Abstract
Specification of the mesodermal lineages requires a complex set of morphogenetic events orchestrated by interconnected signaling pathways and gene regulatory networks. The transcription factor Sox7 has critical functions in differentiation of multiple mesodermal lineages, including cardiac, endothelial, and hematopoietic. Using a doxycycline-inducible mouse embryonic stem cell line, we have previously shown that expression of Sox7 in cardiovascular progenitor cells promotes expansion of endothelial progenitor cells (EPCs). In this study, we show that the ability of Sox7 to promote endothelial cell fate occurs at the expense of the cardiac lineage. Using ChIP-Seq coupled with ATAC-Seq we identify downstream target genes of Sox7 in cardiovascular progenitor cells and by integrating these data with transcriptomic analyses, we define Sox7-dependent gene programs specific to cardiac and EPCs. Furthermore, we demonstrate a protein–protein interaction between SOX7 and GATA4 and provide evidence that SOX7 interferes with the transcriptional activity of GATA4 on cardiac genes. In addition, we show that Sox7 modulates WNT and BMP signaling during cardiovascular differentiation. Our data represent the first genome-wide analysis of Sox7 function and reveal a critical role for Sox7 in regulating signaling pathways that affect cardiovascular progenitor cell differentiation.
Keywords: Sox7, transcriptional regulation, cardiovascular progenitor cells
Introduction
The cardiovascular system is a complex system composed of the heart, blood vessels, and blood. The cell types contributing to these lineages are derived from a common mesodermal progenitor population early during development. Specification and differentiation of the cardiac mesoderm depends on the complex interaction of highly conserved transcriptional regulatory networks and signaling pathways [1]. The developing embryo contains multipotent cardiovascular progenitor cells that are capable of generating cardiac muscle, vascular smooth muscle, and endothelial lineages [2–5]. These cells can be defined by expression of Nkx2–5, Kdr (Flk1), and Isl1 [2]. In vitro differentiation of mouse embryonic stem cells (mESCs) to cardiac cell types occurs through biological processes that recapitulate normal cellular and molecular events occurring during embryonic development [6,7].
The SOX family [Sry-related high-mobility group (HMG) box] of transcription factors have key roles in the regulation of transcription during multiple developmental processes [8]. Sox7 is part of the Sox-F gene family (Sox7, Sox17, and Sox18) that has been shown to play a critical role in hematopoiesis, angiogenesis, cardiovascular development, and formation of endoderm during embryonic development [9–12]. Sox7 is highly expressed in the yolk sac beginning at embryonic day (E)7.5 and the heart tube, vascular endothelial cells, and posterior dorsal aorta beginning at E8.25 [9,13,14]. From E12.5, Sox7 expression becomes restricted to the endocardium and vascular endothelium in the heart [13,14].
Multiple studies have demonstrated that Sox7 plays an important role in vascular development [11,15–17]. Knockout of Sox7 is embryonic lethal from E10.5 with developmentally delayed embryos characterized by dilated pericardial sacs and a failure to remodel yolk sac vasculature, which altogether suggest cardiovascular failure [18]. Furthermore, Sox7 is expressed throughout KDR+ mesodermal cells but is more highly expressed in KDR+ cells that contribute to the vascular lineage indicating that Sox7 may act as a regulatory switch in the decision between cardiac and vascular cell fates [19,20].
Sox7 is located on human chromosome 8p23.1, which also includes the transcription factor Gata4. Previous reports have shown that Gata4 is a dosage-sensitive regulator of embryonic heart development [21–23]. In patients, mutations in Gata4 are associated with a spectrum of cardiac malformations [21,22,24,25]. Microdeletions, duplications, and copy number variants of the 8p23.1 chromosomal region are associated with congenital heart disease, diaphragmatic hernia, developmental delay, and behavioral problems [26–34]. Since mutation of Gata4 alone does not account for the range and severity of cardiovascular defects observed in patients with 8p23.1 microdeletions, it has been hypothesized that haploinsufficiency of Sox7 contributes to the observed phenotypes [28].
Despite the accumulated evidence for the role of Sox7 in development of the cardiovascular system, the specific molecular mechanisms underlying Sox7 function and the interplay with other lineage-specific transcription factors have not been uncovered. In this study, using genome-wide approaches, we show that Sox7 functions in cardiovascular progenitor cells by promoting endothelial lineage commitment at the expense of the cardiac program. We provide evidence that (1) SOX7 selectively represses cardiac gene transcription through interaction with GATA4 and (2) Sox7-dependent cardiac-versus-endothelial cell choice involves interaction with the WNT or BMP signaling pathways during cardiovascular cell differentiation. Our study provides novel mechanistic insights about the function of Sox7 regulatory networks in cardiovascular progenitor cells.
Materials and Methods
Growth and differentiation of mESCs
mESCs (A2loxCre) with doxycycline (dox)-inducible expression [tetracycline-responsive promoter (TRE)] of myc-tagged Sox7 (Sox7myc) were generated as previously described [15,35]. The addition of dox causes the reverse tetracycline transactivator to bind to TRE resulting in Sox7myc expression.
mESCs were cultured on irradiated mouse embryonic fibroblasts (MEFs) in Dulbecco's modified Eagle's medium (DMEM; Gibco, Thermo Fisher) supplemented with 1,000 U/mL leukemia inhibitory factor (LIF) (Millipore), 15% inactivated fetal bovine serum (FBS; Gemini), 0.1 mM nonessential amino acids (Gibco), and 0.1 mM of β-mercaptoethanol (Sigma). For embryoid body (EB) differentiation, following preplating to remove MEFs, ESCs were suspended in hanging drops in EB differentiation medium [IMDM (Thermo Fisher) with 15% FBS, 4.5 mM monothioglycerol (Sigma), 100 mg/mL ascorbic acid (Sigma), and 200 mg/mL iron-saturated transferrin (Sigma)]. After 48 h, EBs were collected, resuspended in 10-cm2 Petri dishes in EB differentiation medium and cultured on a slowly swirling table rotator. To induce Sox7myc expression during EB differentiation, doxycycline (Sigma) was added to the cultures at 1 mg/mL at day 3 for 24 h unless indicated. For WNT or BMP inhibitor studies, 10 μM endo-IWR-1 (Tocris) or 2 μM dorsomorphin (Stemgent) or 0.2 μM LDN193189 (Cayman Chemical) were added to EB cultures at the same time as doxycycline for 24 h. For beating assays, day 6 EBs were plated in individual 96-wells and allowed to adhere to tissue cultured treated plates; beating was assessed at day 10.
Flow cytometry and FACS sorting
EBs were dissociated with 0.25% trypsin (Thermo Fisher) at 37°C for 2 min with gentle shaking and resuspended in phosphate-buffered saline (PBS) supplemented with 10% FBS. Cells were incubated with APC-conjugated KDR (BD Biosciences) and PE-conjugated PDGFRα (Invitrogen) or PE-conjugated TEK (eBioscience) and PECy7-conjugated PECAM1 (eBioscience) antibodies at 0.5 μg per 106 cells for 20 min on ice. Washed and stained cells were analyzed on a FACS Aria (BD Biosciences) after adding propidium iodide (Thermo Fisher) to exclude dead cells. Data were analyzed using FlowJo Version 10 software (TreeStar).
RNA isolation, gene expression, and RNA-Seq
Total EBs were lysed and RNA was extracted using the PureLink RNA Kit following the manufacturer's instructions (Thermo Fisher). For quantitative real-time polymerase chain reaction (qRT-PCR), cDNAs were prepared using Superscript Vilo (Thermo Fisher) and TaqMan gene expression assays (Thermo Fisher) were used. For sequencing, dox (24 h) and no dox (control) samples were processed and sequenced at the University of Minnesota Genomics Core (UMGC).
Single-cell qRT-PCR
Day 5 EBs were prepared using Nkx2–5:eGFP mESCs and processed for flow cytometry as described above. Sorted cells [Nkx2–5 (eGFP)+ and KDR+] were submitted to UMGC for single-cell qRT-PCR using the Fluidigm system and TaqMan gene expression assays (Thermo Fisher). Data were analyzed using the viSNE method [36].
Coimmunoprecipitation experiments
C2C12 cells were cultured in high-glucose DMEM (Thermo Fisher) with 10% FBS (Equitech-Bio). The cells were transfected with Lipofectamine LTX (Thermo Fisher) with Gata4Flag and Sox7myc expression plasmids. Cell lysates were used for coimmunoprecipitation assays using Dynabeads Protein G magnetic beads (Thermo Fisher) following the manufacturer's protocol. Briefly, 50 μL of Dynabeads were washed and incubated with 2 μg of mouse anti-Flag antibody (Sigma) or mouse IgG for 10 min at room temperature. After washing unbound antibody off, the beads/antibody complex was incubated with 1 mL C2C12 cell lysate for 20 min at room temperature with rotation. Following five washes with 0.2% Tween-20/PBS, proteins were eluted with elution buffer and NuPage LDS sample buffer. Western blots using antibodies against SOX7 (R&D Systems), or GATA4 (R&D Systems) were done to detect total proteins and coimmunoprecipitated complexes. Sox7 deletion constructs were generated using the NEB Q5 Mutagenesis Kit following the manufacturer's instructions.
Luciferase reporter assays
C2C12 cells were cultured as described above. The day before transfection, 5 × 104 C2C12 cells were seeded onto 12-well plates. Combinations of Sox7 and Gata4 expression plasmids, along with firefly luciferase reporters (Foxp1, Nppa, Vwf, Vwa7) and TK-Renilla luciferase were transfected into cells using Lipofectamine LTX (Thermo Fisher). Cells were lysed 24 h after transfection and examined using the Dual-Luciferase assay (Promega). The firefly luciferase activity was normalized to that of the Renilla luciferase and fold induction was calculated by comparing to empty vector. All of the experiments were repeated at least three times.
Chromatin immunoprecipitation
Day 4 EBs were prepared for chromatin immunoprecipitation (ChIP) from doxycycline-treated (6 or 24 h) and untreated (control) conditions. ChIP was performed following the protocol described by Young and colleagues [37] with minor modifications. In brief, day 4 EBs were dissociated with Hank's enzyme-free cell dissociation solution (Thermo Fisher) at 37°C for 5 min with gentle shaking and reaction was inhibited by adding 10% FBS/PBS. Washed cells were treated with 1% formaldehyde (Pierce) to crosslink protein–DNA complexes (10 min at room temperature) followed by quenching with glycine. Cell pellets were incubated sequentially in lysis buffers 1, 2, 3 supplemented with protease inhibitors (complete-mini protease inhibitor cocktail; Roche) for 10 min each at 4°C (LB1: 50 mM HEPES KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP40, 0.25% Triton X-100; LB2: 10 mM Tris-HCl, pH 8, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA; and LB3: 10 mM Tris-HCl, pH 8, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% sodium deoxycholate, 0.5% N-lauroylsarcosine) and then sonicated for four cycles with a probe tip sonicator at 18% power for 1 min with intervals of 10 s ON-10 s OFF to reach an average chromatin size of 300 bp. After shearing, samples were centrifuged for 10 min at 16,000 g and snap frozen in liquid nitrogen if not processed immediately. For each ChIP, 25 mg of chromatin was precleared for 4 h at 4°C with 15 μL of bovine serum albumin (BSA)-blocked Protein A-conjugated sepharose beads (GE Healthcare) and then supplemented with 1/10 volume of 10% Triton X-100. Immunoprecipitation was performed by overnight incubation with normal mouse IgG (5 μg; Millipore) or anti-myc antibody (5 μg clone 9E10; Roche). Immune complexes were recovered by incubation with 15 μL of BSA-blocked Protein A-conjugated sepharose beads for 4 h at 4°C and then washed five times with RIPA wash buffer (50 mM HEPES KOH, pH7.5, 500 mM LiCl, 1 mM EDTA, 1% NP40, 0.25% Triton X-100, and 0.7% sodium deoxycholate) and one time with TEN buffer (10 mM Tris-HCl, pH 8, 1 mM EDTA, and 50 mM NaCl). Immunoprecipitated chromatin was recovered by incubating beads with 200 μL of elution buffer (50 mM Tris-HCl, pH 8, 10 mM EDTA, and 1% sodium dodecyl sulfate) for 20 min at 65°C. Chromatin from immunoprecipitation and input (equivalent to 1% of starting material) was reverse crosslinked overnight at 65°C, then diluted 1:1 with TE (10 mM Tris-HCl, pH 8, and 1 mM EDTA) supplemented with 4 μL of RNaseA (20 mg/mL; Thermo Fisher) and incubated for 2 h at 37°C followed by Proteinase K treatment (4 μL of 20 mg/mL) for 30 min at 55°C. DNA was purified using phenol–chloroform–isoamyl alcohol extraction, precipitated, washed, and dissolved in 50 μL H2O.
Assay for Transposase-Accessible Chromatin
Day 4 EBs from dox treated (24 h) or no dox (control) were processed for FACs as described above and sorted for PDGFRα+, KDR+ or PDGFRα−, KDR+ cells. Analysis of chromatin accessibility was performed following the protocol described by Buenrostro et al. [38]. Fifty thousand freshly sorted cells were washed with 200 μL of cold PBS then resuspended in 100 μL of cold lysis buffer (10 mM Tris-HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% IGEPAL CA-630), spun at 500 g for 10 min at 4°C and resuspended in 50 μL of the transposition reaction mix. Transposition occurred at 37°C for 30 min, after which transposed DNA was purified using the Qiagen MinElute Kit (Qiagen) and eluted in 10 μL Elution Buffer.
Motif analysis
DNA sequence 200 bp up and downstream of the MACS v1.4 identified summits of peaks in the ChIP or Assay for Transposase-Accessible Chromatin (ATAC) experiments were used for motif analysis through MEME-ChIP [39]. SOX7-binding sites that are specific to Sox7 overexpression (ChIP-Seq data) or open chromatin regions that are cell type specific (ATAC-Seq data) were used as inputs for individual analysis.
Other analysis and statistics
All Gene Ontology (GO) analysis was performed using the ToppGene Suite (https://toppgene.cchmc.org) [40] with functional associations from GREAT. Data are expressed as the mean ± standard error of the mean using the statistical software GraphPad Prism (GraphPad Software, Inc.). Comparisons of two data sets (no dox and dox treated EBs) were analyzed using Student's t-test. Multiple group comparisons (inhibitor studies and luciferase experiments) were analyzed using one-way ANOVA. Numbers indicate independent experiments. See Supplementary Data for details on sequencing analyses.
Results
Overexpression of Sox7 impairs cardiac differentiation
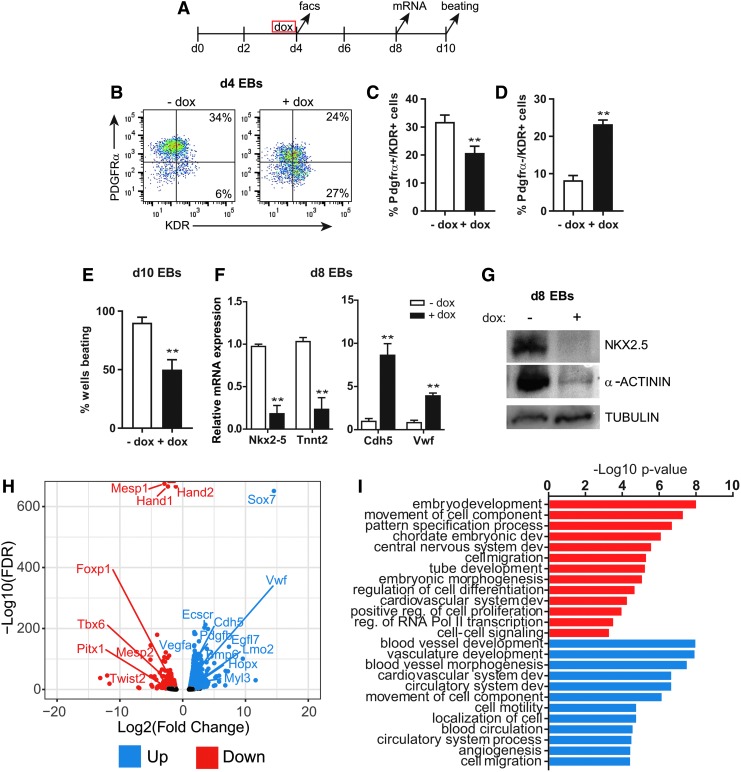
To explore the role of Sox7 during differentiation of the cardiovascular lineages, we used a doxycycline-inducible Sox7 overexpression mESC-EB model [15]. Using this system, we have previously shown that Sox7 is required for endothelial lineage differentiation downstream of Etv2 [15]. In this study, following 24 h of Sox7 induction (day 3–4), the percentage of PDGFRα/KDR double-positive cardiac progenitor cells (CPCs) in day 4 EBs significantly decreased, whereas the percentage of PDGFRα−/KDR+ endothelial progenitor cells (EPCs) increased (Fig. 1A–D). Consistent with a decrease in CPCs, the number of beating EBs decreased at day 10 (Fig. 1E). Furthermore, expression of the cardiac transcription factor, Nkx2–5, and structural components, cardiac Troponin T (Tnnt2), and α-ACTININ (Actn), were also decreased in day 8 EBs (Fig. 1F, G). In contrast, the endothelial genes, Cdh5 and Vwf, were upregulated in day 8 EBs (Fig. 1F). To identify the immediate transcriptional changes driving the impaired cardiac differentiation, we performed RNA-Seq analysis in day 4 EBs following Sox7 induction. Three hundred twenty-eight genes were dysregulated >2-fold following 12 h of Sox7 induction (Fig. 1H and Supplementary Table S1). The known SOX7 target gene Cdh5 was upregulated [41]. GO analysis for biological process indicates upregulated genes are associated with circulatory system development and blood vessel morphogenesis, including specific vasculature-associated genes Vwf, Vegfa, Esccr, and Egfl7. Downregulated genes correspond to genes involved in embryonic development and morphogenesis, regulation of cell differentiation, and cardiovascular system development (Fig. 1I). Specific downregulated genes associated with cardiac mesoderm differentiation include Mesp1, Mesp2, Hand1, Hand2, Twist2, and Tbx6 (Fig. 1H).
FIG. 1.
Overexpression of Sox7 decreases cardiac cells and increases endothelial cells during EB differentiation. (A) Time line of the EB differentiation indicates doxycycline were added at day 3 for 24 h and samples are harvested for analysis at days 4, 8, and 10. (B) Flow cytometry analysis for PDGFRα and KDR demonstrated the percentage of PDGFRα+/KDR+ cells decreased and the percentage of PDGFRα−/KDR+ are increased in Sox7 overexpressing EBs following 24 h dox induction (EB day 3–4). (C, D) Quantification of flow cytometry analysis in (B), n = 6. (E) The percentage of beating EBs at day 10 decreased when Sox7 was induced from EB day 3–4. n = 7. (F) qRT-PCR shows a significant decrease in Nkx2–5 and cardiac Tnnt2 and a significant increase in Cdh5 and Vwf transcripts in day 8 EBs, n = 5. Gene expression was normalized to B2m. (G) Protein levels for NKX2–5 and α-ACTININ are decreased in day 8 EBs. α-TUBULIN was used as a loading control in western blots. (H) Differentially expressed genes following 12 h of Sox7 overexpression (dox vs. no dox) in day 4 EBs are shown. Red, downregulated genes; blue, upregulated genes at an FDR of <0.05. (I) GO term analysis for biological process identified terms enriched in downregulated (red) and upregulated (blue) gene groups. Data are plotted as the −Log10 P-value (using the Bonferroni correction for multiple hypothesis testing). Data in bar graphs are represented as mean ± S.E.M. **P < 0.01. B2m, β-2 microglobin; dox, doxycycline; EB, embryoid body; FDR, false discovery rate; GO, Gene Ontology; qRT-PCR, quantitative real-time polymerase chain reaction; S.E.M., standard error of the mean; Tnnt2, Troponin T.
ChIP-sequencing identifies SOX7-specific binding sites
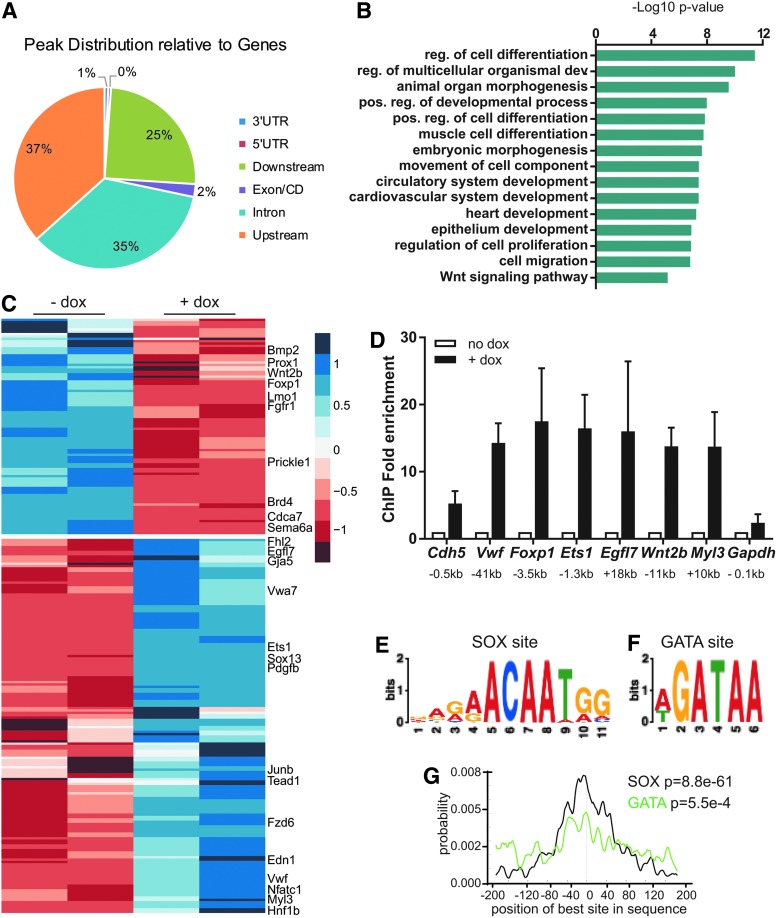
To identify potential SOX7 transcriptional target genes during cardiac differentiation, we performed ChIP-Seq experiments for SOX7 in EBs. We used doxycycline-inducible myc-tagged Sox7 mESCs (Sox7myc) to induce SOX7 expression during EB differentiation and used an α-myc antibody for ChIP. Following doxycycline treatment, SOX7 is robustly expressed in day 4 EBs (Supplementary Fig. S1). From day 4 EBs, we identified 666 SOX7-specific peaks (Supplementary Table S2). Most genomic peaks were distributed in noncoding regulatory regions with slightly more peaks distributed in upstream (37%) versus downstream (25%) regions and additional sites found within intronic regions (35%; Fig. 2A). Analysis of GO terms for biological process indicated SOX7 binding was associated with embryonic morphogenesis, cardiovascular system development, epithelium development, and WNT signaling (Fig. 2B). Of note, SOX7 was bound to regulatory regions associated with multiple WNT pathway genes, specifically Fzd6, Wnt2b, Vangl2, and Wnt5b (Supplementary Table S2).
FIG. 2.
ChIP-sequencing identifies SOX7-specific binding sites. (A) The distribution of SOX7 peaks in relation to genes is shown. (B) GO term analysis for biological process shows enriched terms. Data are plotted as the–Log10 P-value (using the Bonferroni correction for multiple hypothesis testing). (C) Heatmap shows scaled expression of genes that are differentially expressed from day 4 Sox7-induced EBs (dox vs. no dox) and where SOX7 is bound in associated regulatory regions in SOX7 ChIP-Seq experiments. (D) ChIP-qPCR confirms enrichment of SOX7 peaks associated with differentially expressed genes identified in the ChIP-Seq analysis on day 4 EBs (dox vs. no dox treated). Data are represented as mean ± S.E.M., n = 3. Distance from the transcription start site is indicated below gene name. An immediate upstream region for Gapdh used as a control region showed no enrichment. (E, F) SOX- and GATA-binding sites were identified within the ChIP peaks using motif finding. (G) SOX sites are centrally enriched (black line), whereas GATA sites are not centrally enriched. ChIP, chromatin immunoprecipitation.
Comparison of the Sox7-induced day 4 EB RNA-Seq data with the identified SOX7-bound regulatory regions identified 158 genes significantly upregulated and 91 genes significantly downregulated (Fig. 2C) indicating that SOX7 can function as a transcriptional activator or repressor on unique subsets of genes. SOX7 genomic occupancy at selected sites was confirmed by ChIP followed by quantitative PCR (Fig. 2D). Among these, SOX7 was detected at the regulatory regions of the known target gene Cdh5 [41] as well as several genes involved in vascular development, including Vwf, Ets1, and Egfl7. In agreement with a potential role in cardiac differentiation (Fig. 1), SOX7 binding was also observed in proximity of known cardiovascular development genes: Foxp1 and Myl3. These genes are all differentially expressed in Sox7-induced EBs at day 4 (Fig. 2C).
Next, we sought to identify conserved binding motifs within the SOX7 ChIP peaks. As expected, the SOX-binding motif was identified among the top enriched motifs (Fig. 2E) and displayed a centrally enriched distribution in the ChIP peaks (Fig. 2G). Interestingly, this analysis also identified the GATA-binding motif (Fig. 2F), while no enrichment was observed for other motifs associated with endothelial lineage-specific transcription factors, such as ETV2. Although the GATA motif is not centrally located, the distribution shows many sites within the central portion of the SOX7 peak (Fig. 2G). Altogether, these data suggest that a subset of genes could be regulated by SOX7 and GATA factors.
SOX7 interacts with GATA4
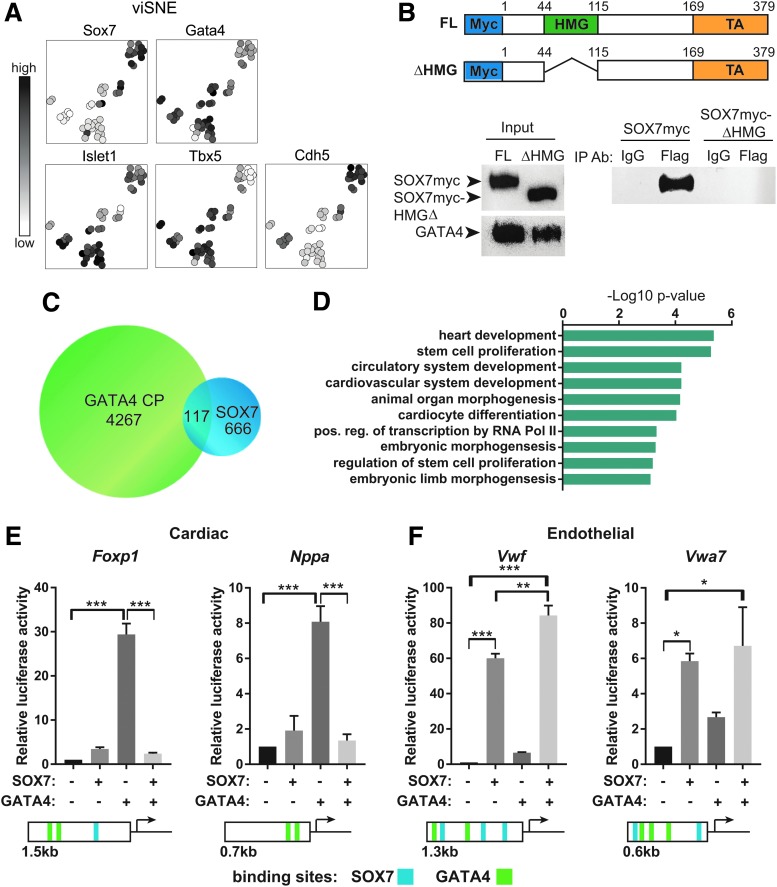
Since our binding site analysis identified the presence of GATA-binding sites within SOX7 ChIP peaks, we tested the possibility of a direct interaction between SOX7 and GATA4. Within the GATA family, we first considered Gata4 due to the possibility of a genetic interaction with Sox7 in humans. We compared the gene expression patterns of Sox7 and Gata4 during EB differentiation. Sox7 was expressed in a similar temporal pattern as Gata4 during EB differentiation with initial Sox7 expression at day 3.5, just after Gata4 expression began (Supplementary Fig. S2A). Expression of both genes peaked at EB day 6 when Nkx2–5 is also highly expressed. To assess whether Sox7 and Gata4 were coexpressed in single cells, we utilized the Nkx2–5:eGFP mESC reporter line [42]. Sox7 was enriched in day 5 eGFP+ KDR+ double-positive cardiovascular progenitor cells when they were isolated by flow cytometry (Supplementary Fig. S2B, C). Among the isolated cardiovascular progenitor cells, all Sox7+ cells coexpressed Gata4 (Fig. 3A). Furthermore, Sox7+ cells coexpressed a combination of Isl1, Tbx5, and Cdh5 (Fig. 3A).
FIG. 3.
SOX7 is coexpressed and interacts with GATA4. (A) Single-cell qRT-PCR of day 5 Nkx2–5+ (eGFP) and KDR+ double-positive cells demonstrates Sox7 expression clusters into two groups: all Sox7+ cells (19 of 47) are Gata4 and Islet1 positive. Some Sox7+ cells also coexpress Tbx5 (12 cells) and/or Cdh5 (16 cells). Using the viSNE method to visualize the distribution of cell types [36], darker cells represent stronger relative expression and white cells no expression. (B) Diagrams show SOX7 full-length and HMG domain deletion constructs used in coimmunoprecipitation experiments. Western blots show total SOX7 (left top) or GATA4 (left bottom) protein expression. GATA4-Flag immunoprecipitates full-length (FL) SOX7 but not SOX7-ΔHMG (right blot). (C) Venn diagram indicates the overlap in GATA4 ChIP peaks (from mESC-derived cardiac progenitor cells as previously published [44]) and SOX7 ChIP peaks. (D) GO term analysis for biological process identified enriched terms associated with heart development and cardiomyocyte differentiation. Data are plotted as the −Log10 P-value (using the Bonferroni correction for multiple hypothesis testing). (E, F) Luciferase reporter assays for Foxp1, Nppa, Vwf, and Vwa7 demonstrate SOX7 and GATA4 transcriptional activity. Gene models indicate the size of the promoter fragment and the relative location of putative GATA4 (green) and SOX7 (blue) DNA-binding sites. Data are represented as mean ± S.E.M., n = 3, *P < 0.05, **P < 0.01, ***P < 0.001. HMG, high-mobility group; mESC, mouse embryonic stem cell.
Since Sox7 and Gata4 were found to be coexpressed in a subset of cardiovascular progenitor cells, we tested whether they could be identified in the same protein complex through coimmunoprecipitation. C2C12 cells were transfected with Gata4-Flag plasmid or Gata4-Flag plus Sox7myc plasmid. We detected SOX7 by western blot when a Flag antibody was used to coimmunoprecipitate GATA4 and associated proteins (Fig. 3B). Furthermore, because the HMG domain of SOX7 is a highly conserved protein–protein and protein–DNA-binding domain [43], we tested whether the SOX7–GATA4 interaction is mediated through the HMG domain. Upon deletion of the HMG domain, the SOX7–GATA4 interaction is abolished, thus demonstrating the HMG domain is required for binding between GATA4 and SOX7.
To determine whether SOX7 and GATA4 bound to shared chromatin regions, we compared our data for SOX7 ChIP peaks with a published GATA4 ChIP data set. The GATA4 ChIP data from the Bruneau group were used since it was generated from mESC-derived cardiac progenitor cells using a comparable differentiation protocol [44]. We identified 117 GATA4 chromatin peaks that overlapped with SOX7 peaks, which represented ∼20% of SOX7 peaks (Fig. 3C and Supplementary Table S2). Genes associated with these regulatory regions are enriched for biological process GO terms specific for heart development, including cardiovascular system development, and cardiocyte differentiation (Fig. 3D). Accordingly, a subset of the SOX7-GATA4 loci can be annotated to genes characterized by differential expression upon SOX7 induction in day 4 EBs (Fig. 2C).
To identify a functional interaction between SOX7 and GATA4, we employed a Luciferase reporter assay to measure the transcriptional activity of these two proteins. For cardiac genes, we used the well-characterized Nppa (atrial natriuretic factor) promoter [45–48] and the novel Foxp1 regulatory element (Fig. 2D). GATA4, along with other cardiac transcription factors, has been previously shown to activate the conserved proximal promoter of Nppa. For endothelial genes, we tested the proximal promoter of Vwf [49] and a novel potential regulatory element for Vwa7 identified in our ChIP-Seq dataset. As shown in Fig. 3E and F, this assay revealed that GATA4 strongly activates both Foxp1 and Nppa regulatory elements while it has minimal effect on the Vwf or Vwa7-dependent reporters. SOX7, instead, displays minimal activity on the Foxp1 and Nppa reporters. It mildly induces the Vwa7 regulatory element and strongly activates the Vwf proximal promoter (Fig. 3E, F). Interestingly, coexpression of these two transcription factors demonstrated that GATA4-mediated transactivation of the cardiac reporters is negatively affected by SOX7 (Fig. 3E). In contrast, SOX7 activity on the endothelial reporters is not affected by GATA4 (Fig. 3F).
ATAC-Seq identifies Sox7-dependent chromatin differences between cardiac and EPCs
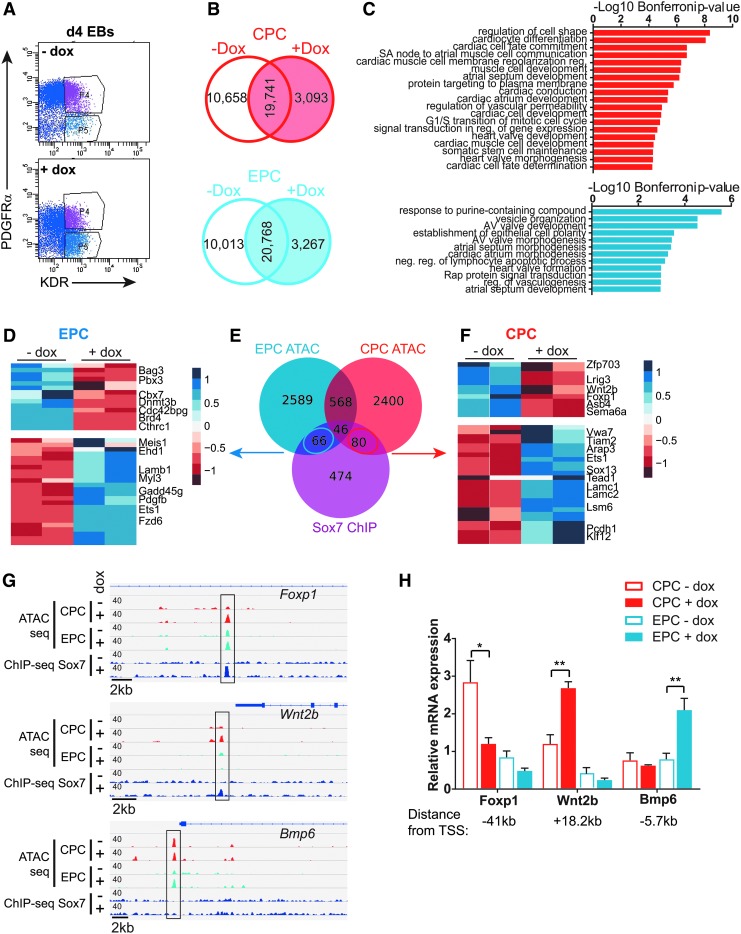
We sought to distinguish the function of Sox7 in promoting the endothelial lineage while repressing the cardiac lineage, therefore, we surveyed chromatin accessibility in CPC and EPC to identify potential genes downstream of Sox7 that were distinct in cardiac and endothelial lineages. We used the Assay for Transposase-Accessible Chromatin coupled with deep sequencing (ATAC-Seq [38]) on SOX7-induced (+ dox) and control (no dox) CPC and EPC populations isolated by FACS from day 4 EBs (Fig. 4A). By comparing the chromatin accessibility profile of SOX7-induced (+ dox) and control (no dox) cells from CPC and EPC fractions, we identified 3,093 chromatin regions in CPCs and 3,267 chromatin regions in EPCs that were remodeled in a SOX7-dependent manner (Fig. 4B and Supplementary Table S3). The SOX7-specific peaks identified in CPC and EPC populations are enriched for distinct sets of GO terms for biological process. Genes associated with SOX7-dependent CPC peaks are specifically enriched for biological process related to cardiac cell fate commitment and muscle cell development while genes associated with SOX7-dependent EPC peaks are associated with atrioventricular valve development, atrial septum morphogenesis, epithelial cell polarity, and regulation of vasculogenesis (Fig. 4C).
FIG. 4.
ATAC-Seq identifies chromatin regions that are regulated by Sox7. (A) PDGFRα+/KDR+ CPCs (gate p4) or PDGFRα−/KDR+ EPCs (gate p5) were isolated by flow cytometry from Sox7-induced (+ dox days 3–4) and control (no dox) day 4 EBs for ATAC-Seq. (B) Venn diagrams show intersection of ATAC-Seq chromatin peaks from control (no dox) and Sox7-induced (plus dox) cells in CPCs (red) and EPCs (blue). (C) GO term analysis for biological process identified distinct terms for CPC (red, top) and EPC (blue, bottom) populations. (D) Heatmap for EPC-specific open chromatin regions, where SOX7 is bound demonstrate gene expression differences in day 4 EBs following Sox7 induction with doxycycline. (E) Venn diagram shows the intersection of open chromatin regions specific to Sox7-induced condition (+ dox) in CPC or EPC with SOX7 ChIP regions. (F) Heatmap for CPC-specific open chromatin regions where Sox7 is bound demonstrate gene expression differences in day 4 EBs following Sox7 induction with doxycycline. (G) IGV view of genomic regions for Foxp1, Wnt2b, and Bmp6 show chromatin accessibility upon Sox7 induction in CPC (red) and EPC (blue) populations and SOX7 ChIP peaks. (H) qRT-PCR on isolated progenitor cell populations (CPC, red and EPC, blue) shows cell type-specific gene expression differences for selected genes. Gene expression was normalized to B2m. Data are represented as mean ± S.E.M., n = 4, *P < 0.05, **P < 0.01. ATAC, Assay for Transposase - Accessible Chromatin; CPC, cardiac progenitor cell; EPC, endothelial progenitor cell.
Interestingly, motif analysis identified an enrichment for SOX- and GATA-binding sites in both CPC and EPC populations. A number of additional motifs could further discriminate between CPCs and EPCs: MEIS-binding sites were enriched specifically in CPCs, whereas ELK-binding sites were enriched in EPCs (Supplementary Fig. S3A). Consistent with the motif enrichment, Meis1 is expressed at higher levels in CPCs, whereas Elk3 is expressed at higher levels in EPCs and Gata4 is expressed at similar levels in both cell populations (Supplementary Fig. S3B).
To determine the specificity of Sox7 gene regulation in cardiac and EPCs, we compared the SOX7 ChIP-Seq data with the ATAC-Seq data generated from CPC and EPC populations. Among the CPC (2,589) and EPC (2,400) unique chromatin regions, we identified 80 regulatory regions in CPCs, 66 regions in EPCs, and 46 regions in both cell populations that SOX7 bound in our ChIP-Seq experiments (Fig. 4E). Comparison to our gene expression analysis (Sox7-induced RNA-Seq) demonstrated that a subset of the genes annotated with these regulatory regions are differentially expressed in day 4 EBs upon SOX7 induction (Fig. 4D, F). We identified SOX7 ChIP peaks and open chromatin peaks specifically in CPC upon SOX7 induction in regulatory regions associated with Foxp1 (in the first intron) and Wnt2b (immediately downstream; Fig. 4G). Chromatin accessibility is increased at the promoter region of Bmp6 in EPCs upon SOX7 induction (Fig. 4G). Slightly further upstream, there is also an ATAC peak of unknown significance detected in CPCs. Consistent with the cell type-specific chromatin accessibility, these genes are differentially regulated in response to Sox7 in a cell type-specific manner: expression of Foxp1 is downregulated and Wnt2b is upregulated specifically in CPCs (Fig. 4H). Bmp6 is specifically upregulated in EPCs (Fig. 4H). The expression of these genes mirrors the chromatin accessibility in cardiac or EPCs.
Inhibition of WNT or BMP signaling rescued Sox7 impaired cardiac differentiation
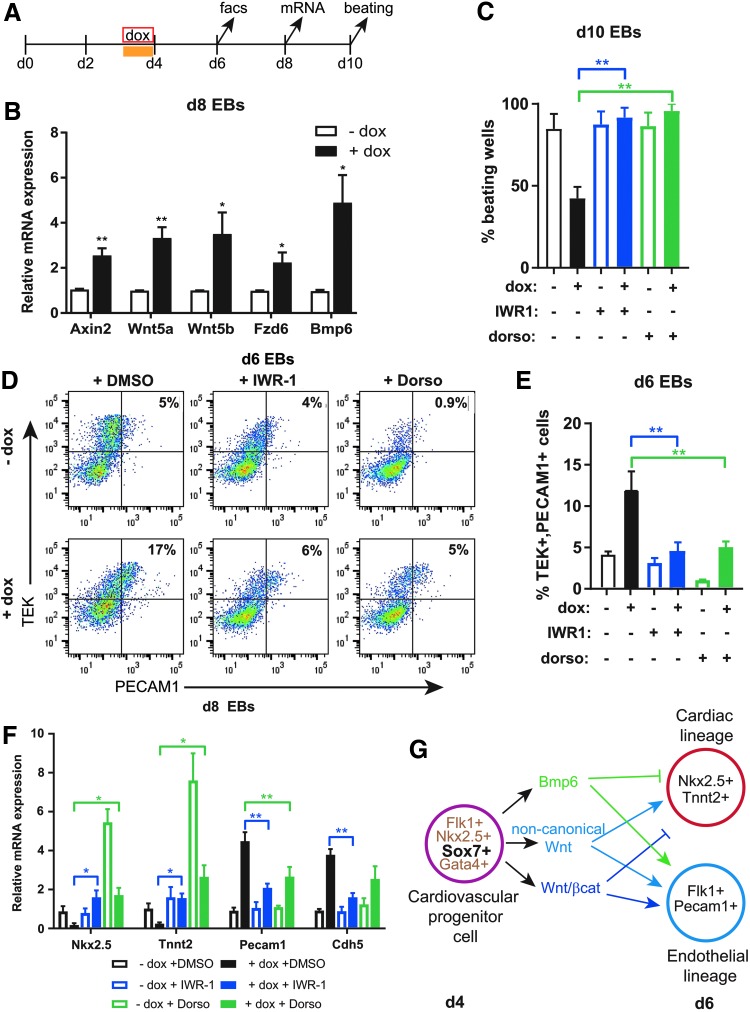
The ATAC-Seq analysis identified chromatin accessibility changes in WNT and BMP signaling pathway components (Supplementary Fig. S4). In addition, SOX7 was bound to multiple WNT pathway genes. We also observed that a significant number of WNT and BMP signaling pathway genes were differentially expressed in day 4 EBs upon SOX7 overexpression (Supplementary Fig. S4 and Supplementary Table S1). Consistent with the possible modulation of WNT signaling by Sox7, the canonical WNT signaling target gene Axin2 is upregulated in Sox7-induced EBs at day 8 (Fig. 5B). Additionally, gene expression of Wnt5a, Wnt5b, and Fzd6, as well as, Bmp6 are increased in day 8 EBs upon Sox7 induction indicating prolonged gene expression differences during cardiac lineage differentiation (Fig. 5B).
FIG. 5.
Inhibition of either WNT or BMP signaling rescues Sox7-impaired cardiac differentiation. (A) Time line of the EB differentiation indicates doxycycline and inhibitors (yellow bar) were added at day 3 for 24 h and samples are harvested for analysis at days 6, 8, and 10. (B) qRT-PCR indicates Axin2, Wnt5a, Wnt5b, Fzd6, and Bmp6 are increased following Sox7 induction in day 8 EBs. Gene expression was normalized to B2m, n = 5. (C) The percentage of beating EBs at day 10 increases in Sox7-induced EBs (+ dox, black) when IWR-1 (blue) or dorsomorphin (green) are added at day 3–4. n = 4. (D) Representative flow cytometry plots for PECAM1 (CD31) and TEK (TIE-2) at day 6 EBs show that the percent double-positive cells are decreased in the presence of IWR-1 (blue) or dorsomorphin (green) indicating endothelial cell differentiation is blocked. (E) Quantitative analysis of flow cytometry shown in (D). n = 3. (F) qRT-PCR on day 8 EBs shows Nkx2–5 and cardiac Tnnt2 transcripts are increased and Pecam1 and Chd5 transcripts are decreased in Sox7-induced EBs in the presence of IWR-1 (blue) or dorsomorphin (green). Gene expression was normalized to B2m. n = 4. Data in all bar graphs are represented as mean ± S.E.M., *P < 0.05, **P < 0.01. (G) Schematic shows a model of how Sox7 expression in cardiac progenitor cells promotes the BMP, canonical, and noncanonical WNT signaling pathways between day 4 and 6 to direct progenitor cell fate toward the endothelial rather than cardiac lineage.
The biphasic role of WNT and BMP signaling in specifying the cardiac lineage is well established; both pathways must be downregulated following formation of the cardiac mesoderm to promote robust cardiomyocyte differentiation [50–52]. Because crosstalk between WNT and BMP signaling pathways have been implicated in the specification of the mesodermal progenitors toward the cardiac and hematopoietic lineages [53], we hypothesized that Sox7 functioned at the interface of WNT and BMP signaling. To test this possibility, we determined whether inhibition of either pathway could rescue the differentiation defects seen in Sox7-induced EBs. The small molecules, endo-IWR-1, a β-catenin inhibitor, and dorsomorphin, a BMP inhibitor, have been previously shown to promote cardiogenesis [50,51,54–56]. Sox7-induced (+ dox) and noninduced (− dox) day 3 EBs were treated with endo-IWR-1 or dorsomorphin (Fig. 5A). The addition of endo-IWR-1 or dorsomorphin to dox-treated EBs significantly increased the number of beating EBs at day 10 (Fig. 5C). In day 6 EBs, the addition of endo-IWR-1 or dorsomorphin to Sox7-induced EBs decreased the percentage of TEK+ (TIE-2), PECAM1+ (CD31), EPCs (Fig. 5D, E). Gene expression of Nkx2–5 and Tnnt2 increased significantly, whereas gene expression of Pecam1 and Cdh5 decreased in inhibitor-treated EBs compared with Sox7-induced EBs at day 8 (Fig. 5F). The effect of inhibiting WNT or BMP signaling in control cells was distinct: addition of dorsomorphin had a robust effect on cardiac differentiation and a greater reduction in endothelial cell specification. Differentiation of the two lineages looked similar to control (no dox) cells when IWR-1 was added alone.
Since dorsomorphin can inhibit additional protein kinases, we tested whether the decreased cardiac differentiation observed upon Sox7 induction can be rescued using the BMP inhibitor LDN-193189 [57]. As expected, addition of LDN-193189 in day 3 cultures promoted cardiac specification at the expense of the endothelial lineage similarly to that observed with dorsomorphin (Supplementary Fig. S5). Altogether, these data indicate that inhibition of either WNT or BMP signaling can counter the effect of Sox7 overexpression during cardiovascular lineage commitment and rescue cardiomyocyte differentiation. We propose a model where Sox7 is expressed in the multipotent cardiovascular progenitor cell and maintains expression of WNT (canonical and noncanonical) and BMP pathway genes thereby promoting prolonged WNT or BMP signaling at a time when these pathways would normally be downregulated. Prolonged WNT or BMP signaling promotes endothelial cell lineage differentiation at the expense of cardiomyocyte differentiation (Fig. 5G).
Discussion
The potential role of Sox7 in regulating cardiovascular development has been appreciated; however, the mechanistic details for Sox7 regulation have been lacking. Previous work has suggested that Sox7 may act as a regulatory switch in the decision between cardiac and vascular cell fates downstream of Kdr [19]. Our study supports this role for Sox7 in specifying the cardiac and endothelial lineages from a common cardiovascular progenitor cell. Using multiple large-scale genomic approaches, we have identified novel genomic regulatory regions bound by SOX7 and identified lineage-specific changes in chromatin accessibility during EB differentiation. Furthermore, we show that Sox7 modulates the WNT and BMP signaling pathways to influence the cell fate of cardiovascular progenitor cells.
Sox7 induces distinct gene regulation in cardiovascular progenitors
The use of ATAC-Seq in addition to ChIP-Seq was valuable in identifying chromatin regions that Sox7 not only bound but were likely accounting for Sox7-dependent gene expression differences specifically in cardiac or EPCs. We observed good correlation between chromatin accessibility and gene expression, thus allowing us to identify distinct activities for Sox7. One limitation to our study is the relatively low number of Sox7-bound genes identified in the ChIP-Seq. This could be due to the low affinity of the antibody for the myc-tagged SOX7 or the heterogeneity of cell types found in EBs. However, given the strength of the ATAC-Seq data in identifying chromatin changes, we believe the regulatory regions we have identified are robust Sox7 targets. Altogether, our genomic data generate a global view of Sox7 regulation during cardiovascular cell specification and provide mechanistic insights into downstream gene regulatory networks.
Genes involved in cardiac cell fate commitment and cardiomyocyte differentiation demonstrate Sox7-dependent chromatin accessibility changes in cardiac progenitor cells. These include structural proteins, transcription factors, and signaling molecules. Foxp1 expression is specifically decreased in CPCs upon Sox7 overexpression. Furthermore, SOX7 is capable of blocking the ability of GATA4 to activate a Foxp1-regulatory element. Foxp1 is an interesting potential downstream target gene since it is expressed in developing myocardium and endocardium and Foxp1 controls cardiomyocyte proliferation and differentiation through multiple mechanisms [58].
As predicted, given the known role of Sox7 in vascular development, this transcription factor binds and mediates changes in chromatin accessibility at the genomic loci associated with known endothelial genes, including Vwf, Ets1, Erg, and Pdgfb. Many of these endothelial genes are upregulated in response to Sox7 induction during EB differentiation. We show that SOX7 strongly activates the proximal promoter region of Vwf and a novel regulatory element of Vwa7. Complete knockout as well as KDR-specific deletion of Sox7 results in widespread vascular defects by e10.5 [12,18]. We previously showed that Sox7 is a direct target of ETV2 in EPC formation [15]. Many of the regulatory regions associated with known endothelial genes demonstrated open chromatin in the absence of Sox7 overexpression. These data are consistent with the idea that Sox7 is not the first factor recruited to these regulatory regions but acts downstream of Etv2 to promote differentiation of the endothelial lineage. The identification of specific endothelial genes downstream of Sox7 provides mechanistic information about how Sox7 promotes endothelial lineage specification.
The ability to interact with other DNA-binding proteins to create a transcriptional activation complex is an important characteristic of the SOX family of transcription factors that provides promoter and cell specificity [43,59]. Our motif analysis suggests that SOX7 interacts with different protein partners in different cellular contexts. An interaction between SOX and GATA family members has been demonstrated in multiple contexts. Binding of SOX4 to GATA3 in T cells impaired the ability of GATA3 to bind DNA and negatively regulated TH2 cell differentiation [60]. Interaction between Gata2, Ets family proteins, and Sox7 (or Sox18) is indispensable for endothelial cell differentiation [61], and in parietal endoderm, SOX7 and GATA4 coregulate the Fgf3 promoter [62]. In this study, we show that SOX7 and GATA4 are found within a protein complex together and that the SOX7 HMG domain is required for this interaction. This is consistent with the critical role the HMG domain plays in the function of SOX family proteins, including binding DNA and mediating the interaction with other transcription factors [43].
Annotation of genomic sites identified as co-occupied by SOX7 and GATA4 displays enrichment for genes involved in heart development, including specific genes known to be involved in endocardial or atrioventricular valve development. On regulatory regions associated with two cardiac genes, we show that SOX7 inhibits GATA4s transcriptional activity. We speculate this represents a possible mechanism by which SOX7 actively represses the cardiac transcriptional program. SOX7 has been previously shown to inhibit the transcriptional activity of RUNX1 by interfering with its ability to bind DNA and its cofactor, thereby blocking differentiation of the hematopoietic lineage [17]. Altogether, these data suggest Sox7 represses lineage-specific transcriptional programs by interfering with a positive regulator of that lineage. This repressive ability of Sox7 favors the endothelial program over either hematopoietic (RUNX1) or cardiac (GATA4) lineages. Sox7 and Sox18 were recently reported to be downstream target genes of Gata4 [63]. The interaction between these two factors may result in a feedback loop, where Gata4 regulates Sox7 expression allowing for an efficient switch between cardiac and endothelial lineage specification. The cellular context and chromatin-specific interactions between Gata4 and Sox7 may explain the spectrum of congenital heart defects associated with 8p23 microdeletions or duplication syndromes in patients [26–28,34].
Sox7 integrates BMP and WNT signaling
The role for BMP and WNT signaling in specification and differentiation of the cardiovascular lineages is well established [50]. The interaction of these signaling pathways is complex and the downstream effect on different lineages depends on the specific cellular context and timing. WNT signaling plays a biphasic role in cardiomyocyte differentiation, where early WNT/β-catenin signaling is required for specification of the mesoderm to a cardiac fate then WNT/β-catenin signaling must be blocked in cardiac mesoderm to promote cardiomyocyte differentiation [50,52]. Additionally, noncanonical WNT signaling, primarily through Wnt5a and Wnt11, plays a role in promoting specification of cardiac mesoderm by interfering with canonical WNT/β-catenin signaling [64–66]. Similar to WNT signaling during cardiogenesis, the timing and dose of BMP signal is crucial for robust cardiomyocyte differentiation. BMP4 is required for efficient specification of cardiac mesoderm and later inhibition of BMP signaling by dorsomorphin amplifies cardiomyocyte differentiation [50,51,54].
There are conflicting reports of the role of WNT signaling in endothelial differentiation. Both activation and inhibition of canonical WNT signaling has been shown to promote endothelial cell differentiation [67–69]. Wnt5a is capable of promoting endothelial differentiation through both canonical and noncanonical WNT signaling pathways [70,71]. In contrast, it is well accepted that BMP signaling positively regulates endothelial cell differentiation and angiogenesis [72]. BMP2, BMP4, and BMP6, all induce endothelial cell proliferation, migration, and vessel assembly [72,73].
Crosstalk between WNT and BMP signaling pathways has been reported during specification of mesoderm. This crosstalk can be mediated at the level of transcription factor interactions or signaling modifiers [53,74]. An example is the ability of BMP2 to induce angiogenesis in vivo occurs through activation of both canonical and noncanonical WNT pathways [72,75]. In our study, inhibition of either WNT signaling or BMP signaling in cardiovascular progenitor cells can rescue the cardiac differentiation defect seen with overexpression of Sox7. Our data suggest that Sox7 positively regulates BMP and both canonical and noncanonical WNT pathways in cardiovascular progenitor cells. We speculate that prolonged signaling through these pathways promotes an endothelial lineage cell fate at the expense of the cardiac lineage. This Sox7 activity appears to be at the level of transcriptional regulation of Wnt ligands, however, the effect on Bmp6 seems to be indirect as SOX7 is not bound to regulatory regions associated with Bmp6, although we cannot exclude that the SOX7-binding site was not detected in our ChIP-Seq.
In conclusion, our study provides novel information about the gene regulatory networks downstream of Sox7, which will allow for further exploration of the role of Sox7 during cardiovascular cell differentiation. Using the power of large-scale genomic approaches, we define a set of genes that Sox7 regulates specifically in endothelial or cardiac progenitors and which results in the promotion of an endothelial cell differentiation program at the expense of cardiac differentiation. Sox7 is a factor that influences the balance of cardiac and endothelial lineage differentiation through positive regulation of WNT and BMP signaling.
Accession Numbers
Analyzed data are provided in supplementary tables. Genome sequencing datasets are deposited at GEO, submission number GSE133899.
Supplementary Material
Acknowledgments
This work was supported by NIH/NHLBI (1K08HL102157) to C.M.M. The authors thank Ann Behrens and Kristen Quaglia for technical assistance. They also thank Dr. Wuming Gong for assistance with the viSNE method to visualize the single-cell qRT-PCR data. They are grateful to the University of Minnesota Genomics Center (UMGC) for sequencing (RNA-Seq, ChIP-Seq and ATAC-Seq) and for the single-cell qRT-PCR analyses.
Author Disclosure Statement
No competing financial interests exist.
Supplementary Material
References
- 1. Kathiriya IS,. Nora EP. and Bruneau BG. (2015). Investigating the transcriptional control of cardiovascular development. Circ Res 116:700–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, et al. (2006). Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell 127:1151–1165 [DOI] [PubMed] [Google Scholar]
- 3. Kattman SJ,. Huber TL. and Keller GM. (2006). Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell 11:723–732 [DOI] [PubMed] [Google Scholar]
- 4. Wu SM,. Fujiwara Y, Cibulsky SM, Clapham DE, Lien CL, Schultheiss TM. and Orkin SH. (2006). Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell 127:1137–1150 [DOI] [PubMed] [Google Scholar]
- 5. Bondue A, Tännler S, Chiapparo G, Chabab S, Ramialison M, Paulissen C, Beck B, Harvey R. and Blanpain C. (2011). Defining the earliest step of cardiovascular progenitor specification during embryonic stem cell differentiation. J Cell Biol 192:751–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kattman SJ,. Adler ED. and Keller GM. (2007). Specification of multipotential cardiovascular progenitor cells during embryonic stem cell differentiation and embryonic development. Trends Cardiovasc Med 17:240–246 [DOI] [PubMed] [Google Scholar]
- 7. Murry CE. and Keller G. (2008). Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell 132:661–680 [DOI] [PubMed] [Google Scholar]
- 8. Lefebvre V, Dumitriu B, Penzo-Méndez A, Han Y. and Pallavi B. (2007). Control of cell fate and differentiation by Sry-related high-mobility-group box (Sox) transcription factors. Int J Biochem Cell Biol 39:2195–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gandillet A, Serrano AG, Pearson S, Lie-A-Ling M, Lacaud G. and Kouskoff V. (2009). Sox7-sustained expression alters the balance between proliferation and differentiation of hematopoietic progenitors at the onset of blood specification. Blood 114:4813–4822 [DOI] [PubMed] [Google Scholar]
- 10. Francois M, Koopman P. and Beltrame M. (2010). SoxF genes: key players in the development of the cardio-vascular system. Int J Biochem Cell Biol 42:445–448 [DOI] [PubMed] [Google Scholar]
- 11. Kim K, Kim IK, Yang JM, Lee E, Koh BI, Song S, Park J, Lee S, Choi C, et al. (2016). SoxF transcription factors are positive feedback regulators of VEGF signaling. Circ Res 119:839–852 [DOI] [PubMed] [Google Scholar]
- 12. Lilly AJ,. Mazan A, Scott DA, Lacaud G. and Kouskoff V. (2017). SOX7 expression is critically required in FLK1-expressing cells for vasculogenesis and angiogenesis during mouse embryonic development. Mech Dev 146:31–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sakamoto Y, Hara K, Kanai-Azuma M, Matsui T, Miura Y, Tsunekawa N, Kurohmaru M, Saijoh Y, Koopman P. and Kanai Y. (2007). Redundant roles of Sox17 and Sox18 in early cardiovascular development of mouse embryos. Biochem Biophys Res Commun 360:539–544 [DOI] [PubMed] [Google Scholar]
- 14. Wat JJ. and Wat MJ. (2014). Sox7 in vascular development: review, insights and potential mechanisms. Int J Dev Biol 58:1–8 [DOI] [PubMed] [Google Scholar]
- 15. Behrens AN,. Zierold C, Shi X, Ren Y, Koyano-Nakagawa N, Garry DJ. and Martin CM. (2014). Sox7 is regulated by ETV2 during cardiovascular development. Stem Cells Dev 23:2004–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cermenati S, Moleri S, Cimbro S, Corti P, Del Giacco L, Amodeo R, Dejana E, Koopman P, Cotelli F. and Beltrame M. (2008). Sox18 and Sox7 play redundant roles in vascular development. Blood 111:2657–2666 [DOI] [PubMed] [Google Scholar]
- 17. Lilly AJ,. Costa G, Largeot A, Fadlullah MZ, Lie-A-Ling M, Lacaud G. and Kouskoff V. (2016). Interplay between SOX7 and RUNX1 regulates hemogenic endothelial fate in the yolk sac. Development 143:4341–4351 [DOI] [PubMed] [Google Scholar]
- 18. Wat MJ,. Beck TF, Hernández-García A, Yu Z, Veenma D, Garcia M, Holder AM, Wat JJ, Chen Y, et al. (2012). Mouse model reveals the role of SOX7 in the development of congenital diaphragmatic hernia associated with recurrent deletions of 8p23.1. Hum Mol Genet 21:4115–4125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nelson TJ,. Chiriac A, Faustino RS, Crespo-Diaz RJ, Behfar A. and Terzic A. (2009). Lineage specification of Flk-1+ progenitors is associated with divergent Sox7 expression in cardiopoiesis. Differentiation 77:248–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Scialdone A, Tanaka Y, Jawaid W, Moignard V, Wilson NK, Macaulay IC, Marioni JC. and Göttgens B. (2016). Resolving early mesoderm diversification through single-cell expression profiling. Nature 535:289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fahed AC,. Gelb BD, Seidman JG. and Seidman CE. (2013). Genetics of congenital heart disease: the glass half empty. Circ Res 112:707–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS, Hirayama-Yamada K, et al. (2003). GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature 424:443–447 [DOI] [PubMed] [Google Scholar]
- 23. Zhou P, He A. and Pu WT. (2012). Regulation of GATA4 transcriptional activity in cardiovascular development and disease. Curr Top Dev Biol 100:143–169 [DOI] [PubMed] [Google Scholar]
- 24. Su W, Zhu P, Wang R, Wu Q, Wang M, Zhang X, Mei L, Tang J, Kumar M, et al. (2017). Congenital heart diseases and their association with the variant distribution features on susceptibility genes. Clin Genet 91:349–354 [DOI] [PubMed] [Google Scholar]
- 25. Rajagopal SK,. Ma Q, Obler D, Shen J, Manichaikul A, Tomita-Mitchell A, Boardman K, Briggs C, Garg V, et al. (2007). Spectrum of heart disease associated with murine and human GATA4 mutation. J Mol Cell Cardiol 43:677–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Devriendt K, Matthijs G, Van Dael R, Gewillig M, Eyskens B, Hjalgrim H, Dolmer B, McGaughran J, Bröndum-Nielsen K, et al. (1999). Delineation of the critical deletion region for congenital heart defects, on chromosome 8p23.1. Am J Hum Genet 64:1119–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Giglio S, Graw SL, Gimelli G, Pirola B, Varone P, Voullaire L, Lerzo F, Rossi E, Dellavecchia C, et al. (2000). Deletion of a 5-cM region at chromosome 8p23 is associated with a spectrum of congenital heart defects. Circulation 102:432–437 [DOI] [PubMed] [Google Scholar]
- 28. Wat MJ,. Shchelochkov OA, Holder AM, Breman AM, Dagli A, Bacino C, Scaglia F, Zori RT, Cheung SW, Scott DA. and Kang SH. (2009). Chromosome 8p23.1 deletions as a cause of complex congenital heart defects and diaphragmatic hernia. Am J Med Genet A 149A:1661–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ballarati L, Cereda A, Caselli R, Selicorni A, Recalcati MP, Maitz S, Finelli P, Larizza L. and Giardino D. (2011). Genotype–phenotype correlations in a new case of 8p23.1 deletion and review of the literature. Eur J Med Genet 54:55–59 [DOI] [PubMed] [Google Scholar]
- 30. Longoni M, Lage K, Russell MK, Loscertales M, Abdul-Rahman OA, Baynam G, Bleyl SB, Brady PD, Breckpot J, et al. (2012). Congenital diaphragmatic hernia interval on chromosome 8p23.1 characterized by genetics and protein interaction networks. Am J Med Genet A 158A:3148–3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Long F, Wang X, Fang S, Xu Y, Sun K, Chen S. and Xu R. (2013). A potential relationship among beta-defensins haplotype, SOX7 duplication and cardiac defects. PLoS One 8:e72515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lalani SR,. Shaw C, Wang X, Patel A, Patterson LW, Kolodziejska K, Szafranski P, Ou Z, Tian Q, et al. (2013). Rare DNA copy number variants in cardiovascular malformations with extracardiac abnormalities. Eur J Hum Genet 21:173–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang Y, Li Y, Wang Y, Shan B. and Duan Y. (2013). 8p23.1 duplication detected by array-CGH with complete atrioventricular septal defect and unilateral hand preaxial hexadactyly. Am J Med Genet A 161A:561–565 [DOI] [PubMed] [Google Scholar]
- 34. Barber JC,. Rosenfeld JA, Foulds N, Laird S, Bateman MS, Thomas NS, Baker S, Maloney VK, Anilkumar A, et al. (2013). 8p23.1 duplication syndrome; common, confirmed, and novel features in six further patients. Am J Med Genet A 161A:487–500 [DOI] [PubMed] [Google Scholar]
- 35. Iacovino M, Hernandez C, Xu Z, Bajwa G, Prather M. and Kyba M. (2009). A conserved role for Hox paralog group 4 in regulation of hematopoietic progenitors. Stem Cells Dev 18:783–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Amir el-AD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, Shenfeld DK, Krishnaswamy S, Nolan GP. and Pe'er D. (2013) viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol 31:545–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Boyer LA,. Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, et al. (2005). Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122:947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Buenrostro JD,. Giresi PG, Zaba LC, Chang HY. and Greenleaf WJ. (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10:1213–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bailey TL,. Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW. and Noble WS. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen J, Aronow BJ. and Jegga AG. (2009). Disease candidate gene identification and prioritization using protein interaction networks. BMC Bioinformatics 10:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Costa G, Mazan A, Gandillet A, Pearson S, Lacaud G. and Kouskoff V. (2012). SOX7 regulates the expression of VE-cadherin in the haemogenic endothelium at the onset of haematopoietic development. Development 139:1587–1598 [DOI] [PubMed] [Google Scholar]
- 42. Hsiao EC,. Yoshinaga Y, Nguyen TD, Musone SL, Kim JE, Swinton P, Espineda I, Manalac C, deJong PJ. and Conklin BR. (2008). Marking embryonic stem cells with a 2A self-cleaving peptide: a NKX2–5 emerald GFP BAC reporter. PLoS One 3:e2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bernard P. and Harley VR. (2010). Acquisition of SOX transcription factor specificity through protein-protein interaction, modulation of Wnt signalling and post-translational modification. Int J Biochem Cell Biol 42:400–410 [DOI] [PubMed] [Google Scholar]
- 44. Luna-Zurita L, Stirnimann CU, Glatt S, Kaynak BL, Thomas S, Baudin F, Samee MA, He D, Small EM, et al. (2016). Complex interdependence regulates heterotypic transcription factor distribution and coordinates cardiogenesis. Cell 164:999–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Durocher D, Chen CY, Ardati A, Schwartz RJ. and Nemer M. (1996). The atrial natriuretic factor promoter is a downstream target for Nkx-2.5 in the myocardium. Mol Cell Biol 16:4648–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Durocher D, Charron F, Warren R, Schwartz RJ. and Nemer M. (1997). The cardiac transcription factors Nkx2–5 and GATA-4 are mutual cofactors. EMBO J 16:5687–5696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Morin S, Charron F, Robitaille L. and Nemer M. (2000). GATA-dependent recruitment of MEF2 proteins to target promoters. EMBO J 19:2046–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Morin S, Paradis P, Aries A. and Nemer M. (2001). Serum response factor-GATA ternary complex required for nuclear signaling by a G-protein-coupled receptor. Mol Cell Biol 21:1036–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Guan J, Guillot PV. and Aird WC. (1999). Characterization of the mouse von Willebrand factor promoter. Blood 94:3405–3412 [PubMed] [Google Scholar]
- 50. Burridge PW,. Keller G, Gold JD. and Wu JC. (2012). Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell 10:16–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kattman SJ,. Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J. and Keller G. (2011). Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell 8:228–240 [DOI] [PubMed] [Google Scholar]
- 52. Cohen ED,. Tian Y. and Morrisey EE. (2008). Wnt signaling: an essential regulator of cardiovascular differentiation, morphogenesis and progenitor self-renewal. Development 135:789–798 [DOI] [PubMed] [Google Scholar]
- 53. Baik J, Magli A, Tahara N, Swanson SA, Koyano-Nakagawa N, Borges L, Stewart R, Garry DJ, Kawakami Y, Thomson JA. and Perlingeiro RC. (2016). Endoglin integrates BMP and Wnt signalling to induce haematopoiesis through JDP2. Nat Commun 7:13101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hao J, Daleo MA, Murphy CK, Yu PB, Ho JN, Hu J, Peterson RT, Hatzopoulos AK. and Hong CC. (2008). Dorsomorphin, a selective small molecule inhibitor of BMP signaling, promotes cardiomyogenesis in embryonic stem cells. PLoS One 3:e2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yu PB,. Hong CC, Sachidanandan C, Babitt JL, Deng DY, Hoyng SA, Lin HY, Bloch KD. and Peterson RT. (2008). Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat Chem Biol 4:33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Willems E, Spiering S, Davidovics H, Lanier M, Xia Z, Dawson M, Cashman J. and Mercola M. (2011). Small-molecule inhibitors of the Wnt pathway potently promote cardiomyocytes from human embryonic stem cell-derived mesoderm. Circ Res 109:360–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sanvitale CE,. Kerr G, Chaikuad A, Ramel MC, Mohedas AH, Reichert S, Wang Y, Triffitt JT, Cuny GD, et al. (2013). A new class of small molecule inhibitor of BMP signaling. PLoS One 8:e62721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang Y, Li S, Yuan L, Tian Y, Weidenfeld J, Yang J, Liu F, Chokas AL. and Morrisey EE. (2010). Foxp1 coordinates cardiomyocyte proliferation through both cell-autonomous and nonautonomous mechanisms. Genes Dev 24:1746–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kondoh H. and Kamachi Y. (2010). SOX-partner code for cell specification: regulatory target selection and underlying molecular mechanisms. Int J Biochem Cell Biol 42:391–399 [DOI] [PubMed] [Google Scholar]
- 60. Kuwahara M, Yamashita M, Shinoda K, Tofukuji S, Onodera A, Shinnakasu R, Motohashi S, Hosokawa H, Tumes D, et al. (2012). The transcription factor Sox4 is a downstream target of signaling by the cytokine TGF-β and suppresses T(H)2 differentiation. Nat Immunol 13:778–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kanki Y, Nakaki R, Shimamura T, Matsunaga T, Yamamizu K, Katayama S, Suehiro JI, Osawa T, Aburatani H, et al. (2017). Dynamically and epigenetically coordinated GATA/ETS/SOX transcription factor expression is indispensable for endothelial cell differentiation. Nucleic Acids Res 45:4344–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Murakami A, Shen H, Ishida S. and Dickson C. (2004). SOX7 and GATA-4 are competitive activators of Fgf-3 transcription. J Biol Chem 279:28564–28573 [DOI] [PubMed] [Google Scholar]
- 63. Afouda BA,. Lynch AT, de Paiva Alves E. and Hoppler S. (2018). Genome-wide transcriptomics analysis identifies sox7 and sox18 as specifically regulated by gata4 in cardiomyogenesis. Dev Biol 434:108–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cohen ED,. Miller MF, Wang Z, Moon RT. and Morrisey EE. (2012). Wnt5a and Wnt11 are essential for second heart field progenitor development. Development 139:1931–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bisson JA,. Mills B, Paul Helt JC, Zwaka TP. and Cohen ED. (2015). Wnt5a and Wnt11 inhibit the canonical Wnt pathway and promote cardiac progenitor development via the Caspase-dependent degradation of AKT. Dev Biol 398:80–96 [DOI] [PubMed] [Google Scholar]
- 66. Gessert S. and Kühl M. (2010). The multiple phases and faces of wnt signaling during cardiac differentiation and development. Circ Res 107:186–199 [DOI] [PubMed] [Google Scholar]
- 67. Wang H, Gilner JB, Bautch VL, Wang DZ, Wainwright BJ, Kirby SL. and Patterson C. (2007). Wnt2 coordinates the commitment of mesoderm to hematopoietic, endothelial, and cardiac lineages in embryoid bodies. J Biol Chem 282:782–791 [DOI] [PubMed] [Google Scholar]
- 68. Lian X, Bao X, Al-Ahmad A, Liu J, Wu Y, Dong W, Dunn KK, Shusta EV. and Palecek SP. (2014). Efficient differentiation of human pluripotent stem cells to endothelial progenitors via small-molecule activation of WNT signaling. Stem Cell Reports 3:804–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Reichman DE,. Park L, Man L, Redmond D, Chao K, Harvey RP, Taketo MM, Rosenwaks Z. and James D. (2018). Wnt inhibition promotes vascular specification of embryonic cardiac progenitors. Development 145:dev159905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yang DH,. Yoon JY, Lee SH, Bryja V, Andersson ER, Arenas E, Kwon YG. and Choi KY. (2009). Wnt5a is required for endothelial differentiation of embryonic stem cells and vascularization via pathways involving both Wnt/beta-catenin and protein kinase Calpha. Circ Res 104:372–379 [DOI] [PubMed] [Google Scholar]
- 71. Masckauchán TN,. Agalliu D, Vorontchikhina M, Ahn A, Parmalee NL, Li CM, Khoo A, Tycko B, Brown AM. and Kitajewski J. (2006). Wnt5a signaling induces proliferation and survival of endothelial cells in vitro and expression of MMP-1 and Tie-2. Mol Biol Cell 17:5163–5172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Le Bras A, Vijayaraj P. and Oettgen P. (2010). Molecular mechanisms of endothelial differentiation. Vasc Med 15:321–331 [DOI] [PubMed] [Google Scholar]
- 73. Ren R, Charles PC, Zhang C, Wu Y, Wang H. and Patterson C. (2007). Gene expression profiles identify a role for cyclooxygenase 2-dependent prostanoid generation in BMP6-induced angiogenic responses. Blood 109:2847–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Jain R, Li D, Gupta M, Manderfield LJ, Ifkovits JL, Wang Q, Liu F, Liu Y, Poleshko A, et al. (2015). HEART DEVELOPMENT. Integration of Bmp and Wnt signaling by Hopx specifies commitment of cardiomyoblasts. Science 348:aaa6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. de Jesus Perez VA,. Alastalo TP, Wu JC, Axelrod JD, Cooke JP, Amieva M. and Rabinovitch M. (2009). Bone morphogenetic protein 2 induces pulmonary angiogenesis via Wnt-beta-catenin and Wnt-RhoA-Rac1 pathways. J Cell Biol 184:83–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.