Abstract
Artificial intelligence (AI) and machine learning (ML) techniques are revolutionizing several industrial and research fields like computer vision, autonomous driving, natural language processing, and speech recognition. These novel tools are already having a major impact in radiology, diagnostics, and many other fields in which the availability of automated solution may benefit the accuracy and repeatability of the execution of critical tasks. In this narrative review, we first present a brief description of the various techniques that are being developed nowadays, with special focus on those used in spine research. Then, we describe the applications of AI and ML to problems related to the spine which have been published so far, including the localization of vertebrae and discs in radiological images, image segmentation, computer‐aided diagnosis, prediction of clinical outcomes and complications, decision support systems, content‐based image retrieval, biomechanics, and motion analysis. Finally, we briefly discuss major ethical issues related to the use of AI in healthcare, namely, accountability, risk of biased decisions as well as data privacy and security, which are nowadays being debated in the scientific community and by regulatory agencies.
Keywords: artificial neural networks, deep learning, ethical implications, outcome prediction, segmentation
1. INTRODUCTION
The last decade has seen a massive increase in the use of artificial intelligence (AI), especially machine learning (ML) technologies, for several applications. For example, personal assistants able to understand vocal natural language and to perform simple tasks such as retrieving information from a calendar, managing home automation devices and place online orders are now being used on millions of smartphones. A notable example of state‐of‐the‐art AI is the self‐driving car, which employs computer vision and other sensors to sense the surrounding environment, and automated control systems to take decisions and move without any human input.
While AI and ML are sometimes used in the generalist press as synonyms, ML constitutes only a branch of AI, the one dealing with methods to give a machine the capability to “learn,” that is to improve the performance in specific tasks, based on previous experience or on provided data.1 Although other AI branches such as symbolic reasoning, heuristics, and evolutionary algorithms have had a tremendous impact on science and technology,2 ML arguably constitutes the most interesting and promising field of AI for applications in medical research (Figure 1).
Figure 1.
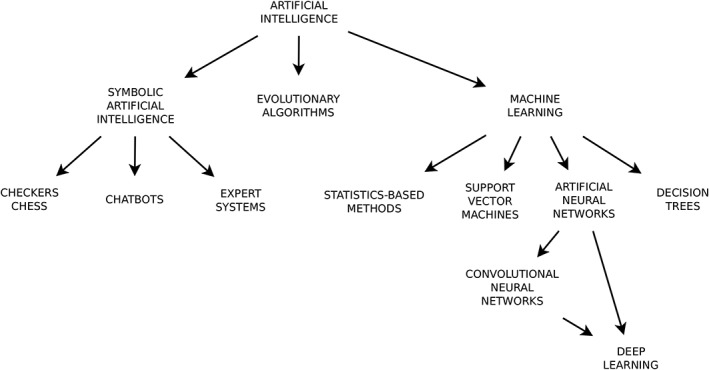
Schematic overview of the main branches of artificial intelligence (AI), including machine learning (ML) methods which are having an impact on spine research
ML is based on the availability of data, which is used to train the machine to perform the desired tasks. Due to its nature, ML lends itself well to applications in which input data are used to generate an output based on some features of the inputs themselves, for example, to perform image classification. Indeed, a research area which was dramatically advanced by ML in recent years is image processing. Thanks to the continuous technical improvements, in 2015, a deep neural network achieved for the first time superhuman performance in a famous image classification contest, the ImageNet Large Scale Visual Recognition Challenge.3 Computer can nowadays perform tasks such as image classification, object detection (eg, face detection and recognition), and landmark localization better than expert human operators. Although the deployment of such powerful technologies to medical imaging is still in its infancy, radiologists generally agree that ML is a truly disruptive technology which can deeply transform how imaging data are interpreted and exploited for treatment planning and follow‐up.4 The impact of ML and AI on other basic medical research fields has been less conspicuous so far; nevertheless, numerous novel applications, for example, in motion analysis and mechanical characterization of tissues, are starting to emerge.
As testified by the sharp increase in the number of published papers in recent years, AI and ML are more and more being used to investigate issues related to the spine, especially in radiological imaging but also in other fields such as the outcome prediction of treatments. The reported results are either promising or already surpassing the previous state of the art in several applications; for example, ML techniques nowadays allow for an accurate and perfectly repeatable grading of intervertebral disc degeneration on magnetic resonance imaging (MRI) scans. Indeed, the current pace of technical improvements is expected to being further benefits in the next future.
With this narrative literature review, we aim at raising the awareness of the current achievements and potential spine‐related applications of AI in the spine science community, including readers working in different fields who are not familiar with the technical aspects of such technologies. To this aim, the paper first presents a brief general overview of AI, with special emphasis on ML and its recent advances which are having a practical or potential impact on spine research. The following paragraphs describe the state of the art of the use of ML and AI in spine science, including diagnostic spine imaging, the prediction of the outcome of therapeutic interventions, clinical decision support systems, information retrieval, biomechanical analysis and characterization of biological tissues, and motion analysis.
2. HISTORICAL PERSPECTIVE
The first steps toward AI date back to the development of general purpose computers, which were pioneered during the Second World War and become available for nonmilitary use in the 1950s. The newly available computing power allowed creating symbolic AI programs, that is, algorithms that apply a set of rules in order to imitate reasoning and to draw decisions.2 Notable examples of such programs are those aimed at checkers1 and chess gaming, which achieved very good performances already in the 1970s,5 and the first chatbots which could simulate to some extent a conversation in natural language.6 In parallel, taking advantage of the recent advances in neurological research which showed that the central nervous system consists of a large network of units communicating via electric signals, research groups started developing the so‐called artificial neural networks (ANNs), that is, networks of artificial neurons mimicking the brain structure (Figure 2A),7 by means of analog systems.8 These networks, such as, for example, the perceptron, 9 showed to be able to perform simple logical functions and to recognize classes of patterns, although with significant limitations.10
Figure 2.
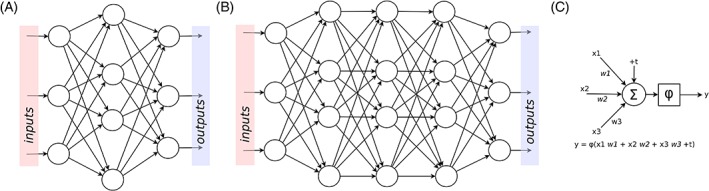
Schematic representation of an artificial neural network (A), a deep network (B), and a unit, also called artificial neuron (C). In each unit, the inputs (“x 1,3 ”) are multiplied by weights (“w 1,3”), summed to a bias term (“+t”), and the total sum is processed by a linear or nonlinear activation function (“φ”)
After the first two decades of research, there was a succession of phases of general skepticism (the so‐called “AI winters”) mainly due to an underestimation of the complexity of the problems to be solved and lack of the necessary computer power, and optimistic phases with larger funding and technological breakthroughs.2 In the 1980s, expert systems, that is, computer programs able to deal with practical problems based on set of rules derived from human expert knowledge, were successfully employed in several research and industrial fields. In the same years, ANNs were revamped by the development of backpropagation, 11 a powerful training algorithm which is still the base for their use nowadays.
In the last two decades, the increases in computer power and its improved accessibility even for small research institutes, made possible by graphics processing units (GPUs) with tremendous parallel computing capabilities, fostered the adoption of AI solutions for many practical applications.12 While the achievement of strong AI, that is, a computer program with a flexible intelligence which can perform any task feasible for humans, remains out of the foreseeable future, narrow AI, that is a machine able to apply AI only to a specific problem, has found a widespread use. Internet search engines and speech recognition software are good showcases of the huge potential of the recent advances.
One of the branches of AI which is seeing the fastest improvements is deep learning 12 (Figure 2B). In most implementations, this ML method is based on deep neural networks, that is, network architectures with several layers, and is revolutionizing research fields such as image processing, voice recognition and natural language processing. In addition to the improved computer power, a key driver for the success of deep learning was the availability of big data, massive datasets collected from various sources, including the Internet and medical institutions (eg, imaging databases), which are extremely valuable for an effective exploitation of deep learning in practical applications. As a matter of facts, most of the scientific papers applying AI to spine research, which are described in the paragraph “Applications of AI and ML in spine research,” are based on deep learning.
3. MACHINE LEARNING
The expression “ML” was introduced by Arthur Samuel in 1959, who defined it as the field of study that gives computers the ability to learn without being explicitly programmed.1 This paragraph summarizes the main concepts of ML, which are presented in deeper details elsewhere.13, 14
The general aim of ML is to make a prediction, that is, to estimate the value of a desired output given an input, based solely on features provided by the model developer or automatically learned from training data. More specifically, common applications of ML include:
(a) Classification: the input is assigned to a specific category among a group of two or more. An example of binary classification is the automated diagnosis of cancer based on histopathological images, in which the machine should decide if an image shows features (eg, texture and color information) depicting a pathological condition. The automation of Pfirrmann grading for disc degeneration exemplifies a multiclass classification problem, in which an MRI scan of the disc should be assigned to a category ranging from 1 (healthy disc) to 5 (severe disc degeneration).15 Image segmentation, in which each pixel is labeled based on its belonging to a specific region or anatomical structure, can also be considered as a subclass of classification problems.
(b) Regression: the output of the task is continuous rather than discrete. An example of a regression problem is the determination of the coordinates of an anatomical landmark in a radiographic image.
(c) Clustering: the provided inputs are divided into groups, based on features learned from the inputs themselves. Cluster analysis is used to classify data when no a priori knowledge about the belonging to a specific class is available. Clustering has been used, for example, to subdivide into groups patients suffering from osteoporotic vertebral fractures based on pain progression.16
Another way to describe the different forms of ML is based on the nature of the tasks to be performed:
(a) Supervised learning: the machine learns to predict the output based on a collection of inputs for which the correct output (ground truth) is known. In most implementations, supervised learning consists in learning the optimal manner to map the inputs to the outputs, by minimizing the value of a loss function representing the difference between the machine predictions and the ground truth. It is the most common type of learning used in medical research.
(b) Unsupervised learning: the machine learns from input data for which there is no ground truth. This type of learning task identifies patterns and features in the inputs, with the aim of extracting new knowledge from the available data. Clustering is an application of unsupervised learning.
(c) Reinforcement learning: instead of having ground truth data available at the beginning of the task, feedback about the correctness of the execution is provided after the task has been completed, thus acting like to a reward or a punishment. Reinforcement learning is typically used in dynamic or interactive environments, for example, in gaming. Clinical decision‐making is rapidly gaining interest as another field of application. Models of reinforcement learning are valuable tools for the investigation of how nonhuman animals and humans learn the causal structure of tasks and phenomena.
Regardless of the task to be performed, the availability of large datasets to be used for training the algorithm and to test its accuracy is essential for a successful implementation of ML. Especially in medical research, this requirement poses serious challenges related to data privacy, ethics, regulation, and liability, which are described in Section 6.
4. METHODS USED IN SUPERVISED LEARNING
The next paragraphs provide a brief summary of the methods used for supervised learning, which play a cardinal role among the ML tasks in medical research, and are described in detail elsewhere.13 The concept of supervised learning is based on the estimation of a function which maps an input, which can be, for example, an image or a collection of clinical data regarding a patient, to an output value. The training data therefore consists of a set of pairs including an input and the relative output, which is known. When the mapping function has been determined, it can be used to process new inputs for which the value of the output is not available. If the number of training examples is sufficient and an appropriate learning algorithm has been chosen, the algorithm itself should be able to generalize well, that is, to provide accurate results for inputs similar but different to those included in the training data. Conversely, the predictions may reveal overfitting, that is, results fitting precisely the input data but not able to make accurate predictions on additional data, or underfitting, which happens when the learning model is not sufficiently complex to capture the features of the input data17 (Figure 3).
Figure 3.
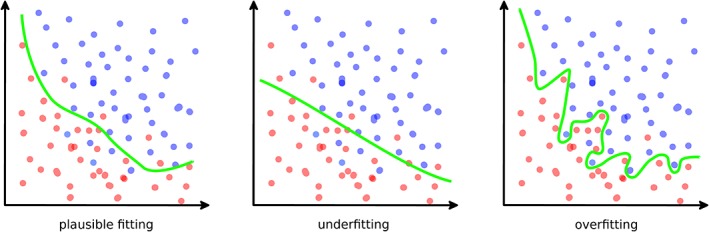
Examples of a plausible good fitting (left), underfitting (center), and overfitting (right) in a binary classification task
4.1. Methods derived from statistics
Although considering linear regression in the realm of ML might be counterintuitive, it constitutes a good example of a simple method to create a function which maps an input (a number, or more frequently a vector of numbers) to an output. Indeed, any form of input can be mathematically formulated as a multidimensional vector of numbers, conventionally named features, which can be processed by linear regression. Features are a set of variables which characterize the data, and can be either simple and human readable (such as, eg, age and sex of a patient) or more difficult to interpret, such as the image features extracted with specialized algorithms like SIFT18 and ORB,19 or with texture and shape analysis. Even without feature extraction, an image such as a radiograph can be also viewed as an array of integer numbers with length equal to the number of pixels in the image; each element of the array would contain the color (gray level) of the specific pixel. From this perspective, the application of linear regression even in case of complex and large inputs is straightforward.
The linear regression function is commonly fitted by means of the least squares method, which therefore acts as the learning algorithm. In this case, performing a linear regression corresponds to minimizing the mean square error (MSE) between the predictions and the inputs; MSE therefore represents the loss function of the algorithm. In ML literature, MSE is also commonly named as L2 loss, whereas the L1 loss is the mean absolute error (MAE) which is also a possibly effective choice for regression problems. Due to its simplicity and its inherent incapability of capturing a nonlinear behavior, linear regression is prone to underfitting, and therefore is not the method of choice for complex ML regression tasks.
Logistic regression can be seen as the equivalent of linear regression for classification problems. In its simplest form, the inputs (one or multiple continuous numbers) are fitted to a binary output (0 or 1) by means of a nonlinear curve, the logistic sigmoid function, which represents the probability that an input is mapped to the “1” output. If the output probability is greater or equal than 0.5, a “1” is predicted, whereas on the contrary the output is “0.” In addition to predicting binary outputs, logistic regression can be effectively generalized to multiclass classification problems. MSE is not the most appropriate choice to act as loss function for logistic regression; specialized functions such as the cross entropy are employed in this respect. Similar to linear regression, logistic regression is outperformed by more complex algorithms for most ML classification tasks.
Another method derived from statistical inference which found its place in ML literature is the Bayes classifier,20 which is based on Bayes' theorem of conditional probability. The naive Bayes classifier, which assumes the independence of the features from each other, is especially simple to implement, fast to train even for very large training datasets and potentially very effective in tasks where the assumption of feature independence is reasonable. In spine research, Bayes classifiers have been used for the classification of vertebral fractures21 and for computer‐aided diagnosis.22
4.2. Support vector machines
Considering each input belonging to the training data as a multidimensional vector and therefore as a point in a multidimensional space, performing a classification task corresponds to determining a partition of the space which divides the points belonging to the various classes. A support vector machine (SVM) is an algorithm which builds the hyperplane, or a number of them, which can divide the space so that the points of the different classes are effectively and optimally partitioned23 (Figure 4).
Figure 4.
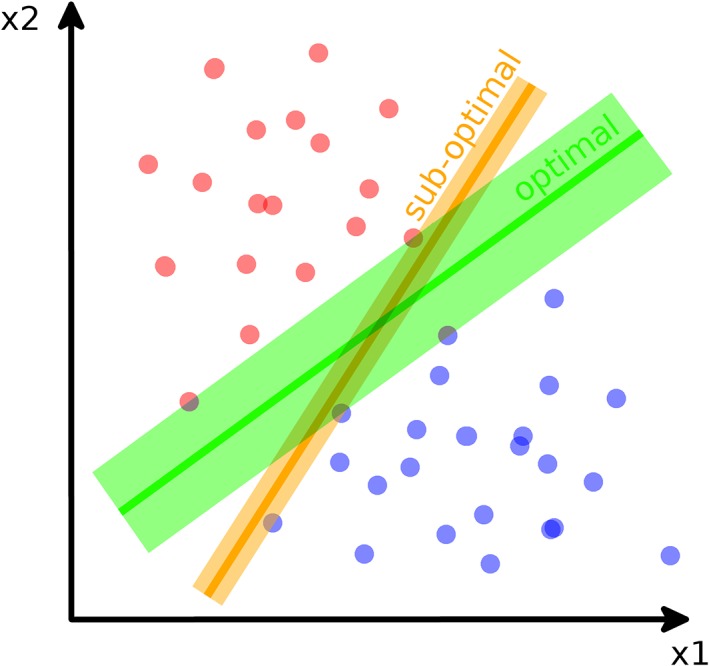
Schematic representation of a simple support vector machine (SVM) used for binary classification. In brief, the SVM builds the optimal hyperplane (in green) which separates the two classes maximizing the gap between them. A non‐optimal hyperplane (in orange) which correctly separates the two classes, but with a smaller gap, is also shown. The SVM operates in the feature space (“x1” and “x2” in the exemplary figure)
SVMs are powerful tools to perform multiclass linear classification tasks, including image segmentation. Although the original publication of the method dates back to 1963,24 SVMs are still widely used nowadays and may outperform the most recent techniques in specific cases, for example, when the dataset available for training has a limited size. In spine science, SVMs have been used, for example, for the grading of disc degeneration25 and for the classification of scoliosis curve types.26 SVMs can be adapted to nonlinear classification and regression problems, as well as to unsupervised learning (eg, for clustering).
4.3. Classification and regression decision trees
The use of tree‐like structures in AI dates back to the pioneering checkers programs by Arthur Samuel.1 Even for classification and regression purposes, decision tree was first employed in the 1950s.27, 28 Nowadays, decision trees are valuable support tools in various fields including economics and military; notably, they are commonly used for the choice of the most appropriate medical treatment in health care.
In ML, a classification and regression decision tree (CART) links the values of the features to the possible outputs, therefore implementing a classification or a regression task, by means of a set of conditions.29 For each condition, the tree splits into branches, which end with terminal nodes representing the outcome of the decision; due to this peculiar structure, CARTs are easier to understand for humans with respect to other ML techniques. CARTs can be trained based on large sets of input data by means of specialized algorithms,30, 31 which are generally not computationally intensive and thus suitable for very large datasets. Regarding downsides, CARTs are prone to overfitting, which can be limited by using special techniques such as pruning, which reduces the size of the tree, and random forests, 32 which exploit multiple decision trees built on random subsets of the features and average their predictions. CARTs and random forests have been used for several applications in spine research. As a clinical decision support system, decision trees have been used for the management of low back pain,33 and for the preoperative selection of patients with adult spinal deformity.34 Other applications include the evaluation of the primary fixation strength of pedicle screws35, 36 (Figure 5), and the prediction of proximal junctional failure.37
Figure 5.
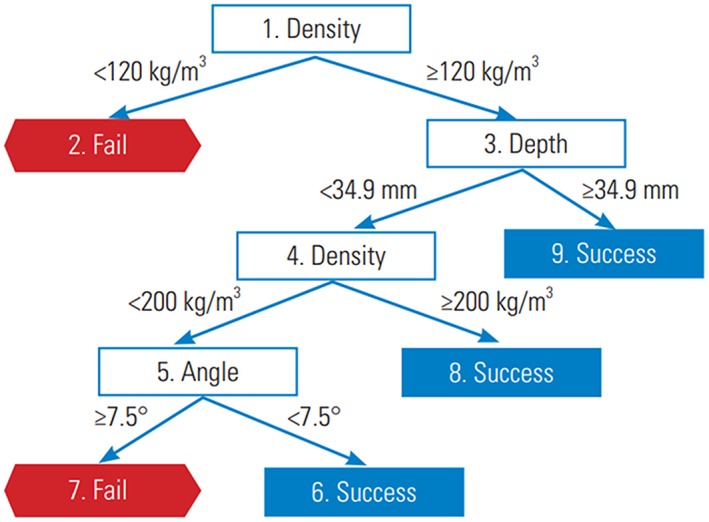
Example of a decision tree trained to predict the risk of failure of pedicle screws. Reproduced with permission from Varghese et al36
4.4. Artificial neural networks
ANNs constitute the branch of ML which has seen the most impressive improvements in recent years, so much that it has been identified by the general public with ML itself. Applications of ANNs in medical research as well as in spine science are countless, and are described in detail in the paragraph “Applications of AI and ML in spine research.”
ANNs are biologically inspired networks which loosely resemble how the neurons are connected and interact in the brain.7 Mimicking the principles of Hebbian learning,38 information flows from the inputs to the outputs through artificial neurons, which are organized in layers and perform simple operations such as making linear combinations of their inputs multiplied by a weight, and then processing the result through a linear or nonlinear activation function (Figure 2C). The networks may include regularization terms, which are aimed at reducing the risk of overfitting by penalizing large values of the weights through a penalty coefficient. Training the ANN consists in finding the optimal values of the weights, so that the inputs belonging to the training data are processed and transmitted through the layers resulting in outputs which fit well the ground truth.
The same loss functions described in the previous paragraphs, that is, MSE, MAE, and cross entropy, are commonly used to train ANNs and as metrics for their performance. In its simplest implementation, the training algorithm, named backpropagation,11 consists in calculating the derivatives of the loss function with respect to each weight, and adjusting the specific weight by the value of the respective derivative multiplied by a coefficient, the learning rate. Iterating the process determines a decrease of the loss function, which would reach a minimum after convergence has been achieved. This gradient descent algorithm has been superseded by more sophisticated methods, such as, for example, the stochastic gradient descent39 and Adam,40 which can generally achieve a faster and more robust convergence.
ANNs are used in several industrial and research fields, for both classification and regression problems. Although the applications of ANNs in spine research are mostly based on supervised learning, these networks are also proficiently employed for unsupervised tasks and reinforcement learning. Starting from the earlier examples such the single layer perceptron,9 a simple linear binary classifier consisting of a single layer of outputs directly connected to the inputs via a series of weights, high‐performance network architectures which are optimized to deal with specific problems have been developed. For example, ANNs are nowadays used to generate new data which share some characteristics with known data by means of the so‐called generative models, 41 and to process data keeping memory of previous inputs, for example, with recurrent neural networks.42 The latter methods found widespread use in speech recognition and automated language translation.
4.5. Convolutional neural networks
Image processing, as well as computer vision in general, are arguably the largest fields of application of ANNs. The design of convolutional neural networks (CNNs or ConvNets) has been inspired by the structure of the animal visual cortex, based on experiments carried out in cats and monkeys.43, 44 In the 1960s, Hubel and Wiesel described that specific groups of neurons in the visual cortex are stimulated only be small areas of the visual field, and extract features and information from those areas. Specific groups of neurons are sensitive to features such as a certain edge orientation, and others to other directions or shapes. Visual perception then results from combining the information coming from the neuron groups and exploiting information about their architecture.
CNNs mimic rather closely such neuronal architecture.45, 46 In a convolutional layer, which is the characterizing component of a CNN, a small filter (having most commonly a size of 3 × 3 × 3 or 5 × 5 × 3) slides, or convolves, on the input image; for each possible position in the image, a number is calculated by element‐wise multiplication of the weights of the filter by the corresponding values of the input of the layer. The collection of all calculated numbers constitutes the so‐called activation map (Figure 6). Since a typical convolutional layer consists of several filters, the convolution process results in a three‐dimensional matrix, each layer of which is an activation map. Convolutional layers are usually combined with pooling layers, 47 which downsample the data and help in reducing the risk of overfitting, and dense (fully connected) layers, which are the standard nonconvolutional layers used in ANNs, to generate an output and thus to perform a classification or a regression task. Dropout layers, in which a predefined fraction of artificial neurons are artificially canceled, force the network to learn different ways of achieving the same output and are frequently integrated in CNNs to reduce the risk of overfitting. Training the convolutional layer consists in finding the optimal values of the weights of the filters, and is performed by means of optimization algorithms similar to those used for standard ANNs.48
Figure 6.
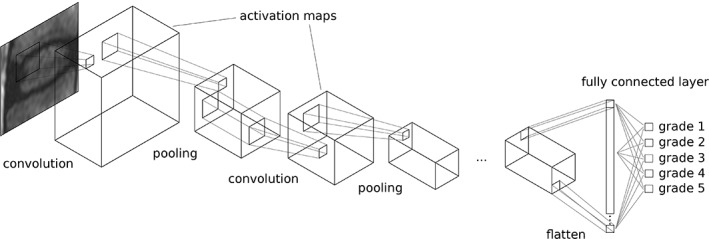
Schematic representation of a convolutional neural network (CNN), here exemplary aimed at performing the grading of disc degeneration on T2‐weighted MRI scans based on the scheme presented by Pfirrmann et al.15 In a convolutional layer, a small filter convolves over the data creating a series of activation maps; these maps can be downsampled by pooling layers, and then processed by another convolutional layer. In the simplest forms of a CNN, one or more fully connected layers perform the final classification or regression decision
4.6. Deep learning
In simple terms, deep learning is the branch of ML which employs methods involving multiple layers of processing units, with the final aim of being able to capture different levels of abstraction. Practically, deep learning is most commonly based on the use of multilayer ANNs, commonly referred as deep neural networks. Although such ANNs with several layers were developed in conjunction with CNNs and have been available already in the 1970s, they never gained widespread use due to the computational resources required for training, and the lack of effective learning algorithms. In 1989, the research group of LeCun introduced the first of a family of networks, LeNet‐1, which featured two convolutional layers and two pooling layers and could be trained with the standard backpropagation.49, 50 LeNet‐1 scored state‐of‐the‐art results in an image classification task; later developments, notably LeNet‐5, showed that increasing the depth of the network, that means adding layers, could drastically improve the accuracy of the predictions.51 These pioneering studies, together with the improved accessibility of computer power, opened the way to deep learning which is nowadays considered as the most advanced frontier in ML. It should be noted that deep learning architectures are not only based on ANNs and aimed at computer vision, but also cover other domains, such as, for example, the deep Boltzmann machines commonly used for making music and movie recommendations on the Internet, and deep recurrent neural networks for speech recognition and natural language understanding.49
Recent developments of deep architectures are continuously raising the bar in image classification tasks. In 2012, AlexNet,52 a CNN having five convolutional layers followed by three dense layers, won several competitions and demonstrated that deep CNNs have more potential for computer vision than any other current ML technique. Among the various designs that were introduced afterward, some are worth of mention. The Visual Geometry Group (VGG) architecture was developed at the University of Oxford and is a large network with 138 million trainable parameters, 13 convolutional layers and two dense layers.53 GoogLeNet, introduced in 2014, has 22 layers (including nine Inception layers, a novel design) but a smaller number of parameters (11 million), benefiting the computational resources necessary for training.54 The ResNet family of networks, presented by Microsoft in 2015, features a large number of layers, up to 152, none of which is fully connected.3 ResNet was the first architecture to achieve superhuman performance in image classification; its foundation innovation, the concept of residual learning, that is, skipping layers in order to make the deep network easier to train, is still exploited in many of the most recent architectures.
A key driver for the widespread diffusion of deep learning is its easy accessibility. In the spirit of knowledge sharing and cooperative work which characterizes computer science and is gaining momentum also in other fields, the vast majority of the recently developed algorithms are publicly available on the Internet. ML frameworks such as Torch (http://torch.ch/), Tensorflow (https://www.tensorflow.org/), and Caffe (http://caffe.berkeleyvision.org/), as well as high‐level libraries such as Keras (https://keras.io/) and PyTorch (https://pytorch.org/) are also freely available, even for commercial use.
Together with the improved accessibility of powerful GPUs and of cloud computing platforms offering AI products and services, the availability of state‐of‐the‐art deep learning software is fostering its use in a wide range of research fields. Although the adoption of deep learning for real‐world problems in spine science is still limited by the short time passed since its first introduction, we expect it to become a disruptive technology in the near future, especially for spine imaging applications.
4.7. Assessing the accuracy and robustness of ML tools
Before any ML tools can be used to address practical problems and deployed to industrial or research environments, their accuracy and robustness need to be proven by performing a proper validation. To do so, in supervised learning, the available data are typically split in two or three datasets, which serve different purposes.55 The first one is the training dataset in the strictest sense of the word, which includes the majority of the available data (typically around 70%‐80%) and is actually used to train the model, that is, to calculate the weights of the artificial neurons in case of ANNs. The second set is named validation dataset and is aimed at tuning the model hyperparameters, such as learning and dropout rates, penalty coefficients in regularization terms or even the number of units or layers, in order to improve the model fit on the training data. The validation dataset might not be present in the simplest ML implementations, when all hyperparameters have been defined by the developer prior to training. The latter set is the test dataset, which includes data which has not been seen by the model, that is, neither used for learning the weights nor for tuning the hyperparameters, and therefore allows for an unbiased assessment of the model accuracy and robustness. The test dataset should be used only when the model is completely trained; if modifications to the model architecture or hyperparameters are performed after testing, for example, to further improve the accuracy or to reduce overfitting, a new test should be performed on another set of data which has not been seen previously by the model.
For a proper assessment of the model performance, it is critical that training, validation, and test datasets do not overlap. Besides, selection bias should be avoided when creating the three datasets from the available data; all sets should be equally representative samples of the data of interest. The quality of ground truth data is a further issue of uttermost importance, especially when the size of the database is limited; noisy ground truth would result in outputs which are inaccurate to some extent, depending on the amount of data available.56 As a matter of fact, there is no precise rule to estimate the minimum size of the training database for a good performance of the model. Heuristics methods as well as naive guesses are sometimes used to this purpose; a more comprehensive evaluation requires training the model on databases of different sizes, and creates a learning curve representing accuracy vs data size. An estimation of the minimum required size can then be extrapolated from the curve.
Test automation is currently widely used in software engineering to execute a large number of tests in a controlled and formalized environment, by means of specifically designed software. Although this technology still has to find its place in the rapidly evolving field of ML, especially regarding medical applications, its adoption for model validation is easy to foresee in the next future. A prerequisite for such advance is the definition of standardized data sets, which shall be used to perform quantitative comparisons between different models.
The validation process may reveal either underfitting, which results in poor performance of the model on all the three datasets, or overfitting, which can be detected when good accuracies are achieved on the training data, but the unbiased evaluation on the test dataset reveals a poor outcome. Whereas addressing underfitting typically involves increasing the complexity of the model, overfitting can be remediated by means of specific techniques such as pooling and dropout layers or regularization, as mentioned above, or by simplifying the model architecture.
5. APPLICATIONS OF AI AND ML IN SPINE RESEARCH
AI technologies are having a major impact in several research fields related to the spine, which is expected to further increase in the future. In the following paragraphs, we summarize the published applications of AI and ML in various domains of spine research, such as diagnostic imaging, prediction of treatment outcomes, and decision support systems. Applications more closely related to basic science such as biomechanics and motion analysis are covered as well.
5.1. Localization and labeling of spinal structures
ML approaches have been employed to extract information such as the location of vertebrae, discs and spinal shape from radiological images like planar radiographs, computed tomography (CT) and MRI scans. As a matter of fact, localizing anatomical structures in an imaging dataset is commonly a first step toward the development of fully automated methods for the detection and classification of pathological features, or to predict the outcome of therapies.
In addition to methods not strictly related to ML, based, for example, on thresholding and heuristic search,57, 58 proper ML techniques have been used for localization tasks. Schmidt used a classification tree to generate a probability map of the location of each intervertebral disc centroid in MRI scans, which were then used by a probabilistic graphical model to infer the most likely location, resulting in an average localization error of 6.2 mm with respect to a human‐created reference.59 Oktay and Akgul trained an SVM for disc localization based on a feature descriptor, the pyramidal histogram of oriented gradients, obtaining mean localization errors ranging between 2.6 and 3.6 mm depending on the disc level.60 In simple words, the method was based on a sliding window, which is a rectangular region which slides over a multiscaled version of the original image; for each position of the window, the value of the feature descriptor is calculated, and passed as input to the SVM to determine if the current window contains an intervertebral disc. When a set of the most likely disc locations have been calculated, a graphical model is used to infer the position of each specific disc. The same authors expanded and improved the method to allow also localizing the vertebrae, achieving average errors lower than 4 mm.25 Glocker et al confronted the challenging topic of localization of vertebrae in CT datasets of pathological spines, including severe scoliosis, sagittal deformity and presence of fixation devices, obtaining mean localization errors between 6 and 8.5 mm61, 62 (Figure 7). The proposed method was based on classification random forests trained to determine the location of the vertebral centroid, and employed novel techniques to generate appropriate training data and to eliminate false positive predictions.
Figure 7.
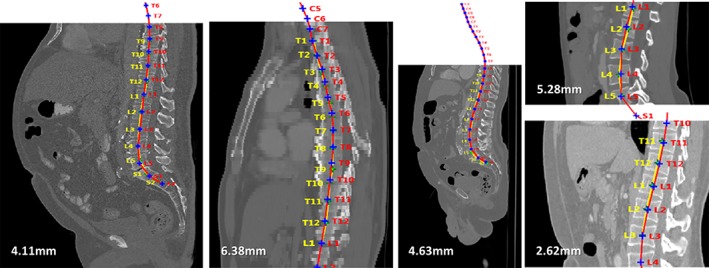
Examples of localization of the vertebral centroids from a literature study,61 dealing with different types of CT images (from left to right: standard, low resolution, noisy, cropped). Manual annotations by an expert operator are shown in yellow, whereas the computer predictions are in red. The numbers indicate the mean absolute error (MAE) with respect to the manual annotations. Reproduced with permission from Glocker et al61
More recently, ANNs and deep learning were also employed for the localization of spinal structures. Chen et al used a hybrid method involving a random forest classifier which performs a first coarse localization used to drive a deep CNN63, 64; this approach allowed for a clear improvement with respect to the previous state of the art not based on deep learning,62 that is, average localization errors for the centroid of the intervertebral disc of 1.6 to 2 mm. The same research group also used CNNs, both based on a 2D convolution, that is, processing separately the single slices, and a novel 3D convolutional layer.65 Suzani et al used a six‐layer neural network to localize the vertebral centroids by means of a regression task: for each voxel in the dataset, the network voted the vector connecting the voxel itself to the centroid. The votes were then used to statistically estimate the most probable location of the vertebral centroid66. An alternative approach was presented by Payer et al, who used 2D and 3D CNNs to build regression heatmaps of the landmark locations67; the method was, however, not applied to spine images. In several papers, after a satisfactory localization of the vertebral or disc centroids has been achieved, the labeling task was performed by fitting a graphical model.68, 69 Recent works achieved high accuracies with complex models able to perform the localization of landmarks and vertebral centroids by taking as inputs the whole 3D dataset, without any preliminary coarse localization or sliding window approach. Yang et al were able to achieve localization errors for the vertebral centroid between 6.9 and 9 mm in CT scans of patients suffering from various pathologies as well as subjected to surgical instrumentation, with strongly variable fields of view as well as image resolution70.
As a matter of fact, state‐of‐the‐art techniques for localizing and labeling spinal structures have achieved high performance comparable to that of expert human observers. Detection and labeling functions are nowadays already integrated in commercial Picture and Archiving Communication System and commercially available clinical imaging software, although technical details about those have not been publicly disclosed.
5.2. Segmentation
A key problem in image analysis is understanding the content of the image, that is, subdividing the image in regions at a pixel level so that each pixel belongs to a specific region. This process is named semantic segmentation and can be conducted either manually or automatically; this topic has been the subject of a vast body of literature, since it is fundamental for applications such as computer vision and autonomous driving.71 In medical imaging, in addition to identifying if a pixel belongs, for example, to a disc, the segmentation algorithm should typically determine to which specific instance it belongs (eg, either L1‐L2 or L2‐L3). This type of segmentation is named instance segmentation, and is the most relevant for spine research.72
Assessing the quality of a segmentation algorithm involves the definition of quantitative metrics, which might be less intuitive than the localization error employed in localization tasks. Among the several metrics which have been introduced in previous studies, the most common ones are the Dice similarity coefficient (DSC), which expresses the amount of spatial overlap between the segmented image and the ground truth, and the mean surface distance (MSD), which describes the mean distance between every surface voxel of the segmented surface from the closest surface voxel in the ground truth.
Many papers introduced methods for spine segmentation not involving ML techniques, which in several cases required the intervention of the user73, 74, 75; fully automated methods were described as well.76 Other methods relied on fitting deformable anatomical models to the images by means of optimization procedures.76, 77, 78 Among many published techniques, the ones based on graphs and the normalized cuts were especially successful,79, 80 as well as methods derived from them.81, 82, 83 For example, by using normalized cuts, Ayed et al79 achieved DSC values of 0.88 and MSD of 2.7 mm. Marginal space learning assumes that the pose and shape of the object to be segmented is quantized in a number of parameters.84, 85 A large number of hypotheses covering the parameter space, that is, describing all the possible poses of the object, are then formulated; the best hypothesis is selected by means of a classifier.
In recent years, CNNs specifically designed for instance segmentation tasks were employed. Chen et al65 used a deep CNN including 3D convolutional layers to generate the probability of belonging to a specific region at the voxel level. Postprocessing techniques including thresholding and smoothing were used to refine the segmentation. Lessmann et al86 introduced a 3D CNN with a memory component in order to remember which vertebrae were already classified. In order to be able to process large datasets, the technique uses a 3D sliding window approach which first determines the position in which the window contains an entire vertebra, and then performs the pixel‐level segmentation with a deep classifier. The memory is then updated so that if a portion of the already segmented vertebrae is detected while looking for the next ones, it is then ignored. This method allowed achieving outstanding accuracies, with an average DSC of 0.94 and MSD of 0.2 mm.
Although promising results have been achieved, the segmentation of the anatomical structures of the spine still appears to have large room for improvements. Indeed, spine segmentation challenges have been proposed even very recently (Computational Methods and Clinical Applications for Spine Imaging (Figure 8), xVertSeg (http://lit.fe.uni-lj.si/xVertSeg/overview.php),87, 88 and databases hosting annotated images to be used for the development of new segmentation methods are currently publicly available (http://spineweb.digitalimaginggroup.ca/spineweb/).
Figure 8.
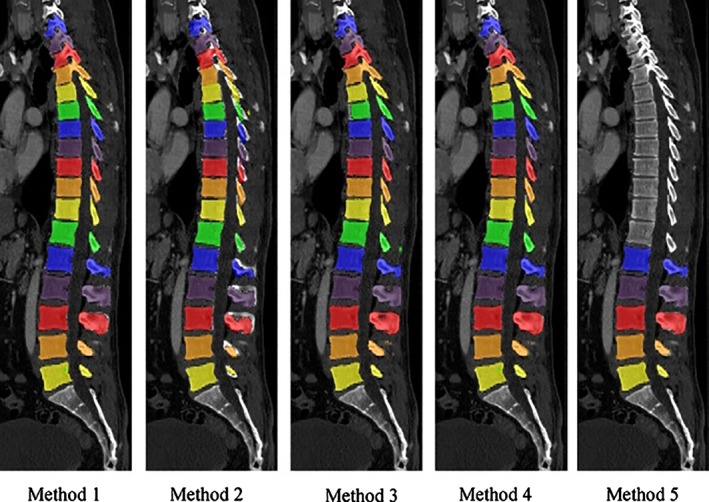
Five automated segmentation methods for CT scans developed in the frame of the grand challenge organized by the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) Workshop on Computational Spine Imaging (CSI 2014). Reprinted with permission from Yao et al87
5.3. Computer‐aided diagnosis and diagnostic imaging
The use of ML for diagnostic purposes dates back to the 1980s. In 1988, Bounds et al89 trained a multilayer perceptron to diagnose low back pain and sciatica, with reported accuracies ranging between 77% and 82%, better than those obtained by human medical doctors (68%‐76%) (. Symptoms and previous medical history, in a standardized form, were used as training data; as output, the ANN classified the back pain in four categories, namely simple back pain, radicular pain, spinal pathology (tumor, inflammation, or infection), and back pain with significant psychological overlay. More recently, most papers exploited the availability of imaging data to perform the automated diagnosis of a spinal disorder. Nowadays, the use of ML for diagnostic imaging of the spine encompasses several types of disorders, such as degenerative diseases, spinal deformities as well as oncology.
Similar to the detection and segmentation of spinal structures, the first published works about computer‐aided diagnosis based on medical imaging employed non‐ML techniques based on classical image processing techniques,90 or simple ML methods such as Bayesian classifiers.91 Shallow ANNs such as perceptrons were also used in the 2000s for various purposes, for example, detecting osteophytes.92 Two automated classification systems for degenerated intervertebral discs on T2‐weighted MRI images were presented in 2009,74, 93 and both provided a binary output (“normal” vs “degenerated”). One study was based on a simple statistical model trained on 30 MRI datasets,93 whereas the other paper employed a Bayesian binary classifier and exploited MRI scans from 34 patients74; both studies took into account information about the signal intensity and the texture of the disc. In 2011, Ghosh et al tested several different classifiers in performing the same task, including an SVM, all trained on 35 MRI stacks,94 obtaining accuracies ranging between 80% and 94%; the SVM resulted to be the most accurate technique. Hao et al95 proposed an SVM‐based method which considered, in addition to the intensity and texture information, the shape of the disc in order to classify it as degenerated or not; accuracies up to 91.6% were achieved. Oktay et al96 further refined such approach by including information from the T1‐weighted MRI scan. A significant advance was provided by the works of Ruiz‐Espana et al97 and Castro‐Mateos et al,98 who classified disc degeneration not on a binary basis but following the classification scheme published by Pfirrmann et al,15 which describes five degeneration degrees and is commonly employed in the clinical practice. Both studies included the extraction of features describing the intensity as well as the shape of the discs which were then passed to a classifier, which was a custom solution in the former paper and a simple ANN in the latter one. Prior to the feature extraction, the discs were segmented automatically in both works. The paper by Jamaludin et al99 introduced several improvements and innovations, such as the collection of a high number of disc images to be used for training and testing, namely 12 018 discs from 2009 patients whereas most previous papers involved less than 100 MRI datasets, and the use of a CNN as a classifier, which obviated the need for a segmentation prior to the classification (Figure 9). The method allowed achieving an agreement with human observations of 70.1%, comparable to the reported inter‐rater agreement between distinct expert radiologists of 70.4%. Furthermore, the same method was used to successfully detect other features such as endplate lesions and marrow changes. Recently, Niemeyer and coworkers used a deep CNN and further increased the size of the training set, setting the state‐of‐the‐art accuracy for automatic degeneration grading with the Pfirrmann classification system at 97%.100
Figure 9.
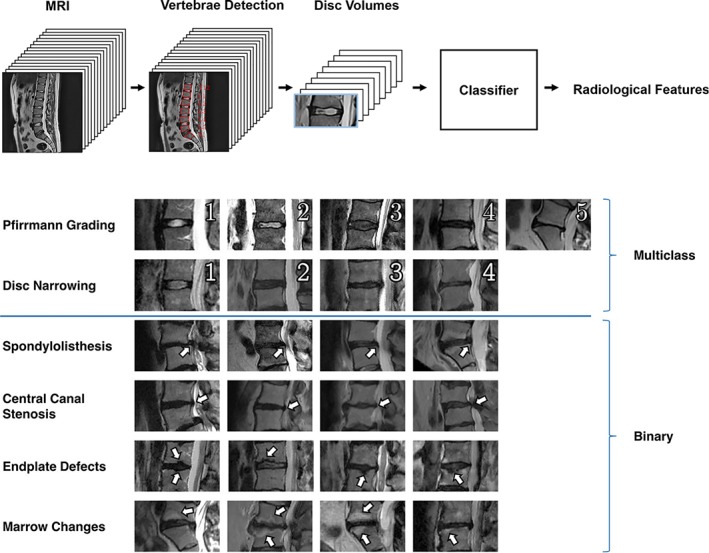
Top: workflow to perform classification tasks on lumbar MRI scans from a literature study.99 First, vertebrae are detected, then the volumes corresponding to the intervertebral discs are extracted and passed to a classifier. Bottom: the various radiological parameters (Pfirrmann grading of disc degeneration15; disc narrowing; spondylolisthesis; central canal stenosis; endplate defects; marrow changes) automatically extracted from the images in the same study. Reproduced from Jamaludin et al99
Aside from the degenerative spine, ML techniques have been also applied to the study of spinal deformities. The research area which has been impacted to the largest extent by ML is the evaluation of the severity of adolescent idiopathic scoliosis by means of noninvasive techniques such as surface topography. As a matter of fact, such techniques do not offer a direct visualization of the spine; the extraction of clinically relevant conclusions can therefore take a decisive advantage from inference tools which can exploit subtle patterns in the data which may not be visible to human observers. Ramirez et al101 classified surface topographies of scoliotic patients in three categories, namely mild, moderate, and severe curves, by means of an SVM, a decision tree, and a technique derived from statistics, the linear discriminant analysis. The authors achieved an accuracy of 85% with the SVM, which outperformed the other classifiers. Bergeron et al102 used a regression SVM to extract the spinal centerline from surface topography, using as ground truth data obtained from biplanar radiographs of 149 scoliotic subjects. The first attempt to predict the curve type, a simplified version of the Lenke classification system distinguishing three types of scoliotic curves,103 was performed by Seoud et al,26 who used an SVM trained on radiographs from 97 adolescent subjects suffering from idiopathic scoliosis, and achieved an overall accuracy of 72.2% with respect to diagnoses based on measurements conducted on planar radiographs. More recently, Komeili et al104 trained a decision tree to classify surface topography data into mild, moderate and severe curves as well as to identify the curve location (thoracic‐thoracolumbar, proximal thoracic, or lumbar), in order to determine the risk of curve progression. The model was able to detect 85.7% of the progression curves and 71.6% of the nonprogression ones.
The analysis of radiographic data of patients suffering from spinal deformities has also been tackled exploiting ML techniques. The challenging automated analysis of the Cobb angle describing the severity of a scoliotic curve has been confronted with various approaches, ranging from non‐ML methods such as the fuzzy Hough transform105 to deep learning techniques. Sun et al106 used a regression SVM to predict the Cobb angle from coronal radiographs, with a very good accuracy (relative root mean squared error of 21.6%) highlighting a potential clinical use. Zhang et al107 trained a deep ANN to predict the vertebral slopes on coronal radiographic images and used the slope data to estimate the Cobb angle, achieving absolute errors lower than 3°. Wu et al108 and Galbusera et al109 exploited the three‐dimensional information contained in biplanar radiographs to perform a more comprehensive assessment of the pathological curvature. Seeing the problem from another perspective, Thong et al110 attempted to use an unsupervised clustering method to obtain a novel classification scheme for adolescent idiopathic scoliosis which effectively describes the variability of the curves among the subjects. Based on 915 biplanar radiographs, the clustering method defined 11 classes differing based on the location of the main curve, in particular of the apical vertebra, as well as kyphosis and lordosis (Figure 10).
Figure 10.
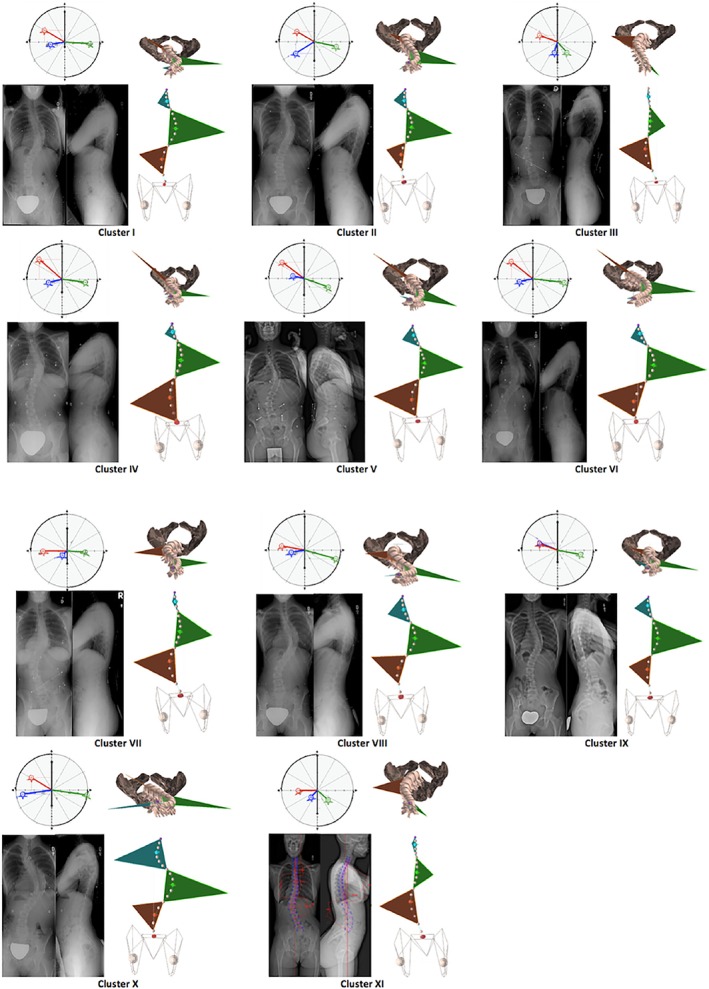
Eleven clusters of spine curves of patients suffering from adolescent idiopathic scoliosis, automatically determined from a large database of biplanar radiographs.110 For each cluster, exemplary radiographs, da Vinci views,111 coronal and top views of the three‐dimensional reconstructions are shown. Reproduced with permission from Thong et al110
Although the definition of computer‐aided detection (CADe) systems is rather general and may cover all the studies which have been summarized in this paragraph, this name is commonly employed in the scientific literature to describe computer programs able to identify and localize relevant features such as lesions and fractures in medical images, with the aim of reducing the risk of missed diagnosis and favoring incidental findings. In the spine field, CADe systems have been used to detect and classify with good success vertebral fractures using either a regression SVM112 or a CNN,113 with accuracies up to 95% for vertebral body compression fractures. CADe systems are also being developed for the detection of spine metastases on CT scans, which has been undertaken by using a classifier trained on a number of features extracted from the image of each single vertebra.114, 115 The developed systems were able to detect both lytic and blastic lesions in real time, with occasional false positives requiring the judgment of a human operator. Burns et al116 developed an alternative approach, in which a watershed segmentation algorithm was used to identify large regions with similar intensities, which were considered as candidate lesions. By means of an SVM classifier processing features extracted from the shape, location, and intensity of the region, the method determined if the candidate region is indeed a tumoral lesion. This method was also rather prone to produce false positives (620 false‐positive detections vs 439 true‐positive lesions), which appear to be an issue requiring further research efforts.
In summary, in light of the tremendous advances which have been observed in recent years, there is no doubt that ML is bringing a revolution to diagnostic imaging, both in general and concerning the study of spine disorders. Although the figure of a human radiologist is not going to be replaced by a computer soon, also taking into account ethics aspects such as the issue of individual responsibility, the potential impact of accurate and reliable automated diagnostic tools is enormous.
5.4. Outcome prediction and clinical decision support
Predictive analytics is a branch of statistics aimed at making predictions about the future based on available data from the past, and has been largely impacted by novel AI technologies and big data sources.117 Healthcare has shown interest in predictive analytics since its early days, due to its large potential in providing improvements to patient care and financial management. Applications of predictive analytics which have been applied to healthcare include the identification of chronic patients at risk of poor health outcome and who may benefit from interventions, the development of personalized medicine and therapies, the prediction of adverse events during the hospital stay, and the optimization of the supply chain.
In the last decade, several studies presented models aimed at predicting various aspects of the outcome of spine surgeries, a selection of those is described below. McGirt et al118 used simple statistics‐derived techniques such as linear and logistic regression to predict values such as the Oswestry Disability Index (ODI)119 1 year after the surgery, the occurrence of complications, readmission to the hospital, and return to work. The prediction model was based on data from 750 to 1200 patients, and scored accuracies between 72% and 84% regarding complications and return to work. The predictors taken into account by the model were more than 40 and included the preoperative ODI, age, ethnicity, body mass index, a detailed description of the symptoms, the possible presence of other spinal disorders as well as various scores describing the health and functional status of the patient. More recently, Kim et al120 used logistic regression and a shallow ANN to specifically predict the occurrence of four types of major complications in patients undergoing spine fusion, namely cardiac complications, wound complications, venous thromboembolism, and mortality, and achieved results largely better than by using the clinical score commonly employed for such applications (Figure 11). A similar approach was used by Lee et al121 who focused on the prediction of surgical site infection. Interestingly, a successive study performed an external validation, that is, based on another sample of patients, of this predictive model, highlighting several limitations and showing a generally poor performance.122 Recently, a large retrospective study123 presented an ensemble of decision trees to predict, with an overall accuracy of 87.6%, major intraoperative or perioperative complications following adult spine deformity surgery. Durand et al investigated a different outcome, the necessity of blood transfusion after adult deformity surgery, which was predicted with good success using single decision trees and a random forest.124
Figure 11.
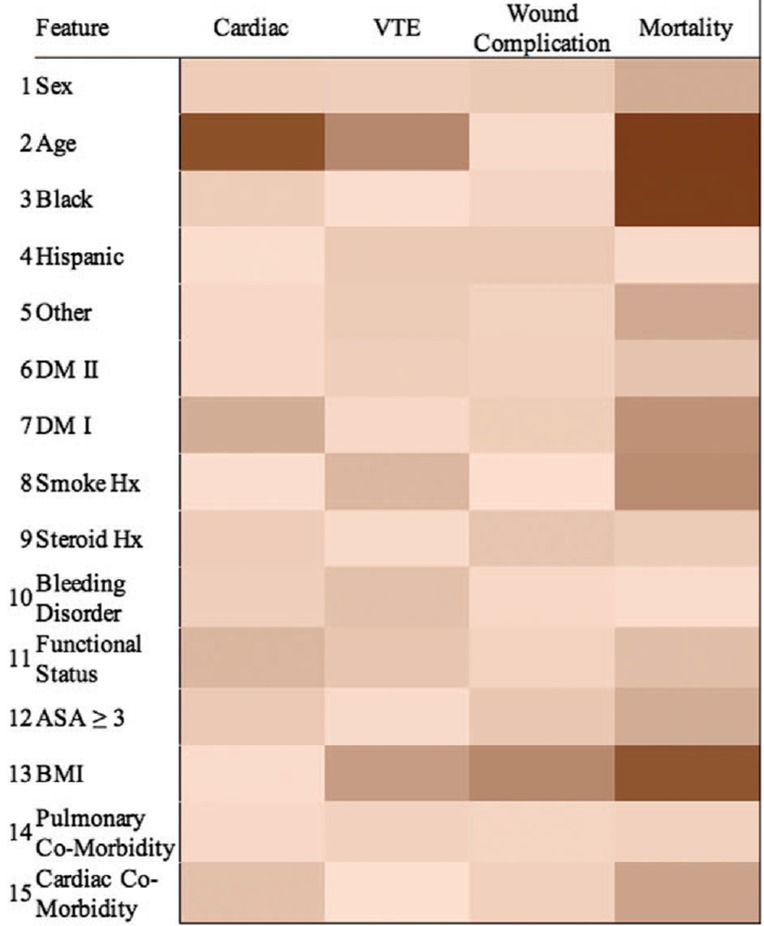
Example of heatmap showing the importance of the various factors (first column) in determining an outcome, namely the risk of complications following posterior lumbar spine fusion, as predicted with machine learning (ML) techniques in a literature study.120 Reproduced with permission from Kim et al120
An application of predictive analytics which is nowadays finding a wide use in the clinical practice is the decision support tool (DST), which exploits the predictive power of the models to support clinical decisions by providing personalized predictions. A recent example of DST in spine care is the Nijmegen Decision Tool for Chronic Low Back Pain,125, 126 which is based on predictors covering various aspects of the patient's health (namely, sociodemographic, pain, somatic, psychological, functioning, and quality of life) to suggest either surgical treatment, conservative care, or no intervention. This DST is still under development, and the technical implementation of the decision has not been finalized yet.
Compared to the other applications of AI and ML in spine research, predictive analytics and clinical decision support currently appear to be at a lower level of development. As a matter of fact, there is no DST based on ML techniques to support the decisions in spine surgery, for example, the length of instrumentation and the choice of the anchoring implants in spine deformity surgery. Imaging data are usually not exploited by predictive models, which are not generally based on state‐of‐the‐art techniques such as deep learning. Indeed, large databases including clinical and imaging data, which would be necessary to train such models, are still lacking, under construction or inaccessible by AI researchers. Nevertheless, the recent proliferation of national and local spine registries, some of these including imaging data, will likely allow for significant advances in the near future also in this field.
5.5. Content‐based image retrieval
The digital imaging databases of large hospitals typically contain several thousands of images for each anatomical district and imaging modality. To facilitate image retrieval for clinical studies or educational purposes, many institutions implement an indexing based on the content of each image, so that the whole imaging database can be easily searched by means of keywords. This indexing process is commonly manually performed, but is a cumbersome, error‐prone and expensive task.127 Automated content‐based image retrieval (CBIR) has become an active area of research in recent years, and is strongly benefiting from the introduction of ML techniques.
Several CBIR frameworks employ the so‐called relevance feedback, which consists in an evaluation of the relevance of each item returned by the query.128 This feedback can be either explicit, that is, the user is asked to grade the relevance of the returned items, or implicit, that is derived automatically from the user behavior, for example, based on which documents are selected by the user for a closer inspection or on the time spent looking at the item. Recent studies introduced ML techniques such as SVMs to implement relevance feedback.129 For the classification of the images, most CBIR systems are based on simple solutions such as SVMs rather than on deep learning architectures.130, 131 Nevertheless, recent studies started to employ deep learning.132, 133
Regarding spine imaging, a few sophisticated algorithms tailored to exploit the features of spine images has been presented. Xu et al134 proposed a novel relevance feedback algorithm for spine radiographs retrieval based on the vertebral contour. The algorithm includes a short‐term memory feature which was able to keep a memory of the human choices between different feedback iterations; the final selection about the relevance of each image is then performed by a decision tree. The same research group presented a CBIR system which also took into account the shape of the intervertebral space.135
5.6. Biomechanics
So far, AI and ML impacted basic biomechanics to a lower extent with respect to applied clinical and radiological research. Nevertheless, in recent years, a few papers describing applications of ANNs for typical biomechanical problems such as the estimation of loads and stresses started to appear. Although studies specifically addressing spine biomechanics are currently not available, we believe that it is worthy to briefly mention here some ML‐based studies investigating other musculoskeletal districts, since the analysis of the state‐of‐the‐art may help in delineating the possible future fields of applications of ML techniques in spine biomechanics.
ML has been used to estimate the material properties of biological tissues. Chande et al136 employed shallow ANNs to estimate the relationship between the stiffness of the ligaments and the kinematics of the foot in patients suffering from adult acquired flatfoot deformity. In order to create the training data, the authors constructed and employed patient‐specific computer models of the foot anatomy. Zadpoor et al137 investigated a related problem, that is, the prediction of the mechanical loads that determine certain mechanical properties of a biological tissue subject to remodeling, namely trabecular bone. The authors employed an existing biomechanical computational model able to predict bone tissue adaptation under mechanical loading based on the local strains, and used it to run a series of simulations in which random loads were applied to a small bone trabecular sample. The outputs of the simulations, that is, the remodeled local bone densities, were used to train the ANN to predict the loads which induced that form of remodeling.
Another field of application of ML is the calculation of stresses in patient‐specific analysis, thus eliminating the need for computationally expensive finite element models. For example, Lu et al138 developed a shallow ANN able to predict the stress in the cartilage of the tibial plateau and femoral condyles of the knee joint. A finite element model of the knee was used to generate a dataset then used for training the ANN, which was able to predict the stress in each element of the articular cartilage with a dramatic reduction in time and cost with respect to creating and solving the finite element model itself.
In general, the use of ML techniques in musculoskeletal biomechanics appears to be still in its infancy; the few published papers did not exploit yet the potential of the latest innovations such as deep learning. Nevertheless, the available papers clearly demonstrate the potential of ML in this field. Computational models that are able to predict the biomechanical response of bones, joints as well as the spine are widely available and could be used for generating large datasets to be used as training data for ML models, as suggested previously.138 This approach would facilitate a more widespread adoption of patient‐specific modeling in bench‐to‐bedside applications where the computational resources and time required for the construction and solution of a traditional biomechanical model may conflict with the clinical demands.
5.7. Motion and gait analysis
The quantitative analysis of human motion, and especially gait, with cameras, optoelectronic systems, wearable inertial devices, electromyography systems, force plates, and pressure sensors is widely employed for the scientific and clinical investigations of several pathologies. Indeed, the study of gait pattern alterations in patients suffering from spinal disorders is a very active area of research.139, 140 Traditional gait analysis aims at the measurement of spatiotemporal parameters such as walking velocity, stride and step lengths, cadence, and duration of the stance and swing phases; kinematic parameters such as the angles of rotation of the various joints; kinetic parameters such as forces and moments in the joints, which typically involve the use of force platforms. The value of these parameters are then compared to reference ranges and used for diagnostic purposes, or to monitor patient recovery. In addition to the study of gait, specific motion analysis protocols have been developed for the investigation of spine motion during common activities such as standing, chair rise sitting, stair climbing, and flexing the trunk.141
In the last two decades, this consolidated approach has been revisited while ML techniques have been gaining a wide use in several research fields.142 Recent papers employed ML techniques such as SVMs143, 144, 145 and ANNs146 for the classification of abnormal gait patterns with good success. However, only a few studies involving ML techniques to investigate spinal disorders have been presented so far; this lack of documentation reflects the technical difficulties in assessing position and motion of the vertebrae due to soft tissue artifacts.147 An example of a pioneering study in this field is offered by Hayashi et al,148 who trained an SVM to distinguish gait patterns associated to either L4 or L5 radiculopathy in patients suffering from lumbar canal stenosis, achieving an accuracy of 80.4%.
ML has also been successfully employed to investigate spine disorders by means of electromyography systems.149 The authors built an SVM to identify patients responding to a functional restoration rehabilitation program for chronic low back pain, based on dynamic surface electromyography readings, with an accuracy of 96% on a sample of 30 patients.
A radically different research field related to gait and ML concerns humanoid or animal‐shaped agents, that is, computer models, learning how to walk and move in a simulated environment, which may be geometrically complex and including obstacles. The process of learning to walk consists in appropriately activating the actuators, which act as the muscles in a human subject, while keeping equilibrium and achieving the locomotion goal, and has been shown to be very challenging to be replicated in a ML framework. Indeed, the implementation of such models requires sophisticated reinforcement algorithms, which typically provide rewards when the model is able to accomplish its goal, that is, reaching the target location, and punishments when the agent fails, for example, if it falls on the ground. A good example of the state of the art is offered by Heess et al150 (https://www.youtube.com/watch?v=hx_bgoTF7bs).
6. ETHICS ISSUES AND REGULATION
The implementation of AI technologies in healthcare, especially regarding tools with a direct clinical impact such as those aimed at supporting diagnosis or clinical decisions, is undoubtedly determining a paradigm shift. Such a change of perspective involves the emergence of several major ethics issues, which are being heatedly discussed both in the scientific community and by regulatory agencies.
Most AI technologies, notably including deep learning networks which now are having a major role, appear as a black box to an external user.151 Although methods to visualize the inner structure and behavior of the AI tools have been presented (eg, 152) and more human‐readable technologies such as decision trees are also being used, AI predictions appears largely to be determined by an obscure logic which cannot be understood or interpreted by a human observer.153 This limitation directly leads to the issue of the accountability of the decisions, which is nowadays being debated at a regulatory level. In other words, if a prediction fails, for example, in case of misdiagnosis, determining if the responsibility is of the radiologists who used the AI system, of the device itself or of the manufacturer is of critical importance. This obscure nature has also severe implications regarding the marketing approval of novel AI tools, which require deeper testing and verification with respect to other technologies, and thus longer time‐to‐market and cost.
A second issue concerns possible biases in the predictions, which may be either intentional, that is, fraudulent, or unintended. Examples of intentional biases are a DST preferably promoting the use of drugs or devices by a specific manufacturer, or a tool designed to maximize a specific quality metrics relevant for the hospital but not necessarily optimizing patients' care.153 Unintended biases may be related to scarce availability of data regarding some rare pathologies or phenotypes, which may be then insufficiently covered in the training dataset with respect to more common conditions, or ethnicities for which datasets are indeed not existing or limited.151 Besides, insufficient data collection efforts, for example, by privileging data sources easier to access, may also lead to unintended biases. To limit the impact of such issues, efforts toward a governance of AI are starting to be undertaken, with the final aim of building a robust public trust.154 It should be noted that cultural differences between the European Union, the United States, and East Asian countries may likely result in dramatically different attitudes from a regulatory and governance point of view.155
The use of AI in healthcare also raises serious concerns about data privacy and security, due to the massive amount of clinical and imaging data required for training and validation of the tools, thus involving issues about data collection, transmission and storage, as well as informed consent. Data anonymization is being commonly used to enhance privacy and security; nevertheless, patients retain rights on their anonymized data, which are subjected to strict regulations about storage, transmission and use, especially when data are used in a for‐profit environment. The recent introduction of the General Data Protection Regulation in the European Union considerably expanded the rights of the patients by adopting an explicit opt‐in policy regarding the permission for data processing; on the other side, it further enlarges the policy differences with the less strict United States, thus possibly strengthening the leading role of this country in AI innovation.155 Due to the large amounts of investments related to AI technologies and their potential economic consequences, policy makers and regulatory agencies need to take into account these aspects as well.
Following in the footsteps of the free software movement, providing open access to ML models and training data would be a possible way to foster public trust, as well as to improve accountability and prediction bias by giving the scientific community the possibility of further testing and developing these technologies. As a matter of facts, source code for most of the recent AI and ML algorithms is publicly available, released by public research institutions as well as companies such as Google (Mountain View, California) and Nvidia (Santa Clara, California). However, due to business and regulatory reasons, the public release of detailed technical information about production‐ready ML software intended to be used for clinical applications is highly unlikely to happen.
7. CONCLUSIONS
AI and ML are emerging disruptive technologies which have nowadays reached a substantial level of development, enabling them to have already a practical impact on several research fields. Computer vision and image processing are especially gaining momentum, due to the latest innovations in deep learning and improved accessibility of computational resources, such as powerful GPUs. Indeed, most recent spine research studies using AI and ML techniques are related to medical imaging, but an increasing impact on other fields such as spine biomechanics should be expected in the near future. Ethics aspects related to accountability, data privacy and security as well as the risk of biased predictions are relevant and are currently under the attention of policy makers and regulatory agencies.
CONFLICTS OF INTEREST
The authors declare that there is no conflict of interest regarding the publication of this article.
Author contributions
F.G.: literature analysis, manuscript preparation and revision; G.C.: literature analysis, manuscript revision; T.B.: literature analysis, manuscript preparation and revision.
Galbusera F, Casaroli G, Bassani T. Artificial intelligence and machine learning in spine research. JOR Spine. 2019;2:e1044. 10.1002/jsp2.1044
REFERENCES
- 1. Samuel AL. Some studies in machine learning using the game of checkers. IBM J Res Dev. 1959;3:210‐229. [Google Scholar]
- 2. Nilsson NJ. The Quest for Artificial Intelligence. Cambridge, UK: Cambridge University Press; 2009. [Google Scholar]
- 3. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; Piscataway, NJ: Institute of Electrical and Electronics Engineers (IEEE); 2016. ArXiv Preprint arXiv:1512.03385. [Google Scholar]
- 4. Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJ. Artificial intelligence in radiology. Nat Rev Cancer. 2018;18:510‐518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Slate DJ, Atkin LR. Chess 4.5—the Northwestern University chess program In: Frey PW, ed. Chess Skill in Man and Machine. New York, NY: Springer; 1983. [Google Scholar]
- 6. Weizenbaum J. ELIZA—a computer program for the study of natural language communication between man and machine. Commun ACM. 1966;9:36‐45. [Google Scholar]
- 7. McCulloch WS, Pitts W. A logical calculus of the ideas immanent in nervous activity. Bull Math Biophys. 1943;5:115‐133. [PubMed] [Google Scholar]
- 8. Minsky M. Neural Nets and the Brain‐Model Problem [doctoral dissertation]. Princeton University, NJ; 1954.
- 9. Rosenblatt F. The perceptron: a probabilistic model for information storage and organization in the brain. Psychol Rev. 1958;65:386‐408. [DOI] [PubMed] [Google Scholar]
- 10. Minsky M, Papert SA. Perceptrons: An Introduction to Computational Geometry. Cambridge, UK: MIT Press; 1969. [Google Scholar]
- 11. Rumelhart DE, Hinton GE, Williams RJ. Learning representations by back‐propagating errors. Nature. 1986;323:533‐536. [Google Scholar]
- 12. Schmidhuber J. Deep learning in neural networks: An overview. Neural Netw. 2015;61:85‐117. [DOI] [PubMed] [Google Scholar]
- 13. Michalski RS, Carbonell JG, Mitchell TM. Machine Learning: An Artificial Intelligence Approach. Berlin, Germany: Springer Science & Business Media; 2013. [Google Scholar]
- 14. Shalev‐Shwartz S, Ben‐David S. Understanding Machine Learning: From Theory to Algorithms. Cambridge, UK: Cambridge University Press; 2014. [Google Scholar]
- 15. Pfirrmann CW, Metzdorf A, Zanetti M, Hodler J, Boos N. Magnetic resonance classification of lumbar intervertebral disc degeneration. Spine. 2001;26:1873‐1878. [DOI] [PubMed] [Google Scholar]
- 16. Toyoda H, Takahashi S, Hoshino M, et al. Characterizing the course of back pain after osteoporotic vertebral fracture: a hierarchical cluster analysis of a prospective cohort study. Arch Osteoporosis. 2017;12:82. [DOI] [PubMed] [Google Scholar]
- 17. Domingos P. A few useful things to know about machine learning. Commun ACM. 2012;55:78‐87. [Google Scholar]
- 18. Lowe DG. Object recognition from local scale‐invariant features Computer Vision, 1999, the Proceedings of the Seventh IEEE International Conference. Piscataway, NJ: Institute of Electrical and Electronics Engineers (IEEE); 1999. 10.1109/ICCV.1999.790410. [DOI] [Google Scholar]
- 19. Rublee E, Rabaud V, Konolige K, Bradski G. ORB: an efficient alternative to SIFT or SURF Computer Vision (ICCV), 2011 IEEE International Conference. Piscataway, NJ: Institute of Electrical and Electronics Engineers (IEEE); 2011. 10.1109/ICCV.2011.6126544. [DOI] [Google Scholar]
- 20. Russell SJ, Norvig P. Artificial Intelligence: A Modern Approach. London, UK: Pearson Education Limited; 2016. [Google Scholar]
- 21. Frighetto‐Pereira L, Rangayyan RM, Metzner GA, de Azevedo‐Marques PM, Nogueira‐Barbosa MH. Shape, texture and statistical features for classification of benign and malignant vertebral compression fractures in magnetic resonance images. Comput Biol Med. 2016;73:147‐156. [DOI] [PubMed] [Google Scholar]
- 22. Unal Y, Polat K, Kocer HE. Pairwise FCM based feature weighting for improved classification of vertebral column disorders. Comput Biol Med. 2014;46:61‐70. [DOI] [PubMed] [Google Scholar]
- 23. Cortes C, Vapnik V. Support‐vector networks. Mach Learn. 1995;20:273‐297. [Google Scholar]
- 24. Vapnik V. Pattern recognition using generalized portrait method. Automat Rem Control. 1963;24:774‐780. [Google Scholar]
- 25. Oktay AB, Akgul YS. Simultaneous localization of lumbar vertebrae and intervertebral discs with SVM‐based MRF. IEEE Trans Biomed Eng. 2013;60:2375‐2383. [DOI] [PubMed] [Google Scholar]
- 26. Seoud L, Adankon MM, Labelle H, Dansereau J, Cheriet F. Prediction of scoliosis curve type based on the analysis of trunk surface topography Biomedical Imaging: From Nano to Macro, 2010 IEEE International Symposium. Piscataway, NJ: Institute of Electrical and Electronics Engineers (IEEE); 2010. 10.1109/ISBI.2010.5490322. [DOI] [Google Scholar]
- 27. Belson WA. Matching and prediction on the principle of biological classification. Applied Statistics. 1959;8(2):65‐75. [Google Scholar]
- 28. Morgan JN, Sonquist JA. Problems in the analysis of survey data, and a proposal. J Am Stat Assoc. 1963;58:415‐434. [Google Scholar]
- 29. Breiman L. Classification and Regression Trees. London, UK: Routlegde; 2017. [Google Scholar]
- 30. Quinlan JR. C4. 5: Programs for Machine Learning. Burlington: Morgan Kaufmann; 2014. [Google Scholar]
- 31. Quinlan JR. Induction of decision trees. Mach Learn. 1986;1:81‐106. [Google Scholar]
- 32. Breiman L. Random forests. Mach Learn. 2001;45:5‐32. [Google Scholar]
- 33. Nijeweme‐d'Hollosy WO, van Velsen LS, Soer R, Hermens HJ. Design of a web‐based clinical decision support system for guiding patients with low back pain to the best next step in primary healthcare Proceedings of the 9th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2016). Cham: Springer International Publishing; 2016. [Google Scholar]
- 34. Oh T, Scheer JK, Smith JS, et al. Potential of predictive computer models for preoperative patient selection to enhance overall quality‐adjusted life years gained at 2‐year follow‐up: a simulation in 234 patients with adult spinal deformity. Neurosurg Focus. 2017;43:E2. [DOI] [PubMed] [Google Scholar]
- 35. Varghese V, Kumar GS, Krishnan V. Effect of various factors on pull out strength of pedicle screw in normal and osteoporotic cancellous bone models. Med Eng Phys. 2017;40:28‐38. [DOI] [PubMed] [Google Scholar]
- 36. Varghese V, Krishnan V, Kumar GS. Evaluating pedicle‐screw instrumentation using decision‐tree analysis based on pulloutaStrength. Asian Spine J. 2018;12(4):611‐621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yagi M, Akilah KB, Boachie‐Adjei O. Incidence, risk factors and classification of proximal junctional kyphosis: surgical outcomes review of adult idiopathic scoliosis. Spine. 2011;36:E60‐E68. [DOI] [PubMed] [Google Scholar]
- 38. Hebb DO. The First Stage of Perception: Growth of the Assembly. London: Psychology Press; 2005:102‐120. [Google Scholar]
- 39. Bottou L, Bousquet O. The tradeoffs of large scale learning Advances in Neural Information Processing Systems 20; Cambridge, MA: MIT Press; 2008. https://papers.nips.cc/paper/3323-the-tradeoffs-of-large-scale-learning. [Google Scholar]
- 40. Kingma DP, Ba J. Adam: a method for stochastic optimization. ArXiv Preprint arXiv. 2014;1412:6980. [Google Scholar]
- 41. Goodfellow I, Pouget‐Abadie J, Mirza M, et al. Generative adversarial nets Advances in Neural Information Processing Systems; Cambridge, MA: MIT Press; 2014. [Google Scholar]
- 42. Lipton ZC, Berkowitz J, Elkan C. A critical review of recurrent neural networks for sequence learning. ArXiv Preprint arXiv. 2015;1506:00019. [Google Scholar]
- 43. Hubel DH, Wiesel TN. Receptive fields and functional architecture of monkey striate cortex. J Physiol. 1968;195:215‐243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hubel DH, Wiesel TN. Receptive fields, binocular interaction and functional architecture in the cat's visual cortex. J Physiol. 1962;160:106‐154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Denker JS, Gardner W, Graf HP, et al. Neural network recognizer for hand‐written zip code digits Advances in Neural Information Processing Systems; Cambridge, MA: MIT Press; 1989. https://papers.nips.cc/paper/107-neural-network-recognizer-for-hand-written-zip-code-digits. [Google Scholar]
- 46. Fukushima K, Miyake S. Neocognitron: A Self‐Organizing Neural Network Model for a Mechanism of Visual Pattern Recognit. 1982;15(6):455‐469. [Google Scholar]
- 47. Scherer D, Müller A, Behnke S. Evaluation of pooling operations in convolutional architectures for object recognition In: Diamantaras K, Duch W, Iliadis LS, eds. Artificial Neural Networks—ICANN 2010. ICANN 2010. Lecture Notes in Computer Science. Vol 6354 Berlin, Heidelberg: Springer; 2010. [Google Scholar]
- 48. LeCun Y, Boser B, Denker JS, et al. Backpropagation applied to handwritten zip code recognition. Neural Comput. 1989;1:541‐551. [Google Scholar]
- 49. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436‐444. [DOI] [PubMed] [Google Scholar]
- 50. LeCun Y, Boser BE, Denker JS, et al. Handwritten digit recognition with a back‐propagation network Advances in Neural Information Processing Systems. Cambridge, MA: MIT Press; 1990. https://papers.nips.cc/paper/293‐handwritten‐digit‐recognition‐with‐a‐back‐propagation‐network. [Google Scholar]
- 51. LeCun Y, Bottou L, Bengio Y, Haffner P. Gradient‐based learning applied to document recognition. Proc IEEE. 1998;86:2278‐2324. [Google Scholar]
- 52. Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks NIPS'12 Proceedings of the 25th International Conference on Neural Information Processing Systems—Volume 1. Cambridge, MA: MIT Press; 2012:1097‐1105. [Google Scholar]
- 53. Simonyan K, Zisserman A. Very deep convolutional networks for large‐scale image recognition. ArXiv Preprint arXiv. 2014;1409:1556. [Google Scholar]
- 54. Szegedy C, Liu W, Jia Y, et al. Going deeper with convolutions. ArXiv Preprint arXiv. 2015;1409:4842. [Google Scholar]
- 55. Ripley BD. Pattern Recognition and Neural Networks. Cambridge, UK: Cambridge University Press; 1996. [Google Scholar]
- 56. Krig S. Ground truth data, content, metrics, and analysis Computer Vision Metrics. Berkeley, CA: Apress; 2014. [Google Scholar]
- 57. Chwialkowski MP, Shile PE, Pfeifer D, Parkey RW, Peshock RM. Automated localization and identification of lower spinal anatomy in magnetic resonance images. Comput Biomed Res. 1991;24:99‐117. [DOI] [PubMed] [Google Scholar]
- 58. Peng Z, Zhong J, Wee W, Lee J. Automated vertebra detection and segmentation from the whole spine MR images Conference Proceedings IEEE Engineering in Medicine and Biology Society. Vol 3 Piscataway, NJ: Institute of Electrical and Electronics Engineers (IEEE); 2006:2527‐2530. [DOI] [PubMed] [Google Scholar]
- 59. Schmidt S, Kappes J, Bergtholdt M, et al. Spine detection and labeling using a parts‐based graphical model. Inf Process Med Imaging. 2007;20:122‐133. [DOI] [PubMed] [Google Scholar]
- 60. Oktay AB, Akgul YS. Localization of the lumbar discs using machine learning and exact probabilistic inference In: Fichtinger G, Martel A, Peters T, eds. Medical Image Computing and Computer‐Assisted Intervention – MICCAI 2011. MICCAI 2011. Lecture Notes in Computer Science. Vol 6893 Berlin, Heidelberg: Springer; 2011. [DOI] [PubMed] [Google Scholar]
- 61. Glocker B, Feulner J, Criminisi A, Haynor DR, Konukoglu E. Automatic localization and identification of vertebrae in arbitrary field‐of‐view CT scans In: Ayache N, Delingette H, Golland P, Mori K, eds. Medical Image Computing and Computer‐Assisted Intervention – MICCAI 2012. MICCAI 2012. Lecture Notes in Computer Science. Vol 7512 Berlin, Heidelberg: Springer; 2012. [DOI] [PubMed] [Google Scholar]
- 62. Glocker B, Zikic D, Konukoglu E, Haynor DR, Criminisi A. Vertebrae localization in pathological spine CT via dense classification from sparse annotations In: Mori K, Sakuma I, Sato Y, Barillot C, Navab N, eds. Medical Image Computing and Computer‐Assisted Intervention – MICCAI 2013. MICCAI 2013. Lecture Notes in Computer Science. Vol 8150 Berlin, Heidelberg: Springer; 2013. [DOI] [PubMed] [Google Scholar]
- 63. Chen C, Belavy D, Yu W, et al. Localization and segmentation of 3D intervertebral discs in MR images by data driven estimation. IEEE Trans Med Imaging. 2015a;34:1719‐1729. [DOI] [PubMed] [Google Scholar]
- 64. Chen H, Shen C, Qin J, et al. Automatic localization and identification of vertebrae in spine CT via a joint learning model with deep neural networks In: Navab N, Hornegger J, Wells W, Frangi A, eds. Medical Image Computing and Computer‐Assisted Intervention—MICCAI 2015. MICCAI 2015. Lecture Notes in Computer Science. Vol 9349 Cham: Springer; 2015b. [Google Scholar]
- 65. Chen H, Dou Q, Wang X, Qin J, Cheng JC, Heng P. 3D fully convolutional networks for intervertebral disc localization and segmentation International Conference on Medical Imaging and Virtual Reality. Cham: Springer; 2016. 10.1007/978-3-319-43775-0_34. [DOI] [Google Scholar]
- 66. Suzani A, Seitel A, Liu Y, Fels S, Rohling RN, Abolmaesumi P. Fast automatic vertebrae detection and localization in pathological ct scans‐a deep learning approach In: Navab N, Hornegger J, Wells W, Frangi A, eds. Medical Image Computing and Computer‐Assisted Intervention – MICCAI 2015. MICCAI 2015. Lecture Notes in Computer Science. Vol 9351 Cham: Springer; 2015. [Google Scholar]
- 67. Payer C, Štern D, Bischof H, Urschler M. Regressing heatmaps for multiple landmark localization using CNNs In: Ourselin S, Joskowicz L, Sabuncu M, Unal G, Wells W, eds. Medical Image Computing and Computer‐Assisted Intervention – MICCAI 2016. MICCAI 2016. Lecture Notes in Computer Science. Vol 9901 Cham: Springer; 2016. [Google Scholar]
- 68. Forsberg D, Sjöblom E, Sunshine JL. Detection and labeling of vertebrae in MR images using deep learning with clinical annotations as training data. J Digit Imaging. 2017;30:406‐412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lootus M, Kadir T, Zisserman A. Vertebrae Detection and Labelling in Lumbar MR Images. Cham: Springer; 2014:219‐230. [Google Scholar]
- 70. Yang D, Xiong T, Xu D, et al. Deep image‐to‐image recurrent network with shape basis learning for automatic vertebra labeling in large‐scale 3D CT volumes In: Descoteaux M, Maier‐Hein L, Franz A, Jannin P, Collins D, Duchesne S, eds. Medical Image Computing and Computer‐Assisted Intervention − MICCAI 2017. MICCAI 2017. Lecture Notes in Computer Science. Vol 10435 Cham: Springer; 2017. [Google Scholar]
- 71. Thoma M. A survey of semantic segmentation. ArXiv Preprint arXiv. 2016;1602:06541. [Google Scholar]
- 72. Romera‐Paredes B, Torr PHS. Recurrent instance segmentation. ArXiv Preprint arXiv. 2016;1511:08250. [Google Scholar]
- 73. Law MW, Tay K, Leung A, Garvin GJ, Li S. Intervertebral disc segmentation in MR images using anisotropic oriented flux. Med Image Anal. 2013;17:43‐61. [DOI] [PubMed] [Google Scholar]
- 74. Michopoulou SK, Costaridou L, Panagiotopoulos E, Speller R, Panayiotakis G, Todd‐Pokropek A. Atlas‐based segmentation of degenerated lumbar intervertebral discs from MR images of the spine. IEEE Trans Biomed Eng. 2009;56:2225‐2231. [DOI] [PubMed] [Google Scholar]
- 75. Neubert A, Fripp J, Engstrom C, et al. Automated detection, 3D segmentation and analysis of high resolution spine MR images using statistical shape models. Phys Med Biol. 2012;57:8357‐8376. [DOI] [PubMed] [Google Scholar]
- 76. Klinder T, Ostermann J, Ehm M, Franz A, Kneser R, Lorenz C. Automated model‐based vertebra detection, identification, and segmentation in CT images. Med Image Anal. 2009;13:471‐482. [DOI] [PubMed] [Google Scholar]
- 77. Korez R, Ibragimov B, Likar B, Pernuš F, Vrtovec T. Deformable model‐based segmentation of intervertebral discs from MR spine images by using the SSC descriptor In: Vrtovec T. et al., eds. Computational Methods and Clinical Applications for Spine Imaging. CSI 2015. Lecture Notes in Computer Science. Vol 9402 Cham: Springer; 2016. [Google Scholar]
- 78. Korez R, Ibragimov B, Likar B, Pernuš F, Vrtovec T. Interpolation‐based shape‐constrained deformable model approach for segmentation of vertebrae from CT spine images In: Yao J, Glocker B, Klinder T, Li S, eds. Recent Advances in Computational Methods and Clinical Applications for Spine Imaging. Lecture Notes in Computational Vision and Biomechanics. Vol 20 Cham: Springer; 2015. [Google Scholar]
- 79. Ayed IB, Punithakumar K, Garvin G, Romano W, Li S. Graph cuts with invariant object‐interaction priors: application to intervertebral disc segmentation. Inf Process Med Imaging. 2011;22:221‐232. [DOI] [PubMed] [Google Scholar]
- 80. Carballido‐Gamio J, Belongie SJ, Majumdar S. Normalized cuts in 3‐D for spinal MRI segmentation. IEEE Trans Med Imaging. 2004;23:36‐44. [DOI] [PubMed] [Google Scholar]
- 81. Egger J, Kapur T, Dukatz T, et al. Square‐cut: a segmentation algorithm on the basis of a rectangle shape. PloS One. 2012;7:e31064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Huang S, Chu Y, Lai S, Novak CL. Learning‐based vertebra detection and iterative normalized‐cut segmentation for spinal MRI. IEEE Trans Med Imaging. 2009;28:1595‐1605. [DOI] [PubMed] [Google Scholar]
- 83. Schwarzenberg R, Freisleben B, Nimsky C, Egger J. Cube‐cut: vertebral body segmentation in MRI‐data through cubic‐shaped divergences. PloS One. 2014;9:e93389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kelm BM, Wels M, Zhou SK, et al. Spine detection in CT and MR using iterated marginal space learning. Med Image Anal. 2013;17:1283‐1292. [DOI] [PubMed] [Google Scholar]
- 85. Zheng Y, Barbu A, Georgescu B, Scheuering M, Comaniciu D. Four‐chamber heart modeling and automatic segmentation for 3‐D cardiac CT volumes using marginal space learning and steerable features. IEEE Trans Med Imaging. 2008;27:1668‐1681. [DOI] [PubMed] [Google Scholar]
- 86. Lessmann N, van Ginneken B, Išgum I. Iterative convolutional neural networks for automatic vertebra identification and segmentation in CT images. ArXiv Preprint arXiv. 2018;1804:04383. [DOI] [PubMed] [Google Scholar]
- 87. Yao J, Burns JE, Forsberg D, et al. A multi‐center milestone study of clinical vertebral CT segmentation. Comput Med Imaging Graph. 2016;49:16‐28. 10.1016/j.compmedimag.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zheng G, Chu C, Belavý DL, et al. Evaluation and comparison of 3D intervertebral disc localization and segmentation methods for 3D T2 MR data: A grand challenge. Med Image Anal. 2017;35:327‐344. [DOI] [PubMed] [Google Scholar]
- 89. Bounds DG, Lloyd PJ, Mathew B, Waddell G. A multilayer perceptron network for the diagnosis of low back pain Proceedings of IEEE International Conference on Neural Networks, San Diego. Piscataway, NJ: Institute of Electrical and Electronics Engineers (IEEE); 1988. 10.1109/ICNN.1988.23963. [DOI] [Google Scholar]
- 90. Tsai M, Jou S, Hsieh M. A new method for lumbar herniated inter‐vertebral disc diagnosis based on image analysis of transverse sections. Comput Med Imaging Graph. 2002;26:369‐380. [DOI] [PubMed] [Google Scholar]
- 91. Koompairojn S, Hua KA, Bhadrakom C. Automatic classification system for lumbar spine X‐ray images Computer‐Based Medical Systems, 2006. CBMS 2006. 19th IEEE International Symposium. Piscataway, NJ: Institute of Electrical and Electronics Engineers (IEEE); 2006. 10.1109/CBMS.2006.54. [DOI] [Google Scholar]
- 92. Cherukuri M, Stanley RJ, Long R, Antani S, Thoma G. Anterior osteophyte discrimination in lumbar vertebrae using size‐invariant features. Comput Med Imaging Graph. 2004;28:99‐108. [DOI] [PubMed] [Google Scholar]
- 93. Raja'S A, Corso JJ, Chaudhary V, Dhillon G. Desiccation diagnosis in lumbar discs from clinical MRI with a probabilistic model Biomedical Imaging: From Nano to Macro, 2009. ISBI'09. IEEE International Symposium. Piscataway, NJ: Institute of Electrical and Electronics Engineers (IEEE); 2009. 10.1109/ISBI.2009.5193105. [DOI] [Google Scholar]
- 94. Ghosh S, Raja'S A, Chaudhary V, Dhillon G. Computer‐aided diagnosis for lumbar MRI using heterogeneous classifiers Biomedical Imaging: From Nano to Macro, 2011 IEEE International Symposium. Piscataway, NJ: Institute of Electrical and Electronics Engineers (IEEE); 2011. 10.1109/ISBI.2011.5872612. [DOI] [Google Scholar]
- 95. Hao S, Jiang J, Guo Y, Li H. Active learning based intervertebral disk classification combining shape and texture similarities. Neurocomputing. 2013;101:252‐257. [Google Scholar]
- 96. Oktay AB, Albayrak NB, Akgul YS. Computer aided diagnosis of degenerative intervertebral disc diseases from lumbar MR images. Comput Med Imaging Graph. 2014;38:613‐619. [DOI] [PubMed] [Google Scholar]
- 97. Ruiz‐España S, Arana E, Moratal D. Semiautomatic computer‐aided classification of degenerative lumbar spine disease in magnetic resonance imaging. Comput Biol Med. 2015;62:196‐205. [DOI] [PubMed] [Google Scholar]
- 98. Castro‐Mateos I, Pozo JM, Lazary A, Frangi AF. 2D segmentation of intervertebral discs and its degree of degeneration from T2‐weighted magnetic resonance images Medical Imaging 2014: Computer‐Aided Diagnosis. Bellingham, WA: The International Society for Optics and Photonics (SPIE); 2014. 10.1117/12.2043755. [DOI] [Google Scholar]
- 99. Jamaludin A, Lootus M, Kadir T, et al. ISSLS PRIZE IN BIOENGINEERING SCIENCE 2017: automation of reading of radiological features from magnetic resonance images (MRIs) of the lumbar spine without human intervention is comparable with an expert radiologist. Eur Spine J. 2017;26:1374‐1383. [DOI] [PubMed] [Google Scholar]
- 100. Niemeyer F, Galbusera F, Kienle A, Wilke H. A Deep Learning System for Consistent Automatic Disc Degeneration Grading. Dublin: World Congress of Biomechanics; 2018. [Google Scholar]
- 101. Ramirez L, Durdle NG, Raso VJ, Hill DL. A support vector machines classifier to assess the severity of idiopathic scoliosis from surface topography. IEEE Trans Inf Technol Biomed. 2006;10:84‐91. [DOI] [PubMed] [Google Scholar]
- 102. Bergeron C, Cheriet F, Ronsky J, Zernicke R, Labelle H. Prediction of anterior scoliotic spinal curve from trunk surface using support vector regression. Eng Appl Artif Intel. 2005;18:973‐983. [Google Scholar]
- 103. Lenke LG, Edwards CC, Bridwell KH. The Lenke classification of adolescent idiopathic scoliosis: how it organizes curve patterns as a template to perform selective fusions of the spine. Spine. 2003;28:S199‐S207. [DOI] [PubMed] [Google Scholar]
- 104. Komeili A, Westover L, Parent EC, El‐Rich M, Adeeb S. Monitoring for idiopathic scoliosis curve progression using surface topography asymmetry analysis of the torso in adolescents. Spine J. 2015;15:743‐751. [DOI] [PubMed] [Google Scholar]
- 105. Zhang J, Lou E, Le LH, Hill DL, Raso JV, Wang Y. Automatic Cobb measurement of scoliosis based on fuzzy Hough transform with vertebral shape prior. J Digit Imaging. 2009;22:463‐472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Sun H, Zhen X, Bailey C, Rasoulinejad P, Yin Y, Li S. Direct estimation of spinal Cobb angles by structured multi‐output regression In: Niethammer M. et al., eds. Information Processing in Medical Imaging. IPMI 2017. Lecture Notes in Computer Science. Vol 10265 Cham: Springer; 2017. [Google Scholar]
- 107. Zhang J, Li H, Lv L, Zhang Y. Computer‐aided cobb measurement based on automatic detection of vertebral slopes using deep neural network. Int J Biomed Imaging. 2017;2017:1‐6. 10.1155/2017/9083916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Wu H, Bailey C, Rasoulinejad P, Li S. Automated comprehensive Adolescent Idiopathic Scoliosis assessment using MVC‐Net. Med Image Anal. 2018;48:1‐11. [DOI] [PubMed] [Google Scholar]
- 109. Galbusera F, Niemeyer F, Wilke HJ, et al. Fully automated radiological analysis of spinal disorders and deformities: a deep learning approach. Eur Spine J. under review. [DOI] [PubMed] [Google Scholar]
- 110. Thong WE, Parent S, Wu J, Aubin CE, Labelle H, Kadoury S. Three‐dimensional morphology study of surgical adolescent idiopathic scoliosis patient from encoded geometric models. Eur Spine J. 2016;15:3104‐3113. [DOI] [PubMed] [Google Scholar]
- 111. Labelle H, Aubin CE, Jackson R, Lenke L, Newton P, Parent S. Seeing the spine in 3D: how will it change what we do? J Pediatr Orthop. 2011;31(1 Suppl):S37‐S45. [DOI] [PubMed] [Google Scholar]
- 112. Burns JE, Yao J, Summers RM. Vertebral Body Compression Fractures and Bone Density: Automated Detection and Classification on CT Images. Radiology. 2017;284:788‐797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Roth HR, Wang Y, Yao J, Lu L, Burns JE, Summers RM. Deep convolutional networks for automated detection of posterior‐element fractures on spine CT. ArXiv Preprint arXiv. 2016;1602:0020. [Google Scholar]
- 114. Hammon M, Dankerl P, Tsymbal A, et al. Automatic detection of lytic and blastic thoracolumbar spine metastases on computed tomography. Eur Radiol. 2013;23:1862‐1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. O'Connor SD, Yao J, Summers RM. Lytic metastases in thoracolumbar spine: computer‐aided detection at CT—preliminary study. Radiology. 2007;242:811‐816. [DOI] [PubMed] [Google Scholar]
- 116. Burns JE, Yao J, Wiese TS, Muñoz HE, Jones EC, Summers RM. Automated detection of sclerotic metastases in the thoracolumbar spine at CT. Radiology. 2013;268:69‐78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Amarasingham R, Patzer RE, Huesch M, Nguyen NQ, Xie B. Implementing electronic health care predictive analytics: considerations and challenges. Health Aff. 2014;33:1148‐1154. [DOI] [PubMed] [Google Scholar]
- 118. McGirt MJ, Sivaganesan A, Asher AL, Devin CJ. Prediction model for outcome after low‐back surgery: individualized likelihood of complication, hospital readmission, return to work, and 12‐month improvement in functional disability. Neurosurg Focus. 2015;39:E13. [DOI] [PubMed] [Google Scholar]
- 119. Fairbank JC, Couper J, Davies JB, O'Brien JP. The Oswestry low back pain disability questionnaire. Physiotherapy. 1980;66:271‐273. [PubMed] [Google Scholar]
- 120. Kim JS, Merrill RK, Arvind V, et al. Examining the ability of artificial neural networks machine learning models to accurately predict complications following posterior lumbar spine fusion. Spine. 2018;43:853‐860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Lee MJ, Cizik AM, Hamilton D, Chapman JR. Predicting surgical site infection after spine surgery: a validated model using a prospective surgical registry. Spine J. 2014;14:2112‐2117. [DOI] [PubMed] [Google Scholar]
- 122. Janssen DM, van Kuijk SM, d'Aumerie BB, Willems PC. External validation of a prediction model for surgical site infection after thoracolumbar spine surgery in a Western European cohort. J Orthop Surg Res. 2018;13:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Scheer JK, Smith JS, Schwab F, et al. Development of a preoperative predictive model for major complications following adult spinal deformity surgery. J Neurosurg Spine. 2017;26:736‐743. [DOI] [PubMed] [Google Scholar]
- 124. Durand WM, DePasse JM, Daniels AH. Predictive modeling for blood transfusion after adult spinal deformity surgery: a tree‐based machine learning approach. Spine. 2018;43:1058‐1066. [DOI] [PubMed] [Google Scholar]
- 125. Coupé VM, van Hooff ML, de Kleuver M, Steyerberg EW, Ostelo RW. Decision support tools in low back pain. Best Pract Res Clin Rheumatol. 2016;30:1084‐1097. [DOI] [PubMed] [Google Scholar]
- 126. van Hooff ML, van Loon J, van Limbeek J, de Kleuver M. The Nijmegen decision tool for chronic low back pain. Development of a clinical decision tool for secondary or tertiary spine care specialists. PLoS One. 2014;9:e104226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Antani SK, Long LR, Thoma GR. Content‐based image retrieval for large biomedical image archives. Stud Health Technol Inform. 2004;107(Pt 2):829‐833. [PubMed] [Google Scholar]
- 128. Schütze H, Manning CD, Raghavan P. Introduction to Information Retrieval. Cambridge, UK: Cambridge University Press; 2008. [Google Scholar]
- 129. Liu R, Wang Y, Baba T, Masumoto D, Nagata S. SVM‐based active feedback in image retrieval using clustering and unlabeled data. Pattern Recognition. 2008;41:2645‐2655. [Google Scholar]
- 130. Hoi SC, Jin R, Zhu J, Lyu MR. Semisupervised SVM batch mode active learning with applications to image retrieval. ACM Trans Inf Syst. 2009;27:16. [Google Scholar]
- 131. Rahman MM, Bhattacharya P, Desai BC. A framework for medical image retrieval using machine learning and statistical similarity matching techniques with relevance feedback. IEEE Trans Inf Technol Biomed. 2007;11:58‐69. [DOI] [PubMed] [Google Scholar]
- 132. Anavi Y, Kogan I, Gelbart E, Geva O, Greenspan H. Visualizing and enhancing a deep learning framework using patients age and gender for chest x‐ray image retrieval Medical Imaging 2016: Computer‐Aided Diagnosis, Proceedings Volume 9785. Bellingham, WA: The International Society for Optics and Photonics (SPIE); 2016. 10.1117/12.2217587. [DOI] [Google Scholar]
- 133. Shah A, Conjeti S, Navab N, Katouzian A. Deeply learnt hashing forests for content based image retrieval in prostate MR images Medical Imaging 2016: Image Processing. Bellingham, WA: The International Society for Optics and Photonics (SPIE); 2016. 10.1117/12.2217162. [DOI] [Google Scholar]
- 134. Xu X, Lee D, Antani SK, Long LR, Archibald JK. Using relevance feedback with short‐term memory for content‐based spine X‐ray image retrieval. Neurocomputing. 2009;72:2259‐2269. [Google Scholar]
- 135. Lee D, Antani S, Chang Y, Gledhill K, Long LR, Christensen P. CBIR of spine X‐ray images on inter‐vertebral disc space and shape profiles using feature ranking and voting consensus. Data Knowl Eng. 2009;68:1359‐1369. [Google Scholar]
- 136. Chande RD, Hargraves RH, Ortiz‐Robinson N, Wayne JS. Predictive behavior of a computational foot/ankle model through artificial neural networks. Comput Math Methods Med. 2017;2017:3602928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Zadpoor AA, Campoli G, Weinans H. Neural network prediction of load from the morphology of trabecular bone. App Math Model. 2013;37:5260‐5276. [Google Scholar]
- 138. Lu Y, Pulasani PR, Derakhshani R, Guess TM. Application of neural networks for the prediction of cartilage stress in a musculoskeletal system. Biomed Signal Process Control. 2013;8:475‐482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Haddas R, Belanger T. Clinical gait analysis on a patient undergoing surgical correction of kyphosis from severe ankylosing spondylitis. Int J Spine Surg. 2017;11:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Mahaudens P, Mousny M. Gait in adolescent idiopathic scoliosis. Kinematics, electromyographic and energy cost analysis. Stud Health Technol Inform. 2010;158:101‐106. [PubMed] [Google Scholar]
- 141. Leardini A, Biagi F, Merlo A, Belvedere C, Benedetti MG. Multi‐segment trunk kinematics during locomotion and elementary exercises. Clin Biomech (Bristol, Avon). 2011;26(6):562‐571. [DOI] [PubMed] [Google Scholar]
- 142. Prakash C, Kumar R, Mittal N. Recent developments in human gait research: parameters, approaches, applications, machine learning techniques, datasets and challenges. Artif Intell Rev. 2018;49:1‐40. [Google Scholar]
- 143. Fukuchi RK, Eskofier BM, Duarte M, Ferber R. Support vector machines for detecting age‐related changes in running kinematics. J Biomech. 2011;44:540‐542. [DOI] [PubMed] [Google Scholar]
- 144. Lai DT, Begg RK, Palaniswami M. Computational intelligence in gait research: a perspective on current applications and future challenges. IEEE Trans Inf Technol Biomed. 2009;13:687‐702. [DOI] [PubMed] [Google Scholar]
- 145. Zhang J, Lockhart TE, Soangra R. Classifying lower extremity muscle fatigue during walking using machine learning and inertial sensors. Ann Biomed Eng. 2014;42:600‐612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Begg R, Kamruzzaman J. A machine learning approach for automated recognition of movement patterns using basic, kinetic and kinematic gait data. J Biomech. 2005;38:401‐408. [DOI] [PubMed] [Google Scholar]
- 147. Stucovitz E, Vitale J, Galbusera F. In vivo measurements: motion analysis Biomechanics of the Spine—Basic Concepts, Spinal Disorders and Treatments. London, UK: Academic Press; 2018. [Google Scholar]
- 148. Hayashi H, Toribatake Y, Murakami H, Yoneyama T, Watanabe T, Tsuchiya H. Gait analysis using a support vector machine for lumbar spinal stenosis. Orthopedics. 2015;38:e959‐e964. [DOI] [PubMed] [Google Scholar]
- 149. Jiang N, Luk KD, Hu Y. A machine learning‐based surface electromyography topography evaluation for prognostic prediction of functional restoration rehabilitation in chronic low back pain. Spine. 2017;42:1635‐1642. [DOI] [PubMed] [Google Scholar]
- 150. Heess N, Sriram S, Lemmon J, et al. Emergence of locomotion behaviours in rich environments. ArXiv Preprint arXiv. 2017;1707:02286. [Google Scholar]
- 151. Pesapane F, Volonté C, Codari M, Sardanelli F. Artificial intelligence as a medical device in radiology: ethical and regulatory issues in Europe and the United States. Insights Imaging. 2018;9(5):745‐753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Simonyan K, Vedaldi A, Zisserman A. Deep inside convolutional networks: Visualising image classification models and saliency maps. ArXiv Preprint arXiv. 2013;1312:6034. [Google Scholar]
- 153. Char DS, Shah NH, Magnus D. Implementing machine learning in health care ‐ addressing ethical challenges. N Engl J Med. 2018;378:981‐983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Winfield AF, Jirotka M. Ethical governance is essential to building trust in robotics and artificial intelligence systems. Philos Trans Royal Soc A. 2018;376:20180085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Thierer, A. D. , Castillo, A. , Russell, R. Artificial Intelligence and Public Policy. Arlington, VA: Mercatus Research, Mercatus Center at George Mason University; 2017. https://www.mercatus.org/system/files/thierer‐artificial‐intelligence‐policy‐mr‐mercatus‐v1.pdf [Google Scholar]


