Summary
The ability to generate T cells from self-renewing pluripotent stem cells (PSC) has the potential to transform the current practice of autologous T cell immunotherapy into universal off-the-shelf products. However, differentiation of human PSCs into mature, conventional T cells has been challenging with existing methods. We report that a 3D artificial thymic organoid (PSC-ATO) system induced efficient differentiation of human embryonic stem cell and induced pluripotent stem cell-derived mesoderm progenitors to mature, functional T cells with a diverse T cell receptor (TCR) repertoire. This continuous culture system supported both hematopoietic specification and terminal differentiation to naïve CD3+CD8αβ+ and CD3+CD4+ conventional T cells. Introduction of an MHC class I-restricted TCR in PSCs produced naïve, antigen-specific cytotoxic CD8αβ+ T cells which lacked endogenous TCR Vβ expression. Functional assays and RNA sequencing aligned PSC-derived T cells with primary naïve CD8+ T cells. The PSC-ATO system presented here is an efficient platform for generating functional, mature T cells from human PSCs.
Introduction
Engineered T cell therapies hold promise for the effective treatment of cancer and chronic viral infections. The ability to generate T cells on demand from self-renewing human pluripotent stem cells (PSC) may substantially advance the field by allowing the production of universal-donor T cells from stably gene-modified PSC lines (Themeli et al., 2015). Although protocols to differentiate PSC into essentially any non-hematopoietic or hematopoietic lineage have been extensively reported, generation of fully functional mature cells that resemble their adult counterparts has been more problematic. Differentiation of mature T cells from human PSCs has been limited on two fronts: the ability to specify hematopoietic progenitor cells with T-lineage potential (Dravid et al., 2011; Kennedy et al., 2012), and the capacity of existing methods to support maturation of T-lineage committed precursors to conventional, naïve T cells (Themeli et al., 2013; Vizcardo et al., 2013). Improved PSC-to-T cell differentiation strategies must therefore integrate T-competent hematopoietic specification with the full span of conventional, thymic-like T cell differentiation.
T cell development from multipotent bone marrow-derived hematopoietic stem/progenitor cells (HSPCs) in the thymus is enforced by spatiotemporal interactions of precursor T cells with signals from thymic epithelial, mesenchymal, and hematopoietic cells (Rothenberg et al., 2008). Of these interactions, the stromal-expressed Notch ligand DLL4 plays a critical role in the onset and maintenance of T-lineage commitment (Hozumi et al., 2008; Koch et al., 2008). In vitro T-lineage commitment from human HSPCs can be induced by co-culture with Notch ligand-expressing stromal cell lines (De Smedt et al., 2004; La Motte-Mohs et al., 2005), however positive selection and thus conventional maturation of T cells using these methods is limited. We recently reported that a 3D artificial thymic organoid (ATO) culture system permits in vitro differentiation of human HSPCs to functional, mature T cells using a standardized Notch ligand-expressing stromal cell line in serum-free conditions (Seet et al., 2017). Notably, we observed that both the medium and the 3D structure were critical for the efficient positive selection of CD4+CD8+ double positive (DP) precursors to conventional CD3+TCRαβ+CD8αβ+ T cells in ATOs.
Separately, we have shown that human hematopoiesis proceeds from PSCs through a human embryonic mesodermal progenitor (hEMP) stage marked by downregulation of CD326 (EpCAM) and upregulation of CD56 (NCAM) (Chin et al., 2016; Evseenko et al., 2010). Hematopoietic specification from hEMPs could be subsequently induced by co-culture with the murine stromal line OP9 in the presence of hematopoietic cytokines (Evseenko et al., 2010). Given their mesodermal restriction and ease of production, we reasoned that hEMPs may serve as a logical substrate for the development of a combined hematopoietic/T cell directed differentiation protocol from PSCs based on the ATO system.
We report here that a modified ATO system (PSC-ATO) permits the differentiation of human embryonic stem cell (ESC) and induced pluripotent stem cell (iPSC)-derived hEMPs to mature, conventional T cells in vitro. Following mesoderm induction, DLL4-expressing organoids supported hematopoietic specification and T cell development in a continuous organoid structure, which was effective across multiple different ESC and iPSC lines. PSC-derived T cells were predominantly of a CD3+TCRαβ+CD8αβ+ naïve T cell phenotype and exhibited a diverse TCR repertoire; CD3+TCRαβ+CD4+ naïve T cells with a diverse repertoire were also generated, particularly in the presence of DLL1. Furthermore, stable introduction of an antigen-specific TCR at the PSC stage resulted in the development of naïve CD8αβ+ T cells with potent antigen-specific cytotoxicity, and which through allelic exclusion lacked endogenous TCR Vβ expression. RNA sequencing of T cells derived from PSCs and TCR-engineered PSCs revealed a transcriptional profile highly similar to that of primary naïve CD8+ T cells from the human thymus. Overall, our findings that PSC-ATOs can support the efficient differentiation of conventional T cells from human PSCs has relevance to both the study of human T cell development, and the generation of potentially universal adoptive T cell immunotherapies for cancer and other diseases.
RESULTS
A modified artificial thymic organoid (ATO) system permits hematopoietic specification and T cell differentiation from human PSCs
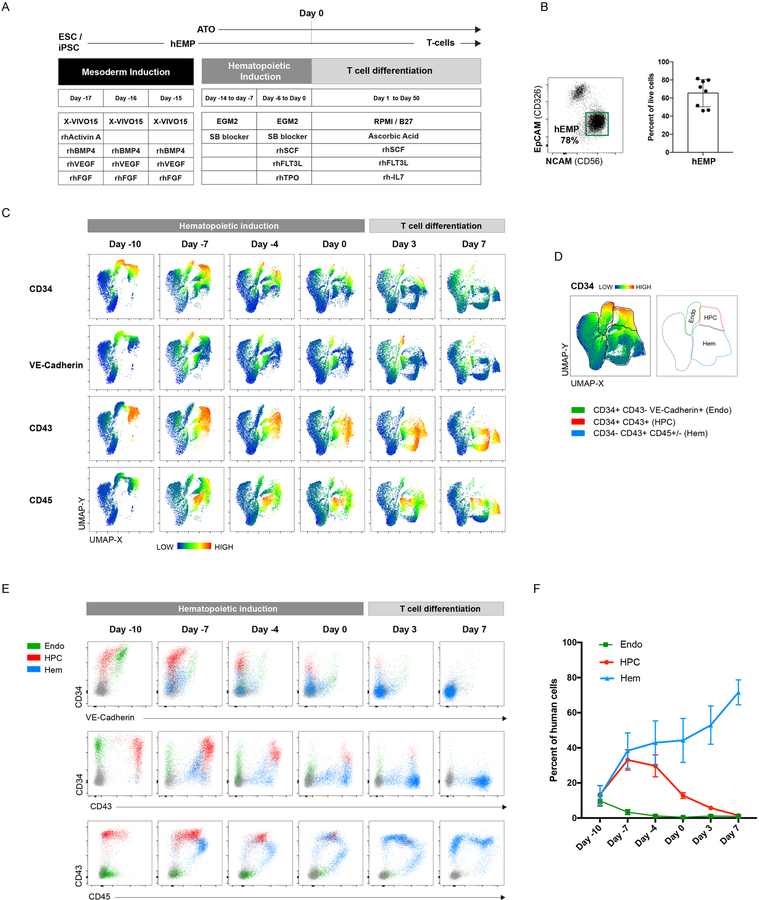
The previously developed artificial thymic organoid (ATO) system (Seet et al., 2017) was modified for PSC differentiation by adopting a simple, three-phase differentiation protocol (Figure 1A). In the first three-day phase (Day −17 to Day-15), mesoderm induction from PSCs in feeder-free conditions generated cultures that comprised 45–80% of hEMPs, defined previously by our group as CD326-CD56+ (Chin et al., 2016; Evseenko et al., 2010) (Figure 1B). In the second phase (hematopoietic induction, Day −14 to Day 0), hEMPs were aggregated into organoids by centrifugation with the mouse bone marrow stromal cell line MS5 (Calvo et al., 2012; Itoh et al., 1989) engineered to express full-length human DLL4 (hereafter referred to as MS5-hDLL4) (Figure 1A). These 3D aggregates (PSC-ATOs, hereafter) were cultured at the air-liquid interface on porous membranes for 14 days in EGM-2 medium in the presence of a TGFβ inhibitor and cytokines to induce hemato-endothelial commitment (Figure 1A). In the third phase, T cell differentiation was induced within the existing organoids by changing the medium at to RPMI supplemented with ascorbic acid and B27 Supplement (“RB27”], with SCF, IL-7 and FLT3L (Figure 1A), as described for primary HSPC ATOs (Seet et al., 2017).
Figure 1: Hematopoietic induction from human pluripotent stem cells (PSCs) in the ATO system.
(A) Schematic of the PSC-ATO differentiation protocol starting from ESC or iPSC. After 3–4 days of mesoderm induction (days −17 to −15), hEMPs are isolated and aggregated with MS5-DLL4 or MS5-DLL1 cells in ATO culture for 2 weeks in hematopoietic induction conditions (days −14 to day 0). T cell differentiation is then initiated within the same ATOs by changing to T cell medium. (B) Representative analysis of hEMP differentiation (n=8) at day −15 after 3.5 days of mesoderm differentiation from H1 ESCs (left panel). hEMP frequency in day −15 cultures across independent experimental replicates (n=8). (C) UMAP analysis of flow cytometry data of different timepoints during hematopoietic induction and T cell differentiation in H1 PSC-ATOs. ATOs were digested to recover all cell populations including adherent cells. Human cells were gated based on negativity for mouse CD29 (expressed by MS5 stromal cells). Heatmaps reflect the mean fluorescence intensity (MFI) of the indicated markers. Data are representative of five independent experiments. (D) Manual gates based on UMAP clusters to broadly categorize cells as endothelial (endo), hematopoietic progenitor (HPC), or differentiated hematopoietic (hem) cells. Gates are shown on a concatenated plot of all timepoints shown in (C) with heatmap coloring based on CD34 MFI (left) or as outlines (right). Extended phenotypes for each population cluster are shown in Supplemental Fig. 1B. (E) Co-expression patterns of CD34, VE-cadherin, CD43, and CD45 on the three population clusters defined in (D). (F) Frequency of endothelial, HPC, and differentiated hematopoietic cells, as defined in (D) in PSC-ATOs at the indicated timepoints (n=5 independent experimental replicates).
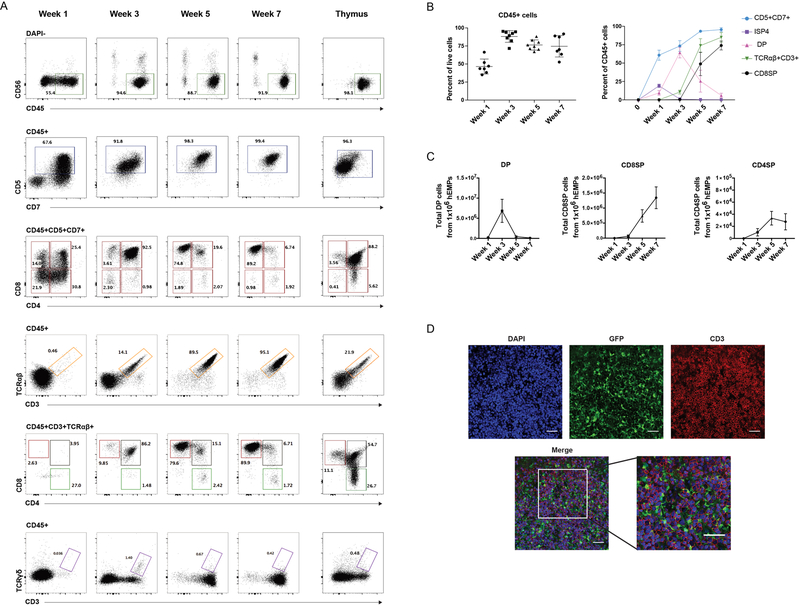
The PSC-ATO system induced rapid hemato-endothelial differentiation from hEMPs derived from the H1 human ESC line (Thomson et al., 1998), (Fig 1C–F and Fig S1). Both endothelial cells (CD34+ CD43- VE-Cadherin+) and hematopoietic progenitor cells (HPC; CD34+CD43+CD45+/−) were detected within 4 days of PSC-ATO culture (i.e. by Day −10) and persisted throughout hematopoietic induction, largely disappearing by the first week in T cell differentiation conditions, and coinciding with a rise in frequency of CD34- hematopoietic cells (Fig 1 C–F). T lineage commitment was seen as early as 1 week from the start of the T cell differentiation phase, as shown by the predominance of CD45+CD5+CD7+ T lineage cells (Figure 2A). T cell differentiation in the PSC-ATO system was thymus-like and orderly, with emergence of CD3-CD4+ immature single positive (CD4ISP) T precursor cells, and CD4+CD8+ (double positive; DP) precursors by week 1, followed by a predominance of DPs by week 3 (Figures 2A and 2B). Of note, a transient population of CD3-CD8+ cells was detected at Week 1 but disappeared by week 3 (Fig 2A); these CD3-CD8+ cells were CD8αα+ cells, consistent with an innate-like phenotype. CD3+TCRαβ+ cells appeared as early as week 3, most of which were initially DP (Figures 2A–C); by week 5, mature CD3+TCRαβ+CD8+CD4- (CD8 single-positive; CD8SP) T cells dominated the cultures (Figures 2A–C), consistent with the occurrence of positive selection within PSC-ATOs. Mature CD3+TCRαβ+CD4+CD8- (CD4 single-positive; CD4SP) T cells were generated to a lesser degree than CD8SP T cells (Fig 2A and 2C), consistent with a CD8+ bias previously observed in ATOs using primary HSPCs (Seet et al., 2017). Low frequencies of CD3+TCRγδ+ T cells were also generated (Figure 2A). On average, a standard small-scale experiment generated 1.5×106 CD8SP T cells from 1×106 hEMPs seeded in 100 ATOs by week 7 (Figure 2C). Immunofluorescence imaging of whole mounted PSC-ATOs demonstrated a dense, tissue-like architecture with abundant T-lineage cells (as shown by CD3 expression) interspersed with MS5-hDLL4 stromal cells (as shown by GFP expression) (Figure 2D).
Figure 2: T cell development from human pluripotent stem cells (PSCs) in the ATO system.
(A) Representative kinetics (n=8 independent experiments) of T cell differentiation in PSC-ATPs from H1 ESCs at the time points shown. Total live cells are shown in the DAPI- gate. Subsequent parent gates are shown above each panel. Human postnatal thymocytes are shown on the far right for comparison. (B) Left: Frequency of hematopoietic (CD45+) cells in PSC-ATOs at the indicated time points (gated on DAPI- cells). Right: frequencies of precursor and mature T cell populations at the indicated time points (gated on CD45+ cells) (mean SD, n=8 independent experiments). (C) Numbers of DP cells, CD8SP cells (defined as CD3+TCR +CD8+CD4-) and CD4SP cells (defined as CD3+TCR +CD4+CD8-) generated from 1×106 hEMP at indicated time points (mean SEM; n=8). (D) Representative whole-mount immunofluorescence analysis (n=3) of PSC-ATOs at week 5. Staining for GFP (marking MS5-DLL4 cells), CD3 (red), and DAPI (nuclear stain). Scale bars, 50 m.
The previously reported ATO system used MS-5 cells expressing DLL1 (Seet et al., 2017), however, studies in mice have suggested that DLL4 may be superior for the support of T cell differentiation (Mohtashami et al., 2010). We directly compared the efficiency of T cell development from H1 ESCs using either hDLL1 or hDLL4-expressing MS5 cells in PSC-ATOs. The total cell numbers generated at week 5 were not significantly different (Figure S2A), and both Notch ligands efficiently supported T cell differentiation (Figure S2B). However, MS5-hDLL4 consistently generated a higher purity of hematopoietic (CD45+) and T lineage (CD56-CD5+CD7+) cells, and significantly enhanced the frequency of mature CD3+TCRαβ+CD8+ (CD8SP) T cells (p<0.01, n=8 hDLL4, n=5 hDLL1) (Figure S2B and S2C); MS5-hDLL4 was thus selected for use in subsequent PSC-ATO experiments. Interestingly, MS5-hDLL1 resulted in the generation of a significantly higher frequency of CD4SP T cells compared to MS5-hDLL4 (p<0.01) (Figure S2C).
Development of naïve conventional T cells and TCR diversity in PSC-ATOs
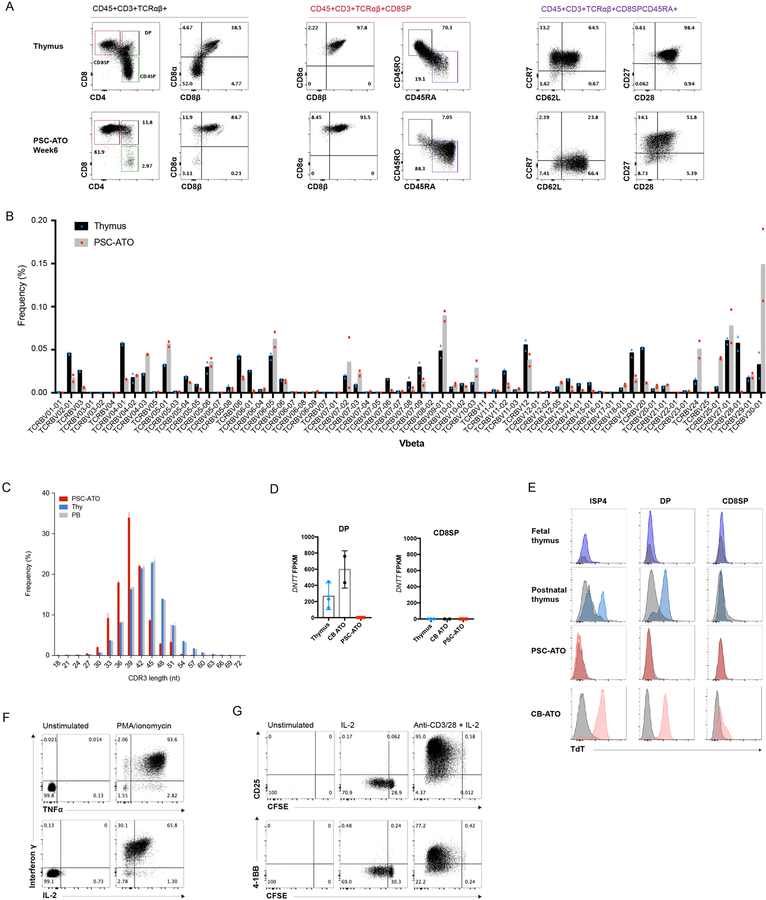
CD8SP T cells produced in PSC-ATOs from H1 ESCs displayed a CD8αβ+ phenotype and the naive markers CD45RA, CD27, CD28, and CD62L (Figure 3A), reflecting the emergence of conventional, naïve T cells phenotypically similar to those in the thymus (Figure 3A) and blood, and those generated from primary HSPCs in ATOs (Seet et al., 2017), however with relatively lower levels of CCR7 expression on PSC-ATO derived cells. A similar pattern of cell surface markers was also seen on CD4SP cells, again suggesting a naïve state (Figure S3A). TCR diversity of H1 PSC-ATO derived CD8SP T cells was assessed by deep sequencing of the TCR Vβ complementary-determining region 3 (CDR3) regions compared to naïve CD8+ T cells isolated from postnatal human thymi (Figure 3B). PSC-ATO-derived T cells displayed a highly diverse distribution of Vβ gene usage comparable to that in thymic CD8SP T cells (Figure 3B). Vβ analysis by flow cytometry of both CD8SP and CD4SP T cells derived from MS5-hDLL1 PSC-ATOs confirmed surface TCR diversity in both subsets (Figure S3B). Interestingly, sequencing analysis revealed shorter CDR3 lengths in mature CD8SP T cells generated in PSC-ATOs compared to CD8SP from the postnatal thymus or peripheral blood (Figure 3C). We postulated that a deficiency of N-nucleotide addition (mediated by terminal deoxynucleotidyl transferase, TdT, during TCR rearrangement) may underlie this difference. Indeed, RNA expression of DNTT (encoding TdT) was absent in DPs from PSC-ATOs, but present as expected in DPs from isolated from the thymus or from ATOs initiated with umbilical cord blood HSPCs (CB-ATOs) (Figure 3D). As DNTT expression has been reported to be lower in fetal compared to postnatal and adult B cell precursors (Rother et al., 2016), we analyzed TdT expression by flow cytometry and found detectable TdT expression at both the ISP4 and DP stages (consistent with stage-specific TCRβ and α chain rearrangement, respectively) from the postnatal thymus and CB-ATOs, but very low and absent TdT expression in the same subsets from the fetal thymus and PSC-ATOs, respectively (Figure 3E), suggesting a developmental correlation between fetal and PSC-ATO T cell development.
Figure 3: Maturation, TCR diversity and function of PSC derived T cells generated in PSC-ATOs.
(A) Representative flow cytometry analysis of T cell maturation markers on CD3+TCR + cells from H1 PSC-ATOs at week 7, demonstrating a conventional T cell phenotype (CD8 ) and generation of mature (CD45RA+CD45RO-) nave T cells. Parent gates are indicated above each panel. Human postnatal thymocytes are shown for comparison (top row). Data are representative of 8 independent experiments. (B) TCR V diversity in nave CD8SP T cells isolated from H1 PSC-ATOs (gray bars, n=2) compared to nave CD8SP from postnatal thymi (black bars, n=2) by deep sequencing of TCR V CDR3 regions. Frequency of cells expressing each V segment is shown. (C) TCR V CDR3 lengths in nave CD8SP T cells isolated from H1 PSC-ATOs (red bars, n=2), postnatal thymi (blue bars, n=2) and adult peripheral blood (gray bars, n=2) assessed by deep sequencing. (D) Expression of DNTT (TdT) by RNA sequencing in DP cells (left) and CD8SP T cells (right) isolated from H1 PSC-ATOs (n=3) compared to the same populations isolated from postnatal thymi (n=3) or ATOs initiated with human cord blood CD34+ HSPCs (CB-ATOs) (n=2). Gene expression is quantified as fragments per kilobase of transcript per million reads (FPKM). (E) Representative intracellular flow cytometry analysis of TdT expression gated on ISP4, DP and CD8SP populations from the human fetal and postnatal thymus (n=1 and n=2, respectively); H1 PSC-ATOs (n=3), and CB-ATOs (n=2). Isotype staining controls are shown in gray for each plot. (F) Polyfunctional cytokine production by H1 PSC-ATO derived CD8SP T cells after treatment with PMA/ionomycin. Data are representative of 3 independent experiments. (G) Proliferation (as measured by the dilution of CFSE) and upregulation of activation markers CD25 and 4–1BB by H1 PSC-ATO derived CD8SP T cells after 5 days of treatment with media only, IL-2, or anti-CD3/CD28 beads plus IL-2. Data are representative of three independent experiments.
Despite this interesting developmental difference, PSC-derived CD8SP T cells were highly functional and exhibited polyfunctional production of interferon (IFN)γ, TNFα and IL-2 in response to phorbol 12-myristate 13-acetate (PMA) and ionomycin (Figure 3F). Both CD8SP and CD4SP proliferated vigorously in response to treatment with anti-CD3/CD28 and IL-2, and upregulated the activation markers CD25 and 4–1BB (Figure 3G and Figure S3C). A low level of proliferation without activation was seen in CD8SP T cells in response to IL-2 alone, consistent with homeostatic proliferation (Figure 3G).
T cell differentiation from multiple ESC and iPSC lines in PSC-ATOs
In addition to the H1 ESC line, T cells were produced in PSC-ATOs from the GMP-compliant ESI-017 cell line (Figure S4A) as well as an induced pluripotent stem cell (iPSC) line derived from a human fibroblast using non-integrating RNA reprogramming (Hazim et al., 2017) (Figure S4B). Both PSC lines produced CD8SP cells that expressed CD8αβ heterodimers and the naïve markers CD45RA, CD27, CD28, CCR7, and CD62L (Figures S4A and S4B), with similar output of total cells and frequency of CD8SP T cells (Figure S4C). Two additional NIH-registered ESC lines (UCLA3 and UCLA6) (Diaz Perez et al., 2012) were tested in ATOs and again produced mature naïve CD8SP cells (Figure S4D), with a diverse TCR Vβ repertoire (Figure S4E).
Generation of conventional TCR-engineered naïve T cells in PSC-ATOs
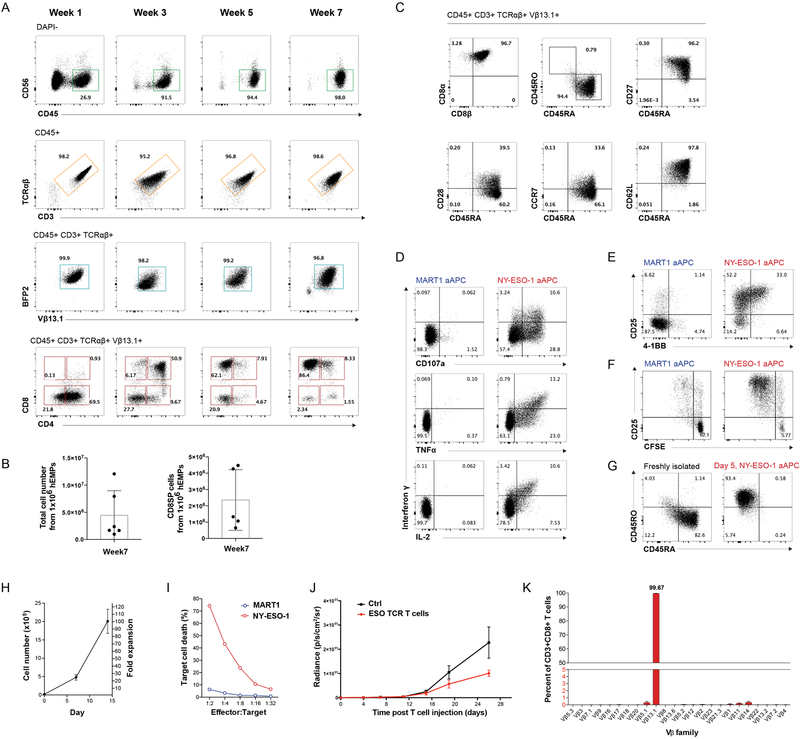
We next investigated whether PSC-ATOs could be adapted to generate naïve antigen-specific T cells from PSCs stably engineered with TCR transgenes. The H1 ESC line was transduced with a lentiviral vector encoding α and β chains of an HLA-A*02:01-restricted TCR specific for the tumor-associated NY-ESO-1157–165 peptide (ESO TCR, hereafter) (Gschweng et al., 2014; Robbins et al., 2008). Stably transduced H1 cells were easily propagated without loss of pluripotency or impairment of hEMP differentiation. TCR-engineered (TCR)PSC-ATOs displayed rapid and orderly T cell development, with unperturbed sequential generation of CD4ISP, DP, and CD8SP developmental stages (Figure 4A). Accelerated CD3/TCRαβ expression (by week 1 of T cell induction) was seen, with virtually all CD3+ cells expressing the transgenic Vβ13.1 TCR chain (Figure 4A). Mature T cell yields tended to be higher in (TCR)PSC-ATOs compared to non-transduced H1 PSC-ATOs, with 1×106 hEMPs in (TCR)PSC-ATOs generating on average of ~2.5×106 CD8SP T cells (Figures 2C and4B). As with non-transduced PSC-ATOs, T cells from (TCR)PSC-ATOs displayed a conventional CD8αβ+ phenotype and naïve T cell markers (Figure 4C). They also showed robust polyfunctional production of IFNγ, TNFα, and IL-2, and CD107a mobilization in response to artificial antigen-presenting cells (aAPCs) expressing a cognate (NY-ESO-1) but not irrelevant (MART1) peptide-MHC (Figure 4D). ESO TCR T cells also underwent antigen-specific proliferation and upregulation of CD25 and 4–1BB (Figures 4E and 4F) and, consistent with a naïve state, switched from a naïve to an effector/memory surface phenotype following prolonged antigen-specific activation (Figure 4G, and Figure S5A). Following 14 days of antigen-specific activation in the presence of IL-2, (TCR)PSC-ATO-derived ESO TCR T cells underwent an approximately 100-fold expansion (Figure 4H), with expanded cells displaying a larger cell size whilst retaining a conventional CD8αβ+ phenotype (Figure S5A) and tetramer staining (Figure S5B). Expanded ESO TCR T cells displayed potent antigen-specific cytotoxicity in vitro against K562 cells expressing NY-ESO-1 but not MART-1 peptide-MHCs at an effector-to-target ratio as low as 1:32 (Figure 4I). Following intravenous injection into immune deficient (NSG) mice, activated ESO TCR T cells were detectable in the peripheral blood and spleen at 48 hours (Figure 5SC and 5SD). They were also able to control the growth of subcutaneously implanted luciferase-expressing K562 tumors expressing the cognate antigen, relative to control mice which did not receive T cells (Figure 4J).
Figure 4: Differentiation, antigen specific function, and V allelic exclusion of TCR-engineered T cells in PSC-ATOs.
(A) Analysis of differentiation kinetics (representative of six independent experiments) of HLA-A*0201/NY-ESO-1157–165-specific (ESO TCR) TCR-engineered T cells in PSC-ATOs initiated from TCR-transduced H1 ESCs ((TCR)PSC-ATOs). H1 ESCs were stably transduced with a lentiviral vector co-expressing the ESO TCR (detectable using an antibody specific for V?13.1) and the mTagBFP2 fluorescent protein. Total live cells are shown in the DAPI- gate. Subsequent parent gates are shown above each panel. (B) Number of total live cells (left) and V 13.1+ (ESO TCR) CD8SP T cells (right) generated in PSC-ATOs at week 7, starting with 1×106 hEMPs (mean SD) (n=6 experiments). (C) Representative (n=6) flow cytometry analysis of maturation markers on ESO TCR CD8SP T cells from (TCR)PSC-ATOs, showing a conventional (CD8 +) and nave (CD45RA+CD45RO-) T cell phenotype including expression of CD27, CD28, CCR7, and CD62L. (D) Cytokine production and CD107a membrane mobilization of TCR-PSC-ATO-derived ESO TCR CD8SP T cells in response to K562 artificial antigen presenting cells (aAPC) expressing an irrelevant (MART-1) or cognate (NY-ESO-1) peptide-MHC single chain trimer. Data are representative of three independent experiments. (E) Upregulation of activation markers CD25 and 4–1BB on ESO TCR CD8SP T cells in response to MART1 or NY-ESO-1 aAPCs for 24h. (F) Proliferation (as measured by dilution of CFSE) of ESO TCR CD8 SP T cells in response to MART1 or NY-ESO-1 aAPCs for 5 days. (G) Evolution of an effector/memory T cell phenotype (CD45RA- CD45RO+) from nave (CD45RA+CD45RO-) ESO TCR CD8SP T cells after 5 days of stimulation with NY-ESO-1 aAPCs and IL-2. (H) Post-ATO expansion of ESO TCR CD8SP T cells isolated from (TCR)PSC-ATOs in response to cognate NY-ESO-1 aAPCs and IL-2. Mean and s.d. of technical triplicates are shown; data are representative of 3 independent experiments. (I) In vitro cytotoxicity of ESO TCR CD8SP T cells against target K562 cells expressing an irrelevant (MART1) or cognate (NY-ESO-1) single chain trimer. T cells were activated/expanded using NY-ESO-1 aAPCs and IL-2 prior to assay. Cell killing is shown as the percentage of target cells positive for annexin V binding at 12h, at the indicated effector to target cell ratios. Data are representative of two independent experiments. (J) In vivo slowing of tumor growth after I.V. administration of (TCR)PSC-ATP derived ESO-TCR CD8SP T cells. NSG mice were subcutaneously implanted with 2105 luciferase-transduced K562-ESO tumor cells 3 days prior to T cell infusion. 5 106 ESO TCR T cells isolated from (TCR)PSC-ATOs and expanded for 10 days, or PBS (control) were injected intravenously into each mouse. Bioluminescence was measured at the indicated timepoints. Mean and SEM for each group is shown (control n=5, ESO TCR T cells n=5). (K) Allelic exclusion of endogenous V expression shown by flow cytometry of CD8SP T cells isolated from (TCR)PSC-ATOs (n=3). Frequencies of T cells expressing the indicated V segments are shown (error bars represent s.d.).
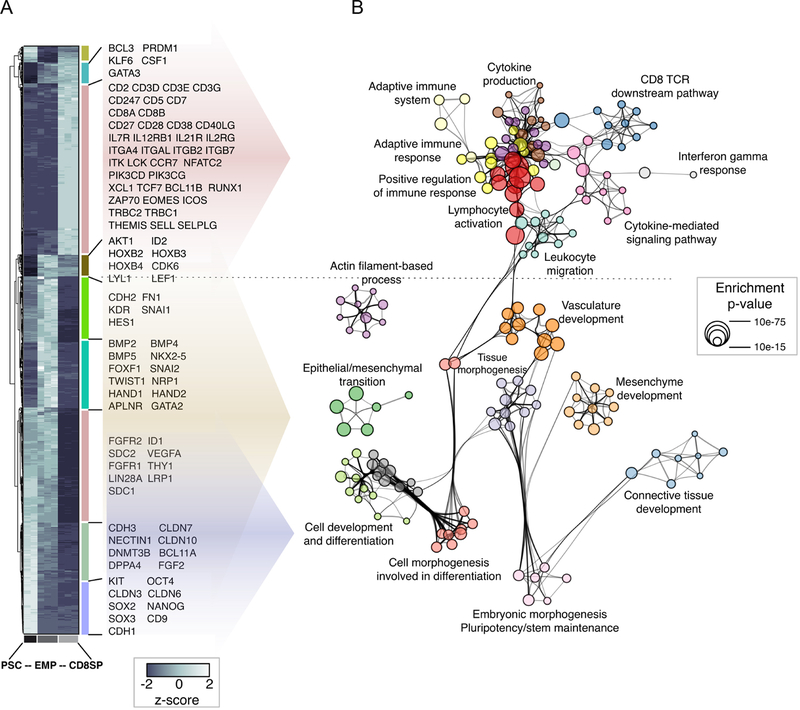
Figure 5. Gene expression changes during the transition from PSC to hEMP to CD8SP T cell.
(A) Hierarchical clustering for the 1981 genes classified as differentially expressed between PSC (H1 ESC), hEMP and PSC-ATO-derived CD8SP cells (pool of three pair-wise comparisons). Data for each biological replicate is shown. Individual clusters are labeled with vertical colored bars, and official symbols for representative genes are shown (full results provided in Table S1).
(B) Functional enrichment network for all genes differentially expressed between PSC, hEMP and PSC-ATO-derived CD8SP cells. Individual gene ontology terms with similar gene members are grouped by categories (node color) and labeled using a representative member. Node size is proportional to statistical significance (Enrichment p-value) as shown. Edge thickness is proportional to between-node similarity and reflects the overlap between genes annotated in both ontology terms. Only edges representing a Kappa similarity score greater than 0.3 are shown. Only significant ontology terms are shown (hypergeometric p-value p<1e-03). The network is oriented to highlight the association between the genes in each ontology term and those in the clusters from (A).
Analysis of the Vβ repertoire of CD8SP T cells from (TCR)PSC-ATOs revealed >99% of cells expressed the exogenous Vβ13.1 chain, with near-complete suppression of endogenous Vβ expression (Figure 4K). These results recapitulated those from TCR-engineered primary HSPC ATOs (Seet et al., 2017), and are consistent with allelic exclusion of endogenous Vβ gene rearrangement mediated by the transgenic TCR during early T cell differentiation, as has also been reported in other systems (Giannoni et al., 2013; Starck et al., 2014; Vatakis et al., 2013).
Transcriptional analysis of T cell differentiation in PSC-ATOs
We next analyzed the transcriptional fidelity of T cell differentiation in PSC-ATOs. RNA sequencing (RNA-Seq) of parental H1 ESCs, hEMP progeny derived from H1, and mature CD8SP T cells isolated from PSC-ATOs revealed the transcriptional topography of PSC-to-T differentiation (Table S1). Unbiased mapping of gene expression changes (Figure 5A) onto functional pathways (Figure 5B) across these key stages revealed the shift from pluripotency to mesodermal gene pathway usage at the PSC-to-hEMP transition, characterized by upregulation of mesenchymal and endothelial programs; down-regulation of pluripotency genes; and induction of epithelial-to-mesenchymal transition, as previously described (Chin et al., 2016; Evseenko et al., 2010). The subsequent hEMP-to-T transition was marked by the loss of mesodermal lineage pathways, and gain of T-lineage transcription factor expression (including BCL11B, TCF7, EOMES) as well as TCR/cytokine signaling pathways associated with a CD8+ T cell function (Figures 5A and 5B, Table S1). Importantly, mature T cells did not retain expression of pluripotency genes, including SOX2, OCT4, and NANOG.
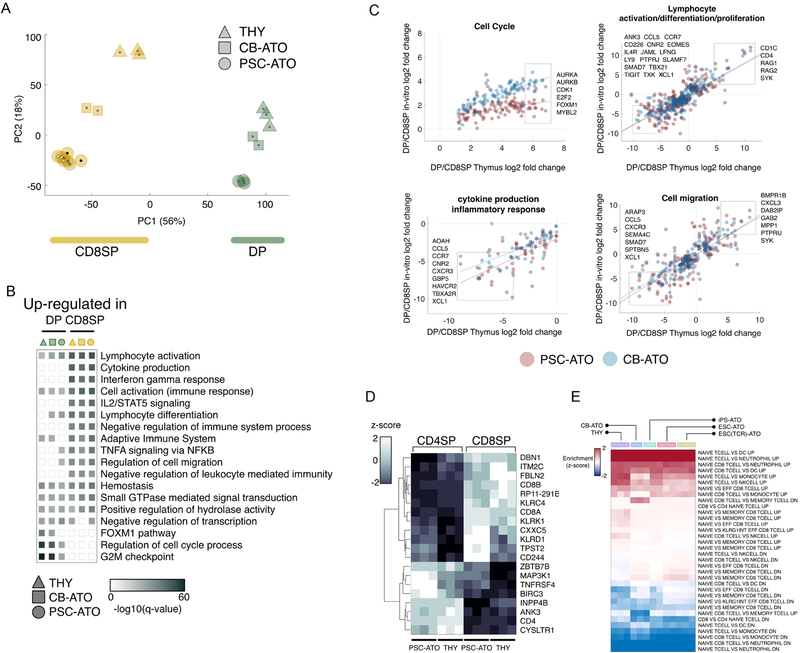
We next examined the DP-to-CD8SP transition in H1 PSC-ATOs, a maturation step associated with positive selection in the thymus (Carpenter and Bosselut, 2010; Rothenberg et al., 2008). DPs and CD8SP T cells from three different sources of PSC-ATOs (H1 ESC, TCR-transduced H1 ESC, and iPSC from RNA-reprogrammed fibroblasts), were compared with their counterparts from two primary sources: postnatal human thymi, and ATOs initiated with primary umbilical cord blood HSPCs (CB-ATOs) (Seet et al., 2017). Principal component analysis using global expression profiles for all populations showed stage-specific clustering of DP and CD8SP cells, while also revealing stage-independent source effects (Figure 6A). T cells from all three types PSC-ATO sources showed minimal transcriptional differences (Figure 6A). Mapping of differential gene expression between DP and CD8SP T cells from PSC-ATOs, CB-ATOs, or thymus onto functional pathways revealed cell cycle genes to be the primary driver of the observed source effect on gene expression, without significant differences in the development of T cell effector pathways (Figures 6B and 6C). To examine ATO-specific differences in cell cycle regulation in developing T cells, we analyzed cell cycling in DP and CD8SP T cells from postnatal thymi, CB-ATOs, and PSC-ATOs by Ki-67 staining. This revealed that DPs from all sources were in cycle significantly more than CD8SP T cells, which were mostly in G0 phase (Fig S6 A–C). However, the frequency of mature CD8SP cells in cycle was higher in CB- and PSC-ATOs than in the thymus (Fig S6D), of which one mechanism may be homeostatic proliferation of naïve T cells in ATOs, possibly in response to continued exposure to IL-7 (Schluns et al., 2000).
Figure 6: RNA-Seq analysis of mature T cell development in PSC-ATOs.
(A) Principal component analysis of gene expression for CD8SP and DP cells from three different sources (THY: normal thymus; CB-ATO: ATOs initiated with cord blood CD34+ HSPCs; and PSC-ATOs). Each symbol shape represents an individual biological replicate for the corresponding cell type; all CD8SP samples are yellow and all DP are green; the PSC-ATO group includes ATOs initated from iPSC (iPSC-ATO; black central dots), H1 ESCs (ESC-ATO; white central dots), and TCR-transduced H1 ESCs (ESC(TCR)-ATO; grey central dots). Shown is the ordination using the first two principal components (PC1 and PC2) and percent of variance explained by each principal component as computed using unfiltered, whole-transcriptome expression levels.
(B) Functional enrichment results for genes differentially expressed between CD8SP and DP cells from three different sources (THY, CB-ATO, and PSC-ATO). For each pairwise comparison, shown are enrichment q-values computed independently for genes upregulated in CD8SP or DP cells for each source. Each functional category summarizes the enrichment of individual ontology terms with similar gene members (full results are provided in Table S1).
(C) Comparison of gene expression fold changes between DP and CD8SP cells derived from different sources, for selected functional categories. The x axis represents the fold change (DP/CD8) for cells isolated from normal thymus. The y axis represents the fold change (DP/CD8) for ATO-derived cells differentiated from CB-ATOs (blue dots) or PSC-ATIs (red dots). For each category, boxes and official symbols highlight representative genes strongly up- or down-regulated between DP and CD8SP cells regardless of the source. Full results are provided in Table S1.
(D) Hierarchical clustering for the 20 genes classified as differentially expressed (Wald adjusted p-value <1–10, fold change >2) between CD4SP and CD8SP cells from both PSC-ATOs and thymic samples. All 3 biological replicates for each cell type are shown.
(E) SaVanT enrichment scores for CD8SP cells using RNA-Seq expression estimates. Enrichment was computed using T cell and CD8 T cell entries in a collection of pair-wise immunologic gene expression signatures (see Methods). Shown are results for CD8SP cells derived from different sources (THY: thymus, CB-ATO, iPS-ATO, ESC-ATO, and TCR-transduced ESC-ATOs). Entries labeled as “UP” include genes more expressed in the first cell type of the signature description. For each pair of cell types, enrichment in both “UP” and “DOWN” (genes more expressed in the second cell type) signatures are shown.
We next compared gene expression in mature CD8SP and CD4SP T cells isolated from both H1 PSC-ATOs and postnatal thymi. In both cases, CD8SP and CD4SP cells were found to be almost identical at the transcriptional level (Table S1) in agreement with previous findings in both mouse and human (Mingueneau et al., 2013; Palmer et al., 2006). The small set of genes which distinguished CD8SP from CD4SP T cells were source-indepependent (Figure 6D), many of which have been previously reported to be differentially expressed between human CD4+ and CD8+ naïve T cells (Ranzani et al., 2015), and included the CD4-lineage regulator ZBTB7B (ThPOK). Finally, we compared the transcriptional profile of naïve phenotype CD8SP T cells isolated from different sources, including PSC-ATOs (derived from H1 ESC, TCR-engineered H1 ESC, or iPSC); CB-ATOs; and primary human postnatal thymi. Gene expressions profiles were compared to a library of immune gene expression signatures (Godec et al., 2016). Unbiased enrichment results revealed the similarity of primary, CB-ATO, and PSC-ATO CD8SP populations using reference gene sets enriched in CD8+ naïve T cells, further confirming PSC-ATO derived T cells as both conventional and naïve (Figure 6E).
Discussion
We have shown here that generation of mature, conventional human T cells from PSCs can be efficiently achieved in a standardized artificial thymic organoid system (PSC-ATO) effective across multiple ESC and iPSC lines. This simplified approach integrates the induction of T lineage-competent hematopoiesis with complete development of conventional, naïve T cells in a continuous 3D structure, representing both qualitative and technical improvements to human T cell generation from PSCs.
Induction of T-lineage competent hematopoietic specification from human PSCs has been historically inefficient. Successful protocols have used either spontaneous embryoid body (Galic et al., 2006; Galic et al., 2009; Kennedy et al., 2012) or OP9 monolayer co-culture methods (Galat et al., 2017; Timmermans et al., 2009; Vizcardo et al., 2013) as an initial step to induce mesodermal and hemato-endothelial specification whilst retaining T potential. Using these methods, limited T cell differentiation was achieved by co-culturing PSC-derived CD34+ cells with monolayers of Notch-ligand transduced OP9 cells (Kennedy et al., 2012; Timmermans et al., 2009). As the hematopoietic potential of PSC-derived CD34+ cells is largely primed for myeloid and erythroid differentiation (Dravid et al., 2011; Kennedy et al., 2012; Kim et al., 2014; Sturgeon et al., 2014; Vodyanik et al., 2005), we propose that the use of hEMPs as the substrate for hematopoiesis in PSC-ATOs improves the efficiency of T cell differentiation by excluding non-mesodermal lineages; capturing all emerging hematopoietic progenitors with T lineage potential; and suppressing non-T hematopoietic lineages via Notch signaling (Rothenberg and Scripture-Adams, 2008). Indeed, PSC-ATOs revealed early generation of CD34+CD43+ hematopoietic progenitor cells (HPC) together with small bystander populations of endothelial and possibly mesenchymal cells; however CD34- hematopoietic cells, comprising mainly T-lineage cells, constituted the majority of ATO cells at later timepoints, consistent with in situ differentiation of HPCs to T-lineage cells, and Notch-mediated suppression of alternate lineages.
Previous methods of T cell generation from human PSCs have permitted limited positive selection of T cell DP precursors to conventional naïve T cells (Kennedy et al., 2012; Themeli et al., 2015; Timmermans et al., 2009; Vizcardo et al., 2013). Notably, human PSC-derived hematopoietic cells cultured on OP9-DL1 monolayers were shown to be largely arrested at the DP precursor stage; subsequent generation of a CD8αβ phenotype has previously been achieved only by isolating DPs from cultures and stimulating them with CD3 antibody or and agonist peptide (Maeda et al., 2016; Vizcardo et al., 2013). Unconventional selection of iPSC-derived T cells to DN innate-like T cells has also been reported (Themeli et al., 2013). In contrast, PSC-ATOs permitted orderly T cell differentiation and spontaneous positive selection of DP precursors to naïve, conventional CD3+TCRαβ+CD8αβ+ T cells without the need for activation-mediated agonist selection. Further studies are required to identify the specific molecular and spatial interactions promoting positive selection in this system, however, as for primary CB-ATOs, we hypothesize that 3D interactions with positively selecting ligands, including self-MHC, on neighboring hematopoietic cells allows for thymic-like signal integration not otherwise possible in monolayer cultures. Supporting this, human fetal thymic implants have been shown to support positive selection of human PSC-derived T cells when implanted into NSG mice (Galic et al., 2006; Galic et al., 2009); and a recent study demonstrated that mouse iPSC-derived T cell precursors generated on OP9-DLL1 monolayers matured to CD8αβ+ T cells only when seeded into mouse fetal thymic organ cultures (FTOCs) (Vizcardo et al., 2018). Importantly, unlike the FTOC or NSG-transplantation models, PSC-ATOs do not contain primary thymic stromal elements, suggesting that 3D interactions rather than thymic-specific stromal signals are sufficient for the positive selection of human CD8αβ+ T cells in our system. It is likely that the higher frequency of CD8SP over CD4SP in ATOs is at least partly due the ubiquitous expression of MHC class I on hematopoietic cells and the rarity of MHC class II-expressing cells; however other factors including Notch signaling strength may also play a role.
We also show that apparently normal T cell differentiation can proceed from PSCs in the absence of DNTT (TdT) expression, albeit resulting in TCRs with shorter CDR3 regions. This observation is consistent with reports that CDR3 length in B and T cells increases progressively during human fetal development (Rechavi et al., 2015; Zemlin et al., 2002). As the PSC-ATO environment is standardized and similar to that of CB-ATOs in which TdT is expressed normally, these findings suggest that control of DNTT expression, and hence TCR CDR3 length, is intrinsic to the PSC-derived T cell program, and furthermore similar to that seen in fetal T cell development.
Our data also validate PSC-ATOs for the in vitro generation of antigen specific T cells from TCR-transduced PSCs. Generation of antigen-specific T cells from human PSCs has been previously achieved using TCR or CAR-transduced iPSCs derived from T cells (T-iPSC) (Maeda et al., 2016; Themeli et al., 2013), or iPSCs derived directly from antigen-specific T cells (Maeda et al., 2016; Vizcardo et al., 2013). As with other human PSC methods, these sources yielded innate-like DN or CD8αα+ T cells in OP9 co-cultures or required activation and agonist selection of DP precursors to generate mature-like T cells. A major advantage of PSC-ATOs is the ability to directly generate antigen-specific naïve T cells without agonist selection and developmental diversion to innate-like T cell types. While innate-like T cells may possess potentially interesting functional properties such as robust cytokine production and antigen-independent cytotoxicity, based on their similarity to intraepithelial lymphocytes, patterns of survival, tissue homing, and peripheral tolerance induction are likely to be very distinct from those of conventional T cells. For example, T-iPSC-derived DN/CD8αα+ T cells derived on OP9-DL1 monolayers exhibited a terminally differentiated TEMRA phenotype, lacked expression of the lymphoid organ homing molecules CCR7 and CD62L, and failed to produce IFNγ in response to antigen (Themeli et al., 2013). In contrast, PSC-ATO-derived CD8αβ+ T cells were transcriptionally similar to primary CD8+ naïve T cells, expressed a CD62L+CCR7+ naïve phenotype, produced IFNγ and underwent conversion to effector/memory T cells. (TCR)PSC-ATO derived T cells exhibited robust in vitro cytotoxicity against NY-ESO-1-expressing K562 cells, as well as in vivo circulation and tumor control which, while modest, may be primarily limited by cytokine and homing incompatibilities across species barriers; indeed other studies of PSC-derived T cell adoptive transfer have circumvented these issued by injecting both tumor cells and T cells into the peritoneal space together with human cytokines (Maeda et al., 2016; Themeli et al., 2013).
Analogous to observations from TCR-transduced HSPCs (Giannoni et al., 2013; Starck et al., 2014; Vatakis et al., 2013), introduction of a fully rearranged TCR at the PSC stage induced allelic exclusion of endogenous TCR Vβ rearrangement during T cell differentiation, which may serve as a built-in mechanism to mitigate graft-versus-host-disease risk upon adoptive transfer of PSC-ATO derived engineered T cells in the allogeneic setting.
PSC-ATOs are thus an efficient and readily adoptable method for modeling both hematopoietic and T cell differentiation from human PSCs. As a platform for therapeutic T cell generation, PSC-ATOs present the opportunity to combine technologies to gene-modify, screen, and expand self-renewing PSC clones engineered for non-alloreactivity and enhanced T cell function, and may serve as the basis for the production of universal, “off-the-shelf” T cell therapies.
STAR methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead Contact, Gay Crooks (gcrooks@mednet.ucla.edu)
EXPERIMENTAL MODELS AND SUBJECT DETAILS
Cell lines
The MS5-hDLL1 cell line was generated in our lab as previously described (Seet et al., 2017). To generate MS5-hDLL4, MS5 cells (Itoh et al., 1989) were transduced with a lentiviral vector encoding full-length human DLL4. The highest 5% DLL4-expressing cells were sorted by FACS using an anti-DLL4 antibody and passaged in DMEM/10% fetal calf serum (FCS). Stable expression was confirmed by flow cytometry for DLL4 expression after several weeks of culture, as well as qRT-PCR and DNA sequencing. For immunofluorescence analysis, we generated MS5-hDLL4-GFP cells by transducing MS5 cells with a lentiviral vector encoding full-length human DLL4 and enhanced green flurorescent protein (eGFP). The top 5% of the cells with the highest expression of GFP were sorted and passaged in DMEM/10% FCS. Stable expression was confirmed by flow cytometry for GFP expression after several weeks of culture.
Artificial antigen presenting cells (aAPCs) were generated using K562 cells (ATCC). Briefly, K562 cells were transduced with lentiviral vectors encoding full-length human CD80, CD83, CD137L, and either HLA-A*02:01/B2M/NY-ESO-1157–165 or MART-126–35 single chain trimers (SCTs; gifts from Dr. David Baltimore, Caltech). Cytotoxicity assay target cells were created by transduction of K562 cells with either NY-ESO-1 or MART-1 SCTs alone.
Human pluripotent cell lines
The hESC lines H1 (Thomson et al., 1998) (WiCell, Madison, WI) , UCLA3, UCLA6 (Diaz Perez et al., 2012), ESI-017 (Crook et al., 2007) (ESI BIO, Alameda, CA), and an iPSC line derived by RNA transfection of healthy female fibroblasts (DMD 1001 R1) (Hazim et al., 2017) were maintained and expanded either on irradiated primary mouse embryonic fibroblasts (MEFs) (EMD Millipore, Billerica, MA) or Matrigel (Growth factor reduced Matrigel matrix, BD Biosciences, Cat. 356231) in mTesSR complete medium (mTeSR medium +5X supplement, Stem Cell Technologies, Cat. 05850).
The TCR-transduced hESC line (H1-opt1G4-mTagBFP2) was generated by transduction of H1 hESCs with a lentiviral vector encoding 1G4 (an MHC Class I restricted NY-ESO-1 specific TCR (described below) and the fluorescent marker mTagBFP2. Briefly, H1 colonies were transferred from MEFs to Matrigel in mTesSR complete medium in 6 well plates. Concentrated lentiviral supernatant was directly added to the wells. Medium was changed the next day. After 72 hours, cells were dissociated with TrypLE™ Express (Gibco Life Technologies, Cat. 12604–013) and isolated by flow cytometry using the following phenotype: TRA-1–81+ mTagBFP2+ and mouse CD29-. Isolated cells were returned to culture on MEFs for expansion and cryopreservation.
Mice
All animal experiments were conducted under a protocol approved by the UCLA Chancellor’s Animal Research Committee. This study used 4–8-week-old NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice from Jackson Laboratory (Bar Harbor, Maine). Mice from both sexes were randomly allocated to experimental groups.
METHOD DETAILS
Generation and isolation of human embryonic mesodermal progenitors (hEMPs)
Mesoderm commitment was induced as previously described (Chin et al., 2016; Evseenko et al., 2010) with certain optimizations. Briefly, hESC colonies were maintained on Matrigel-coated 6-well plates in mTesSR complete medium. hESC/hIPSC cells were then harvested as a single cell suspension after TrypLE™ Express (Gibco Life Technologies Ref 12604–013) treatment for 5 minutes at 37°C, washed, and counted. Cells were resuspended directly in X-VIVO 15 medium (Lonza Ref 04–418Q) supplemented with rhActivin A (10 ng/ml) (R&D Systems, Cat. 338-AC-010), rhBMP4 (10 ng/ml) (R&D Systems, Cat. 314-BP-010), rhVEGF (10 ng/ml) (R&D Systems, Cat. 298-VS-005), rhFGF (10 ng/ml) (R&D Systems, Cat. 233-FB-025), and ROCK inhibitor Y-27632 dihydrochloride (10 uM) (Tocris, Cat. 1254). Cells were plated on Matrigel coated 6-well plates at 3×106 cells per well in 3ml. Medium was then changed daily with X-VIVO 15 supplemented with rhBMP4 (10 ng/ml), rhVEGF (10 ng/ml, and rhFGF (10 ng/ml). At day 3.5, cells were washed 3 times with PBS and incubated with Accutase (Innovative Cell Technologies, Cat. AT-104) (1 ml per well, for 10 min. at 37°C). Cells were harvested using a cell scraper, washed in PBS, and stained with antibodies for flow cytometry. CD326−CD56+ hEMP were isolated by FACS on a FACSARIA instrument (BD Biosciences, San Jose, CA).
Isolation of human cord blood HSPCs for ATO culture
Neonatal umbilical cord blood was obtained from discarded cord and placental units from deliveries at UCLA under UCLA IRB-approved protocols or exemptions, and were enriched for mononuclear cells by Ficoll-Paque (GE Healthcare Life Sciences, Pittsburgh, PA) gradient centrifugation followed by positive selection of CD34+ cells by magnetic cell sorting (MACS) using CD34 MicroBead UltraPure Kit (Miltenyi, Auburn CA). CD34+ enriched fractions were cryopreserved after MACS. Prior to use, cells were thawed, and residual T cells depleted by FACS by sorting for CD34+CD3- cells. Sorted cells were immediately seeded into MS5-hDLL4 ATOs, as previously described (Seet et al., 2017).
Isolation of human thymic and ATO-derived T cells
Postnatal human thymi were obtained under IRB exemption as anonymized, discarded waste from patients undergoing cardiac surgery at Children’s Hospital Los Angeles (CHLA). A fetal thymus sample (17 weeks gestation) was obtained from the UCLA CFAR virology core under their institutionally approved protocol. Thymic fragments were finely dissected in RPMI and disrupted by pipetting to release thymocytes into suspension, followed by passage through a 70 µm nylon strainer. Cells were analyzed fresh on the same or following day. Flow cytometry cell sorting of thymic and ATO-derived T cell populations used the following surface phenotypes: double positive (DP; CD4+CD8+CD3-), naïve CD8 single positive (CD8SP; TCRαβ+CD3+CD8α+CD4-CD45RA+) and naïve CD4 single positive (CD4SP; TCRαβ+CD3+CD8-CD4+CD45RA+). PSC-ATO-derived T cell populations were defined as CD56-CD45+ in combination with the phenotypes described above. PSC-ATO derived mature CD8SP cells were further defined as positive for CD8β.
Cord blood HSPC artificial thymic organoid (CB-ATO) cultures
Cord blood ATOs were generated as previously described (Seet et al., 2017). MS5-hDLL1 cells were harvested by trypsinization and resuspended in serum free ATO culture medium (“RB27”) composed of RPMI 1640 (Corning, Manassas, VA), 4% B27 supplement (ThermoFisher Scientific, Grand Island, NY), 30 µM L-ascorbic acid 2-phosphate sesquimagnesium salt hydrate (Sigma-Aldrich, St. Louis, MO) reconstituted in PBS, 1% penicillin/streptomycin (Gemini Bio-Products, West Sacramento, CA), 1% Glutamax (ThermoFisher Scientific, Grand Island, NY), 5 ng/ml rhFLT3L and 5 ng/ml rhIL-7 (Peprotech, Rocky Hill, NJ). RB27 was made fresh weekly. 1.5×105 MS5-hDLL1 cells were combined with 5×103 purified CD34+CD3- cells per ATO in 1.5 ml Eppendorf tubes (up to 12 ATOs per tube) and centrifuged at 300 g for 5 min at 4˚C in a swinging bucket centrifuge. Supernatants were carefully removed, and the cell pellet was resuspended in 6 µl RB27 per ATO and mixed by brief vortexing. ATOs were plated on a 0.4 µm Millicell transwell insert (EMD Millipore, Billerica, MA; Cat. PICM0RG50) placed in a 6-well plate containing 1 ml RB27 per well. Medium was changed completely every 3–4 days by aspiration from around the cell insert followed by replacement with 1 ml with fresh RB27/cytokines. ATO cells were harvested by adding FACS buffer (PBS/0.5% bovine serum album/2mM EDTA) to each well and briefly disaggregating the ATO by pipetting with a 1 ml “P1000” pipet, followed by passage through a 50 µm nylon strainer.
Pluripotent stem cell-artificial thymic organoid (PSC-ATO) cultures
PSC-ATOs were generated using purified hEMPs and followed a two-step procedure including a hematopoietic induction phase and a T-cell differentiation phase (see Figure 1A). MS5-hDLL4 cells (or MS5-hDLL1 cells, where indicated) were harvested by trypsinization and resuspended in hematopoietic induction medium composed of EGM2 (Lonza Ref CC-4176) supplemented with ROCK inhibitor Y-27632 dihydrochloride (10uM) (Tocris Bioscience, Cat. 1254) and TGF-βRI inhibitor SB-431542 (“SB blocker”) (10 uM) (Tocris Bioscience, Cat. 1614). At day −14, 5×105 MS5-hDLL4 cells were combined with 0.5–1×104 purified hEMP per PSC-ATO in 1.5 ml Eppendorf tubes and centrifuged at 300 g for 5 min at 4˚C in a swinging bucket centrifuge. Multiple (up to 12) PSC-ATOs were prepared per tube. Supernatants were carefully removed and the cell pellet was resuspended by brief vortexing and resuspended in hematopoietic induction medium at a volume of 6 µl per PSC-ATO. 6 µl of cells were plated as PSC-ATOs on a 0.4 µm Millicell transwell insert (EMD Millipore, Billerica, MA; Cat. PICM0RG50) (up to 2 PSC-ATOs per insert were plated) and placed in 6-well plates containing 1 ml hematopoietic induction medium per well. Medium was changed completely every 2–3 days for 1 week, as described above for CB HSPC ATOs, with medium composed of EGM2 with SB blocker (10 µM). At day −7, medium was change to EGM2 + SB blocker (10 µM) with the hematopoietic cytokines rhTPO 5 ng/ml (Peprotech, Cat. 300–18), rhFLT3L 5 ng/ml (Peprotech, Cat. 300–19), and rhSCF 50 ng/ml (Peprotech, Cat. 300–07). This medium was changed every 2–3 days for an additional 7 days. At day 0, T cell differentiation was induced by changing the medium to “RB27” (described above) supplemented with 10 ng/ml rhSCF, 5 ng/ml rhFLT3L, and 5 ng/ml rhIL-7 (Peprotech, Cat. 200–07). Medium was changed completely every 3–4 days.
Isolation of PSC-ATO cells
PSC-ATO T cells were harvested by adding MACS buffer (PBS/0.5% bovine serum album/2mM EDTA) to each well and briefly disaggregating the ATO by pipetting with a 1 ml “P1000” pipet, followed by passage through a 50 µm nylon strainer. For larger numbers of PSC-ATOs, a cell scraper was used to remove PSC-ATOs from culture inserts, and single cell suspensions were prepared by passage through a 50 µm nylon strainer.
A digestion step was performed for the harvest and characterization of progenitor cells during the early stages of differentiation (Figure 1). ATOs were removed from culture insert and incubated in digestion buffer (collagenase type IV solution (StemCell Technologies) supplemented with 0.88mg/ml collagenase/dispase (Roche) and 50U DNAse I (Sigma)) for 10 minutes at 37C, mechanically disrupted by pipetting, and incubated again for 10 minutes at 37°C. After complete disaggregation by pipetting, single cell suspensions were prepared by passage through a 50 µm nylon strainer
Lentiviral vectors and transduction
The full-length coding sequence of human DLL4 was synthesized (Integrated DNA Technologies, Skokie, IL) and cloned into the third-generation lentiviral vector pCCL-c-MNDU3 (gift from Dr. Donald Kohn, UCLA) with IRES-linked eGFP expression.
The codon optimized TCR α and β (including Vβ13.1) chains of a TCR specific for HLA-A*02:01/NY-ESO-1157–165 (derived from the 1G4 TCR; (Robbins et al., 2008) is previously described (Gschweng et al., 2014) (gift from Dr. Antoni Ribas, UCLA). The TCR coding sequences sub-cloned into the third-generation pCCL lentiviral vector downstream of a ubiquitin C (UBC) promoter. A 2A-linked mTagBFP2 fluorescent protein coding sequence (Subach et al., 2011) was added downstream of TCRβ.
Packaging and concentration of lentivirus particles was performed as previously described (Seet et al., 2017). Briefly, 293T cells (ATCC) were co-transfected with a lentiviral vector plasmid, pCMV-∆R8.9, and pCAGGS-VSVG using TransIT 293T (Mirus Bio, Madison, WI) for 17 hours followed by treatment with 20 mM sodium butyrate for 8 hours, followed by generation of cell supernatants in serum-free UltraCulture for 48 hours. Supernatants were concentrated by ultrafiltration using Amicon Ultra-15 100K filters (EMD Millipore, Billerica, MA) at 4000 xg for 40 minutes at 4°C and stored as aliquots at −80C.
TCR Vβ expression analysis by flow cytometry
Total cells isolated from pooled PSC-ATOs or postnatal thymi were stained for CD3, CD4, CD8, and TCRγδ, in conjunction with the IOTest Beta Mark TCR V Kit (Beckman Coulter, Indianapolis, IN). CD3+TCRγδ-CD8+CD4- cells and CD3+TCRγδ-CD8-CD4+ cells were gated for analysis, and Vβ family usage was determined by percent FITC+, PE+, or FITC+PE+ cells, representing 3 different Vβ antibodies per tube, per the manufacturer’s protocol. For Vβ analysis of TCR-transduced PSC-ATOs, total cells from week 6–7 (TCR)PSC-ATOs were additionally pre-labeled with an APC-conjugated HLA-A*02:01/NY-ESO-1157–165 tetramer (MBL International, Woburn, MA) for 10 minutes prior to surface antibody staining, and cells were gated on tetramer+CD3+TCRγδ-CD8+CD4- T cells for Vβ analysis.
TCR repertoire deep sequencing
Mature CD8SP T cells (CD45+CD3+TCRαβ+CD8αβ+CD45RA+) from week 6–7 H1 PSC-ATOs were FACS-sorted from independent duplicate experiments. For comparison, CD8SP T cells from two neonatal thymi and two peripheral blood samples were sorted using based on the same CD8SP surface phenotype. Cell pellets were frozen, and genomic DNA was extracted. Deep sequencing and analysis of TCRβ CDR3 regions was performed by immunoSEQ (Adaptive Biotechnologies, Seattle, WA).
T cell cytokine assays
Mature CD8SP cells from PSC-ATOs were isolated by magnetic negative selection using the CD8+ T Cell Isolation Kit (Miltenyi, Cat. 130–096-495) and sorted by FACS to further deplete CD45RO+ cells (containing immature CD8SP T cells and DN/4ISP T cell precursors). Purified T cell populations were plated in 96-well U-bottom plates in 200 µl AIM V (ThermoFisher Scientific, Grand Island, NY) with 5% human AB serum (Gemini Bio-Products, West Sacramento, CA). PMA/ionomycin/protein transport inhibitor cocktail or control protein transport inhibitor cocktail (eBioscience, San Diego, CA) were added to each well and incubated for 6h. Cells were washed and stained for CD3, CD4, and CD8 (Biolegend, San Diego, CA) and Zombie UV™ Fixable Viability dye (Biolegend, San Diego, CA) prior to fixation and permeabilization with an intracellular staining buffer kit (eBioscience, San Diego, CA) and intracellular staining with antibodies against IFNγ, TNFα, and IL-2 (Biolegend, San Diego, CA).
T cell activation and proliferation assays
For CFSE proliferation assays, PSC-ATO-derived CD8SP and CD4SP T cells were isolated as described above and labeled with 5 µM CFSE (Biolegend, San Diego, CA). Labeled cells were incubated with anti-CD3/CD28 beads per manufacturer’s protocol (ThermoFisher Scientific, Grand Island, NY) in AIM V/5% human AB serum with 20 ng/ml rhIL-2 (Peprotech, Rocky Hill, NJ), and plated in 200 µl per well of 96-well round-bottom plates. At day 3, half the media was changed by gently aspirating and replacing with fresh medium containing 2X rhIL-2. On day 5, cells were washed and stained for CD25 and 4–1BB (Biolegend, San Diego, CA) and analyzed by flow cytometry. For in vitro cell expansion assays, 1×104 (TCR)PSC-ATO-derived ESO TCR CD8SP T cells were isolated as above and plated in triplicate in 96-well round-bottom plates in 200 µl AIM V/5% AB serum with irradiated antigen-expressing K562 aAPCs (K562-CD80/83/137L-1073ESO) in a 1:3 aAPC:T cell ratio in the presence of 20 ng/mL rhIL-2 . Fresh medium was added on day 4, and cells were replated every 2–3 days into larger wells as needed. Cells were counted weekly on a hemacytometer. Flow cytometry analysis was done on expanded cells at the indicated times.
T cell activation/cytokine assays
1×105 total ESO TCR CD8SP T cells were isolated from week 6 (TCR)PSC-ATOs as described above, and co-cultured with cognate (NY-ESO-1) or irrelevant (MART-1) antigen-expressing K562 aAPCs in 96-well round-bottom plates in 200 µl AIM V/5% human AB serum at a 2:1 T cell:aAPC ratio for 6h. CD107a-APC antibody (Biolegend, San Diego, CA) was added to wells at a 1:50 final dilution together with a protein transport inhibitor cocktail (eBioscience, San Diego, CA) for the duration of culture. Cells were then stained for surface markers, fixed, permeabilized, and intracellularly stained for cytokines as described above.
In vitro cytotoxicity assays
ESO TCR CD8SP T cells were isolated from pooled (TCR) PSC-ATOs and were activated in 96 well round-bottom plates in AIM V/5% human AB serum with irradiated cognate aAPCs and 20 ng/ml IL-2, as described above, for 36h. For extended expansions, cells were cultured in 20 ng/ml rhIL-2 for up to 14 days and restimulated with aAPCs between day 7–10. For cytotoxicity assays, 2-fold serial dilutions of activated T cells were plated in 96-well round bottom plates starting at 1×105 cells per well in AIM V/5% human AB serum. K562 target cells transduced with HLA-A*02:01/NY-ESO-1157–165 or HLA-A*02:01/MART-126–35 single chain trimers, were plated at 5×104 cells per well. Apoptotic cell death of target cells was quantified by annexin V/DAPI staining at 8–9h. Target cell death was calculated by subtracting percent annexin V+ target cells in wells receiving no T cells from wells that received T cells.
In vivo tumor assay
4–8 week old NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (Jackson Laboratory, Bar Harbor, Maine) were subcutaneously implanted with 2×105 K562 target cells transduced with a HLA-A*02:01/NY-ESO-1157–165 single chain trimer and firefly luciferase. Mice were imaged for tumor bioluminescence on day 3 by intraperitoneal injection of luciferin. ESO TCR T cells from (TCR)PSC-ATO-derived T cells were isolated and activated/expanded as above for 10 days. 5×106 T cells (verified as a pure population of mTagBFP2+ cells prior to injection) were injected via tail vein on day 3 post-tumor implantation. Injection of PBS into control mice was also performed. Tumor bioluminescence was repeated every 3–4 days for at least 21 days, after which mice were sacrificed based on disease burden criteria.
Short term in vivo analysis of adoptively transferred T cells
TCR engineered PSC-ATO-derived CD8SP T cells were activated/expanded as above for 14 days. 7×106 T cells (100% mTagBFP2+) were then injected via retro-orbital vein in 4–8 week old NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (Jackson Laboratory, Bar Harbor, Maine). Injection of PBS into control mice was also performed. rhIL2 (50000U/animal) was administered to all mice by intraperitoneal injection daily for 3 days. 48 hours after T cell injection, blood was collected and analyzed for the presence of ATO-derived T cells. Some experimental animals were sacrificed 48 hours after T cell injection for splenocyte analysis.
Immunofluorescence imaging of PSC-ATOs
PSC-ATOs were fixed in 4% Formaldehyde (Sigma-Aldrich, St. Louis, MO) for 30 minutes at room temperature followed by 3× 10 min. washes in PBST (0.3% Triton X-100) and a 1 hour block in PBST/BSA (2% BSA). ATOs were stained with anti-GFP (clone FM264G; Biolegend, San Diego, CA) at a 1:100 dilution and anti-CD3 (clone UCHT1; Biolegend, San Diego, CA) at a 1:50 dilution overnight at 4ºC. Secondary antibodies AlexaFluor-594-conjugated anti-mouse IgG (H+L) (Jackson ImmunoResearch, West Grove, PA) and AlexaFluor-488-conjugated anti-rat IgG (H+L) (Jackson ImmunoResearch West Grove, PA) were added at a 1:200 dilution for 2 hours at room temperature. Each ATO was mounted individually in Vectashield Antifade Mounting Medium (Vector Laboratories, England) on a concavity microscope slide (ThermoFisher Scientific, Grand Island, NY). Immunofluorescence images were acquired on a Zeiss LSM 880 confocal microscope equipped with Airyscan and Zen software (Zeiss, Jena, Germany).
Flow cytometry and antibodies
All flow cytometry stains were performed in PBS/0.5% BSA/2 mM EDTA for 30 min on ice. TruStain FcX (Biolegend, San Diego, CA) was added to all samples for 5 min prior to antibody staining. For tetramer co-staining, PE-conjugated HLA-A*02:01/NY-ESO-1157–165 tetramer (MBL International, Woburn, MA) was added to cells at a 1:50 final dilution at room temperature for 20 minutes prior to additional antibody staining for 20 minutes on ice. DAPI was added to all samples prior to analysis.
For intracellular TdT expression analysis, cells were stained for CD5, CD7, CD3, TCRαβ, CD4, and CD8 (Biolegend, San Diego, CA) and Zombie UV™ Fixable Viability dye (Biolegend, San Diego, CA) prior to fixation and permeabilization with the True-Nuclear Transcription Factor Buffer Set (Biolegend, San Diego, CA) followed by intracellular staining with an anti-TdT or an isotype control.
For cell cycle analysis, cells were stained for CD3, TCRαβ, CD4, and CD8 (Biolegend, San Diego, CA) prior to fixation and permeabilization with the FOXP3 staining Buffer Set (eBioscience San Diego, CA), and intracellular staining with the Ki-67 antibody clone (Biolegend, San Diego, CA). DAPI was added to the samples at least 1 hour before analysis.
Analysis was performed on an LSRII Fortessa, and FACS sorting on FACSARIA or FACSARIA-H instruments (BD Biosciences, San Jose, CA) at the UCLA Broad Stem Cell Research Center Flow Cytometry Core. For all analyses (except intracellular staining) DAPI+ cells were gated out, and single cells were gated based FSC-H vs. FSC-W and SSC-H vs. SSC-W. Anti-human antibody clones used for surface and intracellular staining were obtained from Biolegend (San Diego, CA): CD3 (UCHT1), CD4 (RPA-T4), CD5 (UCHT2), CD8a (SK1), CD25 (BC96), CD27 (O323), CD28 (CD28.2), CD326 (EPCAM) (Clone 9C4), CD34 (581), CD45 (HI30), CD45RA (HI100), CD45RO (UCHL1), CD56 (HCD56), CD62L (DREG-56), CD107a (H4A3), CCR7 (G043H7), interferon γ (4S.B3), IL-2 (MQ1–17H12), TCRαβ (IP26), TCRγδ (B1), TNFα (Mab11), Vβ13.1 (H131), 4–1BB (clone 4B4–1), TdT (clone 41A), CD90 (clone 5E10), CD31 (clone WM59), CD38 (Clone HIT2), Isotype control (clone RTK2071), Ki-67 (clone Ki67) and BD Biosciences (San Jose, CA): CD7 (M-T701), CD8b (clone 2ST8.5H7), CD43 (clone 1G10) and CD144 (clone 55–7H1). Anti-mouse CD29 (clone HMβ1–1), CD45 (clone 30-F11) and Ter119 (clone Ter119) were obtained from Biolegend (San Diego, CA). Flow cytometry data were analyzed with FlowJo software (Tree Star Inc.). In some experiments, flow cytometry data were analyzed using the UMAP (Uniform Manifold Approximation and Projection) package (McInnes and Healy, 2018).
RNA sequencing (RNA-seq) and data analysis
RNA was extracted from 3–5×104 cells from each of the indicated PSC (H1 ESC), hEMP, PSC-ATO, CB-ATO, or neonatal thymic populations by FACS, as described above, and total RNA isolated using the RNeasy Micro kit (Qiagen).
| Name of cell population | Number of replicates (n) | Phenotype |
|---|---|---|
| PSC (H1 ESC) | 2 | TRA-1–81+ |
| EMP | 3 | CD56+ Epcam- |
| ESC-ATO DP early | 3 | CD4+ CD8+ CD3- |
| ESC-ATO CD8 SP CD45RA+ | 3 | CD4- CD8+ CD3+ TCRab+ CD45RA+ |
| ESC-ATO CD4 SP CD45RA+ | 3 | CD4+ CD8- CD3+ TCRab+ CD45RA+ |
| ESC (TCR)-ATO CD8 SP CD45RA+ | 3 | CD4- CD8+ CD3+ TCRab+ CD45RA+ |
| ESC (iPSC)-ATO CD8 SP CD45RA+ | 2 | CD4- CD8+ CD3+ TCRab+ CD45RA+ |
| Thymus (THY) DP early | 3 | CD4+ CD8+ CD3- |
| Thymus (THY) CD8 SP CD45RA+ | 3 | CD4- CD8+ CD3+ TCRab+ CD45RA+ |
| Thymus (THY) CD4 SP CD45RA+ | 3 | CD4+ CD8- CD3+ TCRab+ CD45RA+ |
| CB-ATO DP early | 2 | CD4+ CD8+ CD3- |
| CB-ATO CD8 SP CD45RA+ | 2 | CD4- CD8+ CD3+ TCRab+ CD45RA+ |
1.5 ng of total RNA was input to generate sequencing libraries with SMARTer Stranded Total RNA-Seq (Pico) Kit (Clonetech, Cat. 635005). Paired-end 150 bp sequencing was performed on an Illumina HiSeq 3000. A total of 32 libraries were multiplexed and sequenced in 5 lanes. Raw sequence files were obtained, and quality checked using Illumina’s proprietary software, and are available at NCBI’s Gene Expression Omnibus (accession number GSE116015).
The STAR ultrafast universal RNA-seq aligner v2.5.2b (Dobin et al., 2013) was used to generate the genome index and perform paired-end alignments. Reads were aligned to a genome index that includes both the genome sequence (GRCh38 primary assembly) and the exon/intron structure of known gene models (Gencode v26 basic genome annotation). Alignment files were used to generate strand-specific, gene-level count summaries with STAR’s built-in gene counter. Only protein-coding, long-noncoding, anti-sense and T-cell receptor genes in the Gencode v26 annotation were considered (98% of total counts on average). Independent filtering was applied as follows: genes with less than one count per sample on average, count outliers or low mappability were filtered out for downstream analysis (Casero et al., 2015; Love et al., 2014). Counts were normalized per-sample in units of FPKMs after correcting for gene mappable length and sample total counts. The table of expression estimates (FPKM) was used as input for SaVanT (Lopez et al., 2017) to compute enrichment scores on a collection of immunologic gene expression signatures (Godec et al., 2016) (Figure 6E). Non-default parameters for SaVanT were “Convert matrix values to ranks” and “Compute null distribution with 10000 iterations”. Figure 6E shows per-sample z-scores of raw enrichment scores from SaVanT.
Differential expression analysis was performed with DESeq2 (Love et al., 2014). Given the range of phenotypes analyzed in this study, each differential analysis was performed independently to minimize type II error for highly-expressed, condition specific genes (Rapaport et al., 2013). For each analysis, we retrieved both variance-stabilized data and fold-changes for downstream analysis. Additionally, pairwise differential expression was performed to classify genes as differentially expressed between any two cell types (Wald test adjusted p-value < 1e-10, fold change >2). Functional enrichment of selected gene sets (Figures 6C and 6D) was performed with Metascape (http://metascape.org). The network of ontology terms in Figure 5B was computed in-house and visualized with Cytoscape (Shannon et al., 2003). Nodes with the same color are specific ontology terms in the same functional generic class, and are labeled using a representative member (Table S1). Node size is proportional to statistical significance (hypergeometric p-value as provided by Metascape). Edge thickness is proportional to between-node similarity and was computed in-house in Matlab (Release 2017a, The MathWorks, Inc) using Kappa statistics, and reflects the overlap between the gene sets annotated in both ontology terms. Hierarchical clustering (Figures 5A and 6D) was performed and visualized in Matlab using z-scores from variance-stabilized data as input. Principal component analysis (PCA, Figure 6A) was performed with the function prcomp in R (https://www.R-project.org/) using variance-stabilized data as input.
QUANTIFICATION AND STATISTICAL ANALYSIS
In all figures, n represents independent experiments and data are represented as mean ± standard deviation (s.d.) or mean ± standard error of the mean (SEM) as indicated. For Figure S2 and Figure S6, statistical analysis was performed using GraphPad Prism software and p-values were calculated from the two-tailed unpaired t test with Welch correction. The p-values are directly indicated on the figure, above the corresponding graphs. *p< 0.05; **p< 0.01; and ***p< 0.001 were considered statistically significant.
DATA AND SOFTWARE AVAILABILITY
The GEO accession number for the RNA-seq data reported in this paper is GSE116015. A detailed description of data analysis and the softw
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-human CD3 | Biolegend | (clone UCHT1) |
| Anti-human CD4 | Biolegend | (clone RPA-T4) |
| Anti-human CD43 | BD Bioscience | (clone 1G10) |
| Anti-human CD5 | Biolegend | (clone UCHT2) |
| Anti-human CD7 | BD Bioscience | (clone M-T701) |
| Anti-human CD8a | Biolegend | (clone SK1) |
| Anti-human CD8b | BD Bioscience | (clone 2ST8.5H7) |
| Anti-human CD25 | Biolegend | (clone BC96) |
| Anti-human CD27 | Biolegend | (clone O323) |
| Anti-human CD28 | Biolegend | (clone CD28.2) |
| Anti-human CD31 | Biolegend | (clone WM59) |
| Anti-human CD326 (EPCAM) | Biolegend | (Clone 9C4) |
| Anti-human CD34 | Biolegend | (clone 581) |
| Anti-human CD38 | Biolegend | (clone HIT2) |
| Anti-human CD45 | Biolegend | (clone HI30) |
| Anti-human CD45RA | Biolegend | (clone HI100) |
| Anti-human CD45RO | Biolegend | (clone UCHL1) |
| Anti-human CD56 | Biolegend | (clone HCD56) |
| Anti-human CD62L | Biolegend | (clone DREG-56) |
| Anti-human CD90 | Biolegend | (clone 5E10) |
| Anti-human CD107a | Biolegend | (clone H4A3) |
| Anti-human CD144 | BD Bioscience | (clone 55–7H1) |
| Anti-human CCR7 | Biolegend | (clone G043H7) |
| Anti-human interferon γ | Biolegend | (clone 4S.B3) |
| Anti-human IL-2 | Biolegend | (clone MQ1–17H12) |
| Anti-human TCRαβ | Biolegend | (clone IP26) |
| Anti-human TCRγδ | Biolegend | (clone B1) |
| Anti-human TNFα | Biolegend | (clone Mab11) |
| Anti-human Vβ13.1 | Biolegend | (clone H131) |
| Anti-human CD137 (4–1BB) | Biolegend | (clone 4B4–1) |
| Anti-human Ki67 | Biolegend | (clone ki-67) |
| Anti-human TdT | Biolegend | (clone 41A) |
| Anti-mouse CD29 | Biolegend | (clone HMβ1–1) |
| Anti-mouse Ter119 | Biolegend | (clone Ter119) |
| Anti-mouse CD45 | Biolegend | (clone 30-F11) |
| Anti-GFP | Biolegend | (clone FM264G) |
| Rat IgG1, x isotype ctrl | Biolegend | (clone RTK2071) |
| AlexaFluor-594-conjugated donkey anti-mouse IgG (H+L) | Jackson immunoResearch | Cat. 715–515-151 |
| AlexaFluor-488-conjugated donkey anti-rat IgG (H+L) | Jackson immunoResearch | Cat. 712–485-153 |
| Bacterial and Virus Strains | ||
| pCCL-c-MNDU3-hDLL4 (lentivirus) | This paper | N/A |
| pCCL-c-MNDU3-hDLL4-P2A-eGFP (lentivirus) | This paper | N/A |
| pCCL-UBC-opt1G4-mTagBFP2 (lentivirus) | This paper | N/A |
| Biological Samples | ||
| Postnatal human Thymi | CHLA | N/A |
| Cord Blood CD34+ cells | UCLA | N/A |
| Fetal human thymus | UCLA | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| rhIL-2 | Peprotech | Cat. 200–02 |
| rhBMP4 | R&D Systems | Cat. 314-BP-010 |
| rhVEGF | R&D Systems | Cat. 298-VS-005 |
| rhFGF | R&D Systems | Cat. 233-FB-025 |
| ROCK inhibitor Y-27632 dihydrochloride | Tocris Bioscience | Cat. 1254 |
| TGF-βRI inhibitor SB-431542 | Tocris Bioscience | Cat. 1614 |
| rhFLT3L | Peprotech | Cat. 300–19 |
| rhIL-7 | Peprotech | Cat. 200–07 |
| rhTPO | Peprotech | Cat. 300–18 |
| rhSCF | Peprotech | Cat. 300–07 |
| DNAse I | Sigma | Cat. 9003–98-9 |
| Collagenase/dispase | Roche | Cat. 11097113001 |
| Collagenase Type IV | StemCell Technologies | Cat. 07909 |
| B27 supplement | Gibco | Cat. 17504–044 |
| Accutase | Innovative Cell Technologies | Cat. AT-104 |
| TrypLE™ Express | Gibco Life technologies | Cat. 12604–013 |
| Annexin V | Biolegend | |
| TruStain FcX | Biolegend | Cat. 422302 |
| DAPI | Life technologies | Cat. D1306 |
| Formaldehyde | Sigma-Aldrich | Cat. F8775 |
| Vectashield Antifade Mounting Medium | Vector Laboratories | Cat. H1000 |
| Critical Commercial Assays | ||
| CD34 MicroBead Kit UltraPure | Miltenyi | Cat. 130–100-453 |
| IOTest Beta Mark TCR V Kit | Beckman coulter | Cat. IM3497 |
| iTAg Tetramer/APC - HLA-A*02:01 NY-ESO-1 (SLLMWITQC) | MBL International | Cat. T01065 |
| ImmunoSEQ | Adaptive Biotechnologies | https://www.adaptivebiotech.com/immunoseq/products/ |
| CD8+ T Cell Isolation Kit | Miltenyi | Cat. 130–096-495 |
| Cell Stimulation Cocktail (plus protein transport inhibitors) | eBioscience | Cat. 00–4975-03 |
| Intracellular Fixation & Permeabilization Buffer Set | eBioscience | Cat. 88–8824-00 |
| Foxp3 staining Buffer Set | eBioscience | Cat. 00–5523-00 |
| True-Nuclear Transcription Factor Buffer Set | BIolegend | Cat. 424401 |
| CFSE proliferation assay | Biolegend | Cat. 423801 |
| antiCD3/CD38 beads | ThermoFisher Scientific | Cat. 111.61D |
| RNeasy Micro kit | Qiagen | Cat. 74004 |
| SMARTer Stranded Total RNA-Seq (Pico) Kit | Clonetech | Cat. 635005 |
| Zombie UV™ Fixable Viability Kit | Biolegend | Cat. 423107 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GSE116015 |
| Experimental Models: Cell Lines | ||
| H1 ESC line | WiCell | https://www.wicell.org/ |
| UCLA3, UCLA6 ESC lines | UCLA PSC core | Diaz Perez., et al 2012 |
| ESI-017 ESC line | ESI BIO | https://www.esibio.com/media/wysiwyg/esibio/documents/esicells/WEB_Rev_B_ESI-017_Data_Sheet.pdf |
| iPSC (DMD 1001 R1) line | UCLA PSC core | Hazim et al., 2017 |
| H1-opt1G4-mTagBFP2 hESC line | This paper | N/A |
| MS5-hDLL1 | Seet et al., 2017 | N/A |
| MS5-hDLL4 | This paper | N/A |
| MS5-hDLL4-eGFP | This paper | N/A |
| K562-CD80/83/137L-HLA-A*02:01/B2M/NY-ESO-1157–165 aAPC | This paper | N/A |
| K562-HLA-A*02:01/B2M/NY-ESO-1157–165 aAPC | This paper | N/A |
| K562-HLA-A*02:01/B2M/ MART-126–35 aAPC | This paper | N/A |
| Experimental Models: Organisms | ||
| Mouse: NSG: NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ | The Jackson Laboratory | JAX: 005557 |
| Software and Algorithms | ||
| FlowJo | Tree Star Inc. | https://www.flowjo.com/solutions/flowjo |
| UMAP (Uniform Manifold Approximation and Projection for Dimension Reduction | McInnes et al., 2018 | https://umap-learn.readthedocs.io/en/latest/ |
| GraphPad Prism | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| bcl2fastq2 (v2.17) | Illumina | https://support.illumina.com/downloads/bcl2fastq-conversion-software-v2–20.html |
| The STAR ultrafast universal RNA-seq aligner v2.5.2b | Dobin et al., 2013 | N/A |
| DESeq2 | Love et al.,2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Metascape | Metascape | http://metascape.org |
| Cytoscape | Shannon et al.,2003 | http://www.cytoscape.org/ |
| Matlab | The MathWorks, Inc | https://www.mathworks.com/products/matlab.html |
| R | R | https://www.R-project.org/ |
| Zen | Zeiss | https://www.zeiss.com/microscopy/int/products/microscope-software/zen.html |
Acknowledgements
We thank the UCLA Broad Stem Cell Research Center (BSCRC) Flow Cytometry Core for assistance with FACS sorting; Rebecca Chan for assistance with specimen processing; and Dr. Chintan Parekh from Children’s Hospital Los Angeles for generous assistance with thymus samples. This work was supported by NIH/NCATS KL2TR001882 (C.S.S), BSCRC Clinical (C.S.S.) and Predoctoral (S.L.) Fellowships, NIH/NHLBI T32HL066992 (C.S.S., S.T.), and a Tower Cancer Research Foundation Career Development Grant (C.S.S.). Kite Pharma/Gilead Biosciences provided support for the research reported here under a sponsored research agreement with Dr. Gay M. Crooks as Principal Investigator, and holds an exclusive license with UCLA to certain intellectual property relating to the ATO system.
Research reported in this publication included work performed in the Genomics shared resource supported by the National Cancer Institute of the National Institutes of Health under award number P30CA016042, and UCLA Center for AIDS Research Virology Core Lab and UCLA AIDS Institute (grant 5P30 AI028697). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
References
- Calvo J, BenYoucef A, Baijer J, Rouyez MC, and Pflumio F (2012). Assessment of human multi-potent hematopoietic stem/progenitor cell potential using a single in vitro screening system. PloS one 7, e50495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AC, and Bosselut R (2010). Decision checkpoints in the thymus. Nature Immunology 11, 666–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casero D, Sandoval S, Seet C, Scholes J, Zhu Y, Ha V, Luong A, Parekh C, and Crooks G (2015). Long non-coding RNA profiling of human lymphoid progenitor cells reveals transcriptional divergence of B cell and T cell lineages. Nat Immunol 16, 1282–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin CJ, Cooper Aaron R, Lill Georgia R, Evseenko D, Zhu Y, He Chong B, Casero D, Pellegrini M, Kohn Donald B, and Crooks Gay M (2016). Genetic Tagging During Human Mesoderm Differentiation Reveals Tripotent Lateral Plate Mesodermal Progenitors. STEM CELLS 34, 1239–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crook JM, Peura TT, Kravets L, Bosman AG, Buzzard JJ, Horne R, Hentze H, Dunn NR, Zweigerdt R, Chua F, et al. (2007). The generation of six clinical-grade human embryonic stem cell lines. Cell stem cell 1, 490–494. [DOI] [PubMed] [Google Scholar]
- De Smedt M, Hoebeke I, and Plum J (2004). Human bone marrow CD34+ progenitor cells mature to T cells on OP9-DL1 stromal cell line without thymus microenvironment. Blood cells, molecules & diseases 33, 227–232. [DOI] [PubMed] [Google Scholar]
- Diaz Perez SV, Kim R, Li Z, Marquez VE, Patel S, Plath K, and Clark AT (2012). Derivation of new human embryonic stem cell lines reveals rapid epigenetic progression in vitro that can be prevented by chemical modification of chromatin. Human Molecular Genetics 21, 751–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England) 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dravid G, Zhu Y, Scholes J, Evseenko D, and Crooks GM (2011). Dysregulated gene expression during hematopoietic differentiation from human embryonic stem cells. Molecular therapy : the journal of the American Society of Gene Therapy 19, 768–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evseenko D, Zhu Y, Schenke-Layland K, Kuo J, Latour B, Ge S, Scholes J, Dravid G, Li X, MacLellan WR, et al. (2010). Mapping the first stages of mesoderm commitment during differentiation of human embryonic stem cells. Proceedings of the National Academy of Sciences 107, 13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galat Y, Dambaeva S, Elcheva I, Khanolkar A, Beaman K, Iannaccone PM, and Galat V (2017). Cytokine-free directed differentiation of human pluripotent stem cells efficiently produces hemogenic endothelium with lymphoid potential. Stem cell research & therapy 8, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galic Z, Kitchen SG, Kacena A, Subramanian A, Burke B, Cortado R, and Zack JA (2006). T lineage differentiation from human embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America 103, 11742–11747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galic Z, Kitchen SG, Subramanian A, Bristol G, Marsden MD, Balamurugan A, Kacena A, Yang O, and Zack JA (2009). Generation of T lineage cells from human embryonic stem cells in a feeder free system. Stem Cells 27, 100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannoni F, Hardee CL, Wherley J, Gschweng E, Senadheera S, Kaufman ML, Chan R, Bahner I, Gersuk V, Wang X, et al. (2013). Allelic exclusion and peripheral reconstitution by TCR transgenic T cells arising from transduced human hematopoietic stem/progenitor cells. Molecular therapy : the journal of the American Society of Gene Therapy 21, 1044–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godec J, Tan Y, Liberzon A, Tamayo P, Bhattacharya S, Butte AJ, Mesirov JP, and Haining WN (2016). Compendium of Immune Signatures Identifies Conserved and Species-Specific Biology in Response to Inflammation. Immunity 44, 194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gschweng EH, McCracken MN, Kaufman ML, Ho M, Hollis RP, Wang X, Saini N, Koya RC, Chodon T, Ribas A, et al. (2014). HSV-sr39TK positron emission tomography and suicide gene elimination of human hematopoietic stem cells and their progeny in humanized mice. Cancer research 74, 5173–5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazim RA, Karumbayaram S, Jiang M, Dimashkie A, Lopes VS, Li D, Burgess BL, Vijayaraj P, Alva-Ornelas JA, Zack JA, et al. (2017). Differentiation of RPE cells from integration-free iPS cells and their cell biological characterization. Stem cell research & therapy 8, 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hozumi K, Mailhos C, Negishi N, Hirano K, Yahata T, Ando K, Zuklys S, Hollander GA, Shima DT, and Habu S (2008). Delta-like 4 is indispensable in thymic environment specific for T cell development. The Journal of experimental medicine 205, 2507–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K, Tezuka H, Sakoda H, Konno M, Nagata K, Uchiyama T, Uchino H, and Mori KJ (1989). Reproducible establishment of hemopoietic supportive stromal cell lines from murine bone marrow. Experimental hematology 17, 145–153. [PubMed] [Google Scholar]
- Kennedy M, Awong G, Sturgeon Christopher M., Ditadi A, LaMotte-Mohs R, Zúñiga-Pflücker Juan C., and Keller G (2012). T Lymphocyte Potential Marks the Emergence of Definitive Hematopoietic Progenitors in Human Pluripotent Stem Cell Differentiation Cultures. Cell Reports 2, 1722–1735. [DOI] [PubMed] [Google Scholar]
- Kim WS, Zhu Y, Deng Q, Chin CJ, He CB, Grieco AJ, Dravid GG, Parekh C, Hollis RP, Lane TF, et al. (2014). Erythropoiesis from human embryonic stem cells through erythropoietin-independent AKT signaling. Stem Cells 32, 1503–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch U, Fiorini E, Benedito R, Besseyrias V, Schuster-Gossler K, Pierres M, Manley NR, Duarte A, Macdonald HR, and Radtke F (2008). Delta-like 4 is the essential, nonredundant ligand for Notch1 during thymic T cell lineage commitment. The Journal of experimental medicine 205, 2515–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Motte-Mohs RN, Herer E, and Zuniga-Pflucker JC (2005). Induction of T-cell development from human cord blood hematopoietic stem cells by Delta-like 1 in vitro. Blood 105, 1431–1439. [DOI] [PubMed] [Google Scholar]
- Lopez D, Montoya D, Ambrose M, Lam L, Briscoe L, Adams C, Modlin RL, and Pellegrini M (2017). SaVanT: a web-based tool for the sample-level visualization of molecular signatures in gene expression profiles. BMC Genomics 18, 824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T, Nagano S, Ichise H, Kataoka K, Yamada D, Ogawa S, Koseki H, Kitawaki T, Kadowaki N, Takaori-Kondo A, et al. (2016). Regeneration of CD8alphabeta T Cells from T-cell-Derived iPSC Imparts Potent Tumor Antigen-Specific Cytotoxicity. Cancer research 76, 6839–6850. [DOI] [PubMed] [Google Scholar]
- McInnes L, and Healy J (2018). Umap: Uniform manifold approximation and projection for dimension reduction arXiv preprint arXiv:180203426. [Google Scholar]
- Mingueneau M, Kreslavsky T, Gray D, Heng T, Cruse R, Ericson J, Bendall S, Spitzer MH, Nolan GP, Kobayashi K, et al. (2013). The transcriptional landscape of alphabeta T cell differentiation. Nature Immunology 14, 619–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohtashami M, Shah DK, Nakase H, Kianizad K, Petrie HT, and Zuniga-Pflucker JC (2010). Direct comparison of Dll1- and Dll4-mediated Notch activation levels shows differential lymphomyeloid lineage commitment outcomes. Journal of immunology (Baltimore, Md : 1950) 185, 867–876. [DOI] [PubMed] [Google Scholar]
- Palmer C, Diehn M, Alizadeh AA, and Brown PO (2006). Cell-type specific gene expression profiles of leukocytes in human peripheral blood. BMC Genomics 7, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranzani V, Rossetti G, Panzeri I, Arrigoni A, Bonnal RJ, Curti S, Gruarin P, Provasi E, Sugliano E, Marconi M, et al. (2015). The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4. Nature Immunology 16, 318–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport F, Khanin R, Liang Y, Pirun M, Krek A, Zumbo P, Mason CE, Socci ND, and Betel D (2013). Comprehensive evaluation of differential gene expression analysis methods for RNA-seq data. Genome biology 14, R95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechavi E, Lev A, Lee YN, Simon AJ, Yinon Y, Lipitz S, Amariglio N, Weisz B, Notarangelo LD, and Somech R (2015). Timely and spatially regulated maturation of B and T cell repertoire during human fetal development. Science translational medicine 7, 276ra225. [DOI] [PubMed] [Google Scholar]
- Robbins PF, Li YF, El-Gamil M, Zhao Y, Wargo JA, Zheng Z, Xu H, Morgan RA, Feldman SA, Johnson LA, et al. (2008). Single and dual amino acid substitutions in TCR CDRs can enhance antigen-specific T cell functions. Journal of immunology (Baltimore, Md : 1950) 180, 6116–6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg EV, Moore JE, and Yui MA (2008). Launching the T-cell-lineage developmental programme. Nature Reviews Immunology 8, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg EV, and Scripture-Adams DD (2008). Competition and collaboration: GATA-3, PU.1, and Notch signaling in early T-cell fate determination. Seminars in immunology 20, 236–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rother MB, Jensen K, van der Burg M, van de Bovenkamp FS, Kroek R, van Ijcken WFJ, van der Velden VHJ, Cupedo T, Olstad OK, van Dongen JJM, et al. (2016). Decreased IL7Rα and TdT expression underlie the skewed immunoglobulin repertoire of human B-cell precursors from fetal origin. Scientific Reports 6, 33924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluns KS, Kieper WC, Jameson SC, and Lefrancois L (2000). Interleukin-7 mediates the homeostasis of naive and memory CD8 T cells in vivo. Nature Immunology 1, 426–432. [DOI] [PubMed] [Google Scholar]
- Seet CS, He C, Bethune MT, Li S, Chick B, Gschweng EH, Zhu Y, Kim K, Kohn DB, Baltimore D, et al. (2017). Generation of mature T cells from human hematopoietic stem and progenitor cells in artificial thymic organoids. Nature Methods 14, 521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, and Ideker T (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck L, Popp K, Pircher H, and Uckert W (2014). Immunotherapy with TCR-redirected T cells: comparison of TCR-transduced and TCR-engineered hematopoietic stem cell-derived T cells. Journal of immunology (Baltimore, Md : 1950) 192, 206–213. [DOI] [PubMed] [Google Scholar]
- Sturgeon CM, Ditadi A, Awong G, Kennedy M, and Keller G (2014). Wnt signaling controls the specification of definitive and primitive hematopoiesis from human pluripotent stem cells. Nature biotechnology 32, 554–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subach OM, Cranfill PJ, Davidson MW, and Verkhusha VV (2011). An enhanced monomeric blue fluorescent protein with the high chemical stability of the chromophore. PloS one 6, e28674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Themeli M, Kloss CC, Ciriello G, Fedorov VD, Perna F, Gonen M, and Sadelain M (2013). Generation of tumor-targeted human T lymphocytes from induced pluripotent stem cells for cancer therapy. Nature biotechnology 31, 928–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Themeli M, Rivière I, and Sadelain M (2015). New Cell Sources for T Cell Engineering and Adoptive Immunotherapy. Cell stem cell 16, 357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, and Jones JM (1998). Embryonic stem cell lines derived from human blastocysts. Science (New York, NY) 282, 1145–1147. [DOI] [PubMed] [Google Scholar]
- Timmermans F, Velghe I, Vanwalleghem L, De Smedt M, Van Coppernolle S, Taghon T, Moore HD, Leclercq G, Langerak AW, Kerre T, et al. (2009). Generation of T cells from human embryonic stem cell-derived hematopoietic zones. Journal of immunology (Baltimore, Md : 1950) 182, 6879–6888. [DOI] [PubMed] [Google Scholar]
- Vatakis DN, Arumugam B, Kim SG, Bristol G, Yang O, and Zack JA (2013). Introduction of exogenous T-cell receptors into human hematopoietic progenitors results in exclusion of endogenous T-cell receptor expression. Molecular therapy : the journal of the American Society of Gene Therapy 21, 1055–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcardo R, Klemen ND, Islam SMR, Gurusamy D, Tamaoki N, Yamada D, Koseki H, Kidder BL, Yu Z, Jia L, et al. (2018). Generation of Tumor Antigen-Specific iPSC-Derived Thymic Emigrants Using a 3D Thymic Culture System. Cell Rep 22, 3175–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcardo R, Masuda K, Yamada D, Ikawa T, Shimizu K, Fujii S, Koseki H, and Kawamoto H (2013). Regeneration of human tumor antigen-specific T cells from iPSCs derived from mature CD8(+) T cells. Cell stem cell 12, 31–36. [DOI] [PubMed] [Google Scholar]
- Vodyanik MA, Bork JA, Thomson JA, and Slukvin II (2005). Human embryonic stem cell-derived CD34+ cells: efficient production in the coculture with OP9 stromal cells and analysis of lymphohematopoietic potential. Blood 105, 617–626. [DOI] [PubMed] [Google Scholar]
- Zemlin M, Schelonka RL, Bauer K, and Schroeder HW Jr. (2002). Regulation and chance in the ontogeny of B and T cell antigen receptor repertoires. Immunologic research 26, 265–278. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The GEO accession number for the RNA-seq data reported in this paper is GSE116015. A detailed description of data analysis and the softw