glucosidase inhibitor bromoconduritol. Proc. Natl. Acad. Sci. U. S. A. 1982;79:6787–6791. doi: 10.1073/pnas.79.22.6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Miller J.L., Spiro S.G., Dowall S.D., Taylor I., Rule A., Alonzi D.S., Sayce A.C., Wright E., Bentley E.M., Thom R., Hall G., Dwek R.A., Hewson R., Zitzmann N. Minimal in vivo efficacy of iminosugars in a lethal Ebola virus guinea pig model. PLoS One. 2016;11 doi: 10.1371/journal.pone.0167018. e0167018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chang J., Warren T.K., Zhao X., Gill T., Guo F., Wang L., Comunale M.A., Du Y., Alonzi D.S., Yu W., Ye H., Liu F., Guo J.-T., Mehta A., Cuconati A., Butters T.D., Bavari S., Xu X., Block T.M. Small molecule inhibitors of ER α-glucosidases are active against multiple hemorrhagic fever viruses. Antivir. Res. 2013;98:432–440. doi: 10.1016/j.antiviral.2013.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sayce A.C., Alonzi D.S., Killingbeck S.S., Tyrrell B.E., Hill M.L., Caputo A.T., Iwaki R., Kinami K., Ide D., Kiappes J.L., Beatty P.R., Kato A., Harris E., Dwek R.A., Miller J.L., Zitzmann N. Iminosugars inhibit Dengue virus production via inhibition of ER alpha-glucosidases—not glycolipid processing enzymes. PLoS Negl. Trop. Dis. 2016;10 doi: 10.1371/journal.pntd.0004524. e0004524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Miller J.L., Lachica R., Sayce A.C., Williams J.P., Bapat M., Dwek R., Beatty P.R., Harris E., Zitzmann N. Liposome-mediated delivery of iminosugars enhances efficacy against dengue virus in vivo. Antimicrob. Agents Chemother. 2012;56:6379–6386. doi: 10.1128/AAC.01554-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wu S.-F., Lee C.-J., Liao C.-L., Dwek R.A., Zitzmann N., Lin Y.-L. Antiviral effects of an iminosugar derivative on flavivirus infections. J. Virol. 2002;76:3596–3604. doi: 10.1128/JVI.76.8.3596-3604.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Alonzi D.S., Scott K.A., Dwek R.A., Zitzmann N. Iminosugar antivirals: the therapeutic sweet spot. Biochem. Soc. Trans. 2017;45:571–582. doi: 10.1042/BST20160182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kiappes J.L., Hill M.L., Alonzi D.S., Miller J.L., Iwaki R., Sayce A.C., Caputo A.T., Kato A., Zitzmann N. ToP-DNJ, a selective inhibitor of endoplasmic reticulum α-glucosidase II exhibiting antiflaviviral activity. ACS Chem. Biol. 2018;13:60–65. doi: 10.1021/acschembio.7b00870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bagdonaite I., Wandall H.H. Global aspects of viral glycosylation. Glycobiology. 2018;28:443–467. doi: 10.1093/glycob/cwy021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shtyrya Y.A., Mochalova L.V., Bovin N.V. Influenza virus neuraminidase: structure and function. Acta Nat. 2009;1:26–32. [PMC free article] [PubMed] [Google Scholar]
- 97.Bouvier N.M., Palese P. The biology of influenza viruses. Vaccine. 2008;26(Suppl. 4):D49–D53. doi: 10.1016/j.vaccine.2008.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Palese P., Compans R.W. Inhibition of Influenza virus replication in tissue culture by 2-deoxy-2,3-dehydro-N-trifluoroacetylneuraminic acid (FANA): mechanism of action. J. Gen. Virol. 1976;33:159–163. doi: 10.1099/0022-1317-33-1-159. [DOI] [PubMed] [Google Scholar]
- 99.Matrosovich M.N., Matrosovich T.Y., Gray T., Roberts N.A., Klenk H.-D. Neuraminidase is important for the initiation of Influenza virus infection in human airway epithelium. J. Virol. 2004;78:12665–12667. doi: 10.1128/JVI.78.22.12665-12667.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Waxham M.N., Merz D.C., Wolinsky J.S. Intracellular maturation of mumps virus hemagglutinin-neuraminidase glycoprotein: conformational changes detected with monoclonal antibodies. J. Virol. 1986;59:392–400. doi: 10.1128/jvi.59.2.392-400.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xu R., Palmer S.G., Porotto M., Palermo L.M., Niewiesk S., Wilson I.A., Moscona A. Interaction between the Hemagglutinin-Neuraminidase and Fusion glycoproteins of Human Parainfluenza virus type III regulates viral growth in vivo. MBio. 2013;4 doi: 10.1128/mBio.00803-13. e00803-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Connaris H., Takimoto T., Russell R., Crennell S., Moustafa I., Portner A., Taylor G. Probing the sialic acid binding site of the hemagglutinin-neuraminidase of Newcastle disease virus: identification of key amino acids involved in cell binding, catalysis, and fusion. J. Virol. 2002;76:1816–1824. doi: 10.1128/JVI.76.4.1816-1824.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zeltina A., Bowden T.A., Lee B. Emerging Paramyxoviruses: receptor tropism and zoonotic potential. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005390. e1005390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.von Itzstein M. The war against influenza: discovery and development of sialidase inhibitors. Nat. Rev. Drug Discov. 2007;6:967–974. doi: 10.1038/nrd2400. [DOI] [PubMed] [Google Scholar]
- 105.Taylor G., Crennell S., Takimoto T., Portner A. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat. Struct. Biol. 2000;7:1068–1074. doi: 10.1038/81002. [DOI] [PubMed] [Google Scholar]
- 106.Chen Y.-Q., Wohlbold T.J., Zheng N.-Y., Huang M., Huang Y., Neu K.E., Lee J., Wan H., Rojas K.T., Kirkpatrick E., Henry C., Palm A.-K.E., Stamper C.T., Lan L.Y.-L., Topham D.J., Treanor J., Wrammert J., Ahmed R., Eichelberger M.C., Georgiou G., Krammer F., Wilson P.C. Influenza infection in humans induces broadly cross-reactive and protective neuraminidase-reactive antibodies. Cell. 2018;173:417–429.e10. doi: 10.1016/j.cell.2018.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Crowe J.E. Antibody determinants of influenza immunity. J. Infect. Dis. 2019 doi: 10.1093/infdis/jiz010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Desheva Y., Sychev I., Smolonogina T., Rekstin A., Ilyushina N., Lugovtsev V., Samsonova A., Go A., Lerner A. Anti-neuraminidase antibodies against pandemic A/H1N1 influenza viruses in healthy and influenza-infected individuals. PLoS One. 2018;13 doi: 10.1371/journal.pone.0196771. e0196771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Trefry J.C., Wollen S.E., Nasar F., Shamblin J.D., Kern S.J., Bearss J.J., Jefferson M.A., Chance T.B., Kugelman J.R., Ladner J.T., Honko A.N., Kobs D.J., Wending M.Q.S., Sabourin C.L., Pratt W.D., Palacios G.F., Pitt M.L.M. Ebola virus infections in nonhuman primates are temporally influenced by glycoprotein poly-U editing site populations in the exposure material. Viruses. 2015;7:6739–6754. doi: 10.3390/v7122969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Maruyama T., Parren P.W.H.I., Sanchez A., Rensink I., Rodriguez L.L., Khan A.S., Peters C.J., Burton D.R. Recombinant human monoclonal antibodies to Ebola virus. J. Infect. Dis. 1999;179:S235–S239. doi: 10.1086/514280. [DOI] [PubMed] [Google Scholar]
- 111.Druar C., Saini S.S., Cossitt M.A., Yu F., Qiu X., Geisbert T.W., Jones S., Jahrling P.B., Stewart D.I.H., Wiersma E.J. Analysis of the expressed heavy chain variable-region genes of Macaca fascicularis and isolation of monoclonal antibodies specific for the Ebola virus’ soluble glycoprotein. Immunogenetics. 2005;57:730–738. doi: 10.1007/s00251-005-0047-4. [DOI] [PubMed] [Google Scholar]
- 112.Cook J.D., Lee J.E. The secret life of viral entry glycoproteins: moonlighting in immune evasion. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003258. e1003258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Saphire E.O., Schendel S.L., Fusco M.L., Gangavarapu K., Gunn B.M., Wec A.Z., Halfmann P.J., Brannan J.M., Herbert A.S., Qiu X., Wagh K., He S., Giorgi E.E., Theiler J., Pommert K.B.J., Krause T.B., Turner H.L., Murin C.D., Pallesen J., Davidson E., Ahmed R., Aman M.J., Bukreyev A., Burton D.R., Crowe J.E., Davis C.W., Georgiou G., Krammer F., Kyratsous C.A., Lai J.R., Nykiforuk C., Pauly M.H., Rijal P., Takada A., Townsend A.R., Volchkov V., Walker L.M., Wang C.-I., Zeitlin L., Doranz B.J., Ward A.B., Korber B., Kobinger G.P., Andersen K.G., Kawaoka Y., Alter G., Chandran K., Dye J.M. Viral Hemorrhagic Fever Immunotherapeutic Consortium, Systematic analysis of monoclonal antibodies against Ebola virus GP defines features that contribute to protection. Cell. 2018;174 doi: 10.1016/j.cell.2018.07.033. 938–952.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Branco L.M., Garry R.F. Characterization of the Lassa virus GP1 ectodomain shedding: implications for improved diagnostic platforms. Virol. J. 2009;6:147. doi: 10.1186/1743-422X-6-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Branco L.M., Grove J.N., Moses L.M., Goba A., Fullah M., Momoh M., Schoepp R.J., Bausch D.G., Garry R.F. Shedding of soluble glycoprotein 1 detected during acute Lassa virus infection in human subjects. Virol. J. 2010;7:306. doi: 10.1186/1743-422X-7-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cohen-Dvashi H., Cohen N., Israeli H., Diskin R. Molecular mechanism for LAMP1 recognition by Lassa virus. J. Virol. 2015;89:7584–7592. doi: 10.1128/JVI.00651-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hastie K.M., Zandonatti M.A., Kleinfelter L.M., Heinrich M.L., Rowland M.M., Chandran K., Branco L.M., Robinson J.E., Garry R.F., Saphire E.O. Structural basis for antibody-mediated neutralization of Lassa virus. Science. 2017;356:923–928. doi: 10.1126/science.aam7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pryce R., Ng W.M., Zeltina A., Watanabe Y., El Omari K., Wagner A., Bowden T.A. Structure-based classification defines the discrete conformational classes adopted by the arenaviral GP1. J. Virol. 2019;93 doi: 10.1128/JVI.01048-18. e01048-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Israeli H., Cohen-Dvashi H., Shulman A., Shimon A., Diskin R., Chevenet F. Mapping of the Lassa virus LAMP1 binding site reveals unique determinants not shared by other old world arenaviruses. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006337. e1006337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Somnuke P., Hauhart R.E., Atkinson J.P., Diamond M.S., Avirutnan P. N-linked glycosylation of dengue virus NS1 protein modulates secretion, cell-surface expression, hexamer stability, and interactions with human complement. Virology. 2011;413:253–264. doi: 10.1016/j.virol.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Avirutnan P., Zhang L., Punyadee N., Manuyakorn A., Puttikhunt C., Kasinrerk W., Malasit P., Atkinson J.P., Diamond M.S. Secreted NS1 of Dengue virus attaches to the surface of cells via interactions with heparan sulfate and chondroitin sulfate. PLoS Pathog. 2007;3:e183. doi: 10.1371/journal.ppat.0030183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Scaturro P., Cortese M., Chatel-Chaix L., Fischl W., Bartenschlager R. Dengue virus non-structural protein 1 modulates infectious particle production via interaction with the structural proteins. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005277. e1005277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Avirutnan P., Hauhart R.E., Somnuke P., Blom A.M., Diamond M.S., Atkinson J.P. Binding of Flavivirus nonstructural protein NS1 to C4b binding protein modulates complement activation. J. Immunol. 2011;187:424–433. doi: 10.4049/jimmunol.1100750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Thiemmeca S., Tamdet C., Punyadee N., Prommool T., Songjaeng A., Noisakran S., Puttikhunt C., Atkinson J.P., Diamond M.S., Ponlawat A., Avirutnan P. Secreted NS1 protects Dengue virus from mannose-binding lectin–mediated neutralization. J. Immunol. 2016;197:4053–4065. doi: 10.4049/jimmunol.1600323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Beatty P.R., Puerta-Guardo H., Killingbeck S.S., Glasner D.R., Hopkins K., Harris E. Dengue virus NS1 triggers endothelial permeability and vascular leak that is prevented by NS1 vaccination. Sci. Transl. Med. 2015;7:304ra141. doi: 10.1126/scitranslmed.aaa3787. [DOI] [PubMed] [Google Scholar]
- 126.Bukreyev A., Yang L., Fricke J., Cheng L., Ward J.M., Murphy B.R., Collins P.L. The secreted form of Respiratory Syncytial Virus G glycoprotein helps the virus evade antibody-mediated restriction of replication by acting as an antigen decoy and through effects on Fc receptor-bearing leukocytes. J. Virol. 2008;82:12191–12204. doi: 10.1128/JVI.01604-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ressing M.E., van Leeuwen D., Verreck F.A.W., Keating S., Gomez R., Franken K.L.M.C., Ottenhoff T.H.M., Spriggs M., Schumacher T.N., Hutt-Fletcher L.M., Rowe M., Wiertz E.J.H.J. Epstein-Barr Virus gp42 Is posttranslationally modified to produce soluble gp42 that mediates HLA class II immune evasion. J. Virol. 2005;79:841–852. doi: 10.1128/JVI.79.2.841-852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hoebe E.K., Le Large T.Y.S., Tarbouriech N., Oosterhoff D., De Gruijl T.D., Middeldorp J.M., Greijer A.E. Epstein-Barr virus-encoded BARF1 protein is a decoy receptor for macrophage colony stimulating factor and interferes with macrophage differentiation and activation. Viral Immunol. 2012;25:461–470. doi: 10.1089/vim.2012.0034. [DOI] [PubMed] [Google Scholar]
- 129.Moore P.L., Crooks E.T., Porter L., Zhu P., Cayanan C.S., Grise H., Corcoran P., Zwick M.B., Franti M., Morris L., Roux K.H., Burton D.R., Binley J.M. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J. Virol. 2006;80:2515–2528. doi: 10.1128/JVI.80.5.2515-2528.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sattentau Q.J., Moore J.P. Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the gp120 oligomer. J. Exp. Med. 1995;182:185–196. doi: 10.1084/jem.182.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sanders R.W., Derking R., Cupo A., Julien J.-P., Yasmeen A., de Val N., Kim H.J., Blattner C., de la Peña A.T., Korzun J., Golabek M., de los Reyes K., Ketas T.J., van Gils M.J., King C.R., Wilson I.A., Ward A.B., Klasse P.J., Moore J.P. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003618. e1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.de Taeye S.W., Ozorowski G., Torrents de la Peña A., Guttman M., Julien J.-P., van den Kerkhof T.L.G.M., Burger J.A., Pritchard L.K., Pugach P., Yasmeen A., Crampton J., Hu J., Bontjer I., Torres J.L., Arendt H., DeStefano J., Koff W.C., Schuitemaker H., Eggink D., Berkhout B., Dean H., LaBranche C., Crotty S., Crispin M., Montefiori D.C., Klasse P.J., Lee K.K., Moore J.P., Wilson I.A., Ward A.B., Sanders R.W. Immunogenicity of stabilized HIV-1 envelope trimers with reduced exposure of non-neutralizing epitopes. Cell. 2015;163:1702–1715. doi: 10.1016/j.cell.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Helle F., Vieyres G., Elkrief L., Popescu C.-I., Wychowski C., Descamps V., Castelain S., Roingeard P., Duverlie G., Dubuisson J. Role of N-linked glycans in the functions of hepatitis C virus envelope proteins incorporated into infectious virions. J. Virol. 2010;84:11905–11915. doi: 10.1128/JVI.01548-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lavie M., Hanoulle X., Dubuisson J. Glycan shielding and modulation of Hepatitis C virus neutralizing antibodies. Front. Immunol. 2018;9:910. doi: 10.3389/fimmu.2018.00910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Julithe R., Abou-Jaoudé G., Sureau C. Modification of the hepatitis B virus envelope protein glycosylation pattern interferes with secretion of viral particles, infectivity, and susceptibility to neutralizing antibodies. J. Virol. 2014;88:9049–9059. doi: 10.1128/JVI.01161-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bradel-Tretheway B.G., Liu Q., Stone J.A., McInally S., Aguilar H.C. Novel functions of Hendra virus G N-glycans and comparisons to Nipah virus. J. Virol. 2015;89:7235–7247. doi: 10.1128/JVI.00773-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Samal S., Khattar S.K., Kumar S., Collins P.L., Samal S.K. Coordinate deletion of N-glycans from the heptad repeats of the fusion F protein of Newcastle disease virus yields a hyperfusogenic virus with increased replication, virulence, and immunogenicity. J. Virol. 2012;86:2501–2511. doi: 10.1128/JVI.06380-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sodora D.L., Cohen G.H., Eisenberg R.J. Influence of asparagine-linked oligosaccharides on antigenicity, processing, and cell surface expression of herpes simplex virus type 1 glycoprotein D. J. Virol. 1989;63:5184–5193. doi: 10.1128/jvi.63.12.5184-5193.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Machiels B., Lété C., Guillaume A., Mast J., Stevenson P.G., Vanderplasschen A., Gillet L. Antibody evasion by a gammaherpesvirus O-glycan shield. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002387. e1002387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Watanabe Y., Raghwani J., Allen J.D., Seabright G.E., Li S., Moser F., Huiskonen J.T., Strecker T., Bowden T.A., Crispin M. Structure of the Lassa virus glycan shield provides a model for immunological resistance. Proc. Natl. Acad. Sci. 2018;115:7320–7325. doi: 10.1073/pnas.1803990115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhao Y., Ren J., Harlos K., Jones D.M., Zeltina A., Bowden T.A., Padilla-Parra S., Fry E.E., Stuart D.I. Toremifene interacts with and destabilizes the Ebola virus glycoprotein. Nature. 2016;535:169–172. doi: 10.1038/nature18615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lee P.S., Ohshima N., Stanfield R.L., Yu W., Iba Y., Okuno Y., Kurosawa Y., Wilson I.A. Receptor mimicry by antibody F045-092 facilitates universal binding to the H3 subtype of influenza virus. Nat. Commun. 2014;5:3614. doi: 10.1038/ncomms4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.An Y., McCullers J.A., Alymova I., Parsons L.M., Cipollo J.F. Glycosylation analysis of engineered H3N2 Influenza A virus hemagglutinins with sequentially added historically relevant glycosylation sites. J. Proteome Res. 2015;14:3957–3969. doi: 10.1021/acs.jproteome.5b00416. [DOI] [PubMed] [Google Scholar]
- 144.Do Kwon Y., Pancera M., Acharya P., Georgiev I.S., Crooks E.T., Gorman J., Joyce M.G., Guttman M., Ma X., Narpala S., Soto C., Terry D.S., Yang Y., Zhou T., Ahlsen G., Bailer R.T., Chambers M., Chuang G.-Y., Doria-Rose N.A., Druz A., Hallen M.A., Harned A., Kirys T., Louder M.K., O'Dell S., Ofek G., Osawa K., Prabhakaran M., Sastry M., Stewart-Jones G.B.E., Stuckey J., Thomas P.V., Tittley T., Williams C., Zhang B., Zhao H., Zhou Z., Donald B.R., Lee L.K., Zolla-Pazner S., Baxa U., Schön A., Freire E., Shapiro L., Lee K.K., Arthos J., Munro J.B., Blanchard S.C., Mothes W., Binley J.M., McDermott A.B., Mascola J.R., Kwong P.D. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat. Struct. Mol. Biol. 2015;22:522–531. doi: 10.1038/nsmb.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Xu K., Chan Y.-P., Bradel-Tretheway B., Akyol-Ataman Z., Zhu Y., Dutta S., Yan L., Feng Y., Wang L.-F., Skiniotis G., Lee B., Zhou Z.H., Broder C.C., Aguilar H.C., Nikolov D.B. Crystal structure of the pre-fusion Nipah virus fusion glycoprotein reveals a novel hexamer-of-trimers assembly. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005322. e1005322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Crispin M., Ward A.B., Wilson I.A. Structure and immune recognition of the HIV glycan shield. Annu. Rev. Biophys. 2018;47 doi: 10.1146/annurev-biophys-060414-034156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hallenberger S., Bosch V., Angliker H., Shaw E., Klenk H.-D., Garten W. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gpl60. Nature. 1992;360:358–361. doi: 10.1038/360358a0. [DOI] [PubMed] [Google Scholar]
- 148.Ma X., Lu M., Gorman J., Terry D.S., Hong X., Zhou Z., Zhao H., Altman R.B., Arthos J., Blanchard S.C., Kwong P.D., Munro J.B., Mothes W. HIV-1 Env trimer opens through an asymmetric intermediate in which individual protomers adopt distinct conformations. Elife. 2018;7 doi: 10.7554/eLife.34271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ward A.B., Wilson I.A. The HIV-1 envelope glycoprotein structure: nailing down a moving target. Immunol. Rev. 2017;275:21–32. doi: 10.1111/imr.12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wyatt R., Kwong P.D., Desjardins E., Sweet R.W., Robinson J., Hendrickson W.A., Sodroski J.G. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 151.Wei X., Decker J.M., Wang S., Hui H., Kappes J.C., Wu X., Salazar-Gonzalez J.F., Salazar M.G., Kilby J.M., Saag M.S., Komarova N.L., Nowak M.A., Hahn B.H., Kwong P.D., Shaw G.M. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 152.Pritchard L.K., Spencer D.I.R., Royle L., Bonomelli C., Seabright G.E., Behrens A.-J., Kulp D.W., Menis S., Krumm S.A., Dunlop D.C., Crispin D.J., Bowden T.A., Scanlan C.N., Ward A.B., Schief W.R., Doores K.J., Crispin M. Glycan clustering stabilizes the mannose patch of HIV-1 and preserves vulnerability to broadly neutralizing antibodies. Nat. Commun. 2015;6:7479. doi: 10.1038/ncomms8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Behrens A.-J., Vasiljevic S., Pritchard L., Harvey D., Andev R., Krumm S., Struwe W., Cupo A., Kumar A., Zitzmann N., Seabright G., Kramer H., Spencer D.R., Royle L., Lee J., Klasse P., Burton D., Wilson I., Ward A., Sanders R., Moore J., Doores K., Crispin M. Composition and antigenic effects of individual glycan sites of a trimeric HIV-1 envelope glycoprotein. Cell Rep. 2016;14:2695–2706. doi: 10.1016/j.celrep.2016.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Cao L., Diedrich J.K., Kulp D.W., Pauthner M., He L., Park S.-K.R., Sok D., Su C.Y., Delahunty C.M., Menis S., Andrabi R., Guenaga J., Georgeson E., Kubitz M., Adachi Y., Burton D.R., Schief W.R., Yates J.R., III, Paulson J.C. Global site-specific N-glycosylation analysis of HIV envelope glycoprotein. Nat. Commun. 2017;8:14954. doi: 10.1038/ncomms14954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Behrens A.-J., Harvey D.J., Milne E., Cupo A., Kumar A., Zitzmann N., Struwe W.B., Moore J.P., Crispin M. Molecular architecture of the cleavage-dependent mannose patch on a soluble HIV-1 envelope glycoprotein trimer. J. Virol. 2017;91 doi: 10.1128/JVI.01894-16. e01894-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Pritchard L., Vasiljevic S., Ozorowski G., Seabright G., Cupo A., Ringe R., Kim H., Sanders R., Doores K., Burton D., Wilson I., Ward A., Moore J., Crispin M. Structural constraints determine the glycosylation of HIV-1 envelope trimers. Cell Rep. 2015;11:1604–1613. doi: 10.1016/j.celrep.2015.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Behrens A.-J., Crispin M. Structural principles controlling HIV envelope glycosylation. Curr. Opin. Struct. Biol. 2017;44:125–133. doi: 10.1016/j.sbi.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Crispin M., Doores K.J. Targeting host-derived glycans on enveloped viruses for antibody-based vaccine design. Curr. Opin. Virol. 2015;11:63–69. doi: 10.1016/j.coviro.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Burton D.R., Hangartner L. Broadly neutralizing antibodies to HIV and their role in vaccine design. Annu. Rev. Immunol. 2016;34:635–659. doi: 10.1146/annurev-immunol-041015-055515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Doores K.J. The HIV glycan shield as a target for broadly neutralizing antibodies. FEBS J. 2015;282:4679–4691. doi: 10.1111/febs.13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pejchal R., Doores K.J., Walker L.M., Khayat R., Huang P.-S., Wang S.-K., Stanfield R.L., Julien J.-P., Ramos A., Crispin M., Depetris R., Katpally U., Marozsan A., Cupo A., Maloveste S., Liu Y., McBride R., Ito Y., Sanders R.W., Ogohara C., Paulson J.C., Feizi T., Scanlan C.N., Wong C.-H., Moore J.P., Olson W.C., Ward A.B., Poignard P., Schief W.R., Burton D.R., Wilson I.A. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Walker L.M., Huber M., Doores K.J., Falkowska E., Pejchal R., Julien J.-P., Wang S.-K., Ramos A., Chan-Hui P.-Y., Moyle M., Mitcham J.L., Hammond P.W., Olsen O.A., Phung P., Fling S., Wong C.-H., Phogat S., Wrin T., Simek M.D., Principal Investigators P.G., Koff W.C., Wilson I.A., Burton D.R., Poignard P. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wibmer C.K., Moore P.L., Morris L. HIV broadly neutralizing antibody targets. Curr. Opin. HIV AIDS. 2015;10:135–143. doi: 10.1097/COH.0000000000000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhou T., Zheng A., Baxa U., Chuang G.-Y., Georgiev I.S., Kong R., O'Dell S., Shahzad-ul-Hussan S., Shen C.-H., Tsybovsky Y., Bailer R.T., Gift S.K., Louder M.K., McKee K., Rawi R., Stevenson C.H., Stewart-Jones G.B.E., Taft J.D., Waltari E., Yang Y., Zhang B., Shivatare S.S., Shivatare V.S., Lee C.-C.D., Wu C.-Y., Mullikin J.C., Bewley C.A., Burton D.R., Polonis V.R., Shapiro L., Wong C.-H., Mascola J.R., Kwong P.D., Wu X., Benjamin B., Blakesley R., Bouffard G., Brooks S., Coleman H., Dekhtyar M., Gregory M., Guan X., Gupta J., Han J., Hargrove A., Ho S., Legaspi R., Maduro Q., Masiello C., Maskeri B., McDowell J., Montemayor C., Mullikin J., Park M., Riebow N., Schandler K., Schmidt B., Sison C., Stantripop M., Thomas J., Thomas P., Vemulapalli M., Young A. A neutralizing antibody recognizing primarily N-linked glycan targets the silent face of the HIV envelope. Immunity. 2018;48 doi: 10.1016/j.immuni.2018.02.013. 500–513.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chuang G.-Y., Zhou J., Acharya P., Rawi R., Shen C.-H., Sheng Z., Zhang B., Zhou T., Bailer R.T., Dandey V.P., Doria-Rose N.A., Louder M.K., McKee K., Mascola J.R., Shapiro L., Kwong P.D. Structural survey of broadly neutralizing antibodies targeting the HIV-1 Env trimer delineates epitope categories and characteristics of recognition. Structure. 2019;27:196–206.e6. doi: 10.1016/j.str.2018.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Burton D.R. What are the most powerful immunogen design vaccine strategies? Reverse vaccinology 2.0 shows great promise. Cold Spring Harb. Perspect. Biol. 2017;9 doi: 10.1101/cshperspect.a030262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kwong P.D., Mascola J.R. HIV-1 vaccines based on antibody identification, B cell ontogeny, and epitope structure. Immunity. 2018;48:855–871. doi: 10.1016/j.immuni.2018.04.029. [DOI] [PubMed] [Google Scholar]
- 168.Sanders R.W., Moore J.P. Native-like Env trimers as a platform for HIV-1 vaccine design. Immunol. Rev. 2017;275:161–182. doi: 10.1111/imr.12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Seabright G.E., Doores K.J., Burton D.R., Crispin M. Protein and glycan mimicry in HIV vaccine design. J. Mol. Biol. 2019 doi: 10.1016/j.jmb.2019.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Boni M.F. Vaccination and antigenic drift in influenza. Vaccine. 2008;26(Suppl. 3):C8–14. doi: 10.1016/j.vaccine.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Bhatt S., Holmes E.C., Pybus O.G. The genomic rate of molecular adaptation of the human Influenza A virus. Mol. Biol. Evol. 2011;28:2443–2451. doi: 10.1093/molbev/msr044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wu N.C., Wilson I.A. A perspective on the structural and functional constraints for immune evasion: insights from influenza virus. J. Mol. Biol. 2017;429:2694–2709. doi: 10.1016/J.JMB.2017.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Skehel J.J., Stevens D.J., Daniels R.S., Douglas A.R., Knossow M., Wilson I.A., Wiley D.C. A carbohydrate side chain on hemagglutinins of Hong Kong influenza viruses inhibits recognition by a monoclonal antibody. Proc. Natl. Acad. Sci. U. S. A. 1984;81:1779–1783. doi: 10.1073/pnas.81.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Igarashi M., Ito K., Kida H., Takada A. Genetically destined potentials for N-linked glycosylation of influenza virus hemagglutinin. Virology. 2008;376:323–329. doi: 10.1016/J.VIROL.2008.03.036. [DOI] [PubMed] [Google Scholar]
- 175.Sun S., Wang Q., Zhao F., Chen W., Li Z. Glycosylation site alteration in the evolution of influenza A (H1N1) viruses. PLoS One. 2011;6 doi: 10.1371/journal.pone.0022844. e22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Blackburne B.P., Hay A.J., Goldstein R.A. Changing selective pressure during antigenic changes in human influenza H3. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000058. e1000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Cherry J.L., Lipman D.J., Nikolskaya A., Wolf Y.I. Evolutionary dynamics of N-glycosylation sites of influenza virus hemagglutinin. PLoS Curr. 2009;1:RRN1001. doi: 10.1371/CURRENTS.RRN1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Tate M.D., Job E.R., Deng Y.-M., Gunalan V., Maurer-Stroh S., Reading P.C. Playing hide and seek: how glycosylation of the influenza virus hemagglutinin can modulate the immune response to infection. Viruses. 2014;6:1294–1316. doi: 10.3390/v6031294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Das S.R., Puigbò P., Hensley S.E., Hurt D.E., Bennink J.R., Yewdell J.W. Glycosylation focuses sequence variation in the influenza A virus H1 hemagglutinin globular domain. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001211. e1001211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Medina R.A., Stertz S., Manicassamy B., Zimmermann P., Sun X., Albrecht R.A., Uusi-Kerttula H., Zagordi O., Belshe R.B., Frey S.E., Tumpey T.M., García-Sastre A., García-Sastre A. Glycosylations in the globular head of the hemagglutinin protein modulate the virulence and antigenic properties of the H1N1 influenza viruses. Sci. Transl. Med. 2013;5:187ra70. doi: 10.1126/scitranslmed.3005996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lu J., Raghwani J., Pryce R., Bowden T.A., Thézé J., Huang S., Song Y., Zou L., Liang L., Bai R., Jing Y., Zhou P., Kang M., Yi L., Wu J., Pybus O.G., Ke C. Molecular evolution, diversity, and adaptation of Influenza A(H7N9) viruses in China. Emerg. Infect. Dis. 2018;24:1795–1805. doi: 10.3201/eid2410.171063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Thompson C.P., Lourenço J., Walters A.A., Obolski U., Edmans M., Palmer D.S., Kooblall K., Carnell G.W., O'Connor D., Bowden T.A., Pybus O.G., Pollard A.J., Temperton N.J., Lambe T., Gilbert S.C., Gupta S. A naturally protective epitope of limited variability as an influenza vaccine target. Nat. Commun. 2018;9 doi: 10.1038/s41467-018-06228-8. 3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Das S.R., Hensley S.E., David A., Schmidt L., Gibbs J.S., Puigbò P., Ince W.L., Bennink J.R., Yewdell J.W. Fitness costs limit influenza A virus hemagglutinin glycosylation as an immune evasion strategy. Proc. Natl. Acad. Sci. U. S. A. 2011;108:E1417–E1422. doi: 10.1073/pnas.1108754108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kosik I., Ince W.L., Gentles L.E., Oler A.J., Kosikova M., Angel M., Magadán J.G., Xie H., Brooke C.B., Yewdell J.W. Influenza A virus hemagglutinin glycosylation compensates for antibody escape fitness costs. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006796. e1006796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zhang S., Sherwood R.W., Yang Y., Fish T., Chen W., McCardle J.A., Jones R.M., Yusibov V., May E.R., Rose J.K.C., Thannhauser T.W. Comparative characterization of the glycosylation profiles of an influenza hemagglutinin produced in plant and insect hosts. Proteomics. 2012;12:1269–1288. doi: 10.1002/pmic.201100474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Mir-Shekari S.Y., Ashford D.A., Harvey D.J., Dwek R.A., Schulze I.T. The glycosylation of the influenza A virus hemagglutinin by mammalian cells. A site-specific study. J. Biol. Chem. 1997;272:4027–4036. doi: 10.1074/JBC.272.7.4027. [DOI] [PubMed] [Google Scholar]
- 187.Walls A.C., Xiong X., Park Y.-J., Tortorici M.A., Snijder J., Quispe J., Cameroni E., Gopal R., Dai M., Lanzavecchia A., Zambon M., Rey F.A., Corti D., Veesler D. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell. 2019;176 doi: 10.1016/j.cell.2018.12.028. 1026–1–39.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Jardine J., Julien J.-P., Menis S., Ota T., Kalyuzhniy O., McGuire A., Sok D., Huang P.-S., MacPherson S., Jones M., Nieusma T., Mathison J., Baker D., Ward A.B., Burton D.R., Stamatatos L., Nemazee D., Wilson I.A., Schief W.R. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wei C.-J., Boyington J.C., Dai K., Houser K.V., Pearce M.B., Kong W.-P., Yang Z.-Y., Tumpey T.M., Nabel G.J. Cross-neutralization of 1918 and 2009 Influenza viruses: role of glycans in viral evolution and vaccine design. Sci. Transl. Med. 2010;2 doi: 10.1126/scitranslmed.3000799. 24ra21-24ra21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Xu R., Ekiert D.C., Krause J.C., Hai R., Crowe J.E., Wilson I.A. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science. 2010;328:357–360. doi: 10.1126/science.1186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wang L., Shi W., Chappell J.D., Joyce M.G., Zhang Y., Kanekiyo M., Becker M.M., van Doremalen N., Fischer R., Wang N., Corbett K.S., Choe M., Mason R.D., Van Galen J.G., Zhou T., Saunders K.O., Tatti K.M., Haynes L.M., Kwong P.D., Modjarrad K., Kong W.-P., McLellan J.S., Denison M.R., Munster V.J., Mascola J.R., Graham B.S. Importance of neutralizing monoclonal antibodies targeting multiple antigenic sites on MERS-CoV Spike to avoid neutralization escape. J. Virol. 2018;92 doi: 10.1128/JVI.02002-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wang L., Shi W., Joyce M.G., Modjarrad K., Zhang Y., Leung K., Lees C.R., Zhou T., Yassine H.M., Kanekiyo M., Yang Z., Chen X., Becker M.M., Freeman M., Vogel L., Johnson J.C., Olinger G., Todd J.P., Bagci U., Solomon J., Mollura D.J., Hensley L., Jahrling P., Denison M.R., Rao S.S., Subbarao K., Kwong P.D., Mascola J.R., Kong W.-P., Graham B.S. Evaluation of candidate vaccine approaches for MERS-CoV. Nat. Commun. 2015;6 doi: 10.1038/ncomms8712. 7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Pallesen J., Wang N., Corbett K.S., Wrapp D., Kirchdoerfer R.N., Turner H.L., Cottrell C.A., Becker M.M., Wang L., Shi W., Kong W.-P., Andres E.L., Kettenbach A.N., Denison M.R., Chappell J.D., Graham B.S., Ward A.B., McLellan J.S. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Hilleman M.R. Strategies and mechanisms for host and pathogen survival in acute and persistent viral infections. Proc. Natl. Acad. Sci. 2004;101:14560–14566. doi: 10.1073/pnas.0404758101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Du L., Tai W., Yang Y., Zhao G., Zhu Q., Sun S., Liu C., Tao X., Tseng C.-T.K., Perlman S., Jiang S., Zhou Y., Li F. Introduction of neutralizing immunogenicity index to the rational design of MERS coronavirus subunit vaccines. Nat. Commun. 2016;7:13473. doi: 10.1038/ncomms13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Radoshitzky S.R., Bào Y., Buchmeier M.J., Charrel R.N., Clawson A.N., Clegg C.S., DeRisi J.L., Emonet S., Gonzalez J.-P., Kuhn J.H., Lukashevich I.S., Peters C.J., Romanowski V., Salvato M.S., Stenglein M.D., de la Torre J.C. Past, present, and future of arenavirus taxonomy. Arch. Virol. 2015;160:1851–1874. doi: 10.1007/s00705-015-2418-y. [DOI] [PubMed] [Google Scholar]
- 197.Parsy M.-L., Harlos K., Huiskonen J.T., Bowden T.A. Crystal structure of Venezuelan Hemorrhagic Fever Virus fusion glycoprotein reveals a class 1 postfusion architecture with extensive glycosylation. J. Virol. 2013;87:13070–13075. doi: 10.1128/JVI.02298-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Zeltina A., Krumm S.A., Sahin M., Struwe W.B., Harlos K., Nunberg J.H., Crispin M., Pinschewer D.D., Doores K.J., Bowden T.A. Convergent immunological solutions to Argentine hemorrhagic fever virus neutralization. Proc. Natl. Acad. Sci. 2017;114:7031–7036. doi: 10.1073/pnas.1702127114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Bowden T.A., Crispin M., Graham S.C., Harvey D.J., Grimes J.M., Jones E.Y., Stuart D.I. Unusual molecular architecture of the Machupo virus attachment glycoprotein. J. Virol. 2009;83:8259–8265. doi: 10.1128/JVI.00761-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Acciani M., Alston J.T., Zhao G., Reynolds H., Ali A.M., Xu B., Brindley M.A. Mutational analysis of Lassa virus glycoprotein highlights regions required for α-dystroglycan utilization. J. Virol. 2017 doi: 10.1128/JVI.00574-17. JVI.00574-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Li S., Sun Z., Pryce R., Parsy M.-L., Fehling S.K., Schlie K., Siebert C.A., Garten W., Bowden T.A., Strecker T., Huiskonen J.T. Acidic pH-induced conformations and LAMP1 binding of the Lassa virus glycoprotein spike. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005418. e1005418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Lee J.E., Saphire E.O. Ebolavirus glycoprotein structure and mechanism of entry. Futur. Virol. 2009;4:621–635. doi: 10.2217/fvl.09.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.de La Vega M.-A., Wong G., Kobinger G.P., Qiu X. The multiple roles of sGP in Ebola pathogenesis. Viral Immunol. 2015;28:3–9. doi: 10.1089/vim.2014.0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Francica J.R., Varela-Rohena A., Medvec A., Plesa G., Riley J.L., Bates P. Steric shielding of surface epitopes and impaired immune recognition induced by the Ebola virus glycoprotein. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001098. e1001098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Bowden T.A., Aricescu A.R., Gilbert R.J.C., Grimes J.M., Jones E.Y., Stuart D.I. Structural basis of Nipah and Hendra virus attachment to their cell-surface receptor ephrin-B2. Nat. Struct. Mol. Biol. 2008;15:567–572. doi: 10.1038/nsmb.1435. [DOI] [PubMed] [Google Scholar]
- 206.Kirchdoerfer R.N., Cottrell C.A., Wang N., Pallesen J., Yassine H.M., Turner H.L., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Aguilar H.C., Matreyek K.A., Filone C.M., Hashimi S.T., Levroney E.L., Negrete O.A., Bertolotti-Ciarlet A., Choi D.Y., McHardy I., Fulcher J.A., Su S.V., Wolf M.C., Kohatsu L., Baum L.G., Lee B. N-Glycans on Nipah virus fusion protein protect against neutralization but reduce membrane fusion and viral entry. J. Virol. 2006;80:4878–4889. doi: 10.1128/JVI.80.10.4878-4889.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Biering S.B., Huang A., Vu A.T., Robinson L.R., Bradel-Tretheway B., Choi E., Lee B., Aguilar H.C. N-Glycans on the Nipah virus attachment glycoprotein modulate fusion and viral entry as they protect against antibody neutralization. J. Virol. 2012;86:11991–12002. doi: 10.1128/JVI.01304-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Goncalves A.-R., Moraz M.-L., Pasquato A., Helenius A., Lozach P.-Y., Kunz S. Role of DC-SIGN in Lassa virus entry into human dendritic cells. J. Virol. 2013;87:11504–11515. doi: 10.1128/JVI.01893-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Tassaneetrithep B., Burgess T.H., Granelli-Piperno A., Trumpfheller C., Finke J., Sun W., Eller M.A., Pattanapanyasat K., Sarasombath S., Birx D.L., Steinman R.M., Schlesinger S., Marovich M.A. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. J. Exp. Med. 2003;197:823–829. doi: 10.1084/jem.20021840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Geijtenbeek T.B.H., van Kooyk Y. DC-SIGN: a novel HIV receptor on DCs that mediates HIV-1 transmission. Curr. Top. Microbiol. Immunol. 2003;276:31–54. doi: 10.1007/978-3-662-06508-2_2. [DOI] [PubMed] [Google Scholar]
- 212.Regan A.D., Whittaker G.R. Utilization of DC-SIGN for entry of feline coronaviruses into host cells. J. Virol. 2008;82:11992–11996. doi: 10.1128/JVI.01094-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Marzi A., Gramberg T., Simmons G., Moller P., Rennekamp A.J., Krumbiegel M., Geier M., Eisemann J., Turza N., Saunier B., Steinkasserer A., Becker S., Bates P., Hofmann H., Pohlmann S. DC-SIGN and DC-SIGNR interact with the glycoprotein of Marburg virus and the S protein of Severe Acute Respiratory Syndrome Coronavirus. J. Virol. 2004;78:12090–12095. doi: 10.1128/JVI.78.21.12090-12095.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Klimstra W.B., Nangle E.M., Smith M.S., Yurochko A.D., Ryman K.D. DC-SIGN and L-SIGN can act as attachment receptors for alphaviruses and distinguish between mosquito cell- and mammalian cell-derived viruses. J. Virol. 2003;77:12022–12032. doi: 10.1128/JVI.77.22.12022-12032.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Lozach P.-Y., Kühbacher A., Meier R., Mancini R., Bitto D., Bouloy M., Helenius A. DC-SIGN as a receptor for phleboviruses. Cell Host Microbe. 2011;10:75–88. doi: 10.1016/j.chom.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 216.Guo Y., Feinberg H., Conroy E., Mitchell D.A., Alvarez R., Blixt O., Taylor M.E., Weis W.I., Drickamer K. Structural basis for distinct ligand-binding and targeting properties of the receptors DC-SIGN and DC-SIGNR. Nat. Struct. Mol. Biol. 2004;11:591–598. doi: 10.1038/nsmb784. [DOI] [PubMed] [Google Scholar]
- 217.Feinberg H., Mitchell D.A., Drickamer K., Weis W.I. Structural basis for selective recognition of oligosaccharides by DC-SIGN and DC-SIGNR. Science. 2001;294:2163–2166. doi: 10.1126/science.1066371. [DOI] [PubMed] [Google Scholar]
- 218.Londrigan S.L., Turville S.G., Tate M.D., Deng Y.-M., Brooks A.G., Reading P.C. N-Linked glycosylation facilitates sialic acid-independent attachment and entry of Influenza A viruses into cells expressing DC-SIGN or L-SIGN. J. Virol. 2011;85:2990–3000. doi: 10.1128/JVI.01705-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Cormier E.G., Durso R.J., Tsamis F., Boussemart L., Manix C., Olson W.C., Gardner J.P., Dragic T. L-SIGN (CD209L) and DC-SIGN (CD209) mediate transinfection of liver cells by hepatitis C virus. Proc. Natl. Acad. Sci. 2004;101:14067–14072. doi: 10.1073/pnas.0405695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Jeffers S.A., Tusell S.M., Gillim-Ross L., Hemmila E.M., Achenbach J.E., Babcock G.J., Thomas W.D., Thackray L.B., Young M.D., Mason R.J., Ambrosino D.M., Wentworth D.E., Demartini J.C., Holmes K.V., Holmes K.V. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Van Breedam W., Pöhlmann S., Favoreel H.W., de Groot R.J., Nauwynck H.J. Bitter-sweet symphony: glycan–lectin interactions in virus biology. FEMS Microbiol. Rev. 2014;38:598–632. doi: 10.1111/1574-6976.12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Crispin M., Harvey D.J., Bitto D., Halldorsson S., Bonomelli C., Edgeworth M., Scrivens J.H., Huiskonen J.T., Bowden T.A. Uukuniemi Phlebovirus assembly and secretion leave a functional imprint on the virion glycome. J. Virol. 2014;88:10244–10251. doi: 10.1128/JVI.01662-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Hofmann H., Li X., Zhang X., Liu W., Kühl A., Kaup F., Soldan S.S., González-Scarano F., Weber F., He Y., Pöhlmann S. Severe fever with thrombocytopenia virus glycoproteins are targeted by neutralizing antibodies and can use DC-SIGN as a receptor for pH-dependent entry into human and animal cell lines. J. Virol. 2013;87:4384–4394. doi: 10.1128/JVI.02628-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Phoenix I., Nishiyama S., Lokugamage N., Hill T., Huante M., Slack O., Carpio V., Freiberg A., Ikegami T. N-Glycans on the Rift Valley Fever Virus envelope glycoproteins Gn and Gc redundantly support viral infection via DC-SIGN. Viruses. 2016;8:149. doi: 10.3390/v8050149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Lozach P.-Y., Kühbacher A., Meier R., Mancini R., Bitto D., Bouloy M., Helenius A. DC-SIGN as a receptor for phleboviruses. Cell Host Microbe. 2011;10:75–88. doi: 10.1016/j.chom.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 226.Halldorsson S., Li S., Li M., Harlos K., Bowden T.A., Huiskonen J.T. Shielding and activation of a viral membrane fusion protein. Nat. Commun. 2018;9:349. doi: 10.1038/s41467-017-02789-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Léger P., Tetard M., Youness B., Cordes N., Rouxel R.N., Flamand M., Lozach P.-Y. Differential use of the C-type lectins L-SIGN and DC-SIGN for Phlebovirus endocytosis. Traffic. 2016;17:639–656. doi: 10.1111/tra.12393. [DOI] [PubMed] [Google Scholar]
- 228.Halldorsson S., Behrens A.-J., Harlos K., Huiskonen J.T., Elliott R.M., Crispin M., Brennan B., Bowden T.A. Structure of a phleboviral envelope glycoprotein reveals a consolidated model of membrane fusion. Proc. Natl. Acad. Sci. U. S. A. 2016;113:7154–7159. doi: 10.1073/pnas.1603827113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Miller J.L., DeWet B.J.M., Martinez-Pomares L., Radcliffe C.M., Dwek R.A., Rudd P.M., Gordon S. The mannose receptor mediates Dengue virus infection of macrophages. PLoS Pathog. 2008;4:e17. doi: 10.1371/journal.ppat.0040017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Pokidysheva E., Zhang Y., Battisti A.J., Bator-Kelly C.M., Chipman P.R., Xiao C., Gregorio G.G., Hendrickson W.A., Kuhn R.J., Rossmann M.G. Cryo-EM reconstruction of Dengue virus in complex with the carbohydrate recognition domain of DC-SIGN. Cell. 2006;124:485–493. doi: 10.1016/j.cell.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 231.Navarro-Sanchez E., Altmeyer R., Amara A., Schwartz O., Fieschi F., Virelizier J.-L., Arenzana-Seisdedos F., Desprès P. Dendritic-cell-specific ICAM3-grabbing non-integrin is essential for the productive infection of human dendritic cells by mosquito-cell-derived dengue viruses. EMBO Rep. 2003;4:723–728. doi: 10.1038/sj.embor.embor866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Modis Y., Ogata S., Clements D., Harrison S.C. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427:313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 233.Lozach P.-Y., Burleigh L., Staropoli I., Navarro-Sanchez E., Harriague J., Virelizier J.-L., Rey F.A., Desprès P., Arenzana-Seisdedos F., Amara A. Dendritic cell-specific intercellular adhesion molecule 3-grabbing non-integrin (DC-SIGN)-mediated enhancement of dengue virus infection is independent of DC-SIGN internalization signals. J. Biol. Chem. 2005;280:23698–23708. doi: 10.1074/jbc.M504337200. [DOI] [PubMed] [Google Scholar]
- 234.Cheng G., Cox J., Wang P., Krishnan M.N., Dai J., Qian F., Anderson J.F., Fikrig E. A C-Type lectin collaborates with a CD45 phosphatase homolog to facilitate West Nile virus infection of mosquitoes. Cell. 2010;142:714–725. doi: 10.1016/j.cell.2010.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Liu Y., Zhang F., Liu J., Xiao X., Zhang S., Qin C., Xiang Y., Wang P., Cheng G. Transmission-blocking antibodies against mosquito C-type lectins for Dengue prevention. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1003931. e1003931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Baize S., Kaplon J., Faure C., Pannetier D., Georges-Courbot M.-C., Deubel V. Lassa virus infection of human dendritic cells and macrophages is productive but fails to activate cells. J. Immunol. 2004;172:2861–2869. doi: 10.4049/JIMMUNOL.172.5.2861. [DOI] [PubMed] [Google Scholar]
- 237.Geijtenbeek T.B., Kwon D.S., Torensma R., van Vliet S.J., van Duijnhoven G.C., Middel J., Cornelissen I.L.M.H., Nottet H.S.L., KewalRamani V.N., Littman D.R., Figdor C.G., van Kooyk Y. DC-SIGN, a dendritic cell–specific HIV-1-binding protein that enhances trans-infection of T cells. Cell. 2000;100:587–597. doi: 10.1016/S0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 238.Yang Z.-Y., Huang Y., Ganesh L., Leung K., Kong W.-P., Schwartz O., Subbarao K., Nabel G.J. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J. Virol. 2004;78:5642–5650. doi: 10.1128/JVI.78.11.5642-5650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Engering A., Geijtenbeek T.B.H., van Vliet S.J., Wijers M., van Liempt E., Demaurex N., Lanzavecchia A., Fransen J., Figdor C.G., Piguet V., van Kooyk Y. The dendritic cell-specific adhesion receptor DC-SIGN internalizes antigen for presentation to T cells. J. Immunol. 2002;168:2118–2126. doi: 10.4049/jimmunol.168.5.2118. [DOI] [PubMed] [Google Scholar]
- 240.de Witte L., Nabatov A., Pion M., Fluitsma D., de Jong M.A.W.P., de Gruijl T., Piguet V., van Kooyk Y., Geijtenbeek T.B.H. Langerin is a natural barrier to HIV-1 transmission by Langerhans cells. Nat. Med. 2007;13:367–371. doi: 10.1038/nm1541. [DOI] [PubMed] [Google Scholar]
- 241.Sukegawa S., Miyagi E., Bouamr F., Farkašová H., Strebel K. Mannose receptor 1 restricts HIV particle release from infected macrophages. Cell Rep. 2018;22:786–795. doi: 10.1016/J.CELREP.2017.12.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Xiang Y., Karaveg K., Moremen K.W. Substrate recognition and catalysis by GH47 α-mannosidases involved in Asn-linked glycan maturation in the mammalian secretory pathway. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E7890–E7899. doi: 10.1073/pnas.1611213113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Ng K.K.-S., Kolatkar A.R., Park-Snyder S., Feinberg H., Clark D.A., Drickamer K., Weis W.I. Orientation of bound ligands in mannose-binding proteins. Implications for multivalent ligand recognition. J. Biol. Chem. 2002;277:16088–16095. doi: 10.1074/jbc.M200493200. [DOI] [PubMed] [Google Scholar]
- 244.Lee J.H., de Val N., Lyumkis D., Ward A.B. Model building and refinement of a natively glycosylated HIV-1 Env protein by high-resolution cryoelectron microscopy. Structure. 2015;23:1943–1951. doi: 10.1016/j.str.2015.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Calarese D.A., Scanlan C.N., Zwick M.B., Deechongkit S., Mimura Y., Kunert R., Zhu P., Wormald M.R., Stanfield R.L., Roux K.H., Kelly J.W., Rudd P.M., Dwek R.A., Katinger H., Burton D.R., Wilson I.A. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300:2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- 246.Murin C.D., Julien J.-P., Sok D., Stanfield R.L., Khayat R., Cupo A., Moore J.P., Burton D.R., Wilson I.A., Ward A.B. Structure of 2G12 Fab2 in complex with soluble and fully glycosylated HIV-1 Env by negative-stain single-particle electron microscopy. J. Virol. 2014;88:10177–10188. doi: 10.1128/JVI.01229-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Weis W., Brown J.H., Cusack S., Paulson J.C., Skehel J.J., Wiley D.C. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 248.Wilson I.A., Skehel J.J., Wiley D.C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 249.Chen L., Li F. Structural analysis of the evolutionary origins of influenza virus hemagglutinin and other viral lectins. J. Virol. 2013;87:4118–4120. doi: 10.1128/JVI.03476-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Baum L.G., Paulson J.C. Sialyloligosaccharides of the respiratory epithelium in the selection of human influenza virus receptor specificity. Acta Histochem. Suppl. 1990;40:35–38. [PubMed] [Google Scholar]
- 251.Skehel J.J., Wiley D.C. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 252.Yamada S., Suzuki Y., Suzuki T., Le M.Q., Nidom C.A., Sakai-Tagawa Y., Muramoto Y., Ito M., Kiso M., Horimoto T., Shinya K., Sawada T., Kiso M., Usui T., Murata T., Lin Y., Hay A., Haire L.F., Stevens D.J., Russell R.J., Gamblin S.J., Skehel J.J., Kawaoka Y. Haemagglutinin mutations responsible for the binding of H5N1 influenza A viruses to human-type receptors. Nature. 2006;444:378–382. doi: 10.1038/nature05264. [DOI] [PubMed] [Google Scholar]
- 253.Imai M., Kawaoka Y. The role of receptor binding specificity in interspecies transmission of influenza viruses. Curr. Opin. Virol. 2012;2:160–167. doi: 10.1016/j.coviro.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Nicholls J.M., Chan R.W.Y., Russell R.J., Air G.M., Peiris J.S.M. Evolving complexities of influenza virus and its receptors. Trends Microbiol. 2008;16:149–157. doi: 10.1016/j.tim.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 255.Isa P., Arias C.F., López S. Role of sialic acids in rotavirus infection. Glycoconj. J. 2006;23:27–37. doi: 10.1007/s10719-006-5435-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Heggelund J.E., Varrot A., Imberty A., Krengel U. Histo-blood group antigens as mediators of infections. Curr. Opin. Struct. Biol. 2017;44:190–200. doi: 10.1016/j.sbi.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 257.Gee G.V., Dugan A.S., Tsomaia N., Mierke D.F., Atwood W.J. The role of sialic acid in human polyomavirus infections. Glycoconj. J. 2006;23:19–26. doi: 10.1007/s10719-006-5434-z. [DOI] [PubMed] [Google Scholar]
- 258.Li W., Hulswit R.J.G., Widjaja I., Raj V.S., McBride R., Peng W., Widagdo W., Tortorici M.A., van Dieren B., Lang Y., van Lent J.W.M., Paulson J.C., de Haan C.A.M., de Groot R.J., van Kuppeveld F.J.M., Haagmans B.L., Bosch B.-J. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc. Natl. Acad. Sci. 2017;114:E8508–E8517. doi: 10.1073/pnas.1712592114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.de Groot R.J. Structure, function and evolution of the hemagglutinin-esterase proteins of corona- and toroviruses. Glycoconj. J. 2006;23:59–72. doi: 10.1007/s10719-006-5438-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Olofsson S., Bergström T. Glycoconjugate glycans as viral receptors. Ann. Med. 2005;37:154–172. doi: 10.1080/07853890510007340. [DOI] [PubMed] [Google Scholar]
- 261.Izquierdo-Useros N., Lorizate M., McLaren P.J., Telenti A., Kräusslich H.-G., Martinez-Picado J. HIV-1 capture and transmission by dendritic cells: The role of viral glycolipids and the cellular receptor siglec-1. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004146. e1004146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Yoshida-Moriguchi T., Campbell K.P. Matriglycan: a novel polysaccharide that links dystroglycan to the basement membrane. Glycobiology. 2015;25:702–713. doi: 10.1093/glycob/cwv021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Yoshida-Moriguchi T., Willer T., Anderson M.E., Venzke D., Whyte T., Muntoni F., Lee H., Nelson S.F., Yu L., Campbell K.P. SGK196 is a glycosylation-specific O-mannose kinase required for dystroglycan function. Science. 2013;341:896–899. doi: 10.1126/science.1239951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Jae L.T., Raaben M., Herbert A.S., Kuehne A.I., Wirchnianski A.S., Soh T.K., Stubbs S.H., Janssen H., Damme M., Saftig P., Whelan S.P., Dye J.M., Brummelkamp T.R. Lassa virus entry requires a trigger-induced receptor switch. Science (80-.) 2014;344:1506–1510. doi: 10.1126/science.1252480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Cao W., Henry M.D., Borrow P., Yamada H., Elder J.H., Ravkov E.V., Nichol S.T., Compans R.W., Campbell K.P., Oldstone M.B. Identification of α-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science. 1998;282:2079–2081. doi: 10.1126/science.282.5396.2079. [DOI] [PubMed] [Google Scholar]
- 266.Pasqual G., Rojek J.M., Masin M., Chatton J.-Y., Kunz S. Old World arenaviruses enter the host cell via the multivesicular body and depend on the endosomal sorting complex required for transport. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002232. e1002232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Patel M., Yanagishita M., Roderiquez G., Bou-Habib D.C., Oravecz T., Hascall V.C., Norcross M.A. Cell-surface heparan sulfate proteoglycan mediates HIV-1 infection of T-cell lines. AIDS Res. Hum. Retrovir. 1993;9:167–174. doi: 10.1089/aid.1993.9.167. [DOI] [PubMed] [Google Scholar]
- 268.O'Donnell C.D., Shukla D. The importance of heparan sulfate in herpesvirus infection. Virol. Sin. 2008;23:383–393. doi: 10.1007/s12250-008-2992-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Hilgard P., Stockert R. Heparan sulfate proteoglycans initiate dengue virus infection of hepatocytes. Hepatology. 2000;32:1069–1077. doi: 10.1053/jhep.2000.18713. [DOI] [PubMed] [Google Scholar]
- 270.Shukla D., Spear P.G. Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J. Clin. Invest. 2001;108:503–510. doi: 10.1172/JCI13799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Peerboom N., Block S., Altgärde N., Wahlsten O., Möller S., Schnabelrauch M., Trybala E., Bergström T., Bally M. Binding kinetics and lateral mobility of HSV-1 on end-grafted sulfated glycosaminoglycans. Biophys. J. 2017;113:1223–1234. doi: 10.1016/j.bpj.2017.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Altgärde N., Eriksson C., Peerboom N., Phan-Xuan T., Moeller S., Schnabelrauch M., Svedhem S., Trybala E., Bergström T., Bally M. Mucin-like region of Herpes Simplex Virus type 1 attachment protein glycoprotein C (gC) modulates the virus-glycosaminoglycan interaction. J. Biol. Chem. 2015;290:21473–21485. doi: 10.1074/jbc.M115.637363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Green R.S., Stone E.L., Tenno M., Lehtonen E., Farquhar M.G., Marth J.D. Mammalian N-glycan branching protects against innate immune self-recognition and inflammation in autoimmune disease pathogenesis. Immunity. 2007;27:308–320. doi: 10.1016/j.immuni.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 274.Auriti C., Prencipe G., Moriondo M., Bersani I., Bertaina C., Mondì V., Inglese R. Mannose-binding lectin: biologic characteristics and role in the susceptibility to infections and ischemia-reperfusion related injury in critically ill neonates. J Immunol Res. 2017;2017 doi: 10.1155/2017/7045630. 7045630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Kuhlman M., Joiner K., Ezekowitz R.A. The human mannose-binding protein functions as an opsonin. J. Exp. Med. 1989;169:1733–1745. doi: 10.1084/jem.169.5.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Miller A., Phillips A., Gor J., Wallis R., Perkins S.J. Near-planar solution structures of mannose-binding lectin oligomers provide insight on activation of lectin pathway of complement. J. Biol. Chem. 2012;287:3930–3945. doi: 10.1074/jbc.M111.320341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Matsushita M. The lectin pathway of the complement system. Microbiol. Immunol. 1996;40:887–893. doi: 10.1111/j.1348-0421.1996.tb01156.x. [DOI] [PubMed] [Google Scholar]
- 278.Wallis R. Interactions between mannose-binding lectin and MASPs during complement activation by the lectin pathway. Immunobiology. 2007;212:289–299. doi: 10.1016/j.imbio.2006.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Chen C.-B., Wallis R. Two mechanisms for mannose-binding protein modulation of the activity of its associated serine proteases. J. Biol. Chem. 2004;279:26058–26065. doi: 10.1074/jbc.M401318200. [DOI] [PubMed] [Google Scholar]
- 280.Matsushita M., Fujita T. Ficolins and the lectin complement pathway. Immunol. Rev. 2001;180:78–85. doi: 10.1034/j.1600-065X.2001.1800107.x. [DOI] [PubMed] [Google Scholar]
- 281.Fuchs A., Lin T.-Y., Beasley D.W., Stover C.M., Schwaeble W.J., Pierson T.C., Diamond M.S. Direct complement restriction of Flavivirus infection requires glycan recognition by mannose-binding lectin. Cell Host Microbe. 2010;8:186–195. doi: 10.1016/J.CHOM.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Spear G.T., Zariffard M.R., Xin J., Saifuddin M. Inhibition of DC-SIGN-mediated trans infection of T cells by mannose-binding lectin. Immunology. 2003;110:80–85. doi: 10.1046/j.1365-2567.2003.01707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Ji X., Olinger G.G., Aris S., Chen Y., Gewurz H., Spear G.T. Mannose-binding lectin binds to Ebola and Marburg envelope glycoproteins, resulting in blocking of virus interaction with DC-SIGN and complement-mediated virus neutralization. J. Gen. Virol. 2005;86:2535–2542. doi: 10.1099/vir.0.81199-0. [DOI] [PubMed] [Google Scholar]
- 284.Anders E.M., Hartley C.A., Reading P.C., Ezekowitz R.A.B. Complement-dependent neutralization of influenza virus by a serum mannose-binding lectin. J. Gen. Virol. 1994;75:615–622. doi: 10.1099/0022-1317-75-3-615. [DOI] [PubMed] [Google Scholar]
- 285.Zhou Y., Lu K., Pfefferle S., Bertram S., Glowacka I., Drosten C., Pohlmann S., Simmons G. A single asparagine-linked glycosylation site of the severe acute respiratory syndrome Coronavirus spike glycoprotein facilitates inhibition by mannose-binding lectin through multiple mechanisms. J. Virol. 2010;84:8753–8764. doi: 10.1128/JVI.00554-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Pastva A.M., Wright J.R., Williams K.L. Immunomodulatory roles of surfactant proteins A and D: implications in lung disease. Proc. Am. Thorac. Soc. 2007;4:252–257. doi: 10.1513/pats.200701-018AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Hartshorn K.L., White M.R., Shepherd V., Reid K., Jensenius J.C., Crouch E.C. Mechanisms of anti-influenza activity of surfactant proteins A and D: comparison with serum collectins. Am. J. Phys. 1997;273:L1156–L1166. doi: 10.1152/ajplung.1997.273.6.L1156. [DOI] [PubMed] [Google Scholar]
- 288.Crouch E., Hartshorn K., Horlacher T., McDonald B., Smith K., Cafarella T., Seaton B., Seeberger P.H., Head J. Recognition of mannosylated ligands and Influenza A virus by human surfactant protein D: contributions of an extended site and residue 343. Biochemistry. 2009;48:3335–3345. doi: 10.1021/bi8022703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Hillaire M.L.B., van Eijk M., van Trierum S.E., van Riel D., Saelens X., Romijn R.A., Hemrika W., Fouchier R.A.M., Kuiken T., Osterhaus A.D.M.E., Haagsman H.P., Rimmelzwaan G.F. Assessment of the antiviral properties of recombinant porcine SP-D against various Influenza A viruses in vitro. PLoS One. 2011;6 doi: 10.1371/journal.pone.0025005. e25005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Benne C.A., Benaissa-Trouw B., Van Strijp J.A.G., Kraaijeveld C.A., Van Iwaarden J.F.F. Surfactant protein A, but not surfactant protein D, is an opsonin for influenza A virus phagocytosis by rat alveolar macrophages. Eur. J. Immunol. 1997;27:886–890. doi: 10.1002/eji.1830270413. [DOI] [PubMed] [Google Scholar]
- 291.Vigerust D.J., Ulett K.B., Boyd K.L., Madsen J., Hawgood S., McCullers J.A. N-linked glycosylation attenuates H3N2 influenza viruses. J. Virol. 2007;81:8593–8600. doi: 10.1128/JVI.00769-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Funk C.J., Wang J., Ito Y., Travanty E.A., Voelker D.R., Holmes K.V., Mason R.J. Infection of human alveolar macrophages by human coronavirus strain 229E. J. Gen. Virol. 2012;93:494–503. doi: 10.1099/vir.0.038414-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.LeVine A.M., Elliott J., Whitsett J.A., Srikiatkhachorn A., Crouch E., DeSilva N., Korfhagen T. Surfactant protein-D enhances phagocytosis and pulmonary clearance of respiratory syncytial virus. Am. J. Respir. Cell Mol. Biol. 2004;31:193–199. doi: 10.1165/rcmb.2003-0107OC. [DOI] [PubMed] [Google Scholar]
- 294.Levroney E.L., Aguilar H.C., Fulcher J.A., Kohatsu L., Pace K.E., Pang M., Gurney K.B., Baum L.G., Lee B. Novel innate immune functions for galectin-1: galectin-1 inhibits cell fusion by Nipah virus envelope glycoproteins and augments dendritic cell secretion of proinflammatory cytokines. J. Immunol. 2005;175:413–420. doi: 10.4049/jimmunol.175.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Garner O.B., Aguilar H.C., Fulcher J.A., Levroney E.L., Harrison R., Wright L., Robinson L.R., Aspericueta V., Panico M., Haslam S.M., Morris H.R., Dell A., Lee B., Baum L.G. Endothelial galectin-1 binds to specific glycans on Nipah virus fusion protein and inhibits maturation, mobility, and function to block syncytia formation. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000993. e1000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Garner O.B., Yun T., Pernet O., Aguilar H.C., Park A., Bowden T.A., Freiberg A.N., Lee B., Baum L.G. Timing of galectin-1 exposure differentially modulates Nipah virus entry and syncytium formation in endothelial cells. J. Virol. 2015;89:2520–2529. doi: 10.1128/JVI.02435-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Yang M.-L., Chen Y.-H., Wang S.-W., Huang Y.-J., Leu C.-H., Yeh N.-C., Chu C.-Y., Lin C.-C., Shieh G.-S., Chen Y.-L., Wang J.-R., Wang C.-H., Wu C.-L., Shiau A.-L. Galectin-1 binds to influenza virus and ameliorates influenza virus pathogenesis. J. Virol. 2011;85:10010–10020. doi: 10.1128/JVI.00301-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Ouellet M., Mercier S., Pelletier I., Bounou S., Roy J., Hirabayashi J., Sato S., Tremblay M.J. Galectin-1 acts as a soluble host factor that promotes HIV-1 infectivity through stabilization of virus attachment to host cells. J. Immunol. 2005;174:4120–4126. doi: 10.4049/jimmunol.174.7.4120. [DOI] [PubMed] [Google Scholar]
- 299.St-Pierre C., Manya H., Ouellet M., Clark G.F., Endo T., Tremblay M.J., Sato S. Host-soluble galectin-1 promotes HIV-1 replication through a direct interaction with glycans of viral gp120 and host CD4. J. Virol. 2011;85:11742–11751. doi: 10.1128/JVI.05351-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Blattner C., Lee J.H., Sliepen K., Derking R., Falkowska E., de la Peña A.T., Cupo A., Julien J.-P., van Gils M., Lee P.S., Peng W., Paulson J.C., Poignard P., Burton D.R., Moore J.P., Sanders R.W., Wilson I.A., Ward A.B. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity. 2014;40:669–680. doi: 10.1016/j.immuni.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Falkowska E., Le K.M., Ramos A., Doores K.J., Lee J.H., Blattner C., Ramirez A., Derking R., van Gils M.J., Liang C.-H., Mcbride R., von Bredow B., Shivatare S.S., Wu C.-Y., Chan-Hui P.-Y., Liu Y., Feizi T., Zwick M.B., Koff W.C., Seaman M.S., Swiderek K., Moore J.P., Evans D., Paulson J.C., Wong C.-H., Ward A.B., Wilson I.A., Sanders R.W., Poignard P., Burton D.R. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity. 2014;40:657–668. doi: 10.1016/j.immuni.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Huang J., Kang B.H., Pancera M., Lee J.H., Tong T., Feng Y., Imamichi H., Georgiev I.S., Chuang G.-Y., Druz A., Doria-Rose N.A., Laub L., Sliepen K., van Gils M.J., de la Peña A.T., Derking R., Klasse P.-J., Migueles S.A., Bailer R.T., Alam M., Pugach P., Haynes B.F., Wyatt R.T., Sanders R.W., Binley J.M., Ward A.B., Mascola J.R., Kwong P.D., Connors M. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41–gp120 interface. Nature. 2014;515:138–142. doi: 10.1038/nature13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Scanlan C.N., Pantophlet R., Wormald M.R., Saphire E.O., Stanfield R., Wilson I.A., Katinger H., Dwek R.A., Rudd P.M., Burton D.R. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1→2 mannose residues on the outer face of gp120. J. Virol. 2002;76:7306–7321. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Pancera M., Shahzad-ul-Hussan S., Doria-Rose N.A., McLellan J.S., Bailer R.T., Dai K., Loesgen S., Louder M.K., Staupe R.P., Yang Y., Zhang B., Parks R., Eudailey J., Lloyd K.E., Blinn J., Alam S.M., Haynes B.F., Amin M.N., Wang L.-X., Burton D.R., Koff W.C., Nabel G.J., Mascola J.R., Bewley C.A., Kwong P.D. Structural basis for diverse N-glycan recognition by HIV-1–neutralizing V1–V2–directed antibody PG16. Nat. Struct. Mol. Biol. 2013;20:804–813. doi: 10.1038/nsmb.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Watanabe Y., Vasiljevic S., Allen J.D., Seabright G.E., Duyvesteyn H.M.E., Doores K.J., Crispin M., Struwe W.B. Signature of antibody domain exchange by native mass spectrometry and collision-induced unfolding. Anal. Chem. 2018;90:7325–7331. doi: 10.1021/acs.analchem.8b00573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Rouvinski A., Guardado-Calvo P., Barba-Spaeth G., Duquerroy S., Vaney M.-C., Kikuti C.M., Navarro Sanchez M.E., Dejnirattisai W., Wongwiwat W., Haouz A., Girard-Blanc C., Petres S., Shepard W.E., Desprès P., Arenzana-Seisdedos F., Dussart P., Mongkolsapaya J., Screaton G.R., Rey F.A. Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature. 2015;520:109–113. doi: 10.1038/nature14130. [DOI] [PubMed] [Google Scholar]
- 307.Dejnirattisai W., Wongwiwat W., Supasa S., Zhang X., Dai X., Rouvinski A., Jumnainsong A., Edwards C., Quyen N.T.H., Duangchinda T., Grimes J.M., Tsai W.-Y., Lai C.-Y., Wang W.-K., Malasit P., Farrar J., Simmons C.P., Zhou Z.H., Rey F.A., Mongkolsapaya J., Screaton G.R. A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat. Immunol. 2015;16:170–177. doi: 10.1038/ni.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Rouvinski A., Dejnirattisai W., Guardado-Calvo P., Vaney M.-C., Sharma A., Duquerroy S., Supasa P., Wongwiwat W., Haouz A., Barba-Spaeth G., Mongkolsapaya J., Rey F.A., Screaton G.R. Covalently linked dengue virus envelope glycoprotein dimers reduce exposure of the immunodominant fusion loop epitope. Nat. Commun. 2017;8:15411. doi: 10.1038/ncomms15411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Dejnirattisai W., Supasa P., Wongwiwat W., Rouvinski A., Barba-Spaeth G., Duangchinda T., Sakuntabhai A., Cao-Lormeau V.-M., Malasit P., Rey F.A., Mongkolsapaya J., Screaton G.R. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nat. Immunol. 2016;17:1102–1108. doi: 10.1038/ni.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Tokatlian T., Read B.J., Jones C.A., Kulp D.W., Menis S., Chang J.Y.H., Steichen J.M., Kumari S., Allen J.D., Dane E.L., Liguori A., Sangesland M., Lingwood D., Crispin M., Schief W.R., Irvine D.J. Innate immune recognition of glycans targets HIV nanoparticle immunogens to germinal centers. Science. 2019;363:649–654. doi: 10.1126/SCIENCE.AAT9120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Hudak J.E., Bertozzi C.R. Glycotherapy: new advances inspire a reemergence of glycans in medicine. Chem. Biol. 2014;21:16–37. doi: 10.1016/j.chembiol.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Imberty A., Chabre Y.M., Roy R. Glycomimetics and glycodendrimers as high affinity microbial anti-adhesins. Chem. Eur. J. 2008;14:7490–7499. doi: 10.1002/chem.200800700. [DOI] [PubMed] [Google Scholar]
- 313.von Itzstein M., Wu W.-Y., Kok G.B., Pegg M.S., Dyason J.C., Jin B., Van Phan T., Smythe M.L., White H.F., Oliver S.W., Colman P.M., Varghese J.N., Ryan D.M., Woods J.M., Bethell R.C., Hotham V.J., Cameron J.M., Penn C.R. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature. 1993;363:418–423. doi: 10.1038/363418a0. [DOI] [PubMed] [Google Scholar]
- 314.Asano N. Glycosidase inhibitors: update and perspectives on practical use. Glycobiology. 2003;13:93R–104. doi: 10.1093/glycob/cwg090. [DOI] [PubMed] [Google Scholar]
- 315.Copeland R., Balasubramaniam A., Tiwari V., Zhang F., Bridges A., Linhardt R.J., Shukla D., Liu J. Using a 3-O-sulfated heparin octasaccharide to inhibit the entry of herpes simplex virus type 1. Biochemistry. 2008;47:5774–5783. doi: 10.1021/bi800205t. [DOI] [PMC free article] [PubMed] [Google Scholar]