Abstract
Mucopolysaccharidosis (MPS) are a group of rare genetic disorders caused by deficiency in the activity of specific lysosomal enzymes required for the degradation of glycosaminoglycans (GAGs). A defect in the activity of these enzymes will result in the abnormal accumulation of GAGs inside the lysosomes of most cells, inducing progressive cellular damage and multiple organ failure. DNA samples from 70 patients with biochemical diagnosis of different MPSs genotypes confirmed by Sanger sequencing were used to evaluate a Next Generation Sequencing (NGS) protocol. Eleven genes related to MPSs were divided into three different panels according to the clinical phenotype. This strategy led to the identification of several pathogenic mutations distributed across all exons of MPSs-related genes. We were able to identify 96% of all gene variants previously identified by Sanger sequencing, showing high sensitivity in detecting different types of mutations. Furthermore, new variants were not identified, representing 100% specificity of the NGS protocol. The use of this NGS approach for genotype identification in MPSs is an attractive option for diagnosis of patients. In addition, the MPS diagnosis workflow could be divided in a two-tier approach: NGS as a first-tier followed by biochemical confirmation as a second-tier.
Keywords: Lysosomal storage disease, mucopolysaccharidoses, next generation sequencing, target sequence, mutation detection
Introduction
Mucopolysaccharidoses (MPSs) are a group of rare genetic disorders caused by deficiency in the activity of specific lysosomal enzymes required for the degradation of glycosaminoglycans (GAGs). Thus, a defect in the activity of any of the 11 enzymes responsible for the stepwise degradation of GAGs will result in their abnormal accumulation inside the lysosomes of most cells, inducing progressive cellular damage and multiple organ failure, with consequent reduction of the quality of life and life expectancy (Neufeld and Muenzer, 2001).
MPS disorders are inherited in an autosomal recessive manner and affect males and females equally. The exception is MPS II, an X-linked recessive disorder that primarily affects males, but due to autosomal X-chromosomal translocation and non-random X-chromosome inactivation, rare female patients with MPS II have also been reported (Neufeld and Muenzer, 2001). The clinical manifestations of MPSs vary considerably, with a wide spectrum of signs and symptoms in multiple organ systems, being chronic and progressive conditions (Muenzer et al., 2011).
MPS I, II, and VII are characterized by many similar clinical features that include skeletal abnormalities (dysostosis multiplex), coarse facies, corneal opacity, hearing loss, decreased pulmonary function, cardiac disease, umbilical and inguinal hernias, visceromegaly, among other problems. The disease onset, rate of progression, and manifestations vary from mild to severe. In addition to the symptomatic manifestations, patients with severe forms of MPS I, II, and VII may have cognitive impairment, typically appearing in childhood. MPS VI patients present similar somatic manifestations as MPS I, II, and VII, also with a wide spectrum of severity but with absence of cognitive impairment (Valayannopoulos et al., 2005; Martin et al., 2008; Muenzer et al., 2009, 2011; Montaño et al., 2016).
Patients with any form of MPS III present severe central nervous system degeneration with little or no somatic involvement. This disorder may be recognized by a rapid loss of social skills with aggressive behavior and hyperactivity, hirsutism, and coarse facies. The skeletal pathology is relatively mild and often becomes apparent only after a diagnosis is established (Neufeld and Muenzer, 2001; Valstar et al., 2010).
Both forms of MPS IV are characterized by skeletal dysplasia, ligamentous laxity, odontoid hypoplasia, and short stature, without cognitive impairment. Patients with severe phenotype may live into their second or third decade, and those with attenuated disease may live much longer (Northover et al., 1996; Tomatsu et al., 2011).
MPS IX is the rarest form of MPS, with only four patients diagnosed to date, mainly with joint disease, short stature, bifid uvula, submucosal cleft palate, flat nasal bridge, generalized cutaneous swelling, multiple periarticular soft-tissue masses and popliteal cyst (Triggs-Raine et al., 1999).
The purpose of this work was to validate and establish the sensitivity and specificity of NGS panels to identify genetic mutations in MPS patients previously diagnosed and genotyped. Panels were designed to be phenotype-oriented, considering the feasibility of this approach as a first-tier alternative followed by biochemical confirmation as a second-tier.
Subjects and Methods
Patients
Samples from 70 MPS patients (8 MPS I, 12 MPS II, 23 MPS III, 17 MPS IV, 6 MPS VI, and 4 MPS VII) and eight controls were included in this study. All patients had a previous biochemical diagnosis and were already genotyped by Sanger sequencing. The study was approved by the Hospital de Clinicas de Porto Alegre Research Ethics Committee, which is recognized by the Office for Human Research Protections as an Institutional Review Board (IRB0000921).
AmpliSeq gene panels
Ion Torrent semiconductor technology is able to load onto an Ion 314TM, 316TM and 318TM chip, 10 Mb, 100 Mb and 1 Gb of sequence per run, respectively. Targeted NGS can be achieved by Ion AmpliSeq technology (Thermo Fisher Scientific), an ultrahigh-multiplex PCR amplification strategy that uses very low input genomic DNA for a simple and fast library construction of specific human genes or genomic regions (Buermans and den Dunnen, 2014). To set up a fast and comprehensive assay for molecular analysis of MPS based in NGS sequencing using the Ion Torrent Personal Genome MachineTM, we designed three customized AmpliSeqTM panels for sequencing eleven genes (IDUA, IDS, SGSH, NAGLU, HGSNAT, GNS, GALNS, GLB1, ARSB, GUSB, and HYAL1) related with MPSs. These panels were validated in 78 samples (70 MPS patients and 8 controls) previously sequenced by Sanger method.
The 11 genes related with MPS disease were divided into three different customized panels according to the similarity of clinical presentations. Panel 1, first comprised MPS types I, II, VI and VII and was later redesigned to include MPS IX; Panel 2 included all MPS III types, and Panel 3 included the MPS IV types A and B. AmpliSeqTM primer panels were designed by Ion AmpliSeq Designer software (Thermo Fisher Scientific) resulting in a two-pool design for each panel.
DNA isolation
Genomic DNA (gDNA) was extracted from fresh peripheral blood samples using a salting out method or automated extraction using iPrep Purelink gDNA Blood kit (Invitrogen). DNA sample quantity and purity of the nucleic acid samples were assessed using a Nanodrop 1000 (Thermo Fisher Scientific) specrophotometer. All of the gDNAs had 260/280-nm absorbance ratios between 1.8 and 2.0. Patient DNA samples were adjusted to a final concentration of 10 ng/μL.
Library preparation
The customized Ion AmpliSeqTM panel was processed using the Ion AmpliSeq Library Kit 2.0 (Thermo Fisher Scientific) according to the manufacturer’s recommendations, starting from 10 ng of gDNA per pool. The samples were barcoded with the Ion Express Barcode Kit (Thermo Fisher Scientific) to optimize patient pooling on the same sequencing chip. The number of samples by chip was determined taking into account the total length of each panel, aiming for at least 100X coverage (number of reads per amplicon).
Ion Torrent PGM sequencing
Template preparation was performed using an Ion One Touch 2 System (Thermo Fisher Scientific) and an Ion One Touch ES (Thermo Fisher Scientific) following the latest version of the manufacturer’s manuals. The template-positive Ion Sphere Particles (ISP+) were sequenced on an Ion Torrent Personal Genome Machine (PGM) (Thermo Fisher Scientific) using the 314 Chip v2, following the Ion PGM Sequencing 200 Kit v2 manual. We used 500-flow runs, which support a template read length of approximately 200 bp.
Bioinformatics analyses
The raw data was processed with the Torrent Suite Software v5.0 (Thermo Fisher Scientific) using the standard pipeline parameters. Read alignment and variant identification was done with the Torrent Mapping Program (TMAP) v3.4.1 and Torrent Variant Caller (TVC) v5.0 software. We used hg19 (Human Genome version 19, UCSC) as reference for alignment with TMAP, and a BED (Browser Extensible Display, UCSC) file to define our regions of interest on TVC (Merriman et al., 2012). The Integrative Genome Viewer (IGV; Broad Institute) was used for the analysis of depth coverage, sequence quality, and variant visualization (Thorvaldsdóttir et al., 2013).
The Coverage Analysis plugin from Torrent Suite software was also used to establish four different types of coverage charts. For each sample, the amplicon coverage summary file was downloaded, sorted according to the .bed reference file, and the total reads information was used to create a unique spreadsheet for all samples of a given panel. Depth values of each amplicon were divided by the median depth of the batch, following by calculation of the log25 ratio to access comparisons between sequenced samples. To evaluate the intra-run performance of each amplicon, the mean of log25 value and the standard deviation were calculated. In order to analyze the inter-run reproducibility, we concatenated the intra-run results. We also used the normalized total reads mean values to explore the depth of each sequenced gene.
Results
In this study, we aimed to evaluate the sensitivity and specificity of three custom Ampliseq panels designed for amplifying the coding regions of MPS related genes. These designed panels allowed the analysis of 81 out of 90 exons, targeting 90-95% of the genes (Table 1a,b). Libraries of 78 samples, previously sequenced by Sanger sequencing, were re-sequenced by NGS in 12 separated runs: six for Panel 1 and three for Panels 2 and 3. To ensure adequate depth and coverage for variant identification, we sequenced 8-16 barcoded samples on a 314 chip. For this sample set, we obtained an average of mapped reads of 76,000 for Panel 1, 58,000 for Panel 2, and 47,000 for Panel 3; the average of reads on target, depth of coverage, and uniformity varied from gene to gene (Table 1c).
Table 1. Coverage metrics of gene panels.
| a) Panel metrics | ||||
|---|---|---|---|---|
| Size | Number of amplicons | Missed bp | Coverage (%) | |
| Panel 1 | 13.42Kb | 83 | 342 | 95.9 |
| Panel 2 | 7.81Kb | 66 | 435 | 95.1 |
| Panel 3 | 8.61Kb | 49 | 562 | 90.1 |
| b) Target genes coverage and missed regions | |||
|---|---|---|---|
| Gene | Target size (bp) | Missed (bp) | Covered (%) |
| IDUA | 2,256 | 209 | 90.74 |
| IDS | 2,079 | 0 | 100 |
| ARSB | 1,820 | 96 | 94.73 |
| GUSB | 2,208 | 37 | 98.32 |
| SGSH | 1,597 | 0 | 100 |
| NAGLU | 2,298 | 294 | 87.21 |
| HGSNAT | 2,106 | 141 | 93.3 |
| GNS | 1,813 | 0 | 100 |
| GALNS | 2,657 | 541 | 79.64 |
| GLB1 | 3,049 | 21 | 99.31 |
| c) Panel coverage metrics | ||||||
|---|---|---|---|---|---|---|
| Number of samples | Mapped reads | Reads on target | Depth of coverage | Uniformity of coverage | ||
| Panel 1 | ||||||
| IDUA | 8 | 77,000 | 79% | 1160 | 88% | |
| IDS | 12 | 76,000 | 81% | 641 | 87% | |
| ARSB | 6 | 75,000 | 82% | 841 | 86% | |
| GUSB | 4 | 76,000 | 82% | 580 | 86% | |
| Panel 2 | ||||||
| SGSH | 11 | 56,000 | 94% | 693 | 90% | |
| NAGLU | 8 | 61,000 | 94% | 764 | 89% | |
| HGSNAT | 2 | 58,000 | 76% | 477 | 91% | |
| GNS | 2 | - | - | - | - | |
| Panel 3 | ||||||
| GALNS | 13 | 42,000 | 67% | 565 | 87% | |
| GLB1 | 4 | 53,000 | 83% | 800 | 87% | |
We were able to identify 250 variants in the 70 samples. Variants were filtered according to: a) minimum coverage of 100X; b) variant detection in both strands; and c) variant frequency < 1%. After filtration, 65 variants remained, including 48 missense/nonsense mutations and 17 pathogenic indels. Overall, we were able to identify 98% of all variants previously identified by Sanger method, 96% in Panel 1, 100% in Panel 2, and 100% in Panel 3, showing a high sensitivity in detecting different types of mutations using this approach (Table 2). Furthermore, no other known pathogenic mutation was detected, in addition to those found by Sanger sequencing, showing 100% specificity of our assays.
Table 2. Number of variants identified by Sanger and NGS.
| Gene | Pathogenic point mutations | Pathogenic indels | Other variants * | |||
|---|---|---|---|---|---|---|
| Panel 1 | Sanger | NGS | Sanger | NGS | ||
| IDUA | 6 | 3 | 0 | 0 | 44 | |
| IDS | 6 | 6 | 6 | 6 | 10 | |
| ARSB | 4 | 4 | 2 | 2 | 25 | |
| GUSB | 4 | 4 | 1 | 1 | 18 | |
| Panel 2 | ||||||
| SGSH | 6 | 6 | 2 | 2 | 21 | |
| NAGLU | 9 | 9 | 3 | 3 | 19 | |
| GNS | 0 | 0 | 0 | 0 | 14 | |
| HGSNAT | 2 | 2 | 0 | 0 | 11 | |
| Painel 3 | GALNS | 12 | 12 | 1 | 1 | 15 |
| GLB1 | 2 | 2 | 2 | 2 | 8 | |
Non-pathogenic mutations: synonymous or polymorphisms
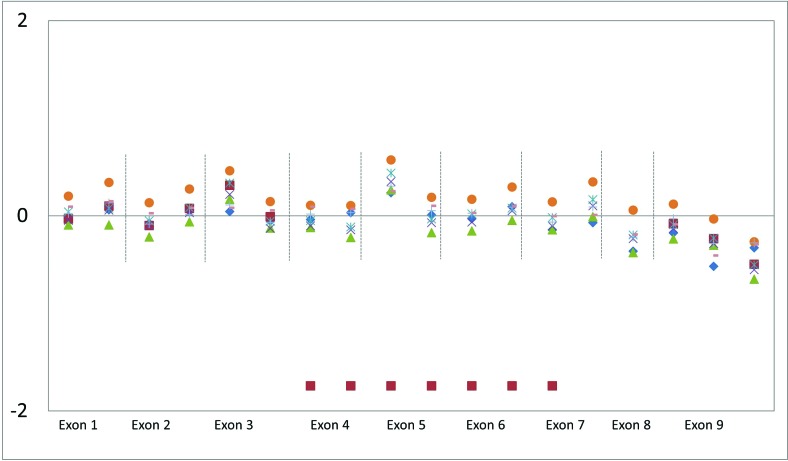
A partial deletion present in two MPS II patients could be inferred from coverage graphs. These events are noticed as decreased coverage of consecutives exons, compared with the median observed for samples sequenced in the same batch (Figure 1). Nevertheless, the exact breakpoints could not be determined, and other technologies, such as array-CGH, should be used for this purpose.
Figure 1. Deletion visualized in the decrease coverage of log25 values. Relative copy number derived from coverage data per target. The red squares mark the probable deletion of the exons IV to VIII of IDS gene.
Exon 1 of some genes (IDUA, ARSB, NAGLU, HGSNAT, and GALNS) and other four exons (exon 10 of IDUA, exon 7 and 11 of GUSB, exon 5 of GLB1), failed in the design of amplicons, probably due to high GC content and/or presence of repeat regions.
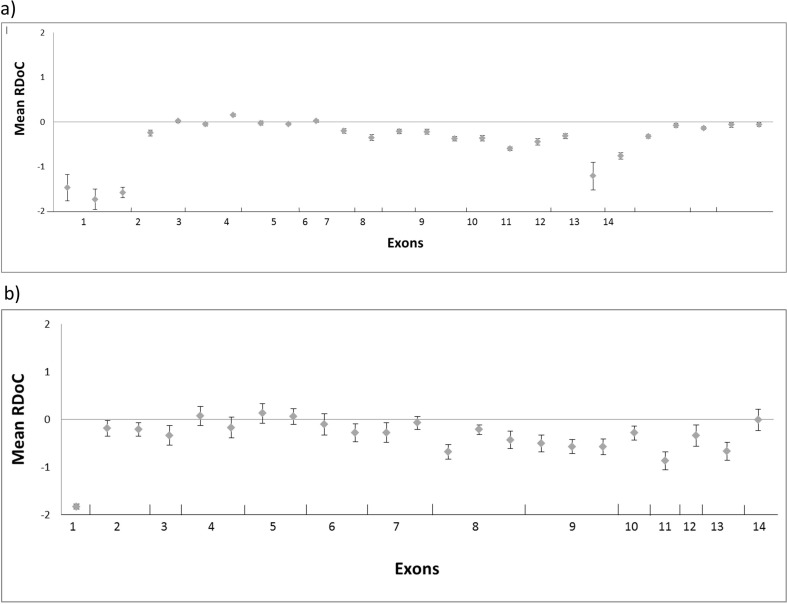
The IDUA gene had low amplification efficiency of some amplicons, as can be seen in Figure 2a, which shows the log25 ratio for eight samples sequenced in different runs. Consistently, all of them fell below the median depth of the batch in most of exons; as MPS I was the third-most frequent MPS in our routine analysis, we decided to re-design Panel 1, improving exon coverage.
Figure 2. IDUA gene amplicon coverage. (a) Intra-run relative depth of coverage (RDoC) at 14 exons corresponding to 8 samples sequenced in 6 runs; (b) IDUA gene amplicon coverage of second design. Intra-run RDoC at 14 exons corresponding to 8 samples sequenced in 1 run.
A run with eight samples was performed in order to evaluate the new panel design for the IDUA gene. We could identify 12 out of 14 mutations previously detected by Sanger sequencing. The other two mutations not detected were in exon 1, and no amplicons were designed by AmpliseqTM design software for this region. The median depth of the batch showed a high coverage depth when compared with the first panel designed (Figure 2b). Calling of variants in highly homologous regions that cannot be accurately detected by NGS can often be solved by other methods, such as Sanger sequencing, through the design of specific primers, or by other methods, such as bioinformatics, through realignment.
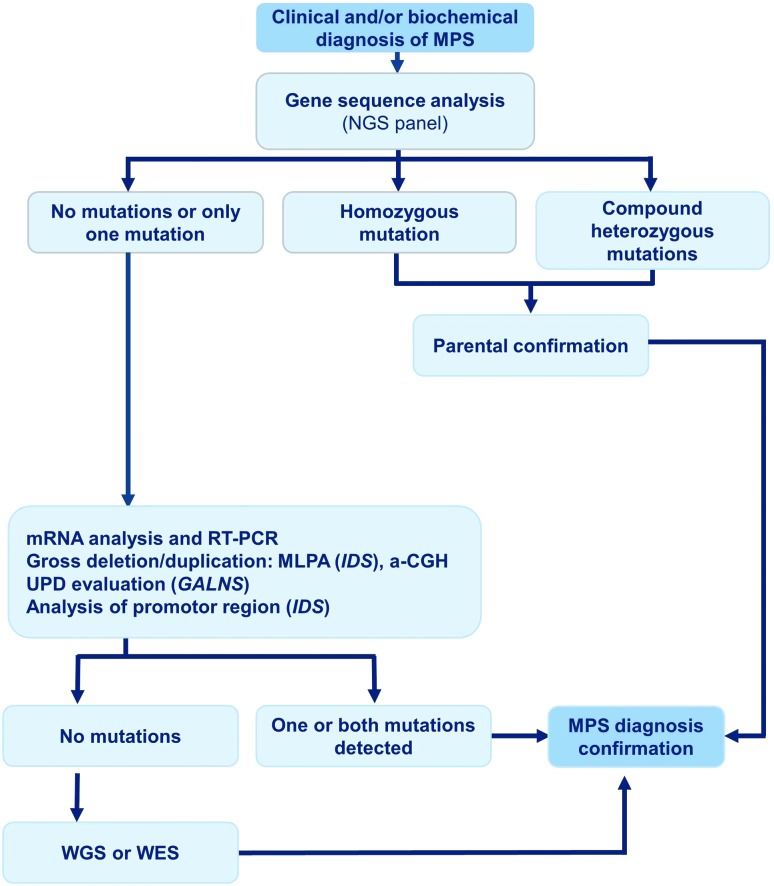
The HYAL1 gene, which is associated with MPS IX, was included in the re-designed panel in order to have a complete coverage of the MPSs genes. This analysis was validated using only the eight normal samples, once this type of MPS is very rare and no positive case was available. The analysis showed a great coverage and depth for all exons. Although the finding of a specific enzyme deficiency in leucocytes or fibroblasts confirms the diagnosis, molecular analysis of the respective gene is recommended, whenever possible (Figure 3).
Figure 3. Flow Chart for MPS investigation.
Discussion
MPSs are genetic metabolic diseases that can be caused by mutations in several distinct genes, each one coding for a specific enzyme in the GAG degradation pathway. Symptoms may be similar in some MPS types, and nowadays definitive diagnosis is achieved by enzyme assays, usually confirmed by genotype analysis.
Once a clinical suspicion of MPS is raised, biochemical studies are usually performed with diagnostic purposes. The first step in this process aims to identify if the levels of total GAGs in urine are increased and which are the GAGs species excreted in urine. Quantitation of urinary GAGs is usually colorimetric (de Jong et al., 1989), and the qualitative pattern can be obtained by thin-layer chromatography (TLC), electrophoresis, or tandem mass spectrometry (TMS) techniques (Hopwood and Harrison, 1982; Dembure et al., 1990; Auray-Blais et al., 2012; Tomatsu et al., 2014). The gold standard diagnosis of MPS is based on determination of the enzyme activity in plasma, leukocytes, or fibroblasts by fluorometric methods (Voznyi et al., 2001). Dried blood spots (DBS) could also be used for most enzyme assays, but it is usually recommended to confirm positive results in leukocytes or fibroblasts. These biochemical tests are often laborious and require specialized personnel and specific substrates. Sometimes, it is hard to obtain viable samples, especially when there is the need to travel long distances and across international borders.
Although the biochemical tests (enzyme assays and GAG analysis) are sufficient to confirm the diagnosis of MPS, genetic analysis of the specific genes involved is necessary for the characterization of the molecular defect and prognosis prediction. This enables the detection of carrier status of relatives (especially important for female relatives of MPS II patients) and accurate genetic counseling. The knowledge of the causative mutation also allows a faster and more precise prenatal diagnosis in future pregnancies in affected families.
Over the past 30 years, Sanger sequencing technology has been regarded as the gold standard to identify sequence alterations in the target region, being an accurate approach for molecular diagnosis. This method relies on analyzing individual genes, usually exon-by-exon, being expensive and time consuming (Sanger et al., 1977).
In this sense, we developed a custom panel for molecular analysis of MPS patients using NGS, thus allowing the simultaneous sequence of multiple genes grouped according to the MPS phenotype. In our study, we chose to group MPS patients according to their clinical symptoms instead of developing a comprehensive panel for all disease types. This allows maximizing the number of patients per run, avoiding unnecessary sequencing of genes that do not fit the clinical hypothesis.
The use of the NGS mutation detection method compared with Sanger sequencing led to the identification of 250 variants and 90% coverage of the 11 genes involved in MPSs etiology. The high sensitivity of our assay could be demonstrated in the detection of 94% mutations, including missense, nonsense, splice sites, in-frame deletions, and large deletions, such as a deletion of four exons in the IDS gene.
On the other hand, in five samples, the genotype could not be defined. This was due to mutations located in regions not covered or with low amplification (< 100X), caused by a high GC content or repeat regions. For these regions, for which Ion Torrent PGM was unable to sequence, or for regions not designed by Ampliseq design software, other methodologies, such as Sanger sequencing must be used for a complete gene amplification.
Moreover, due to the inability of NGS to adequately cover some GC-rich regions, as seen for the IDUA gene (MPS I), we re-designed Panel 1, changing primers for these regions in order to improve read depth, because there are pathogenic mutations previously reported in these regions. We did not redesign the other panels that presented genes without coverage or with low coverage depth due to the presence of high GC content in the segments.
Other studies had used NGS to diagnose lysosomal storage disorders by whole-exome approaches (Fernández-Marmiesse et al., 2014; Prada et al., 2014). Selmer et al. (2012) described a mild form of MPS IIIB and illustrated the diagnostic potential of targeted NGS in Mendelian disease with unknown etiology. Wei et al. (2011), reported a novel disease causing mutation in the IDS gene using this strategy.
NGS is limited by the enormous amounts of genomic data generated after sequencing of large genomic regions, making interpretation a big challenge. We were able to minimize this by our targeted sequencing strategy. Assigning pathogenicity for previously undetected variants can also be difficult (Grada and Weinbrecht, 2013; Barba et al., 2014). In the specific scenario, the existence of validated biochemical tests can help to establish which alterations are disease-causing. However, if more than two variants of unknown significance are found in the same gene (or one in the case of X-linked IDS) there might remain the doubt of which one (if any) is a non-pathogenic variant.
Additionally, complex rearrangements and large gene deletions could not be detected by the Ion Torrent platform, requiring the use of other technologies, including aCGH, MLP, and cDNA analysis by Sanger sequencing, to complement the NGS molecular analysis.
In conclusion, the Ion Torrent PGM was found to be a fast tool with a high capacity of sequencing, allowing mutation detection of multiple genes and patients at the same time, improving the molecular diagnosis approach. This massive parallel sequencing technology allows the sequencing of large genomic regions in a short time and at a low cost. In addition, NGS platforms are becoming more popular and consequently more affordable to smaller centers. Thus, we suggest this NGS protocol as a first-tier for MPS diagnosis followed by a second-tier biochemical confirmation.
Acknowledgments
This study was supported by grants from FAPERGS, CNPq, and FIPE-HCPA funds. ACBF received a governmental postdoctoral scholarship from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) - Finance Code 001. SLS and RG were recipients of CNPq research grants.
Footnotes
Associate Editor: Mariluce Riegel
Conflict of interest
The authors declare no conflict of interest.
Author Contributions
ACBF was involved in the design, experimental execution, laboratory analysis and manuscript preparation; MS was involved in the experimental execution; DL was involved in the bioinformatics analysis and manuscript preparation; DRM was involved in the generation of figures and graphs; GP was involved in MPS I Sanger sequencing; FT was involved with the DNA extractions of biological samples; UM was involved in the experimental design and manuscript review; RG was involved in the manuscript review; SLS is the principal researcher and was involved with experimental design, oversaw all aspects of the project and reviewed the manuscript draft; All authors reviewed and commented on the manuscript during its drafting and approved the final version.
References
- Auray-Blais C, Lavoie P, Zhang H, Gagnon R, Clarke JT, Maranda B, Young SP, An Y, Millington DS. An improved method for glycosaminoglycan analysis by LC-MS/ MS of urine samples collected on filter paper. Clin Chim Acta. 2012;413:771–778. doi: 10.1016/j.cca.2012.01.012. [DOI] [PubMed] [Google Scholar]
- Barba M, Czosnek H, Hadidi A. Historical perspective, development and applications of next-generation sequencing in plant virology. Viruses. 2014;6:106–136. doi: 10.3390/v6010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buermans HP, den Dunnen JT. Next generation sequencing technology: Advances and applications. Biochim Biophys Acta. 2014;1842:1932–1941. doi: 10.1016/j.bbadis.2014.06.015. [DOI] [PubMed] [Google Scholar]
- de Jong JG, Wevers RA, Laarakkers C, Poorthuis BJ. Dimethylmethylene blue-based spectrophotometry of glycosaminoglycans in untreated urine: a rapid screening procedure for mucopolysaccharidosis. Clin Chem. 1989;35:1472–1477. [PubMed] [Google Scholar]
- Dembure PP, Drumheller JE, Barr SM, Elsas LJ. Selective urinary screening for mucopolysaccharidoses. Clin Biochem. 1990;23:91–96. doi: 10.1016/0009-9120(90)90561-8. [DOI] [PubMed] [Google Scholar]
- Fernández-Marmiesse A, Morey M, Pineda M, Eiris J, Couce ML, Castro-Gago M, Fraga JM, Lacerda L, Gouveia S, Pérez-Poyato MS. Assessment of a targeted resequencing assay as a support tool in the diagnosis of lysosomal storage disorders. Orphanet J Rare Dis. 2014;9:59. doi: 10.1186/1750-1172-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grada A, Weinbrecht K. Next-Generation Sequencing: Methodology and application. J Invest Dermatol. 2013;133:e11. doi: 10.1038/jid.2013.248. [DOI] [PubMed] [Google Scholar]
- Hopwood JJ, Harrison JR. High-resolution electrophoresis of urinary glycosaminoglycans: An improved screening test for the mucopolysaccharidoses. Anal Biochem. 1982;119:120–127. doi: 10.1016/0003-2697(82)90674-1. [DOI] [PubMed] [Google Scholar]
- Martin R, Beck M, Eng C, Giugliani R, Harmatz P, Muñoz V, Muenzer J. Recognition and diagnosis of mucopolysaccharidosis II (Hunter syndrome) Pediatrics. 2008;121:377–386. doi: 10.1542/peds.2007-1350. [DOI] [PubMed] [Google Scholar]
- Merriman B, Ion Torrent R&D Team and Rothberg JM Progress in Ion Torrent semicondutor chip based sequencing. Electrophoresis. 2012;33:3397–3417. doi: 10.1002/elps.201200424. [DOI] [PubMed] [Google Scholar]
- Montaño AM, Lock-Hock N, Steiner RD, Graham BH, Szlago M, Greenstein R, Pineda M, Gonzalez-Meneses A, Çoker M, Bartholomew D. Clinical course of Sly syndrome (mucopolysaccharidosis type VII) J Med Genet. 2016;53:403–418. doi: 10.1136/jmedgenet-2015-103322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muenzer J, Wraith JE, Clarke LA, International Consensus Panel on Management and Treatment of Mucopolysaccharidosis I Mucopolysaccharidosis I: Management and treatment guidelines. Pediatrics. 2009;123:19–29. doi: 10.1542/peds.2008-0416. [DOI] [PubMed] [Google Scholar]
- Muenzer J, Beck M, Giugliani R, Suzuki Y, Tylki-Szymanska A, Valayannopoulos V, Vellodi A, Wraith JE. Idursulfase treatment of Hunter syndrome in children younger than 6 years: Results from the Hunter Outcome Survey. Genet Med. 2011;13:102–109. doi: 10.1097/GIM.0b013e318206786f. [DOI] [PubMed] [Google Scholar]
- Neufeld EF, Muenzer J. The mucopolysaccharidoses. In: Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Kinzler KW and Vogelstein B (eds) The Metabolic and Molecular Basis of Inherited Disease. 8th edition. McGraw-Hill, Medical Publishing Division. 2001;3:3421–3452. [Google Scholar]
- Northover H, Cowie RA, Wraith JE. Mucopolysaccharidosis type IVA (Morquio syndrome): A clinical review. J Inherit Metab Dis. 1996;19:357–365. doi: 10.1007/BF01799267. [DOI] [PubMed] [Google Scholar]
- Prada CE, Gonzaga-Jauregui C, Tannenbaum R, Penney S, Lupski JR, Hopkin RJ, Sutton VR. Clinical utility of whole-exome sequencing in rare diseases: Galactosialidosis. Eur J Med Genet. 2014;57:339–344. doi: 10.1016/j.ejmg.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmer KK1, Gilfillan GD, Strømme P, Lyle R, Hughes T, Hjorthaug HS, Brandal K, Nakken S, Misceo D, Egeland T. A mild form of Mucopolysaccharidosis IIIB diagnosed with targeted next-generation sequencing of linked genomic regions. Eur J Hum Genet. 2012;20:58–63. doi: 10.1038/ejhg.2011.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomatsu S, Montaño AM, Oikawa H, Smith M, Barrera L, Chinen Y, Thacker MM, Mackenzie WG, Suzuki Y, Orii T. Mucopolysaccharidosis type IVA (Morquio A disease): Clinical review and current treatment. Curr Pharm Biotechnol. 2011;12:931–945. doi: 10.2174/138920111795542615. [DOI] [PubMed] [Google Scholar]
- Tomatsu S, Shimada T, Mason RW, Kelly J, LaMarr WA, Yasuda E, Shibata Y, Futatsumori H, Montaño AM, Yamaguchi S. Assay for glycosaminoglycans by tandem mass spectrometry and its applications. J Anal Bioanal Tech Suppl. 2014;2:006. doi: 10.4172/2155-9872.S2-006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triggs-Raine B1, Salo TJ, Zhang H, Wicklow BA, Natowicz MR. Mutations in HYAL1, a member of a tandemly distributed multigene family encoding disparate hyaluronidase activities, cause a newly described lysosomal disorder, mucopolysaccharidosis IX. Proc Natl Acad Sci U S A. 1999;96:6296–6300. doi: 10.1073/pnas.96.11.6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valayannopoulos V, Nicely H, Harmatz P. Mucopolysaccharidosis VI. Orphanet J Rare Dis. 2005;5:5. doi: 10.1186/1750-1172-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valstar MJ, Neijs S, Bruggenwirth HT, Olmer R, Ruijter GJ, Wevers RA, van Diggelen OP, Poorthuis BJ, Halley DJ, Wijburg FA. Mucopolysaccharidosis type IIIA: Clinical spectrum and genotype-phenotype correlations. Ann Neurol. 2010;68:876–887. doi: 10.1002/ana.22092. [DOI] [PubMed] [Google Scholar]
- Voznyi YV, Keulemans JL, van Diggelen OP. A fluorimetric enzyme assay for the diagnosis of MPS II (Hunter disease) J Inherit Metab Dis. 2001;24:675–680. doi: 10.1023/a:1012763026526. [DOI] [PubMed] [Google Scholar]
- Wei X, Jin F, Ye Y, Xu C, Qu N, Ju X, Yi X. A novel mutation of IDS gene in a Chinese patient with mucopolysaccharidosis II by next-generation sequencing. Clin Chim Acta. 2011;412:2340–2342. doi: 10.1016/j.cca.2011.08.031. [DOI] [PubMed] [Google Scholar]