Abstract
In flowering plants, gene body methylation (gbM) is associated with a subset of constitutively expressed genes. It has been proposed that gbM modulates gene expression. Here, we show that there are no consistent and direct differences to expression following the loss of gbM. By comparing expression of gbM genes in Arabidopsis thaliana accessions to orthologous genes in two Eutrema salsugineum genotypes, we identified both positive and negative expression differences associated with gbM loss. However, expression is largely unaffected by gbM loss in E. salsugineum. Expression differences between species were within the variation of expression observed within A. thaliana accessions that displayed variation in gbM. Furthermore, experimentally induced loss of gbM did not consistently lead to differences in expression compared to wild type. To date, there is no convincing data to support a direct causal link between the presence/absence of gbM and the modulation of expression in flowering plants.
Keywords: DNA methylation, gene body methylation, gene expression, epigenetics
Gene body methylation (gbM) is characterized by the enrichment of CG methylation levels within coding regions (Tran et al. 2005; Zhang et al. 2006; Zilberman et al. 2007; Cokus et al. 2008; Lister et al. 2008; Takuno and Gaut 2012). Compared to unmethylated (UM) genes, gbM genes tend to be moderately and constitutively expressed (Zhang et al. 2006; Lister et al. 2008; Niederhuth et al. 2016). Many have speculated on the function of gbM, suggesting that its primary role is likely to be homeostatic (Zilberman 2017; Muyle and Gaut 2019). The discovery of species without gbM led to testing hypotheses for its function and evolutionary origin (Bewick et al. 2016; Bewick et al. 2017). No obvious and consistent consequences to gene expression were observed following the loss of gbM in Eutrema salsugineum (Bewick et al. 2016). However, a reanalysis of Bewick, et al. (2016) reported a small, but statistically significant reduction in gene expression associated with genes that have lost gbM in one of the two E. salsugineum accessions (Muyle and Gaut 2019). To say the least, the function of gbM remains enigmatic.
Methods
Datasets
Whole Genome Bisulfite Sequencing (WGBS) and RNA-seq data were downloaded from Gene Expression Omnibus (GEO) or Short Read Archive (SRA) for Arabidopsis thaliana and Eutrema salsugineum. For A. thaliana we selected six additional accessions to Col-0 that spanned the gamut of CG DNA methylation levels for gbM genes (∼30–60%) (Kawakatsu et al. 2016). WGBS and RNA-seq data from these additional accessions represent a single replicate per accession. However, comparison between wild type and met1 epiRIL was performed in triplicate (Bewick et al. 2016). For E. salsugineum we selected the two available genotypes with available WGBS and RNA-seq (Bewick et al. 2016). GEO or SRA accession numbers and mapping statistics are located in Table S7.
Identification of putative orthologs
Basic Local Alignment Search Tool (BLAST) (Altschul et al. 1990) was to identify putative orthologs between A. thaliana and E. salsugineum. We used identical BLAST parameters as Bewick et al. (2016) to recover the same set of putative one-to-one orthologs. Orthologs were then split into Group 1 (putative orthologous pairs that changed gbM status in E. salsugineum) and Group 2 (putative orthologous pairs that are UM in both A. thaliana and E. salsugineum) based on DNA methylation status in each species (see subsection WGBS mapping and analysis of DNA methylation for classification of gbM and UM). For Group 1, genes were required to be gbM across all A. thaliana accessions and UM for both E. salsugineum genotypes. Similarly, for Group 2 genes were required to be UM across all A. thaliana accessions and UM for both E. salsugineum genotypes. The number of genes within each group were lower than previously reported by Bewick et al. (2016) and Muyle and Gaut (2019) because of the DNA methylation requirements across accessions and genotypes, and the incorrect A. thaliana-E. salsugineum one-to-one orthology assignment of 1,301 genes by Muyle and Gaut (2019).
WGBS mapping and analysis of DNA methylation
WGBS data were aligned to each species respective genome assembly using the methylpy pipeline (Schultz et al. 2015). In brief, reads were trimmed of sequencing adapters using Cutadapt v1.9 (Martin and Marcel 2011), and then mapped to both a converted forward strand (cytosines to thymines) and converted reverse strand (guanines to adenines) using bowtie v1.1.1 (Langmead et al. 2009). Reads that mapped to multiple locations, and clonal reads were removed. Additionally, PCR-duplicated reads were identified and removed using Picard v2.16.0 (https://github.com/broadinstitute/picard). Unmethylated lambda phage DNA or plastid genome was used as a control for sodium bisulfite conversion. Non-conversion rates are located in Table S7.
Weighted DNA methylation was calculated for CG sites by dividing the total number of aligned methylated reads by the total number of methylated plus unmethylated reads (Schultz et al. 2012). CG sequence context enrichment for each gene was determined through a binomial test followed by Benjamini-Hochberg false discovery rate following Takuno and Gaut (2012). A background mCG level was determined from all coding sequence, which was used as a threshold in determining significance with a False Discovery Rate (FDR) correction. Genes were classified as gene body methylated (gbM) if they had at least 3 reads mapping to 20 CG sites and a CG q-value ≤ 0.05. Genes were classified as unmethylated (UM) if they had reads mapping to at least 20 reads mapping to 20 CG sites and a CG, CHG, and CHH (H = A, C, or T) q-value > 0.05.
RNA-seq mapping
Raw RNA-seq FASTQ reads were trimmed for adapters and preprocessed to remove low-quality reads using Trimmomatic v0.33 (arguments: default) (Bolger et al. 2014) prior to mapping to the species respective genome assembly. Reads were mapped using HISAT2 v2.1.0 (Pertea et al. 2016) supplied with a reference GTF (General Transfer Format) and splice-site and exon information (arguments: default). Following mapping, RNA-Seq alignments were assembled into potential transcripts using StringTie v1.3.3b (Pertea et al. 2016) (arguments: default).
Analysis of gene expression
Differentially expressed genes between met1 epiRIL and wild-type libraries were determined using edgeR v3.20.1 (Robinson and Oshlack 2010; Robinson et al. 2010) implemented in R v3.2.4 (https://www.r-project.org/). Mutant and wild-type libraries were collectively normalized using the Trimmed mean of M values (TMM) method (Robinson and Oshlack 2010). Genes were retained for DEG analysis if they possessed a count per million (CPM) ≥ 1 in at least ≥ 2 libraries. Significance was determined using the glmQLFTest function, which uses empirical Bayes quasi-likelihood F tests, and an FDR cutoff of 5%. Parameter settings were determined following best practices for DEG analysis as described by (Chen et al. 2016).
A bootstrap test with 1,000 replicates was used to determine significance of gene expression distributions for Group 1 between A. thaliana accessions and E. salsugineum genotypes, E. salsugineum genotypes, and for Group 1 and Group 2 between wild type and met1 epiRIL. A Wilcoxon signed-rank test was also used to compare expression between pairs of genes in Group 1 (Table S8). Paired tests are appropriate when the data are not independent (i.e., shared ancestry) and when the dependency results in a one-to-one match (i.e., single-copy putative orthologous gene pairs). Both the statistical significance (P value) and the substantive difference (effect size) are important in the interpretation of studies (Sullivan and Feinn 2012). Hence, in addition to reporting statistical significance, we used Cohen’s d to assess effect size of difference in gene expression within Groups 1 and 2.
Data Availability
All data are available at GEO or SRA and the accession numbers are listed in Table S7. Supplemental material available at FigShare: https://doi.org/10.25387/g3.8204582.
Results and Discussion
To test for a function of gbM in modulating gene expression (Fragments Per Kilobase of transcript per Million mapped reads [FPKM]), Muyle and Gaut (2019) contrasted gene expression (log10[FPKM+1]) of two groups of genes. Group 1 genes consisted of putative orthologous pairs that changed gbM status in E. salsugineum (i.e., gbM→UM) since its last common ancestor with Arabidopsis thaliana. Group 2 genes consisted of putative orthologous pairs that are UM in both A. thaliana and E. salsugineum. This comparison (i) does not directly test the function of gbM in modulating gene expression and (ii) introduces unlikely assumptions regarding gene expression. First, Muyle and Gaut’s (2019) model is designed to test for an effect of Group on gene expression, rather than a change in gene expression following the loss of gbM. Second, Group 2 is an inappropriate control to test for the function of gbM in modulating gene expression. Using Group 2 as a control incorrectly assumes that gene expression levels of UM genes have remained relatively constant over millions of years. This assumption is contrary to the proposed homeostatic function of gbM, where under this model, it is predicted that UM genes have more variable gene expression compared to orthologous gbM genes (Zilberman 2017).
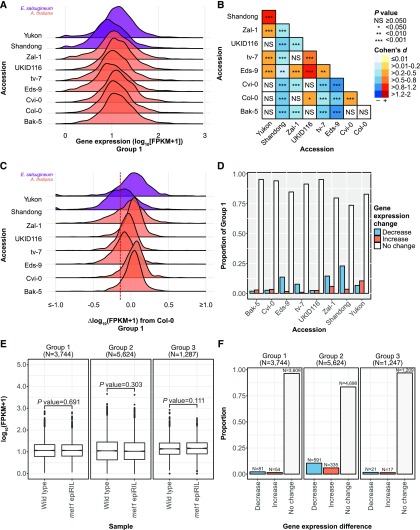
If gbM modulates gene expression, a consistent effect on gene expression would be expected upon loss of gbM. However, only one of the two E. salsugineum genotypes tested in the Muyle and Gaut (2019) study supported the conclusion that gene expression is reduced following loss of gbM. This raises the possibility that the gene expression variation observed by Muyle and Gaut (2019) is within the expected range for expression variation between any group of orthologous genes that have been diverging for millions of years. To test this hypothesis further, we used data from the 1,001 A. thaliana epi/genomes project (1001 Genomes Consortium 2016; Kawakatsu et al. 2016). Expression levels of gbM genes from seven A. thaliana accessions and their putative orthologs from two E. salsugineum genotypes that have lost gbM (N = 1,328) show substantial overlap (Figure 1A and Table S1). Mean gene expression and standard error of the mean (SEM) of E. salsugineum Yukon fall within the range of A. thaliana accessions investigated (Table S2). However, mean gene expression and SEM of E. salsugineum Shandong is the lowest of genotypes and accessions investigated (Table S2). Gene expression consequences following the loss of gbM are inconsistent across E. salsugineum genotypes when compared to A. thaliana accessions (Figure 1B). Gene expression is higher in E. salsugineum Yukon compared to A. thaliana accessions, with several nonsignificant exceptions, whereas gene expression is lower in E. salsugineum Shandong compared to A. thaliana accessions. Differences in gene expression between A. thaliana accessions for gbM genes is also observed, but several comparisons are nonsignificant (Figure 1B). The effect size of gene expression differences between accessions, as measured using Cohen’s d, ranges from small to large for between and within species comparisons. These data fail to support that gene expression is reduced following the loss of gbM and instead suggest that the observed differences between E. salsugineum and A. thaliana are within an expected distribution of natural variation in gene expression.
Figure 1.
No direct and consistent consequences to gene expression following the loss of gbM. (A) Ridge plot of gene expression for the same 1,328 genes across accessions and genotypes. All genes have lost gbM in E. salsugineum Shandong and Yukon, whereas all genes maintain gbM in A. thaliana accessions (Group 1). (B) Pair-wise comparison of gene expression for Group 1 between species (E. salsugineum-A. thaliana) and within species (E. salsugineum-E. salsugineum and A. thaliana-A. thaliana). Significance is represented by asterisks. Color of cell indicates direction and effect size of difference relative to the x-axis. The effect size is represented as Cohen’s d. (C) Ridge plot of gene expression differences for each genotype and accession relative to A. thaliana Col-0. Gene expression differences ≤-1.0 and ≥1.0 log10(FPKM+1) were collapsed to -1.0 and 1.0, respectively. Dashed red line indicates the average difference in gene expression reported by Muyle and Gaut (2019) for Group 1 genes between E. salsugineum and A. thaliana Col-0 (-0.14 log10[FPKM+1]). (D) Bar plot of discrete differences in genes expression for each accession and genotype relative to A. thaliana Col-0. (E) Distribution of gene expression for Group 1, Group 2, and Group 3 genes between wild type and met1 epiRIL. Gene expression was averaged per gene across three wild type and met1 epiRIL RNA-seq libraries, respectively. Boxplot elements: center line, median; upper and lower “hinges”, first and third quartiles (the 25th and 75th percentiles), respectively; whiskers, 1.5× interquartile range; large points, outliers. (F) Bar plot of discrete differences in genes expression for Group 1, Group 2, and Group 3 genes between wild type and met1 epiRIL.
Muyle and Gaut (2019) report an average 0.14 log10(FPKM+1) decrease following the loss of gbM in E. salsugineum. Similarly, to Muyle and Gaut (2019), we compared Group 1 gene expression between E. salsugineum genotypes and A. thaliana Bak-5, Cvi-0, Eds-9, tv-7, UKID116, and Zal-1 to A. thaliana Col-0 using the exact same methods. Compared to A. thaliana Col-0, both increases and decreases to gene expression are observed for genes that lost gbM in either E. salsugineum and/or possess gbM in A. thaliana accessions (Figure 1C). Although E. salsugineum Shandong shows an on average negative difference in gene expression identical to that reported by Muyle and Gaut (2019) (-0.138 log10[FPKM+1]), E. salsugineum Yukon shows an average positive difference in gene expression (+0.017 log10[FPKM+1]). However, in E. salsugineum Shandong and Yukon, 53.24% and 82.53% of genes that have lost gbM show an increase in gene expression greater than the 0.14 log10(FPKM+1) reduction reported by Muyle and Gaut (2019).
Furthermore, requiring a ≥1 (increase) or ≤-1 (decrease) log2 difference in gene expression, more genes that have lost gbM have higher rather than lower expression in E. salsugineum Yukon compared to A. thaliana Col-0 gbM genes (Figure 1D). However, the majority of genes that have lost gbM in E. salsugineum and gbM genes in A. thaliana accessions Bak-5, Cvi-0, Eds-9, tv-7, UKID116, and Zal-1 are not differentially expressed given this threshold compared to A. thaliana Col-0 gbM genes (Figure 1D). Differences in sequencing coverage between A. thaliana Col-0 and E. salsugineum may interfere with gene expression comparisons, particularly if there are heterogeneous transcript distributions between samples (Bullard et al. 2010; Hansen et al. 2010). However, equalizing sequencing coverage of genes between A. thaliana Col-0 and E. salsugineum (e.g., Shandong) has no effect on gene expression levels, suggesting transcript heterogeneity is trivial and is not introducing biases to these comparisons (Fig. S1). Furthermore, the on average negative difference in gene expression between A. thaliana Col-0 and E. salsugineum Shandong for Group 1 is similar to the difference between E. salsugineum Shandong and Yukon for all genes (-0.170 log10[FPKM+1]) (Fig. S2), suggesting an outgroup effect influencing differences in gene expression. Collectively these results demonstrate gene expression differences following the loss of gbM in E. salsugineum are within the range of differences between gbM genes of A. thaliana accessions. These results further support an inconsistent consequence to differences in gene expression following the loss of gbM.
Gene expression comparison between species could be confounded by inherent experimental and biological biases. Hence, to test for direct differences to gene expression following the loss of gbM, while mitigating biases, we took advantage of the availability of eighth-generation met1 epigenetic Recombinant Inbred Lines (epiRILs) (Reinders et al. 2009). The met1 epiRILs contain mosaic methylomes, including genomic regions with normal DNA methylation from the wild-type parent and gbM-free regions from the met1 parent. No difference in gene expression is observed for Group 1 genes between the met1 epiRIL and wild type (Figure 1E and Table S3). Using the expression difference threshold as above, we identified 31/3,744 (0.83%) and 22/3,744 (0.59%) Group 1 genes experiencing a positive and negative difference in gene expression, respectively. To account for the slight possibility of transcript heterogeneity between met1 epiRIL and wild type, we collectively normalized gene expression across samples and replicates using the Trimmed mean of M values (TMM) method prior to the identification of Differentially Expressed Genes (DEGs) in edgeR v3.20.1 (Robinson and Oshlack 2010; Robinson et al. 2010). This more formal approach to compare gene expression differences recovered a total of 1,061 DEGs with 135 (12.72%) belonging to Group 1 and 926 (87.28%) belonging to Group 2 (Table S4). Gene expression differences are not independent of Group (χ2 = 6.255e-06, d.f. = 1, P value = 1.788e-82) with fewer gene expression differences following the loss of gbM being observed than expected by chance (NExpected = 424) (Table S5). Among the 135 DEGs in Group 1, 81 have decreased and 54 have increased gene expression levels in the met1 epiRIL relative to wild type (Figure 1F and Table S6). Similarly, among the 926 DEGs in Group 2 genes, 591 show decreased and 335 show increased expression in the met1 epiRIL relative to wild type (Figure 1F and Table S6). Hence, there are similar negative and positive gene expression differences between Groups 1 and 2 (∼3:2). We also compared gene expression of genes that are gbM in both met1 epiRIL and wild type (Group 3). Group 3 genes are similar to Group 1 genes with respect to gene expression level and the proportion of DEGs (Figure 1E and F). Overall, these results show the loss of gbM does not lead to direct and consistent differences to gene expression. Instead, gene expression differences that are observed for Group 1 genes are likely indirect effects associated with the gain and recovery of MET1.
The association of gbM to gene expression is often interpreted as it is possessing a functional role, however, there is currently no direct evidence to support this role. Evolutionary loss of gbM without clear consequences to gene expression suggests a lack of a role in transcriptional regulation, but this too is an indirect test which requires further support. In contrast, experimentally induced loss of gbM in the met1 epiRIL did not lead to significant differences in gene expression and the few genes with significant differences to gene expression were both negative and positive. These data suggest that the function of gbM is not likely modulating or buffering gene expression. Zilberman (2017) proposes that the primary effect of gbM is likely to be homeostatic, possibly by preventing aberrant transcription within genes or restricting access to histone H2A.Z. However, the pattern of H2A.Z in E. salsugineum Shandong and met1 epiRIL remains comparable to that in wild type (Bewick et al., 2016). If a functional role of gbM exists, it has yet to be convincingly determined. Continued discussions and research on this curious feature of flowering plant genomes will help unravel this mystery.
Acknowledgments
We thank Frank Johannes (Technical University of Munich), Chad Niederhuth (Michigan State University), David Hall (University of Georgia) and Nathan Springer (University of Minnesota) for comments. We additionally thank the National Science Foundation (NSF) IOS-1444567 (Dr. Jim Leebens-Mack, re. AJB), IOS-1546867 and MCB-1856143 (RJS), and IOS-1811694 (JMW) for funding.
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25387/g3.8204582.
Communicating editor: B. Gregory
Literature Cited
- Genomes Consortium , 2016. 1,135 Genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166: 481–491. 10.1016/j.cell.2016.05.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Bewick A. J., Ji L., Niederhuth C. E., Willing E. M., Hofmeister B. T. et al. , 2016. On the origin and evolutionary consequences of gene body DNA methylation. Proc. Natl. Acad. Sci. USA 113: 9111–9116. 10.1073/pnas.1604666113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewick A. J., Niederhuth C. E., Ji L., Rohr N. A., Griffin P. T. et al. , 2017. The evolution of CHROMOMETHYLASES and gene body DNA methylation in plants. Genome Biol. 18: 65 10.1186/s13059-017-1195-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., and Usadel B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard J. H., Purdom E., Hansen K. D., and Dudoit S., 2010. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics 11: 94 10.1186/1471-2105-11-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Lun A. T. L., and Smyth G. K., 2016. From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000 Res. 5: 1438 10.12688/f1000research.8987.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus S. J., Feng S., Zhang X., Chen Z., Merriman B. et al. , 2008. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452: 215–219. 10.1038/nature06745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen K. D., Brenner S. E., and Dudoit S., 2010. Biases in Illumina transcriptome sequencing caused by random hexamer priming. Nucleic Acids Res. 38: e131 10.1093/nar/gkq224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T., Huang S. C., Jupe F., Sasaki E., Schmitz R. J. et al. , 2016. Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell 166: 492–505. 10.1016/j.cell.2016.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., and Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R., O’Malley R. C., Tonti-Filippini J., Gregory B. D., Berry C. C. et al. , 2008. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536. 10.1016/j.cell.2008.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M., and Marcel M., 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17: 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Muyle A., and Gaut B. S., 2019. Loss of gene body methylation in Eutrema salsugineum is associated with reduced gene expression. Mol. Biol. Evol. 204: 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederhuth C. E., Bewick A. J., Ji L., Alabady M. S., Kim K. D. et al. , 2016. Widespread natural variation of DNA methylation within angiosperms. Genome Biol. 17: 194 10.1186/s13059-016-1059-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M., Kim D., Pertea G. M., Leek J. T., and Salzberg S. L., 2016. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11: 1650–1667. 10.1038/nprot.2016.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders J., Wulff B. B., Mirouze M., Marí-Ordóñez A., Dapp M. et al. , 2009. Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev. 23: 939–950. 10.1101/gad.524609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., and Oshlack A., 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11: R25 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., and Smyth G. K., 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz M. D., Schmitz R. J., and Ecker J. R., 2012. ‘Leveling’ the playing field for analyses of single-base resolution DNA methylomes. Trends Genet. 28: 583–585. 10.1016/j.tig.2012.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz M. D., He Y., Whitaker J. W., Hariharan M., Mukamel E. A. et al. , 2015. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature 523: 212–216. 10.1038/nature14465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan G. M., and Feinn R., 2012. Using effect size—or why the P value is not enough. J. Grad. Med. Educ. 4: 279–282. 10.4300/JGME-D-12-00156.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takuno S., and Gaut B. S., 2012. Body-methylated genes in Arabidopsis thaliana are functionally important and evolve slowly. Mol. Biol. Evol. 29: 219–227. 10.1093/molbev/msr188 [DOI] [PubMed] [Google Scholar]
- Tran R. K., Henikoff J. G., Zilberman D., Ditt R. F., Jacobsen S. E. et al. , 2005. DNA methylation profiling identifies CG methylation clusters in Arabidopsis genes. Curr. Biol. 15: 154–159. 10.1016/j.cub.2005.01.008 [DOI] [PubMed] [Google Scholar]
- Zilberman D., Gehring M., Tran R. K., Ballinger T., and Henikoff S., 2007. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 39: 61–69. 10.1038/ng1929 [DOI] [PubMed] [Google Scholar]
- Zilberman D., 2017. An evolutionary case for functional gene body methylation in plants and animals. Genome Biol. 18: 87 10.1186/s13059-017-1230-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yazaki J., Sundaresan A., Cokus S., Chan S. W. et al. , 2006. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126: 1189–1201. 10.1016/j.cell.2006.08.003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are available at GEO or SRA and the accession numbers are listed in Table S7. Supplemental material available at FigShare: https://doi.org/10.25387/g3.8204582.