Abstract
Migratory behaviors such as the timing and duration of migration are genetically inherited and can be under strong natural selection, yet we still know very little about the specific genes or molecular pathways that control these behaviors. Studies in candidate genes Clock and Adcyap1 have revealed that both of these loci can be significantly correlated with migratory behaviors in birds, though observed relationships appear to vary across species. We investigated geographic genetic structure of Clock and Adcyap1 in four populations of blackpoll warblers (Setophaga striata), a Neotropical–Nearctic migrant that exhibits geographic variation in migratory timing and duration across its boreal breeding distribution. Further, we used data on migratory timing and duration, obtained from light‐level geolocator trackers to investigate candidate genotype–phenotype relationships at the individual level. While we found no geographic structure in either candidate gene, we did find evidence that candidate gene lengths are correlated with five of the six migratory traits. Maximum Clock allele length was significantly and negatively associated with spring arrival date. Minimum Adcyap1 allele length was significantly and negatively associated with spring departure date and positively associated with fall arrival date at the wintering grounds. Additionally, we found a significant interaction between Clock and Adcyap1 allele lengths on both spring and fall migratory duration. Adcyap1 heterozygotes also had significantly shorter migration duration in both spring and fall compared to homozygotes. Our results support the growing body of evidence that Clock and Adcyap1 allele lengths are correlated with migratory behaviors in birds.
Keywords: Adcyap1, avian migration, Clock, phenology, pituitary adenylate cyclase‐activating polypeptide
1. INTRODUCTION
Avian migration involves a suite of complex behavioral and physiological responses to exogenous seasonal cues including changes in diet and metabolism, the onset of migratory restlessness (zugunruhe), and precise directional orientation (Bairlein, Eikenaar, & Schmaljohann, 2015; Berthold, 1996; Gwinner, 1990; Jenni & Schaub, 2003). The culmination of these responses allow individual birds to accomplish amazing migratory feats, sometimes flying over ocean for several days at a time or navigating thousands of kilometers through unfamiliar habitat between wintering and breeding grounds (Bairlein et al., 2012; Battley et al., 2012; DeLuca et al., 2019, 2015; Gill et al., 2009; McKinnon, Artuso, & Love, 2017). Each of these behavioral and physiological responses is likely genetically inherited (Berthold & Pulido, 1994; Helbig, 1991; Liedvogel & Lundberg, 2014; Pulido, Berthold, Mohr, & Querner, 2001) and could be under natural selection (Gienapp, Leimu, & Merilä, 2007; Nilsson, Klaassen, & Alerstam, 2013; Pulido & Berthold, 2004, 2010). Yet we still know very little about the specific genes or molecular pathways that control these behaviors (Liedvogel, Åkesson, & Bensch, 2011). Further, it is unknown whether molecular pathways controlling migration vary among animal taxa, or even among migratory bird taxa given that migration is evolutionarily labile and likely arose independently multiple times within birds (Pulido, 2007; Zink, 2011). Investigating the genetic control of migratory behaviors in a diversity of bird taxa will therefore allow us to better understand the evolution of migration within birds, as well as ongoing natural selection on these behaviors in response to environmental change (Pulido & Berthold, 2010).
Recent studies have attempted to identify candidate genes that may be linked to migration in insects, birds, and other vertebrates (Contina, Bridge, & Kelly, 2016; Delmore et al., 2015; Johnston, Paxton, Moore, Wayne, & Smith, 2016; Lemopoulos, Uusi‐Heikkilä, Huusko, Vasemägi, & Vainikka, 2018; Lundberg et al., 2013, 2017; Merlin & Liedvogel, 2019; Mueller, Pulido, & Kempenaers, 2011; Steinmeyer, Mueller, & Kempenaers, 2009; Zhu, Gegear, Casselman, Kanginakudru, & Reppert, 2009). Two genes that have received considerable attention, especially in regard to migratory phenology, are Clock and Adcyap1. Clock plays a central role in regulating the circadian oscillator gene complex, which controls circadian and circannual rhythmicity in response to exogenous cues (Panda, Hogenesch, & Kay, 2002; Yu & Hardin, 2006). Adcyap1 codes for pituitary adenylate cyclase‐activating polypeptide (PACAP) which influences circadian rhythms in part by directly activating Clock and other genes in the circadian oscillator complex (Nagy & Csernus, 2007). PACAP also has multiple effects on physiology and behavior throughout the body (Vaudry et al., 2009), including melatonin production in the pineal gland (Csernus et al., 2004) and a role in processing light signals from the retina into neuronal signals (Hannibal et al., 1997), both of which likely play a role in the photoperiodic control of seasonality in birds (Dawson, King, Bentley, & Ball, 2001). Clock and Adcyap1, therefore, represent two candidate genes that natural selection may act on independently or in concert to shape migratory behaviors in natural populations of migratory organisms.
Several studies have shown a correlation between length polymorphisms in Clock and Adcyap1 and migratory traits in birds (Table 1). For Clock, individuals with a greater number of glutamine repeats in the 3′ polyglutamine tail (i.e., longer alleles) exhibit delayed migratory and breeding phenology (Bazzi et al., 2015; Bourret & Garant, 2015; Caprioli et al., 2012; Liedvogel, Szulkin, & Knowles, 2009; Saino et al., 2015) and longer migratory distance (Peterson et al., 2013), relative to individuals with shorter alleles. For Adcyap1, longer alleles have been shown to be associated with greater migratory restlessness (Mueller et al., 2011; Peterson et al., 2013), earlier spring arrival dates (Mettler, Segelbacher, & Schaefer, 2015), and earlier postnatal dispersal (Chakarov, Jonker, Boerner, Hoffman, & Krüger, 2013). Additionally, in one species, more northerly breeding populations had longer Adcyap1 alleles on average than southerly populations (Bazzi et al., 2016), which may reflect geographic variation in migratory strategies or phenological schedules resulting from local adaptation to environmental cues (Johnsen et al., 2007). However, the relationships between candidate genes and migratory phenotypes may also be influenced by local environmental factors such as temperature, photoperiod, and breeding density (Bourret & Garant, 2015), potentially complicating interpretations of geographic patterns within species. Further complicating the study of candidate genes and migratory traits, an increasing number of studies in birds have found no correlation between Clock or Adcyap1 allele length and migratory behavior (Table 1; Bazzi et al., 2016, 2017; Contina, Bridge, Ross, Shipley, & Kelly, 2018; Dor et al., 2012). The interspecific variation in genotype–phenotype correlations for these candidate migratory‐phenology genes highlights the challenge thus far of generalizing the expected relationship between length polymorphism and migration timing, as well as identifying the mechanism by which length variants affect migratory behaviors.
Table 1.
Studies of association between Clock and Adcyap1 allele lengths, migratory behaviors, and breeding latitude in birds
| Gene | Study | Species | Phenology | Migratory propensity | breeding latitude |
|---|---|---|---|---|---|
| Clock | Johnsen et al. (2007) | Bluethroat, Luscinia svecica | 0 | ||
| Blue tit, Cyanistes caeruleus | + | ||||
| Liedvogel et al. (2009) | Blue tit, Cyanistes caeruleus | +/0a | |||
| Liedvogel and Sheldon (2010) | Great tit, Parus major | 0 | |||
| Dor et al. (2011) | Barn swallow, Hirundo rustica | 0 | |||
| Mueller et al. (2011) | Eurasian blackcaps, Sylvia atricapilla | 0 | |||
| Caprioli et al. (2012) | Barn swallow, Hirundo rustica | +/0a | |||
| Dor et al. (2012) | Swallows, Tachycineta | 0 | 0 | ||
| Chakarov et al. (2013) | Common buzzard, Buteo buteo | 0 | |||
| Peterson et al. (2013) | Dark‐eyed junco, Junco hyemalis | + | |||
| Kuhn et al. (2013) | Pied flycatcher, Ficedula hypoleuca | 0 | 0 | ||
| Bazzi et al. (2015) | Barn swallow, Hirundo rustica | + | |||
| Bourret and Garant (2015) | Tree swallow, Tachycineta bicolor | + | |||
| Saino et al. (2015) | Nightingale, Luscinia megarhynchos | + | |||
| Pied flycatcher, Ficedula hypolecua | 0 | ||||
| Tree pipit, Anthus trivialis | +/0a | ||||
| Winchat, Saxicola ruberta | 0 | ||||
| Bazzi et al. (2016) | WILSON'S warbler, Cardellina pusilla | 0 | 0 | ||
| Bazzi et al. (2017) | Willow warbler, Phylloscopus trochilus | 0 | |||
| Contina et al. (2018) | Painted bunting, Passerina ciris | 0 | 0 | ||
| Romano et al. (2018) | Yellow‐legged gull, Larus michahellis | 0 | |||
| Adcyap1 | Mueller et al. (2011) | Eurasian blackcaps, Sylvia atricapilla | + | ||
| Chakarov et al. (2013) | Common buzzard, Buteo buteo | − | |||
| Peterson et al. (2013) | Dark‐eyed junco, Junco hyemalis | + | |||
| Bourret and Garant (2015) | Tree swallow, Tachycineta bicolor | −/+b | |||
| Mettler et al. (2015) | Eurasian blackcaps, Sylvia atricapilla | −/0a | |||
| Saino et al. (2015) | Nightingale, Luscinia megarhynchos | 0 | |||
| Pied flycatcher, Ficedula hypolecua | 0 | ||||
| Tree pipit, Anthus trivialis | 0 | ||||
| Winchat, Saxicola ruberta | 0 | ||||
| Bazzi et al. (2016) | Wilson's warbler, Cardellina pusilla | 0 | 0/+ | ||
| Bazzi et al. (2017) | willow warbler, Phylloscopus trochilus | 0 | |||
| Contina et al. (2018) | Painted bunting, Passerina ciris | 0 | 0 | ||
| Romano et al. (2018) | Yellow‐legged gull, Larus michahellis | 0 |
Migratory propensity includes studies of migratory restlessness, distance, and duration. Symbols indicate positive (+), negative (−), or no observed relationships (0).
Relationships that varied by sex (F/M).
Relationships that varied by latitude (low latitude/high latitude).
Given the potential evolutionary and ecological importance of these migratory candidate genes, and the variation in observed patterns across species, it is valuable to continue to build evidence to either support or refute a role of these genes in migratory behavior in different avian species, particularly to help understand why patterns vary across species. Here, we contribute to this growing number of studies by examining relationship between Adcyap1 and Clock and migratory behavior in the blackpoll warbler (Setophaga striata), a small (12 g) long distance Neotropical–Nearctic migrant (Figure 1) with geographic variation in migratory behaviors (DeLuca, Holberton, Hunt, & Eliason, 2013; DeLuca et al., 2015; Morris, Covino, Jacobs, & Taylor, 2016).
Figure 1.

Blackpoll warblers breed across the North American boreal forest and winter in tropical South America. Left: adult breeding male captured in 2010 in New Brunswick. Photo credit: J Ralston. Right: adult breeding male captured in Yukon Territories in 2018, and affixed with a light‐level geolocator. Photo credit: H A Cooke
Blackpoll warblers breed across the North American boreal forest from Alaska to Newfoundland and in isolated montane fir forests at the southern periphery of their range in northeastern United States (Figure 2; DeLuca et al., 2013). Annual blackpoll warbler migration between the northern breeding grounds and wintering grounds in the Amazon basin of northern South America is one of the longest migratory routes for any North American songbird (DeLuca et al., 2013; Morris et al., 2016), and was recently tracked from geographically distant breeding populations using light‐level geolocators (DeLuca et al., 2015, 2019; Figure 1). Birds depart the wintering grounds in mid‐April to mid‐May and travel through the Greater Antilles and continental United States to their northern breeding territories, a trip that can vary in duration considerably depending on breeding location (mean 34 days, range 17–49 days; DeLuca et al., 2019). After departing the breeding grounds in August through October, blackpoll warblers first migrate southeastward to the Atlantic coast of United States and Canada, then migrate south over the Atlantic Ocean to their South American wintering grounds, a nonstop trans‐oceanic flight of up to 3,000 km that may take up to three days (DeLuca et al., 2015, 2019).
Figure 2.
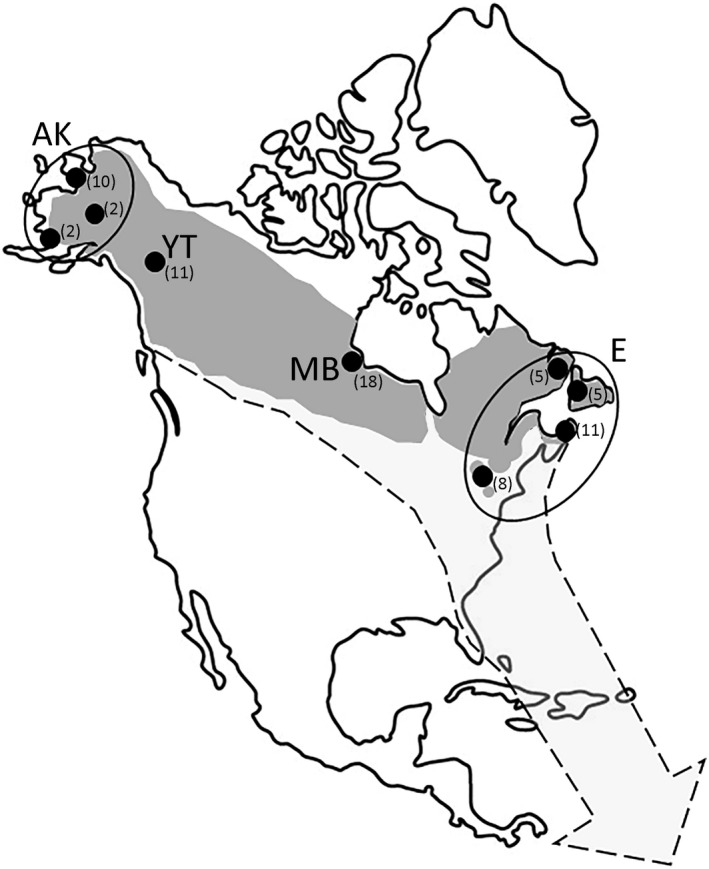
Breeding distribution (dark gray) and fall migratory route (light gray with dashed outline) for blackpoll warblers following DeLuca et al. (2013, 2019, 2015). Sample location is shown with black circles with sample sizes in parentheses. AK, Alaska; YT, Whitehorse, Yukon Territories; MB, Churchill, Manitoba; E, Eastern population. Three sampling sites were grouped together into the AK population: Nome, Denali National Park, and southwest Alaska. Four sampling populations were grouped together into the E population: Adirondack and Catskill Mountains, New York; Cape Breton, Nova Scotia; western Newfoundland; southeastern Labrador
The large breeding range means that blackpoll warblers from different breeding localities vary in the timing, direction, distance, and duration of their migrations. Birds that breed further north and west migrate further and take longer; these populations tend to depart the wintering grounds earlier but arrive on the breeding grounds at similar times compared to birds breeding further east (DeLuca et al., 2015, 2019). Like in the spring, fall migration timing and duration varies across breeding populations with western breeding birds departing earlier and taking longer to arrive at wintering grounds compared to eastern breeding birds (DeLuca et al., 2015, 2019).
We assessed variation in Clock and Adcyap1 across four breeding populations of blackpoll warbler and tested for correlations between allele lengths and the timing of departure, arrival, and duration in both spring and fall migratory periods. Based on published evidence from other species (Table 1), we predicted allele lengths in both Clock and Adcyap1 would be positively associated with migratory distance and duration. We predicted Clock would have a positive relationship with the timing of migration (departure and arrival dates), and Adcyap1 would have a negative relationship with migratory timing.
2. MATERIALS AND METHODS
2.1. Genetic sampling
We collected Blackpoll Warbler blood and tissue samples from throughout the breeding range using mist nets during the May–July breeding seasons from 2009 to 2019 (n = 64). Blood samples taken in the field were collected using brachial venipuncture and were stored in lysis buffer at ambient temperature until they were transported to a genetics laboratory where they were stored at −20°C. Samples collected in the field were supplemented with a small number of frozen tissues from natural history museum collections, also collected from the breeding seasons from 2008 to 2009 (n = 8, Appendix 1). Our total sample (n = 72) included only breeding adults, no nestlings, or known relatives, and was mostly male (64/72, 89% males). We extracted DNA from all samples using a DNeasy Blood and Tissue Extraction Kit (Qiagen, Valencia, CA) following the manufacturer's protocol with one modification: Final elution was conducted in two rounds of 75 μl. All blood samples and DNA extracts used in the current study, including those for geolocator tagged birds, are archived at Saint Mary's College, Notre Dame, Indiana, USA, or at New York State Museum, Albany, New York, USA.
We amplified Clock and Adcyap1 loci in 10 μl polymerase chain reactions (PCRs) using a Qiagen Multiplex PCR Master Mix (HotStar Taq DNA Polymerase, dNTP mix, final concentration of 3.0 mM MgCl2), 0.2 μM reverse and fluorescent‐labeled forward primers (Applied Biosystems, Foster City, CA), and <200 ng of template DNA. Primer sequences were obtained from Johnsen et al. (2007) and Steinmeyer et al. (2009). Our PCR thermal regime included a 15‐min initial denaturation at 95°C; 35 cycles of 30‐s denaturation at 94°C, 90‐s primer annealing at 54°C, and 90‐s extension at 72°C; and a final extension period of 10 min at 72°C. Final PCR products were mixed with LIZ500 size standard (Thermo Fisher Scientific), diluted in Hi‐Di formamide, and sent to the University of Notre Dame Genomics and Bioinformatics Core Facility for genotyping on an Applied Biosystems 3730xl Genetic Analyzer. We used Peak Scanner (Applied Biosystems) to examine electropherograms and estimate the sizes of the alleles (measurement unit = number of base pairs) from each individual at Clock and Adcyap1 loci.
2.2. Analysis of geographic variation
We grouped samples into four populations for analysis: Alaska (n = 14), Yukon (n = 11), Manitoba (n = 18), and Eastern (n = 29). The Alaska population included samples from Nome (n = 10), Denali National Park (n = 2), and southwest Alaska (n = 2). Four sampling locations were grouped together into the Eastern population: Adirondack and Catskill Mountains, New York (n = 8); Cape Breton, Nova Scotia (n = 11); western Newfoundland (n = 5); and southeastern Labrador (n = 5). All samples from Yukon are from Whitehorse, Yukon Territories, and all samples from Manitoba are from Churchill, Manitoba. Previous analysis of neutral loci revealed significant genetic structure in mitochondrial DNA marker between Newfoundland and other Eastern subpopulations (Ralston & Kirchman, 2012), but no significant structure and a large number of shared alleles at neutral microsatellite markers (Ralston & Kirchman, 2013). Due to the small sample size of Newfoundland in the current study, we group these samples within the Eastern population. We used the package hierfstat (Goudet, 2004) in program R version 3.5.5 (R Core Team, 2019) to calculate overall F ST and F IS. We used the program ARLEQUIN version 3.5 (Excoffier & Lischer, 2010) to test for deviation from Hardy–Weinberg equilibrium, evidence of genetic linkage disequilibrium between the two loci, and to assess genetic population differentiation using pairwise F ST and an analysis of molecular variance (AMOVA). Significance for each test was determined using default ARLEQUIN settings for number of permutations and number of steps in Markov chain Monte Carlo (MCMC) algorithms.
We further investigated geographic structure at these loci using the program STRUCTURE version 2.3.4 (Pritchard, Stephens, & Donnelly, 2000), which uses a Bayesian clustering analysis to determine the most likely number of populations (K). We used an admixture model with an assumption of correlated allele frequencies among populations, and population of each sample as a prior. We ran 10 iterations for each K ranging from 1 to 4 with 1,000,000 MCMC steps following a burn‐in of 100,000 steps, and used mean natural log probability to determine the most likely number of populations. We used the program DISTRUCT (Rosenberg, 2004) to visualize population assignment of each individual from the STRUCTURE run with the greatest log probability for each value of K. Additionally, we performed a principal components analysis (PCA) implemented in the R package ade4 (Dray & Dufour, 2007). We visualized clustering of genotypes in ordination space using principal components with eigenvalues greater than 1.0.
While the above genetic analyses assess variation in the frequencies of alleles across populations, they do not directly assess patterns in allele length per se. In other words, in the above analyses, alleles are treated as categorical, while we are also interested in allele length as a continuous variable across geography. We therefore also assessed variation in allele lengths across populations, as well as across latitude and longitude. For these population‐level analyses, we consider latitude and longitude as proxies of migratory behaviors, specifically migratory distance, with the understanding that populations that breed further north and west migrate further each year. We used a series of general linear models (GLMs) with longitude and latitude as the predictor variables to determine whether allele length varied across either sampling latitude or longitude. It is unknown whether there are any allelic interactions or dominance effects at these loci relative to the migratory traits of interest, though previous studies have suggested dominance of the longer allele in Clock (Saino et al., 2015) and other genes with 3′ polyglutamine repeats (Ross, 2002). Due to small sample sizes per genotype, and high variability in Adcyap1, we could not directly compare genotypes for dominance effects (as in Liedvogel et al., 2009; Saino et al., 2015). However, we did run separate GLMs using either minimum allele, maximum allele, or mean allele length for each individual as the dependent variable. If longer Clock alleles are dominant, we would expect to see stronger relationships between individuals' maximum allele length and migratory traits.
2.3. Analysis of candidate genes relative to individual migratory traits
To investigate correlations between candidate genes and migratory behaviors at the individual level, we combined genotype data described above with migratory data obtained from light‐level geolocators. In a separate study specifically addressing migratory routes and phenology, DeLuca et al. (2019) tracked migration in blackpoll warblers breeding in Alaska, Yukon, and Manitoba with Biotrack model MK‐6 light‐level geolocators (Wareham, UK), and took blood samples from tagged individuals for the current study (Alaska n = 5, Yukon n = 4, Manitoba n = 8). All individuals used in this analysis were unrelated breeding adult males. Full details on the methods of geolocator deployment and the analysis of geolocator track data are available in DeLuca et al. (2019). For the current study, we analyze six migratory traits related to phenology and duration of migration: (a) spring departure date from wintering grounds, (b) duration of spring migration, (c) spring arrival date on the breeding grounds, (d) fall departure date from breeding grounds, (e) duration of fall migration, and (f) fall arrival date on the wintering grounds.
We tested for correlations between Clock and Adcyap1 allele lengths and each of the six migratory traits using GLMs. To account for known variation in migratory traits across populations (DeLuca et al., 2019), we include population, coded as a factor, as a predictor variable in the models. Like in the population‐level analyses, we built separate GLMs using either minimum allele, maximum allele, or mean allele length for each individual. For six individuals, geolocator track information was not available from the spring migration period. Therefore, analyses comparing spring departure date, spring duration, or spring arrival date with allele length had lower sample sizes (n = 12, n = 12, n = 11, respectively; n = 17 for all fall migratory traits). We repeated these analyses using longitude and latitude as covariables instead of population to determine whether our results were robust to this selection.
Next, we tested for an interaction between Clock and Adcyap1 on each of the six migratory traits using GLMs with population, Clock allele length, Adcyap1 allele length, and a Clock × Adcyap1 interaction term as the independent variables. So as to not vastly increase our number of tests by comparing all possible allele length combinations of Clock and Adcyap1, we only tested GLMs with interactions between allele lengths that showed significant or marginally significant correlations in the individual gene models described above (see Section 3). If we found a significant Clock × Adcyap1 interaction effect on a migratory trait, we do not further discuss single gene effects for this migratory trait.
Lastly, we compared each of the six migratory traits between homozygous and heterozygous individuals for both Clock and Adcyap1 using GLMs with location and heterozygosity as the predictor variables. If heterozygosity correlates with individual fitness and migratory performance (Mettler et al., 2015), we would expect heterozygotes to have significantly shorter migratory durations and earlier arrival times. While several studies have shown sex‐specific effects of candidate genes on phenology (e.g., Mettler et al., 2015; Bazzi et al., 2017), we could not test for sex effects because all 17 geolocator birds were male.
3. RESULTS
We successfully genotyped 72 Blackpoll Warblers at candidate genes Clock and Adcyap1. We observed four Clock alleles ranging in size from 186 to 195 base pairs (Table 2). We found relatively greater variation in Adcyap1; we detected 13 Adcyap1 alleles ranging from 153 to 169 base pairs, five of which were found in single individuals (Table 2). Observed heterozygosity was fairly high (H o = 0.722 for each locus), with only five individuals being homozygous at both loci. None of the populations deviated from Hardy–Weinberg equilibrium (all p > 0.05, Table 3), and we found no evidence of linkage disequilibrium (p > 0.05).
Table 2.
Allele frequencies for two candidate genes, Clock and Adcyap1, at four populations of blackpoll warblers
| Population | Clock allele lengths | Adcyap1 allele lengths | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 186 | 189 | 192 | 195 | 153 | 155 | 157 | 159 | 160 | 161 | 162 | 163 | 164 | 165 | 166 | 167 | 169 | |
| Alaska | 5 | 12 | 4 | 7 | 0 | 1 | 4 | 13 | 0 | 4 | 0 | 4 | 1 | 1 | 0 | 0 | 0 |
| Yukon | 0 | 11 | 5 | 6 | 1 | 2 | 6 | 7 | 0 | 4 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| Manitoba | 4 | 13 | 13 | 6 | 0 | 3 | 6 | 14 | 0 | 4 | 0 | 4 | 0 | 1 | 1 | 2 | 1 |
| Eastern | 6 | 20 | 15 | 17 | 0 | 1 | 10 | 17 | 1 | 19 | 1 | 5 | 2 | 1 | 0 | 1 | 0 |
Allele numbers represent lengths in number of base pairs.
Table 3.
Summary statistics for each candidate gene, Clock and Adcyap1, for four populations of blackpoll warbler
| Population | n | Clock | Adcyap1 | Pairwise F ST | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| H o | H e | A | H o | H e | A | Alaska | Yukon | Manitoba | ||
| Alaska | 14 | 0.571 | 0.728 | 4 | 0.714 | 0.746 | 7 | |||
| Yukon | 11 | 0.636 | 0.654 | 3 | 0.636 | 0.810 | 6 | 0.041 | ||
| Manitoba | 18 | 0.667 | 0.719 | 4 | 0.722 | 0.806 | 9 | −0.027 | 0.040 | |
| Eastern | 29 | 0.862 | 0.730 | 4 | 0.759 | 0.780 | 10 | −0.001 | 0.049 | −0.016 |
Sample size (n), observed (H o) and expected heterozygosity (H e), number of alleles (A) per locus, and pairwise F ST. None of the H o were statistically different from H e, and no pairwise F ST were significant.
We found no evidence of structured geographic variation in either Clock or Adcyap1. Overall F ST was −0.003, and F IS was 0.070. All pairwise F ST values between populations were nonsignificant (Table 3), and the AMOVA revealed that only 0.72% of the variation in candidate genes was explained by difference among populations, while 99.28% corresponded to variation within populations. The STRUCTURE analysis, similarly, suggested no geographic structure with K = 1 as the most likely number of groups (Figure 3a). For all values of K, the assignment probability of each individual was roughly equal for all populations (i.e., the probability of any individual belonging to any of K populations ≈ 1/K; Figure 3b). The first two principal components from a PCA on genotype data had eigenvalues greater than 1.0 (1.61 and 1.26, respectively) and together explained 71.68% of the variation in genotype data. A PCA plot on these axes revealed no clustering by population. All populations overlapped in ordination space (Figure 3c). We found no significant relationship between allele length for either locus and either longitude or latitude, regardless of whether individual minimum, maximum, or mean allele lengths were considered (all p > 0.05, all R 2 values <0.05; Appendix 2). From our GLMs, we found a significant correlation between population and spring and fall departure, and spring and fall duration, but not with spring and fall arrival. These results are not explored further here, as population variation in migratory timing is more thoroughly discussed in DeLuca et al. (2019).
Figure 3.
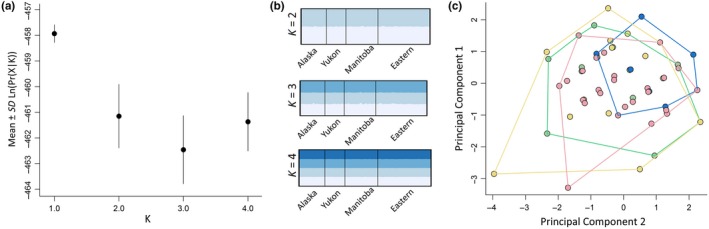
(a) Mean log probability for each number of populations (K) from 10 replicates in program STRUCTURE. Error bars indicate standard deviation. K = 1 was the most likely number of populations indicating no geographic structure. (b) Individual probability assignment to each population when K > 1. For all values of K, the assignment probability of each individual was roughly equal for all populations indicating no geographic structure. (c) Clustering of individuals according to a principal components analysis on genotype data. Colors indicate populations (Alaska = green, Yukon = blue, Manitoba = yellow, and Eastern = pink), and a minimum convex polygon was drawn around each population. All population overlap with no clustering based on population, indicating no geographic structure
Despite finding no evidence of geographic variation in candidate genes, we did find evidence that both Clock and Adcyap1 allele length were correlated with migratory behaviors at an individual level (Table 4). Maximum Clock allele length was significantly and negatively associated with spring arrival date on the breeding ground (GLM: F 3,7 = 9.466, p = 0.007, R 2 = 0.718; β MaxClock = −1.25, p MaxClock = 0.024); individuals with longer Clock alleles tended to arrive to the breeding territories earlier in the spring (Figure 4). We found no other correlations for Clock allele length and the other five migration traits from our single gene models (Appendix 3). We found significant correlation between individual's minimum Adcyap1 allele length and the timing of spring and fall migration. Blackpoll warblers with longer minimum Adcyap1 alleles departed earlier in the spring (F 3,8 = 6.936, p = 0.013, R 2 = 0.618; β MinAdcyap1 = −5.615, p MinAdcyap1 = 0.007) and arrived later to the southern wintering grounds (F 3,13 = 2.418, p = 0.113, R 2 = 0.210; β MinAdcyap1 = 2.122, p MinAdcyap1 = 0.043; Figure 5). A single individual that was homozygous for the longest minimum Adcyap1 allele had by far the earliest spring departure, the longest spring and fall duration, and the latest fall arrival. Removal of this individual from GLM analyses resulted in nonsignificant relationships between minimum Adcyap1 allele length and both spring departure (F 3,7 = 8.531, p = 0.010, R 2 = 0.693; β MinAdcyap1 = −3.239, p MinAdcyap1 = 0.088) and fall arrival (F 3,12 = 2.469, p = 0.112, R 2 = 0.227; β MinAdcyap1 = 0.580, p MinAdcyap1 = 0.535). The correlation between Adcyap1 and fall arrival was also significant when considering individuals' mean allele length (Table 4). Results were consistent when using longitude and latitude as covariates instead of population.
Table 4.
General linear model results for individual‐level analyses of blackpoll warbler candidate gene alleles and migratory traits
| Gene | Migratory trait | Allele | n | β Allele Length | SE | p Allele Length | R 2 Allele Length |
|---|---|---|---|---|---|---|---|
| Clock | Spring arrival | Max | 11 | −1.250 | 0.438 | 0.024 | 0.538 |
| Adcyap1 | Spring departure | Min | 12 | −5.615 | 1.577 | 0.007 | 0.618 |
| Spring duration | Min | 12 | 5.123 | 1.657 | 0.015* | 0.544 | |
| Fall duration | Min | 17 | 1.941 | 0.588 | 0.006* | 0.456 | |
| Mean | 17 | 1.844 | 0.797 | 0.038* | 0.292 | ||
| Fall arrival | Min | 17 | 2.122 | 0.945 | 0.043 | 0.279 | |
| Mean | 17 | 2.889 | 1.053 | 0.017 | 0.367 |
Allele indicates whether an individual's minimum, maximum, or mean allele length was used for each analysis. Slope (β), p‐value, and partial R 2 values are given for allele length and each migratory trait. Population was also used as a predictor variable, though the slopes and p‐values for this factor are not provided. Only models for which allele length was significant (p ≤ 0.05) are shown. Results for all general linear models provided in Appendix 3.
Significant single gene effects that are not further discussed because of a significant genetic interaction effect on this migratory trait (see Table 5).
Figure 4.
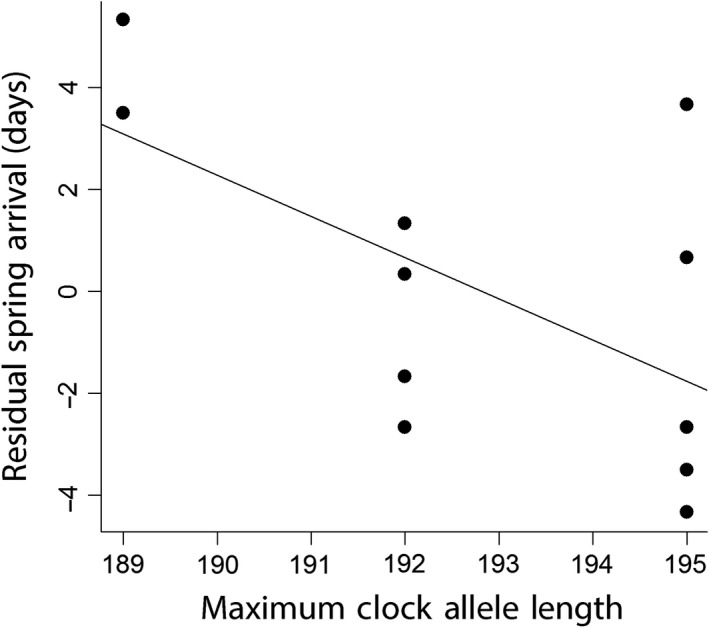
Relationship between individual maximum Clock allele length and spring arrival date residuals after accounting for population. Line represents slope from the general linear models with population and maximum Clock allele as independent variables. Individuals with longer maximum Clock allele lengths arrived earlier for their population compared to individuals with shorter maximum Clock alleles
Figure 5.
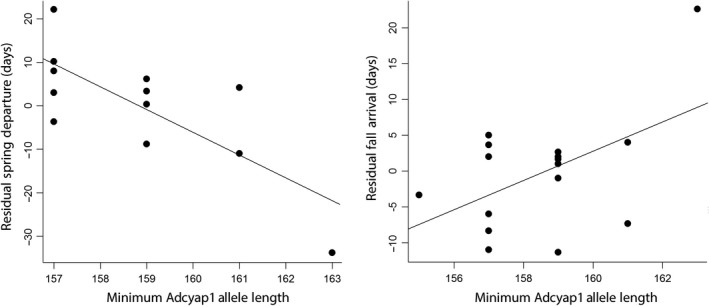
Relationship between individual minimum Adcyap1 allele length and the residuals of spring departure and fall arrival after accounting for population. Lines represent slopes from general linear models with population and minimum Adcyap1 allele length as independent variables. Individuals with longer minimum Adcyap1 allele lengths departed earlier in the spring and arrived earlier in the fall for their population compared to individuals with shorter minimum Adcyap1 alleles
We also found an interaction effect between Clock and Adcyap1 allele lengths on migratory duration (Figure 6). When we used mean Clock allele length and minimum Adcyap1 allele length, this interaction was significantly correlated with both spring duration (F 5,6 = 10.15, p = 0.007, R 2 = 0.806; p MeanClock = 0.020, p MinAdcyap1 = 0.018, p MeanClock × MinAdcyap1 = 0.019) and fall duration (F 5,11 = 22.38, p < 0.001, R 2 = 0.870; p MeanClock = 0.007, p MinAdcyap1 = 0.007, p MeanClock × MinAdcyap1 = 0.007). For individuals with shorter‐than‐average mean Clock allele lengths, both spring duration and fall duration were determined by a significant positive relationship with minimum Adcyap1 allele length. For individuals with longer‐than‐average mean Clock allele lengths, duration was not correlated with Adcyap1 length (Figure 6). When the individual with the longest minimum Adcyap1 allele is removed from the analyses, this relationship becomes nonsignificant for spring duration (F 5,5 = 5.538, p = 0.042, R 2 = 0.694; pMeanClock × MinAdcyap1 = 0.120), but remains significant for fall duration (F 5,10 = 23.37, p < 0.001, R 2 = 0.882; p MeanClock × MinAdcyap1 = 0.011).This Clock × Adcyap1 interaction effect was weaker when using either maximum Clock allele length or mean Adcyap1 allele length (Table 5).
Figure 6.
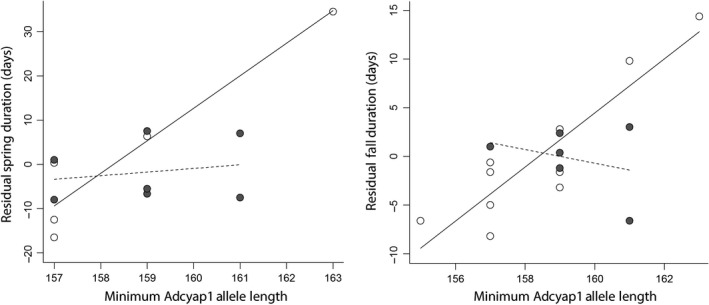
GLM results for an interaction effect between mean Clock and minimum Adcyap1 allele lengths on spring and fall migratory duration. White circles and solid lines represent individuals with shorter‐than‐average mean Clock allele lengths, and gray circles and dashed lines represent individuals with longer‐than‐average mean Clock allele lengths. Gene interactions were significantly correlated (p < 0.05) with both spring and fall duration. Migratory duration shows a significant positive relationship with minimum Adcyap1 allele length in both seasons when Clock alleles are short, but not when Clock alleles are long
Table 5.
General linear model results for interaction effects between candidate genes on spring and fall duration
| Clock allele | Adcyap1 allele | Migratory trait | βClock × Adcyap1 | pClock × Adcyap1 |
|---|---|---|---|---|
| Mean | Min | Spring duration | −1.910 | 0.019 |
| Fall duration | −0.740 | 0.007 | ||
| Mean | Spring duration | −0.143 | 0.934 | |
| Fall duration | −0.720 | 0.029 | ||
| Max | Min | Spring duration | −1.450 | 0.056 |
| Fall duration | −0.605 | 0.042 | ||
| Mean | Spring duration | 0.658 | 0.671 | |
| Fall duration | −0.558 | 0.08 |
Clock and Adcyap1 allele columns indicate whether an individual's minimum, maximum, or mean allele length was used for each analysis. Slope (β) and p‐value are given for the interaction term. Population was also used as a predictor variable, though the slope and p‐values for this factor are not provided. GLMs were only run using mean or max Clock allele lengths and minimum or mean Adcyap1 allele lengths based on results from single gene models. Only models with spring or fall duration as the dependent variable are presented here; no other migratory traits had a significant interaction term. Models with significant interaction effects (p ≤ 0.05) are bolded.
Lastly, we tested for differences in each of the migratory traits between homozygotes and heterozygotes of each locus. We found individuals that were heterozygous at Adcyap1 had significantly shorter spring duration after accounting for population origin in a GLM (mean homozygote residuals = 8.47; mean heterozygote residuals = −8.47, p = 0.030) and significantly shorter fall duration (mean homozygote residuals = 3.26; mean heterozygote residuals = −2.90, p = 0.031) compared to homozygotes.
4. DISCUSSION
We found evidence suggesting allele lengths in candidate genes Clock and Adcyap1 are correlated with spring and fall migratory behavior in blackpoll warblers. Despite no geographic variation across populations in allele frequencies or lengths, variation in candidate gene allele lengths was associated with nearly every migratory trait analyzed at the individual level. It is worth noting that we conducted a large number of statistical tests (N = 78 GLMs), increasing the chance of type I errors. However, with α = 0.05, we would we expect an average of 3.7 of the 78 tests to yield significant results at random. We find significant correlations (p < 0.05) in 12 cases, significantly more than expected at random (binomial test, p < 0.001). Given that the observed relationships are mostly in agreement with predicted direction of relationships, and are consistent with studies in other species, it is likely that our results represent real relationships between the variables and not spurious statistical effects. While sample sizes are low given the low recovery rates of geolocator‐fitted birds (DeLuca et al., 2019), future work that aims to increase sample sizes may allow more rigorous evaluation of links between candidate genes and migration behavior. For example, with more birds, analyses could control for other factors known to influence migratory timing and speed, such as age, sex, or environmental conditions experienced during and leading up to migration, which could obscure genetic contributions to migration phenology.
The only consistent effect we found for Clock alleles was a negative correlation with spring arrival date (Figure 4, Table 4). This was a surprising result as all other published studies that report a significant effect of Clock allele length on spring phenology in birds report a positive correlation (Table 1; Bazzi et al., 2015; Bourret & Garant, 2015; Caprioli et al., 2012; Liedvogel et al., 2009; Saino et al., 2015). Why blackpoll warblers would exhibit an opposite pattern at this locus is unclear, though this highlights the emerging pattern of variation across species in migratory genotype–phenotype relationships and a need for further detailed studies that investigate the functional influence of candidate gene alleles. Previous studies have suggested that longer Clock alleles are dominant over shorter alleles in their influence on phenology (Bazzi et al., 2015; Saino et al., 2015). Our results seem to support this given that we find a stronger relationship between maximum Clock allele length and spring arrival time, compared to minimum Clock alleles.
Our results showed longer minimum Adcyap1 alleles were associated with earlier spring departure and later fall arrival on the wintering grounds. These results are generally consistent with work previously published on other species (Table 1). While no other studies to our knowledge have shown a significant correlation between Adcyap1 and arrival date on the wintering grounds as we show here, these results are consistent with an effect of Adcyap1 on longer fall migration duration (Mueller et al., 2011; Peterson et al., 2013). The strongest relationships between migratory traits and Adcyap1 were with an individual's minimum allele length (Appendix 3). This may suggest dominance of the shortest Adcyap1 allele, a finding not reported in other species. However, the one individual in our sample that was homozygous for the longest Adcyap1 allele (163 bp) had by far the earliest spring departure, the longest spring and fall duration, and the latest fall arrival. This may support that the effects of long Adcyap1 alleles on migratory behavior are additive instead of dominant, as was previously suggested by Bourret and Garant (2015). Future studies with larger sample sizes may be able to more directly assess dominance by comparing migratory traits across genotypes at the individual level (Liedvogel et al., 2009; Saino et al., 2015). Our results that a single individual with a rare genotype differs greatly in migratory behavior are also consistent with studies published on other species that show a few individuals with rare genotypes can show significantly different migratory traits compared to the rest of the population (Bazzi et al., 2015). For example, a single individual barn swallow with the longest observed Clock allele had significantly later migration in both spring and fall compared to the rest of the population (Bazzi et al., 2015).
We found evidence of a significant interaction between Clock and Adcyap1 allele lengths on migratory duration, the first such finding in studies of migratory birds to our knowledge. Previous studies that have investigated the effects of both Clock and Adcyap1 either did not find a significant interaction between genes or did not test for one (Bazzi et al., 2016, 2017; Bourret & Garant, 2015; Contina et al., 2018; Peterson et al., 2013; Saino et al., 2015). In this interaction in our study, Adcyap1 length appears to increase migration duration when corresponding Clock allele length is short, especially in the fall. The peptide product of Adcyap1, PACAP, regulates the expression of Clock in chicken (Gallus gallus domesticus) pineal glands (Nagy & Csernus, 2007), which may suggest the interaction effect we observe is the result of differential expression of Clock as determined by Adcyap1 allele length. However, it is yet unknown how Adcyap1 allele length influences PACAP structure or function, including downstream regulation of Clock. These complex genetic interactions may explain in some cases why previous studies have failed to identify effects in single candidate genes.
Our findings that candidate gene allele lengths are correlated with five of the six migratory traits raise an important question about whether the migratory phenotypic characters assessed in this study and others are evolutionarily independent of one another. For example, is variation in arrival dates a secondary consequence of natural selection on departure date and migratory duration, or might selection be acting on these characters separately and independently? Among the 17 blackpoll warblers used in this study, duration and departure date for both spring and fall migration were strongly and negatively correlated (spring r = −0.948, fall r = −0.677), suggesting that birds who departed later had a shorter duration (faster rate of migration). However, duration and arrival date were not correlated or only moderately so for spring and fall, respectively (spring r = −0.010, fall r = 0.457). While a study with similar sample sizes in another species showed a close relationship between departure and arrival dates (Ouwehand & Both, 2017), surprisingly, departure and arrival dates were weakly correlated in our data for both seasons (spring r = 0.270, fall r = 0.290). These data suggest migratory departure and arrival dates may be independent of one another. Further, variation in duration may be more closely tied to variation in departure date than arrival date, perhaps due to stronger selection on arrival date especially in the spring (Nilsson et al., 2013). Our data also showed a strong positive correlation between spring and fall duration (r = 0.688), and both of these characters were associated with a significant interaction between Clock and Adcyap1 (Figure 6). Whether these genes influence spring and fall duration via a common mechanism, perhaps through increased migratory restlessness (Mueller et al., 2011; Peterson et al., 2013), or whether these characters are independent requires further investigation. Nilsson et al. (2013) examined timing and speed of fall and spring migration in published tracking studies and found evidence for stronger selection on spring migratory phenology compared to fall, potentially suggesting evolutionary independence of these phenotypic characters.
Part of the answer to this question about evolutionary independence depends on the control of these characters at a proximate molecular level. Are there separate molecular pathways that control departure dates and duration, or that control spring and fall duration, or are these all proximately linked? PACAP is known to have broad influence on the physiology and behavior of organisms, acting in the brain and throughout peripheral organs (Mueller et al., 2011; Vaudry et al., 2009). It is therefore plausible that this gene could influence similar behaviors in separate seasons via independent molecular pathways. Conversely, it is perhaps equally plausible that a common molecular pathway is triggered by environmental cues in multiple seasons. This again highlights the need for studies that further investigate the functional role of candidate genes, how they influence migratory behaviors throughout the annual cycle, both independently and interactively with other factors (e.g., age, sex, environmental conditions), and specifically how variation in allele length influences the expression level, structure, and molecule functioning of gene products.
Although we found relationships between candidate genes and migratory traits at the individual level, we found no geographic structure in candidate genes. One possible explanation for this is that geographic variation in migratory behavior is explained by variation in environmental cues and not local adaptation in candidate genes. Geographic variation in behavior may be the result of plastic responses to variable environments, independent of the effects of those environments on selection in candidate genes (Foster, 1999). Therefore, to the extent environmental cues vary across breeding (or wintering) sites, it is possible we might observe behavioral differences in migratory behaviors without underlying geographic differences in genes. For example, during fall migration western breeding blackpoll warblers depart earlier and take longer to arrive at wintering grounds compared to eastern breeding birds (DeLuca et al., 2015, 2019), despite no differences in candidate gene frequencies between those populations. Our results suggest these differences across populations could be the results of plastic responses to differing environmental cues, while candidate genes are still correlated with variation in timing among individuals within each population. Further, Bourret and Garant (2015) point out that gene‐by‐environment interactions are underappreciated in the study of candidate genes. In their study of breeding phenology in tree swallows (Tachycineta bicolor), they found most candidate gene genotype–phenotype relationships were affected by environmental variables such as breeding density, latitude (a proxy of photoperiod), and temperature (Bourret & Garant, 2015). If genotype–phenotype relationships are influenced by environment, again we may observe behavioral differences across populations without underlying population genetic differences. Alternatively, it might simply be that any interpopulation variation in allele frequencies or lengths are undetectable given high levels of intrapopulation variation and our relatively small sample size; however, our result of no geographic structure of candidate genes is consistent with studies in other bird species (Bazzi et al., 2016; Dor et al., 2012, 2011; Johnsen et al., 2007). It is unlikely that the lack of geographic structure in candidate genes in blackpoll warblers is due solely to admixture from gene flow between locally adapted populations. Ralston and Kirchman (2012) found genetic structure due to isolation by distance at the continental scale in this species, despite a relatively high number of shared alleles among populations (Ralston & Kirchman, 2012, 2013). Future studies may benefit from comparing geographic patterns in candidate and neutral loci (Contina et al., 2018).
We observed greater allelic diversity in blackpoll warblers at Adcyap1 than at Clock, a pattern that appears to be typical across birds (Bazzi et al., 2016; Contina et al., 2018; Peterson et al., 2013; Steinmeyer et al., 2009). However, our results differ in that we find relatively high heterozygosity (74.3%) at the Clock locus, despite only observing four alleles. In studies of other bird species, Clock heterozygosity was quite low (0%–9%; Bazzi et al., 2015, 2016; Chakarov et al., 2013; Dor et al., 2011). Low heterozygosity at microsatellite loci may be the result of stabilizing selection or loss of diversity due to inbreeding and therefore useful in studies of individual fitness (Chapman, Nakagawa, Coltman, Slate, & Sheldon, 2009; Dor et al., 2011). While a single microsatellite locus is not sufficient in estimating genome‐wide heterozygosity or inbreeding (Miller et al., 2014), individual heterozygosity at a small number of microsatellite loci can be useful in the study of avian life history traits and fitness (Forstmeier, Schielzeth, Mueller, Ellegren, & Kempenaers, 2012; Lens et al., 2000), especially if those loci are functionally important and under selection (Chapman et al., 2009). Adcyap1 heterozygosity in Eurasian blackcaps was significantly associated with earlier spring arrival (Mettler et al., 2015), perhaps suggesting these individuals were of greater migratory fitness and able to migrate more quickly. We found similar results in blackpoll warblers in that heterozygotes at Adcyap1 had shorter spring and fall duration than homozygotes. We found no associations between heterozygosity at Clock and any migratory trait. Few studies of migratory candidate genes have examined the effect of individual heterozygosity, but our results and those of Mettler et al. (2015) suggest this is a potentially fruitful avenue of further investigation.
The results of our individual‐level analyses demonstrate the value of studies that reveal individual migratory phenotypes, for example, by using light‐level geolocators (McKinnon & Love, 2018). The combination of geolocator data with candidate gene analysis has been an important advancement in the study of migration (Bazzi et al., 2015; Contina et al., 2018; Saino et al., 2017). For example, in the study of barn swallows, individual‐level analyses revealed that rare Clock genotypes can have a significant impact on migratory phenology (Bazzi et al., 2015) and that degree of DNA methylation at the Clock gene can explain individual variation in migratory behavior (Saino et al., 2017). Both of these insights would likely have been missed with analysis of population‐level genetic and migratory variation. Increasing the number of species and individuals with linked genotype–phenotype information will allow richer investigations of migratory candidate genes.
Understanding variation in candidate genes across species is especially important for behaviors that are evolutionarily labile and may have arisen independently in multiple lineages. We therefore encourage future studies of candidate migratory genes to investigate species that are codistributed, share a biogeographic history, or are in the same family as those species that have already been studied. This will allow a better understanding of the influences of environment and history on selection at candidate genes, as well as the degree to which these patterns are conserved within lineages.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
JR, LL, and MM conceived of the project, performed the genetics laboratory work and statistical analyses, and wrote the paper. All other authors contributed to field efforts to collect blood samples or retrieve geolocators, and provided critical feedback on analyses and the manuscript. WVD took the lead on geolocator data analysis.
Supporting information
ACKNOWLEDGMENTS
We thank Christian Artuso, Yousif Attia, John Brett, Jukka Jantunen, Jill Boelsma, George Gress, David Merz, Jason Reppert, Emily Williams, and Ted Murphy‐Kelly for outstanding assistance in the field. We are also grateful to the following organizations for support of this research: Saint Mary's College, Kenneth M. Molson Foundation, Canadian Foundation for Innovation, Natural Sciences and Engineering Research Council of Canada, Wildlife Conservation Society Canada, U.S. National Park Service, Society of Yukon Bird Observatories, Bird Studies Canada, the W. Garfield Weston Foundation, Teslin Tlingit Council, the estate of Fred Bodsworth, Bering Straits Council, Solomon Native Corporations, Churchill Northern Science Centre, CNSC Northern Research Fund, Alaska Geographic.
APPENDIX 1.
Tissue and blood sampling locations, population assignments for the current study, and original tissue sources (population, sample location, source, n)
| Population (n) | Source | Source n | Catalog numbers | Geolocator numbers |
|---|---|---|---|---|
| Alaska (14) | Current study (blood)a | 10 | 3254‐001, 3254‐003, 3254‐008, 3254‐011, 3254‐05 | |
| University of Alaska Museum (frozen tissue) | 3 | UAM7394, UAM20089, UAM20508 | ||
| UCLA Conservation Genetics Resource Center (feather) | 1 | 01N9262 | ||
| Yukon (11) | Current study (blood)a | 11 | blpw12, blpw14, blpw15, blpw25 | |
| Manitoba (18) | Current study (blood)a | 17 | 4105‐002, 4105‐004, 4105‐006, 4105‐008, 4105‐009, 4105‐010, 4105‐016, 4105‐017 | |
| Ralston and Kirchman (2012; blood)b | 1 | |||
| Eastern (29) | Current study (blood)a | 10 | ||
| New York State Museum (frozen tissue) | 4 | NYSM11077, NYSM11078, NYSM11079, NYSM11080 | ||
| Ralston and Kirchman (2012; blood)b | 15 |
Blood samples and DNA extracts archived at Saint Mary's College, Notre Dame, Indiana, USA.
Blood samples and DNA extracts archived at New York State Museum, Albany, New York, USA.
APPENDIX 2.
General linear model results for variation in allele lengths by longitude and latitude. There was no significant relationship between allele length for either locus and either longitude or latitude, regardless of whether individual minimum, maximum, or mean allele lengths were considered
| Gene | Allele | β Longitude | p Longitude | β Latitude | p Latitude | F | df | p |
|---|---|---|---|---|---|---|---|---|
| Clock | Min | 0.005 | 0.720 | 0.004 | 0.544 | 0.220 | 2,69 | 0.804 |
| Clock | Mean | 0.008 | 0.466 | 0.018 | 0.759 | 0.417 | 2,69 | 0.664 |
| Clock | Max | 0.011 | 0.388 | −0.006 | 0.926 | 1.437 | 2,69 | 0.245 |
| Adcyap1 | Min | −0.004 | 0.776 | −0.055 | 0.417 | 0.579 | 2,69 | 0.563 |
| Adcyap1 | Mean | 0.007 | 0.555 | 0.003 | 0.957 | 0.483 | 2,69 | 0.619 |
| Adcyap1 | Max | 0.017 | 0.209 | 0.062 | 0.389 | 0.859 | 2,69 | 0.428 |
APPENDIX 3.
General linear model results for individual‐level analyses of blackpoll warbler candidate gene alleles and migratory traits. Allele indicates whether an individual's minimum, maximum, or mean allele length was used for each analysis. Slope (β) with standard error (SE), p‐value, and partial R 2 are given for the relationship between allele length and each migratory trait. Population was also used as a predictor variable, though the slope and p‐values for this factor are not provided. Models with a significant slope for allele length (p ≤ 0.05) are shown with bold text.
| Gene | Migratory trait | Allele | n | β AlleleLength | SE | p AlleleLength | R 2 AlleleLength |
|---|---|---|---|---|---|---|---|
| Clock | Spring departure | Min | 12 | −0.258 | 1.811 | 0.890 | 0.003 |
| Mean | 12 | −0.488 | 2.156 | 0.827 | 0.006 | ||
| Max | 12 | −0.810 | 2.478 | 0.752 | 0.013 | ||
| Spring duration | Min | 12 | −0.333 | 1.752 | 0.854 | 0.005 | |
| Mean | 12 | −0.353 | 2.09 | 0.870 | 0.004 | ||
| Max | 12 | −0.310 | 2.413 | 0.901 | 0.002 | ||
| Spring arrival | Min | 11 | −0.626 | 0.384 | 0.147 | 0.275 | |
| Mean | 11 | −0.912 | 0.425 | 0.069 | 0.397 | ||
| Max | 11 | −1.250 | 0.438 | 0.024 | 0.538 | ||
| Fall departure | Min | 17 | −0.102 | 0.543 | 0.854 | 0.003 | |
| Mean | 17 | −0.533 | 0.633 | 0.415 | 0.052 | ||
| Max | 17 | −0.923 | 0.599 | 0.147 | 0.154 | ||
| Fall duration | Min | 17 | −0.369 | 0.577 | 0.534 | 0.03 | |
| Mean | 17 | 0.065 | 0.702 | 0.927 | 0.001 | ||
| Max | 17 | 0.662 | 0.679 | 0.347 | 0.068 | ||
| Fall arrival | Min | 17 | −0.556 | 0.804 | 0.502 | 0.035 | |
| Mean | 17 | −0.574 | 0.967 | 0.563 | 0.026 | ||
| Max | 17 | −0.354 | 0.977 | 0.723 | 0.01 | ||
| Adcyap1 | Spring departure | Min | 12 | −5.615 | 1.577 | 0.007 | 0.613 |
| Mean | 12 | −2.646 | 2.575 | 0.334 | 0.117 | ||
| Max | 12 | 0.611 | 1.893 | 0.755 | 0.013 | ||
| Spring duration | Min | 12 | 5.123 | 1.657 | 0.015 | 0.544 | |
| Mean | 12 | 2.435 | 2.509 | 0.360 | 0.105 | ||
| Max | 12 | −0.537 | 1.835 | 0.777 | 0.011 | ||
| Spring arrival | Min | 11 | −0.742 | 0.543 | 0.214 | 0.211 | |
| Mean | 11 | −0.322 | 0.645 | 0.633 | 0.034 | ||
| Max | 11 | 0.106 | 0.462 | 0.826 | 0.007 | ||
| Fall departure | Min | 17 | 0.122 | 0.738 | 0.871 | 0.002 | |
| Mean | 17 | 0.522 | 0.866 | 0.557 | 0.027 | ||
| Max | 17 | 0.470 | 0.631 | 0.470 | 0.041 | ||
| Fall duration | Min | 17 | 1.941 | 0.588 | 0.006 | 0.456 | |
| Mean | 17 | 1.844 | 0.797 | 0.038 | 0.292 | ||
| Max | 17 | 0.512 | 0.681 | 0.465 | 0.042 | ||
| Fall arrival | Min | 17 | 2.122 | 0.945 | 0.043 | 0.279 | |
| Mean | 17 | 2.889 | 1.053 | 0.017 | 0.367 | ||
| Max | 17 | 1.500 | 0.878 | 0.111 | 0.183 |
Ralston J, Lorenc L, Montes M, et al. Length polymorphisms at two candidate genes explain variation of migratory behaviors in blackpoll warblers (Setophaga striata). Ecol Evol. 2019;9:8840–8855. 10.1002/ece3.5436
Data Availability Statement: All data used in analyses in the paper, including Clock and Adcyap1 genotype information, capture coordinates, and migratory trait values are available in Dryad Digital Repository (https://doi.org/10.5061/dryad.d10qb58). An R script is available in supplementary materials (Data S1) and can be used in combination with the dataset on Dryad to perform all analyses in this paper.
DATA ACCESSIBILITY
All data used in analyses in the paper, including Clock and Adcyap1 genotype information, capture coordinates, and migratory trait values are available in Dryad Digital Repository (https://doi.org/10.5061/dryad.d10qb58). An R script is available in supplementary materials (Data S1) and can be used in combination with the dataset on Dryad to perform all analyses in this paper.
REFERENCES
- Bairlein, F. , Eikenaar, C. , & Schmaljohann, H. (2015). Routes to genes: Unravelling the control of avian migration – An integrated approach using Northern Wheatear Oenanthe oenanthe as a model organism. Journal of Ornithology, 156, 3–14. [Google Scholar]
- Bairlein, F. , Norris, D. R. , Nagel, R. , Bulte, M. , Voigt, C. C. , Fox, J. W. , … Schmaljohann, H. (2012). Cross‐hemisphere migration of a 25 g songbird. Biology Letters, 8, 505–507. 10.1098/rsbl.2011.1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battley, P. F. , Warnock, N. , Tibbitts, T. L. , Gill, R. E. , Piersma, T. , Hassell, C. J. , … Riegen, A. C. (2012). Contrasting extreme long‐distance migration patterns in bar‐tailed godwits Limosa lapponica . Journal of Avian Biology, 43, 21–32. 10.1111/j.1600-048X.2011.05473.x [DOI] [Google Scholar]
- Bazzi, G. , Ambrosini, R. , Caprioli, M. , Costanzo, A. , Liechti, F. , Gatti, E. , … Rubolini, D. (2015). Clock gene polymorphism and scheduling of migration: A geolocator study of the barn swallow Hirundo rustica . Nature Scientific Reports, 5, 12443 10.1038/srep12443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzi, G. , Galimberti, A. , Hays, Q. , Bruni, I. , Cecere, J. G. , Gianfranceschi, L. … Rubolini, D. (2016). Adcyap1 polymorphism covaries with breeding latitude in a Nearctic migratory songbird, the Wilson's warbler (Cardellina pusilla). Ecology and Evolution, 6, 3226–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzi, G. , Podofillini, S. , Gatti, E. , Gianfranceschi, L. , Cecere, J. G. , Spina, F. , … Rubolini, D. (2017). Candidate genes have sex‐specific effects on timing of spring migration and moult speed in a long‐distance migratory bird. Current Zoology, 63, 479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold, P. (1996). Control of bird migration. London, UK: Chapman & Hall. [Google Scholar]
- Berthold, P. , & Pulido, F. (1994). Heritability of migratory activity in a natural bird population. Proceedings of the Royal Society of London Series B: Biological Sciences, 257, 311–315. [Google Scholar]
- Bourret, A. , & Garant, D. (2015). Candidate gene‐environment interactions and their relationships with timing of breeding in a wild bird population. Ecology and Evolution, 5, 3628–3641. 10.1002/ece3.1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprioli, M. , Ambrosini, R. , Boncoraglio, G. , Gatti, E. , Romano, A. , Romano, M. , … Saino, N. (2012). Clock gene variation is associated with breeding phenology and maybe under directional selection in the migratory barn swallow. PLoS ONE, 7, e35140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakarov, N. , Jonker, R. M. , Boerner, M. , Hoffman, J. I. , & Krüger, O. (2013). Variation at phenological candidate genes correlates with timing of dispersal and plumage morph in a sedentary bird of prey. Molecular Ecology, 22, 5430–5440. 10.1111/mec.12493 [DOI] [PubMed] [Google Scholar]
- Chapman, J. R. , Nakagawa, S. , Coltman, D. W. , Slate, J. , & Sheldon, B. C. (2009). A quantitative review of heterozygosity‐fitness correlations in animal populations. Molecular Ecology, 18, 2746–2765. 10.1111/j.1365-294X.2009.04247.x [DOI] [PubMed] [Google Scholar]
- Contina, A. , Bridge, E. , & Kelly, J. (2016). Exploring novel candidate genes from the Mouse Genome Informatics database: Potential implications for avian migration research. Integrative Zoology, 11, 240–249. 10.1111/1749-4877.12199 [DOI] [PubMed] [Google Scholar]
- Contina, A. , Bridge, E. S. , Ross, J. D. , Shipley, J. R. , & Kelly, J. F. (2018). Examination of Clock and Adcyap1 gene variation in a neotropical migratory passerine. PLoS ONE, 13, e0190859 10.1371/journal.pone.0190859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csernus, V. , Józsa, R. , Reglodi, D. , Hollósy, T. , Somogyvári‐Vigh, A. , & Arimura, A. (2004). The effect of PACAP on rhythmic melatonin release of avian pineals. General and Comparative Endocrinology, 135, 62–69. 10.1016/S0016-6480(03)00284-3 [DOI] [PubMed] [Google Scholar]
- Dawson, A. , King, V. M. , Bentley, G. E. , & Ball, G. F. (2001). Photoperiodic control of seasonality in birds. Journal of Biological Rhythms, 16, 365–380. 10.1177/074873001129002079 [DOI] [PubMed] [Google Scholar]
- Delmore, K. E. , Hübner, S. , Kane, N. C. , Schuster, R. , Andrew, R. L. , Câmara, F. , … Irwin, D. E. (2015). Genomic analysis of a migratory divide reveals candidate genes for migration and implicates selective sweeps in generating islands of differentiation. Molecular Ecology, 24, 1873–1888. 10.1111/mec.13150 [DOI] [PubMed] [Google Scholar]
- DeLuca, W. V. , Holberton, R. , Hunt, P. D. , & Eliason, B. C. (2013). Blackpoll warbler (Setophaga striata), version 2.0 In Poole A. F. (Ed.), The birds of North America. Ithaca, NY: Cornell Lab of Ornithology; 10.2173/bna.431 [DOI] [Google Scholar]
- DeLuca, W. V. , Woodworth, B. K. , Mackenzie, S. A. , Newman, A. E. M. , Cooke, H. A. , Phillips, L. M. , … Ryan Norris, D. (2019). A boreal songbird's 20,000 km migration across North America and the Atlantic Ocean. Ecology, 100, e02651. [DOI] [PubMed] [Google Scholar]
- DeLuca, W. V. , Woodworth, B. K. , Rimmer, C. C. , Marra, P. P. , Taylor, P. D. , McFarland, K. P. , … Norris, D. R. (2015). Transoceanic migration by a 12 g songbird. Biology Letters, 11, 2014105 10.1098/rsbl.2014.1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dor, R. , Cooper, C. B. , Lovette, I. J. , Massoni, V. , Bulit, F. , Liljesthrom, M. , & Winkler, D. W. (2012). Clock gene variation in Tachycineta swallows. Ecology and Evolution, 2, 95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dor, R. , Lovette, I. J. , Safran, R. J. , Billerman, S. M. , Huber, G. H. , Vortman, Y. , … Winkler, D. W. (2011). Low variation in the polymorphic Clock gene poly‐Q region despite population genetic structure across barn swallow (Hirundo rustica) populations. PLoS ONE, 6, e28843 10.1371/journal.pone.0028843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dray, S. , & Dufour, A. B. (2007). The ade4 package: Implementing the duality diagram for ecologists. Journal of Statistical Software, 22, 1–20. [Google Scholar]
- Excoffier, L. , & Lischer, H. E. L. (2010). Arlequin suite ver 3.5: A new series of programs to perform population genetic analyses under Linux and Windows. Molecular Ecology Resources, 10, 564–567. [DOI] [PubMed] [Google Scholar]
- Forstmeier, W. , Schielzeth, H. , Mueller, J. C. , Ellegren, H. , & Kempenaers, B. (2012). Heterozygosity‐fitness correlations in zebra finches: Microsatellite markers can be better than their reputation. Molecular Ecology, 21, 3237–3249. 10.1111/j.1365-294X.2012.05593.x [DOI] [PubMed] [Google Scholar]
- Foster, S. A. (1999). The geography of behavior: An evolutionary perspective. Trends in Ecology and Evolution, 14, 190–195. [DOI] [PubMed] [Google Scholar]
- Gienapp, P. , Leimu, R. , & Merilä, J. (2007). Responses to climate change in avian migration time – Microevolution versus phenotypic plasticity. Climate Research, 35, 25–35. 10.3354/cr00712 [DOI] [Google Scholar]
- Gill, R. E. , Tibbitts, T. L. , Douglas, D. C. , Handel, C. M. , Mulcahy, D. M. , Gottschalck, J. C. , … Piersma, T. (2009). Extreme endurance flights by landbirds crossing the Pacific Ocean: Ecological corridor rather than barrier? Proceedings of the Royal Society B: Biological Sciences, 276, 447–457. 10.1098/rspb.2008.1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet, J. (2004). HIERFSTAT, a package for R to compute and test hierarchical F‐statistics. Molecular Ecology Notes, 5, 184–186. 10.1111/j.1471-8286.2004.00828.x [DOI] [Google Scholar]
- Gwinner, E. (1990). Bird migration. Heidelberg, Germany: Springer. [Google Scholar]
- Hannibal, J. , Ding, J. M. , Chen, D. , Fahrenkrug, J. , Larsen, P. J. , Gillette, M. U. , & Mikkelsen, J. D. (1997). Pituitary adenylate cyclase‐activating peptide (PACAP) in the retinohypothalamic tract: A potential daytime regulator of the biological clock. The Journal of Neuroscience, 17, 2637–2644. 10.1523/JNEUROSCI.17-07-02637.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbig, A. J. (1991). Inheritance of migratory direction in a bird species: A cross‐breeding experiment with SE‐and SW‐migrating blackcaps (Sylvia atricapilla). Behavioral Ecology and Sociobiology, 28, 9–12. 10.1007/BF00172133 [DOI] [Google Scholar]
- Jenni, L. , & Schaub, M. (2003). Behavioural and physiological reactions to environmental variation in bird migration: A review In Berthold P., Gwinner E., & Sonnenschein E. (Eds.), Avian migration (pp. 155–171). Berlin, Germany: Springer. [Google Scholar]
- Johnsen, A. , Fidler, A. E. , Kuhn, S. , Carter, K. L. , Hoffmann, A. , Barr, I. R. , … Kempenaers, B. (2007). Avian Clock gene polymorphism: Evidence for a latitudinal cline in allele frequencies. Molecular Ecology, 16, 4867–4880. 10.1111/j.1365-294X.2007.03552.x [DOI] [PubMed] [Google Scholar]
- Johnston, R. A. , Paxton, K. L. , Moore, F. R. , Wayne, R. K. , & Smith, T. B. (2016). Seasonal gene expression in a migratory songbird. Molecular Ecology, 25, 5680–5691. 10.1111/mec.13879 [DOI] [PubMed] [Google Scholar]
- Kuhn, K. , Schwenk, K. , Both, C. , Canal, D. , Johansson, U. S. , van der Mije, S. , … Päckert, M. (2013). Differentiation in neutral genes and a candidate gene in the pied flycatcher: Using biological archives to track global climate change. Ecology and Evolution, 3, 4799–4814. 10.1002/ece3.855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemopoulos, A. , Uusi‐Heikkilä, S. , Huusko, A. , Vasemägi, A. , & Vainikka, A. (2018). Comparison of migratory and resident populations of brown trout reveals candidate genes for migration tendency. Genome Biology and Evolution, 10, 1493–1503. 10.1093/gbe/evy102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lens, L. , Van Dongen, S. , Galbusera, P. , Schenck, T. , Matthysen, E. , & Van De Casteele, T. (2000). Developmental instability and inbreeding in natural bird populations exposed to different levels of habitat disturbance. Journal of Evolutionary Biology, 13, 889–896. 10.1046/j.1420-9101.2000.00232.x [DOI] [Google Scholar]
- Liedvogel, M. , Åkesson, S. , & Bensch, S. (2011). The genetics of migration on the move. Trends in Ecology and Evolution, 26, 561–569. 10.1016/j.tree.2011.07.009 [DOI] [PubMed] [Google Scholar]
- Liedvogel, M. , & Lundberg, M. (2014). The genetics of migration In Hansson L. A., & Åkesson S. (Eds.), Animal movement across scales. Oxford, UK: Oxford University Press. [Google Scholar]
- Liedvogel, M. , & Sheldon, B. C. (2010). Low variability and absence of phenotypic correlates of Clock gene variation in a great tit Parus major population. Journal of Avian Biology, 41, 543–550. 10.1111/j.1600-048X.2010.05055.x [DOI] [Google Scholar]
- Liedvogel, M. , Szulkin, M. , Knowles, S. C. , Wood, M. J. , & Sheldon, B. C. (2009). Phenotypic correlates of Clock gene variation in a wild blue tit population: Evidence for a role in seasonal timing of reproduction. Molecular Ecology, 18, 2444–2456. [DOI] [PubMed] [Google Scholar]
- Lundberg, M. , Boss, J. , Canbäck, B. , Liedvogel, M. , Larson, K. W. , Grahn, M. , … Wright, A. (2013). Characterisation of a transcriptome to find sequence differences between two differentially migrating subspecies of the willow warbler Phylloscopus trochilus . BMC Genomics, 14, 330 10.1186/1471-2164-14-330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg, M. , Liedvogel, M. , Larson, K. , Sigeman, H. , Grahn, M. , Wright, A. , … Bensch, S. (2017). Genetic differences between willow warbler migratory phenotypes are few and cluster in large haplotype blocks. Evolution Letters, 1, 155–168. 10.1002/evl3.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon, E. A. , Artuso, C. , & Love, O. P. (2017). The mystery of the missing warbler. Ecology, 98, 1970–1972. 10.1002/ecy.1844 [DOI] [PubMed] [Google Scholar]
- McKinnon, E. A. , & Love, O. P. (2018). Ten years tracking the migrations of small landbirds: Lessons learned in the golden age of bio‐logging. The Auk: Ornithological Advances, 135, 834–856. 10.1642/AUK-17-202.1 [DOI] [Google Scholar]
- Merlin, C. , & Liedvogel, M. (2019). The genetics and epigenetics of animal migration and orientation: Birds, butterflies and beyond. Journal of Experimental Biology, 222, jeb191890 10.1242/jeb.191890 [DOI] [PubMed] [Google Scholar]
- Mettler, R. , Segelbacher, G. , & Schaefer, H. M. (2015). Interactions between candidate gene for migration (ADCYAP1), morphology and sex predict spring arrival in blackcap populations. PLoS ONE, 10, e0144587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J. M. , Malenfant, R. M. , David, P. , Davis, C. S. , Poissant, J. , Hogg, J. T. , … Coltman, D. W. (2014). Estimating genome‐wide heterozygosity: Effects of demographic history and marker type. Heredity, 112, 240–247. 10.1038/hdy.2013.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, S. R. , Covino, K. M. , Jacobs, J. D. , & Taylor, P. D. (2016). Fall migratory patterns of the Blackpoll Warbler at a continental scale. The Auk, 133, 41–51. [Google Scholar]
- Mueller, J. C. , Pulido, F. , & Kempenaers, B. (2011). Identification of a gene associated with avian migratory behavior. Proceedings of the Royal Society of London Series B: Biological Sciences, 278, 2848–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy, A. D. , & Csernus, V. J. (2007). The role of PACAP in the control of circadian expression of clock genes in the chicken pineal gland. Peptides, 28, 1767–1774. 10.1016/j.peptides.2007.07.013 [DOI] [PubMed] [Google Scholar]
- Nilsson, C. , Klaassen, R. H. G. , & Alerstam, T. (2013). Differences in speed and duration of bird migration between spring and autumn. The American Naturalist, 181, 837–845. 10.1086/670335 [DOI] [PubMed] [Google Scholar]
- Ouwehand, J. , & Both, C. (2017). African departure rather than migration speed determines variation in spring arrival in pied flycatchers. Journal of Animal Ecology, 86, 88–97. 10.1111/1365-2656.12599 [DOI] [PubMed] [Google Scholar]
- Panda, S. , Hogenesch, J. B. , & Kay, S. A. (2002). Circadian rhythms from flies to humans. Science, 417, 329–335. [DOI] [PubMed] [Google Scholar]
- Peterson, M. P. , Abolins‐Abols, M. , Atwell, J. W. , Rice, R. J. , Milá, B. , & Ketterson, E. D. (2013). Variation in candidate genes CLOCK and ADCYAP1 does not consistently predict differences in migratory behavior in the songbird genus Junco. F1000 Research, 2, 115 10.12688/f1000research.2-115.v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, J. K. , Stephens, M. , & Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics, 155, 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido, F. (2007). The genetics and evolution of avian migration. BioScience, 57, 165–174. 10.1641/B570211 [DOI] [Google Scholar]
- Pulido, F. , & Berthold, P. (2004). Microevolutionary response to climatic change. Advances in Ecological Research, 35, 151–182. [Google Scholar]
- Pulido, F. , & Berthold, P. (2010). Current selection for lower migratory activity will drive the evolution of residency in a migratory bird population. Proceedings of the National Academy of Sciences of the United States of America, 107, 7341–7346. 10.1073/pnas.0910361107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido, F. , Berthold, P. , Mohr, G. , & Querner, U. (2001). Heritability of the timing of autumn migration in a natural bird population. Proceedings of the Royal Society of London Series B: Biological Sciences, 268, 953–959. 10.1098/rspb.2001.1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2019). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; Retrieved from https://www.R-project.org/ [Google Scholar]
- Ralston, J. , & Kirchman, J. J. (2012). Continent‐scale genetic structure in a boreal forest migrant, the blackpoll warbler (Setophaga striata). The Auk, 129, 467–478. [Google Scholar]
- Ralston, J. , & Kirchman, J. J. (2013). Predicted range shifts in North American boreal forest birds and the effect of climate change on genetic diversity in blackpoll warblers (Setophaga striata). Conservation Genetics, 14, 543–555. 10.1007/s10592-012-0418-y [DOI] [Google Scholar]
- Romano, A. , Possenti, C. D. , Caprioli, M. , Gatti, E. , Gianfranceschi, L. , Rubolini, D. , … Parolini, M. (2018). Circadian genes polymorphism and breeding phenology in a resident bird, the yellow‐legged gull. Current Zoology, 304, 117–123. [Google Scholar]
- Rosenberg, N. A. (2004). DISTRUCT: A program for the graphical display of population structure. Molecular Ecology Notes, 4, 137–138. 10.1046/j.1471-8286.2003.00566.x [DOI] [Google Scholar]
- Ross, C. A. (2002). Polyglutamine pathogenesis: Emergence of unifying mechanisms for Huntington's disease and related disorders. Neuron, 35, 819–822. 10.1016/S0896-6273(02)00872-3 [DOI] [PubMed] [Google Scholar]
- Saino, N. , Ambrosini, R. , Albetti, B. , Caprioli, M. , De Giorgio, B. , Gatti, E. , … Rubolini, D. (2017). Migration phenology and breeding success are predicted by methylation of a photoperiodic gene in the barn swallow. Nature Scientific Reports, 7, 45412 10.1038/srep45412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saino, N. , Bazzi, G. , Gatti, E. , Caprioli, M. , Cecere, J. G. , Possenti, C. D. , … Spina, F. (2015). Polymorphism at the Clock gene predicts phenology of long‐distance migration in birds. Molecular Ecology, 24, 1758–1773. [DOI] [PubMed] [Google Scholar]
- Steinmeyer, C. , Mueller, J. C. , & Kempenaers, B. (2009). Search for informative polymorphisms in candidate genes: Clock genes and circadian behaviour in blue tits. Genetica, 136, 109–117. 10.1007/s10709-008-9318-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaudry, D. , Falluel‐Morel, A. , Bourgault, S. , Basille, M. , Burel, D. , Wurtz, O. , … Vaudry, H. (2009). Pituitary adenylate cyclase‐activating polypeptide and its receptors: 20 years after the discovery. Pharmacological Reviews, 61, 284–357. 10.1124/pr.109.001370 [DOI] [PubMed] [Google Scholar]
- Yu, W. , & Hardin, P. E. (2006). Circadian oscillators of Drosophila and mammals. Journal of Cell Science, 119, 4793–4795. 10.1242/jcs.03174 [DOI] [PubMed] [Google Scholar]
- Zhu, H. , Gegear, R. J. , Casselman, A. , Kanginakudru, S. , & Reppert, S. M. (2009). Defining behavioral and molecular differences between summer and migratory monarch butterflies. BMC Biology, 7, 14 10.1186/1741-7007-7-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink, R. M. (2011). The evolution of avian migration. Biological Journal of the Linnean Society, 104, 237–250. 10.1111/j.1095-8312.2011.01752.x [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in analyses in the paper, including Clock and Adcyap1 genotype information, capture coordinates, and migratory trait values are available in Dryad Digital Repository (https://doi.org/10.5061/dryad.d10qb58). An R script is available in supplementary materials (Data S1) and can be used in combination with the dataset on Dryad to perform all analyses in this paper.


