Abstract
Protein analysis of potential disease markers in blood is complicated by the fact that proteins in plasma show very different abundances. As a result, high-abundance proteins dominate the analysis, which often render the analysis of low-abundance proteins impossible. Depleting high-abundance proteins is one strategy to solve this problem. Here, we present, for the first time, a very simple approach based on selective binding of serum proteins to the surface of nanodiamonds. In our first proof-of-principle experiments, we were able to detect, on average, eight proteins that are present at a concentration of 1 ng/mL (instead of 0.5 ng/mL in the control without sample preparation). Remarkably, we detect proteins down to a concentration of 400 pg/mL after only one simple depletion step. Among the proteins we could analyze are also numerous disease biomarkers, including markers for multiple cancer forms, cardiovascular diseases, or Alzheimer’s disease. Remarkably, many of the biomarkers we find also could not be detected with a state-of-the-art ultrahigh-performance liquid chromatography column (which depletes the 64 most-abundant serum proteins).
It is believed that the majority of disease markers are still unidentified, since they are among the low-abundance proteins in plasma.1 However, recently, several methods have been developed to deplete high-abundance proteins from serum, thus allowing the analysis of low-abundance proteins. For instance, there are commercially available high-pressure liquid chromatography (HPLC) columns, which contain antibodies against high-abundance proteins and thus retain them in the column.2−4 While initially only a few proteins were depleted, now columns are available that deplete several tens of proteins simultaneously. An alternative is extraction with an organic solvent.5 Another approach is to use nanoparticles, which bind to certain proteins. For instance, Liu et al. used several steps of precipitation with polyethylene glycol (PEG) for this purpose, followed by depletion with one of the above-mentioned antibody columns.6 Large amounts of proteins have also been identified. However, the authors used more-complex multistep protocols.7 One alternative that does not require specific antibodies is represented by molecularly imprinted polymer particles.8 To produce these, one must imprint a polymer with the proteins that need to be depleted. However, in order to achieve this, one must know the proteins that should be depleted and have them available. This issue was solved elegantly by Yang et al.;9,10 the authors imprinted with the full bovine serum. By varying the concentration that was used for imprinting, they could tune the amount of proteins that are adsorbed.
An alternative approach for protein enrichment is combinatorial peptide ligand libraries (CPLLs).11 To produce the library, beads are coated with many different covalently attached peptides.12 These bind different proteins in the serum, which are thus removed from the sample. The remaining serum is strongly depleted of all types of proteins, including the most abundant ones. This approach does not require specific antibodies or prior knowledge and has already been successfully applied to several different samples with a complex proteome.13−15
However, despite these efforts, the depletion of high-abundance proteins still remains an issue.16 Here, we show a simple, fast, and cost-effective method to achieve high-abundance protein depletion. To achieve protein depletion, we use the fact that only some proteins bind to nanodiamonds. Our approach works similarly to CPLLs in the sense that there are particles that bind to many different proteins. However, we have the advantage that our particles are slightly simpler and, since there are no biomolecules attached, they are likely more durable. A disadvantage is probably that the surface chemistry is less complex and, thus, probably binds less proteins than the complex surface of CPLLs.
The nanodiamonds in our experiments have traditionally been used as abrasive and are thus readily available commercially. They also recently gained popularity for their magneto-optical properties17 and their use as long-term fluorescent labels,18,19 as well as their use in drug delivery.20 However, their application in depleting high-abundance proteins from plasma is entirely new.
Materials and Methods
To eliminate high-abundance proteins, nanodiamonds and NaCl were added to the serum. As a result, aggregates precipitate. Since several of the high-abundance proteins bind poorly to the diamond surface, one can deplete them by removing the supernatant. When the protein corona on the diamond surface is analyzed via mass spectrometry, we find an increased number of low-abundance proteins. For a schematic representation of the protein depletion, see Figure 1.
Figure 1.
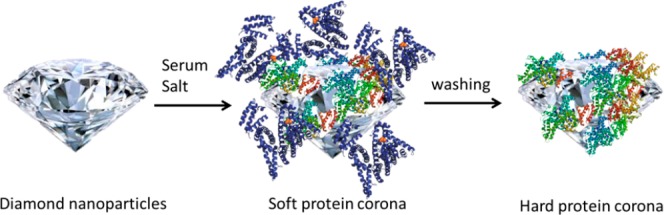
Schematic representation of the experiment: First, nanodiamonds and salts are mixed with the serum samples. Certain proteins (mostly proteins whose biological function is binding to negatively charged molecules) adhere to the diamonds. Analyzing proteins on the diamond particles reveals that high-abundance proteins were successfully depleted. At this point, loosely binding proteins, the so-called “soft corona”, is still adhered. These proteins can be removed by an additional washing step, which further depletes some proteins.
Materials
Throughout this article we used nanodiamonds with a hydrodynamic diameter of 25 nm from Microdiamant and a flakelike structure.21 They are produced by the manufacturer via grinding high-pressure high-temperature diamonds. Since the diamonds are acid-cleaned their surface contains oxygen groups.22 As a result, mostly proteins with positive domains or proteins, which, in nature, bind to negatively charged molecules, adhere to the particles. Human plasma was donated to us from the Bischoff group and stored at −80 °C in aliquots until use.
Sample Preparation
To achieve binding, we added nanodiamonds (25 nm diameter from Microdiamant) and NaCl, which were previously identified to facilitate diamond aggregation, to the serum.23 After aggregation, the samples were centrifuged (13 200 rpm for 21 min) and the supernatant was removed. These aggregates also contain loosely bound proteins, the so-called “soft corona” (which was also found on other nanoparticles24−26). The samples were then either analyzed immediately or washed. To wash the particles, the pellets were resuspended in distilled water once and centrifuged again. Subsequently, the supernatant was removed, leaving only the tightly bound proteins behind in the pellet, followed by freeze-drying. The control sample was the pure serum. To prepare the samples for mass spectrometry, they were subjected to the digesting protocol published in ref (27). Small amounts of the freeze-dried sample (and a few microliters of the control, respectively) were first treated with 20 μL of freshly prepared 10 mM dithiothreitol (DTT) in 100 mM NH4HCO3, to reduce the protein. This was followed by an incubation step at 55–60 °C for 30 min. The alkylation of the cysteines was achieved by adding 10 μL of iodoacetamide in 100 mM NH4HCO3 (incubation for 45 min). Subsequently, a second treatment with DTT followed for 30 min (to remove unreacted iodoacetamide). A trypsin digest followed by adding 20 μL of solution with 10 ng/μL trypsin (sequencing grade, Promega, Madison, WI, USA). An overnight incubation followed at 37 °C. A cleanup using SPE with C-18 cartridges followed, using a 70/30/0.1 acetonitrile/water/formic acid mixture for elution.
Sample Preparation with Carbon Black
Next, we answered whether the protein depletion is specific for diamond nanoparticles. To this end, we prepared our samples in exactly the same way as with FND, except the FNDs was replaced with carbon black.
Protein Analysis
The samples were analyzed via nanoLC–MS/MS on an Ultimate 3000 system (Dionex, Amsterdam, The Netherlands) interfaced online with a Q-ExactivePlus (Orbitrap) mass spectrometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). Peptide mixtures were loaded onto a 5 mm × 300 μm i.d. trapping microcolumn that was packed with C18 PepMAP100 5 μm particles (Dionex) in 2% acetonitrile in 0.1% formic acid at the flow rate of 20 μL/min. After loading and washing for 3 min, peptides were back-flush eluted onto a 15 cm × 75 μm i.d. nanocolumn and packed with C18 PepMAP100 1.8 μm particles (Dionex). The following mobile phase gradient (total run time: 75 min) was delivered at the flow rate of 300 nL/min: 2%–50% of solvent B in 60 min; 50%–90% B in 1 min; 90% B during 13 min, and back to 2% B in 1 min (held for 15 min). Solvent A was 100:0 H2O/acetonitrile (v/v) with 0.1% formic acid, and solvent B was 0:100 H2O/acetonitrile (v/v) with 0.1% formic acid. Peptides were infused into the mass spectrometer via dynamic nanospray probe (Thermo Fisher Scientific, Inc.) with a stainless steel emitter (Thermo Fisher Scientific, Inc.). The typical spray voltage was 1.8 kV with no sheath and auxiliary gas flow; ion transfer tube temperature was 275 °C. Mass spectrometer was operated in data-dependent mode. DDA cycle consisted of the survey scan within m/z 300–1650 at the Orbitrap analyzer with target mass resolution of 70 000 (full width at half-maximum (fwhm) at m/z 200), followed by MS/MS fragmentations of the top 10 precursor ions. Singly charged ions were excluded from MS/MS experiments and m/z of fragmented precursor ions were dynamically excluded for an additional 20 s.
Data Processing
The software PEAKS Studio version 7 (Bioinformatics Solutions, Inc., Waterloo, Canada) was applied to the spectra generated by the Q-Exactive Plus mass spectrometer to search against the protein sequence database UniProtKB/Trembl of the UniProt Knowledgebase (UniProtKB), limited to protein sequences of Homo sapiens (a search including the entire database was performed as well, to rule out the relevance of possible contaminations). Searching for the fixed modification carbamidomethylation of cysteine and the variable post-translational modifications oxidation of methionine was done with a maximum of five post-translational modifications per peptide at a parent mass error tolerance of 10 ppm and a fragment mass tolerance of 0.02 Da. The false discovery rate was set at 0.1%.
From the mass spectrometry, one obtains spectral counts. These reflect how often protein fragments are found that can be attributed to a certain protein. However, larger proteins naturally lead to more fragments. To compensate for this fact, one must calculate normalized spectral counts. These give a semiquantitative measure for the (relative) the concentration of a certain protein in the sample. The normalized spectral counts are calculated by using the following equation:28−30
| 1 |
where NpSpCk is the normalized percentage of spectral count (which is the number of spectra associated with a protein) for protein k, SpC is the spectral count identified, and MW is the molecular weight (in daltons) of the protein k.
Waterfall plots were created by comparing the protein lists with the human proteome project database (HPP-DB). The concentrations in the database reflect the current knowledge from selected references.
Results
When we precipitate proteins together with nanodiamonds in a salt-containing medium, we find that some of the most abundant serum proteins bind poorly to the nanodiamonds surface. We then used liquid chromatography coupled with mass spectrometry (LCMS) analysis to determine which proteins can be found on the diamond surface. We typically find several hundred proteins on our diamond surface. Figure 2 summarizes the depletion that we find for different media.
Figure 2.
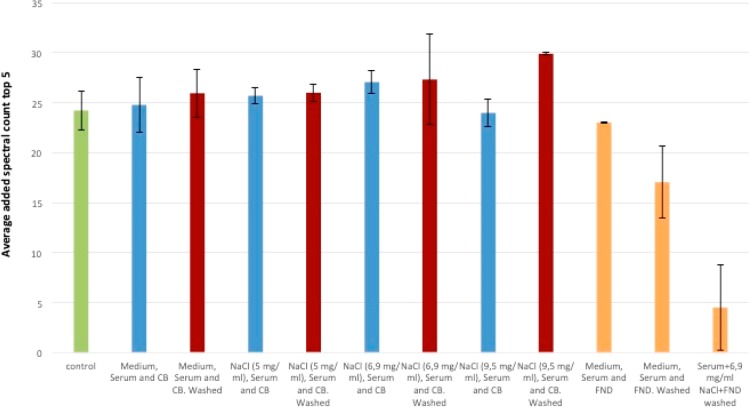
Depletion of high-abundance proteins with nanodiamonds. Compared to the control (serum without any treatment), shown in green, the amount of high-abundance proteins that is found by mass spectrometry is significantly reduced when these were previously depleted with nanodiamonds. Different media are used to precipitate protein-coated diamonds, and the depletion is compared. Error bars are generated from three different independent experiments and represent the standard error of the mean.
To generate the figure, we added the normalized spectral count values (which give a rough estimate for the concentration) for the five most-abundant proteins. The first bar (shown in green in Figure 2) represents the control, where the serum was analyzed without our method. We investigated the depletion after adding Dulbecco Modified Eagle Medium (DMEM), since this is one of the most common cell culture media. In addition, we had first found a similar depletion effect for bovine serum proteins, which are routinely used in mammalian cell cultures.16 However, as we can see here, mainly the salt component of the medium is responsible for the precipitation.
To determine the optimal conditions where most low-abundance proteins bind to the surface while high-abundance proteins remain in the supernatant, we tested different salt concentrations. The concentrations that we chose were near the physiological concentration of 6.9 mg/mL NaCl. In addition to varying the salt concentration, we also investigated the effect of washing in order to differentiate between the hard and soft protein corona. The soft corona (before washing) contains loosely and strongly binding proteins. The hard corona is what remains after washing and only contains strongly binding proteins. For most cases, we do see a small decrease in high-abundance proteins after washing. In addition to quantifying the most abundant proteins, we were also interested in the composition of the protein corona. Figure 3 shows which categories of proteins we find on which sample.
Figure 3.
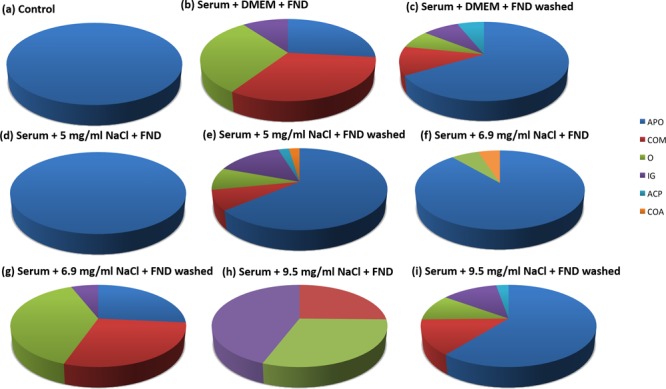
Analyzing the proteins that are found on the diamonds. Depending on the sample (panels (a)–(i)), the most prominent 50% can be assigned to different groups of proteins with different functions. [Legend: APO, apolipoproteins; COM, complement factors; O, other; IG, immunoglubulins; ACP, acute phase proteins; and COA, coagulation factors.]
The categories are chosen based on their biological function. To make this classification, we ranked the proteins from the highest concentration to the lowest concentration. We took into account all proteins in the top 50%. We chose to use the top 50% here, since, for lower-abundance proteins, these classifications are scarce or not available at all. The groups, based on biological functions that we could distinguish, are apolipoproteins (APO), complement factors (COM), other (O), immunoglubulins (IG), acute phase proteins (ACP), and coagulation factors (COA). We found large differences in the corona composition. Whereas, in the control, the top 50% of the corona consists of apolipoproteins, the diamond samples are more diverse. Most likely binding to the diamond surface occurs via electronegative oxygen groups on the diamond surface, which can interact with electropositive groups within proteins. While we could not establish a clear relationship between, for instance, binding and the isoelectric point of the proteins, we do often see proteins binding whose function in biology is to bind to electronegative structures. What we observe is similar to CPLLs, which offer a rich surface chemistry, to which proteins can bind. Similar to CPLLs, we also do not target a specific protein or a number of protein (as an antibody column does) but rather deplete anything that does not bind. Next, we compared the samples based on their ability to detect low-abundance proteins. To this end, we used so-called “waterfall plots”. To construct a waterfall plot, the protein lists are compared with the database. Figure 4 shows one of these waterfall plots, which we obtained for the best condition (serum +6.9 mg/mL NaCl + FND). The proteins in the database are plotted in order of decreasing concentrations. Every protein that is identified in the sample receives a blue dot. To illustrate the improvement, a dotted line is used to indicate the lowest concentrated protein that we could detect with the control. The proteins below that dotted line (marked with a rectangle) are only accessible with the diamond sample preparation step.
Figure 4.
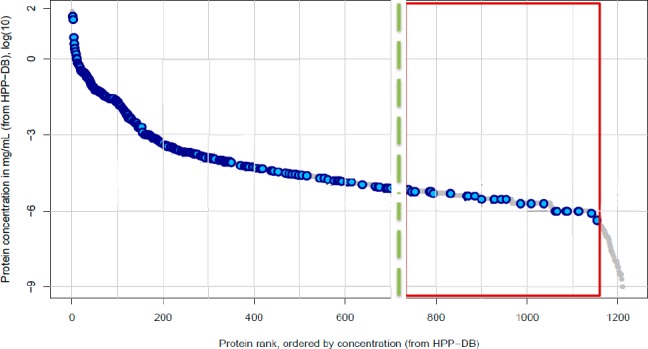
Graphical depiction of the waterfall plot: the waterfall plot lists all proteins, starting with the most-concentrated ones down to the least concentrated ones. Each blue dot indictes that the protein was found in the sample. The waterfall plot shown here is from the condition with serum +6.9 mg/mL NaCl + nanodiamonds. The dotted green line shows the detection limit for the control. All the proteins in the red square are only accessible with our sample preparation method.
Most interesting for proteomics are proteins with concentrations of <1 ng/mL. These are challenging to analyze without specialized sample preparation. In Figure 5, we compare how many of these low-abundance proteins one can find with each sample preparation method. The condition with serum +6.9 mg/mL NaCl + FND, which can reveal eight proteins, on average, gives the best results. For instance, the control only gives 0.5 proteins, on average.
Figure 5.
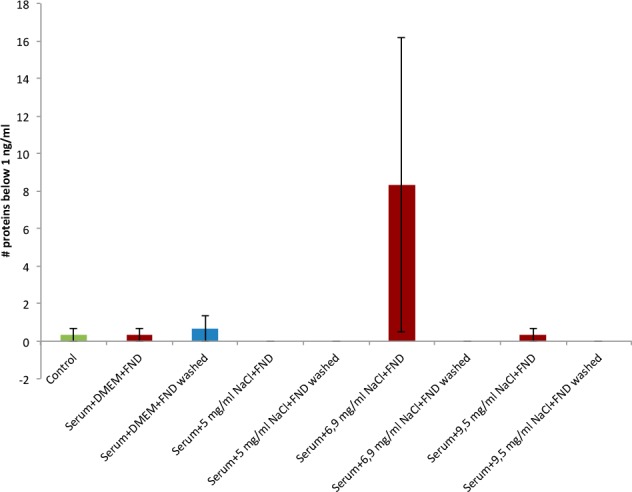
Low-abundance proteins: To demonstrate the abilities of our method, we compare the amount of proteins that were found in the samples that are below 1 ng/mL in the original plasma sample. Error bars are generated from three different independent experiments and represent the standard error of the mean.
As a final assessment of usefulness of our method, we compared the proteins that we could identify with proteins that are already used as biomarkers in the literature. Table 1 gives a few examples, which seemed to be most interesting to us.
Table 1. Examples of Proteins Identified in the Best Sample (Serum +6.9 mg/mL NaCl + FND) That Could Be Detected Neither in the Reference nor with a State-of-the-Art Depletion Column with 64 Antibodies and Their Clinical Relevance.
| protein | clinical relevance | ref |
|---|---|---|
| von Willebrand factor | Willebrand disease, the most common inherited bleeding disorder | (31) |
| Tetranectin | marker for disease activity in patients with rheumatoid arthritis | (32) |
| Proteoglycan 4 | diagnostic biomarker for COPD (chronic obstructive pulmonary disease) | (33) |
| Vitamin D-binding protein | risk factor for colorectal cancer | (34) |
| Fibulin-1 | cardiovascular risk markers in chronic kidney disease and diabetes | (35) |
| Hornerin | aberrantly expressed in breast cancer | (36) |
| Hepatocyte growth factor activator | diagnostic value for numerous diseases, as well as age and pregnancy | (37) |
| Apolipoprotein M | suspected to be a biomarker for certain diabetes types | (38) |
| Endostatin | diagnosing malignant pleural effusions, anti angiogenic agent | (39) |
| Suprabasin | tumor endothelial cell marker | (40) |
| Angiogenin | used in prediction of failure on long-term treatment response and for poor overall survival in non-Hodgkin lymphoma (a certain cancer type) | (41) |
| Desmoplakin | biomarker for Creutzfeldt–Jakob disease | (42) |
| Ribonuclease 4 | diagnosis of pancreatic cancer | (43) |
Sample Preparation with Carbon Black
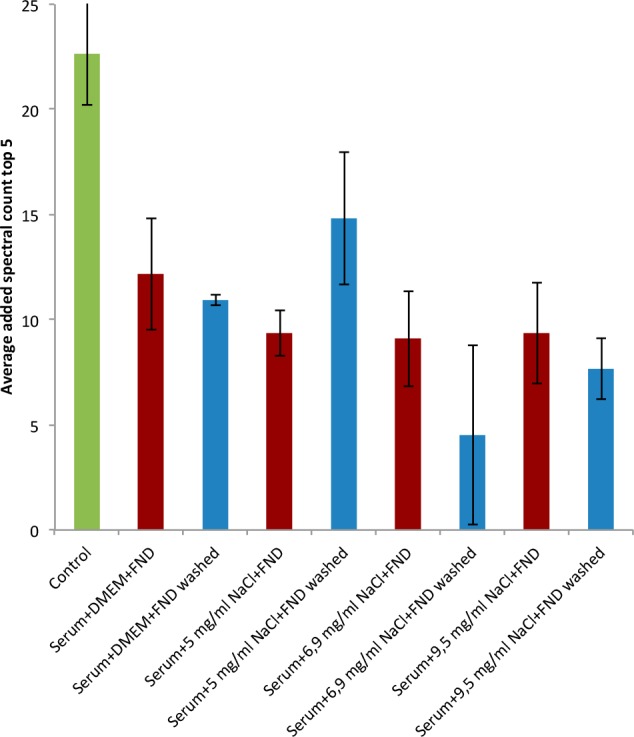
Finally, we wanted to determine if the depletion effect that we see is specific for diamond. When diamond is replaced in the above-mentioned experiments, as shown in Figure 6, we do not observe any depletion effects under any conditions. This finding indicates that the depletion of high-abundance serum proteins is indeed a peculiarity of diamond nanoparticles (or particles that resemble them). The main difference between carbon black and HPHT diamond is the content of SP2 vs SP3. While carbon black contains large amounts of SP2 (carbon black is actually more similar to graphite than it is to diamond), HPHT diamond is almost exclusively SP3 carbon. The consequence is that carbon black can interact with proteins via π–π interactions (which are not available in diamond). If such groups are exposed on the protein surface, they will interact more with carbon black. Oxygen-containing polar groups, on the other hand, are more prominent on the diamond surface. Since graphitic layers are (apart from defects) saturated and give less opportunities for oxygen-containing groups.
Figure 6.
Comparison with carbon black. Compared with the control (green, just serum), we do not observe any significant depletion for any conditions using carbon black (blue). Also, the washing step did not improve the situation (red). We added the FND samples for comparison and for a positive control.
Conclusions
While antibody-based depletion columns are generally quite expensive, nanodiamonds are surprisingly inexpensive, since they are commercially available mass products, which are used as abrasives. In addition, the depletion process is just one fast and straightforward step. While antibodies bind very specifically to a predefined target, here, we use a less-specific approach. We believe that proteins bind to specific groups on the diamond. Diamond particles provide a rich surface chemistry, which provide all types of oxygen-containing groups that (similar to a CPLL) can interact with different proteins. During our experiments, we were able to deplete high-abundance proteins significantly. As a result, we have access to low-abundance proteins for analysis, which would otherwise be undetectable. With this simple method, we were able to detect proteins down to the pg/mL range. The best results (the highest number of low-abundance proteins) that we can achieve were found when salt was added in physiological concentrations. With this approach, we are able to detect several disease biomarkers, including, among others, markers for several cancer types, cardiovascular diseases, or kidney function.
Acknowledgments
Furthermore, we would like to acknowledge support with mass spectrometry and sample preparation from Prof. Bischoff and Dr. Permentier and their team.
Glossary
ABBREVIATIONS
- APO
apolipoproteins
- COM
complement factors
- O
other
- IG
immunoglubulins
- ACP
acute phase proteins
- COA
coagulation factors
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
R.S. acknowledges financial support from FOM via Projectruimte Grant No. 15PR3229 and for an ERC Starting Grant No. 714289—Stress Imaging. F.P.M. acknowledges support from the Chilean government via a CONICYT scholarship (Grant No. 72160222).
The authors declare no competing financial interest.
References
- Polaskova V.; Kapur A.; Khan A.; Molloy M. P.; Baker M. S. High-abundance protein depletion: Comparison of methods for human plasma biomarker discovery. Electrophoresis 2010, 31, 471–482. 10.1002/elps.200900286. [DOI] [PubMed] [Google Scholar]
- Echan L. A.; Tang H.-Y.; Ali-Khan N.; Lee K.; Speicher D. W. Depletion of multiple high-abundance proteins improvesprotein profiling capacities of human serum and plasma. Proteomics 2005, 5, 3292–3303. 10.1002/pmic.200401228. [DOI] [PubMed] [Google Scholar]
- Millioni R.; Tolin S.; Puricelli L.; Sbrignadello S.; Fadini G. P.; Tessari P.; Arrigoni G. High Abundance Proteins Depletion vs Low Abundance Proteins Enrichment: Comparison of Methods to Reduce the Plasma Proteome Complexity. PLoS One 2011, 6 (5), e19603. 10.1371/journal.pone.0019603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govorukhina N. I.; Keizer-Gunnink A.; Van der Zee A. G. J.; De Jong S.; De Bruijn H. W. A.; Bischoff R. Sample preparation of human serum for the analysis of tumor markers: Comparison of different approaches for albumin and γ-globulin depletion. Journal of Chromatography A 2003, 1009 (1–2), 171–178. 10.1016/S0021-9673(03)00921-X. [DOI] [PubMed] [Google Scholar]
- Chertov O.; Biragyn A.; Kwak L. W.; Simpson J. T.; Boronina T.; Hoang V. M.; Prieto D. A.; Conrads T. P.; Veenstra T. D.; Fisher R. J. Organic solvent extraction of proteins and peptides from serum as an effective sample preparation for detection and identification of biomarkers by mass spectrometry. Proteomics 2004, 4 (4), 1195–1203. 10.1002/pmic.200300677. [DOI] [PubMed] [Google Scholar]
- Liu Z.; Fan S.; Liu H.; Yu J.; Qiao R.; Zhou M.; Yang Y.; Zhou J.; Xie P. Enhanced detection of low-abundance human plasma proteins by integrating polyethylene glycol fractionation and immunoaffinity depletion. PLoS One 2016, 11 (11), e0166306 10.1371/journal.pone.0166306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H.; Sun L.; Huang X.; Zhang P.; Zhao X. A proteomic approach for plasma biomarker discovery with 8-plex iTRAQ labeling and SCX-LC-MS/MS. Mol. Cell. Biochem. 2010, 343 (1), 91–99. 10.1007/s11010-010-0502-x. [DOI] [PubMed] [Google Scholar]
- Gao R.; Zhao S.; Hao Y.; Zhang L.; Cui X.; Liu D.; Zhang M.; Tang Y. Synthesis of magnetic dual-template molecularly imprinted nanoparticles for the specific removal of two high-abundance proteins simultaneously in blood plasma. J. Sep. Sci. 2015, 38, 3914. 10.1002/jssc.201500882. [DOI] [PubMed] [Google Scholar]
- Yang C.; Liu Y. R.; Zhang Y.; Wang J.; Tian L. L.; Yan Y. N.; Cao W. Q.; Wang Y. Y. Depletion of abundant human serum proteins by per se imprinted cryogels based on sample heterogeneity. Proteomics 2017, 17 (9), 1600284. 10.1002/pmic.201600284. [DOI] [PubMed] [Google Scholar]
- Yang C.; Zhang Y.; Cao W.-Q.; Ji X.-F.; Wang J.; Yan Y.-N.; Zhong T.-L.; Wang Y. Synthesis of Molecularly Imprinted Cryogels to Deplete Abundant Proteins from Bovine Serum. Polymers 2018, 10, 97. 10.3390/polym10010097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boschetti E.; D’Amato A.; Candiano G.; Righetti P. G. Protein biomarkers for early detection of diseases: The decisive contribution of combinatorial peptide ligand libraries. J. Proteomics 2018, 188, 1–14. 10.1016/j.jprot.2017.08.009. [DOI] [PubMed] [Google Scholar]
- Candiano G.; Santucci L.; Petretto A.; Lavarello C.; Inglese E.; Bruschi M.; Ghiggeri G. M.; Boschetti E.; Righetti P. G. Widening and diversifying the proteome capture by combinatorial peptide ligand libraries via Alcian Blue dye binding. Anal. Chem. 2015, 87 (9), 4814–4820. 10.1021/acs.analchem.5b00218. [DOI] [PubMed] [Google Scholar]
- González-García E.; Marina M. L.; García M. C.; Righetti P. G.; Fasoli E. Identification of plum and peach seed proteins by nLC-MS/MS via combinatorial peptide ligand libraries. J. Proteomics 2016, 148, 105–112. 10.1016/j.jprot.2016.07.024. [DOI] [PubMed] [Google Scholar]
- Van Vaerenbergh M.; Debyser G.; Smagghe G.; Devreese B.; de Graaf D. C. Unraveling the venom proteome of the bumblebee (Bombusterrestris) by integrating a combinatorial peptide ligand library approach with FT-ICR MS. Toxicon 2015, 102, 81–88. 10.1016/j.toxicon.2013.10.002. [DOI] [PubMed] [Google Scholar]
- Pisanu S.; Biosa G.; Carcangiu L.; Uzzau S.; Pagnozzi D. Comparative evaluation of seven commercial products for human serum enrichment/depletion by shotgun proteomics. Talanta 2018, 185, 213–220. 10.1016/j.talanta.2018.03.086. [DOI] [PubMed] [Google Scholar]
- Tu C.; Rudnick P. A.; Martinez M. Y.; Cheek K. L.; Stein S. E.; Slebos R. J. C.; Liebler D. C. Depletion of Abundant Plasma Proteins and Limitations of Plasma Proteomics. J. Proteome Res. 2010, 9 (10), 4982–4991. 10.1021/pr100646w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirhagl R.; Chang K.; Loretz M.; Degen C. L. Nitrogen-vacancy centers in diamond: nanoscale sensors for physics and biology. Annu. Rev. Phys. Chem. 2014, 65, 83–105. 10.1146/annurev-physchem-040513-103659. [DOI] [PubMed] [Google Scholar]
- Tzeng Y. K.; Faklaris O.; Chang B. M.; Kuo Y.; Hsu J. H.; Chang H. C. Superresolution imaging of albumin-conjugated fluorescent nanodiamonds in cells by stimulated emission depletion. Angew. Chem., Int. Ed. 2011, 50 (10), 2262–2265. 10.1002/anie.201007215. [DOI] [PubMed] [Google Scholar]
- Merson T. D.; Castelletto S.; Aharonovich I.; Turbic A.; Kilpatrick T. J.; Turnley A. M. Nanodiamonds with silicon vacancy defects for nontoxic photostable fluorescent labeling of neural precursor cells. Opt. Lett. 2013, 38 (20), 4170–4173. 10.1364/OL.38.004170. [DOI] [PubMed] [Google Scholar]
- Liu K. K.; Zheng W. W.; Wang C. C.; Chiu Y. C.; Cheng C. L.; Lo Y. S.; Chen C.; Chao J. I. Covalent linkage of nanodiamond-paclitaxel for drug delivery and cancer therapy. Nanotechnology 2010, 21 (31), 315106. 10.1088/0957-4484/21/31/315106. [DOI] [PubMed] [Google Scholar]
- Ong S. Y.; Chipaux M.; Nagl A.; Schirhagl R. Shape and crystallographic orientation of nanodiamonds for quantum sensing. Phys. Chem. Chem. Phys. 2017, 19 (17), 10748–10752. 10.1039/C6CP07431F. [DOI] [PubMed] [Google Scholar]
- Nagl A.; Hemelaar S.; Schirhagl R. Improving Surface and Defect Center Chemistry of Fluorescent Nano-Diamonds for Imaging Purposes – A Review. Anal. Bioanal. Chem. 2015, 407 (25), 7521–7536. 10.1007/s00216-015-8849-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemelaar S. R.; Nagl A.; Bigot F.; Rodríguez-García M. M.; de Vries M. P.; Chipaux M.; Schirhagl R. The interaction of fluorescent nanodiamond probes with cellular media. Microchim. Acta 2017, 184 (4), 1001–1009. 10.1007/s00604-017-2086-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzen S.; Schoettler S.; Baier G.; Rosenauer C.; Mailaender V.; Landfester K.; Mohr K. Complementary analysis of the hard and soft protein corona: sample preparation critically effects corona composition. Nanoscale 2015, 7, 2992–3001. 10.1039/C4NR05982D. [DOI] [PubMed] [Google Scholar]
- Tenzer S.; Docter D.; Kuharev J.; Musyanovych A.; Fetz V.; Hecht R.; Schlenk F.; Fischer D.; Kiouptsi K.; Reinhardt C.; Landfester K.; Schild H.; Maskos M.; Knauer S. K.; Stauber R. H. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat. Nanotechnol. 2013, 8, 772–781. 10.1038/nnano.2013.181. [DOI] [PubMed] [Google Scholar]
- Salvati A.; Pitek A. S.; Monopoli M. P.; Prapainop K.; Baldelli Bombelli F.; Hristov D. R.; Kelly P. M.; Aberg C.; Mahon E.; Dawson K. A. Transferrin-functionalized nanoparticles lose their targeting capabilities when a biomolecule corona adsorbs on the surface. Nat. Nanotechnol. 2013, 8 (2), 137–143. 10.1038/nnano.2012.237. [DOI] [PubMed] [Google Scholar]
- Govorukhina N. I.; Reijmers T. H.; Nyangoma S. O.; Van der Zee A. G. J.; Jansen R. C.; Bischoff R. Analysis of human serum by liquid chromatography–mass spectrometry: Improved sample preparation and data analysis. Journal of Chromatography A 2006, 1120 (1–2), 142–150. 10.1016/j.chroma.2006.02.088. [DOI] [PubMed] [Google Scholar]
- Sakulkhu U.; Maurizi L.; Mahmoudi M.; Motazacker M. M.; Vries M.; Gramoun A.; Ollivier Beuzelin M.-G.; Vallee J.-P.; Rezaee F.; Hofmann H. Nanoscale 2014, 6, 11439. 10.1039/C4NR02793K. [DOI] [PubMed] [Google Scholar]
- Sakulkhu U.; Mahmoudi M.; Maurizi L.; Coullerez G.; Hofmann-Amtenbrink M.; Vries M.; Motazacker M.; Rezaee F.; Hofmann H. Significance of surface charge and shell material of superparamagnetic iron oxide nanoparticle (SPION) based core/shell nanoparticles on the composition of the protein corona. Biomater. Sci. 2015, 3 (2), 265–278. 10.1039/C4BM00264D. [DOI] [PubMed] [Google Scholar]
- Zhu W.; Smith J. W.; Huang C. M. Mass spectrometry-based label-free quantitative proteomics. J. Biomed. Biotechnol. 2010, 2010, 2010. 10.1155/2010/840518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yawn B.; Nichols W. L.; Rick M. E. Diagnosis and management of von Willebrand disease: Guidelines for primary care. Am. Fam. Physician 2009, 80 (11), 1261–1268. [PubMed] [Google Scholar]
- Kamper E. F.; Kopeikina L. T.; Koutsoukos V.; Stavridis J. Plasma tetranectin levels and disease activity in patients with rheumatoid arthritis. J. Rheumatol. 1997, 24 (2), 262–268. [PubMed] [Google Scholar]
- Lee K. Y.; Chuang H. C.; Chen T. T.; Liu W. T.; Su C. L.; Feng P. H.; Chiang L. L.; Bien M. Y.; Ho S. C. Proteoglycan 4 is a diagnostic biomarker for COPD. Int. J. Chronic Obstruct. Pulm. Dis. 2015, 10, 1999. 10.2147/COPD.S90926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H. Q.; Sun H. L.; He B. S.; Pan Y. Q.; Wang F.; Deng Q. W.; Chen J.; Liu X.; Wang S. K. Circulating vitamin D binding protein, total, free and bioavailable 25-hydroxyvitamin D and risk of colorectal cancer. Sci. Rep. 2015, 5, 7956. 10.1038/srep07956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholze A.; Bladbjerg E. M.; Sidelmann J. J.; Diederichsen A. C.; Mickley H.; Nybo M.; Argraves W. S.; Marckmann P.; Rasmussen L. M. Plasma concentrations of extracellular matrix protein fibulin-1 are related to cardiovascular risk markers in chronic kidney disease and diabetes. Cardiovasc. Diabetol. 2013, 12 (1), 6. 10.1186/1475-2840-12-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming J. M.; Ginsburg E.; Oliver S. D.; Goldsmith P.; Vonderhaar B. K. Hornerin, an S100 family protein, is functional in breast cells and aberrantly expressed in breast cancer. BMC Cancer 2012, 12 (1), 266. 10.1186/1471-2407-12-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi H.; Nakamura T. Hepatocyte growth factor: from diagnosis to clinical applications. Clin. Chim. Acta 2003, 327 (1–2), 1–23. 10.1016/S0009-8981(02)00302-9. [DOI] [PubMed] [Google Scholar]
- Mughal S. A.; Park R.; Nowak N.; Gloyn A. L.; Karpe F.; Matile H.; Malecki M. T.; McCarthy M. I.; Stoffel M.; Owen K. R. Apolipoprotein M can discriminate HNF 1AMODY from Type 1 diabetes. Diabetic medicine 2013, 30 (2), 246–250. 10.1111/dme.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W. B.; Bai M.; Jin Y. Diagnostic value of vascular endothelial growth factor and endostatin in malignant pleural effusions. Int. J. Tuberc. Lung Dis. 2009, 13 (3), 381–386. [PubMed] [Google Scholar]
- Alam M. T.; Nagao-Kitamoto H.; Ohga N.; Akiyama K.; Maishi N.; Kawamoto T.; Shinohara N.; Taketomi A.; Shindoh M.; Hida Y.; Hida K. Suprabasin as a novel tumor endothelial cell marker. Cancer science 2014, 105 (12), 1533–1540. 10.1111/cas.12549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S.; Repo H.; Joensuu H.; Orpana A.; Salven P. High serum angiogenin at diagnosis predicts for failure on long-term treatment response and for poor overall survival in non-Hodgkin lymphoma. Eur. J. Cancer 2011, 47 (11), 1708–1716. 10.1016/j.ejca.2011.02.018. [DOI] [PubMed] [Google Scholar]
- Gawinecka J.; Ciesielczyk B.; Sanchez-Juan P.; Schmitz M.; Heinemann U.; Zerr I. Desmoplakin as a potential candidate for cerebrospinal fluid marker to rule out 14–3-3 false positive rates in sporadic Creutzfeldt-Jakob disease differential diagnosis. Neurodegener. Dis. 2012, 9 (3), 139–144. 10.1159/000334499. [DOI] [PubMed] [Google Scholar]
- Wang Y. M.; Yu S. Y. Serum ribonuclease and its isoenzymes for diagnosis of pancreatic cancer. Chin. Med. J. 1989, 102 (3), 183–187. [PubMed] [Google Scholar]