Abstract
The advent of mass cytometry (CyTOF®) has permitted simultaneous detection of more than 40 antibody parameters at the single-cell level, although a limited number of metal-labeled antibodies are commercially available. Here we present optimized and scalable protocols for conjugation of lanthanide as well as bismuth ions to immunoglobulin (Ig) using a maleimide-functionalized chelating polymer and for characterization of the conjugate. The maleimide functional group is reactive with cysteine sulfhydryl groups generated through partial reduction of the Ig Fc region. Incubation of Ig with polymer pre-loaded with lanthanide ions, produces metal-labeled IgG without disrupting antigen specificity. Antibody recovery rates can be determined by UV spectrophotometry and frequently exceeds 60%. Each custom-conjugated antibody is validated using positive and negative cellular control populations and is titrated for optimal staining at concentrations ranging from 0.1 to 10 μg/ml. The preparation of metal-labeled antibodies can be completed in 4.5 hrs., and titration requires an additional 3 to 5 hrs.
Keywords: CyTOF, mass cytometry, conjugation, antibody, immunoglobulin, IgG, lanthanide, bismuth, isotope, titration, phenotype, phosphorylation, MaxPAR, chelator, polymer
1. Introduction
1.1. Background
Multicellular biological systems require the interplay of a diversity of cellular phenotypes. The advent of monoclonal antibody technology allowed the precise definition of many cellular phenotypes, first in the immune system, and more recently in solid tissues and tumors. Cellular phenotypes are most often defined by combinations of extracellular surface antigens. Many of these markers, such as those best understood in the immune system, are gained and lost during developmental maturation and in response to environmental stimuli. As the knowledge of cellular roles, particularly among immune cell subtypes, has become increasingly detailed, phenotypic definitions have begun to involve intracellular regulatory proteins or protein modifications that can act as proxies for cellular function: these include transcription factors, cytokines, and post-translational modifications of signaling proteins (e.g., phosphorylation, acetylation, cleavage).
Multiparametric single-cell fluorescence cytometry platforms are unique in their ability to measure multiple features per cell on thousands or millions of cells per experiment—allowing for quantitative capturing of subtle or wholesale shifts in cell subset frequencies and marker expression across diverse cellular phenotypes. The absolute number of cellular components that can be measured simultaneously on each cell is limited in fluorescence-based cytometry by constraints inherent in light-based measurements and emission spectral overlap of available fluorophores. This restriction in the number of simultaneous measurements has limited the scope of inquiry regarding the biological system under study such as human immune system states and cancers.
Recognizing the need for increased simultaneous measurement and quantification on a per cell basis, the Tanner group at the University of Toronto created a new detection modality, in which antibodies were tagged with stable heavy metal isotopes and quantified using a technology called inductively-coupled plasma mass spectrometry (ICP-MS) [1]. By combining sensitive and highly multiparametric ICP-MS immunoassay technology with a single-cell acquisition source, Tanner and colleagues produced the first mass cytometry platform, which was later released commercially as the CyTOF® [2].
To date, this technology has been employed to analyze multiple facets of biology, biochemistry and molecular regulation at the single cell level. Using the protocols described herein our groups have created numerous customized antibody panels, including those for studying: human hematopoiesis and regulatory cell signals [3]; cell and context specific kinase inhibitor activity in a high throughput assay [4]; broad facets of cell cycle across the human hematopoietic compartment [5]; cellular apoptosis and necrosis [6]; activation profiles of virus-specific cytotoxic T cells; and comparison of regulatory phosphorylation kinetics governing T cell receptor activation across different populations [7].
Using CyTOF®, more than 40 antibody-based parameters can be analyzed simultaneously at the single-cell level [3, 4, 8], though reagents for its implementation are not as currently widespread as fluorophore-conjugated materials. Additionally, the pursuit of new biology combined with the enormity of possible combinations of measurement reagents will likely always necessitate the creation of novel, custom-conjugated antibodies. Designing an optimal panel to investigate relevant biological questions of interest requires custom conjugation of purified antibodies with heavy metal ions such that bound antibodies can be detected by inductively coupled plasma time-of-flight mass spectrometry (ICP-TOF-MS). To standardize and optimize this process we have developed a protocol for labeling of purified monoclonal and polyclonal antibodies designed to maximize conjugation efficiency while maintaining desired binding affinity.
This antibody labeling protocol utilizes MaxPar® chelating polymers commercially available from Fluidigm. These water-soluble polymers contain a sulfhydryl-reactive bismaleimide group and a trivalent metal cation-chelating 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid (DOTA) or diethylene triamine pentaacetic acid (DTPA) group. The binding affinities of DOTA and DTPA for lanthanide metal reporters approach 10−16 M [9]. Importantly, an advantage of this platform is that there are 44 mass channels within the optimal mass cytometry measurement range that can be occupied by stable isotopes of transition metals, mainly the lanthanide series. These transition metals readily form stable trivalent salts in solution with an oxidation state of (III) (Table 1). Consequently, antibody reagents for all 44 mass channels can be created using the protocol described herein.
Table 1 –
A summary of compatible trivalent stable elemental isotopic reporters, commercial sources, and relative sensitivities and performance notes and common interferences in mass cytometry assays.
| Isotopic Mass | Element (Symbol)A | Commercial AvailabilityB | Relative SensitivityC | Most Common InterferenceD *Ba Oxide or Hydroxide (Contaminant) |
|---|---|---|---|---|
| 113 | Indium (In) | P | 0.1 | 113In impurity from another In |
| 115 | Indium (In) | P | 0.1 | 115In impurity from another In |
| 139 | Lanthanum (La) | N | 0.3 | 138Ba (contamination) +1 signal bleed |
| 140 | Cerium (Ce) | P | 0.3 | |
| 141 | Praseodymium (Pr) | M, N | 0.3 | |
| 142 | Neodymium (Nd) | M, P | 0.4 | 142Nd impurity from another Nd |
| 143 | Neodymium (Nd) | M, P | 0.4 | 143Nd impurity from another Nd |
| 144 | Neodymium (Nd) | M, P | 0.5 | 144Nd impurity from another Nd |
| 145 | Neodymium (Nd) | M, P | 0.5 | 145Nd impurity from another Nd |
| 146 | Neodymium (Nd) | M, P | 0.5 | 146Nd impurity from another Nd |
| 147 | Samarium (Sm) | M, P | 0.6 | 147Sm impurity from another Sm |
| 148 | Neodymium (Nd) | M, P | 0.6 | 148Sm impurity from an Sm reporter |
| 149 | Samarium (Sm) | M, P | 0.6 | 149Sm impurity from another Sm |
| 150 | Neodymium (Nd) | M, P | 0.7 | 150Sm impurity from an Sm reporter |
| 151 | Europium (Eu) | M, P | 0.7 | * |
| 152 | Samarium (Sm) | M, P | 0.7 | *; 152Gd impurity from a Gd reporter |
| 153 | Europium (Eu) | M, P | 0.8 | * |
| 154 | Samarium (Sm) | M, P | 0.8 | * |
| 155 | Gadolinium (Gd) | M, P | 0.8 | *; 139La Oxide; 155Gd impurity from another Gd |
| 156 | Gadolinium (Gd) | M, P | 0.9 | 140Ce Oxide; 156Gd impurity from another Gd |
| 157 | Gadolinium (Gd) | P | 0.9 | 141Pr Oxide; 157Gd impurity from another Gd |
| 158 | Gadolinium (Gd) | M, P | 0.9 | 142Nd Oxide; 158Gd impurity from another Gd |
| 159 | Terbium (Tb) | M, N | 1 | 143Nd Oxide |
| 160 | Gadolinium (Gd) | M, P | 1 | 144Nd Oxide; 160Dy impurity from a Dy reporter |
| 161 | Dysprosium (Dy) | M, P | 1 | 145Nd Oxide; 161Dy impurity from another Dy |
| 162 | Dysprosium (Dy) | M, P | 1 | 146Nd Oxide; 162Dy impurity from another Dy |
| 163 | Dysprosium (Dy) | M, P | 1 | 147Sm Oxide; 163Dy impurity from another Dy |
| 164 | Dysprosium (Dy) | M, P | 1 | 148Nd Oxide; 164Dy impurity from another Dy |
| 165 | Holmium (Ho) | M, N | 1 | 149Sm Oxide |
| 166 | Erbium (Er) | M, P | 1 | 150Nd Oxide; 166Er impurity from another Er |
| 167 | Erbium (Er) | M, P | 1 | 167Er impurity from another Er |
| 168 | Erbium (Er) | M, P | 1 | 152Sm Oxide; 168Er impurity from another Er |
| 169 | Thulium (Tm) | M, N | 1 | |
| 170 | Erbium (Er) | M, P | 0.9 | 154Sm Oxide; 170Yb impurity from a Yb reporter |
| 171 | Ytterbium (Yb) | M, P | 0.9 | 155Gd Oxide; 171Yb impurity from another Yb |
| 172 | Ytterbium (Yb) | M, P | 0.9 | 156Gd Oxide; 172Yb impurity from another Yb |
| 173 | Ytterbium (Yb) | M, P | 0.8 | 157Gd Oxide; 173Yb impurity from another Yb |
| 174 | Ytterbium (Yb) | M, P | 0.8 | 158Gd Oxide; 174Yb impurity from another Yb |
| 175 | Lutetium (Lu) | M, P | 0.8 | 159Tb Oxide |
| 176 | Ytterbium (Yb) | M, P | 0.8 | 160Gd-oxide; 176Lu impurity from a Lu reporter |
| 197 | Gold (Au) | N | 0.3 | High non-specific binding |
| 203 | Thallium (Tl) | P | 0.5 | 203Tl impurity from another Tl |
| 205 | Thallium (Tl) | P | 0.5 | 205Tl impurity from another Tl |
| 209 | Bismuth (Bi) | N | 0.5 | 193Ir Oxide (DNA intercalator) |
Most abundant elemental isotope with that mass. Other stable elemental isotopes with the same mass may exist.
M: MaxPAR labeling kit; P: source (III) purified chloride or nitrate isotope from supplier (e.g. Trace Sciences International); N: Natural (III) chloride or nitrate element is >99.9% monoisotopic.
Based on the original mass cytometry specifications [2]. Transmission efficiency varies between instruments. This should serve only as a guide.
The process of antibody conjugation has three fundamental steps:
The MaxPar® polymer is incubated in a trivalent metal salt solution to facilitate chelation with high efficiency.
To generate sites for maleimide labeling, the purified, carrier-free immunoglobulin, type G (IgG) is incubated in a low concentration of tris-carboxyethyl phosphine (TCEP) to preferentially reduce disulfide bonds within the Fc region, ideally without compromising the antigen specificity of the Fab region [10, 11].
Lastly, the chelated polymer and partially reduced antibody are mixed and incubated to achieve full conjugation.
Notably, antibodies, as well as many buffer components such as phosphates, have a propensity to precipitate out of solution containing even low concentrations of free lanthanide ion; therefore, it is critical to chelate metal ions and wash the polymer thoroughly prior to addition of the reduced antibody. Furthermore, the tertiary structure of Fab regions of antibodies incubated under harsh reducing conditions may become altered due to disruption of structural disulfide bonds. As such, the concentration of Tris-carboxyethyl phosphine (TCEP) in reducing buffer and the length of incubation are critical, and the protocol below should be followed precisely as described. To maximize conjugation efficiency, we utilize a series of buffers optimized for metal loading, antibody reduction, polymer coupling to the antibody, and subsequent washing. It is also essential to ensure that carrier protein (i.e., BSA) is removed from the antibody solution as the carrier may compete for free maleimide groups of the polymer. If the monoclonal or polyclonal antibody preparation to be used includes carrier protein, these proteins may be removed by Melon gel (Pierce) or Protein G column purification. Using purified antibodies, the coupling reaction of polymer to partially reduced antibody approaches completion, resulting in covalent binding of around six polymers to each IgG.
1.2. Experimental Considerations
The overriding advantage of mass cytometry over other biological analysis platforms is its ability to acquire single-cell, 40–50 parameter data with negligible spectral overlap. Although this cytometric analysis is best known as a method for analyzing immunologic subpopulations [3, 8], the utility of high-parameter single-cell analysis goes well beyond blood and bone marrow—it is useful in virtually any biological system that can be prepared as a single-cell suspension. For example, the multiparametric and quantitative data afforded by mass cytometry is valuable in studies of solid-tissue stem cell differentiation, cellular heterogeneity in tumors [12, 13], coordinated anti-tumor immunity across an entire organism [14]; defining cellular phenotypes in autoimmunity [15]; tumor-stroma interactions, stochasticity and kinetics in cell-culture models of apoptosis and cell cycle [5, 6], network analysis of the interplay between phosphorylation events, and as a highly parallelized drug screening platform capable of monitoring many kinase targets simultaneously [4].
The common theme of most of these applications is the need for simultaneous detection of many cellular components at a single-cell level to enable the fine-grained discrimination of many heterogeneous cell types, to efficiently multiplex many functional assays in a single analysis, or to gain insights into correlated phenomena and emergent properties that can only be mined from quantitative, high-dimensional data [16, 17]. These high dimensional single-cell approaches create a challenge and an opportunity to leverage the mutual information encoded in these experiments to make new models of human systems and take unsupervised approaches to cellular identification and disease classification [12, 18–20].
One minor limitation is the nature of the immunoglobulins used with success in this protocol. IgGs from mouse, rat, rabbit, goat, and sheep have been tested extensively and are compatible with the protocol described here. Attempts to use type E and type M immunoglobulins (Igs) have been less successful presumably due to disulfide bonds in these antibody types which are required to maintain structure and binding activity. We have also occasionally observed that labeling Igs from an Armenian hamster background decreases binding activity.
1.3. Experimental design
When designing a mass cytometry experiment, the researcher must first take into consideration the technological limitations of the current generation of mass cytometry instruments: i) The expected data capture rate is only ~30% of the input number of cells for each sample depending on the staining protocol and cell introduction system. ii) The cell acquisition rate should be limited to 500–1000 cells/second to maximize data quality and avoid double cell events. iii) This is a destructive method. Cells are vaporized as they enter the plasma ionization source, thus no cell recovery is possible. iv) Lastly, investigator-specific reagents must be labeled and tested as detailed in the protocol herein. Although many experiments have been designed to circumvent these limitations, there are some experiments that are simply not well suited for mass cytometry analysis. Mainly, experiments that involve an extremely rare cell population such as hematopoietic progenitors [3] or antigen specific T cells [8] require either very large numbers of cells, some sort of pre-enrichment with the use of carrier cells to be discriminated in silico, or both.
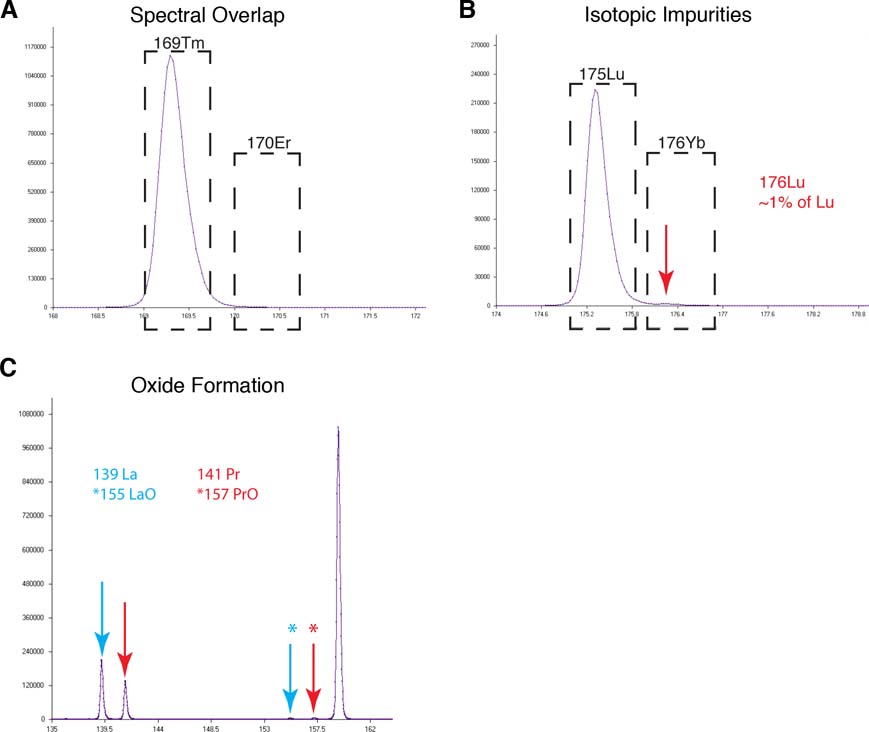
Mass cytometry and the use of elemental isotopic reporters overcome many of the limitations inherent with fluorescent or colorimetric reporter technologies, namely spectral overlap and background signal inherent in biological samples. Still, there are sources of interference that can confound analysis, namely: oxide formation, isotopic contamination, and, to a lesser extent, spectral overlap (‘signal bleed’) when strong signals are adjacent to weaker ones on the mass scale (Figure 1). Isotopic reporter signals that differ by >102 may cause visible spectral bleed from the higher-abundance ion into the lower-abundant mass channel next to it on the mass scale. For example, a small percentage of a strong 169Tm signal may be present in the 170Er measurement window (Figure 1A) despite no expected signal. Because the magnitude of this ‘signal bleed’ is only a very small proportion of the overall interfering signal (169Tm), if the expected signal in the channel targeted by the interference (170Er) is not 100 times (two orders of magnitude) lower than the interfering signal, it is not expected to confound the analysis. Additionally, most of this spectral overlap in time-of-flight mass spectrometry, as in the CyTOF mass cytometer, tails in the M+1 direction (i.e., the 169Tm has a higher propensity to be measured as 170Er rather than the other way around).
Figure 1 –
Sources of signal interference in mass cytometry assays as demonstrated by analyzing a mixture of metal standard (139La, 141Pr, 159Tb, 169Tm, and 175Lu) on a CyTOF mass cytometer. (A) Spectral overlap from the 169 Tm standard into the 170 Er measurement channel. 169 Tm is naturally mono-isotopic with no expectation of a constituent with mass 170. (B) Natural Lu, used to make the standard in this experiment, contains ~99% 175 Lu and 1% 176 Lu. A distinct peak corresponding to the 176 Lu isotopic ‘contaminant’ can be seen in the measurement window that would be used for 176 Yb labeled reagents (red arrow). The dashed black boxes in (A) and (B) represent the measurement window where all signals within that range would be attributed to the indicated elemental isotope. (C) La and Pr oxide formation indicated with the blue and red arrows, respectively. Here the resulting ‘+16Da’ oxide interference peak is indicated with a ‘*’ of a matching color.
The two primary sources of convoluting reporter signal interference are isotopic impurities (Figure 1B) and oxide formation during ionization (Figure 1C). Isotopic impurities are a result of incomplete purification from the naturally occurring element, which can be a mixture of stable isotopes. As a general rule, the most common isotopic contaminants are in those elements that have +1 or −1 mass adjacent isotopes. For example, the contaminants in 97% pure 145Nd are likely 144Nd and 146Nd. In contrast, 151Eu can be obtained in 99% purity with little or no contaminating 153Eu (the only other stable Eu isotope) because the additional mass resolution allows for more efficient purification. A profile of the contaminating isotopes is generally available from the supplier of the isotopically enriched material. Other heavy elements (La, Pr, Tb, Ho, and Tm) are natural or near natural (>99.9% pure) mono-isotopes, therefore isotopic contamination is also not a concern in these cases. These natural mono-isotopes and series of mass adjacent isotopic reporters are also noted in Table 1 and were reviewed in Ornatsky [21].
Oxide, and to a much lesser extent hydroxide, formation occurs during analysis when incomplete vaporization/ionization results in the formation of adducts between the elemental isotopic reporter with oxygen or a hydroxide, thus adding 16 or 17 amu to the reporter mass, respectively [22]. For example, 139La, one of the most likely elements to oxidize, will also have a small signal in the 155Gd channel resulting from the oxidation product (139La+16O) (Figure 1C). While oxidation creates this M+16 artifact for certain metal isotope reporters the mass cytometer is tuned such that this occurrence is typically 3% or less of the total signal. The level of oxidation is dependent on the oxide bond strength of the atomic ion of the reporter element and the local temperature of the plasma. The relative likelihoods of oxide or hydroxide formation for common mass cytometry elements are summarized in Table 2. The most significant sources of interference due to oxide formation combined with common isotopic interferences are summarized in Table 1 and should be considered in the context of expected signal levels when selecting panels of elemental isotopic reporters for mass cytometry analyses. For example, antibodies against low-abundance antigens should not be conjugated to an isotope that will receive considerable oxidation from a reporter metal used to label an antibody targeting a high-abundance, or “bright”, marker.
Table 2 –
The expected relative occurrence of oxide (M+O) and hydroxide (M+OH) ion adducts during ICP-MS analysis of metals commonly present in mass cytometry assays. The frequency of occurrence has been normalized to ratio of LaO/La acquired under the same conditions.
| Element (Symbol) | Relative to LaO OccurrenceA | |
|---|---|---|
| Oxide Level (M+16O) | Hydroxide Level (M+16O1H) | |
| Barium (Ba) | 0.04 | 0.05 |
| Lanthanum (La) | 1.0 | 0.09 |
| Cerium (Ce) | 1.0 | 0.07 |
| Praseodymium (Pr) | 0.7 | 0.05 |
| Neodymium (Nd) | 0.58 | 0.03 |
| Samarium (Sm) | 0.11 | 0.01 |
| Europium (Eu) | 0.02 | - |
| Gadolinium (Gd) | 0.30 | 0.08 |
| Terbium (Tb) | 0.25 | 0.02 |
| Dysprosium (Dy) | 0.11 | 0.01 |
| Holmium (Ho) | 0.10 | 0.00 |
| Erbium (Er) | 0.09 | 0.00 |
| Thulium (Tm) | 0.03 | 0.00 |
| Ytterbium (Yb) | 0.01 | 0.00 |
| Lutetium (Lu) | 0.07 | 0.01 |
Based on the published ratios of oxide (M+O/M) or hydroxide (M+OH/M) measured by ICP-MS analysis [22], normalized to the expected ratio of LaO/La in the same experiment and accounting for the relative mass sensitivities as reported in Table 1. This frequency of LaO occurrence is a common metric in tuning oxidation levels in mass cytometry experiments. For example, if the level of LaO was found to be 2% of the total La signal then the expected GdO level would only be 0.6% of the total Gd signal. Note that the abundance of oxide ions also depends on local plasma temperature and thus may deviate slightly from these values.
Finally, the CyTOF utilizes TOF mass measurement in conjunction with a series of mass filters in order to remove overly abundant ions inherent in biological samples as well as the ions from argon plasma that can be detrimental to the detector. Current mass cytometry instrumentation can be tuned with a mass window of approximately 130 atomic mass units (amu). While this allows the detection of stable isotopic reporters with masses between ~80 and 238 the peak sensitivity typically lies between 160–170 amu and drops off sharply towards the low mass end and more gradually towards the higher end. The relative sensitivity of various isotopic mass reporters is summarized in Table 1 and should be considered in the context of the expected antigen expression level on a cell when assigning a mass reporter to an antibody. Specifically, low-abundance or “weak” antigens are often best measured using reporter isotopes in the peak sensitivity range. However, because these channels can receive bleed from the oxidation of lighter isotopes, effective panel design aims to optimize these factors to minimize confounding signal as is routinely performed for fluorescence-based methods.
2. Materials
2.1. General considerations
Barium is a commonly occurring element in many detergent products and is frequently found at high concentrations in bottles cleaned in laboratory dishwashers. For optimum sensitivity and lifetime of the CyTOF® instrument, wash solutions used in the immunostaining protocol must be nearly free of barium (< 1 ppb). Many commercially available biological reagents (e.g., 500 mL liquid bottles of liquid GIBCO DPBS) contain high amounts of barium and should be avoided in later steps of the staining protocol. The recipes below are specifically designed to avoid barium contamination, but it is recommended that each laboratory test every wash solution in the workflow for barium contamination before running the first set of samples on the CyTOF.
2.2. Reagents
100–500 μg immunoglobulin
Tris-carboxyethyl phosphine (TCEP), neutral pH (0.5 M in 10 μl aliquots) (Thermo Scientific)
- MaxPar X8 or DN3 Antibody Labeling Kit (Fluidigm):
- MaxPar X8 or DN3 Antibody Labeling Reagent (one test per 0.1 mg of Ig)
- L-Buffer (suitable substitution: 20 mM ammonium acetate, pH 6)
- R-Buffer (suitable substitution: 0.1 M phosphate buffer with 2.5mM EDTA, pH 7.2)
- C-Buffer (suitable substitution: tris-buffered salt with 1 mM EDTA, pH 7.5)
- W-Buffer (suitable substitution: tris-buffered salt, pH 7.5)
- Trivalent metal lanthanide solution (0.05 M stocks of XCl3 or X(NO3)3 in L-buffer, where X is the elemental metal isotope)
Modified C-Buffer, 150 mM tris, 150 mM NaCl, 1 mM EDTA, pH 7.5
10x PBS, 320 g of NaCl, 8 g of KCl, 46 g of Na2HPO4•7H20, and 8 g of KH2PO4 in 3 L of ddH20. Bring solution to pH 7.4 using concentrated aqueous NaOH. Bring volume to 4 L with ddH2O. To create a 1X stock mix 1 part of 10X stock with 9 parts ddH2O.
FACS buffer, 500 ml PBS, 2.5 g BSA (final concentration: 0.5% wt/vol), and 100 mg sodium azide NaN3 (final concentration: 0.02% wt/vol). Store at 4 °C for up to 4 months.
Antibody stabilization buffer, 0.1% (wt/vol) (NaN3) in Antibody Stabilizer solution. Store at 4 °C for up to several years.
16% Paraformaldehyde (PFA) ampules (wt/vol in water).
DNA intercalator: 1 ml PBS, 100 μl of 16% PFA, and 0.25 μl of Ir-Intercalator (500 μM stock concentration, Fluidigm).
Methanol, store at 4 °C.
Positive and negative control cells (or cell populations) of interest.
Monensin Solution (1000X, Biolegend).
Brefeldin A Solution (1000X, Biolegend).
2.3. Equipment
Amicon Ultra-0.5 mL Centrifugal Filter Unit with Ultracel-3 membrane (Millipore)
Amicon Ultra-0.5 mL Centrifugal Filter Unit with Ultracel-50 membrane (Millipore)
Amicon Ultrafree Durapore 0.1 μm PVDF 0.5-mL centrifugal filters (Millipore)
Screw-top Eppendorf tubes
Pipettes
Filter pipette tips
Polystyrene round bottom test tubes (FACS tubes)
Pasteur pipettes
Vacuum flask connected to vacuum line
Water bath heated to 37 °C
Room temperature centrifuge with rotor for microtubes
Refrigerated centrifuge with rotor for FACS tubes
A low volume (μl) UV/Vis spectrophotometer
CyTOF mass cytometer
Flow cytometry analysis software (we use Cytobank [23], www.Cytobank.org)
3. Methods
3.1. Antibody conjugation
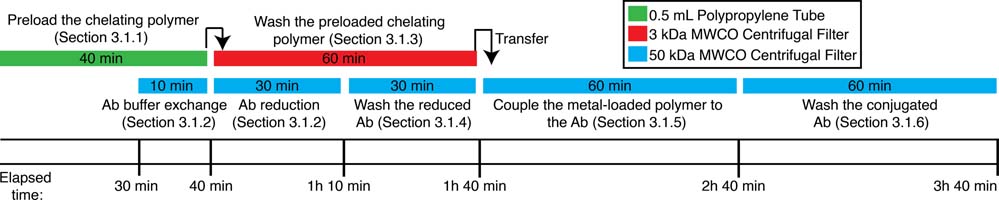
The following conjugation procedure has some critical timing steps that are important to the success of the protocol. These steps are noted in text and summarized in a workflow diagram in Figure 2.
In the main text, we describe the conjugation of 100 μg of immunoglobulin. Due to the inevitable loss of a certain amount of protein during the conjugation procedure, conjugation of <100 μg in one reaction is not recommended. However, conjugation of multiples of 100 μg is possible and might be desirable in cases in which frequent usage is anticipated and where potential batch-effects between different conjugations are to be avoided. In order to conjugate >100 μg (we have tested conjugation of up to 500 μg), the presented protocol has to be slightly modified which will be pointed out and referred to in the Notes section.
Conjugation of antibodies to Bismuth (209Bi) requires slightly adjusted reagents and again, this will be referred to in the Notes section.
Figure 2 –
A workflow summarizing the timing and coordination of steps for the conjugation of a purified immunoglobulin with a sulfhydryl reactive polymer pre-loaded with metal isotope reporters.
3.1.1. Preloading the chelating polymer (40 min)
Per 100 μg of immunoglobulin (see Note 1) to be conjugated, spin one tube containing the MaxPar chelating polymer for 10 s in a microcentrifuge. Polymer is difficult to see by eye and can escape from the tube easily upon opening. Spinning beforehand ensures that the reagent is at the bottom of the tube.
Reconstitute the MaxPar reagent in 95 μl of L-Buffer per labeling reaction using a filter pipette tip (for conjugation of >100 μg of antibody see Note 2 and for conjugation of antibodies to 209Bi see Note 3).
Add 5 μl of 0.05 M stock of metal solution to the polymer solution (final concentration: 2.5 mM). Vortex briefly to mix (for conjugation of >100 μg of antibody see Note 4).
Incubate at room temperature (RT) for 40 min, vortexing every 10 min. In order to ensure that the polymer and antibody are ready for conjugation simultaneously, proceed to section 3.1.2 after approximately 30 min of the incubation has elapsed (see Figure 2).
3.1.2. Buffer exchanging and reducing the antibody (40 min)
Add 300 μl of R-buffer to a 50-kDa MWCO micro-filter device (see Note 5).
Add 100 μg of antibody (see Note 6) to the R-buffer in the 50-kDa MWCO micro-filter device (see Note 7).
Reduce volume by spinning at 12,000 g for 10 min at RT. Discard the flow-through. Final volume should be 20 μl or less before proceeding.
Mix 8 μl of TCEP stock with 992 μl of R-buffer (final concentration: 4 mM TCEP).
Add 100 μl of the diluted TCEP solution to the concentrated antibody in the 50-kDa MWCO micro-filter device. Tap the tube by hand to mix. Mixing too vigorously by vortexing at high speeds can compromise the structural integrity of the antibody when mixed with the mild reducing agent TCEP.
Incubate covered for 30 min at 37 °C. The antibody should not be left in TCEP for more than 30 min; longer incubation may result in full reduction of disulfide bonds necessary for the structural integrity of the protein.
3.1.3. Washing pre-loaded MaxPar labeling reagent (60 min)
Following the 40 min incubation, add 200 μl of C-buffer to the metal-loaded polymer. Pipette the mixture or briefly vortex the column to mix (for conjugation to 209Bi see Note 8).
Transfer the mixture to the 3-kDa MWCO micro-filter device.
Reduce the volume by centrifugation at 12,000 g for 25 min at RT. Discard the flow-through.
Add 300 μl of C-buffer to the 3-kDa MWCO micro-filter. Pipette the mixture or briefly vortex the column to mix.
Reduce the volume by centrifugation at 12,000 g for 30 min at RT. Discard the flow-through. Final volume should not exceed 20 μl. A higher volume could result in excess free metal concentration and induce antibody precipitation.
3.1.4. Washing the partially reduced antibody (30 min)
Following the 30 min incubation (section 3.1.2), collect the partially reduced antibody from the 37 °C incubator. Add 300 μl of C-buffer to the partially reduced antibody in the 50-kDa MWCO micro-filter device. Pipette the mixture or briefly vortex the column to mix.
Reduce the volume by centrifugation at 12,000 g for 10 min at RT. Discard the flow-through.
Add an additional 400 μl of C-buffer to the 3-kDa MWCO micro-filter device. Pipette the mixture or briefly vortex the column to mix.
Reduce the volume by centrifugation at 12,000 g for 10 min at RT. Discard the flow-through.
3.1.5. Coupling metal-loaded polymer to partially reduced antibody (1 h)
Remove all micro-filter devices from the centrifuge. Resuspend the metal-loaded polymer in 60 μl of C-buffer in the 3-kDa MWCO micro-filter device using a pipette equipped with a filter tip.
Transfer the contents of the 3-kDa MWCO micro-filter device into the corresponding 50-kDa MWCO micro-filter device containing the partially reduced antibody of choice. Pipette the mixture or briefly vortex the column to mix.
Incubate at 37 °C for at least 60 min. Incubation time can be extended up to 2 h, though the reaction should approach completion after 60 min.
3.1.6. Washing and recovering the conjugated antibody (1 h)
Add 250 μl of W-buffer to the antibody conjugation mixture in the 50-kDa MWCO micro-filter device. Pipette the mixture or briefly vortex the column to mix.
Centrifuge at 12,000 g for 10 min at RT. Discard the flow-through. Volume should not exceed 20 μl after spin.
Add 400 μl of W-buffer to the antibody conjugation mixture in the 50-kDa MWCO micro-filter device. Pipette or briefly vortex to mix.
Centrifuge at 12,000 g for 10 min at RT. Discard flow-through. Volume should not exceed 20 μl after spin.
Repeat steps 3–4 twice.
Add 50 μl of W-buffer to the 50-kDa MWCO micro-filter device. Pipette to mix and rinse the walls of the column.
Invert the micro-filter device into a new collection tube (supplied with the AMICON filters).
Centrifuge at 1,000 g for 2 min at RT.
Gently remove micro-filter device from collection tube. Add an additional 50 μl of W-buffer to the 50-kDa MWCO micro-filter device. Pipette to mix and rinse the walls of the column.
Invert the micro-filter device into the same collection tube.
Centrifuge at 1,000 g for 2 min at RT.
Using a pipette equipped with a filter tip, transfer the conjugated antibody to a screw-top polypropylene tube (to prevent evaporation) for long-term storage. The antibody can be stored in W-buffer at 4 °C for at least a week. For long-term storage, the antibody should be diluted to the appropriate concentration as determined by titration in antibody stabilization buffer supplemented with NaN3. Ideally, the antibody should be diluted such that 1 to 2 μl is sufficient for a 100 μl cell staining reaction. This facilitates the creation of low volume staining cocktails containing more than 20 antibodies.
3.2. Quantification of conjugated antibody (15 min)
Set up a low volume spectrophotometer according the manufacturer’s instructions. Measure the absorbance of W-buffer at 280 nm and use as a ‘blank’ for subsequent measurements.
Against the blank, measure the absorbance of the conjugated antibody at 280 nm. Calculate the concentration of antibody present in solution. For mammalian IgG, an A280 of 1.38 absorbance units corresponds to a concentration of 1 mg/ml. The expected recovery is > 60% of conjugated antibody. For quantification of Bismuth-conjugated antibodies see Note 9.
Within a week of labeling proceed to antibody validation and titration in order to select the appropriate storage concentration. If in doubt, dilute the conjugated antibody to a concentration of 0.2 mg/ml in antibody stabilization buffer supplemented with NaN3 for long-term storage. The antibody can be stored in antibody stabilization solution at 4 °C for 6 months or more. However, before using the conjugated antibody for an experiment on the mass cytometer, the antibody must be validated and titrated as follows.
3.3. Staining samples for validation and titration of metal conjugated antibody (3–5 h)
We here focus on the validation and titration of antibodies against stable cell-surface molecules. Cases in which conjugated antibodies recognize inducible intracellular modifications or secreted molecules (i.e., cytokines) will be referred to in Note 10.
Obtain a suspension of single cells that are expected to contain the marker of choice (positive control) as well as cells that are not (negative control). Choosing appropriate cell populations is critical to validate that the antigen specificity of the newly conjugation antibody has not been altered. For example, when validating an antibody against anti-human CD3, Jurkat T cells could serve as an appropriate positive control, and Nalm-6 pre-B cells could serve as an appropriate negative control. In this same example, human peripheral blood mononuclear cells (PBMCs) could be used and CD3-positive T cells could be identified with a CD2 or CD5 stain and CD3 negative B cells with a CD19 or CD20 stain. For more examples, see the expected results in Figure 3.
Add 1 ml of cell culture media (user’s choice based on cell line or cell type) or FACS buffer at 37 °C to two FACS tubes labeled positive and negative. Add 10 × 106 positive control cells or 10 × 106 negative control cells to the FACS tubes, respectively. If using a single control (i.e., when using cells containing known positive and negative cell types; e.g., PBMCs) use a single tube. If the recognized epitope is stimulation dependent, see Note 10.
Add 111 μl of 16% paraformaldehyde (1.6% final concentration), and pipette thoroughly to mix.
Incubate for 10 min at RT.
Centrifuge at 500 g for 5 min at 4 °C. Aspirate supernatant using a vacuum equipped Pasteur pipette. Vortex to resuspend cells. Leaving the cells in minimal residual volume greatly enhances the efficiency of these wash steps. We prefer to aspirate the supernatant rather than decanting FACS tubes. This also maximizes cell recovery. Immediately continue to the following washing steps to prevent over-fixation of cells.
Add 3.3 mL of FACS buffer to each tube.
Add 500 μl of the above FACS buffer cell mixture into each of six labeled FACS tubes.
Centrifuge at 500 g for 5 min at 4 °C. Aspirate supernatant using a Pasteur pipette equipped with a fresh pipette tip, leaving cells in 60 μl residual volume.
If surface antibody staining is not being titrated, dilute the appropriate amount of antibody or antibodies for each staining reaction in FACS buffer to a final volume of 40 μl per sample. Transfer the 40 μl to each cell sample and vortex to mix. Then proceed to the incubation (step 18). If titrating the surface antibody staining cocktail proceed as follows: create an antibody staining cocktail by diluting 4 μg of each antibody for titration to a total volume of 200 μl in FACS buffer (final concentration: 20 μg/ml per antibody). If the antibody stock solution was initially diluted to 0.2 mg/ml, then 20 μl of the antibody stock will contain 4 μg.
This dilution is designed to produce a final concentration of 8 μg/ml for each antibody in a total staining volume of 100 μl for the first titration step. Therefore, if staining will be performed in a larger volume or the antibody was diluted to a different initial concentration, adjust these guidelines accordingly. Also, this titration is designed for two series (i.e., positive and negative controls) of six concentrations. If only using one series or more than two series of control cell lines amount of antibody can be scaled and subsequently diluted accordingly.
If titrating some antibodies while holding the concentration of others ‘constant’ in order to identify control cell populations, prepare a separate, low volume (ideally < 10 μl per sample) mixture of the ‘constant’ antibodies that will be added separately to each sample following the addition of the titrated antibodies.
Perform a six-step, two-fold serial dilution.
Add 40 μl of the first diluted antibody solution to the 60 uL in FACS ‘tube 1’ for each of the cellular controls (final concentration: 8 μg/ml antibody).
Add 100 μl of the remaining diluted antibody solution to 100 μl of FACS buffer (final concentration: 10 μg/ml antibody with cells).
Add 40 μl of the new diluted antibody solution to the 60 uL in FACS ‘tube 2’ for each of the cellular controls (final concentration: 4 μg/ml antibody with cells).
Repeat steps 15 & 16 four more times to create a six-step serial dilution with final staining concentrations of 2 μg/ml in ‘tube 3’, 1 μg/ml in ‘tube 4’, 0.5 μg/ml in ‘tube 5’, and 0.25 μg/ml antibody in ‘tubes 6’.
Incubate for 30 min at RT.
Add 3 ml of FACS buffer to each tube.
Centrifuge at 500 g for 5 min at 4 °C. Aspirate supernatant using a Pasteur pipette equipped with a fresh pipette tip.
Vortex to resuspend cells. If the cell pellet is not resuspended the cells will clump upon addition of methanol for cell permeabilization. Add 1 ml of 4 °C methanol to each disrupted cell pellet. Vortex to mix and incubate on ice for 10 min.
Add 2 ml of FACS buffer to each tube.
Centrifuge at 500 g for 5 min at 4 °C. Aspirate supernatant using a Pasteur pipette equipped with a fresh pipette tip. Vortex to resuspend cells.
Add 3 ml of FACS buffer to each tube. Centrifuge at 500 g for 5 min at 4 °C.
Aspirate supernatant, leaving cells in 60 μl residual volume.
If staining and/or titrating intracellular antibodies, prepare an intracellular antibody staining cocktail and stain the cell samples according to steps 11–20.
Add 1 ml of Ir DNA intercalator solution to each FACS tube and mix thoroughly (see Note 11).
Incubate for at least 20 min at RT. Cells can be stored in DNA intercalator solution for up to 3 days at 4°C before acquisition on mass cytometer.
Add 2 ml of FACS buffer to each tube.
Centrifuge at 500 g for 5 min at 4 °C. Aspirate supernatant using a Pasteur pipette equipped with a fresh pipette tip. Vortex to resuspend the cells.
Add 3 ml of ddH2O to each tube.
Centrifuge at 500 g for 5 min at 4 °C. Aspirate supernatant using a Pasteur pipette equipped with a fresh pipette tip. Vortex to resuspend the cells.
Repeat steps 31–32.
Place cell pellets on ice. Just prior to analysis, resuspend cells in ddH2O at a concentration of 1–2 × 106 cells per ml.
Filter with a cell strainer and analyze each tube on the mass cytometer. At this stage, 1x EQ beads can be added to the sample to enable later data normalization. Collect cell events at a rate no faster than 500–1000 cells/s. If the cells are too concentrated the instrument can clog and the data quality may suffer due to overlapping cell boundaries. Adjust cell dilution with ddH2O accordingly.
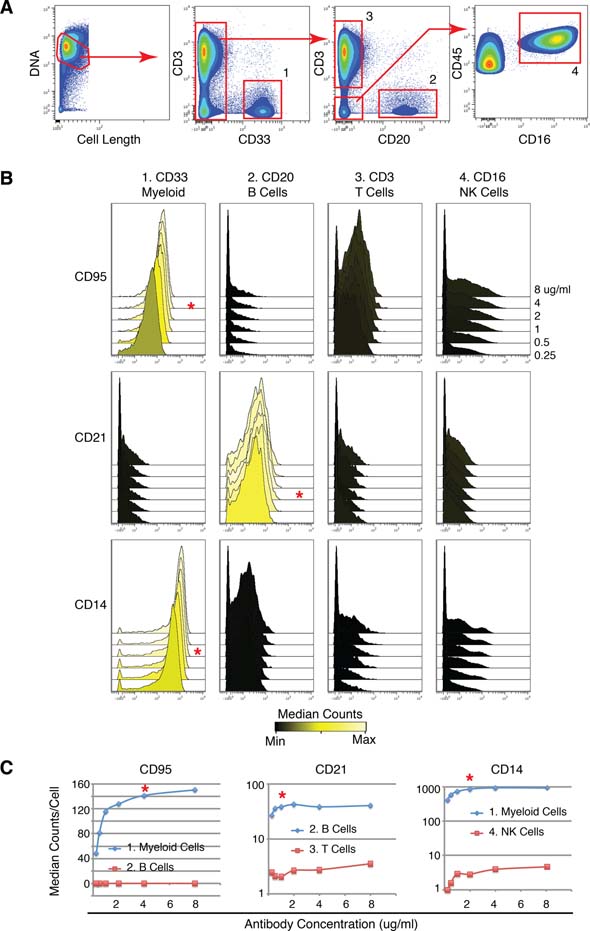
Figure 3 –
Anticipated results for the titration of metal reporter conjugated antibodies by CyTOF mass cytometry. Using the protocol described herein, antibodies against human CD95, CD21, and CD14 were labeled with 164Dy, 152Sm, and 148Nd, respectively. Using the surface staining protocol and a methanol permeabilization, these antibodies were simultaneously titrated from 0.25–8 μg/ml on human peripheral blood mononuclear cells that were counterstained with CD45-154Sm, CD3-170Er, CD20-147Sm, CD33-158Gd, and CD16-165Ho in order to identify positive and negative cell populations. (A) From data analysis at cytobank.org, the following populations were identified: 1) CD33-positive myeloid cells, 2) CD20-positive B cells, 3) CD3-positive T cells, and 4) CD16-positive NK cells. (B) Histogram overlays of CD95, CD21, and CD14 expression levels in the gated populations from (A) were created at cytobank.org. Histogram color scale indicates minimum (black) and maximum (yellow) median counts for each given antibody. (C) Dot plots summarizing the median counts of representative positive (blue) and negative (red) cell controls for each of the titrated antibodies across the range of 0.25–8 μg/ml. In (B) and (C) the red asterisk indicates the ideal antibody concentration with maximum signal-to-noise (the ratio of positive control to negative control signal) and lowest antibody concentration where the positive signal begins to saturate.
3.4. Gating strategy and data analysis
Figure 3 provides an example of anticipated results and analysis of titration data.
In .FCS file browsing software, gate on single cells (parameters: cell_length, Ir-191 or Ir-193) for all samples (Figure 3A). This gate may have to be tailored for each sample as the intensity of the Ir DNA intercalator staining can vary depending on a number of conditions such as cell type and number of cell in the staining reaction.
View different cell populations or control samples as stacked histograms or dot plots to visually validate epitope specificity of the antibody (Figure 3B).
Select the ideal antibody concentration based on the greatest overall signal in the positive control and signal-to-noise when compared to the negative control. This can be accomplished by comparing the channel medians (50th percentile) (Figure 3C) or, if focusing on an outlier population, the 95th percentile of the positive and negative samples at the different concentrations. Alternative approaches such as maximum separation index (SI) could also be employed [24].
3.5. Troubleshooting
Troubleshooting advice regarding antibody conjugation and titration can be found in Table 3. All potential problems will be observed during antibody recovery or during data analysis.
Table 3 –
A troubleshooting guide for common issues observed following custom antibody conjugation.
| Problem | Possible reason | Solution | ||
|---|---|---|---|---|
| Antibody recovery too low post-conjugation (<50%) | Precipitation | Likely induced by excess lanthanide metal exposure or antibody denaturation due to reduction. Ensure complete washes of the polymer and metal with post-centrifuge filter volumes of < 20μl to achieve desired dilution factor. |
||
| Antibody starting concentration |
Measure concentration of starting antibody stock by A280 to ensure concentration is as stated by manufacturer |
|||
| Defective 50kDa MWCO filter |
Repeat labeling protocol with a new filter and fresh antibody preparation. | |||
| Antibody recovery too high post-conjugation (>95%) | Carrier protein | Check manufacturer specifications for carrier protein (BSA, gelatin, ‘protein stabilizer’). Obtain carrier free stock or purify antibody away from carrier. |
||
| Antibody starting concentration |
Measure concentration of starting antibody stock by A280 to ensure concentration is as stated by manufacturer. |
|||
| No antibody staining detected | Poor staining | Repeat staining steps with fresh cells | ||
| Antibody integrity destroyed | After staining cells with conjugated antibody, perform secondary stain with fluorescently-labeled anti-IgG antibody and run on flow cytometer (e.g., LSR II from BD) |
|||
| Mass cytometer malfunction | Check mass cytometer performance using CyTOF Calibration Beads | |||
| Weak antibody staining detected | Poor staining | Repeat staining steps with fresh cells | ||
| Improper titration range | Repeat staining steps using increased antibody concentration | |||
| No difference between positive and negative controls | Improper choice of controls | Check antigen expression using fluorescently-labeled antibodies on flow cytometer (e.g., LSR II from BD) | ||
| Loss of antigen specificity | After staining cells with conjugated antibody, perform secondary stain with fluorescently-labeled anti-IgG antibody and run on flow cytometer (e.g., LSR II from BD) | |||
| No difference between titration steps | Improper titration range | Repeat staining steps using decreased antibody concentration | ||
| High background | Improper titration range |
Repeat staining steps using decreased antibody concentration | ||
| Loss of antigen specificity | After staining cells with conjugated antibody, perform secondary stain with fluorescently-labeled anti-IgG antibody and run on flow cytometer (e.g., LSR II from BD) |
3.6. Anticipated Results
To demonstrate the utility of the protocol described herein and provide representative data for a mass cytometry antibody titration, antibodies against human CD95, CD21, and CD14 were conjugated to 164Dy, 152Sm, and 148Nd, respectively (Figure 3). When choosing negative and positive control cell populations for an antibody titration, we prefer to utilize cells from a similar source as those to be interrogated experimentally, which, in this case, were human peripheral blood mononuclear cells (PBMC). The expectation was that CD21, complement component receptor 2, would be uniformly expressed on peripheral B cells, and CD14, the LPS co-receptor, would be expressed on the majority of monocytes. Similarly, we expected that CD95, the FAS receptor, would be expressed by most monocytes, at lower levels on some T cells and NK cells, but not by resting B cells, which constitute the majority of those present in unstimulated healthy human PBMC [25, 26]. The staining protocol with sample fixation and methanol permeabilization was used to stain human PBMC, which was expected to contain both positive and negative cell types for these antibody targets. A mixture of CD95, CD21, and CD14 were titrated according to the above described method. A separate mixture with CD45-154Sm, CD3-170Er, CD20-147Sm, CD33-158Gd, and CD16-165Ho was prepared and added to each sample after the completion of step 17 in section 3.3 in order to identify control cell populations while simultaneously titrating CD95, CD21, and CD14. Six different antibody concentrations were evaluated using six samples, as the lack of crosstalk between reporter signals allowed counterstaining all significant cell populations while titrating all three antibodies at the same time. Approximately, 105 cell events were acquired per sample on the CyTOF mass cytometer, and all data was uploaded to and analyzed at cytobank.org. Single cells were first identified by visualizing a biaxial plot of the iridium DNA intercalator signal versus cell length, a metric of the number of individual mass scans integrated to form the cell event as previously described [3]. Based on the expression of the counterstaining antibodies, the following cell populations were subsequently identified and gated: 1) CD33-positive myeloid cells, 2) CD20-positive B cells, 3) CD3-positive T cells, and 4) CD16-positive NK cells (Figure 3A). To assess the titration of CD95, CD21, and CD14 for each control cell populations, histogram overlays were created for each control population (Figure 3B). To more quantitatively visualize the differences between positive and negative control cell populations, selected based on known biology, dot plots summarizing the median counts for each antibody concentration from 0.25–8 μg/ml were made (Figure 3C). Depending on the antibody, linear (CD95) or log (CD21 and CD14) scaled plots best revealed differences across different concentrations. In Figure 3B and C, the red asterisks indicate the optimal antibody concentrations selected for these particular preparations. These concentrations were selected based on signal-to-noise ratio (the ratio of positive control to negative control signal) and on the lowest antibody concentration where the positive signal was beginning to saturate (plateau at higher concentrations). Alternative mechanisms of titration analysis could include using the 5th or 95th percentile, as opposed to the median, to compare situations where outlier cell events in each population are a primary concern when assessing resolution of positive and negative controls. Separation index is a good example of this and is described by Bigos et al. [24].
Although not completely necessary, it is helpful to have the antibody at close to saturating concentration in order to buffer the effects of staining volume differences as well as differences in cell staining numbers. For example, for CD95 (Figure 3C) at the lower concentrations of antibody there was a linear relationship between the measured counts per cell and the concentration of the antibody. If the antibody were applied at one of these lower concentrations, changes in cell staining volumes, a common source of experimental variation, would have inversely proportional effects on the resulting stain intensity. Variations in cell numbers have a similar effect. Both of these situations are particularly problematic when attempting to perform comparative single-cell measurements in a relatively quantitative fashion. Also of note in Figure 3C is that all three antibodies had selected titration concentrations that were below the maximum possible positive signal. In the cases shown here, the signal-to-noise ratio was not significantly higher at the selected concentration than at higher concentrations and the positive signal had begun to saturate, evidenced by the titration curve plateau. For these reasons, and to conserve custom-labeled antibody, the lower titration point in each case was selected.
4. Notes
Immunoglobulin should be free of a cysteine-containing carrier protein in solution (e.g., BSA). Notably, the cysteine content of gelatin is variable, from 10 to 100-fold lower than that of BSA by mass to completely absent. We have conjugated antibodies in the presence of Porcine gelatin though it is not recommended as it makes the Ig recovery difficult to quantify. If a carrier is present, a purification procedure should be performed before attempting the conjugation. Small molecule preservatives (e.g., trehalose, glycerol and sodium azide) or standard buffer salts are compatible additives.
For the conjugation of > 100 μg of the same immunoglobulin to a given heavy metal isotope use 1 tube of MaxPar polymer per 100 ug of antibody. You will need to add 5 μl of metal stock per tube of polymer. Resuspend the first polymer in a volume ‘X’ of L-buffer such that ‘X’ = 100 μl – ((Amount of antibody in μg / 100 μg) * 5 μl). E.g. for the conjugation of 500 μg, the first polymer tube is to be resuspended in 75 μl of L buffer. Subsequently, resuspend the next 4 tubes of polymer with the L-buffer from the first. Complete the pre-loading step by adding 25 μl of metal stock.
If antibodies are to be conjugated with 209Bi, replace L buffer with 1% HNO3 in this step. Note that the Bi stock will also be in 1–10% HNO3 to maintain solubility.
For the conjugation of > 100 μg of immunoglobulin, add 5 μl of stock metal solution per 100 μg of protein.
To confirm the integrity of the filter column, add 400 μl of ddH2O and centrifuge at 12,000 g for 30 sec. The volume of the flow-through should be around 150 μl. Spin for 5 min to get rid of excess ddH2O and discard flow-through. During the conjugation procedure, make sure to never touch the column membrane with the pipet tip to avoid scratches which can lead to lower antibody recovery.
Optional: To ensure that the volume of antibody solution corresponds to the intended quantity (within a tolerance of +/− 10%), confirm the protein concentration of the antibody stock solution by measuring the absorbance at 280 nm as described in section 3.2.
MWCO micro-filter devices are designed to hold up to 500 μl. If the desired amount of antibody requires addition of more than 200 μl of solution, pre-concentrate the antibody first in the same 50-kDa MWCO micro-filter device by spinning at 12,000 g for 8 min at RT until all of the antibody solution has been added to the column and the final volume is 200 μl. If these additional steps are required to pre-concentrate the antibody, this step should begin earlier (see Figure 2).
If antibodies are to be conjugated with 209Bi, replace C-buffer with modified C-buffer in this step.
Bismuth-ion chelator complexes interfere with the measured absorbance at 280 nm and thus cannot easily quantified using this method. Alternatively, bicinchoninic acid assays (BCA) can be used to determine the protein content.
In case the recognized epitope is stimulation dependent, proceed as follows: add 1 ml of cell culture media at 37 °C to two FACS tubes, labeled stimulated and unstimulated. Add stimulation cocktail of choice to one tube. Selecting the appropriate stimulation condition is essential for validating and titrating the conjugated antibody. The stimulation should result in selective induction of the activated form of the signaling molecule of interest. Please refer to these for additional information on antibody targets, appropriate stimulation, and timing – reviewed in [27]. For secreted molecules such as cytokines, add 1X brefeldin A and monensin to the cell culture media prior to addition of cells to medium. More details on cytokine production and measurement by mass cytometry can be found in Newell et al. [8]. Add 10 × 106 cells to each FACS tube. Incubate cells at 37 °C for the appropriate time for monitoring of the selected cellular target. Because distinct signaling pathways are activated at different times following stimulation, the incubation time should be adjusted accordingly. Some cellular signaling events are very time sensitive. Be ready to proceed to fixation immediately.
The reagent should be made fresh for each use. The amount of Ir intercalating reagent can also be reduced (to 0.15–0.2 μl per ml) for more sensitive instruments and larger cells, or increased (to 0.5 μl per ml) for staining cells that were not permeabilized (not described herein).
5. Acknowledgements
We are grateful to Scott Tanner for providing useful feedback on this manuscript and to Fluidigm for supplying the chelating polymer that was used to optimize the protocol described here. F.J.H received support from the Swiss National Science Foundation (SNF Early Postdoc. Mobility), the Novartis Foundation for medical-biological Research (Research Fellowship) and EMBO (Long-Term Fellowship). Also, DP5OD023056, R21AG057224 from the NIH and the Parker Institute for Cancer immunotherapy to M.H.S. and S10OD018040 Shared Instrumentation Grant to UCSF. E.F.S. is supported by a Damon Runyon Cancer Research Foundation Fellowship (DRG-2190–14). G.P.N. is supported by the NIH grants R01CA184968, 1R01GM10983601, 1R01NS08953301, 1R01CA19665701, R01HL120724, 1R21CA183660, R33CA0183692, 1R33CA183654–01, U19AI057229, 1U19AI100627, U54-UCA149145A, N01-HV-00242 HHSN26820100034C, and 5UH2AR067676; the NIH Northrop-Grumman Corporation Subcontract 7500108142; FDA grant HHSF223201210194C; DOD grants OC110674 and W81XWH-14-1-0180; NWCRA Entertainment Industry Foundation; Bill and Melinda Gates Foundation grant OPP1113682. S.C.B. is supported by the Damon Runyon Cancer Research Foundation Fellowship (DRG-2017–09), the NIH grants 1DP2OD022550–01, 1R01AG056287–01, 1R01AG057915–01, 1-R00-GM104148–01, 1U24CA224309–01, 5U19AI116484–02, 1U24CA224309–01, The Bill and Melinda Gates Foundation, and a Translational Research Award from the Stanford Cancer Institute.
Footnotes
Competing Financial Interests
S.C.B. and E.F.S. have been paid consultants for the company Fluidigm Sciences, the manufacturers that produced some of the reagents and instrumentation described in this manuscript.
7. References
- 1.Baranov VI, Quinn Z, Bandura DR, Tanner SD (2002) A Sensitive and Quantitative Element-Tagged Immunoassay with ICPMS Detection. Anal Chem 74:1629–1636. doi: 10.1021/ac0110350 [DOI] [PubMed] [Google Scholar]
- 2.Bandura DR, Baranov VI, Ornatsky OI, et al. (2009) Mass Cytometry: Technique for Real Time Single Cell Multitarget Immunoassay Based on Inductively Coupled Plasma Time-of-Flight Mass Spectrometry. Anal Chem 81:6813–6822. doi: 10.1021/ac901049w [DOI] [PubMed] [Google Scholar]
- 3.Bendall SC, Simonds EF, Qiu P, et al. (2011) Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332:687–96. doi: 10.1126/science.1198704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodenmiller B, Zunder ER, Finck R, et al. (2012) Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat Biotechnol 30:858–67. doi: 10.1038/nbt.2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behbehani GK, Bendall SC, Clutter MR, et al. (2012) Single-cell mass cytometry adapted to measurements of the cell cycle. Cytom Part A 81 A:552–566. doi: 10.1002/cyto.a.22075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fienberg HG, Simonds EF, Fantl WJ, et al. (2012) A platinum-based covalent viability reagent for single-cell mass cytometry. Cytom Part A 81 A:467–475. doi: 10.1002/cyto.a.22067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mingueneau M, Kreslavsky T, Gray D, et al. (2013) The transcriptional landscape of alphabeta T cell differentiation. Nat Immunol 14:619–632. doi: 10.1038/ni.2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newell EW, Sigal N, Bendall SC, et al. (2012) Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity 36:142–52. doi: 10.1016/j.immuni.2012.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lou X, Zhang G, Herrera I, et al. (2007) Polymer-based elemental tags for sensitive bioassays. Angew Chemie - Int Ed 46:6111–6114. doi: 10.1002/anie.200700796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seegan GW, Smith CA, Schumaker VN (1979) Changes in quaternary structure of IgG upon reduction of the interheavy-chain disulfide bond. Immunology 76:907–911. doi: 10.1073/pnas.76.2.907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu H, Chumsae C, Gaza-Bulseco G, et al. (2010) Ranking the susceptibility of disulfide bonds in human IgG1 antibodies by reduction, differential alkylation, and LC-MS analysis. Anal Chem 82:5219–5226. doi: 10.1021/ac100575n [DOI] [PubMed] [Google Scholar]
- 12.Amir ED, Davis KL, Tadmor MD, et al. (2013) viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol 31:545–52. doi: 10.1038/nbt.2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibbs KD, Jager A, Crespo O, et al. (2012) Decoupling of tumor-initiating activity from stable immunophenotype in HoxA9-Meis1-driven AML. Cell Stem Cell 10:210–217. doi: 10.1016/j.stem.2012.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spitzer MH, Carmi Y, Reticker-Flynn NE, et al. (2017) Systemic Immunity Is Required for Effective Cancer Immunotherapy. Cell 168:487–502.e15. doi: 10.1016/j.cell.2016.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartmann FJ, Bernard-Valnet R, Quériault C, et al. (2016) High-dimensional single-cell analysis reveals the immune signature of narcolepsy. J Exp Med jem.20160897. doi: 10.1084/jem.20160897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arrell DK, Terzic a (2010) Network systems biology for drug discovery. Clin Pharmacol Ther 88:120–125. doi: 10.1038/clpt.2010.91 [DOI] [PubMed] [Google Scholar]
- 17.Sachs K (2005) Causal Protein-Signaling Networks Derived from Multiparameter Single-Cell Data. Science (80- ) 308:523–529. doi: 10.1126/science.1105809 [DOI] [PubMed] [Google Scholar]
- 18.Aghaeepour N, Jalali A, O’Neill K, et al. (2012) RchyOptimyx: Cellular hierarchy optimization for flow cytometry. Cytom Part A 81 A:1022–1030. doi: 10.1002/cyto.a.22209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu P, Simonds EF, Bendall SC, et al. (2011) Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol 29:886–91. doi: 10.1038/nbt.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mair F, Hartmann FJ, Mrdjen D, et al. (2016) The end of gating? An introduction to automated analysis of high dimensional cytometry data. Eur J Immunol 46:34–43. doi: 10.1002/eji.201545774 [DOI] [PubMed] [Google Scholar]
- 21.Ornatsky OI, Kinach R, Bandura DR, et al. (2008) Development of analytical methods for multiplex bio-assay with inductively coupled plasma mass spectrometry. J Anal At Spectrom 23:463. doi: 10.1039/b710510j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dulski P (1994) Interferences of oxide, hydroxide and chloride analyte species in the determination of rare earth elements in geological samples by inductively coupled plasma-mass spectrometry. Fresenius J Anal Chem 350:194–203. doi: 10.1007/BF00322470 [DOI] [Google Scholar]
- 23.Kotecha N, Krutzik PO, Irish JM (2010) Web-based analysis and publication of flow cytometry experiments. Curr Protoc Cytom Chapter: 10 Unit1017. doi: 10.1002/0471142956.cy1017s53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bigos M (2007) Separation index: an easy-to-use metric for evaluation of different configurations on the same flow cytometer. Curr Protoc Cytom Chapter:1 Unit121. doi: 10.1002/0471142956.cy0121s40 [DOI] [PubMed] [Google Scholar]
- 25.Medvedev AE, Johnsen AC, Haux J, et al. (1997) Regulation of Fas and Fas-ligand expression in NK cells by cytokines and the involvement of Fas-ligand in NK/LAK cell-mediated cytotoxicity. Cytokine 9:394–404. doi: 10.1006/cyto.1996.0181 [DOI] [PubMed] [Google Scholar]
- 26.Garrone P, Neidhardt EM, Garcia E, et al. (1995) Fas ligation induces apoptosis of CD40-activated human B lymphocytes. J Exp Med 182:1265–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krutzik PO, Trejo A, Schulz KR, Nolan GP (2011) Phospho flow cytometry methods for the analysis of kinase signaling in cell lines and primary human blood samples. Methods Mol Biol 699:179–202. doi: 10.1007/978-1-61737-950-5_9 [DOI] [PubMed] [Google Scholar]