Abstract
Background
Individuals can be infected with multiple strains of Helicobacter pylori. However, the differences among co-infecting strains have not been well analyzed yet. This study aimed to investigate whether the virulence factors and antibiotic resistance patterns of H. pylori differ between strains isolated from different locations of the stomach in the same patient.
Methods
H. pylori isolates were obtained from the antrum and body of the stomach. Genetic differences were examined by random amplified polymorphic DNA (RAPD) fingerprinting. Antibiotic resistance was assessed using the agar dilution method. Virulence factors were identified by polymerase chain reaction and DNA sequencing.
Results
Among 80 patients, co-infection by two H. pylori strains was detected in 10 patients. Among the 10 pairs of H. pylori strains, differences in antibiotic resistance patterns were detected in 7 pairs (clarithromycin, 1 patient; quinolone, 3 patients; metronidazole, 4 patients) and differences in virulence factors were detected in 5 pairs. The cagA virulence gene was detected in all 10 patients, and 2 patients had H. pylori strains with different EPIYA motifs. Differences in vacA genotypes were detected in 4 patients.
Conclusions
Co-infection by two H. pylori strains was confirmed by RAPD fingerprinting. Frequently, two H. pylori strains obtained from a single host differed in their virulence factors and antibiotic resistance patterns. Co-infection by multiple H. pylori strains could undermine the success of eradication therapy and should be considered when interpreting the results of antimicrobial susceptibility tests.
Electronic supplementary material
The online version of this article (10.1186/s12876-019-1062-5) contains supplementary material, which is available to authorized users.
Keywords: Random amplified polymorphic DNA, Helicobacter pylori, Virulence factor, Antibiotic resistance
Background
Helicobacter pylori is a spiral microaerophilic gram-negative bacterium that colonizes the human gastric mucosa. H. pylori is a major cause of gastric and duodenal diseases, and it has been estimated to currently infect half of the world’s population [1, 2]. It is already known that many virulence factors of H. pylori are related to the occurrence of various gastrointestinal disorders [3, 4]. The effect of H. pylori eradication therapy on gastrointestinal disorders has been vigorously studied. There is evidence that H. pylori eradication can cure gastritis, delay or prevent progression to long-term complications, or prevent the recurrence of disease. Even some extragastric diseases are thought to be related to H. pylori infection and could potentially be cured with eradication therapy.
Despite these benefits of H. pylori eradication therapy, an increasing resistance of the organism to antibiotics is becoming a major concern. Since the antibiotic resistance of H. pylori is the most important cause of eradication failure in many regions of the world, the role of antimicrobial susceptibility testing is becoming more important, especially in populations from regions with a high antibiotic resistance or after experiencing eradication failure [5, 6]. In addition, co-infection by more than one H. pylori strain in a single patient should be considered when antibiotic resistance is suspected [7–10]. Multiple H. pylori strains isolated from the same host might differ in certain characteristics such as their virulence factors or antibiotic resistance patterns. Such co-infections could affect the success of eradication therapy, thereby influencing the treatment course and prognosis of patients with H. pylori infection. However, the differences among H. pylori strains obtained from the same host in terms of their characteristics have not been well investigated yet.
In our study, infection by two H. pylori strains in a single patient was identified using random amplified polymorphic DNA (RAPD) fingerprinting, and the differences in virulence factors and antibiotic resistance between the co-infecting strains were investigated.
Methods
Patients and bacterial strains
We retrospectively enrolled 80 patients. Enrolled patients were those who visitied Chung-Ang University Yong-san Hospital between January 2005 and December 2009 due to upper gastrointestinal symptoms and underwent esophagogastroduodenoscopy with biopsies from the antrum and body of the stomach, with successful H. pylori isolation from both antrum and body. The patients with positive H. pylori culture only from one site, either antrum or body, were not eligible for this study. Totally, 80 pairs of H. pylori isolates were obtained from the enrolled subjects. The Institutional Review Board of Chung-Ang University Hospital approved this study [IRB number: C2014247(1444)].
Histopathologic evaluation of biopsy samples from gastric mucosa
Two gastric mucosal tissues, one from the lesser curvature of the antrum and the other from the lesser curvature of the body, were taken for histologic evaluation. These tissue samples were histologically examined using the updated Sydney system: H. pylori density, inflammatory activity (neutrophil infiltration), atrophy, and intestinal metaplasia were assessed and scored as 0 (none), 1 (mild), 2 (moderate), and 3 (marked) [11].
H. pylori culture and genomic DNA isolation
H. pylori isolates were cultured at 37 °C on Brucella agar plates (Becton Dickinson, Franklin Lakes, NJ, USA) containing 5% defibrinated sheep blood (Hanil Komed, Seongnam, Republic of Korea) under microaerobic conditions (5% O2, 10% CO2, 85% N2) for 3–5 days. The H. pylori isolates subcultured less than 3 times were used in all experiments, and subculture was carried out on blood brucella agar plates under the same conditions. Organisms were identified as H. pylori by colony morphology, rapid urease test, H. pylori-selective media [Oxoid™ SR 147 supplement (Thermo Fisher Scientific, Waltham, MA) and 5% defibrinated sheep blood], and polymerase chain reaction (PCR) to detect ureA.
Genomic DNA extraction was performed by harvesting H. pylori subcultured for 3–5 days and using a HiYield™ genomic DNA mini kit (Real Biotech Corporation, Taipei, Taiwan). The isolated genomic DNA was stored at − 20 °C until required for PCR amplification.
PCR for virulence factors and RAPD fingerprinting
PCR was used to detect the H. pylori-specific ureA gene, CagA EPIYA motif type, vacA genotype, oipA status, and cagA, dupA, and iceA genes. RAPD fingerprinting was used to analyze H. pylori isolated from the stomach to determine whether there were multiple strains co-infecting a single patient [10, 12–14]. All the primer sets used were selected from previously published literature (Additional file 1: Table S1).
The Ex Taq™ DNA polymerase (Takara Bio, Otsu, Shiga, Japan) was used for PCR amplification, which was performed in a volume of 50 μL, containing 5 μL 10× Ex Taq™ buffer including MgCl2, 4 μL dNTP mixture (2.5 mM each), 10 pmole of each primer, and 1.25 U Ex Taq™ DNA polymerase, following previously described methods [12, 15–24]. A GeneAmp® PCR system 2700 (Applied Biosystems, Foster City, CA, USA) was used for amplification.
Sequencing of CagA EPIYA, oipA, V domain of 23S rRNA gene, gyrA and gyrB
PCR and DNA sequencing were used to confirm whether the cagA genes of the isolated H. pylori strains had the Western or East Asian type of EPIYA motif [4]. The EPIYA-containing region of amplified cagA was confirmed by agarose gel electrophoresis and gel extraction was performed. The electrophoretic bands were cut out and PCR products were extracted from the gel using a HiYield™ gel/PCR DNA mini kit (Real Biotech Corporation). The gel extraction products thus obtained were submitted for sequencing (Macrogen Corporation, Seoul, Republic of Korea). Sequences of the EPIYA-containing regions thus obtained were converted to protein sequences using CLC Main Workbench 5 (Qiagen, Hilden, Germany) and aligned. The sequence of the amplification product of the primers targeting the oipA gene was aligned through CLC Main Workbench 5 and the presence of the CT repeat motif was confirmed. Additionally, the mutations of 23S rRNA gene and gyrA/gyrB sequence were analyzed to clarify the mechanism of antibiotic resistance for clarithromycin and quinolone, respectively.
Determination of minimum inhibitory concentrations (MICs) of antibiotics
In our study, we selected the internationally recognized agar dilution method, which can accurately measure MICs and is widely used for antibiotic resistance testing (Clinical and Laboratory Standards Institute document M07-A10). The antibiotics used in the MIC tests were clarithromycin (0.0625–16 μg/mL), levofloxacin (0.125–16 μg/mL), amoxicillin (0.125–16 μg/mL), metronidazole (2–128 μg/mL), moxifloxacin (0.125–16 μg/mL), and tetracycline (0.125–16 μg/mL).
H. pylori were inoculated on a Mueller–Hinton agar plate (Becton Dickinson) containing each antibiotic and 5% defibrinated sheep blood. After H. pylori were cultured at 37 °C in a 10% CO2 environment for 5–7 days, the plate was examined for visible growth and the lowest concentration of antibiotics that completely inhibited growth was determined as the MIC point. MIC testing was performed three times for each strain. Only the MIC point obtained using agar dilution method in previous literature was used to avoid confusion over the interpretation of antibiotic resistance results determined using other MIC methods [25]. Differences of MIC for an antibiotic between two strains cultured from a single patient were considered significant when the MICs differed by 4 times or more. When there was a difference in the presence of resistance even for one antibiotic between two strains cultured from one patient, the strains were defined as having different antibiotic resistance patterns.
Results
RAPD fingerprinting and clinical information
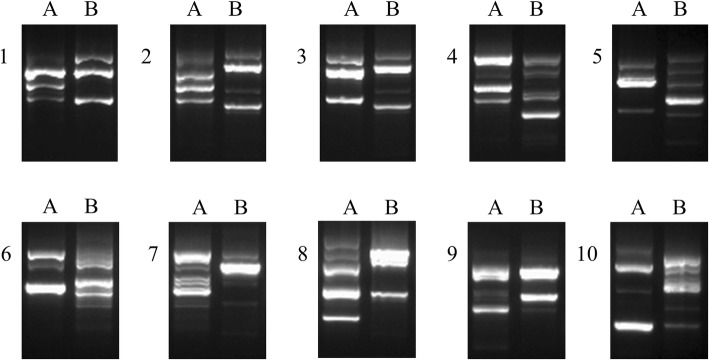
Among 80 patients with successful H. pylori isolation from the antrum and body of the stomach, the co-existence of different H. pylori strains was detected in 10 patients by RAPD fingerprinting of H. pylori (Fig. 1). Furthermore, PCR for the ureA gene was performed to confirm the H. pylori isolates from 10 patients (6 females, 4 males). The median age of the 10 patients was 57 years (range 40–69). All of the 10 patients were diagnosed with gastritis, and none had gastric dysplasia, gastric cancer or peptic ulcer. They all showed chronic atrophic gastritis, and only one of them had intestinal metaplasia. The degree of atrophy was mild in most of the cases. Detailed clinical information of these 10 patients is shown in Table 1.
Fig. 1.
RAPD fingerprinting patterns of H. pylori isolates from patients obtained by RAPD analysis with primer 1281. Numbers 1–10 indicate individual patients; A, stomach antrum; B, stomach body
Table 1.
Clinical information of 10 patients
| Patient | Pathology | |||
|---|---|---|---|---|
| Inflammatoryactivity | H. pylori | Intestinal metaplasia | Atrophy | |
| 1A | 1 | 2 | 0 | 1 |
| 1B | 1 | 2 | 1 | |
| 2A | 1 | 1 | 0 | 1 |
| 2B | 0 | 1 | 1 | |
| 3A | 1 | 2 | 0 | 1 |
| 3B | 1 | 2 | 1 | |
| 4A | 1 | 2 | 0 | 1 |
| 4B | 0 | 1 | 1 | |
| 5A | 1 | 1 | 0 | 1 |
| 5B | 0 | 1 | 1 | |
| 6A | 1 | 2 | 0 | 1 |
| 6B | 0 | 2 | 1 | |
| 7A | 2 | 2 | 0 | 2 |
| 7B | 2 | 2 | 1 | |
| 8A | 1 | 1 | 0 | 1 |
| 8B | 0 | 1 | 1 | |
| 9A | 1 | 1 | 0 | 1 |
| 9B | 0 | 1 | 1 | |
| 10A | 1 | 1 | 1 | 1 |
| 10B | 1 | 2 | 1 | |
Numbers 1–10 indicate individual patients; A, stomach antrum; B, stomach body
Differences in virulence factors
The positivity and subtypes of several virulence factors are described in Table 2. Overall, 5 of the 10 patients (50%) showed differences in the profiles of virulence factors between their two co-infecting H. pylori strains. All the H. pylori strains obtained from the 10 patients had an s1-type vacA s-region. In 4 patients (40%), the co-infecting H. pylori strains differed in their vacA s1 subtypes. Regarding the vacA m-region and i-region, co-infecting H. pylori strains with different patterns were isolated in 2 of the 10 patients (20%) (Additional file 2: Figure S1).
Table 2.
Positivity of vacA, CagA EPIYA, iceA, oipA, and dupA of H. pylori in 10 patients
| Strain no. | vacA | iceA | CagA EPIYA | oipA | dupA | ||
|---|---|---|---|---|---|---|---|
| s-region | m-region | i-region | |||||
| 1A | s1c | m1 | i1 | iceA1 | ABD | 3 + 1 on | – |
| 1B | s1a | m1 | i1 | iceA1 | ABD | 3 + 1 on | – |
| 2A | s1c | m1 | i1 | iceA1 | ABD | 3 + 1 on | – |
| 2B | s1a | m1 | i1 | iceA1 | ABD | 3 + 1 on | – |
| 3A | s1a | m1 | i1 | iceA1 | ABD | 3 + 1 on | – |
| 3B | s1a | m1 | i1 | iceA1 | ABD | 3 + 1 on | – |
| 4A | s1c | m1 | i1 | iceA1 | ABD | 2 + 1 + 1 + 1 on | – |
| 4B | s1c | m1 | i1 | iceA1 | ABD | 3 + 1 on | – |
| 5A | s1c | m2 | i2 | iceA1 | ABC | 3 + 1 on | – |
| 5B | s1a | m1 | i1 | iceA1 | ABD | 3 + 1 on | + |
| 6A | s1c | m1 | i1 | iceA1 | ABD | 2 + 1 + 3 on | – |
| 6B | s1c | m1 | i1 | iceA1 | ABD | 2 + 1 on | – |
| 7A | s1c | m1 | i1 | iceA1 | ABD | 3 + 1 on | – |
| 7B | s1c | m1 | i1 | iceA1 | ABD | 3 + 1 on | – |
| 8A | s1c | m1 | i1 | iceA1 | ABC | 5 + 3 on | + |
| 8B | s1a | m2 | i2 | iceA1 | ABC | 5 + 3 on | + |
| 9A | s1c | m2 | i1 | iceA1 | ABD | 2 + 1 + 1 + 1 on | – |
| 9B | s1c | m2 | i1 | iceA1 | ABD | 2 + 3 + 1 on | – |
| 10A | s1c | m1 | i1 | iceA1 | ABD | 2 + 2 + 1 on | + |
| 10B | s1c | m1 | i1 | iceA1 | ABC | 3 + 1 on | – |
Numbers 1–10 indicate individual patients; A, stomach antrum; B, stomach body; oipA status is expressed as CT repeat number and on/off status. The expression of OipA is regulated by the slipped-strand repair mechanism based on the number of CT dinucleotide repeats in the 5′ signal-sequence coding region the oipA gene (“on” = functional; “off” = nonfunctional)
The cagA gene was positively detected in all the strains from the 10 patients, and 2 of 10 patients (20%) were found to have H. pylori strains with different EPIYA motifs (Additional file 2: Figure S2) and dupA statuses (Additional file 2: Figure S3). In all the strains, the oipA gene had ‘on’ status (Additional file 2: Table S2) and the iceA1 allele of iceA was detected (Additional file 2: Figure S3).
Antibiotic resistance of H. pylori strains
The antibiotic resistance patterns of all the H. pylori strains from the 10 patients are described in Table 3. Among the 10 pairs of H. pylori strains, 9 pairs (90%) showed significantly different MIC results, and 7 pairs (70%) showed different profiles of antibiotic resistance. H. pylori strains with different antibiotic resistance patterns to quinolone, metronidazole, and clarithromycin were detected in 3, 4, and 1 patients (30, 40, and 10%), respectively. (Additional file 3: Table S3)
Table 3.
Minimum inhibitory concentration of H. pylori in 10 patients
| Strain no. | Clarithromycin (> 1.0) | Levofloxacin (> 1.0) | Amoxicillin (≥0.5) | Metronidazole (> 8) | Moxifloxacin (> 1.0) | Tetracycline (> 4.0) |
|---|---|---|---|---|---|---|
| ATCC 43504 | 0.0625 | 0.25 | 0.125 | 128 | 0.25 | 0.125 |
| 1A | 0.0625 (S) | 16 (R) | 0.125 (S) | 8 (S) | 8 (R) | 0.25 (S) |
| 1B | 0.0625 (S) | 0.125 (S) | 0.125 (S) | 4 (S) | 0.125 (S) | 0.125 (S) |
| 2A | 0.0625 (S) | 8 (R) | 0.125 (S) | 16 (R) | 8 (R) | 0.5 (S) |
| 2B | 0.0625 (S) | 0.5 (S) | 0.125 (S) | 128 (R) | 0.5 (S) | 1 (S) |
| 3A | 0.0625 (S) | 0.25 (S) | 0.125 (S) | 64 (R) | 0.25 (S) | 0.25 (S) |
| 3B | 0.0625 (S) | 1 (S) | 0.125 (S) | 128 (R) | 1 (S) | 1 (S) |
| 4A | 0.0625 (S) | 0.5 (S) | 0.125 (S) | 4 (S) | 0.5 (S) | 0.5 (S) |
| 4B | 0.0625 (S) | 0.5 (S) | 0.125 (S) | 4 (S) | 0.25 (S) | 0.5 (S) |
| 5A | 0.0625 (S) | 0.25 (S) | 0.125 (S) | 4 (S) | 0.125 (S) | 0.5 (S) |
| 5B | 0.0625 (S) | 0.5 (S) | 0.125 (S) | 16 (R) | 0.25 (S) | 1 (S) |
| 6A | 0.0625 (S) | 0.5 (S) | 0.125 (S) | 128 (R) | 0.25 (S) | 0.5 (S) |
| 6B | 0.0625 (S) | 0.5 (S) | 0.125 (S) | 4 (S) | 0.25 (S) | 1 (S) |
| 7A | 0.0625 (S) | 0.5 (S) | 0.125 (S) | 4 (S) | 0.5 (S) | 0.125 (S) |
| 7B | 0.0625 (S) | 1 (S) | 0.125 (S) | 2 (S) | 1 (S) | 1 (S) |
| 8A | 16 (R) | 8 (R) | 0.25 (S) | 128 (R) | 8 (R) | 0.25 (S) |
| 8B | 16 (R) | 0.25 (S) | 0.125 (S) | 128 (R) | 0.25 (S) | 0.125 (S) |
| 9A | 0.0625 (S) | 0.25 (S) | 0.125 (S) | 4 (S) | 0.25 (S) | 0.5 (S) |
| 9B | 0.0625 (S) | 0.5 (S) | 0.125 (S) | 128 (R) | 0.25 (S) | 2 (S) |
| 10A | 0.0625 (S) | 0.5 (S) | 0.125 (S) | 16 (R) | 0.25 (S) | 2 (S) |
| 10B | 16 (R) | 1 (S) | 0.125 (S) | 16 (R) | 1 (S) | 1 (S) |
Numbers 1–10 indicate individual patients; A, stomach antrum; B, stomach body; S, susceptible; R, resistant
Discussion
In our study, co-infecting H. pylori strains obtained from two different gastric locations in a single patient showed different RAPD fingerprinting patterns, and co-infections by two strains were detected in 12.5% of all our tested cases. In addition, half of them had H. pylori strains with different profiles of virulence factors. These findings show that patients may be affected by multiple different H. pylori strains with varying virulence factors. Similarly, the antibiotic resistance patterns of the co-infecting strains differed in the majority of cases. These results suggest that co-infecting H. pylori strains frequently have different antibiotic resistance patterns as well as different virulence factors.
Though there have been some reports showing co-infection with multiple H. pylori strains in a single patient, the results have not been consistent among studies [7–10]. In a previous study conducted in Japan, it was suggested that infection with single H. pylori strain was very common, while infection with multiple strains was rare (detected in 0 of 30 patients) [26]. The difference in the prevalence of infection by multiple strains detected in our study compared with previous studies might be mainly due to regional differences in the overall prevalence of H. pylori infection. Most importantly, the prevalence of H. pylori infection in South Korea (54.4%, 1998–2011) [27] appears to be higher than that in Japan (39.3%, 1974–1994) [28]. In regions where the prevalence of H. pylori infection is high, the chance of multiple infection with different strains is relatively higher than that in regions with low prevalence of H. pylori infection [29]. The prevalence of multiple infection was generally lower in developed countries (0–10%) than the prevalence in developing countries (> 20–35%) [8, 9, 30, 31]. It could be inferred that low H. pylori infection rate due to improved hygiene, sanitation, and eradication with antibiotic therapy in developed countries contributed to the low rate of multiple infection in a single host. Although being a developed country, Korea is also well-known for the highest incidence rates of gastric cancer in the world with high prevalence of H. pylori infection. Recently, the H. pylori prevalence has been steadily decreasing especially among the young population in Korea [32]. Considering these factors, the multiple infection rate of 12% in our study seems a feasible value considering the rates in other developed countries. In addition, the differences of study design, such as type of samples and biopsy locations, could also account for the differences in the multiple infection rates among studies from different regional areas.
Our study not only confirmed the occurrence of co-infections by multiple strains of H. pylori using RAPD fingerprinting, but also identified differences between the co-infecting strains in their virulence factors and antibiotic resistance patterns. These are important factors that influence the natural course of H. pylori-related diseases, as well as the treatment results and prognosis of the infected individuals.
It has been reported that more than 90% of East Asian H. pylori strains are cagA positive [33], and the East Asian-type CagA can show increased virulence as compared to the Western-type CagA [34]. In our study, all the tested H. pylori strains were cagA-positive and almost all of them had the East Asian-type cagA, which is similar to the results from previous studies [33]. In addition, almost all the cagA-positive strains had the s1/m1-type vacA, which presents the most cytotoxic profile among various vacA subtypes [18, 35]. Our results, like those of the previous study, showed that all the H. pylori strains with cagA had the s1-type vacA. Some researchers suggest that multiple infection could affect the histologic changes of gastric mucosa [31, 36]. Previous study has shown that there are differences in histological changes in the antrum and the body of the stomach between the patients with mixed H. pylori infections and those with single strain infection. Mixed infection was associated with significant higher rate of presence of intestinal metaplasia in the antrum [31]. In another study, mixed H. pylori infection was significantly frequent in patients with duodenal ulcer than in those with chronic gastritis [36].
Recently, since clarithromycin has become widely used for various indications including respiratory infections or nontuberculous mycobacterial infections, the prevalence of H. pylori resistance to clarithromycin has been increasing, resulting in a worldwide reduction of eradication rates [37, 38]. Resistance to other antibiotics has also been increasing, leading to a steady decrease in eradication rates. This problem is especially a concern in local areas with high clarithromycin, quinolone, or metronidazole resistance. Eradication rates with previously efficacious regimens have been decreasing worldwide [5, 6, 39, 40].
The decreasing eradication rate and increasing resistance to antibiotics support the importance of antimicrobial susceptibility testing and the need for new efficacious eradication regimens. Given that co-infecting H. pylori strains from a single patient frequently have different antibiotic resistance patterns, infection by multiple H. pylori strains may make it more difficult to interpret the results of antibiotic susceptibility tests and select appropriate eradication regimens. It has been suggested that heteroresistance of H. pylori in a single patient can contribute to treatment failure by conventional therapeutic regimens [10]. This implies that the presence of multiple infections should be considered as a possible cause of failure in the treatment of H. pylori infection. Besides, there are previous reports that show H. pylori isolates from different gastric sites in a single host presented different profiles of antibiotic resistance even though the isolates had similar or same genotype [41, 42]. This means that antibiotic resistance of H. pylori can also develop from pre-existing susceptible strains in a single host with topical variations. Therefore, it should be remembered that the etiology of heteroresistant antibacterial phenotypes is complex, and not only confined to infection with multiple strains.
Considering these points, it might be a reasonable strategy to obtain specimen from different gastric sites for H. pylori culture at the discretion of the clinician in certain clinical situations. These situations may include when multiple infection rate is not negligible and high antibiotic resistance is anticipated, for example, in areas with both high H. pylori prevalence and high antibiotic resistance. This strategy may also be useful when rescue therapy is considered.
There are some limitations in this study. First, only one type of primer was used. Although H. pylori strains can be distinguished by RAPD fingerprinting using a single primer, some recent studies have used several primers in combination to more accurately distinguish between different strains [10, 26]. However, antimicrobial susceptibility testing and the analysis of virulence factors supported the robustness of our RAPD fingerprinting results in identifying different H. pylori strains in this study.
Second, clinical records of subsequent eradication therapy and results in the patients were not available. Third, as the number of patients included was small, and the patients with single H. pylori infection were not included in the analysis, the results should be carefully interpreted. According to the enrollment criteria in our study, we only included patients with successful H. pylori isolation from both antrum and body. So this study does not give information comparing the clinical characteristics of H. pylori from patients with single infection and those with multiple infections. As the cases eventually showing only one positive culture in either antrum or body were not enrolled in this study, the true prevalence of multiple infection among the whole population with H. pylori infection may thus be lower.
Conclusions
In conclusion, the presence of two H. pylori strains in a single patient was confirmed by RAPD fingerprinting. In the majority of cases, two H. pylori strains from a single host showed different virulence factors and antibiotic resistance patterns. Co-infection by multiple strains of H. pylori can increase the failure rate of eradication therapy and should be considered when interpreting the results of antimicrobial susceptibility tests.
Additional files
A list of primer sets. Contains primer sets used for genotyping H. pylori by PCR. (DOCX 21 kb)
Primary data for virulence factor analysis. Contains agarose gel electrophoresis of the PCR-based vacA subtype, iceA, and dupA genotyping, amino acid sequences analysis of the cagA and oipA gene. (DOCX 1342 kb)
Additional mechanistic study for antibiotic resistance. Contains mechanistic analysis of antibiotics resistance for clarithromycin and quinolone. (DOCX 17 kb)
Acknowledgments
The authors thank all the medical staff who collected the specimens used in this study. A part of this work has been presented at conference (Asian Pacific Digestive Week (APDW). Connecting Excellence on Gastroenterology and Hepatology in Asia-Pacific, Seoul, Korea, 15–18 November 2018).
Abbreviations
- MIC
Minimum inhibitory concentration
- PCR
Polymerase chain reaction
- RAPD
Random amplified polymorphic DNA
Authors’ contributions
JWS, JYP, TSS analyzed and processed the data. JWS, JYP drafted the manuscript. JWS did the specimen culture. JWS, TSS did the PCR and antibiotic assays. JGK helped clinical coordination and obtaining the specimen from the patients. JYP, TSS, JGK reviewed and edited the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported by Biomedical Science, Department of Medicine Research Scholarship Grants, Chung-Ang University in 2014. The funding body played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The Institutional Review Board of Chung-Ang University Hospital approved this study [IRB number: C2014247(1444)]. Written informed consent forms were obtained from either the patients, or their authorized representatives.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Marshall B, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;323(8390):1311–1315. doi: 10.1016/S0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- 2.Hussein NR. Helicobacter pylori and gastric cancer in the Middle East: a new enigma? World J Gastroenterol. 2010;16(26):3226. doi: 10.3748/wjg.v16.i26.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suerbaum S, Michetti P. Helicobacter pylori infection. N Engl J Med. 2002;347(15):1175–1186. doi: 10.1056/NEJMra020542. [DOI] [PubMed] [Google Scholar]
- 4.Yamaoka Y. Mechanisms of disease: Helicobacter pylori virulence factors. Nat Rev Gastroenterol Hepatol. 2010;7(11):629–641. doi: 10.1038/nrgastro.2010.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graham DY. Antibiotic resistance in Helicobacter pylori: implications for therapy. Gastroenterology. 1998;115(5):1272–1277. doi: 10.1016/S0016-5085(98)70100-3. [DOI] [PubMed] [Google Scholar]
- 6.Megraud F, Coenen S, Versporten A, Kist M, Lopez-Brea M, Hirschl AM, Andersen LP, Goossens H, Glupczynski Y. Helicobacter pylori resistance to antibiotics in Europe and its relationship to antibiotic consumption. Gut. 2013;62(1):34–42. doi: 10.1136/gutjnl-2012-302254. [DOI] [PubMed] [Google Scholar]
- 7.Jorgensen M, Daskalopoulos G, Warburton V, Mitchell HM, Hazell SL. Multiple strain colonization and metronidazole resistance in Helicobacter pylori-infected patients: identification from sequential and multiple biopsy specimens. J Infect Dis. 1996;174(3):631–635. doi: 10.1093/infdis/174.3.631. [DOI] [PubMed] [Google Scholar]
- 8.Norazah A, Rasinah WZ, Zaili Z, Aminuddin A, Ramelah M. Analysis of PCR-RAPD DNA and antibiotic susceptibility profiles of antrum and corpus isolates of Helicobacter pylori from Malaysian patients. Malays J Pathol. 2009;31(1):29–34. [PubMed] [Google Scholar]
- 9.Hua J, Ng HC, Yeoh KG, Ho B. Predominance of a single strain of Helicobacter pylori in gastric antrum. Helicobacter. 1999;4(1):28–32. doi: 10.1046/j.1523-5378.1999.09043.x. [DOI] [PubMed] [Google Scholar]
- 10.Lee YC, Lee SY, Pyo JH, Kwon DH, Rhee JC, Kim JJ. Isogenic variation of Helicobacter pylori strain resulting in heteroresistant antibacterial phenotypes in a single host in vivo. Helicobacter. 2005;10(3):240–248. doi: 10.1111/j.1523-5378.2005.00316.x. [DOI] [PubMed] [Google Scholar]
- 11.Dixon MF, Genta RM, Yardley JH, Correa P. Classification and grading of gastritis. The updated Sydney system. International workshop on the histopathology of gastritis, Houston 1994. Am J Surg Pathol. 1996;20(10):1161–1181. doi: 10.1097/00000478-199610000-00001. [DOI] [PubMed] [Google Scholar]
- 12.Akopyanz N, Bukanov NO, Westblom TU, Kresovich S, Berg DE. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 1992;20(19):5137–5142. doi: 10.1093/nar/20.19.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Occhialini A, Urdaci M, Doucet-Populaire F, Bébéar CM, Lamouliatte H, Mégraud F. Macrolide resistance in Helicobacter pylori: rapid detection of point mutations and assays of macrolide binding to ribosomes. Antimicrob Agents Chemother. 1997;41(12):2724–2728. doi: 10.1128/AAC.41.12.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marshall D. Molecular analysis of Helicobacter pylori populations in antral biopsies from individual patients using randomly amplified polymorphic DNA (RAPD) fingerprinting. FEMS Immunology and Medical Microbiology. 1995;10(3-4):317–323. doi: 10.1111/j.1574-695X.1995.tb00050.x. [DOI] [PubMed] [Google Scholar]
- 15.van Doorn LJ, Figueiredo C, Sanna R, Plaisier A, Schneeberger P, de Boer W, Quint W. Clinical relevance of the cagA, vacA, and iceA status of Helicobacter pylori. Gastroenterology. 1998;115(1):58–66. doi: 10.1016/S0016-5085(98)70365-8. [DOI] [PubMed] [Google Scholar]
- 16.Miyabayashi H, Furihata K, Shimizu T, Ueno I, Akamatsu T. Influence of oral Helicobacter pylori on the success of eradication therapy against gastric Helicobacter pylori. Helicobacter. 2000;5(1):30–37. doi: 10.1046/j.1523-5378.2000.00004.x. [DOI] [PubMed] [Google Scholar]
- 17.Panayotopoulou EG, Sgouras DN, Papadakos K, Kalliaropoulos A, Papatheodoridis G, Mentis AF, Archimandritis AJ. Strategy to characterize the number and type of repeating EPIYA phosphorylation motifs in the carboxyl terminus of CagA protein in Helicobacter pylori clinical isolates. J Clin Microbiol. 2007;45(2):488–495. doi: 10.1128/JCM.01616-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Atherton JC, Cao P, Peek RM, Tummuru MK, Blaser MJ, Cover TL. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270(30):17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 19.Yamaoka Y, Kodama T, Gutierrez O, Kim JG, Kashima K, Graham DY. Relationship between Helicobacter pylori iceA, cagA, and vacA status and clinical outcome: studies in four different countries. J Clin Microbiol. 1999;37(7):2274–2279. doi: 10.1128/jcm.37.7.2274-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamaoka Y, Kodama T, Kita M, Imanishi J, Kashima K, Graham DY. Relationship of vacA genotypes of Helicobacter pylori to cagA status, cytotoxin production, and clinical outcome. Helicobacter. 1998;3(4):241–253. doi: 10.1046/j.1523-5378.1998.08056.x. [DOI] [PubMed] [Google Scholar]
- 21.Rhead JL, Letley DP, Mohammadi M, Hussein N, Mohagheghi MA, Hosseini ME, Atherton JC. A new Helicobacter pylori vacuolating cytotoxin determinant, the intermediate region, is associated with gastric cancer. Gastroenterology. 2007;133(3):926–936. doi: 10.1053/j.gastro.2007.06.056. [DOI] [PubMed] [Google Scholar]
- 22.Kauser F, Hussain MA, Ahmed I, Ahmad N, Habeeb A, Khan AA, Ahmed N. Comparing genomes of Helicobacter pylori strains from the high-altitude desert of Ladakh. India J Clin Microbiol. 2005;43(4):1538–1545. doi: 10.1128/JCM.43.4.1538-1545.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arachchi H, Kalra V, Lal B, Bhatia V, Baba C, Chakravarthy S, Rohatgi S, Sarma PM, Mishra V, Das B. Prevalence of duodenal ulcer-promoting gene (dupA) of Helicobacter pylori in patients with duodenal ulcer in north Indian population. Helicobacter. 2007;12(6):591–597. doi: 10.1111/j.1523-5378.2007.00557.x. [DOI] [PubMed] [Google Scholar]
- 24.Gerhard M, Lehn N, Neumayer N, Borén T, Rad R, Schepp W, Miehlke S, Classen M, Prinz C. Clinical relevance of the Helicobacter pylori gene for blood-group antigen-binding adhesin. Proc Natl Acad Sci U S A. 1999;96(22):12778–12783. doi: 10.1073/pnas.96.22.12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim JG, Kim BJ. Antibiotic resistance in Helicobacter pylori infection. Korean J Helicobacter Up Gastrointest Res. 2011;11(1):13–20. doi: 10.7704/kjhugr.2011.11.1.13. [DOI] [Google Scholar]
- 26.Toita N, Yokota S-i, Fujii N, Konno M. Clonality analysis of Helicobacter pylori in patients isolated from several biopsy specimens and gastric juice in a Japanese urban population by random amplified polymorphic DNA fingerprinting. Gastroenterol Res Pract. 2013;2013:721306. doi: 10.1155/2013/721306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim SH, Kwon J-W, Kim N, Kim GH, Kang JM, Park MJ, Yim JY, Kim HU, Baik GH, Seo GS. Prevalence and risk factors of Helicobacter pylori infection in Korea: nationwide multicenter study over 13 years. BMC Gastroenterol. 2013;13(1):104. doi: 10.1186/1471-230X-13-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujisawa T, Kumagai T, Akamatsu T, Kiyosawa K, Matsunaga Y. Changes in seroepidemiological pattern of Helicobacter pylori and hepatitis a virus over the last 20 years in Japan. Am J Gastroenterol. 1999;94(8):2094–2099. doi: 10.1111/j.1572-0241.1999.01283.x. [DOI] [PubMed] [Google Scholar]
- 29.Ben Mansour K, Fendri C, Battikh H, Garnier M, Zribi M, Jlizi A, Burucoa C. Multiple and mixed helicobacter pylori infections: comparison of two epidemiological situations in Tunisia and France. Infect Genet Evol. 2016;37:43–48. doi: 10.1016/j.meegid.2015.10.028. [DOI] [PubMed] [Google Scholar]
- 30.Selgrad M, Tammer I, Langner C, Bornschein J, Meissle J, Kandulski A, Varbanova M, Wex T, Schluter D, Malfertheiner P. Different antibiotic susceptibility between antrum and corpus of the stomach, a possible reason for treatment failure of helicobacter pylori infection. World J Gastroenterol. 2014;20(43):16245–16251. doi: 10.3748/wjg.v20.i43.16245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheu SM, Sheu BS, Lu CC, Yang HB, Wu JJ. Mixed infections of helicobacter pylori: tissue tropism and histological significance. Clin Microbiol Infect. 2009;15(3):253–259. doi: 10.1111/j.1469-0691.2008.02666.x. [DOI] [PubMed] [Google Scholar]
- 32.Kim SY. Time trends in the prevalence of helicobacter pylori infection and future directions in Korea. Korean J Helicobacter Up Gastrointest Res. 2016;16(3):123–128. doi: 10.7704/kjhugr.2016.16.3.123. [DOI] [Google Scholar]
- 33.Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, Massone A, Papini E, Xiang Z, Figura N. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci U S A. 1993;90(12):5791–5795. doi: 10.1073/pnas.90.12.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Higashi H, Tsutsumi R, Fujita A, Yamazaki S, Asaka M, Azuma T, Hatakeyama M. Biological activity of the Helicobacter pylori virulence factor CagA is determined by variation in the tyrosine phosphorylation sites. Proc Natl Acad Sci U S A. 2002;99(22):14428–14433. doi: 10.1073/pnas.222375399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamaoka Y. Pathogenesis of Helicobacter pylori-related gastroduodenal diseases from molecular epidemiological studies. Gastroenterol Res Pract. 2012;2012:371503. doi: 10.1155/2012/371503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai CH, Huang JC, Chiang-Ni C, Li JP, Wu LT, Wu HS, Sun YC, Lin ML, Lee JF, Lin HJ. Mixed infections of helicobacter pylori isolated from patients with gastrointestinal diseases in Taiwan. Gastroenterol Res Pract. 2016;2016:7521913. doi: 10.1155/2016/7521913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uygun A, Kadayifci A, Yesilova Z, Safali M, Ilgan S, Karaeren N. Comparison of sequential and standard triple-drug regimen for Helicobacter pylori eradication: a 14-day, open-label, randomized, prospective, parallel-arm study in adult patients with nonulcer dyspepsia. Clin Ther. 2008;30(3):528–534. doi: 10.1016/j.clinthera.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 38.Fujioka T, Aoyama N, Sakai K, Miwa Y, Kudo M, Kawashima J, Matsubara Y, Miwa J, Yakabi K. A large-scale nationwide multicenter prospective observational study of triple therapy using rabeprazole, amoxicillin, and clarithromycin for Helicobacter pylori eradication in Japan. J Gastroenterol. 2012;47(3):276–283. doi: 10.1007/s00535-011-0487-6. [DOI] [PubMed] [Google Scholar]
- 39.Lee JW, Kim N, Kim JM, Nam RH, Chang H, Kim JY, Shin CM, Park YS, Lee DH, Jung HC. Prevalence of primary and secondary antimicrobial resistance of Helicobacter pylori in Korea from 2003 through 2012. Helicobacter. 2013;18(3):206–214. doi: 10.1111/hel.12031. [DOI] [PubMed] [Google Scholar]
- 40.Heo J, Jeon SW. Changes in the eradication rate of conventional triple therapy for Helicobacter pylori infection in Korea. Korean J Gastroenterol. 2014;63(3):141–145. doi: 10.4166/kjg.2014.63.3.141. [DOI] [PubMed] [Google Scholar]
- 41.Kao CY, Lee AY, Huang AH, Song PY, Yang YJ, Sheu SM, Chang WL, Sheu BS, Wu JJ. Heteroresistance of helicobacter pylori from the same patient prior to antibiotic treatment. Infect Genet Evol. 2014;23:196–202. doi: 10.1016/j.meegid.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 42.Kim JJ, Kim JG, Kwon DH. Mixed-infection of antibiotic susceptible and resistant helicobacter pylori isolates in a single patient and underestimation of antimicrobial susceptibility testing. Helicobacter. 2003;8(3):202–206. doi: 10.1046/j.1523-5378.2003.00145.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A list of primer sets. Contains primer sets used for genotyping H. pylori by PCR. (DOCX 21 kb)
Primary data for virulence factor analysis. Contains agarose gel electrophoresis of the PCR-based vacA subtype, iceA, and dupA genotyping, amino acid sequences analysis of the cagA and oipA gene. (DOCX 1342 kb)
Additional mechanistic study for antibiotic resistance. Contains mechanistic analysis of antibiotics resistance for clarithromycin and quinolone. (DOCX 17 kb)
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.