Abstract
Amphibians are highly vulnerable and diverse vertebrates for which we still have modest genomic resources. Amphibian larvae are key components of continental wetlands, where they have strong influences on energy fluxes, nutrient cycling, and community structure. Amphibian larvae are highly sensitive to environmental conditions and can often alter their physiology, behavior and even morphology in response to the local conditions experienced, although we still know relatively little about the transcriptomic changes that enable such plasticity. Here we contribute the larval transcriptomes of two spadefoot toad species with divergent developmental rates and degree of developmental plasticity in response to pond drying.
Keywords: Amphibian development, developmental rate, amphibian transcriptomes, developmental plasticity, cross-species comparisons
Most amphibian species exhibit a complex life-cycle including two or more life stages separated by an ontogenetic switch point such as hatching or metamorphosis. Adaptations to divergent environments can require the modification of the timing of such switch points and the relative investment in growth and differentiation between subsequent stages (Moran 1994). Such alterations of developmental trajectories, however, often have substantial repercussions at several organismal levels, from physiology to morphology and even genomic structure. Adaptive divergence in developmental rate tracking aquatic habitats of different duration in spadefoot toads is a well-known example of this. Spadefoot toads from Europe and northern Africa typically have tadpoles that grow to be quite large over a long larval period, but can otherwise accelerate development and precipitate metamorphosis if at risk of pond drying, whereas north American spadefoot toads tend to have smaller tadpoles that develop faster and are less capable of further developmental acceleration (Buchholz and Hayes 2002). At opposite ends of this spectrum, we find Pelobates cultripes, distributed throughout most of the Iberian Peninsula and southern France, and Scaphiopus couchii, distributed across southwestern USA to northern Mexico. Pelobates cultripes larvae grow quite large (> 16 g) and can take up to 6 months to reach metamorphosis, whereas S. couchii tadpoles are much smaller (1.5-2 g) and can develop to metamorphosis in as little as eight days. Such developmental acceleration is rather energetically demanding and requires a substantial increase in metabolic activity (Kulkarni et al. 2017), hence causing oxidative stress (Gomez-Mestre et al. 2013). Precipitating metamorphosis alters growth and developmental trajectories, and as these are non-isometrical for different parts of the body, the emerging metamorphs are not only smaller but also have relatively shorter limbs (Gomez-Mestre et al. 2010; Kulkarni et al. 2011; Johansson and Richter-Boix 2013). Developmental acceleration is achieved through neuroendocrine regulation mainly resulting in increased corticosterone and thyroid hormone levels (Denver 2009; Kulkarni et al. 2017), as well as through differential expression of hormone receptors (Gomez-Mestre et al. 2013). Interestingly, the canalized fast development of S. couchii to a large extent mirrors the environmentally-induced accelerated state of the more plastic P. cultripes (Kulkarni et al. 2017).
At the genomic level, evolutionary divergences in developmental rate seem to leave a big imprint on whole genomes with some studies showing that fast developmental rates are often associated with smaller genome sizes (Wyngaard et al. 2005; Alfsnes et al. 2017). The rule also holds true for amphibians, whether at a large macroevolutionary scale (Liedtke et al. 2018) or focused on specific species groups (Jockusch 1997). Spadefoot toads present broad differences in developmental rate across species, which are consequently also reflected in large differences in genome size (Zeng et al. 2014): slow developing P. cultripes has a large genome (∼3.9 Gbp), whereas fast developing S. couchii has only about one third its size (∼1.5 Gbp). Here we present a first description of the transcriptomes of these species at the onset of metamorphosis to explore the potential consequences of such dramatic divergence in their genomes and to uncover the transcriptomic basis of their differences in developmental rate.
The NCBI Transcriptome Shotgun assemblies database currently lists transcriptome assemblies for 26 species of amphibians and of those, only four are larval phase transcriptomes: Rhinella marina (Richardson et al. 2018), Microhyla fissipes (Zhao et al. 2016), Rana (Lithobates) catesbeiana and Xenopus laevis (Birol et al. 2015). The addition of transcriptomes for the larval phases of two more species, especially as they represent a distinct evolutionary lineage, is therefore a significant contribution to the current knowledgebase.
Methods
Sample collection, total RNA extraction and sequencing
The experimental procedures described below were approved by the CSIC IACUC committee (permit #17_01 CEEA-EBD). Three egg clutches of P. cultripes were collected from a natural pond in Doñana National Park, southwestern Spain, brought to a walk-in chamber in the laboratories of Doñana Biological Station (EBD-CSIC) and placed in a plastic tray with carbon-filtered dechlorinated tap water with aerators to ensure adequate oxygenation. Three clutches of S. couchii were spawned in the laboratory at EBD-CSIC by adult pairs obtained from a captive colony at Daniel Buchholz’s laboratory at the University of Cincinnati (USA), and originally collected in SE Arizona. Adults were hormonally stimulated to breed by intraperitoneally injecting 20–100 μL of 1 μg/100 μL GnRH agonist (des-Gly, [D-His(Bzl)]-luteinizing hormone releasing hormone ethylamide, Sigma). Upon hatching, we transferred tadpoles from each clutch of each species to 3 L plastic containers with dechlorinated tap water where they were individually kept under standard conditions of 24°, 12:12 L:D photoperiod, and ad libitum food supply consisting of finely powdered rabbit chow. Tadpoles were stage-matched so that when they advanced in development to Gosner stage 35, as determined by the individualization of all five toes (Gosner 1960), we euthanized twelve individuals per species via MS-222 overdose, eviscerated them to avoid interferences from fecal material, and snap-froze them in liquid nitrogen. We extracted whole-body total RNA from each tadpole using Trizol reagent following the manufacturer’s protocol (Invitrogen). Total RNA was assayed for quantity and quality using Qubit RNA HS Assay (Life Technologies) and RNA 6000 Nano Assay on a Bioanalyzer 2100 (Agilent).
The RNASeq libraries were prepared from total RNA using the TruSeqStranded mRNA LT Sample Prep Kit (Illumina Inc., Rev.E, October 2013). Briefly, 500ng of total RNA was used as the input material and was enriched for the mRNA fraction using oligo-dT magnetic beads. The mRNA was fragmented in the presence of divalent metal cations. The second strand cDNA synthesis was performed in the presence of dUTP instead of dTTP, this allowed to achieve the strand specificity. The blunt-ended double stranded cDNA was 3′adenylated and Illumina indexed adapters were ligated. The ligation product was enriched with 15 PCR cycles and the final library was validated on an Agilent 2100 Bioanalyzer with the DNA 7500 assay.
Each library was sequenced using TruSeq SBS Kit v3-HS, in paired end mode with the read length 2x76bp. We generated on average 38 million paired-end reads for each sample in a fraction of a sequencing lane on HiSeq2000 (Illumina) following the manufacturer’s protocol. Images analysis, base calling and quality scoring of the run were processed using the manufacturer’s software Real Time Analysis (RTA 1.13.48) and followed by generation of FASTQ sequence files by CASAVA 1.8.
Assembling de novo transcriptomes of P. cultripes and S. couchii
Quality of raw reads was inspected using FASTQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and MULTIQC (Ewels et al. 2016). Assembly was performed using Trinity v2.4.0 (Haas et al. 2013) for the two species separately. Reads from all samples per species were combined, trimmed (using default Trimmomatic settings SLIDINGWINDOW:4:5 LEADING:5 TRAILING:5 MINLEN:25)(Bolger et al. 2014) and normalized using in silico normalization with default Trinity settings (flags used:–trimmomatic–normalize_max_read_cov 50).
Assessment of transcriptome quality and completeness
Transcriptome quality in terms of read representation was evaluated by mapping the normalized reads (pairs only) back onto the transcriptome using Bowtie2 v2.3.2 (Langmead and Salzberg 2012). Completeness in terms of gene content was assessed using BUSCO v3.0.2 (Simão et al. 2015) with the tetrapoda-odb9 database as a reference as well as by running blastx (E-value cut off E ≤ 1e-20) against both the SwissProt database (downloaded on 01.11.2017) and the Xenopus tropicalis proteome (Ensemble JGI 4.2; downloaded on 03.11.2017) with a stringent Evalue criteria of ≤ 1e-20. The count of full-length transcripts with blastx hits was based on grouped high scoring segment pairs per transcript to avoid multiple fragments per transcript aligning to a single protein sequence.
Functional annotation
We used Trinotate v3.0 (https://trinotate.github.io/) to annotate the transcriptome. This involves finding similarities to known proteins by querying transcripts against the Swissprot database (accessed in June 2018) (The UniProt Consortium 2017) (blastx with a cut-off of E-value ≤ 1e-5). Moreover, likely coding regions were detected with TransDecoder (https://github.com/TransDecoder) and resulting protein products (coding sequence; CDS) were matched against both the complete Swissprot database and a subset including only vertebrate genes, using blastp (E-value ≤ 1e-5), and a conserved protein domain search was conducted using HMMER (http://hmmer.org/) on the Pfam database (Finn et al. 2016). SignalP v4.1 (Petersen et al. 2011) and TmHMM v2.0 (http://www.cbs.dtu.dk/services/TMHMM/) were used to predict signal peptides and transmembrane regions respectively. Finally, gene ontology identifiers were assigned to transcripts based on available annotations from best-matching Swissprot entries. Trinotate also provides KEGG (Kyoto Encycopedia of Genes and Genomes; http://www.genome.jp/kegg/) and EggNOG (Huerta-Cepas et al. 2016) annotations. Exploring the Trinotate output was facilitated using the TrinotateR R package (https://github.com/cstubben/trinotateR).
Expression-based filtering and cross-species comparisons
Transcripts with low expression levels have questionable biological relevance. Because of this, abundances were estimated using Kallisto v0.4.3.1 (Bray et al. 2016), and the transcriptome was filtered to retain only transcripts with more than 1 Transcript Per Million (TPM). This threshold roughly coincides with a notable influx of lowly expressed transcripts when plotting the number of transcripts as a function of minimum TPM thresholds. In addition, any transcripts flagged by NCBI as containing potential vector, adaptor or primer contaminations were also filtered out.
The PANTHER classification scheme (Mi et al. 2013) for Xenopus tropicalis was used to organize gene function and ontology. The filtered transcriptomes were annotated using the Xenopus Ensembl protein identifiers recovered from the blastx search (see above) and using the PANTHER web server, we performed both functional classifications and a statistical overrepresentation test (Fisher’s exact test with a false discovery rate correction) to investigate which genes are significantly (P < 0.05) over or under represented in the filtered transcriptomes compared to the X. tropicalis reference. Further functional classification and gene pathway enrichment was performed using gProfiler (g:GOST; Raudvere et al. 2019) using the accompanying R package gprofiler2 v0.1.3. The expression-filtered Xenopus protein identifiers were queried against the Gene Ontology – Biological Process, KEGG and Reactome databases (Fabregat et al. 2017). An overrepresentation test was performed to calculate the probability that the intersection of query and a functional category has arisen by chance, using X. tropicalis as the reference organism, limiting the gene domain to only annotated genes and excluding GO electronic annotations. The gene list was ranked in order of decreasing expression (using the mean TPM per gene across all biological replicates) and treated as an ordered query to perform an incremental enrichment analysis, giving more importance to higher expressed terms. P-values were adjusted for multiple testing using the g:SCS algorithm. Functional groupings and gene networks of significantly overrepresented/enriched terms and pathways were further explored using the EnrichmentMap add-on in Cytoscape v3.7.1 (Shannon et al. 2003; Merico et al. 2010).
Finally, OrthoFinder v2.2.3 (Emms and Kelly 2015) was used to find orthologous genes across the two species. OrthoFinder was run with default settings, taking the TransDecoder predicted CDS of both P. cultripes and S. couchii as the input, as well as the proteome of X. tropicalis to provide context (as an ‘outgroup’).
Data availability
The data sets supporting the results of this article are available as supporting information. Specifically, we provide quality assessment results of both BUSCO and BowTie2, the unfiltered transcriptome assemblies plus their transdecoded CDS files and annotations in the form of Trinotate summary tables and Panther annotation lists. In addition, all raw reads as well as the expression filtered transcriptome assemblies are deposited on the NCBI’s Sequence Read Archive [SRA; SRP161446] and Transcriptome Shotgun Assembly database [TSA; P. cultripes: GHBH01000000; S. couchii: GHBO01000000], under BioProject [PRJNA490256]. Supplemental material available at FigShare: https://doi.org/10.25387/g3.8201825.
Results and Discussion
Transcriptome comparison and quality assessment
The twelve P. cultripes samples each consisted of 30.5-43.3 million, 101bp paired-end reads (888.3 million reads in total) pooling to 84.2 million post-normalization pair-end reads used for the assembly (10.5% of total). Trinity generated 753,223 transcript contigs with median length 362, of which 428,406 clustered into ‘genes’ (transcript clusters with shared sequence content; Table 1). Bowtie2 mapped 83.96% of the reads back onto the transcriptome (Supporting Data 1). In comparison, the S. couchii samples consisted of 32.1-53.9 million, 101bp reads (958.8 million reads in total) with 84.4 million post-normalization pair-end reads used for the final assembly (9.19%). 657,280 transcripts were generated by Trinity with a median length of 432bp clustering into 381,135 ‘genes’ (Table 1). Bowtie2 mapped 90.71% of the reads back onto the transcriptome. The Trinity-assembled, unfiltered transcriptomes for both species are available as Supporting Data 2.
Table 1. Transcriptome assembly statistics for both tadpole species. Summaries for Trinity outputs are given both at the transcript and at the ‘gene’ level.
| P. cultripes | S. couchii | |
|---|---|---|
| Total number of raw reads | 888,265,444 | 958,782,922 |
| Number of in silico normalized reads | 84,209,684 | 84,420,786 |
| Number of read pairs aligned to assembly | 419,274,288 (94.4%) | 453,683,501 (94.6%) |
| Number of proper pair reads aligned to assembly | 359,039,281 (85.6%) | 425,833,848 (93.9%) |
| N50 of transcripts | longest isoform per ‘gene’ | 1,496bp | 731bp | 2,057bp | 872bp |
| Number of Trinity transcripts | ‘genes’ | 753,223 | 428,406 | 657,280 | 381,135 |
| Size of transcript | longest isoform per ‘gene’: | ||
| Total | 581,464,720bp | 237,111,496bp | 644,907,581bp | 232,600,864bp |
| Median | 362bp | 313bp | 432bp | 331bp |
| Average | 771.97bp | 553.47bp | 981.18bp | 610.28bp |
The BUSCO results support near-complete gene sequence information for 89.7% of genes in the P. cultripes transcriptome with only 7.4% of the genes being fragmented and 2.9% missing. The quality of the S. couchii assembly was similar with 86.6% complete sequence information, 10.5% fragmented genes and 2.9% missing (Supporting Data 3).
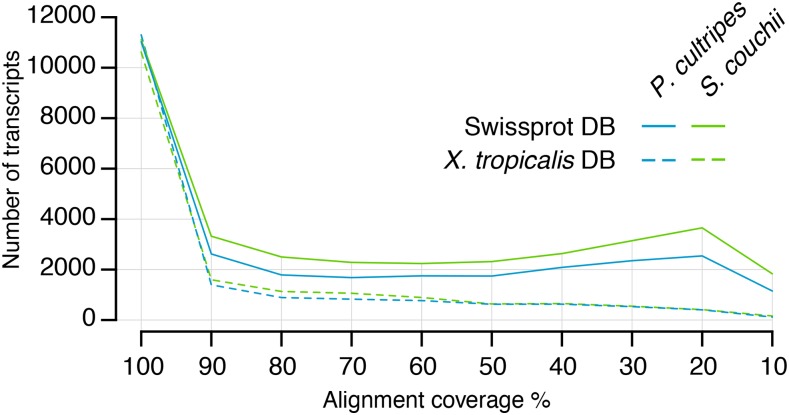
Querying the Trinity assembly against both the Swissprot database and the X. tropicalis proteome (using blastx) revealed large numbers of fully reconstructed coding transcripts, with 13,645 Swissprot proteins and 12,715 X. tropicalis proteins represented by nearly full-length transcripts (>80% alignment coverage) in the P. cultripes assembly, and 14,429 Swissprot proteins and 12,216 X. tropicalis proteins in the S. couchii assembly (Figure 1; Supporting Data 4).
Figure 1.
Number of transcripts (using grouped highest scoring segment pairs) per alignment coverage bins when querying against the SwissProt and Xenpus tropicalis proteome sequence databases.
Functional annotation
Gene annotation via the Trinotate pipeline is useful for providing biological context to the assembled transcriptomes (Trinotate tables available as Supporting Data 5). Querying (using blastx) the SwissProt database with the Trinity assembly allowed for the annotation of 162,031 P. cultripes and 204,646 S. couchii transcripts. Gene Ontology (GO) derived from these hits resulted in 18,585 unique (out of a total of 1,626,015) GO annotations for P. cultripes and 19,917 unique (out of a total 2,155,849) GO annotations for S. couchii.
TransDecoder recovered fewer candidate-coding regions for P. cultripes (154,906) than for S. couchii (175,331; Table 2). This corresponds to 36.2% and 46.1% of the Trinity-identified ‘genes’ for P. cultripes and S. couchii, respectively. Homology searches using blastp against the entire Swissprot database were able to annotate 108,881 and 132,578 of the trasdecoded CDS, and 107,199 and 130,847 when searching the vertebrates-only database (Table 2). Of the sequences with vertebrate gene hits, 24,327 and 27,309 unique vertebrate swissprot proteins were identified for P. cultripes and S. couchii (genes with unique UniProtKB-IDs). Of these, the two species share 56.5% (18,651 proteins), with 17.2% being unique to P. cultripes (5,676 proteins) and 26.2% unique to S. couchii (8,658 proteins). Similarly, the number of hits of candidate coding regions against other databases including pfam, signalP, tmHMM, KEGG and EggNOG was greater for S. couchii than P. cultripes (Table 2).
Table 2. Number of unique | total TransDecoder-predicted candidate genes with annotations via different search tools and databases (summary of Trinotate results).
| P. cultripes | S. couchii | |
|---|---|---|
| TransDecoder predicted coding regions (ORFs) | 154,906 | 175,331 |
| Protein hits (blastp - SwissProt) | 79,504 | 108,881 | 91,337 | 132,578 |
| Protein hits (blastp – SwissProt vertebrates only) | 77,924 | 107,199 | 89,704 | 130,847 |
| pfam hits (HMMER search) | 65,597 | 91,929 | 76,741 | 112,740 |
| signalP predicted peptides | 3,943 | 10,981 | 4,114 | 13,097 |
| tmHMM predicted transmembrane proteins | 17,990 | 25,833 | 19,881 | 29,991 |
| GO Pfam | 2,471 | 57,308 | 2,583 | 71,966 |
| KEGG | 31,313 | 127,707 | 40,765 | 174,712 |
| EggNOG | 8,125| 117,983 | 8,681| 156,494 |
Expression-based filtering and cross-species comparisons
Expression-based filtering results in only 20.8% of the P. cultripes transcripts and 26.8% S. couchii transcripts to be retained. Post-NCBI screening, the transcriptomes consisted of 153,520 transcripts for P. cultripes and 175,261 for S. couchii corresponding to 42,949 and 60,662 transdecoded peptide sequences respectively. The number of predicted coding sequences post expression filtering is in the range of published amphibian larval transcriptomes [R. catesbeiana: 51,720 CDS (Birol et al. 2015), M. fissipes 51,506 CDS (Zhao et al. 2016), R. marina 62, 365 CDS (Richardson et al. 2018)].
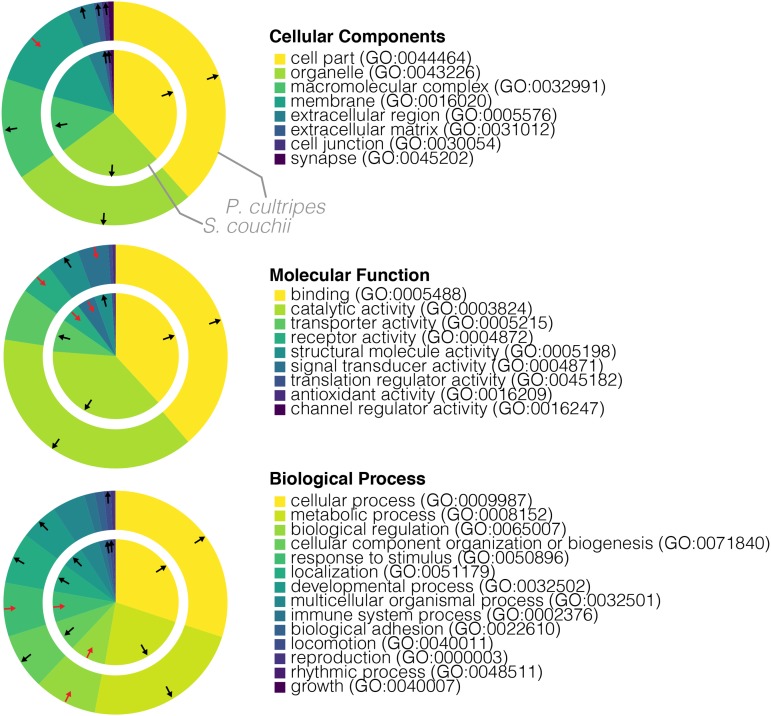
The PANTHER GO-slim classification system designed for X. tropicalis provides a curated, functional classification scheme of GO terms and allows for relative over or underrepresentation of terms to be assessed in relation to the reference (in this case X. tropicalis) database. For P. cultripes, 10,666 out of 13,421unique annotations could be mapped to PANTHER genes, and 11,598 out of 14,811 annotations for S. couchii. The relative abundances of GO terms, as well as their statistical representation in relation to X. tropicalis, are highly comparable across the two species (Figure 2). Cell parts and organelles (cellular components; CC), binding and catalytic activity (molecular function; MF) and cellular and metabolic processes (biological processes; BP) make up more than half of the terms for each ontology, in each case being significantly over represented in both species compared to in X. tropicalis. Underrepresented terms are less common, restricted only to genes related to receptor and signal transducer activity (MF) and biological regulation and response to stimulus (BP) in both species, as well as membrane genes (CC) in P. cultripes only. Bar charts showing the over and underrepresentation of each PANTHER term per species per ontology are provided as supporting data (Supporting Data 6).
Figure 2.
PANTHER functional classification of transcriptomes. Wedge size reflect number of unique genes per category and out/in, black/red arrow annotations specify significant over/under representation of the GO-slim term compared to the X. tropicalis reference database.
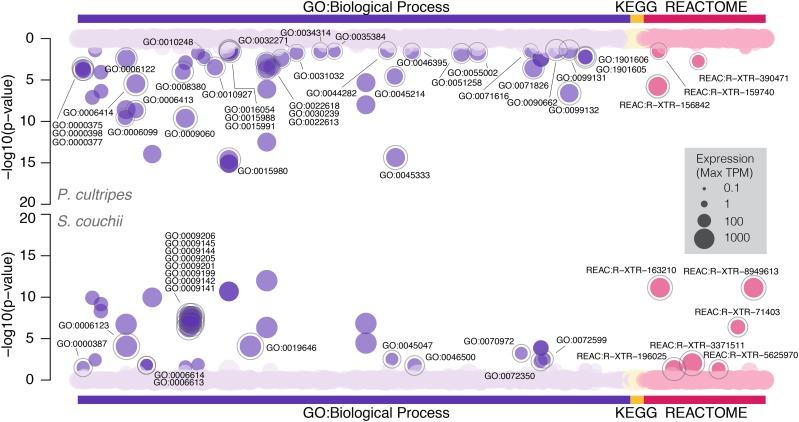
gProfiler could map 12,391 genes out of a total query of 13,421 to 8,229 functional terms for P. cultripes, of which 54 are significantly enriched (adjusted p-value < 0.05). In comparison, for S. couchii, gProfiler could map 13,315 out of 14,811 queried genes to 8,455 functional terms, of which 40 are significantly enriched (Supporting Data 7). For both species, the majority of enriched terms were GO terms, with the addition of nine Reactome pathways (three for P. cultripes and six for S. couchii). No KEGG pathways were enriched for either species (Figure 3; Table 3). Sixty-two of the significantly enriched terms and pathways were unique to each species, with only 16 shared terms, all within the GO term domain space (Figure 3; Table 3). When exploring this subset of significant terms and pathways further using Cytoscape, the most extensively represented clusters include ribonucleoside triphosphate metabolic processes (both species, though primarily S. couchii), mitochondrial electron transport (both species) and cytoplasmic initiation complex (both species), with the most represented species-specific terms being actomyosin structure organization (P. cultripes) and protein targeting to endoplasmic reticulum (S. couchii; Supporting Data 8). Genes related to thyroid hormone regulated development or oxidative stress were not recovered as significantly overrepresented in either species. Interestingly, however, the enriched annotations for S. couchii showed a notable representation of nucleoside triphosphate metabolism related processes (Supporting Data 8), which may be related to the species’ need for higher rates of DNA replication linked to its faster development and higher metabolic rates (Kulkarni et al. 2017).
Figure 3.
gProfiler gene pathway enrichment showing symmetrical Manhattan plots of annotated terms and gene pathways for P. cultripes (above) and S. couchii (below). Color code reflects annotation domains (Gene Ontology - Biological Processes, KEGG and Reactome gene pathways) with darker shading highlighting overrepresented terms (adj. P < 0.05). Term size reflects mean expression (TPM) and overrepresented terms that are unique to each species are outlined in gray and labeled.
Table 3. gProfiler enriched Gene Ontology (Biological Processes only) terms and Reactome pathways for P. cultripes and S. couchii. Terms and pathways match highlighted annotations in Figure 3.
| Domain | Term/Pathway | Term/Pathway description | Species | adj. p value |
|---|---|---|---|---|
| GO: BP | GO:0000375 | RNA splicing, via transesterification reactions | P. cultripes | <0.001 |
| GO: BP | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile | P. cultripes | <0.001 |
| GO: BP | GO:0000398 | mRNA splicing, via spliceosome | P. cultripes | <0.001 |
| GO: BP | GO:0001732 | Formation of cytoplasmic translation initiation complex | P. cultripes; S. couchii | <0.001;<0.001 |
| GO: BP | GO:0002181 | Cytoplasmic translation | P. cultripes; S. couchii | <0.001;<0.001 |
| GO: BP | GO:0002183 | cytoplasmictranslationalinitiation | P. cultripes; S. couchii | <0.001;<0.001 |
| GO: BP | GO:0006099 | tricarboxylic acid cycle | P. cultripes | <0.001 |
| GO: BP | GO:0006119 | oxidative phosphorylation | P. cultripes; S. couchii | <0.001;<0.001 |
| GO: BP | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen | S. couchii | <0.001 |
| GO: BP | GO:0006413 | translational initiation | P. cultripes | <0.001 |
| GO: BP | GO:0006414 | translational elongation | P. cultripes | <0.001 |
| GO: BP | GO:0006754 | ATP biosynthetic process | P. cultripes; S. couchii | <0.001;<0.001 |
| GO: BP | GO:0008380 | RNA splicing | P. cultripes | <0.001 |
| GO: BP | GO:0009060 | aerobic respiration | P. cultripes | <0.001 |
| GO: BP | GO:0009141 | nucleoside triphosphate metabolic process | S. couchii | <0.001 |
| GO: BP | GO:0009142 | nucleoside triphosphate biosynthetic process | S. couchii | <0.001 |
| GO: BP | GO:0009144 | purine nucleoside triphosphate metabolic process | S. couchii | <0.001 |
| GO: BP | GO:0009145 | purine nucleoside triphosphate biosynthetic process | S. couchii | <0.001 |
| GO: BP | GO:0009199 | ribonucleoside triphosphate metabolic process | S. couchii | <0.001 |
| GO: BP | GO:0009201 | ribonucleoside triphosphate biosynthetic process | S. couchii | <0.001 |
| GO: BP | GO:0009205 | purine ribonucleoside triphosphate metabolic process | S. couchii | <0.001 |
| GO: BP | GO:0009206 | purine ribonucleoside triphosphate biosynthetic process | S. couchii | <0.001 |
| GO: BP | GO:0010927 | cellular component assembly involved in morphogenesis | P. cultripes | <0.001 |
| GO: BP | GO:0015980 | energy derivation by oxidation of organic compounds | P. cultripes | <0.001 |
| GO: BP | GO:0015985 | energy coupled proton transport, down electrochemical gradient | P. cultripes; S. couchii | <0.001;<0.001 |
| GO: BP | GO:0015986 | ATP synthesis coupled proton transport | P. cultripes; S. couchii | <0.001;<0.001 |
| GO: BP | GO:0019646 | aerobic electron transport chain | S. couchii | <0.001 |
| GO: BP | GO:0022618 | ribonucleoprotein complex assembly | P. cultripes | <0.001 |
| GO: BP | GO:0022900 | electron transport chain | P. cultripes; S. couchii | <0.001;<0.001 |
| GO: BP | GO:0022904 | respiratory electron transport chain | P. cultripes; S. couchii | <0.001;<0.001 |
| GO: BP | GO:0030239 | myofibril assembly | P. cultripes | <0.001 |
| GO: BP | GO:0042773 | ATP synthesis coupled electron transport | P. cultripes; S. couchii | <0.001;<0.001 |
| GO: BP | GO:0042775 | mitochondrial ATP synthesis coupled electron transport | P. cultripes; S. couchii | <0.001;<0.001 |
| GO: BP | GO:0045214 | sarcomere organization | P. cultripes | <0.001 |
| GO: BP | GO:0045333 | cellular respiration | P. cultripes | <0.001 |
| GO: BP | GO:0071826 | ribonucleoprotein complex subunit organization | P. cultripes | <0.001 |
| GO: BP | GO:0099132 | ATP hydrolysis coupled cation transmembrane transport | P. cultripes | <0.001 |
| GO: BP | GO:0009063 | cellular amino acid catabolic process | P. cultripes; S. couchii | 0.001;<0.001 |
| GO: BP | GO:0022613 | ribonucleoprotein complex biogenesis | P. cultripes | 0.001 |
| GO: BP | GO:0070972 | protein localization to endoplasmic reticulum | S. couchii | 0.001 |
| GO: BP | GO:0045047 | protein targeting to ER | S. couchii | 0.003 |
| GO: BP | GO:0072599 | establishment of protein localization to endoplasmic reticulum | S. couchii | 0.003 |
| GO: BP | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c | P. cultripes | 0.004 |
| GO: BP | GO:0031032 | actomyosin structure organization | P. cultripes | 0.004 |
| GO: BP | GO:0072376 | protein activation cascade | P. cultripes; S. couchii | 0.004;0.004 |
| GO: BP | GO:0072378 | blood coagulation, fibrin clot formation | P. cultripes; S. couchii | 0.004;<0.001 |
| GO: BP | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient | P. cultripes | 0.005 |
| GO: BP | GO:0072350 | tricarboxylic acid metabolic process | S. couchii | 0.005 |
| GO: BP | GO:1901605 | alpha-amino acid metabolic process | P. cultripes | 0.006 |
| GO: BP | GO:1901606 | alpha-amino acid catabolic process | P. cultripes | 0.006 |
| GO: BP | GO:0006613 | cotranslational protein targeting to membrane | S. couchii | 0.016 |
| GO: BP | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | S. couchii | 0.016 |
| GO: BP | GO:0051258 | protein polymerization | P. cultripes | 0.016 |
| GO: BP | GO:0009620 | response to fungus | P. cultripes; S. couchii | 0.018;0.014 |
| GO: BP | GO:0046500 | S-adenosylmethionine metabolic process | S. couchii | 0.018 |
| GO: BP | GO:0032271 | regulation of protein polymerization | P. cultripes | 0.019 |
| GO: BP | GO:0055002 | striated muscle cell development | P. cultripes | 0.021 |
| GO: BP | GO:0016054 | organic acid catabolic process | P. cultripes | 0.025 |
| GO: BP | GO:0046395 | carboxylic acid catabolic process | P. cultripes | 0.025 |
| GO: BP | GO:0034314 | Arp2/3 complex-mediated actin nucleation | P. cultripes | 0.028 |
| GO: BP | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient | P. cultripes | 0.029 |
| GO: BP | GO:0035384 | thioester biosynthetic process | P. cultripes | 0.029 |
| GO: BP | GO:0071616 | acyl-CoA biosynthetic process | P. cultripes | 0.029 |
| GO: BP | GO:0000387 | spliceosomal snRNP assembly | S. couchii | 0.031 |
| GO: BP | GO:0044282 | small molecule catabolic process | P. cultripes | 0.036 |
| GO: BP | GO:0001878 | response to yeast | P. cultripes; S. couchii | 0.038;0.028 |
| GO: BP | GO:0015991 | ATP hydrolysis coupled proton transport | P. cultripes | 0.04 |
| GO: BP | GO:0090662 | ATP hydrolysis coupled transmembrane transport | P. cultripes | 0.04 |
| GO: BP | GO:0099131 | ATP hydrolysis coupled ion transmembrane transport | P. cultripes | 0.04 |
| Reactome | REAC:R-XTR-156842 | Eukaryotic Translation Elongation | P. cultripes | <0.001 |
| Reactome | REAC:R-XTR-163210 | Formation of ATP by chemiosmotic coupling | S. couchii | <0.001 |
| Reactome | REAC:R-XTR-71403 | Citric acid cycle (TCA cycle) | S. couchii | <0.001 |
| Reactome | REAC:R-XTR-8949613 | Cristae formation | S. couchii | <0.001 |
| Reactome | REAC:R-XTR-390471 | Association of TriC/CCT with target proteins during biosynthesis | P. cultripes | 0.002 |
| Reactome | REAC:R-XTR-3371511 | HSF1 activation | S. couchii | 0.009 |
| Reactome | REAC:R-XTR-159740 | Gamma-carboxylation of protein precursors | P. cultripes | 0.03 |
| Reactome | REAC:R-XTR-196025 | Formation of annular gap junctions | S. couchii | 0.047 |
| Reactome | REAC:R-XTR-5625970 | RHO GTPases activate KTN1 | S. couchii | 0.049 |
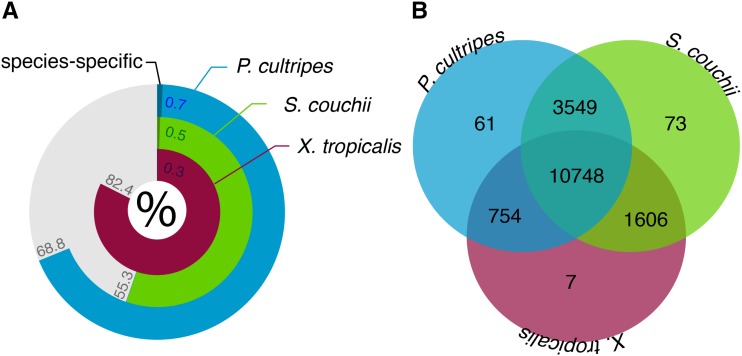
OrthoFinder assigned 81,833 genes (64.8% of total) to 16,798 orthogroups. 82.4% of X. tropicalis genes could be assigned to orthogroups, compared to 68.8% of P. cultripes genes and 55.3% of S. couchii genes (Figure 4a). Of the assigned genes, only small fractions of genes were in species-specific orthogroups (X. tropicalis: 0.3%, P. cultripes: 0.7%, S. couchii 0.5%; Figure 3a). Fifty percent (G50) of all genes were in orthogroups with 4 or more genes and were contained in the largest 9,842 orthogroups (O50). There were 10,748 orthogroups (the largest share) with all species present (Figure 4b) and 2,123 of these consisted entirely of single-copy genes. Pelobates cultripes and S. couchii shared more orthogroups than either did with X. tropicalis with 61 and 73 orthogroups being unique to each species respectively, compared to only 7 being unique to X. tropicalis.
Figure 4.
OrthoFinder results showing a) the percentage of genes that could be assigned to orthogroups per species (darker shading represents percentage of genes in species-specific orthogroups) and b) the number of species-specific and shared orthogroups recovered.
Conclusion
De novo transcriptome assemblies of the larval phase of two amphibians with vastly differing environmental sensitivity in developmental rate are presented and annotated. Despite having drastically different sized genomes (with that of Pelobates cultripes being 2.6 times larger than that of Scaphiopus couchii; Liedtke et al. 2018), the assemblies are of similar sizes (0.58Gbp vs. 0.64Gbp). The assemblies are of high quality, with 83–90% of raw reads mapping onto the transcriptomes, and both transcriptome assemblies consist of >86% full length BUSCO matches with only 2.9% of the assemblies having no corresponding match.
The PANTHER and gProfile overrepresentation tests suggest the two transcriptomes (post express-filtering) are largely comparable in terms of their annotation composition and how they differ to the proteome of X. tropicalis. Similarly, the largest share of OrthoFinder orthogroups contains all three species and not unexpectedly (due to their closer phylogenetic affinity; Pyron and Wiens 2011), P. cultripes and S. couchii share more orthogroups with each other than either does with X. tropicalis.
Approximately 40% of the unfiltered assemblies were predicted to be protein coding sequences allowing for extensive annotation and here we provide information on SwissProt proteins (and their GO terms), protein family proteins (Pfam; and their GO terms), protein orthologous groups (eggnog), biological pathways (KEGG database), signal peptide cleave sites (SignalP) and transmembrane protein predictions (TMHMM). The herein provided transcriptomes should therefore serve as an important resource for the advancement in the understanding of amphibian larval transcriptomics.
Acknowledgments
We thank D. Buchholz for providing adult toads for us to breed for this project. Informatic resources were provided by ICTS-RBD. This project was funded by Ministerio de Economía, Industria y Competitividad (MINECO) through the grant CGL2014-59206-P awarded to the EBD group. The CNAG team was funded by grant PT17/0009/0019 from MINECO, as well as Fondo Europeo de Desarrollo Regional (FEDER).
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25387/g3.8201825.
Communicating editor: D. Grunwald
Literature Cited
- Alfsnes K., Leinaas H. P., and Hessen D. O., 2017. Genome size in arthropods; different roles of phylogeny, habitat and life history in insects and crustaceans. Ecol. Evol. 44: 498–499. 10.1002/ece3.3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birol I., Behsaz B. Hammond S. A., Kucuk E., Veldhoen N. et al. , 2015. De novo transcriptome assemblies of Rana (Lithobates) catesbeiana and Xenopus laevis tadpole livers for comparative genomics without reference genomes. PLoS One 10: e0130720 10.1371/journal.pone.0130720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., and Usadel B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N. L., Pimentel H., Melsted P., and Pachter L., 2016. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34: 525–527. 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- Buchholz D. R., and Hayes T. B., 2002. Evolutionary patterns of diversity in spadefoot toad metamorphosis (Anura: Pelobatidae). Copeia 2002: 180–189. 10.1643/0045-8511(2002)002[0180:EPODIS]2.0.CO;2 [DOI] [Google Scholar]
- Denver R. J., 2009. Stress hormones mediate environment-genotype interactions during amphibian development. Gen. Comp. Endocrinol. 164: 20–31. 10.1016/j.ygcen.2009.04.016 [DOI] [PubMed] [Google Scholar]
- Emms D. M., and Kelly S., 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16: 157 10.1186/s13059-015-0721-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels P., Magnusson M., Lundin S., and Käller M., 2016. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32: 3047–3048. 10.1093/bioinformatics/btw354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M. et al. , 2017. The Reactome pathway knowledgebase. Nucleic Acids Res. 46: D649–D655. 10.1093/nar/gkx1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Coggill P., Eberhardt R. Y., Eddy S. R., Mistry J. et al. , 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44: D279–D285. 10.1093/nar/gkv1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Mestre I., Kulkarni S., and Buchholz D. R., 2013. Mechanisms and consequences of developmental acceleration in tadpoles responding to pond drying. PLoS One 8: e84266 10.1371/journal.pone.0084266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Mestre I., Sacoccio V. L., Iijima T., Collins E. M., Rosenthal G. G. et al. , 2010. The shape of things to come: linking developmental plasticity to post-metamorphic morphology in anurans. J. Evol. Biol. 23: 1364–1373. 10.1111/j.1420-9101.2010.02016.x [DOI] [PubMed] [Google Scholar]
- Gosner K. L., 1960. A simplified table for staging anuran embryos and larvae with notes on identification. Herpetologica 16: 183–190. [Google Scholar]
- Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D. et al. , 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8: 1494–1512. 10.1038/nprot.2013.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J., Szklarczyk D., Forslund K., Cook H., Heller D. et al. , 2016. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 44: D286–D293. 10.1093/nar/gkv1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jockusch E. L., 1997. An evolutionary correlate of genome size change in plethodontid salamanders. P. Roy. Soc. B-Biol. Sci. 264: 597–604. 10.1098/rspb.1997.0085 [DOI] [Google Scholar]
- Johansson F., and Richter-Boix A., 2013. Within-population developmental and morphological plasticity is mirrored in between-population differences: linking plasticity and diversity. Evol. Biol. 40: 494–503. 10.1007/s11692-013-9225-8 [DOI] [Google Scholar]
- Kulkarni S. S., Denver R. J., Gomez-Mestre I., and Buchholz D. R., 2017. Genetic accommodation via modified endocrine signalling explains phenotypic divergence among spadefoot toad species. Nat. Commun. 8: 993 10.1038/s41467-017-00996-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni S. S., Gomez-Mestre I., Moskalik C. L., Storz B. L., and Buchholz D. R., 2011. Evolutionary reduction of developmental plasticity in desert spadefoot toads. J. Evol. Biol. 24: 2445–2455. 10.1111/j.1420-9101.2011.02370.x [DOI] [PubMed] [Google Scholar]
- Langmead B., and Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liedtke H. C., Gower D. J., Wilkinson M., and Gomez-Mestre I., 2018. Macroevolutionary shift in the size of amphibian genomes and the role of life history and climate. Nat. Ecol. Evol. 2: 1792–1799. 10.1038/s41559-018-0674-4 [DOI] [PubMed] [Google Scholar]
- Merico D., Isserlin R., Stueker O., Emili A., and Bader G. D., 2010. Enrichment Map: A network-based method for gene-set enrichment visualization and interpretation. PLoS One 5: e13984 10.1371/journal.pone.0013984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., Casagrande J. T., and Thomas P. D., 2013. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8: 1551–1566. 10.1038/nprot.2013.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran N. A., 1994. Adaptation and constraint in the complex life cycles of animals. Annu. Rev. Ecol. Syst. 25: 573–600. 10.1146/annurev.es.25.110194.003041 [DOI] [Google Scholar]
- Petersen T. N., Brunak S., von Heijne G., and Nielsen H., 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8: 785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- Pyron R. A., and Wiens J. J., 2011. A large-scale phylogeny of Amphibia including over 2800 species, and a revised classification of extant frogs, salamanders, and caecilians. Mol. Phylogenet. Evol. 61: 543–583. 10.1016/j.ympev.2011.06.012 [DOI] [PubMed] [Google Scholar]
- Raudvere U., Kolberg L., Kuzmin I., Arak T., Adler P. et al. , 2019. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 44: W90–W98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson M. F., Sequeira F., Selechnik D., Carneiro M., Vallinoto M. et al. , 2018. Improving amphibian genomic resources: a multitissue reference transcriptome of an iconic invader. Gigascience 7: 1–7. 10.1093/gigascience/gix114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T. et al. , 2003. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13: 2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão F. A., Waterhouse R. M., Panagiotis I., and Kriventseva E. V., 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- The UniProt Consortium , 2017. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45: D158–D169. 10.1093/nar/gkw1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyngaard G. A., Rasch E. M., Manning N. M., Gasser K., and Domangue R., 2005. The relationship between genome size, development rate, and body size in copepods. Hydrobiologia 532: 123–137. 10.1007/s10750-004-9521-5 [DOI] [Google Scholar]
- Zeng C., Gomez-Mestre I., and Wiens J. J., 2014. Evolution of rapid development in spadefoot toads is unrelated to arid environments. PLoS One 9: e96637 10.1371/journal.pone.0096637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Liu L., Wang S., Wang H., and Jiang J., 2016. Transcriptome profiles of metamorphosis in the ornamented pygmy frog Microhyla fissipes clarify the functions of thyroid hormone receptors in metamorphosis. Sci. Rep. 6: 1–11. 10.1038/srep27310 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets supporting the results of this article are available as supporting information. Specifically, we provide quality assessment results of both BUSCO and BowTie2, the unfiltered transcriptome assemblies plus their transdecoded CDS files and annotations in the form of Trinotate summary tables and Panther annotation lists. In addition, all raw reads as well as the expression filtered transcriptome assemblies are deposited on the NCBI’s Sequence Read Archive [SRA; SRP161446] and Transcriptome Shotgun Assembly database [TSA; P. cultripes: GHBH01000000; S. couchii: GHBO01000000], under BioProject [PRJNA490256]. Supplemental material available at FigShare: https://doi.org/10.25387/g3.8201825.