Abstract
Background
Epithelial ovarian cancer (EOC) is the malignant tumor of the female reproductive system with the highest fatality rate. Tolerance of chemotherapeutic drugs like cisplatin (DDP) occurring in very early stage is one of the important factors of the poor prognosis of epithelial ovarian cancer. Here we aim to study the dysregulation of a particular long noncoding RNA, lncRNA GAS5, and its role in EOC progression.
Methods
The low expression of lncRNA GAS5 in EOC tissues and OC cell lines was determined by microarray analyses and Real-Time qPCR. Flow cytometer assays were used to detect cell cycle and apoptosis of OC cells. CCK8 assay were performed to investigate the DDP sensitivity of OC cells. Western blot was carried out to detect cell growth markers, apoptotic markers, PARP1, E2F4, MAPK pathway protein expression and other protein expression in OC cell lines. The binding of GAS5 and E2F4 were proved by RNA pull-down and RIP assay. The effect of E2F4 on PARP1 were determined by CHIP-qPCR assay and luciferase reporter assay. The effect of lncRNA GAS5 on OC cells was assessed in vitro and in vivo.
Results
By microarray (3 EOC tissues νs. 3 normal ovary tissues) and RT- qPCR (53 EOC tissues νs. 10 normal ovary tissues) we identified lncRNA GAS5 to be dramatically low expressed in EOC samples and correlated with prognosis. Compared with sensitive cell lines, GAS5 was also low expressed in DDP resistant OC cell lines, and over-expression of GAS5 significantly enhanced the sensitivity of OC cells to DDP in vivo and in vitro. Meanwhile the over-expression of GAS5 also caused OC cells G0/G1 arrest and apoptosis increase. Mechanistically, GAS5 might regulate PARP1 expression by recruiting the transcription factor E2F4 to its promoter, and then affect the MAPK pathway activity. Due to the 5’TOP structure, GAS5 could be regulated by transcription inhibitor rapamycin in OC cells.
Conclusion
Here we explored the specific mechanisms of EOC cisplatin resistance and tumor progress due to lncRNA-GAS5, presented the GAS5-E2F4-PARP1-MAPK axis and its role in OC drug-sensitivity and progression for the first time, and the results may provide experimental basis for clinical application.
Keywords: lncRNA GAS5, PARP1, E2F4, MAPK, Ovarian cancer
Background
Ovarian cancer (OC) is considered to be the lethal gynecological cancer with the highest mortality rate worldwide, 5-year survival rate maintains at 25–30% [1, 2]. Epithelial ovarian cancer (EOC) accounts for 90% of OC cases in Asian populations [3]. In clinical practice, early metastasis and drug resistance result in poor prognosis of EOC [4]. At present, the clinical strategy for EOC is still cytoreductive surgery supplemented by platinum-based chemotherapy, while statistical studies have shown that about 15 to 25% of EOC patients appeared primary resistant to platinum-based chemotherapy, and at least 80% of all patients eventually develop secondary resistance to platinum [5]. Therefore, exploring the mechanism and regulation pathway of EOC drug resistance is essential to improve the survival benefit of EOC patients. In addition to the reduction of intracellular drug accumulation, increased drug efflux and drug inactivation caused by multi-drug resistance-related genes and their protein products such as P-glycoprotein (MDR1/P-gp), lung resistance associated protein (LRP), glutathione transferase (GST), etc.; and the classical resistance mechanisms such as DNA damage response pathway (DDR) activation; drug targets abnormalities or drug failure caused by abnormal regulation of apoptosis, metastasis or proliferation-related pathways are also new directions of recent researches on molecular mechanisms of EOC drug resistance. The epigenetic regulation of key genes and proteins on the above drug resistance related pathways has also become a hot spot in the study of tumor resistance interventions.
With the wide regulatory range, diverse regulatory pathways and specific targets, non-coding RNA (non-coding RNA) has become one important part of epigenetic researches. The aberrant expression of long non-coding RNA (lncRNA), a generic term for non-coding RNAs over 200 nucleotides in length, is involved in many disease processes [6], and is closely related to tumorigenesis, tumor progression and drug resistance [7–10]. In this study, we identified growth inhibition associated lncRNA GAS5 to be dramatically lower expressed in EOC and correlated with poor prognosis. LncRNA GAS5 is transcribed from the non-protein-coding snoRNA host gene GAS5, which is also known as the growth arrest-specific transcript 5 [11]. As a tumor suppressor-like lncRNA, its abnormal low expression has been found in various tumors including lung cancer [12], liver cancer [13], breast cancer [14], cervical cancer [15], etc. and participates in affecting tumor development process, immune regulation [16] and drug resistance [14]. Recent years, lncRNA-GAS5 has been reported to influence the cisplatin sensitivity in non-small cell lung cancer [17] and cervical cancer [18], and affected the adriamycin sensitivity in bladder transitional cell carcinoma as well [19]. But its influence and related mechanisms on progression or drug resistence in ovarian cancer are still unclear.
In this study, we confirmed the low-expression of lncRNA GAS5 in EOC tissues and OC cell lines, ascertained its tumor suppressor gene like role and discovered its sensitization function of cisplatin in ovarian cancer. And for the first time, we presented that GAS5 may regulate PARP1 expression by recruiting the transcription factor E2F4 to its promoter, and then affect MAPK pathway activity as well, thereby it may enhanced chemosensitivity by promoting apoptosis and causing cell cycle arrest of OC cells. In addition, because of its 5’TOP structure, the expression of GAS5 in cytoplasm could be elevated by translation inhibitor Rapamycin, and this intervention may be used to offset the low expression of GAS5 in ovarian cancer.
Methods
Cell culture
The human ovarian cancer cell lines HEY, A2780, A2780/DDP, HO8910, HO8910PM, SKOV3, SKOV3/DDP, and normal human ovarian epithelial cell line IOSE were maintained by our laboratory. Cells were cultured in RPMI− 1640 medium (Gibco, Grand Island, NY, USA), supplemented with 10% FBS (HyClone, Logan, UT, USA). All cells were cultured in a humidified atmosphere, at 37 °C with 5% CO2.
Patients and specimens
Fifty three EOC tissues and 10 normal ovarian tissues were obtained from surgical specimens at Department of Obstetrics and Gynecology, Ren Ji hospital, School of Medicine, Shanghai Jiao Tong University (Shanghai, China) after informed consent during January 2013 to December 2015. All the specimens were staged according to FIGO classification of malignant Tumours-8th edition, and graded according to the 2018 world health
The inclusion and exclusion criteria of participants are as followed:
EOC group
Inclusion criteria:
Pathology criteria: Epithelial ovarian cancer, confirmed by two pathologists.
Staging criteria: I-IV (FIGO,2018).
Surgical method: Radical resection of ovarian cancer.
Medical history:
① No treatment with radiotherapy or chemotherapy before surgery.
② No death occurring within 1 month after surgery.
③ Clinicopathological characteristics and follow-up information available.
Requirements for the tissue samples:
① Derived from the first surgery.
② RNA concentration more than 200 ng/μl.
Exclusion criteria:
① Death within 1 month after operation.
② Lost cases.
③ RNA quality is not up to standard.
Normal group
Inclusion criteria:
Pathology criteria: Normal ovarian cancer, confirmed by two pathologists.
Surgical method: Gynecological malignancies or benign diseases except ovarian cancer undergoing total hysterectomy and adnexal resection.
Requirements for the tissue samples:
① Derived from the first surgery.
② RNA concentration more than 200 ng/μl.
Exclusion criteria:
① Lesion invade ovaries.
② RNA quality is not up to standard.
Sample collection
All samples derived from surgery were cut into two parts, each of which was no less than 0.5 cm*0.5 cm*0.5 cm. One part was immediately placed in a test tube containing RNA later for RNA extraction, and the other part is immediately stored in an empty test tube in a − 80 °C refrigerator. Cutting and transferring were done on ice.
The Institutional Review Board (IRB) approval number of this work is [2018]114 (Issuance of Ethics Approval by Ethics Committee of Renji Hospital).
Microarray analysis
LncRNAs in the RNA samples were profiled using human SBC Human (4*180 K) ceRNA microarray analysis (Bio-tech, Shanghai, China). Quantile normalization and data processing were performed using the GeneSpring GX v11.5.1 software package (Agilent Technologies). Differentially expressed lncRNAs were identified by Heatmap. Up-regulated or down-regulated lncRNAs were selected based on changes ≥2 fold threshold and P < 0.05.
Real-time reverse transcription polymerase chain reaction
The relative quantity of lncRNA and mRNA were measured by RT-qPCR. The gene-specific primers were designed by Primer Premier 5.0 software (Premier Biosoft International, Palo Alto,CA, USA).
Western blot
Total protein was separated by 10% SDS-PAGE and transferred into polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA). After that, the membranes were blocked by 5% nonfat milk, and incubated with primary antibodies (anti-PARP1, anti-E2F4, anti-CDK4, anti-CDK6, anti-CyclinD, anti-CASP3, anti-cleaved CASP3, anti-CASP7, anti-cleaved CASP7, anti-GAPDH, anti-β-actin [Abcam, USA], anti-p-ERK, anti-p-ERK, anti-p-JNK and anti-P38MAPK [Cell Signaling Technology, USA]), at 4 °C overnight. The membranes were then incubated with anti-rabbit/mouse IgG secondary antibody (CST, USA). The protein bands were visualized using enhanced chemiluminescent substrate (ECL, Pierce, USA).
Flow cytometry
Cell apoptosis was evaluated using the Annexin-V/Propidium Iodide Detection Kit (Key GEN, China). Cells were analyzed by FACS cytometry (BD Biosciences Inc.)
Luciferase reporter assay
The PARP1 sequence containing predicted E2F4 binding site was sub-cloned and inserted into pmirGLO vector (Promega, Madison, WI, USA). After that, SKOV3/HEY cells were incubated in a 96-well plate and co-transfected with recombinant plasmids (50 nM) or empty pmirGLO vector (200 ng), After incubation for 48 h, the cells were lysed in 1x Passive lysis. The Dual-Luciferase Reporter Assay System (Promega) was applied to measure the Renilla luciferase activity, with firefly luciferase serving as a transfection control.
RNA immunoprecipitation (RIP)
RIP experiments were performed using Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA) according to the manufacturer’s instructions. The co-precipitated RNAs were detected by reverse transcription PCR. To demonstrate that the detected signals were from the RNA that was specifically bound, total RNA (input controls) and corresponding species IgG controls were performed simultaneously (n = 3). For the anti-E2F4 RIP experiment, GAS5 were transiently transfected into SKOV3 and HEY cells. Cells were harvested and lysed for RIP using anti-E2F4 antibody after 48 h of transfection
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed using the EZ-ChIP chromatin immunoprecipitation kit (Millipore, Billerica, MA, USA) according to the manufacturer’s guidelines. Immunoselections of cross-linked protein-DNA were performed using anti-PARP1 antibody together agarose beads A/G, at 4 °C for overnight. The anti-rabbit IgG was used as a negative control. The purified DNAs were analyzed by PCR, the forward and reverse primers were: 5′- CTGATGTTGCAGGAAAAGCCC-3′ and 5′- AATAAAACACCGCCACCCAGA-3′ for human PARP1 promoter.
RNA pull-down assay
Cellular nuclear protein was extracted by using the NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific, Inc., USA), and then incubated with biotin-labeled GAS5 truncation probe and streptavidin agarose beads (Invitrogen). Finally, the retrieved protein was detected by Western-blot.
Tumorigenicity assays in nude mice
All animal studies were conducted in accordance with institutional guidelines for animal care and were approved by the committee for the use and care of animals of Renji hospital (Shanghai, China). The Institutional Review Board (IRB) approval number of this work is [2018]114 (Issuance of Ethics Approval by Ethics Committee of Renji Hospital). Briefly, SKOV3 cells were first transfected with the GAS5 expression vector or with empty vector for 72 h. The cells were then harvested at the exponential growth stage when they reached 90% confluence. Approximately 2.5 × 106 cells in 50% Matrigel were injected subcutaneously into the right flanks of 4- to 6-week-old nude mice. Tumor growth was monitored, and tumor sizes were measured every other day using caliper. The tumor volume (V) was calculated using the formula: V = 1/2(length × width2).In the DDP treatment groups, 5 mg/kg DDP was injected through tail vein every 3 days. When the maximum tumor size ≥1000 mm3, end monitor and execute mice.
Immunohistochemistry
Tissue samples were embedded in paraffin. Expression of CDK4, Ki67 and PARP1 were detected by immunohistochemical staining. Sections were visualized under a microscope (400× or 200×) (Olympus, Japan). The results were graded according to the percentage of positive cells.
RNA-fish
Cy3-labeled GAS5 and DAPI-labeled U6 probes were obtained from RiboBio (Guangzhou, China). RNA FISH were performed using fluorescent in situ hybridization kit (RiboBio) according to manufacturer’s instructions.
Statistical analysis
Statistical analysis was performed using the statistical program SPSS 13.0. Quantitative results were expressed as the mean ± standard error of mean (SEM) and were analyzed using Student’s t test. p value < 0.05 were considered statistically significant.
Results
LncRNA GAS5 was down-regulated in EOC tissues and OC cell lines, and associated with EOC prognosis
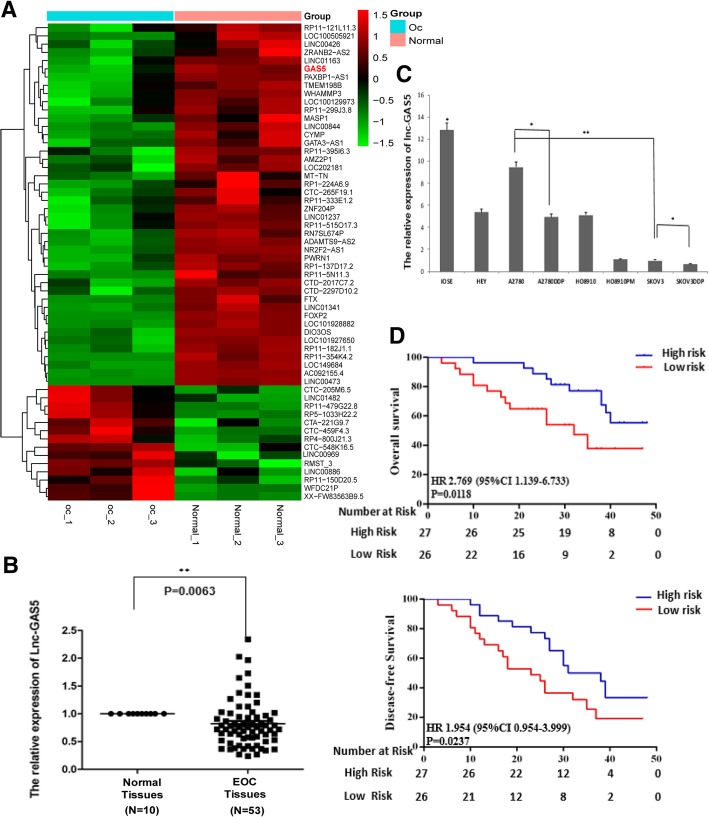
Microarray (3 EOC tissues νs. 3 normal ovary tissues) results determined that the expression of lncRNA GAS5 was significantly decreased in EOC tissues compared with normal ovarian tissues (OC νs. Normal fold change = 0.003, p value = 0.028) (Fig. 1a). RT-qPCR also showed that compared with 10 normal ovarian tissues, GAS5 was remarkably low-expressed in 53 EOC tissues (p value = 0.006) (Fig. 1b). And the expression level of GAS5 in seven OC cell lines (HEY, A2780, A2780/DDP, HO8910, HO8910PM, SKOV3, SKOV3/DDP) were also dramatically lower than that in normal human ovarian epithelial cell line IOSE (Fig. 1c). Furthermore, Kaplan-Meier survival analysis revealed that the low expression of GAS5 was associated with low overall survival rate (p value = 0.012) and disease-free survival rate (p value = 0.024) of OC patients (Fig. 1d), and as shown in Table 1, as an independent prognostic factor, GAS5 expression was significantly correlated with advanced FIGO stage and histological type. This result suggested that GAS5 downregulation might be vital in the progression of ovarian cancer.
Fig. 1.
LncRNA GAS5 was down-regulated in EOC tissues and OC cell lines, and associated with EOC prognosis. a Hierarchical clustering of Agilent lncRNA array result showing abnormal expressed lncRNAs in normal ovarian or EOC tumor tissues. GAS5 is one of the most low-expressed lncRNAs in EOC tissues. b Total RNA was extracted from human normal ovarian tissues (Normal) of 10 participants and EOC tumor tissues (OC Tissues) of 53 patients, GAS5 expression levels were then detected by RT-qPCR assay. c GAS5 expression levels in OC cell lines and normal human ovarian epithelial cell line were detected by RT-qPCR assay. d Kaplan–Meier overall survival curves (OS) and disease-free survival curves (DFS) was observed based on GAS5 level. Data were the means±SD of triplicate determinants. *P < 0.05 versus control groups. **P < 0.005 versus control groups
Table 1.
Clinical characteristics of EOC patients with high and low GAS5 risk scores
| Case(n) | GAS5 | P | ||
|---|---|---|---|---|
| Low-risk (n) | High-risk (n) | |||
| Age | ||||
| ≥ mean | 33 | 17 | 16 | 0.269 |
| < mean | 20 | 8 | 12 | |
| FIGO Stage | ||||
| I- II | 22 | 16 | 6 | 0.007* |
| III-IV | 31 | 10 | 21 | |
| Histological Type | ||||
| I | 11 | 7 | 4 | 0.024* |
| II- III | 42 | 16 | 26 | |
| Residual tumor (cm) | ||||
| < 1 cm | 34 | 18 | 16 | 0.332 |
| ≥ 1 cm | 19 | 8 | 11 | |
| Histology | ||||
| Mucinous | 7 | 3 | 4 | 0.402 |
| Serous | 41 | 17 | 24 | |
| Endometrioid | 3 | 1 | 2 | |
| Clear cell | 2 | 1 | 1 | |
| Lymphatic metastasis | ||||
| No | 20 | 8 | 12 | 0.257 |
| Yes | 33 | 14 | 19 | |
| Ascites | ||||
| No | 21 | 14 | 7 | 0.096 |
| Yes | 32 | 13 | 19 | |
P Values are calculated by X2 test or Fisher’s exact test
GAS5 over-expression caused cell cycle arrest and promoted apoptosis of OC cells
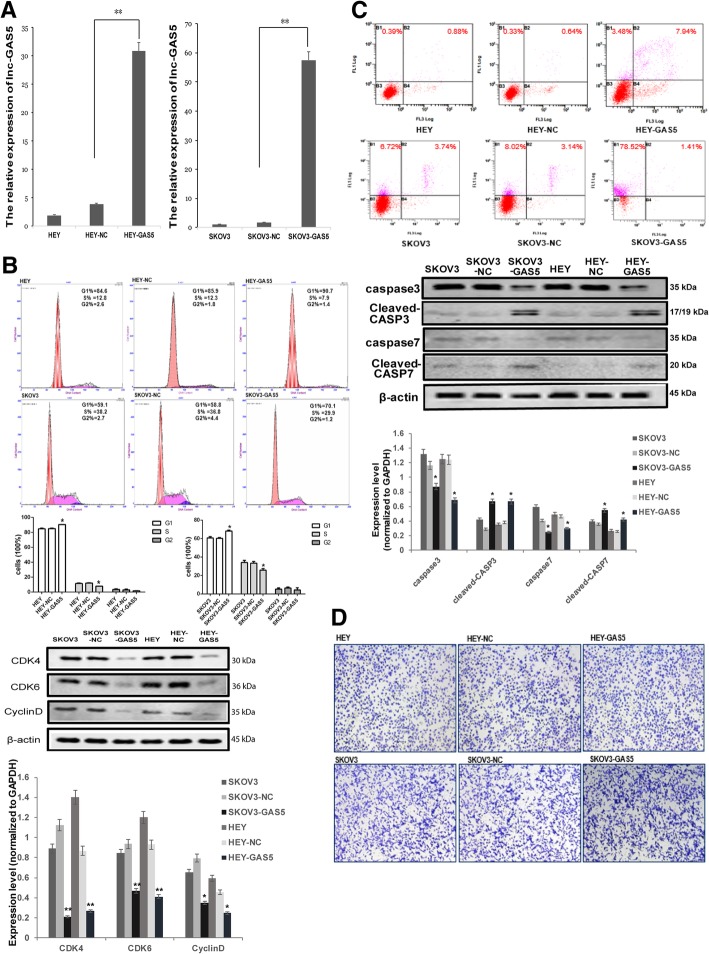
After constructed a retroviral stable expression of GAS5 in HEY and SKOV3 cells, qRT-PCR were performed to validate the over-expression levels of GAS5 in these two cell lines (Fig. 2a). As shown by the results of the flow cytometry, over-expression of GAS5 caused G0/G1 cell cycle arrest and apoptosis increase. In the meantime, Western-blot showed that GAS5 overexpression affects the expression of cell cycle and apoptosis-related proteins (Fig. 2b and c). However, overexpression of GAS5 had little impact on OC cells migration rate (Fig. 2d). These results indicated that GAS5 worked as a tumor suppressor gene, and played a specific role in regulating the cell cycle and apoptosis but not the migration of OC cells in vitro.
Fig. 2.
GAS5 over-expression caused cell cycle arrest and promoted apoptosis of OC cells. a LV5-GAS5 or the empty LV5 vector plasmids were transfected into OC cells. 72 h later, overexpression efficient of GAS5 was evaluated by RT-qPCR. b 72 h after transfection of LV5-GAS5 or the empty vector, flow cytometry assay were performed to detect the cell cycle impacted by overexpression of GAS5 in OC cell lines SKOV3 and HEY, and cell cycle-related protein levels were then assessed by Western-blot assay. β-actin was served as the internal control. c After transfection, cells were harvested and stained by Annexin V-PE and 7-AAD, and apoptosis rates were analyzed by flow cytometry, and apoptosis-related protein levels were then assessed by Western-blot assay. β-actin was served as the internal control. d Transwell assay were performed to detect the migration rate of SKOV3 and HEY. *P < 0.05 versus control groups
GAS5 over-expression enhanced sensitivity to DDP of OC both in vitro and in vivo
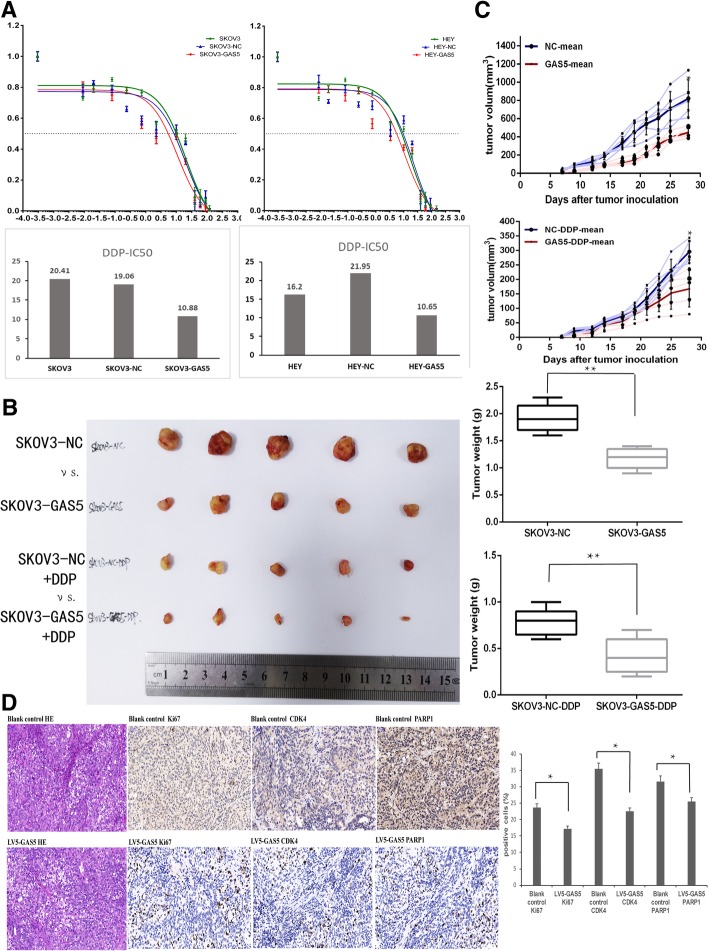
As shown in the qRT-PCR assay, GAS5 expression in DDP-resistant OC cell lines was significantly lower than in DDP-sensitive cells (A2780DDP vs. A2780, SKOV3DDP vs. SKOV3, A2780 vs. SKOV3) (Fig.1c). We then meditated that GAS5 might be correlated with DDP resistant in OC. As expected, HEY and SKOV3 cells stably expressing GAS5 showed to be more sensitive to DDP compared with control group in vitro (Fig.3a). When injected into nude mice, the tumor size and weight of stably expressing GAS5 groups were all remarkably lessened than the control groups 4 weeks after injection, and GAS5 overexpression groups seems more sensitive to DDP compared with control group (Fig. 3b and c). Furthermore, the immunohistochemical staining of Ki67, CDK4 and PARP1 showed that the proportion of positive cells of these protein in GAS5 over-expression group were lower than those in NC group (Fig.3d). Taken together, our data showed that GAS5 suppressed tumor growth and enhanced the sensitivity of cisplatin (DDP) in ovarian cancer.
Fig. 3.
GAS5 over-expression enhanced sensitivity to DDP in OC cells both in vitro and in vivo. a DDP sensitivity were detected by CCK8 assay and IC50 in OC cell lines SKOV3 and HEY. b The tumor tissues of nude mice were presented. c The volume of tumor was calculated on the 7, 9, 12, 14, 17, 19, 21, 23, 25 and 28 days. And the tumor weight were measured when tumors were harvested. d Immunohistochemical staining were performed to detect the Ki67, CDK4 and PARP1 expression in animal tumor tissues. *P < 0.05 versus control groups
GAS5 regulated the expression of PARP1 and effected MAPK pathway in OC cells
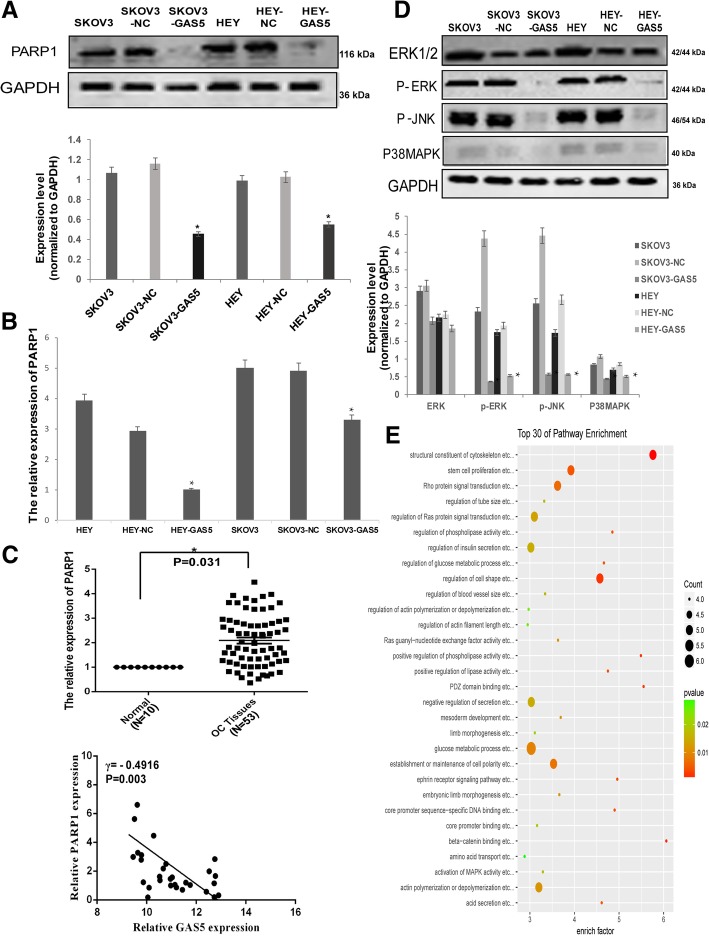
Poly (ADP-ribose) Polymerase1 (PARP1) promotes tumor progress via diversified biological functions such as cell cycle regulation, DNA repair, apoptosis inhibition, and maintaining MAPK activity, etc. [20] Clinically, PARP1 inhibitors have shown their potential in treating breast and ovarian cancers [21]. In this study, the clustering analysis of whole genome microarray data predicted that GAS5 may affect PARP1 Bind Protein (PARPBP), we then found that forced expression of GAS5 markedly down-regulated the protein and mRNA level of PARP1 in OC cells (Fig. 4a and b). As shown in RT-qPCR assay, PARP1 was high expressed in 53 EOC patient samples (P = 0.031), negatively related with GAS5 expression level (γ = − 0.4916, P = 0.003) (Fig. 4c). PARP1 has been reported to be indispensible for maintaining MAPK activity. Western-blot showed that GAS5 overexpression indeed affected the phosphorylation level of MAPK pathway members (Fig. 4d), as assumed by GO analysis of microarray data (Fig. 4e).
Fig. 4.
GAS5 regulated the expression of PARP1 and effected MAPK pathway in OC cells. a The protein level of PARP1 were assessed by Western-blot assay. GAPDH was served as the internal control. b The mRNA level of PARP1 were assessed by RT-qPCR assay. c PARP1 transcriptional activities of OC cells stably transfected with LV5-GAS5 or the empty vector were assessed by luciferase reporter assay. d The protein level of MAPK pathway members were assessed by Western-blot assay. GAPDH was served as the internal control. e GO analysis showed the top 30 pathway enrichment affected by GAS5 over-expression. *P < 0.05 versus control groups
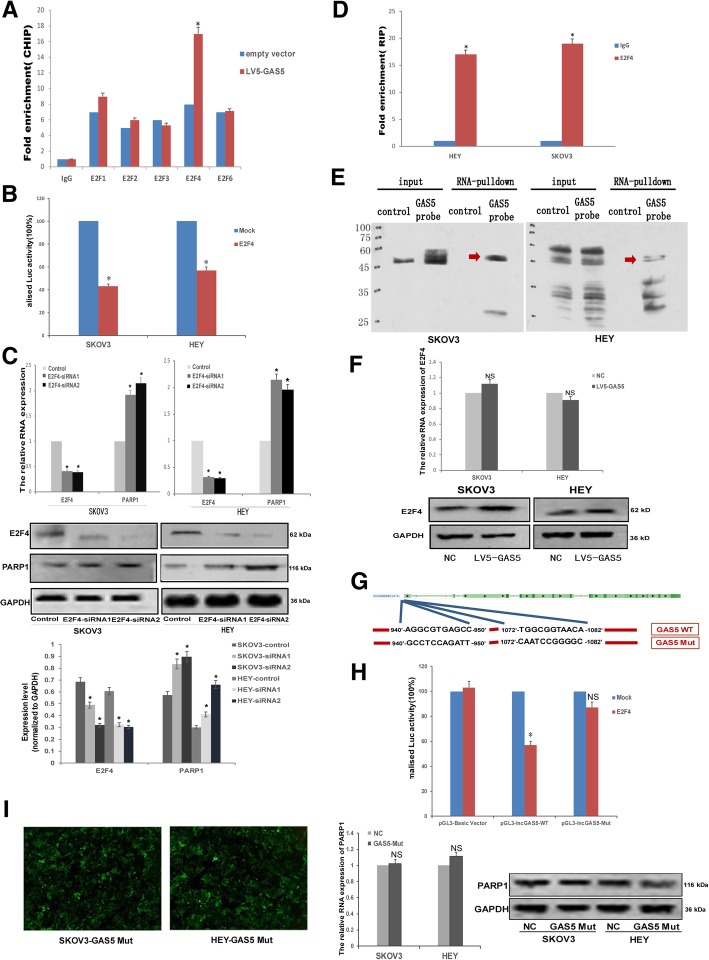
GAS5 regulated the PARP1 transcriptional by recruiting the transcription factor E2F4 to its promoter in OC cells
While PARP1 transcription has been reported to be repressed by E2Fs complex in human monocytes [22], in hypoxic cancer cells [23], and in bladder cancer [24]. We found that there were 69 E2Fs-binding sites in PARP1 promoters by Jaspar matrix models (http://jaspar.genereg.net/) (Table 2). The E2F family is mainly comprised of 8 members (E2F1–8). Though both E2F1, E2F2, E2F3, E2F4 and E2F6 could bind to PARP1 promoter, the CHIP-qPCR assay showed that in OC cells, only E2F4’s binding capacity was improved evidently with GAS5 over-expression (Fig. 5a). And there are 13 E2F4-binding sites in PARP1 promoters predicted by Jaspar (Table 3). Therefore we focus E2F4 as the target of our study. To illustrate the mechanism of the regulation of PARP1 expression by E2F4, human PARP1 luciferase reporter plasmid was used for luciferase reporter assay, and co-transfected E2F4 resulted in a decreased promoter activity of PARP1 in HEY and SKOV3 cells (Fig. 5b). E2F4 was then knockdown by siRNA, and the results of RT-qPCR and western blot revealed that PARP1 expression was significantly upregulated (Fig. 5c). And then RNA immunoprecipitation (RIP) assay was performed, which demonstrated that E2F4 protein could bind to GAS5 (Fig. 5d) as well. The direct interaction between E2F4 and GAS5 was further verified by applying total protein to GAS5 pull-down assay (Fig. 5e). In addition, we found GAS5 over-expression did not impact the expression of E2F4 protein and mRNA (Fig. 5f). To explored whether the combination of GAS5 with E2F4 is essential for regulating PARP1 expression, we checked the possible binding sites for E2F4 in GAS5 promoter predicted by Jaspar (Table 4) and constructed mutation type (Mut) GAS5 retroviral plasmid and luciferase plasmid (Fig. 5g). As shown by luciferase reporter assay, mutation type GAS5 (GAS5 Mut) plasmid co-transfected with E2F4 manifested no changes in GAS5 promoter activity in 293 cells, indicated that the mutation of the binding sites abolish the combination of GAS5 with E2F4 (Fig. 5h). While stable transfect mutation type GAS5 (GAS5 Mut) plasmid in SKOV3 and HEY cell lines (Fig. 5i, left), RT-qPCT and Western blot assay indicated that mutation type GAS5 transfection lost the effect on PARP1 expression (Fig. 5i, middle and right). Taken together, we concluded that GAS5 inhibited PARP1 transcriptional activity by recruiting transcription factor E2F4 to its promoter.
Table 2.
Possible binding sites of E2Fs in PARP1 promoter
| Model ID | Model name | Score | Relative score | Start | End | Strand | Predicted site sequence |
|---|---|---|---|---|---|---|---|
| MA0024.2 | E2F1 | 3.427 | 0.820 | 35 | 45 | −1 | TGAGCTGCAGC |
| MA0024.1 | E2F1 | 8.832 | 0.870 | 86 | 93 | 1 | TTTGGGGC |
| MA0024.2 | E2F1 | 5.271 | 0.847 | 102 | 112 | 1 | TGGGTGCCAGG |
| MA0471.1 | E2F6 | 8.684 | 0.889 | 375 | 385 | −1 | AAGTGGGAGGA |
| MA0024.2 | E2F1 | 4.624 | 0.838 | 376 | 386 | −1 | GAAGTGGGAGG |
| MA0470.1 | E2F4 | 4.217 | 0.812 | 388 | 398 | −1 | ACTCGGGAGGC |
| MA0471.1 | E2F6 | 4.490 | 0.827 | 388 | 398 | −1 | ACTCGGGAGGC |
| MA0024.2 | E2F1 | 5.067 | 0.844 | 389 | 399 | −1 | TACTCGGGAGG |
| MA0471.1 | E2F6 | 2.914 | 0.804 | 415 | 425 | −1 | TGGTGGCAGGT |
| MA0024.2 | E2F1 | 5.276 | 0.847 | 416 | 426 | −1 | GTGGTGGCAGG |
| MA0469.1 | E2F3 | 7.755 | 0.840 | 909 | 923 | −1 | CTCCCACCTCGACTT |
| MA0024.2 | E2F1 | 6.718 | 0.869 | 914 | 924 | 1 | GAGGTGGGAGG |
| MA0470.1 | E2F4 | 5.788 | 0.836 | 915 | 925 | 1 | AGGTGGGAGGA |
| MA0471.1 | E2F6 | 10.763 | 0.920 | 915 | 925 | 1 | AGGTGGGAGGA |
| MA0024.2 | E2F1 | 6.352 | 0.864 | 1238 | 1248 | 1 | TGGACGGCAGG |
| MA0471.1 | E2F6 | 5.290 | 0.839 | 1269 | 1279 | 1 | AGGAGGGTGGA |
| MA0024.2 | E2F1 | 5.156 | 0.846 | 1299 | 1309 | −1 | TTGGCCCGAGG |
| MA0469.1 | E2F3 | 5.736 | 0.813 | 1319 | 1333 | − 1 | CCCCCGCCTCGGGAA |
| MA0024.2 | E2F1 | 5.886 | 0.857 | 1324 | 1334 | 1 | GAGGCGGGGGC |
| MA0470.1 | E2F4 | 7.463 | 0.863 | 1325 | 1335 | 1 | AGGCGGGGGCC |
| MA0471.1 | E2F6 | 5.390 | 0.841 | 1325 | 1335 | 1 | AGGCGGGGGCC |
| MA0471.1 | E2F6 | 6.073 | 0.851 | 1364 | 1374 | 1 | GGGAGAGAGGA |
| PB0112.1 | E2F2_2 | 7.546 | 0.807 | 1384 | 1400 | −1 | ACGCCGGCCCCAAACTC |
| MA0024.1 | E2F1 | 8.832 | 0.870 | 1387 | 1394 | 1 | TTTGGGGC |
| MA0469.1 | E2F3 | 5.222 | 0.806 | 1434 | 1448 | −1 | TTCACGCCTCAGCCT |
| MA0024.2 | E2F1 | 7.150 | 0.876 | 1439 | 1449 | 1 | GAGGCGTGAAG |
| MA0470.1 | E2F4 | 6.980 | 0.855 | 1440 | 1450 | 1 | AGGCGTGAAGA |
| MA0471.1 | E2F6 | 7.460 | 0.871 | 1440 | 1450 | 1 | AGGCGTGAAGA |
| MA0024.2 | E2F1 | 2.878 | 0.811 | 1487 | 1497 | −1 | GTCTCGCCAAG |
| MA0024.2 | E2F1 | 5.067 | 0.844 | 1560 | 1570 | 1 | TACTCGGGAGG |
| MA0470.1 | E2F4 | 4.217 | 0.812 | 1561 | 1571 | 1 | ACTCGGGAGGC |
| MA0471.1 | E2F6 | 4.490 | 0.827 | 1561 | 1571 | 1 | ACTCGGGAGGC |
| MA0469.1 | E2F3 | 5.644 | 0.811 | 1568 | 1582 | −1 | CTCCCACCTCAGCCT |
| MA0024.2 | E2F1 | 6.718 | 0.869 | 1573 | 1583 | 1 | GAGGTGGGAGG |
| MA0470.1 | E2F4 | 5.788 | 0.836 | 1574 | 1584 | 1 | AGGTGGGAGGA |
| MA0471.1 | E2F6 | 10.763 | 0.920 | 1574 | 1584 | 1 | AGGTGGGAGGA |
| PB0112.1 | E2F2_2 | 8.488 | 0.826 | 1619 | 1635 | −1 | GCAGTGCCGCCATCATG |
| PB0112.1 | E2F2_2 | 7.730 | 0.810 | 1620 | 1636 | 1 | ATGATGGCGGCACTGCA |
| PB0113.1 | E2F3_2 | 7.920 | 0.808 | 1620 | 1636 | 1 | ATGATGGCGGCACTGCA |
| MA0470.1 | E2F4 | 3.684 | 0.803 | 1635 | 1645 | −1 | GCGCTGGAGTG |
| MA0471.1 | E2F6 | 3.063 | 0.806 | 1635 | 1645 | −1 | GCGCTGGAGTG |
| MA0469.1 | E2F3 | 5.667 | 0.812 | 1637 | 1651 | 1 | CTCCAGCGCGGTGAG |
| MA0024.2 | E2F1 | 4.515 | 0.836 | 1669 | 1679 | 1 | AAAGGGGGAGG |
| MA0471.1 | E2F6 | 5.461 | 0.842 | 1670 | 1680 | 1 | AAGGGGGAGGG |
| PB0112.1 | E2F2_2 | 9.187 | 0.840 | 1738 | 1754 | −1 | GACCCGGCGCCACCCCT |
| PB0113.1 | E2F3_2 | 10.273 | 0.856 | 1738 | 1754 | −1 | GACCCGGCGCCACCCCT |
| MA0024.2 | E2F1 | 4.195 | 0.831 | 1742 | 1752 | 1 | GTGGCGCCGGG |
| MA0024.2 | E2F1 | 4.281 | 0.832 | 1821 | 1831 | 1 | CACCCGGCAGG |
| MA0024.2 | E2F1 | 5.062 | 0.844 | 1827 | 1837 | −1 | CGGGCGCCTGC |
| MA0024.2 | E2F1 | 4.402 | 0.834 | 1828 | 1838 | 1 | CAGGCGCCCGG |
| MA0024.2 | E2F1 | 6.411 | 0.864 | 1832 | 1842 | 1 | CGCCCGGGAAA |
| MA0470.1 | E2F4 | 7.805 | 0.868 | 1833 | 1843 | 1 | GCCCGGGAAAC |
| MA0471.1 | E2F6 | 5.723 | 0.845 | 1833 | 1843 | 1 | GCCCGGGAAAC |
| MA0024.1 | E2F1 | 7.993 | 0.841 | 1835 | 1842 | −1 | TTTCCCGG |
| MA0470.1 | E2F4 | 3.661 | 0.803 | 1847 | 1857 | −1 | GGCCGGGGGGC |
| MA0024.2 | E2F1 | 6.975 | 0.873 | 1853 | 1863 | 1 | CGGCCGGCAGG |
| MA0470.1 | E2F4 | 3.553 | 0.801 | 1854 | 1864 | 1 | GGCCGGCAGGG |
| PB0008.1 | E2F2_1 | 8.763 | 0.811 | 1860 | 1874 | 1 | CAGGGGGCGCGCGCG |
| PB0009.1 | E2F3_1 | 9.191 | 0.826 | 1860 | 1874 | 1 | CAGGGGGCGCGCGCG |
| MA0024.2 | E2F1 | 6.297 | 0.863 | 1863 | 1873 | 1 | GGGGCGCGCGC |
| PB0008.1 | E2F2_1 | 8.245 | 0.800 | 1863 | 1877 | −1 | CGGCGCGCGCGCCCC |
| MA0470.1 | E2F4 | 3.869 | 0.806 | 1864 | 1874 | 1 | GGGCGCGCGCG |
| MA0024.2 | E2F1 | 5.095 | 0.845 | 1865 | 1875 | 1 | GGCGCGCGCGC |
| MA0024.2 | E2F1 | 5.095 | 0.845 | 1866 | 1876 | −1 | GGCGCGCGCGC |
| MA0470.1 | E2F4 | 3.825 | 0.805 | 1879 | 1889 | −1 | GGGCGGGGCCG |
| MA0470.1 | E2F4 | 12.687 | 0.946 | 1911 | 1921 | −1 | CCGCGGGAACG |
| MA0471.1 | E2F6 | 8.773 | 0.890 | 1911 | 1921 | −1 | CCGCGGGAACG |
| MA0024.2 | E2F1 | 6.807 | 0.870 | 1912 | 1922 | -1 | GCCGCGGGAAC |
| MA0469.1 | E2F3 | 13.658 | 0.920 | 1913 | 1927 | 1 | TTCCCGCGGCCAGGC |
69 putative sites were predicted with these settings (80%) in sequence named NC_000001.11:c226408100–226360691
Fig. 5.
GAS5 affected the PARP1 transcriptional expression by recruiting the transcription factor E2F4 to its promoter in OC cells. a LV5-GAS5 or the empty vector was transfected into OC cells, and chrome immunoprecipitations (CHIP) were performed by using specific anti-E2F1, anti-E2F2, anti-E2F3, anti-E2F4 or anti-E2F6 antibodies. b PARP1 luciferase reporter plasmid was used for luciferase reporter assay, and co-transfected E2F4 resulted in a decreased promoter activity of PARP1 in HEY and SKOV3 cells. c E2F4 siRNAs (siE2F4–1, 2) or the control siRNA were transfected into OC cells for 36 h, E2F4 and PARP1 mRNA and protein levels were then assessed by RT-qPCR or Western-blot assay. GAPDH was served as the internal control. d RNA immunoprecipitations (RIP) were performed in OC cells, and the relative quantities of GAS5 were detected by RT-qPCR assay, normalized to the input groups. IgG and E2F4 represented for the groups coprecipitation with IgG protein and anti-E2F4 antibody respectively. e Total proteins were extracted from SKOV3 and HEY cells, and then lncRNA GAS5 pull-down assay was performed. The E2F4 protein levels were evaluated by Western-blot. GAS5 probe represented the biotin-labeled GAS5 probe group and control stood for the oligo probe group. f After forced over-expression of GAS5, the mRNA and protein level of E2F4 were assessed by RT-qPCR and Western-blot assay. g Sketch map of GAS5 mutated E2F4 binding sites. h luciferase reporter assay: mutation type GAS5 (GAS5 Mut) plasmid co-transfected with E2F4 manifested no changes in GAS5 promoter activity in 293 cells. i After stable transfect mutation type GAS5 (GAS5 Mut) plasmid, the mRNA and protein level of PARP1 were assessed by RT-qPCR and Western-blot assay in SKOV3 and HEY cell lines. *P < 0.05 versus control groups
Table 3.
Possible binding sites of E2F4 in PARP1 promoter
| Model ID | Model name | Score | Relative score | Start | End | Strand | Predicted site sequence |
|---|---|---|---|---|---|---|---|
| MA0470.1 | E2F4 | 4.217 | 0.812 | 388 | 398 | -1 | ACTCGGGAGGC |
| MA0470.1 | E2F4 | 5.788 | 0.836 | 915 | 925 | 1 | AGGTGGGAGGA |
| MA0470.1 | E2F4 | 7.463 | 0.863 | 1325 | 1335 | 1 | AGGCGGGGGCC |
| MA0470.1 | E2F4 | 6.980 | 0.855 | 1440 | 1450 | 1 | AGGCGTGAAGA |
| MA0470.1 | E2F4 | 4.217 | 0.812 | 1561 | 1571 | 1 | ACTCGGGAGGC |
| MA0470.1 | E2F4 | 5.788 | 0.836 | 1574 | 1584 | 1 | AGGTGGGAGGA |
| MA0470.1 | E2F4 | 3.684 | 0.803 | 1635 | 1645 | -1 | GCGCTGGAGTG |
| MA0470.1 | E2F4 | 7.805 | 0.868 | 1833 | 1843 | 1 | GCCCGGGAAAC |
| MA0470.1 | E2F4 | 3.661 | 0.803 | 1847 | 1857 | -1 | GGCCGGGGGGC |
| MA0470.1 | E2F4 | 3.553 | 0.801 | 1854 | 1864 | 1 | GGCCGGCAGGG |
| MA0470.1 | E2F4 | 3.869 | 0.806 | 1864 | 1874 | 1 | GGGCGCGCGCG |
| MA0470.1 | E2F4 | 3.825 | 0.805 | 1879 | 1889 | -1 | GGGCGGGGCCG |
| MA0470.1 | E2F4 | 12.687 | 0.946 | 1911 | 1921 | -1 | CCGCGGGAACG |
13 putative sites were predicted with these settings (80%) in sequence named NC_000001.11:c226408100–226360691
Table 4.
Possible binding sites of E2F4 in GAS5 promoter
| Model ID | Model name | Score | Relative score | Start | End | Strand | Predicted site sequence |
|---|---|---|---|---|---|---|---|
| MA0470.1 | E2F4 | 5.773 | 0.836 | 789 | 799 | -1 | AGGCAGGAGAA |
| MA0470.1 | E2F4 | 4.619 | 0.818 | 940 | 950 | 1 | AGGCGTGAGCC |
| MA0470.1 | E2F4 | 4.206 | 0.811 | 1049 | 1059 | -1 | AGGAGCGAAAG |
| MA0470.1 | E2F4 | 6.297 | 0.845 | 1072 | 1082 | 1 | TGGCGGTAACA |
4 putative sites were predicted with these settings (80%) in sequence named NC_000001.11:c226408100–226360691
The expression of GAS5 in cytoplasm could be elevated by rapamycin
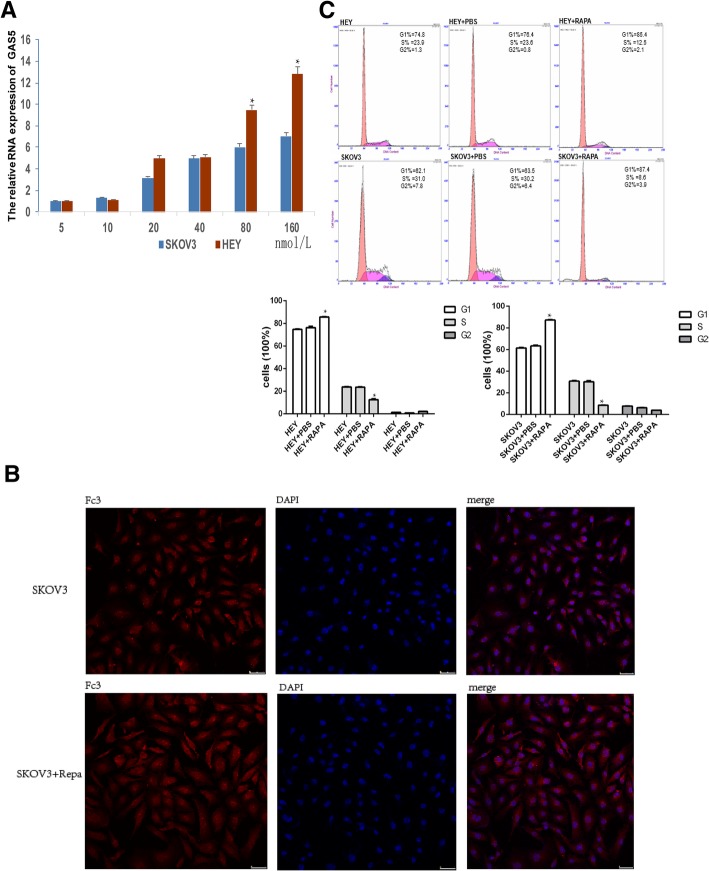
Since lncRNA-GAS5 is a 5′ TOP (5′-terminal oligonucleotide) gene [11], translational inhibitors such as rapamycin, cycloheximide, and miconazole can increase the accumulation of GAS5 spliced mRNA [25, 26]. Our RT-qPCR assay confirmed that GAS5 mRNA levels increased in a dose-dependent manner after rapamycin treatment (Fig. 6a), and FISH experiments confirmed that rapamycin treatment caused GAS5 to aggregate in SKOV3 cells (Fig. 6b). And as shown by the results of the flow cytometry, rapamycin treatment caused G0/G1 cell cycle arrest as well (Fig.6c). Here rapamycin might be considered as an intervention therapy against ovarian cancer progression by regulating GAS5 expression, and our findings may lay a theoretical foundation for the development of combination therapy of ovarian cancer.
Fig. 6.
The expression of GAS5 in cytoplasm could be elevated by Rapamycin. a The mRNA and protein level of GAS5 were assessed by RT-qPCR in OC cell lines treated by Rapamycin. b Cy3-labeled GAS5 and DAPI-labeled U6 probes were obtained from RiboBio. RNA FISH were performed using fluorescent in situ hybridization kit (RiboBio) according to manufacturer’s instructions. c Flow cytometry assay were performed to detect the cell cycle impacted by Rapamycin treating in OC cell lines SKOV3 and HEY. *P < 0.05 versus control groups
Discussion
In recent years, research on tumor drug sensitivity-related lncRNA has been receiving increasing attention. Among them, the studies on tumor metastasis-associated lncRNA MALAT1 is relatively mature. Some studies reported that MALAT1 inhibited E-cadherin expression by binding to highly expressed EZH2 at its 3′ end in colorectal cancer, thereby caused tumor cell epithelial-mesenchymal transition (EMT) and oxaliplatin resistance [27]. Meanwhile, MALAT1 caused tumor cell infiltration and drug resistance in glioblastoma multiforme (GBM), combining standard TMZ treatment with targeted nanocomplex carrying siRNA against MALAT1 substantially enhanced the very poor prognosis for GBM patients [28]. And the use of small interfering RNA knockdown MALAT1 in the treatment of enzalutamide (Enz)-resistant prostate cancer has also entered preclinical studies stage [29]. HOTAIR is another lncRNA that has been studied a lot. Teschendorff AE, etc. [30] reported that HOTAIR is highly expressed in ovarian cancer and is associated with poor prognosis and carboplatin resistance. Özeş AR, etc [31] showed that HOTAIR could affects platinum resistance in ovarian cancer by down-regulating NF-κB inhibitor Iκ-Bα, prolonging NF-κB activity, activating its downstream protein interleukin-6 expression to activate DNA damage response pathway (DDR), ultimately maintained tumor cell genome stability, maintain cell survival and avoided apoptosis.
In addition to HOTAIR, more and more lncRNAs affecting ovarian cancer drug sensitivity have been discovered and studied. Wang F, etc [32] found that lncRNA UCA1 affected platinum resistance in ovarian cancer by targeting the SRPK1/PI3K-Akt signaling pathway. Ji An, etc [33] reported that lncRNA NEAT1 affected paclitaxel resistance in ovarian cancer by recruiting miR − 194 to target ZEB1 and then regulating P-gp and GST. However, compared with other tumors, the function and mechanism study of lncRNA in ovarian cancer is relatively weak. In this study, we selected lncRNA-GAS5 to be dramatically down-regulated in ovarian cancer by microarray assay, and then confirmed the low-expression of GAS5 in EOC tissues and OC cell lines. When detected the expression level of GAS5 in OC cell lines we found that the loss of GAS5 in drug-resistant cell lines was much more evidently than in drug-sensitive cells. These findings leaded us to speculate whether GAS5 could affects OC drug sensitivity. Drug sensitivity tests in vitro and in vivo confirmed our conjecture, exogenous overexpression of GAS5 by retroviral plasmids significantly increased the susceptibility of OC to DDP in vivo and in vitro. The results of the flow cytometry and Western-blot showed that overexpression of GAS5 caused G0/G1 cell cycle arrest, apoptosis increase, and affected the expression of cell cycle and apoptosis-related proteins including PARP1.
Poly-ADP-ribose polymerase 1 (PARP1) is a multitasking enzyme that regulates many intracellular processes, including DNA repair, metabolism, signaling and transcription, by direct interaction with other proteins and DNA [34, 35]. High levels of PARP1 in cancer cells promote cell cycle progression, and may in response to proliferation arrest thereby sensitize cells to agents that challenge redox homeostasis [21]. As for epithelial ovarian cancers, the inhibition of PARP1 even became a promising targeted therapies [36]. PARP1 transcription was reported to be repressed by lncRNAs in lung cancer [37], hepatocellular carcinoma [38], neuroblastoma [39], thyroid cancer [40], multiple myeloma [41], lung morphogenesis [42] and angiogenesis [43]. While in this study, based on our microarray, RT-qPCT and WB data, we found that PARP1 is high-expressed in EOC samples, negatively related with GAS5 expression level, and forced expression of GAS5 markedly down-regulated PARP1 both on protein and mRNA level. The repression of PARP1 transcription by E2F4 complex in human monocytes was reported by Wiśnik E, etc. [22] and Robaszkiewicz A, etc [44] Our luciferase reporter assay showed that E2F4 could decreased promoter activity of PARP1 in OC cells. 13 E2F4-binding sites in PARP1 promoters were predicted by Jaspar matrix models, CHIP assay showed that E2F4’s binding capacity to PARP1 promote was improved evidently with GAS5 overexpression. RNA immunoprecipitation (RIP) assay was performed to demonstrate the E2F4 binding to GAS5. The direct interaction between E2F4 and GAS5 was further verified by GAS5 pull-down assay. While when mutated the binding sites with E2F4, GAS5 lost the regulation capacity on PARP1 expression. Taken together, we concluded that GAS5 inhibited PARP1 transcriptional activity by recruiting transcription factor E2F4 to PARP1 promoter (Fig. 7).
Fig. 7.
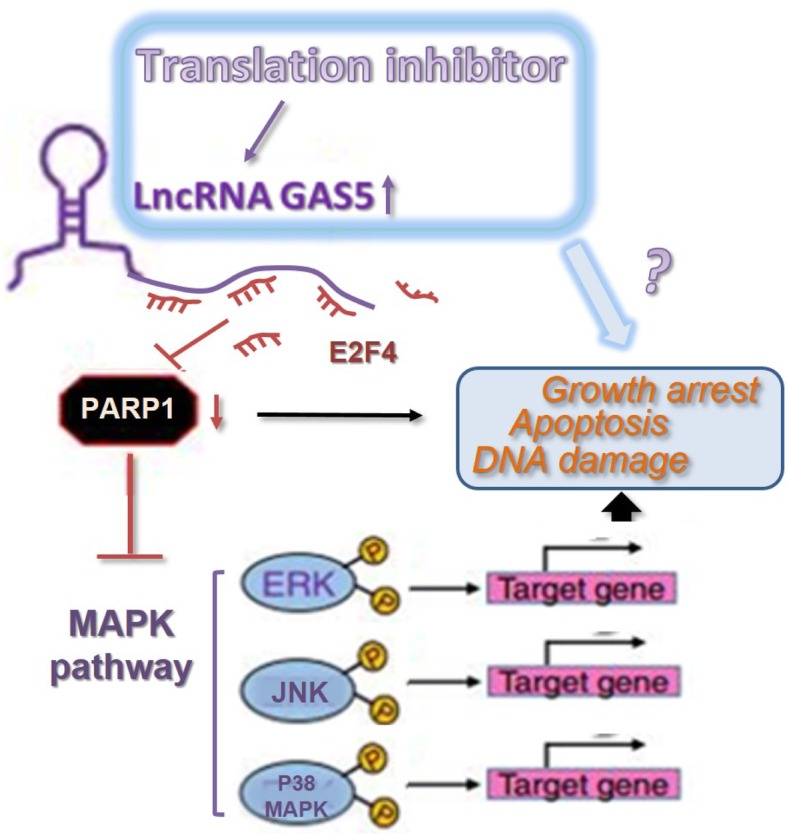
Schematic diagram for GAS5/E2F4/PARP1/MAPK axis in ovarian cancer
Since lncRNA-GAS5 is a 5′ TOP (5′-terminal oligonucleotide) gene [11], translational inhibitors such as rapamycin, cycloheximide, and miconazole can increase the accumulation of GAS5 spliced mRNA [25, 26]. Here with RT-qPCR and FISH, we confirmed the up-regulation of GAS5 by rapamycin in OC cells. As an intervention therapy against the low-expression of GAS5, rapamycin may take part in the combination therapy of ovarian cancer, but need more investigation.
There are some limitations of our study, first of all, whether rapamycin can affect the apoptosis of ovarian cancer cells by targeting GAS5 expression and how to participate in GAS5 target gene (E2F4 or PARP1) regulation need to be further studied. Secondly, the clinical sample size in this study can be enlarged, especially in the normal group, and the effect of GAS5 expression level on prognosis in different stages of ovarian cancer can be lucubrated. Finally, the function and molecular mechanism of GAS5 can be further verified in GAS5 knockdown OC cell lines.
Conclusion
In conclusion, we assessed the lncRNA expression profile of EOC, proved that GAS5 worked as a tumor suppressor like gene in OC and might effected DDP sensitivity. We presented the GAS5-E2F4-PARP1-MAPK axis and explored its role in OC progression and drug-sensitivity for the first time. And we proposed that Rapamycin might be used to offset the low expression of GAS5 in ovarian cancer.
Acknowledgements
Thanks all support from Department of Obstetrics and Gynecology, Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University. The authors are grateful to all staffs who contributed to this study.
Abbreviations
- CHIP
Chromatin Immunoprecipitation
- DDP
Cisplatin
- EOC
Epithelial ovarian cancer
- FISH
Fluorescence in situ hybridization
- lncRNA
Long non-coding RNA
- OC
Ovarian cancer
- RIP
RNA immunoprecipitation
- RT-qPCT
Quantificational real-time polymerase chain reaction
- WB
Western blot
Authors’ contributions
XRL and KQS designed the experiments, performed experiments, analyzed data and wrote the paper. HH helped to analyze the experimental data and amend the writing of this article. QT and WJW helped analyzed data. QD helped in cell culture. XY helped in providing tissue samples. WD initiated and organized the study and critically revised the manuscript. All authors read and approved the final version of the manuscript.
Funding
This work was supported by: the 63th batch of China’s Postdoctoral Science Fund projects (No.195380), Key Discipline Project of Shanghai Municipal Commission of Health and Family Planning (No.15GWZK0701), National Natural Science Foundation of China (No. 81772770), Key Clinical Disciplines of Shanghai (No.2017ZZ02016) and National Key R&D Program of China (No.2016YFC1302900).
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
This study was conducted in accordance with the Declaration of Helsinki principles. It was approved by the Medical Research Ethics Committee of Renji Hospital, School of Medicine, Shanghai Jiao Tong University.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiaoran Long, Keqi Song and Hao Hu contributed equally to this work.
Contributor Information
Xiaoran Long, Email: longxr@mail2.sysu.edu.cn.
Keqi Song, Email: songkeqi@hotmail.com.
Hao Hu, Email: 18262639612@163.com.
Qi Tian, Email: tianqizaifei@sina.com.
Wenjing Wang, Email: 1090212224@qq.com.
Qian Dong, Email: dongqian95fy@163.com.
Xia Yin, Email: yinxia2@aliyun.com.
Wen Di, Phone: +86-13826075261, Email: diwen163@163.com.
References
- 1.Coward JI, Middleton K, Murphy F. New perspectives on targeted therapy in ovarian cancer. Int J Women's Health. 2015;7:189–203. doi: 10.2147/IJWH.S52379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 4.Bowtell DD. The genesis and evolution of high-grade serous ovarian cancer. Nat Rev Cancer. 2010;10:803–808. doi: 10.1038/nrc2946. [DOI] [PubMed] [Google Scholar]
- 5.Pinato DJ, Graham J, Gabra H, Sharma R. Evolving concepts in the management of drug resistant ovarian cancer: dose dense chemotherapy and the reversal of clinical platinum resistance. Cancer Treat Rev. 2013;39:153–160. doi: 10.1016/j.ctrv.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Harries LW. Long non-coding RNAs and human disease. Biochem Soc Trans. 2012;40:902–906. doi: 10.1042/BST20120020. [DOI] [PubMed] [Google Scholar]
- 7.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 9.Orom UA, Shiekhattar R. Long noncoding RNAs usher in a new era in the biology of enhancers. Cell. 2013;154:1190–1193. doi: 10.1016/j.cell.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31:4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith CM, Steitz JA. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol. 1998;18(12):6897–6909. doi: 10.1128/MCB.18.12.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong S, Qu X, Li W, Zhong X, Li P, Yang S, Chen X, Shao M, Zhang L. The long non-coding RNA, GAS5, enhances gefitinib-induced cell death in innate EGFR tyrosine kinase inhibitor-resistant lung adenocarcinoma cells with wide-type EGFR via downregulation of the IGF−1R expression. J Hematol Oncol. 2015;8:43. [DOI] [PMC free article] [PubMed]
- 13.Tao R, Hu S, Wang S, Zhou X, Zhang Q, Wang C, Zhao X, Zhou W, Zhang S, Li C, et al. Association between indel polymorphism in the promoter region of lncRNA GAS5 and the risk of hepatocellular carcinoma. Carcinogenesis. 2015;36:1136–1143. doi: 10.1093/carcin/bgv099. [DOI] [PubMed] [Google Scholar]
- 14.Pickard MR, Williams GT. Regulation of apoptosis by long non-coding RNA GAS5 in breast cancer cells: implications for chemotherapy. Breast Cancer Res Treat. 2014;145:359–370. doi: 10.1007/s10549-014-2974-y. [DOI] [PubMed] [Google Scholar]
- 15.Yang W, Hong L, Xu X, Wang Q, Huang J, Jiang L. LncRNA GAS5 suppresses the tumorigenesis of cervical cancer by downregulating miR−196a and miR-205. Tumour Biol. 2017;39:1010428317711315. doi: 10.1177/1010428317711315. [DOI] [PubMed] [Google Scholar]
- 16.Sun D, Yu Z, Fang X, Liu M, Pu Y, Shao Q, Wang D, Zhao X, Huang A, Xiang Z, et al. LncRNA GAS5 inhibits microglial M2 polarization and exacerbates demyelination. EMBO Rep. 2017;18:1801–1816. doi: 10.15252/embr.201643668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang NYG, Shao XM, Wei L. GAS5 modulated autophagy is a mechanism modulating cisplatin sensitivity in NSCLC cells. Eur Rev Med Pharmacol Sci. 2016;20(11):2271–2277. [PubMed] [Google Scholar]
- 18.Wen Q, Liu Y, Lyu H, Xu X, Wu Q, Liu N, Yin Q, Li J, Sheng X. Long noncoding RNA GAS5, which acts as a tumor suppressor via microRNA 21, regulates cisplatin resistance expression in cervical Cancer. Int J Gynecol Cancer. 2017;27:1096–1108. doi: 10.1097/IGC.0000000000001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H, Guo Y, Song Y, Shang C. Long noncoding RNA GAS5 inhibits malignant proliferation and chemotherapy resistance to doxorubicin in bladder transitional cell carcinoma. Cancer Chemother Pharmacol. 2017;79:49–55. doi: 10.1007/s00280-016-3194-4. [DOI] [PubMed] [Google Scholar]
- 20.Pietrzak J, Spickett CM, Ploszaj T, Virag L, Robaszkiewicz A. PARP1 promoter links cell cycle progression with adaptation to oxidative environment. Redox Biol. 2018;18:1–5. doi: 10.1016/j.redox.2018.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain Priyancy G., Patel Bhumika D. Medicinal chemistry approaches of poly ADP-Ribose polymerase 1 (PARP1) inhibitors as anticancer agents - A recent update. European Journal of Medicinal Chemistry. 2019;165:198–215. doi: 10.1016/j.ejmech.2019.01.024. [DOI] [PubMed] [Google Scholar]
- 22.Wiśnik E, Płoszaj T, Robaszkiewicz A. Downregulation of PARP1 transcription by promoter-associated E2F4-RBL2-HDAC1-BRM complex contributes to repression of pluripotency stem cell factors in human monocytes. Sci Rep. 2017;7(1):9483. [DOI] [PMC free article] [PubMed]
- 23.Hegan DC, Lu Y, Stachelek GC, Crosby ME, Bindra RS, Glazer PM. Inhibition of poly(ADP-ribose) polymerase down-regulates BRCA1 and RAD51 in a pathway mediated by E2F4 and p130. Proc Natl Acad Sci U S A. 2010;107:2201–2206. doi: 10.1073/pnas.0904783107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Léger K, Hopp AK, Fey M, Hottiger MO. ARTD1 regulates cyclin E expression and consequently cell-cycle re-entry and G1/S progression in T24 bladder carcinoma cells. Cell Cycle. 2016;15(15):2042–2052. doi: 10.1080/15384101.2016.1195530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lucafò M, Bravin V, Tommasini A, Martelossi S, Rabach I, Ventura A, Decorti G, De Iudicibus S. Differential expression of GAS5 in rapamycin-induced reversion of glucocorticoid resistance. Clin Exp Pharmacol Physiol. 2016;43:602–605. doi: 10.1111/1440-1681.12572. [DOI] [PubMed] [Google Scholar]
- 26.Yacqub-Usman K, Pickard MR, Williams GT. Reciprocal regulation of GAS5 lncRNA levels and mTOR inhibitor action in prostate cancer cells. Prostate. 2015;75(7):693–705. doi: 10.1002/pros.22952. [DOI] [PubMed] [Google Scholar]
- 27.Li P, Zhang X, Wang H, Wang L, Liu T, Du L, Yang Y, Wang C. MALAT1 is associated with poor response to oxaliplatin-based chemotherapy in colorectal cancer patients and promotes Chemoresistance through EZH2. Mol Cancer Ther. 2017;16(4):739–751. doi: 10.1158/1535-7163.MCT-16-0591. [DOI] [PubMed] [Google Scholar]
- 28.Kim SS, Harford JB, Moghe M, Rait A, Pirollo KF. Chang EH targeted nanocomplex carrying siRNA against MALAT1 sensitizes glioblastoma to temozolomide. Nucleic Acids Res. 2018;46(3):1424–1440. doi: 10.1093/nar/gkx1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang R, Sun Y, Li L, Niu Y, Lin W, Lin C, Antonarakis ES, Luo J, Yeh S, Chang C. Preclinical study using Malat1 small interfering RNA or androgen receptor splicing variant 7 degradation enhancer ASC-J9((R)) to suppress enzalutamide-resistant prostate Cancer progression. Eur Urol. 2017;72:835–844. doi: 10.1016/j.eururo.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teschendorff AE, Lee SH, Jones A, Fiegl H, Kalwa M, Wagner W, Chindera K, Evans I, Dubeau L, Orjalo A, et al. HOTAIR and its surrogate DNA methylation signature indicate carboplatin resistance in ovarian cancer. Genome Med. 2015;7:108. doi: 10.1186/s13073-015-0233-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozes AR, Miller DF, Ozes ON, Fang F, Liu Y, Matei D, Huang T, Nephew KP. NF-kappaB-HOTAIR axis links DNA damage response, chemoresistance and cellular senescence in ovarian cancer. Oncogene. 2016;35:5350–5361. doi: 10.1038/onc.2016.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.WANG F., ZHOU J., XIE X., HU J., CHEN L., HU Q., GUO H., YU C. Involvement of SRPK1 in cisplatin resistance related to long non-coding RNA UCA1 in human ovarian cancer cells. Neoplasma. 2015;62(03):432–438. doi: 10.4149/neo_2015_051. [DOI] [PubMed] [Google Scholar]
- 33.An J, Lv W, Zhang Y. LncRNA NEAT1 contributes to paclitaxel resistance of ovarian cancer cells by regulating ZEB1 expression via miR-194. Onco Targets Ther. 2017;10:5377–5390. doi: 10.2147/OTT.S147586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hottiger MO. Nuclear ADP-Ribosylation and its role in chromatin plasticity, cell differentiation, and epigenetics. Annu Rev Biochem. 2015;84:227–263. doi: 10.1146/annurev-biochem-060614-034506. [DOI] [PubMed] [Google Scholar]
- 35.Rosado MM, Bennici E, Novelli F, Pioli C. Beyond DNA repair, the immunological role of PARP−1 and its siblings. Immunology. 2013;139:428–437. doi: 10.1111/imm.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dal Molin GZ, Omatsu K, Sood AK, Coleman RL. Rucaparib in ovarian cancer: an update on safety, efficacy and place in therapy. Ther Adv Med Oncol. 2018;10:1758835918778483. doi: 10.1177/1758835918778483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo H, Sun Y, Wei G, Luo J, Yang X, Liu W, Guo M, Chen R. Functional characterization of long noncoding RNA Lnc_bc060912 in human lung carcinoma cells. Biochemistry. 2015;54:2895–2902. doi: 10.1021/acs.biochem.5b00259. [DOI] [PubMed] [Google Scholar]
- 38.Qi Heqiang, Lu Yuyan, Lv Jie, Wu Huita, Lu Jing, Zhang Changmao, Zhang Sheng, Bao Qing, Zhang Xiuming, Xie Chengrong, Yin Zhenyu. The long noncoding RNA lncPARP1 contributes to progression of hepatocellular carcinoma through up-regulation of PARP1. Bioscience Reports. 2018;38(3):BSR20180703. doi: 10.1042/BSR20180703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao X, Li D, Huang D, Song H, Mei H, Fang E, Wang X, Yang F, Zheng L, Huang K, Tong Q. Risk-associated long noncoding RNA FOXD3-AS1 inhibits neuroblastoma progression by repressing PARP1-mediated activation of CTCF. Mol Ther. 2018;26:755–773. doi: 10.1016/j.ymthe.2017.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Xiang C, Zhang ML, Zhao QZ, Xie QP, Yan HC, Yu X, Wang P, Wang Y. LncRNA-SLC6A9-5:2: a potent sensitizer in 131I-resistant papillary thyroid carcinoma with PARP−1 induction. Oncotarget. 2017;8(14):22954–22967. doi: 10.18632/oncotarget.14578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu Y, Lin J, Fang H, Fang J, Li C, Chen W, Liu S, Ondrejka S, Gong Z, Reu F, et al. Targeting the MALAT1/PARP1/LIG3 complex induces DNA damage and apoptosis in multiple myeloma. Leukemia. 2018;32:2250–2262. doi: 10.1038/s41375-018-0104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Banerjee P, Surendran H, Bharti K, Morishita K, Varshney A, Pal R. Long noncoding RNA RP11-380D23.2 drives distal-proximal patterning of the lung by regulating PITX2 expression. Stem Cells. 2018;36:218–229. doi: 10.1002/stem.2740. [DOI] [PubMed] [Google Scholar]
- 43.Man HSJ, Sukumar AN, Lam GC, Turgeon PJ, Yan MS, Ku KH, Dubinsky MK, Ho JJD, Wang JJ, Das S, et al. Angiogenic patterning by STEEL, an endothelial-enriched long noncoding RNA. Proc Natl Acad Sci U S A. 2018;115:2401–2406. doi: 10.1073/pnas.1715182115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robaszkiewicz A, Wisnik E, Regdon Z, Chmielewska K, Virag L. PARP1 facilitates EP300 recruitment to the promoters of the subset of RBL2-dependent genes. Biochim Biophys Acta Gene Regul Mech. 2017;1874–9399. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.