Abstract
Oil rapeseed (Brassica napus L.) is a typical winter biennial plant, with high cold tolerance during vegetative stage. In recent years, more and more early-maturing rapeseed varieties were planted across China. Unfortunately, the early-maturing rapeseed varieties with low cold tolerance have higher risk of freeze injury in cold winter and spring. Little is known about the molecular mechanisms for coping with different low-temperature stress conditions in rapeseed. In this study, we investigated 47,328 differentially expressed genes (DEGs) of two early-maturing rapeseed varieties with different cold tolerance treated with cold shock at chilling (4°) and freezing (−4°) temperatures, as well as chilling and freezing stress following cold acclimation or control conditions. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis indicated that two conserved (the primary metabolism and plant hormone signal transduction) and two novel (plant-pathogen interaction pathway and circadian rhythms pathway) signaling pathways were significantly enriched with differentially-expressed transcripts. Our results provided a foundation for understanding the low-temperature stress response mechanisms of rapeseed. We also propose new ideas and candidate genes for genetic improvement of rapeseed tolerance to cold stresses.
Keywords: Transcriptome, rapeseed, cold acclimation, cold shock, chilling, freezing
Low-temperature is a major environmental stress that adversely affects plant growth and development, limiting crop productivity. Plants evolutionarily developed a wide variety of mechanisms to cope with low-temperature stress. The response to low-temperature in plants is a complex process that involves morphological, physiological, biochemical and molecular processes, including gene expression, enzyme activity and metabolic homeostasis (Theocharis et al. 2012; Nievola et al. 2017). Chilling stress (0-15°) causes the membrane to rigidify, destabilizes protein complexes and impairs photosynthesis, whereas freezing stress (< 0°) causes more serious injuries to the plant (Shi et al. 2018; Ruelland et al. 2009).
Many plants showed increased freezing tolerance upon exposure to low non-freezing temperatures in a phenomenon known as cold acclimation (Thomashow 1999). Cold acclimation involved an array of physiological and biochemical modifications, and these altered processes involved changes in gene expression patterns via phytohormone and the ICE-CBF-COR transcriptional regulatory cascade (Shi et al. 2018).
C-repeat binding factors (CBF) transcription factors, also known as dehydration responsive element binding factor 1 (DREB1) proteins, could bind directly to the CRT/DRE cis-element in the promoters of cold regulated (COR) genes, and subsequently induced the expression of COR genes (Jaglo-Ottosen et al. 1998; Liu et al. 1998). The COR genes protected plant cells against cold-induced damage, repaired cold-rigidified membranes and stabilized cellular osmotic potential by encoding cryoprotective proteins and key enzymes for osmolyte biosynthesis (Chinnusamy et al. 2007). In Arabidopsis, the basic helix-loop-helix (bHLH) transcription factor ICE1 was shown to bind directly to the CANNTG cis-elements in the promoters of CBF and upregulated CBF expression under cold stress (Chinnusamy et al. 2003; Kim et al. 2015). Apart from CBFs, various transcriptional factors including HSFC1, ZAT12, ZF, ZAT10 and RAV1 were also involved in modulating COR expression by bypassing CBF signaling (Park et al. 2015).
Abscisic acid (ABA) is a vital plant hormone that plays a key role in stress resistance during plant growth and development (Vishwakarma et al. 2017). It was reported that OST1/SnRK2E, a serine-threonine protein kinase in ABA core signaling pathway, acted upstream of CBFs to positively regulate freezing tolerance via phosphorylating ICE1 to prevent its 26S proteasome-mediated degradation by HOS1 (Ding et al. 2015). SnRK2E phosphorylated basic transcription factors 3 (BTF3) and BTF3-like factors, and facilitated their interactions with CBFs to promote CBF stability under cold stress (Ding et al. 2018).
Jasmonic acid (JA) is a lipid-derived plant hormone that plays an important role in the plant abiotic and biotic stresses (Wasternack and Song 2017; Wasternack and Strnad 2018). It was reported that JA signaling positively regulated the plant responses to freezing stress via the interaction between the repressors of jasmonate signaling proteins (TIFY/JAZ) and ICE1/2, thus suppressing the transcriptional activity of ICE proteins, thereby attenuating CBF genes expression (Hu et al. 2013).
Oil rapeseed is a winter biennial oil-seed crop, responds to vernalization and shows an excellent tolerance to cold stresses during vegetative stage. In recent years, more and more early-maturing rapeseed varieties were planted across China. Unfortunately, the early-maturing rapeseed varieties with low cold tolerance have higher risk of freeze injury in cold winter and spring. Hence, it is vital to compare early-maturing rapeseed varieties tolerant to cold and evaluate molecular mechanisms that adapt to different low-temperature stress conditions.
In this study, RNA-seq technology was used to compare differentially expressed genes (DEGs) of two early-maturing rapeseed varieties with different cold tolerance, that were treated with cold shock (4° and −4°) temperatures, as well as chilling (4°) and freezing (−4°) temperatures following cold acclimation and control conditions. Analyses identified conserved and novel signaling pathways and genes. Our results provided a foundation for understanding the low-temperature stress response mechanisms of rapeseed. We also propose new ideas and candidate genes for genetic improvement of rapeseed tolerance to cold stresses.
Materials and Methods
Plant materials and treatments
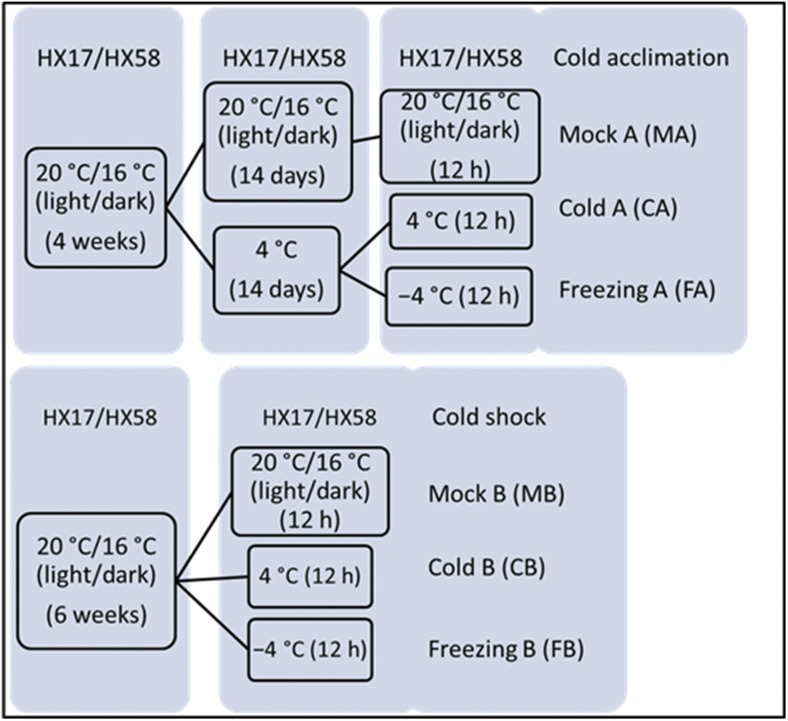
The seedlings of two early-maturing semi-winter rapeseed varieties (HX17 and HX58) were cultured in incubators under 20° (14 h light: am6:00-pm8:00)/16° (10 h dark: pm8:00-am6:00) 4 weeks, then treated with 4° (14 days) →4° (12 h) or - 4° (12 h), 20°/16° (light/dark) 6 weeks → 4° (12 h), 20° (14 h light: am6:00-pm8:00)/16° (10 h dark: pm8:00-am6:00) 6 weeks →- 4° (12 h). For the acclimation condition, after the 14 days at 4°, 4°/-4° (12h) mean a treatment with 4° or -4° at pm8:00-am8:00 (10 h dark and 2 h light). Then the third leaves from the top were collected at am8:00 after cold treatment and stored at - 80° immediately until RNA extraction.
Total RNA extraction and sequencing
The samples were sent to the sequencing cooperation of Novogene Co. Ltd (Bejing, China) for RNA isolation, examination and sequencing. HiSeq PE150 of Illumina was selected for the sample transcriptome sequencing.
Transcriptome assembly and annotation
Raw data (raw reads) of fastq format were first processed through in-house perl scripts. In this step, clean data/reads were obtained by removing reads containing adapter or, reads containing ploy-N and low quality reads from raw data. At the same time, Q20, Q30 and GC content for the clean data were calculated. All the downstream analyses were based on the clean data with high quality.
Reference genome and gene model annotation files were downloaded from genome website directly. Index of the reference genome was built using Hisat2 v2.0.4 and paired end reads were aligned to the reference genome using Hisat2 v2.0.4. Hisat2 was selected as the mapping tool because it can generate a database of splice junctions based on the gene model annotation file, thus providing better mapping result than other non-splice mapping tools.
HTSeq v0.9.1 was used to count the reads mapped to each gene. FPKM of each gene was calculated based on the length of the gene and read counts mapped to the gene. FPKM considers the combined effects of sequencing depth and gene length for the read counts, and is currently the most commonly used method for estimating gene expression levels (Trapnell et al. 2009).
Differential expression analyses of transcripts
Differential expression analysis of two conditions/groups (three biological replicates per condition) was performed using the DESeq R package (1.18.0). DESeq provide statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution (Kij ∼ NB(μij,σij2)). The resulting P-values were adjusted using the Multiple Hypothesis Testing for controlling false discovery rate. Genes with an adjusted P-value < 0.05 calculated by DESeq were assigned as differentially expressed.
GO and KEGG enrichment analyses of differentially expressed genes (DEGs)
Gene ontology (GO) enrichment analysis of DEGs was implemented by the GOseq R package, in which gene length bias was corrected. GO terms with corrected P-value < 0.05 were considered significantly enriched in DEGs.
KEGG is a database resource for understanding high-level functions and utilities of the biological system, such as the cell, the organism and the ecosystem, at molecular level, especially large-scale datasets generated by genome sequencing and other high throughput technologies (http://www.genome.jp/kegg/). KOBAS software was used to test the statistical significance of enrichment of DEGs in KEGG pathways.
qRT-PCR
To confirm the validity of the RNA-Sequencing data, we randomly selected nine DEGs for qRT-PCR analysis. 3 µg total RNAs/sample was used for cDNA biosynthesis with Superscript III reverse transcriptase (Invitrogen, San Diego, USA). The qRT-PCR was performed using the CFX96 Real-Time System (Bio-Rad, USA) with SYBR green (Bio-Rad, USA). The relative changes were calculated with 2-∆Ct and the BnaActin (BnaC05g34300D) from B. napus was used as the endogenous reference gene. The relative transcript level was determined and normalized using the reference level and averaged over the three technical replicates. Primers for the qRT-PCR (Table S10) were designed according to the cDNAs sequences using Primer Premier 5 (http://www.premierbiosoft.com/crm/jsp/com/pbi/crm/clientside/ProductList.jsp).
Data Availability
The sequencing rawdata are accessible through GEO (GSE129220): https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE129220. Supplemental material available at Figshare: https://10.6084/m9.figshare.7379858.
Results and discussion
Transcriptomic analyses
Two early-maturing semi-winter rapeseed varieties (HX17 and HX58) with different cold tolerance (Figure S1) were selected for transcriptomic investigation by RNA-Seq following different low-temperature treatments.
The responsive mechanisms were different between chilling and freezing temperatures, as well as cold shock and cold acclimation in plants (Ruelland et al. 2009; Guy 1999; Nievola et al. 2017). A short-term exposure to low, non-freezing temperatures sufficiently induced cold acclimation resulting in increased tolerance to freezing conditions [5]. Accordingly, 20°/16° (light/dark) →cold-acclimation (4°, 14 days) → 4°/−4° (12 h), 20°/16° (light/dark) → 4°/−4° (12 h), were selected as the treatment conditions (Figure 1).
Figure 1.
The processes of different low-temperature treatments on two rapeseed (Brassica. napus L. cv. HX17 and HX58) seedlings.
Total RNA was extracted from the seedlings of HX17 and HX58 treated with different low-temperature conditions and sequenced using the Illumina HiSeq 2500 platform. A total of 296.67 Gb clean data (1,977,933,478 clean reads) were acquired after removal of adaptor sequences, ambiguous nucleotides and low-quality sequences (Table 1). Approximately 43.1-78.1 million clean reads were obtained from the libraries. The Q-score for 88.17–93.99% of reads was Q30, and 82.57–86.51% of the total reads were uniquely mapped to the reference genome (Table S1). The mapped sequences were assembled with Cufflinks, referenced against the annotated genome sequence of the tetraploid species B. napus provided by Index of /brassicanapus/data (http://www.genoscope.cns.fr/brassicanapus/data/). RNA-Seq revealed that there was a total of 172,412 transcripts and 110,367 unigenes, and 10,578 genes on average for each sample, including 78,945 known genes and 26,233 novel genes on average (Table S2). Gene expression levels were estimated by fragments per kilo base of transcript per million fragments mapped (FPKM) (Langmead et al. 2009). Approximately 8.51–11.97% of total genes’ FPKM was more than 15.0, and 59.26–65.50% of total genes’ FPKM was lower than 1.0 (Table S3).
Table 1. Quality filtering and statistics of raw reads obtained in transcriptome libraries of B. napus exposed to low-temperature treatments.
| Sample name | Raw reads | Clean reads | clean bases | Error rate (%) | Q20 (%) | Q30 (%) | GC content (%) |
|---|---|---|---|---|---|---|---|
| HX17_MA1 | 61608094 | 58966572 | 8.84G | 0.03 | 97.36 | 92.93 | 44.06 |
| HX17_MA2 | 46842038 | 45252272 | 6.79G | 0.03 | 96.98 | 92.17 | 44.39 |
| HX17_MA3 | 51620482 | 50405900 | 7.56G | 0.03 | 97.11 | 92.01 | 45.91 |
| HX17_CA1 | 45028428 | 44096512 | 6.61G | 0.03 | 96.97 | 91.66 | 45.83 |
| HX17_CA2 | 53540226 | 52633174 | 7.89G | 0.03 | 96.72 | 91.49 | 46.35 |
| HX17_CA3 | 49776538 | 47962390 | 7.19G | 0.03 | 96.70 | 91.54 | 43.39 |
| HX17_FA1 | 46114476 | 45390992 | 6.81G | 0.03 | 96.66 | 91.37 | 44.84 |
| HX17_FA2 | 66471914 | 64442766 | 9.67G | 0.03 | 97.11 | 92.40 | 45.85 |
| HX17_FA3 | 49756338 | 49191330 | 7.38G | 0.03 | 97.20 | 92.53 | 45.64 |
| HX17_MB1 | 45873106 | 45028940 | 6.75G | 0.03 | 95.42 | 88.17 | 44.09 |
| HX17_MB2 | 47889494 | 47067140 | 7.06G | 0.03 | 96.75 | 91.57 | 45.00 |
| HX17_MB3 | 46228026 | 45344394 | 6.80G | 0.03 | 96.94 | 92.04 | 44.25 |
| HX17_CB1 | 70149774 | 68467860 | 10.27G | 0.03 | 97.85 | 94.12 | 44.40 |
| HX17_CB2 | 54975576 | 53977612 | 8.10G | 0.03 | 97.12 | 92.04 | 44.50 |
| HX17_CB3 | 55136028 | 53840712 | 8.08G | 0.03 | 97.02 | 91.82 | 44.74 |
| HX17_FB1 | 79518680 | 78163306 | 11.72G | 0.03 | 97.70 | 93.70 | 44.76 |
| HX17_FB2 | 48126142 | 47147508 | 7.07G | 0.03 | 96.75 | 91.28 | 44.80 |
| HX17_FB3 | 52485614 | 51527762 | 7.73G | 0.03 | 97.77 | 93.79 | 44.19 |
| HX58_MA1 | 67391320 | 65727088 | 9.86G | 0.03 | 97.16 | 92.16 | 44.28 |
| HX58_MA2 | 66677164 | 64776152 | 9.72G | 0.03 | 97.09 | 91.98 | 43.86 |
| HX58_MA3 | 47017928 | 44892330 | 6.73G | 0.03 | 97.07 | 92.07 | 43.88 |
| HX58_CA1 | 44491830 | 43128444 | 6.47G | 0.03 | 96.66 | 91.39 | 46.46 |
| HX58_CA2 | 47877742 | 46243718 | 6.94G | 0.03 | 96.73 | 91.40 | 45.62 |
| HX58_CA3 | 49056666 | 47594184 | 7.14G | 0.03 | 97.85 | 93.99 | 45.77 |
| HX58_FA1 | 77825480 | 76080646 | 11.41G | 0.03 | 97.92 | 94.08 | 44.84 |
| HX58_FA2 | 72714882 | 70675296 | 10.6G | 0.03 | 97.72 | 93.61 | 45.33 |
| HX58_FA3 | 62524530 | 60787534 | 9.12G | 0.03 | 97.71 | 93.59 | 45.10 |
| HX58_MB1 | 55247676 | 54098192 | 8.11G | 0.03 | 96.74 | 91.23 | 44.28 |
| HX58_MB2 | 57837196 | 56764648 | 8.51G | 0.03 | 96.82 | 91.34 | 45.16 |
| HX58_MB3 | 49353542 | 48225898 | 7.23G | 0.03 | 97.34 | 92.54 | 44.69 |
| HX58_CB1 | 61754716 | 60061840 | 9.01G | 0.03 | 97.88 | 93.95 | 43.42 |
| HX58_CB2 | 66709668 | 64672348 | 9.70G | 0.03 | 97.69 | 93.60 | 42.76 |
| HX58_CB3 | 52648416 | 51400508 | 7.71G | 0.03 | 96.73 | 91.28 | 43.77 |
| HX58_FB1 | 58213716 | 56978060 | 8.55G | 0.03 | 97.04 | 91.89 | 46.07 |
| HX58_FB2 | 59190884 | 57729982 | 8.66G | 0.03 | 97.11 | 92.37 | 46.39 |
| HX58_FB3 | 60769766 | 59189468 | 8.88G | 0.03 | 97.10 | 92.36 | 45.06 |
| Assembly Steps | Total assembled transcripts | Number of transcripts (>1000bp) | Percent of transcripts | Maximum transcript length | Average transcript length | N50 value (bp) | Coverage (X) |
|---|---|---|---|---|---|---|---|
| Assembler SOAPdenovotrans (65 k-mer) | 91,765 | 32,509 | 35.43 | 12,805 | 903.39 | 1,427 | 160 |
| After gap filling and hierarchical clustering | 53,806 | 24,429 | 45.40 | 12,674 | 1,096.96 | 1,499 | 218 |
| After removing mis-assembled transcripts | 46,556 | 19,347 | 41.55 | 9,811 | 1,013.66 | 1,393 | 226 |
| BLASTX best group representatives (unigenes) | 25,400 | 12,588 | 49.56 | 9,811 | 1,153.30 | 1,502 | 240 |
For the evaluation of DEGs’ reliability and the filter of abnormal samples, Pearson correlation coefficient (PCC) analysis was conducted. The correlation analysis indicated that all three technical replicates replication samples showed more than 0.929 (R2) of similarities (Table S4).
Differentially expressed genes (DEGs) in rapeseed under different low-temperature treatments
Differential expression analysis of treatments and control group was performed using the DESeq (Anders and Huber 2010). A threshold of fold change of ≥2 and p-adjusted q-value of < 0.005 was used for identifying DEGs.
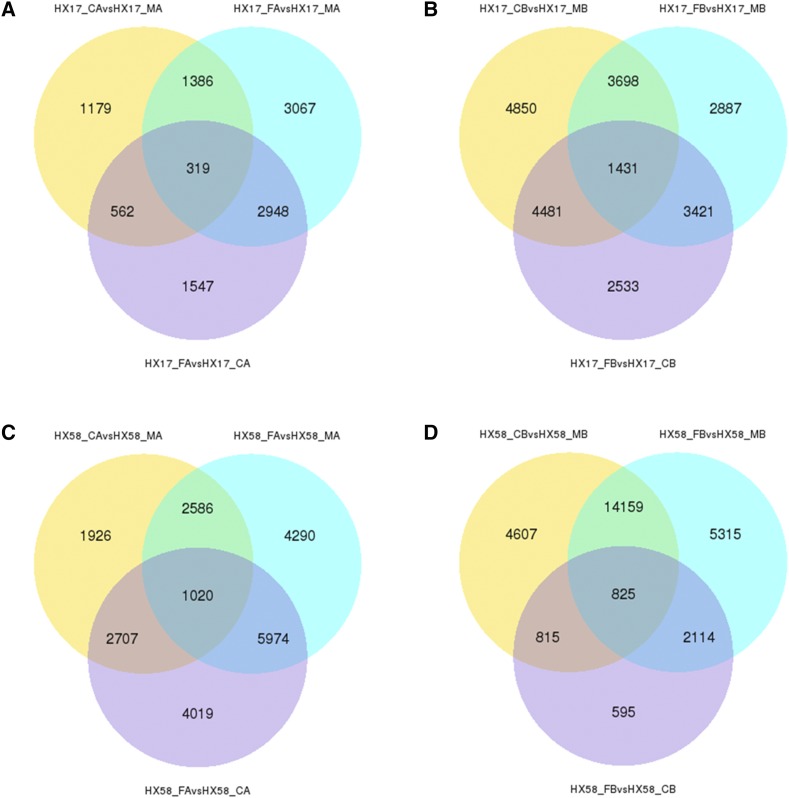
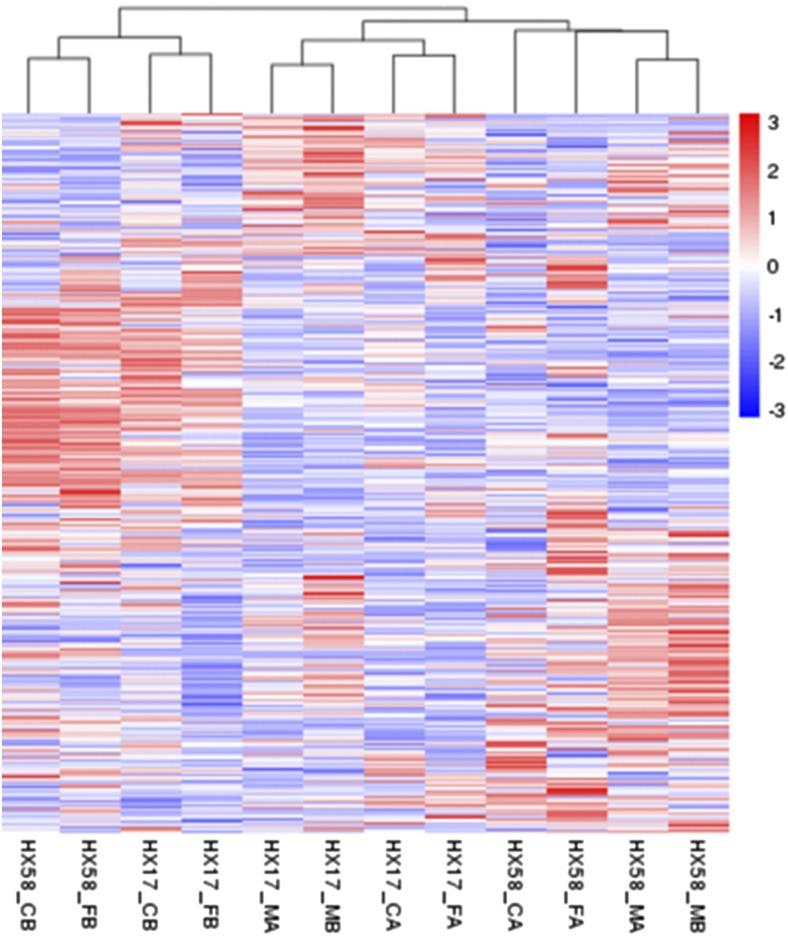
A total of 47,328 DEGs (HX17_CA vs. HX17_MA: 3446; HX17_FA vs. HX17_MA: 7738; HX17_FA vs. HX17_CA: 5378; HX17_CB vs. HX17_MB: 16802; HX17_FB vs. HX17_MB: 22271; HX17_FB vs. HX17_CB: 3704; HX58_CA vs. HX58_MA: 10340; HX58_FA vs. HX58_MA: 16505; HX58_FA vs. HX58_CA: 18280; HX58_CB vs. HX58_MB: 21902; HX58_FB vs. HX58_MB: 25279; HX58_FB vs. HX58_CB: 4381) were identified in the different low-temperature treatments (Figure 2 and Table S5). Statistically, DEGs in both HX17 and HX58 showed that FB had the most DEGs and CA had the least, in the order of FB > CB > FA> CA. HX58 presented with more DEGs than HX17 (Figure 2), consistent with the cluster analysis of DEGs (Figure 3 and Figure S2). In addition, the lower temperature correlated with more injuries, and HX58 was more susceptible to low-temperature stress (Figure S1). DEGs under different stresses in both HX17 and HX58 were presented as CA vs. FA > CB vs. FB (Figure 2), confirming that the cold acclimation altered the responses to freezing stress in plant.
Figure 2.
Venn diagram of DEGs between HX17_MA, HX17_CA, HX17_FA (A); HX17_MB, HX17_CB, HX17_FB (B); HX58_MA, HX58_CA, HX58_FA (C); HX58_MA, HX58_CA, HX58_FB (D).
Figure 3.
Hierarchical cluster analysis of differentially expressed genes (DEGs) among all the 12 samples. Heatmap of DEGs among the 12 samples. Red indicates high expression, and blue indicates low expression. Color from red to blue represents descending log10 (FPKM + 1).
Pathway analyses of responses to different low-temperature stresses in rapeseed
Pathway analyses of DEGs against the Kyoto Encyclopedia of Genes and Genomes (KEGG) contributed to the understanding of gene functions (Kanehisa et al. 2008). As shown in Figure S3 and Table S6, there was no enriched pathway in both HX17 and HX58 treated with cold shock (CB and FB) compared to control condition (MB), indicating that cold shock stress caused serious injuries to the plant. Interestingly, there was no enriched pathway in HX58 acclimatized to cold (CA and FA), similar to cold shock stress. However, the plant hormone signal transduction pathway was enriched in HX17 acclimatized to cold (CA and FA), indicating that the plant hormone signal transduction was activated by cold acclimation in cold-tolerant rapeseed. Further analyses of the upregulated and downregulated DEGs enriched pathways are reported in Table S7. Pathways involved in primary metabolism and plant hormone signal transduction were enriched under different low-temperature stresses in both HX17 and HX58 (Fig. S3, Table S6). Previous studies also reported changes in these two pathways in various crops and plants exposed to low-temperature stresses (Nievola et al. 2017; Qu et al. 2015; Zhang et al. 2017; Li et al. 2017; Bai et al. 2015; Zhang et al. 2014), suggested that low-temperature stresses were conservative to affect the normal metabolism of plants and the plant hormone signal transduction.
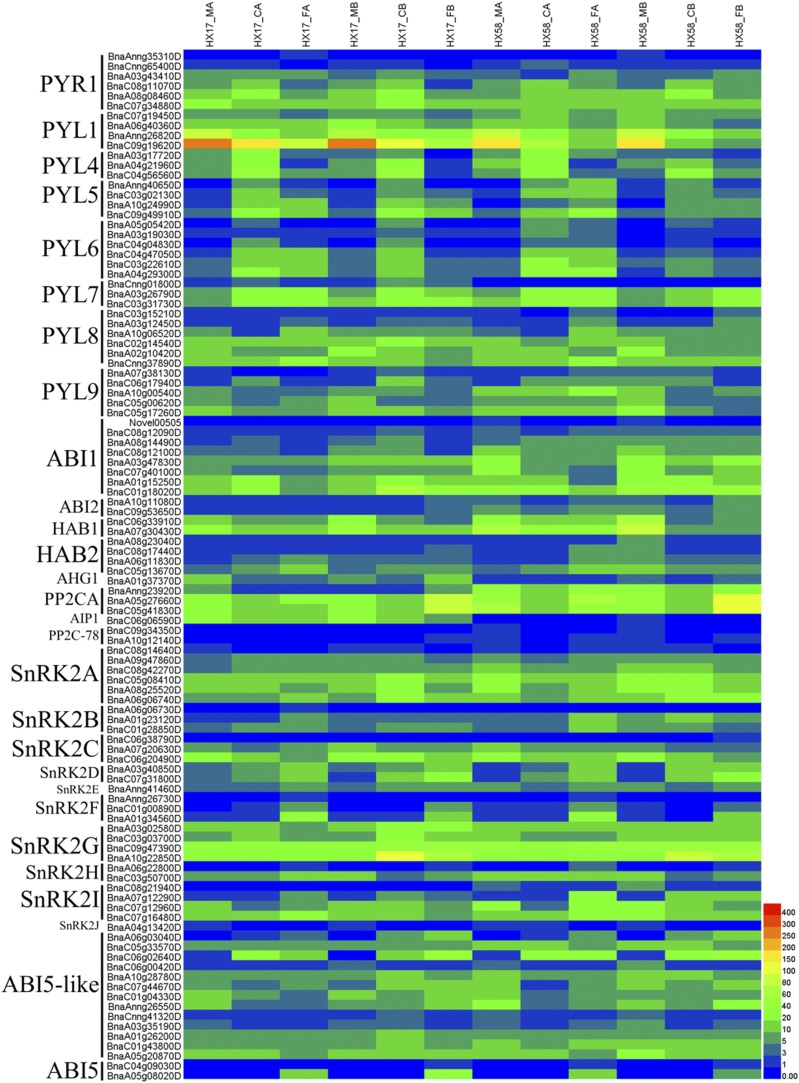
There were many reports that the plant hormone signal pathways played important roles in the plant responses to abiotic stresses (Colebrook et al. 2014; Kazan 2015; Gururani et al. 2015; Bielach et al. 2017; Verma et al. 2016; de Zelicourt et al. 2016; Khan et al. 2015; Sharma et al. 2017). The ABA and JA biosynthesis and signaling were induced by various abiotic stresses, and positively regulated the low-temperature tolerance in plants (Yoshida et al. 2014; Huang et al. 2017; Hu et al. 2017; Shinkawa et al. 2013; Hu et al. 2013; Du et al. 2013; Wang et al. 2016a). Hence, we analyzed the expression patterns of ABA and JA signaling genes. The results showed that among the 503 DEGs of the plant hormone signaling transduction (total of 1148 genes), 94 of them were the ABA signaling genes (total of 139 genes) and 43 of them were JA signaling genes (total of 61 genes)(Table S7). Most of the ABA signaling genes were changed under cold acclimation and cold shock treatments in both HX17 and HX58 but they presented different expression profiles (Figure 4). The ABA receptors pyrabactin resistance proteins/PYR-like proteins/regulatory components of ABA receptors (PYR/PYL/RCAR) PYL5 and PYL7 were both induced by cold and freezing treatments in HX17 and HX58, while the PYL1 and PYL9 were suppressed in all treatments. The PYR1 and PYL4 were only induced by cold treatment but not by freezing treatment. The PYL6 were induced by cold treatment and freezing treatment following cold acclimation. ABA co-receptor phosphatase 2C (PP2C) ABI1 and HAB1 were suppressed by all treatments, while HAB2 was upregulated. The Snf1-related protein kinase 2 (SnRK2) SnRK2B and SnRK2D were induced by all low-temperature treatments, while SnRK2C was suppressed. Interestingly, SnRK2F was induced only by freezing treatment, similar to ABI5 expression, indicating that they played important roles in response to freezing in rapeseed. The Arabidopsis ABI5 was a bZIP transcription factor phosphorylated by SnRK2D and SnRK2I, and participated in ABA-regulated gene expression during seed development and subsequent vegetative stage by acting as the major mediator of ABA repression of growth (Skubacz et al. 2016; Nakashima et al. 2009; Brocard et al. 2002). It is possible that SnRK2F directly phosphorylated ABI5 in rapeseed. The SnRK2E/OST1 was induced only in HX17 but not in HX58, which may be a part reason for the better cold-tolerance of HX17 than HX58.
Figure 4.
Heatmap analysis of the genes in ABA signal pathway. PYR1 (PYRABACTIN RESISTANCE 1), Abscisic acid receptor; PYL (PYR1-like protein), Abscisic acid receptor; ABI1/2 (ABSCISIC ACID-INSENSITIVE 1/2), Protein phosphatase 2C; HAB1/2 (HYPERSENSITIVE TO ABA 1/2), Protein phosphatase 2C; AHG1 (ABA-HYPERSENSITIVE GERMINATION 1), Protein phosphatase 2C; PP2C, Protein phosphatase 2C; AIP1 (AKT1-INTERACTING 1), Protein phosphatase 2C; SnRK2 (SNF1-related kinase 2), Serine/threonine-protein kinase; ABI5 (ABSCISIC ACID-INSENSITIVE 5), bZIP transcription factor. The expression levels of each gene (FPKM values) in each sample, as indicated by different color rectangles. Red means high expression; Blue means low expression.
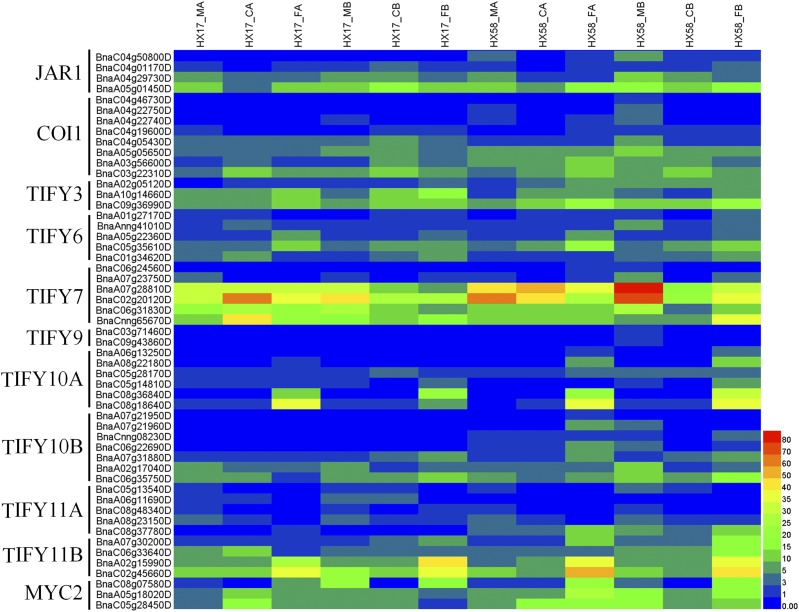
It was reported that exogenous application of JA enhances plant freezing tolerance with or without cold acclimation, and overexpression of JAZ1/TIFY10A and JAZ4/TIFY6 suppressed the transcriptional function of ICE1, thereby attenuating the ICE-CBF-COR signaling pathway and freezing responses in Arabidopsis (Hu et al. 2013). As shown in Figure 5, the expressions of many JA signaling genes were altered in both HX17 and HX58 under low-temperature conditions. TIFY3 and TIFY6 were induced in both HX17 and HX58 under all low-temperature conditions. Interestingly, TIFY7 was suppressed only under cold shock treatments, while the TIFY10A and TIFY11B were upregulated only by freezing treatments, similar to SnRK2F and ABI5 (Figure 4 and Figure 5). These changes suggest that they may be important regulators in the freezing response in rapeseed. Additionally, MYC2 was only induced by cold acclimation, but not by cold shock.
Figure 5.
Heatmap analysis of the genes in JA signal pathway. JAR1 (JASMONATE RESISTANT 1), Jasmonic acid-amido synthetase; COI1 (Coronatine-insensitive protein 1), Jasmonic acid receptor; TIFY, Jasmonate ZIM domain-containing protein; MYC2, bHLH transcription factor MYC2. The expression levels of each gene (FPKM values) in each sample, as indicated by different color rectangles. Red means high expression; Blue means low expression.
To test whether exogenous ABA and methyl-jasmonic acid (MeJA) could improve the rapeseed cold tolerance, 4-week-old rapeseed seedlings were sprayed with ABA (100 μM), MeJA (100 μM) and water before cold shock at -6°. The survival rate results demonstrated that exogenous ABA significantly improved the rapeseed freezing tolerance. On the contrary, exogenous MeJA reduced the rapeseed freezing tolerance (Figure 6). The concentration of exogenous MeJA might be too high, affecting the rapeseed freezing tolerance. Other studies reported that treatment with exogenous JA induced leaf senescence and expression of senescence associated genes, while exogenous high concentration MeJA caused plant injures (Zhu et al. 2015). All the results indicated that a complex gene regulatory network composed of plant hormones signaling affected the low-temperature stresses in rapeseed, and appropriate concentrations of exogenous plant hormones could increase rapeseed’s cold tolerance.
Figure 6.
Effect of ABA and MeJA on the freezing tolerance of ZS11 (Brassica. napus L. cv. Zhongshuang 11) seedlings. ZS11 seedlings were grown for 4-week under normal conditions at 20 °C, and then sprayed with ABA (100 μM), MeJA (100 μM) or water (Mock) for 1 day. Then the seedlings were treated with -6 °C for 2 days and recovered under normal conditions at 20 °C for 7 days. Survival rates of seedlings were shown in brackets.
Identification of the transcriptional regulatory network responding to low-temperature stress in rapeseed
Transcription factors (TF) play a central role in the gene regulatory networks that mediate various aspects of plant developmental processes and responses to environmental changes (Hoang et al. 2017; Riechmann and Ratcliffe 2000). The program iTAK was used to analyze DEGs and identified 7633 TFs. Of these, 3670 (48.1%) belong to 76 transcription factors and transcriptional regulator families. The 10 top families were MYB, AP2-EREBP, bHLH, Orphans, HB, bZIP, NAC, C2H2, WRKY and C3H (Table S8).
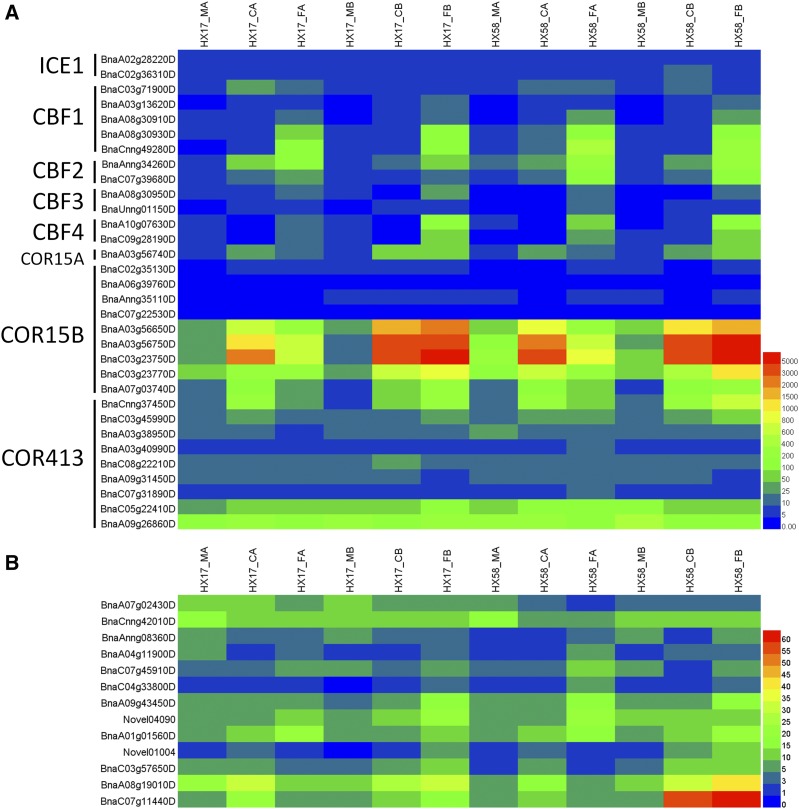
It was reported that CBFs were the core regulator in the cold-response signaling in plants (Shi et al. 2018; Guo et al. 2018; Ruelland et al. 2009; Wang et al. 2017). CBFs belong to the DREB1 proteins, including DRE1A/CBF3, DRE1B/CBF1, DRE1C/CBF2 and DRE1D/CBF4 (Liu et al. 1998; Jaglo-Ottosen et al. 1998; Sakuma et al. 2002). We analyzed the ICE1, CBFs and some COR genes, and observed that COR15A and COR15B were significantly induced by all low-temperature treatments (Figure 7A). Apart from BnaC03g71900D, all the CBF genes were induced by freezing stress more than chilling stress, regardless of cold acclimation (Figure 7A), highlighting the importance of CBFs in rapeseed freezing tolerance. However, ICE1 was not induced by low-temperature treatments, as opposed to AtICE1 (Chinnusamy et al. 2003).
Figure 7.
Heatmap analysis of the ICE-CBF-COR signal pathway (A) and cold shock domain-containing proteins (B). ICE1 (Inducer of CBF expression 1), bHLH transcription factor; CBF (C-repeat-binding factor), dehydration-responsive element-binding protein; COR, cold-regulated. The expression levels of each gene (FPKM values) in each sample, as indicated by different color rectangles. Red means high expression; Blue means low expression.
The cold shock domain-containing proteins (CSP) act as RNA chaperones that destabilize mRNA secondary structures at low temperatures, and they are regulated by low-temperature stress (Sasaki and Imai 2011; Choi et al. 2015; Radkova et al. 2014; Chaikam and Karlson 2008). Overexpression of AtCSP2 resulted in decreased freezing tolerance despite cold acclimation (Sasaki et al. 2013). CSP functions are associated mainly with cold adaptation, but they are also involved in other biological processes under normal growth conditions (Sasaki and Imai 2011; Nakaminami et al. 2009; Park et al. 2009). All the 13 cold CSPs were regulated by the low-temperature treatments (Figure 7B). As shown in Figure 7B, three CSP genes (BnaA07g02430D, BnaCnng42010D, and BnaAnng08360D) were suppressed by low-temperature treatments, and the others were induced by at least one treatment. BnaA01g01560D was induced by all low-temperature treatments. Four CSP (Novel01004, BnaC03g57650D, BnaA08g19010D, and BnaC07g11440D) were induced by cold shock (CB and FB) and chilling after cold acclimation (CA), but not freezing after acclimation (FA). BnaA09g43450D and Novel04090 were induced by cold shock (CB and FB) and chilling after cold acclimation (FA), but not freezing after acclimation (CA). All the results indicated that the CSP genes have a complex expression pattern in response to different low-temperature conditions.
Heat stress transcription factors (HSFs) play a crucial role in plant responses to high-temperature by regulating the expression of stress-responsive genes, such as heat shock proteins (Guo et al. 2016). Approximately half of HSFs were affected under low-temperature condition (Table S8), suggesting that HSFs responded to high-temperature and low-temperature changes.
Plant-pathogen interaction pathways play important roles in low-temperature stresses in rapeseed
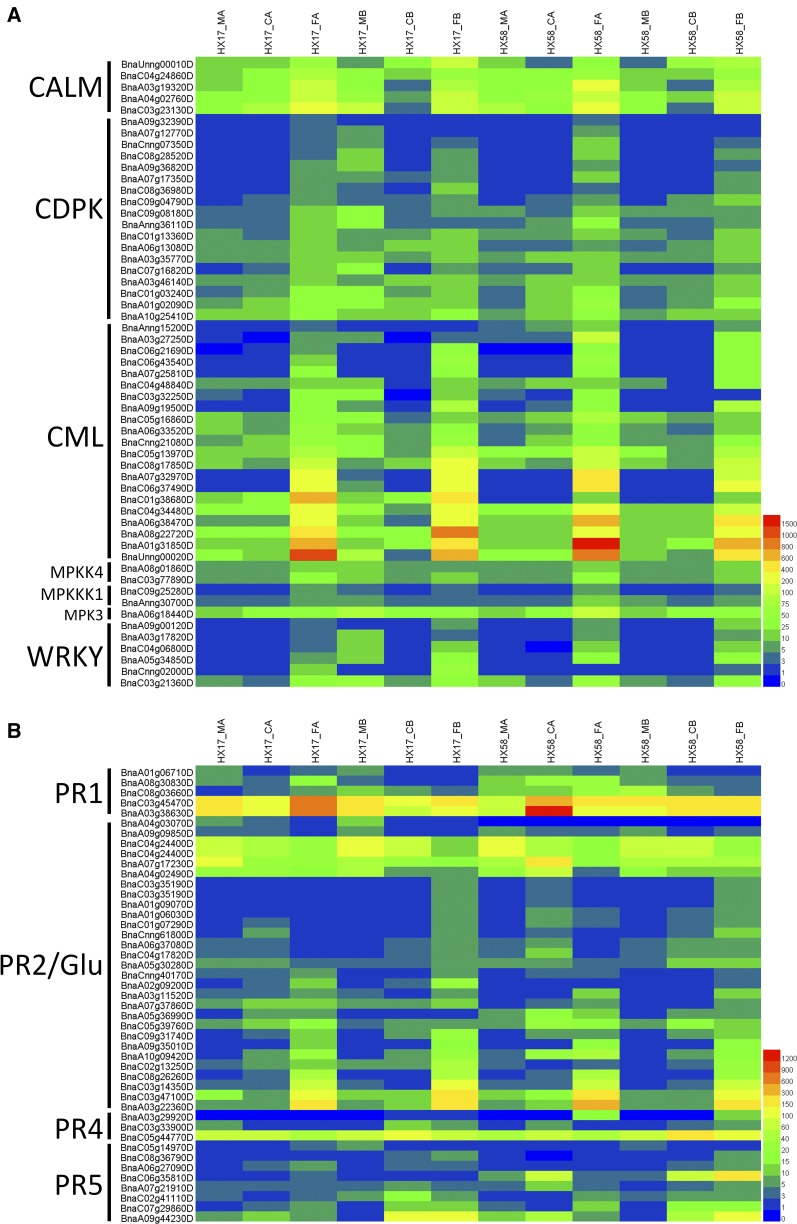
As shown in Table S6-S7, the plant-pathogen interaction pathway was enriched in both HX17 and HX58 under low-temperature conditions, especially under the freezing stresses, consistent with expression of the plant-pathogen interaction pathway genes (Figure 8). Even though the plant-pathogen interaction pathway was consistently reported to be enriched under low-temperature treatments in various plants (Zhang et al. 2017; Qu et al. 2015; Yang and Huang 2018; Xu et al. 2016; Wang et al. 2016b; Tian et al. 2013; Du et al. 2015; Wang et al. 2015), the functions of this pathway in the plant cold responsiveness were neglected by researchers. The pathogenesis-related proteins (PR) were induced by cold acclimation (Kuwabara and Imai 2009; Hincha et al. 1997). Pre-treatment of tomato fruit with MeJA or MeSA induced the synthesis of PR proteins, which led to increased chilling tolerance and resistance to pathogens (Ding et al. 2002). Overexpression of 3 PR genes (PR2, PR4, Glu) in Arabidopsis enabled the plant to tolerate freezing temperatures (Cabello et al. 2012). As shown in Figure 8b, PR1 and PR5 were induced by cold acclimation and cold shock. Overexpression of a rice PR5 gene in rapeseed enhanced the resistance to Sclerotinia sclerotiorum, a fungal pathogen that caused Sclerotinia stem rot in rapeseed (Aghazadeh et al. 2017). We propose that overexpression of PR genes in rapeseed might produce rapeseed germplasm with tolerance to cold and S. sclerotiorum.
Figure 8.
Heatmap analysis of the regulator genes (A) and PR genes (B) in the plant-pathogen interaction pathway. CAML, Calmodulin, CDPK; Calcium-dependent protein kinase; CML, calcium-binding protein; MPK3, Mitogen-activated protein kinase 3; MPKK4, Mitogen-activated protein kinase kinase 4; MPKKK1, Mitogen-activated protein kinase kinase kinase 1; WRKY, WRKY transcription factor; PR, Pathogenesis-related protein; Glu, Glucan endo-1,3-beta-glucosidase. The expression levels of each gene (FPKM values) in each sample, as indicated by different color rectangles. Red means high expression; Blue means low expression.
Circadian rhythms play an important role in low-temperature stresses in rapeseed
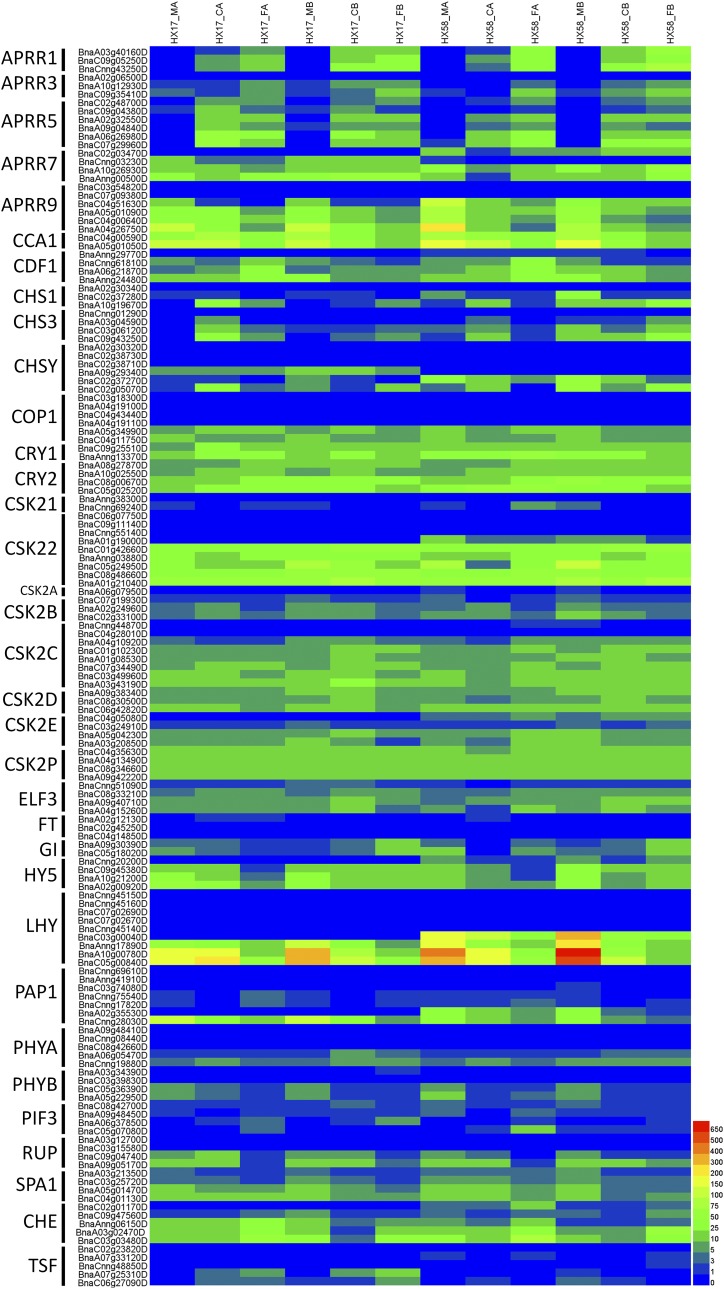
As shown in Table S6-S7, the circadian rhythms pathway was enriched in the DEGs for both HX17 and HX58 under low-temperature conditions. It was also shown to respond to low-temperature conditions in several plants (Li et al. 2017; Sobkowiak et al. 2014; Liu et al. 2018; Abeynayake et al. 2015; Barah et al. 2013), and regulated CBFs and CORs gene expression (Edwards et al. 2006; Bieniawska et al. 2008). Almost all the genes associated with circadian rhythms showed differential expression (Figure 9). These included induction of the pseudo-response regulators (APRR) APRR1/TOC1, APRR3 and APRR5 by chilling and freezing stresses in both HX17 and HX58, and suppression of APRR7 and APRR9, consistent with the reports on Arabidopsis (Lee et al. 2005). A prr7-3 prr9-1 double mutant failed to entrain to temperature cycles (Salome and McClung 2005), while prr5/7/9 triple mutant plant was more tolerant to cold stresses through upregulation of CBF expression (Nakamichi et al. 2009). The Cholcone synthase (CHS), twin sister of FT (TSF), phytochrome A (PHYA) and transcription factor PIF3 were induced by chilling and freezing stresses. The circadian clock associated 1 (CCA1), early flowering 3 (ELF3), late elongated hypocotyl (LHY), phytochrome B (PHYB), suppressor of PHYA-105 1 (SPA1), repressor of UV-B photomorphogenesis 1 (RUP) and transcription factors HY5 and MYB75 were suppressed by chilling and freezing stresses. Cyclic dof factor 1 (CDF1) and cytochrome 1/2 (CRY1/2) were induced by cold acclimation, but were suppressed by cold shock. Gigantea (GI) and CHE/TCP21 were induced only by freezing stress.
Figure 9.
Heatmap analysis of the genes in circadian rhythms pathway. APRR (pseudo response regulator), Two-component response regulator-like; CCA1 (CIRCADIAN CLOCK ASSOCIATED 1), MYB-related transcription factor; CDF1 (Cyclic dof factor 1), Dof zinc finger protein; CHS (Chalcone synthase); COP1 (Constitutive photomorphogenesis protein 1), E3 ubiquitin-protein ligase; CRY1/2 (Cryptochrome 1/2), Blue light photoreceptor; CSK2, Casein kinase II subunit; ELF3 (EARLY FLOWERING 3); FT (FLOWERING LOCUS T), Phosphatidylethanolamine-binding protein; GI (GIGANTEA); HY5 (ELONGATED HYPOCOTYL5), bZIP transcription factor; LHY (LATE ELONGATED HYPOCOTYL), MYB-related transcription factor; PAP1(Production of anthocyanin pigment 1 protein), MYB-related transcription factor; PHYA/B (Phytochrome A/B), ; PIF3 (Phytochrome-interacting factor 3), bHLH transcription factor; RUP1, WD repeat-containing protein; SPA1 (SUPPRESSOR OF PHYA-105 1), WD repeat-containing protein; CHE (CCA1 HIKING EXPEDITION), TCP transcription factor; TSF (TWIN SISTER of FT). The expression levels of each gene (FPKM values) in each sample, as indicated by different color rectangles. Red means high expression; Blue means low expression.
Two phytochrome mutants phyB and phyD showed increased cold tolerance via the upregulation of CBF regulon (Franklin and Whitelam 2007). During the warm long-day condition growing season, the CBF pathway is actively suppressed by PHYB, PIF4, and PIF7 (Lee and Thomashow 2012). PHYA and PHYB function antagonistically to regulate cold tolerance via abscisic acid-dependent jasmonate signaling in tomato (Wang et al. 2016a). PIF3 acts as a negative regulator of plant cold acclimation by direct suppression of CBF expression (Jiang et al. 2017).
HY5 transcription was regulated by low temperature through a CBF- and ABA-independent pathway, and mutant hy5-1 provoked a significant reduction in the ability of Arabidopsis to cold acclimate (Catala et al. 2011). GI increased freezing tolerance via a CBF-independent pathway (Cao et al. 2005). CCA1/LHY bound to promoter of CBFs and promoted cold acclimation (Dong et al. 2011). COR27 and COR28 were direct targets of CCA1, meanwhile they bind to the chromatin of APRR1 and APRR5 and repress their expression (Li et al. 2016). Therefore, we could infer that the circadian rhythm pathways play a role in freezing tolerance, through regulation of the circadian rhythm genes in rapeseed.
Vernalization in rapeseed
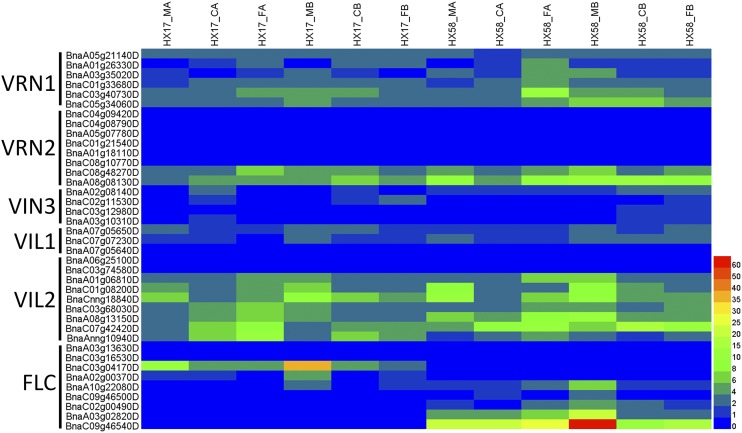
Although both cold acclimation and vernalization are responses to low temperature, the durations of cold exposure required to initiate these responses are quite distinct. The most upstream event of the vernalization pathway identified is the induction of vernalization insensitive 3 (VIN3) in response to low temperatures. Together with VIL1, VIN3 was required during vernalization for the modifications of FLC and FLM chromatin, it led to an epigenetically silenced state and acquisition of competence to flower (Sung and Amasino 2004).
We analyzed the expression pattern of all rapeseed VIN and FLC genes under different low-temperature conditions. As shown in Figure 10, only two VIN genes (BnaC07g42420D and BnaAnng10940D) were induced by cold acclimation and cold shock. BnaC01g08200D and BnaCnng18840D were suppressed by both cold acclimation and cold shock. BnaC03g68030D and BnaA08g13150D were induced only by cold acclimation but not by cold shock. The induction of a few VIN genes in response to low temperatures suggested that the vernalization pathway in early-maturing rapeseed was attenuated.
Figure 10.
Heatmap analysis of the genes in the vernalization pathway. VRN1 (VERNALIZATION 1), B3 domain-containing transcription factor; VRN2 (VERNALIZATION 2), Zinc finger protein; VIN3 (VERNALIZATION INSENSITIVE 3), PHD finger protein; VIL, VIN3-like protein; FLC (FLOWERING LOCUS C), MADS-box protein. The expression levels of each gene (FPKM values) in each sample, as indicated by different color rectangles. Red means high expression; Blue means low expression.
The expression patterns of nine rapeseed FLCs were different between HX17 and HX58 (Figure 10). Two FLC genes were suppressed only in HX17 under cold-acclimation and cold-shock, while three FLC genes were suppressed only in HX58. BnaA10g22080D was suppressed in both HX17 and HX58 under cold shock stresses. All the results suggest that FLC genes vary extensively among the rapeseed populations.
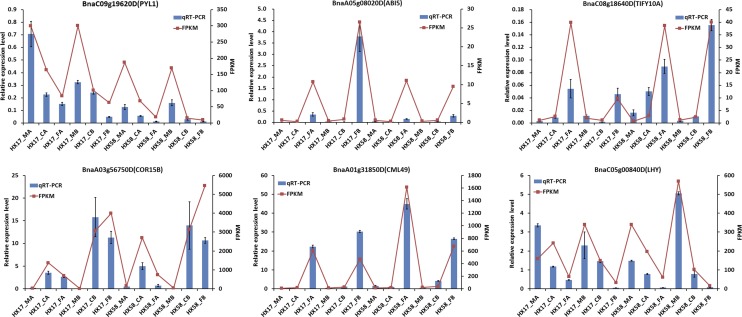
Validation of gene expression patterns by qRT-PCR
As shown in Figure 11, all the 4 candidate genes were induced by freezing stress strongly, with or without cold acclimation. Although the fold-changes in their expression detected by sequencing did not exactly match those detected by qRT-PCR, the detected expression patterns were mostly consistent for all the selected genes, confirming the reliability of the RNA-Seq results.
Figure 11.
qRT-PCR verification of 6 selected DEGs. Comparison of qRT-PCR (blue bar) with FPKM data (red line). The relative changes were calculated with 2-∆Ct and the rapeseed BnaActin gene was amplified as the reference gene. The relative qRT-PCR expression level (selected DEG/ BnaActin) is shown on the left y-axis. The relative transcript level was determined and normalized using the reference level and averaged over the three technical replicates. The FPKM from the RNA-Seq data are indicated on the right y-axis.
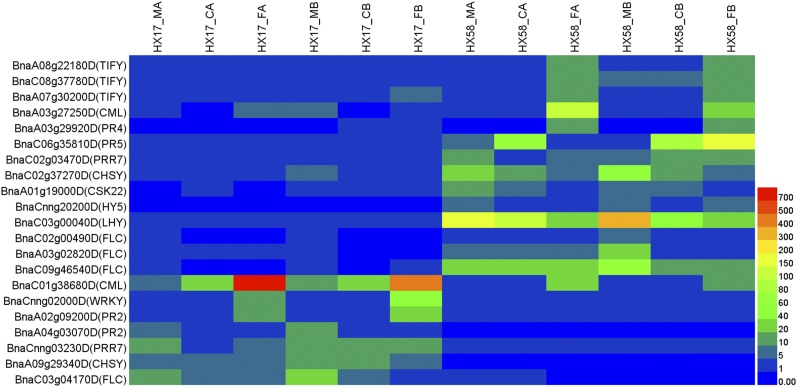
The differences in cold tolerance between HX58 and HX17
As shown in Figure 12, there are some genes involved in JA signaling, Ca2+-signaling, plant-pathogen interaction pathway, circadian rhythms pathway and flowering pathway with different expression patterns and levels between HX58 and HX17, and this may suggest that those genes’ expression extent and amount lead the difference in cold tolerance between HX58 and HX17.
Figure 12.
Heatmap analysis of the DEG genes between HX58 and HX17. The expression levels of each gene (FPKM values) in each sample, as indicated by different color rectangles. Red means high expression; Blue means low expression.
Conclusions
This study is the first report of the transcriptome data of two early-maturing rapeseeds under different low-temperature stresses (cold acclimation and cold shock; chilling and freezing) using HiSeq PE150 of Illumina. A total of 47,328 DEGs were identified in rapeseed treated with different low temperatures. Further analysis of these DEGs showed that the low-temperature response was a complex process in rapeseed. Many KEGG pathways were enriched following low-temperature treatments, including the primary metabolism, plant hormone signal transduction, plant-pathogen interaction pathway, circadian rhythms and so on. These results provide a reference for our understanding of the rapeseed adaptation to different low-temperature environments and provide new insights into the molecular mechanisms of different low-temperature tolerance in rapeseed.
Acknowledgments
This work was supported by Institutions of higher learning innovation ability enhancement (30552-520190100004) and National Natural Science Foundation of China (31801398).
Footnotes
Supplemental material available at Figshare: https://10.6084/m9.figshare.7379858.
Communicating editor: D. J. de Koning
Literature Cited
- Abeynayake S. W., Byrne S., Nagy I., Jonaviciene K., Etzerodt T. P. et al. , 2015. Changes in Lolium perenne transcriptome during cold acclimation in two genotypes adapted to different climatic conditions. BMC Plant Biol. 15: 250 10.1186/s12870-015-0643-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghazadeh R., Zamani M., Motallebi M., and Moradyar M., 2017. Agrobacterium-Mediated Transformation of the Oryza sativa Thaumatin-Like Protein to Canola (R Line Hyola308) for Enhancing Resistance to Sclerotinia sclerotiorum. Iranian J. Biotechnol. 15: 201–207. 10.15171/ijb.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., and Huber W., 2010. Differential expression analysis for sequence count data. Genome Biol. 11: R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai B., Wu J., Sheng W. T., Zhou B., Zhou L. J. et al. , 2015. Comparative Analysis of Anther Transcriptome Profiles of Two Different Rice Male Sterile Lines Genotypes under Cold Stress. Int. J. Mol. Sci. 16: 11398–11416. 10.3390/ijms160511398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barah P., Jayavelu N. D., Rasmussen S., Nielsen H. B., Mundy J. et al. , 2013. Genome-scale cold stress response regulatory networks in ten Arabidopsis thaliana ecotypes. BMC Genomics 14: 722 10.1186/1471-2164-14-722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielach A., Hrtyan M., and Tognetti V. B., 2017. Plants under Stress: Involvement of Auxin and Cytokinin. Int. J. Mol. Sci. 18: E1427 10.3390/ijms18071427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniawska Z., Espinoza C., Schlereth A., Sulpice R., Hincha D. K. et al. , 2008. Disruption of the Arabidopsis Circadian Clock Is Responsible for Extensive Variation in the Cold-Responsive Transcriptome. Plant Physiol. 147: 263–279. 10.1104/pp.108.118059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard I. M., Lynch T. J., and Finkelstein R. R., 2002. Regulation and role of the Arabidopsis abscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol. 129: 1533–1543. 10.1104/pp.005793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello J. V., Arce A. L., and Chan R. L., 2012. The homologous HD-Zip I transcription factors HaHB1 and AtHB13 confer cold tolerance via the induction of pathogenesis-related and glucanase proteins. Plant J. 69: 141–153. 10.1111/j.1365-313X.2011.04778.x [DOI] [PubMed] [Google Scholar]
- Cao S., Ye M., and Jiang S., 2005. Involvement of GIGANTEA gene in the regulation of the cold stress response in Arabidopsis. Plant Cell Rep. 24: 683–690. 10.1007/s00299-005-0061-x [DOI] [PubMed] [Google Scholar]
- Catala R., Medina J., and Salinas J., 2011. Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 16475–16480. 10.1073/pnas.1107161108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaikam V., and Karlson D., 2008. Functional characterization of two cold shock domain proteins from Oryza sativa. Plant Cell Environ. 31: 995–1006. 10.1111/j.1365-3040.2008.01811.x [DOI] [PubMed] [Google Scholar]
- Chinnusamy V., Ohta M., Kanrar S., Lee B. H., Hong X. et al. , 2003. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17: 1043–1054. 10.1101/gad.1077503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V., Zhu J., and Zhu J. K., 2007. Cold stress regulation of gene expression in plants. Trends Plant Sci. 12: 444–451. 10.1016/j.tplants.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Choi M. J., Park Y. R., Park S. J., and Kang H., 2015. Stress-responsive expression patterns and functional characterization of cold shock domain proteins in cabbage (Brassica rapa) under abiotic stress conditions. Plant Physiol. Biochem. 96: 132–140. 10.1016/j.plaphy.2015.07.027 [DOI] [PubMed] [Google Scholar]
- Colebrook E. H., Thomas S. G., Phillips A. L., and Hedden P., 2014. The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 217: 67–75. 10.1242/jeb.089938 [DOI] [PubMed] [Google Scholar]
- de Zelicourt A., Colcombet J., and Hirt H., 2016. The Role of MAPK Modules and ABA during Abiotic Stress Signaling. Trends Plant Sci. 21: 677–685. 10.1016/j.tplants.2016.04.004 [DOI] [PubMed] [Google Scholar]
- Ding C. K., Wang C. Y., Gross K. C., and Smith D. L., 2002. Jasmonate and salicylate induce the expression of pathogenesis-related-protein genes and increase resistance to chilling injury in tomato fruit. Planta 214: 895–901. 10.1007/s00425-001-0698-9 [DOI] [PubMed] [Google Scholar]
- Ding Y., Jia Y., Shi Y., Zhang X., Song C. et al. , 2018. OST1-mediated BTF3L phosphorylation positively regulates CBFs during plant cold responses. EMBO J. 37: e98228 10.15252/embj.201798228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Li H., Zhang X., Xie Q., Gong Z. et al. , 2015. OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev. Cell 32: 278–289. 10.1016/j.devcel.2014.12.023 [DOI] [PubMed] [Google Scholar]
- Dong M. A., Farre E. M., and Thomashow M. F., 2011. Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 7241–7246. 10.1073/pnas.1103741108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du F., Xu J. N., Li D., and Wang X. Y., 2015. The identification of novel and differentially expressed apple-tree genes under low-temperature stress using high-throughput Illumina sequencing. Mol. Biol. Rep. 42: 569–580. 10.1007/s11033-014-3802-5 [DOI] [PubMed] [Google Scholar]
- Du H., Liu H., and Xiong L., 2013. Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front. Plant Sci. 4: 397 10.3389/fpls.2013.00397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K. D., Anderson P. E., Hall A., Salathia N. S., Locke J. C. et al. , 2006. FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell 18: 639–650. 10.1105/tpc.105.038315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin K. A., and Whitelam G. C., 2007. Light-quality regulation of freezing tolerance in Arabidopsis thaliana. Nat. Genet. 39: 1410–1413. 10.1038/ng.2007.3 [DOI] [PubMed] [Google Scholar]
- Guo M., Liu J.-H., Ma X., Luo D.-X., Gong Z.-H. et al. , 2016. The Plant Heat Stress Transcription Factors (HSFs): Structure, Regulation, and Function in Response to Abiotic Stresses. Front. Plant Sci. 7: 114 10.3389/fpls.2016.00114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X. Y., Liu D. F., and Chong K., 2018. Cold signaling in plants: Insights into mechanisms and regulation. J. Integr. Plant Biol. 60: 745–756. 10.1111/jipb.12706 [DOI] [PubMed] [Google Scholar]
- Gururani M. A., Mohanta T. K., and Bae H., 2015. Current Understanding of the Interplay between Phytohormones and Photosynthesis under Environmental Stress. Int. J. Mol. Sci. 16: 19055–19085. 10.3390/ijms160819055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy C., 1999. Molecular responses of plants to cold shock and cold acclimation. J. Mol. Microbiol. Biotechnol. 1: 231–242. [PubMed] [Google Scholar]
- Hincha D. K., Meins F. Jr, and Schmitt J. M., 1997. [beta]-1,3-Glucanase Is Cryoprotective in Vitro and Is Accumulated in Leaves during Cold Acclimation. Plant Physiol. 114: 1077–1083. 10.1104/pp.114.3.1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang X. L. T., Nhi D. N. H., Thu N. B. A., Thao N. P., and Tran L. S. P., 2017. Transcription Factors and Their Roles in Signal Transduction in Plants under Abiotic Stresses. Curr. Genomics 18: 483–497. 10.2174/1389202918666170227150057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Jiang L., Wang F., and Yu D., 2013. Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 25: 2907–2924. 10.1105/tpc.113.112631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Jiang Y., Han X., Wang H., Pan J. et al. , 2017. Jasmonate regulates leaf senescence and tolerance to cold stress: crosstalk with other phytohormones. J. Exp. Bot. 68: 1361–1369. 10.1093/jxb/erx004 [DOI] [PubMed] [Google Scholar]
- Huang X. B., Shi H. Y., Hu Z. R., Liu A., Amombo E. et al. , 2017. ABA Is Involved in Regulation of Cold Stress Response in Bermudagrass. Front. Plant Sci. 8: 1613 10.3389/fpls.2017.01613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglo-Ottosen K. R., Gilmour S. J., Zarka D. G., Schabenberger O., and Thomashow M. F., 1998. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280: 104–106. 10.1126/science.280.5360.104 [DOI] [PubMed] [Google Scholar]
- Jiang B., Shi Y., Zhang X., Xin X., Qi L. et al. , 2017. PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 114: E6695–E6702. 10.1073/pnas.1706226114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M. et al. , 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36: D480–D484. 10.1093/nar/gkm882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K., 2015. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 20: 219–229. 10.1016/j.tplants.2015.02.001 [DOI] [PubMed] [Google Scholar]
- Khan M. I., Fatma M., Per T. S., Anjum N. A., and Khan N. A., 2015. Salicylic acid-induced abiotic stress tolerance and underlying mechanisms in plants. Front. Plant Sci. 6: 462 10.3389/fpls.2015.00462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. S., Lee M., Lee J. H., Lee H. J., and Park C. M., 2015. The unified ICE-CBF pathway provides a transcriptional feedback control of freezing tolerance during cold acclimation in Arabidopsis. Plant Mol. Biol. 89: 187–201. 10.1007/s11103-015-0365-3 [DOI] [PubMed] [Google Scholar]
- Kuwabara C., and Imai R., 2009. Molecular Basis of Disease Resistance Acquired through Cold Acclimation in Overwintering Plants. J. Plant Biol. 52: 19–26. 10.1007/s12374-008-9006-6 [DOI] [Google Scholar]
- Langmead B., Trapnell C., Pop M., and Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. H., Henderson D. A., and Zhu J. K., 2005. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17: 3155–3175. 10.1105/tpc.105.035568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. M., and Thomashow M. F., 2012. Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 109: 15054–15059. 10.1073/pnas.1211295109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Liu H., Xia W., Mu J., Feng Y. et al. , 2017. De Novo Transcriptome Sequencing and the Hypothetical Cold Response Mode of Saussurea involucrata in Extreme Cold Environments. Int. J. Mol. Sci. 18: E1155 10.3390/ijms18061155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Ma D., Lu S. X., Hu X., Huang R. et al. , 2016. Blue Light- and Low Temperature-Regulated COR27 and COR28 Play Roles in the Arabidopsis Circadian Clock. Plant Cell 28: 2755–2769. 10.1105/tpc.16.00354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S. et al. , 1998. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406. 10.1105/tpc.10.8.1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Cheng C., Lin Y., XuHan X., and Lai Z., 2018. Genome-wide identification and characterization of mRNAs and lncRNAs involved in cold stress in the wild banana (Musa itinerans). PLoS One 13: e0200002 10.1371/journal.pone.0200002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N., Kusano M., Fukushima A., Kita M., Ito S. et al. , 2009. Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 50: 447–462. 10.1093/pcp/pcp004 [DOI] [PubMed] [Google Scholar]
- Nakaminami K., Hill K., Perry S. E., Sentoku N., Long J. A. et al. , 2009. Arabidopsis cold shock domain proteins: relationships to floral and silique development. J. Exp. Bot. 60: 1047–1062. 10.1093/jxb/ern351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K., Fujita Y., Kanamori N., Katagiri T., Umezawa T. et al. , 2009. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 50: 1345–1363. 10.1093/pcp/pcp083 [DOI] [PubMed] [Google Scholar]
- Nievola C. C., Carvalho C. P., Carvalho V., and Rodrigues E., 2017. Rapid responses of plants to temperature changes. Temperature (Austin) 4: 371–405. 10.1080/23328940.2017.1377812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Lee C. M., Doherty C. J., Gilmour S. J., Kim Y. et al. , 2015. Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J. 82: 193–207. 10.1111/tpj.12796 [DOI] [PubMed] [Google Scholar]
- Park S. J., Kwak K. J., Oh T. R., Kim Y. O., and Kang H., 2009. Cold shock domain proteins affect seed germination and growth of Arabidopsis thaliana under abiotic stress conditions. Plant Cell Physiol. 50: 869–878. 10.1093/pcp/pcp037 [DOI] [PubMed] [Google Scholar]
- Qu Y., Zhou A., Zhang X., Tang H., Liang M. et al. , 2015. De Novo Transcriptome Sequencing of Low Temperature-Treated Phlox subulata and Analysis of the Genes Involved in Cold Stress. Int. J. Mol. Sci. 16: 9732–9748. 10.3390/ijms16059732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radkova M., Vitamvas P., Sasaki K., and Imai R., 2014. Development- and cold-regulated accumulation of cold shock domain proteins in wheat. Plant Physiol. Biochem. 77: 44–48. 10.1016/j.plaphy.2014.01.004 [DOI] [PubMed] [Google Scholar]
- Riechmann J. L., and Ratcliffe O. J., 2000. A genomic perspective on plant transcription factors. Curr. Opin. Plant Biol. 3: 423–434. 10.1016/S1369-5266(00)00107-2 [DOI] [PubMed] [Google Scholar]
- Ruelland E., Vaultier M. N., Zachowski A., and Hurry V., 2009. Cold Signalling and Cold Acclimation in Plants. Adv. Bot. Res. 49: 35–150. 10.1016/S0065-2296(08)00602-2 [DOI] [Google Scholar]
- Sakuma Y., Liu Q., Dubouzet J. G., Abe H., Shinozaki K. et al. , 2002. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290: 998–1009. 10.1006/bbrc.2001.6299 [DOI] [PubMed] [Google Scholar]
- Salome P. A., McClung C. R., 2005. PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17: 791–803. 10.1105/tpc.104.029504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K., and Imai R., 2011. Pleiotropic roles of cold shock domain proteins in plants. Front. Plant Sci. 2: 116 10.3389/fpls.2011.00116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K., Kim M. H., and Imai R., 2013. Arabidopsis COLD SHOCK DOMAIN PROTEIN 2 is a negative regulator of cold acclimation. New Phytol. 198: 95–102. 10.1111/nph.12118 [DOI] [PubMed] [Google Scholar]
- Sharma I., Kaur N., and Pati P. K., 2017. Brassinosteroids: A Promising Option in Deciphering Remedial Strategies for Abiotic Stress Tolerance in Rice. Front. Plant Sci. 8: 2151 10.3389/fpls.2017.02151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Ding Y., and Yang S., 2018. Molecular Regulation of CBF Signaling in Cold Acclimation. Trends Plant Sci. 23: 623–637. 10.1016/j.tplants.2018.04.002 [DOI] [PubMed] [Google Scholar]
- Shinkawa R., Morishita A., Amikura K., Machida R., Murakawa H. et al. , 2013. Abscisic acid induced freezing tolerance in chilling-sensitive suspension cultures and seedlings of rice. BMC Res. Notes 6: 351 10.1186/1756-0500-6-351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skubacz A., Daszkowska-Golec A., and Szarejko L., 2016. The Role and Regulation of ABI5 (ABA-Insensitive 5) in Plant Development, Abiotic Stress Responses and Phytohormone Crosstalk. Front. Plant Sci. 7: 1884 10.3389/fpls.2016.01884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobkowiak A., Jonczyk M., Jarochowska E., Biecek P., Trzcinska-Danielewicz J. et al. , 2014. Genome-wide transcriptomic analysis of response to low temperature reveals candidate genes determining divergent cold-sensitivity of maize inbred lines. Plant Mol. Biol. 85: 317–331. 10.1007/s11103-014-0187-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S., and Amasino R. M., 2004. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427: 159–164. 10.1038/nature02195 [DOI] [PubMed] [Google Scholar]
- Theocharis A., Clement C., and Barka E. A., 2012. Physiological and molecular changes in plants grown at low temperatures. Planta 235: 1091–1105. 10.1007/s00425-012-1641-y [DOI] [PubMed] [Google Scholar]
- Thomashow M. F., 1999. PLANT COLD ACCLIMATION: Freezing Tolerance Genes and Regulatory Mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 571–599. 10.1146/annurev.arplant.50.1.571 [DOI] [PubMed] [Google Scholar]
- Tian D. Q., Pan X. Y., Yu Y. M., Wang W. Y., Zhang F. et al. , 2013. De novo characterization of the Anthurium transcriptome and analysis of its digital gene expression under cold stress. BMC Genomics 14: 827 10.1186/1471-2164-14-827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., and Salzberg S. L., 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma V., Ravindran P., and Kumar P. P., 2016. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16: 86 10.1186/s12870-016-0771-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishwakarma K., Upadhyay N., Kumar N., Yadav G., Singh J. et al. , 2017. Abscisic Acid Signaling and Abiotic Stress Tolerance in Plants: A Review on Current Knowledge and Future Prospects. Front. Plant Sci. 8: 161 10.3389/fpls.2017.00161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D. Z., Jin Y. N., Ding X. H., Wang W. J., Zhai S. S. et al. , 2017. Gene regulation and signal transduction in the ICE-CBF-COR signaling pathway during cold stress in plants. Biochemistry (Mosc.) 82: 1103–1117. 10.1134/S0006297917100030 [DOI] [PubMed] [Google Scholar]
- Wang F., Guo Z., Li H., Wang M., Onac E. et al. , 2016a Phytochrome A and B Function Antagonistically to Regulate Cold Tolerance via Abscisic Acid-Dependent Jasmonate Signaling. Plant Physiol. 170: 459–471. 10.1104/pp.15.01171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Zhang X. N., and Liu J. H., 2015. Deep sequencing-based characterization of transcriptome of trifoliate orange (Poncirus trifoliata (L.) Raf.) in response to cold stress. BMC Genomics 16: 555 10.1186/s12864-015-1629-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Liu N., Yang X., Tu L., and Zhang X., 2016b Small RNA-mediated responses to low- and high-temperature stresses in cotton. Sci. Rep. 6: 35558 10.1038/srep35558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C., and Song S., 2017. Jasmonates: biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 68: 1303–1321. 10.1093/jxb/erw443 [DOI] [PubMed] [Google Scholar]
- Wasternack C., and Strnad M., 2018. Jasmonates: News on Occurrence, Biosynthesis, Metabolism and Action of an Ancient Group of Signaling Compounds. Int. J. Mol. Sci. 19: E2539 10.3390/ijms19092539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Zhang M., Liu G., Yang X., and Hou X., 2016. Comparative transcriptome profiling of chilling stress responsiveness in grafted watermelon seedlings. Plant Physiol. Biochem. 109: 561–570. 10.1016/j.plaphy.2016.11.002 [DOI] [PubMed] [Google Scholar]
- Yang T., and Huang X. S., 2018. Deep sequencing-based characterization of transcriptome of Pyrus ussuriensis in response to cold stress. Gene 661: 109–118. 10.1016/j.gene.2018.03.067 [DOI] [PubMed] [Google Scholar]
- Yoshida T., Mogami J., and Yamaguchi-Shinozaki K., 2014. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 21: 133–139. 10.1016/j.pbi.2014.07.009 [DOI] [PubMed] [Google Scholar]
- Zhang S., Song G., Gao J., Li Y., Guo D. et al. , 2014. Transcriptome characterization and differential expression analysis of cold-responsive genes in young spikes of common wheat. J. Biotechnol. 189: 48–57. 10.1016/j.jbiotec.2014.08.032 [DOI] [PubMed] [Google Scholar]
- Zhang T., Huang L. Y., Wang Y. X., Wang W. S., Zhao X. Q. et al. , 2017. Differential transcriptome profiling of chilling stress response between shoots and rhizomes of Oryza longistaminata using RNA sequencing. PLoS One 12: e0188625 10.1371/journal.pone.0188625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Chen J., Xie Z., Gao J., Ren G. et al. , 2015. Jasmonic acid promotes degreening via MYC2/3/4- and ANAC019/055/072-mediated regulation of major chlorophyll catabolic genes. Plant J. 84: 597–610. 10.1111/tpj.13030 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequencing rawdata are accessible through GEO (GSE129220): https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE129220. Supplemental material available at Figshare: https://10.6084/m9.figshare.7379858.