Summary
Cold temperature during the reproductive stage often causes great yield loss of grain crops in subtropical and temperate regions. Previously we showed that the rice transcription factor bZIP73Jap plays an important role in cold adaptation at the seedling stage. Here we further demonstrate that bZIP73Jap also confers cold stress tolerance at the reproductive stage. bZIP73 Jap was up‐regulated under cold treatment and predominately expressed in panicles at the early binucleate and flowering stages. bZIP73Jap forms heterodimers with bZIP71, and co‐expression of bZIP73 Jap and bZIP71 transgenic lines significantly increased seed‐setting rate and grain yield under natural cold stress conditions. bZIP73Jap:bZIP71 not only repressed ABA level in anthers, but also enhanced soluble sugar transport from anthers to pollens and improved pollen grain fertility, seed‐setting rate, and grain yield. Interestingly, bZIP73Jap:bZIP71 also regulated the expression of qLTG3‐1 Nip, and qLTG3‐1 Nip overexpression lines greatly improved rice tolerance to cold stress during the reproductive stage. Therefore, our work establishes a framework for rice cold stress tolerance through the bZIP71‐bZIP73Jap‐qLTG3‐1Nip‐sugar transport pathway. Together with our previous work, our results provide a powerful tool for improving rice cold stress tolerance at both the seedling and the reproductive stages.
Keywords: cold tolerance, bZIP73Jap, ABA level, sugar transport, reproductive stage
Introduction
As rice originated from tropical and subtropical regions, it is vulnerable to cold stress at all growth stages, which is especially critical during the reproductive stage (booting and flowering stage), because it adversely affects grain yield and quality (Espe et al., 2017; Pan et al., 2015; Zhang et al., 2014, 2017). During different booting stages, including the transition of the tetrad to early uni‐nucleate stage (also called young microspore or YM stage), and the early binucleate (EB) stage, cold stress may cause sterile pollen grains or reduced number of mature pollen grains, especially at the YM stage, thus leading to spikelet sterility (Oliver et al., 2005; Satake, 1976; Shimono et al., 2016; Suh et al., 2010). During the flowering stage, cold stress may affect pollen germination, pollen tube elongation, fertilization, also leading to spikelet sterility (Shinada et al., 2013, 2014). Rice is widely planted in China, from Hainan island (18°90′N) to the Mohe River (53°27′N) in Heilongjiang, and from the eastern coastal areas to the Yunnan‐Guizhou Plateau and Xinjiang in Western China, and almost all rice production areas are highly vulnerable to cold injury, leading to an estimated annual loss of about 3–5 million tons in China (Li et al., 2018; Liu et al., 2018b; Zhang et al., 2017; Zhu et al., 2015). Therefore, breeding cold‐tolerant varieties is an effective method to maintain high and stable yields in those rice cultivation regions.
Cold tolerance is a quantitative trait that is controlled by multiple loci and also affected by the environment (Li et al., 2018; Sun et al., 2018; Zhang et al., 2017). Over the past decades, only three QTL, CTB4a, Ctb1 and qPSR10, have been cloned and functionally characterized (Saito et al., 2010; Xiao et al., 2018; Zhang et al., 2017). CTB4a encodes a conserved leucine‐rich repeat receptor‐like kinase that interacts with AtpB, thus increasing ATP synthase activity and ATP content and enhancing seed‐setting rate and improved yield under cold stress conditions (Zhang et al., 2017). Ctb1 encodes an F‐box protein, suggesting that a ubiquitin‐proteasome pathway is involved in chilling tolerance at this stage (Saito et al., 2010). qPSR10 was identified by GWAS using a 1033‐accession diversity panel and confers cold tolerance both at the seedling and reproductive stages (Xiao et al., 2018). However, little is known about the molecular mechanisms of rice cold tolerance at the reproductive stage.
Morphological and histological investigations indicated that cold results in reduced number of mature pollen grains, thus increasing the rate of male sterility (Nishiyama, 1976, 1982; Sakata et al., 2014). The proposed mechanism for cold‐induced pollen sterility (CIPS) is that cold leads to an absence of starch accumulation in mature pollen grains and concurrent accumulation of sugar in anthers due to repression of anther cell wall invertase and monosaccharide transporter genes, thus resulting in reduced pollen sink strength (Ji et al., 2011; Koonjul et al., 2005; Oliver et al., 2005). In agreement with this, expression of cell wall invertase and monosaccharide transporter genes is negatively affected under cold stress conditions (Ji et al., 2011; Oliver et al., 2007; Sakata et al., 2014).
Beijing is located in Northern China (39.9°N) where low temperature stress during the rice growing season often occurs. Previously, we reported that bZIP73 Jap, the japonica version of bZIP73, a bZIP transcription factor‐coding gene with only one functional polymorphism (+511 G>A) between the two subspecies japonica and indica, significantly improved rice tolerance to cold stress at the seedling stage when co‐expressed with bZIP71 (Liu et al., 2018a). Here, we further demonstrate that bZIP71 and bZIP73Jap also improve rice cold tolerance at the reproductive stage by mediating ABA and sugar partitioning in flowers, which indicates that these factors can improve cold tolerance throughout the entire growth period of rice.
Results
Expression patterns of bZIP73 Jap and bZIP71
Previously we demonstrated that the bZIP71:bZIP73Jap heterodimer plays an important role in rice cold adaptation at the seedling stage (Liu et al., 2018a). To further elucidate bZIP71:bZIP73Jap functions, the expression of bZIP71 and bZIP73 Jap in different tissues was examined. bZIP73 Jap was predominately expressed in panicles at the early binucleate (EB) and flowering stages (Figure S1), while bZIP71 was constitutively expressed in almost all tissues and organs examined and its expression was higher in seedlings (shoots and roots) than in other tissues. Further examination of its expression profile under cold stress at the reproductive stage revealed that bZIP73 Jap was strongly up‐regulated both in panicles and flag leaves following 3 h of cold treatment (Figure 1) and reached the highest expression at 1 h (EB stage) or 2 h (YM stage) after cold treatment (Figure 1). At the flowering stage, bZIP73 Jap was up‐regulated in panicles during a 24 h time course and reached the highest expression at 12 h after cold treatment. However, bZIP71 was unchanged after cold treatment at the YM, EB, and flowering stages (Figure S2). These results indicated that bZIP73 Jap may also participate in cold stress tolerance during the reproductive stage of rice development.
Figure 1.
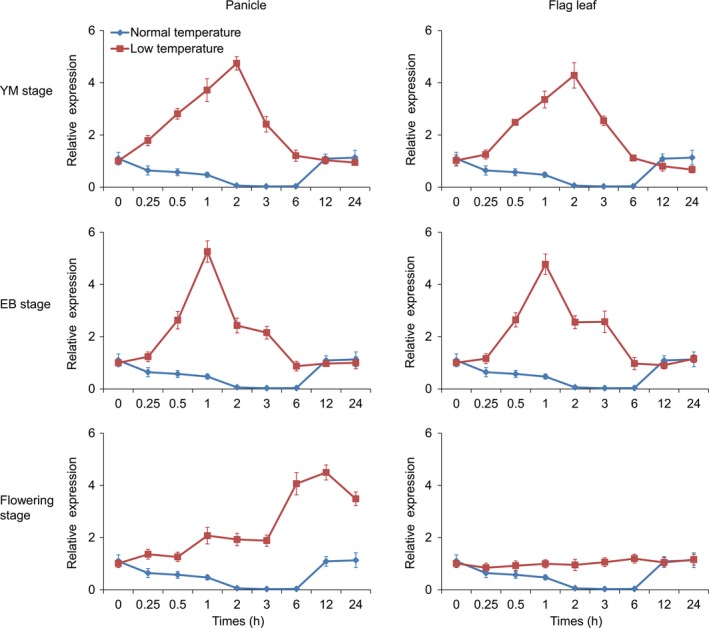
bZIP73 Jap expression patterns in flag leaves and panicles of wild‐type ZH11 under cold treatment during the reproductive stage. YM, young microspore stage. EB, early binucleate stage. Plants grown under normal warm temperature conditions were used as untreated control. Error bar, standard deviation from three independent experiments.
bZIP73 Jap and bZIP71 co‐expression lines significantly improve cold tolerance during the reproduction stage
To examine low temperature stress during the reproductive stage under natural growth conditions in Beijing (116.4°E/39.9°N), we adopted a batch sowing approach with different T3 homozygous transgenic lines, including bZIP73 Jap/Ind overexpression plants (73JapOE, 73IndOE), bZIP71 overexpression plants (71OE), bZIP73 Jap RNAi plants (73JapRi), bZIP71 RNAi plants (71Ri), and bZIP73 Jap and bZIP71 co‐expression plants (71‐73JapOE), and wild‐type Zhonghua 11 (ZH11) as a control. The late sowing rice plants started booting in the time ranging from late August to late September, when the typical night terrestrial temperature was below 20 °C (Figure S3), and agronomic traits were investigated and compared between normal and cold conditions in field plots. In 2012, the late planting transgenic lines, which headed on September 13, met the low temperature stress requirement, experiencing a minimum terrestrial temperature of less than 20 °C during the reproductive stage (Figure S3). The 73IndOE, 73JapRi, and 71Ri transgenic lines were more sensitive to cold stress with decreased seed‐setting rates and yield per plant compared to wild‐type ZH11, that is, the seed‐setting rate decreased by 40.4%–52.6%, and the yield per plant decreased by 40.8%–55.9% (Figure 2, Table S1). However, the seed‐setting rate and yield per plant of 73JapOE and 71OE lines were not significantly different from those of wild‐type ZH11 plants under natural cold stress. On the other hand, the seed‐setting rate of 71‐73JapOE lines improved by 12%–20.7%, and the yield per plant improved by 40.8%–63.2% (Table S1). Transgenic plants that were planted much later headed on September 20 and also met the low temperature stress requirement, experiencing even lower minimum terrestrial temperatures during the reproductive stage (Figure S3), and the seed‐setting rate of 71‐73JapOE lines improved by 30.7%–42%, while the yield per plant improved by 37.9%–52.1% compared to wild‐type ZH11 (Figure 2, Table S2). Moreover, we found that seed‐setting rate and yield per plant was positively correlated with co‐overexpression levels of bZIP71 and bZIP73 Jap (Figure 2, Figure S4). However, we did not observe significant differences for all other agronomic traits between transgenic lines and wild‐type controls under normal temperature conditions (Table S3). These results indicated that co‐overexpression of bZIP71 and bZIP73 Jap significantly improves rice tolerance to cold stress during the reproductive period.
Figure 2.
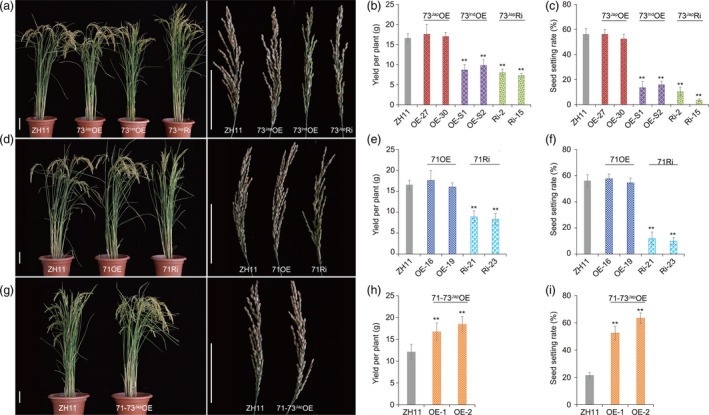
Cold tolerance phenotypes and agronomic traits of bZIP71 and bZIP73 transgenic rice plants during the reproductive stage. (a‐c) Overexpression lines bZIP73 Jap (73Jap OE), bZIP73 Ind (73Ind OE), and RNAi lines bZIP73 Jap (73JapRi) under natural cold stress conditions in Beijing during the autumn of 2012 (heading date: September 13). (b) Yield per plant, (c) Seed‐setting rate. (d–f) bZIP71 overexpression lines (71OE) and bZIP71 RNAi (71Ri) lines under natural cold stress conditions in Beijing during the autumn of 2012 (heading date: September 13), (e) Yield per plant, (f) Seed‐setting rate. (g–i) Co‐overexpression lines of bZIP71‐bZIP73 Jap (71‐73Jap OE) under natural cold stress conditions in Beijing during the autumn of 2012 (heading date: September 20), (h) Yield per plant. (i) Seed‐setting rate. Scale bars = 10 cm. All data were collected using two independent homozygous transgenic lines. Three biological replicates (n = 30 each) were used for each line. Error bar, standard deviation. **P < 0.01, two‐tailed t‐test.
71‐73JapOE lines increase fertility by increasing the amount of mature pollen grains under cold stress
Rice plants experiencing temperatures lower than 20 °C at the YM stage for only 2–4 successive nights will suffer irreversible pollen sterility and a reduction in pollen number, resulting in reduced fertilization (Mamun et al., 2010; Sakata et al., 2014; Sharma and Nayyar, 2016). 73JapRi, 71Ri, and 73IndOE lines had decreased seed‐setting rates, while 71‐73JapOE lines had increased seed‐setting rates compared to wild type ZH11 under cold stress. However, starch staining showed that the rate of pollen sterility in 73JapRi, 71Ri, and 73IndOE lines was not significantly different compared to wild‐type ZH11 pollen under cold stress (Figure 3a), but compared to wild‐type ZH11, the number of mature pollen grains in those lines was significantly reduced by 46.5%–60.4% under cold stress (Figure 3c). In contrast, the number of mature pollen grains in 71‐73JapOE lines was 28.9%–37.1% higher than in wild‐type under cold stress (Figure 3c), and no differences in the rate of pollen sterility and pollen number between 71‐73JapOE lines and wild‐type was observed under normal growth temperature (Figure 3a,b).
Figure 3.
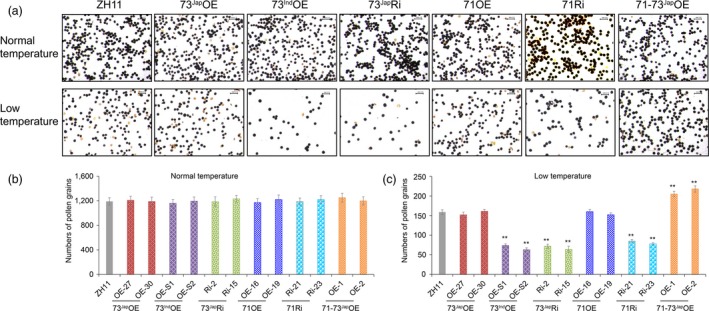
Effects of cold treatment on starch pollen viability and pollen numbers of transgenic lines and ZH11 control plants. (a) KI/I2 staining of pollen grains from panicles of transgenic lines and wild‐type ZH11 control plants grown in Beijing, one day before heading either under normal warm temperature conditions (heading date: August 17) or natural cold stress conditions (heading date: September 13) with average terrestrial temperatures below 20 °C (Figure S3). Scale bars = 0.1 mm. Number of pollen grains in anthers of plants grown at warm temperatures (b) and natural cold stress conditions (c). For each plant, three biological replicates, n = 3 flowers, were tested. Error bar, standard deviation. **P < 0.01, one way anova test, P‐value in Table S8A,B. [Correction added on 16 April 2019, after first online publication. Information regarding Figure 3 was previously incorrect and is updated in this version.]
bZIP71 enhances bZIP73Jap by repressing the expression of ABA synthesis genes and reducing ABA levels in anthers under cold stress
Cold stress leads to abscisic acid (ABA) accumulation in rice anthers (Ji et al., 2011; Oliver et al., 2007). Our previous work showed that bZIP71 enhances bZIP73Jap by repressing the ABA biosynthesis genes OsNCED3 and OsNCED5, leading to reduced ABA levels in rice seedlings under cold stress (Liu et al., 2018a). To test whether bZIP71 and bZIP73 Jap also play a role in modulating ABA levels at the reproductive stage, ABA levels in anthers of transgenic lines grown both under normal and low temperature conditions were measured. ABA levels in anthers of 71Ri, 73JapRi, and 73IndOE lines were 23.1%–34.5% higher than that of wild‐type ZH11 under low temperature conditions (Figure 4a). In contrast, ABA levels in anthers of 71‐73JapOE lines were 22.0%–27.4% lower than in wild‐type ZH11 (Figure 4a). However, ABA levels in anthers of all transgenic lines were not significantly different under normal condition (Figure S5A).
Figure 4.
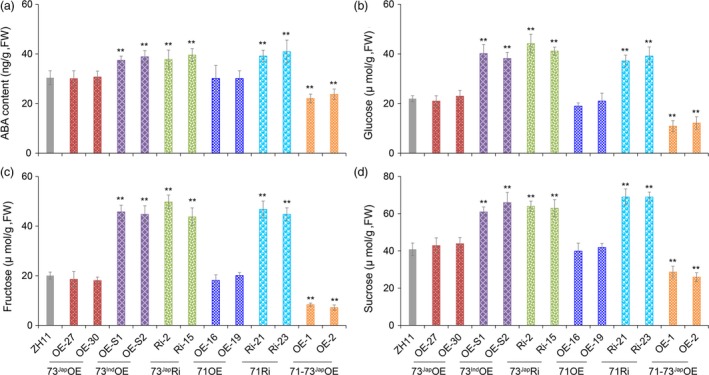
ABA and soluble sugar contents in transgenic lines and wild‐type plants under natural cold stress conditions. Shown are ABA (a), glucose (b), fructose (c), and sucrose (d) contents in transgenic lines and wild‐type ZH11 plants under cold stress conditions. FW, fresh weight. All experiments were conducted using flowers one day before heading. Values are the mean of ten independent biological replicates. Error bars indicate SD. **P < 0.01, one way anova test, P‐value in Table S8C–F.
Chromatin immunoprecipitation (ChIP) assays using flowers of bZIP73Jap::Flag overexpression lines moreover showed that the anti‐Flag antibody specifically precipitated promoter sequences of the ABA biosynthesis genes OsNCED3 and OsNCED5 (Figure 5a), indicating that bZIP73Jap binds to the promoters of these two genes during the reproductive stage. In addition, expression analyses showed that OsNCED3 and OsNCED5 were up‐regulated in anthers of 73JapRi, 71Ri and 73IndOE lines (Figure S6B,D,E), which was consistent with the observed higher ABA levels in anthers of those lines under cold stress conditions (Figure 4a). In contrast, OsNCED3 and OsNCED5 genes were down‐regulated in anthers of 71‐73JapOE lines (Figure S6F), resulting in the observed lower ABA levels in anthers of 71‐73JapOE lines under cold stress conditions (Figure 4a).
Figure 5.
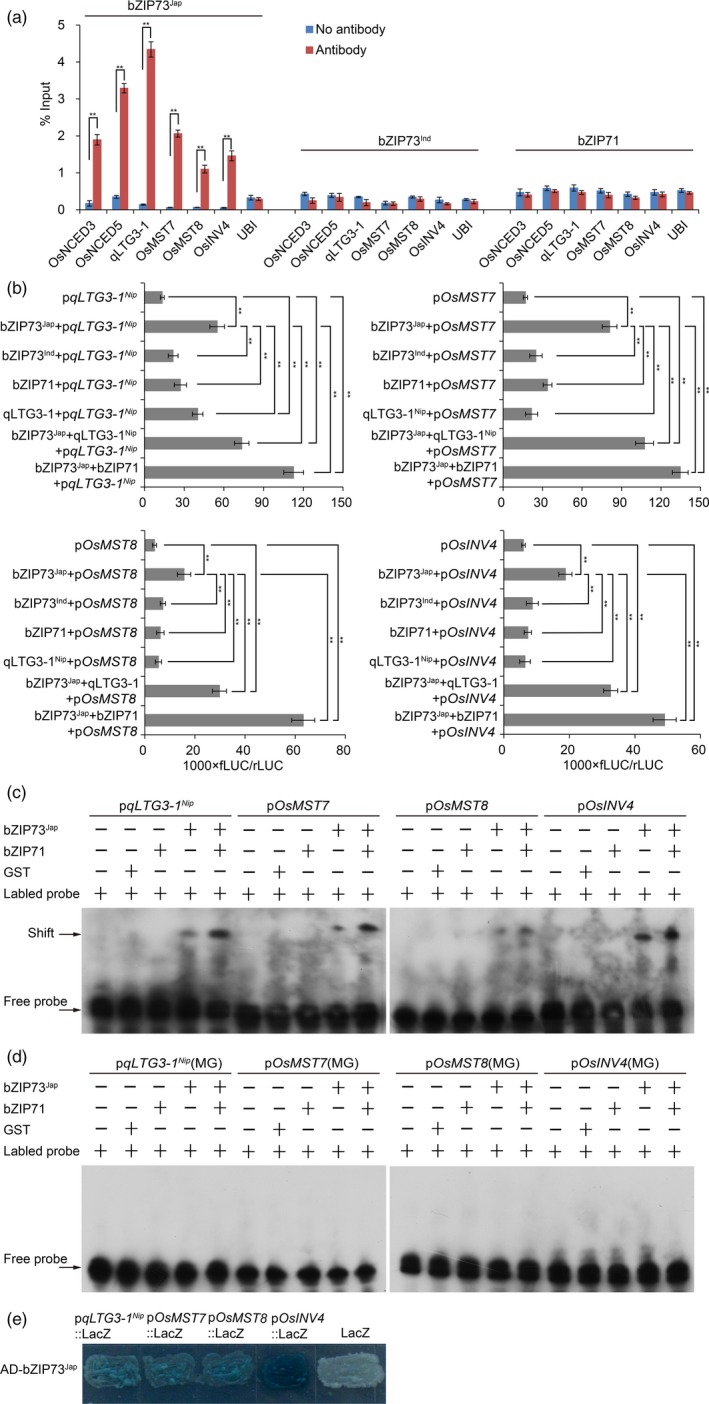
bZIP71 enhances bZIP73Jap trans‐activates the expression of qLTG3‐1 Nip, OsMST7, OsMST8 and OsINV4. (a) ChIP‐qPCR assays indicating that bZIP73Jap directly binds in vivo to promoters of OsNCED3, OsNCED5,qLTG3‐1 Nip, OsINV4, OsMST7, and OsMST8. Flowers of bZIP73Jap::Flag, bZIP73Ind::Flag, and bZIP71::Flag overexpression lines were harvested for ChIP analysis using three biological replicates. The anti‐Flag antibody was used to precipitate DNA sequences interacting with different bZIP fusion proteins. Precipitated DNA was amplified with primers overlapping with G‐box motifs. ChIP, chromatin immunoprecipitation. Error bar, standard deviation. **P < 0.01, two‐tailed t‐test. (b) Transient trans‐activation expression assays in rice protoplasts showing that bZIP71 enhances bZIP73Jap in trans‐activating the promoters of qLTG3‐1 Nip, OsINV4, OsMST7 and OsMST8. Moreover, qLTG3‐1Nip also enhances bZIP73Jap in trans‐activating these downstream genes expression. The relative fLUC/rLUC ratio (see Materials and Methods) shown on the x‐axes represents the relative promoter activity. Constructs used for each transfections into rice protoplasts are shown on the y‐axes. Promoters of qLTG3‐1 Nip, OsINV4, OsMST7, and OsMST8 were fused to LUC as the reporter gene. Values are the mean of five independent replicates and error bars indicate SD. **P < 0.01, one way anova test. (c) In vitro EMSA showing that bZIP71 promotes bZIP73Jap binding to promoters of qLTG3‐1 Nip, OsINV4, OsMST7, and OsMST8, which contains G‐box sequences. EMSA, electrophoretic mobility shift assay. GST was used as a negative control. (d) In vitro EMSA showing that bZIP73Jap could not bind to promoters of qLTG3‐1 Nip, OsINV4, OsMST7, and OsMST8, which contains mutated G‐box (MG) sequences. GST was used as a negative control. (e) Yeast one‐hybrid assay showing that bZIP73Jap activated the expression of pqLTG3‐1 Nip::LacZ, pO sINV4::LacZ, pO sMST7::LacZ, and pO sMST8::LacZ. While, bZIP73Jap could not activate the expression of empty LacZ.
bZIP71 enhances bZIP73Jap activity of reducing sugar accumulation in spikelets under cold stress
ABA plays an important role in integrating sugar signaling, sugar metabolism, and sugar distribution (Sharma and Nayyar, 2016). It has been shown that increased ABA levels regulate expression of the tapetum cell wall‐bound invertase gene OsINV4, which cleaves sucrose in rice anthers and plays a key role in sugar regulation under cold stress conditions (Oliver et al., 2005, 2007). ABA also regulates the monosaccharide transporter genes OsMST7 and OsMST8, which transport glucose and fructose from anther to tapetum and developing pollen grains, and they are critical for pollen development (Mamun et al., 2010; Oliver et al., 2005, 2007; Sharma and Nayyar, 2016). Yeast one‐hybrid (Y1H) assays showed that the GAL4 activation domain (AD), fusion protein AD‐bZIP73Jap binds to the promoters of OsINV4, OsMST7, and OsMST8, and activates the expression of the LacZ reporter gene (Figure 5e). Furthermore, transient expression assays in rice protoplasts showed that bZIP73Jap trans‐activates these promoters and their activities were enhanced by the addition of bZIP71 (Figure 5b). Promoter analyses revealed that the promoters of these genes contain bZIP binding G‐box motifs (Figure S7). To further investigate binding of bZIP73Jap to the promoter regions of these three genes, in vitro electrophoretic mobility shift assays (EMSA) were done with recombinant protein GST‐bZIP73Jap. Shifted bands were clearly detected when probes containing G‐box elements in the promoter regions of these three genes were incubated with the GST‐bZIP73Jap fusion protein (Figure 5c). Furthermore, we found that bZIP71 enhanced such interactions in vitro (Figure 5c). Similarly, ChIP assays showed that bZIP73Jap also specifically precipitated these promoter sequences in flowers of bZIP73Jap::Flag overexpression lines (Figure 5a). Thus, our data demonstrated that bZIP73Jap is an upstream transcriptional regulator of OsINV4, OsMST7 and OsMST8.
Cold stress during pollen development was shown to result in an accumulation of sucrose in spikelets and depletion of starch in mature pollen grains (Oliver et al., 2007). bZIP73Jap can regulate genes encoding a cell wall invertase and monosaccharide transporters, leading to a change of soluble sugar content in anthers under cold stress. Therefore, we examined the soluble sugar content in anthers of transgenic lines and wild‐type ZH11 plants under both normal and natural cold stress conditions, indicating that the abundance of glucose, fructose, and sucrose in 71Ri, 73JapRi and 73IndOE lines was increased dramatically compared to ZH11 under cold stress (Figure 4b–d). In contrast, levels of glucose, fructose, and sucrose in the 71‐73JapOE lines were significantly decreased compared to ZH11 under cold stress (Figure 4b–d), while all transgenic lines and wild‐type ZH11 had no significant differences in levels of those sugars under warm temperature growth conditions (Figure S5B‐D). The expression levels of OsINV4, OsMST7 and OsMST8 in flowers of those transgenic lines were well correlated with changes in soluble sugars, which were down‐regulated in the anthers of 73JapRi, 71Ri and 73IndOE lines (Figure S6B,D,E), but up‐regulated in the anthers of 71‐73JapOE lines (Figure S6F).
bZIP73Jap interacts with the qLTG3‐1Nip protein and activates its downstream genes
Using a yeast two‐hybrid (Y2H) screen, we previously identified 23 bZIP73Jap interacting proteins (Liu et al., 2018a). One of them was qLTG3‐1, which was shown to be involved in rice low‐temperature germinability (Fujino et al., 2008). Interestingly, our analysis of the three different qLTG3‐1 haplotypes: a Nipponbare haplotype (Nip); a Kasalath haplotype (Ka, which is identical to the allele from Italica Livorno); and a Hejiang19 haplotype (Hj, a 71‐bp deletion in the qLTG3‐1 gene, which is identical to the allele from Hayamasari). To examine interactions between bZIP73Jap and different haplotypes of qLTG3‐1, we fused the full‐length CDS of bZIP73 Jap to the GAL4 DNA‐binding domain (BD) as bait, while the full‐length CDSs of the three different qLTG3‐1 alleles (Figures 6a, S8) from Nip, Ka, and Hj were fused to the GAL4 DNA‐activation domain (AD) as prey. In the Y2H assay, only the transformants AD‐qLTG3‐1Nip (aa: 1‐184) + BD‐bZIP73Jap could grow on the quadruple dropout medium SD/Trp‐Leu‐His‐Ade‐ plates, while the transformants AD‐qLTG3‐1Ka (aa: 1‐184) + BD‐bZIP73Jap and AD‐qLTG3‐1Hj (aa: 1‐116) + BD‐bZIP73Jap could not grow on quadruple dropout medium (Figure 6b). These results demonstrated that bZIP73Jap interacts directly with qLTG3‐1Nip, but not qLTG3‐1Ka or qLTG3‐1Hj. The interaction between bZIP73Jap and qLTG3‐1Nip was further confirmed in vivo using bimolecular fluorescence complementation (BiFC) assays and co‐immunoprecipitation (Co‐IP) experiments in Nicotiana benthamiana by agroinfiltration (Figure 6c,d).
Figure 6.
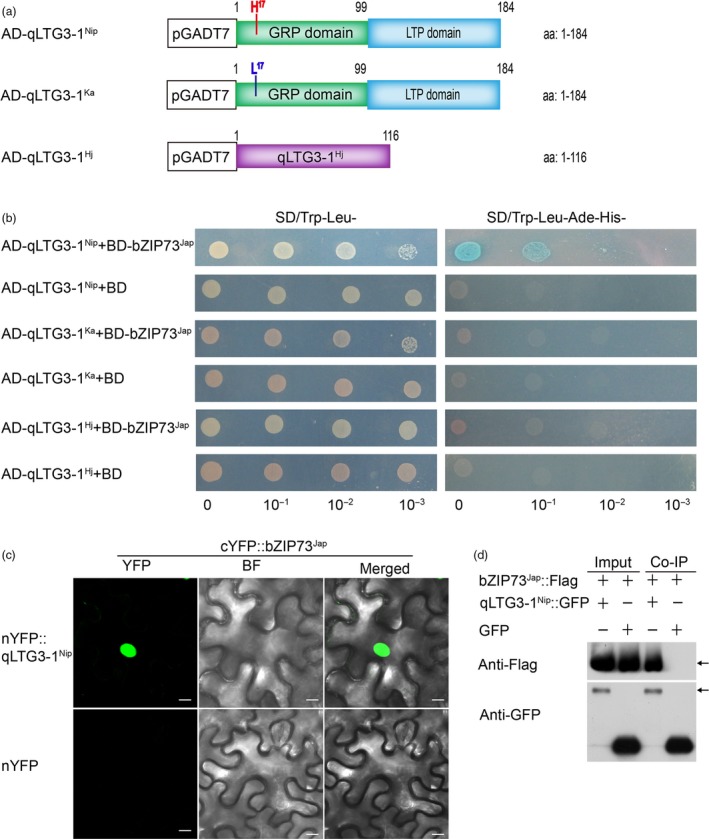
Protein‐protein interactions among bZIP73Jap and qLTG3‐1. (a) Graphic representation of the qLTG3‐1 protein structure. (b) bZIP73Jap interacts with qLTG3‐1Nip/ka/Hj in yeast‐two hybrid (Y2H) assay. bZIP73 Jap was fused to the GAL4 binding domain (BD) as bait; qLTG3‐1 Nip, qLTG3‐1 Ka, or qLTG3‐1 Hj was fused to the GAL4 activation domain (AD) as prey, the empty BD as negative control, prey and bait co‐transformated into yeast Y2HGOLD cells. Shown are growth phenotypes of yeast transformants on selective media of SD/Trp‐Leu‐ (left panel) and SD/Trp‐Leu‐His‐Ade‐ (right panel). Ka: Kasalath; ZH11: Zhonghua11; Hj: Hejiang19. (c) bZIP73Jap interacts with qLTG3‐1Nip in a BiFC assay. bZIP73Jap was fused to the C‐terminal region of vernus fluorescent protein, qLTG3‐1Nip was fused to the N‐terminal region of vernus fluorescent protein. Co‐expression of the cYFP‐bZIP73Jap and nYFP‐qLTG3‐1Nip in N. benthamiana leaves. Fluorescence observed by confocal microscopy. YFP fluorescence (left), bright field (middle) and bright field overlay images (right) are depicted. Scale bars = 10 μm. (d) Detection of bZIP73Jap and qLTG3‐1Nip interaction by co‐immunoprecipitation analysis. GFP‐tagged bZIP73Jap and Flag‐tagged bZIP73Jap were expressed in N. benthamiana leaves for the analysis. Left panel (Input), before precipitation; Right panel (Co‐IP), Anti‐Flag and anti‐GFP antibodies were used to detect Flag and GFP peptides, respectively.
Y1H, EMSA and ChIP assays showed that the bZIP73Jap protein specifically binds to the promoter sequence of qLTG3‐1 Nip (Figure 5a,c–e). Transient expression assays in rice protoplasts also showed that bZIP73Jap trans‐activates the qLTG3‐1 promoter and this activity was enhanced by the addition of bZIP71 (P < 0.01, one way anova test) (Figure 5b). Correspondingly, the expression of qLTG3‐1 Nip was down‐regulated in 73JapRi lines (Figure S6E). Furthermore, bZIP73Jap activates qLTG3‐1 Nip, OsMST7, OsMST8 and OsINV4, and addition of the qLTG3‐1Nip protein enhances this activation in transient expression assays of rice protoplasts (P < 0.01, one way anova test; Figure 5b). Thus, our data demonstrated that bZIP73Jap interacts with qLTG3‐1Nip, and this protein‐protein interaction enhances bZIP73Jap activation of downstream target gene expression.
qLTG3‐1 Nip overexpression transgenic lines improve cold tolerance
It was previously shown that qLTG3‐1 is specifically expressed in panicles and embryos of germinating seeds (Fujino and Matsuda, 2010; Fujino et al., 2008). Our expression analyses revealed that qLTG3‐1 Nip is predominantly expressed in panicles at the EB and flowering stages (Figure S1) and up‐regulated by cold stress (Figure S9). These results suggested that qLTG3‐1 Nip might play an important role in cold stress tolerance during the reproductive stage. To further characterize qLTG3‐1 function, three different alleles of qLTG3‐1 were introduced into the indica‐type cultivar Kasalath via Agrobacterium‐mediated transformation. Analyses of the transgenic plants harboring qLTG3‐1 Nip demonstrated that the seed‐setting rate of qLTG3‐1 Nip overexpression (OE) lines improved by 30.6%–41%, and yield per plant improved by 52.7%–82.5% compared to wild‐type Kasalath under natural cold stress conditions (Figure 7, Table S4). Seed‐setting rate and yield per plant were positively correlated with expression levels of qLTG3‐1 Nip (Figure 7, Figure S10). Moreover, qLTG3‐1 Nip OE lines reduced soluble sugar (sucrose, glucose and fructose) accumulation in flowers (Figure S11), but compared to wild‐type Kasalath, increased pollen fertility under natural cold stress conditions (Figure 8). Similar results were obtained with another batch of field trial under natural cold stress conditions (Table S5). Surprisingly, both qLTG3‐1 Ka OE and qLTG3‐1 Hj OE lines showed no differences compared to Kasalath under natural cold stress conditions (Tables S4 and S5). All agronomic traits of transgenic lines harboring the three different alleles of qLTG3‐1 were not different compared to Kasalath under normal growth conditions (Table S6). These results indicated that qLTG3‐1 Nip OE lines significantly improve rice tolerance to cold stress during the reproductive stage.
Figure 7.
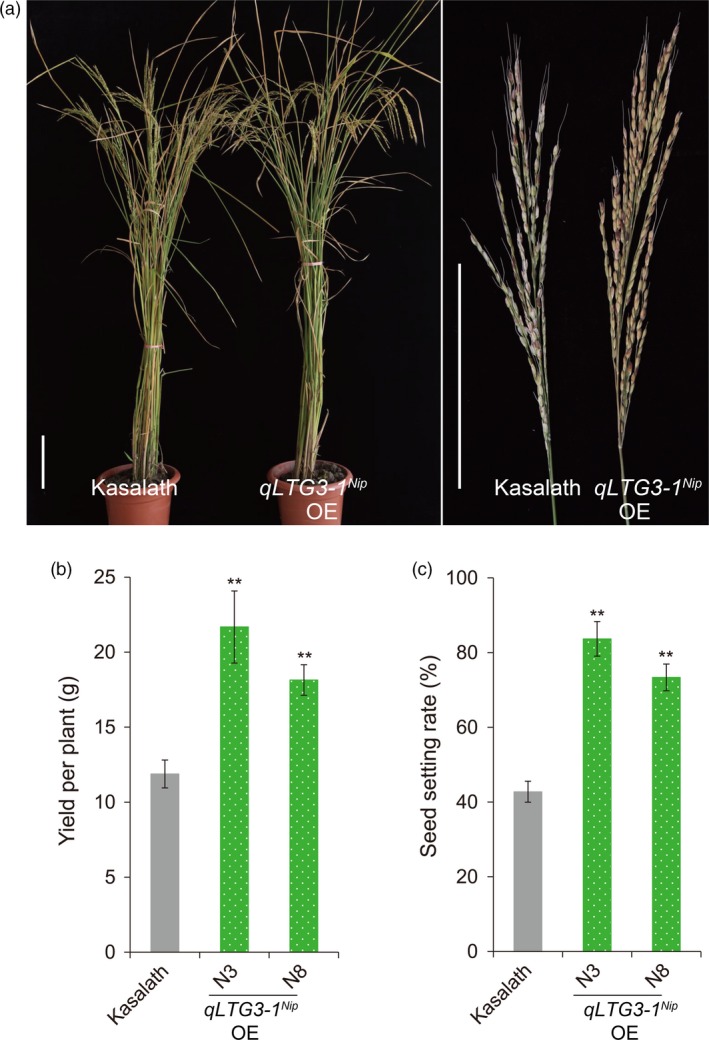
Cold tolerance phenotypes and agronomic traits of qLTG3‐1 Nip overexpression transgenic lines experiencing natural cold stress conditions during the reproductive stage grown in the autumn of 2012 in Beijing (heading date: September 12). (a) Phenotypes of qLTG3‐1 Nip overexpression (qLTG3‐1 Nip OE) lines and Kasalath wild‐type control plants, (b) Yield per plant, (c) Seed‐setting rate. Scale bars = 15 cm. Three biological replicates (n = 30 each) were used per line. Error bar, standard deviation. **P < 0.01, two‐tailed t‐test in comparison to ZH11.
Figure 8.
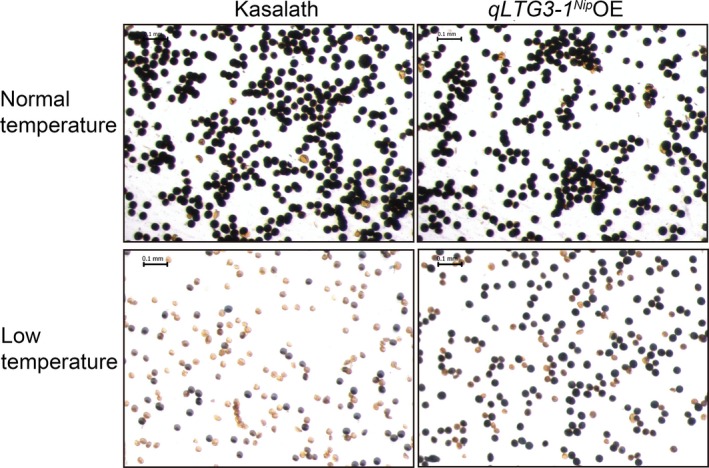
KI/I2 staining phenotype of pollen grains from qLTG3‐1 Nip overexpression lines and Kasalath control plants under natural cold stress conditions as a measure of the effect of cold treatment on pollen viability. Shown are staining phenotypes of pollen grains of plants grown under normal warm temperature conditions in Beijing (heading date: August 13), and natural cold stress conditions with average terrestrial temperatures below 20 °C (Figure S3; heading date: September 12). Scale bars = 0.1 mm. Microscopic observation shows an increased amount of starch (dark blue color) in pollen grains from qLTG3‐1 Nip overexpression lines compared to pollen grains from Kasalath control plants after experiencing natural cold stress conditions.
Discussion
ABA plays a critical role in mediating cold stress responses in plants (Mittler and Blumwald, 2015). Rice mutants deficient in carotenoid biosynthesis genes, the mutants deficient in ABA biosynthetic precursors, and plants overexpressing the ABA‐catabolic gene OsABA8ox1 have reduced ABA levels and are more tolerant to cold stress than wild‐type plants at the seedling stage (Du et al., 2013; Mega et al., 2015). It was also shown that cold‐induced accumulation of ABA was higher in anthers of cold‐sensitive cultivars than cold‐tolerant cultivars (Oliver et al., 2007). Overexpression of the wheat ABA catabolic gene TaABA8′OH1 resulted in reduced ABA levels in anthers and improved cold tolerance of rice (Ji et al., 2011). Our work revealed that 71‐73JapOE transgenic lines had lower ABA levels and were more tolerant to cold stress at the seedling stage (Liu et al., 2018a), and as shown here, also at the reproductive stage, while 71Ri, 73JapRi, 73IndOE lines had higher ABA levels and were more sensitive to cold stress than wild‐type plants, indicating that a reduction of ABA levels improved cold tolerance both at the seedling and reproductive stage in rice.
ABA also plays an important role in regulating sugar signaling and enhances the ability of plant tissues to respond to subsequent sugar signals and repression of OsINV4 and OsMST8 (Rook et al., 2001; Wang et al., 2015). The invertase OsINV4 catalyzes the hydrolysis of sucrose to glucose and fructose, which plays a critical role in sugar partitioning and regulating source‐sink interactions under cold stress conditions (Mamun et al., 2006; Oliver et al., 2005, 2007; Tymowskalalanne and Kreis, 1998). Under cold stress, reduced expression of the monosaccharide transporter gene OsMST8 and the invertase gene OsINV4 in chilling sensitive rice cultivars results in perturbed carbohydrate metabolism and reduced sugar transport to the tapetum and developing pollen grains, thus causing pollen abortion and lower pollen number (Oliver et al., 2005, 2007; Sakata et al., 2014). bZIP73Jap trans‐activates the expression of OsINV4, OsMST7, and OsMST8, and bZIP71 enhances this activity. High expression level of bZIP71 and bZIP73 Jap in 71‐73JapOE lines leads to up‐regulation of OsINV4, OsMST7 and OsMST8. Meanwhile, 71‐73JapOE lines have lower ABA levels in anthers during cold stress than wild‐type controls, which attenuates repression of OsINV4, OsMST8, resulting in enough soluble sugars transported to pollen in 71‐73JapOE lines, which improves pollen grain fertility, seed‐setting rate, and overall yield under cold stress conditions. In contrast, expression levels of bZIP73 Jap and bZIP71 in 71Ri, 73JapRi, and 73IndOE lines are lower than in wild‐type ZH11, leading to down‐regulated expression of OsINV4, OsMST7 and OsMST8. Because cold stress causes ABA accumulation in anthers of those lines, increased ABA levels would feedback inhibit the expression of OsINV4 and OsMST8, leading to reduced soluble sugars transported to the pollen grains. Therefore, sugars accumulate in anthers instead of pollens and pollen grains become sterile, leading to low seed‐setting rate and reduced yield under cold stress conditions.
Both bZIP73 and bZIP71 belong to bZIP transcription factor group S (Liu et al., 2014). In Arabidopsis, Group S members of bZIP transcription factors such as AtbZIP1, AtbZIP2/GBF5, AtbZIP11/ATB2, AtbZIP44, and AtbZIP53 play important roles in sugar signaling (Kang et al., 2010; Ma et al., 2011; Wiese et al., 2004). bZIP71 enhances bZIP73Jap in regulating soluble sugar allocation between source (anthers) and sink (pollen) under cold stress, suggesting that bZIP73 Jap has a conserved function in Group S and plays a role in the cross‐talk of ABA and sugar signaling.
Interestingly, 73IndOE lines are as cold sensitive as 73JapRi lines with a loss‐of‐function phenotype. A possible explanation for this is that the 73IndOE lines were made by overexpressing bZIP73 Ind in a japonica cultivar, Zhonghua 11, which contains an endogenous allele of bZIP73 Jap. Since both bZIP73Jap and bZIP73Ind can form heterodimer with bZIP71 in vitro and in vivo, there is competition between bZIP73Ind and bZIP73Jap for interaction with bZIP71, which reduces the endogenous amount of productive bZIP73Jap‐bZIP71 complexes in transgenic lines and thus might have a dominant‐negative effect (Liu et al., 2018a). This suggests that reduction of endogenous bZIP73Jap‐bZIP71 complexes leads to the down‐regulation of qLTG3‐1 Nip, monosaccharide transporter genes, and cell wall invertase genes (Figure S6), and thus reduced soluble sugars transport from anthers to pollens and reduced pollen grain number (Figure 3a,c), seed‐setting rate, and grain yield in 73IndOE lines (Figure 2a–c). The phenotype of 73IndOE lines at the reproductive stage was similar to that at the seedling stage (Liu et al., 2018a).
The qLTG3‐1 protein has two conserved domains, a glycine‐rich protein (GRP) motif and a lipid transfer protein (LTP) motif (Fujino et al., 2008). qLTG3‐1 and its orthologs are conserved among rice, Brachypodium, sorghum, maize, and dicots such as Arabidopsis, and orthologous genes have a conserved function during seed germination (Fujino et al., 2014; Xu et al., 2011). GRP domains are functionally conserved in plants, and many genes coding for those domains are involved in cold signaling. AtGRP2, AtGRP4, AtGRP7 and AtRZ‐1b in Arabidopsis exhibit RNA chaperone activity during the cold adaptation process (Kim et al., 2007, 2010b; Kwak et al., 2011). In rice, plants overexpressing OsGRP1, OsGRP4, and OsGRP6 were more tolerant to low temperature stress (Kim et al., 2010a). In Nicotiana tabacum, NtGRP1, NtGRP‐1a and NtGRP3 transcripts were up‐regulated after cold treatment (Chen et al., 2010; Czolpinska and Rurek, 2018). It was previously shown that the Italica Livorno allele of qLTG3‐1, which is identical to the Kasalath allele, qLTG3‐1 Ka, has a much stronger activity than the Nipponbare allele of qLTG3‐1, qLTG3‐1 Nip, in conferring low‐temperature germinability and pre‐harvest sprouting resistance (Fujino et al., 2008; Hori et al., 2010; Iwata and Fujino, 2010). In our study, however, only the Nipponbare allele qLTG3‐1 Nip conferred cold stress tolerance in rice at the reproductive stage, while the Kasalath and Hejiang 19 alleles qLTG3‐1 Ka and qLTG3‐1 Hj were non‐functional in improving cold stress tolerance at that stage. Compared to the Kasalath allele qLTG3‐1 Ka, qLTG3‐1 Nip has only one nucleotide change leading to a nonsynonymous substitution in the 17th amino acid, resulting in the novel function of cold tolerance at the reproductive stage. Uncovering in future work the molecular mechanism of how this single amino‐acid change results in a novel functional allele that improves cold stress tolerance during the reproductive stage will be extremely interesting for understanding mechanisms of cold stress tolerance in rice. One possibility is that the amino acid change affects the subcellular localization of qLTG3‐1. In agreement with this, the subcellular localization of fluorescent signals of the qLTG3‐1Ka::GFP fusion protein was stronger than that of the qLTG3‐1Nip::GFP fusion, (Figure S12), possibly because the amino acid substitution is within the presumed N‐terminal signal sequence of the protein.
Although qLTG3‐1Nip cannot activate OsMST7, OsMST8 or OsINV4 expression (Figure 5b), qLTG3‐1 Nip OE lines nonetheless have a reduced level of soluble sugars (sucrose, glucose and fructose) in flowers under natural cold stress conditions (Figure S11). A possible explanation for this is that the physical interaction of qLTG3‐1Nip with bZIP73Jap enhances the expression of OsMST7, OsMST8 and OsINV4, thus causing qLTG3‐1 Nip OE lines to transport more soluble sugars than wild‐type plants from the source (flowers) to the sink (pollen), leading to reduced soluble sugar accumulation in flowers (Figure S11), improved pollen fertility (Figure 8), seed‐setting rates, and yield per plant (Figure 7).
Previous ChIP‐qPCR analysis showed that bZIP73Jap binds to the promoter regions of peroxidase precursors (POXs, LOC_Os01g22249, LOC_Os03g02920, LOC_Os03g32050 and LOC_Os04g59210) in seedlings of bZIP73Jap::Flag OE lines, and up‐regulated their expression in bZIP71‐bZIP73 Jap OE (71‐73JapOE) lines (Liu et al., 2018a). However, the factor cannot bind to the promoters of qLTG3‐1 Nip, OsINV4, OsMST7, and OsMST8 at the seedling stage (Figure S13B), and there is no significant difference in expression levels of those genes between 71‐73JapOE lines and wild‐type ZH11 (Figure S13D). In contrast, ChIP‐qPCR showed that bZIP73Jap binds to the promoters of qLTG3‐1 Nip, OsMST7, OsMST8, and OsINV4, in the panicles of bZIP73Jap::Flag OE lines (Figure 5a), but not to the promoters of the four POX genes (Figure S13A), nor does it change their expression in panicles of the 71‐73JapOE lines (Figure S13C). qLTG3‐1 Nip is predominantly expressed in panicle at EB and flowering stages (Figure S1), while OsMST7 OsMST7, OsMST8 and OsINV4 were expressed predominantly in pollen, anther wall, or tapetum from the YM until the pollen maturity stage (Oliver et al., 2005, 2007). A possible explanation for these tissue specific expression differences is that the chromatin structure at loci of qLTG3‐1 Nip, OsMST7, OsMST8, and OsINV4 might be open and accessible for bZIP73Jap or bZIP71:bZIP73Jap at the reproductive stage, while it might be closed and inaccessible for those factors at the seedling stage. This means that a combination of chromatin accessibility and possible interactions with different binding partners allows transcription factors such as bZIP73Jap or bZIP71:bZIP73Jap to regulate different target genes at different development stages. On the other hand, expression levels of bZIP71 and bZIP73 Jap in response to cold were also different in vegetative seedling and at the reproductive stage. In shoots of two‐week seedlings, the expression of bZIP71 was much higher than that of bZIP73 Jap (Figure S14), while expression levels of those two genes were similar in panicles at the reproductive stage (Figure S14). Moreover, the expression of bZIP73 Jap did not change during cold exposure in shoots at the seedling stage (Liu et al., 2018a), while it was cold induced in flag leaves and panicles at the reproductive stage (Figure 1). At the same time, expression levels of bZIP71 did not change in response to cold conditions both at the seedling (Liu et al., 2014) and reproductive stages (Figure S2). Therefore, the abundance of bZIP71:bZIP73Jap heterodimers is most likely different at the seedling and reproductive stages, thus activation of downstream genes is different. Furthermore, because bZIP73Jap binds to the promoter of qLTG3‐1 Nip, which is preferentially expressed in panicles at EB and flowering stages (Figure S1), and also physically interacts with qLTG3‐1Nip (Figure 6), there might be a feedback regulation of qLTG3‐1 Nip on the transcriptional activity of bZIP73Jap as follows: bZIP73Jap upregulates qLTG3‐1 Nip at the rice reproductive stage, thus promoting a physical interaction between qLTG3‐1Nip and bZIP73Jap, which in turn enhances expression of reproductive stage specific bZIP73Jap target genes such as qLTG3‐1 Nip, OsMST7, OsMST8 and OsINV4 (Figure 5b). Alternatively, interaction of qLTG3‐1Nip and bZIP73Jap may sequester the bZIP71‐bZIP73Jap heterodimer mainly to those four target genes, thus preventing the bZIP71‐bZIP73Jap heterodimer from activating the expression of POX genes at the reproductive stage. Moreover, the effect of transcription factors on their target genes usually varies depending on particular developmental or cellular contexts (Lu et al., 2013; Spitz and Furlong, 2012), and genes associated with bZIP73Jap binding sites might be differently expressed at the seedling and reproductive stages. This would suggest a developmental or cellular context dependent regulation of bZIP73Jap.
Based on our results, we propose the following model for rice cold tolerance at the reproductive stage: when rice encounters cold stress, bZIP73Jap is induced which recruits bZIP71 to form heterodimers, leading to repressed ABA biosynthesis and activated peroxidase expression, and ultimately improved cold stress tolerance at the seedling stage (Liu et al., 2018a). During the reproductive stage, bZIP71:bZIP73Jap forms heterodimers and activates transcription of qLTG3‐1 Nip, monosaccharide transporter genes, and cell wall invertase genes, which enhances soluble sugar transport from anthers to pollen, thus improving the rice seed‐setting rate under cold stress conditions (Figure 9).
Figure 9.
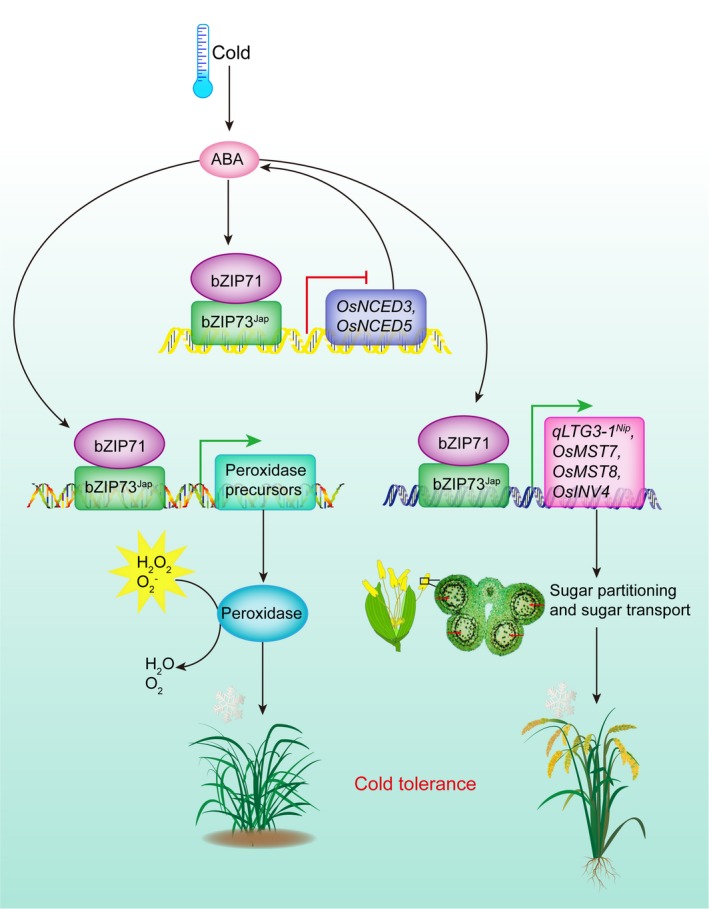
Proposed model for the molecular mechanisms of bZIP73Jap‐mediated cold tolerance in rice plants at both the seedling and the reproductive stage.
Materials and methods
Plant materials and growth conditions
The Japonica cultivar (cv.) Zhonghua 11 (ZH11) was used to generate bZIP71 (LOC_Os09g13570), bZIP73 Jap (LOC_Os09g29820), bZIP73 Ind overexpression (71OE, 73JapOE, 73IndOE), bZIP71 and bZIP73 Jap co‐expression (71‐73JapOE), and bZIP71, bZIP73 Jap RNAi transgenic lines (71Ri, 73JapRi). The Indica type cv. Kasalath was used to generate qLTG3‐1 Nip, qLTG3‐1 Ka, and qLTG3‐1 Hj overexpression transgenic lines. Rice plants were grown in a paddy field under natural conditions in Beijing (116.4°E/39.9°N).
Expression analyses with real‐time PCR
For analysis of gene expression patterns in response to cold stress during the reproductive stage, anther developmental stages were determined based on the AD method [AD: the distance in cm between the nodes of the flag leaf and the penultimate leaf (Oliver et al., 2005)]. The YM stage was AD 0 to +2.5 cm; the EB stage was AD +6 to +8 cm; and the flowering stage was AD: +16 to 18 cm. Cold treatment was carried out in a controlled growth chamber (14 h light/10 h dark) for 1 day at a constant temperature of 12 °C, and the panicles and flag leaves of ZH11 were collected 0, 0.25, 0.5, 1, 3, 6, 12, and 24 h after cold treatment. For tissue expression pattern analysis, different ZH11 tissues including shoots (two‐week‐old), roots (two‐week‐old), stems (YM stage), flag leaves (YM stage), panicles (YM and EB stages), and flowers (1 day before flowering) were used for RNA extraction and quantitative RT‐PCR.
Total RNA was prepared from rice tissues using the Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. Total RNA was reverse transcribed to cDNA using the ReverTra Ace qPCR RT Master Mix with gDNA remover Kit (Toyobo, Osaka, Japan) according to the manufacturer's instruction. cDNA templates were used for real‐time PCR analysis. Real‐time PCR was done using the SYBR® Premix Ex TaqTM Kit (TaKaRa, Kusatsu, Japan) protocol in an ABI PRISM 7900HT (Applied Biosystems, Foster City, CA, USA). The rice Ubiquitin gene (LOC_Os03g13170) was used as internal control. Primers for real‐time PCR are listed in Table S7. Data analysis was conducted as described by Pfaffl (Pfaffl, 2001).
Vector construction and plant transformation
To avoid negative agronomic traits in transformed plants caused by fusion tags of binary expression vectors, overexpression vectors were constructed without fusion tags. Three different alleles of qLTG3‐1 (LOC_Os03g01320) coding sequences (CDS) were amplified by RT‐PCR from Nipponbare, Kasalath, and Hejiang19, and subsequently cloned into the binary vector pCambia1390‐Ubiquitin to generate the pCAMBIA1390‐Ubiquitin‐ qLTG3‐1 Nip/Ka/Hj overexpression constructs. The CDS of bZIP71/bZIP73 Jap/Ind was cloned into pENTER‐TOPO vectors (Invitrogen, Carlsbad, CA, USA), then they were cloned into the binary vector pH2GW7 (Karimi et al., 2002) (Invitrogen) using the LR reaction of the pENTER‐TOPO vectors, to generate the pH2GW7‐bZIP71/bZIP73 Jap/Ind overexpression vectors. All binary constructs were introduced into Agrobacterium tumefaciens strain AGL1 and subsequently transformed into the Indica type cultivar Kasalath. Primer sequences for vector construction are listed in Table S7.
Stress tolerance assays
For field test of cold resistance at the reproductive stage, transgenic lines were planted in the temperate zone of Beijing (39.9°N) in Northern China, where temperatures in late August and early September are relatively low, with average daily temperatures usually below 20 °C. Exposure of rice to temperatures below 20 °C at the booting stage causes pollen sterility or fertilization failure. Different planting date experiments using transgenic lines were conducted in Beijing every fifteen days from May 10th to July 9th, which ensured cold temperature would hit at the rice booting stage in late August or early September. Because the constitutive bZIP71 overexpression lines (71OE) flowered 7 days later than wild‐type ZH11 (Liu et al., 2014), we sowed 71OE lines 7 days earlier than wild‐type and other transgenic lines to ensure that the heading date of each batch was the same. Thirty plants of each line were used for each plot, and the experiments were replicated at least three times. Agronomic traits including plant height, tiller number, seed‐setting rate, and yield per plant were statistically analyzed.
Microscopy
Starch staining assays of pollen grains (Nelson, 1968) were performed just one day before flowering. For the morphological observations of anthers, samples were fixed in a Formalin‐Aceto‐Alcohol (FAA) solution. To determine pollen fertility, fixed anthers were ground to release pollen grains, stained with I2‐KI solution, then observed microscopically and photographs taken with an Olympus SEX16. Blue stained pollen grains were counted as a measure to determine pollen fertility. Pollen or male sterility is defined as an absence of or presence of non‐functional pollen grains in plants, or as the inability of plants to produce or release functional pollen grains. Immature pollen is sterile. One way avova test, P‐value in Table S8.
ABA quantification
Quantification of endogenous ABA was performed as described previously (Li et al., 2011). Rice flowers (approximately 3 g of fresh weight) of transgenic lines and wild‐type plants were harvested one day before flowering both under normal (heading at August 17, 2012, Beijing) and natural cold conditions (heading at September 13, 2012, Beijing). Each series of experiments were performed using biological triplicates.
Sugar measurements
Samples were harvested one day before flowering both under normal (heading on August 17, 2012, Beijing) and natural cold conditions (heading on September 13, 2012, Beijing). Anthers were ground in liquid nitrogen to a fine powder, and 10 replicates for each line were used. 0.7 mL of 80% ethanol was added to each sample and immediately incubated at 80 °C for 60 min. After cooling, samples were centrifuged at 4 °C and 16 873 g for 5 min. The supernatant was transferred to a new Eppendorf tube, and the remaining pellet was washed twice with 0.7 mL of 80% ethanol. The supernatant was evaporated in a speed vacuum dryer at 50 °C for around 60–90 min and the residue resuspended in 200–250 μL of water and used directly for measurements. The content of glucose, fructose, and sucrose were measured using enzyme‐based methods as described previously (Spackman and Cobb, 2002). One way avova test, P‐value in Table S8.
EMSA
The CDS of bZIP71 and bZIP73 Jap were amplified and cloned into the pGEX4T‐3 vector to generate GST fusion proteins, and were purified as previously reported (Liu et al., 2018a). For promoter binding experiments, complementary single‐stranded oligonucleotides derived from 70 bp of the G‐box region of downstream gene (qLTG3‐1 Nip, OsMST7, OsMST8 and OsINV4) promoters were synthesized as DNA probes. To obtain double stranded G‐Box motif‐containing fragments, two complementary oligonucleotides were mixed in a water bath at 95 °C for 5 min and cooled to room temperature for annealing. A mutated G‐box sequence was amplified from wild type using mutated primers and was used as negative control. EMSA was performed using a chemiluminescent EMSA kit (Thermo, Massachusetts, USA). Probe sequences are shown in Table S7.
ChIP‐qPCR
ChIP assays were performed as described previously, with some modifications (Fiil et al., 2008). Briefly, about 2 g of flowers from T3 homozygous lines of bZIP73Jap::Flag, bZIP73Ind::Flag, or bZIP71::Flag transgenic lines in the ZH11 background were cross‐linked in 1% formaldehyde under a vacuum, and cross‐linking was stopped with 0.125 m glycine. Samples were ground to a powder in liquid nitrogen, and nuclei were isolated. An antibody against bZIP fused Flag tag was used at a 1:5000 dilution to immuno‐precipitate protein‐DNA complexes. Immunoprecipitation was performed with anti‐Flag antibody (anti‐Flag) or without antibody (No Ab) as a negative control. Fragments consisting of a 1000 bp sequence upstream of the ATG were chosen as the promoter (Figure S6). This promoter fragment contained G‐box element regions used for designing primers (Table S7). The Ubiquitin gene was used as a negative control. The amount of precipitated DNA was calculated relative to the total input of chromatin and expressed as a percentage of the total according to the formula: % input = 2▵Ct × 100%, where ▵Ct = Ct (input) − Ct (IP) and Ct is the mean threshold cycle of the corresponding PCR reaction (Chernukhin et al., 2011). Three independent biological replicates were used.
Transient expression in rice protoplasts
For determination of subcellular localization, the full‐length CDSs of qLTG3‐1 Nip/Ka were inserted into pCambia2300‐35S‐GFP, creating qLTG3‐1 Nip::GFP and qLTG3‐1 Ka::GFP fusion vectors. Plasmid DNA was prepared using the Plasmid Midi Kits (Qiagen, Germany) according to the manufacturer's instructions. qLTG3‐1 Nip/Ka::GFP fusion vectors were transformed into rice (ZH11) leaf protoplasts using the polyethylene glycol method (Zhang et al., 2011). After 12 h of incubation in the dark at 28 °C, transient expression of green fluorescent protein (GFP) fluorescence was recorded using a confocal laser‐scanning microscope (Carl Zeiss, LSM510, Germany).
For the Dual‐Luciferase Assay, promoters of qLTG3‐1 Nip, OsMST7 (LOC_Os01g38680), OsMST8 (LOC_Os01g58670), and OsINV4 (LOC_Os04g33720) (~2000 bp upstream of ATG) were amplified and cloned into the pGreenII 0800‐LUC vector containing the firefly luciferase (fLUC) gene and the Renilla LUC gene (rLUC) as reporters (Hellens et al., 2005), while the CDS of bZIP71, bZIP73 Jap/Ind and qLTG3‐1 Nip were cloned into the pCambia1300‐Flag vector as effectors. Empty pCambia1300‐Flag vector was used as negative control for the effector. Rice (ZH11) shoot protoplasts were prepared and transfected and followed by a 12 h incubation to allow transient expression (Zhang et al., 2011). Protoplasts were collected by centrifugation at 450 g for 3 min and immediately utilized for luciferase assays. Luciferase activity was quantified using a dual‐luciferase reporter assay kit (Promega, E1910, Madison, WI, USA). Five independent transformations for each sample were performed, and the relative luciferase activity was calculated as the ratio of fLUC to rLUC (fLUC/rLUC).
Yeast assays
To prepare constructs for the yeast one‐hybrid (Y1H) assay, the promoter region of qLTG3‐1 Nip, OsMST7, OsMST8 and OsINV4 (2 kb upstream of ATG) was amplified and ligated into the EcoRI‐SalI sites of the pLacZi2μ vector (Lin et al., 2007) to generate pqLTG3‐1 Nip /pOsMST7/pOsMST8/pOsINV4::LacZ. To generate AD‐bZIP73Jap, the full‐length CDS of OsbZIP73 Jap was ligated into the EcoRI‐XhoI sites of the pJG4‐5 vector (Clontech). The Y1H assay was performed according to the Yeast Protocols Handbook (Clontech). Briefly, the AD fusion constructs were co‐transformed with various LacZ reporter plasmids into yeast strain EGY48. Transformants were grown on SD/Trp‐Ura‐ dropout plates containing 5‐bromo‐4‐chloro‐3‐indolyl‐ß‐D‐galactopyranoside (X‐Gal) for blue color development.
Yeast two‐hybrid (Y2H) assays were conducted using the Matchmaker Two‐Hybrid System 3 (Clontech). The activation domain (AD) vector pGADT7, the DNA‐binding domain (BD) vector pGBDT7, and the yeast strain Y2HGold were utilized. The full‐length CDSs of qLTG3‐1 Nip (aa: 1‐184), qLTG3‐1 Ka (aa: 1‐184), or qLTG3‐1 Hj (aa: 1‐116) were cloned respectively into the vector pGADT7 as prey, and the full‐length CDS of bZIP73 Jap was cloned into the vector pGBDT7 as bait. The amplification primers used are shown in Table S7. The prey and bait vectors were co‐transformed into Y2HGold cells and the transformants were identified according to the manufacturer's instructions.
BiFC and Co‐IP
To generate BiFC constructs, the CDSs of bZIP73 Jap and qLTG3‐1 Nip were PCR amplified and ligated into VYCE(R) and VYNE(R) (Waadt et al., 2008), resulting in bZIP73Jap::cYFP and qLTG3‐1Nip::nYFP, respectively. Empty vectors of BiFC constructs were used as negative controls. To generate Co‐IP constructs, the CDSs of bZIP73 Jap and qLTG3‐1 Nip were PCR amplified and ligated into pCambia1300‐Flag and pCambia2300‐35S‐GFP, resulting in bZIP73Jap::Flag and qLTG3‐1Nip::GFP, respectively. Primers used for generating fusion constructs are listed in Table S7. All the constructs were transformed into the Agrobacterium tumefaciens GV3101 strain and used for BiFC and Co‐IP experiments.
Agrobacterium solutions were adjusted to OD600 = 0.8 and then equally mixed before Agroinfiltration. An Agrobacterium strain carrying the viral suppressor p19 was used to suppress gene silencing in transformed tobacco leaves (Guan et al., 2017) in this experiment. Middle leaves of three‐week‐old juvenile N. bethamiana plants were used for Agroinfiltration. The detailed procedures are described in previous report (Sparkes et al., 2006). The infiltrated N. bethamiana leaves were imaged using a Carl Zeiss (LSM510, Germany) confocal microscope 3 days after agroinfiltration.
Co‐IP analysis was performed based on a previous report (Lin and Lai, 2017). Briefly, proteins from 250 mg of tobacco leaves, which continued growth 3 days after agroinfiltration, were extracted in 500 μL of IP buffer, then centrifuged and mixed with 20 μL of anti‐GFP magnetic beads (Sigma). After a 1‐h incubation, the beads were washed five times with wash buffer. Beads were then boiled in 25 μL of 2 × SDS sample buffer and subjected to immunoblotting analysis. The original blot scans are shown in Figure S15.
Author contributions
C.L. performed most experiments. M.S and A.W. performed vector constructions of qLTG3‐1. B.M. and W.W. performed investigation of agronomic traits in the field. C.L. and C.C. designed experiments and wrote the manuscript. M.S. edited the manuscript.
Conflict of interest
The authors declare no competing financial and non‐financial interests.
Supporting information
Figure S1 bZIP71, bZIP73 Jap, qLTG3‐1 Nip expression patterns in different organs of wild type ZH11.
Figure S2 bZIP71 expression patterns in flag leaves and panicles of wild‐type ZH11 under cold treatment during the reproductive stage.
Figure S3 Temperature profile at the experimental station of IGDB at Changping, Beijing, during the periods of August, September, and October in 2012.
Figure S4 Relative expression of bZIP71 and bZIP73 in transgenic lines.
Figure S5 ABA and soluble sugar contents in transgenic lines and wild‐type plants under normal warm conditions.
Figure S6 Relative expression levels of ABA biosynthetic genes, invertase gene, monosaccharide transporter genes in (A) bZIP71 overexpression (71OE) lines, (B) bZIP71 RNAi (71Ri) lines, (C) bZIP73 Jap overexpression (73JapOE) lines, (D) bZIP73 Ind overexpression (73IndOE) lines, (E) bZIP73 Jap RNAi (73JapRi) lines, and (F) bZIP73 Jap and bZIP71 co‐overexpression (71‐73JapOE) lines as detected by qPCR.
Figure S7 Distribution of G‐box elements in the promoters of OsNCED3, OsNCED5, qLTG3‐1, OsMST7, OsMST8, and OsINV4 genes.
Figure S8 Sequence alignment of three alleles of qLTG3‐1.
Figure S9 qLTG3‐1 Nip expression patterns in flag leaves and panicles of wild‐type ZH11 under cold stress conditions during the reproductive stage.
Figure S10 Relative expression levels of qLTG3‐1 in flowers of Kasalath and qLTG3‐1 Nip/Ka/Hj transgenic lines measured by real‐time PCR under normal warm conditions.
Figure S11 Soluble sugar contents in qLTG3‐1 Nip overexpression lines and wild‐type plants under natural cold conditions.
Figure S12 Subcellular localization of qLTG3‐1Nip and qLTG3‐1Ka protein in rice protoplasts.
Figure S13 bZIP73Jap could not bind downstream genes both at seedling and reproductive stages.
Figure S14 bZIP71 and bZIP73 Jap expression patterns in different organs of ZH11 (japonica rice cultivar).
Figure S15 Original blotting images of co‐IP assay (Figure 6D).
Table S1 Agronomic traits of bZIP71, bZIP73 transgenic lines and wild‐type ZH11 grown in the field under natural cold stress conditions for plants heading on September 13, 2012, Beijing.
Table S2 Agronomic traits of bZIP71 and bZIP73 Jap co‐overexpression transgenic lines and wild‐type ZH11 grown in the field under natural cold stress conditions for plants heading on September 20, 2012, Beijing.
Table S3 Agronomic traits of bZIP71, bZIP73 transgenic lines and wild‐type ZH11 grown in the field under normal warm condition for plants heading on August 17, 2012, Beijing.
Table S4 Agronomic traits of qLTG3‐1 Nip/Ka/Hj overexpression transgenic lines and wild‐type Kasalath grown in the field under natural cold stress conditions for plants heading on September 3, 2012, Beijing.
Table S5 Agronomic traits of qLTG3‐1 Nip/Ka/Hj overexpression transgenic lines and wild‐type Kasalath grown in the field under natural cold stress conditions, for plants heading on September 12, 2012, Beijing.
Table S6 Agronomic traits of qLTG3‐1 Nip/Ka/Hj overexpression transgenic lines and wild‐type Kasalath grown in the field under normal warm conditions for plants heading on August 22, 2012, Beijing.
Table S7. Primers used in real‐time PCR analyses and vector constructions.
Table S8 One way anova test for P‐value.
Acknowledgements
We thank Mr. Gupo Li for plant management in the field, and Mrs. Shouyun Cao for generating transgenic rice plants. This work was supported by grants from the National Natural Science Foundation of China (#31501283 and #31271680) and in part by USDA‐AFRI‐NIFA grant #2016‐67013‐24587 and #2018‐67014‐27470 (to M.S.).
References
- Chen, X. , Zeng, Q. , Xiuping, L.U. , Diqiu, Y.U. and Wenzheng, L.I. (2010) Characterization and expression analysis of four glycine‐rich RNA‐binding proteins involved in osmotic response in tobacco (Nicotiana tabacum cv. Xanthi). Agric. Sci. China, 9, 1577–1587. [Google Scholar]
- Chernukhin, I. , Kang, S.Y. , Brown, S. , Gretton, S. , Mendez‐Catala, C.F. , Cowieson, D. and Klenova, E. (2011) BioVyon Protein A, an alternative solid‐phase affinity matrix for chromatin immunoprecipitation. Anal. Biochem. 412, 183–188. [DOI] [PubMed] [Google Scholar]
- Czolpinska, M. and Rurek, M. (2018) Plant glycine‐rich proteins in stress response: an emerging, still prospective story. Front. Plant Sci. 9, 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, H. , Wu, N. , Chang, Y. , Li, X. , Xiao, J. and Xiong, L. (2013) Carotenoid deficiency impairs ABA and IAA biosynthesis and differentially affects drought and cold tolerance in rice. Plant Mol. Biol. 83, 475–488. [DOI] [PubMed] [Google Scholar]
- Espe, M.B. , Hill, J.E. , Hijmans, R.J. , McKenzie, K. , Mutters, R. , Espino, L.A. , Leinfelder‐Miles, M. et al (2017) Point stresses during reproductive stage rather than warming seasonal temperature determine yield in temperate rice. Glob. Chang. Biol. 23, 4386–4395. [DOI] [PubMed] [Google Scholar]
- Fiil, B.K. , Qiu, J.‐L. , Petersen, K. , Petersen, M. and Mundy, J. (2008) Coimmunoprecipitation (co‐IP) of nuclear proteins and chromatin immunoprecipitation (ChIP) from Arabidopsis . Cold Spring Harb. Protoc. 2008, pdb.prot5049. [DOI] [PubMed] [Google Scholar]
- Fujino, K. and Matsuda, Y. (2010) Genome‐wide analysis of genes targeted by qLTG3‐1 controlling low‐temperature germinability in rice. Plant Mol. Biol. 72, 137–152. [DOI] [PubMed] [Google Scholar]
- Fujino, K. , Sekiguchi, H. , Matsuda, Y. , Sugimoto, K. , Ono, K. and Yano, M. (2008) Molecular identification of a major quantitative trait locus, qLTG3‐1, controlling low‐temperature germinability in rice. Proc. Natl. Acad. Sci. USA, 105, 12623–12628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino, K. , Obara, M. and Sato, K. (2014) Diversification of the plant‐specific hybrid glycine‐rich protein (HyGRP) genes in cereals. Front. Plant Sci. 5, 489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, P. , Ripoll, J.J. , Wang, R. , Vuong, L. , Bailey‐Steinitz, L.J. , Ye, D. and Crawford, N.M. (2017) Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc. Natl. Acad. Sci. USA, 114, 2419–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens, R.P. , Allan, A.C. , Friel, E.N. , Bolitho, K. , Grafton, K. , Templeton, M.D. , Karunairetnam, S. et al (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods, 1, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori, K. , Sugimoto, K. , Nonoue, Y. , Ono, N. , Matsubara, K. , Yamanouchi, U. , Abe, A. et al (2010) Detection of quantitative trait loci controlling pre‐harvest sprouting resistance by using backcrossed populations of japonica rice cultivars. Theor. Appl. Genet. 120, 1547–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata, N. and Fujino, K. (2010) Genetic effects of major QTLs controlling low‐temperature germinability in different genetic backgrounds in rice (Oryza sativa L.). Genome, 53, 763–768. [DOI] [PubMed] [Google Scholar]
- Ji, X. , Dong, B. , Shiran, B. , Talbot, M.J. , Edlington, J.E. , Hughes, T. , White, R.G. et al (2011) Control of abscisic acid catabolism and abscisic acid homeostasis is important for reproductive stage stress tolerance in cereals. Plant Physiol. 156, 647–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, S.G. , Price, J. , Lin, P.C. , Hong, J.C. and Jang, J.C. (2010) The Arabidopsis bZIP1 transcription factor is involved in sugar signaling, protein networking, and DNA binding. Mol. Plant, 3, 361–373. [DOI] [PubMed] [Google Scholar]
- Karimi, M. , Inze, D. and Depicker, A. (2002) GATEWAY vectors for Agrobacterium‐mediated plant transformation. Trends Plant Sci. 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Kim, J.S. , Park, S.J. , Kwak, K.J. , Kim, Y.O. , Kim, J.Y. , Song, J. , Jang, B. et al (2007) Cold shock domain proteins and glycine‐rich RNA‐binding proteins from Arabidopsis thaliana can promote the cold adaptation process in Escherichia coli . Nucleic Acids Res. 35, 506–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.Y. , Kim, W.Y. , Kwak, K.J. , Oh, S.H. , Han, Y.S. and Kang, H. (2010a) Glycine‐rich RNA‐binding proteins are functionally conserved in Arabidopsis thaliana and Oryza sativa during cold adaptation process. J. Exp. Bot. 61, 2317–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, W.Y. , Kim, J.Y. , Jung, H.J. , Oh, S.H. , Han, Y.S. and Kang, H. (2010b) Comparative analysis of Arabidopsis zinc finger‐containing glycine‐rich RNA‐binding proteins during cold adaptation. Plant Physiol. Biochem. 48, 866–872. [DOI] [PubMed] [Google Scholar]
- Koonjul, P. , Minhas, J. , Nunes, C. , Sheoran, I. and Saini, H. (2005) Selective transcriptional down‐regulation of anther invertases precedes the failure of pollen development in water‐stressed wheat. J. Exp. Bot. 56, 179–190. [DOI] [PubMed] [Google Scholar]
- Kwak, K.J. , Park, S.J. , Han, J.H. , Kim, M.K. , Oh, S.H. , Han, Y.S. and Kang, H. (2011) Structural determinants crucial to the RNA chaperone activity of glycine‐rich RNA‐binding proteins 4 and 7 in Arabidopsis thaliana during the cold adaptation process. J. Exp. Bot. 62, 4003–4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Jiang, J. , Qian, Q. , Xu, Y. , Zhang, C. , Xiao, J. , Du, C. et al (2011) Mutation of rice BC12/GDD1, which encodes a kinesin‐like protein that binds to a GA biosynthesis gene promoter, leads to dwarfism with impaired cell elongation. Plant Cell, 23, 628–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Pan, Y. , Guo, H. , Zhou, L. , Yang, S. , Zhang, Z. , Yang, J. et al (2018) Fine mapping of QTL qCTB10‐2 that confers cold tolerance at the booting stage in rice. Theor. Appl. Genet. 131, 157–166. [DOI] [PubMed] [Google Scholar]
- Lin, J.S. and Lai, E.M. (2017) Protein‐Protein Interactions: Co‐Immunoprecipitation. Methods Mol. Biol. (Clifton, NJ), 1615, 211–219. [DOI] [PubMed] [Google Scholar]
- Lin, R. , Ding, L. , Casola, C. , Ripoll, D.R. , Feschotte, C. and Wang, H. (2007) Transposase‐derived transcription factors regulate light signaling in Arabidopsis . Science, 318, 1302–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C. , Mao, B. , Ou, S. , Wang, W. , Liu, L. , Wu, Y. , Chu, C. et al (2014) OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol. Biol. 84, 19–36. [DOI] [PubMed] [Google Scholar]
- Liu, C. , Ou, S. , Mao, B. , Tang, J. , Wang, W. , Wang, H. , Cao, S. et al (2018a) Early selection of bZIP73 facilitated adaptation of japonica rice to cold climates. Nat. Commun. 9, 3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C. , Wang, W. , Mao, B. and Chu, C. (2018b) Cold stress tolerance in rice: physiological changes, molecular mechanism, and future prospects. Hereditas (Beijing), 40, 171–185. [DOI] [PubMed] [Google Scholar]
- Lu, Z. , Yu, H. , Xiong, G. , Wang, J. , Jiao, Y. , Liu, G. , Jing, Y. et al (2013) Genome‐wide binding analysis of the transcription activator ideal plant architecture1 reveals a complex network regulating rice plant architecture. Plant Cell, 25, 3743–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J. , Hanssen, M. , Lundgren, K. , Hernandez, L. , Delatte, T. , Ehlert, A. , Liu, C.M. et al (2011) The sucrose‐regulated Arabidopsis transcription factor bZIP11 reprograms metabolism and regulates trehalose metabolism. New Phytol. 191, 733–745. [DOI] [PubMed] [Google Scholar]
- Mamun, E.A. , Alfred, S. , Cantrill, L.C. , Overall, R.L. and Sutton, B.G. (2006) Effects of chilling on male gametophyte development in rice. Cell Biol. Int. 30, 583–591. [DOI] [PubMed] [Google Scholar]
- Mamun, E. , Cantrill, L.C. , Overall, R.L. and Sutton, B.G. (2010) Mechanism of low‐temperature‐induced pollen failure in rice. Cell Biol. Int. 34, 469–476. [DOI] [PubMed] [Google Scholar]
- Mega, R. , Meguro‐Maoka, A. , Endo, A. , Shimosaka, E. , Murayama, S. , Nambara, E. , Seo, M. et al (2015) Sustained low abscisic acid levels increase seedling vigor under cold stress in rice (Oryza sativa L.). Sci. Rep. 5, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler, R. and Blumwald, E. (2015) The roles of ROS and ABA in systemic acquired acclimation. Plant Cell, 27, 64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, O.E. (1968) The waxy locus in maize. II. The location of the controlling element alleles. Genetics, 60, 507–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama, I. (1976) Male sterility caused by cooling treatment at the young microspore stage in rice plants: XIII. Ultrastructure of tapetal hypertrophy without primary wall. Proc. Crop. Sci. Soc. Jpn, 45, 270–278. [Google Scholar]
- Nishiyama, I. (1982) Male sterility caused by cooling treatment at the young microspore stage in rice plants. XXIII. Anther length, pollen number and the difference in susceptibility to coolness among spikelets on the panicle. Jpn J. Crop. Sci. 51, 462–469. [Google Scholar]
- Oliver, S.N. , Van Dongen, J.T. , Alfred, S.C. , Mamun, E.A. , Zhao, X. , Saini, H.S. , Fernandes, S.F. et al (2005) Cold‐induced repression of the rice anther‐specific cell wall invertase gene OsINV4 is correlated with sucrose accumulation and pollen sterility. Plant Cell Environ. 28, 1534–1551. [Google Scholar]
- Oliver, S.N. , Dennis, E.S. and Dolferus, R. (2007) ABA regulates apoplastic sugar transport and is a potential signal for cold‐induced pollen sterility in rice. Plant Cell Physiol. 48, 1319–1330. [DOI] [PubMed] [Google Scholar]
- Pan, Y. , Zhang, H. , Zhang, D. , Li, J. , Xiong, H. , Yu, J. , Li, J. et al (2015) Genetic analysis of cold tolerance at the germination and booting stages in rice by association mapping. PLoS ONE, 10, e0120590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl, M.W. (2001) A new mathematical model for relative quantification in real‐time RT–PCR. Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook, F. , Corke, F. , Card, R. , Munz, G. , Smith, C. and Bevan, M.W. (2001) Impaired sucrose‐induction mutants reveal the modulation of sugar‐induced starch biosynthetic gene expression by abscisic acid signalling. Plant J. 26, 421–433. [DOI] [PubMed] [Google Scholar]
- Saito, K. , Hayano‐Saito, Y. , Kuroki, M. and Sato, Y. (2010) Mapbased cloning of the rice cold tolerance gene Ctb1 . Plant Sci. 179, 97–102. [Google Scholar]
- Sakata, T. , Oda, S. , Tsunaga, Y. , Shomura, H. , Kawagishi‐Kobayashi, M. , Aya, K. , Saeki, K. et al (2014) Reduction of gibberellin by low temperature disrupts pollen development in rice. Plant Physiol. 164, 2011–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satake, T. (1976) Determination of the most sensitive stage to sterile‐type cool injury in rice plants. Res. Bull. Hokkaido Nat. Agric. Exp. Stn., 113, 1–43. [Google Scholar]
- Sharma, K.D. and Nayyar, H. (2016) Regulatory networks in pollen development under cold stress. Front. Plant Sci. 7, 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono, H. , Abe, A. , Aoki, N. , Koumoto, T. , Sato, M. , Yokoi, S. , Kuroda, E. et al (2016) Combining mapping of physiological quantitative trait loci and transcriptome for cold tolerance for counteracting male sterility induced by low temperatures during reproductive stage in rice. Physiol. Plant. 157, 175–192. [DOI] [PubMed] [Google Scholar]
- Shinada, H. , Iwata, N. , Sato, T. and Fujino, K. (2013) Genetical and morphological characterization of cold tolerance at fertilization stage in rice. Breed. Sci. 63, 197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinada, H. , Iwata, N. , Sato, T. and Fujino, K. (2014) QTL pyramiding for improving of cold tolerance at fertilization stage in rice. Breed. Sci. 63, 483–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spackman, V.M.T. and Cobb, A.H. (2002) An enzyme‐based method for the rapid determination of sucrose, glucose and fructose in sugar beet roots and the effects of impact damage and postharvest storage in clamps. J. Sci. Food Agric. 82, 80–86. [Google Scholar]
- Sparkes, I.A. , Runions, J. , Kearns, A. and Hawes, C. (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025. [DOI] [PubMed] [Google Scholar]
- Spitz, F. and Furlong, E.E. (2012) Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 13, 613–626. [DOI] [PubMed] [Google Scholar]
- Suh, J.P. , Jeung, J.U. , Lee, J.I. , Choi, Y.H. , Yea, J.D. , Virk, P.S. , Mackill, D.J. et al (2010) Identification and analysis of QTLs controlling cold tolerance at the reproductive stage and validation of effective QTLs in cold‐tolerant genotypes of rice (Oryza sativa L.). Theor. Appl. Genet. 120, 985–995. [DOI] [PubMed] [Google Scholar]
- Sun, J. , Yang, L. , Wang, J. , Liu, H. , Zheng, H. , Xie, D. , Zhang, M. et al (2018) Identification of a cold‐tolerant locus in rice (Oryza sativa L.) using bulked segregant analysis with a next‐generation sequencing strategy. Rice, 11, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tymowskalalanne, Z. and Kreis, M. (1998) The plant invertases: physiology, biochemistry and molecular biology. Adv. Bot. Res. 28, 71–117. [Google Scholar]
- Waadt, R. , Schmidt, L.K. , Lohse, M. , Hashimoto, K. , Bock, R. and Kudla, J. (2008) Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56, 505–516. [DOI] [PubMed] [Google Scholar]
- Wang, Z. , Xu, Y. , Chen, T. , Zhang, H. , Yang, J. and Zhang, J. (2015) Abscisic acid and the key enzymes and genes in sucrose‐to‐starch conversion in rice spikelets in response to soil drying during grain filling. Planta, 241, 1091–1107. [DOI] [PubMed] [Google Scholar]
- Wiese, A. , Elzinga, N. , Wobbes, B. and Smeekens, S. (2004) A conserved upstream open reading frame mediates sucrose‐induced repression of translation. Plant Cell, 16, 1717–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, N. , Gao, Y. , Qian, H. , Gao, Q. , Wu, Y. , Zhang, D. , Wang, Z. et al (2018) Identification of genes related to cold tolerance and a functional allele that confers cold tolerance. Plant Physiol. 177, 1108–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, D. , Huang, X. , Xu, Z.Q. and Schläppi, M. (2011) The HyPRP gene EARLI1 has an auxiliary role for germinability and early seedling development under low temperature and salt stress conditions in Arabidopsis thaliana . Planta, 234, 565–577. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Su, J. , Duan, S. , Ao, Y. , Dai, J. , Liu, J. , Wang, P. et al (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast‐related processes. Plant Methods, 7, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q. , Chen, Q. , Wang, S. , Hong, Y. and Wang, Z. (2014) Rice and cold stress: methods for its evaluation and summary of cold tolerance‐related quantitative trait loci. Rice, 7, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Li, J. , Pan, Y. , Li, J. , Zhou, L. , Shi, H. , Zeng, Y. et al (2017) Natural variation in CTB4a enhances rice adaptation to cold habitats. Nat. Commun. 8, 14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y. , Chen, K. , Mi, X. , Chen, T. , Ali, J. , Ye, G. , Xu, J. et al (2015) Identification and fine mapping of a stably expressed QTL for cold tolerance at the booting stage using an interconnected breeding population in rice. PLoS ONE, 10, e0145704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 bZIP71, bZIP73 Jap, qLTG3‐1 Nip expression patterns in different organs of wild type ZH11.
Figure S2 bZIP71 expression patterns in flag leaves and panicles of wild‐type ZH11 under cold treatment during the reproductive stage.
Figure S3 Temperature profile at the experimental station of IGDB at Changping, Beijing, during the periods of August, September, and October in 2012.
Figure S4 Relative expression of bZIP71 and bZIP73 in transgenic lines.
Figure S5 ABA and soluble sugar contents in transgenic lines and wild‐type plants under normal warm conditions.
Figure S6 Relative expression levels of ABA biosynthetic genes, invertase gene, monosaccharide transporter genes in (A) bZIP71 overexpression (71OE) lines, (B) bZIP71 RNAi (71Ri) lines, (C) bZIP73 Jap overexpression (73JapOE) lines, (D) bZIP73 Ind overexpression (73IndOE) lines, (E) bZIP73 Jap RNAi (73JapRi) lines, and (F) bZIP73 Jap and bZIP71 co‐overexpression (71‐73JapOE) lines as detected by qPCR.
Figure S7 Distribution of G‐box elements in the promoters of OsNCED3, OsNCED5, qLTG3‐1, OsMST7, OsMST8, and OsINV4 genes.
Figure S8 Sequence alignment of three alleles of qLTG3‐1.
Figure S9 qLTG3‐1 Nip expression patterns in flag leaves and panicles of wild‐type ZH11 under cold stress conditions during the reproductive stage.
Figure S10 Relative expression levels of qLTG3‐1 in flowers of Kasalath and qLTG3‐1 Nip/Ka/Hj transgenic lines measured by real‐time PCR under normal warm conditions.
Figure S11 Soluble sugar contents in qLTG3‐1 Nip overexpression lines and wild‐type plants under natural cold conditions.
Figure S12 Subcellular localization of qLTG3‐1Nip and qLTG3‐1Ka protein in rice protoplasts.
Figure S13 bZIP73Jap could not bind downstream genes both at seedling and reproductive stages.
Figure S14 bZIP71 and bZIP73 Jap expression patterns in different organs of ZH11 (japonica rice cultivar).
Figure S15 Original blotting images of co‐IP assay (Figure 6D).
Table S1 Agronomic traits of bZIP71, bZIP73 transgenic lines and wild‐type ZH11 grown in the field under natural cold stress conditions for plants heading on September 13, 2012, Beijing.
Table S2 Agronomic traits of bZIP71 and bZIP73 Jap co‐overexpression transgenic lines and wild‐type ZH11 grown in the field under natural cold stress conditions for plants heading on September 20, 2012, Beijing.
Table S3 Agronomic traits of bZIP71, bZIP73 transgenic lines and wild‐type ZH11 grown in the field under normal warm condition for plants heading on August 17, 2012, Beijing.
Table S4 Agronomic traits of qLTG3‐1 Nip/Ka/Hj overexpression transgenic lines and wild‐type Kasalath grown in the field under natural cold stress conditions for plants heading on September 3, 2012, Beijing.
Table S5 Agronomic traits of qLTG3‐1 Nip/Ka/Hj overexpression transgenic lines and wild‐type Kasalath grown in the field under natural cold stress conditions, for plants heading on September 12, 2012, Beijing.
Table S6 Agronomic traits of qLTG3‐1 Nip/Ka/Hj overexpression transgenic lines and wild‐type Kasalath grown in the field under normal warm conditions for plants heading on August 22, 2012, Beijing.
Table S7. Primers used in real‐time PCR analyses and vector constructions.
Table S8 One way anova test for P‐value.


