Abstract
Metabolic disorders present a public health challenge of staggering proportions. In diabetes, there is an urgent need to better understand disease heterogeneity, clinical trajectories, and related comorbidities. A pressing and timely question is whether we are ready for precision medicine in diabetes. Some biological insights that have emerged during the last decade have already been used to direct clinical decision making, especially in monogenic forms of diabetes. However, much work is necessary to integrate high-dimensional explorations into complex disease architectures, less penetrant biological alterations, and broader phenotypes, such as type 2 diabetes. In addition, for precision medicine to take hold in diabetes, reproducibility, interpretability, and actionability remain key guiding objectives. In this review, we examine how mounting data sets generated during the last decade to understand biological variability are now inspiring new venues to clarify diabetes nosology and ultimately translate findings into more effective prevention and treatment strategies.
Keywords: precision medicine, diabetes, diabetes heterogeneity, omics
Precision medicine
The concept of precision medicine has evolved from an initial focus on individualized preventive strategies and patient care (personalized medicine) to a wider and more realistic notion that intends to convey the principle that, although therapeutics are rarely developed for single individuals, subgroups of individuals with unique features may be increasingly defined and treated in more efficient ways. This is now possible owing to the comprehensive capture of multiple data points across orthogonal axes of information, the development of analytical methods that permit the interpretation of complex data and enable the construction of more refined categories, and concomitant advances in targeted therapeutics.
Precision medicine is not a new construct—but our ability to implement it in a sophisticated and rational manner has finally come of age. Indeed, the science of medicine has been historically focused around notions of subgroups and categorization. In ancient Greek civilization, health (“a gift from the gods”) and illness (“divine punishment”) were presumed to be affected by a few categories, such as gender, geographic localization, social class, humors, diet, trauma, beliefs, and mindset.1 From that initial understanding, the concept of heath and disease has progressed, and the number of categories (now considered disease risk factors) has increased exponentially. This is due to the widespread adoption of the scientific method, by which clinical investigators observe, predict, test, and generalize.2,3 Evidence-based medicine is the application of the scientific method in biomedicine and has become the gold standard to advance knowledge and make decisions about patient care. The continuous updating of the scientific method to maximize preventive strategies and patient care has launched thriving research initiatives to explore the ways in which several axes of biological information and environmental characteristics drive disease pathogenesis and influence responses to therapy.3 However, as the scientific method is systematically deployed across all areas of medicine, it has become apparent that large disease heterogeneity and response variability exist. Thus, the generalizability of diagnostic groups and the extrapolation of average responses from clinical trials, predicated on the assumption that medical interventions should work ubiquitously in individuals sharing a limited set of similar characteristics, can often be misleading.4,5
The mounting troves of data generated during the last decade to understand biological variability are now inspiring new venues for complex disease prevention, treatment, and cure through a deeper understanding and characterization of underlying molecular processes.6,7 One early step toward the historical change in the global reach of biological inquiry started just a few decades ago with the Human Genome Project, whose major contribution was to hand the world a complete resource of detailed information about the structure, organization, and function of the full set of human genes.8 Since then, technological and analytical advances have enabled the design of genome-wide association studies (GWASs), which have generated the discovery of hundreds of associated loci across the human genome for many complex diseases of large public health impact.9–11 Led by genomics, other high-throughput technologies have emerged recently, providing comprehensive information about the epigenome, transcriptome, proteome, metabolome, and microbiome. This information is allowing investigators to explore complementary biological axes in greater breadth and depth.12–17 Although each layer of information provides relevant insights into disease nosology, the interplay between diverse biological layers, even within the same tissue or metabolic condition, hinders the translation of the newly generated knowledge, given that it cannot be assessed by a simplified reduction approach.18 In addition, tissue-specific regulation and cross talk between tissues orchestrating the same molecular process in response to different environmental or physiological triggers presents an extra level of difficulty in unraveling the biological basis of complex diseases like diabetes.
Some areas of biomedicine are more amenable to the integration of these features. In cancer, each tumor can be seen as a unique and localized set of genetic and epigenetic changes resulting in a definite molecular signature. Successful examples of personalized medicine in cancer often rest on the direct detection of penetrant somatic mutations in the diseased tissue. For example, somatic mutations in dominant melanoma oncogenes, such as BRAF, have been shown to be relevant in some nonmelanoma cancers, making BRAF a generally targetable oncogene and practically redefining cancer types by their primary molecular pathogenic defects rather than their tissue of origin.19 A potential source of heterogeneity in cancer cell response may be attributable to the specific tumor microenvironment, in part captured through chromatin regulation and catalyzing the emergence of chromatin regulation as a new druggable objective for resistant cancers.20 Other approaches for resistant cancers include personalized vaccines that take advantage of differences in DNA sequence between tumor and healthy cells to re-engineer more efficient T cells.21 For example, two recent phase I clinical trials showed that a personalized T cell–based vaccine to treat people with skin cancer successfully enhanced the immune response, and no signs of tumor recurrence were reported in the majority of participants after a follow-up period of up to 32 months after vaccination.22,23
Whether knowledge generated in cancer is ready to be implemented in other metabolic complex diseases, such as diabetes, is less clear. The genetic architectures of diabetes and cancer are quite different: both type 1 and type 2 diabetes (T1D and T2D, respectively) are caused by the combination of genetic predisposition and environmental triggers, with most germline variants (with the exception of the HLA region in T1D) conferring only modest effects on risk; whereas in cancer a specific high-penetrance somatic mutation in a given gene in a particular cell type is likely to start cancer initiation, in some cases also triggered by an external insult and modified by additional gene variants.11,24
Diabetes is a heterogeneous disease
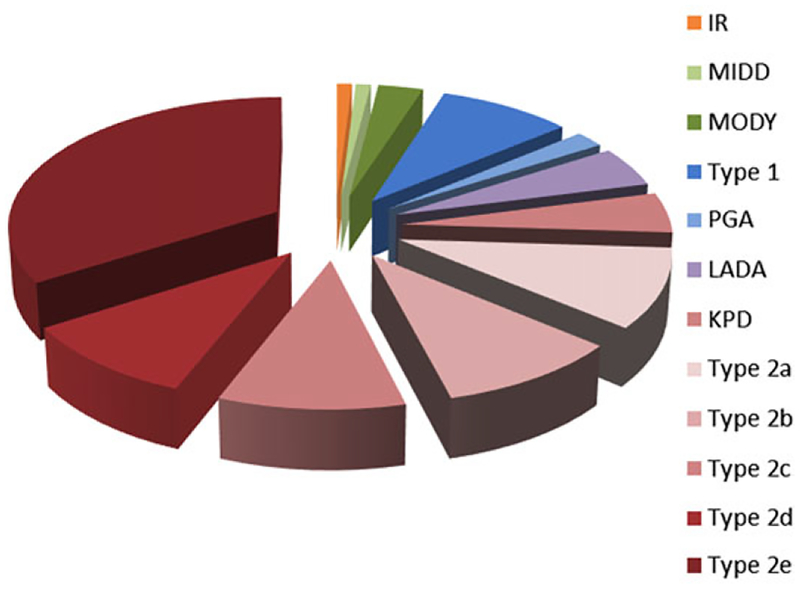
Diabetes, albeit with the common denominators of relative insulin deficiency and consequent elevated blood glucose, is a much more heterogeneous disease than the present classification into T1D and T2D suggests.25,26 T1D, with a typical onset at an early age, develops as a result of autoimmune destruction of the insulin-producing β cells, whereas T2D is due to a combination of insulin resistance superimposed on an insulin-secretory defect. However, the spectrum of diabetes has broadened in the past few decades with the realization that several different overlapping mechanisms can lead to diabetes, suggesting that individuals might have features of different biological alterations (Fig. 1). For example, latent autoimmune diabetes of adults (LADA; also known as autoimmune diabetes in adults, type 1 and 1/2 diabetes, and slow-onset diabetes in adults) might constitute a category of diabetes on its own, since patients share features of both T1D (autoimmunity, eventual insulin dependence) and T2D (onset at later ages, insidious presentation).27,28 Recent work suggests that the genetic architecture of LADA is closer to T1D than to T2D.29 Ketosis-prone diabetes (KPD) in adults is another hybrid form of diabetes with features of both T1D and T2D, but without the autoimmune characteristics of LADA.30 These individuals, often of Asian or Afro-Caribbean ancestry, are characterized by relative insulin deficiency and are prone to developing diabetic ketoacidosis.31
Figure 1.
Heterogeneity of diabetes. The pie chart represents the multiple ways people can develop hyperglycemia and reach the diagnosis of diabetes. The size of each piece only represents an approximate proportion of prevalence in the population. IR, rare genetic forms of insulin resistance; MIDD, maternally inherited diabetes and deafness; MODY, maturity-onset diabetes of the young; type 1, type 1 diabetes; PGA, diabetes caused in the setting of polyglandular autoimmune syndrome; LADA, latent autoimmune diabetes of adults; KPD, ketosis-prone diabetes; type 2a–2e, hypothetical subgroups of type 2 diabetes.
Despite these hybrid forms of diabetes, T2D is the major cause of diabetes worldwide and accounts for nearly 90–95% of diabetes cases. T2D by itself is also a heterogeneous disease, as patients can range from those with a predominantly insulin-resistance phenotype but with sufficient β cell reserve to remain insulin independent to those who may require insulin treatment early in the course of their disease.26 The need for escalation of therapy with additional agents differs across individuals, as does the likelihood of progression to specific macrovascular or microvascular complications. In addition, the influence of obesity on T2D risk varies greatly across populations, with individuals of East Asian descent developing T2D at much lower indices of adiposity.32,33 As more diverse populations are examined, it is becoming apparent that differences in allele frequencies across populations may explain disparities in prevalence.34–36
Subclassification of type 2 diabetes
Given the heterogeneity in diabetes phenotype, several attempts to define molecular subgroups of particular forms of diabetes through clinical features have been implemented (Fig. 1). For example, up to four different phenotypes have been described in KPD, depending on the presence or absence of islet cell autoantibodies (A− or A+) and β cell functional reserve (β− or β+).37 This classification, based on both immunologic and β cell function criteria, has the highest accuracy and predictive value in classifying patients with KPD with regard to clinical outcomes and pathophysiologic subtypes.38 In addition, a long-term longitudinal follow-up study including KPD A−β+ patients has revealed that this phenotype comprises two distinct subtypes distinguished by whether T1D-associated HLA susceptibility is present or not.39 In gestational diabetes (GDM), about half of women with GDM had predominant insulin-sensitivity defects with hyperinsulinemia, a phenotype that was linked with altered adipokine profiles, larger infants, and greater risk of GDM-related complications; these were not observed among women with GDM due to predominant insulin-secretion defects, suggesting uneven characteristics within GDM.40 Finally, a recent data-driven cluster analysis of six simple clinical variables (age, body mass index (BMI), GAD antibody status, hemoglobin A1c, and homeostasis model assessments of β cell function and insulin resistance) measured at baseline in patients with newly diagnosed diabetes identified five replicable clusters of patients with different clinical presentations. The five diabetes subtypes (severe autoimmune diabetes, severe insulin-deficient diabetes, severe insulin-resistant diabetes, mild obesity-related diabetes, and mild age-related diabetes) also showed varying degrees of risk of diabetic complications. For example, individuals with severe insulin-resistant diabetes had significantly higher risk of diabetic kidney disease, while those with severe insulin-deficient diabetes had the highest risk of retinopathy.41
The revolution of omics profiling technologies can help identify subgroups of individuals with diabetes sharing unique biological features. Among all available approaches for more detailed personalized profiling, assaying genetic variation has taken the lead and made rapid progress. This is the case for a number of reasons, including (1) the ability to query millions of variants across the human genome in a single experiment;42,43 (2) the development of accurate analytical methods and stringent statistical standards to interpret results with appropriate statistical rigor;42 (3) the unique feature that germline genetic variation is fixed in the individual and thus needs to be measured only once in the person’s lifetime;44 and (4) the potential to derive causal inference, since genetic variation is free from conventional confounding owing to the unidirectional arrow of time (the variant always precedes the phenotype and is unaffected by the disease process or its treatment) and the random independent assortment of alleles at meiosis.45
To date, well over 100 genetic loci have been identified in successive waves of GWAS meta-analyses as robustly associated with T2D and/or related traits.46–52 Comprehensive sequencing studies that capture both common and rare variation suggest that most genetic variation influencing T2D appears to reside at common variant sites.11 Although genetic risk variants at these loci have modest effects on disease predisposition (collectively accounting for 10–15% of overall disease risk),11,50 the knowledge gained has paved the way to elucidate the molecular taxonomy of the disease and the potential identification of novel therapeutic approaches.42,53 To highlight disease heterogeneity, ethnic-specific alleles are emerging, thanks to genotyping and sequencing experiments in diverse populations. For example, a nonsense polymorphism in the TBC1D4 locus (with a minor allele frequency (MAF) of 17% in Inuit populations, but almost nonexistent in other groups), raises 2-h glucose and increases T2D risk 10-fold.54 As TBC1D4 is implicated in transducing the insulin signal in skeletal muscle, it is believed that these individuals suffer from a type of T2D mostly defined by muscle insulin resistance and might benefit preferentially from treatment with an insulin sensitizer, a hypothesis that can be tested in a pharmacogenetic clinical trial.55 Similarly, a risk haplotype in the SLC16A11 locus is common (MAF < 40–50%) among people of Mexican or Latin American descent but rare among Europeans and absent in Africa; together with other such variants, it might explain some portion of the increased T2D prevalence in Mexico.34 A recent functional study demonstrated that lower levels of monocarboxylate transporter 11 (the protein encoded by SLC16A11) in the plasma membrane of primary human hepatocytes are associated with T2D-relevant changes in fatty acid and lipid metabolism.56 Though the mechanism of action is incompletely understood, therapies targeting this monocarboxylate transporter and enhancing its function in hepatocytes may be particularly effective in people whose risk of developing T2D is driven by this disrupted mechanism.57 In another example, a missense polymorphism in HNF1A (MAF of 2% in Mexicans with T2D) increases the risk of T2D fivefold.36 Because carriers of loss-of-function mutations in this gene experience a more favorable response to sulfonylureas, it is possible that these patients might be better treated with those agents as well, at least early in their disease course. Finally, whole-exome sequencing data have uncovered a single coding variant in PAX4 that was strongly associated with T2D, but only in people from East Asian countries, including Korea, China, and Singapore.11
Another approach to better define particular subtypes of T2D is to use T2D-associated genetic variants as biomarkers in unsupervised classification methods or aggregate them into biologically relevant polygenic risk scores (GRS). Using unsupervised clustering analysis of 37 established T2D susceptibility loci, it was shown that T2D risk loci may fall into different groups related to (1) insulin sensitivity, (2) insulin secretion, (3) insulin processing, and (4) insulin processing and secretion without a detectable change in fasting glucose levels.58 Hierarchical clustering analysis using 19 common genetic variants associated with fasting insulin–based measures identified 11 variants associated with a metabolic profile consistent with a mild form of lipodystrophy.59 A GRS composed of these 11 prioritized risk alleles was paradoxically associated with lower BMI but increased visceral to-subcutaneous adipose tissue ratio and caused metabolic alterations, such as higher triglycerides and elevated transaminases or hepatic steatosis and lower HDL or adiponectin. In a follow-up reciprocal analysis, a GRS of 11 favorable adiposity variants was associated with lower T2D relative risk and higher body fat percentage, with greater subcutaneous storage capacity.60 Once variants are aggregated into physiologically meaningful clusters, the next step is to verify whether GRS based on such a classification can also group individuals in clinically meaningful categories, particularly at the extreme ends of the distribution.
As the number of T2D-associated variants continues to mount, the construction of GRS allows for a continuous and quantitative measure of T2D genetic susceptibility that can help to identify relevant subgroups. For example, with rising populationobesity, distinguishing T1D and T2D in patients with new-onset diabetes has proven difficult, particularly in clinical scenarios, because more young people are developing T2D, and many individuals with T1D will be obese.61 Accordingly, up to 15% of young adults with diabetes are estimated to be wrongly classified and consequently incorrectly treated, which may have consequences in poor glycemic control, inappropriate insulin regimens, and the risk of life-threatening ketoacidosis.62 A study assessing a T1D GRS based on 30 T1D-associated risk variants provided evidence that the GRS is highly discriminative and indicative of T1D, especially in young adults, where the T1D GRS alone predicted progression to insulin deficiency.63 In aseparate study, a similar T1D GRS improved the discrimination of monogenic diabetes from T1D,64 confirming the relevance of genetics to correctly classify individuals based on divergent pathophysiological processes.
Finally, more comprehensive methods based on high-dimensional data from electronical medical records combined with genetic information have been implemented to attempt to characterize the heterogenic complexity of T2D and its complications.65–67 A recent study suggested thata topological analysis of many clinical features gives rise to three distinct subgroups of T2D:67 subtype 1 was characterized by T2D microvascular complications, including diabetic nephropathy and diabetic retinopathy; subtype 2 was enriched for cancer malignancy and cardiovascular diseases; and subtype 3 was associated most strongly with cardiovascular diseases, neurological diseases, allergies, and HIV infections. Distinct sets of genetic variants could be mapped to these subtypes. However, it is difficult to replicate these types of high-dimensional explorations, as their biological relevance is not obvious, and no clear clinical decision-making implications have emerged. For precision medicine to take hold in diabetes, reproducibility, interpretability, and actionability remain key guiding objectives.
Another angle to characterize diabetes into distinct categories and provide biological insights into early metabolic alterations is via particular metabolomic profiling.68,69 During the last decade, metabolomics has emerged as an integrative tool for biological states through the global measurement of chemical endophenotypes that lie downstream of genomic, transcriptomic, and proteomic variability.70 One key advantage of metabolomics is that the measured entity is closer to the organismal phenotype than genetic variation and integrates a number of biological processes; thus, the effect size on the trait of interest is typically larger. Disadvantages include the complex and incompletely characterized nature of the metabolome, the unknown nature of many measured metabolites, the diversity of technologies and data-analysis techniques used, and the correlational nature of most analyses where causal inference is challenging. As a result, generalizability and replication has been difficult, though standardized methods for metabolite identification and reporting are beginning to emerge.71
Despite intrinsic limitations, particular attention has focused on metabolomics of insulin resistance and T2D. Several large prospective analyses using either targeted or untargeted metabolomics approaches have validated the association between branched-chain amino acids (BCAAs) and aromatic amino acids with insulin resistance and T2D.72,73 Beyond BCAA, downstream BCAA metabolic products, such as branched-chain ketoacids and acylcarnitines, were significantly elevated in both individuals with impaired fasting glucose and subjects with T2D compared with control subjects.74 Other products of amino acids catabolism, such as 2-aminoadipic acid or α-hydroxybutyrate, have been found to be strongly correlated with incident T2D.75,76 These findings are consistent with a model in which excess of BCAAs contributes to impaired efficiency of fatty acid oxidation, resulting in the accumulation of incompletely oxidized lipid species, perhaps of particular relevance in insulin resistance.70 Whether these metabolite alterations are common to all patients with T2D or serve to identify specific subgroups requires further exploration.
Recently, GWASs have been integrated with high-throughput metabolomic profiling to provide biological insights into how genetic variation influences metabolism and how such metabolic differences in plasma can help to identify relevant genes within genomic regions associated with complex diseases.77 In addition, the integration of genomics with metabolomics can help place specific metabolites on causal pathways. For instance, a study that pooled data from four European cohorts found that CYP7A1, which encodes the rate-limiting enzyme in bile acid synthesis, was associated with lower concentrations of deoxycholic acid and higher T2D risk.78 In addition, this study also identified variants in or near the genes encoding sphingosine-1-phosphate phosphatase 1 (SGPP1), glucokinase regulator (GCKR), and fatty acid desaturase 1 and 2 (FADS1/2) that were associated with diabetes-associated phospholipids and T2D risk. Finally, using Mendelian randomization in combination with plasma metabolomics suggested a causal role for lower levels of palmitoleic acid and oleic acid on insulin resistance.79 Two recent Mendelian randomization studies (see Ref. 45 for methodological details) have implicated BCAAs in the pathogenesis of T2D and suggested that genetically raised insulin resistance drives higher circulating fasting BCAA levels.80,81 In a recent analysis of 1622 nondiabetic participants from the Framingham Heart Study, the combination of genetics, metabolomics, and clinical factors increased the prediction of future T2D.82 In brief, a 62-variant GRS showed an area under the curve (AUC) of 64%; addition of metabolites increased the AUC to 82%, and the combination of genetics, metabolomics and clinical factors achieved an AUC of 88%. The results from this study suggest that metabolite and genetic traits also provide complementary information to each other for the prediction of future T2D. This emerging information may help classify individuals at high risk for different forms of T2D and potentially translate findings into more personalized prevention or treatment strategies.
The observation that the great majority of GWAS variants identified for T2D do not affect protein-coding sequence suggests that gene regulation has a central role in the development of the disease.83 New analytical methods exploiting available data sets that describe gene expression patterns and epigenetic marks obtained from cells and tissues at different stages of development and disease states have become an extremely useful resource.84 In diabetes, one of the most salient findings is the discovery that T1D GWAS signals localize to enhancer sequences active in the thymus, T and B cells, and CD34+ stem cells, confirming the regulatory disruption of the immune component in the etiology of T1D.85 A landmark study using integrative analysis was conducted for the fat-mass obesity (FTO) locus, the strongest genome-wide association signal for obesity.86,87 In brief, using a variety of data sets, the authors predicted the cell type (preadipocyte) and regulatory element (enhancer) disrupted by the causal variant and linked the predicted enhancer (ARID5B motif) to two target genes (IRX3 and IRX5) involved in early adipocyte differentiation. Finally, the investigators were able to restore the correct expression of the affected target genes in cells isolated from patients and a mouse model using CRISPR–Cas9 genome editing and to demonstrate major allele-dependent effects on thermogenesis in adipocytes.87 Though additional mechanisms could be at play, these integration efforts illustrate how resource building can result in major biological insights into the functional consequences of genetic alterations, uncovering novel pathways that could be harnessed for therapeutic development. In addition, improved mechanistic understanding is a first step in determining whether such processes are uniformly operational across the entire phenotypic spectrum of diabetes or could serve to describe specific subtypes.
Convincing evidence suggests that the dysbiotic state conferred by gut microbiota composition is associated with metabolic diseases.88 Previous profiles of the gut microbiome in T2D have found compositional changes between patients and healthy controls, showing increased capacity for oxidative stress resistance and a decreased capacity for flagellar assembly and riboflavin metabolism.89,90 However, the human microbiome contains vast numbers of uncharacterized enzymes, limiting our functional understanding of this community and its effects on host health and disease. A possible mechanism linking the gut microbiome and insulin resistance may be due to specific microbiota species (Prevotella copri and Bacteroides vulgatus), which have been reported to drive the association between biosynthesis of BCAAs and insulin resistance in ~200 individuals with insulin resistance.91 However, given the large number of microbiota species and interconnected metabolic pathways between species, efforts to distinguish which bacterial species increases diabetes risk in a causal manner or to define diabetes subgroups according to metagenome characterization are still largely unrealized.
Does omic information make a difference for treatment?
The clinical management of diabetes is currently based on reducing plasma glucose to levels that are associated with a low risk of developing long-term complications.92 However, significant variability exists in response to these interventions, indicating that treatment heterogeneity may reflect underlying biological differences. For example, genetic factors can influence the glycemic response to metformin, explaining from 21% to 34% of its variance depending on how response is defined.93 A better understanding of the underlying causes for differential pharmacological response of subgroups of diabetic individuals may catalyze the delivery of the most accurate intervention strategy to a given individual based on his or her unique characteristics.
For monogenic diabetes, the implementation of precision medicine can be considered an early success, given that there are discrete subgroups that are easily defined by molecular genetics. For example, neonatal diabetes is defined by an extreme phenotype (onset of hyperglycemia within the first 6 months of life), often caused by penetrant mutations in a well-understood locus (ABCC8, which encodes the sulfonylurea receptor SUR1 and the adjacent gene KCNJ11 encoding its associated ATP-dependent potassium channel).94 This biological understanding has led directly to genetically driven individualized therapy (high-dose sulfonylureas).95 Other monogenic diabetes forms, such as maturity-onset diabetes of the young (MODY), also provide proof of concept that genetic information can guide therapy. The most common causes of MODY include mutations in the genes encoding the hepatocyte nuclear factor 1α (HNF1A) and the enzyme glucokinase (GCK) and HNF4A.96,97 Individuals with loss-of-function HNF1A mutations, which cause MODY 3, are extremely sensitive to the hypoglycemic effects of sulfonylureas. This knowledge has key translational implications when the diagnosis of MODY 3 is genetically confirmed: in patients who were mistakenly diagnosed as having T1D (on the basis of the early onset of disease and lean body habitus), the clinician can discontinue insulin therapy and initiate treatment with sulfonylureas, whereas in those who were mistakenly diagnosed as having T2D (on the basis of absence of autoimmunity and a nonketotic presentation), the clinician can substitute metformin for low-dose sulfonylureas.98 Individuals with a heterozygous, inactivating mutation in GCK (MODY 2) have a defect in glucose sensing; hence, glucose homeostasis is maintained at a higher set point, resulting in mild, asymptomatic fasting hyperglycemia (5.5–8.0 mmol/L) that shows little deterioration with age, does not require escalation of therapy, and is not associated with cardiovascular complications.99 The general consensus is that treatment is not recommended outside of pregnancy.100 Insulin treatment might be required during pregnancy to prevent excess fetal growth only if the fetus has not inherited the GCK mutation.101 Similar to individuals with MODY 3, carriers of mutations in HNF4A (MODY 1) display progressive β cell dysfunction; hence, low-dose sulfonylureas is the preferred tailored intervention.102 However, a subgroup of phenotypically different patients with MODY 1 due to a common mutation in HNF4A(p.R114W) was recently described.103 These individuals showed reduced sensitivity to low-dose sulfonylurea treatment, reduced penetrance, and no effect on birth weight and therefore may need high-dose sulfonylurea treatment.
T2D is considerably different from monogenic diabetes. The genetic architecture of T2D is mostly composed of small-effect common variants that hinder the use of any of these variants as a handle to reverse the disease. However, the precise combination of risk and protective variants carried by any given individual is likely to be unique, offering potential translational opportunities.104 Accordingly, research efforts are now moving to determine where the boundaries of risk lie and how to eventually predict whether a patient is likely to develop T2D in his/her lifetime or respond differently to conventional treatments.105 As a critical factor in T2D pathogenesis seems to be early β cell dysfunction, tailored interventions might be administered to individuals with specific β cell risk factors early in the disease course, when insulin secretion has not yet markedly deteriorated. Future T2D pharmacotherapy may focus on preserving normal glucose homeostasis by β cell–based interventions from different angles, including β cell genetic reprogramming, differentiation, or enhancement of activity.
Metformin is currently recommended as a first-line drug for the treatment of T2D: it is effective at reducing hyperglycemia primarily by inhibiting hepatic glucose production and secondarily by modestly increasing insulin sensitivity.106 Whether metformin has different effects on β cell function preservation is a matter of debate. For example, the ADOPT study showed that the durability of metformin monotherapy was better than that of sulfonylureas, but it still resulted in a 21% failure rate at 5 years in participants with recently diagnosed T2D.107
Genetic studies for metformin response may help to prioritize which individuals are more likely to respond better to metformin and achieve greater β cell preservation. Initial candidate gene approaches focused on SLC22A1 (encoding the organic cation transporter 1) or SLC47A1 (encoding the multidrug and toxin extrusion 1) have failed to produce a definite picture of the genetic determinants of metformin response.108
GWASs have identified two metformin response loci (ATM and SLC2A2). The ATM (ataxia telangiectasia mutated) locus, located in a large linkage dis-equilibrium block on chromosome 11 that includes a total of seven potential candidate genes, has been associated with glycemic response to metformin.109 A functional study to comprehensively identify genes and regulatory elements associated with metformin treatment has showed that variants in linkage disequilibrium with the ATM GWAS lead SNP (rs11212617) had increased enhancer activity. Expression quantitative trait locus analysis and CRISPR–Cas9 activation suggest that this enhancer haplotype could be regulating ATM in the liver and activating transcription factor 3, leading to gluconeogenesis repression.110 A noncoding variant in SLC2A2 (encoding the facilitated glucose transporter GLUT2) represents another identified genome-wide signal for metformin response, and it was associated with reduction in hemoglobin A1c in 10,577 participants of European ancestry.111 This regulatory variant influences GLUT2 expression in the human liver, identifying hepatic GLUT2 as an effector of metformin action. However, the increased response to metformin in carriers of this variant did not prevent diabetes in participants from the Diabetes Prevention Program,112 indicating that perhaps genetic influences on drug response are also dependent on the metabolic state of the individual, with differential interactions occurring at diverse stages of disease progression.
In terms of sulfonylurea response, a limited number of polymorphisms in sulfonylurea drug target genes and T2D risk genes has been studied, and most of the results have been limited to small, observational studies. Prior research has found that individuals carrying homozygous loss-of-function mutations in CYP2C9 (encoding the cytochrome P450 2C9 enzyme responsible for liver sulfonylurea metabolism) improved glycemic control after sulfonylurea therapy.113 It should be noted that, in all of these pharmacogenetic studies, while the effects seen thus far are interesting mechanistically, their magnitude is too small to underwrite individualization of therapy.
Conclusions: a working model
Clinical decision making is, by necessity, dichotomous: on the basis of complex and often continuous information, the practitioner needs to decide whether to act or not to act, to intervene or to merely observe. One course must be taken among several possible options, and the key question is whether modern omics technologies will be able to capture enough biological variation to enable the construction of sensible discrete categories to facilitate rational decision analysis, or this will remain the province of “boutique” rare forms of diabetes.
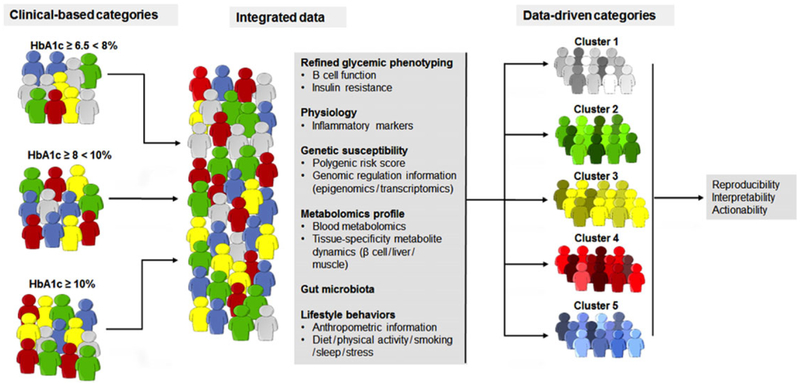
The answer to that question will depend, to a great extent, on the underlying biology. In T2D, the emerging picture is one in which a large constellation of genetic factors (from many hundreds to a few thousands, according to some empirical estimates50) work in concert with environmental and demographic factors to increase T2D risk. The number of potential variations in these interactions can be linked to the colors on a painter’s palette,7 where hues and tones are mixed, and individual primary colors might be difficult to discern. There may still be sections of the “McCarthy palette,” where a particular color (or genetic variant or environmental exposure) may predominate, in which case a targeted intervention may be feasible; and specific subtypes might be defined by the extremes along empirical scores that combine genetic and other variables. Where that is possible, at least some individuals may be placed in strata in which specific surveillance, prevention, lifestyle, pharmacological, and/or surgical strategies might be deployed. Figure 2 hypothetically illustrates how precision medicine might deconstruct traditional symptom-based categories through the study and integration of several biological axes of information to parse current heterogeneous syndromes into homogeneous clusters.
Figure 2.
Implementing precision medicine in diabetes. A hypothetical example illustrating how precision medicine might deconstruct traditional clinical-based categories through the study and integration of the many axes of biological information that can serve to parse current heterogeneous syndromes into homogeneous clusters. The example suggests that the way to prevent new cases of diabetes or treat individuals with diabetes should be tailored to the specific molecular event or pathway that raises glycemia.
As new treatments are introduced, it will be crucial to verify whether they are equally effective across all subtypes; if comparatively greater effectiveness is demonstrated for a specific segment of the population, this information may be used in a public health setting to prioritize the subgroups more likely to benefit. The body of knowledge that will guide these decisions must be developed, and the experiments designed to answer this question ought to be reproducible, interpretable, and actionable. Decision-making support tools must be implemented at the point of care, so clinicians can easily act on available information in a seamless fashion. The new technologies that generate data relevant to health outcomes will need be scaled up and made accessible in community settings, so that they help us understand, rather than deepen, existing health disparities.
Acknowledgments
J.M. is supported by a postdoctoral fellowship funded by the European Commission; Horizon 2020 program; Marie Skłodowska-Curie actions (H2020-MSCA-IF-2015-703787). J.C.F. is a Massachusetts General Hospital Research Scholar and is supported by NIDDK K24 DK110550. We thank the members of the Florez research group and the larger diabetes genetics community for fruitful and engaging discussions.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Edelstein SL 1967. The Hippocratic physician In Ancient Medicine. Selected Papers of Ludwig Edelstein Temkin O &Temkin C, Eds.: 87–110. Baltimore: Johns Hopkins Press. [Google Scholar]
- 2.Feinstein AR & Horwitz RI. 1982. Double standards, scientific methods, and epidemiologic research. N. Engl. J. Med 307: 1611–1617. [DOI] [PubMed] [Google Scholar]
- 3.Gabriel SE & Normand S-LT. 2012. Getting the methods right—the foundation of patient-centered outcomes research. N. Engl. J. Med 367: 787–790. [DOI] [PubMed] [Google Scholar]
- 4.Lillie EO, Patay B, Diamant J, et al. 2011. The n-of-1 clinical trial: the ultimate strategy for individualizing medicine? Per. Med 8: 161–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schork NJ 2015. Personalized medicine: time for one-person trials. Nature 520: 609–611. [DOI] [PubMed] [Google Scholar]
- 6.Calkin AC & Tontonoz P. 2010. Genome-wide association studies identify new targets in cardiovascular disease. Sci. Transl. Med 2: 48ps46. [DOI] [PubMed] [Google Scholar]
- 7.McCarthy MI 2017. Painting a new picture of person-alised medicine for diabetes. Diabetologia 60: 793–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins FS & McKusick VA. 2001. Implications of the Human Genome Project for medical science. JAMA 285: 540–544. [DOI] [PubMed] [Google Scholar]
- 9.Lander ES 2011. Initial impact of the sequencing of the human genome. Nature 470: 187–197. [DOI] [PubMed] [Google Scholar]
- 10.Visscher PM, Brown MA, McCarthy MI, et al. 2012. Five years of GWAS discovery. Am. J. Hum. Genet 90: 7–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuchsberger C, Flannick J, Teslovich TM, et al. 2016. The genetic architecture of type 2 diabetes. Nature 536: 41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cravatt BF, Simon GM & Yates JR III. 2007. The biological impact of mass-spectrometry-based proteomics. Nature 450: 991–1000. [DOI] [PubMed] [Google Scholar]
- 13.Nicholson JK & Lindon JC. 2008. Systems biology: metabonomics. Nature 455: 1054–1056. [DOI] [PubMed] [Google Scholar]
- 14.Wang Z, Gerstein M & Snyder M. 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet 10: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clemente JC, Ursell LK, Parfrey LW, et al. 2012. The impact of the gut microbiota on human health: an integrative view. Cell 148: 1258–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ardlie KG, Deluca DS, Segre AV, et al. 2015. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348: 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kundaje A, Meuleman W, Ernst J, et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mias GI & Snyder M. 2013. Personal genomes, quantitative dynamic omics and personalized medicine. Quant. Biol 1: 71–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hyman DM, Puzanov I, Subbiah V, et al. 2015. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. N. Engl. J. Med 373: 726–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller TE, Liau BB, Wallace LC, et al. 2017. Transcription elongation factors represent in vivo cancer dependencies in glioblastoma. Nature 547: 355–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melief CJM 2017. Cancer: precision T-cell therapy targets tumours. Nature 547: 165–167. [DOI] [PubMed] [Google Scholar]
- 22.Ott PA, Hu Z, Keskin DB, et al. 2017. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547: 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sahin U, Derhovanessian E, Miller M, et al. 2017. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 547: 222–226. [DOI] [PubMed] [Google Scholar]
- 24.Hindorff LA, Gillanders EM & Manolio TA. 2011. Genetic architecture of cancer and other complex diseases: lessons learned and future directions. Carcinogenesis 32: 945–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tuomi T, Santoro N, Caprio S, et al. 2014. The many faces of diabetes: a disease with increasing heterogeneity. Lancet 383: 1084–1094. [DOI] [PubMed] [Google Scholar]
- 26.American Diabetes Association. 2017. 2. Classification and diagnosis of diabetes. Diabetes Care 40(Suppl. 1): S11–S24. [DOI] [PubMed] [Google Scholar]
- 27.Groop LC, Eriksson J, Ekstrand A, et al. 1991. Metabolic characteristics of autoimmune diabetes mellitus in adults. Diabetologia 34: 46–51. [DOI] [PubMed] [Google Scholar]
- 28.Chatzianagnostou K, Iervasi G & Vassalle C. 2016. Challenges of LADA diagnosis and treatment. Am. J. Ther 23: e1270–e1274. [DOI] [PubMed] [Google Scholar]
- 29.Mishra R, Chesi A, Cousminer DL, et al. 2017. Relative contribution of type 1 and type 2 diabetes loci to the genetic etiology of adult-onset, non-insulin-requiring autoimmune diabetes. BMC Med 15: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karalliedde J & Gnudi L. 2016. Diabetes mellitus, a complexand heterogeneousdisease, and theroleofinsulin resistance as a determinant of diabetic kidney disease. Nephrol. Dial. Transplant 31: 206–213. [DOI] [PubMed] [Google Scholar]
- 31.Umpierrez GE 2006. Ketosis-prone type 2 diabetes: time to revise the classification of diabetes. Diabetes Care 29: 2755–2757. [DOI] [PubMed] [Google Scholar]
- 32.Yoon K-H, Lee J-H, Kim J-W, et al. 2006. Epidemic obesity and type 2 diabetes in Asia. Lancet 368: 1681–1688. [DOI] [PubMed] [Google Scholar]
- 33.Ma RCW & Chan JCN. 2013. Type 2 diabetes in East Asians: similarities and differences with populations in Europe and the United States. Ann. N.Y. Acad. Sci 1281: 64–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams AL, Jacobs SBR, et al. ; SIGMA Type 2 Diabetes Consortium. 2014. Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature 506: 97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fumagalli M, Moltke I, Grarup N, et al. 2015. Greenlandic Inuit show genetic signatures of diet and climate adaptation. Science 349: 1343–1347. [DOI] [PubMed] [Google Scholar]
- 36.Estrada K, Aukrust I, et al. ; SIGMA Type 2 Diabetes Consortium. 2014. Association of a low-frequency variant in HNF1A with type 2 diabetes in a Latino population. JAMA 311: 2305–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oak S, Gaur LK, Radtke J, et al. 2014. Masked and overt autoantibodies specific to the DPD epitope of 65-kDa glutamate decarboxylase (GAD65-DPD) are associated with preserved β-cell functional reserve in ketosis-prone diabetes. J. Clin. Endocrinol. Metab 99: E1040–E1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balasubramanyam A, Garza G, Rodriguez L, et al. 2006. Accuracy and predictive value of classification schemes for ketosis-prone diabetes. Diabetes Care 29: 2575–2579. [DOI] [PubMed] [Google Scholar]
- 39.Brooks-Worrell BM, Iyer D, Coraza I, et al. 2013. Islet-specific T-cell responses and proinflammatory monocytes define subtypes of autoantibody-negative ketosis-prone diabetes. Diabetes Care 36: 4098–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Powe CE, Allard C, Battista M-C, et al. 2016. Heterogeneous contribution of insulin sensitivity and secretion defects to gestational diabetes mellitus. Diabetes Care 39: 1052–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ahlqvist E, Storm P, Karajamaki A, et al. 2017. Clustering of adult-onset diabetes into novel subgroups guides therapy and improves prediction of outcome. bioRxiv 10.1101/186387. [DOI] [Google Scholar]
- 42.Florez JC 2016. Leveraging genetics to advance type 2 diabetes prevention. PLoS Med 13: e1002102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Telenti A, Pierce LCT, Biggs WH, et al. 2016. Deep sequencing of 10,000 human genomes. Proc. Natl. Acad. Sci. USA 113: 11901–11906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seoighe C & Scally A. 2017. Inference of candidate germline mutator loci in humans from genome-wide haplotype data. PLoS Genet 13: e1006549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lawlor DA, Harbord RM, Sterne JAC, et al. 2008. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat. Med 27: 1133–1163. [DOI] [PubMed] [Google Scholar]
- 46.Zeggini E, Scott LJ, Saxena R, et al. 2008. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet 40: 638–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dupuis J, Langenberg C, Prokopenko I, et al. 2010. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet 42: 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Voight BF, Scott LJ, Steinthorsdottir V, et al. 2010. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat. Genet 42: 579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scott RA, Lagou V, Welch RP, et al. 2012. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat. Genet 44: 991–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morris AP, Voight BF, Teslovich TM, et al. 2012. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet 44: 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Manning AK, Hivert M-F, Scott RA, et al. 2012. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat. Genet 44: 659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scott RA, Scott LJ, Mägi R, et al. 2017. An expanded genome-wide association study of type 2 diabetes in Europeans. Diabetes 66: 2888–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scott RA, Freitag DF, Li L, et al. 2016. A genomic approach to therapeutic target validation identifies a glucose-lowering GLP1R variant protective for coronary heart disease. Sci. Transl. Med 8: 341ra76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moltke I, Grarup N, Jørgensen ME, et al. 2014. A common Greenlandic TBC1D4 variant confers muscle insulin resistance and type 2 diabetes. Nature 512: 190–193. [DOI] [PubMed] [Google Scholar]
- 55.Manousaki D, Kent JW, Haack K, et al. 2016. Toward precision medicine: TBC1D4 disruption is common among the Inuit and leads to underdiagnosis of type 2 diabetes. Diabetes Care 39: 1889–1895. [DOI] [PubMed] [Google Scholar]
- 56.Rusu V, Hoch E, Mercader JM, et al. 2017. Type2diabetes variantsdisruptfunctionofSLC16A11throughtwodistinct mechanisms. Cell 170: 199–212.e20.28666119 [Google Scholar]
- 57.Stadler LKJ & Farooqi IS. 2017. A new drug target for type 2 diabetes. Cell 170: 12–14. [DOI] [PubMed] [Google Scholar]
- 58.Dimas AS, Lagou V, Barker A, et al. 2014. Impact of type 2 diabetes susceptibility variants on quantitative glycemic traits reveals mechanistic heterogeneity. Diabetes 63: 2158–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yaghootkar H, Scott RA, White CC, et al. 2014. Genetic evidence for a normal-weight metabolically obese phenotype linking insulin resistance, hypertension, coronary artery disease, and type 2 diabetes. Diabetes 63: 4369–4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yaghootkar H, Lotta LA, Tyrrell J, et al. 2016. Genetic evidence for a link between favorable adiposity and lower risk of type 2 diabetes, hypertension, and heart disease. Diabetes 65: 2448–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hamman RF, Bell RA, Dabelea D, et al. 2014. The SEARCH for diabetes in youth study: rationale, findings, and future directions. Diabetes Care 37: 3336–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Farmer A & Fox R. 2011. Diagnosis, classification, and treatment of diabetes. BMJ 342: d3319. [DOI] [PubMed] [Google Scholar]
- 63.Oram RA, Patel K, Hill A, et al. 2016. A type 1 diabetes genetic risk score can aid discrimination between type 1 and type 2 diabetes in young adults. Diabetes Care 39: 337–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patel KA, Oram RA, Flanagan SE, et al. 2016. Type 1 diabetes genetic risk score: a novel tool to discriminate monogenic and type 1 diabetes. Diabetes 65: 2094–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jensen PB, Jensen LJ & Brunak S. 2012. Mining electronic health records: towards better research applications and clinical care. Nat. Rev. Genet 2: 395–405. [DOI] [PubMed] [Google Scholar]
- 66.Simmons M, Singhal A & Lu Z. 2016. Text mining for precision medicine: bringing structure to EHRs and biomedical literature to understand genes and health. Adv. Exp. Med. Biol 939: 139–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li L, Cheng W-Y, Glicksberg BS, et al. 2015. Identification of type 2 diabetes subgroups through topological analysis of patient similarity. Sci. Transl. Med 7: 311ra174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bain JR, Stevens RD, Wenner BR, et al. 2009. Metabolomics applied to diabetes research: moving from information to knowledge. Diabetes 58: 2429–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gooding JR, Jensen MV & Newgard CB. 2016. Metabolomics applied to the pancreatic islet. Arch. Biochem. Biophys 589: 120–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Newgard CB 2017. Metabolomics and metabolic diseases: where do we stand? Cell Metab 25: 43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Salek RM, Arita M, Dayalan S, et al. 2015. Embedding standards in metabolomics: the Metabolomics Society data standards task group. Metabolomics 11: 782–783. [Google Scholar]
- 72.Newgard CB, An J, Bain JR, et al. 2009. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab 9: 311–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang TJ, Larson MG, Vasan RS, et al. 2011. Metabolite profiles and the risk of developing diabetes. Nat. Med 17: 448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Menni C, Fauman E, Erte I, et al. 2013. Biomarkers for type 2 diabetes and impaired fasting glucose using a non-targeted metabolomics approach. Diabetes 62: 4270–4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang TJ, Ngo D, Psychogios N, et al. 2013. 2-Aminoadipic acid is a biomarker for diabetes risk. J. Clin. Invest 123: 4309–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ferrannini E, Natali A, Camastra S, et al. 2013. Early metabolic markers of the development of dysglycemia and type 2 diabetes and their physiological significance. Diabetes 62: 1730–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shin S-Y, Fauman EB, Petersen A-K, et al. 2014. An atlas of genetic influences on human blood metabolites. Nat. Genet 46: 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fall T, Salihovic S, Brandmaier S, et al. 2016. Non-targeted metabolomics combined with genetic analyses identifies bile acid synthesis and phospholipid metabolism as being associated with incident type 2 diabetes. Diabetologia 59: 2114–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nowak C, Salihovic S, Ganna A, et al. 2016. Effect of insulin resistance on monounsaturated fatty acid levels: a multi-cohort non-targeted metabolomics and Mendelian randomization study. PLoS Genet 12: e1006379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lotta LA, Scott RA, Sharp SJ, et al. 2016. Genetic predisposition to an impaired metabolism of the branched-chain amino acids and risk of type 2 diabetes: a Mendelian randomisation analysis. PLoS Med 13: e1002179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mahendran Y, Jonsson A, Have CT, et al. 2017. Genetic evidence of a causal effect of insulin resistance on branched-chain amino acid levels. Diabetologia 60: 873–878. [DOI] [PubMed] [Google Scholar]
- 82.Walford GA, Porneala BC, Dauriz M, et al. 2014. Metabolite traits and genetic risk provide complementary information for the prediction of future type 2 diabetes. Diabetes Care 37: 2508–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Maurano MT, Humbert R, Rynes E, et al. 2012. Systematic localization of common disease-associated variation in regulatory DNA. Science 337: 1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stricker SH, Köferle A & Beck S. 2016. From profiles to function in epigenomics. Nat. Rev. Genet 18: 51–66. [DOI] [PubMed] [Google Scholar]
- 85.Onengut-Gumuscu S, Chen W-M, Burren O, et al. 2015. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat. Genet 47: 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Frayling TM, Timpson NJ, Weedon MN, et al. 2007. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316: 889–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Claussnitzer M, Dankel SN, Kim K-H, et al. 2015. FTO obesity variant circuitry and adipocyte browning in humans. N. Engl. J. Med 373: 895–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yassour M, Lim MY, Yun HS, et al. 2016. Sub-clinical detection of gut microbial biomarkers of obesity and type 2 diabetes. Genome Med 8: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Qin J, Li Y, Cai Z, et al. 2012. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490: 55–60. [DOI] [PubMed] [Google Scholar]
- 90.Karlsson FH, Tremaroli V, Nookaew I, et al. 2013. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498: 99–103. [DOI] [PubMed] [Google Scholar]
- 91.Pedersen HK, Gudmundsdottir V, Nielsen HB, et al. 2016. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 535: 376–381. [DOI] [PubMed] [Google Scholar]
- 92.American Diabetes Association. 2011. Standards of medical care in diabetes—2011. Diabetes Care 34(Suppl. 1): S11–S61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou K, Donnelly L, Yang J, et al. 2014. Heritability of variation in glycaemic response to metformin: a genome-wide complex trait analysis. Lancet Diabetes Endocrinol 2: 481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Florez JC, Burtt N, de Bakker PIW, et al. 2004. Haplotype structure and genotype–phenotype correlations of the sulfonylurea receptor and the islet ATP-sensitive potassium channel gene region. Diabetes 53: 1360–1368. [DOI] [PubMed] [Google Scholar]
- 95.Gloyn AL, Pearson ER, Antcliff JF, et al. 2004. Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. N. Engl. J. Med 350: 1838–1849. [DOI] [PubMed] [Google Scholar]
- 96.McCarthy MI & Hattersley AT. 2008. Learning from molecular genetics: novel insights arising from the definition of genes for monogenic and type 2 diabetes. Diabetes 57: 2889–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hattersley AT & Patel KA. 2017. Precision diabetes: learning from monogenic diabetes. Diabetologia 60: 769–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pearson ER, Starkey BJ, Powell RJ, et al. 2003. Genetic cause of hyperglycaemia and response to treatment in diabetes. Lancet 362: 1275–1281. [DOI] [PubMed] [Google Scholar]
- 99.Steele AM, Shields BM, Wensley KJ, et al. 2014. Prevalence of vascular complications among patients with glucokinase mutations and prolonged, mild hyperglycemia. JAMA 311: 279–286. [DOI] [PubMed] [Google Scholar]
- 100.Stride A, Shields B, Gill-Carey O, et al. 2014. Cross-sectional and longitudinal studies suggest pharmacological treatment used in patients with glucokinase mutations does not alter glycaemia. Diabetologia 57: 54–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chakera AJ, Steele AM, Gloyn AL, et al. 2015. Recognition and management of individuals with hyperglycemia because of a heterozygous glucokinase mutation. Diabetes Care 38: 1383–1392. [DOI] [PubMed] [Google Scholar]
- 102.Pearson ER, Pruhova S, Tack CJ, et al. 2005. Molecular genetics and phenotypic characteristics of MODY caused by hepatocyte nuclear factor 4alpha mutations in a large European collection. Diabetologia 48: 878–885. [DOI] [PubMed] [Google Scholar]
- 103.Laver TW, Colclough K, Shepherd M, et al. 2016. The common p.R114W HNF4A mutation causes a distinct clinical subtype of monogenic diabetes. Diabetes 65: 3212–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Flannick J, Johansson S & Njølstad PR. 2016. Common and rare forms of diabetes mellitus: towards a continuum of diabetes subtypes. Nat. Rev. Endocrinol 12: 394–406. [DOI] [PubMed] [Google Scholar]
- 105.Collins FS & Varmus H. 2015. A new initiative on precision medicine. N. Engl. J. Med 372: 793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tahrani AA, Bailey CJ, Del Prato S, et al. 2011. Management of type 2 diabetes: new and future developments in treatment. Lancet 378: 182–197. [DOI] [PubMed] [Google Scholar]
- 107.Kahn SE, Haffner SM, Heise MA, et al. 2006. Glycemic durability of rosiglitazone, metformin, or glyburide monotherapy. N. Engl. J. Med 355: 2427–2443. [DOI] [PubMed] [Google Scholar]
- 108.Florez JC 2017. Pharmacogenetics in type 2 diabetes: precision medicine or discovery tool? Diabetologia 60: 800–807. [DOI] [PubMed] [Google Scholar]
- 109.Zhou K et al. ; GoDARTS and UKPDS Diabetes Pharmacogenetics Study Group; Wellcome Trust Case Control Consortium 2. 2011. Common variants near ATM are associated with glycemic response to metformin in type 2 diabetes. Nat. Genet 43: 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Luizon MR, Eckalbar WL, Wang Y, et al. 2016. Genomic characterization of metformin hepatic response. PLoS Genet 12: e1006449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhou K, Yee SW, Seiser EL, et al. 2016. Variation in the glucose transporter gene SLC2A2 is associated with glycemic response to metformin. Nat. Genet 48: 1055–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Florez JC, Jablonski KA, McAteer JB, et al. 2012. Effects of genetic variants previously associated with fasting glucose and insulin in the Diabetes Prevention Program. PLoS One 7: e44424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhou K, Donnelly L, Burch L, et al. 2010. Loss-of-function CYP2C9 variants improve therapeutic response to sulfonylureas in type 2 diabetes: a Go-DARTS Study. Clin. Pharmacol. Ther 87: 52–56. [DOI] [PubMed] [Google Scholar]