Abstract
Drug-resistant strains of malaria parasites have emerged for most of antimalarial medications. A new chemotherapeutic compound is needed for malarial therapy. Antimalarial activity against both drug-sensitive and drug-resistant P. falciparum has been reported for an isocryptolepine derivative, 8-bromo-2-fluoro-5-methyl-5H-indolo[3,2-c]quinoline (ICL-M), which also showed less toxicity to human cells. ICL-M has indoloquinoline as a core structure and its mode of action remains unclear. Here, we explored the mechanisms of ICL-M in P. falciparum by assessing the stage-specific activity, time-dependent effect, a proteomic analysis and morphology. Since human topo II activity inhibition has been reported as a function of isocryptolepine derivatives, malarial topo II activity inhibition of ICL-M was also examined in this study. The ICL-M exhibited antimalarial activity against both the ring and trophozoite stages of P. falciparum. Our proteomics analysis revealed that a total of 112 P. falciparum proteins were differentially expressed after ICL-M exposure; among these, 58 and 54 proteins were upregulated and downregulated, respectively. Proteins localized in the food vacuole, nucleus, and cytoplasm showed quantitative alterations after ICL-M treatment. A bioinformatic analysis revealed that pathways associated with ribosomes, proteasomes, metabolic pathways, amino acid biosynthesis, oxidative phosphorylation, and carbon metabolism were significantly different in P. falciparum treated with ICL-M. Moreover, a loss of ribosomes was clearly observed by transmission electron microscopy in the ICL-M-treated P. falciparum. This finding is in agreement with the proteomics data, which revealed downregulated levels of ribosomal proteins following ICL-M treatment. Our results provide important information about the mechanisms by which ICL-M affects the malaria parasite, which may facilitate the drug development of isocryptolepine derivatives.
Introduction
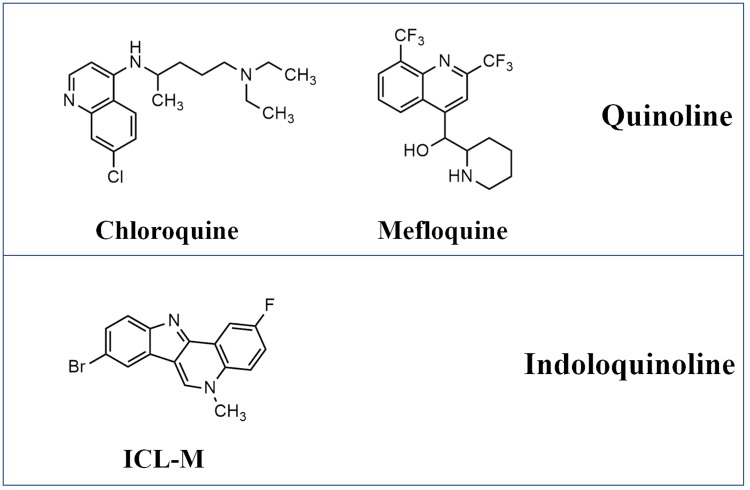
Over the last decades, antimalarial drug resistance in Plasmodium falciparum has presented an obstacle for the malaria elimination program of the World Health Organization (WHO) [1]. Many studies have attempted to discover a new generation of antimalarial drugs. Herbal medicine has been the most common source of new candidate antimalarial compounds. Cryptolepis sanguinolenta is a Ghanaian traditional medicine used for malarial treatment. A report in 1989 showed that the aqueous root extract of this plant can successfully treat uncomplicated malaria in Ghana [2]. Isocryptolepine was first isolated from the roots of C. sanguinolenta in 1995, and its structure was determined to be 5-methyl-5H-indolo[3,2-c]quinoline. Isocryptolepine was later synthesized and tested for potential antimalarial properties. The results demonstrated that isocryptolepine possesses activity against both drug-sensitive and drug-resistance strains of P. falciparum [3–5]. Several strategies have been used to design and construct isocryptolepine derivatives with improved antimalarial activity [3–6]. In 2015, an isocryptolepine derivative containing fluorine and bromine, 8-bromo-2-fluoro-5-methyl-5H-indolo[3,2-c]quinoline (ICL-M), was reported to display an improved antimalarial activity against both drug-sensitive and drug-resistant strains and to induce less toxicity in human cells compared with the original isocryptolepine. The half maximal inhibitory concentration (IC50) of ICL-M in normal Homo sapiens lung cells was 121,944 nM, whereas the IC50 of chloroquine-resistant and mefloquine-resistant strains of P. falciparum were 132 and 211 nM, respectively [5]. Additionally, furfural, the agricultural byproduct, was found to possess the ability to be converted to isocryptolepine [7], suggesting the possibility of a low-cost synthesis process for this compound. Although ICL-M has indoloquinoline as its core structure, it could kill quinoline-resistant P. falciparum such as chloroquine-resistant and mefloquine-resistant strains [5]. Therefore, ICL-M may have different mechanisms for killing malaria parasite. The structure of chloroquine, mefloquine and ICL-M are shown in Fig 1. Isocryptolepine derivatives have been reported to target human topoisomerase II (topo II) in several different types of cancer cells [8]. However, the targets and modes of action of ICL-M in malaria parasites remain unclear. Understanding the molecular mechanism of ICL-M against malaria parasites may provide important insight into the mode of action for isocryptolepine derivatives as well as guidance for further structure development.
Fig 1. The structures of chloroquine, mefloquine and ICL-M.
Proteomics is a high-throughput protein analysis that can be used to reveal the biological activity in parasites. Quantitative proteomics is a useful tool with which to study parasite gene expression during both the asexual and sexual stages [9]. This method has been applied to several studies of the host response to parasites [10, 11], the parasite response to its host [12–14], and drug target identification [15, 16]. In drug development, the differential protein expression between treated and non-treated parasites is commonly used as part of the pharmacodynamics information for a compound on parasite cells. Polyacrylamide gel electrophoresis (PAGE) coupled with mass spectrometry (MS) is a crucial method for identifying and quantifying proteins from large-scale antimalarial drug studies [17–19]. In malarial proteomics research, label-free quantification, such as the exponentially modified protein abundance index [20], has been applied for various types of samples, spanning the cultural blood stage [21], clinical blood stage [22], and mosquito stage [23]. Here, we aimed to apply a proteomics approach to investigating the mechanism of ICL-M against P. falciparum. The protein profile resulting from ICL-M exposure may provide crucial information for understanding the mechanism of isocryptolepine derivatives against malaria parasites and further facilitate antimalarial drug development.
Methods
1. Cultivation of P. falciparum strain 3D7
P. falciparum 3D7 was cultured using O+ human erythrocytes in culture medium containing RPMI 1640 (Gibco, New York, USA) supplemented with 10% human serum, 2% sodium bicarbonate (Merck, New Jersey, USA), and 40 mg/L gentamycin sulphate (GPO, Bangkok, Thailand). Parasite culture was maintained at 37 °C under mixed gas (5% CO2, 5% O2, and 90% N2). The culture media was changed daily. Parasite growth was monitored by Giemsa-stained thin blood smears. Culture with mixed stages of P. falciparum was treated with 5% sorbitol (Amresco, Pennsylvania, USA) for 30 min at 37 °C for synchronization. After RPMI 1640 was used to wash the treated parasites, the parasite culture was continued [24]
2. Antimalarial activity assay
ICL-M was synthesized following the protocol developed by Aroonkit and colleagues in 2015 [5]. The compound was dissolved in dimethyl sulfoxide (DMSO) (Merck) to prepare a 10 mM stock concentration. The IC50 and ninety percent inhibitory concentration (IC90) were assessed to evaluate the antimalarial activity of ICL-M. Briefly, the compound was prepared at various concentrations in a 96-well plate. Synchronized parasites at 1% parasitemia (ring form) and 2% hematocrit were then loaded into the wells. All tests were performed in triplicate. The plate was further cultured for 24 h prior to [3H]hypoxanthine (PerkinElmer, Massachusetts, USA) labelling. After incubation for another 24 h, the surviving parasites were measured by radiolabeling. The inhibitory concentration was determined by an analysis of the dose-response curves calculated using GRAFIT software (Erithacus Software, Kent, UK).
3. Stage-specific antimalarial activity
Stage-specific antimalarial activity was analyzed. Ring (6–16 h after invasion) and trophozoite (30–34 h after invasion) stages were identified based on the criteria previously described by Silamut and colleagues [25]. Untreated and 450 nM ICL-M (three-fold the IC50)-treated parasites at 1% parasitemia in 2% hematocrit were incubated at 37 °C for 24 or 48 h. After incubation, the parasitemia percentages were estimated by Giemsa-stained thin blood smear visualized by microscopy.
4. Parasite topoisomerase II activity assay
Red blood cells (RBCs) infected with the trophozoite stage of P. falciparum were washed with 10 ml of RPMI 1640. Parasites were released from the RBCs by treatment with 0.15% saponin (Amresco). Free parasites were then washed three times with 10 ml of cold phosphate-buffered saline (PBS) (Merck). The resulting pellets were mixed with 2 ml of lysis buffer containing 20 mM HEPES [pH 7.8], 10 mM potassium chloride (KCl), 1 mM EDTA, 1 mM dithiothreitol (DTT), 1 mM phenylmethanesulfonyl fluoride (PMSF), and 1% Triton X-100 (all components from Merck). The mixture was incubated on ice for 5 min, after which the parasite nuclei were isolated using a modified protocol from Voss and colleagues [26]. Nuclear proteins were extracted using a Topoisomerase Type IIα Nuclear Extraction/Assay Kit (TopoGen Inc., Colorado, USA). The concentration of nuclear proteins was measured by a Bradford Assay (BioRAD Inc., California, USA) [27]. The solution used for the P. falciparum topoisomerase II (topo II) activity assay was prepared from 10 μg of nuclear proteins, 100 ng of kinetoplast DNA, 1× TopoII reaction buffer, and the test compounds (ICL-M or pyronaridine) at a 100 μM final concentration. The reaction was incubated for 30 min at 37 °C. After incubation, each sample was mixed with Tris-Borate-EDTA (TBE) buffer and GelRed solution (Biotium Inc., California, USA). The topo II reaction samples were loaded into a 1% agarose gel and subjected to electrophoresis, after which the gel was visualized by Gel Documentary (BioRAD Inc.).
5. Time-dependent effect of antimalarial activity
Synchronized trophozoite stage P. falciparum was prepared at 2% parasitemia and 2% hematocrit. The culture was separated into three groups: untreated, <0.01% DMSO-treated, and 250 nM of ICL-M (IC90)-treated parasites. The cultures were started using 5 ml, and 400-μl samples were taken at each timepoint. After 24 h, the infected RBCs were washed three times with RPMI 1640 and resuspended in 400 μl of culture medium. A 100-μl aliquot of each sample was taken and used for continued culture in 96-well plates, individually (in triplicate). Surviving parasites were quantified by [3H]hypoxanthine labelling. Parasitic growth was determined by measurement of ionizing radiation.
6. Protein preparation
The <0.01% DMSO- and 250 nM of ICL-M (IC90)-treated trophozoite stage parasites were cultured for 4 h (in three biological replicates). Total RBCs were collected, washed with RPMI 1640, and centrifuged at 500 ×g at room temperature for 5 min. Parasites were released from the RBCs by the addition of 0.15% saponin in PBS buffer followed by an incubation at 4 °C for 5 min. Free parasites were washed three times with ice-cold PBS, then centrifuged at 2,500 ×g at 4 °C for 10 min. The resulting parasite pellets were kept at −80 °C until use.
7. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE)
Parasite proteins were extracted by adding lysis buffer (1% SDS, 1% Triton-X, and 0.5% NaCl [all components from Merck]) and rotated at 4 °C for 1 h. The lysate was centrifuged at 10,000 ×g at 4 °C for 10 min, after which the resulting supernatant was collected. The protein concentration of each sample was measured using a Bradford assay (BioRAD Inc.) [27]. Proteins (30 μg per sample) were separated by 12% SDS-PAGE. The gel was stained by Coomassie blue R (BioRAD Inc.) and de-stained in 45% methanol (Merck) and 10% acetic acid (Merck). Each lane was sliced into 16 pieces and subjected to tryptic digestion.
8. In-gel digestion
Gel pieces were de-stained with 50% acetonitrile in 50 mM ammonium bicarbonate (Merck). The disulfide bonds in the proteins were reduced by treatment with 4 mM DTT in 50 mM ammonium bicarbonate at 60 °C for 15 min. Subsequently, the proteins were alkylated by incubation with 250 mM iodoacetamide at room temperature in the dark for 30 min. The reaction was quenched by 4 mM DTT in 50 mM ammonium bicarbonate for 5 min at room temperature. All solutions were removed, and the gel pieces were dehydrated by acetonitrile. The gel pieces were then air dried and resuspended in 0.1 μg/μl of Trypsin Proteomics Grade (Sigma Aldrich, Missouri, USA) in 50 mM ammonium bicarbonate. The digestion was performed overnight at 37 °C. The digested peptides were extracted by acetonitrile and dried using a centrifugal evaporator.
9. Mass spectrometry
Peptide mixtures were resuspended in 0.1% formic acid. Each sample was injected into the UltiMate 3000 nano-liquid chromatography (nano-LC) system (Dionex, Surrey, UK). Peptide separation was performed using a C18 column at a flow rate of 300 nL/min. Mobile phase A was 0.1% formic acid in water. Mobile phase B was 80% acetonitrile in 0.1% formic acid. The elution occurred during the 30-min gradient from 4% mobile phase B to 50% mobile phase A and infused to a micrOTOF-Q (Bruker Daltonics, Bremen, Germany). The mass spectra from the mass spectrometry (MS) and tandem mass spectrometry (MS/MS) covered mass ranges of m/z 400–2000 and m/z 50–1500, respectively. A mascot generic file (.mgf) was generated using DataAnalysis 3.4 version software. Mascot daemon version 2.3.2 (Matrix Science, London, UK) was used to merge the .mgf files and identify the proteins. Identification and quantification of the proteins were performed against a NCBInr database (24 October 2018) specific to P. falciparum 3D7. The protein abundance was determined by a peptide count analysis using the emPAI value. Three biological replications were performed. Differential expression in at least in two of the biological replicates was reported as protein alteration during ICL-M treatment. The UniProt database (www.uniprot.org) was used for indicating gene ontology and protein localization. STRINGS software (https://string-db.org/) was used to analyze protein–protein interactions [28].
10. Transmission electron microscopy (TEM)
RBCs infected with the trophozoite stage of P. falciparum were treated with <0.01% DMSO (buffer control) and 250 nM of ICL-M (IC90) for 4 h. The infected RBCs were then centrifuged at 500 ×g for 5 min, and the resulting pellets were washed with RPMI 1640. The sample preparation protocol used for TEM analysis was modified from that described by Ampawong and colleagues [29]. Infected RBCs were fixed overnight by 2.5% glutaraldehyde in 0.1 M sucrose phosphate buffer (SPB) at 4 °C, then washed three times with 0.1 M SPB. The cells were incubated in 1% osmium tetroxide (Electron Microscopy Sciences [EMS], Pennsylvania, USA) in 0.1 M SPB for 1 h. Afterwards, the cells were washed three times with 0.1 M SPB, dehydrated with ethanol at room temperature, and infiltrated by a series of LR white resin (EMS). Finally, the cells were embedded with 100% LR white resin in a capsule overnight at 65 °C. Each sample was cut into thin sections and stained with 2% uranyl acetate (Sigma Aldrich, Missouri, USA) for 1 min followed by staining with lead citrate for 3 min, then washed with water. The samples were examined by a transmission electron microscope (HT7700–6610LV, Hitachi, Japan).
Results
1. The activity of ICL-M against P. falciparum 3D7
Antimalarial assays for ICL-M were performed using P. falciparum 3D7. The IC50 and IC90 of ICL-M were found to be 148.31 ± 22.87 and 243.39 ± 17.99 nM, respectively (Table 1). To assess the stage-specific antimalarial properties of ICL-M, the ring and trophozoite stages of P. falciparum 3D7 were each incubated with ICL-M. After 24 h of treatment, 99% of the ring-stage parasites were killed, and the total population was killed after 48 h of incubation (Fig 2). In contrast, ICL-M-treated trophozoite-stage parasites displayed 86% and 99% growth inhibition after 24 h and 48 h of incubation, respectively. Thus, ICL-M has the ability to kill both the ring and trophozoite stages of P. falciparum, but the trophozoite stage is slightly less sensitive to treatment with ICL-M compared with the early stage.
Table 1. Antimalarial activity of ICL-M against P. falciparum 3D7.
| Compound | IC50 | IC90 |
|---|---|---|
| ICL-M | 148.31 ± 22.87 | 243.39 ± 17.99 |
Mean (nM) ± SD
Fig 2. Stage-specific antimalarial activity of ICL-M.
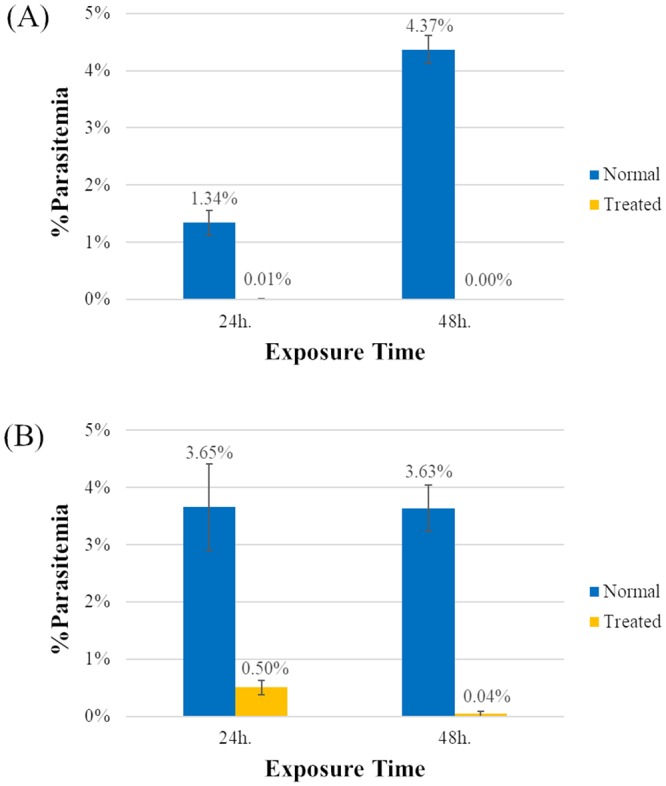
Synchronized P. falciparum in the ring or trophozoite stages were exposed to treatment with ICL-M at 3× IC50 for 24 or 48 h. (A–B) Antimalarial activity on the ring stage (A) and the trophozoite stage (B).
2. Inhibition of P. falciparum topoisomerase II activity
Isocryptolepine derivatives have been reported to target topoisomerase II (topo II) in several different types of cancer cells [8]. Here, we tested the inhibitory activity of ICL-M against P. falciparum topo II. Pyronaridine is an antimalarial drug with documented parasite topo II inhibiting activity, so it was used as a positive control in this experiment (Fig 3). Topo II can break catenated DNA into decatenated DNA. In parasites treated with the negative control (DMSO, compound solvent), P. falciparum topo II activity was observed as the cutting of substrate kinetoplast DNA (catenated DNA) into circular DNA (decatenated DNA). As expected, an inhibition of P. falciparum topo II activity was observed in the presence of the positive control pyronaridine. Specifically, the resulting band of circular DNA was faint compared with that of the negative control-treated sample. When P. falciparum parasites were treated with ICL-M, a band of circular DNA with an intensity similar to that of the negative control was observed, suggesting that ICL-M treatment did not inhibit the activity of P. falciparum topo II in this assay.
Fig 3. Effect of ICL-M on P. falciparum 3D7 topoisomerase II activity.
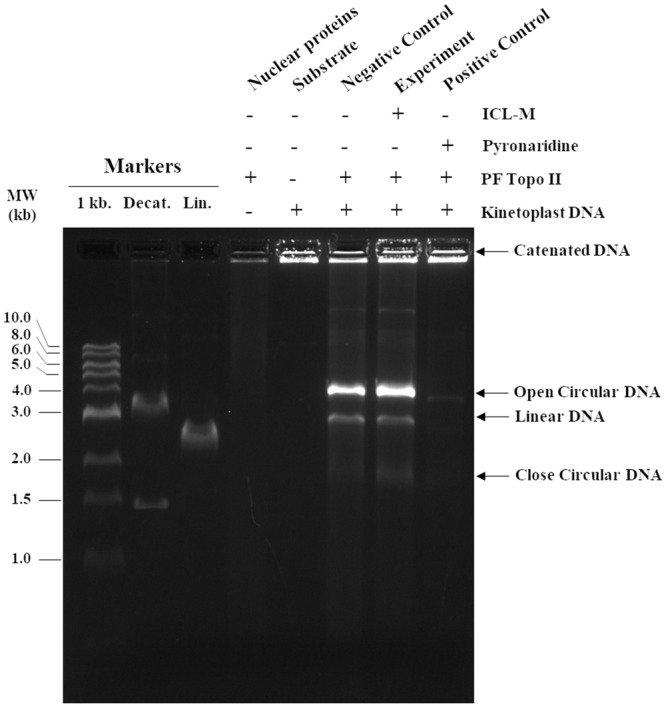
Each reaction contained P. falciparum 3D7 topoisomerase II, kinetoplast DNA (substrate), and pyronaridine (positive control), DMSO (negative control), or ICL-M (experimental reaction).
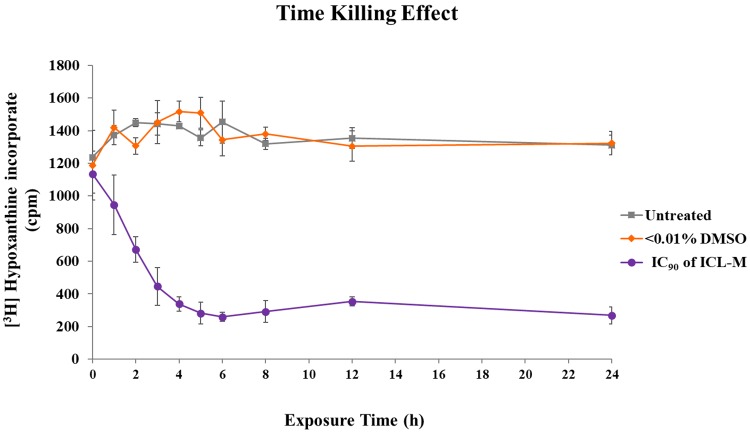
3. Time course of P. falciparum growth rate inhibition by ICL-M
To assess the growth rate inhibition against P. falciparum by ICL-M, three groups of trophozoite stage P. falciparum were prepared: the untreated, <0.01% DMSO-treated (solvent control), and 250 nM ICL-M (IC90)-treated groups. For the growth rate assay, a [3H] hypoxanthine supplement in the parasite culture was used to measure the growth of the parasite. The amount of parasite growth at each timepoint is shown in Fig 4. The growth rate of the P. falciparum in the <0.01% DMSO-treated group was similar to that of the untreated group, which suggests that the presence of the DMSO solvent did not affect the parasite growth rate. In contrast, the ICL-M treatment (IC90) initially caused detectable interference with parasite growth after 1 h of treatment and gradually reached the maximum inhibition rate after 4 h of exposure, maintaining this rate until at least 24 h of exposure.
Fig 4. Time-dependent effects of treatment with ICL-M at IC90 on P. falciparum 3D7.
The untreated, <0.01% DMSO-treated (solvent control), and 250 nM ICL-M (IC90)-treated groups were collected at different timepoints and assayed the growth of the parasite by [3H] hypoxanthine method.
4. Proteomic profile
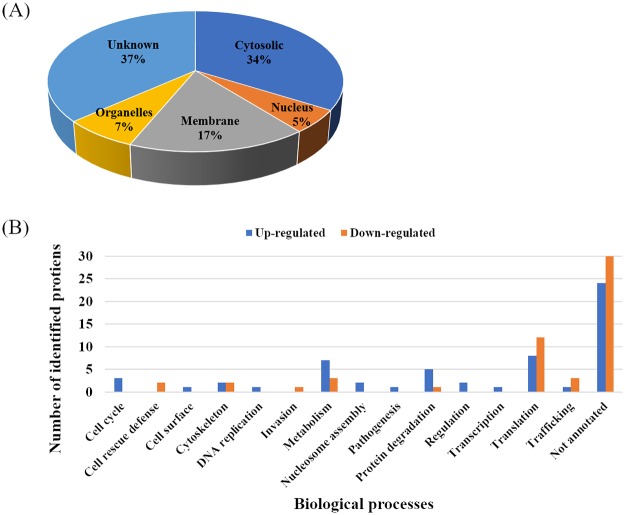
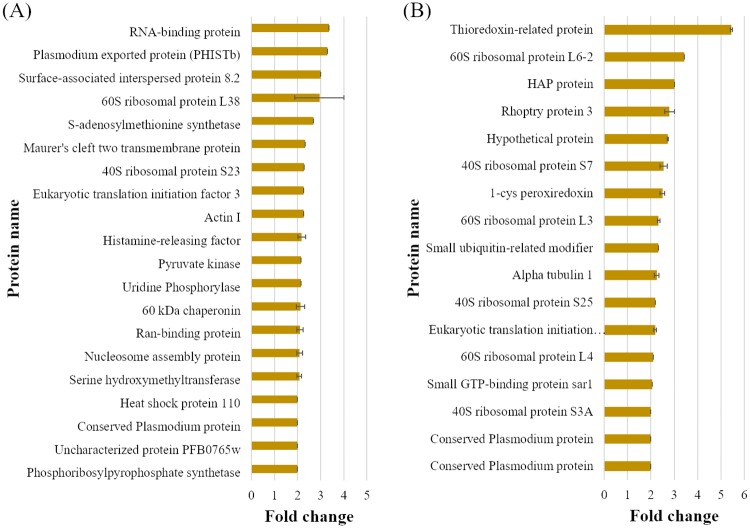
For studying differential protein expression, a proteomic approach was applied to P. falciparum exposed to ICL-M. Two groups of trophozoite stage P. falciparum parasites were prepared: <0.01% DMSO-treated (solvent control) and 250 nM ICL-M (IC90)-treated groups. Proteins were extracted from each experimental group after 4 h of treatment (the minimum exposure time needed to induce irreversible parasite death). The proteins were then separated by 1D-SDS-PAGE, and each lane was cut into 16 pieces (Fig 5) prior to in-gel trypsin digestion. The digested peptides were identified by LC-MS/MS, and each protein was quantified using a label-free (emPAI) spectral counting technique. A total of 668 unique proteins were identified (Table 2), with 511 and 514 proteins in the control and ICL-M-treated groups, respectively. Among these, 112 proteins were differentially expressed in the ICL-M-treated group when compared with the control group; there were 58 upregulated proteins (Table 3) and 54 downregulated proteins (Table 4) in the ICL-M-treated group. According to the Uniprot database, the differentially expressed proteins were localized to the cytosol (34%), membrane (17%), nucleus (5%), other organelles (7%), or had an unknown cellular location (37%) (Fig 6A). The upregulated proteins were specifically involved in several biological processes, including the cell cycle, cell surface, cytoskeleton, DNA replication, metabolism, nucleosome assembly, pathogenesis, protein degradation, regulation, transcription, translation, and trafficking. The downregulated proteins were linked to seven biological processes: cell rescue defense, cytoskeleton, invasion, metabolism, protein degradation, translation, and trafficking (Fig 6B). The most upregulated and downregulated proteins with their fold changes calculated by the semi-quantitative data were demonstrated in Fig 7.
Fig 5. Coomassie blue-stained gel of DMSO- or ICL-M-treated P. falciparum 3D7 proteins.
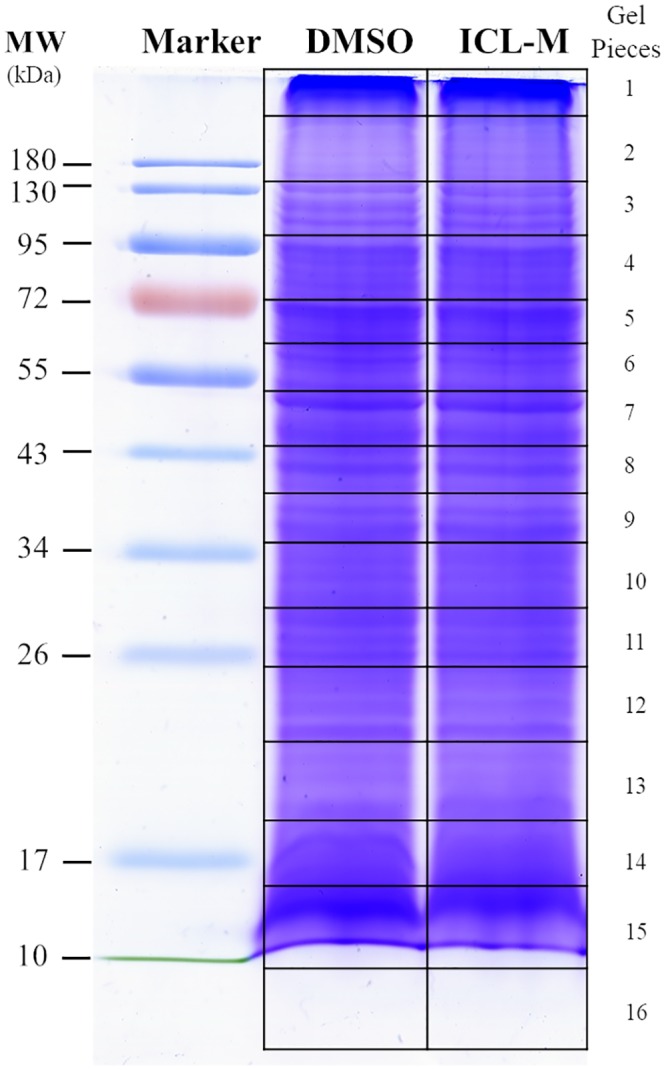
P. falciparum 3D7 parasites were exposed to DMSO (<0.01%) or ICL-M (IC90) for 4 h. Each gel lane was cut into 16 pieces for in-gel tryptic digestion.
Table 2. Protein detection of P. falciparum 3D7 after 4 h of treatment with IC90 ICL-M.
| Compound | All Detection | All Change | Up | Down |
|---|---|---|---|---|
| DMSO | 511 | - | - | - |
| ICL-M | 514 | 112 | 58 | 54 |
Total detected proteins = 668
Table 3. Upregulated proteins in ICL-M-treated P. falciparum 3D7.
| Biological Process | NCBI Accession Number | Protein Description | Protein Mass | PI | Protein Score | Percent Coverage | Localization |
|---|---|---|---|---|---|---|---|
| Cell Cycle | gi|23504595| | Histamine-releasing factor | 19967 | 4.58 | 210 | 23.4 | Cytoplasm |
| gi|46362290| | DNA repair protein RAD50, putative | 267786 | 8.78 | 39 | 17.6 | Nucleus | |
| gi|356624409| | Chain A, Translationally-controlled Tumor Protein Homolog | 21594 | 4.9 | 26 | 19.7 | Cytoplasm | |
| Cell Surface | gi|23505252| | Cytoadherence linked asexual protein 9 | 160313 | 8.88 | 42 | 17.1 | Membrane |
| Cytoskeleton | gi|74929507| | Actin I | 41844 | 5.21 | 78 | 24.7 | Cytoplasm |
| gi|124806724| | GAS8-like protein, putative | 54772 | 7.98 | 42 | 8.9 | Golgi Apparatus | |
| DNA Replication | gi|17148533| | Ran-binding protein | 33176 | 4.92 | 57 | 16.4 | Cytoplasm |
| Metabolism | gi|46361220| | Pyruvate kinase | 55625 | 7.5 | 405 | 44.8 | Cytoplasm |
| gi|10129955| | S-adenosylmethionine synthetase | 44816 | 6.28 | 250 | 31.1 | Unknown | |
| gi|47169189| | Chain A, Uridine Phosphorylase, Putative | 27745 | 6.32 | 188 | 44.3 | Unknown | |
| gi|23615408| | Phosphoribosylpyrophosphate synthetase | 49352 | 9.39 | 151 | 26.3 | Unknown | |
| gi|322812543| | Chain A, Glucose-6-phosphate isomerase | 69517 | 6.9 | 144 | 24.3 | Unknown | |
| gi|124806534| | Serine hydroxymethyltransferase | 49749 | 8.29 | 103 | 31 | Cytoplasm | |
| gi|124801366| | ATP synthase F1, alpha subunit | 61731 | 8.72 | 33 | 19.6 | Mitochondria | |
| Nucleosome Assembly | gi|23505090| | Nucleosome assembly protein | 31807 | 4.17 | 131 | 24.2 | Cytoplasm |
| gi|124806302| | WD repeat-containing protein, putative | 378674 | 9.02 | 45 | 12.8 | Nucleus | |
| Pathogenesis | gi|258597440| | Antigen 332, DBL-like protein | 688870 | 3.86 | 45 | 5.2 | Membrane |
| Protein degradation | gi|124810483| | Proteasome subunit alpha type-1, putative | 28819 | 5.51 | 192 | 24 | Cytoplasm |
| gi|23615667| | Proteasome subunit alpha type-4, putative | 27930 | 5.85 | 47 | 20.3 | Cytoplasm | |
| gi|23615691| | Ubiquitin-conjugating enzyme, putative | 22869 | 5.32 | 40 | 17.3 | Cytoplasm | |
| gi|258597535| | 60 kDa chaperonin | 81434 | 4.97 | 30 | 22.8 | Apicoplast | |
| gi|23505203| | Peptidyl-prolyl cis-trans isomerase | 72507 | 9.11 | 25 | 15.8 | Nucleus | |
| Regulation | gi|23504638| | Phosphatidylinositol 3-kinase | 255758 | 9.27 | 16 | 11.2 | Food Vacuole |
| gi|23498743| | tRNA m5C-methyltransferase, putative | 141140 | 6.35 | 30 | 17 | Unknown | |
| Transcription | gi|124802200| | PRE-binding protein | 131545 | 9.19 | 30 | 14.3 | Nucleus |
| Translation | gi|124804373| | 60S ribosomal protein L38 | 10307 | 10.71 | 100 | 58.6 | Cytoplasm |
| gi|7672213| | Eukaryotic translation initiation factor 3 subunit K, putative | 28017 | 5.61 | 92 | 31.1 | Cytoplasm | |
| gi|23615551| | 60S ribosomal protein L18, putative | 21733 | 10.62 | 88 | 45.1 | Cytoplasm | |
| gi|23499061| | RNA-binding protein, putative | 32421 | 9.11 | 81 | 15.4 | Unknown | |
| gi|4493905| | 40S ribosomal protein S23, putative | 16120 | 10.83 | 66 | 33.8 | Cytoplasm | |
| gi|46361134| | 60S ribosomal protein L27a, putative | 16712 | 10.54 | 59 | 31.8 | Cytoplasm | |
| gi|124802168| | Eukaryotic translation initiation factor 2 subunit beta, putative | 25306 | 9.23 | 42 | 43.2 | Cytoplasm | |
| gi|124808549| | NOT family protein, putative | 519499 | 6.88 | 27 | 11.8 | Cytoplasm | |
| Trafficking | gi|124805983| | Clathrin heavy chain, putative | 232803 | 6 | 38 | 12.3 | Cytoplasm |
| Unknown | gi|225632011| | Heat shock protein 110, putative | 108119 | 5.5 | 95 | 16.8 | Apicoplast |
| gi|23498308| | Plasmodium exported protein (PHISTb), unknown function | 35939 | 8.75 | 88 | 32 | Unknown | |
| gi|8439487| | hypothetical protein, partial | 23455 | 5.87 | 61 | 14.6 | Membrane | |
| gi|23505032| | Elongation factor 1-beta | 32007 | 4.94 | 50 | 25.4 | Unknown | |
| gi|23504974| | ATP-dependent protease ATPase subunit ClpY | 106396 | 8.42 | 48 | 17.4 | Cytoplasm | |
| gi|3694805| | Cytoadherence linked asexual protein, partial | 160994 | 8.98 | 42 | 20.1 | Membrane | |
| gi|225631696| | Conserved Plasmodium protein, unknown function | 1116481 | 9.37 | 39 | 11.1 | Membrane | |
| gi|23498992| | Surface-associated interspersed protein 8.2 (SURFIN 8.2) | 248324 | 5.35 | 38 | 17.7 | Membrane | |
| gi|124808162| | Conserved Plasmodium protein, unknown function | 240981 | 9.51 | 36 | 13 | Unknown | |
| gi|23505262| | Plasmodium exported protein (PHISTc), unknown function | 45472 | 9.71 | 33 | 16.4 | Membrane | |
| gi|23505053| | Conserved Plasmodium protein, unknown function | 85353 | 9.1 | 32 | 12.3 | Unknown | |
| gi|258597726| | Conserved Plasmodium protein, unknown function | 248348 | 9.01 | 30 | 10.2 | Unknown | |
| gi|258549210| | Conserved Plasmodium protein, unknown function | 30341 | 7.64 | 29 | 18.6 | Unknown | |
| gi|124804341| | Parasitophorous vacuolar protein 1 | 51919 | 4.97 | 29 | 24.3 | Cytoplasm | |
| gi|23499096| | Conserved Plasmodium protein, unknown function | 170170 | 6.4 | 27 | 12.1 | Unknown | |
| gi|46362309| | Conserved Plasmodium protein, unknown function | 324818 | 6.62 | 25 | 10.5 | Unknown | |
| gi|296005130| | Pfmc-2TM Maurer~s cleft two transmembrane protein | 27380 | 9.4 | 25 | 31.2 | Membrane | |
| gi|74862993| | RecName: Full = Uncharacterized protein PFB0765w | 166903 | 6.19 | 25 | 17.1 | Unknown | |
| gi|258597812| | Conserved Plasmodium protein, unknown function | 70994 | 9.15 | 24 | 19 | Unknown | |
| gi|46362277| | Conserved Plasmodium protein, unknown function | 334204 | 8.49 | 22 | 18.4 | Unknown | |
| gi|23498914| | Mitochondrial import inner membrane translocase subunit TIM14, putative | 13044 | 10.09 | 21 | 22.6 | Unknown | |
| gi|23615369| | Conserved Plasmodium membrane protein, unknown function | 404731 | 8.76 | 21 | 14.6 | Membrane | |
| gi|23498252| | Regulator of chromosome condensation, putative | 236211 | 9.23 | 21 | 9.2 | Unknown | |
| gi|23505265| | Plasmodium exported protein, unknown function | 31267 | 9.69 | 20 | 8.9 | Membrane |
Table 4. Downregulated proteins in ICL-M-treated P. falciparum 3D7.
| Biological Process | NCBI Accession Number | Protein Description | Protein Mass | PI | Protein Score | Percent Coverage | Localization |
|---|---|---|---|---|---|---|---|
| Cell Rescue Defence | gi|23499261| | 1-cys peroxiredoxin | 25148 | 6.31 | 155 | 47.3 | Cytoplasm |
| gi|23615654| | Thioredoxin-related protein, putative | 23972 | 9.44 | 134 | 26.9 | Membrane | |
| Cytoskeleton | gi|23504938| | Alpha tubulin 1 | 50264 | 4.93 | 234 | 23.8 | Cytoplasm |
| gi|23504954| | Dynein heavy chain, putative | 720134 | 6.18 | 32 | 10.6 | Cytoplasm | |
| Invasion | gi|23504955| | High molecular weight rhoptry protein 3 | 104789 | 6.25 | 150 | 21.4 | Rhoptry |
| Metabolism | gi|237640532| | HAP protein | 37376 | 4.97 | 141 | 32.2 | Food Vacuole |
| gi|46361188| | Hexokinase | 55226 | 6.72 | 53 | 31.2 | Cytoplasm | |
| gi|23504543| | Small ubiquitin-related modifier | 11053 | 4.74 | 52 | 38 | Nucleus | |
| Protein degradation | gi|124804234| | Autophagy-related protein 7, putative | 156530 | 6.05 | 29 | 18.2 | Cytoplasm |
| Translation | gi|23504556| | 60S ribosomal protein L4 | 46183 | 10.5 | 268 | 34.3 | Cytoplasm |
| gi|23615172| | 40S ribosomal protein S7, putative | 22467 | 9.81 | 266 | 41.8 | Cytoplasm | |
| gi|23615526| | 60S ribosomal protein L6-2, putative | 25516 | 10.1 | 222 | 26.7 | Cytoplasm | |
| gi|225631740| | 60S ribosomal protein L19 | 21566 | 11.32 | 168 | 14.3 | Cytoplasm | |
| gi|23499154| | 60S ribosomal protein L13-2, putative | 25425 | 10.78 | 123 | 30.2 | Cytoplasm | |
| gi|124802670| | 60S ribosomal protein L3 | 44193 | 10.21 | 89 | 29.3 | Cytoplasm | |
| gi|4494003| | 40S ribosomal protein S3A, putative | 30028 | 9.8 | 55 | 33.2 | Cytoplasm | |
| gi|23504862| | Eukaryotic translation initiation factor 3 subunit E, putative | 61379 | 7.08 | 49 | 23.4 | Cytoplasm | |
| gi|258597702| | 40S ribosomal protein S25 | 11656 | 10.12 | 44 | 55.2 | Cytoplasm | |
| gi|23498839| | Eukaryotic translation initiation factor 3 subunit I, putative | 37261 | 6.43 | 36 | 19.6 | Cytoplasm | |
| gi|124804772| | 60S ribosomal protein L35ae, putative | 16255 | 10.55 | 25 | 26.4 | Cytoplasm | |
| gi|23498787| | 60S ribosomal protein L34 | 17340 | 10.77 | 24 | 10.7 | Cytoplasm | |
| Trafficking | gi|23498233| | Small GTP-binding protein sar1 | 22006 | 6.75 | 72 | 31.3 | Endoplasmic Reticulum |
| gi|13375179| | Putative GTPase | 22872 | 6.18 | 47 | 16 | Membrane | |
| gi|23615179| | Sodium/hydrogen exchanger, Na+, H+ antiporter | 225940 | 8.68 | 29 | 11.4 | Membrane | |
| Unknown | gi|8247298| | Hypothetical protein, partial | 4755 | 9.23 | 159 | 60 | Unknown |
| gi|124801947| | RNA-binding protein, putative | 30033 | 10.07 | 55 | 35.1 | Unknown | |
| gi|23504857| | Hsc70-interacting protein | 51092 | 4.67 | 45 | 14.6 | Unknown | |
| gi|225632182| | Conserved Plasmodium protein, unknown function | 126994 | 5.28 | 39 | 12 | Unknown | |
| gi|23498915| | Conserved Plasmodium protein, unknown function | 36570 | 8.79 | 39 | 22 | Unknown | |
| gi|225632293| | Plasmodium exported protein, unknown function | 36390 | 5.74 | 35 | 22.2 | Unknown | |
| gi|14530178| | Krueppel-like protein | 151550 | 7.89 | 33 | 11.5 | Membrane | |
| gi|23498950| | Zinc finger, C3HC4 type, putative | 253816 | 8.32 | 32 | 15.5 | Unknown | |
| gi|23615182| | Conserved Plasmodium protein, unknown function | 1111079 | 9.12 | 30 | 15.6 | Membrane | |
| gi|23504877| | Conserved Plasmodium protein, unknown function | 225390 | 6.01 | 30 | 16 | Unknown | |
| gi|3649757| | Conserved Plasmodium protein, unknown function | 202018 | 8.3 | 29 | 16.5 | Cytoplasm | |
| gi|225631936| | MORN repeat protein, putative | 519893 | 9.15 | 29 | 10.3 | Membrane | |
| gi|124808756| | Conserved Plasmodium protein, unknown function | 192484 | 9.67 | 29 | 16.8 | Membrane | |
| gi|124806636| | Conserved Plasmodium protein, unknown function | 212627 | 4.96 | 29 | 12.8 | Unknown | |
| gi|23504575| | Conserved Plasmodium protein, unknown function | 369953 | 5.29 | 29 | 9 | Unknown | |
| gi|124809084| | Conserved Plasmodium protein, unknown function | 152167 | 8.42 | 29 | 12.9 | Unknown | |
| gi|124805631| | Conserved Plasmodium protein, unknown function | 208541 | 8.3 | 29 | 15.2 | Unknown | |
| gi|124802600| | Conserved Plasmodium protein, unknown function | 190279 | 8.29 | 29 | 10.9 | Unknown | |
| gi|124804432| | Conserved Plasmodium protein, unknown function | 133324 | 5.18 | 29 | 17.4 | Unknown | |
| gi|124804384| | Conserved Plasmodium protein, unknown function | 118257 | 8.84 | 29 | 14.6 | Unknown | |
| gi|225632254| | rRNA-processing protein FCF1, putative | 23145 | 9.63 | 29 | 23.2 | Nucleus | |
| gi|23505139| | RNA-binding protein, putative | 22965 | 9.51 | 27 | 20.3 | Unknown | |
| gi|124808373| | Conserved Plasmodium protein, unknown function | 390446 | 9.23 | 25 | 11.7 | Unknown | |
| gi|23615433| | Conserved Plasmodium protein, unknown function | 319935 | 7.32 | 25 | 15.5 | Unknown | |
| gi|23498752| | Conserved Plasmodium protein, unknown function | 118270 | 8.1 | 23 | 9 | Unknown | |
| gi|225631798| | Conserved protein, unknown function | 296598 | 9.02 | 22 | 15.4 | Membrane | |
| gi|124804419| | Conserved Plasmodium protein, unknown function | 330099 | 6.16 | 21 | 12.1 | Unknown | |
| gi|225631940| | Conserved Plasmodium protein, unknown function | 737235 | 9.13 | 21 | 11.3 | Membrane | |
| gi|124804142| | Leucine-rich repeat protein | 93275 | 8.97 | 18 | 13.9 | Unknown | |
| gi|225631849| | Conserved Plasmodium protein, unknown function | 102515 | 8.83 | 18 | 11.7 | Unknown |
Fig 6. Gene ontology of differential Plasmodium falciparum proteins after ICL-M treatment.
(A) Protein localization (B) Biological process classification. Blue bar: number of upregulated proteins, Orange bar: number of downregulated proteins.
Fig 7. The most upregulated and downregulated proteins effect by ICL-M treatment ranked by emPAI values.
(A) Upregulated proteins after ICL-M treatment. (B) Downregulated proteins after ICL-M treatment.
The predicted protein–protein interaction network identified the ribosome pathway as the major affected pathway in both the upregulated and downregulated proteins (Fig 8). In contrast, the proteasomal, amino acid biosynthesis, metabolic, oxidative phosphorylation, and carbon metabolism pathways were identified as minor pathways, and the proteins linked to them were detected in only the group of upregulated proteins.
Fig 8. STRINGS protein–protein interaction analysis of differentially expressed proteins following ICL-M treatment.

(A) Upregulated proteins after ICL-M treatment. (B) Downregulated proteins after ICL-M treatment. Red: Ribosome, Blue: Proteasome, Dark green: Metabolic pathways, Light green: Biosynthesis of amino acids, Yellow: Oxidative phosphorylation, and Pink: Carbon metabolism.
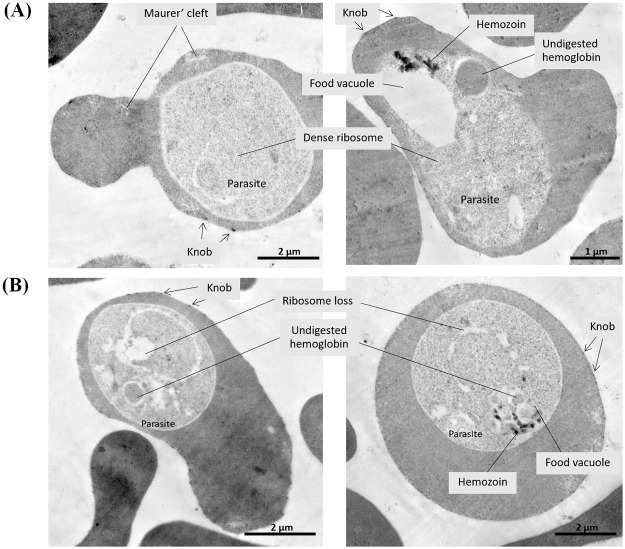
5. Ultrastructural effect of ICL-M
To examine the effect of ICL-M treatment on parasite ultrastructure, the morphology of ICL-M-treated P. falciparum was investigated. Differences were observed between the trophozoite stage parasites within RBCs in the control culture (Fig 9A) and the ICL-M treatment (Fig 9B) groups. Numerous Maurer’s clefts and knobs were seen in the infected RBCs of both groups. Internally, both the control and ICL-M-treated parasites displayed normal food vacuoles with hemozoin, undigested hemoglobin, and dense ribosomes. Interestingly, the loss of ribosome area could be observed in the ICL-M-treated group.
Fig 9. Transmitted electron micrograph of trophozoite stage P. falciparum 3D7 parasites after a 4-h compound exposure.
(A) The control group treated with DMSO. (B) The experimental group treated with ICL-M.
Discussion
The present study aimed to explore the molecular mechanisms of ICL-M. We studied the effects of ICL-M on P. falciparum by assessing its stage-specific activity, topo II activity inhibition, and time-dependent effect as well as by performing a proteomic analysis and a TEM morphology analysis. The stage-specific activity results indicate that ICL-M has antimalarial activity against both the ring and trophozoite stages of P. falciparum. This result corresponds to those of quinine-, artemisinin-, artesunate-, and chloroquine-treated parasites [30, 31]. After drug exposure, the ICL-M demonstrated a stronger inhibitory effect on the ring stage of P. falciparum than on the trophozoite stage, which is similar to the results of reports on chloroquine and artemisinin [30, 31]. In contrast, quinine and artesunate have stronger inhibitory effects on trophozoite stage parasites than they do on ring stage parasites. We also found that the trophozoite stage of P. falciparum was irreversibly damaged by ICL-M (IC90) treatment by 4 h after exposure. Thus, ICL-M causes irreversible damage faster than do either artemether (IC90) or lumefantrine (IC90), which display irreversible killing parasite at 5 h or 8 h after exposure, respectively [17].
In previous studies, several isocryptolepine derivatives have been tested for use in anti-cancer chemotherapy, where they demonstrated high efficiency supporting their further development as novel drugs. Indoloquinoline, which forms the core of ICL-M, has demonstrated activity against the human cancer topo II enzyme in melanoma, leukemia, and non-melanoma skin cancer [32–34]. In addition, the inhibition activity of another isocryptolepine derivative, 3-methoxy-11H-pyrido[3',4':4,5]pyrrolo[3,2-c]quinoline-1,4-dione, has been studied on mouse topo I and topo II [8]. Moreover, etoposide, a well-known anticancer targeting human topo IIα, induced the same inhibitory activity on P. falciparum 3D7 topo II [35]. These previous findings raised a concern that ICL-M might inhibit P. falciparum 3D7 topo II. However, our results indicate that ICL-M has no inhibitory activity against P. falciparum topo II, although indoloquinoline was reported to accumulate in the nucleus of P. falciparum [36]. In addition, many studies have provided evidence of isocryptolepine analogues interrupting the macromolecules that interfere with the replication and transcription processes in both eukaryotic and prokaryotic cells [37–39]. However, P. falciparum topo II might not be the direct target of this compound. The compound accumulation in nucleus may result from DNA intercalation observed in other indoloquinoline derivatives [37]. On the other hands, ICL-M may interact with other topoisomerases or other target proteins. To investigate whether ICL-M affects protein targets associated with antimalarial mechanisms, we applied a large-scale proteomics approach using mass spectrometry. The control and ICL-M-treated groups were identified 511 and 514 proteins, respectively. An proteomic study performed on Plasmodium using in-solution digestion by endoprotease Lys-C and trypsin could yield 714 protein identification in asexual blood stage [9]. The different number of protein identification may cause by the completeness of protease digestion to prepare peptide mixture for mass spectrometric analysis.
Fifty-eight P. falciparum nuclear proteins were identified in our study. Among these, six proteins showed differential expression; DNA repair protein RAD50 (RAD50), peptidyl-prolyl cis-trans isomerase (PPIase), PRE-binding protein (PREBP), and WD repeat-containing protein (WDR) were upregulated after ICL-M exposure, whereas rRNA-processing protein FCF1 (FCF1) and small ubiquitin-related modifier (SUMO) were downregulated. Interestingly, PPIase, PREBP, and SUMO have been reported as drug or vaccine targets in P. falciparum [40], [41], [42]. These mechanisms may be involved in the inhibition of P. falciparum by ICL-M. PPIase accelerates protein folding by catalyzing the cis–trans isomerization of peptide bonds that are N-terminal to proline residues in polypeptide chains. This enzyme has been reported as a target of several antimalarial drugs, such as cyclosporine A, FK506, and rapamycin [40]. PREBP is involved in DNA-binding transcription factor activity. This protein is expressed in the Plasmodium merozoite stage and has been reported as a target of monoclonal antibodies that inhibit parasite invasion [41]. In addition, PREBP has also been reported as a potential candidate for P. falciparum vaccine development [42]. SUMO plays an important role in the oxidative stress response during the P. falciparum intra-erythrocyte developmental cycle. It is a promising target for the development of P. falciparum sumoylation inhibitors and parasite replication [43]. The other three differentially expressed proteins detected in our study, RAD50, WDR, and FCF1, have not been reported to have an association with existing antimalarial drugs. Based on our results, it is possible that ICL-M might have some effects on parasite nuclear proteins as described above.
In addition to affecting nuclear proteins, one isocryptolepine derivative was reported to affect activity in the parasite food vacuole [44]. A number of studies suggested that quinoline antimalarial drugs, such as chloroquine, quinine, and mefloquine, could inhibit hemozoin formation in the food vacuole of parasites [45]. Since isocryptolepine also contains part of quinoline in its core structure, this drug has been investigated for its ability to inhibit heme detoxification. The result revealed that β-hematin detoxification was inhibited by 3 fold less isocryptolepine than chloroquine [4]. Therefore, isocryptolepine may contribute to other actions in the parasite food vacuole. Our proteomics data identified nine P. falciparum food vacuole proteins in our samples. Among these, histo-aspartic protease (HAP) protein was 3-fold downregulated and phosphatidylinositol 3-kinase (PI3K) was only identified in the treated parasite. HAP is a type of P. falciparum aspartic protease and plays a role in hemoglobin degradation. [46]. An alternative name for HAP is plasmepsin (PM) III. In P. falciparum, four PMs, specifically PM I, II, III, and IV, are key mediators for hemoglobin degradation [47]. All four of these PMs have received considerable attention as potential antimalarial drug targets. The absence of PM III led to the most significant hyper-susceptibility in response to antimalarial compounds such as chloroquine [48] and piperaquine [49]. In our study, PM III was downregulated under ICL-M treatment. The low level of HAP may play a role in increasing the susceptibility of parasites to ICL-M, thus improving the antimalarial efficiency of ICL-M. PI3K was up-regulated in P. falciparum parasites exposed to ICL-M. PI3K is involved in trafficking of host hemoglobin to the parasite food vacuole by endocytosis [50]. While, the downregulation of PM III may reduce the hemoglobin degradation. An upregulation of PI3K may be a compensatory effect to the increased hemoglobin uptake that occurs when hemoglobin digestion is altered.
The protein–protein interactions of all the differentially expressed proteins detected by our proteomics analysis were assessed using the STRING server. Following ICL-M treatment, the significantly enriched pathways included ribosomal, proteasomal, metabolic, amino acid biosynthesis, oxidative phosphorylation, and carbon metabolism pathways. Ribosomes are the major machinery in the translational process; thus, a mutation or alteration of ribosomal proteins leads to abnormal function in cell proliferation [51], [52], [53]. Several ribosomal proteins have been reported as drug targets. Tetracycline, an antibiotic, is able to bind to 70S ribosome and inhibit protein synthesis in bacteria [54]. For P. falciparum, cryogenic electron microscopy revealed that the ribosome GTPase-associated center reacts with mefloquine. Correspondingly, the inhibition of translational processes is one of modes of action for mefloquine [55]. Our proteomics data identified a total of 58 ribosomal proteins; four and ten proteins were upregulated and downregulated, respectively. Interestingly, 60S ribosomal protein L18 and 60S ribosomal protein L27a were both upregulated after ICL-M treatment. This result corresponds to a similar finding following artemisinin treatment of parasites [12]. ICL-M and artemisinin might share some modes of action relating to interference with protein synthesis.
The proteasomal pathway was significantly enriched following ICL-M exposure of P. falciparum. The proteasome is the major protein degradation regulatory network. When homeostasis between protein synthesis and degradation is unbalanced, cell survival is not possible [56]. In Plasmodium, proteasomal proteins have been suggested as potential antimalarial drug targets that could act to disrupt parasite protein homeostasis [57]. Our data identified a total of 11 proteasomal proteins, among which there were two upregulated after ICL-M treatment: proteasome subunit alpha type-1 and proteasome subunit alpha type-4. The differential expression of either of these two proteins has not been reported in studies of other antimalarial drugs. However, other proteins in the proteasome pathway, such as 20S proteasome beta subunit, proteasome regulatory subunit, and proteasome subunit beta type 1, were found to be upregulated after artemisinin treatment of parasites [12]. Additionally, the downregulation of 26S proteasome regulatory subunit was observed in doxycycline-treated parasites [13]. Based on our findings, creating an imbalance in P. falciparum protein homeostasis might be one mode of action for ICL-M.
The glycolytic metabolic pathway was also affected in P. falciparum after ICL-M exposure. Glycolysis is an important metabolic pathway that produces energy for the parasite. Several studies have explored the effect of antimalarial drugs in the expression of proteins involved in this pathway [17, 58] [13, 18]. Our study detected three proteins in this pathway that were differentially expressed in P. falciparum after ICL-M exposure: hexokinase, glucose-6-phosphate isomerase, and pyruvate kinase. Hexokinase and pyruvate kinase have also been reported to have altered expression levels in parasites following doxycycline treatment [13]. Given that ICL-M altered the expression level of proteins in the glycolysis pathway, ICL-M may influence parasite energy production.
In our electron microscopy analysis, we observed a clear loss of ribosomes in the ICL-M-treated P. falciparum. This result supports the finding from our proteomics analysis that ribosomal proteins were highly downregulated in ICL-M-treated P. falciparum. Similarly abnormal parasite morphology was observed by previous work testing other antimalarial drugs, such as artesunate, quinine, and piperaquine [59]. Ribosome depletion leads to a lack of necessary protein functions for parasite biological processes. Moreover, the expression of ribosomal proteins is crucial for the development of several organisms. For example, the ribosomal protein L19 expression differs throughout the growth phases of Leishmania spp. during their lifecycle [60]. Additionally, a study in female Schistosoma japonicum reported that the ribosomal protein expression profiles were changed during sexual maturation [61]. Thus, the downregulation of ribosomal proteins after ICL-M treatment may affect the development of P. falciparum.
Conclusion
The effect of ICL-M, a derivative of isocryptolepine, on malarial parasites was studied in vitro using P. falciparum 3D7 as a model. This compound had an antimalarial effect on both the ring and trophozoite stages of P. falciparum. Our proteomics analysis revealed that ICL-M disrupts several P. falciparum biological processes; ribosomal, proteasomal, and metabolism pathways were the main pathways found to be affected by ICL-M treatment.
Supporting information
Total proteins were identified and quantified by Mascot daemon version 2.3.2 software. The protein database was collected from an NCBInr database (24 October 2018) specific to P. falciparum 3D7.
(PDF)
Total proteins were identified and quantified by Mascot daemon version 2.3.2 software. The protein database was collected from an NCBInr database (24 October 2018) specific to P. falciparum 3D7.
(PDF)
Total proteins were identified and quantified by Mascot daemon version 2.3.2 software. The protein database was collected from an NCBInr database (24 October 2018) specific to P. falciparum 3D7.
(PDF)
The experiment was performed in three biological replicates. Proteins with altered expression levels were selected based on different expression levels in at least two biological replicates.
(PDF)
Acknowledgments
We thank Katie Oakley, PhD, from Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This study was supported by FTM2013 of Faculty of Tropical Medicine, Mahidol university and Thailand Toray Science Foundation.
References
- 1.World Health Organization. World Malaria Report 2018. Geneva: World Health Organization; 2018. [Google Scholar]
- 2.Willcox ML, Bodeker G. Traditional herbal medicines for malaria. BMJ. 2004;329(7475):1156–9. 10.1136/bmj.329.7475.1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grellier P, Ramiaramanana L, Millerioux V, Deharo E, Schrével J, Frappier F, et al. Antimalarial Activity of Cryptolepine and Isocryptolepine, Alkaloids Isolated fromCryptolepis sanguinolenta. Phytother Res. 1996;10(4):317–21. [Google Scholar]
- 4.Van Miert S, Hostyn S, Maes BU, Cimanga K, Brun R, Kaiser M, et al. Isoneocryptolepine, a synthetic indoloquinoline alkaloid, as an antiplasmodial lead compound. J Nat Prod. 2005;68(5):674–7. 10.1021/np0496284 [DOI] [PubMed] [Google Scholar]
- 5.Aroonkit P, Thongsornkleeb C, Tummatorn J, Krajangsri S, Mungthin M, Ruchirawat S. Synthesis of isocryptolepine analogues and their structure-activity relationship studies as antiplasmodial and antiproliferative agents. Eur J Med Chem. 2015;94(0):56–62. [DOI] [PubMed] [Google Scholar]
- 6.Whittell LR, Batty KT, Wong RP, Bolitho EM, Fox SA, Davis TM, et al. Synthesis and antimalarial evaluation of novel isocryptolepine derivatives. Bioorg Med Chem. 2011;19(24):7519–25. 10.1016/j.bmc.2011.10.037 [DOI] [PubMed] [Google Scholar]
- 7.Uchuskin MG, Pilipenko AS, Serdyuk OV, Trushkov IV, Butin AV. From biomass to medicines. A simple synthesis of indolo[3,2-c]quinolines, antimalarial alkaloid isocryptolepine, and its derivatives. Org Biomol Chem. 2012;10(36):7262–5. 10.1039/c2ob25836f [DOI] [PubMed] [Google Scholar]
- 8.Riou JF, Helissey P, Grondard L, Giorgi-Renault S. Inhibition of eukaryotic DNA topoisomerase I and II activities by indoloquinolinedione derivatives. Mol Pharmacol. 1991;40(5):699–706. [PubMed] [Google Scholar]
- 9.Lasonder E, Ishihama Y, Andersen JS, Vermunt AM, Pain A, Sauerwein RW, et al. Analysis of the Plasmodium falciparum proteome by high-accuracy mass spectrometry. Nature. 2002;419(6906):537–42. 10.1038/nature01111 [DOI] [PubMed] [Google Scholar]
- 10.Kassa FA, Shio MT, Bellemare MJ, Faye B, Ndao M, Olivier M. New inflammation-related biomarkers during malaria infection. PLoS One. 2011;6(10):e26495 10.1371/journal.pone.0026495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ray S, Kamath KS, Srivastava R, Raghu D, Gollapalli K, Jain R, et al. Serum proteome analysis of vivax malaria: An insight into the disease pathogenesis and host immune response. J Proteomics. 2012;75(10):3063–80. 10.1016/j.jprot.2011.10.018 [DOI] [PubMed] [Google Scholar]
- 12.Prieto JH, Koncarevic S, Park SK, Yates J 3rd, Becker K. Large-scale differential proteome analysis in Plasmodium falciparum under drug treatment. PLoS One. 2008;3(12):e4098 10.1371/journal.pone.0004098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Briolant S, Almeras L, Belghazi M, Boucomont-Chapeaublanc E, Wurtz N, Fontaine A, et al. Plasmodium falciparum proteome changes in response to doxycycline treatment. Malar J. 2010;9:141 10.1186/1475-2875-9-141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morita M, Sanai H, Hiramoto A, Sato A, Hiraoka O, Sakura T, et al. Plasmodium falciparum endoplasmic reticulum-resident calcium binding protein is a possible target of synthetic antimalarial endoperoxides, N-89 and N-251. J Proteome Res. 2012;11(12):5704–11. 10.1021/pr3005315 [DOI] [PubMed] [Google Scholar]
- 15.Yang KS, Budin G, Tassa C, Kister O, Weissleder R. Bioorthogonal approach to identify unsuspected drug targets in live cells. Angew Chem Int Ed Engl. 2013;52(40):10593–7. 10.1002/anie.201304096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ismail HM, Barton V, Phanchana M, Charoensutthivarakul S, Wong MH, Hemingway J, et al. Artemisinin activity-based probes identify multiple molecular targets within the asexual stage of the malaria parasites Plasmodium falciparum 3D7. Proc Natl Acad Sci U S A. 2016;113(8):2080–5. 10.1073/pnas.1600459113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makanga M, Bray PG, Horrocks P, Ward SA. Towards a proteomic definition of CoArtem action in Plasmodium falciparum malaria. Proteomics. 2005;5(7):1849–58. 10.1002/pmic.200401076 [DOI] [PubMed] [Google Scholar]
- 18.Segura C, Cuesta-Astroz Y, Nunes-Batista C, Zalis M, von Kruger WM, Mascarello Bisch P. Partial characterization of Plasmodium falciparum trophozoite proteome under treatment with quinine, mefloquine and the natural antiplasmodial diosgenone. Biomedica. 2014;34(2):237–49. 10.1590/S0120-41572014000200010 [DOI] [PubMed] [Google Scholar]
- 19.Aly NS, Hiramoto A, Sanai H, Hiraoka O, Hiramoto K, Kataoka H, et al. Proteome analysis of new antimalarial endoperoxide against Plasmodium falciparum. Parasitol Res. 2007;100(5):1119–24. 10.1007/s00436-007-0460-8 [DOI] [PubMed] [Google Scholar]
- 20.Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, et al. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics. 2005;4(9):1265–72. 10.1074/mcp.M500061-MCP200 [DOI] [PubMed] [Google Scholar]
- 21.Reamtong O, Srimuang K, Saralamba N, Sangvanich P, Day NP, White NJ, et al. Protein profiling of mefloquine resistant Plasmodium falciparum using mass spectrometry-based proteomics. Int J Mass Spectrom. 2015;391:82–92. 10.1016/j.ijms.2015.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertin GI, Sabbagh A, Argy N, Salnot V, Ezinmegnon S, Agbota G, et al. Proteomic analysis of Plasmodium falciparum parasites from patients with cerebral and uncomplicated malaria. Sci Rep. 2016;6:26773 10.1038/srep26773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lasonder E, Janse CJ, van Gemert GJ, Mair GR, Vermunt AM, Douradinha BG, et al. Proteomic profiling of Plasmodium sporozoite maturation identifies new proteins essential for parasite development and infectivity. PLoS Pathog. 2008;4(10):e1000195 10.1371/journal.ppat.1000195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lambros C, Vanderberg JP. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J Parasitol. 1979;65(3):418–20. [PubMed] [Google Scholar]
- 25.Silamut K, Phu NH, Whitty C, Turner GD, Louwrier K, Mai NT, et al. A quantitative analysis of the microvascular sequestration of malaria parasites in the human brain. Am J Pathol. 1999;155(2):395–410. 10.1016/S0002-9440(10)65136-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voss TS, Mini T, Jenoe P, Beck HP. Plasmodium falciparum possesses a cell cycle-regulated short type replication protein A large subunit encoded by an unusual transcript. J Biol Chem. 2002;277(20):17493–501. 10.1074/jbc.M200100200 [DOI] [PubMed] [Google Scholar]
- 27.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72(1):248–54. [DOI] [PubMed] [Google Scholar]
- 28.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–52. 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ampawong S, Isarangkul D, Reamtong O, Aramwit P. Adaptive effect of sericin on hepatic mitochondrial conformation through its regulation of apoptosis, autophagy and energy maintenance: a proteomics approach. Sci Rep. 2018;8(1):14943 10.1038/s41598-018-33372-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skinner TS, Manning LS, Johnston WA, Davis TM. In vitro stage-specific sensitivity of Plasmodium falciparum to quinine and artemisinin drugs. Int J Parasitol. 1996;26(5):519–25. [DOI] [PubMed] [Google Scholar]
- 31.Le Manach C, Scheurer C, Sax S, Schleiferbock S, Cabrera DG, Younis Y, et al. Fast in vitro methods to determine the speed of action and the stage-specificity of anti-malarials in Plasmodium falciparum. Malar J. 2013;12:424 10.1186/1475-2875-12-424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonjean K, De Pauw-Gillet MC, Defresne MP, Colson P, Houssier C, Dassonneville L, et al. The DNA intercalating alkaloid cryptolepine interferes with topoisomerase II and inhibits primarily DNA synthesis in B16 melanoma cells. Biochemistry. 1998;37(15):5136–46. 10.1021/bi972927q [DOI] [PubMed] [Google Scholar]
- 33.Dassonneville L, Bonjean K, De Pauw-Gillet MC, Colson P, Houssier C, Quetin-Leclercq J, et al. Stimulation of topoisomerase II-mediated DNA cleavage by three DNA-intercalating plant alkaloids: cryptolepine, matadine, and serpentine. Biochemistry. 1999;38(24):7719–26. 10.1021/bi990094t [DOI] [PubMed] [Google Scholar]
- 34.Pal HC, Katiyar SK. Cryptolepine, a Plant Alkaloid, Inhibits the Growth of Non-Melanoma Skin Cancer Cells through Inhibition of Topoisomerase and Induction of DNA Damage. Molecules. 2016;21(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mudeppa DG, Kumar S, Kokkonda S, White J, Rathod PK. Topoisomerase II from Human Malaria Parasites: EXPRESSION, PURIFICATION, AND SELECTIVE INHIBITION. J Biol Chem. 2015;290(33):20313–24. 10.1074/jbc.M115.639039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lavrado J, Cabal GG, Prudencio M, Mota MM, Gut J, Rosenthal PJ, et al. Incorporation of basic side chains into cryptolepine scaffold: structure-antimalarial activity relationships and mechanistic studies. J Med Chem. 2011;54(3):734–50. 10.1021/jm101383f [DOI] [PubMed] [Google Scholar]
- 37.Marquez VE, Cranston JW, Ruddon RW, Kier LB, Burckhalter JH. Mechanism of action of amodiaquine. Synthesis of its indoloquinoline analog. J Med Chem. 1972;15(1):36–9. 10.1021/jm00271a010 [DOI] [PubMed] [Google Scholar]
- 38.Ibrahim E-S, Montgomerie AM, Sneddon AH, Proctor GR, Green B. Synthesis of indolo[3,2-c]quinolines and indolo[3,2-d]benzazepines and their interaction with DNA. Eur J Med Chem. 1988;23(2):183–8. [Google Scholar]
- 39.Ghosh P, Purkayastha P. Selective interaction of methylindoloquinolines with DNA. Rsc Advances. 2014;4(43):22442–8. [Google Scholar]
- 40.Bell A, Wernli B, Franklin RM. Roles of peptidyl-prolyl cis-trans isomerase and calcineurin in the mechanisms of antimalarial action of cyclosporin A, FK506, and rapamycin. Biochem Pharmacol. 1994;48(3):495–503. 10.1016/0006-2952(94)90279-8 [DOI] [PubMed] [Google Scholar]
- 41.Epping RJ, Goldstone SD, Ingram LT, Upcroft JA, Ramasamy R, Cooper JA, et al. An epitope recognised by inhibitory monoclonal antibodies that react with a 51 kilodalton merozoite surface antigen in Plasmodium falciparum. Mol Biochem Parasitol. 1988;28(1):1–10. [DOI] [PubMed] [Google Scholar]
- 42.Sanasam BD, Kumar S. PRE-binding protein of Plasmodium falciparum is a potential candidate for vaccine design and development: An in silico evaluation of the hypothesis. Med Hypotheses. 2019;125:119–23. 10.1016/j.mehy.2019.01.006 [DOI] [PubMed] [Google Scholar]
- 43.Reiter K, Mukhopadhyay D, Zhang H, Boucher LE, Kumar N, Bosch J, et al. Identification of biochemically distinct properties of the small ubiquitin-related modifier (SUMO) conjugation pathway in Plasmodium falciparum. J Biol Chem. 2013;288(39):27724–36. 10.1074/jbc.M113.498410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang N, Wicht KJ, Imai K, Wang MQ, Anh Ngoc T, Kiguchi R, et al. Synthesis, beta-haematin inhibition, and in vitro antimalarial testing of isocryptolepine analogues: SAR study of indolo[3,2-c]quinolines with various substituents at C2, C6, and N11. Bioorg Med Chem. 2014;22(9):2629–42. 10.1016/j.bmc.2014.03.030 [DOI] [PubMed] [Google Scholar]
- 45.Kumar S, Guha M, Choubey V, Maity P, Bandyopadhyay U. Antimalarial drugs inhibiting hemozoin (beta-hematin) formation: a mechanistic update. Life Sci. 2007;80(9):813–28. 10.1016/j.lfs.2006.11.008 [DOI] [PubMed] [Google Scholar]
- 46.Bhaumik P, Xiao H, Hidaka K, Gustchina A, Kiso Y, Yada RY, et al. Structural insights into the activation and inhibition of histo-aspartic protease from Plasmodium falciparum. Biochemistry. 2011;50(41):8862–79. 10.1021/bi201118z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Francis SE, Gluzman IY, Oksman A, Knickerbocker A, Mueller R, Bryant ML, et al. Molecular characterization and inhibition of a Plasmodium falciparum aspartic hemoglobinase. EMBO J. 1994;13(2):306–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moura PA, Dame JB, Fidock DA. Role of Plasmodium falciparum digestive vacuole plasmepsins in the specificity and antimalarial mode of action of cysteine and aspartic protease inhibitors. Antimicrob Agents Chemother. 2009;53(12):4968–78. 10.1128/AAC.00882-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mukherjee A, Gagnon D, Wirth DF, Richard D. Inactivation of Plasmepsins 2 and 3 Sensitizes Plasmodium falciparum to the Antimalarial Drug Piperaquine. Antimicrob Agents Chemother. 2018;62(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vaid A, Ranjan R, Smythe WA, Hoppe HC, Sharma P. PfPI3K, a phosphatidylinositol-3 kinase from Plasmodium falciparum, is exported to the host erythrocyte and is involved in hemoglobin trafficking. Blood. 2010;115(12):2500–7. 10.1182/blood-2009-08-238972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pelchovich G, Nadejda S, Dana A, Tuller T, Bravo IG, Gophna U. Ribosomal mutations affecting the translation of genes that use non-optimal codons. Febs j. 2014;281(16):3701–18. 10.1111/febs.12892 [DOI] [PubMed] [Google Scholar]
- 52.Guo X, Shi Y, Gou Y, Li J, Han S, Zhang Y, et al. Human ribosomal protein S13 promotes gastric cancer growth through down-regulating p27(Kip1). J Cell Mol Med. 2011;15(2):296–306. 10.1111/j.1582-4934.2009.00969.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen B, Zhang W, Gao J, Chen H, Jiang L, Liu D, et al. Downregulation of ribosomal protein S6 inhibits the growth of non-small cell lung cancer by inducing cell cycle arrest, rather than apoptosis. Cancer Lett. 2014;354(2):378–89. 10.1016/j.canlet.2014.08.045 [DOI] [PubMed] [Google Scholar]
- 54.Jenner L, Starosta AL, Terry DS, Mikolajka A, Filonava L, Yusupov M, et al. Structural basis for potent inhibitory activity of the antibiotic tigecycline during protein synthesis. Proc Natl Acad Sci U S A. 2013;110(10):3812–6. 10.1073/pnas.1216691110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wong W, Bai X-C, Sleebs BE, Triglia T, Brown A, Thompson JK, et al. Mefloquine targets the Plasmodium falciparum 80S ribosome to inhibit protein synthesis. Nature Microbiology. 2017;2:17031 10.1038/nmicrobiol.2017.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cromm PM, Crews CM. The Proteasome in Modern Drug Discovery: Second Life of a Highly Valuable Drug Target. ACS central science. 2017;3(8):830–8. 10.1021/acscentsci.7b00252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ng CL, Fidock DA, Bogyo M. Protein Degradation Systems as Antimalarial Therapeutic Targets. Trends Parasitol. 2017;33(9):731–43. 10.1016/j.pt.2017.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nirmalan N, Sims PF, Hyde JE. Quantitative proteomics of the human malaria parasite Plasmodium falciparum and its application to studies of development and inhibition. Mol Microbiol. 2004;52(4):1187–99. 10.1111/j.1365-2958.2004.04049.x [DOI] [PubMed] [Google Scholar]
- 59.Sachanonta N, Chotivanich K, Chaisri U, Turner GD, Ferguson DJ, Day NP, et al. Ultrastructural and real-time microscopic changes in P. falciparum-infected red blood cells following treatment with antimalarial drugs. Ultrastruct Pathol. 2011;35(5):214–25. 10.3109/01913123.2011.601405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Almeida-Bizzo JH, Alves LR, Castro FF, Garcia JBF, Goldenberg S, Cruz AK. Characterization of the pattern of ribosomal protein L19 production during the lifecycle of Leishmania spp. Exp Parasitol. 2014;147:60–6. 10.1016/j.exppara.2014.08.015 [DOI] [PubMed] [Google Scholar]
- 61.Sun J, Li C, Wang S. The Up-Regulation of Ribosomal Proteins Further Regulates Protein Expression Profile in Female Schistosoma japonicum after Pairing. PLoS One. 2015;10(6):e0129626–e. 10.1371/journal.pone.0129626 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Total proteins were identified and quantified by Mascot daemon version 2.3.2 software. The protein database was collected from an NCBInr database (24 October 2018) specific to P. falciparum 3D7.
(PDF)
Total proteins were identified and quantified by Mascot daemon version 2.3.2 software. The protein database was collected from an NCBInr database (24 October 2018) specific to P. falciparum 3D7.
(PDF)
Total proteins were identified and quantified by Mascot daemon version 2.3.2 software. The protein database was collected from an NCBInr database (24 October 2018) specific to P. falciparum 3D7.
(PDF)
The experiment was performed in three biological replicates. Proteins with altered expression levels were selected based on different expression levels in at least two biological replicates.
(PDF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.