Abstract
Poly(ADP-ribose) is a posttranslational modification and signaling molecule that regulates many aspects of human cell biology, and it is synthesized by enzymes known as poly(ADP-ribose) polymerases, or PARPs. A diverse collection of domain structures dictates the different cellular roles of PARP enzymes and regulates the production of poly(ADP-ribose). Here we primarily review recent structural insights into the regulation and catalysis of two family members: PARP-1 and Tankyrase. PARP-1 has multiple roles in the cellular response to DNA damage and the regulation of gene transcription, and Tankyrase regulates a diverse set of target proteins involved cellular processes such as mitosis, genome integrity, and cell signaling. Both enzymes offer interesting modes of regulating the production and the target site selectivity of the poly(ADP-ribose) modification.
The ADP-ribose modification and the PARP family of enzymes
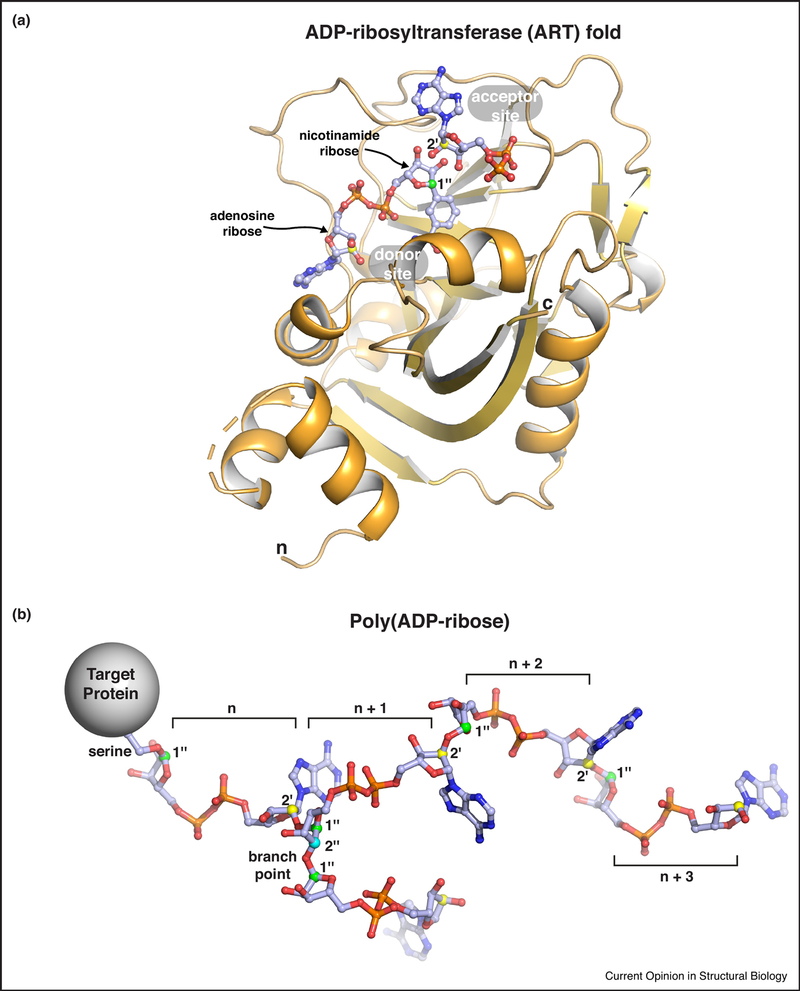
ADP-ribose is a posttranslational modification and signaling molecule produced by the ADP-ribosyltransferase (ART) activity of PARP enzymes using NAD+ as a substrate (Fig. 1a). ADP-ribose is primarily attached to proteins, but can also be found on DNA and small chemical groups [1]. The ADP-ribose modification is important for the regulation of multiple cellular processes including DNA repair, transcription, cell fate decisions, and the stress response [2]. The poly(ADP-ribose) polymerase (PARP) family of proteins is responsible for catalyzing the production of the ADP-ribose modification in cells. The PARP family has 17 members that all share a conserved catalytic domain fold that is homologous to the diphteria toxin ART fold [3](Fig. 1a). Hence, the PARP family of proteins are also referred to as ART diphteria toxin-like (ARTD) enzymes [4]. Among the 17 PARP family members, PARP-1, PARP-2 and PARP-5a/PARP-5b (the Tankyrases) synthesize polymers of ADP-ribose (Fig. 1b), while other PARPs catalyze a mono ADP-ribose posttranslational modification [5]. In addition to linear chains of ADP-ribose, poly(ADP-ribose) can also include branch points (Fig. 1b) [6,7]. Like other posttranslational modifications, poly(ADP-ribose) can alter the biochemical properties and activities of proteins targeted for modification. Moreover, there are a number of protein modules that recognize and bind to poly(ADP-ribose), and an assortment of enzymes that can trim and/or reverse the complex structure of poly(ADP-ribose) [8,9].
Figure 1. Poly(ADP-ribose) and the ADP-ribosyltransferase (ART) fold.
(a) The ADP-ribosyltransferase (ART) fold of PARP family enzymes. The minimal catalytic region of human PARP-1 is shown bound to a non-hydrolyzable NAD+ analog (PDB code 6bhv; [32]). Substrate NAD+ binds to the “donor site.” The adenosine ribose and the nicotinamide ribose are indicated. Adenosime ribose atoms are labeled as single prime (‘) and nicotinamde ribose atoms are labeled as double prime (“). Protein side chains to be modified with ADP-ribose attack the 1 ” carbon of nicotinamide ribose (labeled and colored green), thus releasing nicotinamide. The current model for the extension of the primary ADP-ribose group is based on an “acceptor site” identified in the crystal structure of chicken PARP-1 (PDB code la26; [76]). The ADP group from the chicken PARP-1 structure was overlayed on the structure of human PARP-1. Extension of the ADP-ribose chain is proposed to arise from the acceptor site adenosine ribose 2’ hydroxyl (2’ carbon is labeled and colored yellow) attacking the donor site NAD+ on the nicotinamide ribose 1” carbon (creating a 1” to 2’ linkage; see panel b). Branches are proposed to arise from an ADP-ribose that binds in a reversed orientation in the “acceptor site,” thus instead placing the nicotinamide ribose 2” hydroxyl in position to attack the donor site NAD+ on the nicotinamide ribose 1” carbon (creating a 1” to 2” linkage; see panel b). Structures that capture reaction intermediates are required to fully understand the catalysis of poly(ADP-ribose). The termini of the ART fold are labeled (n and c).
(b) The poly(ADP-ribose) posttranslational modification. Four linear units of ADP-ribose (denoted n, n+1, …) are shown attached to a serine side chain on a target protein. A single branch point is shown extending from the n + 1 ADP-ribose of the linear chain. The serine sidechain oxygen is attached to the 1” carbon of the nicotinamide ribose (colored green as above). Linear chains of ADP-ribose units are formed through a ribose-ribose linkage: nicotinamide ribose 1” carbon linked to the adenosine ribose 2’ carbon (colored yellow as above). A branch in the poly(ADP-ribose) chain forms a different ribose-ribose linkage: nicotinamide ribose 1” carbon linked to a nicotinamide ribose 2” carbon (colored cyan on the n + 1 ADP-ribose unit).
The PARP family members are NAD+ consumers and have the capacity to dramatically influence cellular levels of NAD+. PARP enzyme activities are thus tightly regulated to control the activation of cell signaling pathways but also in order to avoid depletion of the cellular energy supplies. This review presents recent structural insights into the regulation and catalysis of PARP enzymes involved in the production of poly(ADP-ribose): PARP-1, PARP-2, and the Tankyrases.
PARP regulation and catalysis in response to DNA damage
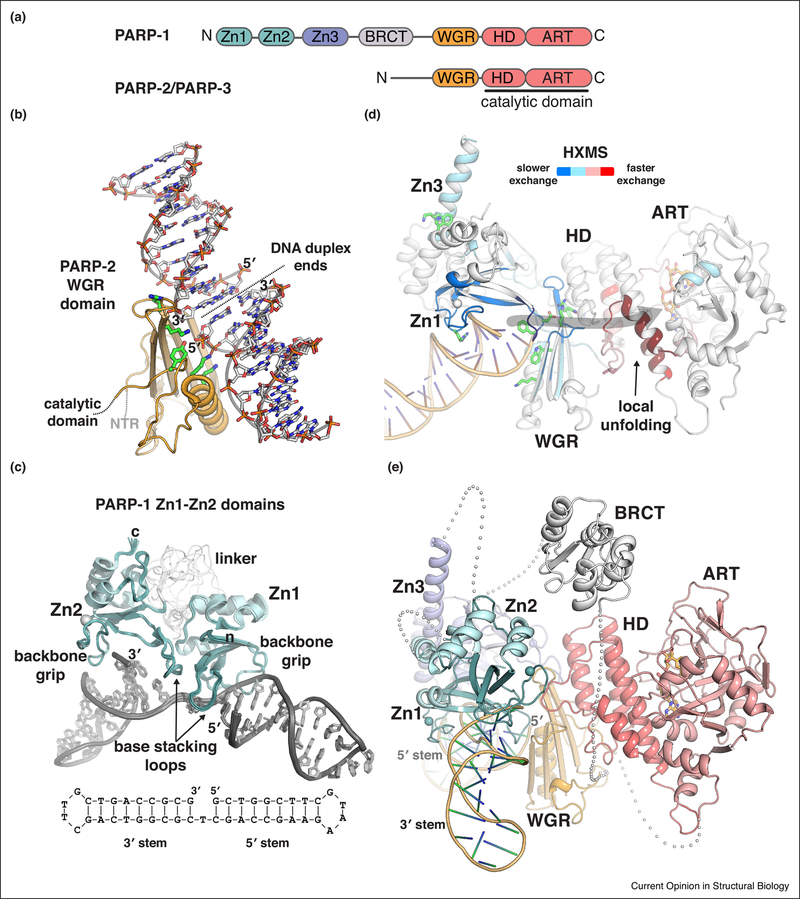
Poly(ADP-ribose) catalysis by PARP-1, PARP-2 and PARP-3 is robustly activated through binding to DNA strand breaks. These enzymes are thereby involved in various pathways of the cellular response to DNA damage [10–12]. The catalytic domain of these PARP enzymes includes a regulatory helical domain (HD) that is closely coupled to the ART fold (Fig. 2a). Outside of the catalytic domain, PARP-1, PARP-2 and PARP-3 share a common Trp-Gly-Arg (WGR) domain that interacts with DNA and acts as a key regulator of catalytic activity in response to DNA damage [13]. In addition, PARP-1 bears three zinc finger domains (Zn1, Zn2, Zn3) and a BRCA-C-terminus (BRCT) domain, whereas PARP-2 and PARP-3 have more compact structures with only short N-terminal extensions from the WGR domain. The N-terminal extensions of PARP-2 and PARP-3 contribute to overall DNA binding affinity, but are not strictly required for catalytic activity [14] [15]. Thus, the WGR domain serves as the central regulatory feature of PARP-2 and PARP-3. Notably, the WGR domains of PARP-2 and PARP-3 are sensitive to the nature of the DNA break. For example, both PARP-2 and PARP-3 are preferentially activated by DNA breaks carrying a 5’ phosphate group (5’P) [15–17], and PARP-3 activity is most responsive to 5’ phosphorylated single strand nicks [15]. In contrast, PARP-1 is relatively insensitive to the phosphorylation state of the DNA breaks that it binds and is activated by. A recent crystal structure of the WGR domain of PARP-2 bound to DNA has provided the first insights into the structural basis for selectivity toward 5’ phosphorylated DNA and for the detection of DNA breaks (Fig. 2b) [18]. Interestingly, the WGR domain of PARP-2 spans two DNA ends in the crystal structure (Fig. 2b), leading to the proposal that PARP-2 might serve a role in bridging DNA ends in pathways of DNA repair [18].
Figure 2. Structural insights into PARP-1 and PARP-2 DNA damage detection.
(a) Schematic representation of human PARP-1, PARP-2, and PARP-3 domain organization. Zinc fingers: Zn1, Zn2, and Zn3; BRCT – BRCA C-terminus; WGR: Trp-Gly-Arg domain; HD –helical domain; ART – ADP-ribosyltransferase.
(b) Structure of the human PARP-2 WGR domain bound to 5’ phosphorylated DNA (PDB code 6f5b; [18]). The crystal asymmetric unit contains two DNA duplexes. The WGR binds where the two DNA ends meet, which mimics a DNA strand break. Thus, the WGR spans the DNA break, engaging the 5’ phosphorylated end of one DNA duplex (labeled 5’), and the 3ΌH terminated end of the second duplex (labeled 3’). Some of the key esidues involved in binding the 5’ phosphate, or the second duplex, are shown as sticks and colored green. A second WGR domain is present in the crystal asymmetric unit, but has not been illustrated for clarity. The second WGR domain is bound to the opposite face of the DNA in the same manner as shown.
(c) Structure of the Zn1-Zn2 fragment of human PARP-1 detecting a DNA single-strand break (PDB code 2n8a; [21]). A schematic of the DNA is drawn below the complex structure, illustrating the single strand break with a one nucleotide gap. The two sides of the DNA break are denoted 3’ stem and 5’ stem, based on the nature of the DNA terminus. Zn1 engages nucleotide bases on the 5’ stem, and Zn2 engages nucleotide bases on the 3’ stem. The N-terminus of Zn1 is labeled (n), and the C-terminus of Zn2 is labeled (c). The extended linker region connecting Zn1 to Zn2 adopts multiple conformations, and is likely to allow the two zinc fingers to engage a variety of damaged DNA structures in which the relative positioning of the ends could vary [21]. (c) Changes in PARP-1 dynamics upon detecting DNA damage were revealed by HXMS [31]. PARP-1 peptides experiencing slower amide hydrogen exchange in the presence of a DNA strand break are indicative of newly formed protein-DNA contacts and/or domain-domain contacts, and the peptides exhibiting slower exchange (colored blue) are consistent with contacts observed in the PARP-1 complex with a DNA double-strand break (shown in cartoon with domains labeled) [13]. Remarkably, several PARP-1 peptides grouped in the HD experienced much faster amide hydrogen exchange in the presence of DNA (colored red). The rate of exchange was much faster than possible for amide hydrogens involved in forming a helix, thus these helical regions of the HD were proposed to unfold in the presence of DNA, or to rapidly sample the unfolded state [31]. Several of the key PARP-1 residues essential for DNA-dependent poly(ADP-ribose) catalysis [13,19,20,29] are drawn as sticks, and they highlight the allosteric network that connects PARP-1 DNA damage detection to the catalytic active site (indicated by grey arrow).
(d) A composite model for full-length PARP-1 detecting a single-strand DNA break. The crystal structure of Zn1-Zn3-WGR-CAT was aligned to the NMR structure of the Zn1-Zn2 fragment bound to a single-strand break [13,21]. The BRCT domain was manually positioned between the C-terminus of the Zn3 domain and the N-terminus of the WGR domain. The linker residues connecting PARP-1 domains are shown as grey spheres, with each sphere representing an amino acid residue. The DNA break site is noted by the labeling of the 5’ terminus adjacent to the WGR domain.
PARP-1 detection of DNA strand breaks
PARP-1 binds in a sequence-independent manner to a variety of DNA strand break structures, including nicked and gapped single strand breaks (SSBs), and overhang and blunt ended double strand breaks (DSBs). Zinc fingers Zn1 and Zn2 serve as the primary sensors of DNA strand breaks. Crystal structures of the individual Zn1 and Zn2 domains bound to blunt-ended, double-strand DNA breaks indicated similar modes of interaction, with the zinc fingers contacting a continuous segment of the DNA backbone (“the backbone grip”) and the nucleobases that were exposed at the DNA end (“the base-stacking loop”) [19](Fig. 2c). A crystal structure containing the domains of PARP-1 that are essential for DNA-dependent catalysis (Zn1, Zn3, WGR and CAT) in complex with a DNA double-strand break indicated how multiple domains of PARP-1 could assemble around the Zn1 domain bound to the end of the DNA [13](Fig. 2d). The essential domains of PARP-1 exhibit mutually compatible DNA binding contacts and collectively provide high affinity interaction with a DNA break. Moreover, the contact points between the domains are critical for poly(ADP-ribose) production in response to DNA double-strand breaks [13,20].
The structural data described above for PARP-1 interaction with DNA double-strand breaks established a model for PARP-1 activation, but there were still questions regarding PARP-1 interaction with other types of DNA damage, and whether the same assembly of domains and mode of activation would be utilized. Given PARP-1 involvement in SSB repair pathways [10], the mode of activation by single-strand break DNA damage was of particular interest and importance. Recently, the Neuhaus group determined the NMR structure of the Zn1-Zn2 fragment of PARP-1 detecting a single-strand DNA break [21] (Fig. 2c), indicating how the PARP-1 zinc fingers are utilized to specifically engage this type of DNA damage. Briefly, the NMR study indicated that the two zinc fingers are distributed to separate sides of the DNA break site, with Zn1 engaging the 5′ stem, and Zn2 engaging the 3′ stem (Fig. 2c). Moreover, NMR analysis with the Zn1–Zn2–Zn3 fragment bound to the SSB, and with the WGR domain titrated into the Zn1–Zn2–Zn3/DNA complex, indicated that the same domain interfaces formed in the PARP-1 complex with a DSB are also utilized in the complex with an SSB (e.g. Zn3 contacts with Zn1, and WGR contacts with Zn1 and Zn3). Notably, the specific positioning of the Zn1 domain on the 5′ stem of the SSB allows the other domains of PARP-1 to collectively assemble on a single-strand break in the same manner observed on a double-strand break, thus providing a unified model for PARP-1 detection of DNA damage (Fig. 2e).
The structural data summarized above together with other recent studies involving sedimentation analysis, small-angle x-ray scattering, single molecule tracking, and cell-based functional assays [13,21–25] indicate a monomeric, intramoleculer mechanism for PARP-1 detection of DNA strand breaks and subsequent activation. Collectively, these data argue strongly against studies that had suggested a dimeric interaction of PARP-1 with DNA [26–28]. One feature of the monomeric, intramolecular activation mechanism is that it positions PARP-1 for self-modification (automodification) in cis, as recently demonstrated biochemically [21]. The biochemical analysis of DNA strand-break activation of PARP-1 has highlighted the critical importance of the interfaces that form between PARP-1 domains after detecting DNA damage [13,19–21,29]. More recently, cell-based CRISPR-Cas9 screens have identified PARP-1 point mutations that lie at these same domain interfaces and give rise to PARP inhibitor resistance, as well as a patient-derived example of a clinical resistance mutation that targets the WGR interface with Zn1 [30].
Opening the HD for NAD+
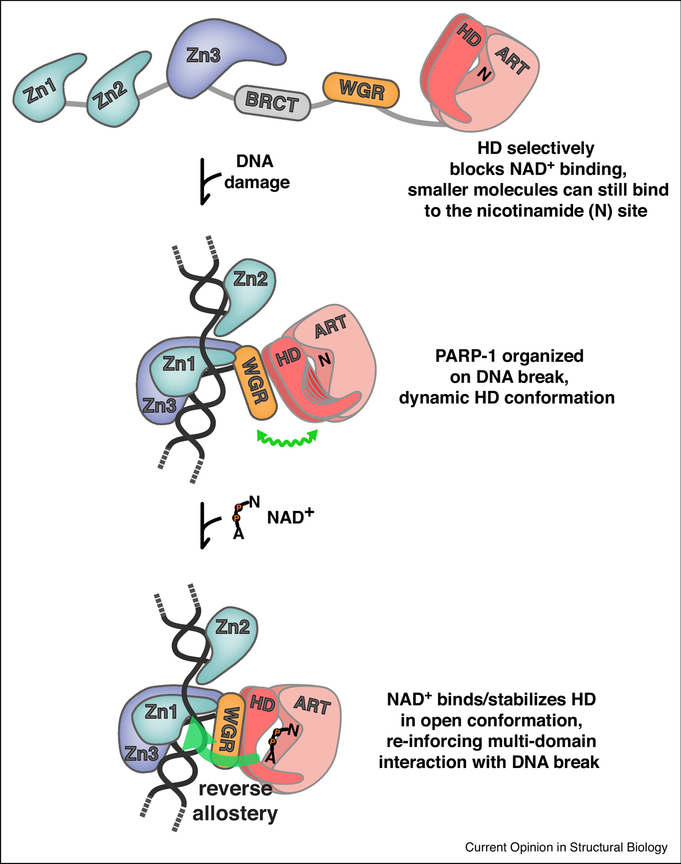
The crystal structure of PARP-1 bound to a DNA double-strand break founded an allosteric activation mechanism in which the detection of DNA damage organized PARP-1 domains into a configuration that imposed a de-stabilizing structural transition in the HD domain [13]. However, the structural transition in the HD observed in the crystal structure did not offer an obvious mechanistic explanation for 1000-fold increase in poly(ADP-ribose) production in response to DNA breaks. A critical advance in the model of PARP-1 allosteric activation came from the analysis of PARP-1 dynamics using hydrogen/deuterium exchange coupled to mass spectrometry (HXMS) [31](Fig. 2d). HXMS identified that specific regions of the HD that form helices in the absence of DNA become unfolded when PARP-1 binds to DNA damage. Deletion of the HD led to constitutive PARP-1 activity at a level that matched DNA break-induced activation, thus revealing an inhibitory function for the HD [31]. The HD inhibitory mechanism was proposed either (i) to force bound NAD+ into a conformation that was incompatible with efficient catalysis, or (ii) to completely block NAD+ binding to the active site. This next critical element of the allosteric activation mechanism was recently provided from a study using the non-hydrolyzable NAD+ analog called BAD (benzamide adenine dinucleotide)[32]. This study revealed that the HD fully blocks binding of substrate NAD+ to the active site of PARP-1, thereby inhibiting poly(ADP-ribose) production by restricting access to substrate (Fig. 3). The HD provides a selective steric block of the active site, since smaller compounds such as benzamide and nicotinamide are still able to bind to the catalytic active site in the presence of a folded HD structure. Only larger compounds like NAD+ that extend toward the HD are prevented from binding (Fig. 3). When PARP-1 binds to DNA damage, the block on substrate NAD+ is relieved through local unfolding of the HD that is induced through contacts with the regulatory domains bound to DNA. In the absence of DNA, PARP-1 can only bind to NAD+ when the HD transiently samples the unfolded conformation, thus explaining the very low basal activity of PARP-1.
Figure 3. Allosteric regulation of PARP-1.
Regulation of NAD+ access and reverse allostery. The HD regulates PARP-1 catalytic activity through a substrate-blocking mechanism [32]. In the absence of DNA, the domains of PARP-1 exist in an open configuration. In this state, small molecules can bind to the nicotinamide site (N) of the ART; however, NAD+ binding is completely blocked. Upon detecting DNA damage, PARP-1 domains are organized around the DNA break, and an allosteric network of contacts destabilizes the HD, leading to a dynamic HD structure, illustrated as multiple potential HD conformations, and accented by a wavy green line that indicates a flexible conformation. NAD+ is now able to access the catalytic active site, engaging the nicotinamide site (N) and the adenosine site (A) that was previously blocked by the HD. NAD+ binding pushes the distribution of HD conformations toward the unfolded state, thus promoting the PARP-1 assembly of domains on DNA. Thus, NAD+ binding can influence PARP-1 interaction with DNA through a reverse allostery mechanism from catalytic active site to DNA binding domain (large green arrow).
The same allosteric mechanism of activation exists in PARP-2 and PARP-3, where the HD also serves an inhibitory function that blocks NAD+ binding [31,32]. The need to tightly control the activity of PARP-1, PARP-2 and PARP-3 is likely to be related to the fact that they are NAD+ consumers with the capacity to globally effect the cellular energy pool and metabolic state, and with the potential for dramatic cellular consequences [33]. Indeed, it was shown that the PARP-1 HD mutant L713F, with elevated constitutive activity, leads to NAD+ depletion and apoptosis when expressed in human cells [34]. Importantly, the HD does not influence the NAD+ binding site conformation, but instead provides a blockage that can be reversibly controlled in order to achieve different levels of activation that allow PARP-1 to perform cellular housekeeping functions as well as acute responses to cellular stress.
Active site communication to the DNA binding domains – allostery in reverse
PARP-1 DNA binding domains communicate the detection of a DNA strand break to the catalytic active site (Fig. 2d). Analysis of PARP-1 binding to the non-hydrolyzable NAD+ analog BAD indicated that active site occupancy can be communicated back to the DNA binding domains, thereby influencing PARP-1 persistence on DNA [32](Fig. 3). BAD binding to the catalytic domain increased the affinity of PARP-1 for a DNA strand break and the retention of PARP-1 on a DNA probe in a competition experiment. At the structural level, a decrease in hydrogen/deuterium exchange was observed in PARP-1 peptides involved in domain-domain and DNA contacts, indicating that these interactions were strengthened in the presence of BAD [32], This effect was specific to a compound like BAD with a binding mode that extends to the HD and can therefore influence the distribution of HD conformations; the smaller compound benzamide was not able to influence active site communication to the DNA binding domains.
Inhibition of PARP enzymes has emerged as a strategy for targeting the inherent genomic instabilities of certain cancer cells [35,36]. A reverse allostery mechanism has been proposed to explain why some PARP-1 inhibitors are more efficient at trapping PARP-1 on DNA and exhibit correspondingly greater cytotoxicity, despite having similar inhibitory potential [37,38]. However, some studies have suggested that reverse allostery does not play a significant role for current clinical inhibitors in PARP-1 trapping on DNA and that the observed differences in trapping abilities might solely be explained by differences in inhibitory potential and inhibitor dissociation rates [3 9–41]. Moving forward, a deeper understanding of inhibitor influence on PARP-1 allostery is necessary and could provide important insights into the continued development and clinical use of PARP inhibitors.
In addition to a better understanding of the intramolecular activation mechanism of PARP-1, it will be important to understand how other cellular factors influence the PARP-1 catalytic output. An interesting recent example is histone PARylation factor 1 (HPF1), a binding partner of PARP-1 (and PARP-2) that modulates the output of PARP-1 catalytic activity [42], HPF1 interacts with the catalytic domain of PARP-1 and shifts the distribution of the poly(ADP-ribose) modification toward heteromodification of histones (rather than just PARP-1 automodification). Moreover, HPF1 steers PARP-1 to create serine-linked ADP-ribose modifications over other residues such as glutamic acid [43]. Structural studies will be necessary to determine how exactly HPF1 affects PARP-1/PARP-2 conformation and/or their interaction with histones to operate this switch in target residues. Notably, recent studies have highlighted an underappreciated prevalence of serine-linked ADP-ribosylation in cells [44,45]. These studies underscore the need to better understand the specificity underlying the modification of serine residues, and to understand the differences in biological outcomes associated with serine-linked ADP-ribosylation versus other types of modification, such as glutamate-linked ADP-ribosylation.
Tankyrase enzymes
The PARP enzymes known as Tankyrases (also referred to as PARP5a/PARP5a and PARP5b/PARP6) participate in diverse cellular functions, for example telomere maintenance, mitosis, and β-catenin/Wnt signaling [46–49]. There are two human Tankyrase enzymes, Tankyrase-1 and Tankyrase-2 (here we will use the term Tankyrase to refer to both enzymes). Tankyrase modification of target proteins with poly(ADP-ribose) frequently marks these proteins for processing by the ubiquitin ligase/proteosomal degradation system [50,51], thus allowing Tankyrase to regulate cellular protein levels and thereby influence biological pathways. In a notable example, Tankyrase catalytic activity regulates the canonical Wnt signaling pathway by binding and modifying Axin with poly(ADP-ribose) [49,52–54]. Axin is the limiting factor of a multicomponent assembly of proteins referred to as the “destruction complex,” [55,56], which degrades β-catenin in the cytoplasm to prevent its movement to the nucleus where it activates gene transcription [56]. Tankyrase modification of Axin with poly(ADP-ribose) lowers Axin levels, protects β-catenin from destruction, and thereby increases β-catenin dependent transcription [49].
Tankyrase-1 and Tankyrase-2 have the same overall domain organization composed of an ankyrin repeat region, a sterile alpha motif (SAM) fold, and a catalytic domain with the ART fold (Fig. 4a). The N-terminus of Tankyrase-1 additionally bears a region of low complexity sequence with runs of histidine, proline, and serines (HPS). In contrast to the autoinhibited catalytic domain of PARP-1, Tankyrase catalytic domains do not contain an HD and are largely accessible to bind NAD+ substrate. The principal Tankyrase regulatory mechanisms appear to be ankyrin repeat selection of binding partners, and SAM domain modulation of Tankyrase polymer formation. Consistent with the diverse cellular function of Tankyrase, the ankyrin repeats interact with a variety of partners proteins: e.g. telomere repeat factor 1 (TRF1), nuclear mitotic apparatus protein (NuMA), the β-catenin/Wnt signaling protein Axin, and DNA repair factor MERIT40.
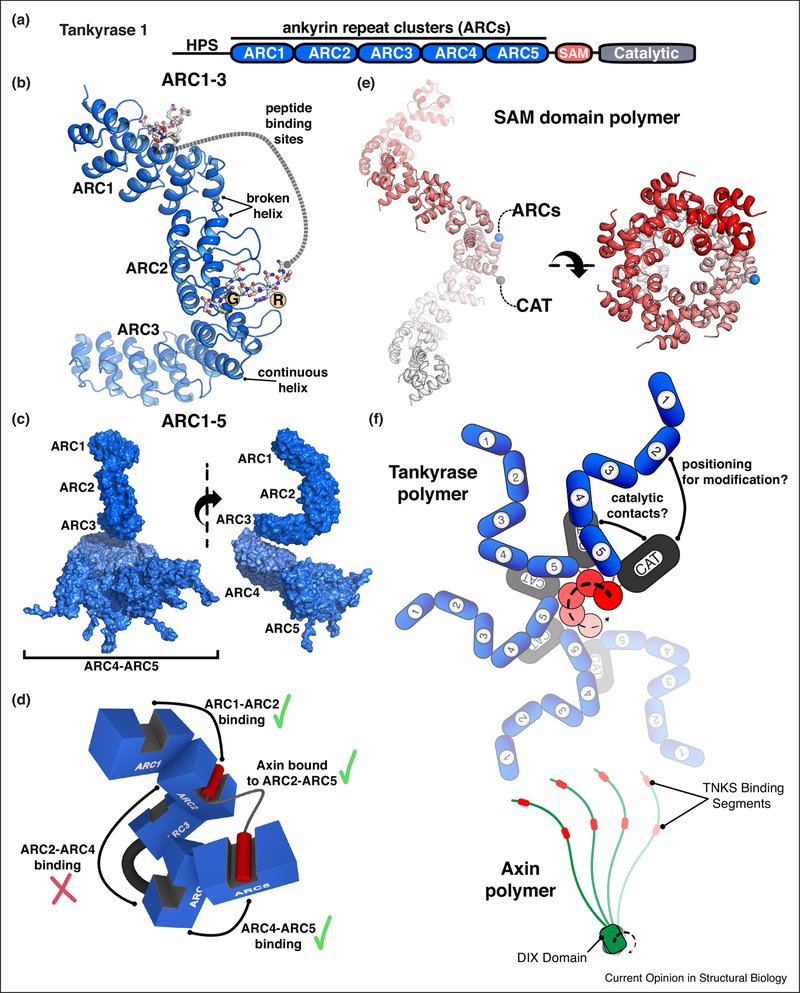
Figure 4. Structural biology of Tankyrase regulation.
(a) Schematic representation of human Tankyrase-1 domain architecture. ARC – ankyrin repeat cluster (ARC); SAM – sterile alpha motif; HPS – histidine/proline/serine-rich region of unknown function. Human Tankyrase-2 has the same domain architecture, but lacks the HPS region.
(b) Crystal structure of the ARC1–ARC2–ARC3 fragment of human Tankyrase-1 bound to a peptide derived from IRAP (insulin regulated aminopeptidase) [59]. Peptide bound to ARC1 and ARC2 are drawn as sticks. The peptide bound to ARC2 indicates the essential contact points – an Arg (R) at position 1 and a Gly (G) at position 6. ARC3 lacks the amino acids necessary for peptide binding. The structure indicated two types of transitions between consecutive ARCs: a “broken helix” connecting ARC1 to ARC2, and a “continuous helix” connecting ARC2 to ARC3. The relative positioning of the two peptide binding sites is fixed. The ARC1–ARC2–ARC3 conformation was confirmed by SAXS analysis [59].
(c) SAXS analysis of the entire ankyrin repeat region, ARC1–5. In contrast to the rigid conformation of ARC1–3, ARC4 and ARC5 are more flexibly connected. An ensemble of structures was used to model the SAXS data [59], and a selection of ARC4-ARC5 conformations are shown.
(d) Axin interaction with ARC1–5. A cartoon representation of ARC1–5 based on SAXS-based modeling in panel (c). Axin contains two peptide regions that bind to Tankyrase [60] (shown here as red cylinders). Combinatorial mutagenesis of ARC peptide binding sites and Axin binding affinity analysis indicated that specific ARC pairs can function together to bind Axin: ARC1:ARC2, ARC4:ARC5, ARC2:ARC5 (shown as green check mark). Non-binding ARC3 separates ARC2 and ARC4 and prevents them from simultaneously engaging Axin (red check mark.
(e) Tankyrase SAM domain polymer. The crystal structure of the wild-type human Tankyrase-1 SAM domain is shown from two views related by a 90° rotation (PDB code 5kni [69]; see also [68]). For one SAM domain in the polymer, the N-terminus (blue sphere) and C-terminus (grey sphere) are indicated to highlight that the ARCs and the catalytic domain (CAT) will extend from the outside surface of the SAM polymer.
(f) A model for the polymeric from of Tankyrase based on current structural information. A juxtaposed cartoon model of the Axin polymer [72] illustrates the potential for avidity-dependent interaction of these proteins [51,68].
Tankyrase ankyrin repeats – a flexible, multivalent binding platform
The ankyrin repeat region mediates interaction with Tankyrase binding partners. It consists of 25 copies of the ankyrin motif, a ~33 amino acid structure composed of a helix-loop-helix that can form extended solenoid-like configurations. The N-terminal repeats of Tankyrase are segmented into 5 ankyrin repeat clusters (ARCs) owing to the regularly-spaced positioning of variant ankyrin repeats that form N-terminus-capping and C-terminus-capping structures, and thereby disrupt the otherwise continuous extension of the repeats [57,58](Fig. 4a, b). The ARCs form the basic unit for recognizing peptide motifs in Tankyrase binding partners. ARCI, ARC2, ARC4, and ARC5 are capable of interacting with Tankyrase peptide motifs, whereas ARC3 lacks key peptide-interacting residues and therefore does not bind to Tankyrase peptide motifs [58–60]. Tankyrase-interacting peptide motifs generally span a segment of 8-amino acids and exhibit degenerate sequence requirements in most positions, with the exception of a strict requirement for an arginine at position 1 and a glycine at position 6 [58](Fig. 4b).
X-ray crystallography has provided a firm structural basis for ARC interaction with peptides derived from binding partners [58–63]. In contrast, much less is understood about Tankyrase interaction with these peptide motifs in the context of the complete tertiary/quaternary structures of diverse binding partners. Most, if not all, Tankyrase binding partners exists as multimers (e.g. dimeric TRF1, polymeric Axin), and thus present multiple peptide motifs with different spatial positioning. Moreover, some binding partners present consecutive peptide motifs, such as the two motifs identified in Axin [60], which provide the potential for multivalent binding. Likewise, the relative positioning of the five ARCs will also play a role in the mode of Tankyrase interaction with binding partners. Together, these structural determinants are likely to play a major role in the overall affinity and stability of Tankyrase interaction with binding partners.
A crystal structure of the ARC1-ARC2-ARC3 (ARC1–3) segment of Tankyrase-1 represents the largest portion of the ankyrin repeats analyzed to date and provides new insights into the overall organization of the ankyrin repeat region [59] (Fig. 4b). The X-ray structure and solution structure from small-angle X-ray scattering (SAXS) indicated a rigid conformation for the ARC1–3 section of the ankyrin repeat region, with defined transitions from ARC1 to ARC2 (“broken helix”) and ARC2 to ARC3 (“continuous helix”) giving rise to a distinctive C-shape (Fig. 4b). The positioning of the peptide binding pockets on ARC1 and ARC2 are thus held in a fixed relative conformation. A SAXS-derived model for the full ankyrin repeat region, ARC1–5, indicated that ARC4 and ARC5 are flexibly positioned relative to the more rigid ARC1–3 segment (Fig. 4c). Binding analysis with a divalent peptide derived from Axin was used to test the importance of the relative positioning of the two Axin motifs, and the ability of ARCs to simultaneously engage the two motifs. Consistent with the fixed relative spacing of ARC1 and ARC2 observed in the crystal structure, the binding analysis indicated that the affinity of the Tankyrase-1/Axin interaction is sensitive to the spacing between the two peptide motifs in Axin1, in that a shortened linker between the motifs lowered the affinity of interaction [59]. Tankyrase-1 mutagenesis indicated that specific pairs of ARCs can bind simultaneously to the two motifs in Axin: ARC1/ARC2, ARC4/ARC5, and ARC1/ARC5, but not ARC2/ARC4 (Fig. 4d). ARC3 likely serves as a fixed structural element that prevents ARC2 and ARC4 from simultaneously engaging Axin1. The pairing of ARC1 and ARC5 is quite interesting since it suggests a high level of overall flexibility, since ARC1 and ARC5 are located at opposite ends of the ankyrin repeat region (Fig. 4d). Unlike the pairing of ARC1/ARC2 and ARC4/5, the pairing of ARC1/ARC5 was not sensitive to the shortened linker between the two peptide motifs in Axin. The SAXS analysis also indicated that the conformation of the ankyrin repeats is altered upon binding Axin, suggesting that Tankyrase is an adaptable binding platform that responds to the nature of the interacting partner.
Structural analysis of Tankyrase bound to complete binding partners will ultimately be required to answer questions regarding the specificity of ankyrin repeat interactions with proteins of varied structures, and the potential for the binding interaction to be coupled to delivery to the catalytic domain for ADP-ribose modification. It is noteworthy that not all Tankyrase binding partners are modified with ADP-ribose [64,65], potentially indicating that the mode of interaction could influence catalysis, for example by imposing an inactive Tankyrase conformation.
SAM domain regulation of Tankyrase catalysis and polymer formation
The SAM domain is a ~70 amino acid helical protein module that is commonly involved in homo- and hetero-dimerization/oligomerization, but is also used to mediate other functions such as RNA binding [66]. Tankyrase-1 and Tankyrase-2 polymerize through their SAM domains, and SAM domain polymerization is required for full catalytic activity [67]. Recent X-ray crystallographic studies and structural modeling have provided detailed insights into the interfaces that form in the isolated SAM domain polymer [68–70](Fig. 3e), allowing structure-based mutagenesis to illustrate the essential role of Tankyrase polymerization in promoting β-catenin/Wnt signaling [68,69]. Polymerization-deficient mutants were not able to stimulate Wnt signaling and exhibited greatly reduced catalytic activity. In contrast to the disruption of SAM domain polymerization, neither Tankyrase catalytic inhibitor treatment nor catalytic active site mutations were able to completely abolish Tankyrase stimulation of Wnt signaling, at least under Tankyrase overexpression conditions [68]. These results suggest that Tankyrase plays a protein scaffolding role that supports Wnt signaling, in addition to the role that catalytic activity plays in regulating Wnt signaling. The components of the Wnt signaling destruction complex reversibly and dynamically associate into cytoplasmic signaling complexes [71]. Axin self-polymerizes through a Dishevelled and Axin (DIX) domain (Fig. 4f) and interacts with destruction complex components, thereby playing a pivotal role in regulating the assembly and stability of the destruction complex [72]. The polymeric structure of Tankyrase could play a similar role in organizing cytoplasmic signaling complexes through its proposed scaffolding function (Fig. 4f)[68].
A complete understanding of Tankyrase regulation and catalysis will clearly require further structural insights into its multidomain architecture. There are several key questions to be addressed. For example, what is the mechanism by which Tankyrase oligomerization contributes to catalytic activity? The linker connecting the SAM domain to the CAT domain is fairly short, perhaps allowing an interaction between these domains that could influence catalytic output, and possibly be reversed through SAM polymerization. Zhang and colleagues have recently suggested that dimerization of the Tankyrase catalytic domain regulates catalytic output [73]. In this model, catalytic domain dimerization depends on SAM-mediated polymerization, thus providing a potential basis for SAM polymerization to regulate poly(ADP-ribose) production. Another plausible mechanism is that the oligomeric form of Tankyrase serves to optimally position the CAT relative to a target protein(s) bound to the ankyrin repeats. Indeed, the N- and C-termini of the SAM domain both face the outside surface of the polymeric structure, thus supporting this type of communication between the ARCs and the CAT (Fig. 4e, f).
Conclusion
The PARP family has a variety of regulatory domains and structural features that matches the diverse aspects of cellular biology that PARP enzymes regulate. As highlighted here, PARP-1 and Tankyrase offer a glimpse of the interesting mechanisms that regulate PARP enzymes. PARP inhibitors have gained considerable interest for targeting cancers with genomic instabilities that depend on PARP-1 function and are sensitive to PARP-1 being trapped on DNA [74,75], and also for treating cancers with abnormal Wnt signaling, where Tankyrase can be targeted for its substantial regulatory role in this pathway [49–51]. Thus, the structural analysis of PARP-1 and Tankyrase will not only yield interesting insights into unique modes of enzyme regulation and catalysis, but could also contribute to the development of inhibitors that specifically target these unique regulatory mechanisms.
Acknowledgements
The work described here was in part supported by grants from the Canadian Institute of Health Research (BMA342854 and PJT374609 to J.M.P.). Efforts to apply crystallography to characterize eukaryotic pathways relevant to human cancers are supported in part by National Cancer Institute grant Structural Biology of DNA Repair (SBDR) CA92584.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
None declared.
REFERENCES
- 1.Palazzo L, Mikoč A, Ahel I: ADP-ribosylation: new facets of an ancient modification. FEBS J 2017, doi: 10.1111/febs.14078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupte R Liu Z, Kraus WL: PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes Dev 2017, 31:101–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vyas S, Chang P: New PARP targets for cancer therapy. Nat Rev Cancer 2014, 14:502–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hottiger MO, Hassa PO, Lüscher B, Schüler H, Koch-Nolte F: Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci 2010, 35:208–219. [DOI] [PubMed] [Google Scholar]
- 5.Vyas S, Matic I, Uchima L, Rood J, Zaja R Hay RT, Ahel I, Chang P: Family-wide analysis of poly(ADP-ribose) polymerase activity. 2014, 5:4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miwa M, Saikawa N, Yamaizumi Z, Nishimura S, Sugimura T: Structure of poly(adenosine diphosphate ribose): identification of 2’-[1”-ribosyl-2”-(or 3”-)(1’”-ribosyl)]adenosine-5’”,5”,5’”-tris(phosphate) as a branch linkage. Proc Natl Acad Sci USA 1979, 76:595–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miwa M, Ishihara M, Takishima S, Takasuka N, Maeda M, Yamaizumi Z, Sugimura T, Yokoyama S, Miyazawa T: The branching and linear portions of poly(adenosine diphosphate ribose) have the same alpha(l leads to 2) ribose-ribose linkage. J Biol Chem 1981, 256:2916–21. [PubMed] [Google Scholar]
- 8.Barkauskaite E, Jankevicius G, Ladurner AG, Ahel I, Timinszky G: The recognition and removal of cellular poly(ADP-ribose) signals. FEBS J 2013, 280:3491–3507. [DOI] [PubMed] [Google Scholar]
- 9.Crawford K, Bonfiglio JJ, Mikoč A, Matic I, Ahel I: Specificity of reversible ADP-ribosylation and regulation of cellular processes. Crit Rev Biochem Mol Biol 2018, 53:64–82. [DOI] [PubMed] [Google Scholar]
- 10.Ray Chaudhuri A, Nussenzweig A: The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat Rev Mol Cell Biol 2017,18:610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Vos M, Schreiber V, D F: The diverse roles and clinical relevance of PARPs in DNA damage repair: current state of the art. Biochem Pharmacol 2012,84:137–46. [DOI] [PubMed] [Google Scholar]
- 12.Pascal JM: The comings and goings of PARP-1 in response to DNA damage. DNA Repair (Amst) 2018, doi: 10.1016/j.dnarep.2018.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langelier M, Planck JL, Roy S, Pascal JM: Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science 2012, 336:728–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riccio AA, Cingolani G, Pascal JM: PARP-2 domain requirements for DNA damage-dependent activation and localization to sites of DNA damage. Nucleic Acids Res 2015, 44:1691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langelier M-FM-F, Riccio AA, Pascal JM: PARP-2 and PARP-3 are selectively activated by 5’ phosphorylated DNA breaks through an allosteric regulatory mechanism shared with PARP-1. Nucleic Acids Res 2014,42:7762–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Obaji E, Haikarainen T, Lehtiö L: Characterization of the DNA dependent activation of human ARTD2/PARP2. Sci Rep 2016, 6:34487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grundy GJ, Polo LM, Zeng Z, Rulten SL, Hoch NC, Paomephan P, Xu Y, Sweet SM, Thorne AW, Oliver AW, et al. : PARP3 is a sensor of nicked nucleosomes and monoribosylates histone H2BGlu2. Nat Commun 2016, 7:12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Obaji E, Haikarainen T, Lehtiö L: Structural basis for DNA break recognition by ARTD2/PARP2. Nucleic Acids Res 2018, doi: 10.1093/nar/gky927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langelier M-F, Planck JL, Roy S, Pascal JM: Crystal structures of poly(ADP-ribose) polymerase-1 (PARP-1) zinc fingers bound to DNA: structural and functional insights into DNA-dependent PARP-1 activity. J Biol Chem 2011,286:10690–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steffen JD, Tholey RM, Langelier M-F, Planck JL, Schiewer MJ, Lai S, Bildzukewicz NA, Yeo CJ, Knudsen KE, Brody JR, et al. : Targeting PARP-1 allosteric regulation offers therapeutic potential against cancer. Cancer Res 2014, 74:31–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eustermann S, Wu W-F, Langelier M-F, Yang J-C, Easton LE, Riccio AA, Pascal JM, Neuhaus D: Structural Basis of Detection and Signaling of DNA Single-Strand Breaks by Human PARP-1. Mol Cell 2015, 60:742–54. [DOI] [PMC free article] [PubMed] [Google Scholar]; ••Reveals the structure of human PARP-1 zinc fingers Zn1 and Zn2 bound to a single-strand DNA break, and the interdomain contacts formed on this type of DNA damage. Biochemical evidence for in cis PARP-1 automodification on defined DNA damage.
- 22.Lilyestrom W, van der Woerd MJ, Clark N, Luger K: Structural and biophysical studies of human PARP-1 in complex with damaged DNA. J Mol Biol 2010, 395:983–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eustermann S, Videler H, Yang J-C, Cole PT, Gruszka D, Veprintsev D, Neuhaus D: The DNA-binding domain of human PARP-1 interacts with DNA single-strand breaks as a monomer through its second zinc finger. J Mol Biol 2011,407:149–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu L, Kong M, Gassman NR, Freudenthal BD, Prasad R, Zhen S, Watkins SC, Wilson SH, Van Houten B: PARP1 changes from three-dimensional DNA damage searching to one-dimensional diffusion after auto-PARylation or in the presence of APE1. Nucleic Acids Res 2017, 45:12834–12847. [DOI] [PMC free article] [PubMed] [Google Scholar]; •Insights into PARP-1 identification of strand breaks using single molecule tracking.
- 25.Steffen JD, McCauley MM, Pascal JM: Fluorescent sensors of PARP-1 structural dynamics and allosteric regulation in response to DNA damage. Nucleic Acids Res 2016,44:9771–9783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pion E, Bombarda E, Stiegler P, Ullmann GM, Mély Y, De Murcia G, Gérard D: Poly(ADP-ribose) Polymerase-1 Dimerizes at a 5′ Recessed DNA End in Vitro: A Fluorescence Study. Biochemistry 2003, doi: 10.1021/bi0344432. [DOI] [PubMed] [Google Scholar]
- 27.Mendoza-Alvarez H, Alvarez-Gonzalez R: Poly(ADP-ribose) polymerase is a catalytic dimer and the automodification reaction is intermolecular. J Biol Chem 1993, [PubMed] [Google Scholar]
- 28.Ali AAE, Timinszky G, Arribas-Bosacoma R, Kozlowski M, Hassa PO, Hassler M, Ladurner AG, Pearl LH, Oliver AW: The zinc-finger domains of PARP1 cooperate to recognize DNA strand breaks. Nat Struct Mol Biol 2012, doi: 10.1038/nsmb.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langelier M-F, Ruhl DD, Planck JL, Kraus WL, Pascal JM: The Zn3 domain of human poly(ADP-ribose) polymerase-1 (PARP-1) functions in both DNA-dependent poly(ADP-ribose) synthesis activity and chromatin compaction. J Biol Chem 2010,285:18877–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pettitt SJ, Krastev DB, Brandsma I, Dréan A, Song F, Aleksandrov R, Harrell MI, Menon M, Brough R, Campbell J, et al. : Genome-wide and high-density CRISPR-Cas9 screens identify point mutations in PARP1 causing PARP inhibitor resistance. Nat Commun 2018, 9:1849. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• PARP inhibitor resistance mutations identified through an in vivo screen. The mutations are interpreted in the context of the PARP-1 structure bound to a DNA double strand break [14] and the zinc fingers bound to a single strand break [16].
- 31.Dawicki-McKenna JM, Langelier M-F, DeNizio JE, Riccio AA, Cao CD, Karch KR, McCauley M, Steffen amin D, Black BE, Pascal JM: PARP-1 Activation Requires Local Unfolding of an Autoinhibitory Domain. Mol Cell 2015, 60:755–68. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• HXMS analysis of the changes in PARP-1 dynamics upon binding DNA damage. Identification of an autoinhibitory domain in PARP-1, PARP-2, and PARP-3.
- 32.Langelier M-F, Zandarashvili L, Aguiar PM, Black BE, Pascal JM: NAD+ analog reveals PARP-1 substrate-blocking mechanism and allosteric communication from catalytic center to DNA-binding domains. Nat Commun 2018, 9:844. [DOI] [PMC free article] [PubMed] [Google Scholar]; ••A non-hydrolyzable NAD+ substrate analog is used to establish a substrate blocking mechanism for the HD of PARP-1. Analog binding to the catalytic active site influences PARP-1 interaction with DNA damge, indicating reverse allostery.
- 33.Andrabi SA, Umanah GKE, Chang C, Stevens DA, Karuppagounder SS, Gagne J-P, Poirier GG, Dawson VL, Dawson TM: Poly (ADP-ribose) polymerase-dependent energy depletion occurs through inhibition of glycolysis. Proc Natl Acad Sci 2014, 111:10209–10214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rank L, Veith S, Gwosch EC, Demgenski J, Ganz M, Jongmans MC, Vogel C, Fischbach A, Buerger S, Fischer JMF, et al. : Analyzing structure-function relationships of artificial and cancer-associated PARP1 variants by reconstituting TALEN-generated HeLa PARP1 knock-out cells. Nucleic Acids Res 2016,44:gkw859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T: Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005,434:913–7. [DOI] [PubMed] [Google Scholar]
- 36.Farmer H, McCabe N, Lord CJ, Tutt ANJ, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. : Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005,434:917–21. [DOI] [PubMed] [Google Scholar]
- 37.Murai J, Huang S-YN, Renaud A, Zhang Y, Ji J, Takeda S, Morris J, Teicher B, Doroshow JH, Pommier Y: Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol Cancer Ther 2014,13:433–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murai J, Huang SN, Das BB, Renaud A, Zhang Y, Doroshow JH, Ji J, Takeda S, Pommier Y: Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res 2012, 72:5588–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hopkins TA, Shi Y, Rodriguez LE, Solomon LR, Donawho CK, DiGiammarino EL, Panchal SC, Wilsbacher JL, Gao W, Olson AM, et al. : Mechanistic Dissection of PARP1 Trapping and the Impact on In Vivo Tolerability and Efficacy of PARP Inhibitors. Mol Cancer Res 2015,13:1465–1477. [DOI] [PubMed] [Google Scholar]
- 40.Shen Y, Rehman FL, Feng Y, Boshuizen J, Bajrami I, Elliott R, Wang B, Lord CJ, Post LE, Ashworth A: BMN 673, a Novel and Highly Potent PARP1/2 Inhibitor for the Treatment of Human Cancers with DNA Repair Deficiency. Clin Cancer Res 2013, doi: 10.1158/1078-0432.CCR-13-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rudolph J, Mahadevan J, Dyer P, Luger K: Poly(ADP-ribose) polymerase 1 searches DNA via a ‘monkey bar’ mechanism. Elife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]; •Evidence that PARP-1 uses segmental transfer to move efficiently througout the nucleus.
- 42.Gibbs-Seymour I, Fontana P, Rack JGM, Ahel I: HPFl/C4orf27 Is a PARP-1-Interacting Protein that Regulates PARP-1 ADP-Ribosylation Activity. Mol Cell 2016,62:432–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bonfiglio JJ, Fontana P, Zhang Q, Colby T, Gibbs-Seymour I, Atanassov I, Bartlett E, Zaja R, Ahel I, Matic I: Serine ADP-Ribosylation Depends on HPF1. Mol Cell 2017, 65:932–940.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palazzo L, Leidecker O, Prokhorova E, Dauben H, Matic I, Ahel I: Serine is the major residue for ADP-ribosylation upon DNA damage. Elife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larsen SC, Hendriks IA, Lyon D, Jensen LJ, Nielsen ML: Systems-wide Analysis of Serine ADP-Ribosylation Reveals Widespread Occurrence and Site-Specific Overlap with Phosphorylation. Cell Rep 2018, 24:2493–2505.e4. [DOI] [PubMed] [Google Scholar]
- 46.Smith S, Giriat I, Schmitt A, de Lange T: Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 1998,282:1484–1487. [DOI] [PubMed] [Google Scholar]
- 47.Hsiao SJ, Smith S: Tankyrase function at telomeres, spindle poles, and beyond. Biochimie 2008, 90:83–92. [DOI] [PubMed] [Google Scholar]
- 48.Chang P, Coughlin M, Mitchison TJ: Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat Cell Biol 2005, 7:1133–9. [DOI] [PubMed] [Google Scholar]
- 49.Huang S-M a, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud G a, Charlat O, Wiellette E, Zhang Y, Wiessner S, et al. : Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 2009,461:614–620. [DOI] [PubMed] [Google Scholar]
- 50.Haikarainen T, Krauss S, Lehtio L: Tankyrases: structure, function and therapeutic implications in cancer. Curr Pharm Des 2014,20:6472–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mariotti L, Pollock K, Guettler S: Regulation of Wnt/β-catenin signalling by tankyrase-dependent poly(ADP-ribosyl)ation and scaffolding. Br J Pharmacol 2017,174:4611–4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y, Liu S, Mickanin C, Feng Y, Charlat O, Michaud GA, Schirle M, Shi X, Hild M, Bauer A, et al. : RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat Cell Biol 2011,13:623–9. [DOI] [PubMed] [Google Scholar]
- 53.DaRosa PA, Wang Z, Jiang X, Pruneda JN, Cong F, Klevit RE, Xu W: Allosteric activation of the RNF146 ubiquitin ligase by a polyfADP-ribosyllation signal. Nature 2015, 517:223–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Callow MG, Tran H, Phu L, Lau T, Lee J, Sandoval WN, Liu PS, Bheddah S, Tao J, Lili JR, et al. : Ubiquitin ligase RNF146 regulates tankyrase and Axin to promote Wnt signaling. PLoS One 2011,6:e22595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kimelman D, Xu W: beta-catenin destruction complex: insights and questions from a structural perspective. Oncogene 2006, 25:7482–91. [DOI] [PubMed] [Google Scholar]
- 56.MacDonald BT, Tamai K, He X: Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell 2009,17:9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Rycker M, Venkatesan RN, Wei C, Price CM: Vertebrate tankyrase domain structure and sterile alpha motif (SAM)-mediated multimerization. Biochem J 2003,372:87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guettler S, Larose J, Petsalaki E, Gish G, Scotter A, Pawson T, Rottapel R, Sicheri F: Structural basis and sequence rules for substrate recognition by tankyrase explain the basis for cherubism disease. Cell 2011,147:1340–1354. [DOI] [PubMed] [Google Scholar]
- 59.Eisemann T, McCauley M, Langelier M-F, Gupta K, Roy S, Van Duyne GD, Pascal JM: Tankyrase-1 Ankyrin Repeats Form an Adaptable Binding Platform for Targets of ADP-Ribose Modification. Structure 2016, doi: 10.1016/j.str.2016.07.014. [DOI] [PubMed] [Google Scholar]; ••Crystallographic and SAXS analysis of the ankyrin repeat region of Tankyrase-1 and its interaction with Axin.
- 60.Morrone S, Cheng Z, Moon RT, Cong F, Xu W: Crystal structure of a Tankyrase-Axin complex and its implications for Axin turnover and Tankyrase substrate recruitment. Proc Natl Acad Sci USA 2012,109:1500–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li B, Qiao R, Wang Z, Zhou W, Li X, Xu W, Rao Z: Crystal structure of a tankyrase 1 -telomere repeat factor 1 complex research communications. 2016, 2:320–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao F, Vermeer B, Lehmann U, Kreipe H, Manns MP, Greten TF: Identification of a novel murine pancreatic tumour antigen, which elicits antibody responses in patients with pancreatic carcinoma. 2009, doi: 10.1111/j.1365-2567.2009.03090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu D, Liu J, Fu T, Shan B, Qian L, Pan L, Yuan J: USP25 regulates Wnt signaling by controlling the stability of tankyrases. Genes Dev 2017, 31:1024–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bisht KK, Dudognon C, Chang WG, Sokol ES, Ramirez A, Smith S: GDP-mannose-4,6-dehydratase is a cytosolic partner of tankyrase 1 that inhibits its poly(ADP-ribose) polymerase activity. Mol Cell Biol 2012, 32:3044–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bae J, Donigian JR, Hsueh AJW: Tankyrase 1 interacts with Mcl-1 proteins and inhibits their regulation of apoptosis. J Biol Chem 2003, 278:5195–5204. [DOI] [PubMed] [Google Scholar]
- 66.Qiao F, Bowie JU: The many faces of SAM. Sci STKE 2005, 2005:re7. [DOI] [PubMed] [Google Scholar]
- 67.De Rycker M, Price CM: Tankyrase polymerization is controlled by its sterile alpha motif and poly(ADP-ribose) polymerase domains. Mol Cell Biol 2004, 24:9802–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mariotti L, Templeton CM, Ranes M, Paracuellos P, Cronin N, Beuron F, Morris E, Guettler S: Tankyrase Requires SAM Domain-Dependent Polymerization to Support Wnt-β-Catenin Signaling. Mol Cell 2016,63:498–513. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Structure-based mutagenesis of the SAM domain indicates the importance of Tankyrase-1/2 polymerization in Wnt signaling. A scaffolding function for Tankyrase 1/2 is proposed to support Wnt signaling, in addition to a role for catalytic activity.
- 69.Riccio AA, McCauley M, Langelier M-FF, Pascal JM: Tankyrase Sterile Alpha Motif Domain Polymerization Is Required for Its Role in Wnt Signaling. Structure 2016, 24:1573–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]; ••Crystal structure for the wild-type SAM domain of Tankyrase-1. Together with [58], the importance of Tankyrase-1 polymerization in Wnt signaling is established.
- 70.DaRosa PA, Ovchinnikov S, Xu W, Klevit RE: Structural insights into SAM domain-mediated tankyrase oligomerization. Protein Sci 2016, 25:1744–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bienz M: Signalosome assembly by domains undergoing dynamic head-to-tail polymerization. Trends Biochem Sci 2014, 39:487–95. [DOI] [PubMed] [Google Scholar]
- 72.Schwarz-Romond T, Fiedler M, Shibata N, Butler PJG, Kikuchi A, Higuchi Y, Bienz M: The DIX domain of Dishevelled confers Wnt signaling by dynamic polymerization. Nat Struct Mol Biol 2007,14:484–92. [DOI] [PubMed] [Google Scholar]
- 73.Fan C, Yarravarapu N, Chen H, Kulak O, Dasari P, Herbert J, Yamaguchi K, Lum L, Zhang X: Regulation of tankyrase activity by a catalytic domain dimer interface. Biochem Biophys Res Commun 2018, 503:1780–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lord CJ, Ashworth A: PARP inhibitors: Synthetic lethality in the clinic. Science (80-) 2017, 355:1152–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pommier Y, O’Connor MJ, de Bono J: Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci Transi Med 2016,8:362ps17. [DOI] [PubMed] [Google Scholar]
- 76.Ruf a, Rolli V, de Murcia G, Schulz GE: The mechanism of the elongation and branching reaction of poly(ADP-ribose) polymerase as derived from crystal structures and mutagenesis. J Mol Biol 1998, 278:57–65. [DOI] [PubMed] [Google Scholar]