Summary
The directed migration of cells sculpts the embryo, contributes to homeostasis in the adult and, when dysregulated, underlies many diseases [1, 2]. During these processes, cells move singly or as a collective. In both cases, they follow guidance cues which direct them to their destination [3–6]. In contrast to single cells, collectively migrating cells need to coordinate with their neighbors to move together in the same direction. Recent studies suggest that leader cells in the front sense the guidance cue, relay the directional information to the follower cells in the back and can pull the follower cells along [7–19]. In this manner, leader cells steer the collective and set the collective′s overall speed. However, whether follower cells also participate in steering and speed setting of the collective is largely unclear. Using chimeras, we analyzed the role of leader and follower cells in the collectively migrating zebrafish posterior lateral line primordium. This tissue expresses the chemokine receptor Cxcr4 and is guided by the chemokine Cxcl12a [20–23]. We find that leader and follower cells need to sense the attractant Cxcl12a for efficient migration, are coupled to each other through cadherins, and require coupling to pull Cxcl12a-insensitive cells along. Analysis of cell dynamics in chimeric and protein-depleted primordia shows that Cxcl12a-sensing and cadherin-mediated adhesion contribute jointly to direct migration at both single-cell and tissue levels. These results suggest that all cells in the primordium need to sense the attractant and adhere to each other to coordinate their movements and migrate with robust directionality.
Ghaphical abstract
Colak-Champollion et al. combine chimeric analysis with live imaging to examine how collectively migrating cells coordinate their migration in the zebrafish lateral line primordium. They show that the cells require cadherins and chemokine sensing for polarity, cell coupling and efficient directional migration of the primordium.
Results and discussion
Cxcr4-mediated Cxcl12a signaling is required in all primordium cells for efficient directional migration
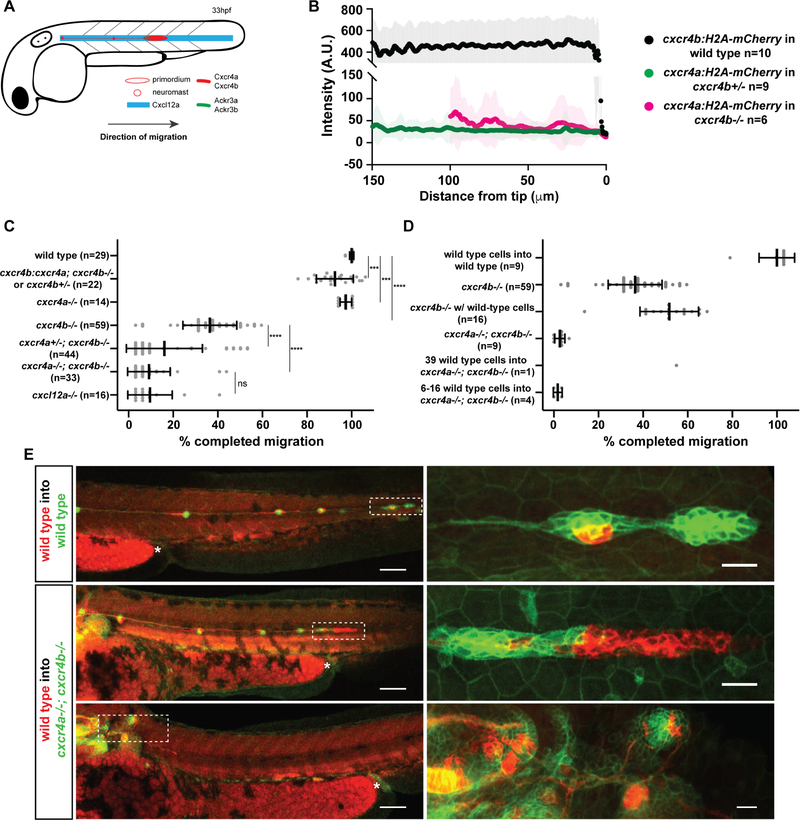
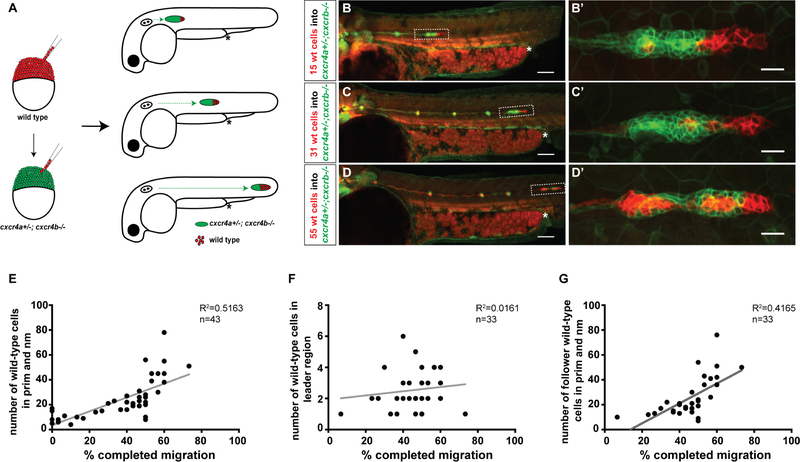
The primordium expresses the chemokine receptor Cxcr4b and migrates along a stripe of constant Cxcl12a expression [20–22]. It expresses the Cxcl12a clearance receptor Ackr3b in its back [24, 25], where Ackr3b removes the chemokine to generate a local Cxcl12a gradient across the primordium to direct it forward [22, 23] (Figure 1A). Chimeric analysis indicates that Cxcr4b is only required in the leader cells of the primordium (cells located in the front 15 μm of the primordium for directed migration [21]), suggesting that the leader cells guide the follower cells in the back of the primordium. Since cells in the first 120 μm of the primordium (about 80% of the cells) sense a Cxcl12a-gradient across themselves [22, 23], we hypothesized that the follower cells (cells located behind the front 15 μm of the primordium) could also contribute to directed migration. To explore this idea, we first analyzed the expression levels of the two Cxcl12a receptors Cxcr4a and Cxcr4b using transcriptional reporters. This analysis showed that the primordium expresses cxcr4a and cxcr4b as previously reported [20, 23, 26] (Figure S1A, S1B) but at very different levels; cxcr4b expression is 19-fold higher than cxcr4a expression in the primordium (Figure 1B). The difference in expression was lost when we expressed cxcr4a from the cxcr4b promoter (Figure S1C, S1D). Second, we asked to what degree the two Cxcr4 receptors contribute to the migration of the primordium. Quantification of the speed, directionality and traveled distance in embryos with progressively lower overall Cxcr4 activity showed that Cxcr4a contributes little and Cxcr4b majorly to the directional sensing of the primordium (Figures 1C, S1E). Loss of Cxcr4b function did not result in the upregulation of cxcr4a expression, indicating that the absence of Cxcr4b activity is not compensated for by increased cxcr4a transcription (Figure S1B). Importantly, expressing cxcr4a from the cxcr4b promoter in cxcr4b−/− embryos restored primordium migration (Figure 1C), suggesting that the different receptor expression levels underlie the different contributions to directional sensing during primordium migration rather than different affinities for Cxcl12a as observed in other contexts [27, 28]. Third, we assessed whether a few wild-type cells can also restore directional migration of primordia completely lacking Cxcr4 activity. Using chimeric analysis [29, 30], we found that 16 or fewer wild-type cells did not restore the migration of cxcr4a−/−; cxcr4b−/− primordia (Figure 1D, E). Increasing the number of wild-type cells to 29 (this corresponds to 54 % of the total cells in this primordium) partly restored migration of a cxcr4a−/−; cxcr4b−/− primordium (Figure 1E). Similarly, and consistent with published work [21], placing as few as seven wild-type cells into cxcr4b−/− primordia that lack Cxcr4b activity but retained Cxcr4a activity partly restored primordium migration (Figure S2). Placing wild-type cells into wild-type primordia did not affect primordium migration (Figure 1D, E). Thus, a few wild-type cells are not sufficient to direct the migration of cxcr4-deficient neighbors in the primordium, suggesting that both the leader cells in the front and the follower cells in the back of the primordium need to sense the attractant Cxcl12a for directional migration. To determine the contribution of the follower cells to the directed migration of the primordium, we generated cxcr4a−/+; cxcr4b−/− primordia which harbored an increasing number of wild-type cells (Figure 2A). This analysis showed that the distance traversed by the chimeric primordia increased in a fairly linear fashion with the initial number of wild-type cells in the primordium (Figure 2B–E). In contrast, there was little to no correlation between the traversed distance and the number of wild-type cells when only considering the cells in the front of the primordium (Figure 2F, G). This suggests that all the cells in the primordium - leader and follower cells - contribute remarkably equally to the directed migration of the collective.
Figure 1. A small number of Cxcr4-expressing cells cannot restore migration of primordia lacking Cxcr4 activity.
(A) Schematic of primordium migration and chemokine signaling system in primordium.
(B) Fluorescent intensity quantification of cxcr4b transcriptional reporter in wild-type primordia (black) and cxcr4a transcriptional reporter in cxcr4b heterozygous (green) and cxcr4b mutant primordia (pink) in 33–36 hpf embryos. Fluorescent intensities were averaged along the front-to-back axis of the primordia. Mean, SD, and n are indicated.
(C) Quantification of primordium migration in 48 hpf embryos of indicated genotypes. Note that the cxcr4b:cxcr4a embryos originated from a cross that yielded on average 50 % cxcr4b−/− and 50% cxcr4b+/− embryos. Grey dots are individual data points. Sample size is as indicated. Vertical and horizontal black lines indicate mean and SD, respectively. *** = p<0.001, **** = p < 0.0001, ns=not significant (Mann-Whitney test).
(D) Quantification of the migration defects in cxcr4 mutants and degree of migration restoration in chimeric primordia. Note, the number of wild-type cells for the chimeras includes wild-type cells in the neuromasts and the primordium.
(E) Sum projections of fixed chimeric embryos of indicated genotypes at 48 hpf. Magnifications of primordia in left panels indicated by dashed white rectangles are shown in the panels on the right. The asterisks mark the yolk extension. Scale bars in left and right panels correspond to 100 μm and 20 μm, respectively. (See also Figure S1 and Methods S2).
Figure 2. Cxcr4-deficient primordia migrate further with increasing number of wild-type cells.
(A) Experimental design and predictions for migratory behavior of cxcr4-deficient primordia with increasing number of wild-type cells.
(B-D) Sum projections of 48 hpf embryos with chimeric primordia. Wild-type donor-derived cells in primordia labeled in red with prim:lyn2mCherry and cxcr4a+/−; cxcr4b−/− host-derived cells labeled in green with cldnB:lyn2GFP (B-D) and magnifications of chimeric primordia outlined with rectangles in B-D (B′-D′). Chimeric primordia migrate further as the number of wild-type donor cells increases in the cxcr4a+/−; cxcr4b−/− primordia. The chimeric primordia are outlined with dashed rectangles and asterisks mark the end of the yolk extension in B-D. Scale bars in B-D and B′-D′ correspond to 100 μm and 20 μm, respectively.
(E) Graph of the migration distance of cxcr4-deficient primordia versus the total number of wild-type cells in the chimeric primordia (total number of wild-type cells include neuromast (nm) cells and primordium (prim) cells).
(F) Graph of wild-type cell number in the leader cell region (first 15 microns of the front of the primordium as defined in [31]) versus migration distance of cxcr4-deficient chimeric primordia.
(G) Graph of the number of wild-type cells in the follower cell region (past 15 microns from the front of the primordium as defined in [31]) versus migration distance of cxcr4-deficient chimeric primordia. (See also Figure S2 and Methods S2).
n indicates number of chimeric primordia in E-G.
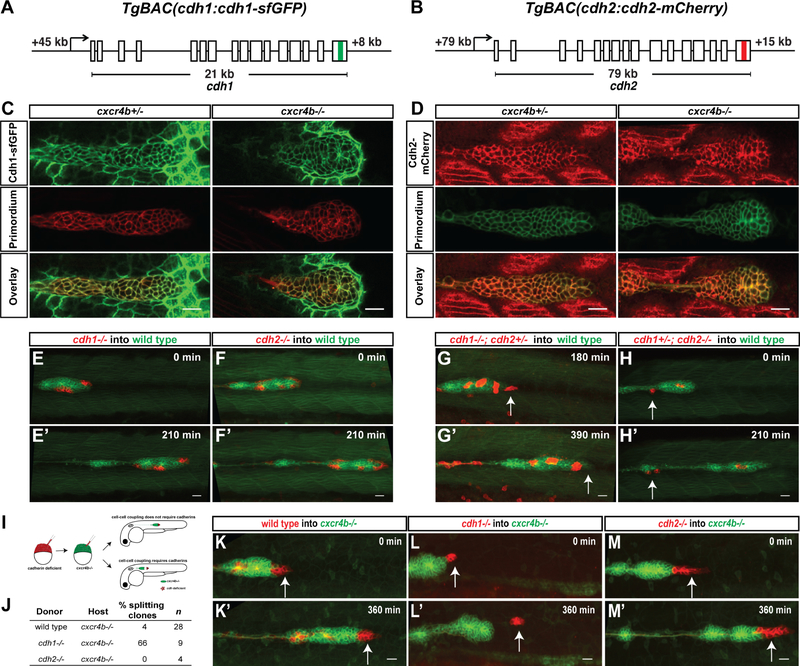
Cadherins mediate cell-cell adhesion between primordium cells
Wild-type cells can direct the migration of neighboring cells in primordia with reduced Cxcr4 activity. Since the cells in the primordium adhere tightly to each other [31], wild-type cells could direct the migration of Cxcr4-deficient cells in the primordium by pulling their neighbors along. The primordium expresses the two cell-cell adhesion receptors Cadherin-1 (E-Cadherin, Cdh1) and Cadherin-2 (N-Cadherin, Cdh2) [30–33]. To characterize the role of Cdh1 and Cdh2 in primordium migration, we first assessed their expression using functional BAC transgenes that drive expression of Cdh1-superfolderGFP (Cdh1-sfGFP) and Cdh2-mCherry from the cdh1 and cdh2 genomic architecture (cdh1:cdh 1-sfGFP and cdh2:cdh2-mCherry, Figure 3A, B). Consistent with previous reports [30–33], we find that Cdh1 and Cdh2 are expressed in the migrating primordium (Figure 3C, D and Video S1). The front cells express 1.7-fold more Cdh1-sfGFP and the rear cells express 1.6-fold mor6e Cdh2-mCherry than the rear and front cells, respectively (Figure S3A–S3C). The graded Cadherin expression pattern in the primordium is independent of Cxcr4b-mediated Cxcl12a and Fgf3/10 signaling (Figures 3C, D, S3B–S3F). Second, we asked whether Cdh1 and Cdh2 are required for cell-cell adhesion in the primordium. While cdh1−/− embryos disintegrate during gastrulation [34] - precluding the analysis of the migration of the primordium, cdh2−/− embryos are viable until 4 dpf but malformed [35, 36]. As previously reported [37, 38], we find that the primordium only completes on average 70 % of its migration in cdh2−/− embryos (Figure S3L). This is probably due to the reduced or absent stripe of cxcH2a expression along the migratory route in cdh2−/− embryos (Figure S3G). We therefore used chimeric analysis to assess the role of Cdh1 and Cdh2 in primordium migration. As reported for cdh1 morphant cells [30], cdh1−/− and cdh2−/− cells migrate together with their wild-type neighbors (Figure 3E, F and Video S2). Since Cdh1 and Cdh2 might act redundantly, we analyzed the behavior of cdh1−/−; cdh2−/+ and cdh1−/+; cdh2−/− cells in wild-type primordia. Indeed, cells lacking three of the four cdh1 and cdh2 copies detached and fell behind when they were located at the primordium′s edge (3 of 3 clones) and co-migrated with their wild-type neighbors only when they were located in the center of the primordium (Figure 3G, H and Video S2). Importantly, we never obtained cdh1−/−; cdh2−/− double mutant clones in the primordium (Figure S3J), suggesting that cdh1−/−; cdh2−/− cells are not incorporated into the primordium or fail to co-migrate with wild-type primordium cells. Also, Cdh1-sfGFP was not upregulated in the absence of Cdh2 activity (Figure S3H, S3I). Thus, while Cdh1 and Cdh2 are graded in their expression across the primordium, both proteins work together to mediate adhesion between the cells in the primordium.
Figure 3. Cadherin-mediated adhesion couples the cells in the primordium.
(A, B) Schematic diagrams of BAC transgenes that drive expression of Cdh1-sfGFP and Cdh2-mCherry.
(C) Single confocal section through primordium in live prim:lyn2-mCherry, cdh1:cdh1-sfGFP embryos heterozygous (left) and mutant for cxcr4b (right) at 36 hpf.
(D) Single confocal section through primordium in live cldnB:lyn2-GFP, cdh2:cdh2-mCherry embryos heterozygous (left) and mutant for cxcr4b (right) at 36 hpf. Scale bar corresponds to 20 μm. See also Video S1.
(E-H) Sum projections of time lapse videos of wild-type primordia (green) with clones of cdh1−/− (E), cdh2−/− (F), cdh1−/−; cdh2−/+ (G) and cdh1−/+; cdh2−/− (H) primordium cells (red). The time lapse videos start at 33 hpf (E), 28 hpf (F,H), 31 hfp (G) and stop at the indicated lapsed times. Arrows in G and H indicate mutant cells on the edge of the primordium that fail to co-migrate with their wild-type neighbors. Scale bar is 20 μm. Images correspond to frames 1 and 8 (E, F and H) and frames 7 and 14 (G) in Video S2.
(I) Experimental design and predictions for the cadherin mutant cells′ ability to rescue the cxcr4b mutant primordia migration defect.
(J) Table summarizing the chimeric analysis of cadherin deficient cells in cxcr4b mutant primordia. Genotypes of donor clones and host primordia, percentage of chimeric primordia in which donor-cells split from the host-cells and sample number are indicated.
(K-M) Sum projections of time lapse videos of cxcr4b mutant primordia (green) with clones of wild-type control (K), cdh1−/− (L) and cdh2−/− (M) primordium cells (red). The time lapse videos start at 33–34 hpf and stop at the indicated lapsed times. Arrows indicate wild-type (K) and mutant (L-M) cells in the front of the primordium. Scale bar is 20 μm. Images correspond to frames 1 and 13 in Video S2. (See also Figure S3, Videos S1 and S2 and Methods S1)
Cadherin-1 is required in wild-type cells to restore the migration of cxcr4b mutant primordial
Since wild-type cells restore directed migration and increase the speed of cxcr4-deficient primordia with increasing cell number (Figure 2E), we hypothesized that wild-type cells physically adhere to and pull cxcr4-deficient cells in the correct direction. We tested this idea by placing cdh1−/− and cdh2−/− cells into cxcr4b−/− primordia (Figure 3I). Similar to wild-type cells, cdh2−/− cells restored the directed migration of cxcr4b−/− primordia (4 of 4 chimeric primordia, Figure 3J, K, M and Video S2). In contrast, cdh1−/− cells mostly failed to restore cxcr4b−/− primordium migration. cdh1−/− cells occupied the front region, initiated migration but then separated from the cxcr4b−/− primordium (6 of 9 chimeric primordia, Figure 3J, L and Video S2). Thus, Cdh1, and not Cdh2, is important for cells to remain attached to and restore the migration of primordium cells with reduced directional sensing. This observation suggests that reducing directional sensing and cell-cell adhesion in the same cells should also cause them to separate from the primordium. However, when we placed cdh1−/−; cxcr4b−/− or cdh2−/−; cxcr4b−/− in wild-type primordia, cells of both genotypes co-migrated with their wild-type neighbors and did not separate from the primordium or caused the primordium to split (Figure S3M, S3N). Consistent with the increased levels of Cdh1 in the primordium′s front, these observations suggest that Cdh1 physically couples cells in the front of the primordium to one another.
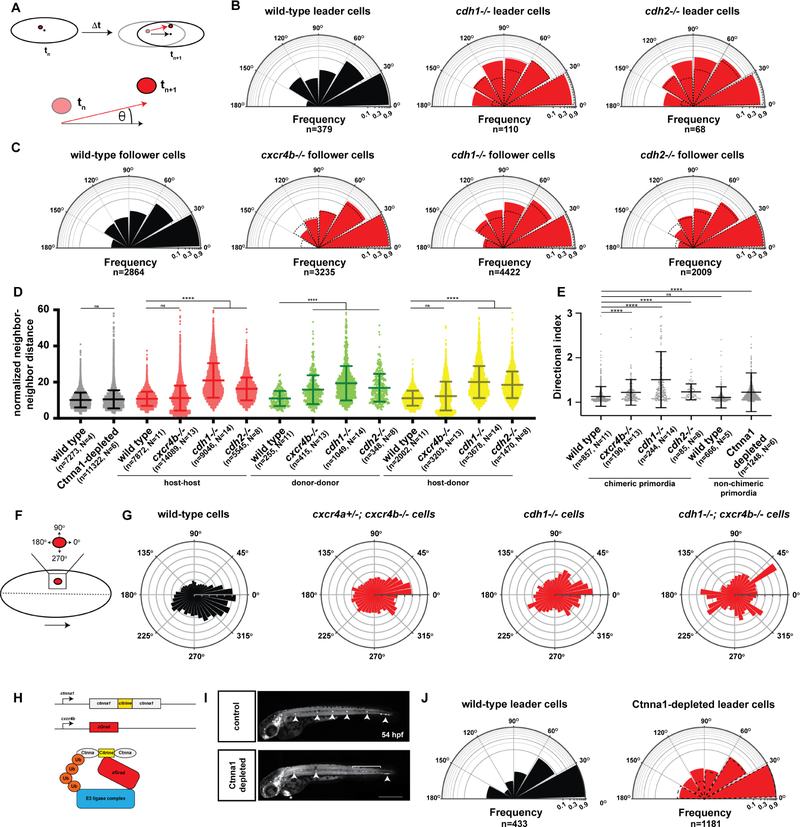
Directional migration of cells in the primordium requires Cadherins
Our observations indicate that Cxcr4-mediated Cxcl12a sensing and Cadherin-mediated cell-cell interactions coordinate directional migration of the cells in the primordium. To assess the contribution of these two aspects to migration, we first characterized the behavior of wild-type cells in different locations of the primordium. We placed wild-type cells labeled with nuclear H2A-GFP into wild-type primordia whose cell nuclei were labeled with H2A-mCherry and tracked the nuclear movements of the donor and host primordium cells over time. Using these measurements, we determined the directionality and neighbor relations of the donor and host cells at different positions in the primordium. As expected, the directionality angle of the cells (Figure 4A) compared to the overall directionality of the primordium varied slightly with the cell′s position in the primordium. Leader cells in the primordium moved slightly more directional than follower cells (Figure 4B, C). Also, cells closer to the midline of the primordium aligned more with the overall directionality of the primordium than cells further outwards (Figure S4C, S4D). To assess the contribution of attractant sensing, we placed cxcr4b−/− cells labeled with H2A-GFP into wild-type primordia labeled with H2A-mCherry. As expected [21], cxcr4b−/− cells occupied follower positions in the rear of the primordia (Figure S4E) and aligned less with the primordium′s direction of migration than wild-type cells (Figure 4C). This resulted in less directional migration of individual cxcr4b−/− cells as judged by their averaged directional indices - a measure of how close to a straight path a cell moves (Figure 4E). As a consequence, neighboring cxcr4b−/− cells separated from each other more over time than neighboring wild-type cells (Figure 4D). This also caused their wild-type host neighbors to move in a slightly less coordinate manner (Figure S4B). Next, we placed cdh1−/− cells into wild-type primordia. In contrast to cxcr4b−/− cells, cdh1−/− cells were not excluded from the front of the primordium (Figure S4E). However, the directional migration of cdh1−/− cells was impaired (Figure 4E). cdh1−/− leader cells moved 3-times more often sideways or backwards than wild-type cells causing the cells behind them to also slow down (Figure 4B and Video S3). cdh1−/− follower cells were also impaired in their directionality but to a lesser degree than cdh1−/− leader cells (Figure 4C). Similar to cxcr4b−/− cells but more severely, cdh1−/− cells also perturbed the directional migration of their wild-type neighbor cells (Figure S4A, S4B, S4D). This caused pairs of neighboring cdh1−/− cells and - surprisingly also pairs of neighboring wild-type cells - to separate over time (Figure 4D). As a consequence, these chimeric primordia migrated in an accordion-like manner (Video S3). In contrast to cdh1−/− cells, cdh2−/− cells in the front or back of otherwise wild-type primordia were less impaired in their ability to migrate directionally (Figures 4B, C, E, S4A–S4C). Also, cdh2−/− cells perturbed the neighbor relations across the entire tissue similar to cdh1−/− cells but to a lesser extent (Figure 4D). Nuclear movements reflect the combined effects of a cell′s own movement and the influence of the neighbors on a cell′s movement. To assess whether impaired attractant sensing or reduced cell-cell adhesion affect the cell′s own movement similarly to its movement in a group, we used the protrusive activity of cell clones as a measure of their preferred direction of migration (Figure 4F and Video S3). Recording the extensions and retractions of membrane-labeled clones in wild-type primordia over time, we found that the protrusive activity of wild-type cell clones is biased towards the front and the midline of the primordium (Figure 4G). This bias is slightly reduced in cxcr4a−/+; cxcr4b−/− cell clones and strongly reduced in cdh1−/− cell clones (Figure 4G). Combined reduction of attractant sensing and cell-cell adhesion in cxcr4b−/−; cdh1−/− cell clones did not reduce the bias of the protrusive activity further (Figure 4G). This suggests that attractant signaling and cadherin activity act on a cellular level to provide directional information and on a tissue level to coordinate cell movements for efficient directional migration.
Figure 4. Chemokine signaling and cadherin-mediated adhesion provide directionality to the cells in the primordium.
(A) Schematic diagrams of nuclear tracking of primordium cells. The directional angle (θ) was calculated as the angle between the averaged displacement vector of the primordium (black arrow) and the displacement vector of each cell nucleus in the primordium (red arrow) between two consecutive time points tn and tn+1 in 3-dimensional space. For perfectly aligned cells, the directional angle will be zero and for cells migrating in the opposite direction of the primordium, the directional angle will be 180 degrees.
(B) Semi-circular histogram plots of the directional angle frequencies (binned in 30 degree intervals) of wild-type (left), cdh1 mutant (middle) and cdh2 mutant leader cells (right) in wild-type primordia. For the wild-type directional angles, donor and host leader cell directional angles were pooled. Note that cxcr4b mutant cells do not occupy leader cell positions in wild-type primordia and directional angles cannot be determined. Dashed lines indicate directional angle frequencies of the wild-type leader cells. The frequencies on the radial axes are plotted on a log10-scale. n indicates the number of directional angles. The directional angle distributions for wild-type and cdh1 mutant leader cells (p = 0.0003) and wild-type and cdh2 mutant leader cells (p = 0.0015) are significantly different (Kolmogorov-Smirnov test).
(C) Semi-circular histogram plots of the directional angle frequencies (binned in 30 degree intervals) of wild-type (far left), cxcr4b mutant (middle left), cdh1 mutant (middle right) and cdh2 mutant follower cells (far right) in wild-type primordia. Dashed lines indicate directional angle frequencies of the wild-type follower cells. The frequencies on the radial axes are plotted on a log10-scale. n indicates the number of directional angles. The directional angle distributions for wild-type and cxcr4b mutant follower cells (p = 0.0075), wild-type and cdh1 mutant follower cells (p < 0.0001), and wild-type and cdh2 mutant follower cells (p =0.0032) are significantly different (Kolmogorov-Smirnov test).
(D) Normalized neighbor-neighbor distances for wild-type and Ctnna1-depleted primordia (black) and host-host (red), donor-donor (green) and host-donor (yellow) chimeric primordia. For chimeric primordia, all host cells were wild-type and the genotypes of the donor cells are indicated. Cells were defined as neighbors if the initial distance between a pair of cells was smaller than 15 μm. The distance between such pairs was tracked for 11 time intervals (22 min). The neighbor-neighbor distances were normalized to the average speed of the primordium cells. Mean and standard deviation are indicated. n is the number of cell pairs and N is the number of embryos analyzed. ns indicates p > 0.05 and **** indicates p < 0.0001 (Kruskal-Wallis one-way analysis of variance).
(E) Directional indices of donor cells of indicated genotypes in wild-type host primordia and directional indices of wild-type cells and Ctnna1-depleted cells. Note that the directional indices of the host and donor cells are pooled for chimeric primordia with wild-type donor and wild-type host cells. Directional indices were calculated for a track length of 18 time intervals (36 min). Mean and standard deviation are indicated. n is the number of cell tracks and N is the number of embryos analyzed. ns indicates p > 0.05 and **** indicates p < 0.0001 (Kruskal-Wallis one-way analysis of variance).
(F) Schematic diagram of donor cell clone (red) location and protrusive activity angle distribution with respect to the primordium midline and direction of migration. Dashed line indicates the primordium midline in AP axis, and large arrow indicates the direction of migration. Protrusive activity towards 0 degrees is in the direction of migration and protrusive activity towards 270 degree is towards the midline of the primordium and the underlying cxcl12a-expressing cells.
(G) Circular histogram plots of protrusive activity frequencies of donor-derived wild-type cells (far left, n=8), cxcr4a+/−; cxcr4b−/− cells (middle left, n=8), cdh1−/− cells (middle right, n=5), and cdh1−/−; cxcr4b−/− cells (far right, n=2) in wild-type primordia. The distributions of the protrusive activities for wild-type cells and the mutant (cxcr4a+/−; cxcr4b−/−, cdh1−/− or cdh1−/−; cxcr4b−/−) cells are significantly different (p < 0.0001, Kolmogorov-Smirnov test).
(H) Schematic depiction of ctnna1:ctnna1-Citrine locus, cxcr4b:zGrad transgene and principle of zGrad-mediated Ctnna1-Citrine depletion.
(I) Tiled and sum-projected Z-stacks of ctnna1:ctnna1-Citrine/ctnna1:ctnna1-Citrine, prim:lyn2-mCherry embryo (top) and ctnna1:ctnna1-Citrine/ctnna1:ctnna1-Citrine, prim:/yn2-mCherry cxcr4b:zGrad embryo (bottom). For clarity, only the mCherry signal is shown. Arrowheads, bracket and asterisk indicate neuromasts, shed primordium cells and mScarlet expression in the heart from the transgenesis marker on the cxcr4b:zGrad BAC transgene, respectively. Scale bar 0.5 mm.
(J) Semi-circular histogram plots of the directional angle frequencies (binned in 30 degree intervals) of leader cells in wild-type (left) and Ctnna1-depleted primordia (right). Dashed lines indicate directional angle frequencies of the wild-type leader cells. The frequencies on the radial axes are plotted on a log10-scale. n indicates the number of directional angles. The directional angle distributions for wild-type and Ctnna-depleted leader cells are not significantly different (p = 0.3454, Kolmogorov-Smirnov test). (See also Figure S4, Videos S3 and S4 and Methods S3 and S4)
Alpha E-catenin coordinates movements between cells in the primordium
Our observations suggest that Cadherin-mediated cell-cell adhesion coordinates the movement of the cells in the primordium. To directly test this idea, we depleted alpha E-catenin (Ctnna1) protein in the primordium using a knock-in line that expresses functional Ctnna1-Citrine (ctnna1-citrine) from the endogenous promoter [39] and zGrad from the cxcr4b promoter (cxcr4b:zGrad, Figure 4H). zGrad is a fusion protein of an F-box to an anti-GFP nanobody that recognizes Citrine-tagged proteins and targets them for degradation [40, 41]. Since Ctnna1 connects Cadherins on the plasma membrane to the actin filaments inside the cell, its depletion should impair Cadherin-mediated cell-cell adhesion [42]. Mostly consistent with in vitro studies [17, 43, 44], time lapse analysis indicated that Ctnna1-Citrine-depleted primordium cells separated from neighbors and migrated less directional than cells in control primordia (Figures 4D, E, J, S4F, S4G and Video S4). This resulted in primordia with gaps in the cluster, uncoordinated movements, cell shedding and overall defective neuromast deposition (Figure 4I and Video S4). Although Ctnna1 is thought to connect Cdh1 and Cdh2 to the actin meshwork [45–47], accordion-like migration displayed by primordia with cdh1−/− cells was not observed in Ctnna1-Citrine-depleted primordia, possibly because depletion of Ctnna1 was not complete or because of Ctnna1-independent functions of Cadherins. This corroborates the idea that cell-cell coupling coordinates cell movements and enhances the directionality of the primordium cells.
In summary, our observations suggest a two-layered model for primordium migration. On the cellular level, each cell derives directional information from the Cxcl12a attractant gradient and through Cdh1, possibly by Cdh1-mediated local tugging from neighbors as observed among mesoderm cells [48]. On a tissue level, cells are mechanically coupled to their neighbors through Cadherins. These mechanisms ensure that the directed migration of the group is the result of locally sampled directional information by the cells and averaged motions of the individual cells. In effect, this changes the behavior of the individual cells from less directional, more single cell-like to more directional and coordinated movements [49–51] and allows the primordium to tolerate perturbations and increase its directional robustness.
Our findings parallel observations in cell culture where coordinated movements through cell-cell adhesion between leader and follower cells is necessary to propel the collective forward [17, 48, 52–57], ex vivo studies where Cdh1 and Cdh2 mark cell-cell contacts orient the polarity of protrusions and suppress or mediate contact inhibition of locomotion and thus cell dispersal, respectively [58–61], and in vivo studies where Cdh1 coordinates the movement of individual cells in a group and thereby affects the group′s directionality [62, 63]. In some scenarios, these effects are mediated by cells pulling on the cadherins of their neighbors across adherens junctions, eliciting signals that provide the neighbors with directional information and maintain leader-follower cell behavior [19, 48, 63–65], a mechanism that may also be employed by cells in the primordium to coordinate cell movements. Future studies will have to unravel to what degree cell-cell adhesion couples the movements of cells and provides polarity information to neighbors to provide coordination and robustness to migrating collectives.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for reagents and resources should be communicated to and will be fulfilled by the Lead Contact, Holger Knaut (Holger.Knaut@nyulangone.org). There are restrictions to the availability of the BAC transgenes (cdh2:cdh2-mCherry, cxcr4a:H2A-mCherry and cxcr4b:cxcr4a) due to the lack of an external centralized repository for its distribution and our need to maintain the bacterial stocks. We are glad to share the bacterial strains carrying the BAC transgenes upon request with compensation by requestor for shipping. Zebrafish lines TgBAC(cdh2:cdh2-mCherry), TgBAC(cxcr4a:H2A-mCherry) and Tg(cxcr4b:cxcr4a) are not available via central repositories due to their criteria for accepting lines and will be maintained as live stocks in our laboratory for at least two years. We will share the zebrafish lines upon request with compensation by requestor for shipping.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Zebrafish strains
Zebrafish care and the use of live fish for experiments were approved and overseen by the New York University School of Medicine Institutional Animal Care and Use Committee. Zebrafish were raised and maintained at 28.5°C. Zebrafish embryos between 5 dpf and 21 dpf were fed on Cylop-eeze (Argent Laboratories). Zebrafish older than 21 dpf were raised on brine shrimp (Brine Shrimp Direct, USA) and at a consistent light-dark cycle (14 hrs light-10 hrs dark). Embryos were collected 15 min past egg laying and raised at 28.5 °C. Unfertilized eggs were discarded. Embryos were staged as described previously [66]. At the developmental stages presented in this study, the gender is not been determined yet and cannot be distinguished.
The cxcr4bt26035 allele contains a nonsense mutation that results in a premature stop codon [67], the cxcr4aum21 allele contains a 29 nt deletion and 4 nt insertion [68], the cxcl12at30516 allele contains a nonsense mutation resulting in a premature stop codon [25], the cdh1tx230 (weg) has a nonsense mutation resulting in a premature stop codon [34], the cdh2m117 (glass onion) [69, 70] allele has a missense mutation that results in an amino acid change [36], the fgf3t24149 (lia) allele contrains a missense mutation (E138K) [71] and the fgf10atbvbo allele has a nonsense mutation that results in a premature stop codon at amino acid 5 [72]. cxcr4a, cdh1, and cdh2 homozygous embryos were generated by inbreeding heterozygous adults. Mutant, heterozygous, and wild-type embryos were identified through PCR amplification of the mutated locus followed by investigation of the size difference of the amplicons for cxcr4a, and restriction digests for cdh1 and cdh2. Additionally, cdh1 mutants were identified by gross morphological phenotype and sequencing of the PCR product. Primers used for genotyping are as follows; cxcr4a. forward outer 5′CCAACTTTGAGGTCCCGTGTGATG 3′ reverse outer 5′ CTGTGGACACGGATGACATTCCTG 3′, forward inner same as forward outer, reverse inner 5′ GAATGGCAGAGTGAGCAC 3′, cdh1: forward outer 5′CAATATAACAGGCTCTGGGCAGAT 3′, reverse outer 5′ CAGACCAATTTTTACTGGTATCACC 3′, forward inner 5′TGTCCGTTATAGAGAGAAGC 3′, reverse inner 5′ GTCAAGAATGCTTTGGATCG 3′, cdh2. forward outer 5′ TGCAGTGTGTCTGTATGCGTAT 3′, reverse outer 5′ AGACTGGATCTGAAACACACAC 3′, forward inner 5′AGTGATACTGTTTCCTCGGCAC3′, reverse inner 5′ TCTGACTGCACTTCACATGATG 3′, fgf3: forward outer 5′ ATCCCGCCATGCCACAAT 3′, reverse outer 5′ TCTCGTACCCCACATAAAACTGAC 3′, forward inner 5′CTGCTCTTGTTGTTACTGAGC 3′, reverse inner 5′ CTCAAATATCAAACGGTTTACTCAC 3′, fgf10: forward outer 5′ TGCATCACCCTTTCTCCCATCCAG 3′, reverse outer 5′ TCGTCCTTGCTTTTGGTGCCATTG 3′, forward inner 5′GCTCTTCCCAGTTTTCCGAGCTCCAGGACAATGTGCAAATCG 3′, reverse inner 5′ TCCGTTCTTATCGATCCTGAG 3′. Restriction enzymes used for cdh1, cdh2, fgf3 and fgf10 are DdeI (New England Biolabs), BsrI (New England Biolabs), Apol (New England Biolabs) and Taqal (New England Biolabs), respectively. Tg(cldnB:lyn2GFP) [21], prim:lyn2mCherry [73], TgBAC(cxcr4b:H2A-GFP) [30], TgBAC(cxcr4a:H2A-mCherry) (this study), TgBAC(cdh1:cdh1-sfGFP) [41], TgBAC(cxcr4b:zGrad) [41], Gt(ctnna-citrine)Ct3a [39], TgBAC(cxcr4b:cxcr4a) (this study) and TgBAC(cdh2:cdh2-mCherry) (this study) embryos were generated by crossing heterozygous transgenic adults to wild-type adults. Transgenic embryos were identified by GFP or RFP fluorescence.
METHOD DETAILS
Generation of transgenic strains
cdh2:cdh2-mCherry
For the cdh2:cdh2-mCherry transgene, we used the BAC clone CHORI-211–75O1 which was obtained from BACPAC Resources, Children′ Hospital Oakland Research Institute, CA, USA (bacpacorders@chori.org). This BAC clone spans 172 kb (172343 bp) of genomic region and contains the cdh2 locus which we modified in two ways by recombineering. First, the Tol2 sites and the cryaa:dsRed transgenesis marker were inserted into the BAC backbone [74]. Second, a cassette consisting of mCherry-FRT-galK-FRT, flanked by 206 bp and 497 bp of homology upstream of the stop codon in cdh2 exon 16, and downstream of the stop codon in cdh2 exon 16, respectively, was inserted before the cdh2 stop codon using galK-mediated recombineering [75]. The GalK was removed using Flippase-mediated recombination. This transgene expresses full length Cdh2 fused to mCherry from the cdh2 promoter and its final version was characterized using EcoRI (New England Biolabs) restriction digestion and sequencing of the PCR amplicons of the modified locus. The final BAC was purified using nucleobond BAC 100 kit (Clontech). We co-injected 1 nl of 40 ng/μl Tol2 mRNA and 50–250 ng/μl of the cdh2:cdh2-mCherry transgene DNA into one-cell stage embryos. The Tol2 mRNA was transcribed using the mMessage mMachine SP6 Transcription Kit (Thermo Fisher) from the pCS2FA-transposase plasmid [76]. Stable transgenic lines were generated by outcrossing the adult fish injected with the transgene and raising embryos with red fluorescence in the eye lens at 4 days post fertilization. The full name of this transgenic line is TgBAC(cdh2:cdh2-mCherry)p3.
cxcr4a:H2A-mCherry
For the cxcr4a:H2a-mCherry transgene, the BAC clone CHORI73–268G8 was modified in two ways by recombineering. First, the Tol2 sites and the cryaa:Cerulean transgenesis marker were inserted into the BAC backbone [74]. Second, a cassette consisting of H2A-mCherry-FRT-kanR-FRT flanked by 737 bp and 753 bp of homology upstream of cxcr4a exon 2, and downstream of the cxcr4a stop codon, respectively, was inserted to replace the cxcr4a coding sequence in cxcr4a exon 2 (amino acid 6–361, the last amino acid before the stop codon) using kanR-mediated recombineering [75]. The kanR cassette was removed using Flippase-mediated recombination. This transgene expresses the first five amino acids from cxcr4a exon 1 fused to H2A-mCherry from the cxcr4a promoter. The final BAC transgene was characterized by SpeI and EcoRI (New England Biolabs) restriction digestion and sequencing of PCR amplicons of the modified locus. The final BAC was purified using nucleobond BAC 100 kit (Clontech). We co-injected 1 nl of 40 ng/μl Tol2 mRNA and 50–250 ng/μl of the cdh2:cdh2-mCherry transgene DNA into one-cell stage embryos. The Tol2 mRNA was transcribed using the mMessage mMachine SP6 Transcription Kit (Thermo Fisher) from the pCSFA-transposase plasmid [76]. Stable transgenic lines were generated by out-crossing the adult fish injected with the transgene and raising embryos with red fluorescence in the cxcr4a expression domains since the expression of Cerulean from the cryaa promoter is too weak to detect in the lens at 4 days post fertilization. The full name of this transgenic strain is TgBAC(cxcr4a:H2A-mCherry)p1.
cxcr4b:cxcr4a
For the cxcr4b:cxcr4a transgene, we modified the BAC clone DKEY169F10 (ImaGenes GmbH, Germany, sales@imagenes-bio.de) which contains a 67 kb genomic DNA fragment that spans the cxcr4b locus. First, we inserted the targeting cassette pIndigo-356-Tol2-kanR-myl7-mScarlet into the BAC backbone through recombineering [41, 74]. This modification adds tol2 sites and a myocardium-specific transgenesis marker to the transgene. Second, a targeting cassette containing the coding sequence for the first five amino acids (MAYYE) of cxcr4a fused to GalK flanked by homology arms 429 bp upstream of the cxcr4b start codon and 345 bp downstream of cxcr4b exon 1 was inserted to replace the coding sequence for the first five amino acids of zebrafish cxcr4b in exon 1. The GalK sequence was removed by seamless recombineering using a PCR template identical to the one described above but lacking the GalK sequence [75]. Next, a targeting cassette containing cxcr4a-FRT-GalK-FRT flanked by homology arms 413 bp upstream and 433 bp downstream of the coding sequence for cxcr4b in exon 2 replaced the coding sequence of zebrafish cxcr4b in exon 2 with the coding sequence of cxcr4a (codons 5–355). The GalK cassette was removed by flippase-mediated recombination. This transgene expresses cxcr4a from the cxcr4b promoter. The full name of this transgenic line is Tg(cxcr4b:cxcr4a)p1.
Whole-mount in situ hybridization
RNA probe preparation and in situ hybridization were carried out as previously described [77]. RNA probes against cxc12a and cxcr4a were labeled with a DIG RNA labeling mix (Roche, Cat. No. 11277073910) and detected with anti-DIG-AP antibody (1:5000; Sigma-Aldrich, Cat. No. 11093274910) and NBT/BCIP stain (1:1000; Sigma-Aldrich, Cat. No. 11681451001). During in situ hybridization procedure, cdh2 mutant embryos and their wild-type siblings and cxcr4b:cxcr4b embryos and their non-transgenic wild-type siblings were processed in the same tubes to reduce variability in staining. Embryos were mounted in 3% methly cellulose (Sigma-Aldrich, Cat. No. 64670) and imaged with a 10× (NA 0.5) objective on a Axioplan Microscope (Zeiss) equipped with an Axiocam (Zeiss). The probes are complementary to nucleotides 32–1013 and 119–1138 of the cxcl12a and cxcr4a mRNAs, respectively [78].
Chimeric analysis
For chimeric analysis, we transplanted 20–50 cells from donor embryos at the 1000–8000 cell stage into host embryos of the same stage. Donor and host embryos were transgenic for one of the following transgenes: Tg(cldnB:lyn2GFP), Tg(prim:lyn2mCherry), TgBAC(cxcr4b:H2A-GFP), or TgBAC(cxcr4b:H2A-mCherry), which label the membranes (first two transgenes) and the nuclei (last two transgenes) of the primordium cells. These transgenes also label other structures. The cldnB:lyn2-GFP transgene labels the primordium and the skin cells. The prim:lyn2-mCherry transgene is an enhancer trap line that labels the primordium and the muscle cells. The cxcr4b:H2A-GFP and the cxcr4b:H2A-mCherry transgenes label the primordium and - more dimly - the skin cells and the muscle cells. Therefore, donor-derived cells in the muscle and skin-depending on the transgene used to mark the donor cells - are mostly also tracked. Since the donor cells were targeted to regions of the host embryo that give rise to neural crest, placodal and neuronal tissue, donor-derived skin and muscle cells were very infrequent in the chimeric embryos. At 26 to 30 hpf, primordia with donor-derived cells were identified using a Zeiss Axio Zoom V16 stereo microscope. Embryos were either used to record time lapse videos or fixed in 4% PFA at 48 hpf and imaged. To determine the genotype of the donor embryos, the donor embryos were digested with Proteinase K (Sigma-Aldrich, Cat. No. P2308) for DNA extraction at 90% epiboly. Additionally, the cdh1 mutant donors were also identified based on the cdh1 mutant phenotype which manifests around gastrulation. Genotyping of donor embryos for cxcr4a, cdh1, and cdh2 mutations by PCR was performed as described above.
Confocal imaging
Still images
Fixed and live embryos were mounted in 0.5% low melt agarose/Ringer′s solution (HEPES 5mM, NaCl 111mM, KCl 5mM, CaCl2 1mM, MgSO4 0.6mM) on No 1.5 coverslips with ring spacers and sealed with glass slides. For live embryos the Ringer′s solution was supplemented with 0.4 mg/ml MS-222 anesthetic. The embryos were imaged using a Leica SP5 II confocal microscope equipped with HyD detectors with a Leica 10× (NA 0.3) objective for overview images and a Leica 40× (NA 1.1) water immersion objective for high spatial resolution images.
The pinhole was set to 1.0 airy unit.
For quantification of the Cdh1-sfGFP, Cdh2-mCherry, cxcr4a:H2A-mCherry and cxcr4b:H2A-mCherry fluorescence intensities, the embryos were mounted in 0.5% low melt agarose/Ringer′s solution supplemented with 0.4 mg/ml MS-222 anesthetic on a glass coverslips as described above. The images were collected using a 40× water immersion objective (HC PL APO 40×/1.10 W CORR CS2) on a Leica SP5 II laser scanning confocal microscope equipped with HyD hybrid detectors set to photon counting mode with exception of the quantification of the Cdh1-sfGFP fluorescence intensity measurements in cdh2 mutant embryos, which were collected on a Leica SP8 scanning confocal microscope equipped with HyD hybrid detectors. For Cdh1-sfGFP image acquisition, the pinhole was set to 2.1 airy unit. The 488 nm laser was set to 293 μW and the 561 nm laser was set to 198 μW. For Cdh2-mCherry image acquisition, the pinhole was set to 3.1 airy units. The 488 nm laser was set to 23 μW and the 561 nm laser was set to 677 μW. For documenting the co-expression of Cdh1-sfGPF and Cdh2-mCherry, the pinhole was set to 3.1 airy units. The 488 laser was set to 160 μW and the 561 laser was set to 672 μW. For cxcr4a:H2A-mCherry and cxcr4b:H2A-mCherry image acquisition, the pinhole was set to 3.8 airy units. The 488 laser was set to 14 μW and the 561 laser was set to 1.30 mW.
The laser power was calibrated before imaging sessions using an X-Cite Power Meter Model XR2100 (Lumen Dynamics), which measures the power of laser light emitted from the objective onto the stage.
For whole cxcr4a:H2A-mCherry; cldnB:lyn2GFP embryo imaging, embryos were mounted in 0.5% low melt agarose/Ringer′s solution supplemented with 0.4 mg/ml MS-222 anesthetic on a glass coverslips as described above. The embryos were imaged with a Leica SP8 laser scanning confocal microscope using the LAS X Navigator software.
Time lapse videos
Chimeric embryos with membrane-labeled primordium cells (wild-type, cdh1−/−, cdh2−/−, cdh1+/−; cdh2−/− and cdh1−/−; cdh2+/− donors in wild-type or cxcr4b−/− hosts) were mounted in 0.5% low melt agarose/Ringer′s solution (HEPES 5mM, NaCl 111mM, KCl 5mM, CaCl2 1mM, MgSO4 0.6mM) supplemented with 0.4 mg/ml MS-222 anesthetic in a 35 mm petri dish and covered with Ringer′s solution supplemented with 0.4 mg/ml MS-222 anesthetic or on No 1.5 coverslips with ring spacers and sealed with glass slides using Vaseline Petroleum Jelly as a sealant. The embryos were imaged on a Leica SP5 II or Leica SP8 laser scanning confocal microscope equipped with HyD hybrid detectors and a heated stage (Warner Instruments). We used a Leica 40× NA 0.8 objective (HCX APO L 40×/0.80 W U-V-I) to document the migration of chimeric primordia (Video S2) or a Leica 40× NA 1.1 water immersion objective (HC PL APO 40×/1.10 W CORR CS2) to document the nucelar movements and protrusive activity of clones in chimeric primordia (Figure 4G and Videos S3 and S4). The heated stage was set to 28 °C. Images were acquired every 0.5, 15, 20 or 30 min using the HyD detectors in photon counting mode. For wild-type donors in cxcr4b mutant hosts, the pinhole was set to 2.73 airy units. The 488 laser was set to 10% and the 561 laser was set to 25%. For cdh1−/− cdh2−/−, cdh1+/−; cdh2−/−; cdh1−/−; cdh2+/− donors in cxcr4b mutant hosts the pinhole was set to 2.17 airy units. To record the protrusive activity of clones the pinhole was set to 1 airy unit.
Chimeric embryos with labeled primordium nuclei (wild-type, cdh1−/−, cdh2−/− and cxcr4b−/− donors in wild-type hosts) were mounted in 0.5% low melt agarose/Ringer′s solution (HEPES 5mM, NaCl 111mM, KCl 5mM, CaCl2 1mM, MgSO4 0.6mM) supplemented with 0.4 mg/ml MS-222 anesthetic on No 1.5 coverslips with ring spacers and sealed with glass slides. The embryos were imaged on a Leica SP5 II laser scanning confocal microscope equipped with HyD hybrid detectors set to photon counting mode and a 40× NA 1.1 water immersion objective (HC PL APO 40×/1.10 W CORR CS2). Images were acquired every 2 min for a duration of 60 to 120 min starting at 33 to 35 hpf. The pinhole was set to 2.1 airy unit.
For low magnification time lapse of Ctnna1-Citrine-depleted primordium cells, ctnna1:ctnna1-Citrine/ctnna1:ctnna1-Citrine, cxcr4b:zGrad, prim:lyn2mCherry and Ctnna1:ctnna1-Citrine/Ctnna1:ctnna1-Citrine, prim:lyn2mCherry control embryos were mounted in 0.5% low melt agarose/Ringer′s solution (HEPES 5mM, NaCl 111mM, KCl 5mM, CaCl2 1mM, MgSO4 0.6mM) supplemented with 0.4 mg/ml MS-222 anesthetic on a 35 mm petri dish. The embryos were imaged on a Leica SP8 laser scanning confocal microscope equipped with HyD hybrid detectors set to photon counting mode and a Leica 20× NA 0.5 water dipping objective (L 20×/0.5 W U-V-I). Images were acquired every 4 min for a duration of 12 hours. The pinhole was set to 1 airy unit.
For cell tracking analysis of Ctnna1-Citrine-depleted primordium cells, ctnna1:ctnna1-Citrine/ctnna1:ctnna1-Citrine, cxcr4b:zGrad, cxcr4b:H2A-mCherry embryos and ctnna1:ctnna1-Citrine/Ctnna1:ctnna1-Citrine, cxcr4b:H2A-mCherry and ctnna1:ctnna1-Citrine/+; cxcr4b:zGrad, cxcr4b:H2A-mCherry control embryos were mounted in 0.5% low melt agarose/Ringer′s solution (HEPES 5mM, NaCl 111mM, KCl 5mM, CaCl2 1mM, MgSO4 0.6mM) supplemented with 0.4 mg/ml MS-222 anesthetic on No 1.5 coverslips with ring spacers and sealed with glass slides. The embryos were imaged on a Leica SP8 laser scanning confocal microscope equipped with HyD hybrid detectors set to photon counting mode and a Leica 40× NA 1.1 water immersion objective (HC PL APO 40×/1.10 W CORR CS2). Images were acquired every 2 min for a duration of 30 to 200 min. The pinhole was set to 1 airy unit.
For the cell protrusion analysis, embryos with chimeric primordia were mounted in 0.5% low melt agarose/Ringer′s solution (HEPES 5mM, NaCl 111mM, KCl 5mM, CaCl2 1mM, MgSO4 0.6mM) supplemented with 0.4 mg/ml MS-222 anesthetic on No 1.5 coverslips with ring spacers and sealed with glass slides. The embryos were imaged on a Leica SP5 II (wild-type and cxcr4a+/−; cxcr4b−/− donor cells) or a Leica SP8 (cdh1−/− and cdh1−/−; cxcr4b−/− donor cells) laser scanning confocal microscope equipped with HyD detectors set to photon counting mode and a 40× NA 1.1 water immersion objective (HC PL APO 40×/1.10 W CORR CS2). Images were acquired every 30 seconds for a duration of 15 to 30 minutes starting at 33 to 35 hpf. The pinhole was set to 1 or 1.5 airy units.
Image analysis
Migration distance and donor cell number
The somite number where the primordium tip was located was determined based on the overview images collected with a Leica 10× objective (HCX PL NA 0.3). The percent of completed migration was calculated by dividing the somite number where the mosaic primordium′s tip was located by the average of the somite numbers where the wild-type primordium tips were located. The total number of donor cells in the posterior lateral line (neuromasts, interneuromasts and the primordium) was counted using the high resolution images collected with a Leica 40× objective (HC PL APO 40×/1.10 W CORR CS2).
To determine the migration distance of primordia in cdh2−/−; cldnB:lyn2GFP and cxcr4b:cxcr4a, cxcr4b−/−; cldnB:lyn2GFP embryos, the embryos were imaged between 48 and 50 hpf using a Leica M165 FC stereo microscope. The percent of completed migration was determined by dividing the distance between the ear and the primordium′s front by the distance between the ear and the end of the tail. For cdh2−/− embryos and their siblings, the values were normalized to the average migration distance of primordia in wild-type embryos. For primordia in cxcr4b:cxcr4a embryos and their non-transgenic siblings, the values were normalized to the average migration distance of primordia in wild-type embryos.
Cdh1-sfGFP and Cdh2-mCherry fluorescence quantification
For Cdh1-sfGFP fluorescence intensity quantification, we generated a mask of the primordium by thresholding the prim:lyn2mCherry channel and filling the holes to encompass the cytoplasm in addition to the membranes. We multiplied the GFP channel to this mask, removed the zeroes, and plotted the average GFP intensity at each anterior-posterior position against the distance from the front of the primordium. For Cdh2-mCherry fluorescence intensity quantification, we used the same procedure but generated a mask based on the cldnB:lyn2GFP channel. These procedures were automated using a custom-written imageJ script (Methods S1).
cxcr4a and cxcr4b expression quantification
For the quantification of H2A-mCherry expressed from the cxcr4a and cxcr4b promoters, we generated a mask of the primordium by thresholding the cldnB:lyn2GFP channel and filling the holes to encompass the cytoplasm in addition to the membranes. We multiplied the mCherry channel to this mask, removed the zeroes, and plotted the average mCherry intensity at each anterior-posterior position against the distance from the front of the primordium. These procedures were automated using a custom-written imageJ script (Methods S2).
Quantification of cell protrusive activity
For the quantification of the protrusive activity of cell clones in the chimeric primordia, individual Z-stacks of the time lapse videos were maximum projected and subjected to a median filter of one to remove noise and improve edge detection. The time lapse videos were then thresholded and the position of the cell clone′s centroid was registered to the image frame. Extensions and retractions of a cell clone were defined as an increase or a decrease in area compared to the cell clone in the previous image (time interval 30 seconds). Protrusive activity was defined as the sum of the absolute area change of a cell clone between two consecutive time points. The protrusive activity was summed over the entire time lapse and radially resliced from the centroid of the cell clone outward with 0 degrees pointing in the direction of primordium migration (posterior), 90 degrees pointing dorsal, 180 degree pointing anterior and 270 degree pointing ventral [79]. The protrusive activity per degree was summed and normalized to the time length of the video, averaged over all clones and binned in 10 degree intervals and normalized to the total protrusive activity of the cell clones of a given genotype for visualization as a rose diagram (Figure 4G). Since the attractant Cxcl12a is secreted underneath the midline of the primordium, dorsal and ventral primordium cells are oriented differently towards the chemokine. To correct for this, cell clones in the ventral half of the primordium were mirror-imaged along the midline of the primordium to make them comparable to dorsal clones. This procedure was automated using a custom-written imageJ script (Methods S3).
Tracking of the cell nuclei in the primordium
To track the cell nuclei in the primordium, we imported the time lapse videos of chimeric primordia with H2A-GFP labeled donor cells (wild-type, cxcr4b−/−, cdh1−/− or cdh2−/− cells) and H2A-mCherry labeled wild-type host cells into the software Imaris Version 8.0 (Bitplane, Oxford Instruments). As in the original recordings, the dimensions were set to × = 0.378 μm, y = 0.378 μm, z = 1.00 μm and t = 120 s. To track the cell nuclei the spot tool in the Imaris software was used with a specified cell diameter of 3.78 μm and default background subtraction activated. The histogram of the spot quality was adjusted such that about 70 to 80 % of the cells were selected. The cell nuclei were tracked using the autoregressive motion method with a maximum distance that an object can move between two consecutive time points set to 20 μm for the red channel (H2A-mCherry wild-type host cell nuclei) and 10 μm for the green channel (H2A-GFP donor cell nuclei). The maximum gap size - maximum number of consecutive time points that are allowed to be missing in order to join track fragments - was set to two for the red channel and 1 for the green channel. After running the tracking algorithm the filter for track length was set to select 75 to 85 % of the longest tracks and the tracks were manually curated to remove tracked nuclei that do not belong to the primordium or represent dying cells and to delete or correct track segments. For dividing cells, only one daughter cell was tracked. To correct for stage drift a muscle cell nucleus was tracked and chosen as a reference point. The x, y, z, t and track IDs were exported and the position of the reference cells was subtracted from all tracked cells at each time point. This analysis resulted in a set of vectors in three-dimensional space where the vector Pt denotes the xyz-coordinates of each cell nucleus at time point t.
Calculation of normalized average speed
We calculated the velocity vt for t = 1 to n where n denotes the last time point of the cell track according to the following relations:
where ‖·‖ denotes the Euclidean norm in 3D space and Δt denotes the time interval (120 s for all time lapse videos).
Calculation of directionality indices
Since the directionality index depends on the track length [80], we only included tracks with the median track length of 18 time intervals for the calculation of the directionality indices. For tracks with a length of 18 or more time intervals, we selected the first 18 time intervals of the track and treated the remaining track as a new track. The directionality indices of such tracks by normalizing the total distance traveled to the actual distance between start (P1) and end position (P18) which is given by:
Calculation of directional angles
To calculate the directional angle of each cell nucleus with a track length of 2 or more time intervals, we first determined the average displacement (Dt) of all wild-type host cell nuclei (total number of nuclei = n) in the primordium at each time interval (t), which is given by:
The displacement of a single nucleus is given by:
The angular deviation of the displacement of each cell nucleus i at time t from the average wild-type host cell nuclei′s displacement is given by:
and ranges from 0 to π.
The position of a cell in respect to the primordium′s front-back (posterior-anterior) and medial-lateral axes was determined based on the center of mass, Mt, and the overall displacement Dt. The center of mass and the overall displacement define the front-back axis with Mt at the origin and Dt pointing towards the front of the primordium. For each cell i at time t, the distance of the cell to the axis is calculated by
using the function “fminsearch” in MATLAB (Mathworks). The corresponding coordinate on axis of cell i at time t is defined as c‖Dt‖.
Calculation of neighbor-neighbor distances
Cells were defined as neighbors if the distance from the center of mass of their nuclei was smaller than 15 · m at first time point that both cell nuclei were tracked. The distance of neighboring cells was then tracked for 11 time intervals and recorded. Tracks spanning less than 11 time intervals were excluded from the analysis and tracks spanning more than 11 time intervals were cut to include only the first 11 time intervals. If a track spanned 22 time intervals or more the track was cut in two and treated as two independent cell tracks. We chose 11 time intervals for the analysis because this corresponds to the mean track length. In case of chimeric primordia, this analysis was done on the host-host cells, donor-donor cells and host-donor cells. The custom-written MATLAB (Mathworks) and python scripts are available as Methods S4.
Experimental design
All data were obtained from at least two independent experiments. No statistical method was used to predetermine sample size. The experiments were not randomized and the data analysis was not performed blindly. Embryos that were not healthy during data acquisition were excluded from the analysis.
QUANTIFICATION AND STATISTICAL ANALYSIS
The sample size and statistical tests used for each experiment are indicated in the figure panels and legends. The Mann-Whitney test was used to assess if the migration distances of primordia between two different conditions was significantly different. The Kolmogorov-Smirnov test was used to assess if the angle distributions among different genotypes/conditions was significantly different. Kruskal-Wallis one-way analysis of variance was used to asses if neighbor-neighbor distances among different genotypes were significantly different. These analyses were performed using Prism software, version 8 (GraphPad).
DATA AND CODE AVAILABILITY
The codes generated during this study are included in the online version of this report (Methods S1–S4).
Supplementary Material
Methods S4. Zip file containing MatLab and Python scripts for the extraction of the directional angle, neighbor-neighbor distance, speed and directionality index of primordium cells labeled with H2A-fluorescent protein and tracked over time, Related to Figure 4 and STAR Methods.
Video S1. Z-stacks of Cdh1 and Cdh2 protein localization in wild-type and cxcr4b mutant primordia, Related to Figure 3. Z-stack of the primordium from lateral (apical side of primordium) to medial (basal side of primordium) in wild-type (left) and cxcr4b−/− primordia (right) transgenic for cdh1:cdh1-GFP; prim:lyn2mCherry (top), cdh2:cdh2-mCherry; cldnB:lyn2-GFP (middle) and cdh1:cdh1-GFP; cdh2:cdh2-mCherry (bottom). Scale bar = 10 μm, Z-slice = 1.01 μm (top and middle) and 1.51 μm (bottom).
Video S2. Time lapse of cadherin mutant cells in wild-type and cxcr4b mutant primordia, Related to Figure 3. Time lapse of wild-type primordia labeled with the prim:lyn2mCherry transgene (false-colored in green) containing cdh1−/− cells (top row left), cdh2−/− cells (top row right), cdh1−/−; cdh2−/+ cells (second row left) and cdh1−/+; cdh2−/− cells (second row right) labeled with the cldnB:iyn2-GFP transgene (false-colored in red), cxcr4b−/− primordia labeled with the prim:lyn2mCherry transgene (false-colored in green) containing cdh1−/− cells labeled with the cldnB:lyn2-GFP transgene (false-colored in red) (third row left), cxcr4b−/− primordia labeled with the cldnB:lyn2-GFP transgene containing cdh1−/− (third row right), cdh2−/− (bottom row left) and wild-type (bottom row right) cells labeled with the prim:lyn2mCherry transgene. Arrows indicate cadherin deficient cells that are detaching from the primordium. Scale bar = 20 μm, time stamp in min. Each time frame is a maximum projection of an individual Z-stack. The videos start at 33 hpf (top row left), 28 hpf (top and second row right), 31 hpf (second row left) and at 33–34 hpf (third row left, bottom row left, third row right and bottom row right).
Video S3. Time lapse of nuclear movements of wild-type, cxcr4b−/−, cdh1−/− and cdh2−/− cells in wild-type primordia and protrusive activity of wild-type, cxcr4a+/−; cxcr4b−/−, cdh1−/− and cdh1−/−; cxcr4b−/− cell clones in wild-type primordia, Related to Figure 4. Top and middle row. Time lapse videos of mosaic primordia consisting of wild-type host cells (red, top two rows) and wild-type (top row left), cxcr4b−/− (middle row left), cdh1−/− (top row right) and cdh2−/− (middle row right) donor cells (green). H2A-mCherry and H2A-GFP mark host and donor cells, respectively. Each time frame is a maximum projection of an individual Z-stack. Z-stacks were collected every 2 min. Scale bar indicates 20 μm, time stamp is in min. The second set of videos shows another set of examples of chimeric primordia of the same genotypes as the first set of videos.
Bottom row. Time lapse videos of donor-derived wild-type (bottom row left), cxcr4a+/−; cxcr4b−/− (bottom row middle left), cdh1−/− (bottom row middle right) and cdh1−/−; cxcr4b−/− (bottom row right) cells labeled with the cldnB:lyn2-GFP transgene (false-colored in grey) in wild-type host primordia (not shown). The second set of videos in the bottom row are identical to the first set but show the masked cells (blue) registered in space with their extensions (green) and retractions (red) in relation to the previous time point. Scale bar indicates 10 μm. Each frame is a maximum projection of an individual Z-stack. Z-stacks were collected every 30 seconds. The videos start between 33 to 36 hpf.
Video S4. Time lapses of wild-type and Ctnna1 protein-depleted primordia, Related to Figure 4. Time lapse of wild-type (top row left) and Ctnna1-Citrine depleted primordia (top row right, second row right and left) labeled with the prim:lyn2mCherry transgene and ctnna1:ctnna1-Citrine (false-colored in cyan). The second set of videos are identical to the first set but only show the prim:lyn2mCherry transgene (false-colored in grey). All embryos are homozygous for ctnna1:ctnna1-Citrine and embryos carrying the cxcr4b:zGrad transgene are indicated. Scale bar = 100 μm, time stamp in min. Each time frame is a sum projection of an individual Z-stack. The videos start at 27 hpf.
Time lapse of wild-type (third row) and Ctnna1-Citrine depleted primordia (bottom row) whose nuclei are labeled by the cxcr4b:H2A-mCherry transgene (red) and ctnna1:ctnna1-Citrine (false-colored in cyan). The embryo shown in the third row left does not carry the cxcr4b:zGrad transgene and the embryo in the third row right expresses zGrad from the cxcr4b promoter and is heterozygous for ctnna1:ctnna1-Citrine such that it expresses both Citrine-tagged Ctnna1 and untagged Ctnna1. The second set of videos in the bottom two rows are identical to the first set but only show the cxcr4b:H2A-mCherry transgene (false-colored in grey). Scale bar = 50 μm, time stamp in min. Each time frame is a sum projection of an individual Z-stack. The videos start at 36 hpf.
Methods S1. Zip file containing ImageJ-based macros for the extraction of Cdh1-sfGFPand Cdh2-mCherry fluorescence intensities across the primordium, Related to Figure 3 and STAR Methods.
Methods S2. ImageJ-based macro for the extraction of H2A-mCherry fluorescence intensities across the primordium expressed from the cxcr4a and cxcr4b promoters, Related to Figure 2 and STAR Methods.
Methods S3. ImageJ-based macro for the extraction of the protrusive activity of membrane-labeled cell clones in the primordium, Related to Figure 4 and STAR Methods.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-DIG AP | Sigma Aldrich | SKU 11093274910 |
| Bacterial and Virus Strains | ||
| NEB 5-alpha | New England Biolabs | Cat No C2987H |
| SW105 | [75] | N/A |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| NBT | Sigma Aldrich | SKU 11383213001 |
| BCIP | Sigma Aldrich | SKU 11383221001 |
| Digoxigenin-labeled nucleotides | Sigma Aldrich | SKU 11277073910 |
| Paraformaldehyde | Sigma Aldrich | SKU P6148–500G |
| Proteinase K | Sigma Aldrich | SKU P6556–1G |
| Bovine Serum Albumin | Sigma Aldrich | SKU A8022–50g |
| Methycellulose | Sigma Aldrich | SKU 1424506–1G |
| Glycerol | Sigma Aldrich | SKU G7893–1L |
| Tricaine (Ethyl 3-aminobenzoate methanesulfonate) | Sigma Aldrich | SKU E10521–50G |
| Low melt agarose (AquaPor HR) | National Diagnostics | Cat No EC-205 (100g) |
| BsrI | New England Biolabs | R0527S |
| DdeI | New England Biolabs | R0175S |
| Critical Commercial Assays | ||
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| cxcr4bt26035 | [67] | N/A |
| cxcr4aum2 | [68] | N/A |
| cldnb:lyn2GFP | [21] | N/A |
| prim:lyn2mCherry | [73] | N/A |
| cxcl12at30516 | [25] | N/A |
| cxcr4a:H2A-mCherry | This study | N/A |
| cxcr4b:H2A-mCherry | [73] | N/A |
| cxcr4b:cxcr4a | This study | N/A |
| fgf10atbvbo | [72] | N/A |
| fgf3t24149 | [71] | N/A |
| cdh1tx230 | [34] | N/A |
| cdh2m117 | [36] | N/A |
| cdh1:cdh1-sfGFP | [41] | N/A |
| cdh2:cdh2-mCherry | This study | N/A |
| cxcr4b:H2A-GFP | [30] | N/A |
| cxcr4b:zGrad | [41] | N/A |
| Gt(ctnna-citrine)Ct3a | [39] | N/A |
| Oligonucleotides | ||
| For primer sequences used in this study, see Table S1 | Integrated DNA Technologies | Custom synthesis |
| Recombinant DNA | ||
| CHORI-211–75O1 (cdh2 BAC) | BACPAC Resources Center | N/A |
| CHORI73–268G8 (cxcr4a BAC) | BACPAC Resources Center | N/A |
| DKEY169F10 (cxcr4b BAC) | Source Bioscience | N/A |
| Software and Algorithms | ||
| Prism 8 (Version 8.1.2) | Graphpad | |
| FIJI | N/A | https://fiji.sc/ |
| MatLab | Mathworks | https://www.mathworks.com/ |
| Imaris 8.0 | Oxford Instruments | https://www.oxinst.com/ |
| Python 2.7 | N/A | https://www.python.org/ |
| Data S1–S4 | This study | N/A |
| Other | ||
Highlights.
Primordium leader and follower cells need to sense Cxcl12a for efficient migration
Cadherins mediate adhesion between primordium cells
Wild-type cells require Cdh1 to pull attractant-blind neighboring cells
Cadherins and Cxcl12a-sensing mediate cell polarity and primordium directionality
Acknowledgements
We thank S. Lau for critical comments; D. Kane, L. Trinh, D. Gilmour, A. Feitzinger, and the Zebrafish International Resource Center at the University of Oregon, Eugene, OR 97403–5274 (https://zebrafish.org) for reagents; F. Fuentes and T. Gerson for fish care; and M. Goerner and L. Champollion for help with data analysis. The Leica TCS SP5 and SP8 confocal microscopes were supported in part by grant S10 RR024708 from the National Center for Research Resources, NIH. This work was supported by NIH grants R21HD088779 (H.K.) and R01NS102322 (H.K.), New York State Stem Cell Science grant C322560GG (N.Y.) and in part by the intramural program of the National Institute of Allergy and Infectious Diseases (M.M-S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- 1.Friedl P, and Gilmour D (2009). Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol 10, 445–457. [DOI] [PubMed] [Google Scholar]
- 2.Sonnemann KJ, and Bement WM (2011). Wound Repair: Toward Understanding and Integration of Single-Cell and Multicellular Wound Responses. Annu. Rev. Cell Dev. Biol 27, 237–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pocha SM, and Montell DJ (2014). Cellular and Molecular Mechanisms of Single and Collective Cell Migrations in Drosophila: Themes and Variations. Annu. Rev. Genet 48, 295–318. [DOI] [PubMed] [Google Scholar]
- 4.Swaney KF, Huang C-H, and Devreotes PN (2010). Eukaryotic Chemotaxis: A Network of Signaling Pathways Controls Motility, Directional Sensing, and Polarity. Annu. Rev. Biophys 39, 265–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scarpa E, and Mayor R (2016). Collective cell migration in development. J. Cell Biol 212, 143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richardson BE, and Lehmann R (2010). Mechanisms guiding primordial germ cell migration: strategies from different organisms. Nat. Rev. Mol. Cell Biol 11, 37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu P, Ewald AJ, Martin GR, and Werb Z (2008). Genetic mosaic analysis reveals FGF receptor 2 function in terminal end buds during mammary gland branching morphogenesis. Dev. Biol 321, 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shakya R, Watanabe T, and Costantini F (2005). The role of GDNF/Ret signaling in ureteric bud cell fate and branching morphogenesis. Dev. Cell 8, 65–74. [DOI] [PubMed] [Google Scholar]
- 9.Cabernard C, and Affolter M (2005). Distinct Roles for Two Receptor Tyrosine Kinases in Epithelial Branching Morphogenesis in Drosophila. Dev. Cell 9, 831–842. [DOI] [PubMed] [Google Scholar]
- 10.Ghabrial AS, and Krasnow MA (2006). Social interactions among epithelial cells during tracheal branching morphogenesis. Nature 441, 746–749. [DOI] [PubMed] [Google Scholar]
- 11.Duchek P (2001). Guidance of Cell Migration by EGF Receptor Signaling During Drosophila Oogenesis. Science 291, 131–133. [DOI] [PubMed] [Google Scholar]
- 12.Duchek P, Somogyi K, Jékely G, Beccari S, and Rorth P (2001). Guidance of cell migration by the Drosophila PDGF/VEGF receptor. Cell 107, 17–26. [DOI] [PubMed] [Google Scholar]
- 13.Prasad M, and Montell DJ (2007). Cellular and molecular mechanisms of border cellmigration analyzed using time-lapse live-cell imaging. Dev. Cell 12, 997–1005. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, He L, Wu YI, Hahn KM, and Montell DJ (2010). Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat. Cell Biol 12, 591–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vishwakarma M, Russo J, Probst D, Schwarz US, Das T, and Spatz JP (2018). Mechanical interactions among followers determine the emergence of leaders in migrating epithelial cell collectives. Nat. Commun 9, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellison D, Mugler A, Brennan MD, Lee SH, Huebner RJ, Shamir ER, Woo LA, Kim J, Amar P, Nemenman I, et al. (2016). Cell-cell communication enhances the capacity of cell ensembles to sense shallow gradients during morphogenesis. Proc. Natl. Acad. Sci. USA 113, E679–E688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vitorino P, and Meyer T (2008). Modular control of endothelial sheet migration. Genes Dev 22, 3268–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reffay M, Parrini MC, Cochet-Escartin O, Ladoux B, Buguin A, Coscoy S, Amblard F, Camonis J, and Silberzan P (2014). Interplay of RhoA and mechanical forces in collective cell migration driven by leader cells. Nat. Cell Biol 16, 217–223. [DOI] [PubMed] [Google Scholar]
- 19.Riahi R, Sun J, Wang S, Long M, Zhang DD, and Wong PK (2015). Notch1-Dll4 signalling and mechanical force regulate leader cell formation during collective cell migration. Nat. Commun 6, 6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.David NB, Sapède D, Saint-Etienne L, Thisse C, Thisse B, Dambly-Chaudière C, Rosa FM, and Ghysen A (2002). Molecular basis of cell migration in the fish lateral line: role of the chemokine receptor CXCR4 and of its ligand, SDF1. Proc. Natl. Acad. Sci. USA 99, 16297–16302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haas P, and Gilmour D (2006). Chemokine signaling mediates self-organizing tissue migration in the zebrafish lateral line. Dev. Cell 10, 673–680. [DOI] [PubMed] [Google Scholar]
- 22.Venkiteswaran G, Lewellis SW, Wang J, Reynolds E, Nicholson C, and Knaut H (2013). Generation and Dynamics of an Endogenous, Self-Generated Signaling Gradient across a Migrating Tissue. Cell 155, 674–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doná E, Barry JD, Valentin G, Quirin C, Khmelinskii A, Kunze A, Durdu S, Newton LR, Fernandez-Minan A, Huber W, et al. (2013). Directional tissue migration through a self-generated chemokine gradient. Nature 503, 285–289. [DOI] [PubMed] [Google Scholar]
- 24.Dambly-Chaudière C, Cubedo N, and Ghysen A (2007). Control of cell migration in the development of the posterior lateral line: antagonistic interactions between the chemokine receptors CXCR4 and CXCR7/RDC1. BMC Dev. Biol 7 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valentin G, Haas P, and Gilmour D (2007). The chemokine SDF1a coordinates tissue migration through the spatially restricted activation of Cxcr7 and Cxcr4b. Curr. Biol 17, 1026–1031. [DOI] [PubMed] [Google Scholar]
- 26.Siekmann AF, Standley C, Fogarty KE, Wolfe SA, and Lawson ND (2009). Chemokine signaling guides regional patterning of the first embryonic artery. Genes Dev 23, 2272–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boldajipour B, Doitsidou M, Tarbashevich K, Laguri C, Yu SR, Ries J, Dumstrei K, Thelen S, Dörries J, Messerschmidt E-M, et al. (2011). Cxcl12 evolution--subfunctionalization of a ligand through altered interaction with the chemokine receptor. Development 138, 2909–2914. [DOI] [PubMed] [Google Scholar]
- 28.Malhotra D, Shin J, Solnica-Krezel L, and Raz E (2018). Spatio-temporal regulation of concurrent developmental processes by generic signaling downstream of chemokine receptors. eLife 7, 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mcgraw HF, Drerup CM, Culbertson MD, Linbo T, Raible DW, and Nechiporuk AV (2011). Lef1 is required for progenitor cell identity in the zebrafish lateral line primordium. Development 138, 3921–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozlovskaja-Gumbrienė A, Yi R, Alexander R, Aman A, Jiskra R, Nagelberg D,Knaut H, McClain M, and Piotrowski T (2017). Proliferation-independent regulation of organ size by Fgf/Notch signaling. eLife 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Revenu C, Streichan S, Dona E, Lecaudey V, Hufnagel L, and Gilmour D (2014). Quantitative cell polarity imaging defines leader-to-follower transitions during collective migration and the key role of microtubule-dependent adherens junction formation. Development 141, 1282–1291. [DOI] [PubMed] [Google Scholar]
- 32.Matsuda M, and Chitnis AB (2010). Atoh1a expression must be restricted by Notch signaling for effective morphogenesis of the posterior lateral line primordium in zebrafish. Development 137, 3477–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Q, Ensign RD, and Azodi E (2003). Cadherin-1, −2 and −4 expression in the cranial ganglia and lateral line system of developing zebrafish. Gene Expr. Patterns 3, 653–658. [DOI] [PubMed] [Google Scholar]
- 34.Kane DA, McFarland KN, and Warga RM (2005). Mutations in half baked/E-cadherin block cell behaviors that are necessary for teleost epiboly. Development 132, 1105–1116. [DOI] [PubMed] [Google Scholar]
- 35.Lele Z, Folchert A, Concha M, Rauch G-J, Geisler R, Rosa F, Wilson SW, Hammerschmidt M, and Bally-Cuif L (2002). parachute/n-cadherin is required for morphogenesis and maintained integrity of the zebrafish neural tube. Development 129, 3281–3294. [DOI] [PubMed] [Google Scholar]
- 36.Malicki J, Jo H, and Pujic Z (2003). Zebrafish N-cadherin, encoded by the glass onion locus, plays an essential role in retinal patterning. Dev. Biol 259, 95–108. [DOI] [PubMed] [Google Scholar]
- 37.Kerstetter AE, Azodi E, Marrs JA, and Liu Q (2004). Cadherin-2 function in the cranial ganglia and lateral line system of developing zebrafish. Dev. Dyn 230, 137–143. [DOI] [PubMed] [Google Scholar]
- 38.López-Schier H, Starr CJ, Kappler JA, Kollmar R, and Hudspeth AJ (2004). Directional cell migration establishes the axes of planar polarity in the posterior lateral-line organ of the zebrafish. Dev. Cell 7, 401–412. [DOI] [PubMed] [Google Scholar]
- 39.Trinh LA, Hochgreb T, Graham M, Wu D, Ruf-Zamojski F, Jayasena CS, Saxena A, Hawk R, Gonzalez-Serricchio A, Dixson A, et al. (2011). A versatile gene trap to visualize and interrogate the function of the vertebrate proteome. Genes Dev 25, 2306–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caussinus E, Kanca O, and Affolter M (2011). Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat. Struct. Mol. Biol 19, 117–121. [DOI] [PubMed] [Google Scholar]
- 41.Yamaguchi N, colak-Champollion T, and Knaut H (2019). zGrad is a nanobody-based degron system that inactivates proteins in zebrafish. eLife 8, 4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leckband DE, and de Rooij J (2014). Cadherin Adhesion and Mechanotransduction. Annu. Rev. Cell Dev. Biol 30, 291–315. [DOI] [PubMed] [Google Scholar]
- 43.Vedula SRK, Leong MC, Lai TL, Hersen P, Kabla AJ, Lim CT, and Ladoux B (2012). Emerging modes of collective cell migration induced by geometrical constraints. Proc. Natl. Acad. Sci. USA 109, 12974–12979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsuzawa K, Himoto T, Mochizuki Y, and Ikenouchi J (2018). α-Catenin Controls the Anisotropy of Force Distribution at Cell-Cell Junctions during Collective Cell Migration. Cell Rep 23, 3447–3456. [DOI] [PubMed] [Google Scholar]
- 45.Chen C-S, Hong S, Indra I, Sergeeva AP, Troyanovsky RB, Shapiro L, Honig B, and Troyanovsky SM (2015). α-Catenin-mediated cadherin clustering couples cadherin and actin dynamics. J. Cell Biol 210, 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Desai R, Sarpal R, Ishiyama N, Pellikka M, Ikura M, and Tepass U (2013). Monomeric α-catenin links cadherin to the actin cytoskeleton. Nat. Cell Biol 15, 261–273. [DOI] [PubMed] [Google Scholar]
- 47.Kim T-J, Zheng S, Sun J, Muhamed I, Wu J, Lei L, Kong X, Leckband DE, and Wang Y (2015). Dynamic Visualization of alpha;-Catenin Reveals Rapid, Reversible Conformation Switching between Tension States. Curr. Biol 25, 218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weber GF, Bjerke MA, and DeSimone DW (2012). A Mechanoresponsive Cadherin-Keratin Complex Directs Polarized Protrusive Behavior and Collective Cell Migration. Dev. Cell 22, 104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petrie RJ, Doyle AD, and Yamada KM (2009). Random versus directionally persistent cell migration. Nat. Rev. Mol. Cell. Biol 10, 538–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rørth P (2011). Whence directionality: guidance mechanisms in solitary and collective cell migration. Dev. Cell 20, 9–18. [DOI] [PubMed] [Google Scholar]
- 51.Malet-Engra G, Yu W, Oldani A, Rey-Barroso J, Gov NS, Scita G, and Dupré L (2015). Collective Cell Motility Promotes Chemotactic Prowessand Resistance to Chemorepulsion. Curr. Biol 25, 242–250. [DOI] [PubMed] [Google Scholar]
- 52.Trepat X, Wasserman MR, Angelini TE, Millet E, Weitz DA, Butler JP, and Fredberg JJ (2009). Physical forces during collective cell migration. Nat. Phys 5, 426–430. [Google Scholar]
- 53.Kim JH, Serra-Picamal X, Tambe DT, Zhou EH, Park CY, Sadati M, Park J-A, Krishnan R, Gweon B, Millet E, et al. (2013). Propulsion and navigation within the advancing monolayer sheet. Nat. Mater 12, 856–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Camand E, Peglion F, Osmani N, Sanson M, and Etienne-Manneville S (2012). N-cadherin expression level modulates integrin-mediated polarity and strongly impacts on the speed and directionality of glial cell migration. J. Cell. Sci 125, 844–857. [DOI] [PubMed] [Google Scholar]
- 55.Ng MR, Besser A, Danuser G, and Brugge JS (2012). Substrate stiffness regulates cadherin-dependent collective migration through myosin-II contractility. J. Cell Biol 199, 545–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bazellières E, Conte V, Elosegui-Artola A, Serra-Picamal X, Bintanel-Morcillo M, Roca-Cusachs P, Muñoz JJ, Sales-Pardo M, Guimerà R, and Trepat X (2015). Control of cell-cell forces and collective cell dynamics by the intercellular adhesome. Nature 17, 409–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seddiki R, Narayana GHNS, Strale P-O, Balcioglu HE, Peyret G, Yao M, Le AP, Teck Lim C, Yan J, Ladoux B, et al. (2018). Force-dependent binding of vinculin to α-catenin regulates cell-cell contact stability and collective cell behavior. Mol. Biol. Cell 29, 380–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scarpa E, Szabó A, Bibonne A, Theveneau E, Parsons M, and Mayor R (2015). Cadherin Switch during EMT in Neural Crest Cells Leads to Contact Inhibition of Locomotion via Repolarization of Forces. Dev. Cell 34, 421–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuriyama S, Theveneau E, Benedetto A, Parsons M, Tanaka M, Charras G, Kabla A, and Mayor R (2014). In vivo collective cell migration requires an LPAR2-dependent increase in tissue fluidity. J. Cell Biol 206, 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Theveneau E, Marchant L, Kuriyama S, Gull M, Moepps B, Parsons M, and Mayor R (2010). Collective chemotaxis requires contact-dependent cell polarity. Dev. Cell 19, 39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Theveneau E, Steventon B, Scarpa E, Garcia S, Trepat X, Streit A, and Mayor R (2013). Chase-and-run between adjacent cell populations promotes directional collective migration. Nat. Cell Biol 15, 763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arboleda-Estudillo Y, Krieg M, StUhmer J, Licata NA, Muller DJ, and Heisenberg C-P (2010). Movement Directionality in Collective Migration of Germ Layer Progenitors. Curr. Biol 20, 161–169. [DOI] [PubMed] [Google Scholar]
- 63.Cai D, Chen S-C, Prasad M, He L, Wang X, Choesmel-Cadamuro V, Sawyer JK, Danuser G, and Montell DJ (2014). Mechanical Feedback through E-Cadherin Promotes Direction Sensingduring Collective Cell Migration. Cell 157, 1146–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hayer A, Shao L, Chung M, Joubert L-M, Yang HW, Tsai F-C, Bisaria A, Betzig E, and Meyer T (2016). Engulfed cadherin fingers are polarized junctional structures between collectively migrating endothelial cells. Nat. Cell Biol 18, 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Das T, Safferling K, Rausch S, Grabe N, Boehm H, and Spatz JP (2015). A molecular mechanotransduction pathway regulates collective migration of epithelial cells. Nat. Cell Biol 17 276–287. [DOI] [PubMed] [Google Scholar]
- 66.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, and Schilling TF (1995). Stages of embryonic development of the zebrafish. Dev. Dyn 203, 253–310. [DOI] [PubMed] [Google Scholar]
- 67.Knaut H, Werz C, Geisler R, Nüsslein-Volhard C, and Tübingen 2000 Screen Consortium (2003). A zebrafish homologue of the chemokine receptor Cxcr4 is a germ-cell guidance receptor. Nature 421, 279–282. [DOI] [PubMed] [Google Scholar]
- 68.Bussmann J, Wolfe SA, and Siekmann AF (2011). Arterial-venous network formation during brain vascularization involves hemodynamic regulation of chemokine signaling. Development 138, 1717–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jiang YJ, Brand M, Heisenberg C-P, Beuchle D, Furutani-Seiki M, Kelsh RN, Warga RM, Granato M, Haffter P, Hammerschmidt M, et al. (1996). Mutations affecting neurogenesis and brain morphology in the zebrafish, Danio rerio. Development 123, 205–216. [DOI] [PubMed] [Google Scholar]
- 70.Malicki J, Neuhauss SC, Schier AF, Solnica-Krezel L, Stemple DL, Stainier DY, Abdelilah S, Zwartkruis F, Rangini Z, and Driever W (1996). Mutations affecting development of the zebrafish retina. Development 123, 263–273. [DOI] [PubMed] [Google Scholar]
- 71.Herzog W, Sonntag C, Hardt, von der S, Roehl HH, Varga ZM, and Hammerschmidt M (2004). Fgf3 signaling from the ventral diencephalon is required for early specification and subsequent survival of the zebrafish adenohypophysis. Development 131, 3681–3692. [DOI] [PubMed] [Google Scholar]
- 72.Norton WHJ, Ledin J, Grandel H, and Neumann CJ (2005). HSPG synthesis by zebrafish Ext2 and Extl3 is required for Fgf10 signalling during limb development. Development 132, 4963–4973. [DOI] [PubMed] [Google Scholar]
- 73.Wang J, Yin Y, Lau S, sankaran J, Rothenberg E, Wohland T, Meier-Schellersheim M, and Knaut H (2018). Anosmin1 Shuttles Fgf to Facilitate Its Diffusion, Increase Its Local Concentration, and Induce Sensory Organs. Dev. Cell 46, 751–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fuentes F, Reynolds E, Lewellis SW, Venkiteswaran G, and Knaut H (2016). A Plasmid Set for Efficient Bacterial Artificial Chromosome (BAC) Transgenesis in Zebrafish. G3. 6(4):829–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Warming S, Costantino N, Court DL, Jenkins NA, and Copeland NG (2005). Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Research 33, 24;33(4):e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, and Chien C-B (2007). The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn 236, 3088–3099. [DOI] [PubMed] [Google Scholar]
- 77.Thisse C, and Thisse B (2008). High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc 3, 59–69. [DOI] [PubMed] [Google Scholar]
- 78.Knaut H, Blader P, Strähle U, and Schier AF (2005). Assembly of trigeminal sensory ganglia by chemokine signaling. Neuron 47, 653–666. [DOI] [PubMed] [Google Scholar]
- 79.Soll DR, Wessels D, Heid PJ, and Voss E (2003). Computer-assisted reconstruction and motion analysis of the three-dimensional cell. Sci. World J 3, 827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gorelik R, and Gautreau A (2014). Quantitative and unbiased analysis of directional persistence in cell migration. Nat. Protoc 9, 1931–1943. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Methods S4. Zip file containing MatLab and Python scripts for the extraction of the directional angle, neighbor-neighbor distance, speed and directionality index of primordium cells labeled with H2A-fluorescent protein and tracked over time, Related to Figure 4 and STAR Methods.
Video S1. Z-stacks of Cdh1 and Cdh2 protein localization in wild-type and cxcr4b mutant primordia, Related to Figure 3. Z-stack of the primordium from lateral (apical side of primordium) to medial (basal side of primordium) in wild-type (left) and cxcr4b−/− primordia (right) transgenic for cdh1:cdh1-GFP; prim:lyn2mCherry (top), cdh2:cdh2-mCherry; cldnB:lyn2-GFP (middle) and cdh1:cdh1-GFP; cdh2:cdh2-mCherry (bottom). Scale bar = 10 μm, Z-slice = 1.01 μm (top and middle) and 1.51 μm (bottom).
Video S2. Time lapse of cadherin mutant cells in wild-type and cxcr4b mutant primordia, Related to Figure 3. Time lapse of wild-type primordia labeled with the prim:lyn2mCherry transgene (false-colored in green) containing cdh1−/− cells (top row left), cdh2−/− cells (top row right), cdh1−/−; cdh2−/+ cells (second row left) and cdh1−/+; cdh2−/− cells (second row right) labeled with the cldnB:iyn2-GFP transgene (false-colored in red), cxcr4b−/− primordia labeled with the prim:lyn2mCherry transgene (false-colored in green) containing cdh1−/− cells labeled with the cldnB:lyn2-GFP transgene (false-colored in red) (third row left), cxcr4b−/− primordia labeled with the cldnB:lyn2-GFP transgene containing cdh1−/− (third row right), cdh2−/− (bottom row left) and wild-type (bottom row right) cells labeled with the prim:lyn2mCherry transgene. Arrows indicate cadherin deficient cells that are detaching from the primordium. Scale bar = 20 μm, time stamp in min. Each time frame is a maximum projection of an individual Z-stack. The videos start at 33 hpf (top row left), 28 hpf (top and second row right), 31 hpf (second row left) and at 33–34 hpf (third row left, bottom row left, third row right and bottom row right).
Video S3. Time lapse of nuclear movements of wild-type, cxcr4b−/−, cdh1−/− and cdh2−/− cells in wild-type primordia and protrusive activity of wild-type, cxcr4a+/−; cxcr4b−/−, cdh1−/− and cdh1−/−; cxcr4b−/− cell clones in wild-type primordia, Related to Figure 4. Top and middle row. Time lapse videos of mosaic primordia consisting of wild-type host cells (red, top two rows) and wild-type (top row left), cxcr4b−/− (middle row left), cdh1−/− (top row right) and cdh2−/− (middle row right) donor cells (green). H2A-mCherry and H2A-GFP mark host and donor cells, respectively. Each time frame is a maximum projection of an individual Z-stack. Z-stacks were collected every 2 min. Scale bar indicates 20 μm, time stamp is in min. The second set of videos shows another set of examples of chimeric primordia of the same genotypes as the first set of videos.
Bottom row. Time lapse videos of donor-derived wild-type (bottom row left), cxcr4a+/−; cxcr4b−/− (bottom row middle left), cdh1−/− (bottom row middle right) and cdh1−/−; cxcr4b−/− (bottom row right) cells labeled with the cldnB:lyn2-GFP transgene (false-colored in grey) in wild-type host primordia (not shown). The second set of videos in the bottom row are identical to the first set but show the masked cells (blue) registered in space with their extensions (green) and retractions (red) in relation to the previous time point. Scale bar indicates 10 μm. Each frame is a maximum projection of an individual Z-stack. Z-stacks were collected every 30 seconds. The videos start between 33 to 36 hpf.
Video S4. Time lapses of wild-type and Ctnna1 protein-depleted primordia, Related to Figure 4. Time lapse of wild-type (top row left) and Ctnna1-Citrine depleted primordia (top row right, second row right and left) labeled with the prim:lyn2mCherry transgene and ctnna1:ctnna1-Citrine (false-colored in cyan). The second set of videos are identical to the first set but only show the prim:lyn2mCherry transgene (false-colored in grey). All embryos are homozygous for ctnna1:ctnna1-Citrine and embryos carrying the cxcr4b:zGrad transgene are indicated. Scale bar = 100 μm, time stamp in min. Each time frame is a sum projection of an individual Z-stack. The videos start at 27 hpf.
Time lapse of wild-type (third row) and Ctnna1-Citrine depleted primordia (bottom row) whose nuclei are labeled by the cxcr4b:H2A-mCherry transgene (red) and ctnna1:ctnna1-Citrine (false-colored in cyan). The embryo shown in the third row left does not carry the cxcr4b:zGrad transgene and the embryo in the third row right expresses zGrad from the cxcr4b promoter and is heterozygous for ctnna1:ctnna1-Citrine such that it expresses both Citrine-tagged Ctnna1 and untagged Ctnna1. The second set of videos in the bottom two rows are identical to the first set but only show the cxcr4b:H2A-mCherry transgene (false-colored in grey). Scale bar = 50 μm, time stamp in min. Each time frame is a sum projection of an individual Z-stack. The videos start at 36 hpf.
Methods S1. Zip file containing ImageJ-based macros for the extraction of Cdh1-sfGFPand Cdh2-mCherry fluorescence intensities across the primordium, Related to Figure 3 and STAR Methods.
Methods S2. ImageJ-based macro for the extraction of H2A-mCherry fluorescence intensities across the primordium expressed from the cxcr4a and cxcr4b promoters, Related to Figure 2 and STAR Methods.
Methods S3. ImageJ-based macro for the extraction of the protrusive activity of membrane-labeled cell clones in the primordium, Related to Figure 4 and STAR Methods.
Data Availability Statement
The codes generated during this study are included in the online version of this report (Methods S1–S4).