Abstract
Efforts to develop treatments for diseases caused by nonsense mutations have focused on identification of small molecules that promote translational read-through of messenger RNAs (mRNAs) harboring nonsense stop codons to produce full-length proteins. However, to date, no small molecule read-through drug has received FDA approval, probably because of a lack of balance between efficacy and safety. Depletion of translation termination factors eukaryotic release factor (eRF) 1 and eRF3a in cells was shown to promote translational read-through of a luciferase reporter gene harboring a nonsense mutation. In this study, we identified antisense oligonucleotides (ASOs) targeting translation termination factors and determined if ASO-mediated depletion of these factors could be a potentially effective and safe therapeutic approach for diseases caused by nonsense mutations. We found that ASO-mediated reduction of either eRF1 or eRF3a to 30%–40% of normal levels in the mouse liver is well tolerated. Hemophilia mice that express a mutant allele of human coagulation factor IX (FIX) containing nonsense mutation R338X were treated with eRF1- or eRF3a-ASO. We found that although eRF1- or eRF3a-ASO alone only elicited a moderate read-through effect on hFIX-R338X mRNA, both worked in synergy with geneticin, a small molecule read-through drug, demonstrating significantly increased production of functional full-length hFIX protein to levels that would rescue disease phenotypes in these mice. Overall our results indicate that modulating the translation termination pathway in the liver by ASOs may provide a novel approach to improving the efficacy of small molecule read-through drugs to treat human genetic diseases caused by nonsense mutations.
Keywords: translational read-through, nonsense mutation, therapeutic, antisense oligonucelotides
Introduction
Genetic diseases affect ∼30 million people in the United States and ∼300 million people worldwide (http://rarediseases.info.nih.gov). An extensive meta-analysis study using the Human Gene Mutation Database has revealed that 11% of all genetic disease-causing mutations are nonsense mutations that generate translation premature termination codons (PTCs) [1]. These mutations commonly inactivate gene function because of the production of truncated proteins and may also lead to reduced messenger RNA (mRNA) levels because of rapid mRNA turnover by the nonsense-mediated mRNA decay (NMD) pathway [2–5]. Efforts to develop treatments for these diseases have focused on developing small molecule translational read-through drugs that induce the translation machinery to record a PTC as a sense codon such that translation continues in the correct reading frame to complete the synthesis of a full-length, potentially functional protein [6–8].
Translation has been shown to be a tightly regulated, high-fidelity process [9,10]. Translation termination occurs when the ribosome reads a stop codon and recruits translation termination factors, instead of aminoacyl-transfer RNAs (tRNAs), to the A-site of the ribosome. Occasionally, a near-cognate aminoacyl-tRNA is recruited to the stop codon that leads to translational read-through [6–8]. The basal level of translational read-through for a normal stop codon occurs at a frequency of 0.001%–0.1% of mRNAs [11]; however, the read-through at nonsense mutations is 10 times higher and occurs in 0.01%–1% of mRNAs [12–14]. These differences in basal translational read-through frequencies indicate that premature translation termination is different, and possibly less regulated, compared with the normal translation termination process, suggesting that pharmacological approaches may be effective when applied to harness translation termination at PTCs to further enhance read-through efficiency.
In fact, it has been demonstrated that small molecules, such as aminoglycosides and Ataluren, are able to further promote translational read-through at PTCs without affecting translation termination at normal stop codons [7,8,15]. The most utilized read-through compounds, aminoglycosides and their derivatives, have been tested in preclinical and clinical studies, demonstrating varying levels of efficacy in promoting translational read-through. However, the ototoxicity, nephrotoxicity, and retinal toxicity of aminoglycosides hamper chronic use of these drugs to treat genetic diseases caused by nonsense mutations [6–8,14,16]. Several studies using in vitro and in vivo systems provided strong evidence that these deleterious effects are not caused by their ability to bind cytoplasmic ribosomal RNA (rRNA), which promotes translational read-through, but rather by their ability to also bind mitochondrial rRNA, resulting in inhibition of mitochondrial protein synthesis. In fact, aminoglycosides have a higher affinity for mitochondrial rRNA than cytoplasmic rRNA and consequently inhibit mitochondrial protein synthesis at concentrations that promote cytoplasmic translational read-through [16–19]. Therefore, safer approaches to promoting cytoplasmic translational read-through of mRNAs harboring PTCs are desperately needed.
In eukaryotes, two translation termination factors are required for cytoplasmic translation termination, eukaryotic release factor (eRF) 1 and eRF3. eRF1 recognizes all three stop codons (UAA, UAG, and UGA) through direct interaction at the ribosome decoding A site and activates the peptidyl transferase center to trigger hydrolysis of the peptidyl-tRNA, releasing the newly synthesized polypeptide [20,21]. Translational read-through efficiency depends on competition between stop codon recognition by eRF1 and decoding of the stop codon by a near-cognate tRNA [7,8]. In fact, it has been shown that small interfering RNA (siRNA) or antisense oligonucleotides (ASOs) targeting eRF1 promote translational read-through of a luciferase reporter in HEK293 cells [22]. eRF3 is a GTPase that binds to and stabilizes eRF1, assisting with stop codon recognition by eRF1 [23]. eRF3 also binds to Poly(A)-binding protein to stimulate translation termination and facilitates ribosome recycling [24]. Mammals have two eRF3 genes, eRF3a and eRF3b. eRF3b is mostly expressed in the brain, whereas eRF3a is ubiquitously expressed [25]. An siRNA targeting eRF3a, which also leads to downregulation of eRF1 at the protein level, promotes translational read-through of a luciferase reporter in HEK293 cells [25].
These in vitro reporter assay results suggest that reduction of translation termination factors could be an approach to promote translational read-through at PTCs. It remains to be tested, however, if we can safely reduce the expression of translation termination factors to promote translational read-through at PTCs without perturbing translation termination at normal stop codons. Furthermore, it is unknown whether these approaches will lead to translational read-through in animals and whether the level of read-through is sufficient to ameliorate disease phenotypes. Therefore, we sought to determine if reduced levels of translation termination factors are tolerated in animals, and whether increased read-through will produce sufficient functional protein to rescue the disease phenotype in a hemophilia mouse model with nonsense mutations.
Antisense technology is a clinically validated platform for drug discovery. Ionis ASOs are short chemically modified oligonucleotides that bind specifically to their RNA targets through Watson–Crick base pairing and trigger endogenous RNase H1-mediated degradation of their targeted mRNAs [26,27]. ASOs have proven to be a specific, potent, and well-tolerated therapeutic approach for cardiovascular, metabolic, neurological, and severe genetic diseases, as well as cancer [28]. In this study, we initiated an effort to use ASOs to target the translation termination factors to determine if ASO-mediated reduction of eRF1 or eRF3a could be a potentially effective and safe therapeutic approach for diseases caused by a nonsense mutation.
Materials and Methods
Antisense oligonucleotides
ASOs used in this study were chemically modified with phosphorothioate in the backbone and 2′-methoxyethyl (MOE) modification in the wings with a central 10-nucleotide deoxy gap (5-10-5 gapmer). Oligonucleotides were synthesized using an Applied Biosystems 380B automated DNA synthesizer (PerkinElmer Life and Analytical Sciences) and purified as previously described [27,29]. Lyophilized ASOs were dissolved in sterile Dulbecco's phosphate-buffered saline (DPBS; without calcium or magnesium) and quantified by ultraviolet spectrometry, diluted to the desired concentration, sterilized through a 0.2 μm filter.
Animals
All the animals of wild-type (C57BL/6; purchased from JAX) and hemophilia mice [30] (licensed from The Children's Hospital of Philadelphia and maintained in Taconic) were housed under standard condition in a pathogen-free mouse facility. All animal procedures were performed in accordance with National Institution of Health guidelines and approved by the Institutional Animal Care and Use Committee at Ionis Pharmaceuticals. ASOs were administrated subcutaneously at a volume of 10 μL/g. Geneticin (G418; 5.6 mg/mL) was administrated subcutaneously at a volume 5 μL/g.
Plasma chemistry analysis
Blood samples were collected by cardiac puncture at time of sacrifice. Plasma chemistry values were measured on the AU480 Clinical Chemistry Analyzer (Beckman Coulter).
RNA analysis
Animal tissues were homogenized in guanidine isothiocyanate solution (Invitrogen) supplemented with 8% 2-mercaptoethanol (Sigma-Aldrich). Total RNA was prepared using the RNeasy Mini Kit (Qiagen). Quantitative real-time polymerase chain reaction (qRT-PCR) was performed using Express One-Step Superscript qRT-PCR kits on an ABI StepOne Plus Real-Time PCR system. Taqman primer probe sets: eRF1 forward ATTTCCAGGGAATGGAGTACCA, reverse GGGTATGCTCCTTGGGTTGA, probe CGGCAACCGTGCCTCACCCT; eRF3a forward CGGAACCTGTAGAGTCCTCTCAA, reverse CATTTCTGTCTCTCCATTTTCTACAACA, probe TCGTGTGAAGGTTCAAA; hFIX forward TGAGGAAGAATTCAACAGTGTGTCT, reverse CCTCTGGTCTAGGCAACTTCAAC, probe CAGCAGTGTTCAGAGCCAAGCAAGA; C/EBP homologous protein (CHOP) forward TGAGCCTAACACGTCGATTATATCA, reverse TCTGGAACACTCTCTCCTCAGGTT, probe CAGCGACAGAGCCAGAATAACAGCCG; glyceradehyde 3-phosphate dehydrogenase (Gapdh) forward GGCAAATTCAACGGCACAGT, reverse GGGTCTCGCTCCTGGAAGAT, probe AAGGCCGAGAATGGGAAGCTTGTCATC.
Protein analysis
Protein levels were measured using western blot. Animal tissues were homogenized in RIPA buffer (Thermo Fisher Scientific) containing Halt Protease Inhibitor Cocktail (Life Technologies). Protein concentrations were determined using the BioRad DC protein assay, and protein was loaded at 40 μg onto a 4%–15% Criterion™ TGX™ Precast Midi Protein Gel (BioRad). Western blot membranes were probed with primary anti-eRF1 antibody (ab31799; Abcam) or anti-eRF3a antibody (ab49878; Abcam). An antibody against β-actin (A5316; Sigma) was used as a loading control. Membranes were then incubated with IRDye secondary antibodies (Li-COR) and scanned using an Odyssey infrared system (Li-COR). Images were quantified using Image Studio (Li-COR).
hFIX protein level in mouse plasma was measured by enzyme-linked immunosorbent assay (ELISA) using Human Factor IX (FIX) ELISA Kit (ab188393; Abcam) following manufacture instructions.
FIX activity assay
FIX activity assay was performed at UCSD Murine Hematology and Coagulation Core Laboratory. In brief, clotting times were determined in duplicate with an ST4 semi-automated mechanical coagulation instrument (Diagnostica Stago). Thirty microliters of citrated sample plasma diluted 1/10 in HN/BSA buffer was incubated with 30 μL of APTT reagent and 30 μL of human citrated plasma deficient of FIX at 37°C for 5 min, followed by the addition of 30 μL of 25 mM 37°C CaCl2 to initiate clotting. Time until clot formation was measured and interpolated on a standard curve of serial dilutions citrated normal (BL/6 pool) mouse plasma tested as described. Final result was reported in % hFIX-WT mice.
Results
ASO-mediated reduction of eRF1 and eRF3a is well tolerated in mice
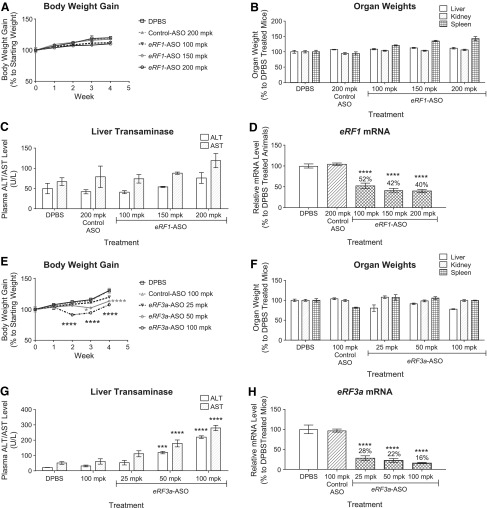
We first designed and tested ∼350 ASOs targeting mouse eRF1 and eRF3a mRNAs in mouse cell lines (data not shown). The top 15 ASOs that achieved the best target reduction for each of the translation termination factors were then evaluated for in vivo tolerability at a single-dose level in mice (data not shown). The best tolerated ASOs were selected for subsequent experiments. To determine the threshold of translation termination factor reduction that is well tolerated in animals, we performed ASO dose–response experiments in C57Bl/6J mice. ASOs targeting mouse eRF1 or eRF3a were dosed twice a week for 4 weeks at the indicated doses. Weekly body weights and also terminal organ weights and blood chemistry were measured to determine if the ASO treatments were tolerated. As liver is the most sensitive organ for ASO-mediated target knockdown with second-generation ASO chemistry, liver RNA was extracted and measured for target reduction and plasma levels of the liver enzymes; alanine transaminase (ALT) and aspartate transaminase (AST) were measured to monitor liver injury. Our results demonstrated that reduction of eRF1 to ∼40% of normal levels in mouse liver with 200 mg/kg/week ASO treatment is well tolerated. ASO-treated mice showed normal body weight gain, organ weights, and liver transaminase levels compared with the DPBS-treated control group (Fig. 1A–D). It is possible that reduction of eRF1 to levels <40% of normal levels could also be well tolerated; however, we could not achieve further target reduction with ASOs targeting eRF1 (data not shown). Moreover, we observed that reduction of eRF3a to ∼28% of normal levels in mouse liver with 25 mg/kg/week ASO treatment is well tolerated (Fig. 1E–H). Animals exhibited normal body weight gain, organ weights, and liver transaminase levels compared with the DPBS-treated control group (Fig. 1E–H). However, when eRF3a was depleted to ∼22% or 16% of control levels with either 50 or 100 mg/kg/week ASO treatments, respectively, the mice showed less body weight gain and elevated plasma transaminase levels (Fig. 1E–H), indicating that reducing eRF3a to <28% of the normal levels in mouse liver is not well-tolerated. These results were confirmed using two ASOs for each translation termination factor (Fig. 1; data not shown).
FIG. 1.
Reduction of eRF1 to ∼40% and eRF3a to ∼30% of normal levels in mouse liver is well tolerated. (A–D) Male C57BL/6 mice 7 weeks of age (n = 4) were treated with eRF1-ASO at 100, 150, or 200 mg/kg/week. DPBS and a scrambled ASO dosed at 200 mg/kg/week were used as controls. Animals were dosed twice a week for a total of eight doses in a 4-week period. Necropsy was performed 48 h after the last dose of ASO. Results are presented as mean ± standard error. (A) Body weights measured once a week. (B) Liver, kidney, and spleen weights measured at necropsy. (C) Plasma ALT and AST levels measured by clinical analyzer at the time of necropsy. (D) qPCR analysis of eRF1 mRNA levels in mouse liver samples. Mouse Gapdh mRNA was used as endogenous control. eRF1 mRNA level in DPBS-treated animals was set as 100%. (E–H) Male C57BL/6 mice 7 weeks of age (n = 4) were treated with eRF3a-ASO at 25, 50, or 100 mg/kg/week. DPBS and a scrambled ASO dosed at 100 mg/kg/week were used as controls. Animals were dosed twice a week for a total of eight doses in a 4-week period. Necropsy was performed 48 h after the last dose of ASO. Results are presented as mean ± standard error. (E) Body weights measured once a week. (F) Liver, kidney, and spleen weights measured at necropsy. (G) Plasma ALT and AST levels measured by clinical analyzer at the time of necropsy. (H) qPCR analysis of eRF3a mRNA levels in mouse liver samples. Mouse Gapdh mRNA was used as endogenous control. eRF3a mRNA level in DPBS-treated animals was set as 100%. Statistical significance was determined using a one-way ANOVA and Dunnett's multiple comparison test in Prism. All groups were compared with DPBS-treated group. ***P < 0.001; ****P < 0.0001. mpk, mg/kg/week; ALT, alanine transaminase; AST, aspartate transaminase; eRF, eukaryotic release factor; mRNA, messenger RNA; qPCR quantitative polymerase chain reaction; DPBS, Dulbecco's phosphate buffered saline; ANOVA, analysis of variance.
ASO targeting eRF3a moderately improves translational read-through in hFIX-R338X mice
It has been reported that siRNA-mediated depletion of eRF3a also reduces the level of eRF1 protein by affecting its stability [25]; therefore, we postulated that eRF3a could be a more effective target to promote translational read-through. To evaluate this hypothesis, we tested ASO-mediated reduction of eRF3a in a hemophilia B mouse model carrying a nonsense mutation. Hemophilia B is an X-linked bleeding disorder that results from a defect in the gene encoding coagulation FIX, a serine protease that is critical for blood clotting [31]. Patients with severe hemophilia B have functional FIX levels that are <1% of normal values and have frequent bleeding episodes that are associated with crippling arthropathy and early death [32]. Increasing circulating FIX to 1% of normal levels can substantially ameliorate the bleeding phenotype [33]. In this hemophilia B mouse model, a human FIX minigene, either wild-type (hFIX-WT as control) or with a nonsense mutation R338X (hFIX-R338X), is expressed in mice that lack the endogenous mouse FIX gene [30]. hFIX-WT mice contain two copies of the transgene and express hFIX mRNA and protein at similar levels to that of FIX expression in human liver and plasma, respectively. hFIX-R338X mice contain six copies of the transgene and express hFIX mRNA approximately four-fold higher than human FIX mRNA level in liver. Truncated protein expressed from the hFIX-R338X transgene is detected in mouse liver but not in plasma. These mice effectively recapitulate the phenotype of patients carrying the R338X mutation (CGA to TGA) demonstrating severe hemophilia with no detectable circulating FIX protein [30]. The hFIX transgenes are driven by the human transthyretin promoter and thus are primarily expressed in mouse hepatocytes, which are the main cell type for endogenous FIX production [30]. Therefore, it is an optimal model for demonstrating proof-of-concept for ASO-mediated translational read-through approach.
As the hFIX-R338X transgene is primarily expressed in hepatocytes, we evaluated whether we could further deplete eRF3a in hepatocytes using a triantennary N-acetyl galactosamine (GalNAc)-conjugated eRF3a ASO to improve the read-through efficacy while reducing any unwanted toxic effects in other cell types. GalNAc is a high-affinity ligand for the hepatocyte-specific asialoglycoprotein receptor (ASGPR) [34]. GalNAc conjugation results in preferential ASO delivery to hepatocytes relative to extrahepatic cells, resulting in 10-fold higher ASO potency in mouse liver than unconjugated ASOs of the same sequence [35]. This enables lower doses of ASO to be administered to reduce hepatocyte targets while largely sparing target reduction in extrahepatic tissues.
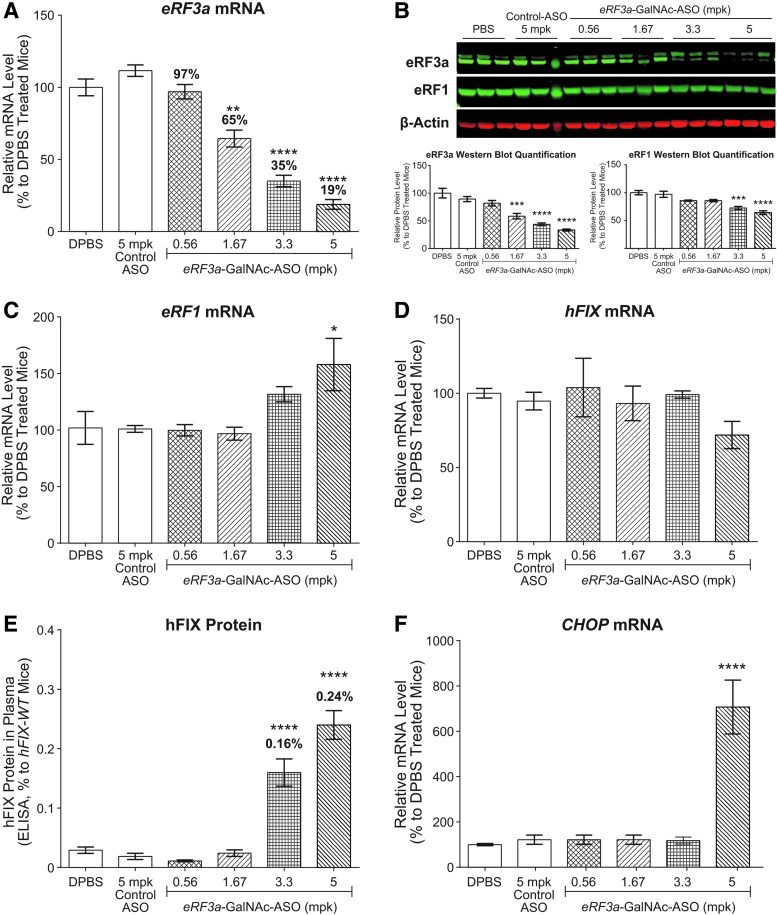
Dose–response experiments were performed with GalNAc-conjugated eRF3a ASO to determine the optimal level of target reduction to achieve a balance between efficacy and safety in hemophilia mice. Male hemophilia mice between ages 6 and 12 weeks were dosed once a week with ASOs at 0.56, 1.67, 3.3 and 5 mg/kg/week for 4 weeks. eRF3a-GalNAc-ASO treatments led to dose-dependent liver target reduction in hemophilia mice, reducing eRF3a mRNA levels to ∼97%, 65%, 35%, and 19% with respect to the levels of DPBS-treated mice (Fig. 2A). Western blot analysis indicated similar eRF3a target reduction at the protein level (Fig. 2B). In contrast to data previously reported in HEK293 cells whereby siRNA-mediated depletion of eRF3a also significantly reduced eRF1 protein levels [25], reduction of eRF3a in hemophilia mouse livers only caused a minor reduction in eRF1 protein levels (Fig. 2B). Of interest, we observed an upregulation of eRF1 mRNA levels in mouse livers treated with eRF3a ASO (Fig. 2C), indicating that a feedback regulation might exist to increase eRF1 expression in compensation for the loss of eRF3a expression.
FIG. 2.
ASO targeting eRF3a moderately improves translational read-through in hFIX-R338X mice. hFIX-R338X male mice 6–12 weeks of age (n = 4–6) were treated weekly with four total doses of DPBS, GalNAc-control ASO, and eRF3a-GalNAc-ASO at indicated doses. Necropsy was performed 48 h after the last dose of ASO. Results are presented as means ± standard errors. (A) qPCR analysis of eRF3a mRNA levels in mouse liver samples. (B) Western blot analysis of eRF1 and eRF3a protein levels in mouse liver samples. (C) qPCR analysis of eRF1 mRNA levels in mouse liver samples. (D) qPCR analysis of hFIX mRNA levels in mouse liver samples. (E) Mouse plasma hFIX protein levels as measure by ELISA. Plasma hFIX protein level in hFIX-WT mice was set as 100%. (F) qPCR analysis of CHOP mRNA levels in mouse liver samples. For qPCR analysis, Gapdh mRNA was used as an endogenous control. The mRNA expression levels in DPBS-treated mouse livers were set as 100%. For western blot analysis, β-actin was used as endogenous control. Statistical significance was determined using a one-way ANOVA and Dunnett's multiple comparison test in Prism. All groups were compared with DPBS-treated group. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. mpk: mg/kg/week; CHOP, C/EBP homologous protein; hFIX, human Factor IX; ELISA, enzyme-linked immunosorbent assay; GalNAc, N-acetyl galactosamine; WT, wild type.
Without affecting hFIX-R338X mRNA levels (Fig. 2D), 3.3 and 5 mg/kg/week eRF3a-GalNAc-ASO treatments led to the detection of full-length hFIX protein in mouse plasma, indicating that reduction of eRF3a to <35% of normal levels induces translational read-through of hFIX-R338X mRNAs (Fig. 2E). However, in contrast to other treatment groups, mice that were treated with eRF3a-GalNAc-ASO at 5 mg/kg/week exhibited less body weight gain during the study and slightly elevated liver transaminase levels (data not shown), confirming that reducing eRF3a RNA level to <28% of normal levels in liver is not well tolerated in mice (Fig. 1E–H) even with GalNAc-conjugated ASO that restricts target reduction in the liver primarily to hepatocytes [35]. In line with these observations, an upregulation of CHOP mRNA was also observed in mice treated with 5 mg/kg/week eRF3a-GalNAc-ASO (Fig. 2F), indicating induction of the unfolded protein response (UPR). This effect is likely because of the impairment of global translation accuracy, which is in agreement with a previous report that high concentrations of aminoglycoside treatment cause endoplasmic reticulum stress and activation of the UPR [36].
Therefore, we conclude that a minimum of ∼28% eRF3a is needed to avoid effects on global translation. Although eRF3a-GalNAc-ASO treatment at a well-tolerated dose significantly increased the production of otherwise undetectable full-length hFIX protein, it did not reach the level required (1% of normal levels) to improve the hemophilia disease phenotype [33].
ASO targeting eRF1 moderately improves translational read-through in hFIX-R338X mice
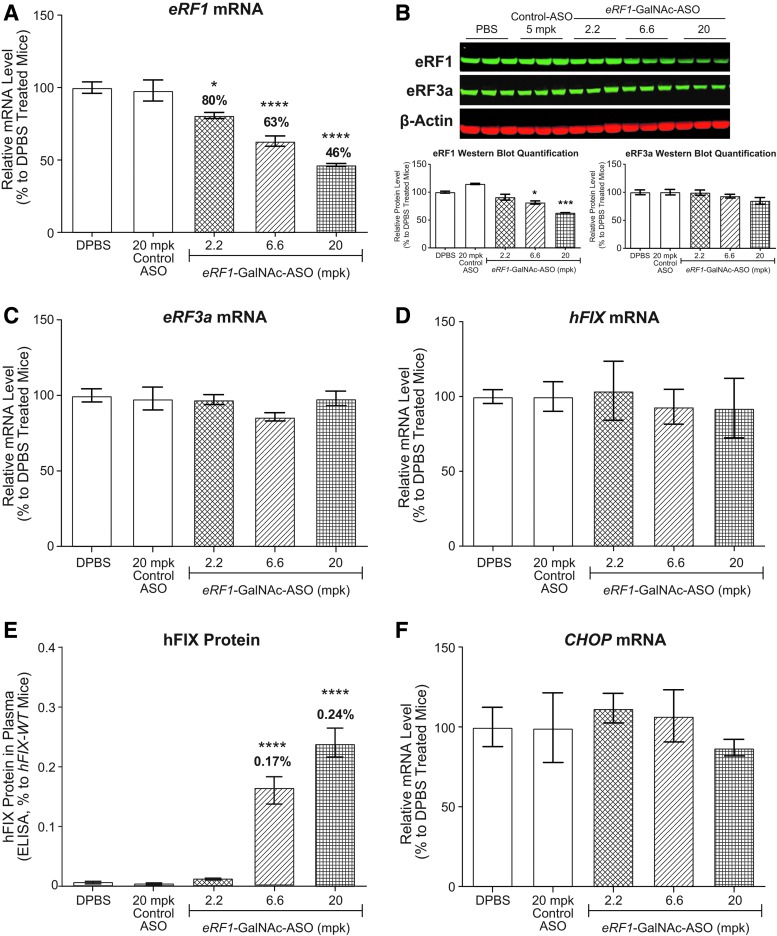
As reduction of eRF3a did not significantly reduce eRF1 protein levels as originally predicted, we evaluated the effects of ASO-mediated reduction of eRF1 on promoting translational read-through in hFIX-R338X hemophilia mice, because it is the translation termination factor that directly recognizes and binds to the stop codon. To deplete eRF1 more efficiently in hepatocytes, we generated a GalNAc-conjugated eRF1 ASO. Male hemophilia mice between ages 6 and 12 weeks were dosed once a week with ASOs at 2.2, 6.6 and 20 mg/kg/week for 4 weeks. eRF1-GalNAc-ASO treatment led to dose-dependent liver target reduction in hemophilia mice and reduced eRF1 mRNA levels to ∼80%, 63%, and 46% of DPBS-treated mice (Fig. 3A). Western blot analysis indicated similar eRF1 target reduction at the protein level (Fig. 3B). All treatment groups demonstrated normal body weight gain, and terminal organ weights and blood chemistry, indicating that all treatments were well tolerated (data not shown). Reduction of eRF1 to these levels did not affect eRF3a expression at the mRNA or protein level (Fig. 3B, C).
FIG. 3.
ASO targeting eRF1 moderately improves translational read-through in hFIX-R338X mice. hFIX-R338X male mice 6–12 weeks of age (n = 4–6) were treated weekly with four total doses of DPBS, GalNAc-control ASO, and eRF1-GalNAc-ASO at the indicated doses. Necropsy was performed 48 h after the last dose of ASO. Results are presented as mean ± standard error. (A) qPCR analysis of eRF1 mRNA levels in mouse liver samples. (B) Western blot analysis of eRF1 and eRF3a protein levels in mouse liver samples. (C) qPCR analysis of eRF3a mRNA levels in mouse liver samples. (D) qPCR analysis of hFIX mRNA levels in mouse liver samples. (E) Mouse plasma hFIX protein levels as measured by ELISA. Plasma hFIX protein level in hFIX-WT mice was set as 100%. (F) qPCR analysis of CHOP mRNA levels in mouse liver samples. For qPCR analysis, Gapdh mRNA was used as an endogenous control. The mRNA expression levels in DPBS-treated mouse liver were set as 100%. For western blot analysis, β-actin was used as endogenous control. Statistical significance was determined using a one-way ANOVA and Dunnett's multiple comparison test in Prism. All groups were compared with DPBS-treated group. *P < 0.05; ***P < 0.001; ****P < 0.0001.
Although reduction of eRF1 also did not affect hFIX-R338X mRNA levels (Fig. 3D), full-length hFIX protein was detected in plasma of mice treated with eRF1-GalNAc-ASO at 6.6 and 20 mg/kg/week, indicating that reduction of eRF1 to 63% of normal levels induces translational read-through of hFIX-R338X mRNA (Fig. 3E). Based on these results, it seems that, in contrast to eRF3a, eRF1 is a haploinsufficient gene in its function in translation termination at PTC. Of interest, we did not detect upregulation of CHOP mRNA in any of the treatment groups in this study (Fig. 3F), indicating a lack of UPR induction. These results suggest that ASO-mediated reduction of eRF1 to 46–63% of normal levels promotes translational read-through at PTC without significantly affecting global translation accuracy.
Therefore, eRF1 seems to be a safer and preferable target for promoting translational read-through at PTCs in comparison with eRF3a. Despite this encouraging data, however, the highest average level of full-length hFIX protein detected in eRF1-GalNAc-ASO-treated mice was only ∼0.24% of hFIX-WT mice (Fig. 3E), which would not result in meaningful clinical outcomes for hemophilia disease.
ASOs targeting eRF1 and eRF3a demonstrate synergistic effects with the aminoglycoside geneticin, significantly increasing translational read-through in hFIX-R338X mice
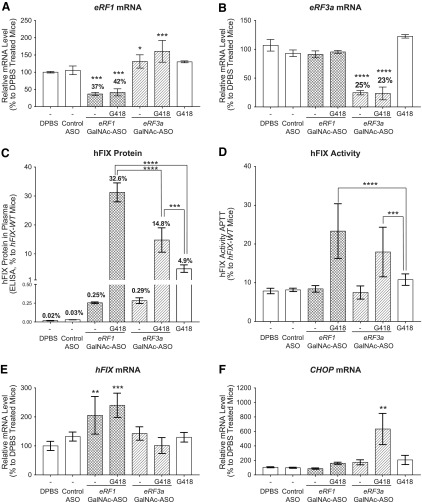
Because GalNAc-ASO-mediated reduction of eRF1 or eRF3a demonstrated limited efficacy in promoting translational read-through and seems less likely to be effective as a single therapy, we wanted to test if ASO-mediated reduction of eRF1 or eRF3a will improve the efficacy of the aminoglycoside geneticin (G418), which was previously shown to promote translational read-through in hFIX-R338X mice [37]. We treated hemophilia mice with the highest tolerable dose of either eRF1-GalNAc-ASO (20 mg/kg/week) or eRF3a-GalNAc-ASO (3.3 mg/kg/week) in combination with G418. Male hFIX-R338X mice between ages 6 and 12 weeks were dosed once a week with either eRF1- or eRF3a-GalNAc-ASO for 4 weeks. Seven days before the end of study, ASO-treated cohorts were divided and half were administered 28 mg/kg G418 daily for 7 days [37]. A separate cohort of mice were treated with G418 alone daily for the last 7 days of the study. Animals were sacrificed 48 h after the last ASO treatment and 9 h after the last dose of G418. All groups of animals demonstrated normal body weight gain throughout the study, as well as normal organ weights and plasma transaminase levels as measured at the time of necropsy (data not shown).
In this study, both eRF1-GalNAc-ASO and eRF3a-GalNAc-ASO demonstrated similar efficacy in promoting translational read-through when administered alone without G418 compared with previous studies (hFIX protein levels in the plasma of eRF1- and eRF3a-GalNAc-ASO treated mice were 0.25% and 0.29% of that of hFIX-WT mice, respectively) (Fig. 4A–C). Of interest, when combined with G418 treatment, both eRF1- and eRF3a-GalNAc-ASO demonstrated synergistic effects with G418 in promoting translational read-through. eRF3a-GalNAc-ASO treatment increased G418 activity by ∼three-fold, increasing plasma hFIX protein levels from ∼4.9% to ∼14.8% compared with untreated hFIX-WT mice (Fig. 4C). eRF1-GalNAc-ASO improved G418 efficacy even further by more than six-fold, reaching ∼32% of untreated hFIX-WT levels (Fig. 4C). Furthermore, both eRF1-ASO/G418 and eRF3a-ASO/G418 treatments significantly improved FIX activity in an activated partial thrombo-plastin time-based coagulation assay (APTT) compared with G418 alone, with eRF1-ASO/G418 demonstrating a trend of greater improvement (Fig. 4D). We also observed an approximately two-fold increase in hFIX-R338X mRNA levels in eRF1-GalNAc-ASO-treated mouse liver, which we did not observe in the previous dose–response study where slightly less eRF1 target reduction was achieved (Fig. 4E). As a measurement of the UPR, CHOP mRNA level did not significantly change in any treatment groups except in the eRF3a-GalNAc-ASO/G418-treated group (Fig. 4F), probably because of additive effects of eRF3a-GalNAc-ASO and G418 on global translation accuracy.
FIG. 4.
ASOs targeting eRF1 and eRF3a demonstrate synergistic effects with the aminoglycoside geneticin, significantly increasing translational read-through in hFIX-R338X mice. hFIX-R338X male mice 6–12 weeks of age (n = 3) were treated weekly with four total doses of DPBS, GalNAc-control ASO (20 mg/kg/dose), eRF1-GalNAc-ASO (20 mg/kg/dose), and eRF3a-GalNAc-ASO (3 mg/kg/dose). G418 (28 mg/kg) was administered daily during the final 7 days of the study either alone or in combination with ASO treatments as described. Necropsy was performed 48 h after the last dose of ASO and 9 h after the last dose of geneticin. Results are presented as mean ± standard error. (A, B) qPCR analysis of eRF1 and eRF3a mRNA levels in mouse liver samples. (C) Mouse plasma hFIX protein levels as measured by ELISA. Plasma hFIX protein level in hFIX-WT mice was set as 100%. (D) APTT FIX activity assay in plasma from treated mice. FIX activity in hFIX-WT mice was set as 100%. (E, F) qPCR analysis of hFIX and CHOP mRNA levels in mouse liver samples. For qPCR analysis, mouse Gapdh mRNA was used as endogenous control. The mRNA expression levels in DPBS-treated animals were set as 100%. Statistical significance was determined using a one-way ANOVA and Dunnett's multiple comparison test in Prism. (A, B, D and E) All groups were compared with DPBS-treated hFIX-R338X group. (C, D) Dunnett's multiple comparison test was performed between ASO/G418 combo groups and G418-treated group *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. APTT, activated partial thrombo-plastin time.
Overall, in hemophilia mice, GalNAc-conjugated ASOs targeting eRF1 and eRF3a worked in synergy with the small molecular read-through drug geneticin, significantly increasing production of full-length hFIX protein, which results in improved plasma FIX activity. GalNAc-ASO-mediated reduction of eRF1 seems to be safer and more effective in promoting the translational read-through of hFIX-R338X mRNA than reduction of eRF3a when combined with G418.
Discussion
It has been decades since the discovery that aminoglycoside antibiotics can promote translational read-through at PTCs in both yeast and human [38,39]. Despite various efforts to therapeutically develop aminoglycosides and their derivatives for the treatment of diseases with nonsense mutations, none have been approved by the FDA [6–8]. Ataluren, a small molecule promoting translational read-through by targeting the ribosome to promote specific near-cognate tRNA selection [15,40], is in clinical development in United States and approved in European Union Member States for the treatment of Duchene muscular dystrophy caused by a nonsense mutation. However, it has failed to significantly improve respiratory function in cystic fibrosis (CF) patients with nonsense mutations, according to results of a Phase 3 clinical trial that has recently completed. The high unmet medical need for various genetic diseases caused by nonsense mutations calls for novel strategies to promote translational read-through at PTCs.
In this study, we investigated the therapeutic potential of ASO-mediated reduction of the translation termination factors for the treatment of diseases with nonsense mutations. We demonstrated that ASO-mediated reduction of the translation termination factors eRF1 and eRF3a to ∼40% and ∼30% of normal levels in mouse liver, respectively, is well tolerated (Fig. 1). Using a hemophilia B disease mouse model, we demonstrated that reducing eRF1 to ∼40% and eRF3a to ∼30% of normal levels induced translational read-through of hFIX-R338X mRNA, leading to the production of an otherwise undetectable full-length hFIX-protein to the level of ∼0.2% of that in hFIX-WT mice (Figs. 2 and 3). Moreover, we discovered that although ASO-mediated reduction of eRF1 or eRF3a alone did not produce sufficient hFIX protein to rescue the disease phenotype, the ASO treatments demonstrated synergistic effects with the aminoglycoside geneticin in promoting translational read-through and produced significant amounts of hFIX protein leading to improved plasma FIX activity that is sufficient for rescuing the disease phenotypes in these mice (Fig. 4). These results suggest that although ASO-mediated reduction of the translation termination factors may not be an effective standalone read-through therapy, it could be a potentially effective therapy if utilized to improve the efficacy of small molecule read-through compounds. It will be interesting to test if the read-through improvement observed by ASO-mediated reduction of translation termination factors in combination with G418 also applies to other aminoglycosides or other small molecule read-through drugs such as Ataluren [15,41].
One note of caution is that the incorporation of certain amino acids at a PTC by near-cognate aminoacyl-tRNAs could lead to reduced protein function [42]. Moreover, the identity of the incorporated amino acid is sequence-context dependent [43] and also can be different depending on the small molecule translational read-through reagent used [40]. Therefore, it will be important to characterize the identity of the amino acid inserted by the treatment of ASOs targeting the translation termination factors in an mRNA-specific manner.
We also performed a side-by-side comparison to determine which translation termination factor is a better ASO target to safely induce efficient translational read-through at PTCs. Our results suggest that eRF1 is a better and more robust target than eRF3a for this approach. First, the reduction of eRF1 to 63% of normal levels can induce translational read-through of hFIX-R338X mRNA (Fig. 3), whereas eRF3a needs to be depleted to 35% of normal levels to induce translational read-through of the same transcript (Fig. 2). Second, although reduction of eRF1 and eRF3a elicits a similar magnitude of translational read-through without G418, eRF1 reduction demonstrated much greater activity in improving the efficacy of G418 in generating stable full-length hFIX protein (Fig. 4). Third, although eliciting similar levels of read-through, eRF1 reduction seems to be better tolerated than eRF3a reduction, because the UPR is only triggered upon eRF3a depletion as demonstrated by upregulation of CHOP mRNA (Fig. 4). Therefore, we conclude that eRF1 is a better potential therapeutic target to be utilized in combination with small molecule read-through compounds.
It is not clear why eRF1 reduction seems to promote translational read-through more efficiently compared with eRF3a reduction without inducing the UPR. It has been suggested that eRF1 binding to the stop codon is the rate-limiting step for translation termination [44]. It is possible, since translation termination at PTCs is less efficient compared with normal stop codons, that a higher concentration of eRF1 is required for efficient translation termination at PTCs. Therefore, a slight reduction of eRF1 to 63% of normal levels can lead to increased translational read-through at PTCs without significantly impacting normal translation termination efficiency. Of interest, we observed an approximately two-fold increase in hFIX-R338X mRNA levels in eRF1-GalNAc-ASO-treated mouse liver (Fig. 4D), which probably contributed to the significant elevation of plasma hFIX protein levels in these mice. It remains to be investigated how the reduction of eRF1 leads to the upregulation of hFIX mRNA and whether it affects mRNA stability globally.
We also found that eRF1 mRNA level is upregulated in eRF3a-ASO-treated mice (Fig. 3), indicating that a feedback regulation might exist to ensure efficient translation termination. It was recently demonstrated that expression of eRF1 in plants is controlled by an autoregulatory circuit involving read-through and nonsense-mediated decay [45]. It will be interesting to determine if this mechanism is conserved in other eukaryotes.
It is surprising to discover that reducing mouse translation termination factors only triggered a minor translational read-through of hFIX-R338X mRNA, whereas mutations in eRF1 and eRF3a in yeast induced significant translational read-through [46–48]. These results indicate there might be other proteins that can serve as translation termination factors or additional mechanisms to govern translation termination in higher eukaryotes. Alternatively, reducing eRF1 and eRF3a expression simultaneously might lead to greater translational read-through. More studies will need to be performed to provide further understanding of translation termination at normal and premature stop codons.
The efficiency of translation termination depends on the context of the stop codon and its surrounding sequences [49]. The PTC and its surrounding sequences of hFIX-R338X are UGU CUU UGA UCU ACA, a sequence context with high termination efficiency [50]. It was demonstrated that the weakest termination contexts were most affected by increases or decreases in the concentration of eRF1 protein [50]. Therefore, it is possible that in a weaker termination context, ASO-mediated reduction of eRF1 or eRF3a could be effective as a single treatment to induce efficient translational read-through and produce sufficient full-length protein to rescue the disease phenotype.
It was found in a clinical study that the aminoglycoside gentamicin shows variable read-through efficiency in CF patients carrying the W1282X nonsense mutation [51,52]. Further evaluation revealed that the variation in efficacy corresponded with the level of CF transmembrane conductance regulator (CFTR) mRNA, leading to the discovery that the efficiency of NMD might vary in these patients and hence affect the response to read-through treatments [53,54]. We previously demonstrated that inhibition of NMD by ASO-mediated reduction of a branch-specific NMD factor, Upf3b [55,56], significantly stabilized hFIX mRNA harboring an early nonsense mutation (R29X) and significantly improved the efficacy of read-through therapy in this mouse hemophilia model [57]. These results underlined the need for a safe and effective read-through therapy and suggested that a combination strategy to simultaneously modulate both the NMD pathway and translation termination process is required to treat genetic diseases caused by nonsense mutations. Overall, our data indicate that modulating the translation termination pathway by ASOs may provide a novel therapeutic approach for human genetic diseases that are caused by nonsense mutations.
Acknowledgments
The authors thank Dr. Stanley T. Crooke and Dr. C. Frank Bennett from Ionis Pharmaceuticals for their helpful discussions.
Author Disclosure Statement
All authors are employees and shareholders of Ionis Pharmaceuticals.
References
- 1. Mort M, Ivanov D, Cooper DN. and Chuzhanova NA. (2008). A meta-analysis of nonsense mutations causing human genetic disease. Hum Mutat 29:1037–1047 [DOI] [PubMed] [Google Scholar]
- 2. Jaffrey SR. and Wilkinson MF. (2018). Nonsense-mediated RNA decay in the brain: emerging modulator of neural development and disease. Nat Rev Neurosci 19:715–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kurosaki T. and Maquat LE. (2016). Nonsense-mediated mRNA decay in humans at a glance. J Cell Sci 129:461–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Karousis ED, Nasif S. and Muhlemann O. (2016). Nonsense-mediated mRNA decay: novel mechanistic insights and biological impact. Wiley Interdiscip Rev RNA 7:661–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang L. and Wilkinson MF. (2012). Regulation of nonsense-mediated mRNA decay. Wiley Interdiscip Rev RNA 3:807–828 [DOI] [PubMed] [Google Scholar]
- 6. Dabrowski M, Bukowy-Bieryllo Z. and Zietkiewicz E. (2018). Advances in therapeutic use of a drug-stimulated translational readthrough of premature termination codons. Mol Med 24:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nagel-Wolfrum K, Moller F, Penner I, Baasov T. and Wolfrum U. (2016). Targeting nonsense mutations in diseases with translational read-through-inducing drugs (TRIDs). BioDrugs 30:49–74 [DOI] [PubMed] [Google Scholar]
- 8. Keeling KM, Xue X, Gunn G. and Bedwell DM. (2014). Therapeutics based on stop codon readthrough. Annu Rev Genomics Hum Genet 15:371–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zaher HS. and Green R. (2009). Fidelity at the molecular level: lessons from protein synthesis. Cell 136:746–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Prabhakar A, Choi J, Wang J, Petrov A. and Puglisi JD. (2017). Dynamic basis of fidelity and speed in translation: coordinated multistep mechanisms of elongation and termination. Protein Sci 26:1352–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Parker J. (1989). Errors and alternatives in reading the universal genetic code. Microbiol Rev 53:273–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bonetti B, Fu L, Moon J. and Bedwell DM. (1995). The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J Mol Biol 251:334–345 [DOI] [PubMed] [Google Scholar]
- 13. Manuvakhova M, Keeling K. and Bedwell DM. (2000). Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system. RNA 6:1044–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Keeling KM, Wang D, Conard SE. and Bedwell DM. (2012). Suppression of premature termination codons as a therapeutic approach. Crit Rev Biochem Mol Biol 47:444–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Welch EM, Barton ER, Zhuo J, Tomizawa Y, Friesen WJ, Trifillis P, Paushkin S, Patel M, Trotta CR, et al. (2007). PTC124 targets genetic disorders caused by nonsense mutations. Nature 447:87–91 [DOI] [PubMed] [Google Scholar]
- 16. Xie J, Talaska AE. and Schacht J. (2011). New developments in aminoglycoside therapy and ototoxicity. Hear Res 281:28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hobbie SN, Akshay S, Kalapala SK, Bruell CM, Shcherbakov D. and Bottger EC. (2008). Genetic analysis of interactions with eukaryotic rRNA identify the mitoribosome as target in aminoglycoside ototoxicity. Proc Natl Acad Sci U S A 105:20888–20893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huth ME, Ricci AJ. and Cheng AG. (2011). Mechanisms of aminoglycoside ototoxicity and targets of hair cell protection. Int J Otolaryngol 2011:937861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shulman E, Belakhov V, Wei G, Kendall A, Meyron-Holtz EG, Ben-Shachar D, Schacht J. and Baasov T. (2014). Designer aminoglycosides that selectively inhibit cytoplasmic rather than mitochondrial ribosomes show decreased ototoxicity: a strategy for the treatment of genetic diseases. J Biol Chem 289:2318–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Frolova L, Le Goff X, Rasmussen HH, Cheperegin S, Drugeon G, Kress M, Arman I, Haenni AL, Celis JE, et al. (1994). A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature 372:701–703 [DOI] [PubMed] [Google Scholar]
- 21. Bulygin KN, Graifer DM, Hountondji C, Frolova LY. and Karpova GG. (2017). Exploring contacts of eRF1 with the 3′-terminus of the P site tRNA and mRNA stop signal in the human ribosome at various translation termination steps. Biochim Biophys Acta Gene Regul Mech 1860:782–793 [DOI] [PubMed] [Google Scholar]
- 22. Carnes J, Jacobson M, Leinwand L. and Yarus M. (2003). Stop codon suppression via inhibition of eRF1 expression. RNA 9:648–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Frolova L, Le Goff X, Zhouravleva G, Davydova E, Philippe M. and Kisselev L. (1996). Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA 2:334–341 [PMC free article] [PubMed] [Google Scholar]
- 24. Ivanov A, Mikhailova T, Eliseev B, Yeramala L, Sokolova E, Susorov D, Shuvalov A, Schaffitzel C. and Alkalaeva E. (2016). PABP enhances release factor recruitment and stop codon recognition during translation termination. Nucleic Acids Res 44:7766–7776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chauvin C, Salhi S, Le Goff C, Viranaicken W, Diop D. and Jean-Jean O. (2005). Involvement of human release factors eRF3a and eRF3b in translation termination and regulation of the termination complex formation. Mol Cell Biol 25:5801–5811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Crooke ST. (2017). Molecular mechanisms of antisense oligonucleotides. Nucleic Acid Ther 27:70–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bennett CF. and Swayze EE. (2010). RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol 50:259–293 [DOI] [PubMed] [Google Scholar]
- 28. Bennett CF, Baker BF, Pham N, Swayze E. and Geary RS. (2017). Pharmacology of antisense drugs. Annu Rev Pharmacol Toxicol 57:81–105 [DOI] [PubMed] [Google Scholar]
- 29. Zong X, Huang L, Tripathi V, Peralta R, Freier SM, Guo S. and Prasanth KV. (2015). Knockdown of nuclear-retained long noncoding RNAs using modified DNA antisense oligonucleotides. In: Springer Protocols: Nuclear Bodies and Noncoding RNAs Methods in Molecular Biology, Vol. 1262 Humana Press, New York, NY: [DOI] [PubMed] [Google Scholar]
- 30. Sabatino DE, Armstrong E, Edmonson S, Liu YL, Pleimes M, Schuettrumpf J, Fitzgerald J, Herzog RW, Arruda VR. and High KA. (2004). Novel hemophilia B mouse models exhibiting a range of mutations in the Factor IX gene. Blood 104:2767–2774 [DOI] [PubMed] [Google Scholar]
- 31. DeLoughery TG. (2015). Hemostasis and Thrombosis. Vol Book, whole. Springer, Cham [Google Scholar]
- 32. Nathwani AC. and Tuddenham EG. (1992). Epidemiology of coagulation disorders. Baillieres Clin Haematol 5:383–439 [DOI] [PubMed] [Google Scholar]
- 33. Nathwani AC, Reiss UM, Tuddenham EG, Rosales C, Chowdary P, McIntosh J, Della Peruta M, Lheriteau E, Patel N, et al. (2014). Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N Engl J Med 371:1994–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Spiess M. (1990). The asialoglycoprotein receptor: a model for endocytic transport receptors. Biochemistry 29:10009–10018 [DOI] [PubMed] [Google Scholar]
- 35. Prakash TP, Graham MJ, Yu J, Carty R, Low A, Chappell A, Schmidt K, Zhao C, Aghajan M, et al. (2014). Targeted delivery of antisense oligonucleotides to hepatocytes using triantennary N-acetyl galactosamine improves potency 10-fold in mice. Nucleic Acids Res 42:8796–8807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lopez-Novoa JM, Quiros Y, Vicente L, Morales AI. and Lopez-Hernandez FJ. (2011). New insights into the mechanism of aminoglycoside nephrotoxicity: an integrative point of view. Kidney Int 79:33–45 [DOI] [PubMed] [Google Scholar]
- 37. Yang C, Feng J, Song W, Wang J, Tsai B, Zhang Y, Scaringe WA, Hill KA, Margaritis P, et al. (2007). A mouse model for nonsense mutation bypass therapy shows a dramatic multiday response to geneticin. Proc Natl Acad Sci U S A 104:15394–15399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Palmer E, Wilhelm JM. and Sherman F. (1979). Phenotypic suppression of nonsense mutants in yeast by aminoglycoside antibiotics. Nature 277:148–150 [DOI] [PubMed] [Google Scholar]
- 39. Bedwell DM, Kaenjak A, Benos DJ, Bebok Z, Bubien JK, Hong J, Tousson A, Clancy JP. and Sorscher EJ. (1997). Suppression of a CFTR premature stop mutation in a bronchial epithelial cell line. Nat Med 3:1280–1284 [DOI] [PubMed] [Google Scholar]
- 40. Roy B, Friesen WJ, Tomizawa Y, Leszyk JD, Zhuo J, Johnson B, Dakka J, Trotta CR, Xue X, et al. (2016). Ataluren stimulates ribosomal selection of near-cognate tRNAs to promote nonsense suppression. Proc Natl Acad Sci U S A 113:12508–12513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Du M, Liu X, Welch EM, Hirawat S, Peltz SW. and Bedwell DM. (2008). PTC124 is an orally bioavailable compound that promotes suppression of the human CFTR-G542X nonsense allele in a CF mouse model. Proc Natl Acad Sci U S A 105:2064–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pranke I, Bidou L, Martin N, Blanchet S, Hatton A, Karri S, Cornu D, Costes B, Chevalier B, et al. (2018). Factors influencing readthrough therapy for frequent cystic fibrosis premature termination codons. ERJ Open Res 4 00080–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xue X, Mutyam V, Thakerar A, Mobley J, Bridges RJ, Rowe SM, Keeling KM. and Bedwell DM. (2017). Identification of the amino acids inserted during suppression of CFTR nonsense mutations and determination of their functional consequences. Hum Mol Genet 26:3116–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tate WP, Cridge AG. and Brown CM. (2018). “Stop” in protein synthesis is modulated with exquisite subtlety by an extended RNA translation signal. Biochem Soc Trans 46: 1615–1625 [DOI] [PubMed] [Google Scholar]
- 45. Nyiko T, Auber A, Szabadkai L, Benkovics A, Auth M, Merai Z, Kerenyi Z, Dinnyes A, Nagy F. and Silhavy D. (2017). Expression of the eRF1 translation termination factor is controlled by an autoregulatory circuit involving readthrough and nonsense-mediated decay in plants. Nucleic Acids Res 45:4174–4188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Akhmaloka , Susilowati PE, Subandi and Madayanti F. (2008). Mutation at tyrosine in AMLRY (GILRY like) motif of yeast eRF1 on nonsense codons suppression and binding affinity to eRF3. Int J Biol Sci 4:87–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Urakov VN, Valouev IA, Kochneva-Pervukhova NV, Packeiser AN, Vishnevsky AY, Glebov OO, Smirnov VN. and Ter-Avanesyan MD. (2006). N-terminal region of Saccharomyces cerevisiae eRF3 is essential for the functioning of the eRF1/eRF3 complex beyond translation termination. BMC Mol Biol 7:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stansfield I, Eurwilaichitr L, Akhmaloka and Tuite MF. (1996). Depletion in the levels of the release factor eRF1 causes a reduction in the efficiency of translation termination in yeast. Mol Microbiol 20:1135–1143 [DOI] [PubMed] [Google Scholar]
- 49. Dabrowski M, Bukowy-Bieryllo Z. and Zietkiewicz E. (2015). Translational readthrough potential of natural termination codons in eucaryotes—the impact of RNA sequence. RNA Biol 12:950–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cridge AG, Crowe-McAuliffe C, Mathew SF. and Tate WP. (2018). Eukaryotic translational termination efficiency is influenced by the 3′ nucleotides within the ribosomal mRNA channel. Nucleic Acids Res 46:1927–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wilschanski M, Yahav Y, Yaacov Y, Blau H, Bentur L, Rivlin J, Aviram M, Bdolah-Abram T, Bebok Z, et al. (2003). Gentamicin-induced correction of CFTR function in patients with cystic fibrosis and CFTR stop mutations. N Engl J Med 349:1433–1441 [DOI] [PubMed] [Google Scholar]
- 52. Wilschanski M, Famini H, Strauss-Liviatan N, Rivlin J, Blau H, Bibi H, Bentur L, Yahav Y, Springer H, et al. (2001). Nasal potential difference measurements in patients with atypical cystic fibrosis. Eur Respir J 17:1208–1215 [DOI] [PubMed] [Google Scholar]
- 53. Linde L, Boelz S, Nissim-Rafinia M, Oren YS, Wilschanski M, Yaacov Y, Virgilis D, Neu-Yilik G, Kulozik AE, et al. (2007). Nonsense-mediated mRNA decay affects nonsense transcript levels and governs response of cystic fibrosis patients to gentamicin. J Clin Invest 117:683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Linde L, Boelz S, Neu-Yilik G, Kulozik AE. and Kerem B. (2007). The efficiency of nonsense-mediated mRNA decay is an inherent character and varies among different cells. Eur J Hum Genet 15:1156–1162 [DOI] [PubMed] [Google Scholar]
- 55. Chan WK, Huang L, Gudikote JP, Chang YF, Imam JS, MacLean JA., 2nd and Wilkinson MF. (2007). An alternative branch of the nonsense-mediated decay pathway. EMBO J 26:1820–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Huang L, Shum EY, Jones SH, Lou CH, Dumdie J, Kim H, Roberts AJ, Jolly LA, Espinoza JL, et al. (2017). A Upf3b-mutant mouse model with behavioral and neurogenesis defects. Mol Psychiatry 23, 1773–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Huang L, Low A, Damle SS, Keenan MM, Kuntz S, Murray SF, Monia BP. and Guo S. (2018). Antisense suppression of the nonsense mediated decay factor Upf3b as a potential treatment for diseases caused by nonsense mutations. Genome Biol 19:4. [DOI] [PMC free article] [PubMed] [Google Scholar]