Abstract
SNP-heritability is a fundamental quantity in the study of complex traits. Recent works have shown that existing methods to estimate genome-wide SNP-heritability yield biases when their assumptions are violated. While various approaches have been proposed to account for frequency- and LD-dependent genetic architectures, it remains unclear which estimates reported in the literature are reliable. Here we show that genome-wide SNP-heritability can be accurately estimated from biobank-scale data irrespective of genetic architecture, without specifying a heritability model or partitioning SNPs by allele frequency and/or LD. We show analytically and through extensive simulations starting from real genotypes (UK Biobank, N = 337K) that, unlike existing methods, our closed-form estimator is robust across a wide range of architectures. We provide estimates of SNP-heritability for 22 complex traits in the UK Biobank and show that, consistent with our results in simulations, existing biobank-scale methods yield estimates up to 30% different from our theoretically-justified approach.
Editorial Summary:
The authors use theoretical justifications coupled with extensive simulations to accurately estimate SNP-heritability for 22 complex traits and diseases from the UK Biobank data irrespective of the underlying genetic architecture of the trait.
SNP-heritability, the proportion of phenotypic variance attributable to the additive effects of a given set of SNPs, is a fundamental quantity in genetics1; it provides an upper bound on risk prediction from a linear model2 and, when defined as a function of all SNPs on an array, yields insights into the “missing heritability” of complex traits3–5. Traditionally, SNP-heritability is estimated by fitting variance components models with REML3,6–9. With some exceptions8, REML-based methods are not scalable to biobanks that assay hundreds of thousands of individuals (e.g., UK Biobank10). SNP-heritability can also be estimated by assessing the deviation in marginal association statistics as a function of LD scores11–14; such methods can scale to millions of individuals. More recently, a randomized extension of Haseman-Elston regression15 was shown to estimate a single genetic variance component from individual-level data as accurately as REML methods but in a fraction of the run-time16.
To facilitate inference, all existing methods for genome-wide SNP-heritability inference make assumptions on genetic architecture, which is typically parametrized by polygenicity (the number of variants with effects larger than some small constant δ) and MAF/LD-dependence (the coupling of effects with minor allele frequency (MAF), local linkage disequilibrium (LD), or other functional annotations)17. Since the true genetic architecture of any given trait is unknown, existing methods are susceptible to bias and often yield vastly different estimates even when applied to the same data9,14,18. Although multi-component methods that stratify SNPs by MAF/LD ameliorate some of these robustness issues7,18,19, fitting multiple variance components to biobank-scale data with REML is highly resource-intensive8 and it is unclear whether multi-component methods based on summary statistics produce accurate estimates of total SNP-heritability. Alternate methods that explicitly model MAF/LD-dependency6,9,14 are also sensitive to model misspecification6,9,14,18,19. In addition, genetic architecture varies across traits and populations due to, for example, variable degrees of negative selection acting on different traits in different populations17,20–25. Methods that jointly infer SNP-heritability and parameters such as the strength of negative selection or polygenicity14,23,26 are computationally intensive and/or sensitive to LD-dependency. Thus, it remains unclear which estimates of SNP-heritability computed from biobank-scale data are reliable.
In this work, we investigate whether genome-wide SNP-heritability can be accurately estimated under a generalized random effects (GRE) model that makes minimal assumptions on genetic architecture. Under this model, every causal effect has an arbitrary SNP-specific variance, and SNP-heritability is defined as the sum of the SNP-specific variances (Methods). To the best of our knowledge, all existing methods make additional assumptions on top of the GRE model (Table 1). For example, GREML3 (and several other methods8,16,27) imposes an inverse relationship between MAF and allelic effect size whereas LDAK assumes that each SNP-specific variance is inversely proportional to both MAF and LD tagging6,9. We derive a closed-form estimator for SNP-heritability as a function of marginal association statistics and in-sample LD and show that this estimator is consistent (approaches the true SNP-heritability as sample size increases) and unbiased (its expectation is equal to the true SNP-heritability) when the number of individuals exceeds the number of SNPs. Most importantly, the accuracy of this estimator is invariant to genetic architecture. While the GRE estimator is similar in form to previously proposed “fixed effect estimators,”28,29 our approach differs from previous work in two main ways. First, SNP-heritability defined under a fixed effect model is different from the estimand of interest here (Methods). Second, previous work applied the estimator locally to identify regions contributing disproportionately to the genome-wide signal28,29; here we define a different genome-wide estimator (Equation 1) that requires large-scale genotype data. In addition, previous work applied an SVD-based regularization to account for errors in LD estimation from reference panels29 which was unnecessary in this work (Methods).
Table 1.
Existing methods to estimate SNP-heritability impose additional assumptions on top of the generalized random effects (GRE) model. Under the GRE model, the causal effects at any two SNPs are assumed to be independent ( for all ) and genome-wide SNP-heritability is defined as , where each can be an arbitrary nonnegative real number as long as (Methods). All existing methods make assumptions on the distribution of and/or the form of that can be subsumed under the GRE model. To simplify notation, we assume for each model that phenotypes are standardized in the population (i.e. for every individual ).
| Model | Assumptions on | Description |
|---|---|---|
| Generalized random effects | , , | Each SNP has a nonnegative SNP-specific variance . Total SNP-heritability is . |
| GREML-SC 3,8,16 | Each SNP explains an equal portion of . In other words, for all . | |
| GREML-MC 7,8,18,42,43 | is partitioned by a set of disjoint SNP partitions that span all SNPs. Partition contains SNPs that have per-SNP variances . Total SNP-heritability is . | |
| LDAK6,9 | , | Each SNP-specific variance is proportional to a function of (the MAF of SNP ) and to (a SNP-specific weight that is a function of the inverse of the LD score of SNP ). controls the relationship between and . The most recent recommendation by ref.9 is to assume . |
| LDSC11 | , | Each SNP explains an equal portion of (similar to the GREML-SC model when is defined with respect to the same set of SNPs). |
| S-LDSC12,13,30 | , | Each SNP-specific variance is a linear function of a set of annotations where each represents a binary or continuous-valued annotation. is the value of annotation at SNP . is the expected contribution of a one-unit increase in annotation to each SNP-specific variance. |
| SumHer14 | , | An extension of the LDAK model to operate on summary-level data; can also efficiently partition by multiple annotations. The most recent recommendations by refs.9,14 is to set . |
Through extensive simulations across a range of MAF/LD-dependent architectures starting from real genotypes from the UK Biobank10 (337K individuals, 593K SNPs), we find that the GRE estimator is nearly unbiased across all architectures whereas existing methods are sensitive to model misspecification. For example, across 126 distinct architectures, the maximum bias of the GRE estimator is 2% of the simulated SNP-heritability whereas stratified LD score regression (S-LDSC)12,13 and SumHer14 yield biases between −64% and 28%. For completeness, we also contrast the GRE estimator with several REML-based methods in simulations at lower sample sizes (due to the computational burden of most REML methods) and find that, consistent with recent reports18, all REML-based methods are biased when their model assumptions are violated, and multi-component REML methods that stratify SNPs by MAF and LD score (GREML-LDMS-I18) are more accurate than single-component REML methods. The performance of the GRE estimator is similar to that of GREML-LDMS-I, thereby confirming that SNP-heritability can be accurately estimated without stratifying SNPs or specifying a heritability model6,9,14.
Finally, we use marginal association statistics and in-sample LD from 290K unrelated British individuals and 460K SNPs (MAF > 1%) to estimate SNP-heritability for 22 complex traits in the UK Biobank10. Consistent with simulations, estimates from S-LDSC and SumHer differ from the GRE estimates by a median of −9% and 11%, respectively, across the 18 traits with SNP-heritability estimates exceeding 0.05. For example, for height, estimates from S-LDSC (0.56) and SumHer (0.63) are approximately 7% lower and 5% higher, respectively, than our estimate of 0.60. Similarly, for hypertension, estimates from S-LDSC (0.14) and SumHer (0.18) are ±12.5% different from our estimate of 0.16. Taken together, our results demonstrate that SNP-heritability can be accurately estimated from biobank-scale data without prior knowledge of the genetic architecture the trait, motivating the development of scalable methods with fewer modeling assumptions.
Results
Overview of the approach
We investigate the utility of an estimator derived under a model that makes minimal assumptions on genetic architecture. We model the standardized phenotype of an individual as where is an M-vector of standardized genotypes, is the corresponding vector of standardized effects, and is environmental noise (Methods). The effect size of each SNP is assumed to have mean zero and a finite SNP-specific variance ( for SNP ) that is allowed to be 0; the covariance between all pairs of effects is assumed to be zero. We term this model the “generalized random effects” (GRE) model as, to the best of our knowledge, all existing methods impose additional assumptions on top of this model. For example, the single-component GREML model3 assumes for whereas the most recent LDAK model9 assumes (where is a SNP-specific LD weight and is MAF) (Table 1). Under the GRE model, the SNP-heritability explained by the SNPs is the sum of the SNP-specific variances: (Methods).
Given genotype measurements across individuals at SNPs and assuming , the estimator , where is the vector of estimated marginal effects, is the pseudoinverse of the in-sample LD matrix, and is the rank of the in-sample LD, is an unbiased estimator of SNP-heritability under the GRE model. That is, (Methods). Unfortunately, even the largest biobanks currently have (i.e. UK Biobank has genotyped SNPs in unrelated British individuals), which limits the utility of the above estimator. We therefore extend our approach by partitioning the genome by chromosome:
| #(1) |
where for chromosome with SNPs, is the of estimated effects, is the pseudoinverse of the in-sample LD matrix, and is the rank of the in-sample LD. Although this estimator introduces bias, we show through extensive simulations that the magnitude of the bias is extremely small when is sufficiently larger than .
The GRE estimator is robust in simulations
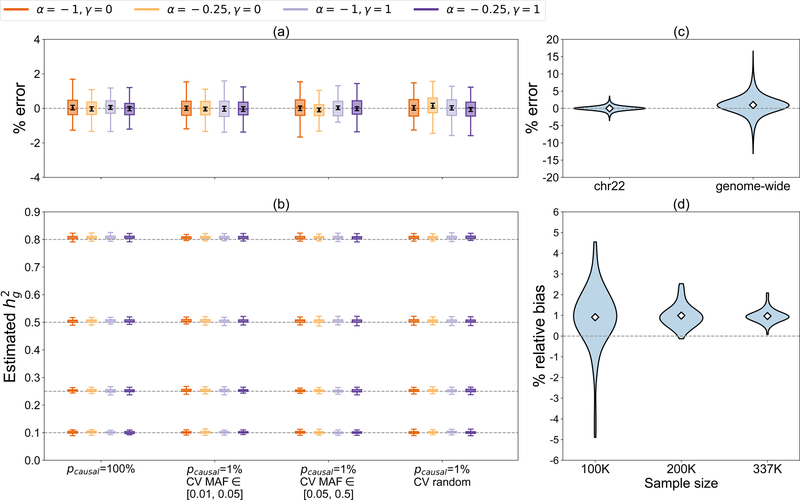
To investigate the bias and variance of , we perform simulations starting from real genotypes , UK Biobank10). First, we simulate 64 MAF/LD-dependent quantitative trait architectures from chromosome 22 typed SNPs) by varying the SNP-heritability , proportion of causal variants , distribution of causal variant MAF (CV MAF), and strength of coupling between effect size and MAF/LD; we use “LDAK-LD-dependent” to describe causal effects that are coupled with “LDAK weights” (Methods). To compare estimates across different values of , we assess bias as a percentage of the simulated value of (relative bias). Errors of individual estimates are also expressed as percentages of . Consistent with analytical derivations, the GRE estimator restricted to chromosome 22 is unbiased across the 64 architectures (bias p-value < 0.05/16 is considered significant in order to correct for 16 tests (architectures) at each value of ; Methods) (Figure 1ac, Supplementary Table 1). The average relative bias across the 64 architectures is and the largest bias under any single architecture is (Supplementary Figure 1a, Supplementary Table 1). In simulations of unascertained case-control studies (Methods), the GRE estimator is approximately unbiased across a range of disease prevalences (for , relative bias range is [−0.20%, 0.30%]) and has larger variance for lower prevalences (Supplementary Figure 2a, Supplementary Table 2). For ascertained case-control studies, estimates are downward-biased but invariant to architecture (when , prevalence = 0.10, and , relative bias is approximately −4%) (Supplementary Table 3). Masking 0%, 50%, or 100% of causal SNPs from the observed summary statistics induces downward-bias when CV MAF = [0.01, 0.05] due to lower average LD between the observed SNPs and masked causal SNPs (Supplementary Figure 3). The analytical estimator of the standard error (Methods) is well-calibrated (Supplementary Figure 4a, Supplementary Table 4). As expected, partitioning chromosome 22 into disjoint, non-independent blocks induces upward bias that increases as block size decreases (Supplementary Figure 5, Supplementary Table 5).
Figure 1.
Simulations under 64 distinct MAF/LD-dependent architectures (N = 337205). For each value of , phenotypes were drawn according to one of 16 genetic architectures defined by , CV MAF, , and (Methods). (a) Distribution of errors (as a percentage of ), where is the estimate from the -th simulation under a given genetic architecture, in simulations on chromosome 22 (M = 9654 SNPs). was computed with 1 chromosome-wide LD block. Black points and error bars represent the mean and standard errors of the mean (s.e.m.) which were used to test whether the bias under a single architecture is significant (Methods). (b) Distribution of in genome-wide simulations (M = 593300 SNPs) where was computed with 22 chromosome-wide LD blocks. In (a) and (b), each boxplot represents estimates from 100 simulations. Boxplot whiskers extend to the minimum and maximum estimates located within from the first and third quartiles, respectively. (c) Distribution of errors for chromosome 22 and genome-wide simulations. Each violin plot represents the errors of 6400 estimates (64 genetic architectures × 100 simulation replicates). (d) Distribution of relative bias (as a percentage of ) as a function of sample size (N = 100K, 200K, or 337K) in genome-wide simulations. Each violin plot represents 64 estimates of relative bias. In (c) and (d), the white diamonds mark the mean of each distribution.
Next, we perform genome-wide simulations individuals, SNPs) to assess with the 22-block approximation (Equation 1). Despite the approximation, is highly accurate and robust across all 64 MAF- and LDAK-LD-dependent quantitative trait architectures (Figure 1b, 1c). Across the 64 architectures, the bias ranges from 0.07% to (Supplementary Figure 1b, Supplementary Table 6). Across all 6400 simulations (64 genetic architectures × 100 simulation replicates), the largest error of any single estimate is approximately (Figure 1c). As increases, the variance of decreases while the relative bias appears to be approximately fixed, ranging between 0.91% and 0.99% (Figure 1d). These trends hold for a range of (Supplementary Figure 6, Supplementary Table 6), for unascertained case-control studies (Supplementary Figure 2b, Supplementary Table 7), and in a smaller set of simulations with individuals of South Asian ancestry and SNPs (Supplementary Table 8; Methods). Most importantly, the accuracy of the GRE estimator is invariant to the underlying architecture (Figure 1b). The analytical estimator for the standard error is downward-biased (and invariant to genetic architecture) with respect to the empirical standard deviation of estimates (Supplementary Figure 4b, Supplementary Table 9). For example, across 16 architectures where , the empirical standard deviation of 100 independent estimates ranges from 0.0049 to 0.0064, whereas our estimated standard errors are approximately 0.0036 across all architectures (Supplementary Figure 4b, Supplementary Table 9).
We investigate the effects of unmodeled substructure and/or cryptic relatedness by filtering individuals at different kinship coefficient thresholds (Methods) and find that using stricter relatedness thresholds increases the variance of the estimates (due to smaller sample size) while reducing bias, albeit not significantly (Supplementary Figure 7, Supplementary Table 10). To assess the impact of population stratification, we simulated an effect of the first genetic principal component (PC) on phenotype and computed OLS association statistics both with and without adjusting for the first PC (Methods). As expected, OLS without PC adjustment yields inflated estimates while OLS with PC adjustment yields approximately unbiased estimates (Supplementary Figure 8, Supplementary Table 11). However, even when a relatively large proportion of phenotypic variance is explained by the first PC (e.g., , ), the maximum bias we observe using unadjusted association statistics is (). Together, these results indicate that the GRE estimator is robust to modest amounts of unmodeled substructure and/or stratification. In all subsequent analyses, we compute with the 22-block approximation as this provides sufficiently accurate estimates and a fair comparison to other methods.
Comparison of methods to estimate SNP-heritability
We compare with existing state-of-the-art methods that are easily scalable to the full UK Biobank data : LD score regression (LDSC), which assumes and no coupling of effects with LD11; stratified LD score regression (S-LDSC), which partitions by a set of annotations of interest12,13; and SumHer, a scalable extension of LDAK which explicitly models MAF/LD-dependency through a specific form of the SNP-specific variances14 (Table 1). To ensure a fair comparison, LD scores for all methods are computed using in-sample LD among the M SNPs, and in all simulations we aim to estimate the SNP-heritability explained by the same M SNPs (Methods).
As expected, is robust across all architectures while LDSC, S-LDSC, and SumHer are sensitive to model misspecification. For example, when (Figure 2), LDSC is approximately unbiased under the “single-component GREML model” (, ) but is sensitive to CV MAF and the degree of coupling between effect size and MAF/LD (e.g., when , relative bias ranges from −44% to 50%) (Supplementary Table 12). Similarly, SumHer is accurate under the “LDAK model” ()but highly sensitive to other architectures (when , relative bias ranges from −19% to 22%) (Figure 2, Supplementary Table 13). S-LDSC (MAF), which partitions by 10 MAF bins (Supplementary Table 14; Methods), is less biased than LDSC when effects are coupled with only MAF, but is significantly downward-biased when effects are also coupled with LDAK weights (for , relative bias range is [1.9%, 7.0%] when and [−58%, −37%] when ) (Figure 2, Supplementary Table 15). S-LDSC with 10 MAF bins and an additional “level of LD” annotation, denoted S-LDSC (MAF+LLD) (Methods), produces similar results (for , relative bias range is [1.8%, 6.5%] when and [−80%, −33%] when ) (Supplementary Table 16). In contrast, the relative bias of ranges from 0.45% to 1.3% across the same 16 architectures where and (Figure 2, Supplementary Table 6). These trends hold for a range of and : across 112 LDAK-LD- and/or MAF-dependent architectures, the average and range of the relative bias of each method are 0.96% [−0.06%, 2.1%] (GRE), −2.2% [−71%, 70%] (LDSC), −22% [−62%, 8.7%] (S-LDSC (MAF)), −29% [−89%, 9.0%] (S-LDSC (MAF+LLD)), and 2.8% [−27%, 28%] (SumHer) (Figure 1b, Figure 2, Supplementary Figures 9–12, Supplementary Tables 6,12,13,15,16). Across 14 alternative LD-dependent architectures where SNP-specific variances are coupled with inverse LD scores instead of LDAK weights (“LD-score-dependent” architectures; Methods, Supplementary Figure 13), remains nearly unbiased (relative bias range [0.52%, 1.3%]) whereas S-LDSC (MAF), S-LDSC (MAF+LLD), and SumHer are generally downward-biased (Supplementary Figure 14, Supplementary Table 17).
Figure 2.
Comparison of with LDSC, S-LDSC (MAF), and SumHer in genome-wide simulations (N = 337205, M = 593300, ). Left: Phenotypes were drawn under one of 16 MAF- and/or LDAK-LD-dependent architectures by varying , , , and CV MAF (Methods). Each boxplot contains estimates of from 100 simulations. Right: Relative bias of each method (as a percentage of ) across 112 distinct MAF- and LDAK-LD-dependent architectures (Methods). Each boxplot contains 112 points; each point is the relative bias estimated from 100 simulations under a single genetic architecture. The white diamonds mark the average of each distribution. Boxplot whiskers extend to the minimum and maximum estimates located within from the first and third quartiles, respectively.
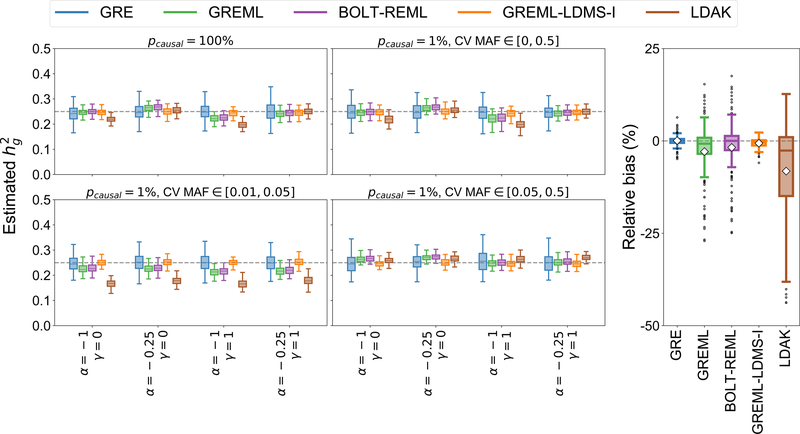
For completeness, we compare to four widely used REML-based methods: GREML, which assumes and no coupling of effects with LD3; GREML-LDMS-I, a multi-component extension of GREML that partitions SNPs by MAF and LD score18; BOLT-REML, a computationally efficient variance components estimation method with assumptions similar to those of GREML8; and LDAK, which assumes a specific form of the SNP-specific LD weights and recommends setting 6,9 (Table 1). Because it is computationally intractable to apply the REML-based methods to thousands of genome-wide simulations with 337K individuals, we perform simulations using a reduced number of individuals and SNPs (Methods). As expected, the single-component methods (GREML, BOLT-REML, and LDAK) are sensitive to MAF/LD-dependency whereas the GRE estimator is robust across all architectures. For example, when (Figure 3), GREML and BOLT-REML are accurate under the GREML model (GREML: , , Supplementary Table 18; BOLT-REML: , , Supplementary Table 19) and LDAK is approximately unbiased under the LDAK model (, , Supplementary Table 20), but all three are sensitive to CV MAF, and . Across 12 architectures where (Figure 3), the relative biases are within [−15%, 7.9%] (GREML), [−14%, 9.1%] (BOLT-REML), and [−34%, 8.2%] (LDAK) (Supplementary Tables 18–20). In contrast, for the same 12 architectures, yields relative biases in the range [−2.1%, 1.7%], which is comparable to the relative bias of GREML-LDMS-I (range [−2.9%, 1.5%]) using 8 GRMs (4 LD quartiles × 2 MAF bins) that align with CV MAF (Figure 3, Supplementary Tables 21, 22). These trends hold over a range of and : across 112 LDAK-LD- and/or MAF-dependent architectures (Supplementary Figures 15–19), the average and range of the relative bias are 0.09% [−4.9%, 6.4%] (GRE), −0.6% [−5.9%, 2.3%] (GREML-LDMS-I), −2.9% [−27%, 15%] (GREML), −1.8% [−25%, 18%] (BOLT-REML), and −8.2% [−44%, 13%] (LDAK) (Supplementary Tables 18–22). Similar trends are observed for LD-score-dependent architectures (Supplementary Figure 20, Supplementary Table 23). In an extreme example where CV MAF is tightly concentrated near 1%, GREML-LDMS-I with the same 8 GRMs as before is downward-biased whereas the GRE estimator remains robust (Supplementary Figure 21, Supplementary Tables 18–22). While the variance of our estimator is larger than the variances of the REML-based methods (Figure 3), our approach is designed for sample sizes several orders of magnitude larger than what we used in these simulations. In summary, our results confirm that it is possible to accurately estimate under the GRE model.
Figure 3.
Comparison of with GREML, BOLT-REML, GREML-LDMS-I, and LDAK in small-scale simulations (N = 8430, M = 14821 SNPs). Left: Phenotypes were drawn under one of 16 MAF- and/or LDAK-LD-dependent architectures by varying , , , and CV MAF (Methods). Each boxplot contains estimates of from 100 simulations. Right: Relative bias of each method (as a percentage of the true ) across 112 distinct MAF- and LDAK-LD-dependent architectures (Methods). Each boxplot represents the distribution of 112 points; each point is the relative bias estimated from 100 simulations under a single genetic architecture. The white diamonds mark the average of each distribution. Boxplot whiskers extend to the minimum and maximum estimates located within from the first and third quartiles, respectively.
SNP-heritability of 22 complex traits in the UK Biobank
Finally, we compute for 22 complex traits in the UK Biobank (290K unrelated British individuals, 460K SNPs; Methods)10. For comparison, we also provide estimates from LDSC, S-LDSC (controlling for the baseline-LD model13,30), and SumHer. Of the 22 traits analyzed (6 quantitative, 16 binary), we focus on 18 traits for which (Table 2). For the 6 quantitative traits, ranges from 0.12 (smoking status) to 0.60 (height). Across the 12 binary traits, ranges from 0.064 (autoimmune disorders) to 0.16 (hypertension) (Table 2). These estimates are robust to filtering of individuals based on relatedness (Supplementary Table 24). We also computed from two additional sets of SNPs (MAF > 0.1% and MAF > 0.01%) and found that the estimates increase slightly for lower MAF thresholds (Supplementary Table 25), which is expected due to the increased number of SNPs. To enable a direct comparison between and the quantities estimated by LDSC, S-LDSC, and SumHer, we run the summary-statistics-based methods with LD scores and regression weights computed from in-sample LD and estimate defined as a function of the same set of SNPs (Methods). Across the 18 traits, S-LDSC (baseline-LD/in-sample) and SumHer (in-sample) differ from by a median of −9% and 11%, respectively (expressed as a percentage of ) (Figure 4, Table 2). As expected11, LDSC (in-sample) yields inflated estimates.
Table 2.
Estimates of from the GRE approach, LDSC (in-sample), S-LDSC (baseline-LD/in-sample), and SumHer (in-sample) for 22 complex traits and diseases in the UK Biobank (N = 290K unrelated British individuals, M = 460K typed SNPs).
| Trait | GRE | S.E. | LDSC | S.E. | S-LDSC | S.E. | SumHer | S.E. |
|---|---|---|---|---|---|---|---|---|
| Smoking Status | 0.122 | 3.90E-03 | 0.178 | 7.70E-03 | 0.110 | 8.50E-03 | 0.132 | 4.30E-03 |
| Height | 0.602 | 4.70E-03 | 0.730 | 2.70E-02 | 0.555 | 3.10E-02 | 0.634 | 2.70E-02 |
| BMI | 0.285 | 4.20E-03 | 0.436 | 1.20E-02 | 0.289 | 1.70E-02 | 0.315 | 9.00E-03 |
| WHR | 0.173 | 4.00E-03 | 0.256 | 1.20E-02 | 0.184 | 1.60E-02 | 0.198 | 9.40E-03 |
| Systolic Blood Pressure | 0.159 | 4.20E-03 | 0.243 | 9.00E-03 | 0.134 | 9.70E-03 | 0.177 | 5.70E-03 |
| Diastolic Blood Pressure | 0.154 | 4.20E-03 | 0.233 | 8.60E-03 | 0.130 | 9.70E-03 | 0.170 | 6.40E-03 |
| Eczema | 0.116 | 4.20E-03 | 0.165 | 1.10E-02 | 0.107 | 1.20E-02 | 0.130 | 8.80E-03 |
| Asthma | 0.116 | 4.90E-03 | 0.163 | 1.20E-02 | 0.116 | 1.70E-02 | 0.131 | 1.20E-02 |
| Hypertension | 0.162 | 4.00E-03 | 0.244 | 9.40E-03 | 0.142 | 1.10E-02 | 0.180 | 6.10E-03 |
| High Cholesterol | 0.082 | 5.10E-03 | 0.127 | 1.30E-02 | 0.138 | 5.80E-02 | 0.088 | 8.30E-03 |
| Diabetes (Any) | 0.070 | 3.70E-03 | 0.093 | 5.90E-03 | 0.062 | 8.70E-03 | 0.074 | 5.00E-03 |
| Type 2 Diabetes | 0.071 | 3.80E-03 | 0.090 | 6.10E-03 | 0.057 | 8.80E-03 | 0.071 | 4.00E-03 |
| Hypothyroidism | 0.088 | 5.20E-03 | 0.142 | 1.30E-02 | 0.078 | 1.20E-02 | 0.110 | 1.70E-02 |
| Thyroid Disorders | 0.084 | 5.20E-03 | 0.141 | 1.30E-02 | 0.080 | 1.20E-02 | 0.110 | 2.00E-02 |
| Endocrinopathies | 0.069 | 5.10E-03 | 0.084 | 7.00E-03 | 0.058 | 9.90E-03 | 0.068 | 5.00E-03 |
| Cardiovascular Diseases | 0.143 | 5.30E-03 | 0.228 | 1.10E-02 | 0.140 | 1.40E-02 | 0.164 | 6.00E-03 |
| Respiratory and ENT Diseases | 0.086 | 5.20E-03 | 0.120 | 1.20E-02 | 0.079 | 1.40E-02 | 0.090 | 9.50E-03 |
| Psoriasis | 0.019 | 5.00E-03 | 0.071 | 3.10E-02 | 0.035 | 1.20E-02 | 0.059 | 4.20E-02 |
| Dermatologic Disorders | 0.023 | 5.00E-03 | 0.049 | 1.40E-02 | 0.034 | 9.90E-03 | 0.031 | 1.10E-02 |
| Rheumatoid Arthritis | 0.008 | 5.00E-03 | 0.041 | 2.10E-02 | 0.010 | 7.90E-03 | 0.021 | 1.20E-02 |
| Autoimmune Disorders (Broad) | 0.063 | 5.10E-03 | 0.105 | 1.20E-02 | 0.050 | 9.50E-03 | 0.079 | 1.70E-02 |
| Autoimmune Disorders (Certain) | 0.015 | 5.00E-03 | 0.052 | 2.60E-02 | 0.005 | 7.60E-03 | 0.047 | 3.40E-02 |
Figure 4.
Percent difference of estimates from LDSC (in-sample), S-LDSC (baseline-LD/in-sample), and SumHer (in-sample) with respect to for 18 complex traits and diseases in the UK Biobank for which (N = 290K unrelated British individuals, M = 460K typed SNPs; Methods). Each bar represents the difference between the estimated from one of the methods (LDSC, S-LDSC, or SumHer) and as a percentage of . Black bars mark standard errors.
To compare to estimates reported in the literature, we also run the summary-statistics methods with their recommended parameter settings11,12,14,30 and with LD scores and regression weights computed from the 1000 Genomes Phase 3 reference panel (489 Europeans)31 – we note that when running these methods as recommended, their estimands are not equivalent to our definition of (see Methods and refs.11,12,14,19 for details). Across the 18 traits for which , the median differences with respect to are −11% for LDSC (1KG), −14% for S-LDSC (baseline-LD/1KG), and 38% for SumHer (1KG) (Supplementary Figure 22, Supplementary Table 26). For 9 of these traits, a previous study reported single-component BOLT-REML estimates (computed from a similar UK Biobank cohort27) that differ from our estimates by a median of 8% (Supplementary Table 26).
Runtime and memory requirement
We report the runtime and memory requirements for computing with the 22-block approximation from 337K individuals and 593K SNPs. First, computing chromosome-wide LD has complexity for chromosome with SNPs. In practice, this step does not impose a computational bottleneck because the computations can be parallelized over SNPs. Second, the pseudoinverse of each LD matrix is computed via truncated SVD, which has complexity for chromosome . For 50K typed SNPs this takes about 3 hours and 60GB of memory. Lastly, given the pseudoinverse LD matrices and OLS association statistics, computing has complexity For any of the traits analyzed in this work, this takes less than 1 hour and requires 24GB of memory; most of this time is spent loading the data into memory. For comparison, running LDSC, S-LDSC, or SumHer consists of precomputing LD scores and SNP-specific weights and performing linear regression to estimate the variance parameters. Precomputing LD scores and SNP-specific weights can be parallelized over blocks of SNPs. The second step (least squares regression) is for M SNPs in the regression and C variance parameters.
Discussion
In this work, we show that SNP-heritability can be accurately estimated under minimal assumptions on genetic architecture. Our proposed estimator allows the SNP-specific variances to capture arbitrary relationships between effect size and MAF/LD, and we demonstrate through simulations that its accuracy is invariant to genetic architecture. We show that all existing methods impose additional assumptions on the GRE model, and we confirm through simulations that these methods can be sensitive to model misspecification. One practical advantage of our approach over summary-statistics methods is that the estimand of our approach is always the same for a given genotype matrix, whereas the definitions and interpretations of the estimands of LDSC, S-LDSC, and SumHer depend on which SNPs are used in each step of inference (e.g., the SNPs used to compute LD scores need not be the same SNPs defining the estimand)11,12,19. Overall, our results show that while existing methods can yield biases, for the purpose of estimating total SNP-heritability, most methods are relatively robust.
We conclude with several caveats and future directions. First, the utility of critically depends on the ratio between the number of SNPs (M) and the number of individuals (N) – as increases, the eigenstructure of the in-sample LD matrix becomes increasingly distorted (larger eigenvalues are overestimated; smaller eigenvalues are underestimated)32. We mitigate this by assuming that chromosomes are approximately independent; as long as N exceeds the number of array SNPs per chromosome, provides meaningful estimates of SNP-heritability. While the utility of our approach is limited by the availability of individual-level biobank-scale data, this concern will abate as more biobanks are established33–35. A major limitation remains with respect to imputed/sequencing data as M will continue to be orders of magnitude larger than N for the foreseeable future. We defer an investigation of regularized estimation of LD in high-dimensional settings to future work.
Second, the theoretical guarantees of rely on the assumption that OLS association statistics and LD are estimated from the same genotypes. While summary statistics have been made publicly available for hundreds of large-scale GWAS, in-sample LD is usually unavailable for these studies since most are meta-analyses36. In addition, summary statistics are often computed using linear mixed models to control for confounding, and previous works have noted that the LD computation must be adjusted to accommodate mixed model association statistics36,37. Thus, the sensitivity of to reference panel LD (with or without regularized LD estimation) and/or mixed model association statistics remains unclear29,38. Furthermore, we simulate phenotypes from typed SNPs because imputed genotypes have highly irregular LD patterns9,18. Although it would be more realistic to simulate from sequencing data18, our simulation design required individual-level genotype measurements in biobank-scale sample sizes.
Third, does not correct for population structure/stratification. In real data, we mitigate this by considering only unrelated individuals (> 3rd degree relatives) and including age, sex, and the top 20 PCs as covariates when computing association statistics. While recent work has found evidence of assortative mating for some traits in the UK Biobank (e.g., height)39, our estimates are robust to different relatedness thresholds, suggesting that adjusting for the top 20 PCs sufficiently controls for population stratification. Still, it remains unclear how to quantify the bias of our genome-wide estimator due to structure or assortative mating in real data. Future work is needed to extend the GRE approach to control for ascertainment bias15,16,40,41.
Finally, while previous works applied similar estimators (defined under fixed effects models) to estimate local SNP-heritability within small regions28,29, additional work is needed to extend our approach to perform partitioning of SNP-heritability by functional annotations. Existing methods for partitioning SNP-heritability make various assumptions on genetic architecture8,12–14,30, motivating the development of new methods in this area.
Methods
The generalized random effects model
We model the phenotype for an individual n randomly sampled from the population as , where is a vector of standardized genotypes measured at M SNPs for individual n, is an M-vector of the corresponding standardized SNP effects, and is environmental noise. We assume and that the genotype at each SNP is centered and scaled in the population such that and ; i.e. , where is the number of copies of the effect allele at SNP for individual , and is the population frequency of the effect allele at SNP . We define the population LD between two SNPs and to be for all . The population LD matrix among the SNPs is therefore . For simplicity, we use “SNP effects” in lieu of “standardized SNP effects” to refer to . We assume that and are independent given allele frequencies and .
Under the generalized random effects (GRE) model, the first two moments of are and , where can be any arbitrary nonnegative finite number. We assume the covariance between the effects of different SNPs is 0 (i.e. for all ). Because the SNP-specific variances can capture any degree of polygenicity and any relationship between genomic features (e.g., MAF and LD) and effect size, the GRE model encompasses most realistic genetic architectures (Table 1).
We define total SNP-heritability to be the proportion of phenotypic variance attributable to the additive effects of a set of M SNPs whose genotypes are directly measured:
| #(2) |
Thus, is defined with respect to a given population and a given set of SNPs. By definition, Similarly, we define regional SNP-heritability to be the proportion of phenotypic variance due to the additive effects of the genotyped SNPs in region We assume that the set of SNPs that defines is a subset of the SNPs that define (thus, ). If region is the whole genome, .
Estimating SNP-heritability under the GRE model
We are interested in estimating under the GRE model (Equation 2). In a GWAS with individuals genotyped at SNPs, let be the matrix of standardized genotypes (each column of has been standardized to have mean 0 and variance 1), be the -vector of standardized phenotypes, and be the in-sample LD matrix (an estimate of population LD, ) with rank , where . Let be the genotype matrices for independent regions spanning all SNPs (e.g., chromosomes). For region containing SNPs, is the standardized genotype matrix and is the corresponding in-sample LD matrix with rank . We propose the following estimator for genome-wide SNP-heritability:
where is the -vector of marginal SNP effects estimated by ordinary least squares (OLS) for region and is the pseudoinverse of . Detailed derivations for can be found in the Supplementary Note.
Analytical variance of
Following quadratic form theory29,44, the variance of in the single-block case is
| #(3) |
When using the K-block approximation, which assumes that the blocks are independent, we approximate Equation 3 as the sum of the variances of the local SNP-heritabilities:
| #(4) |
Equation 3 is estimated by plugging in and Equation 4 is estimated by plugging in , the estimates of the regional SNP-heritabilities.
Simulation Framework
We simulated quantitative phenotypes from real genotype array data (UK Biobank10) under a range of genetic architectures. We obtained a set of unrelated British individuals by extracting individuals with self-reported British ancestry who are > 3rd degree relatives (pairs of individuals with kinship coefficient 10 and excluding individuals with putative sex chromosome aneuploidy. In all simulations, we standardize the genotypes before drawing phenotypes. That is, for each SNP and individual , we compute where is the number of minor alleles and is the in-sample minor allele frequency (MAF).
Simulations of quantitative traits with no population stratification
Given and a fixed value of , phenotypes are drawn according to the following model. The proportion of causal variants, , is set to 1, 0.01, or 0.001. Let be the causal status of SNP . If , for . If , we draw SNPs from the set of SNPs with MAF in one of three ranges: (0, 0.5], (0.01, 0.05], or (0.05, 0.5]. We use “CV MAF” to refer to the MAF range from which the causal variants are drawn. Standardized effects and phenotypes are then drawn according to the model
| #(5) |
| #(6) |
| #(7) |
where controls the coupling of MAF and effect size, is a SNP-specific LD weight, and specifies whether effects are coupled with the LD weights. We simulate two types of LD-dependent architectures by defining to be either (1) the default “LDAK weights” computed by the LDAK software6, or (2) the inverse unpartitioned “LD score” of each SNP computed within a 2-Mb window ( where indexes the set of SNPs within a 2-Mb window centered on SNP )11. When both the LDAK weights and inverse LD score weights cause SNPs in regions of higher LD to have smaller effects than do SNPs in regions of lower LD. We set to one of two values: (a relatively strong inverse relationship between MAF and effect size) or (a weaker inverse relationship between MAF and effect size). Each per-SNP variance is multiplied by a scaling factor so that . Note that if .
Finally, given phenotypes and genotypes , we compute marginal association statistics through ordinary least squares (OLS): .
Simulations of case-control phenotypes with no population stratification
To simulate case-control studies, we first draw each individual’s continuous liability ( for individual n) according to Equation 7. For a given population prevalence we compute the corresponding liability threshold , where is the CDF of the standard normal distribution. Each is then converted into a case-control status: if or if . For unascertained case-control studies, we assume that the proportion of cases in the study is equal to the population prevalence . For ascertained case-control studies , we set and select a random set of controls to satisfy .
We compute association statistics by regressing the binary case-control statuses on genotypes. The GRE estimator produces an estimate of SNP-heritability on the observed scale . Assuming we know the population prevalence, we convert to the liability scale with the transformation , where is the standard normal probability density function45.
Simulations with population stratification
To simulate GWAS with population stratification, we draw phenotypes from a model where a covariate that is correlated to genotypes has a nonzero effect on phenotype. To this end, we simulate an effect of the first genetic principal component . Letting be the proportion of total phenotypic variance explained by , phenotypes are drawn from the model
where . We compute association statistics from one of two models: , which ignores population stratification and other sources of confounding, or , which controls for the effect of .
Comparison of methods in simulations
Unless otherwise specified, in all genome-wide simulations, we use real genotypes of N = 337205 unrelated British individuals measured at M = 593300 array SNPs to draw causal effects for all M SNPs and phenotypes for all N individuals. OLS summary statistics are computed for all M SNPs using the simulated phenotypes and real genotypes of all N individuals. We compare to three methods that operate on summary statistics and are computationally tractable for these simulations: LD score regression (LDSC)11, stratified LD score regression (S-LDSC)12,13, and SumHer14.
For LDSC and S-LDSC, we compute the unpartitioned LD score of each SNP as a function of its LD to all other SNPs in a 2-Mb window centered on the SNP. For each annotation included in S-LDSC, the partitioned LD score of each SNP is a function of its LD to all SNPs within a 2-Mb window that are in the annotation. For both LDSC and S-LDSC, LD scores are computed with the LDSC software (https://github.com/bulik/ldsc/) from a random sample of 40K individuals to reduce the amount of memory required by the LDSC software. We run the regression with an unconstrained intercept, using all M SNPs as observations in the response variable. Each SNP is weighted to account for heteroscedasticity and correlations between association statistics11. For both methods, is estimated as a function of all M SNP-specific variances by using the flags --not-M-5–50 and --chisq-max 99999 (the latter option prevents the LDSC software from dropping high-effect SNPs).
We run S-LDSC in two ways to account for MAF/LD-dependent architectures. S-LDSC (MAF) refers to S-LDSC with 10 binary MAF bin annotations (each bin contains exactly 10% of the typed SNPs), which is intended to mirror the 10 MAF annotations in the “baseline-LD model”13 (see Supplementary Table 14 for precise MAF bin ranges for the UK Biobank Axiom Array). S-LDSC (MAF+LLD) refers to S-LDSC with the same 10 MAF bins and an additional continuous “level of LD” (LLD) annotation computed by quantile-normalizing the unpartitioned LD scores within each MAF bin to a standard normal distribution13. While our definition of LLD is intended to mirror the LLD annotation in the baseline-LD model, we do not set the LLD of variants with MAF < 0.05 to 0 because our estimand of interest includes the effects of SNPs with MAF < 0.0513.
To run SumHer, we use the LDAK software (https://dougspeed.com/ldak/) to compute the default “LDAK weights” using in-sample LD6,9,14. We then compute “LD tagging” (i.e. LD scores) using 1-Mb windows centered on each SNP and setting as recommended14. The LDAK software is memory-efficient, allowing us to use all 337K individuals to compute LDAK weights and LD tagging. Unless otherwise specified, all default parameter settings are used to run SumHer in simulations.
We also perform simulations with N = 8430 unrelated individuals at M = 14821 array SNPs. These individuals and SNPs are a subset of the data used in the genome-wide simulations, chosen by selecting approximately 2.5% of individuals and the first 2.5% of SNPs from the beginning of each chromosome in order to preserve the LD structure among the SNPs. We run single-component GREML3,46 (GCTA software: https://cnsgenomics.com/software/gcta/) and single-component BOLT-REML8 (https://data.broadinstitute.org/alkesgroup/BOLT-LMM/) with default parameters. We run GREML-LDMS-I18,46 using 8 GRMs created from 2 MAF bins ( and MAF > 0.05) and 4 LD score quartiles; LD scores were computed using the GCTA software with the default window size of 200-kb. We run LDAK using the default LDAK weights, setting as recommended6,9.
A third set of simulations was performed using 7,685 individuals of South Asian ancestry in the UK Biobank. This group was composed of individuals of Indian (n = 5,716), Pakistani (n = 1,748), and Bangladeshi (n = 221) ancestry. Due to the small sample size, we used a reduced set of 803 SNPs from chromosome 21 and 839 SNPs from chromosome 22 (1,642 SNPs in total) which were chosen so that for each chromosome k was similar to in the “white British” cohort.
For a given genetic architecture, we generate 100 simulation replicates and obtain 100 estimates of from each method. We estimate the bias of an estimator under a given architecture as bias where is the estimate from the i-th simulation. To test whether the bias is statistically significant (null hypothesis: bias ), we assess the z-score of the bias , where is the standard error of the mean of the 100 estimates) which follows a distribution under the null hypothesis. The p-value of the bias is computed with a two-tailed test. To enable a comparison of estimators across different values of , we assess the relative bias of an estimator under a single architecture as a percentage of . In Figure 1a and 1c, we compute the error of a single estimate as ; errors are also reported as percentages of .
Analysis of UK Biobank phenotypes
We estimate SNP-heritability for 22 complex traits (6 quantitative, 16 binary) in the UK Biobank10. We use PLINK47 (https://www.cog-genomics.org/plink2) to exclude SNPs with MAF < 0.01 and genotype missingness > 0.01 as well as SNPs that fail the Hardy-Weinberg test at significance threshold . We keep only the individuals with self-reported British white ancestry and no kinship (i.e. > 3rd degree relatives, defined as pairs of individuals with kinship coefficient )10. After removing individuals who are outliers for genotype heterozygosity and/or missingness, we obtain a set of N = 290,641 individuals to use in the real data analyses. For all traits, marginal association statistics are computed through OLS in PLINK, using age, sex, and the top 20 genetic principal components (PCs) as covariates in the regression; these 20 PCs were precomputed by UK Biobank from a superset of 488,295 individuals. Additional covariates were used for waist-to-hip ratio (adjusted for BMI) and diastolic/systolic blood pressure (adjusted for cholesterol-lowering medication, blood pressure medication, insulin, hormone replacement therapy, and oral contraceptives). We compute for each trait using in-sample LD estimated from all N individuals.
When using LDSC, S-LDSC, or SumHer to estimate SNP-heritability, it is necessary to define and distinguish between the following sets of SNPs: the set of SNPs containing all possible causal SNPs of interest (used to compute LD scores and LDAK weights), the set of SNPs used as observations in the regression, and the set of SNPs that defines the SNP-heritability estimand of interest. We run two versions of LDSC, S-LDSC (controlling for the most recent baseline-LD model12,13,30), and SumHer14. First, to enable a direct comparison between and the estimands of LDSC, S-LDSC, and SumHer, we run an “in-sample LD” version of each method where the M typed SNPs are used to compute LD scores and LDAK weights, perform the regression, and define the SNP-heritability estimand of interest. We refer to these as LDSC (in-sample), S-LDSC (baseline-LD/in-sample), and SumHer (in-sample). To run LDSC (in-sample) and S-LDSC (baseline-LD/in-sample), we use the LDSC software to compute LD scores and regression weights within 2-Mb windows centered on each SNP, using a random sample of 40K individuals to reduce the memory requirement. To run SumHer (in-sample), we use the LDAK software to compute LD tagging from the genotypes of all N individuals, using 1-Mb windows centered on each SNP and setting as recommended9,14. Unless otherwise specified, all other parameters were set to the default settings.
To enable comparisons between and estimates reported in the literature, we also run each method with its recommended parameter settings and LD estimated from reference panel sequencing data. We refer to these methods as LDSC (1KG), S-LDSC (baseline-LD/1KG), and SumHer (1KG) to indicate that LD is estimated from 489 Europeans in the 1000 Genomes Phase 3 reference panel31. We run LDSC (1KG) and S-LDSC (baseline-LD/1KG) with LD scores and regression weights (1-cM windows) from 9,997,231 SNPs with minor allele count greater than 5 in the reference panel, and we define the SNP-heritability estimand to be a function of the array SNPs with MAF > 0.0511,12. We run SumHer (1KG) using 8,569,062 SNPs with MAF > 0.01 in the reference panel to compute LDAK weights and LD tagging (1-cM windows) and to define the SNP-heritability estimand; we control for a multiplicative inflation of test statistics as recommended14. See refs.11,12,14,19 for details about the definitions and interpretations of the estimands of LDSC, S-LDSC, and SumHer.
Life Sciences Reporting Summary
Additional information on experimental design can be found in the Life Sciences Reporting Summary.
Supplementary Material
Acknowledgments
This research was conducted using the UK Biobank Resource under applications 33297 and 33127. We thank the participants of UK Biobank for making this work possible. We also thank R. Johnson, M. Freund, M. Major, S. Gazal, A. Price and D. Balding for helpful discussions. This work was funded in part by the National Institutes of Health (NIH) under awards R01HG009120, R01MH115676, R01HG006399, U01CA194393, T32NS048004, T32MH073526, and T32HG002536.
Footnotes
Competing interests
The authors declare no competing interests.
Data availability
The baseline-LD annotations used in Fig. 4 are available at https://data.broadinstitute.org/alkesgroup/LDSCORE/. All individual-level genotypes and phenotypes were obtained from the UK Biobank (https://www.ukbiobank.ac.uk); we do not have permission to release this data. The 1000 Genomes Phase 3 reference panel can be downloaded at http://www.internationalgenome.org/data.
Code availability
Open source code implementing the GRE estimator and our simulation framework is available on Github at https://github.com/bogdanlab/h2-GRE.
References
- 1.Visscher PM, Hill WG & Wray NR Heritability in the genomics era — concepts and misconceptions. Nat. Rev. Genet 9, 255 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Wray NR et al. Pitfalls of predicting complex traits from SNPs. Nat. Rev. Genet 14, 507 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang J et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet 42, 565 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Visscher PM, Brown MA, McCarthy MI & Yang J Five Years of GWAS Discovery. Am. J. Hum. Genet 90, 7–24 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Visscher PM et al. 10 Years of GWAS Discovery: Biology, Function, and Translation. Am. J. Hum. Genet 101, 5–22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Speed D, Hemani G, Johnson MR & Balding DJ Improved Heritability Estimation from Genome-wide SNPs. Am. J. Hum. Genet 91, 1011–1021 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang J et al. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat. Genet 47, 1114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loh P-R et al. Contrasting genetic architectures of schizophrenia and other complex diseases using fast variance-components analysis. Nat. Genet 47, 1385 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Speed D et al. Reevaluation of SNP heritability in complex human traits. Nat. Genet 49, 986 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bycroft C et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bulik-Sullivan BK et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet advance on, 291–295 (2015). [DOI] [PMC free article] [PubMed]
- 12.Finucane HK et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet 47, 1228–1235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gazal S et al. Linkage disequilibrium–dependent architecture of human complex traits shows action of negative selection. Nat. Genet 49, 1421 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Speed D & Balding DJ SumHer better estimates the SNP heritability of complex traits from summary statistics. Nat. Genet (2018). doi: 10.1038/s41588-018-0279-5 [DOI] [PMC free article] [PubMed]
- 15.Haseman JK & Elston RC The investigation of linkage between a quantitative trait and a marker locus. Behav. Genet 2, 3–19 (1972). [DOI] [PubMed] [Google Scholar]
- 16.Wu Y & Sankararaman S A scalable estimator of SNP heritability for biobank-scale data. Bioinformatics 34, i187–i194 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Timpson NJ, Greenwood CMT, Soranzo N, Lawson DJ & Richards JB Genetic architecture: the shape of the genetic contribution to human traits and disease. Nat. Rev. Genet 19, 110 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Evans LM et al. Comparison of methods that use whole genome data to estimate the heritability and genetic architecture of complex traits. Nat. Genet 50, 737–745 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gazal S, Marquez-Luna C, Finucane HK & Price AL Reconciling S-LDSC and LDAK models and functional enrichment estimates. bioRxiv 256412 (2018). doi: 10.1101/256412 [DOI] [PMC free article] [PubMed]
- 20.Eyre-Walker A Genetic architecture of a complex trait and its implications for fitness and genome-wide association studies. Proc. Natl. Acad. Sci 107, 1752 LP–1756 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lohmueller KE The Impact of Population Demography and Selection on the Genetic Architecture of Complex Traits. PLOS Genet 10, e1004379 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schoech A et al. Quantification of frequency-dependent genetic architectures and action of negative selection in 25 UK Biobank traits. bioRxiv 188086 (2017). doi: 10.1101/188086 [DOI] [PMC free article] [PubMed]
- 23.Zeng J et al. Signatures of negative selection in the genetic architecture of human complex traits. Nat. Genet 50, 746–753 (2018). [DOI] [PubMed] [Google Scholar]
- 24.O’Connor LJ et al. Polygenicity of complex traits is explained by negative selection. bioRxiv 420497 (2018). doi: 10.1101/420497 [DOI] [PMC free article] [PubMed]
- 25.Uricchio LH, Kitano HC, Gusev A & Zaitlen NA An evolutionary compass for detecting signals of polygenic selection and mutational bias. Evol. Lett 3, 69–79 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Qi G, Park J-H & Chatterjee N Estimation of complex effect-size distributions using summary-level statistics from genome-wide association studies across 32 complex traits. Nat. Genet 50, 1318–1326 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Loh P-R, Kichaev G, Gazal S, Schoech AP & Price AL Mixed-model association for biobank-scale datasets. Nat. Genet 50, 906–908 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gamazon ER, Cox NJ & Davis LK Structural Architecture of SNP Effects on Complex Traits. Am. J. Hum. Genet 95, 477–489 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi H, Kichaev G & Pasaniuc B Contrasting the Genetic Architecture of 30 Complex Traits from Summary Association Data. Am. J. Hum. Genet 99, 139–153 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gazal S et al. Functional architecture of low-frequency variants highlights strength of negative selection across coding and non-coding annotations. Nat. Genet 50, 1600–1607 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Consortium, T. 1000 G. P. et al. A global reference for human genetic variation. Nature 526, 68 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ledoit O & Wolf M A well-conditioned estimator for large-dimensional covariance matrices. J. Multivar. Anal 88, 365–411 (2004). [Google Scholar]
- 33.Nagai A et al. Overview of the BioBank Japan Project: Study design and profile. J. Epidemiol 27, S2–S8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leitsalu L et al. Cohort Profile: Estonian Biobank of the Estonian Genome Center, University of Tartu. Int. J. Epidemiol 44, 1137–1147 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Gaziano JM et al. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J. Clin. Epidemiol 70, 214–223 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Pasaniuc B & Price AL Dissecting the genetics of complex traits using summary association statistics. Nat. Rev. Genet 18, 117 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hormozdiari F, Kichaev G, Yang W-Y, Pasaniuc B & Eskin E Identification of causal genes for complex traits. Bioinformatics 31, i206–i213 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi H, Mancuso N, Spendlove S & Pasaniuc B Local Genetic Correlation Gives Insights into the Shared Genetic Architecture of Complex Traits. Am. J. Hum. Genet 101, 737–751 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yengo L et al. Imprint of assortative mating on the human genome. Nat. Hum. Behav 2, 948–954 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Golan D, Lander ES & Rosset S Measuring missing heritability: inferring the contribution of common variants. Proc. Natl. Acad. Sci. U. S. A 111, E5272–81 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weissbrod O, Flint J & Rosset S Estimating SNP-Based Heritability and Genetic Correlation in Case-Control Studies Directly and with Summary Statistics. Am. J. Hum. Genet 103, 89–99 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee SH et al. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat. Genet 44, 247 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee SH et al. Estimation of SNP Heritability from Dense Genotype Data. Am. J. Hum. Genet 93, 1151–1155 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elman RS, Karpenko N & Merkurjev A The algebraic and geometric theory of quadratic forms 56, (American Mathematical Soc, 2008). [Google Scholar]
- 45.Lee SH, Wray NR, Goddard ME & Visscher PM Estimating Missing Heritability for Disease from Genome-wide Association Studies. Am. J. Hum. Genet 88, 294–305 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang J, Lee SH, Goddard ME & Visscher PM GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet 88, 76–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Purcell S et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.