SUMMARY
N6-methyladenosine (m6A) modification of mRNA is emerging as a vital mechanism regulating RNA function. Here, we show that fragile X mental retardation protein (FMRP) reads m6A to promote nuclear export of methylated mRNA targets during neural differentiation. Fmr1 knockout (KO) mice show delayed neural progenitor cell cycle progression and extended maintenance of proliferating neural progenitors into postnatal stages, phenocopying methyltransferase Mettl14 conditional KO (cKO) mice that have no m6A modification. RNA-seq and m6A-seq reveal that both Mettl14cKO and Fmr1KO lead to the nuclear retention of m6A-modified FMRP target mRNAs regulating neural differentiation, indicating that both m6A and FMRP are required for the nuclear export of methylated target mRNAs. FMRP preferentially binds m6A-modified RNAs to facilitate their nuclear export through CRM1. The nuclear retention defect can be mitigated by wild-type but not nuclear export-deficient FMRP, establishing a critical role for FMRP in mediating m6A-dependent mRNA nuclear export during neural differentiation.
Graphical Abstract
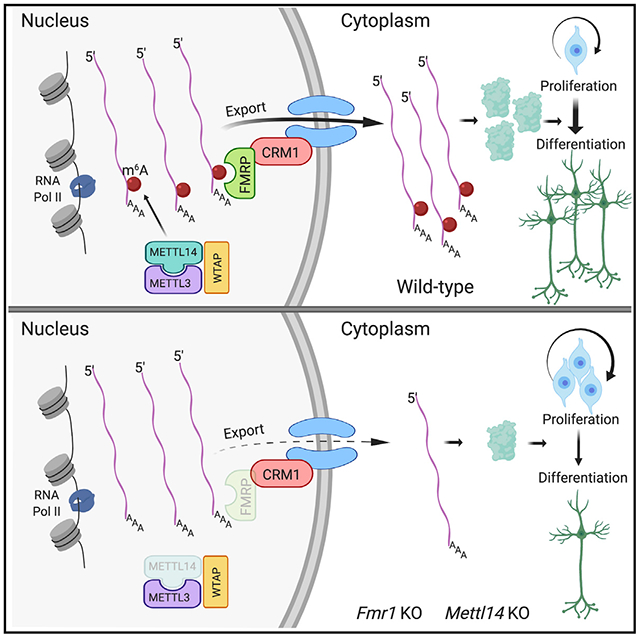
In Brief
Edens et al. reveal fragile X mental retardation protein (FMRP) as an m6A reader that promotes the nuclear export of methylated mRNAs during neural differentiation. Loss of either Fmr1 or the m6A methyltransferase Mettl14 results in the nuclear accumulation of neural differentiation-related mRNAs, causing delayed neural progenitor differentiation in mice.
INTRODUCTION
Methylation of RNA on N6-adenosine (m6A) is emerging as a critical mechanism regulating different aspects of RNA metabolism and function, including stability (Du et al., 2016; Ke et al., 2017; Wang et al., 2014), localization (Roundtree et al., 2017), and translation (Lin et al., 2016; Meyer et al., 2015; Wang et al., 2015). In mammals, methyltransferases METTL3 (methyltransferase-like 3) and METTL14 form a complex that mediates the addition of methyl groups to adenosines in target RNAs (Liu et al., 2014). Readers such as the YTH (YT521-B homology) domain containing proteins bind and interpret m6A in a sequence-dependent manner (Dominissini et al., 2012; Wang et al., 2014; Xu et al., 2014). The presence of m6A erasers has also been revealed, suggesting the complex and dynamic regulation of m6A (Jia et al., 2011; Wei et al., 2018). The emerging biological functions regulated by m6A include the proliferation and differentiation of embryonic (Batista et al., 2014; Geula et al., 2015) and neural stem cells (Wang et al., 2018; Yoon et al., 2017), as well as complex behaviors and processes such as circadian rhythms (Fustin et al., 2013), stress response (Engel et al., 2018), and learning (Koranda et al., 2018).
The RNA-binding protein fragile X mental retardation protein (FMRP) is encoded by the fragile X mental retardation gene (FMR1), mutations in which result in fragile X syndrome, the leading genetic cause of intellectual disability. FMRP contains both a nuclear localization sequence (NLS) and a nuclear export sequence (NES), and it is localized in both the nucleus and the cytoplasm (Eberhart et al., 1996; Kim et al., 2009). The best-studied function of FMRP is the negative regulation of mRNA translation; thus, fragile X syndrome may result in part from the aberrant expression of FMRP target genes (Darnell et al., 2011; Richter et al., 2015). Functionally, loss of Fmr1 has been associated with abnormalities in cortical development and dendritic spine formation, which may contribute to aberrant learning and behavior in fragile X syndrome (Castrén et al., 2005; La Fata et al., 2014; Saffary and Xie, 2011; Shen et al., 2019; Tervonen et al., 2009).
Here, we report that FMRP binds m6A-modified mRNAs and promotes their nuclear export to regulate neural differentiation. Fmr1 knockout (Fmr1KO) mice show delayed cell cycle progression and neural differentiation, phenocopying methyltransferase Mettl14 conditional KO (Mettl14cKO) mice that are devoid of m6A modification (Yoon et al., 2017). Both Mettl14cKO and Fmr1KO lead to the nuclear retention of m6A-modified FMRP target mRNAs regulating neural differentiation, including components of Notch and Hedgehog signaling pathways. FMRP preferentially binds m6A-modified target mRNAs and cooperates with the nuclear export protein CRM1 to facilitate nuclear export. Nuclear retention of methylated FMRP target mRNAs in Fmr1KO can be mitigated by wild-type Fmr1 but not NES-lacking Fmr1ΔNES, suggesting that FMRP is sufficient to drive the nuclear export of m6A-tagged FMRP target mRNAs, and this mechanism requires the NES-nuclear export. Our findings establish a role for FMRP in regulating m6A-dependent mRNA nuclear export during neural differentiation.
RESULTS
Genetic KO of Fmr1 Leads to Delayed Neural Progenitor Cell Cycle Progression
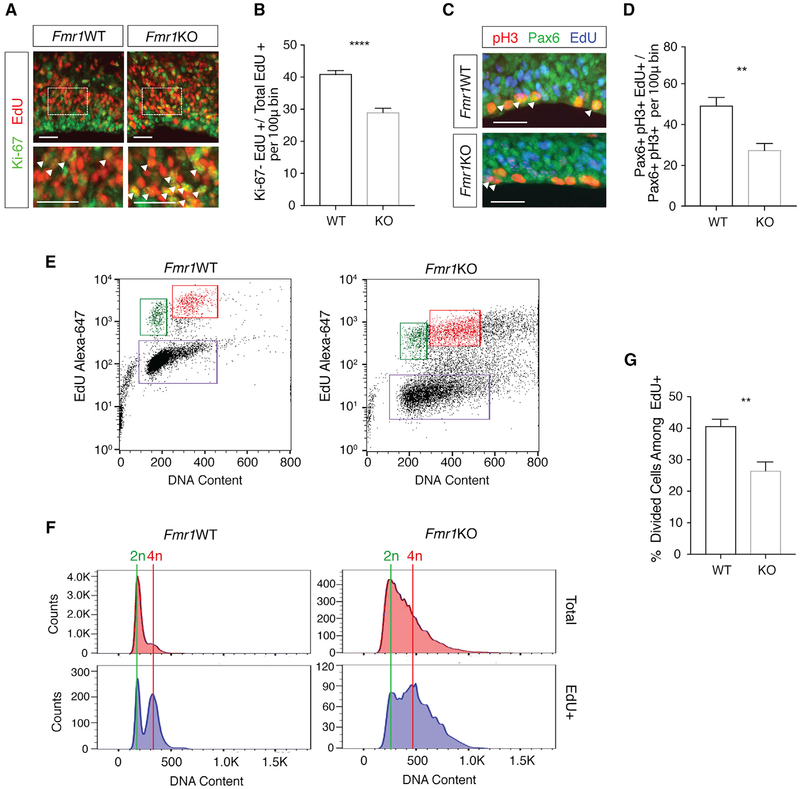
Target binding site analysis suggests that FMRP may bind to consensus RNA methylation motifs (Ascano et al., 2012; Fu et al., 2014). To test whether the genetic removal of Fmr1 affects m6A-related functions during development in vivo, we assessed cortical neural progenitor cell cycle progression, which is regulated by m6A (Yoon et al., 2017). We exposed embryonic day 17.5 (E17.5) wild-type (WT) and Fmr1KO mouse embryos to 5-ethynyl-2′-deoxyuridine (EdU) for 24 h and then measured the cell cycle exit by assessing the colocalization of EdU with Ki-67, a marker of proliferation (Figure 1A). The number of cells that had exited the cell cycle within the 24 h of EdU exposure (Ki-67−EdU+) was significantly decreased in Fmr1KO mice compared to WT mice (Figure 1B), suggesting delayed neural progenitor cell cycle exit in Fmr1KO mice.
Figure 1. Genetic Knockout of Fmr1 Leads to Delayed Neural Progenitor Cell Cycle Progression.
(A) Analysis of cell cycle exit in WT and Fmr1KO E17.5 cortical progenitors. EdU (24 h) staining is shown in red and Ki-67 in green. Enlarged sections (bottom) highlight Ki-67+EdU+ cells (arrows). Scale bars, 25 μm.
(B) Significantly fewer EdU+ cells exit the cell cycle during the 24-h EdU exposure in Fmr1KO cortex (****p ≤ 0.0001; n = 5 WT, 6 KO mice). Data are presented as mean + SEM.
(C) Analysis of M phase entry in WT and Fmr1KO cortical progenitors. pH3 staining is shown in red, Pax6 in green, and EdU in blue. Scale bar, 25 μm.
(D) Significantly fewer EdU+ RGCs entered mitosis within the 2-h EdU exposure in Fmr1KO cortex (**p = 0.0017; n = 6 WT, 6 KO mice). Data are presented as mean + SEM.
(E) FACS analysis of cell cycle dynamics in WT and Fmr1KO NPCs. Cells labeled by EdU are shown in the upper quadrants. Cells that divided during the 5-h incubation are shown in green and cells remaining in G2/M are in red.
(F) Histograms comparing 2n (divided) and 4n (undivided) NPCs between WT and Fmr1KO.
(G) Significantly fewer Fmr1KO EdU+ NPCs completed division by the end of 5 h compared to EdU+ WT NPCs (**p = 0.0018; n = 6 WT, 6 KO mice). Data are presented as mean + SEM.
To explore progenitor cell cycle dynamics further, we exposed E17.5 mouse embryos to EdU for 2 h, followed by labeling with the radial glial cell (RGC) marker Pax6 and the mitotic marker phospho-histone H3 (pH3) (Figure 1C). Triple-labeled cells (pH3+Pax6+EdU+) are RGCs that have progressed from S phase to M phase within the 2-h span of EdU exposure. Comparison of this population to the total Pax6+EdU+ population allows for the assessment of relative rates of cell cycle progression in RGCs. There was a significant reduction in the number of EdU+ RGCs that had entered M phase (EdU+PH3+Pax6+) in the Fmr1KO mouse cortex (Figure 1D), suggesting that cell cycle progression to M phase is delayed by the loss of Fmr1 in vivo.
Further analysis of cell cycle dynamics using flow cytometry revealed prolonged G2/M in cultured Fmr1KO neural precursor cells (NPCs) (Figures 1E and 1F). Following a 30-min EdU pulse and a 5-h incubation, there were significantly fewer divided Fmr1KO NPCs (2n DNA content), accompanied by a greater number of undivided NPCs (4n DNA content), compared to WT (Figure 1G). No difference was observed in EdU incorporation (Figure S1). These data indicate a delay in mitotic exit in Fmr1KO NPCs. Observations from these three experiments in Fmr1KO mice phenocopy altered cell cycle dynamics reported in methyltransferase Mettl14cKO mice that have no m6A (Yoon et al., 2017), suggesting that FMRP mediates m6A function during neural progenitor differentiation.
Genetic Deletion of Fmr1 in Mice Results in Prolonged Maintenance of Proliferating Neural Progenitors in the Postnatal Mouse Cortex
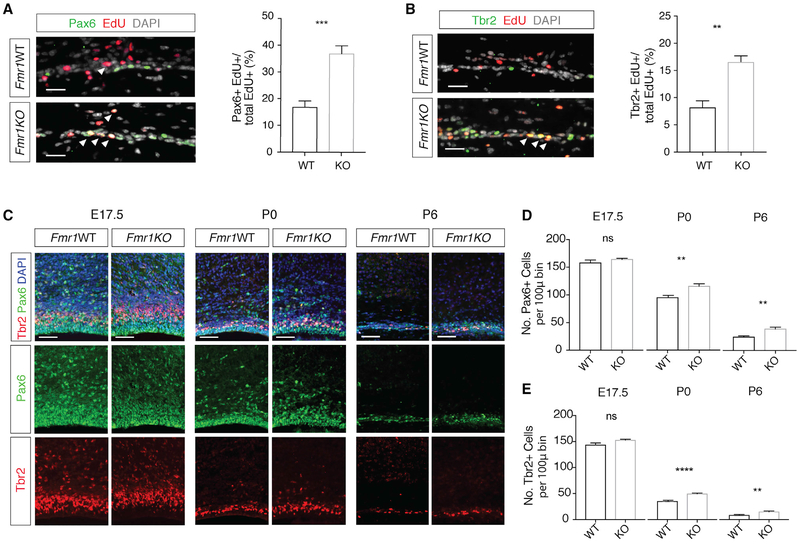
Delayed cell cycle exit may lead to the extended maintenance of neural progenitors. During mouse cortical development, multipotent Pax6+ RGCs and fate-restricted Tbr2+ intermediate progenitors (INPs) are largely depleted by postnatal stages (Dwyer et al., 2016; Yoon et al., 2017). To test whether neural progenitors are maintained postnatally in Fmr1KO mice, we pulsed animals with EdU at postnatal day 5 (P5) and analyzed 2 days later at P7. A significant number of EdU+Pax6+ proliferating RGCs and EdU+Tbr2+ INPs were present in Fmr1KO mice, but very few were present in WT littermates (Figures 2A and 2B). In addition, we examined the maintenance of glial fibrillary acidic protein (GFAP)-expressing radial fibers, which are required for interkinetic nuclear migration and neural progenitor division (Solecki et al., 2004). These radial fibers are normally not maintained postnatally after the completion of cortical neurogenesis. Whereas there were few radial fibers spanning the cortical wall in WT mouse cortex, by P7 many were present in the Fmr1KO cortex (Figure S2A). These data indicate that Fmr1KO mice maintain proliferating neural progenitors into postnatal stages.
Figure 2. Genetic Deletion of Fmr1 in Mice Results in Extended Maintenance of Proliferating Neural Progenitors in the Postnatal Mouse Cortex.
(A) Immunostaining of Pax6 (green) and EdU (red) in P7 cortex from WT and Fmr1KO mice exposed to EdU (48 h). The number of Pax6+EdU+ cells, marked by arrowheads, is increased in KO cortex (***p = 0.0008; n = 5 WT, 4 KO mice). Scale bar, 25 μm. Data are presented as mean + SEM.
(B) Immunostaining of Tbr2 (green) and EdU (red) in P7 cortex from WT and Fmr1KO mice exposed to EdU (48 h). The number of Tbr2+EdU+ cells, marked by arrowheads, is increased in KO cortex (**p = 0.0018; n = 5 WT, 4 KO mice). Scale bar, 25 μm. Data are presented as mean + SEM.
(C) Immunostaining of Pax6 (green) and Tbr2 (red) in WT and Frm1KO cortex at E17.5, P0, and P6. Scale bar, 50 μm.
(D and E) Pax6+ (D) and Tbr2+ (E) cells are increased in Fmr1KO cortex: E17.5, Pax6+ (p = 0.1996; n = 3WT, 4 KO mice) and Tbr2+ (p = 0.053; n = 3 WT, 4 KO mice); P0, Pax6+ (**p = 0.0029; n = 7 WT, 7 KO mice) and Tbr2+ (****p < 0.0001; n = 7 WT, 7 KO mice); P6, Pax6+ (**p = 0.0058; n = 5WT, 6KO mice) and Tbr2+ (**p = 0.0059; n = 5 WT, 6 KO mice). Data are presented as mean + SEM.
To further characterize the impact of Fmr1 deletion on cortical development, we examined the number of Pax6+ RGCs and Tbr2+ INPs in WT and Fmr1KO mouse cortices at E17.5, P0, and P6. While differences in the numbers of RGCs and INPs were undetectable at E17.5, both populations showed significant increases in the Fmr1KO mouse cortex at P0 and P6 (Figures 2C-2E). No differences in the gross morphology of the cortex at embryonic or postnatal stages were detected (Figure S2B). These findings are very similar to the extended maintenance of the proliferating neural progenitors reported in Mettl14cKO mice, which are devoid of m6A (Yoon et al., 2017). The neural differentiation abnormalities we identified in Frm1KO mice phenocopy the m6A-dependent defects found in Mettl14cKO mice, suggesting a functional relation between FMRP and m6A in vivo.
FMRP Preferentially Binds m6A-Modified RNAs to Promote Their Nuclear Export during Neural Differentiation
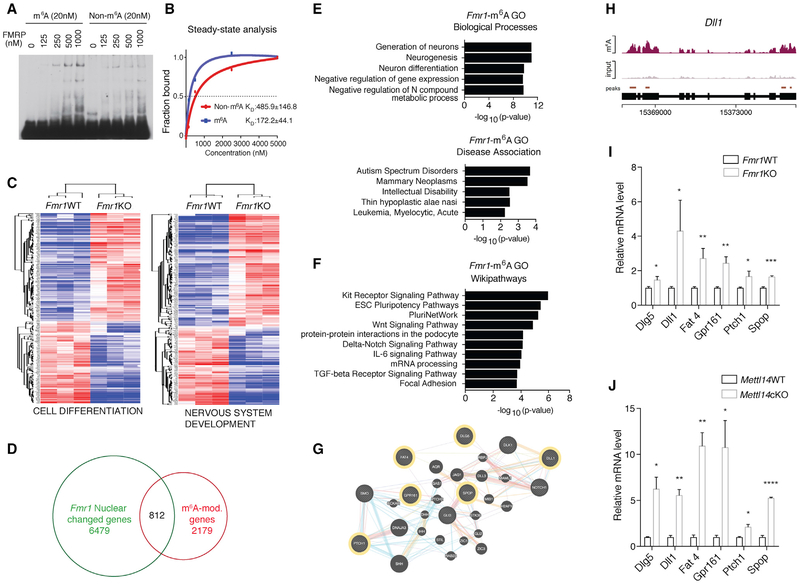
As previous analyses have suggested that FMRP may bind to consensus RNA methylation motifs (Ascano et al., 2012; Edupuganti et al., 2017; Fu et al., 2014), we considered that FMRP could serve as an m6A reader to bind and interpret m6A to regulate RNA function. To test this possibility, we performed electrophoretic mobility shift assay (EMSA) and bio-layer interferometry analysis to assess the binding affinity of purified FMRP to RNA oligos containing an endogenous m6A-modified sequence from mouse Dll1 mRNA or a consensus m6A motif. In both analyses, FMRP binding to RNA probes was significantly enhanced by m6A methylation, suggesting that FMRP preferentially binds to methylated RNAs compared to non-methylated RNAs (Figures 3A, 3B, S3A, and S3B).
Figure 3. FMRP Preferentially Binds m6A-Modified RNAs to Promote Their Nuclear Export in Regulating Neural Differentiation.
(A) EMSA comparing FMRP binding to non-methylated (left) or methylated (right) RNA.
(B) Steady-state analysis of FMRP binding to methylated or non-methylated RNA using bio-layer interferometry. Results are averaged from three independent experiments.
(C) Heatmaps comparing WT and Fmr1KO NPC nuclear expression of genes related to cell differentiation (left) and nervous system development (right).
(D) Venn diagram showing RNAs that are m6A modified and differentially expressed in Fmr1KO nucleus.
(E) Gene Ontology (GO) analysis of RNAs that are both differentially expressed in Fmr1KO nucleus and m6A modified. Biological processes (top) and disease associations (bottom) are shown.
(F) Pathway analysis of RNAs that are both differentially expressed in Fmr1KO nucleus and m6A modified.
(G) Gene interaction network of Hedgehog- and Notch-related signaling components and their cross-talk. m6A-tagged FMRP targets are outlined in yellow.
(H) Coverage plot of m6A modification of Dll1 mRNA.
(I) RNAs of Hedgehog- and Notch-related components are retained in Fmr1KO nucleus quantified by qRT-PCR: Dlg5 (*p = 0.0324), Dll1 (*p = 0.0433), Fat4 (**p = 0.0065), Gpr161 (**p = 0.0020), Ptch1 (*p = 0.0262), and Spop (***p = 0.0006); n = 5 WT, 3 KO biological replicates. Normalization is to U1. Data are presented as mean + SEM.
(J) RNAs of Hedgehog- and Notch-related components are retained in Mettl14cKO nucleus quantified by qRT-PCR: Dlg5 (*p = 0.0180), Dll1 (**p = 0.0016), Fat4 (**p = 0.0021), Gpr161 (*p = 0.0361), Ptch1 (*p = 0.0164), and Spop (****p < 0.0001); n = 3 WT, 4 cKO biological replicates. Normalization is to U1. Data are presented as mean + SEM.
As an RNA-binding protein, FMRP localizes to both the nucleus and the cytoplasm (Eberhart et al., 1996; Kim et al., 2009). Therefore, we hypothesized that FMRP could mediate nuclear RNA export. To test this, we performed RNA sequencing (RNA-seq) to compare mRNA levels in WT and Fmr1KO NPC nuclear fractions (Figures S4A and S4B). We found that genes involved in cell differentiation, neural development (Figure 3C), and embryonic development (Figure S3C) were enriched in Fmr1KO nuclear fractions, suggesting nuclear retention of these mRNAs upon the loss of Fmr1.
To understand how the binding of FMRP to m6A affects neural differentiation, we identified all of the mRNAs that are both m6A tagged and differentially expressed in Fmr1KO NPC nucleus by comparing the above RNA-seq with m6A-seq of mouse embryonic neural progenitors (Figure 3D). Gene Ontology analysis revealed enrichment in processes such as neurogenesis, neural differentiation, and regulation of gene expression (Figure 3E), as well as chromatin and DNA binding (Figure S3D). Furthermore, pathway analysis yielded terms related to pluripotency and developmental signaling pathways (Figure 3F). Among the mRNAs that are both m6A tagged and differentially expressed in Fmr1KO NPC nucleus, components of the Notch and Hedgehog pathways (Figures 3G and 3H) are particularly interesting, as they are involved in regulating the balance between neural stem cell maintenance and differentiation and have been genetically associated with intellectual disability (Chaudhry et al., 2015; Noor et al., 2010). We focused on six of the m6A-modified, differentially expressed Notch- and Hedgehog-related mRNAs (Ptch1, Dll1, Dlg5, Fat4, Gpr161, and Spop), all of which are also validated FMRP targets (Ascano et al., 2012). We quantified the levels of these six targets in WT and Fmr1KO NPCs by qRT-PCR. Each mRNA showed increased nuclear retention in Fmr1KO NPCs (Figure 3I), suggesting a critical role for FMRP in the nuclear export of these methylated target mRNAs. None of these mRNAs showed increased whole-cell levels, indicating that the increase in nuclear levels was not caused by increased general transcription (Figure S3E). We next tested the effect of the loss of m6A on the nuclear levels of these mRNAs in Mettl14cKO NPCs. The elevated nuclear retention of the FMRP target mRNAs observed in Fmr1KO NPCs was largely phenocopied in Mettl14cKO NPCs (Figure 3J), suggesting a nuclear export mechanism that requires both FMRP and m6A.
FMRP Mediates the Nuclear Export of m6A-Tagged Target mRNAs through CRM1
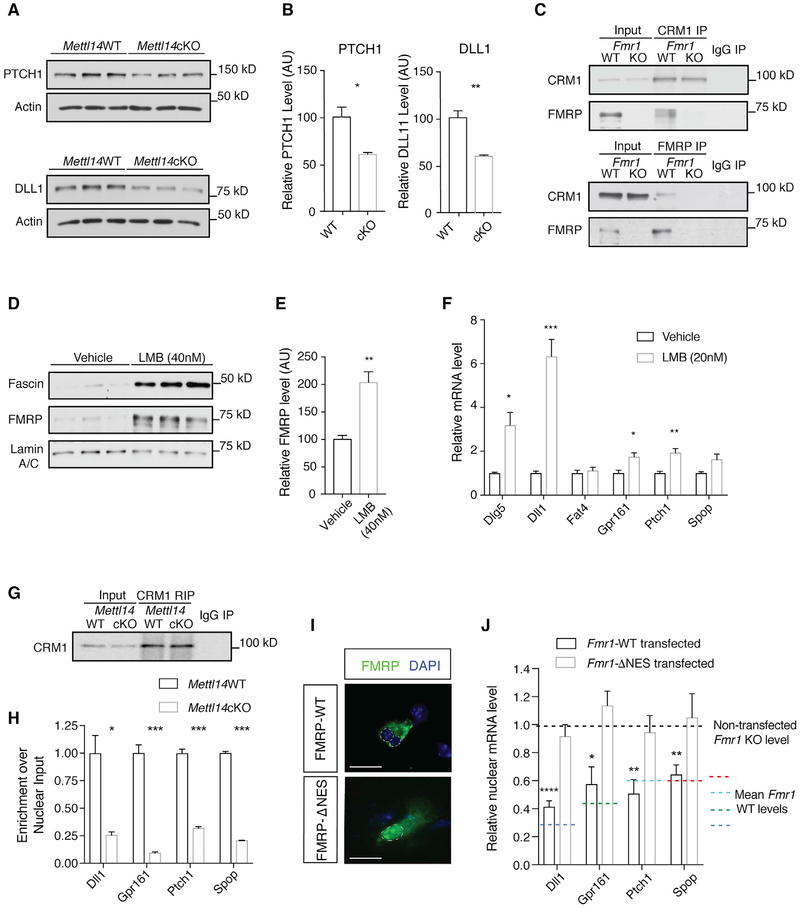
Our results indicate that FMRP can read m6A and that loss of either Fmr1 or m6A results in the reduced nuclear export of methylated FMRP target mRNAs. Nuclear export of mRNA has been shown to be a critical mechanism regulating gene expression (Prasanth et al., 2005; Wegener and Muller-McNicoll, 2018). Therefore, nuclear export defects in Mettl14cKO NPCs may indicate the altered protein expression of methylated FMRP targets. Western blot analysis revealed a significant reduction in PTCH1 and DLL1 protein levels in Mettl14cKO NPCs compared to WT (Figures 4A and 4B). This finding further supports the functional importance of FMRP-m6A-mediated nuclear export of mRNA in neural differentiation.
Figure 4. FMRP Mediates the Nuclear Export of m6A-Tagged FMRP Target mRNAs through CRM1.
(A) Western blot of PTCH1 and DLL1 in WT and Mettl14cKO NPC whole-cell lysates.
(B) PTCH1 (*p = 0.0178; n = 3 WT, 3 cKO biological replicates) and DLL1 (**p = 0.0045; n = 3 WT, 3 cKO biological replicates) protein levels are significantly decreased in Mettl14cKO NPCs. Data are presented as mean + SEM.
(C) Immunoprecipitation of CRM1 (top) or FMRP (bottom) co-precipitates FMRP or CRM1, respectively, in the WT mouse cortex.
(D) Western blot of CRM1 inhibition by LMB in WT NPC nuclear fractions. Lamin A-C serves as loading control and fascin as a positive control.
(E) The nuclear FMRP level is elevated in WT NPCs following LMB treatment (40 nM) (**p = 0.0069; n = 3 biological replicates). Data are presented as mean + SEM.
(F) Notch- and Hedgehog-related RNAs show nuclear retention in WT NPCs following LMB treatment (20 nM) quantified by qRT-PCR: Dlg5 (*p = 0.0170), Dll1 (***p = 0.0006), Fat4 (p = 0.5815), Gpr161 (*p = 0.0154), Ptch1 (**p = 0.0048), and Spop (p = 0.0518); n = 4 vehicle, 6 LMB biological replicates. Normalization is to Snora3. Data are presented as mean + SEM.
(G) CRM1 RIP from WT or Mettl14cKO NPCs.
(H) CRM1 binding to Notch- and Hedgehog-related RNAs is reduced in Mettl14cKO NPCs quantified by qRT-PCR: Dll1 (**p = 0.01), Gpr161 (***p = 0.00029), Ptch1 (***p = 0.0002), and Spop (****p < 0.0001); n = 3 biological replicates. Normalization is to U1. Data are presented as mean + SEM.
(I) Cytoplasmic localization of FMRP-WT and nuclear localization of FMRPΔNES in Fmr1KO NPCs. Scale bar, 20 μm.
(J) Nuclear retention of target mRNAs in Fmr1KO NPCs is rescued by the expression of WT (Dll1 [****p < 0.0001], Gpr161 [*p = 0.026], Ptch1 [**p = 0.008], and Spop [**p = 0.006]; n = 3 biological replicates) but not ΔNES Fmr1 (Dll1 [p = 0.372], Gpr161 [p = 0.253], Ptch1 [p = 0.672], Spop [p = 0.78]; n = 3 biological replicates). Data are presented as mean + SEM.
mRNAs can be exported from the nucleus through either NXF1 or CRM1 (Hutten and Kehlenbach, 2007). CRM1-dependent export is selective, requiring that target mRNAs first bind a CRM1-interacting RNA-binding protein. We identified specific CRM1-FMRP interaction by co-immunoprecipitation (Figure 4C). We also found nuclear accumulation of FMRP following CRM1 inhibition by leptomycin B (LMB) (Figures 4D, 4E, and S4A), suggesting that FMRP is exported from the nucleus through a CRM1-mediated mechanism. To determine whether m6A-tagged FMRP target mRNAs related to Notch and Hedgehog signaling are exported through CRM1, we tested the nuclear levels of these mRNAs in LMB-treated NPCs. Following CRM1 inhibition, there was an increase in the nuclear levels of a majority of these targets (Figure 4F), suggesting that nuclear export of m6A-tagged FMRP target mRNAs is mediated through CRM1. To determine the role of m6A in this process, we performed RNA immunoprecipitation (RIP) of CRM1 in WT and Mettl14cKO NPCs, followed by quantification using qRT-PCR. There was a significant reduction in CRM1 binding to FMRP target mRNAs in Mettl14cKO RIP samples (Figures 4G and 4H), suggesting that RNA methylation is required for CRM1 binding to m6A-modified FMRP target mRNAs and their nuclear export. To further establish the functional link between FMRP and the nuclear export of m6A-tagged target mRNAs, we reintroduced either WT Fmr1 or the NES-lacking Fmr1ΔNES (Figure 4I) into Fmr1KO NPCs and assessed the levels of methylated FMRP target mRNAs by qRT-PCR. While the expression of WT FMRP significantly mitigated the nuclear retention of the methylated FMRP target mRNAs, expression of the nuclear-restricted FMRPΔNES produced no rescue effect (Figure 4J). These data demonstrate a critical role for FMRP in mediating the nuclear export of m6A-tagged target mRNAs to regulate neural differentiation.
DISCUSSION
RNA methylation on N6-adenosine is emerging as a vital mechanism regulating RNA metabolism and function (Dominissini et al., 2012; Meyer et al., 2012; Peer et al., 2017). Here, we report that FMRP regulates neural differentiation through m6A-dependent mRNA nuclear export. We also elucidated the underlying mechanism by which FMRP preferentially binds m6A-modified mRNAs and facilitates their nuclear export through CRM1 to regulate neural differentiation.
In Fmr1KO mice, we discovered delayed cell cycle progression and extended maintenance of proliferating neural progenitors into postnatal stages, which are also m6A-dependent phenotypes found in methyltransferase Mettl14cKO mice that have no m6A. These findings suggest a functional relation between FMRP and m6A in neural differentiation. Our RNA-seq and m6A-seq show that both Mettl14cKO and Fmr1KO lead to nuclear retention of m6A-modified FMRP target mRNAs regulating neural differentiation, indicating that both m6A methylation and FMRP are required for the nuclear export of target mRNAs. To establish the link between FMRP and m6A, we first tested the role of m6A in FMRP target mRNA nuclear export by CRM1 RIP in WT and Mettl14cKO NPCs. We found significantly reduced CRM1 binding to FMRP target mRNAs in Mettl14cKO NPCs that have no m6A (Figures 4G and 4H). This finding highlights the importance of m6A in CRM1-mediated nuclear export of FMRP targets. In addition, we also tested whether the reintroduction of FMRP could rescue the nuclear retention of m6A-tagged FMRP target mRNAs, including components of Notch and Hedgehog signaling pathways that have well-established roles in regulating neural proliferation and differentiation. We found that only WT FMRP, not the NES-lacking and nuclear restricted FMRPΔNES, alleviated nuclear retention deficits in Fmr1KO NPCs (Figures 4I and 4J). Thus, FMRP is sufficient to drive the nuclear export of m6A-tagged FMRP target mRNAs, and this mechanism requires the NES-nuclear export. Our findings establish a critical role for FMRP in regulating m6A-dependent mRNA nuclear export during neural differentiation.
Recently, in a large-scale screen to establish a global m6A interactome using mass spectrometry, FMRP was found to be one of more than 20 candidate proteins that may interact with an m6A-modified probe of 4 GGACU repeats with higher affinity in cell lines (Edupuganti et al., 2017). However, a separate study found no preferential binding of FMRP to m6A-modified RNA probes (Zhang et al., 2018), although specific enrichment of m6A in FMRP targets was discovered. The discrepancy could be due to differences in experimental conditions, design of RNA probes, or the quality of purified FMRP protein used. We used full-length FMRP iso1 purified from Sf9 cells and probe RNA sequences from mouse Dll1 mRNA containing both consensus FMRP-binding sites and RNA methylation motifs to show that FMRP preferentially binds to methylated RNA in both EMSA and bio-layer interferometry analysis. Our data establish the role of FMRP as an m6A reader both in vivo and in vitro.
The nuclear export of mRNAs related to Notch and Hedgehog signaling was found in our study to require both FMRP and m6A. Delta-like (Dll) binding to the Notch receptor drives the expression of Hes1 to maintain stem cell pluripotency and suppress differentiation (Ishibashi et al., 1995). Notch has also been shown to cross-talk with additional pathways, including Hedgehog, to regulate neural stem cell proliferation and differentiation. Hedgehog-induced maintenance of stem cell pluripotency relies in part on the activation of Notch to balance proliferation with differentiation (Dave et al., 2011; Kong et al., 2015). We found increased nuclear retention of m6A-modified FMRP targets related to Notch and Hedgehog pathways in both Fmr1KO and Mettl14cKO NPCs. Our findings suggest that a disruption in the balance between proliferation and differentiation regulated by FMRP-m6A-dependent nuclear export may underlie the aberrant neural differentiation phenotypes observed in Fmr1KO and Mettl14cKO mice, eventually leading to defects in neuronal function.
We discovered the extended maintenance of neural progenitors into postnatal stages, accompanied by delayed cell cycle progression and neural differentiation in Fmr1KO mice. Cell cycle phase and duration are tightly coupled to neurogenic potential (McConnell and Kaznowski, 1991; Pilaz et al., 2016). Moreover, there is a strong association between neuronal birth-date and identity (Rakic, 1988). Therefore, the temporal aberrations in neural progenitor cell cycle and differentiation observed in Fmr1KO mice may influence cortical development by affecting neuron subtype specification, contributing to functional deficits in fragile X syndrome. Consistent with our observations, alterations of layer-specific neuron migration and localization have been found in Fmr1KO mice (La Fata et al., 2014; Tervonen et al., 2009). In addition, delayed cell cycle exit in Fmr1KO progenitors may deregulate the excitatory-inhibitory balance in the developing neural circuitry, as inhibitory interneuron integration into the cortex follows and depends upon pyramidal neuron differentiation and patterning (Bartolini et al., 2013). Defects in these processes in Fmr1KO mice could drive the altered synaptic landscape that is characteristic of fragile X syndrome. Our study elucidates a mechanism by which FMRP reads and facilitates the nuclear export of m6A-modified mRNAs to regulate neural differentiation, defects in which may contribute to functional deficits in fragile X syndrome.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Yongchao C. Ma (ma@northwestern.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All animal experiments conducted within this study have been approved by the Institutional Animal Care and Use Committee (Protocol #IS6359), and performed in accordance with federal regulations governing the use of animals in laboratory research. All animals were maintained in standard housing conditions with 12-hour light/dark cycle and food and water available ad libitum. Fmr1 mice (B6.129P2-Fmr1tm1Cgr/J Jackson Laboratory #003025), originally generated by the Dutch-Belgian Fragile X Consortium (1994), were obtained through Dr. Anis Contractor from Dr. David Nelson’s laboratory, where the original knockout line was backcrossed onto C57/BL6 background. Heterozygous Fmr1 female mice were bred with C57/BL6 males to yield wild-type and knockout littermates for all FMRP experiments at indicated time points (E17.5, P0, P5, or P7), or utilized for generation of neural precursor cells (NPCs) at E13.5. Because Fmr1 is an X-linked gene, only males were used for knockout analysis with the mating scheme described.
METHOD DETAILS
Cell Culture
Dorsal forebrains from timed-pregnant E13.5 mouse embryos were digested with Accutase (Fisher) to yield dissociated cortical neural precursor cells (NPCs) for culture. NPCs were carried on plates coated with Matrigel (Corning) at 80 μg/ml and maintained in DMEM-F12 medium (GIBCO) supplemented with B27 (GIBCO), N2 (GIBCO), and Glutamax (GIBCO). A growth factor cocktail containing EGF (PeproTech) (20ng/ml) and basic FGF (PeproTech) (20ng/ml) in Heparin (5 μg/ml) was added to the medium fresh. Cells were carried at densities not exceeding 80%, and all experiments were performed on density- and passage-matched NPC cultures. Cells were incubated in standard conditions: 37°C with 5% CO2. Multiple lines of NPCs were generated from independent litters of Fmr1 wild-type and knockout embryos.
Immunohistochemistry
Brains harvested from embryonic stages were fixed in fresh 4% PFA (Sigma) for six hours at 4°C, then washed extensively in PBS prior to embedding. Postnatal mice were subjected to transcardial perfusion with PBS, then PFA before brains were harvested and fixed in fresh 4% PFA for either six hours (P0), or overnight (P5, P7). Coronal sections (18 μm) were prepared using a Leica CM 3050S cryostat. Slides were baked at 65°C for two hours. For antigen retrieval, slides were submerged in Antigen Unmasking Solution (DAKO) at 95°C for twenty minutes, then rehydrated in PBS for five minutes. Sections were blocked for one hour at room temperature in 0.01% PBS-TritonX containing 3% BSA and 5% goat and donkey serum. Sections were incubated with primary antibodies overnight, and secondary antibodies for one hour at room temperature. All antibodies were diluted in blocking solution as follows: rat anti-Ki-67 (DAKO, 1:10), rabbit anti-GFAP (DAKO, 1:200), rabbit anti-phospho-Histone H3 (CST, 1:400), rabbit anti-Tbr2 (Abcam, 1:400), and mouse anti-Pax6 (BD Biosciences, 1:400). For embryonic EdU experiments, timed-pregnant dames were injected with EdU at a concentration of 150mg/kg for the indicated durations (2 hours or 24 hours). For postnatal EdU experiments, pups were given an injection of EdU at a concentration of 75mg/kg for the indicated duration (forty-eight hours). For EdU detection, the Click-iT Plus EdU Alexa Fluor 647 Imaging Kit (Invitrogen) was used according to manufacturer’s protocol. Images were acquired on a Leica DM 2500 epifluorescence microscope outfitted with a RETIGA 4000R camera (Q-Imaging). Cell counts represent the total number of labeled cells (as indicated) within 100 μm bins.
Flow Cytometry
NPCs were exposed to EdU for a duration of thirty minutes, and then either fixed immediately or incubated for five hours and then fixed, and incorporation was detected with the Click-iT Plus EdU Alexa Fluor 647 Flow Cytometry Assay Kit (Invitrogen) according to manufacturer’s protocol. DNA was labeled with 7-AAD (Thermo/Fisher). Data were collected using a BD LSR II Flow cytometer (BD Biosciences). Flow cytometry data were visualized with FlowJo software.
Nuclear RNA Extraction and Quality Control
NPCs were harvested at ~80% confluency with Accutase (Fisher), and nuclear fractions were prepared as described in the NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo/Fisher) manufacturer’s protocol, with minor modifications. 0.5U/μl of RNase inhibitor (Ambon/Fisher) was added to the CERI reagent to prevent degradation. After separation from the cytoplasmic fraction, the nuclear pellet was washed twice with PBS (prepared in DEPC water), and resuspended in an appropriate volume of TRIzol reagent (Invitrogen). The nuclear pellet was homogenized with a Polytron PT 1200E (Kinematica). RNA was purified using the TRIzol reagent manufacturer’s protocol, and quantified using the Quant-iT RNA HS Assay Kit (Invitrogen). The efficiency of separation was determined by measuring nuclear and cytoplasmic RNA fractions by qRT-PCR for the relative quantities of nuclear (U1) or cytoplasmic (beta-actin) control genes. Only nuclear RNA preparations with high U1 expression (~8-fold nuclear increase over cytoplasm) and low beta-actin expression (~7-fold cytoplasmic increase over nucleus) were utilized (Figure S4). For qPCR analysis of Leptomycin B (LMB) treated NPCs, a concentration of 20nM was used for 20 hours.
qRT-PCR
RNAs, prepared as described, were reverse-transcribed to produce cDNAs using the QuantaBio qScript cDNA SuperMix (VWR) according to manufacturer’s protocol, and the QuantaBio PerfeCTa SYBR® Green SuperMix (VWR) was used for quantitative real-time PCR (qRT-PCR). All reactions were run on the Applied Biosystems 7500 Fast Real-Time PCR System. The Delta-Delta-Ct method was used to compare relative transcript levels between groups. Unaffected control genes used for normalization are noted in the figure legends for each experiment.
RNA Sequencing and Analysis
RNAs were first subjected to Bioanalyzer analysis (Agilent) to assess RNA quality and concentration prior to sequencing. Libraries of nuclear fraction RNAs were prepared according to the TruSeq RNA-Seq Library protocol (Illumina). Sequencing was performed using the Illumina NextSeq500, 75SE High Output (100 million reads). Gene ontology analysis was performed using the TOPPFUN module of TOPPGENE (Chen et al., 2009). Pathway analysis was performed using Consensus PathDB (Herwig et al., 2016). The gene interaction network schematic was generated using GENEmania (Warde-Farley et al., 2010). Gene sets displayed in heatmaps were obtained from the GO term gene list as indicated within the figures. m6A sequencing data used to generate the list of overlapping Fmr1/m6A-tagged genes was reported previously (Yoon et al., 2017).
Immunoprecipitation
Cortices were harvested from Fmr1 wild-type and KO mice and homogenized with a Kinematica Polytron PT 2500E electric homogenizer in IP buffer (20mM Tris HCl (pH8), 137mM NaCl, 1% Triton-X, 2mM EDTA) supplemented with protease inhibitor cocktail (Roche). Equal amounts of wild-type and KO lysates were used for each IP reaction, and an equal amount of wild-type lysate was used for the IgG control IP reaction. Lysates were precleared with Protein A/G PLUS Agarose beads (SCBT) for one hour at 4°C, followed by a three-hour incubation with antibody (2 μg of CRM1 or 2.75 μg of FMRP) at 4°C. Protein A/G PLUS beads were then added for an overnight 4°C incubation. Beads were washed extensively (10mM Tris (pH7.4), 1mM EDTA, 1mM EGTA, 150mM NaCl, 1% Triton-X, 0.2mM NaVO3) before elution of bound IP complexes for western blot analysis. IP reactions were repeated at least three times, each time with consistent results. One representative experiment is shown. Inputs shown are 1%.
RNA Immunoprecipitation (RIP)
Starting material for each RIP reaction was ~16 million NPCs. Cells were cross-linked with ~1% EM-grade formaldehyde (Polysciences) for 15 minutes with gentle shaking. Glycine was added to a concentration of ~0.2M and shaken gently for five minutes to stop the reaction. Cells were collected, centrifuged to pellet, and resuspended in iced PBS (prepared in DEPC water). Cells were centrifuged to pellet, and washed twice more with PBS. The pellet was next resuspended in nuclear isolation buffer (0.256M sucrose, 8mM Tris HCl (pH7.5), 4mM MgCl2, 1% Triton-X) and incubated on ice for twenty minutes with mixing at regular intervals. Cells were then pelleted by centrifugation, resuspended in 1 mL of RIP buffer (150mM KCl, 25mM Tris HCl (pH7.4), 5mM EDTA, 0.5mM DTT, 0.5% NP40) supplemented with protease inhibitor cocktail (Roche) and RNase inhibitors (Fisher/Ambion). Cells were sheared with twenty strokes of a glass dounce homogenizer (Wheaton), then incubated on a rotator overnight at 4°C with 4 μg of CRM1 antibody (Bethyl). Protein A Dynabeads (Invitrogen) were then added and rotated for 3 hours at 4°C. Dynabeads were pelleted using a magnetic rack and washed four times with RIP buffer, then once with PBS. Beads were resuspended in 200nM RIP buffer containing 40U/ml RNase inhibitor and 20 μg of Proteinase K (VWR), and incubated at 42°C for one hour followed by one hour at 65°C. TRIzol was then added to the beads and RNA was purified as described.
Electrophoretic Mobility Shift Assay (EMSA)
Full-length FMRP isoform 1 protein, tagged with six histidine on the amino-terminus, was purified from codon-optimized baculovirus-infected Sf9 cells by Dr. Yang Xiang at ABclonal. Biotinylated oligos were synthesized containing mouse Dll1mRNA sequence, 5′-GAACACCAACAAGAAGGCGGXCUUUCACGGGGXCCAUGGAGCCGA-3′ or a consensus RNA methylation sequence, 5′-CGUGGXCUGGCU-3′ (X = A or m6A). In addition, oligos containing a mutated consensus sequence were synthesized 5′-GAUACXGAGAAG-3′. RNA probes were resuspended in DEPC-treated water and denatured by heating to 65°C for ten minutes. Recombinant FMRP protein was diluted in binding buffer (10mM HEPES, pH 8.0; 50mM KCl; 1mM EDTA; 0.05% Triton X-100; 5% glycerol; 10 μg/ml salmon sperm DNA; 1mM DTT; 40U/ml RNasin) to produce graded concentrations. One part RNA probe (20nM final concentration) was then mixed with one part protein (0nM, 125nM, 250nM, 500nM, and 1000nM final concentrations) and incubated at room temperature for thirty minutes. A rabbit anti-FMRP antibody (Cell Signaling Technology) or control rabbit IgG was added to binding reactions to induce supershift. The RNA probe-protein mixtures were run on a 4%–20% TBE gradient gel (Novex) and transferred onto a BioDYNE B nylon membrane (Fisher) using a Trans-Blot Turbo semi-dry transfer system (BioRad). The membrane was crosslinked using a UV Staratlinker 2400 (Stratagene) set to 180mg/cm2 for 45 to 60 s. Binding was detected using the Chemiluminescent Nucleic Acid Detection Module (Fisher). Densitometry measurements of all bands were made using the measurement log feature in Adobe Photoshop to calculate the dissociation constant (Kd). Results are averaged from three independent experiments.
Bio-Layer Interferometry Analysis
Bio-layer interferometry analysis of FMRP binding to the short m6A-modified and non-modified RNA oligos was assessed using the BLItz system (Forte Bio). Purified His-tagged FMRP (isoform 1) was loaded onto penta-His biosensors. Binding to m6A-modified and non-modified RNA probes was recorded at the following RNA concentrations: 5000nM, 2500nM, 500nM, 150nM, and 0nM. Reagents were prepared in binding buffer containing 100mM KCl, 20mM HEPES, 1mM EDTA, 0.05% Triton-X, 5% glycerol, and 1mM DTT. Each reaction was performed using 30 s for all baselines, 120 s for loading, 120 s for association, and 120 s for dissociation. KD was analyzed using MATLAB’s nonlinear curve-fit function as described above, and reported data represent three replicates.
Western Blot
Nuclear and cytoplasmic fractions were prepared for western blot analysis according to the NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo/Fisher) protocol and supplemented with protease inhibitor cocktail (Roche). Samples were run on SDS-PAGE, followed by transfer onto PVDF membranes. Membranes were blocked for one hour at room temperature in 5% nonfat milk, incubated with primary antibodies overnight at 4°C, and incubated with secondary antibodies for one hour at room temperature. Primary antibodies were diluted in 0.01% TBS-Tween containing 5% BSA as follows: Rabbit anti-FMRP (Cell Signaling Technology, 1:1000), Rabbit anti-PTCH1 (Proteintech, 1:1000), Rat anti-DLL1 (Millipore-Sigma, 1:500), rabbit anti-CRM1 (Bethyl Labs, 1:2500), mouse anti-Fascin (Santa Cruz Biotechnology, 1:1000), mouse anti-Lamin A/C (Cell Signaling Technology, 1:500), mouse anti-beta-actin (Sigma, 1:2000), and mouse anti-beta tubulin (Fisher, 1:2000). For western blot analysis of Leptomycin B (LMB)-treated NPCs, a concentration of 40nM LMB was used for 20 hours.
Nucleofection of NPCs
Fmr1KO NPCs were cultured as described. Dr. Stephanie Ceman kindly provided pEGFP-Flag-mFmr1 WT and pEGFP-Flag-mFmr1ΔNES plasmids (Kim et al., 2009). Nucleofection was performed using the Amaxa P3 Primary Cell 4D-Nucelofector X kit (Lonza) according to manufacturer’s protocol. Per reaction, about six million cells and 5 μg of plasmid DNA were used. The program “DS113” was utilized for all reactions. Following nucleofection, NPCs were cultured for 24 hours prior to harvesting nuclear and cytoplasmic fractions as described. All samples used for analysis showed comparable nuclear and cytoplasmic separation based on qRT-PCR analysis, and comparable nucleofection efficiency based on GFP fluorescence.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis was performed using Prism7 software (Graphpad). Non-linear curve fitting, used to determine the dissociation constant in Figure 3B, was performed using MATLAB (MathWorks). Specific details regarding the statistical test used, number of samples (n), and number of experimental replicates are in the figure legends. All graphs present mean + SEM. For all experiments, an alpha of 0.05 was used and significance level is indicated as follows: ns p > 0.05, * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, **** p ≤ 0.0001. The distributions of all datasets were analyzed prior to analysis to confirm the applicability of the statistical approach (i.e., verify Normal distribution of data subjected to t test).
DATA AND CODE AVAILABILITY
Sequencing data have been deposited to the Gene Expression Database (GEO) at NCBI. Accession: GSE121809
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-Pax6 (clone 018-1330) | BD Biosciences | Cat# 561462; RRID:AB_10715442 |
| Rabbit polyclonal anti-Tbr2 | Abcam | Cat# ab23345; RRID:AB_778267 |
| Rabbit polyclonal anti-GFAP | DAKO | Cat# Z0334; RRID:AB_10013382 |
| Rat monoclonal anti-Ki-67 | DAKO | Cat# M7249; RRID:AB_2250503 |
| Rabbit polyclonal anti-phospho Histone H3 | CST | Cat# 9701; RRID:AB_331535 |
| Rabbit polyclonal anti-FMRP | CST | Cat# 4317S; RRID:AB_1903978 |
| Rabbit polyclonal anti-CRM1 | Bethyl Labs | Cat# A300-469A; RRID:AB_451004 |
| Rabbit polyclonal anti-Lamin A/C | CST | Cat# 2032; RRID:AB_2136278 |
| Mouse monoclonal anti-Beta-tubulin | Fisher | Cat# A2228; RRID:AB_476697 |
| Mouse monoclonal anti-Fascin | SCBT | Cat# sc-21743; RRID:AB_627580 |
| Rabbit polyclonal anti-PTCH1 | Proetintech | Cat# 17520-1-AP; RRID:AB_2176561 |
| Rat monoclonal anti-DLL1 | Millipore/Sigma | Cat# MABN2284 |
| Peroxidase AffiniPure Goat Anti-Rabbit IgG (H+L) | Jackson Immunoresearch | Cat# 111035003 |
| Peroxidase AffiniPure Goat Anti-Mouse IgG (H+L) | Jackson Immunoresearch | Cat# 115035003 |
| Cy3 AffiniPure Donkey Anti-Rabbit IgG (H+L) | Jackson Immunoresearch | Cat# 711165152 |
| Cy2 AffiniPure Donkey Anti-Mouse IgG (H+L) | Jackson Immunoresearch | Cat# 715225150 |
| Cy2 AffiniPure Donkey Anti-Rat IgG (H+L) | Jackson Immunoresearch | Cat# 712225153 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| FMRP Recombinant Protein (isoform 1) | AB Clonal | N/A |
| EdU (5-ethynyl-2-deoxyuridine) | Invitrogen | Cat# A10044 |
| Paraformaldehyde | Sigma | Cat# P6148-1kg CAS 30525-89-4 |
| PFA, 16% ultrapure EM grade | Polysciences | Cat# 18814-20 |
| Target Retrieval Solution | Agilent/DAKO | Cat# S1699 |
| 7-AAD | Thermo Fisher Scientific | Cat# A1310 |
| StemPro Accutase | Fisher Scientific | Cat# A1110501 |
| SUPER Rase In RNase inhibitor | Fisher Scientific/Ambion | Cat# AM2696 |
| Matrigel | Corning | Cat# #354234 |
| DMEM-F12 | Fisher Scientific/GIBCO | Cat# 11320-033 |
| N-2 Supplement | Fisher Scientific/GIBCO | Cat# 17502048 |
| B27 serum free supplement | Fisher Scientific/GIBCO | Cat# 17504-044 |
| Glutamax | Fisher Scientific/GIBCO | Cat# 35050061 |
| Recombinant Human bFGF | PeproTech | Cat# 100-18B |
| recombinant murine EGF | PeproTech | Cat# 315-09 |
| Heparin | J.T. Baker/Avantor | Cat # M916-00 |
| Complete, Mini, EDTA-free Protease Inhibitor Cocktail | Roche | Cat# 11836170001 |
| Proteinase K | VWR | Cat# 97062-670 |
| Normal Goat Serum | Jackson Immunoresearch | Cat# 005000121 |
| Normal Donkey Serum | Jackson Immunoresearch | Cat# 017000121 |
| Bovine Serum Albumin, Fraction V, Heat Shock Treated | Fisher Scientific | Cat# BP1600100 |
| Leptomycin B | SCBT | Cat# sc-358688, CAS 87081-35-4 |
| TRIzol | Invitrogen | Cat# 15-596-026 |
| Critical Commercial Assays | ||
| Click-iT Plus EdU Alexa Fluor 647 Flow Cytometry Assay Kit | Invitrogen | Cat# C10635 |
| Click-iT Plus EdU Alexa Fluor 647 Imaging Kit | Invitrogen | Cat# C10640 |
| Chemiluminescent Nucleic Acid Detection Module | Fisher Scientific/Pierce | Cat# PI89880 |
| NE-PER Nuclear and Cytoplasmic Extraction Kit | Thermo Fisher Scientific | Cat# PI78835 |
| QuantaBio PerfeCTa SYBR® Green SuperMix with Low ROX | VWR | Cat# 101414-162 |
| QuantaBio qScript cDNA SuperMix | VWR | Cat# 101414-106 |
| Quant-iT RNA HS Assay Kit | Invitrogen | Cat# Q32852 |
| TruSeq RNASeq Library Prep Kit | Illumina | Cat# RS-122-2001 |
| Deposited Data | ||
| Fmr1 Nuclear Fraction RNA Seq data | This paper | GSE121809 |
| Experimental Models: Cell Lines | ||
| Nestin-Cre+/+; Mettl14f/f –derived NPCs | Yoon et al., 2017 | N/A |
| Experimental Models: Organisms/Strains | ||
| B6.129P2-Fmr1tm1Cgr/J | Jackson Laboratory | RRID:IMSR_JAX:003025 |
| Oligonucleotides | ||
| m6A Dll1 oligo: 5′-GAACACCAACAAGAAGGCGGXCUUUCACGGGG m6ACCAUGGAGCCGA-3′ | This paper | N/A |
| Non-m6A Dll1 oligo: 5′-GAACACCAACAAGAAGGCGGXCUUUCACGGGGACCAUGGAGCCGA-3′ | This paper | N/A |
| m6A short oligo: 5′-CGUGG m6ACUGGCU-3′ | This paper | N/A |
| Non-m6A short oligo: 5′-CGUGGACUGGCU-3′ | This paper | N/A |
| m6A mutated oligo: 5′-GAUAC m6AGAGAAG-3′ | This paper | N/A |
| Non-m6A mutated oligo: 5′-GAUACAGAGAAG-3′ | This paper | N/A |
| Primers for qPCR, see Table S1 | This paper | N/A |
| Software and Algorithms | ||
| FlowJo | FlowJo, LLC | RRID:SCR_008520, https://www.flowjo.com |
| TOPPGENE | Chen et al., 2009 | RRID:SCR_005726, https://toppgene.cchmc.org |
| GeneMANIA | Warde-Farley et al., 2010 | RRID:SCR_005709, http://genemania.org |
| Consensus Path-DB | Herwig et al., 2016 | http://cpdb.molgen.mpg.de |
| R | R Project for Statistical Computing | RRID:SCR_001905, https://www.r-project.org |
| MATLAB | MathWorks | RRID:SCR_001622, https://www.mathworks.com/products/matlab.html |
| Prism7 | GraphPad | SCR_002798; https://www.graphpad.com |
| Other | ||
| Protein A Dynabeads | Invitrogen | Cat# 10-001-D |
| Protein A/G PLUS-Agarose | SCBT | Cat# sc-2003; RRID:AB_10201400 |
| Kinematica Polytron PT 1200E Electric Homogenizer | Fisher Scientific | Cat# 05-400-261 |
| 5 Prime Phase Lock Gel Heavy 2 ml | VWR | Cat #10847-802 |
| BioDYNE B nylon membrane | Fisher Scientific/Pierce | Cat #77016 |
| Dip and Read Anti-Penta-HIS (HIS1K) Biosensors | Forte Bio | Cat #18-5120 |
Highlights.
FMRP reads m6A to promote mRNA nuclear export in neural differentiation
Neural progenitor differentiation is delayed by knockout (KO) of Fmr1 or Mettl14
Both Mettl14KO and Fmr1KO lead to nuclear retention of m6A-tagged FMRP target mRNAs
FMRP preferentially binds m6A-tagged mRNAs to facilitate nuclear export through CRM1
ACKNOWLEDGMENTS
This research was supported by grants from the NIH (R01NS094564 and R21NS106307 to Y.C.M.; R37NS047344, U19MH106434, and P01NS097206 to H. Song; RM1HG008935 to C.H.; and R01MH105128, R35NS097370, and U19AI131130 to G.-l.M.); the Simons Foundation Autism Research Initiative (SAFRI) to H. Song (575050); Cure SMA and The Hartwell Foundation (to Y.C.M.); and the Chicago Biomedical Consortium (to Y.C.M. and C.H.). C.V. was partially supported by an NSF predoctoral fellowship and NIH T32GM007445. We thank Dr. Stephanie Ceman for providing WT and ΔNES Fmr1 constructs, Dr. Anis Contractor for providing Fmr1 KO mice, and Dr. Xiaoxi Zhuang for sharing Mettl14f/f mice. We thank Dr. Tian Shao for technical assistance in validating anti-FMRP antibodies. The research reported in this manuscript was made possible in part by the services of the Keck Biophysics Facility and the NUSeq Core Facility, which is supported by the Northwestern University Center for Genetic Medicine, the Feinberg School of Medicine, and the Shared and Core Facilities of Northwestern University’s Office for Research. C.H. is a Howard Hughes Medical Institute Investigator. Y.C.M. is the Ann Marie and Francis Klocke M.D. Research Scholar supported by the Joseph and Bessie Feinberg Foundation.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.06.072.
DECLARATION OF INTERESTS
C.H. is a scientific founder and scientific advisory board member of Accent Therapeutics, Inc.
REFERENCES
- Ascano M Jr., Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, Hafner M, et al. (2012). FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature 492, 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolini G, Ciceri G, and Marín O (2013). Integration of GABAergic interneurons into cortical cell assemblies: lessons from embryos and adults. Neuron 79, 849–864. [DOI] [PubMed] [Google Scholar]
- Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, Bouley DM, Lujan E, Haddad B, Daneshvar K, et al. (2014). m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15, 707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrén M, Tervonen T, Kärkkäinen V, Heinonen S, Castrén E, Larsson K, Bakker CE, Oostra BA, and Akerman K (2005). Altered differentiation of neural stem cells in fragile X syndrome. Proc. Natl. Acad. Sci. USA 102, 17834–17839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry A, Noor A, Degagne B, Baker K, Bok LA, Brady AF, Chitayat D, Chung BH, Cytrynbaum C, Dyment D, et al. ; DDD Study (2015). Phenotypic spectrum associated with PTCHD1 deletions and truncating mutations includes intellectual disability and autism spectrum disorder. Clin. Genet 88, 224–233. [DOI] [PubMed] [Google Scholar]
- Chen J, Bardes EE, Aronow BJ, and Jegga AG (2009). ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37, W305–W311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. (2011). FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave RK, Ellis T, Toumpas MC, Robson JP, Julian E, Adolphe C, Bartlett PF, Cooper HM, Reynolds BA, and Wainwright BJ (2011). Sonic hedgehog and notch signaling can cooperate to regulate neurogenic divisions of neocortical progenitors. PLoS One 6, e14680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. [DOI] [PubMed] [Google Scholar]
- Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, Ma J, and Wu L (2016). YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun 7, 12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutch-Belgian Fragile X Consortium (1994). Fmr1 knockout mice: a model to study fragile X mental retardation. Cell 78, 23–33. [PubMed] [Google Scholar]
- Dwyer ND, Chen B, Chou SJ, Hippenmeyer S, Nguyen L, and Ghashghaei HT (2016). Neural Stem Cells to Cerebral Cortex: Emerging Mechanisms Regulating Progenitor Behavior and Productivity. J. Neurosci 36, 11394–11401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhart DE, Malter HE, Feng Y, and Warren ST (1996). The fragile X mental retardation protein is a ribonucleoprotein containing both nuclear localization and nuclear export signals. Hum. Mol. Genet 5, 1083–1091. [DOI] [PubMed] [Google Scholar]
- Edupuganti RR, Geiger S, Lindeboom RGH, Shi H, Hsu PJ, Lu Z, Wang SY, Baltissen MPA, Jansen PWTC, Rossa M, et al. (2017). N6-methyladenosine (m6A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol 24, 870–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel M, Eggert C, Kaplick PM, Eder M, Roh S, Tietze L, Namendorf C, Arloth J, Weber P, Rex-Haffner M, et al. (2018). The Role of m(6)A/m-RNA Methylation in Stress Response Regulation. Neuron 99, 389–403.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Dominissini D, Rechavi G, and He C (2014). Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet 15, 293–306. [DOI] [PubMed] [Google Scholar]
- Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I, and Okamura H (2013). RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155, 793–806. [DOI] [PubMed] [Google Scholar]
- Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, et al. (2015). Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347, 1002–1006. [DOI] [PubMed] [Google Scholar]
- Herwig R, Hardt C, Lienhard M, and Kamburov A (2016). Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nat. Protoc 11, 1889–1907. [DOI] [PubMed] [Google Scholar]
- Hutten S, and Kehlenbach RH (2007). CRM1-mediated nuclear export: to the pore and beyond. Trends Cell Biol. 17, 193–201. [DOI] [PubMed] [Google Scholar]
- Ishibashi M, Ang SL, Shiota K, Nakanishi S, Kageyama R, and Guillemot F (1995). Targeted disruption of mammalian hairy and enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 9, 3136–3148. [DOI] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, and He C (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol 7, 885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Pandya-Jones A, Saito Y, Fak JJ, Vågbø CB, Geula S, Hanna JH, Black DL, Darnell JE Jr., and Darnell RB (2017). m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 31, 990–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Bellini M, and Ceman S (2009). Fragile X mental retardation protein FMRP binds mRNAs in the nucleus. Mol. Cell. Biol 29, 214–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong JH, Yang L, Dessaud E, Chuang K, Moore DM, Rohatgi R, Briscoe J, and Novitch BG (2015). Notch activity modulates the responsiveness of neural progenitors to sonic hedgehog signaling. Dev. Cell 33, 373–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koranda JL, Dore L, Shi H, Patel MJ, Vaasjo LO, Rao MN, Chen K, Lu Z, Yi Y, Chi W, et al. (2018). Mettl14 Is Essential for Epi-transcriptomic Regulation of Striatal Function and Learning. Neuron 99, 283–292.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Fata G, Gärtner A, Domínguez-Iturza N, Dresselaers T, Dawitz J, Poorthuis RB, Averna M, Himmelreich U, Meredith RM, Achsel T, et al. (2014). FMRP regulates multipolar to bipolar transition affecting neuronal migration and cortical circuitry. Nat. Neurosci 17, 1693–1700. [DOI] [PubMed] [Google Scholar]
- Lin S, Choe J, Du P, Triboulet R, and Gregory RI (2016). The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol. Cell 62, 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol 10, 93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell SK, and Kaznowski CE (1991). Cell cycle dependence of laminar determination in developing neocortex. Science 254, 282–285. [DOI] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, and Jaffrey SR (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3′UTRs and near stop codons. Cell 149, 1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, and Jaffrey SR (2015). 5′ UTR m(6)A Promotes Cap-Independent Translation. Cell 163, 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor A, Whibley A, Marshall CR, Gianakopoulos PJ, Piton A, Carson AR, Orlic-Milacic M, Lionel AC, Sato D, Pinto D, et al. ; Autism Genome Project Consortium (2010). Disruption at the PTCHD1 Locus on Xp22.11 in Autism spectrum disorder and intellectual disability. Sci. Transl. Med 2, 49ra68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer E, Rechavi G, and Dominissini D (2017). Epitranscriptomics: regulation of mRNA metabolism through modifications. Curr. Opin. Chem. Biol 41, 93–98. [DOI] [PubMed] [Google Scholar]
- Pilaz LJ, McMahon JJ, Miller EE, Lennox AL, Suzuki A, Salmon E, and Silver DL (2016). Prolonged Mitosis of Neural Progenitors Alters Cell Fate in the Developing Brain. Neuron 89, 83–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, and Spector DL (2005). Regulating gene expression through RNA nuclear retention. Cell 123, 249–263. [DOI] [PubMed] [Google Scholar]
- Rakic P (1988). Specification of cerebral cortical areas. Science 241, 170–176. [DOI] [PubMed] [Google Scholar]
- Richter JD, Bassell GJ, and Klann E (2015). Dysregulation and restoration of translational homeostasis in fragile X syndrome. Nat. Rev. Neurosci 16, 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y, Sha J, Huang X, Guerrero L, Xie P, et al. (2017). YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. eLife 6, e31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffary R, and Xie Z (2011). FMRP regulates the transition from radial glial cells to intermediate progenitor cells during neocortical development. J. Neurosci 31, 1427–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M, Wang F, Li M, Sah N, Stockton ME, Tidei JJ, Gao Y, Korabelnikov T, Kannan S, Vevea JD, et al. (2019). Reduced mitochondrial fusion and Huntingtin levels contribute to impaired dendritic maturation and behavioral deficits in Fmr1-mutant mice. Nat. Neurosci 22, 386–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solecki DJ, Model L, Gaetz J, Kapoor TM, and Hatten ME (2004). Par6alpha signaling controls glial-guided neuronal migration. Nat. Neurosci 7, 1195–1203. [DOI] [PubMed] [Google Scholar]
- Tervonen TA, Louhivuori V, Sun X, Hokkanen ME, Kratochwil CF, Zebryk P, Castrén E, and Castrén ML (2009). Aberrant differentiation of glutamatergic cells in neocortex of mouse model for fragile X syndrome. Neurobiol. Dis 33, 250–259. [DOI] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, and He C (2015). N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 161, 1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li Y, Yue M, Wang J, Kumar S, Wechsler-Reya RJ, Zhang Z, Ogawa Y, Kellis M, Duester G, and Zhao JC (2018). N6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat. Neurosci 21, 195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. (2010). TheGeneMANIA production server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 38, W214–W220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener M, and Muller-McNicoll M (2018). Nuclear retention of mRNAs - quality control, gene regulation and human disease. Semin. Cell Dev. Biol 79, 131–142. [DOI] [PubMed] [Google Scholar]
- Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC, Shi H, Cui X, Su R, Klungland A, et al. (2018). Differential m(6)A, m(6)Am, and m(1)A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol. Cell 71, 973–985.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y, Lu Z, He C, and Min J (2014). Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol 10, 927–929. [DOI] [PubMed] [Google Scholar]
- Yoon KJ, Ringeling FR, Vissers C, Jacob F, Pokrass M, Jimenez-Cyrus Q, Su Y, Kim NS, Zhu Y, Zheng L, et al. (2017). Temporal Control of Mammalian Cortical Neurogenesis by m(6)A Methylation. Cell 171, 877–889.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Kang Y, Wang M, Li Y, Xu T, Yang W, Song H, Wu H, Shu Q, and Jin P (2018). Fragile X mental retardation protein modulates the stability of its m6A-marked messenger RNA targets. Hum. Mol. Genet 27, 3936–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data have been deposited to the Gene Expression Database (GEO) at NCBI. Accession: GSE121809