Introduction
Antimicrobial susceptibility testing (AST) is a fundamental mission of the clinical microbiology laboratory. AST provides an in vitro measure of bacterial response to an antimicrobial agent that predicts therapeutic efficacy. Standard AST methods require isolated organisms and effectively take a day to perform. Thus, a minimum of two days is required to obtain susceptibility information from a clinical sample. With emerging resistance, this delay may lead to a prolonged antimicrobial susceptibility testing gap during which patients are treated with suboptimal or ineffective therapy. Rapid AST methods are needed to close this gap. In this review, we summarize new and evolving approaches to rapid AST. Importantly, if a rapid AST method provides a timely result and no one is aware, we rhetorically query (akin to the proverbial fallen tree in the forest), whether it is truly rapid. Therefore, we suggest that rapid AST must go hand in hand with new decision support technology to enable an equally rapid therapeutic response.
From Reference to Commercial Methods: Current State
AST methods based on broth or agar dilution are used to determine the minimal inhibitory concentration (MIC), defined as the lowest concentration of antimicrobial inhibiting growth in vitro. Organizations such as the Clinical and Laboratory Standards Institute (CLSI) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) correlate MICs with pharmacokinetic/pharmacodynamics (PK/PD) studies and clinical outcome to derive categorical interpretive criteria (sensitive, resistant, etc.) upon which antimicrobial therapeutic selections are made. The gold standard dilution-based methods - broth macrodilution and microdilution, and agar dilution - have remained relatively unchanged for >40 years.1
These methods serve as reference comparators for clearance of all commercial AST testing systems. However, they are labor intensive and complex so they are rarely if ever performed outside a reference laboratory setting. Instead, most clinical laboratories use automated platforms for routine AST with generally comparable performance characteristics. AST may also be performed with commercially prepared broth dilution panels, or using antimicrobial disk or gradient strip diffusion methods. Practically, results from all of these methods are not available until the next day, and therapeutic corrections in response to results may be delayed even further. All of these methods require inoculation with isolated bacterial colonies, although direct testing of positive culture broth without colony isolation (decreasing turn around by a day) has proven reasonably reliable using disk diffusion.2
With emerging antimicrobial resistance, timeliness becomes important
Until recently, predictable susceptibility profiles allowed for reliable selection of effective empiric antimicrobial therapy, making a one to two day AST gap clinically acceptable (Fig. 1). However, emerging antimicrobial resistance has led to a higher incidence of empiric therapy failure. In particular, for bloodstream infections, mortality correlates with the time needed to get patients on active therapy.3 Delay may have less impact on localized infections such as uncomplicated cystitis and pyelonephritis.4 Nevertheless, patients suffer in the short-term from symptom prolongation and are at increased risk of complications while on suboptimal therapy.
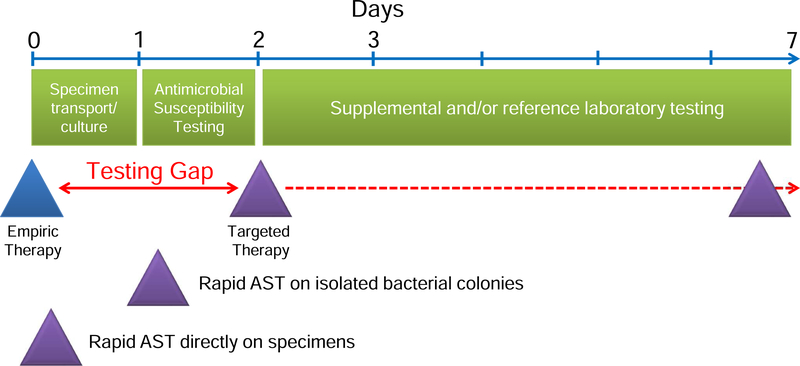
Figure 1. Reducing the antimicrobial testing gap.
Rapid AST will reduce the time between the start of empiric therapy and definitive therapy based on AST data by at least one day for methods that test isolated bacterial growth and by at least two days for methods that test primary specimens directly. Rapid AST data will allow life-saving therapeutic corrections, and tailoring of therapy to foster stewardship goals and avoid complications from extended broad-spectrum empiric therapy.
Therefore, rapid AST will support several clinical goals. First, with emerging antimicrobial resistance, rapid AST will identify active therapy in time to optimize outcome.5 Second, rapid AST will support antimicrobial stewardship goals by allowing timely transition from increasingly broad-spectrum regimens to pathogen-specific therapy. This narrowing of coverage reduces (1) exposure to potentially toxic broad-spectrum single and combination antimicrobial regimens,6,7 (2) complications such as C. difficile colitis; and (3) selection for antimicrobial resistant pathogens in the patient’s colonizing flora, a setup for future multidrug-resistant infection. Based on these considerations, rapid AST methods are not surprisingly an area of intense investigation.
Rapid Susceptibility Landscape Overview
There are two broad classes of AST technology: genotypic and phenotypic, each of which has advantages and disadvantages. The most impactful rapid susceptibility methods would yield results in a few hours. However, holistically, “rapid” must also take into account time required for all pre- and post-analytic components leading to therapeutic adjustments.
Genotypic Testing
Genotypic AST detects presence of specific resistance genes, which may encode enzymes that degrade antibiotic (e.g., β-lactamases), modify antibiotic target (e.g., ribosomal methylases) or alter the target via mutation or substitution (e.g., acquisition of penicillin binding protein 2a by S. aureus). Genotypic susceptibility testing can provide rapid results since neither growth nor metabolism is required for readout. Further, genotypic testing, unlike traditional AST methods, can be performed directly on primary specimens and positive blood culture broth, greatly reducing time to diagnosis.
However, genotypic testing only detects what it is designed to detect, and conversely won’t detect what we don’t yet know about. Additionally, it provides no information about penetrance - to what degree a resistance element is phenotypically expressed – and so may overcall resistance. For example, low levels of serine carbapenemase expression may result in carbapenem MICs (susceptible or intermediate) that should be treatable based on PK/PD considerations.8,9 Of course, it may be argued whether phenotypic MICs are sufficiently reliable based on the steep inoculum effect observed at bacterial concentrations used for AST testing;10 whether sufficient PK/PD exploration has been performed to justify use of carbapenems for carbapenem-producing strains, when other options are available; and thus whether a more conservative approach based on gene detection is preferred.10 However, it is certain that targeted genetic methods lack nuance.
The Gram-Positive/Negative Genotypic Divide
The predictive value and utility of genotypic AST is strongest for Gram-positive pathogens. Here, a relatively small number of resistance elements (mecA/C, vanA/B) contribute to the preponderance of phenotypically observed resistance to therapeutics of major interest.
However, the Gram-positive, mecA target itself also illustrates the not necessarily apparent complexity of genotypic testing. The mecA gene exists within a mobile genetic element (SCCmec) that is heterogeneous.11 Therefore, a large number of primer sets have been incorporated into nucleic acid amplification tests (NAATs) to ensure reliable detection. This is especially important as the clinical consequences of calling a methicillin-resistant blood isolate inappropriately susceptible to β-lactams can be devastating, i.e., a switch to ineffective therapy for an aggressive infection. Furthermore, this genetic heterogeneity is not static. As an exemplar, emergence of mecC in S. aureus led to missed detection of MRSA in mecA assays.12 Therefore, microbial evolution necessitates vigilance and diagnostic product evolution with associated regulatory, technical, and economic hurdles, ultimately limiting NAAT assays to only the most common, relatively stable targets.
In contrast, Gram-negative pathogens frequently exhibit resistance mechanisms that are polygenic and reflect combinatorial effects of permeability, efflux, target modification, enzymatic degradation, and potentially other mechanisms as well. For example, predicting how the hundreds of distinct β-lactamases, which may be expressed in different combinations and at different levels, would affect the MIC against a variety of β-lactam agents seems a daunting task. With this complexity, an inclusive, fully predictive assay may be an elusive goal.
Nevertheless, platforms that rapidly detect presence or absence of high consequence resistance elements such as KPC, VIM, IMP, OXA, and NDM carbapenemases will allow therapeutic adjustment and/or potential reach for agents not typically used for empiric therapy. Genotypic resistance detection within syndromic panels is further discussed in chapter 8, “Direct-from-Specimen Pathogen Identification: Evolution of Syndromic Panels”.
With total analytical times likely trend to under 10 minutes with advanced PCR techniques and related technologies, these types of panels will offer very rapid, targeted direction for a subset of pathogens and resistance types.13 The potential application of whole genome sequencing for AST, the logical extreme of multiplexed genotypic testing, is reviewed in chapter 6, “Next-Generation Sequencing from Strain Typing to Identification to Antimicrobial Susceptibility Prediction: Will It Supplant Traditional Methods in the Clinical Microbiology Laboratory, When and at What Cost?”
Challenges of Rapid Phenotypic AST
A wide body of PK/PD data have been used to set categorical breakpoints based on phenotypic MICs determined by reference methods. Broadly, relation of pharmacokinetic profiles to phenotypic MIC predicts treatment efficacy based on well-established parameters that appear thus far agnostic to underlying resistance gene repertoire. For example, time over the MIC is predictive of a successful response to β-lactam therapy, reviewed in chapter 10, “What the clinical microbiologist should know about PK/PD in the era of emerging multidrug-resistance - focusing on β-lactam / β-lactamase inhibitor combinations.”
Therefore, rapid determination of the phenotypic MIC would provide significant value.
Currently, all reference AST methods are based on direct visual observation of an organism’s response to antibiotics. The threshold for visual detection requires more than a > 200-fold increase in bacterial numbers from the initial inoculum. Accordingly, such bulk growth AST assays require an extended incubation period (16–20 hours) to reach reliable detection thresholds.
Earlier assessment of growth is complicated by both biological and technical factors. A major biological factor is the “lag phase”, during which organisms metabolically adapt to a new environment, but do not yet reproduce.14 Measurements made before organisms exit this lag phase may result in erroneous categorization of organisms as antibiotic susceptible. Lag phase can last for 1–2 hours under optimal conditions and may be extended in the presence of stressors including exposure to subinhibitory levels of antibiotics during MIC testing.15
Even during the active growth phase, genes required for antimicrobial resistance may take a finite time to be expressed. Further, measurements may be confounded by altered morphology of organisms in the presence of antibiotics. For example, Gram-negative cells exposed to inhibitory concentrations of β-lactams or fluoroquinolones filament,16 potentially making it difficult to distinguish between increased volume and true replication.
Biological limitations notwithstanding, rapid phenotypic AST methods must detect organism growth and/or metabolism with very low organism burden. Therefore, the most significant requirement for rapid phenotypic AST is sufficiently sensitive measurement techniques. Several innovative technologies have emerged to address these challenges, broken down by general category below.
Microscopy-Based Rapid Phenotypic AST
The conceptually simplest way to determine whether an organism’s replication is inhibited by an antibiotic is to count bacterial cell numbers by microscopy. However, such measurements are complicated by the need to confine bacteria to a focal plane within a sufficiently optically clear medium to allow microscopic image analysis. Several different strategies have been explored including electrophoretic or centrifugal immobilization onto an adherent surface covered by liquid growth medium; inkjet printing of organisms onto a solid growth surface; or confinement to a microfluidic droplet or channel, several examples of which are discussed in turn. This is not a comprehensive listing, but illustrates the diversity of approaches being pursued. The evolution and natural selection of the most propitious methods will play out over the next decades.
One commercialized system electrophoreses organisms from positive blood culture broth (presumably applicable to primary samples as well) onto a solid surface. An automated microscope records transition from individual organisms to microcolonies in the presence or absence of a panel of antibiotics. MICs are determined based on a machine-learning algorithm that evaluates growth effects of a single concentration of antibiotic during a several hour incubation. These extrapolated MICs have been reported to be accurate.17
Both the elegance and complexity of this system is emblematic of the tradeoffs that must be reconciled as the field matures. For example, the existing one sample per shift per instrument throughput is a concern in high volume laboratories. The underlying complexity also necessarily comes at a price point that will not address the needs of resource limited (see chapter 3, “Clinical Microbiology in Under-Resourced Settings,” for further discussion) and perhaps even not so resource limited settings as well.
Our own group is developing a technology that utilizes inkjet printing of pathogen and antibiotics onto a solidified Mueller-Hinton-based growth surface.18 The system allows testing of any antimicrobial at any concentration at will using commercially available microplates, consumables, and reagents.19 Automated microscopy is combined with a trained convolutional neural network-based algorithm to allow for highly automated MIC determination. Although still early in development, this system has shown >95% agreement for testing standard quality control strains with representative classes of antibiotics in a 2 hour time frame.18 In its current form, it would need to be combined with a separate bacterial identification system, for example, matrix assisted laser desorption time of flight mass spectrometry (MALDI-TOF), to allow appropriate interpretation. Another recently described microscopic technology follows growth of bacteria immobilized in agarose in a customized 96-well plate format from positive blood culture broth with AST determinations in 4 hours.20
Microscopy Plus Microfluidics
A variety of microfluidic-based methods for AST have been described in which bacteria are confined within channels or nanodroplets and assayed by traditional or fluorescence microscopy.21 Microfluidic devices are custom manufactured and may require pumping and sample loading methods that increase device complexity, cost, and scalability. However, implementation of microfluidics in infectious disease sample-to-answer nucleic acid amplification tests suggests that these technologies can be successfully commercialized.
Alternative Optical Detection Methods
A fundamental challenge for direct microscopy-based AST methods is the need for magnification (typically ≥400x), which demands relatively complex and potentially costly optics and automation (for example, an autofocus system coupled to a mechanized stage). In one alternative strategy, organisms from primary samples after exposure to antibiotic and an incubation phase are tethered to magnetic beads and labeled with FISH probes unique for each species, and then pulled by a magnet through a light blocking dye-cushion thereby optically separating labeled bacteria from specimen matrix and unbound reporter.22 Organisms are counted without magnification using a CMOS digital camera chip allowing species-specific quantitative analysis.
An alternative method uses forward laser light scatter, in a way similar to flow cytometry, but in bulk bacterial growth suspension. This biophysical bacterial enumeration method, already commercialized for quantifying bacteria in urine specimens, has been applied to direct AST on urine specimens and positive blood culture broth.23,24
Spectrometry-Based Detection
Raman spectroscopy-based methods rely on spectral scattering of an incident monochromatic laser beam to detect potentially very early physiological changes in bacteria exposed to antibiotics and deposited on a solid surface for analysis.25 To date, no Raman spectroscopy-based AST methods have been comprehensively evaluated and the potential for cost-effective commercialization of these complex technologies remain unclear.
MALDI-TOF has become commonplace in the clinical microbiology laboratory as a qualitative technique for species identification. There has been some progress towards using peak intensity as a proxy for bacterial growth which can in turn be applied to phenotypic AST.26 However, to transition this technology into clinical practice, significant additional levels of automation will be required. Further, applicability to direct specimen AST seems a steep hurdle and the cost of underlying instrumentation will make deployment at point of care unfeasible. MALDI-TOF is discussed in more depth in chapter 5, “MALDI-TOF: The Revolution in Progress.”
Signal amplification
Detection of small numbers of bacteria can also be addressed through signal amplification techniques using either physical or molecular detection methods. For example, after standard incubation for several hours in 96-well format, one platform uses robotics to introduce cationic, europium-cryptate-diamine chelate into microplates that non-specifically coat and bind to natively anionic bacterial surfaces.27 After a washing step to remove unbound chelate, time-resolved fluorescence signal from the chelate correlates with total bacterial surface area and accurately predicts MIC values.
Several strategies use nucleic acid amplification technology (NAAT) to quantify organism number or transcriptional activity and provide a much earlier readout of growth and viability than bulk growth assays.28 A variation uses replication incompetent phage particles to introduce a reporter (e.g., luciferase gene) into bacteria in a species-specific manner for both identification and AST. Luminescence, an ATP-dependent process, provides a proxy for bacterial viability. The technique is potentially applicable to direct testing of patient samples with low organism burden.29 Notably, all of the signal amplification assays discussed are endpoint determinations with a fixed, single incubation time for analysis. These fixed determinations will need to confront lag time and resistance induction issues that can be more easily addressed by real-time phenotypic measures.
Decision Support Must Go Hand in Hand with Rapid AST Implementation
Faster AST results should lead to reduced morbidity and mortality, shorter hospital stays, and cost savings. However, in practice, several factors prevent full realization of these benefits. These include inadequate communication and therapeutic inertia -- reluctance to switch from broad empiric broad-spectrum therapy, if active, to directed therapy.
These factors likely contribute to the less than impressive, but nevertheless instructive, findings in the rapid diagnostics literature. These studies primarily examined the impact of rapid diagnostic molecular testing performed on positive blood culture broth in comparison to traditional methods. They further focused almost exclusively on pathogen identification rather than rapid susceptibility testing, and therapeutic changes were primarily seen for Gram-positive infections. Yet, two important lessons can still be distilled. First, even without AST guidance there was significant improvement in time to effective therapy (5 hours) and reduced length of hospital stay (2.5 days).30 Second, significant improvement in mortality was only demonstrated in the presence of an antimicrobial team on call to respond to “rapid data” and assist in therapeutic adjustment. With some exceptions, it is doubtful that significant change in therapy would have been made in advance of classic AST for Gram-negative pathogens. Therefore, the power of rapid AST is recognized but has not yet been fully demonstrated in the literature.31
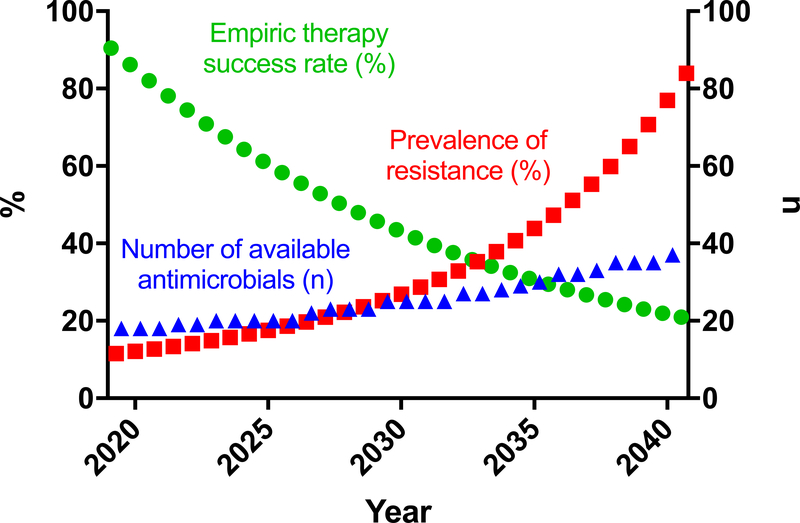
Over the next twenty years, the number of therapeutic choices will certainly increase as will prevalence of antimicrobial resistance (Fig. 2). Therefore, the likely efficacy of empiric therapy will decrease. Further, with increasing number of potential agents, new dosing strategies, and insights from personalized metabolomics, it will be more difficult for caregivers to master optimal use of these agents (optimal dosing, drug interactions, contraindications, adjustments for disease conditions) and render timely, informed therapeutic decisions to take full advantage of rapid AST. Yet such rapid therapeutic adjustments will become critical as empiric therapies become unreliable.
Figure 2. Loss of reliable empiric therapy.
Over the next two decades, antimicrobial resistance will increase significantly. The number of available antimicrobials will also increase, but not at the same pace. As a result, the likelihood that any given empiric therapy will be effective will decrease. Therefore, rapid AST will become increasingly necessary to ensure patients are on active therapy in time to make a difference.
Ultimately, impact of rapid AST technologies will rely on care providers’ willingness and ability to rapidly respond. It is clear that reliance on intervention from specially trained teams, although effective, is only a partial, resource intensive, and not fully sustainable solution.
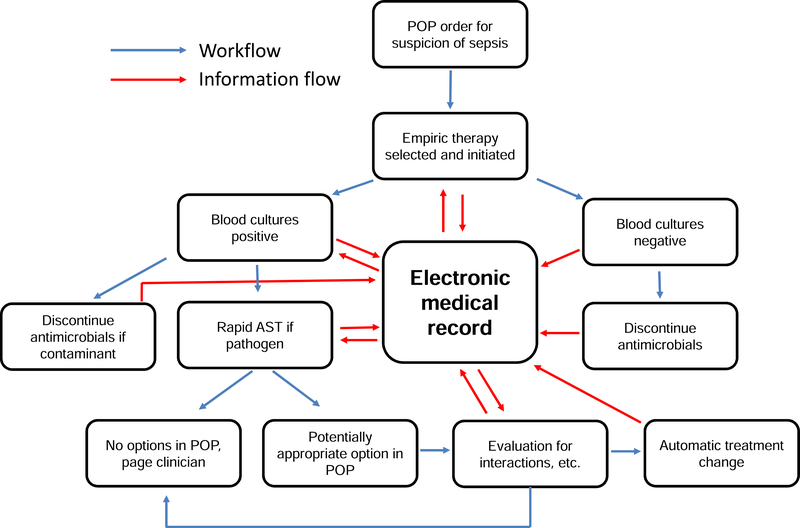
Therefore, we envision that rapid AST must be inextricably linked to a robust and ultimately autonomous decision support system. Such a system, which we call AST-ASSIST, will incorporate bundled orders, rapid identification and susceptibility input; integration of the medical record; and automated flags and real-time therapeutic decisions or suggestions. Each component is discussed in turn as outlined in Fig. 3. This system is designed to provide the most appropriate, timely antimicrobial therapy, while still enlisting direction from primary care givers who are best aware of history, clinical considerations, and patient preferences not reflected in the medical record.
Figure 3. AST-ASSIST.
Rapid AST is only truly rapid if results can be acted upon quickly. To this end, we envision a decision support system, AST-ASSIST, that will leverage use of provider other pathways (POPs), which contain instructions for initial empiric antibiotics and options for reflexed directed antimicrobial therapy populated based on patient medical and microbiological history and local antibiogram. AST-ASSIST will implement therapeutic changes included within the POP based on organism identity and rapid AST results through interface with pharmacy and nursing systems and/or notify clinicians through pager or text message for additional timely input as needed.
We envision leveraging clinicians’ insights by allowing up-front decisions for multiple potential antimicrobial treatment options, or physician order pathways (POPs). Specifically, when infection is suspected, a POP would consist of a single order including both microbiological cultures and treatment. Constituted within this order would be broad-spectrum empiric antibiotics and a series of potential targeted antibiotic regimens that would be reflexed depending on definitive (rapid) AST results. Modifications to the POP would be suggested by AST-ASSIST through query of the electronic medical record (EMR) to consider patient allergies, interacting medications, underlying medical conditions, laboratory values (creatinine, liver function tests, cytopenias) and potential for resistance based on a current antibiogram, prior cultures, clinical service, and length and history of hospitalizations.
If organisms were detected, they would be subject to rapid antimicrobial identification and susceptibility testing using fully interfaced instrumentation. Therefore, data would be available in real-time in the EMR. We envision that AST-ASSIST middleware would evaluate updated microbiological, laboratory and clinical data from the medical record in real-time to detect relevant changes since the original POP. For example, it would identify contraindications not detected in the initial POP evaluation such as interactions with recently instituted therapies. If an appropriate targeted antimicrobial were initially included in the POP, an automatic update to pharmacy and nursing records would result in immediate change in therapy. If appropriate therapy were not included in the POP, clinicians would be alerted by pager/text message with a recommendation for an appropriate antimicrobial.
As an example of this process, if a patient were suspected to have sepsis, a POP would include blood cultures and specify empiric vancomycin/piperacillin-tazobactam as a broad-spectrum antimicrobial regimen based on the local antibiogram and patient history. If methicillin-susceptible S. aureus (MSSA) were isolated, the AST-ASSIST system would evaluate the POP for optimal targeted therapy. If no contraindications were found, coverage would automatically be narrowed to a β-lactam such as cefazolin at the next dose, thereby minimizing all possible delay. However, if coagulase negative staphyloccocci were isolated, the EMR would be automatically queried for previous positive cultures, elevated white cell count, fever, and biomarkers such as procalcitonin (see chapter 9, “Biomarkers - predicting bacterial versus viral infection, or none of the above: current and future prospects”) and lactate levels to predict the probability of a true infection versus contamination. Depending on the results and what was defined in the POP order, therapy may be stopped or the clinician paged to intervene.
Alternatively, if a carbapenem-resistant Enterobacteriaceae expressing a serine carbapenemase (with an MIC = 16 μg/mL by a phenotypic method) were isolated, an alert would be sent to nursing staff and pharmacy for immediate dosing of an appropriate active agent, for example, plazomicin, ceftazidime-avibactam, or meropenem-vaborbactam. Here, dose optimization of active agents may be particularly important owing to inherent toxicity or desire to optimize drug exposure based on PK/PD principles. Therefore, AST-ASSIST dosing algorithms would take into account body mass index, liver and kidney function, genetic metabolic knowledge, drug interactions with other onboard therapies, specific MIC of organism and known PK/PD relationships to optimize dose and dosing frequency for particular agents, as applicable.
Conclusion
Rapid AST represents a critical component in addressing the challenges posed by emerging antimicrobial resistance. A variety of phenotypic and genotypic methods are already FDA-cleared or in development that promise to reduce the time to definitive AST results from days hours. However, to optimize impact, those results must be shared with and acted upon by healthcare providers just as rapidly. Therefore, rapid AST must be supported by a robust LIS, EMR, and decision support infrastructure (POPs and AST-ASSIST). We look forward to fully integrated rapid AST systems and the advantages they will provide at all levels in our healthcare systems.
Key Points.
Emerging antimicrobial resistance makes empiric therapy unreliable. Therefore, rapid antimicrobial susceptibility testing will provide early, definitive therapeutic guidance to optimize patient outcome.
Genotypic rapid AST methods are fast but can only identify what we know about. Phenotypic rapid AST methods provide a nuanced integrated assessment of resistance that can be used to pick the most active therapies.
Early detection by phenotypic AST methods requires very sensitive technology such as microscopic detection of replicating organisms, biophysical assessment, or signal amplification techniques.
For the potential of rapid AST platforms to be fully realized, results must be linked with robust, autonomous decision support solutions that will implement therapeutic changes in real time.
Synopsis.
With emerging antimicrobial resistance, rapid antimicrobial susceptibility testing (AST) is needed to provide early definitive therapeutic guidance to optimize patient outcome. Genotypic methods are fast, but can identify only a subset of known resistance elements. Phenotypic methods determine clinically predictive minimal inhibitory concentrations and include very sensitive optical and biophysical methods to detect changes in replication or physiology of pathogens in response to antibiotics. For the potential of rapid AST to be fully realized, results must be linked with robust decision support solutions that will implement therapeutic changes in real time.
Acknowledgements
Based on space limitations, it was not possible for us to reference and cite all of the relevant literature in the rapidly growing rapid AST field. We apologize to authors, scientists, and companies, large and small, whose innovative and important work we could not specifically identify in this review. We have purposefully not mentioned the name of commercialized products to keep our focus on underlying principles.
Footnotes
disclosure statement
JEK is a member of the Clinical Advisory Board of First Light Biosciences, Chelmsford, MA. TECAN (Morrisville, NC) provided an HP D300 digital dispenser and associated consumables used by JEK’s research group during development of the MAST platform. Neither First Light nor TECAN had a role in manuscript preparation or decision to publish.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brennan-Krohn T, Smith KP, Kirby JE. The Poisoned Well: Enhancing the Predictive Value of Antimicrobial Susceptibility Testing in the Era of Multidrug Resistance. J Clin Microbiol. 2017;55(8):2304–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chandrasekaran S, Abbott A, Campeau S, et al. Direct-from-Blood-Culture Disk Diffusion To Determine Antimicrobial Susceptibility of Gram-Negative Bacteria: Preliminary Report from the Clinical and Laboratory Standards Institute Methods Development and Standardization Working Group. J Clin Microbiol. 2018;56(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rhodes A, Evans LE, Alhazzani W, et al. Surviving Sepsis Campaign: International Guidelines for Management of Sepsis and Septic Shock: 2016. Intensive Care Med. 2017;43(3):304–377. [DOI] [PubMed] [Google Scholar]
- 4.Eliakim-Raz N, Babitch T, Shaw E, et al. Risk Factors for Treatment Failure and Mortality Among Hospitalized Patients With Complicated Urinary Tract Infection: A Multicenter Retrospective Cohort Study (RESCUING Study Group). Clin Infect Dis. 2019;68(1):29–36. [DOI] [PubMed] [Google Scholar]
- 5.Liu VX, Fielding-Singh V, Greene JD, et al. The Timing of Early Antibiotics and Hospital Mortality in Sepsis. Am JRespir Crit Care Med. 2017;196(7):856–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mullins BP, Kramer CJ, Bartel BJ, Catlin JS, Gilder RE. Comparison of the Nephrotoxicity of Vancomycin in Combination With Cefepime, Meropenem, or Piperacillin/Tazobactam: A Prospective, Multicenter Study. Ann Pharmacother. 2018;52(7):639–644. [DOI] [PubMed] [Google Scholar]
- 7.Rutter WC, Cox JN, Martin CA, Burgess DR, Burgess DS. Nephrotoxicity during Vancomycin Therapy in Combination with Piperacillin-Tazobactam or Cefepime. Antimicrob Agents Chemother. 2017;61(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamma PD, Huang Y, Opene BN, Simner PJ. Determining the Optimal Carbapenem MIC That Distinguishes Carbapenemase-Producing and Non-Carbapenemase-Producing Carbapenem-Resistant Enterobacteriaceae. Antimicrob Agents Chemother. 2016;60(10):6425–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neuner EA, Gallagher JC. Pharmacodynamic and pharmacokinetic considerations in the treatment of critically Ill patients infected with carbapenem-resistant Enterobacteriaceae. Virulence. 2017;8(4):440–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith KP, Kirby JE. The Inoculum Effect in the Era of Multidrug Resistance: Minor Differences in Inoculum Have Dramatic Effect on MIC Determination. Antimicrob Agents Chemother. 2018;62(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Becker K, Ballhausen B, Kock R, Kriegeskorte A. Methicillin resistance in Staphylococcus isolates: the “mec alphabet” with specific consideration of mecC, a mec homolog associated with zoonotic S. aureus lineages. Int J Med Microbiol. 2014;304(7):794–804. [DOI] [PubMed] [Google Scholar]
- 12.Lee GH, Pang S, Coombs GW. Misidentification of Staphylococcus aureus by the Cepheid Xpert MRSA/SA BC Assay Due to Deletions in the spa Gene. J Clin Microbiol. 2018;56(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farrar JS, Wittwer CT. Extreme PCR: efficient and specific DNA amplification in 15–60 seconds. Clin Chem. 2015;61(1): 145–153. [DOI] [PubMed] [Google Scholar]
- 14.Rolfe MD, Rice CJ, Lucchini S, et al. Lag phase is a distinct growth phase that prepares bacteria for exponential growth and involves transient metal accumulation. J Bacteriol. 2012;194(3):686–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Belkum A, Dunne WM Jr., Next-generation antimicrobial susceptibility testing. J Clin Microbiol. 2013;51(7):2018–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorian V, Ernst J, Amaral L. The post-antibiotic effect defined by bacterial morphology. JAntimicrob Chemother. 1989;23(4):485–491. [DOI] [PubMed] [Google Scholar]
- 17.Pancholi P, Carroll KC, Buchan BW, et al. Multicenter Evaluation of the Accelerate PhenoTest BC Kit for Rapid Identification and Phenotypic Antimicrobial Susceptibility Testing Using Morphokinetic Cellular Analysis. J Clin Microbiol. 2018;56(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith KP, Richmond DL, Brennan-Krohn T, Elliott HL, Kirby JE. Development of MAST: A Microscopy-Based Antimicrobial Susceptibility Testing Platform. SLAS Technol. 2017;22(6):662–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith KP, Kirby JE. Verification of an Automated, Digital Dispensing Platform for At-Will Broth Microdilution-Based Antimicrobial Susceptibility Testing. J Clin Microbiol. 2016;54(9):2288–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi J, Jeong HY, Lee GY, et al. Direct, rapid antimicrobial susceptibility test from positive blood cultures based on microscopic imaging analysis. Sci Rep. 2017;7(1): 1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campbell J, McBeth C, Kalashnikov M, Boardman AK, Sharon A, Sauer-Budge AF. Microfluidic advances in phenotypic antibiotic susceptibility testing. Biomed Microdevices. 2016;18(6):103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gite S, Archambault D, Cappillino MP, et al. A Rapid, Accurate, Single Molecule Counting Method Detects Clostridium difficile Toxin B in Stool Samples. Sci Rep. 2018;8(1):8364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Idelevich EA, Hoy M, Knaack D, et al. Direct determination of carbapenem-resistant Enterobacteriaceae and Pseudomonas aeruginosa from positive blood cultures using laser scattering technology. Int J Antimicrob Agents. 2018;51(2):221–226. [DOI] [PubMed] [Google Scholar]
- 24.Montgomery S, Roman K, Ngyuen L, et al. Prospective Evaluation of Light Scatter Technology Paired with Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry for Rapid Diagnosis of Urinary Tract Infections. J Clin Microbiol. 2017;55(6):1802–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Novelli-Rousseau A, Espagnon I, Filiputti D, et al. Culture-free Antibiotic-susceptibility Determination From Single-bacterium Raman Spectra. Sci Rep. 2018;8(1):3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jung JS, Hamacher C, Gross B, et al. Evaluation of a Semiquantitative Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry Method for Rapid Antimicrobial Susceptibility Testing of Positive Blood Cultures. J Clin Microbiol. 2016;54(11):2820–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flentie K, Spears BR, Chen F, et al. Microplate-based surface area assay for rapid phenotypic antibiotic susceptibility testing. Sci Rep. 2019;9(1):237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andini N, Hu A, Zhou L, et al. A “Culture” Shift: Broad Bacterial Detection, Identification, and Antimicrobial Susceptibility Testing Directly from Whole Blood. Clin Chem. 2018;64(10):1453–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roche gobbles Smarticles. Nat Biotechnol. 2015;33(10):1012. [DOI] [PubMed] [Google Scholar]
- 30.Timbrook TT, Morton JB, McConeghy KW, Caffrey AR, Mylonakis E, LaPlante KL. The Effect of Molecular Rapid Diagnostic Testing on Clinical Outcomes in Bloodstream Infections: A Systematic Review and Meta-analysis. Clin InfectDis. 2017;64(1):15–23. [DOI] [PubMed] [Google Scholar]
- 31.Henig O, Kaye KS, Chandramohan S, et al. The Hypothetical Impact of Accelerate Pheno (ACC) on Time to Effective Therapy and Time to Definitive Therapy for bloodstream infections due to drug-resistant Gram-negative bacilli. Antimicrob Agents Chemother. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]