Abstract
Introduction:
X-ray crystallography has made important contributions to modern drug development but its application to many important drug targets has been extremely challenging. The recent emergence of X-ray free electron lasers (XFELs) and advancements in serial femtosecond crystallography (SFX) have offered new opportunities to overcome limitations of traditional crystallography to accelerate the structure-based drug discovery (SBDD) process.
Areas covered:
In this review, the authors describe the general principles of X-ray generation and the main properties of XFEL beams, outline details of SFX data collection and processing, and summarize the progress in the development of associated instrumentation for sample delivery and X-ray detection. An overview of the applications of SFX to various important drug targets such as membrane proteins is also provided.
Expert opinion:
While SFX has already made clear advancements towards the understanding of the structure and dynamics of several major drug targets, its robust application in SBDD still needs to develop in terms of the high-throughput techniques for sample production, automation of crystal delivery and data collection, as well as in terms of the optimization of the processing and storage of large amounts of data. The expansion of the available XFEL beamtime is a key to the success of SFX in SBDD.
Keywords: X-ray Free Electron Laser (XFEL), Serial Femtosecond Crystallography (SFX), structure based drug discovery (SBDD), membrane protein, Lipidic Cubic Phase (LCP), G Protein Coupled Receptor (GPCR)
1. Introduction
Over the last century, rational drug discovery efforts have contributed profoundly towards the better understanding and treatment of many devastating and deadly diseases. Yet traditional drug discovery has been extremely time and labor consuming and very costly. According to analytic reports, it takes on average over 13 years along with about $2 billion associated cost to bring a new therapeutic product on market [1]. The cost of developing a drug have been steadily increasing at an annual rate of ~13% [2]. Although a record number of 59 new drugs have been approved by the US Food and Drug Administration (FDA) in 2018, the projected sales are in decline with an average annual peak sales of $720 million per drug [3]. Furthermore, the main hurdles standing in front of the pharmaceutical industry include challenges in the identification and validation of new targets, as well as high attrition rates, with about 90% of drug candidates reaching Phase I clinical trials being destined to fail [4].
Since late 1970-s, structural biology has been making important contributions in various aspects of the drug discovery process, including target validation, hit identification, lead optimization, and selection of the most promising candidates for subsequent animal and clinical trials [5]. While initial structure-based efforts have been severely hampered, there have been remarkable advancements in recombinant protein expression, purification, and crystallization, along with technological breakthroughs in automation, X-ray sources, detectors, and data processing during over the past two decades, which have dramatically expanded the number of accessible protein targets for structural determination and advanced the role of structure-based drug discovery (SBDD) in the successful development of new therapeutics.
Despite the proven success of SBDD with a variety of soluble protein targets, such as kinases and proteases, its application to more challenging protein families including G protein-coupled receptors (GPCRs) and other hard-to-crystallize membrane and soluble proteins has been almost imperceptible. Many of these targets are difficult to express, purify, or crystallize. Even when successful, the crystals are often too small, or their diffraction resolution is poor. The emergence of unique X-ray sources called X-ray free electron lasers (XFELs), about a decade ago, brought with it the possibility of overcoming radiation damage and enabling high-resolution room temperature structure determination from very small or heterogeneous crystals, and, most importantly, provided access to studying conformational transitions in proteins with a sub-picosecond time resolution [6]. Due to extreme brightness of XFEL pulses that can completely annihilate small crystals, crystallographic data are typically collected from thousands of crystals sequentially delivered into the focal point of the XFEL beam in random orientations. This data collection method, known as serial femtosecond crystallography (SFX), has matured over the past few years and started delivering significant new results that overcome the shortcomings of traditional crystallography [7]. In this review we describe the current status of XFEL instrumentation, summarize the most important achievements in structure determination relevant for drug design, and discuss the future perspectives of applying SFX to SBDD.
2. Structure determination in drug discovery
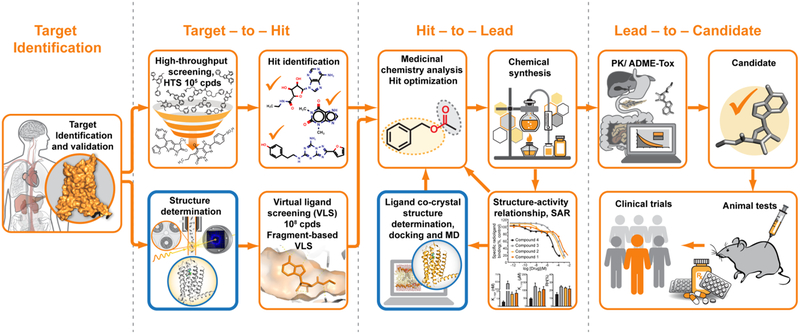
Modern drug discovery is a complex process that, in general, consists of several major steps, including initial target selection and validation, hit identification, hit-to-lead optimization, candidate selection, animal and clinical trials (Figure 1). Traditionally, initial hit identification relies on high-throughput screening (HTS), which is typically limited to assaying libraries of less than a few million compounds out of possible virtual chemical space consisting of over 1063 drug-like small molecules [8]. Combinatorial chemistry along with fragment-based screening bring promise to increase potential coverage of the chemical space, however, these approaches come with additional challenges related to low binding affinity for fragments and to the need for more extensive optimization of fragment-based hits [9]. After initial hits are identified, their optimization is performed iteratively via multiple rounds of structure-activity relationship (SAR) studies [10]. Overall, it takes from several months to a few years to select the most promising candidate for clinical trials.
Figure 1.
Typical drug discovery pipeline. X-ray crystallography contributes to this process mainly by providing initial crystal structure templates for VLS and co-crystal structures during hit-to-lead optimization (outlined in blue). Structural information may also help with target validation and selection of the most promising drug candidates for animal and clinical trials.
X-ray crystallography can make critical contributions to most stages of the described above drug discovery process. First, the knowledge of a 3-dimensional structure of the target protein may shed light on its mechanism of action helping with target validation. Second, available 3D structural templates of the target protein can be used for virtual ligand screening (VLS) of much larger libraries of compounds with substantially less associated cost, time, and effort, compared to HTS [11,12]. For example, one of the largest publicly available library, ZINC [13], contains over 230 million purchasable compounds in ready to dock 3D format. Fragment-based VLS can further expand the covered chemical space to over 1014 feasible for synthesis compounds, significantly increasing the chance of finding more diverse and potent initial hits for further optimization [14]. Third, the lead optimization process can be greatly enhanced by complementing SAR analysis with structural mapping of the ligand-binding pocket. Commonly, hundreds of different ligand co-crystal structures are determined during this stage to better understand specific ligand-protein interactions and guide ligand optimization in an iterative manner.
One of the first examples of using structure to design a new compound that stabilizes the deoxygenated conformation of hemoglobin was reported in 1976 [15]. Designing of antihypertensive renin inhibitors in the 1980-s was initially guided by a homology model of human renin based on crystal structures of fungal pepsins [16] and later by co-crystal structures of mouse and human renins in complex with their inhibitors [17]. Several comprehensive SBDD campaigns during the last 30 years led to the development of high-profile drugs in diverse therapeutic areas, with some of the most prominent examples including HIV protease inhibitors for the treatment of AIDS [18], the neuramidase inhibitor oseltamavir (Tamiflu) against influenza [19], and a number of small-molecule inhibitors for cancer drug targets [20].
Currently, most of the leading pharmaceutical companies employ SBDD in their drug discovery pipelines using either in-house efforts or outsourcing to specialized CRO companies. Recent analysis of over 1,000 drug discovery projects revealed substantially lower attrition rates for SBDD projects compared to project where structural information was not available [21]. Despite the wide acceptance and proven success, many targets are not routinely amenable to SBDD. An important example is the superfamily of G protein-coupled receptors (GPCRs), which represent the targets of about 40% of drugs on the market, however, their members are exceedingly challenging for SBDD applications due to their inherent low expression and high conformational dynamics. Structural studies of GPCRs bound to diffusible ligands were enabled in 2007 [22] by multiple advancements and breakthroughs in their heterologous expression, stabilization in one of their conformational states, crystallization in membrane environment, and microcrystallography. By the end of 2018, more than 300 structures of 59 unique receptors out of over 800 GPCR members that express in human cells have been published [23]. These structures revealed an amazing diversity of shapes and sizes of their orthosteric binding pockets, uncovered a variety of allosteric binding sites, and helped to decipher general signaling mechanisms in GPCRs [24]. Most GPCRs, however, were crystallized only in a single conformational state in complex with a single ligand. Solving the first structure of a new GPCR takes on average several years, and obtaining high-resolution multiple ligand co-crystal structures, required for lead optimization in SBDD, represents a major feat.
XFEL sources that emerged about a decade ago and new serial crystallography data collection technologies that they stimulated bring promise to overcome some of the challenges related to structure determination of difficult targets and transform the SBDD process.
3. Principles of XFEL and SFX
The concept of a free electron laser was proposed in 1971 by John Madey at Stanford [25], based on a previously described process of photon generation by free electrons moving in a periodical array of magnets, known as an undulator, by Vitalii Ginzburg [26] and Hans Motz [27]. The first demonstration of FEL-generated infrared radiation was achieved by Madey’s group in 1976, stimulating further intense research work that culminated in commissioning of the first soft XFEL source FLASH (X-ray photon energy < 0.2 keV) at DESY in Hamburg in 2005 [28]. A few years later, in 2009, the first hard XFEL (X-ray photon energy up to 10 keV) Linac Coherent Light Source (LCLS) was opened for user experiments at the SLAC National Accelerator Laboratory in Menlo Park, California [29,30].
Coherent X-rays are generated in an XFEL by a process called Spontaneous Self-Amplified Emission (SASE) [31], in which a bunch of free electrons is accelerated to near relativistic velocity by a linear accelerator and compressed by magnetic chicanes. The compressed bunch then enters a long undulator, consisting of periodically arranged alternating magnets, where electrons start spontaneously emit incoherent X-rays. As electrons co-propagate and interact with X-ray photons along the undulator, they become arranged in microbunches with the periodicity equal to the wavelength of the emitted radiation. By the end of the long undulator, electrons in microbunches emit X-rays coherently, achieving a power amplification by 9–10 orders of magnitude. Finally, the electrons are dumped, and the XFEL pulse is focused by X-ray optics, such as Kirkpatrick-Baez (K-B) mirrors or beryllium compound refractive lens, to a spot ranging from 0.1 to 10 μm depending on the experimental requirements.
Therefore, XFELs produce extremely bright pulses of coherent X-rays with extremely short duration of only few femtoseconds. Such unique properties enable new applications in many scientific fields including structural biology, in which short XFEL pulses can outrun radiation damage by the diffraction-before-destruction principle [32] and allow for time-resolved crystallographic studies of proteins in action with sub-picosecond time resolution [33,34].
The first successful generation of a hard XFEL beam and initial groundbreaking results achieved at LCLS in physics, chemistry, and structural biology [35] had motivated scientific community to build additional XFEL facilities around the world (Table 1). By the end of 2018, five hard XFEL facilities were available for users: Linac Coherent Light Source (LCLS) in Menlo Park, USA; SPring-8 Angstrom Compact Free Electron Laser (SACLA) in Harima, Japan; Pohang Accelerator Laboratory (PAL)-XFEL in Pohang, South Korea; European XFEL (EuXFEL) in Hamburg, Germany; and SwissFEL at Paul Scherrer Institute in Villigen, Switzerland. Further expansion of XFEL facilities is anticipated via commissioning of new sources such as Shanghai HIgh repetition rate XFEL aNd Extreme light facility (SHINE) in China [36] and planned upgrades such as LCLS-II HE [37].
Table 1.
Operational parameters of currently available hard XFEL facilities. Planned parameters are shown in parentheses.
| XFEL First user experiments |
Photon energy, keV |
Max pulse repetition rate, Hz |
Pulse length, fs (FWHM) |
Photons per pulse, ×1012 |
SFX Beamlines |
Sample environment |
Detector | Beam focus, μm |
Sample delivery |
|---|---|---|---|---|---|---|---|---|---|
| LCLS 2009 |
5-10 | 120 | 30-300 | 1 | CXI | Vacuum | 2 CSPADs | 1.3 0.1 |
liquid/LCP injectors, fixed-target stage |
| MFX (XPP)a | Helium, Cryo, Humidity | Rayonix MX170-HS, CSPAD | 3 (0.7) | liquid/LCP injectors, MESH/coMESH, Roadrunner, drop-on-demand, goniometer | |||||
| SACLA 2012 |
4-20 | 30 (60) | 2-10 | 0.25 | BL3/BL2 | Helium | MPCCD, Rayonix MX225-HS | 1 | fixed-target stage, liquid/LCP injectors, acoustic systems |
| EuXFEL 2017 |
6-10 (3-16) | 1.1×106 (4.5×106)b | 25 (3-300) | 0.6 | SPB/SFX | Vacuum | AGIPD 1 Mpx | 5 (1) 0.5 (0.1) |
liquid/LCP injectors, fixed-target stage |
| SwissFEL 2019 |
1.8-8 (1.8-12.4) | 25 | 100-200 (50-200) | 0.7 | Alvra | Vacuum Helium | 16M JUNGFRAU | 5 | liquid/LCP injectors, fixed-target stage |
| Bernina | Helium Cryo | 1.5M JUNGFRAU | 5 | goniometer | |||||
| PAL-XFEL 2017 |
8-10 | 30 (60) | 25 | 0.1 | NCI | Helium, Vacuum | Rayonix MX225-HS, MPCCD | 5 | liquid/LCP injectors, fixed-target stage |
Many early SFX experiments in ambient environment have been performed at XPP. These types of experiments are currently mainly redirected to a recently commissioned beamline MFX.
EuXFEL has a unique non-uniform pulse sequence consisting of pulse trains generated at 10 Hz with up to 1.1 MHz (4.5 MHz planned) pulse repetition rate within each train.
Since ultrabright focused XFEL pulses can annihilate any material they interact with, structural data are typically acquired via a Serial Femtosecond Crystallography (SFX) approach [38], in which diffraction images are collected from hundreds of thousands of microcrystals delivered to the intersection with the XFEL beam in random orientations, ideally, matched or synchronized with the XFEL pulse repetition rate. The data are then assembled using Monte Carlo integration or other more sophisticated methods. Thus, SFX enables radiation damage-free high-resolution structure determination from micrometer and submicrometer-sized crystals at room temperature, making it an ideal technique for structural studies of extremely radiation-sensitive samples, for example, metalloenzymes, difficult to crystallize proteins such as GPCRs and large protein complexes, as well as for assessing protein dynamics at room temperature and for following ultrafast conformational transitions.
4. An overview of the experimental setup and current status of SFX
In this section we will describe major developments and state-of-the-art related to sample preparation, data collection, and data processing for an SFX experiment. The layout of a typical SFX beamline includes beam focusing and conditioning modules, a sample chamber with an integrated sample delivery system, and an X-ray detector (Figure 2). To reduce background scattering due to a strong XFEL beam, the sample chamber is kept under vacuum or filled with helium at atmospheric pressure. Helium chambers offer simplicity and flexibility in exchange for a moderately increased background. They also can accommodate a broader variety of sample delivery methods along with optical laser systems for time-resolved pump-probe experiments, as well as allow for diverse sample environments such as pressure, temperature, humidity, etc. X-ray detectors for SFX have special requirements due to an extremely large number of photons arriving at the detector within a few femtosecond pulse duration and a high pulse repetition rates at modern XFELs. Finally, SFX data processing requires development of special software for assembling a dataset using static diffraction snapshots captured from thousands of crystals in random orientations.
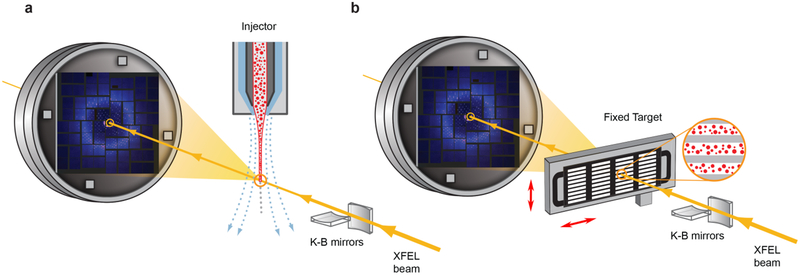
Figure 2.
Schematic layout of an SFX experiment. a) Injector-based and b) fixed target crystal delivery systems. XFEL beam is focused by beamline optics (K-B mirrors in this illustration) and diffracts from microcrystals delivered by an injector (a) or deposited on a solid support and rastered by a fast stage (b). Sample delivery is typically performed in vacuum or helium environment to reduce scattering from the direct beam.
4.1. Crystal preparation and delivery
Since each XFEL pulse destroys the crystals that it interacts with, crystal preparation and delivery are among the most critical aspects of an SFX experiment. An ideal crystal delivery system should be reliable, efficient (minimize sample waste), fast (keep up with the maximum available XFEL pulse repetition rate), compatible with diverse crystallization methods, suitable for fast triggering of conformational transitions (optical lasers, mixing, temperature or pressure jumps), and it should allow for a fast switching between samples. Since it is practically impossible to combine all these characteristics in one system, many sample delivery instruments have been developed, which could be grouped in two main categories: a) injectors that stream crystals across an XFEL beam inside of their native crystallization medium or a specially designed carrier matrix, and b) fixed-target, where crystals are deposited on a solid support that is quickly rastered by the beam (Figure 2).
The first and still one of the most successful injector to date, a gas dynamic virtual nozzle (GDVN), was introduced in 2008 [39]. A simple and robust construction of two coaxial capillaries produces a stream of crystals flowing in their liquid crystallization medium focused to a diameter of only few micrometers by a sheath of gas. GDVN was successfully used for the first SFX demonstration with microcrystals of photosystem I [38] and many other SFX experiments, however, its major drawback of a high sample consumption (flow rates above 10 μl/min) prompted development of modified and alternative injectors. For example, one such modification, a double flow focusing nozzle (DFFN), allowed for sample reduction by up to eight times [40].
An entirely different type of injectors based on the electrospinning principle was designed to substantially reduce the crystal flow rate and thus sample consumption. The first version, called a microfluidic electrokinetic sample holder (MESH) [41], could achieve flow rates down to 0.14 μl/min but required addition of a cryoprotectant, which was remedied by an optimized construction of a concentric-flow microfluidic electrokinetic sample holder (coMESH)_[42]. Another very important injector, known as a lipidic cubic phase (LCP) or viscous media injector [43], offers a wide range of flow rates down to 1 nl/min and compatibility with crystallization of membrane proteins in a membrane-like environment of LCP [44]. An LCP injector was used to obtain about a dozen GPCR structures using less than 0.3 mg of crystallized protein per structure [45] and to record molecular movies of light-induced conformational changes in bacteriorhodopsin [46]. Apart from membrane proteins crystallized in LCP, an LCP injector can be used to highly efficiently deliver crystals of soluble proteins upon mixing them with a viscous carrier matrix, such as LCP, agarose, grease, etc [47].
While crystal injectors have many advantages and are convenient to use, the fixed-target approach [48] can bring some additional benefits such as lower sample consumption, higher crystal hit rates, in situ crystallography, and increased versatility in triggering reactions for time-resolved studies. Major challenges for this approach are to keep crystals hydrated during data collection and to implement a fast and precise scanning and synchronization between crystal positions and XFEL pulse arrivals. A promising fast scanning goniometer, aptly named Roadrunner, partially solved these problems and enabled challenging virus crystallography at a full 120 Hz pulse repetition rate available at LCLS [49]. Another approach for crystal delivery, called drop-on-demand, combines acoustic droplet ejection with a conveyor belt drive to facilitate simultaneous spectroscopic and crystallographic data collection for observation of chemically-triggered or photoinduced reactions [50].
Different crystal delivery systems impose specific requirements on sample preparation. For SFX data collection, crystals, preferably, should be uniform in size, highly isomorphous with respect to cell parameters, well ordered, free of defects, possess low mosaicity, and be prepared in sufficient quantities (typically, 0.1 to 100 mg of crystallized protein depending on the delivery system). Several crystallization techniques have been employed to achieve these requirements, for example, batch crystallization with vigorous mixing [40,51], free-interface diffusion [52], and seeding [53]. In cellulo crystallization has also recently emerged as a promising approach for certain protein targets [54-56] as well as viruses [57]. Crystals prepared in solution can be pre-filtered through a wire mesh to select a more homogeneous in size population and concentrated by centrifugation to optimize crystal hit rates. On the other hand, pre-filtering and concentration are not readily applicable to membrane protein crystals grown in LCP, and, therefore, their preparation and delivery require special considerations [58]. To increase the efficiency of SFX data collection, all prepared samples should be inspected and characterized by various techniques, such as visual and UV-light microscopy, electron microscopy, dynamic light scattering (DLS), and SONICC (Second-Order Nonlinear Imaging of Chiral Crystals) [59].
4.2. XFEL detectors
An accurate detection of X-ray photons diffracted by a crystal is another essential requirement of any crystallographic experiment. Recent development of photon counting hybrid pixel array detectors (HPADs) radically transformed synchrotron crystallography providing determination of diffraction intensities at the level of individual photons over an extremely wide dynamic range [60]. These detectors are called “hybrid” because they consist of two principal parts fabricated separately: a pixelated semiconductor sensor and a pixelated readout chip, also called an Application-Specific Integrated Circuit (ASIC). Two parts are connected electrically pixel-to-pixel by a bump bonding process [60]. The sensor part (typically, a doped silicon chip) converts incoming X-rays into clouds of electrons and holes. Charges are directed pixel-by-pixel into the ASIC layer, where integrated electronics generates a signal in response to each incoming X-ray photon.
Photon counting HPADs can detect sequentially arriving X-rays separated by a minimal time delay mainly dictated by the signal processing electronics; generally, it takes 10−7 - 10−8 s to process one photon count. At XFEL sources X-ray photons arrive in pulses of <10−13 s and therefore individual photons can not be distinguished. Therefore, photon counting HPADs are not suitable for XFELs and integrating analogs are used instead. Integrating HPADs sum all charges arriving at each pixel over the exposure interval, after which it is digitized by ASICs to provide a signal proportional to the charge and therefore to the total number of absorbed photons.
Due to an extremely high X-ray intensity and pulse rate produced by XFELs, integrating HPADs should be able to provide a reasonably low noise for reliable single photon detection over a high dynamic range, and fast readout rates. In most cases detectors are developed individually to match their design and operational characteristics to the parameters of the particular XFEL (Table 1). Cornell-SLAC Pixel Array Detector (CSPAD) was the first detector specifically developed for data collection at LCLS [61]. A 2.3 megapixel CSPAD consists of 64 ASICs assembled into 4 movable tiles with a variable hole in the middle for the passage of a direct XFEL beam. This detector has low noise, high sensitivity, and can acquire data at a full 120 Hz pulse repetition rate of LCLS. However, it suffers from a relatively low for crystallography dynamic range, which can partly be mitigated by selecting one of the two available gain levels for each pixel. A more advanced technique for extending the dynamic range based on a dynamically switched 3-level gain on a per-pixel per-frame basis is implemented in AGIPD (Adaptive Gain Integrating Pixel Detector) at EuXFEL [62] and JUNGFRAU (adJUstiNg Gain detector FoR the Aramis User station) at SwissFEL [63].
Modern XFEL sources based on superconducting LINACs (EuXFEL, future LCLS-II) are capable of producing extremely high pulse repetition rates in a MHz range, which could dramatically shorten data collection time, however, impose great challenges for crystal delivery and data acquisition. The typical achievable detector readout rates are orders of magnitude slower compared to such pulse repetition rates, necessitating a temporary in-ASIC frame storage [64]. EuXFEL has a special timing structure consisting of a 4.5 MHz pulse rate during 600 μs followed by 99.4 ms of idle time, during which the detector is read out, data are digitized and transferred to a mass storage. AGIPD has 352 storage cells inside its pixel circuitry and can operate at up to 6.5 MHz rate, which is compatible with the EuXFEL parameters, but requires an additional storage capacity or on-fly rejection of empty frames to take advantage of the maximum available 27,000 pulses per second.
4.3. SFX data processing software
Compared to traditional goniometer-based crystallography, for which data processing has been well established, SFX represents a relatively young branch with quickly progressing developments of new algorithms and tools. In general, SFX data have two distinct features: i) the high XFEL pulse repetition rate yields large amounts of 2D detector images (hundreds of thousands) that require rapid and automated processing, and ii) only single diffraction snapshots are recorded from each crystal intersecting the beam at random orientations, and neither the crystal orientation nor the full intensity of diffraction spots is known. Consequently, SFX data processing can be divided into two major tasks: i) rapid identification of images (tens of thousands) containing diffraction spots (crystal hits), and ii) indexing, integration, scaling, and merging of crystal hit data. For the first task, a number of algorithms have been proposed to date: Cheetah [65], NanoPeakCell [66], neural network [67], Zaef [68], Dials [69], nXDS [70], cctbx.spotfinder [71]. All of them attempt to quickly scan through the images to identify those that contain at least a certain number of spots with given parameters and also perform preliminary data reduction (rejecting empty images) and beamline-independent data conversion for further off-line processing. Some of these algorithms have been incorporated into real-time monitoring systems, such as OnDA [72], SACLA’s pipeline [73], CASS [74], MICOSS [75], on several XFEL beamlines, helping to estimate sample diffraction quality and hit rate to optimize data collection.
For the second task, several packages, e.g. CrystFEL [76], ccpxfel [77], cctbx.xfel [71], DIALS [69], and nXDS [70], are available, which incorporate both already established (Mosflm, XDS, DirAx) and recently-developed, such as TakeTwo [78], SPIND [79], xgandalf [80], and FELIX [81], indexing algorithms. The problem of partial intensity treatment from single snapshots has been approached by different methods: simple averaging using Monte-Carlo integration in CrystFEL [76], post-refinement partiality estimate implemented in both cctbx.xfel and Crystfel [82-84], PRIME [85] or EVAL [86], improved spot-shape model with no partiality estimate [87], and an Ewald-offset correction in nXDS [70], many of which are still under further development. Notably, during indexing and integration steps, all SFX diffraction images in a dataset can be treated independently, thus allowing for a high level parallelization of data processing to as many CPU-cores as the number of images, compensating for the high volume of data acquired in an SFX experiment.
4.4. Experimental phasing of SFX data
Most SFX structures to date have been solved using molecular replacement. Solving structures of proteins, for which no suitable model is available, however, necessitates experimental phasing that requires higher accuracy of structure factors determination and, in some cases, special sample treatments. Since the first demonstration of SFX experimental phasing five years ago [88] most standard phasing techniques used in traditional crystallography have been applied to SFX experiments (SAD [88], SIRAS [89], SIR [90], and MIRAS [91]) on a number of targets, including GPCR sulfur-SAD phasing [92]. Additionally, the unique properties of XFEL radiation enable new phasing approaches, such as high intensity phasing [93], off-Bragg phasing for nanometer-sized [94] or imperfect crystals [95], as well as offer possibilities for two-color MAD phasing [96].
5. SFX highlights
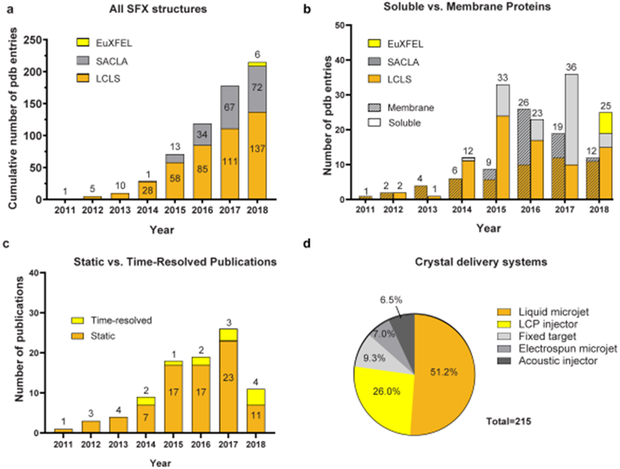
Since its first successful demonstration in 2011 [38], SFX approach has quickly matured from proof-of-principle demonstrations to starting delivering important results in structural biology, limited only by the availability of scarce XFEL beamtime. By the end of 2018, a total of 215 Protein Data Bank (PDB) entries have been published using SFX data collected at XFELs (137 from LCLS, 72 from SACLA, and 6 from EuXFEL) (Figure 3); first results are soon expected from PAL-XFEL [97,98] and SwissFEL. Among them are 69 PDB entries for membrane proteins and 95 (51 unique) for important drug targets (GPCRs, enzymes, viruses). The best resolution so far (1.2 Å) was obtained for Proteinase K (PDB ID 5KXU).
Figure 3.
Statistics of SFX structure determination. a) Cumulative number of PDB entries produced on different XFELs. b) Number of soluble vs membrane protein PDB entries produced per year on different XFELs. c) Number of publications per year reporting static vs time-resolved SFX studies. d) Contributions of different crystal delivery systems used in SFX experiments.
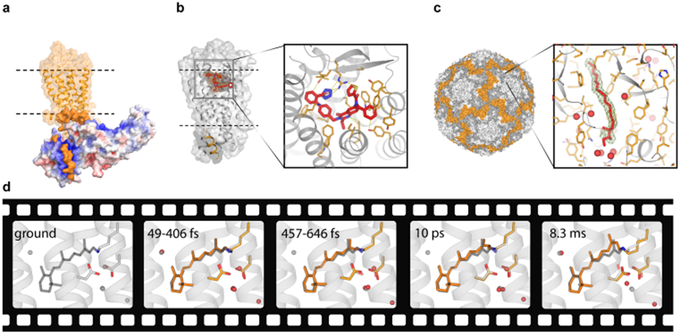
Among some of the most important SFX highlights are structural studies of GPCRs resulting in publications of over a dozen of different receptor structures during the last 5 years [45]. SFX allows to overcome the most significant challenges of GPCR crystallography, such as a low receptor expression and typically small, weakly diffracting crystals. GPCR structure determination by SFX was enabled by the development of an LCP injector [43], which made it possible to deliver crystals within their crystallization matrix and reduced protein consumption on average to less than 0.3 mg of crystallized protein per dataset. In case of GPCRs, SFX helps to substantially shorten crystal optimization time, bypass crystal harvesting and cryocooling, and deliver higher resolution structures captured in close to their native environment without any perceivable radiation damage [99]. The most prominent examples of GPCR work at XFELs include structures elucidating receptor-ligand interactions between the smoothened receptor and the teratogen cyclopamine [100] and between the δ-opioid receptor and a bi-functional peptide acting as a potent and non-addictive pain-killer [101], the structure of a major signaling complex between rhodopsin and arrestin [102,103] (Figure 4a), the full length structures of the Class B glucagon GCGR receptor [104] and Class F smoothened receptor [105], the novel structures of GPCR drug targets against hypertension – angiotensin II receptor type 1 [106], neuropathic pain – angiotensin II receptor type 2 [107] (Figure 4b), and postpartum bleeding – prostaglandin E2 receptor 3 [108]. These structures shed light on the function of corresponding receptors and provide structural templates for rational discovery of new efficient drug candidates.
Figure 4.
Several SFX highlights. a) Structure of a signaling complex between the visual rhodopsin (orange) and arrestin (red-white-blue surface colored by charge density) (PDB ID 5W0P). b) Structure of the angiotensin II receptor type 2 in complex with an antagonist (PDB ID 5UNG). c) Crystal structure of BEV2 virus with a sphingosine molecule (red sticks) bound in a hydrophobic pocket (insert) (PDB ID 5OSN). Ligand omit 2mFo-DFc electron density map contoured at 1 sigma is shown as a green mesh. Waters are shown as red spheres. d) Molecular movie of retinal isomerization in bacteriorhodopsin (PDB IDs 6G7H-6G7L). The ground state retinal conformation is shown in gray sticks in all frames for reference.
Development of antiviral drugs and vaccines can benefit from the knowledge of high-resolution structural information, however, virus crystallography has been very challenging due to the large unit cells, limiting crystal sizes, and radiation damage effects. SFX brings promise of overcoming these problems. Thus, fixed-target SFX with the high-speed Roadrunner goniometer was employed to determine the crystal structures of the picornavirus bovine enterovirus 2 (BEV2) and the cytoplasmic polyhedrosis virus type 18 polyhedrin (CPV18) within just several minutes of data collection and using only few micrograms of crystals [49]. The structure of BEV2 revealed a sphingosine molecule bound in a hydrophobic pocket that can be targeted by antiviral agents (Figure 4c). In another study, potential of SFX for in cellulo virus crystallography has been explored using naturally occurring sub-micron crystallites of bacteriophage phiX174 inside living E. coli cells [57].
Compared with traditional crystallography at synchrotron sources, which is mostly performed with cryo-cooled crystals to minimize radiation damage, SFX provides access to ambient temperature structures, which could reveal essential features that become hidden or lost upon cryo-cooling. For example, room temperature SFX structures of the M2 channel from influenza A virus helped to observe pH-dependent ordering of water inside the channel shedding light on proton transport across the membrane [109]. Other examples include applications of ambient temperature SFX to discover temperature sensitive conformational states of the 30S ribosome induced by interactions with antibiotics [110] and to reveal conformational flexibility of the acyltransferase from the disorazole polyketide synthase that is important for its catalytic activity [111].
While static structures provide a wealth of important information about protein function, one of the most anticipated promises of XFELs is to enable recording molecular movies of proteins in action, capturing ultrafast processes and irreversible conformational transitions, as permitted by crystal lattice. The first time-resolved SFX experiment demonstrating feasibility of pump-probe data collection at an XFEL was published in 2012 [112], followed two years later by capturing the first time-resolved structural snapshots of photosystem II (PSII) [34,113] and photoactive yellow protein (PYP) [51]. The next milestone of observing ultrafast reactions with a sub-picosecond resolution was achieved in experiments on photodisassociation of carbon monooxide from myoglobin [114] and on cis/trans isomerization of the chromophore in PYP [115]. The last two years were marked by a strong expansion of both protein targets and methods of triggering conformational transitions in time-resolved studies (Figure 3c). In addition to exploiting direct activation of photosensitive proteins, such as PSII [116], the reversibly photoswitchable fluorescent protein rsEGFP2 [117], and the light-driven proton pump bacteriorhodopsin [46,118] (Figure 4d), other reaction-triggering methods included the use of a photosensitive caged-NO compound for capturing initial intermediates during enzymatic reaction of a NO reductase [119] and mix-and-inject SFX to observe intermediates during the catalytic reaction of the Mycobacterium tuberculosis β-lactamase with an antibiotic ceftriaxone [120]. It is anticipated that the next boost in time-resolved studies may come from harnessing the extremely high pulse repetition rates available at EuXFEL [121], which could significantly shorten experimental time required for capturing a series of frames for a time-resolved movie.
6. Conclusions
Recently structural biology has undergone a quantum leap propelled by the resolution revolution in cryoelectron microscopy (CryoEM) [122] and breakthroughs in the development of SFX applications at XFEL sources [123]. The field is experiencing a renaissance with an avalanche of new high-resolution structures of highly important targets that previously been considered unattainable. Both CryoEM and SFX have their unique strengths and limitations and can provide complementary contributions towards drug discovery [124,125].
The last decade has been marked by an incredible progress achieved in all aspects of the SFX approach, including development of new instrumentation for sample delivery and data acquisition, new crystallization and data collection methods, as well as new computational algorithms for data processing. These developments enabled room temperature crystallography of extremely radiation sensitive and difficult-to-crystallize proteins and time-resolved studies of ultrafast and irreversible conformational transitions in macromolecules. Important highlights include structural studies of GPCRs, advancements in enzyme and virus crystallography, molecular movies of light-triggered transitions in photosensitive proteins, and capturing transient enzyme intermediates.
7. Expert opinion
SFX offers multiple advantages over traditional goniometer-based cryo-crystallography and carries the strong potential of transforming SBDD. These advantages include the ability to work with challenging targets that have low expression and are difficult to crystallize. Small crystal size often translates into faster crystal optimization, lower mosaicity and better diffraction quality, and can facilitate ligand soaking and exchange, which are critical procedures for the generation of co-crystals with a large number of compounds during the hit-to-lead optimization process. SFX obviates laborious crystal harvesting and necessity for cryo-protection, lending itself suitable for automation. Finally, room temperature structures provide a more realistic view of the strength of water molecule interactions inside the ligand binding pocket, which could inform the drug design process [126].
For robust SBDD applications, the following further developments and considerations are needed. First of all, new high-throughput methods should be established for crystal screening, optimization, ligand co-crystallization and soaking or exchange that would be fully compatible with SFX data collection. Equally important are further optimizations of crystal delivery systems to minimize sample consumption, maintain compatibility with fast pulse repetition rates, enable quick sample change, reduce background scattering, and increase their efficiency and reliability. Next, detector upgrades are critical to optimize data acquisition at high pulse frequency rates with low noise levels, high dynamic range, and tolerance to extreme levels of X-ray radiation. Lastly, data processing algorithms should be further improved to maximize the indexing rates and minimize the number of images required for an accurate measurement of structure factors. The large number of images in a typical SFX dataset require special considerations for data transfer and storage as well as for computing power and architecture that should allow for high parallelization of data processing tasks. Dissemination of expertise in SFX data processing, which is substantially more complicated than conventional crystallography, via publications, workshops, and webinars should be considered as an important component for training future users of this methodology.
While high-resolution static structures are currently the working horses for rational drug design applications, time-resolved molecular movies, enabled by XFEL radiation, can provide important additional insights into functional mechanisms of the target proteins and help to visualize details of drug binding and disassociation, including potential implications of induced fit re-arrangements [127]. Time-resolved studies of a drug binding to the target protein can be carried out using the mix-and-inject approach [120] or the pump-probe technique with either photocaged [119] or photo-switchable ligands. Designing photoswitchable ligands is a promising approach for enabling precise spatial and temporal control over the function of the target protein [128,129].
Other relevant for drug design applications of XFEL radiation include small molecule nanocrystallography [130] and coherent diffraction imaging of amyloid fibrils [131] and drug-delivery particles [132]. One of the most exciting prospects is that of a single molecule imaging, which could revolutionize structural biology enabling studies of the structure and dynamics of macromolecules without the need for crystallization [133].
All these future developments and their applications to SBDD would not be possible to realize without securing adequate access to XFEL beamtime for both academic and industrial teams, which at the moment is scarce and highly competitive, compared to the available beamtime at synchrotron and CryoEM facilities. Currently, most XFELs can accommodate only one experiment at a time. Multiplexing experiments by implementing beam kicking, splitting, switching, and reusing can quickly and significantly expand the available resources [134-136]. In terms of the beam usage efficiency, collection of an SFX dataset typically takes from several minutes to few hours, which seems relatively long compared to reported cases of a full dataset collection within 1 s at synchrotron sources [137]. However, in case of difficult targets, obtaining a high-resolution dataset at a synchrotron requires screening of hundreds or thousands of crystals for diffraction, which can take hours to days. For such targets SFX is already more efficient, and the amount of time to collect a dataset can be further reduced by orders of magnitude by taking advantage of faster pulse repetition rates and optimized data processing protocols. Since construction of large XFEL facilities is very expensive, several groups are exploring designs of less expensive compact XFELs that operate in the attosecond pulse regime [138,139]. If successful, such compact XFELs may help to alleviate the shortage of XFEL beamtime. Additionally, instrumentation and technologies developed for SFX can be translated to serial crystallography experiments [140] at new generation diffraction-limited storage rings (DSLRs) with substantially increased brightness [141] that have recently started to appear around the world, and to which most current third generation synchrotrons soon will be upgraded. Therefore, the prospect for serial crystallography to make impactful and long-lasting contributions to SBDD appears to be very strong.
Article highlights.
Over the last three decades, X-ray crystallography has contributed profoundly to successes in rational drug discovery; however, many important drug targets are not amenable to traditional SBDD approaches.
The recent emergence of XFELs brings the promise of revolutizing structural biology and accelerating SBDD applications through overcoming radiation damage and providing access to difficult-to-crystallize proteins, room temperature structures, and protein dynamics.
Five hard XFEL facilities are currently available for users worldwide: LCLS in USA, SACLA in Japan, EuXFEL in Germany, PAL-XFEL in South Korea, SwissFEL in Switzerland. Several new sources are on the way: LCLS II HE in USA, SHINE in China.
Data at XFEL sources are collected following the “diffraction-before-destruction” principle implemented as an SFX approach that required the development of new instrumentation for crystal delivery and data acquisition, crystal preparation technologies, and data processing software.
SFX technology has quickly matured and started to show high efficiency in the structure determination of various important drug targets, such as GPCRs, channels, enzymes, ribosomes, riboswitches, toxins, and viruses. Time-resolved SFX has been established using pump-probe and mix-and-inject approaches.
Applications of SFX in SBDD will be contingent on further developments focused on the automation of sample preparation and delivery, optimization of data collection efficiency, and the expansion of available XFEL beam time.
Acknowledgments
Funding
This manuscript was support by grants 18–02-40020 and 18–54-00030 from the Russian Foundation for Basic Research (RFBR) and from R35 GM127086 from the National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS).
Footnotes
Declaration of Interest
All authors are supported by the non-profit academic-industrial GPCR consortium as well as by the Russian Foundation for Basic Research. A Mishin, A Gusach, A Luginina, E Marin and V Borshchevskiy are also supported by the Russian Science Foundation and the Ministry of Science and Higher Education of the Russian Federation. V Cherezov also has received grant support from the National Institutes of General Medical Sciences and the National Institute on Drug Abuse. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Reviewer Disclosures:
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
References
Papers of special note have been highlighted as either of interest (*) or of considerable interest (**) to readers.
- [1].Paul SM, Mytelka DS, Dunwiddie CT, et al. How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nat. Rev. Drug Discov. 2010;9:203–214. [DOI] [PubMed] [Google Scholar]
- [2].Munos B Lessons from 60 years of pharmaceutical innovation. Nat. Rev. Drug Discov. 2009;8:959–968. [DOI] [PubMed] [Google Scholar]
- [3].Mullard A 2018 FDA drug approvals. Nat. Rev. Drug Discov. 2019;18:85–89. [DOI] [PubMed] [Google Scholar]
- [4].Smietana K, Siatkowski M, Møller M. Trends in clinical success rates. Nat. Rev. Drug Discov. 2016;15:379–380. [DOI] [PubMed] [Google Scholar]
- [5].Montfort Van RLM, Workman P. Structure-based drug design: aiming for a perfect fit. Essays Biochem. 2017;61:431–437.*Recent editorial on SBDD covering recent advancements and current status of the field
- [6].Spence JCH. XFELs for structure and dynamics in biology. IUCrJ. 2017;4:322–339.**Comprehensive review on applications of XFEL-based techniques in structural biology
- [7].Johansson LC, Stauch B, Ishchenko A, et al. A Bright Future for Serial Femtosecond Crystallography with XFELs. Trends Biochem. Sci. 2017;42:749–762.**Recent review on SFX describing latest developments, current applications, and future insights
- [8].Bohacek RS, McMartin C, Guida WC. The Art and Practice of Structure-Based Drug Design: A Molecular Modeling Perspective. Med. Res. Rev. 1996;16:3–50. [DOI] [PubMed] [Google Scholar]
- [9].Murray CW, Rees DC. The rise of fragment-based drug discovery. Nat. Chem. 2009;1:187–192. [DOI] [PubMed] [Google Scholar]
- [10].Peltason L, Bajorath J. SAR Index : Quantifying the Nature of Structure - Activity Relationships. J. Med. Chem. 2007;50:5571–5578. [DOI] [PubMed] [Google Scholar]
- [11].Ekins S, Mestres J, Testa B. In silico pharmacology for drug discovery : methods for virtual ligand screening and profiling. Br. J. Pharmacol. 2007;152:9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lyu J, Wang S, Balius TE, et al. Ultra-large library docking for discovering new chemotypes. Nature. 2019;566:224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sterling T, Irwin JJ. ZINC 15 - Ligand Discovery for Everyone. J. Chem. Inf. Model. 2015;55:2324–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Keserű GM, Erlanson DA, Ferenczy GG, et al. Design Principles for Fragment Libraries: Maximizing the Value of Learnings from Pharma Fragment-Based Drug Discovery (FBDD) Programs for Use in Academia. J. Med. Chem. 2016;59:8189–8206. [DOI] [PubMed] [Google Scholar]
- [15].Beddell CR, Goodford PJ, Norrington FE, et al. COMPOUNDS DESIGNED TO FIT A SITE OF KNOWN STRUCTURE IN HUMAN HAEMOGLOBIN. Br. J. Pharmacol. 1976;57:201–209.*Early application of SBDD for designing compounds that stabilize the deoxy conformation of human haemoglobin
- [16].Blundell T, Sibanda BL, Pearl L. Three-dimensional structure, specificity and catalytic mechanism of renin. Nature. 1983;304:273–275. [DOI] [PubMed] [Google Scholar]
- [17].Dhanaraj V, Dealwis CG, Frazao C, et al. X-ray analyses of peptide-inhibitor complexes define the structural basis of specificity for human and mouse renins. Nature. 1992;357:466–472. [DOI] [PubMed] [Google Scholar]
- [18].Navia MA, Fitzgerald PMD, McKeever BM, et al. Three-dimensional structure of aspartyl protease from human immunodeficiency virus HIV-1. Nature. 1989;337:615–620. [DOI] [PubMed] [Google Scholar]
- [19].Kim CU, Lew W, Williams MA, et al. Influenza Neuraminidase Inhibitors Possessing a Novel Hydrophobic Interaction in the Enzyme Active Site : Design, Synthesis, and Structural Analysis of Carbocyclic Sialic Acid Analogues with Potent Anti-Influenza Activity. J. Am. Chem. Soc. 1997;119:681–690. [DOI] [PubMed] [Google Scholar]
- [20].Montfort Van RLM, Workman P. Structure-based design of molecular cancer therapeutics. Trends Biotechnol. 2009;27:315–328. [DOI] [PubMed] [Google Scholar]
- [21].Borshell N, Papp T, Congreve M. Deal watch: Valuation benefits of structure-enabled drug discovery. Nat. Rev. Drug Discov. 2011;10:166. [DOI] [PubMed] [Google Scholar]
- [22].Cherezov V, Rosenbaum DM, Hanson MA, et al. High-Resolution Crystal Structure of an Engineered Human b2-Adrenergic G Protein-Coupled Receptor. Science. 2007;318:1258–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Pándy-Szekeres G, Munk C, Tsonkov TM, et al. GPCRdb in 2018: adding GPCR structure models and ligands. Nucleic Acids Res. 2018;46:440–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Katritch V, Cherezov V, Stevens RC. Structure-Function of the G Protein–Coupled Receptor Superfamily. Annu. Rev. Pharmacol. Toxicol. 2013;53:531–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pellegrini C The history of X-ray free-electron lasers. Eur. Phys. J. H. 2012;37:659–708. [Google Scholar]
- [26].Ginzburg VL. On the radiation of microradiowaves and their absorbtion in the air. Izv. Akad. Nauk SSSR, Ser. Fiz. 1947;11:165. [Google Scholar]
- [27].Motz H Applications of the Radiation from Fast Electron Beams. J. Appl. Phys. 1951;22:527–535. [Google Scholar]
- [28].Tiedtke K, Azima A, von Bargen N, et al. The soft x-ray free-electron laser FLASH at DESY: beamlines, diagnostics and end-stations. New J. Phys. 2009;11:023029. [Google Scholar]
- [29].McNeil B Free electron lasers: First light from hard X-ray laser. Nat. Photonics. 2009;3:375–377. [Google Scholar]
- [30].Emma P, Akre R, Arthur J, et al. First lasing and operation of an ångstrom-wavelength free-electron laser. Nat. Photonics. 2010;4:641–647. [Google Scholar]
- [31].McNeil BWJ, Thompson NR. X-ray free-electron lasers. Nat. Photonics. 2010;4:814–821. [Google Scholar]
- [32].Neutze R, Wouts R, Spoel Van Der D, et al. Potential for biomolecular imaging with femtosecond X-ray pulses. Nature. 2000;406:752–757.*Theoretical study proposing the concept of diffraction-before-destruction
- [33].Miller RJD. Femtosecond Crystallography with Ultrabright Electrons and X-rays: Capturing Chemistry in Action. Science. 2014;343:1108–1117. [DOI] [PubMed] [Google Scholar]
- [34].Kupitz C, Basu S, Grotjohann I, et al. Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser. Nature. 2014;513:261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bostedt C, Boutet S, Fritz DM, et al. Linac Coherent Light Source: The first five years. Rev. Mod. Phys. 2016;88:015007. [Google Scholar]
- [36].Lv H, Leng Y, Yan Y, et al. The high level application architecture of the control system for SHINE. Nucl. Instruments Methods Phys. Res. Sect. A Accel. Spectrometers, Detect. Assoc. Equip. 2018;908:167–171. [Google Scholar]
- [37].Raubenheimer TO. The LCLS-II-HE, A High Energy Upgrade of the LCLS-II. In: Chin YH, Zhao Z, Petit-Jean-Genaz C, Schaa VRW, editors. Proceedings of the 60th ICFA Advanced Beam Dynamics Workshop on Future Light Sources; 2018 March 5–9; Shanghai, China: JACoW Publishing; p. 6–11. doi: 10.18429/JACoW-FLS2018-MOP1WA02. [DOI] [Google Scholar]
- [38].Chapman HN, Fromme P, Barty A, et al. Femtosecond X-ray protein nanocrystallography. Nature. 2011;470:73–77.*Pioneering work establishing SFX data collection approach at XFELs using Photosystem I as a test protein
- [39].DePonte DP, Weierstall U, Schmidt K, et al. Gas dynamic virtual nozzle for generation of microscopic droplet streams. J. Phys. D. Appl. Phys. 2008;41:195505.*Development of the Gas Dynamic Virtual Nozzle (GDVN), the first and most popular crystal injector used for SFX
- [40].Oberthuer D, Knoška J, Wiedorn MO, et al. Double-flow focused liquid injector for efficient serial femtosecond crystallography. Sci. Rep. 2017;7:44628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Sierra RG, Laksmono H, Kern J, et al. Nanoflow electrospinning serial femtosecond crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012;68:1584–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Sierra RG, Gati C, Laksmono H, et al. Concentric-flow electrokinetic injector enables serial crystallography of ribosome and photosystem II. Nat. Methods. 2016;13:59–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Weierstall U, James D, Wang C, et al. Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography. Nat. Commun. 2014;5:3309.*Design and operation of an LCP injector that facilitates SFX of membrane proteins
- [44].Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 2009;4:706–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Stauch B, Cherezov V. Serial Femtosecond Crystallography of G Protein–Coupled Receptors. Annu. Rev. Biophys. 2018;47:377–397.**Recent review on structure determination of GPCRs by SFX
- [46].Nango E, Royant A, Kubo M, et al. A three-dimensional movie of structural changes in bacteriorhodopsin. Science. 2016;354:1552–1557.*Time-resolved study of light-driven conformational changes in bacteriorhodopsin using crystals delivered with an LCP injector
- [47].Fromme R, Ishchenko A, Metz M, et al. Serial femtosecond crystallography of soluble proteins in lipidic cubic phase. IUCrJ. 2015;2:545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hunter MS, Segelke B, Messerschmidt M, et al. Fixed-target protein serial microcrystallography with an x-ray free electron laser. Sci. Rep. 2015;4:6026.*One of the first demonstrations of fixed-target SFX
- [49].Roedig P, Ginn HM, Pakendorf T, et al. High-speed fixed-target serial virus crystallography. Nat. Methods. 2017;14:805–810.*Development of high-speed fixed-target SFX and its application to virus crystallography
- [50].Fuller FD, Gul S, Chatterjee R, et al. Drop-on-demand sample delivery for studying biocatalysts in action at X-ray free-electron lasers. Nat. Methods. 2017;14:443–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Tenboer J, Basu S, Zatsepin N, et al. Time-resolved serial crystallography captures high-resolution intermediates of photoactive yellow protein. Science. 2014;346:1242–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kupitz C, Grotjohann I, Conrad CE, et al. Microcrystallization techniques for serial femtosecond crystallography using photosystem II from Thermosynechococcus elongatus as a model system. Philos. Trans. R. Soc. B Biol. Sci. 2014;369:20130316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Dods R, Båth P, Arnlund D, et al. From Macrocrystals to Microcrystals: A Strategy for Membrane Protein Serial Crystallography. Structure. 2017;25:1461–1468. [DOI] [PubMed] [Google Scholar]
- [54].Redecke L, Nass K, DePonte DP, et al. Natively Inhibited Trypanosoma brucei Cathepsin B Structure Determined by Using an X-ray Laser. Science. 2013;339:227–230.*Pioneering work on in cellulo SFX reporting a structure of the natively inhibited cathepsin B crystallized inside insect cells
- [55].Jakobi AJ, Passon DM, Knoops K, et al. In cellulo serial crystallography of alcohol oxidase crystals inside yeast cells. IUCrJ. 2016;3:88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Sawaya MR, Cascio D, Gingery M, et al. Protein crystal structure obtained at 2.9 A resolution from injecting bacterial cells into an X-ray free-electron laser beam. Proc. Natl. Acad. Sci. 2014;111:12769–12774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Duyvesteyn HME, Ginn HM, Pietilä MK, et al. Towards in cellulo virus crystallography. Sci. Rep. 2018;8:3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Liu W, Ishchenko A, Cherezov V. Preparation of microcrystals in lipidic cubic phase for serial femtosecond crystallography. Nat. Protoc. 2014;9:2123–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Kissick DJ, Wanapun D, Simpson GJ. Second-Order Nonlinear Optical Imaging of Chiral Crystals. Annu. Rev. Anal. Chem. 2011;4:419–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Brönnimann C, Trüb P. Hybrid Pixel Photon Counting X-Ray Detectors for Synchrotron Radiation In: Jaeschke E, Khan S, Schneider J, Hastings J, editors. Synchrotron Light Sources Free-Electron Lasers. Springer, Cham; 2015. p. 1–29. [Google Scholar]
- [61].Hart P, Boutet S, Carini G, et al. The Cornell-SLAC pixel array detector at LCLS. IEEE Nucl. Sci. Symp. Conf. Rec. 2012; 538–541. [Google Scholar]
- [62].Henrich B, Becker J, Dinapoli R, et al. The adaptive gain integrating pixel detector AGIPD a detector for the European XFEL. Nucl. Instruments Methods Phys. Res. Sect. A Accel. Spectrometers, Detect. Assoc. Equip. 2011;633:11–14. [Google Scholar]
- [63].Mozzanica A, Andrä M, Barten R, et al. The JUNGFRAU Detector for Applications at Synchrotron Light Sources and XFELs. Synchrotron Radiat. News. 2018;31:16–20. [Google Scholar]
- [64].Allahgholi A, Becker J, Delfs A, et al. The Adaptive Gain Integrating Pixel Detector at the European XFEL. J. Synchrotron Radiat. 2019;26:74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Barty A, Kirian RA, Maia FRNC, et al. Cheetah : software for high-throughput reduction and analysis of serial femtosecond X-ray diffraction data. J. Appl. Crystallogr. 2014;47:1118–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Coquelle N, Brewster AS, Kapp U, et al. Raster-scanning serial protein crystallography using micro- and nano-focused synchrotron beams. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015;71:1184–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Ke TW, Brewster AS, Yu SX, et al. A convolutional neural network-based screening tool for X-ray serial crystallography. J. Synchrotron Radiat. 2018;25:655–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Zaefferer S New developments of computer-aided crystallographic analysis in transmission electron microscopy. J. Appl. Crystallogr. 2000;33:10–25. [Google Scholar]
- [69].Winter G, Waterman DG, Parkhurst JM, et al. DIALS : implementation and evaluation of a new integration package. Acta Crystallogr. Sect. D Struct. Biol. 2018;74:85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Kabsch W Processing of X-ray snapshots from crystals in random orientations. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014;70:2204–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Sauter NK, Hattne J, Grosse-Kunstleve RW, et al. New Python-based methods for data processing. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013;69:1274–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Mariani V, Morgan A, Yoon CH, et al. OnDA : online data analysis and feedback for serial X-ray imaging. J. Appl. Crystallogr. 2016;49:1073–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Nakane T, Joti Y, Tono K, et al. Data processing pipeline for serial femtosecond crystallography at SACLA. J. Appl. Crystallogr. 2016;49:1035–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Foucar L CFEL–ASG Software Suite (CASS): usage for free-electron laser experiments with biological focus. J. Appl. Crystallogr. 2016;49:1336–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Park J, Kim S, Kim S, et al. Multifarious injection chamber for molecular structure study (MICOSS) system: Development and application for serial femtosecond crystallography at Pohang Accelerator Laboratory X-ray Free-Electron Laser. J. Synchrotron Radiat. 2018;25:323–328. [DOI] [PubMed] [Google Scholar]
- [76].White TA, Kirian RA, Martin A V., et al. CrystFEL : a software suite for snapshot serial crystallography. J. Appl. Crystallogr. 2012;45:335–341.*Description of CrystFEL - a popular SFX data processing software suite
- [77].Ginn HM, Evans G, Sauter NK, et al. On the release of cppxfel for processing X-ray free-electron laser images. J. Appl. Crystallogr. 2016;49:1065–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Ginn HM, Roedig P, Kuo A, et al. TakeTwo : an indexing algorithm suited to still images with known crystal parameters. Acta Crystallogr. Sect. D Struct. Biol. 2016;72:956–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Li C, Li X, Kirian R, et al. SPIND : a reference-based auto-indexing algorithm for sparse serial crystallography data. IUCrJ. 2019;6:72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Gevorkov Y xgandalf. 2019. Available from: https://stash.desy.de/users/gevorkov/repos/xgandalf/browse.
- [81].Beyerlein KR, White TA, Yefanov O, et al. FELIX: an algorithm for indexing multiple crystallites in X-ray free-electron laser snapshot diffraction images. J. Appl. Crystallogr. 2017;50:1075–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].White TA. Post-refinement method for snapshot serial crystallography. Philos. Trans. R. Soc. B Biol. Sci. 2014;369:20130330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Ginn HM, Brewster AS, Hattne J, et al. A revised partiality model and post-refinement algorithm for X-ray free-electron laser data. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015;71:1400–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Sauter NK. XFEL diffraction: developing processing methods to optimize data quality. J. Synchrotron Radiat. 2015;22:239–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Uervirojnangkoorn M, Zeldin OB, Lyubimov AY, et al. Enabling X-ray free electron laser crystallography for challenging biological systems from a limited number of crystals. Elife. 2015;4:e05421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Kroon-Batenburg LMJ, Schreurs AMM, Ravelli RBG, et al. Accounting for partiality in serial crystallography using ray-tracing principles. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015;71:1799–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Hattne J, Echols N, Tran R, et al. Accurate macromolecular structures using minimal measurements from X-ray free-electron lasers. Nat. Methods. 2014;11:545–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Barends TRM, Foucar L, Botha S, et al. De novo protein crystal structure determination from X-ray free-electron laser data. Nature. 2014;505:244–247.*First demonstration of de novo phasing of SFX data
- [89].Yamashita K, Pan D, Okuda T, et al. An isomorphous replacement method for efficient de novo phasing for serial femtosecond crystallography. Sci. Rep. 2015;5:14017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Nakane T, Hanashima S, Suzuki M, et al. Membrane protein structure determination by SAD, SIR, or SIRAS phasing in serial femtosecond crystallography using an iododetergent. Proc. Natl. Acad. Sci. 2016;113:13039–13044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Colletier J-P, Sawaya MR, Gingery M, et al. De novo phasing with X-ray laser reveals mosquito larvicide BinAB structure. Nature. 2016;539:43–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Batyuk A, Galli L, Ishchenko A, et al. Native phasing of x-ray free-electron laser data for a G protein–coupled receptor. Sci. Adv. 2016;2:e1600292.*Native sulfur-SAD phasing of SFX data for a GPCR
- [93].Galli L, Son S-K, Barends TRM, et al. Towards phasing using high X-ray intensity. IUCrJ. 2015;2:627–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Millane RP, Chen JPJ. Aspects of direct phasing in femtosecond nanocrystallography. Philos. Trans.R. Soc. B Biol. Sci. 2014;369:20130498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Morgan AJ, Ayyer K, Barty A, et al. Ab initio phasing of the diffraction of crystals with translational disorder. Acta Crystallogr. Sect. A Found. Adv. 2019;75:25–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Gorel A, Motomura K, Fukuzawa H, et al. Multi-wavelength anomalous diffraction de novo phasing using a two-colour X-ray free-electron laser with wide tunability. Nat. Commun. 2017;8:1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Lomelino CL, Kim JK, Lee C, et al. Carbonic anhydrase II microcrystals suitable for XFEL studies. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2018;74:327–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Marin EV, Gusach AY, Luginina AP, et al. Successful GPCR structure determination using PAL XFEL. J. Bioenerg. Biomembr. 2018;50:467–603.30543032 [Google Scholar]
- [99].Liu W, Wacker D, Gati C, et al. Serial Femtosecond Crystallography of G Protein-Coupled Receptors. Science. 2014;342:1521–1524.*First demonstration of SFX with GPCR crystals grown and delivered in LCP
- [100].Wang C, Wu H, Evron T, et al. Structural basis for Smoothened receptor modulation and chemoresistance to anticancer drugs. Nat. Commun. 2014;5:4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Fenalti G, Zatsepin NA, Betti C, et al. Structural basis for bifunctional peptide recognition at human δ-opioid receptor. Nat. Struct. Mol. Biol. 2015;22:265–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Kang Y, Zhou XE, Gao X, et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature. 2015;523:561–567.**Structure of a major signaling complex between GPCR and arrestin obtained by SFX
- [103].Zhou XE, He Y, de Waal PW, et al. Identification of Phosphorylation Codes for Arrestin Recruitment by G Protein-Coupled Receptors. Cell. 2017;170:457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Zhang H, Qiao A, Yang D, et al. Structure of the full-length glucagon class B G-protein-coupled receptor. Nature. 2017;546:259–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Zhang X, Zhao F, Wu Y, et al. Crystal structure of a multi-domain human smoothened receptor in complex with a super stabilizing ligand. Nat. Commun. 2017;8:15383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Zhang H, Unal H, Gati C, et al. Structure of the Angiotensin Receptor Revealed by Serial Femtosecond Crystallography. Cell. 2015;161:833–844.*Structure of the first novel GPCR obtained by SFX
- [107].Zhang H, Han GW, Batyuk A, et al. Structural Basis for Selectivity and Diversity in Angiotensin II Receptors. Nature. 2017;544:327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Audet M, White KL, Breton B, et al. Crystal structure of misoprostol bound to the labor inducer prostaglandin E2 receptor. Nat. Chem. Biol. 2019;15:11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Thomaston JL, Woldeyes RA, Nakane T, et al. XFEL structures of the influenza M2 proton channel: Room temperature water networks and insights into proton conduction. Proc. Natl. Acad. Sci. 2017;114:13357–13362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].O’Sullivan ME, Poitevin F, Sierra RG, et al. Aminoglycoside ribosome interactions reveal novel conformational states at ambient temperature. Nucleic Acids Res. 2018;46:9793–9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Mathews II, Allison K, Robbins T, et al. Conformational Flexibility of the Acyltransferase from the Disorazole Polyketide Synthase Is Revealed by an X-ray Free-Electron Laser Using a Room-Temperature Sample Delivery Method for Serial Crystallography. Biochemistry. 2017;56:4751–4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Aquila A, Hunter MS, Doak RB, et al. Time-resolved protein nanocrystallography using an X-ray free-electron laser. Opt. Express. 2012;20:2706–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Kern J, Tran R, Alonso-Mori R, et al. Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy. Nat. Commun. 2014;5:4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Barends TRM, Foucar L, Ardevol A, et al. Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation. Science. 2015;350:445–450.*First demonstration of ultrafast time-resolved SFX with sub-picosecond time resolution revealing photodissasociation of CO from myoglobin
- [115].Pande K, Hutchison CDM, Groenhof G, et al. Femtosecond structural dynamics drives the trans/cis isomerization in photoactive yellow protein. Science. 2016;352:725–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Kern J, Chatterjee R, Young ID, et al. Structures of the intermediates of Kok’s photosynthetic water oxidation clock. Nature. 2018;563:421–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Coquelle N, Sliwa M, Woodhouse J, et al. Chromophore twisting in the excited state of a photoswitchable fluorescent protein captured by time-resolved serial femtosecond crystallography. Nat. Chem. 2018;10:31–37. [DOI] [PubMed] [Google Scholar]
- [118].Nogly P, Weinert T, James D, et al. Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser. Science. 2018;361:eaat0094. [DOI] [PubMed] [Google Scholar]
- [119].Tosha T, Nomura T, Nishida T, et al. Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate. Nat. Commun. 2017;8:1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Olmos JL, Pandey S, Martin-Garcia JM, et al. Enzyme intermediates captured “on the fly” by mix-and-inject serial crystallography. BMC Biol. 2018;16:59.*Development of a mix-and-inject SFX method for capturing enzyme catalysis in action.
- [121].Wiedorn MO, Oberthür D, Bean R, et al. Megahertz serial crystallography. Nat. Commun. 2018;9:4025.*First structure determination by megahertz SFX
- [122].Kuhlbrandt W The Resolution Revolution. Science. 2014;343:1443–1444. [DOI] [PubMed] [Google Scholar]
- [123].Martin-Garcia JM, Conrad CE, Coe J, et al. Serial femtosecond crystallography: A revolution in structural biology. Arch. Biochem. Biophys. 2016;602:32–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Renaud J-P, Chari A, Ciferri C, et al. Cryo-EM in drug discovery: achievements, limitations and prospects. Nat. Rev. Drug Discov. 2018;17:471–492. [DOI] [PubMed] [Google Scholar]
- [125].Cheng RKY, Abela R, Hennig M. X-ray free electron laser: opportunities for drug discovery. Essays Biochem. 2017;61:529–542. [DOI] [PubMed] [Google Scholar]
- [126].Spyrakis F, Ahmed MH, Bayden AS, et al. The Roles of Water in the Protein Matrix: A Largely Untapped Resource for Drug Discovery. J. Med. Chem. 2017;60:6781–6827. [DOI] [PubMed] [Google Scholar]
- [127].Grebner C, Iegre J, Ulander J, et al. Binding Mode and Induced Fit Predictions for Prospective Computational Drug Design. J. Chem. Inf. Model. 2016;56:774–787. [DOI] [PubMed] [Google Scholar]
- [128].Hauwert NJ, Mocking TAM, Da Costa Pereira D, et al. Synthesis and Characterization of a Bidirectional Photoswitchable Antagonist Toolbox for Real-Time GPCR Photopharmacology. J. Am. Chem. Soc. 2018;140:4232–4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Kienzler MA, Isacoff EY. Precise modulation of neuronal activity with synthetic photoswitchable ligands. Curr. Opin. Neurobiol. 2017;45:202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Dilanian RA, Streltsov V, Coughlan HD, et al. Nanocrystallography measurements of early stage synthetic malaria pigment. J. Appl. Crystallogr. 2017;50:1533–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Seuring C, Ayyer K, Filippaki E, et al. Femtosecond X-ray coherent diffraction of aligned amyloid fibrils on low background graphene. Nat. Commun. 2018;9:1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Huang C-F, Liang KS, Hsu T-L, et al. Free-electron-laser coherent diffraction images of individual drug-carrying liposome particles in solution. Nanoscale. 2018;10:2820–2824. [DOI] [PubMed] [Google Scholar]
- [133].Barty A Single molecule imaging using X-ray free electron lasers. Curr. Opin. Struct. Biol. 2016;40:186–194. [DOI] [PubMed] [Google Scholar]
- [134].MacArthur JP, Lutman AA, Krzywinski J, et al. Microbunch Rotation and Coherent Undulator Radiation from a Kicked Electron Beam. Phys. Rev. X. 2018;8:04036. [Google Scholar]
- [135].Feng Y, Alonso-Mori R, Barends TRM, et al. Demonstration of simultaneous experiments using thin crystal multiplexing at the Linac Coherent Light Source. J. Synchrotron Radiat. 2015;22:626–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Boutet S, Foucar L, Barends TRM, et al. Characterization and use of the spent beam for serial operation of LCLS. J. Synchrotron Radiat. 2015;22:634–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Casanas A, Warshamanage R, Finke AD, et al. EIGER detector: application in macromolecular crystallography. Acta Crystallogr. Sect. D Struct. Biol. 2016;72:1036–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Graves WS, Berggren KK, Carbajo S, et al. Compact XFEL Light Source Proc. FEL2013, New York, NY, USA: 2013:757–761. [Google Scholar]
- [139].Kartner FX. Terahertz driven linear accelerators and photon sources. 2016 41st Int. Conf Infrared, Millimeter, Terahertz waves. IEEE; 2016:1–2. [Google Scholar]
- [140].Martin-Garcia JM, Conrad CE, Nelson G, et al. Serial millisecond crystallography of membrane and soluble protein microcrystals using synchrotron radiation. IUCrJ. 2017;4:439–454.*Injector-based serial crystallography at synchrotrons
- [141].Eriksson M, van der Veen JF, Quitmann C. Diffraction-limited storage rings – a window to the science of tomorrow. J. Synchrotron Radiat. 2014;21:837–842. [DOI] [PubMed] [Google Scholar]