Abstract
Objective
Ovarian cancer (OC) was one of the deadliest gynecological malignancy among women in global. Serum microRNAs (miRNAs) could serve as promising diagnostic biomarkers for patients with OC.
Study design
Using quantitative reverse transcription polymerase chain reaction (qRT-PCR) based Exiqon panel, we identified 27 differentially expressed miRNAs from one normal control (NC) pool and two OC pool samples in the initial screening stage. We further verified the miRNAs in the training (30 OC VS. 36 NCs) and validation stages (80 OC VS. 80 NCs) based on qRT-PCR. Later, the expression levels of the identified miRNAs were also evaluated in exosomes and tissues.
Results
We found a serum microRNA signature including five overexpressed miRNAs (miR-200c-3p, miR-346, miR-127-3p, miR-143-3p and miR-205-5p) in OC in comparison with NCs. The areas under the receiver operating characteristic (ROC) curve (AUC) of the five-miRNA panel were 0.783 for the training stage and 0.745 for the validation stage. The diagnostic sensitivity and specificity of the combined five-miRNA panel was 0.818 and 0.609 when the cut-off value was 0.636. The levels of miR-200c-3p, miR-346 and miR-127-3p in serum were related to tumor grade and distant metastasis of OC. The expression levels of the five miRNAs were also significantly up-regulated in serum exosomes (32 OC VS. 32 NCs). Furthermore, miR-200c-3p was significantly elevated in OC tissues (22 OC VS. 22 NCs). But the levels of the miR-346 and miR-143-3p were significantly lower in OC tissues.
Conclusion
Our findings showed a five-miRNA panel in serum for the detection of OC. Moreover, serum expression levels of miR-200c-3p, miR-346 and miR-127-3p were concerned with tumor grade and distant metastasis of OC.
Keywords: microRNA, Ovarian cancer, Biomarker, Detection, Prediction, qRT-PCR
Introduction
The ovarian cancer remained the same to be the main cause for women deaths of gynecological malignant tumor [1]. According to the 2017 cancer statistics’ report [2], OC was the fifth leading cause of cancer deaths among American women. As well as, the most of patients were diagnosed as OC at advanced stage with massive ascites and peritoneal dissemination. Therefore, the final survival rate only remained 30% though surgery combined with chemotherapy [2]. The reason why the mortality of OC was high was that it was difficult to diagnose early and lacking for effective treatments for patients with an advanced or recurrent disease [3]. Indeed, using the treatment of combination of surgery and chemotherapy, the 5-year survival rates were as follows: Stage IV (25%), Stage III (37%), Stage II (70%) and Stage I (93%) [1]. The pelvic examination, imaging examination, ultrasound examination and serum CA125 level test were carried out in the routine diagnosis process, but early lesions were still difficult to be found [4]. Therefore, developing the novel diagnostic and prognostic biomarkers to detect it and to help optimize treatment were eagerly desired.
MiRNAs were a class of non-coding RNA with length of 20–25 nucleotides. Combined with the principle of complementary base pairing in the target mRNA 3’untranslated region, the miRNAs influenced the translation levels of gene expressions, resulting in inhibition or degradation of the target mRNA [5]. MiRNAs were associated with cellular functions, including cell migration, invasion, proliferation, differentiation and apoptosis [6]. There was a large body of evidence to suggest that aberrant expression levels of miRNAs were relevant to tumorigenesis, progression, and metastases [6]. MiRNAs had been demonstrated to be deregulated in OC, such as overexpression or down-regulation [7]. For example, the increased levels of miR-200 mainly observed in advanced OC suggested that these miRNAs may be closely associated with development and progression of OC [8]. Currently, the academic circles have not yet reached consensus on the appropriate small RNA reference genes used for clinical testing. Based on the spiked-in normalization method, we used a relatively quantification approach to normalize miRNA in body fluid samples (those came from systemic circulation included) [9,10].
Hence, in this study, the serum miRNA profile was analyzed by the qRT-PCR based on miRCURY platform [11]. Then we used relative quantification based on qRT-PCR to verity it [12]. We concentrated on the different expression levels and clinical significance of miRNAs in serum from OC patients, and further evaluated the value of candidate miRNAs as latent markers for OC’s detection and outcome.
Materials and methods
Patients and clinical samples
226 in all serum samples (110 OC patients VS. 116 healthy volunteers) were collected from the patients hospitalized in the First Affiliated Hospital of Nanjing Medical University from 2015 to 2016. Similarly, a total of 64 exosomes from serum samples were collected between 2015 and 2016, and 44 tissue samples were collected between 2015 and 2016. The clinical staging criteria of OC was assessed by the Federation International of Gynecology and Obstetrics (FIGO) stage I to IV [13].
Study design
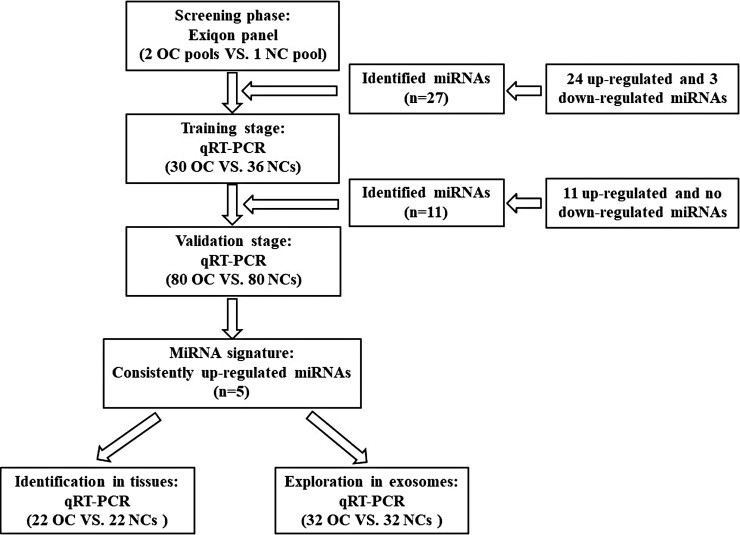
The study was separated into three steps (Fig. 1). At the initial screening stage, we randomly selected one NC pool sample and two OC pool samples (ten serum samples were merged into one pool sample). The differentially expressed miRNAs were assessed by the qRT-PCR based Exiqon panel. On the basis of the manufacturer’s protocol, 20–25 ng RNA in all was extracted from every pooled sample, and then the absorbance was measured by NanoDrop 1000 spectrophotometer used for the determination of RNA’s concentration and purity. The reverse transcribed was conducted using the miRCURY Locked Nucleic Acid (LNA™) Universal Reverse Transcription (RT) microRNA PCR, Polyadenylation and cDNA synthesis kit. Next, based on 7900HT real-time PCR system, 168 miRNAs in serum were detected to recognize miRNAs with different expressions using the Exiqon miRCURY-Ready-to-Use PCR-Human-panel-I+II-V1.M. According to the data from 7900HT real-time PCR system, the values of Ct less than 37 were included in our study. We performed the DNA spike-in (Sp3) and the RNA spike-in (UniSp6) in the panel for quality monitoring to examine whether all samples had similar technical performance. As mentioned above, all experiments were performed with proper melting curves detection, and the melting temperature (Tm) was controlled in the range of known parameters. In this study, normalized Ct (ΔCt) = average Ct (assay) – average Ct (normalizer assays). Use the method of 2−ΔΔCt to evaluate the relative expression between OC patients and NCs [12].
Fig. 1.
Overview of the experiment design. OC: ovarian cancer; NC: normal control.
At the training phase, we recruited 36 NCs and 30 OC patients, and used qRT-PCR to confirm these miRNAs discovered in initial screening stage. At the validation phase, the validated miRNA through training phase were further examined in 80 OC samples and 80 NCs. The identified miRNAs were further validated in 22 normal ovarian tissues and 22 OC tissue samples. Serum exosomal miRNAs were also assessed in 32 OC patients and 32 NCs.
Sample preparation
Tissue samples of surgical patients without preoperative radiotherapy and chemotherapy were collected and stored in liquid nitrogen. Blood samples of OC patients and healthy controls were collected before initial treatment. The whole blood was separated into serum and cellular fractions by centrifugation at 1500 RCF for 10 min and 12,000 RCF for 2 min within 12 h after collection. The serum sample was kept in −80 °C for subsequent processing.
Exosome isolation
To isolate serum exosome, 200 μl of serum was processed with ExoQuick™ based on the manufacturer’s protocol. All serum samples were incubated with ExoQuick exosome precipitation solutions about 30–60 min at 2–8 °C, and then centrifugation at 13,000 RCF about 2 min. After removing the supernatants, the exosome pellets were remained and RNA was further extracted.
RNA extraction
We used the mirVana PARIS Kit (Ambion, Austin, TX, USA) to extract total RNA from 200 μl serum or exosome sample following the manufacturer’s protocol. Then 5 μl synthetic C.elegans miR-39 (5 nM/L, RiboBio, Guangzhou, China) was spiked into every sample after adding the denatured solution (Ambion, Austin, TX, USA) to normalize the variation among samples. Total was extracted by using RNA Trizol (Invitrogen, Carlsbad, CA, USA) from tissue samples. Total RNA was dissolved in 100 μl RNase-free water, and then kept in −80 °C for further use.
Quantitative RT-PCR and data normalization
Using the specific RT primers and PCR from Bulge-Loop™ miRNA qRT-PCR Primer Set conducted amplification of miRNAs. The level of fluorescence emitted by SYBR Green (SYBR® Premix Ex Taq™ II, TaKaRa) was used to evaluate the quantification of PCR production. RT reactions were conducted at 42 °C for 60 min, and then at 70 °C for 10 min. Using the LightCycler® 480 Real-Time PCR System carried out the qRT-PCR on 384-well plates at the temperature of 95 °C for 20 s, followed by 40 cycles with the temperature of 95 °C for 10 s, 60 °C for 20 s and 70 °C for 10 s. The specificity of PCR products was evaluated by the melting curve analysis.
When normalization was based on the combination of exogenous and endogenous reference miRNAs, differences in miRNAs recovery and differences in cDNA synthesis between samples were compensated [14]. The method combined the advantages of exogenous and endogenous reference miRNAs, which not only ensured the quality of samples, but also helped correct the differences in the recovery rates of different samples [14]. So we used the method of 2−△△Ct to calculate the expression of miRNAs in serum or exosome combined with endogenous reference miRNA (miR-103) and exogenous reference miRNA (cel-miR-39), ΔCt = CtmiRNA – 0.5*(Ctcel-miR-39 + CtmiR-103). Similarly, we also used the method of 2-ΔΔCt to assess the relative expression of miRNAs in tissue samples by adding RNU6B (U6) as reference miRNA, ΔCt = CtmiRNA – CtU6.
Statistical analysis
The statistical data were processed by the SPSS 20.0 software. The continuous variable was taken as mean ± SD or median with interquartile range and the categorical variable was described as percentage or count. The categorical variable was evaluated by the chi-square test. The comparison between two groups of continuous variables used the Mann–Whitney U tests or independent sample t-tests. To compare the correlations of two groups continuous variables used the Spearman’s rank order correlations. The probability of the miRNA panel was calculated by binary logistic regression and then using the above value of probability carried out the analysis of ROC curve [15]. The diagnostic value of the identified miRNAs was assessed by ROC curve. ROC curve analysis and comparison of the derived AUC were conducted to evaluated miRNAs as predictors used to distinguish OC patients from healthy controls. The P < 0.05 was considered statistically significant.
Results
Characteristics of subjects
There were 226 in total subjects included in our research to assess the miRNAs of differential expressions in the peripheral serum samples. The experiment was divided into two stages after the screening phase: the training phase and the validation phase (Fig. 1). The demographics and clinical characteristics of the OC patients and NCs were showed in Table 1. In each stage, there was no significant difference in age and gender distribution between OC patients and NCs.
Table 1.
Characteristics of 110 OC patients and 116 normal controls enrolled in the training and the testing stage.
| Variables | Training stage (n = 66) |
Testing stage (n = 160) |
||
|---|---|---|---|---|
| Cases | Controls | Cases | Controls | |
| Number | 30 | 36 | 80 | 80 |
| Age (years) | ||||
| ≤45 | 11 | 14 | 32 | 34 |
| >45 | 19 | 22 | 48 | 46 |
| Histological type | ||||
| Mucinous | 1 | – | 8 | – |
| Serous | 13 | – | 20 | – |
| Papillary | 2 | – | 12 | – |
| Adenocarcinomatous | 5 | – | 15 | – |
| Others | 9 | – | 25 | – |
| Tumor size (cm) | ||||
| ≤1 | 8 | – | 16 | – |
| 1–2 | 17 | – | 44 | – |
| >2 | 5 | – | 20 | – |
| Lymph node status | ||||
| Absent | 26 | – | 58 | – |
| Present | 4 | – | 22 | – |
| Distant metastasis | ||||
| Absent | 15 | – | 24 | – |
| Present | 15 | – | 56 | – |
| FIGO stage | ||||
| I | 14 | – | 15 | – |
| II | 1 | – | 7 | – |
| III | 13 | – | 38 | – |
| IV | 2 | – | 20 | – |
| Menopause | 16 | – | 37 | – |
| Nulliparity | 3 | – | 10 | – |
| Serum CA125 (U/ml) | 113 ± 421 | – | 428 ± 756 | – |
Continuous data are presented as mean ± SD; categorical data are presented as counts.
FIGO, Federation International of Gynecology and Obstetrics; OC, ovarian cancer.
MiRNA profiling in the screening phase
We used the qRT-PCR platform to carried out the Exiqon miRCURY-Ready-to-Use PCR-Human-panel-I + II-V1.M for verifying candidate serum miRNAs for OC diagnosis. 168 in all miRNAs were screened out initially from one NC and two OC pooled serum samples, and 24 up-regulated and 3 down-regulated miRNAs showed at least a 1.5-fold change (Supplementary Table S1). These miRNAs were selected for further validation in the following experiments.
Identified miRNAs of OC by qRT-PCR
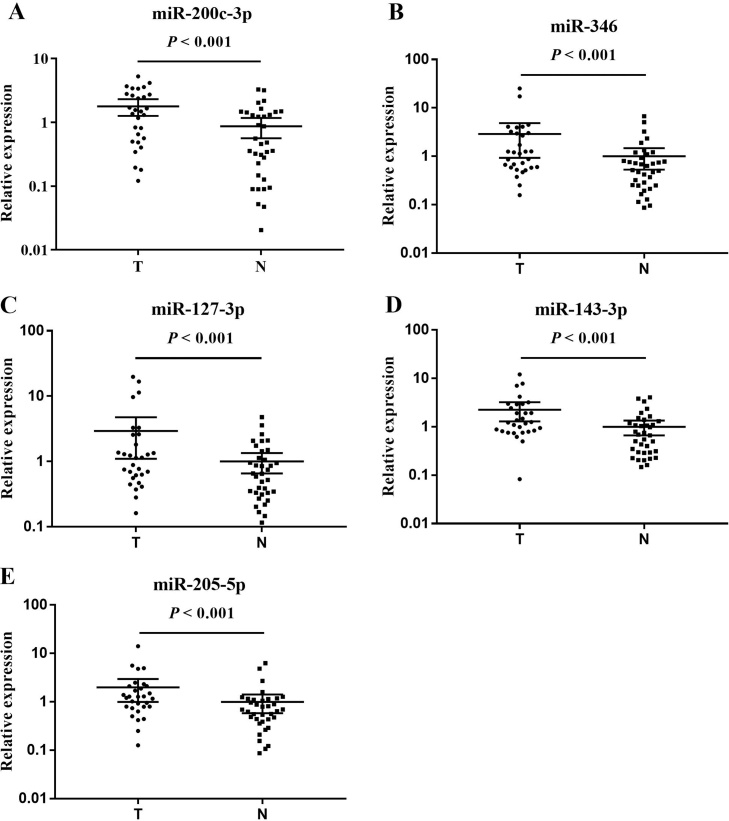
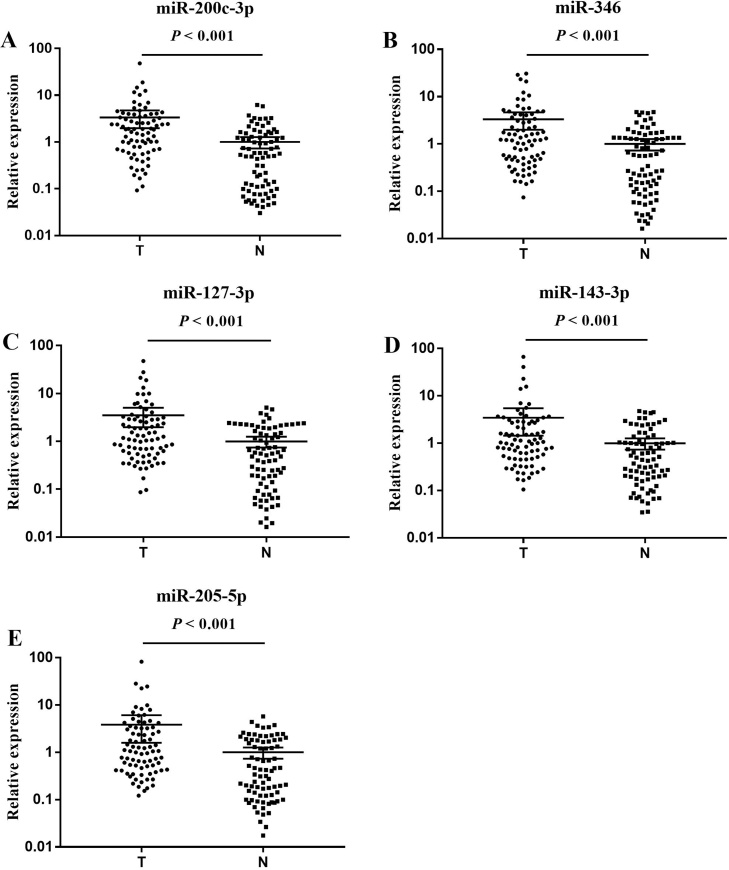
Through qRT-PCR analysis, the screened miRNAs were verified in the training stage including 36 NCs and 30 OC patients. After that, eleven differentially expressed miRNAs were identified and then were confirmed with a larger sample set during the validation phase. Ultimately, five miRNAs (miR-200c-3p, miR-346, miR-127-3p, miR-143-3p and miR-205-5p) were significantly increasing in OC patients and were selected for the following analysis. During the training and validation phase, the differential expression patterns of the five miRNAs between NCs and OC patients were consistent (Table 2; the other miRNAs were shown in the Supplementary Tables S2 and S3). Furthermore, the expression levels of the five miRNAs in serum of OC were significantly higher than that of NCs in the training and validation stages (Fig. 2, Fig. 3).
Table 2.
Realtive expression levels of the 5 miRNAs in the peripheral serum in the training and validation stages (presented as mean ± SD; 2−△△Ct).
| miRNA | Training stage |
validation stage |
Combined |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Controls | Cases | FC | P value | Controls | Cases | FC | P value | FC | P value | |
| miR-200c-3p | 3.11 ± 4.09 | 5.43 ± 5.17 | 1.74 | 0.006 | 1.54 ± 1.91 | 7.53 ± 0.67 | 4.88 | <0.001 | 3.43 | <0.001 |
| miR-346 | 0.42 ± 0.58 | 1.22 ± 2.20 | 2.89 | 0.005 | 2.88 ± 3.54 | 13.20 ± 3.79 | 4.58 | <0.001 | 4.69 | <0.001 |
| miR-127-3p | 0.83 ± 0.85 | 2.44 ± 4.08 | 2.92 | 0.028 | 2.13 ± 2.42 | 7.46 ± 20.86 | 3.50 | <0.001 | 3.52 | <0.001 |
| miR-143-3p | 1.50 ± 1.51 | 3.39 ± 3.86 | 2.26 | 0.002 | 1.47 ± 1.77 | 5.09 ± 6.17 | 3.46 | <0.001 | 3.13 | <0.001 |
| miR-205-5p | 1.47 ± 1.82 | 2.91 ± 3.89 | 1.98 | 0.003 | 1.70 ± 2.04 | 6.57 ± 4.03 | 3.86 | <0.001 | 3.42 | <0.001 |
FC: Realtive fold change to normal control; ΔCt = CtmiRNA − 0.5 * (Ctcel-miR-39 + CtmiR-103).
Fig. 2.
Realtive expression levels of the five miRNAs in the serum of 30 OC patients and 36 NCs (in the training phases; data was presented as 2−△△Ct). A: miR-200c-3p; B: miR-346; C: miR-127-3p; D: miR-143-3p; E: miR-205-5p; N: normal controls; T: tumor. Horizontal line: mean with 95% CI.
Fig. 3.
Realtive expression levels of the five miRNAs in the serum of 80 OC patients and 80 NCs (in the validation phases; data was presented as 2−△△Ct). A: miR-200c-3p; B: miR-346; C: miR-127-3p; D: miR-143-3p; E: miR-205-5p; N: normal controls; T: tumor. Horizontal line: mean with 95% CI.
Diagnostic value of the identified miRNAs
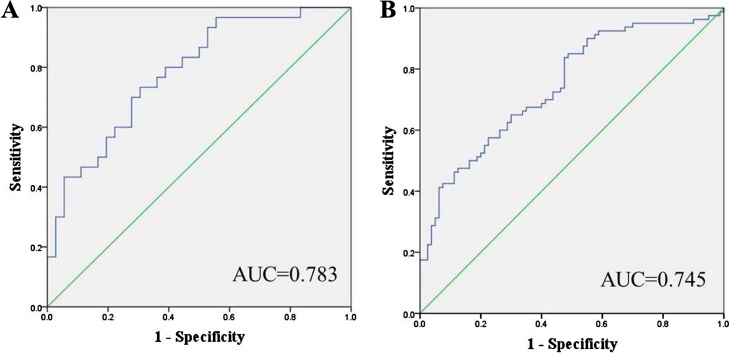
ROC curve analysis was utilized to evaluate the diagnostic value of the 5 miRNAs. The AUCs were 0.726, 0.693, 0.698, 0.687 and 0.689 for miR-200c-3p, miR-346, miR-127-3p, miR-143-3p and miR-205-5p, respectively (Supplementary Fig. S1). According to Jordan index, we calculated the diagnostic sensitivity and specificity of above five miRNAs. The sensitivity and specificity of miR-200c-3p, miR-346, miR-127-3p, miR-143-3p and miR-205-5p were 0.809 and 0.517 at cut-off value of 0.616, 0.718 and 0.603 at cut-off value of 0.592, 0.891 and 0.532 at cut-off value of 0.617, 0.845 and 0.466 at cut-off value of 0.602, and 0.764 and 0.526 at cut-off value of 0.596, respectively. When the five miRNAs were combined as a panel, the diagnostic sensitivity and specificity of the five-miRNAs was higher than single miRNA (0.818 and 0.609 at cut-off value of 0.636). When the five miRNAs were combined as a panel, it was more accurate than any single miRNA in terms of differentiation of OC patients from NCs (AUC: 0.737; 95% CI: 0.673–0.800; Supplementary Fig. S2). In the meantime, the diagnostic value of the five-miRNA panel was also evaluated separately in the training and validation phases, and then the AUCs were 0.783 (95% CI: 0.674–0.892; Fig. 4A) and 0.745 (95% CI: 0.670–0.821; Fig. 4B), respectively. The sensitivity and specificity of the five-miRNAs panel in the training and validation phases were 0.733 and 0.694 when the cut-off value was 0.643, and 0.838 and 0.605 when the cut-off value was 0.633, respectively.
Fig. 4.
Receiver-operating characteristic (ROC) curve analyses of the five-miRNA signature to discriminate OC patients from NCs. A: training phase (30 OC VS. 36 NCs); B: validation phase (80 OC VS. 80 NCs). AUC: areas under the curve; OC: ovarian cancer; NC: normal control.
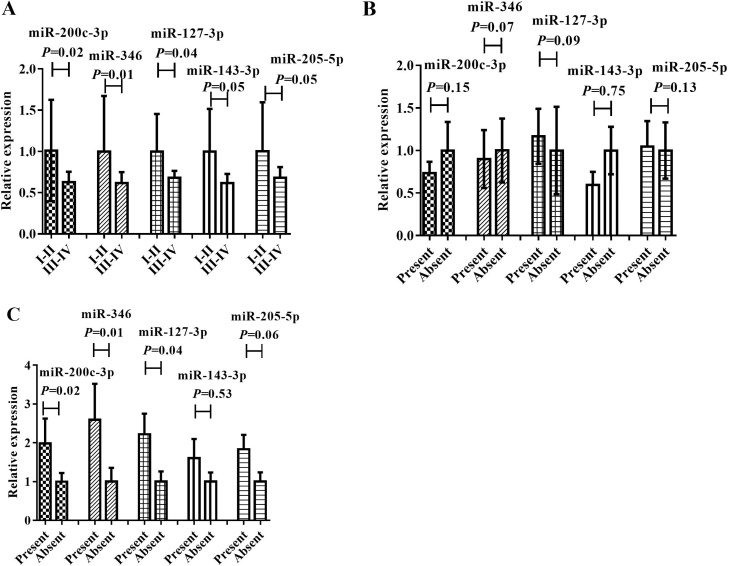
Correlations between identified miRNAs and clinical parameters
The association between the levels of the 5 serum miRNAs and clinical parameters was also evaluated for all 110 OC patients. In comparison with those with stage I + II, the expression levels of miR-200c-3p, miR-346 and miR-127-3p were significantly different in OC patients with stage III + IV (Fig. 5A). Similarly, the levels of miR-200c-3p, miR-346 and miR-127-3p were related with distant metastasis of OC patients (Fig. 5C). However, there was no statistical difference of the rest of the two miRNAs was found between OC patients with stage I + II and stage III + IV, and distant metastasis with present and absent (Fig. 5A; C). The levels of the five miRNAs were not correlated with lymph node status (Fig. 5B). We further analyzed the relationship between five miRNAs in OC patients and age, menopause and nulliparity. As a result, the five miRNAs showed no significant difference between the two cohorts (Supplementary Fig. S3). And there was only miR-205-5p in the five miRNAs significantly related to the level of serum CA125 in patients (P = 0.02; r = 0.87).
Fig. 5.
The differential expression of the five miRNAs among tumor grade, lymph node status and distant metastasis (data was presented as 2−△△Ct). A: tumor grade; B: lymph node status; C: distant metastasis.
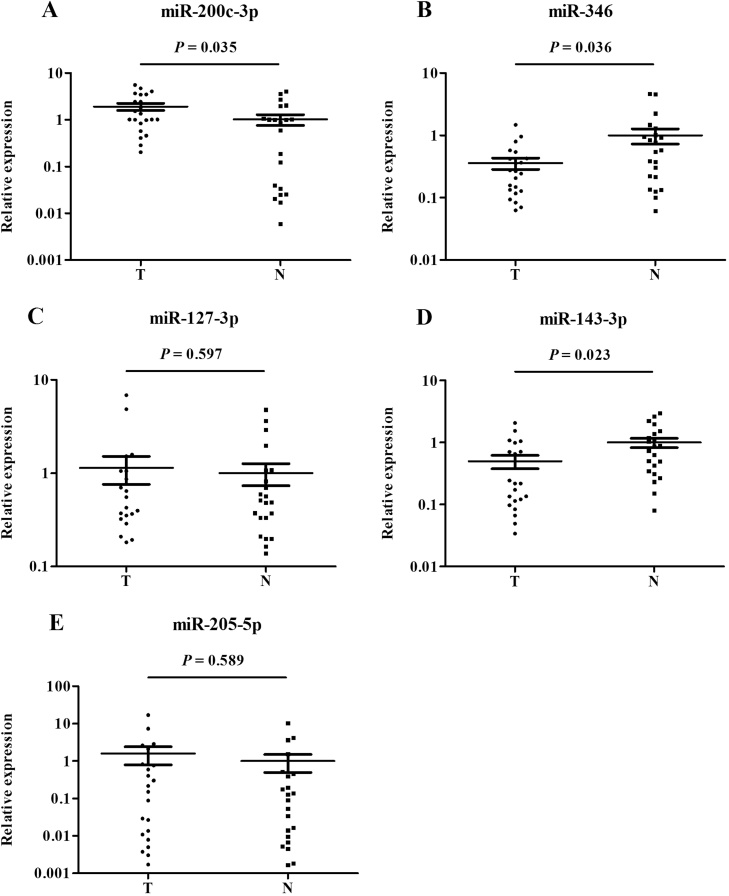
The identified miRNAs in tissue samples
To validate the consistency of the five miRNAs in serum and tissue, we detected the expression levels of the five miRNAs in 22 OC tissue specimens and 22 normal ovarian tissues. The clinical characteristics of these patients were listed in Supplementary Table S4. As shown in Fig. 6, the expression of the miR-346 and miR-143-3p was significantly lower in tumor samples than that in normal tissues. Then the serum level of miR-200c-3p was obviously higher in OC samples than that in normal tissues. The remaining two miRNAs (miR-127-3p and miR-205-5p) were not statistically significant between the two groups.
Fig. 6.
Expression of the five miRNAs in the tumor tissues of 22 OC patients and 22 NCs (data was presented as 2−△△Ct). A: miR-200c-3p; B: miR-346; C: miR-127-3p; D: miR-143-3p; E: miR-205-5p; N: normal controls; T: tumor.
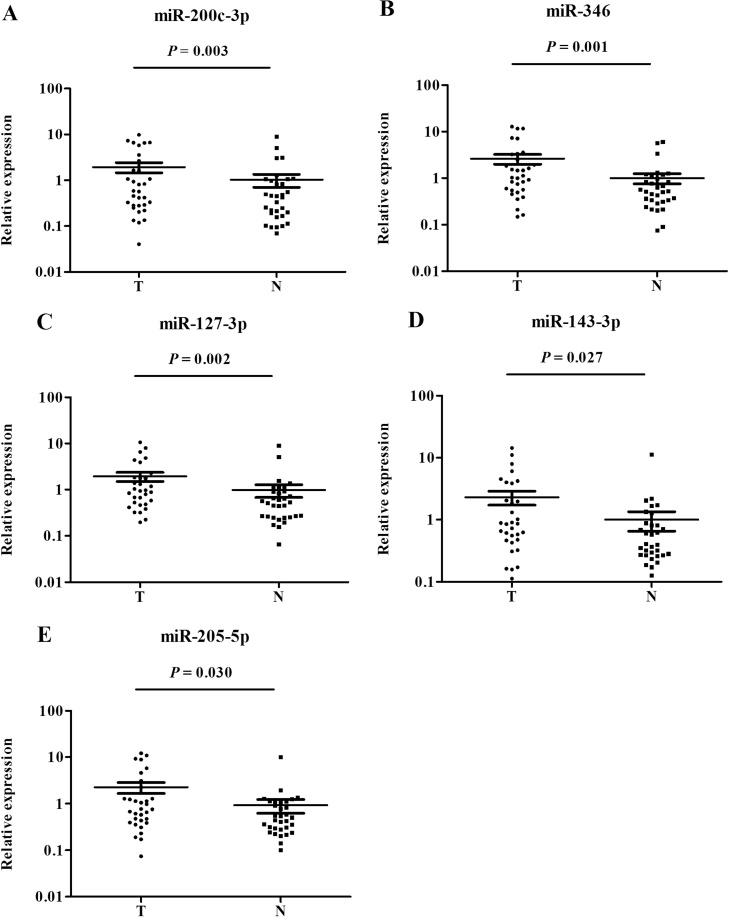
Verification of miRNAs in serum exosomes
For further study, esoxomal miRNAs were extracted from 32 NCs and 32 OC serum samples to evaluate the potential form of these identified miRNAs. In comparison with NCs, the 5 serum miRNAs were over-expressed in OC exosomes, and the results were statistically significant (Fig. 7). The clinical characteristics of these patients listed in Supplementary Table S5.
Fig. 7.
Expression of the five miRNAs in the serum exosomes of 32 OC patients and 32 NCs (data was presented as 2−△△Ct). A: miR-200c-3p; B: miR-346; C: miR-127-3p; D: miR-143-3p; E: miR-205-5p; N: normal controls; T: tumor.
Bioinformatics analysis of identified miRNAs
Experimental verification of the target genes of each confirmed miRNAs was carried out by DIANA-TarBase v7.0. The DIANA-miRPath v3.0 (a pathway analysis web-server for miRNA biological targets) was used to decipher the potential function of above miRNAs (Table S6). We finally found that viral carcinogenesis signaling pathway involved in cell proliferation, apoptosis, differentiation, and development in patients of OC, which was regulated collectively by these identified miRNAs. However, the underlying pathophysiological mechanisms of the miRNAs in OC were still needed to be further explored.
Discussion
There was mounting evidence that miRNAs took part in the regulation of genes expression in the occurrence, development and transformation of multiple tumors [16,17]. By silencing a large number of target genes, miRNAs regulated the downstream signaling pathways to affect the behaviors of human malignant tumor cells [18]. The deregulation of miRNA in OC indicated that miRNA was involved in the occurrence and progression of this disease [6,[19], [20], [21]]. In the current study, we explored expression analysis associated with OC to identify unique tumor expression features that could reliably identify OC [22]. The analysis of serum miRNAs expression level had the potential to be used as diagnostic biomarkers for patients with OC.
In this study, we at last found that the expression levels of the five miRNAs in the serum showed significant difference between OC patients and NCs. The results suggested that the five-serum miRNAs could be specific markers for OC. ROC curve analysis suggested that the five-serum miRNAs had good sensitivity and specificity for diagnosing OC. We further investigated the expression levels of above five miRNAs in exosomes and tissue samples. The result was that the five over-expressed miRNAs in exosomes were the same as in serum. However, the expression the five miRNAs in tissue of OC was inconsistent with the expression in serum. The levels of miR-346 and miR-143-3p were significantly lower in OC samples than that in NCs. Some circulating miRANs were inconsistently expressed in tumor tissues. These miRNAs were produced by tumor-induced systemic responses [[23], [24], [25]]. Zhou et al. [12] described in their research “circulating miRNAs were believed to be passively leaked or actively transported from cells during tumorigenesis and packaged into small membranous vesicles or protected by the formation of a protein-miRNA complex, and the miRNAs with low expression levels in tumor tissue might be broken down in tissues and then released into the bloodstream”. However, the exact mechanism is needed to be further studied in the future.
In addition, we also studied the clinical characteristics of the five miRNAs. The levels of miR-200c-3p, miR-346 and miR-127-3p were concerned with tumor grade and distant metastasis of OC. And the expression level of miR-205-5p significantly related to the level of serum CA125. The results demonstrated that miR-200c-3p, miR-346, miR-127-3p and miR-205-5p were promising as a progressive and prognostic biomarker in malignant OC. Of course, since the small sample size was relatively small, further verification was needed.
By controlling epithelial mesenchymal transition (EMT), miRNA-200 family (including miR-200c) was found to be related to tumor metastasis [26,27]. Gao et al. [28] subsequently verified that microRNA-200c could be a potential biomarker for diagnosis and prognosis of OC. Several studies had shown that miR-346 as a diagnostic marker was identified in different cancer detection, such as nasopharyngeal carcinoma [29], cervical cancer [30], B-cell lymphoma [31] and so on. The miR-127 had been reported to be involved in regulating the expression levels of genes of lung development, placental formation and apoptosis [[32], [33], [34]]. There were several other studies that identified the crucial role of miR-127-3p in malignant tumor [[35], [36], [37]]. For example, in the study of Bi et al. [35], they found that miRNA-127-3p acted as a tumor suppressor in epithelial OC by regulating the BAG5 gene. The function of miR-143 was thought be involved in cardiac morphogenesis but had also been implicated in cancer [33,38]. By directly targeting genes, miR-143-3p could regulate tumor growth, migration and invasion in gastric cancer [39], breast cancer [40] and so on [41]. Besides, Zhang et al. [42] proved that down-regulation of the miR-143-3p was associated with a decreased risk of OC. MiR-205-5p was also demonstrated that could play key roles in various types of cancers by targeting various genes [21]. All these results suggested that above five serum miRNAs had the potential to be a promising biomarker for future clinical OC detection.
Our bioinformatics analysis found that viral carcinogenesis signaling pathway involved in cell proliferation, apoptosis, differentiation, and development in patients of OC, was regulated by above-mentioned miRNAs in common. It suggested that these miRNAs could relate to invasion and metastasis of OC cells.
The circulating miRNAs was closely related to the occurrence and development of tumors and could be used as a new biomarker for the detection of tumors. Therefore, the study of circulating miRNA as tumor markers has been a hotspot in recent years. However, the results of current studies on circulating miRNAs of various tumors were not consistent, among which, in addition to different sample sources, methodological inconsistency was also one of the important reasons. This study was a part of the systematic work of our research group to study the circulating miRNAs of various tumors. We will collect more samples, including benign ovarian tumor samples, as well as more clinical data, including serum CA125, HE4 and imaging data, to evaluate the clinical diagnostic value of miRNAs verified by us.
In conclusion, the findings of this study provided evidence for the clinical value of the five serum miRNAs (miR-200c-3p, miR-346, miR-127-3p, miR-143-3p and miR-205-5p) as non-invasive diagnostic biomarkers in OC. Furthermore, the levels of miR-200c-3p, miR-346, miR-127-3p and miR-205-5p in serum could reflect the progression and prognosis of OC. Without doubt, the precise regulation mechanisms behind these findings suggested that more researches should be done in the future.
Conflict of interest
The authors have no conflicts of interest to declare.
Acknowledgment
This study was supported by the National Natural Science Foundation of China (Grant Nos.: 81672400; 81672788; 81770212).
Contributor Information
Xuemei Jia, Email: xmjia@njmu.edu.cn.
Yan Chen, Email: chenyandoc@163.com.
Wei Zhu, Email: zhuwei@njmu.edu.cn.
References
- 1.Wan W.N., Zhang Y.Q., Wang X.M., Liu Y.J., Zhang Y.X., Que Y.H. Down-regulated miR-22 as predictive biomarkers for prognosis of epithelial ovarian cancer. Diagn Pathol. 2014;9:178. doi: 10.1186/s13000-014-0178-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Kinose Y., Sawada K., Nakamura K., Kimura T. The role of microRNAs in ovarian cancer. Biomed Res Int. 2014;2014:249393. doi: 10.1155/2014/249393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buys S.S., Partridge E., Black A., Johnson C.C., Lamerato L., Isaacs C. Effect of screening on ovarian cancer mortality: the prostate, lung, colorectal and ovarian (PLCO) cancer screening randomized controlled trial. JAMA. 2011;305(22):2295–2303. doi: 10.1001/jama.2011.766. [DOI] [PubMed] [Google Scholar]
- 5.Kloosterman W.P., Plasterk R.H. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11(4):441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Yang J., Li G., Zhang K. MiR-125a regulates ovarian cancer proliferation and invasion by repressing GALNT14 expression. Biomed Pharmacother. 2016;80:381–387. doi: 10.1016/j.biopha.2015.12.027. [DOI] [PubMed] [Google Scholar]
- 7.Langhe R. microRNA and ovarian cancer. Adv Exp Med Biol. 2015;889:119–151. doi: 10.1007/978-3-319-23730-5_8. [DOI] [PubMed] [Google Scholar]
- 8.Meng X., Muller V., Milde-Langosch K., Trillsch F., Pantel K., Schwarzenbach H. Diagnostic and prognostic relevance of circulating exosomal miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer. Oncotarget. 2016;7(13):16923–16935. doi: 10.18632/oncotarget.7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sellars M.J., Vuocolo T., Leeton L.A., Coman G.J., Degnan B.M., Preston N.P. Real-time RT-PCR quantification of Kuruma shrimp transcripts: a comparison of relative and absolute quantification procedures. J Biotechnol. 2007;129(3):391–399. doi: 10.1016/j.jbiotec.2007.01.029. [DOI] [PubMed] [Google Scholar]
- 10.Kroh E.M., Parkin R.K., Mitchell P.S., Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR) Methods. 2010;50(4):298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jensen S.G., Lamy P., Rasmussen M.H., Ostenfeld M.S., Dyrskjot L., Orntoft T.F. Evaluation of two commercial global miRNA expression profiling platforms for detection of less abundant miRNAs. BMC Genomics. 2011;12:435. doi: 10.1186/1471-2164-12-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou X., Zhu W., Li H., Wen W., Cheng W., Wang F. Diagnostic value of a plasma microRNA signature in gastric cancer: a microRNA expression analysis. Sci Rep. 2015;5:11251. doi: 10.1038/srep11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meva J., Chaudhary R.K., Bhaduri D., Bhatia M., Hatti S., Ba R. Lacunae in International Federation of Gynecology and Obstetrics (FIGO) classification for cervical carcinoma: observational study using TNM classification as comparator. Int J Gynecol Cancer. 2013;23(6):1071–1077. doi: 10.1097/IGC.0b013e31829783c4. [DOI] [PubMed] [Google Scholar]
- 14.Sourvinou I.S., Markou A., Lianidou E.S. Quantification of circulating miRNAs in plasma: effect of preanalytical and analytical parameters on their isolation and stability. J Mol Diagn. 2013;15(6):827–834. doi: 10.1016/j.jmoldx.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 15.Wu J., Zhang H., Wang W., Zhu M., Qi L.W., Wang T. Plasma microRNA signature of patients with IgA nephropathy. Gene. 2018;649:80–86. doi: 10.1016/j.gene.2018.01.050. [DOI] [PubMed] [Google Scholar]
- 16.Zhao X., Zhou Y., Chen Y.U., Yu F. miR-494 inhibits ovarian cancer cell proliferation and promotes apoptosis by targeting FGFR2. Oncol Lett. 2016;11(6):4245–4251. doi: 10.3892/ol.2016.4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu S., Gao G., Yan D., Chen X., Yao X., Guo S. Effects of miR-145-5p through NRAS on the cell proliferation, apoptosis, migration, and invasion in melanoma by inhibiting MAPK and PI3K/AKT pathways. Cancer Med. 2017;6(4):819–833. doi: 10.1002/cam4.1030. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Phuah N.H., Nagoor N.H. Regulation of microRNAs by natural agents: new strategies in cancer therapies. Biomed Res Int. 2014;2014:804510. doi: 10.1155/2014/804510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang T., Sun Q., Liu T., Chen J., Du S., Ren C. MiR-451 increases radiosensitivity of nasopharyngeal carcinoma cells by targeting ras-related protein 14 (RAB14) Tumour Biol. 2014;35(12):12593–12599. doi: 10.1007/s13277-014-2581-x. [DOI] [PubMed] [Google Scholar]
- 20.Zhu X., Wang Y., Sun Y., Zheng J., Zhu D. MiR-155 up-regulation by LMP1 DNA contributes to increased nasopharyngeal carcinoma cell proliferation and migration. Eur Arch Oto-Rhino-Laryngol. 2014;271(7):1939–1945. doi: 10.1007/s00405-013-2818-0. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Z., Ma J., Luan G., Kang L., Su Y., He Y. MiR-506 suppresses tumor proliferation and invasion by targeting FOXQ1 in nasopharyngeal carcinoma. PLoS One. 2015;10(4):e0122851. doi: 10.1371/journal.pone.0122851. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Yanaihara N., Noguchi Y., Saito M., Takenaka M., Takakura S., Yamada K. MicroRNA gene expression signature driven by miR-9 overexpression in ovarian clear cell carcinoma. PLoS One. 2016;11(9):e0162584. doi: 10.1371/journal.pone.0162584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nowak M., Janas L., Stachowiak G., Stetkiewicz T., Wilczynski J.R. Current clinical application of serum biomarkers to detect ovarian cancer. Prz Menopauzalny. 2015;14(4):254–259. doi: 10.5114/pm.2015.55887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davidson B., Trope C.G., Reich R. The clinical and diagnostic role of microRNAs in ovarian carcinoma. Gynecol Oncol. 2014;133(3):640–646. doi: 10.1016/j.ygyno.2014.03.575. [DOI] [PubMed] [Google Scholar]
- 25.Ayaz L., Cayan F., Balci S., Gorur A., Akbayir S., Yildirim Yaroglu H. Circulating microRNA expression profiles in ovarian cancer. J Obstet Gynaecol (Lahore) 2014;34(7):620–624. doi: 10.3109/01443615.2014.919998. [DOI] [PubMed] [Google Scholar]
- 26.Lee J.W., Park Y.A., Choi J.J., Lee Y.Y., Kim C.J., Choi C. The expression of the miRNA-200 family in endometrial endometrioid carcinoma. Gynecol Oncol. 2011;120(1):56–62. doi: 10.1016/j.ygyno.2010.09.022. [DOI] [PubMed] [Google Scholar]
- 27.Kan C.W., Hahn M.A., Gard G.B., Maidens J., Huh J.Y., Marsh D.J. Elevated levels of circulating microRNA-200 family members correlate with serous epithelial ovarian cancer. BMC Cancer. 2012;12:627. doi: 10.1186/1471-2407-12-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao Y.C., Wu J. MicroRNA-200c and microRNA-141 as potential diagnostic and prognostic biomarkers for ovarian cancer. Tumour Biol. 2015;36(6):4843–4850. doi: 10.1007/s13277-015-3138-3. [DOI] [PubMed] [Google Scholar]
- 29.Yan H.L., Li L., Li S.J., Zhang H.S., Xu W. miR-346 promotes migration and invasion of nasopharyngeal carcinoma cells via targeting BRMS1. J Biochem Mol Toxicol. 2016;30(12):602–607. doi: 10.1002/jbt.21827. [DOI] [PubMed] [Google Scholar]
- 30.Guo J., Lv J., Liu M., Tang H. miR-346 up-regulates argonaute 2 (AGO2) protein expression to augment the activity of other microRNAs (miRNAs) and contributes to cervical cancer cell malignancy. J Biol Chem. 2015;290(51):30342–30350. doi: 10.1074/jbc.M115.691857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braun F.K., Mathur R., Sehgal L., Wilkie-Grantham R., Chandra J., Berkova Z. Inhibition of methyltransferases accelerates degradation of cFLIP and sensitizes B-cell lymphoma cells to TRAIL-induced apoptosis. PLoS One. 2015;10(3):e0117994. doi: 10.1371/journal.pone.0117994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Afanasyeva E.A., Hotz-Wagenblatt A., Glatting K.H., Westermann F. New miRNAs cloned from neuroblastoma. BMC Genomics. 2008;9:52. doi: 10.1186/1471-2164-9-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altuvia Y., Landgraf P., Lithwick G., Elefant N., Pfeffer S., Aravin A. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33(8):2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bi L., Yang Q., Yuan J., Miao Q., Duan L., Li F. MicroRNA-127-3p acts as a tumor suppressor in epithelial ovarian cancer by regulating the BAG5 gene. Oncol Rep. 2016;36(5):2563–2570. doi: 10.3892/or.2016.5055. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J., Hou W., Chai M., Zhao H., Jia J., Sun X. MicroRNA-127-3p inhibits proliferation and invasion by targeting SETD8 in human osteosarcoma cells. Biochem Biophys Res Commun. 2016;469(4):1006–1011. doi: 10.1016/j.bbrc.2015.12.067. [DOI] [PubMed] [Google Scholar]
- 37.Lu M., Ju S., Shen X., Wang X., Jing R., Yang C. Combined detection of plasma miR-127-3p and HE4 improves the diagnostic efficacy of breast cancer. Cancer Biomark. 2017;18(2):143–148. doi: 10.3233/CBM-160024. [DOI] [PubMed] [Google Scholar]
- 38.Koh W., Sheng C.T., Tan B., Lee Q.Y., Kuznetsov V., Kiang L.S. Analysis of deep sequencing microRNA expression profile from human embryonic stem cells derived mesenchymal stem cells reveals possible role of let-7 microRNA family in downstream targeting of hepatic nuclear factor 4 alpha. BMC Genomics. 2010;11(Suppl. 1):S6. doi: 10.1186/1471-2164-11-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang F., Liu J., Zou Y., Jiao Y., Huang Y., Fan L. MicroRNA-143-3p, up-regulated in H. Pylori-positive gastric cancer, suppresses tumor growth, migration and invasion by directly targeting AKT2. Oncotarget. 2017;8(17):28711–28724. doi: 10.18632/oncotarget.15646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li D., Hu J., Song H., Xu H., Wu C., Zhao B. miR-143-3p targeting LIM domain kinase 1 suppresses the progression of triple-negative breast cancer cells. Am J Transl Res. 2017;9(5):2276–2285. [PMC free article] [PubMed] [Google Scholar]
- 41.He Z., Yi J., Liu X., Chen J., Han S., Jin L. MiR-143-3p functions as a tumor suppressor by regulating cell proliferation, invasion and epithelial-mesenchymal transition by targeting QKI-5 in esophageal squamous cell carcinoma. Mol Cancer. 2016;15(1):51. doi: 10.1186/s12943-016-0533-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Zhang H., Li W. Dysregulation of micro-143-3p and BALBP1 contributes to the pathogenesis of the development of ovarian carcinoma. Oncol Rep. 2016;36(6):3605–3610. doi: 10.3892/or.2016.5148. [DOI] [PubMed] [Google Scholar]