Abstract
In many cancers, high proliferation rates correlate with elevation of rRNA and tRNA levels, and nucleolar hypertrophy. However, the underlying mechanisms linking increased nucleolar transcription and tumorigenesis are only minimally understood. Here we show that IMP dehydrogenase-2 (IMPDH2), the rate-limiting enzyme for de novo guanine nucleotide biosynthesis, is overexpressed in the highly lethal brain cancer, glioblastoma (GBM). This leads to increased rRNA and tRNA synthesis, stabilization of the nucleolar GTP-binding protein, Nucleostemin, and enlarged, malformed nucleoli. Pharmacological or genetic inactivation of IMPDH2 in GBM reverses these effects and inhibits cell proliferation, whereas untransformed glia cells are unaffected by similar IMPDH2 perturbations. Impairment of IMPDH2 activity triggers nucleolar stress and growth arrest of GBM cells even in the absence of functional p53. Our results reveal that upregulation of IMPDH2 is a prerequisite for aberrant nucleolar function and increased anabolic processes in GBM, which constitutes a primary event in gliomagenesis.
Enlargement and irregular shapes of the nucleolus in tumors have been known since at least 1836, but the mechanistic importance to malignancy has remained obscure1–3. In the 1960’s, extensive research identified that the nucleolus is the site for ribosomal RNA transcription and assembly4–6. In addition, a clue to more than a epiphenomenal connection between altered nucleolar morphology and malignancy was the notion that ribonucleic acid (RNA) to deoxyribonucleic acid (DNA) ratio is increased in growing cells and can be an index of overall biosynthetic capacity and cell growth activity from bacteria to humans7,8. RNA is assembled as a chain of four ribonucleotides, ATP, UTP, CTP and GTP, and rRNA and tRNA—both synthesized in the nucleolus6,9,10—account together for over 90% of all RNA molecules. Interestingly and providing a key insight prompting our mechanistic studies, GTP, but not ATP, concentrations are elevated in several types of tumors11, raising the possibility that cells regulate the biosynthesis of ATP and GTP by distinct pathways and that tumors may specifically utilize GTP for processes fundamental to their malignancy.
Increased de novo GTP biosynthesis in glioblastoma but not in primary glia
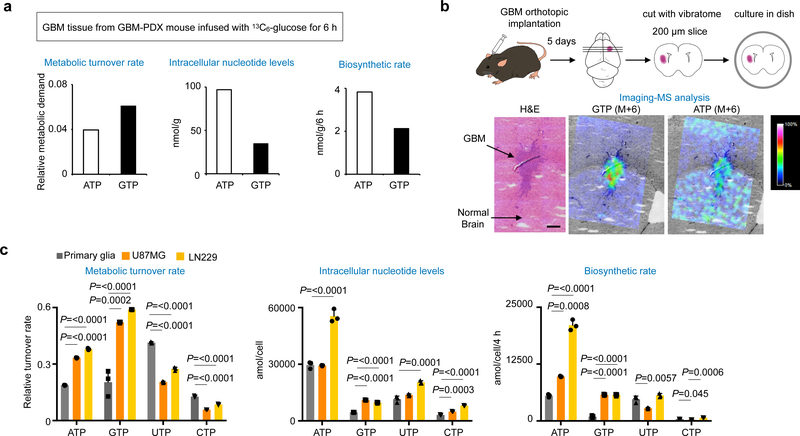
Two evolutionarily conserved purine biosynthesis pathways exist. De novo purine synthesis is an energy-demanding pathway essential for survival in a purine-free environment and embryogenesis. The purine salvage pathway is an energy efficient process that is favored in the adult brain12 and associated with mental disorders and spasticity when deficient13,14. Using patient-derived xenograft (PDX) glioma mice infused with [U-13C]glucose, we found that GBM tissues increase metabolic turnover of GTP (Fig. 1a and Supplementary Fig. 1a). Since GTP is an end product, its high turnover rate may represent the increased metabolic demand, and infused glucose could be metabolized to salvage metabolites before the uptake by GBM. Organotypic GBM brain explant assays, which can directly assess glucose metabolism, revealed significant elevation of de novo GTP biosynthesis in GBM tissue. Interestingly, de novo ATP biosynthesis was comparably active in both GBM and normal brain tissues (Fig. 1b).
Fig. 1 |. Upregulation of GTP biosynthesis in human and murine GBM.
a, Metabolic turnover of GTP is higher than the other nucleotides in GBM-PDX mouse model. Metabolic turnover rate of the indicated nucleotides is calculated based on intracellular pool size (middle) and biosynthetic rate (right) of ribonucleotides in the GBM tissues infused with [U-13C]glucose for 6 h.
b, Murine GBM brain section was treated with [U-13C]glucose and subjected to imaging-mass spectrometry (MS) analysis. M+6 GTP was detected in organotypic GBM but not in normal brain tissue, while M+6 ATP was detected both in GBM and normal brain tissue. Representative images were shown from 2 independent experiments. Scale bar indicates 300 μm.
c, Metabolic turnover of GTP is higher than the other nucleotides in GBM cells, not in primary glia. Metabolic turnover rate of the indicated nucleotides is calculated based on intracellular pool size (middle) and biosynthetic rate (bottom) of ribonucleotides in the indicated cells labeled with [U-13C]glucose for 4 h. Data are presented as mean+s.d. n=3 biologically independent samples. One-way ANOVA.
Consistent with the results from GBM tissues, in cultured GBM cells, the pool size and the turnover rate of GTP were elevated compared to those of ATP, UTP and CTP (Fig. 1c and Supplementary Fig. 1b, c). In contrast, the turnover rate of GTP in less proliferative primary glia is comparable to ATP and CTP and less than that of UTP. Both GBM and primary glia allocate a substantial amount of ATP for biosynthesis of nicotinamide adenine dinucleotide (NAD) and S-adenosylmethionine (SAM) (Supplementary Fig. 1d). Similar results were observed in neural stem cells (NSC) and the isogenic GBM stem cells (GSC) that is responsible for therapeutic resistance15–18 (Supplementary Fig. 1e). Detailed quantification showed that the biosynthetic rate of SAM and NAD+ from ATP pathways was more than two-fold higher in U87MG and LN229 than in primary glia, consistent with increased growth demand in GBM19,20. We also found that primary glia and NSC, compared with GBM and GSC, possess not only more active uptake of purine metabolites from media but also greater active salvage GTP biosynthesis (Supplementary Fig. 1f–j). These results suggest that glucose metabolism in GBM is reprogrammed to increase de novo GTP biosynthesis.
Upregulation of IMPDH2 in human and murine GBM
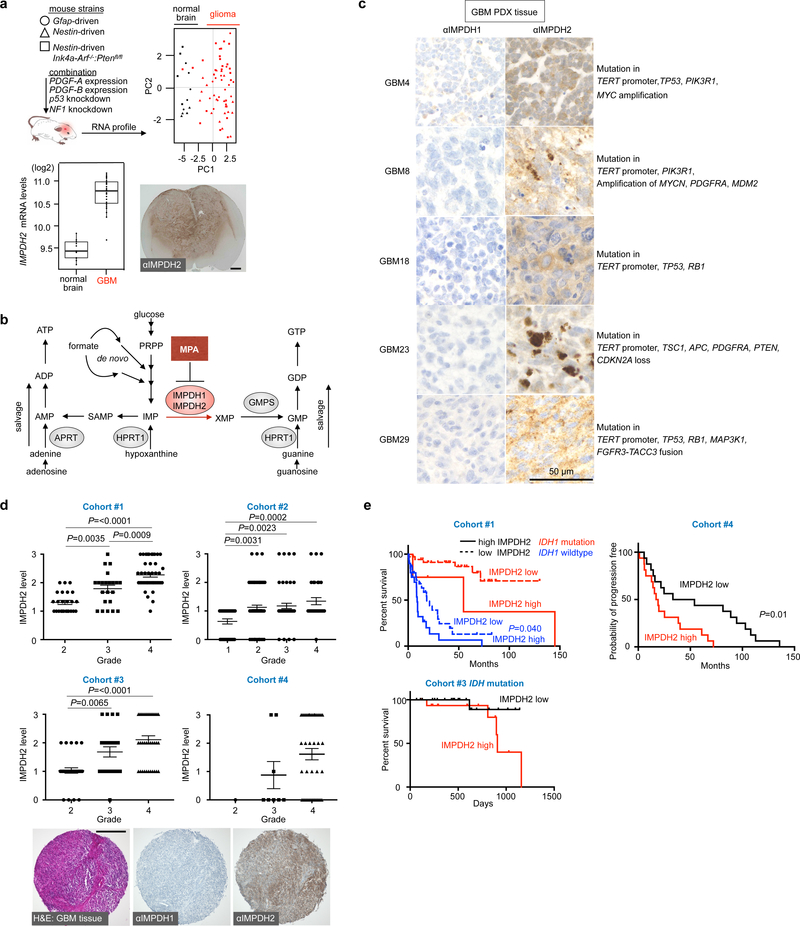
To investigate the molecular mechanism underlying the increased GTP biosynthesis in GBM, we analyzed gene expression in ten genetically different mouse gliomas (Fig. 2a and Supplementary Fig. 2a). Expression of Impdh2, encoding the enzyme that commits IMP for GTP biosynthesis (Fig. 2b), displayed the largest increase relative to control brains (about 2-fold, P<0.05). In contrast, the expression levels of the isoenzyme Impdh1, which has indistinguishable substrate affinities and catalytic activities21–23, was minimally affected or even decreased.
Fig. 2 |. Upregulation of IMPDH2 in human and murine GBM.
a, Principal component analysis in ten clinically relevant glioma mouse models by Illumina cDNA microarray (upper). Black dots are control normal brain samples, red dots are glioma samples. The experiment contains the gene expression profiles from 13 normal and 59 glioma mice (n=72). Impdh2 expression at mRNA levels (lower left) and immunohistochemical (IHC) analysis of IMPDH2 in the Nestin-driven Pdgf-B in lnk4a-arf−/−;Ptenfl/fl mouse brain (lower right). Boxplots follow a Turkey style, in which lower and upper hinges correspond to the first and third quartiles. The IHC image is a representative one from 4 independent animals. Scale bar indicates 1000 μm.
b, A schematic diagram of purine biosynthesis pathway.
c, GBM tissue from the GBM PDX mouse expresses IMPDH2, while IMPDH1 protein was undetectable level. n=1 experiment.
d, Analysis of four cohorts showed the increased IMPDH2 expression in human glioma specimens with different WHO grades (bottom). H&E staining of GBM and representative IHC for IMPDH1 and IMPDH2. Data are presented as mean±s.e.m. n=91 for Cohort#1, n=191 for Cohort#2, n=91 for Cohort#3, n=53 for Cohort#4. One-way ANOVA. The IHC image is the representative one from Cohort#4 (n=53). Scale bar indicates 300 μm.
e, Kaplan-Meier survival curves shown for three cohorts of glioma patients on the basis of IDH mutational status (Cohort #1 (n=90), #3 (n=38)) and the relative strength of cytoplasmic IMPDH2 expression. Cohort #4 (n=32) is progression-free survival and the others are overall survival. Log-rank tests (two-sided) were used for the statistical analysis.
Although the both the association of increased IMPDH activity with tumors24 and the anti-tumor activity of IMPDH inhibition has been noted for over 50 years ago25,26,27–30, the primary role of IMPDH in promoting tumor growth has remained unclear. To investigate the potential primary role for IMPDH, we first surveyed the expression of IMPDH2 in human GBM patients. Five human GBM databases revealed a more than 2-fold upregulation of IMPDH2 mRNA (Supplementary Fig. 2b). Another key enzyme for de novo GTP biosynthesis, GMPS, was also upregulated and showed a high correlation with IMPDH2 expression levels (Supplementary Fig. 2b, c). Interestingly, expression of HPRT1 mRNA, an essential gene for the salvage GTP pathway, was downregulated, while APRT and ADK mRNA, critical genes for the salvage ATP pathway, were reciprocally upregulated showing significant correlation with IMPDH2 levels (Supplementary Fig. 2b, c). Immunodetection confirmed overexpressed IMPDH2 in murine glioma, human PDX GBM tissues, established GBM cell lines and GBM stem cells (Fig. 2c and Supplementary Fig. 3). Four glioma patient cohorts (total 426 specimens) showed a positive correlation of IMPDH2 with increased glioma malignancy (Fig. 2d) and negative correlation with patient survival (Fig. 2e). While the mutational status of isocitrate dehydrogenase (IDH) is one of the most critical biomarkers for molecular classification and patients are in general expected to survive longer16–18,31, high IMPDH2 expression appeared to confer poor survival even in IDH mutated glioma patients. IMPDH1 levels were marginal in the tested glioma specimens (Fig. 2c, d and Supplementary Fig. 3d–g). These results indicate that gliomagenesis is associated with upregulation of IMPDH2, but not IMPDH1. In summary, the upregulation of IMPDH2 and its association with systemic changes of genes critical for de novo and salvage GTP synthesis in GBM is consistent with a direct and primary effect of metabolic reprogramming of GTP biosynthesis on GBM.
IMPDH2 reprograms GTP metabolism in GBM
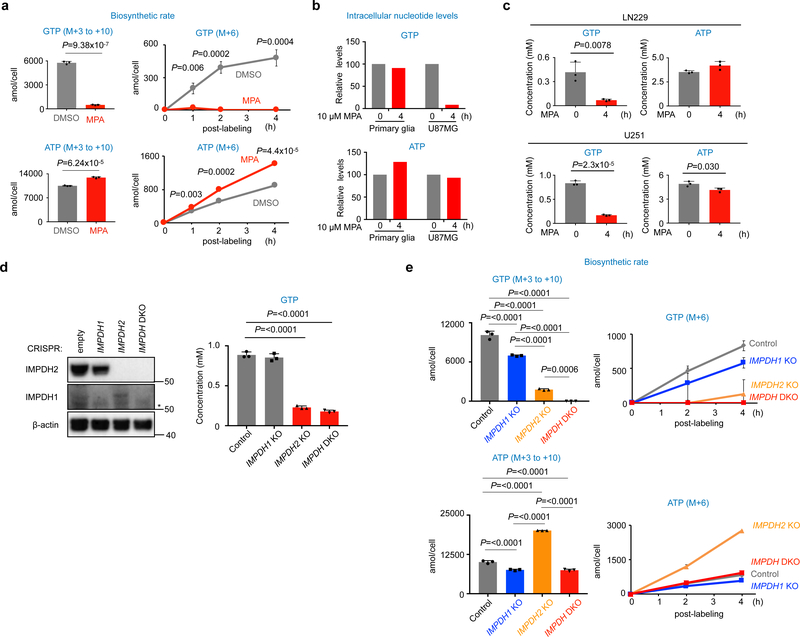
Next, we assessed the impact of pharmacological and genetic inhibition of IMPDH2. Treatment with mycophenolic acid (MPA), a pan IMPDH inhibitor23,32, dramatically decreased de novo GTP biosynthesis in GBM cells, but not in primary glia (Fig. 3a–c). GTP biosynthesis was moderately decreased in IMPDH1 knockout (KO) U87MG cells, whereas IMPDH2 KO markedly decreased GTP biosynthesis. Consequently, double knockout of both isotypes (DKO) led to a greater decrease in GTP biosynthesis and concentrations (Fig. 3d, e). The results suggest that upregulation of IMPDH2 reprograms the nucleotide metabolism towards de novo GTP biosynthesis in GBM.
Fig. 3 |. IMPDH2 reprograms GTP-metabolism in GBM.
a, Biosynthetic rate of GTP, but not ATP, is decreased by pharmacological inhibition of IMPDH in U87MG cells. Isotopomer distribution (M+6) of GTP and ATP from [U-13C]glucose is shown in right. Data are presented as mean+s.d. n=3 biologically independent samples. Unpaired two-sided Student’s t-test.
b, GTP levels were decreased by 4 h of MPA treatment in U87MG cells. n=1 experiment.
c, Pharmacological inhibition of IMPDH activity leads to acute decrease of GTP concentration, but not ATP in GBM cells. The indicated cells were treated with 10 μM MPA for 4 h and GTP and ATP concentrations were quantified by HPLC for U251 cells and CE-MS for LN229 cells. Data are presented as mean+s.d. n=3 biologically independent samples. Unpaired two-sided Student’s t-test.
d, The indicated IMPDH KO U87MG cells were generated by the CRISPR/Cas9 system as in the Method. Western blot shows the upregulation of IMPDH1 in IMPDH2 KO U87MG and LN229 cells. Cells were maintained with 100 μM guanosine supplemented media, which was replaced to DMEM/10% dialyzed FBS media 24 h before the assay. GTP levels were quantified by HPLC. Data are presented as mean+s.d. n=3 biologically independent samples. One-way ANOVA. Western blot analysis was performed at least twice.
e, IMPDH2 KO and IMPDH DKO U87MG decreased biosynthetic rate of GTP. The assay was performed as in (a). Data are presented as mean+s.d. n=3 biologically independent samples. One-way ANOVA.
IMPDH2 upregulation promotes GBM proliferation in vitro
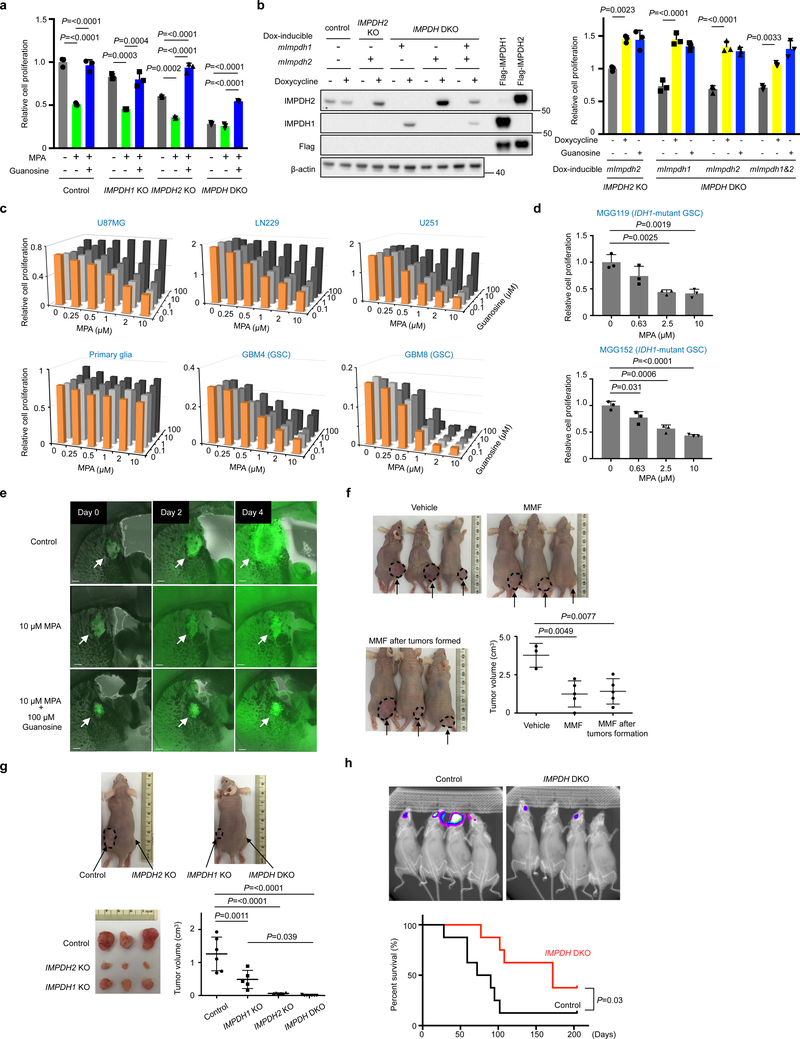
Although IMPDH2 KO decreased proliferation, the cells were still responsive to MPA, presumably owing to the compensation by IMPDH1 (Fig. 4a). In support of this notion, IMPDH DKO further decreased cell proliferation and sensitivity to MPA treatment (Fig. 4a, and Supplementary Fig. 4a). To test this directly, we compared doxycycline (Dox)-dependent ectopic expression of IMPDH1 or IMPDH2 in IMPDH DKO. Either equivalently rescued cell proliferation (Fig. 4b). These data suggest that GBM cells compensate for IMPDH2 loss by increasing IMPDH1 and, thus, pan-inhibition of IMPDH would be critical to suppress GBM growth.
Fig. 4 |. IMPDH2 promotes growth of GBM cells in vitro and in vivo.
a, IMPDH2 KO and IMPDH DKO U87MG cells decrease cell proliferation and MPA sensitivity. Data are presented as mean+s.d. n=3 biologically independent samples. One-way ANOVA. Western blot showing inducible expression of mouse (m)Impdh1 and mImpdh2, which are insensitive to IMPDH sgRNA-mediated targeting.
b, Reconstitution by doxycycline (Dox)-inducible mouse (m)Impdh1 and mImpdh2, which are insensitive to IMPDH sgRNA-mediated targeting, rescue the cell proliferation defect of IMPDH2 KO and IMPDH DKO U87MG cells. Data are presented as mean+s.d. n=3 biologically independent samples. One-way ANOVA. n=1 experiment for Western blot analysis.
c, MPA treatment suppresses GBM cell proliferation, but not primary glia. Data are shown as an average of 2 biologically independent samples.
d, MPA treatment suppresses IDH1-mutated GBM stem cell proliferation. Data are presented as mean+s.d. n=3 biologically independent samples. One-way ANOVA.
e, Growth of murine GSC in brain tissue is inhibited by MPA. GFP-expressing murine GSC brain explant slices in immunocompetent mice were cultured with or without 10 μΜ MPA and 100 μM guanosine. Scale bar indicates 300 μm. n=1 experiment. Similar MPA responses were confirmed in Supplementary Figure 5a.
f, Clinically used IMPDH inhibitor (mycophenolate mofetil, MMF23,27,32) suppresses GBM growth in vivo. MMF was orally administrated (120 mg/kg) to immuno-compromised mice after subcutaneous implantation of U87MG cells. Vehicle (n=3), MMF treatment (n=5). In one group, MMF treatment was initiated after tumor sizes reached 200 mm3. Mice were assessed at day 35. Arrows and dot circles indicate tumors. Data are presented as mean±s.d. One-way ANOVA.
g, IMPDH2 KO and IMPDH DKO significantly decrease in vivo tumorigenic activity. The indicated U87MG cells were injected subcutaneously in immunocompromised mice and assessed at day 37. Control (n=6), IMPDH1 KO (n=5), IMPDH2 KO (n=6), IMPDH DKO (n=7). Arrows and dot circles indicate tumors. Data are presented as mean±s.d. One-way ANOVA.
h, Orthotopic xenografts derived from IMPDH DKO U87MG cells were less progressive and mice survived longer than those with wildtype U87MG (upper). Bioluminescence imaging was performed 4 weeks after the implantation. Data are from Control (n=8) and IMPDH DKO (n=8). Log-rank test (two-sided) was used for the statistical analysis.
Moreover, IMPDH inhibitors, such as MPA, VX-49733 and Mizoribine34 decreased proliferation of all the tested GBM cells and GBM neurospheres, regardless of IDH1 mutational status (Fig. 4c, d and Supplementary Fig. 4b, c). In contrast, MPA treatment did not affect proliferation of primary glia. It is noteworthy that anti-GBM effect of Mizorbine, a pro-drug requiring phosphorylation by adenosine kinase34, required much higher concentration than MPA and those observed in the other type of cancers 28, presumably due to the differential drug metabolism in glioma.
The effect of IMPDH inhibitors was abrogated by high exogenous guanosine concentrations above 10 μM, two orders of magnitude higher than normal blood or cerebrospinal fluid concentrations35,36. To assess the contribution of the salvage pathway to cell proliferation, we generated HPRT1-deficient GBM cells. The HPRT1 KO U87MG, LN229 and A172 cells were completely resistant to 6-thioguanine (6-TG), while high concentration of 6-TG suppressed growth of the parental cells, indicating that these cells have the residual HPRT1 activity (Supplementary Fig. 4d). However, the proliferation rate of HPRT1 KO U87MG, LN229 and A172 cells was comparable to their parental cells (Supplementary Fig. 4d) suggesting that the salvage GTP biosynthesis is dispensable for GBM cells in contrast to the de novo GTP biosynthesis. Interestingly, in line with the restriction in salvage GTP biosynthesis in vitro (Supplementary Fig. 1j), guanosine rescue worked marginally for GSCs in culture dish and brain explant (Fig. 4c, e and Supplementary Fig. 5a), implying a potential metabolic vulnerability of GSC.
IMPDH requirement of in vivo GBM growth and malignancy
In accordance with the in vitro and ex vivo analyses, pharmacological inhibition of IMPDH suppressed tumor formation and progression of subcutaneously xenografted U87MG cells (Fig. 4f and Supplementary Fig. 5b). IMPDH2 KO U87MG cells displayed severely decreased tumorigenic activity and IMPDH DKO failed to form tumors (Fig. 4g and Supplementary Fig. 5c). However, in an orthotopic GBM model, IMPDH2 KO U87MG could form tumors that grow even faster than parental U87MG cells (Supplementary Fig. 5d). Subsequently, we observed significant upregulation of IMPDH1 in IMPDH2 KO U87MG, suggesting a mechanism explaining the tumor growth (Supplementary Fig. 5d). In contrast, IMPDH DKO significantly impaired tumor growth in the brain (Fig. 4h), corroborating the compensatory expression of IMPDH1 upon IMPDH2 depletion. Taken together, these results reveal the essential role of IMPDH activity in the maintenance and proliferation of GBM cells in vivo.
IMPDH2-mediated GTP biosynthesis fuels rRNA and tRNA synthesis
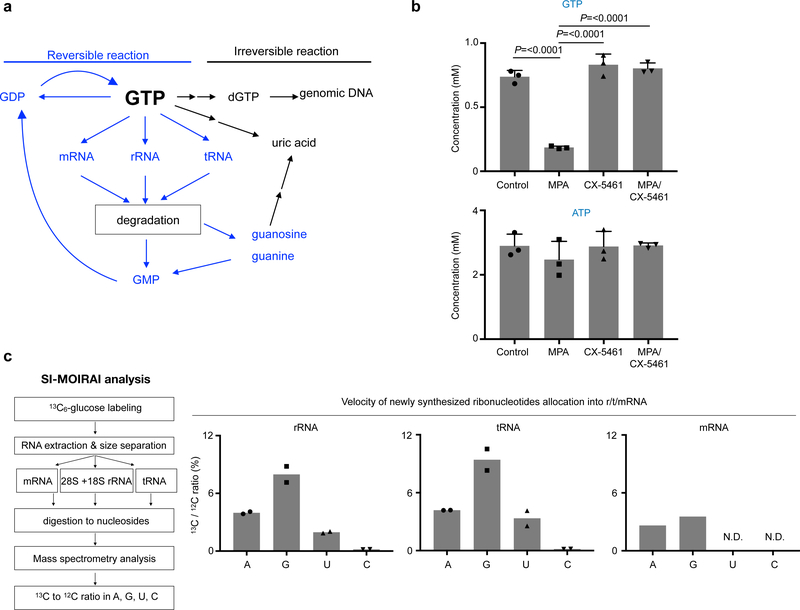
The primary metabolic fates of GTP are degradation to uric acid, deoxygenation to dGTP for DNA synthesis, or substrate for RNA synthesis (Fig. 5a). In GBM, we found that GTP is rapidly turned over in contrast to other ribonucleotides(Fig. 1c, Supplementary Fig. 1c). Inhibition of IMPDH pathway acutely depleted GTP, despite nearly 80% of cells being in a non-S-phase (Fig. 3a–c, Supplementary Fig. 6a, b). Using [U-13C]glucose flux analysis, 13C-labeled dGTP as well as uric acid—a catabolic product of GTP—were almost undetectable (Supplementary Fig. 6c). This suggests that the IMPDH pathway-derived GTP is used in some process(es) other than DNA synthesis or catabolism in the GBM cells. Importantly, treatment with specific inhibitors of rRNA synthesis—CX-546137 and BMH-2138—completely blocked the MPA-induced decrease of GTP, indicating that RNA polymerase I (Pol I)-dependent transcription drives GTP consumption (Fig. 5b, Supplementary Fig. 6d).
Fig. 5 |. SI-MOIRAI analysis identified metabolic fate of GTP for rRNA and tRNA synthesis.
a, A schematic diagram of reversible and irreversible reaction of GTP metabolism.
b, GTP depletion by IMPDH inhibition is abolished by Pol I inhibition. GTP and ATP concentrations quantified by HPLC in 10 μM MPA (4 h) and 1 μM CX-5461 (CX) (5 h including 1 h pretreatment)-treated U87MG cells. Data are presented as mean+s.d. n=3 biologically independent samples. One-way ANOVA.
c, A schematic flow of SI-MOIRAI analysis (left). Metabolic utility of newly synthesized nucleotides into r/m/tRNA. Ratio of the newly synthesized 13C-labeled nucleosides to total nucleosides in indicated RNA species are shown as percentile (right). n=2 biologically independent samples for r/tRNA, n=1 sample for mRNA. N.D. stands for not detectable.
To extend this observation, we established a tracking method for the metabolic fate of ribonucleotides termed Stable-Isotope Measure of Influxed Ribonucleic Acid Index (SI-MOIRAI) (Supplementary Fig. 6e). The SI-MOIRAI is a combination of the [U-13C]glucose flux assay and mass-spectrometry, enabling quantification of the newly synthesized A, U, G, C into RNA under steady state condition. Strikingly, our SI-MOIRAI-based steady state analysis showed that the utilization rate of 13C-labeled GTP is two-fold higher than that of ATP in rRNA and tRNA, but not in mRNA (Fig. 5c). Furthermore, 13C-labeled pyrimidine nucleotides exhibited a significantly lower utilization rate (Fig. 5c). These results suggest that GBM cells require IMPDH2-dependent GTP biosynthesis to support primarily rRNA and tRNA synthesis, whereas mRNA synthesis is not dependent on this change in nucleotide metabolism.
IMPDH2-mediated GTP biosynthesis is required for Pol I and Pol III transcription
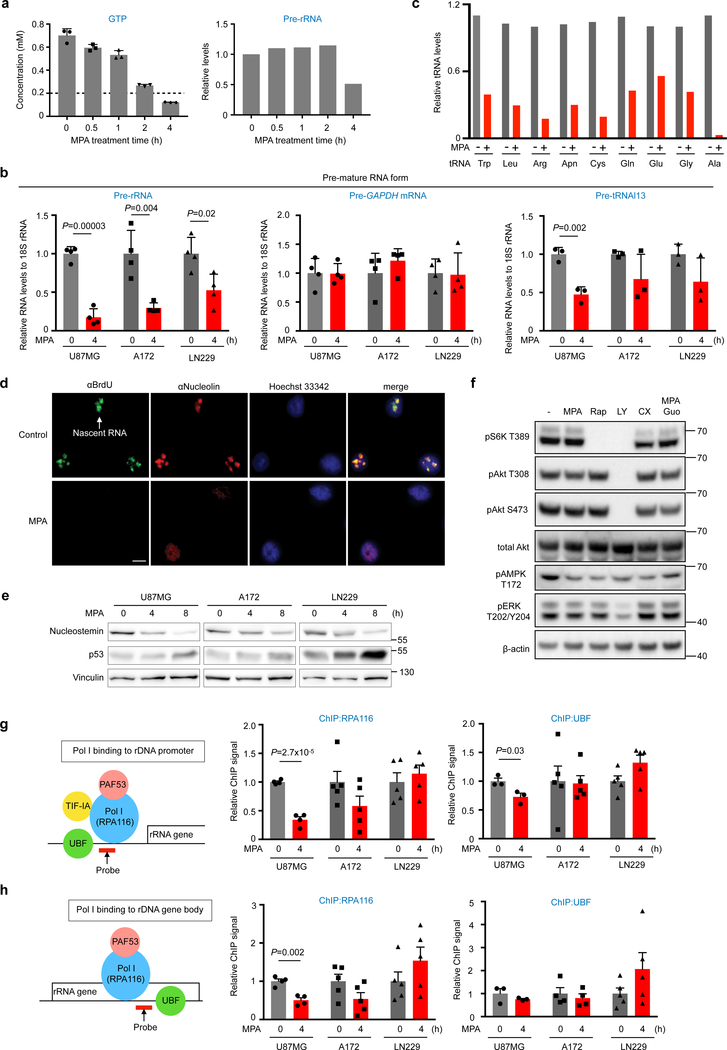
Many tumor cells increase Pol I activity6,10,39 and GTP-dependent Pol I activation has been shown in several types of tumors40–42. Consistent with these previous studies, Pol I activity is upregulated in U87MG compared to primary glia (Supplementary Fig. 7a). Furthermore, we found an additional link between GTP and nucleolar transcription in GBM. When cellular GTP concentrations decreased to below 100 μM, pre-rRNA (Pol I transcript) and pre-tRNAl13 (Pol III transcript) expression levels in GBM cells were diminished, while pre-GAPDH mRNA (Pol II transcript) was unaffected (Fig. 6a, b). Mature tRNAs were also downregulated (Fig. 6c and Supplementary Fig. 7b), suggesting stress-dependent tRNA cleavage43.
Fig. 6 |. De novo GTP biosynthesis is critical for rRNA and tRNA transcription in GBM.
a, GTP concentrations were quantified by HPLC and pre-rRNA levels were analyzed by quantitative (Q)-PCR in 10 μM MPA-treated U87MG cells. Data are presented as mean+s.d. n=3 biologically independent samples for GTP measurements, n=1 biologically independent sample normalized from three technical replicates for Q-PCR.
b, Nascent transcripts of the indicated genes were analyzed by Q-PCR. Data are presented as mean+s.d. Biologically independent samples for Pre-rRNA (n=4), Pre-GAPDH mRNA (n=4), and Pre-tRNAl13 (n=3). Unpaired two-sided Student’s t-test.
c, Mature form of the indicated genes was analyzed by Q-PCR. n=1 biologically independent sample normalized from three technical replicates.
d, IMPDH inhibition diminishes nucleolar transcriptions and evokes nucleolar stress response. U87MG cells were treated with MPA for 4 h and 5-fluorouridine for 20 min. Nascent RNA and nucleolus were visualized by anti-BrdU and anti-nucleolin antibodies, respectively. Data are representatives from n=2 independent experiments. Scale bar indicates 20 μm.
e, Acute inhibition of IMPDH activity in the indicated GBM cells decrease nucleostemin levels, one of the nucleolar stress responses, with concomitant increase of p53 protein. Data are representatives from n=3 independent experiments.
f, Acute inhibition of IMPDH activity has marginal effect on the PI3K, mTOR and ERK/RSK signaling pathways. Western blotting of lysates from U87MG cells, treated with 10 μM MPA, 100 μM guanosine (Guo), 10 nM Rapamycin (Rap), 50 μM LY294002 (LY), 1 μM CX-5461(CX) for 4 h. MPA responses were confirmed in two independent experiments.
g, h, Coordinated suppression of RNA Pol I upon IMPDH inhibition in GBM. The indicated GBM cells were treated with or without MPA for 4 h. Binding of RPA116, a subunit of Pol I and UBF, an architectural factor to promoter (f) and gene body (g) of rRNA gene was assessed by ChIP assay. Data are presented as mean+s.e.m. n=4 biologically independent samples for RPA116 of U87MG (g, h) and UBF of A172 (h), n=3 for UBF of U87MG (g, h), n=5 for RPA116 (g, h) and UBF (g) of A172, RPA116 and UBF of LN229 (g, h). Unpaired two-sided Student’s t-test.
Indeed, under conditions of low Pol I and III expression, MPA treatment triggered the nucleolar stress response44,45, i.e. delocalization of nucleolin, induction of p53 pathway and degradation of nucleostemin, a nucleolar marker of stem and cancer cells whose stability requires GTP binding (Fig. 6d, e and Supplementary Fig. 7c–f). However, no gross changes in the PI3K, mTOR and ERK/RSK signaling were detected (Fig. 6f).
Chromatin immunoprecipitation (ChIP) analyses revealed that, whereas gene occupancy of the architectural factor, UBF, was unaffected by MPA regardless of the cell line, MPA treatment decreased promoter and gene body binding of Pol I in U87MG and A172, but not in LN229 cells (Fig. 6g, h). MPA-treated U87MG and A172 cells, both p53 wildtype, respond with p53-mediated inhibition of Pol I transcription initiation46, whereas in p53-mutated LN229 cells, elongation is blocked, leading to stalling of Pol I. In line with this notion, a Pol I minigene assay showed severely impaired transcription initiation in response to MPA in U87MG cells, but not in LN229 cells (Supplementary Fig. 7g). These data show that inhibition of IMPDH represses rRNA synthesis and induces nucleolar stress, even in GBM cells lacking functional p53.
IMPDH2 upregulation is critical for nucleolar transformation in GBM
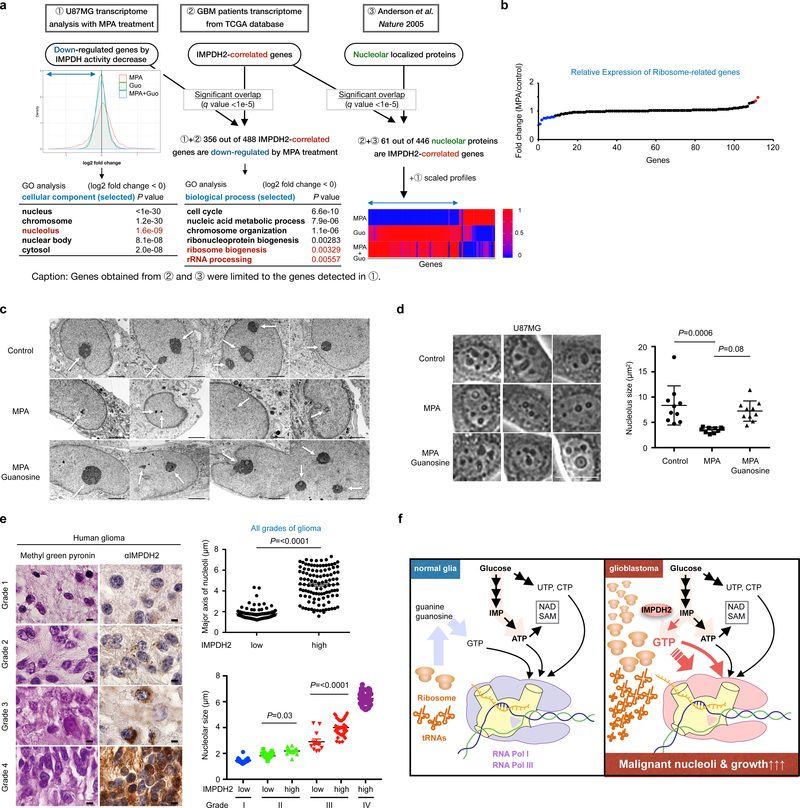
Next, we performed two-dimensional transcriptome screening for IMPDH2-dependent gene expression networks (Fig. 7a). In the first dimension, we set out to identify genes regulated by IMPDH activity. We analyzed the transcriptome of U87MG cells and identified 3,675 genes whose expression were down-regulated after IMPDH inhibition. Interestingly, although the IMPDH inhibition showed a negligible effect on ribosomal protein coding genes (Fig. 7b), a significant change was observed in a subset of nucleolar localizing proteins, including ribosome processing factors, and nucleolar architecture regulators (Fig. 7a). In the second dimension, we sought genes correlated with IMPDH2 expression. We extracted 721 genes whose expression are correlated with IMPDH2 mRNA expression from the transcriptome of GBM patients. 356 genes are overlapped with the IMPDH-associated down-regulated genes (Fig. 7a). The 61 genes correlated with IMPDH2 expression in GBM patients as well as responded to MPA treatment in U87MG cells are implicated in nucleolar function and morphology (Fig. 7a).
Fig. 7 |. IMPDH2 upregulation is critical for nucleolar transformation in GBM.
a, Two-dimensional (2D) transcriptome screening reveals IMPDH2-network and its association with nucleolar activity in GBM. The Gene Ontology (GO) analysis of IMPDH activity-associated genes (left) and the genes associated with both IMPDH2-expression and IMPDH-activity (right) are shown in the tables. Bottom right show the expression of genes encoding nucleolar localizing proteins that are correlated with IMPDH2 mRNA expression in the GBM patient transcriptome. b, 10 μM MPA treatment (24 h) marginally affect the expression of genes encoding ribosomal subunits in U87MG. Red color is for >1.3-fold increased genes, while blue for the >1.3-fold decreased genes. n=1 experiment.
c, U87MG cells were treated with or without 10 μM MPA, guanosine for 72 h and analyzed by transmission electron microscope (x 6,000). Scale bar indicates 2 μm. n=1 experiment.
d, IMPDH inhibition leads to decrease nucleolar size in U87MG. The area of nucleoli from 10 cells was assessed at 24 h of 10 μM MPA, 100 μM guanosine. Data are presented as mean±s.d. Average nucleolus sizes from 10 different cells in each group were indicated. The results were confirmed in two independent experiments. One-way ANOVA. Scale bar indicates 50 μm.
e, IMPDH2 upregulation and glioma malignancy are positively correlated with nucleolar hypertrophy in glioma patients (left). Methyl green pyronin and IMPDH2 staining on human glioma tissue array. Upper right: major axis of nucleoli in human glioma tissue array (all grades of glioma) according to IMPDH2 expression levels (low: n=115, high: n=109 specimens). Lower right: major axis of nucleoli in grade I to IV glioma specimens according to IMPDH2 expression levels (Grade I, low: n=21, Grade II, low: n=61, high: n=16, Grade III, low: n=12, high: n=41, Grade IV, high: n=41 specimens). Data are presented as mean±s.e.m., Mann-Whitney test (two-sided) (upper right), one-way ANOVA (lower right). Scale bar indicates 50 μm.
f, A schematic working model. Among four ribonucleotides, GTP biosynthesis from glucose is elevated in glioblastoma cells to fuel rRNA and tRNA via Pol I and Pol III transcription for their malignant growth. Less proliferative glial cells recycle ribonucleotides.
Remarkably, pharmacological inhibition of IMPDH2 activity led to decreased nucleolar density and size, and more rounded, ring-shape nucleoli as compared to the control cells. Guanosine supplementation and ectopic expression of IMPDH2 reverted the phenotype back to enlarged and irregular shaped nucleoli (Fig. 7c, d and Supplementary Fig. 7h, i). Furthermore, the nucleolar size in IMPDH2 KO U87MG tumor specimens was significantly decreased compared to wildtype U87MG tumors in vivo (Supplementary Fig. 7j) and ex vivo (Supplementary Fig. 7k). In human glioma patient specimens, IMPDH2 expression was significantly correlated with nucleolar enlargement in the grade II and grade III gliomas (Fig. 7e). Together, these data suggest that IMPDH-mediated GTP production directly contributes to the formation of enlarged, irregular nucleoli in GBM cells.
Discussion
Increased rRNA synthesis47 and nucleolar hypertrophy1,2 have both long been recognized as features of malignant transformation. Our data, by revealing that upregulation of IMPDH2 in GBM cells drives de novo GTP biosynthesis, enhances rRNA and tRNA synthesis, and stimulates nucleolar hypertrophy, connects these two long observed features of cancer (Fig. 7f).
Our primary result establishing a mechanistic connection between GTP metabolism, IMPDH2, nucleolar hypertrophy and malignancy prompts a reappraisal of the published literature on IMPDH. The anti-tumor effect of IMPDH inhibitors has been known for over 50 years25,26, but the primary effect of IMPDH inhibition on tumor cell growth has been poorly understood. There are contradictory models27 including integrin dysfunction, caspase-3 activation, PPARγ-mediated differentiation, decreased signaling of Ras/ERK48,49 and mTOR50. The most widely accepted model is that IMPDH inhibition suppresses DNA synthesis and mitotic progression of T cells that enter in S-phase51. However, IMPDH inhibition causes cell cycle arrest at G1-phase in many tumors, and the progression to S-phase and the following mitotic suppression under GTP-depletion depends on cell type or cellular context (e.g. mutation TSC2)28. In the present study, using a newly developed and previously unavailable method, SI-MOIRAI, which overcomes the issue of pleiotropic effect caused by GTP depletion, we have unveiled the metabolic fate of newly synthesized GTP to rRNA and tRNA. The results of SI-MOIRAI explain why inhibition of Pol I activity, by two Pol I inhibitors, completely suppressed the MPA-dependent GTP depletion (Fig. 5b and Supplementary Fig. 6d). Decreased GTP concentration by MPA suppresses both Pol I and Pol III transcription, while Pol II transcription is unaffected. Taken together, our studies provide the direct evidence elucidating the primary role of IMPDH2 in providing metabolic substrates supporting de novo-synthesized GTP to nucleolar rRNA and tRNA synthesis in GBM.
The results also highlight the qualitative differences in rRNA synthesis between GBM and primary glia. In GBM cells, Pol I transcription requires IMPDH2-dependent de novo GTP biosynthesis, while primary glia use the salvage pathway for Pol I transcription. All differentiated, less proliferative, or quiescent cells, such as primary glia, have a nucleolus and continuously produce ribosomes. This observation has at least two major implications: 1) insight into how metabolically inactive cells maintain ribosome biogenesis, one of the most anabolic cellular processes, and 2) the therapeutic potential of IMPDH pathway to suppress ribosome biogenesis in tumor cells but not in normal tissue.
Our study raises a number of important questions. The mechanism of IMPDH2 upregulation in GBM is of particular basic importance. Previous reports show that cMyc increases IMPDH1 and IMPDH229,30,52. But, cMyc itself may not explain the rather selective upregulation of IMPDH2 in GBM, and, therefore, there are likely additional levels of regulation for IMPDH2 upregulation. Other questions include what is mechanism regulating GTP-dependent Pol I and Pol III transcription, whether nucleolar enlargement is a passive structural manifestation, whether the nucleolar changes have an active role in directly promoting tumor growth, and could existing or more selective IMPDH inhibitors benefit and prolong the GBM patients’ survival while minimizing their strong immunosuppressive effects and other toxicities32,53,54? Further studies understanding tumor-specific regulation of IMPDH2 coupled with improved design of currently available IMPDH inhibitors32,55 may open new avenues developing effective therapy for GBM and other types of cancers.
METHODS
Materials
Anti-IMPDH1 (WH0003614M1), BrdU (BMC9318) antibodies (Sigma-Aldrich); anti-IMPDH2 (ab131158), β-actin (ab6276), p21 (ab109199), GAPDH (ab9485), HPRT1 (ab10479) antibodies (Abcam); anti-β-actin (#3700), p-Akt (T308, #4056), (S473, #4060), p-S6K (T389) (#9234), p-ERK (#4370), p-AMPK (#2535), p-p53 (#9284), Nucleostemin (#9495) antibodies (Cell Signaling Technology); anti-pan-Akt (sc-81434), p53 (sc-126), Nucleolin (sc-13057) antibodies (Santa Cruz Biotechnology); anti-GMPS antibody (A302–417A)(Bethyl laboratory); anti-HPRT1 (VMA00483) antibody (Bio-Rad); anti-Nucleostemin (MAB4311) antibody (Millipore); anti-Vinculin (BZL03106) antibody (Biozol Diagnostica).
Cell Culture
U87MG (ATCC line), LN229, U251, and A172 cells were cultured in Dulbecco’s modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS). GBM patient-derived neurospheres and GSC were isolated and cultured as previously described as previously described56–59. For primary glia isolation, the cerebral cortex from new born mice (2 or 3 days-old) was collected, homogenized and harvested with Leibovitz’s L-15 medium. After washing with PBS, homogenates were treated with DNase and Trypsin. Cells were cultured in Minimum Essential Medium (MEM/EBSS, GE Healthcare) containing 10% horse serum (HS). Medium was changed every three days. Human astrocyte (ScienCell Research Laboratories, #1800) were maintained in Astrocyte medium (#1801), which enriches growth factors enabling high proliferation, and habituated in MEM/EBSS/10%HS for two weeks before the assay.
Analysis of Gene Expression in RCAS/tv-a Murine GBM Models
Using Gene Expression Omnibus (the accession number is GSE45874), which harbors total 72 murine samples produced by the RCAS/tv-a system60, we extracted signal intensity data of Illumina BeadChip after normalization by the lumi R package. With knowledge-based search about gene symbols and ids, we obtained 28 probes of 17 genes annotated as a gene in purine synthetic pathway after manual curation (Supplementary Table 8). We applied the prcomp R package for PCA analyses. To detect the genes, which were differentially expressed between normal and tumor samples, we used the P values calculated by the Wilcoxon rank sum test implemented in R (Wilcox.test) after Bonferroni correction.
Quadruple conditional knockout glioma mouse model
Gfap-CreER;Ptenfl/fl;Rb1fl/fl;Trp53fl/fl;Rbl1−/− mice were obtained and induced for gliomagenesis as in59. The brain was analyzed at 8–12 weeks after injection. Mice were maintained under pathogen-free conditions.
IMPDH protein quantification by Western Blot Analysis
For the quantification of IMPDH amounts, His-tagged recombinant human IMPDH proteins were purified in E.coli using Talon Metal Affinity Resin (Clontech). These proteins were subjected to western blot analysis together with cell lysates and cellular IMPDH amounts were calculated by the standard curves. To calculate the concentration, cell numbers were counted and cell volumes were measured by Packed Cell Volume tubes (Techno Plastic Products).
Virus Production and Transduction
Human wildtype and IMPDH1 and IMPDH2 cDNA were cloned into the pLenti CMV Hygro DEST vector (Addgene, #17454), and the virus was produced by co-transfection of pMD2.G (Addgene, #12259) and psPAX2 (Addgene, #12260) to HEK293T. For transduction, 1×105 cells per 12-well plate were treated with virus-containing media and 8 μg/ml polybrene, and centrifuged at 1,200 g for 20 min. The medium was changed with the appropriate fresh medium on the next day. Cells were selected with hygromycin B for at least one week.
Nucleotide measurement by high performance liquid chromatography
Nucleotide measurement was performed as described previously with some modifications61. To calculate the concentration, cell numbers were counted and cell volumes were measured by Packed Cell Volume tubes.
Metabolome analysis
All stable isotopes were purchased from Cambridge Isotope Laboratories, Inc. Cells were seeded on a 100 mm dish and cultured overnight. On the next day, culture media were replaced to [U-13C] glucose containing culture media (DMEM/10% FBS/4.5 g/L [U-13C] glucose for U87MG, MEM/10% HS/1 g/L [U-13C] glucose for primary glia) or 15N5-guanosine-containing culture media (DMEM/10% dialyzed FBS or MEM/10% HS/1 μM guanosine). For neurospheres, neural stem cell-supporting culture medium (NSM) containing 17.5 mM [U-13C] glucose, 2.5 mM 13C5, 15N2-glutamine was used. Cells were harvested at indicated times. Extraction of metabolites was performed as previously described62. Capillary electrophoresis-time-of-flight mass spectrometry (CE-TOFMS) experiments were performed using an Agilent G7100A Capillary Electrophoresis instrument (Agilent technologies), an Agilent 6224 TOF LC/MS system (Agilent technologies), an Agilent 1200 series isocratic HPLC pump, a G1603A Agilent CE-MS adapter kit and a G1607A Agilent CE-electrospray ionization (ESI)-MS sprayer kit. For CE-TOFMS system control and data acquisition, we used an Agilent MassHunter software. Detailed conditions of CE-TOFMS-based metabolome analysis were described63,64. Capillary ion chromatography (IC)-MS analysis was performed using a Dionex ICS-5000+ system equipped with a Q Exactive Orbitrap MS system (Thermo Fisher Scientific).
Cell Proliferation Analysis
To measure cell proliferation, relative cell numbers were measured using crystal violet staining for adherent cells. Cells were fixed with 4% paraformaldehyde in PBS, stained with 0.04% crystal violet aqueous solution for 15 min and dissolved in 1% SDS. The absorbance was measured at 595 nm. For glioblastoma stem cells, cells were assayed using CellTiter 96 Aqueous One Solution Cell Proliferation assay (MTS) (Promega) according to the manufacturer’s instructions.
Establishment of IMPDH knockout cells by CRISPR/Cas9 system and mouse IMPDH-expressing cells
For CRISPR/Cas9-mediated gene knockout, gRNAs were designed by CRISPR design program (http://crispr.mit.edu/) (GTGTCGTCGGGTGTGTAGCG for IMPDH1, ACATCCCGCACGCGATCCTT or GATACCGCAGAAACCATGCC for IMPDH2, TCTTGCTCGAGATGTGATGA for HPRT1). gRNAs were subcloned into lentiCRISPRv2 (Addgene plasmid #52961). U87MG, LN229 and A172 cells were transduced with the lentivirus and selected by puromycin. To avoid compensatory responses, IMPDH KO cells were cultured with 100 μM guanosine. Single clones of IMPDH2 and IMPDH DKO U87MG cells were isolated by FACSAria sorting (BD Biosciences). To establish doxycycline-inducible gene-expressing cells, cDNA fragments of mImpdh1, 2 were subcloned into pTRIPZ (GE Healthcare). Transduced cells were treated with 1 μg/ml doxycycline for the ectopic gene expression.
Xenograft Assay
2.5 × 106 cells were subcutaneously injected into athymic Nude-Foxn1nu homozygous mice (Harlan) around 8 weeks of age with matrigel (Corning). Tumor volumes were measured twice a week and calculated using the equation of D x d x d x π/6 (D=larger diameter, d=smaller diameter). For Mycophenolate mofetil (MMF) treatment, mice were treated with MMF by oral gavage from the next day of injection. For the delayed treatment, mice were treated with MMF by oral gavage after the tumor size reached 200 mm3. MMF was dissolved in 0.5% methylcellulose/0.1% Tween 80 solution and 120 mg/kg of amounts were administrated orally. 0.5% methylcellulose/0.1% Tween 80 solution was used for vehicle treatment. When mice did not form tumors, those data were excluded from the statistical analysis. All of the animal experiments in this study is compliant with all relevant ethical regulations regarding animal research.
Orthotopic implantation
Implantation of cells into the brains of female athymic Nude-Foxn1nu homozygous mice for U87MG/GFP-luciferase cells, C57BL/6J mice for GSC cells was performed as described previously56. Carestream (Kodak) MultiSpectral FX (2D) was used to visualize tumors for U87MG cells after intravenous injection of Luciferin (Gold Biotechnology). For the metabolite quantification experiments, 2 × 106 viable GFP-NSCs were implanted contralaterally. All animal experiments were approved by the Animal Care and Use Committees of University of Cincinnati, Keio University and Hiroshima University.
Ex vivo GBM analysis
For metabolite quantification experiments, brains were removed 5 days after implantation and 200-μm coronal slices were prepared with the use of a LeicaVS1200 vibratome (Leica). Explants were placed on Millicell-CM culture inserts (Merck Millipore) in glass-bottom plates, allowed to recover for 1 h in NSM at 37˚C under a humidified atmosphere of 5% CO265. Images of the slices were acquired with a Fluoview FV10i inverted confocal microscope (Olympus), after which the explants were switched to either fresh NSM or glucose-free NSM supplemented with [U-13C] glucose up to 4.5 g/l. After a 3 h incubation, explants were frozen and stored at −80˚C until analysis. For drug treatment experiments, 150 μm explants were prepared 8 days after implantation, allowed to stabilize for 1 h and then switched to fresh NSM or NSM containing 10 μM MPA, 10 μM MPA and 1 μM guanosine or 10 μM MPA and 10 μM guanosine. Medium with or without drugs was replenished daily. At the end of the study, slices were fixed in fixed overnight with 4% paraformaldehyde, embedded in paraffin, and sectioned at a thickness of 3 μm.
Matrix-assisted laser desorption/ionization-MS imaging acquisition
Matrix-assisted laser desorption/ionization (MALDI) imaging analyses were performed as described previously66,67. Briefly, thin sections (8 μm) of the brain sections were prepared with a cryomicrotome (CM3050, Leica). Sections were attached onto indium tin oxide–coated glass slides (Bruker Daltonics) and were coated with 9-aminoacridine as the matrix (10 mg/mL, dissolved in 80% ethanol) by manually spraying with an artistic-brush (Mr. Hobby). The matrix was simultaneously applied to multiple sections compared in order to maintain consistent analyte extraction and co-crystallization conditions. MALDI imaging was performed using an Ultraflextreme MALDI-TOF/TOF mass spectrometer equipped with an Nd:YAG laser and a linear ion trap MS with a MALDI source (MALDI LTQ XL, Thermo Fisher Scientific) equipped with a nitrogen laser (337 nm; 60 Hz). Data were acquired in the negative reflectron mode with raster scanning at a pitch distance of 50 μm. For the TOF/TOF measurement, signals between m/z 50 and 1000 were collected and image reconstruction for both procedures was performed using FlexImaging 4.1 software (Bruker Daltonics). For the ion-trap instrument, the obtained spectral data were transformed to image data using ImageQuest 1.0.1 software (Thermo Fisher Scientific).
Immunohistochemistry
After deparaffinized, sections were pretreated in Cell Conditioning Solution (CC2) (Roche) for 44 min at 91˚C, then incubated with anti-IMPDH2 Ab (1:100 or 200), anti-IMPDH1 Ab (1:100) for 32 min at room temperature. After incubation signals were detected using UltraView DAB detection kit (Roche). For counterstaining, slides were treated in hematoxylin for 4 min and bluing reagent (Roche) for 4 min. These processes were run on the BenchMark ULTRA (Roche). Human brain glioma tissue arrays were obtained from three independent institutes (anonymous for blind review) and US Biomax (BS17016a, NGL961, GL482). IMPDH1 and IMPDH2 immunostaining intensity was scored by board-certified pathologists on a semiquantitative scale from 0 (negative) to 3 (strong). In figures, low expression group corresponds to the scale 0 and 1, high expression group corresponds to the scale 2 and 3.
Microarray
U87MG cells were treated with 10 μM of MPA or 100 μM guanosine for 24 h, then RNA was extracted with ISOGEN (Nippon gene). The Genechip Primeview Target Synthesis and Labeling Assay was used to create biotin-labeled target cDNA. Samples were subjected to Affymetrix Genechip Primeview Human Gene Expression Array. The microarray data were deposited in the Gene Expression Omnibus (GEO)(GSE124727).
IMPDH2 network analysis
By comparison of the signal strengths between MPA-treated and control U87MG microarray data, down-regulated genes were extracted as the genes affected by the inhibition of IMPDH activity. Gene expression profiles of 10133 genes with a single replicate were applied to the analyses. For GO analyses, p values obtained by a Fisher’s exact test (one-sided) using topGO were shown without any adjustment. For the significance of overlaps, p values obtained by a hypergeometric test after Bonferroni multiple correction were shown as q values. <1e-30 was used as the minimum value in the output of R topGO library due to the underflow problem. P values for the probability of overlaps (shows as q value <1e-5 in Fig.7) also contains the zero p values due to the underflow problem. To avoid an inappropriate description, we put caps of 1e-5 on q values, which at least can be computed precisely. The candidates of IMPDH2-correlated and nucleolar localized genes were obtained from The Cancer Genome Atlas (TCGA) GBM via GlioVis68 and the nucleolar proteome data69, respectively. The overlapped genes between each gene list were extracted for GO enrichment analysis using topGO (Bioconductor, R package version 2.32.0) and P value computation by hypergeometric test.
Chromatin immunoprecipitation (ChIP)
After formaldehyde-fixation (1% formaldehyde, 10 min at room temperature) and quenching with 125 mM glycine (5 min), cells were harvested in PBS and nuclei were prepared by successive incubation in buffer A (100 mM Tris-HCl [pH 8.1], 10 mM DTT), buffer B (10 mM HEPES [pH 7.5], 10 mM EDTA, 0.05 mM EGTA, 0.25% Triton X-100) and buffer C (10 mM HEPES [pH 7.5], 10 mM EDTA, 0.05 mM EGTA, 200 mM NaCl). Nuclei were disrupted in buffer D (50 mM Tris-HCl [pH 8.0], 10 mM EDTA, 1% SDS) and chromatin was sonicated in a Bioruptor Pico (Diagenode) to an average length of 200–500 bp. Chromatin was diluted with 5 volumes of ChIP-buffer (15 mM Tris-HCl [pH 8.0], 180 mM NaCl, 1.2 mM EDTA, 1.2% Triton X-100) and pre-cleared for 1 h at 4˚C on Protein A/G-Sepharose (GE Healthcare), that has been blocked with 1 mg/ml BSA and 1% gelatin from cold water fish skin (Sigma-Aldrich). Per ChIP reaction, approximately 20 μg chromatin were incubated with 1 to 2 μL of polyclonal antisera overnight at 4˚C followed by incubation with Protein A/G-Sepharose for 1 h. Immobilized complexes were washed two times in low salt buffer (20 mM Tris-HCl [pH 8.0], 150 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100) high salt buffer (20 mM Tris-HCl [pH 8.0], 500 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100), LiCl buffer (10 mM Tris-HCl [pH 8.0], 250 mM LiCl, 1 mM EDTA, 1% sodium deoxycholate, 1% NP-40) and TE buffer. DNA was eluted from the beads by incubation in 100 μL of 0.1 M NaHCO3/1% SDS for 10 min at room temperature. The crosslink was reversed by heating at 65˚C for 6 h and the DNA was purified using the ChIP DNA clean and concentrator kit (Zymo Research). Purified DNA was analyzed by qPCR (see Supplementary Table 10 for primer sets).
Real-time PCR analysis
Total RNAs were extracted using TRI REAGENT (Molecular Research Center) or TriFast Reagent (Peqlab) according to the manufacturer’s instruction. High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific) or SuperScript IV Reverse Transcriptase (Life Technologies) was used to prepare cDNA solution. For real-time PCR analyses, Maxima SYBR Green/ROX qPCR Master Mix (2X) (Thermo Fisher Scientific) was used according to the manufacturer’s instruction. Reactions were performed using the pre-set program of ABI 7500 Fast Real-Time PCR system (Thermo Fisher Scientific). For Pol I reporter gene assays, cells were transfected with plasmid pHrP2-BH, which harbors an artificial ribosomal minigene containing a 5’-terminal human rDNA fragment (from −411 to −375) and two rDNA terminator elements. After RNA isolation and cDNA synthesis, the reporter gene was detected with forward and reverse primers matching rDNA and plasmid sequences, respectively. All primers used are found in Supplementary Table 10.
Immunofluorescence and nascent RNA analysis
Cells grown on coverslips were fixed with 1% para-formaldehyde for 5 min, permeabilized with 0.5% Triton X-100 for 5 min and incubated with primary antibodies overnight at 4˚C. For labeling of nascent RNA, cells were pulse-labeled with 2 mM 5-fluoro-uridine (FUrd) for 20 min before fixation. Nucleolar proteins were immunostained with the indicated antibodies and labeled RNA was detected with anti-BrdU antibodies. After incubation overnight at 4˚C and washing with PBS, appropriate secondary antibodies coupled to fluorophores were added for 45 min at room temperature. Coverslips were stained with Hoechst 33342 to visualize nuclear DNA, mounted and pictures were taken with a Nikon fluorescence microscope and digitally recorded.
Tissue Microarray
Human glioma tissue array was constructed at the University of Kentucky, the University of California Los Angeles, Osaka City University after written informed consent. The experimental protocol was approved by the Institutional Review Board of each institute. We also purchased commercially available human glioma tissue arrays from US Biomax, Inc. (BS17016a, NGL961 and GL482). The study is compliant with all relevant ethical regulations regarding research involving human participants.
Patient-derived xenograft (PDX) model
Preparation of PDX models and infusion of [U-13C] glucose were performed as described previously70. Human orthotopic tumor samples were obtained from patients at the University of Texas Southwestern Medical Center after written informed consent and Massachusetts General Hospital under a protocol approved by the Institutional Review Boards. The study is compliant with all relevant ethical regulations regarding research involving human participants.
SI-MOIRAI
Total RNAs were extracted using miRNeasy kit (QIAGEN) and electrophoresed by 0.8% Native-Agarose gel for 40 min with 100V in TBE buffer with ssRNA lader and low range ssRNA ladder RNA size markers (Bio-Rad). RNAs (mRNA (>10,000 mer), 28S rRNA, 18S rRNA and tRNA) were excised with a razor blade, and extracted from the gel using NucleoSpin Gel and PCR Clean-up Kit (MACHEREY-NAGEL). RNA (20 μL) was heat (92˚C for 5 min), then chilled in the ice-water slush. To digest, the RNA samples were treated with 1 μL of antarctic phosphatase buffer (pH 7.0, New England BioLabs) and 1 μL (2 units) of nuclease P1 (Sigma), for 3 h at 50˚C. Then, the samples were treated with 1 μL (0.2 units) of venom phosphodiesterase (Worthington Biochemical) for 2 h at 37˚C, following the treatment with 10 μL (0.5 units, diluted by dH2O) of antarctic phosphatase (New England BioLabs) for 16 h at 37˚C. For mass spectrometry analysis, samples were mixed with 200 μL of 100% MeOH containing 25 μM each of internal standards (L-methionine sulfone, 2-(N-morpholino) ethanesulfonic acid and D-Camphor-10 sulfonic acid), filtered for 3 h at 4˚C with Ultra-free MC-PLHCC5K column (Merck Millipore), and evaporated.
Electron Microscopy
Specimens were immersed in 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (100 mL; pH 7.4) for 2 h at 4˚C. Specimens were then treated with 1% OsO4 for 2 h at 4˚C, and then with saturated uranyl acetate for 3 h at room temperature. Thereafter, specimens were dehydrated in a graded series of ethanol and embedded in epoxy resin. Ultrathin sections (70 nm thick) were made, counterstained with saturated uranyl acetate followed by lead citrate, and observed using a JEM-1400Plus electron microscope (JEOL).
Nucleolus morphology
Cells were cultured as indicated, then pictures were taken. At least 10 cells were assessed by Image J software (National Institutes of Health). For methyl green pyronin (MGP) staining, sections were rinsed in pH 4.8 buffer (acetic acid: 0.08 M, sodium acetate: 0.12 M) and incubated for 60 min in MGP solution (0.2% methyl green, 0.2% pyronin Y). Sections were again rinsed in pH 4.8 buffer, then dehydrated and mounted. After histological evaluation and taking pictures, at least 20 nucleoli were calculated by Image J.
Statistics and Reproducibility
The statistical analyses were performed using the unpaired two-sided Student’s t-test, one-way ANOVA and Mann-Whitney test using Microsoft Excel and GraphPad Prism 6 or 7 (GraphPad Software). ANOVA was used to compare more than two groups. Data were considered statistically significant at *P<0.05, **P<0.01. No statistical method was used to determine sample size. The experiments are repeated independently with similar results. For Fig.1a, a tumor sample was split into two and analyzed by IC-MS in two institutes, showing the similar results. For the flux analyses, nucleotide measurements by HPLC, cell proliferation assays and animal experiments, biologically independent samples were used, showing similar results. For SI-MOIRAI experiment (Fig. 6c), one of the data sets from duplicates is shown. For Fig. 7c, the electron microscope analysis was performed once but the similar results were obtained under the light microscope.
Code Availability
There is no custom code used in this study.
Data Availability
The information of metabolomics data (Supplementary Table 1–6), patient cohorts (Supplementary Table 7) and microarray data (Supplementary Table 9) are available in the online version of the paper. Microarray data that support the findings of this study have been also deposited in the Gene Expression Omnibus (GEO) under accession codes GSE124727. Gene expression data for Supplementary Fig 2b and c are obtained from GlioVis website (http://gliovis.bioinfo.cnio.es/). Probe ID and gene symbols for Supplementary Fig. 2a are provided in Supplementary Table 8. Previously published microarray and sequencing data that were re-analysed here are available under accession code GSE45874. The primer list used in this study is provided in Supplementary Table 10. Source data for Figs. 1, 2, 3, 4, 5, 6, 7, Supplementary Figs. 1, 2, 3, 4, 5, 6, 7 have been provided in Supplementary Table 11. The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgements
We thank the members of the Sasaki Lab, Drs. Carol Mercer, David Plas, Tom Cunnigham, Riko Hatakeyama, Keisuke Ito, Akinori Kawakami, Keiko Kono, Takahisa Nakamura, Kensuke Sasaki, Ryo Kamata, Yoshihisa Hirota, Toshiya Senda, Koh Takeuchi, Yuxiang Zheng, Kaoru Hazeki, Kiyomi Nigorikawa and the Izayoi Society for feedback. We thank Ms. Emily Dobbs for excellent editing. S.Kofuji is supported, in part, by Home for Innovative Researchers and Academic Knowledge Users (HIRAKU), JSPS KAKENHI Grant Number JP18K07233 and the Kanae Foundation. R.K. is supported by a Grant-in-Aid for JSPS Fellows Grant Number JP17J01882. H.Y. was supported by the Uehara Memorial Foundation. K.S. was supported, in part, by the American Association of Neurological Surgeons. Work in DA’s lab was funded by the Francis Crick Institute, which receives its core funding from Cancer Research UK, the UK Medical Research Council and the Wellcome Trust to DA (FC001033). R.J.D. is supported by grants from the NIH (R35CA220449). The infrastructure of imaging MS was supported by JST ERATO Suematsu Gas Biology Project for M.S.(~March 2015). H.B. was supported by the Thuringian state program ProExzellenz (RegenerAging—FSU-I-03/14) of the Thuringian Ministry for Research. The work is supported in part by UC College of Medicine Research Innovation grant, MTP UC-Brain Tumor Center grant, MERF grant, Marlene Harris Ride Cincinnati grant, ABTA Discovery grant, B*Cured research grant, Ohio Cancer Research grant, R21NS100077, and R01NS089815 (A.T.S.).
Footnotes
Competing interest
R.J.D. is an advisor for Agios Pharmaceuticals. P.S.M. is a cofounder, has equity and serves as a consultant for Pretzel Therapeutics, Inc.
References for the main text
- 1.Pianese G, Teuscher R & Ziegler E Beitrag zur Histologie und Aetiologie des Carcinoms. (1896).
- 2.Ruggero D Revisiting the nucleolus: from marker to dynamic integrator of cancer signaling. Science Signaling 5, pe38–pe38 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hara A et al. Correlation between nucleolar organizer region staining and Ki-67 immunostaining in human gliomas. Surg Neurol 33, 320–324 (1990). [DOI] [PubMed] [Google Scholar]
- 4.Busch H, Byvoet P & Smetana K The nucleolus of the cancer cell: a review. Cancer Research 23, 313–339 (1963). [PubMed] [Google Scholar]
- 5.Tsai RYL & Pederson T Connecting the nucleolus to the cell cycle and human disease. FASEB J. 28, 3290–3296 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Grummt I The nucleolus—guardian of cellular homeostasis and genome integrity. Chromosoma 122, 487–497 (2013). [DOI] [PubMed] [Google Scholar]
- 7.WOESE CR & FORRO JR Correlations between ribonucleic acid and deoxyribonucleic acid metabolism during spore germination. Journal of Bacteriology 80, 811–817 (1960). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Srulijes LK, Israeli E & Barzilai D Brief communication: Rna:Dna ratios in human tumors. JNCI J Natl Cancer Inst 58, 769–770 (1977). [DOI] [PubMed] [Google Scholar]
- 9.Thompson M, Haeusler RA, Good PD & Engelke DR Nucleolar clustering of dispersed tRNA genes. Science 302, 1399–1401 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drygin D, Rice WG & Grummt I The RNA Polymerase I Transcription Machinery: An Emerging Target for the Treatment of Cancer. Annu. Rev. Pharmacol. Toxicol 50, 131–156 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Traut T Physiological concentrations of purines and pyrimidines. Molecular and cellular biochemistry 140, 1–22 (1994). [DOI] [PubMed] [Google Scholar]
- 12.Allsop J & Watts RW Purine synthesis and salvage in brain and liver. Adv. Exp. Med. Biol 165 Pt B, 21–26 (1984). [DOI] [PubMed] [Google Scholar]
- 13.Seegmiller JE, Rosenbloom FM & Kelley WN Enzyme Defect Associated with a Sex-Linked Human Neurological Disorder and Excessive Purine Synthesis. Science 155, 1682–1684 (1967). [DOI] [PubMed] [Google Scholar]
- 14.Nyhan WL Behavior in the Lesch-Nyhan syndrome. J Autism Child Schizophr 6, 235–252 (1976). [DOI] [PubMed] [Google Scholar]
- 15.Gerweck LE & Wakimoto H At the Crossroads of Cancer Stem Cells, Radiation Biology, and Radiation Oncology. Cancer Research 76, 994–998 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cloughesy TF, Cavenee WK & Mischel PS Glioblastoma: From Molecular Pathology to Targeted Treatment. Annu. Rev. Pathol. Mech. Dis 9, 1–25 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Nakano I Stem cell signature in glioblastoma: therapeutic development for a moving target. J. Neurosurg 14, 324–330 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CLL & Rich JN Cancer stem cells in glioblastoma. Genes & Development 29, 1203–1217 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hosios AM & Vander Heiden MG The redox requirements of proliferating mammalian cells. Journal of Biological Chemistry jbc.TM117.000239 (2018). doi: 10.1074/jbc.TM117.000239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pavlova NN & Thompson CB The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 23, 27–47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carr S, Papp E, Wu J & Natsumeda Y Characterization of human type I and type II IMP dehydrogenases. Journal of Biological Chemistry 268, 27286 (1993). [PubMed] [Google Scholar]
- 22.Hager PW, Collart FR, Huberman E & Mitchell BS Recombinant human inosine monophosphate dehydrogenase type I and type II proteins Purification and characterization of inhibitor binding. Biochem. Pharmacol 49, 1323–1329 (1995). [DOI] [PubMed] [Google Scholar]
- 23.Hedstrom L IMP dehydrogenase: structure, mechanism, and inhibition. Chemical reviews 109, 2903–2928 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson RC, Weber G & Morris HP IMP dehydrogenase, an enzyme linked with proliferation and malignancy. Nature 256, 331–333 (1975). [DOI] [PubMed] [Google Scholar]
- 25.Carter SB et al. Mycophenolic acid: an anti-cancer compound with unusual properties. Nature 223, 848–850 (1969). [DOI] [PubMed] [Google Scholar]
- 26.Williams RH, Lively DH, DeLong DC, Cline JC & Sweeny MJ Mycophenolic acid: antiviral and antitumor properties. J. Antibiot 21, 463–464 (1968). [DOI] [PubMed] [Google Scholar]
- 27.Majd N et al. A Review of the Potential Utility of Mycophenolate Mofetil as a Cancer Therapeutic. Journal of Cancer Research 2014, 1–12 (2014). [Google Scholar]
- 28.Valvezan AJ et al. mTORC1 Couples Nucleotide Synthesis to Nucleotide Demand Resulting in a Targetable Metabolic Vulnerability. Cancer Cell 32, 624–638.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X et al. Purine synthesis promotes maintenance of brain tumor initiating cells in glioma. Nat. Neurosci 20, 661–673 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang F et al. Inosine Monophosphate Dehydrogenase Dependence in a Subset of Small Cell Lung Cancers. Cell Metab. 28, 369–382.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horbinski C What do we know about IDH1/2 mutations so far, and how do we use it? Acta Neuropathol 125, 621–636 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bentley R Mycophenolic Acid: A One Hundred Year Odyssey from Antibiotic to Immunosuppressant. Chemical reviews 100, 3801–3826 (2000). [DOI] [PubMed] [Google Scholar]
- 33.Sintchak MD & Nimmesgern E The structure of inosine 5’-monophosphate dehydrogenase and the design of novel inhibitors. Immunopharmacology 47, 163–184 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Yokota S Mizoribine: mode of action and effects in clinical use. Pediatr Int 44, 196–198 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Dudzinska W, Lubkowska A, Dolegowska B, Safranow K & Jakubowska K Adenine, guanine and pyridine nucleotides in blood during physical exercise and restitution in healthy subjects. Eur J Appl Physiol 110, 1155–1162 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jové M, Portero-Otín M, Naudí A, Ferrer I & Pamplona R Metabolomics of human brain aging and age-related neurodegenerative diseases. J. Neuropathol. Exp. Neurol 73, 640–657 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Drygin D et al. Targeting RNA Polymerase I with an Oral Small Molecule CX-5461 Inhibits Ribosomal RNA Synthesis and Solid Tumor Growth. Cancer Research 71, 1418 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Peltonen K et al. A Targeting Modality for Destruction of RNA Polymerase I that Possesses Anticancer Activity. Cancer Cell 25, 77–90 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pelletier J, Thomas G & Volarevic S Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat Rev Cancer 18, 51–63 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Grummt I & Grummt F Control of nucleolar RNA synthesis by the intracellular pool sizes of ATP and GTP. Cell 7, 447–453 (1976). [DOI] [PubMed] [Google Scholar]
- 41.Huang M, Ji Y, Itahana K, Zhang Y & Mitchell B Guanine nucleotide depletion inhibits pre-ribosomal RNA synthesis and causes nucleolar disruption. Leuk. Res 32, 131–141 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen LXT et al. Regulation of ribosomal RNA synthesis in T cells: requirement for GTP and Ebp1. Blood 125, 2519–2529 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson DM & Parker R Stressing out over tRNA cleavage. Cell 138, 215–219 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Tsai RYL & McKay RDG A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes & Development 16, 2991–3003 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lo D, Dai MS, Sun XX, Zeng SX & Lu H Ubiquitin- and MDM2 E3 Ligase-independent Proteasomal Turnover of Nucleostemin in Response to GTP Depletion. Journal of Biological Chemistry 287, 10013–10020 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhai W & Comai L Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol Cell Biol 20, 5930–5938 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liebhaber SA, Wolf S & Schlessinger D Differences in rRNA metabolism of primary and SV40-transformed human fibroblasts. Cell 13, 121–127 (1978). [DOI] [PubMed] [Google Scholar]
- 48.Gu JJ Induction of apoptosis in IL-3-dependent hematopoietic cell lines by guanine nucleotide depletion. Blood 101, 4958–4965 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Park J et al. Mycophenolic Acid Inhibits Platelet-Derived Growth Factor-Induced Reactive Oxygen Species and Mitogen-Activated Protein Kinase Activation in Rat Vascular Smooth Muscle Cells. Am J Transplant 4, 1982–1990 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Emmanuel N et al. Purine Nucleotide Availability Regulates mTORC1 Activity through the Rheb GTPase. CellReports 19, 2665–2680 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Eugui EM, Almquist SJ, Muller CD & Allison AC Lymphocyte-selective cytostatic and immunosuppressive effects of mycophenolic acid in vitro: role of deoxyguanosine nucleotide depletion. Scand. J. Immunol 33, 161–173 (1991). [DOI] [PubMed] [Google Scholar]
- 52.Liu Y-C et al. Global Regulation of Nucleotide Biosynthetic Genes by c-Myc. PLoS ONE 3, e2722 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mourad M et al. Correlation of mycophenolic acid pharmacokinetic parameters with side effects in kidney transplant patients treated with mycophenolate mofetil. Clinical Chemistry 47, 88–94 (2001). [PubMed] [Google Scholar]
- 54.Nguyen T, Park JY, Scudiere JR & Montgomery E Mycophenolic acid (cellcept and myofortic) induced injury of the upper GI tract. Am. J. Surg. Pathol 33, 1355–1363 (2009). [DOI] [PubMed] [Google Scholar]
- 55.Chen L & Pankiewicz KW Recent development of IMP dehydrogenase inhibitors for the treatment of cancer. Curr Opin Drug Discov Devel 10, 403–412 (2007). [PubMed] [Google Scholar]
References for the Methods
- 56.Sampetrean O et al. Invasion precedes tumor mass formation in a malignant brain tumor model of genetically modified neural stem cells. Neoplasia 13, 784–791 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kwon C-H et al. Pten haploinsufficiency accelerates formation of high-grade astrocytomas. Cancer Research 68, 3286–3294 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chow LML et al. Cooperativity within and among Pten, p53,and Rb Pathways Induces High-Grade Astrocytoma in Adult Brain. Cancer Cell 19, 305–316 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Steed TC et al. Differential localization of glioblastoma subtype: implications on glioblastoma pathogenesis. Oncotarget 7, 24899–24907 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ozawa T et al. Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell 26, 288–300 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sumita K et al. The Lipid Kinase PI5P4Kβ Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis. Mol Cell 61, 187–198 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krycer JR et al. Dynamic Metabolomics Reveals that Insulin Primes the Adipocyte for Glucose Metabolism. CellReports 21, 3536–3547 (2017). [DOI] [PubMed] [Google Scholar]
- 63.Soga T et al. Metabolomic profiling of anionic metabolites by capillary electrophoresis mass spectrometry. Anal. Chem. 81, 6165–6174 (2009). [DOI] [PubMed] [Google Scholar]
- 64.Hirayama A et al. Effects of processing and storage conditions on charged metabolomic profiles in blood. Electrophoresis 36, 2148–2155 (2015). [DOI] [PubMed] [Google Scholar]
- 65.Minami N et al. Organotypic brain explant culture as a drug evaluation system for malignant brain tumors. Cancer Med 6, 2635–2645 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sugiura Y, Honda K & Suematsu M Development of an Imaging Mass Spectrometry Technique for Visualizing Localized Cellular Signaling Mediators in Tissues. Mass Spectrometry 4, A0040–A0040 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shiota M et al. Gold-nanofève surface-enhanced Raman spectroscopy visualizes hypotaurine as a robust anti-oxidant consumed in cancer survival. Nature Communications 9, 1561 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bowman RL, Wang Q, Carro A, Verhaak RGW & Squatrito M GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro-oncology 19, 139–141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Andersen JS et al. Nucleolar proteome dynamics. Nature 433, 77–83 (2005). [DOI] [PubMed] [Google Scholar]
- 70. Marin-Valencia I et al. Analysis of Tumor Metabolism Reveals Mitochondrial Glucose Oxidation in Genetically Diverse Human Glioblastomas in the Mouse Brain In Vivo. Cell Metab. 15, 827–837 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The information of metabolomics data (Supplementary Table 1–6), patient cohorts (Supplementary Table 7) and microarray data (Supplementary Table 9) are available in the online version of the paper. Microarray data that support the findings of this study have been also deposited in the Gene Expression Omnibus (GEO) under accession codes GSE124727. Gene expression data for Supplementary Fig 2b and c are obtained from GlioVis website (http://gliovis.bioinfo.cnio.es/). Probe ID and gene symbols for Supplementary Fig. 2a are provided in Supplementary Table 8. Previously published microarray and sequencing data that were re-analysed here are available under accession code GSE45874. The primer list used in this study is provided in Supplementary Table 10. Source data for Figs. 1, 2, 3, 4, 5, 6, 7, Supplementary Figs. 1, 2, 3, 4, 5, 6, 7 have been provided in Supplementary Table 11. The data that support the findings of this study are available from the corresponding author upon reasonable request.